Highlights
-
•
2-DE is robust and suitable for comparing the GM soybean with its non-GM counterpart; its technical variability is lower than the biological variability.
-
•
Main source of variability is the gels so 3–4 gel replicates should be used.
-
•
Other sources of variability are minor, which gives some experimental flexibility, i.e. study can be run over several days, several operators.
Abbreviations: 2-DE, two-dimensional gel electrophoresis; CV, coefficient of variation expressed as percentage; GM, genetically modified; IEF, isoelectric focusing; IPG, immobilized pH gradient; SDS-PAGE, sodium dodecyl sulfate polyacrylamide gel electrophoresis
Keywords: False positive rate, Soybean/allergen, Technical variability, Electrophoresis, Genetically modified
Abstract
Two-dimensional gel electrophoresis (2-DE) technique is used as a performing technique to assess the variability of protein expression in crops, and especially soybean endogenous food allergens, which are a subset of proteins of interest for assessing whether genetically modified (GM) soybean has a different allergenic profile compared to its non-GM counterpart.
On top of the biological variability of the 2-DE, which has already been studied by several laboratories, technical variability has to be evaluated. In this study, several sources of variability (number of gel replicates, protein extracts, study timings and operators) were assessed qualitatively and quantitatively on all detectable polypeptide spots as well as on food allergen spots. Results showed that the major source of variability was the number of gel replicates. Other sources were minor.
This has a direct practical impact on the laboratory work as this supports the utilization of three or four gel replicates to get robust results. Furthermore, this implies that the study can be run over several days, and be performed by several trained operators, without impacting its reproducibility.
Furthermore, 2-DE could detect a 2-fold change between two samples with an acceptable rate of false positives (below 7%). This level of sensitivity is acceptable in the context of safety assessment of GM soybean as the biological variability of proteins in soybean is higher than the technical variability shown in this study.
Overall, the 2-DE technique is suitable for investigating endogenous food allergen variability between several soybean seeds, including GM and non-GM counterpart.
1. Introduction
2-DE is a technique for studying protein expression in biological systems such as crops. Several thousands of proteins can be visualized and quantified simultaneously, which allows comparisons under various experimental conditions [17]. For example, influence on protein expression of crop variety (genetic background), growing conditions (e.g. latitude, weather conditions, diseases, soil, exposure to sun) and/or maturity stage can be assessed [1], [18], [40], [42]. Among these proteins, endogenous allergens are a subset of proteins of interest for assessing the genetically modified (GM) crop safety. Evaluation of the level of endogenous allergens in GM soybean is one of the regulatory requirements before marketing them in order to insure they are safe for human consumption [13]. Indeed, one of the concerns involving GM soybean is the potential change in the levels of such endogenous allergens compared to those obtained with conventional breeding methods and subsequent enhancement in their sensitization or elicitation capacities.
The principle of the 2-DE is to separate proteins according to their isoelectric point in the first dimension and according to their molecular weight in the second dimension. Coupled with Coomassie blue staining and image analysis, 2-DE allows for the simultaneous detection and the relative quantification of several hundreds of individual proteins [35].
Technically, 2-DE involves multiple experimental steps (e.g. protein extraction, isoelectric focusing (IEF), sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), gel staining, image acquisition and protein spot quantification), each step representing a potential source of technical variability. Today, the challenge for routine application of 2-DE is to standardize the technique and to ensure its reproducibility. Consequently, it is important to identify the major sources of technical variability and to estimate false positive rates, in order to define an appropriated experimental design [8], [20], [21].
The literature referring to 2-DE is rich but only a few publications investigated technical variability. Most of them were focused on between-gels variability [4], [9], [15], [16], [37]. Some publications assessed the contribution of specific experimental steps, e.g., protein extraction [3], IEF and SDS-PAGE [11], staining procedures [24] or image analysis [43]. A few publications compared image analysis softwares [44], [46] or post-experimental variability, software-dependent and/or operator-dependent [27]. When several sources of variability were examined, the authors focused on variability between-gels and between-extracts [41], between-extracts and between-assays [28], or between-gels and between-operators/laboratory equipment [38]. However, none of these studies evaluated the respective importance of all potential sources of technical variability along the process.
Therefore, the present study aimed at evaluating them using a soybean seed protein extracts, the sources of variability inherent to the 2-DE technique: between-gels variability (variability between gel replicates, also called repeatability), between-extracts variability (inherent to protein extraction procedure), between-assays variability (variability of the whole experiment, also called reproducibility), and between-assays/operators variability. In particular, this study aimed at estimating the false positive rates in order to discriminate significant changes from the experimental background.
The evaluation was conducted on all detectable spots (corresponding to all detectable polypeptides) as well as on a subset of spots corresponding to the known endogenous soybean allergens. Two types of analyses were performed: a qualitative analysis, which referred to the presence or absence of spots, and a quantitative analysis, which referred to the optical intensity of spots.
This variability evaluation is mandatory for describing potential differences in protein allergen levels between GM and non-GM soybean varieties when the 2-DE technique is used [12].
2. Materials and methods
2.1. Study design
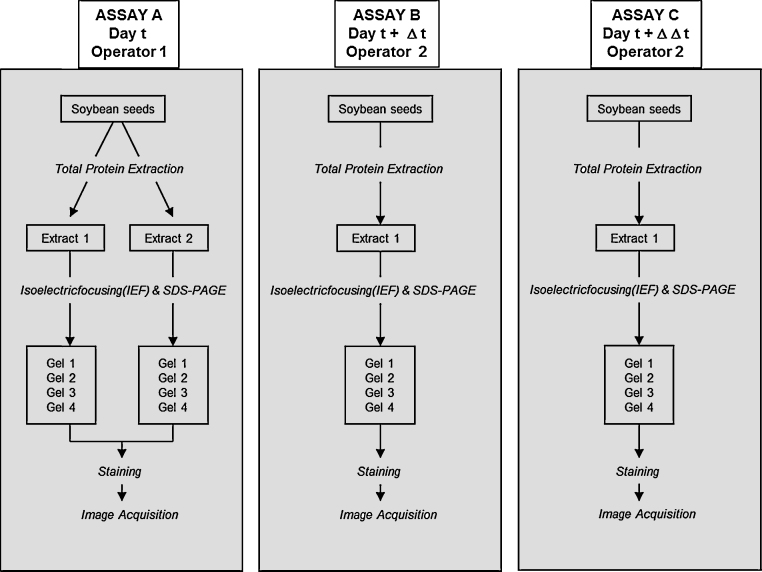
The study design is shown in Fig. 1.
Fig. 1.
Study design.
A first experiment (assay A) was conducted to assess the between-extracts variability and between-gels variability. Two protein extracts were prepared from a unique soybean seed sample. For each extract, 2-DE was performed in quadruplet. Two other experiments (assays B and C), performed at two different periods by a second operator, were set up to assess between-assays variability (assay B versus assay C) and the potential variability induced by the operator (assay A versus assay B).
2.2. Reagents and chemicals
Reagents and chemicals were mainly from Sigma–Aldrich (St Quentin Fallavier, France). Precast IPG ReadyStrips™ (pH 5–8, 17 cm) were purchased from Bio–Rad (Marnes-la-Coquette, France).
2.3. Sample preparation
Mature soybean seeds (non-transgenic Glycine max, Bayer CropScience, Research Triangle Park, USA) were grounded in liquid nitrogen using a mortar and a pestle. Frozen soybean seed powder was homogenized with an Ultraturax in urea/thiourea buffer (7 M urea, 2 M thiourea, 4% 3-[(3-cholamidopropyl) dimethylammonio]-1-propanesulfonate (CHAPS) (w/v), 0.24% Triton X100 (v/v)), supplemented with 20 mM spermine, and 50 mM dithiothreitol [14]. After incubating for 1 h at room temperature, 100 mM iodoacetamide was added to sample, and the mixture was incubated for a further 3 h at room temperature in the dark. After centrifugation (15,000 rpm for 30 min at 20 °C), supernatant was removed, aliquoted and kept frozen at −80 °C until used. Protein concentration was determined by the modified Bradford protein assay [34] using bovine serum albumin as the standard. The protein sample and bovine serum albumin were diluted in urea/thiourea buffer.
2.4. Two-dimensional gel electrophoresis technique
Precast IPG ReadyStrips™ were rehydrated with 330 μl urea/thiourea buffer supplemented with 100 mM 2-hydroxyethyl disulfide (Acros Organics, Geel, Belgium) [33] and 0.001% bromophenol blue as a tracking dye (9 h of passive rehydration at 20 °C). Using a cup-loading system (Bio-Rad), 600 μg of total soybean seed protein in 75 μl urea/thiourea buffer supplemented with 100 mM 2-hydroxyethyl disulfide, were loaded onto IPG strips. The polypeptides were focused successively for 15 min at 250 V, followed by a slow voltage ramping to 10,000 V (15 h), and focusing was continued at 10,000 V up to 300,000 V h. IEF steps were carried out using the Protean IEF Cell (Bio-Rad) at 20 °C under low viscosity oil. Prior to SDS-PAGE, Immobilized pH gradient (IPG) strips were incubated at room temperature for 30 min in an equilibration buffer (0.375 M Tris–HCl [pH 8.8], 6 M urea, 20% glycerol, 4% sodium dodecyl sulfate [SDS]) containing 65 mM dithiothreitol. A second equilibration step was carried out for 40 min under the same conditions, except that dithiothreitol was replaced by 135 mM iodoacetamide and 0.001% bromophenol blue was added as a tracking dye. Equilibrated strips were then loaded onto homemade 12.5% SDS-polyacrylamide gels, with 4% SDS-polyacrylamide gels on top as a stacking gel (2 cm). IPG strips were sealed with 1% low melting point agarose to ensure good contact between the IPG strips and the gel. SDS-PAGE was carried out using the Ettan™ Daltsix Electrophoresis Unit (Amersham, Orsay, France) connected to a PowerPac 1000 Power Supply (Bio-Rad). The electrophoresis was performed at approximately 10 °C with Tris–glycine buffer (25 mM trizma base, 192 mM glycine, 0.1% SDS) at a constant power of 5 watts per gel for 30 min followed by 12.5 W per gel until the tracking dye reached the bottom of the gel. Gels were then stained with colloidal Coomassie blue stain as recommended by the manufacturer (SafeStain, Invitrogen, Cergy Pontoise, France).
2.5. Image acquisition and analysis
Gels were digitalized using the GS-800™ calibrated densitometer (Bio-Rad), high-resolution scanner. Scanned images of 2-DE gels were analyzed with the PDQuest image analysis system (version 7.2, Bio-Rad). Briefly, after automatic spot detection allowing the detection of the majority of protein spots, images were edited manually, e.g., adding, splitting and removing spots. To compare spots across gels in each assay, a match set was obtained with images from all gels. Among them, one gel was selected as the master gel (based on gel and spot quality), against which all other gels were matched. Spots, present in a match set member but absent in the master gel, were manually added to the master gel. Automatic matching of spots on each gel was then performed. Spots matching across all gels were manually verified. For each match set, each gel was normalized to minimize the variability due to slight variation in protein load per gel, staining efficiency or image capture. This normalization was obtained by dividing the raw intensity of each spot in a gel by the total intensity of all spots in that gel that have been included in the master gel. Data were exported to Excel (Microsoft) and from there to the statistical package Statistical Analysis System (Version 8.2, SAS Institute Inc., Cary, NC, USA). All analyses were performed on normalized quantities.
Analyses were conducted independently on all the proteins of the gels and on a subset of 20 spots corresponding to known soybean food allergens, present in the publicly available, peer-reviewed allergen database Allergenonline compiled by the Food Allergy Research and Resource Program (FARRP) (www.allergenonline.org). The identity of the spots was confirmed by mass spectrometry [36]. These spots are shown in Fig. 2.
Fig. 2.
Representative 2D map of soybean seed proteins.
3. Results and discussion
3.1. Between-gels variability
Between-gels variability refers to the comparison between 4 replicate gels of the same extract as described in Fig. 1 extract 1 versus extract 2, in assay A.
3.1.1. Qualitative variability
The total number of spots detected on each gel ranged from 645 to 672 in extract 1 and from 655 to 676 in the extract 2 (Table 1a). A spot was considered to be reproducible when present in the 4 replicate gels. This criterion was very stringent for the qualitative assessment as recommended by Ruebelt et al. [37] to limit false positives or false negatives. The number of reproducible spots was 623 for extract 1 and 642 for extract 2. These numbers represented 91.6% and 94.4%, respectively, of the total of spots detected on any gels.
Table 1a.
Qualitative between-gels variability for all polypeptides.
| Gel 1 | Gel 2 | Gel 3 | Gel 4 | Reproducible spots | All gels | % matching | |
|---|---|---|---|---|---|---|---|
| Extract 1 | 672 | 645 | 649 | 666 | 623 | 680 | 91.6 |
| Extract 2 | 665 | 672 | 676 | 655 | 642 | 680 | 94.4 |
Number of reproducible spots in assay A.
The spots that were not reproducible (8.4% for extract 1 and 5.6% for extract 2) were due to intensities close to the limit of detection, explaining they were not detected in all gels. These undetected spots represent missing values for the quantitative assessment. These missing values can reduce the statistical power.
Although very stringent criteria used, the results showed a low variability. Being more flexible in the definition of reproducible spots would have improved the matching percentages, i.e. when considering the spots matched in 3 out of 4 gels, the percentage of reproducible spots would have increased from 91.6% to 94.1% and from 94.4% to 97.2%, for extracts 1 and 2 respectively (results not presented). This increase was in line with results of Choe and Lee [9] who observed a total number of matched spots present in either 2 (815 spots), 3 (793 spots) or 4 (771 spots) out of 4 gels. In addition, the more replicate gels were used, the less percentage of matching spots was recorded, which limits the statistical benefits of having a large number of replicates [10], [43]. In summary, the experimental design has to include a sufficient number of replicate 2-DE gels. Three or four replicates were often sufficient to take into account the variability and to provide the best results [6], [16].
3.1.2. Quantitative variability
Between-gels variability was assessed using spots matching to all 4 replicate gels (623 or 642 spots in extracts 1 or 2 respectively). Mean coefficient of variation (CV = standard deviation/mean × 100) was calculated per spot. Mean CV of all spots ranged from 35.2% (extract 2) to 41.1% (extract 1) (Table 1b). These mean CV values were a little higher than values reported in literature as follows: 17% [32], 18.7–26.4% [28], 20–28% [5], 24.8% [37] and, 32% [25]. Nevertheless, Zhan and Desiderio [46] reported mean CV of 35.7%, which was comparable to the results generated in this study.
Table 1b.
Quantitative between-gels variability for all polypeptides.
| Reproducible spots in 4 replicate gels | Mean CV (%) | |
|---|---|---|
| Extract 1 | 623 | 41.1 |
| Extract 2 | 642 | 35.2 |
Mean CV of intensities of reproducible spots in assay A.
3.2. Between-extract variability
Between-extract variability refers to the comparison between extracts prepared in parallel as described in Fig. 1 (extract 1 versus extract 2 in assay A).
3.2.1. Qualitative variability
The number of reproducible spots (spots present in the 8 replicate gels of both extracts) is reported in Table 2a. A total of 609 spots were present in both extracts. In addition, 47 spots were present only in one extract. These 47 spots represent false positives, i.e., spots that could be interpreted as different in a study where the two extracts would represent two experimental conditions (e.g., GM crop versus non-GM counterpart, or different locations of growth). These 47 spots trigger a false positive rate of 7.2% of the total of spots.
Table 2a.
Qualitative between-extracts variability for all polypeptides.
| Reproducible spots in 4 replicate gels | Number of spots |
False positive rate (%) | ||
|---|---|---|---|---|
| Present in both extracts | Only present in one extract | |||
| Extract 1 | 623 | 609 | 47 | 7.2 |
| Extract 2 | 642 | |||
Number of reproducible spots in assay A.
3.2.2. Quantitative variability
Mean intensity of each spot was calculated from the 4 replicate gels of each extract. For each spot, the ratio mean intensity in extract 1/mean intensity in extract 2 was calculated. To compare the ratio of mean intensities of each spot between extracts, one of these 2 ranges of ratio [0.8–1.2[or [0.5–2.0[was used. If the ratio was within either one of these ranges, the intensity of the spot was considered homogenous between both extracts. If the ratio was out of either one of these ranges, the intensity of the spot was considered to be non-homogeneous.
When the range [0.8–1.2[was used, 292 spots out of 609 were non-homogenous (Table 2b). This meant that for 292 spots, the mean intensity in one extract was at least 1.2-fold higher than in the other extract. These 292 spots represented false positives, with a false positive rate of 47.9% of the total of spots. As this rate was considered unacceptably high, the same approach was used with the range [0.5–2.0[. In this case, only 33 spots out of 609 were non-homogenous (Table 2b). Therefore, for 33 spots, the mean intensity in one extract was at least 2-fold higher than in the other extract. These 33 spots represented a false positive rate of 5.4% of the total of spots. This less stringent cut-off range of [0.5–2.0[, which defined homogenous intensities was in line with other publications [4], [46]. For example, Zhan and Desiderio [46] found that the mean ratio between two sets of gels, loaded similarly, ranged between 2.35 and 2.48 for the matched spots.
Table 2b.
Quantitative between-extracts variability for all polypeptides.
| Ratio mean intensities in extract 1/extract 2 | Fold difference | Number of spots | False positive rate (%) |
|---|---|---|---|
| Ratio <0.8 or ratio ≥1.2 | 1.2 | 292 | 47.9 |
| Ratio <0.5 or ratio ≥2.0 | 2 | 33 | 5.4 |
Number of spots in assay A.
Therefore, the quantitative evaluation of false positive rates showed that the 2-DE technique could detect a 2-fold change between two samples with an acceptable rate of false positives, but had a limited sensitivity for detecting a 1.2-fold change. Depending on the nature of samples and the goal of experiment, this can be a limitation of the 2-DE technique. However, this level of sensitivity must also be regarded in the context of biological variability of proteins in crops. Biological variability can be caused by crop variety (genetic background and/or genetic modification), growing conditions (e.g. latitude, weather conditions, diseases, soil, and exposure to sun) and/or maturity stage. For example, by comparing the profiles of the two transgenic lines of maize grown in the same location, Barros et al. [2] found a distinct separation between the three growing seasons. A highnumber of data suggested that biological variability of proteins in crops was high. In potato, differences were found among genotypes with 1077 of 1111 protein spots analyzed showing statistical significant differences [23]. In maize, median differences of 5–6-fold, and up to 55-fold, were observed among genotypes and growing locations [1]. In Arabidopsis thaliana, 95% of the spots present in all geographic variants (ecotypes) varied in spot quantity from 2- to 53-fold [37]. These data suggested that the biological variability was higher than the technical variability of 2-DE.
3.3. Between-assays variability
Between-assays variability refers to the comparison between two extracts prepared at different days as descripted in Fig. 1 (Extract 1 in assay B versus extract 1 in assay C).
3.3.1. Qualitative variability
A total of 623 spots were present in both assay B and assay C. In addition, 52 spots were present only in one assay (Table 3a). These 52 spots represented false positives, with a false positive rate of 7.7% of the total of spots. This false positive rate was very similar to that of the between-extract variability (7.2%).
Table 3a.
Qualitative between-assays variability for all polypeptides.
| Number of reproducible spots | Number of spots |
False positive rate (%) | ||
|---|---|---|---|---|
| Present in one assay | Only present in both assays | |||
| Assay B | 672 | 623 | 52 | 7.7 |
| Assay C | 626 | |||
3.3.2. Quantitative variability
When the range of ratio [0.8–1.2[was used, 308 spots out of 623 were non-homogenous (Table 3b). This meant that for 308 spots, the mean intensity in one extract was at least 1.2-fold higher than in the other extract. These 308 spots represented a false positive rate of 49.4% of the total of spots. As this rate was considered unacceptably high, the same approach was used with the range [0.5–2.0[. In this case, only 38 spots out of 623 were non-homogenous (Table 3b). This meant that for 38 spots, the mean intensity in one extract was at least 2-fold higher than in the other extracts. These 38 spots represented a false positive rate of 6.1% of the total of spots. These false positive rates were very similar to those of the between-extracts variability.
Table 3b.
Quantitative between-assays variability for all polypeptides.
| Ratio mean intensities in assay B/assay C | Fold difference | Number of spots | False positive rate (%) |
|---|---|---|---|
| Ratio <0.8 or ratio ≥1.2 | 1.2 | 308 | 49.4 |
| Ratio <0.5 or ratio ≥2.0 | 2 | 38 | 6.1 |
Estimations of false positive rates were similar for between-extracts and between-assays; the variability of the number of reproducible spots and their quantities was not higher between assays than within assay. In other words, two assays conducted during different days did not introduce more variability than if they were conducted during the same day. This has practical consequences, as it indicates that an experiment combining several different assays can be split over several days if needed.
3.4. Between-assays/operators variability
Between-assays/operators variability refers to the comparison between two extracts prepared at different days by different operators (extract 1 in assay A versus extract 1 in assay B) (Fig. 1). This comparison gives the opportunity to assess between-assay/operator variability but without knowing the relative contribution of each factor.
3.4.1. Qualitative variability
A total of 616 spots were present in both assays. In addition, 63 spots were present only in one assay (Table 4a). These 63 spots represented false positives, with a false positive rate of 9.3% of the total of spots. Therefore, this false positive rate was very similar to that of the between-assays variability (7.7%).
Table 4a.
Qualitative between-assays/operators variability for all polypeptides.
| Number of reproducible spots | Number of spots |
False positive rate (%) | ||
|---|---|---|---|---|
| Present in both assays | Only present in one assay | |||
| Assay A | 623 | 616 | 63 | 9.3 |
| Assay B | 672 | |||
3.4.2. Quantitative variability
When the range of ratio [0.8–1.2[was used, 294 spots out of 616 were non-homogenous (Table 4b). This meant that for 294 spots, the mean intensity in one extract was at least 1.2-fold higher than in the other extract. These 294 spots represented a false positive rate of 47.7% of the total of spots. As this rate was considered unacceptably high, the same approach was used with the range [0.5–2.0[. In this case, only 41 spots out of 616 were non-homogenous (Table 4b). This meant that for 41 spots, the mean intensity in one extract was at least 2-fold higher than in the other extract. These 41 spots represented a false positive rate of 6.7% of the total of spots. These false positive rates were very similar to those of the between-assays variability.
Table 4b.
Quantitative between-assays/operators variability for all polypeptides.
| Ratio mean intensities in assay A/assay B | Fold difference | Number of spots | False positive rate (%) |
|---|---|---|---|
| Ratio <0.8 or ratio ≥1.2 | 1.2 | 294 | 47.7 |
| Ratio <0.5 or ratio ≥2.0 | 2 | 41 | 6.7 |
Between-assay/operator false positive estimations were comparable to between-extract and between-assay false positive rate values showing that the variable “operator” did not contribute to strongly increase the technical variability. These results were not consistent with several publications that showed that the operator variable was the major source of variability in 2-DE [22], [39]. However, they also showed that the proper training of operators had a major impact on variability. Some authors [7], [26] showed that involving an untrained operator in 2-DE resulted in a lack of reproducibility. Similarly, Schröder et al. [39] compared gels prepared from the same sample by trained and untrained operators. The variability of low intensity spots in gels generated by the untrained operator was always significantly higher. In our study, a standardized training of both operators and the use of precise standard operating procedures (Good Laboratory Practices environment) might explain that the operator was not a major source of variability.
3.5. Specific analysis of soybean allergens
3.5.1. Between-gels variability
After having compared all spots, the experiment focused on spots which typically represented soybean food allergens. Out of 20 spots corresponding to known allergens, 19 or 20 were detected in each gel from either extract 1 or extract 2. Therefore, the number of reproducible spots used was 19 for each extract. Quantitatively, the mean CV of intensity of these 19 spots ranged from 40.5% (extract 2) to 51.3% (extract 1) (Table 5).
Table 5.
Between-gels variability for allergens.
| Gel 1 | Gel 2 | Gel 3 | Gel 4 | Spots matched in 4 out of 4 gels | Mean CV (%) | |
|---|---|---|---|---|---|---|
| Extract 1 | 20 | 19 | 19 | 19 | 19 | 51.3 |
| Extract 2 | 19 | 20 | 19 | 19 | 19 | 40.5 |
Number of reproducible spots in assay A.
3.5.2. Between-extracts variability
Nineteen out of 20 spots were reproducibly present in the eight gels from extracts 1 and 2 (Table 6). Quantitative analysis showed that, when using the range of ratio [0.8–1.2[, 8 spots out of 19 were non-homogenous (Table 6). These 8 spots represented a false positive rate of 42.1% of the allergen spots, for which the mean intensity in one extract was at least 1.2-fold higher than in the other extract. Using the range [0.5–2.0[, no spots were non-homogenous, meaning that the rate of false positive was 0% (Table 6).
Table 6.
Between-extracts variability for allergens.
| Ratio mean intensities in extract 1/extract 2 | Fold difference | Number of spots | False positive rate (%) |
|---|---|---|---|
| Ratio <0.8 or Ratio ≥1.2 | 1.2 | 8 | 42.1 |
| Ratio <0.5 or Ratio ≥2.0 | 2 | 0 | 0.0 |
Number of spots in assay A.
3.5.3. Between-assay variability
All allergen spots (20 in total) were detected in the eight gels of both assays B and C. When using the range of ratio of [0.8–1.2[, 10 spots out of 20 were non-homogenous (Table 7). These 10 spots represented a false positive rate of 50.0% of the allergen spots, for which the mean intensity in one extract was at least 1.2-fold higher than in the other extract. Using the range [0.5–2.0[, no spots were non-homogenous, i.e. no false positives (Table 7).
Table 7.
Between-assays variability for allergens.
| Ratio mean intensities | Fold difference in assay B/assay C | Number of spots | False positive rate (%) |
|---|---|---|---|
| Ratio <0.8 or ratio ≥1.2 | 1.2 | 10 | 50.0 |
| Ratio <0.5 or ratio ≥2.0 | 2 | 0 | 0.0 |
3.5.4. Between-assays/operators variability
Nineteen out of 20 allergen spots were reproducibly present in the 8 gels of both assays A and B. Quantitatively, when using the range of ratio [0.8–1.2[, 8 spots out of 19 were non-homogenous (Table 8). These 8 spots represented a false positive rate of 42.1% of the allergen spots, for which the mean intensity in one extract was at least 1.2-fold higher than in the other extract. Using the range [0.5–2.0[, one spot was non-homogenous and the false positive rate was 5.2% (Table 8).
Table 8.
Between-assays/operators for allergens.
| Ratio mean intensities | Fold difference in assay A/assay B | Number of spots | False positive rate (%) |
|---|---|---|---|
| Ratio <0.8 or ratio ≥1.2 | 1.2 | 8 | 42.1 |
| Ratio <0.5 or ratio ≥2.0 | 2 | 1 | 5.2 |
3.5.5. Conclusion
Overall the results obtained with all detectable allergen spots (20 spots) were comparable to the results obtained with all detectable polypeptide spots, as expected. In addition, the results showed that the 2-DE technique could detect a 2-fold change between two samples with an acceptable rate of false positives. This level of sensitivity had to be put in perspective with the biological variability data on soybean food allergens. A number of scientific publications investigated qualitatively the biological variability of allergens [31], [30], [29]. However, few data were available on the magnitude of biological variability of soybean allergens. Houston et al. [19] showed that out of 13 polypeptides, corresponding to soybean food allergens, 4 showed at least a 2-fold change in abundance (and up to 7-fold) among 20 soybean varieties. In addition, Yaklich et al. [45] showed that in 18 commercial lines of soybean, the content of β-conglycinin and glycin subunits could be found statistically different, with a difference ranging from 47% to 157%. Overall, these data strongly suggested that the biological variability of soybean food allergens was higher than the technical variability of 2-DE observed in this study. So this 2-DE technique and laboratory procedures are suitable for assessing the soybean food allergen content and compare several soybean varieties, including GM soybean and its non-GM counterpart.
4. General conclusion
Based on this study design, the major source of variability inherent to the 2-DE technique was the number of gel replicates. In comparison, other sources of variability (extracts, assay timings, and operators) were low. This has a direct practical impact on the laboratory work as this supports the utilization of three or four replicates when comparing several experimental conditions. Furthermore, this implies that the study can be run over several days, and be performed by several trained operators, without impacting its reproducibility.
In addition, this study assessed the false positive rates, which helped to characterize the sources of variability in order to better control them in the future studies. The 2-DE technique could detect a 2-fold change between two samples with an acceptable rate of false positives (below 7%). This level of sensitivity is acceptable in the context of safety assessment of GM soybean as the biological variability of proteins in soybean, which can be due to crop variety (genetic background), growing conditions (e.g. latitude, weather conditions, diseases, soil, exposure to sun) and/or maturity stage, is higher than the technical variability shown in this study. Therefore, the 2-DE technique is suitable for detecting biologically significant differences in protein expression levels in different soybean varieties, including the GM and its non-GM counterpart.
Overall, the 2-DE technique is suitable for investigating endogenous food allergen variability between several soybean seeds, including GM and non-GM counterpart.
Conflict of interest
None declared.
Transparency document
Acknowledgement
The authors acknowledge Dr. David Rouquié for its contribution in coordinating the laboratory work and for its detailed review of the article.
Footnotes
Available online 16 September 2014
References
- 1.Anttonen M.J., Lehesranta S., Auriola S., Röhlig R.M., Engel K.H., Kärenlampi S.O. Genetic and environmental influence on maize kernel proteome. J. Proteome Res. 2010;9(12):6160–6168. doi: 10.1021/pr100251p. [DOI] [PubMed] [Google Scholar]
- 2.Barros E., Lezar S., Anttonen M.J., van Dijk J.P., Röhlig R.M., Kok E.J., Engel K.H. Comparison of two GM maize varieties with a near-isogenic non-GM variety using transcriptomics, proteomics and metabolomics. Plant Biotechnol. J. 2010;8(4):436–451. doi: 10.1111/j.1467-7652.2009.00487.x. [DOI] [PubMed] [Google Scholar]
- 3.Bland A.M., D’Eugenio L.R., Dugan M.A., Janech M.G., Almeida J.S., Zile M.R., Arthur J.M. Comparison of variability associated with sample preparation in two-dimensional gel electrophoresis of cardiac tissue. J. Biomol. Tech. 2006;17(3):195–199. [PMC free article] [PubMed] [Google Scholar]
- 4.Bland A.M., Janech M.G., Almeida J.S., Arthur J.M. Sources of variability among replicate samples separated by two-dimensional gel electrophoresis. J. Biomol. Tech. 2010;21(1):3–8. [PMC free article] [PubMed] [Google Scholar]
- 5.Blomberg A., Blomberg L., Norbeck J., Fey S.J., Larsen P.M., Larsen M., Roepstorff P., Degand H., Boutry M., Posch A. Interlaboratory reproducibility of yeast protein patterns analyzed by immobilized pH gradient two-dimensional gel electrophoresis. Electrophoresis. 1995;16(10):1935–1945. doi: 10.1002/elps.11501601320. [DOI] [PubMed] [Google Scholar]
- 6.Burton E.O., Hickey W.J. Assessing variability in gel-based proteomic analysis of Nitrosomonas europaea. Methods Enzymol. 2011;496:435–463. doi: 10.1016/B978-0-12-386489-5.00018-X. [DOI] [PubMed] [Google Scholar]
- 7.Challapalli K.K., Zabel C., Schuchhardt J., Kaindl A.M., Klose J., Herzel H. High reproducibility of large-gel two-dimensional electrophoresis. Electrophoresis. 2004;25(17):3040–3047. doi: 10.1002/elps.200405979. [DOI] [PubMed] [Google Scholar]
- 8.Chich J.F., David O., Villers F., Schaeffer B., Lutomski D., Huet S. Statistics for proteomics: experimental design and 2-DE differential analysis. J. Chromatogr. B. Analyt. Technol. Biomed. Life Sci. 2007;849(1–2):261–272. doi: 10.1016/j.jchromb.2006.09.033. [DOI] [PubMed] [Google Scholar]
- 9.Choe L.H., Lee K.H. Quantitative and qualitative measure of intralaboratory two-dimensional protein gel reproducibility and the effects of sample preparation, sample load, and image analysis. Electrophoresis. 2003;24(19–20):3500–3507. doi: 10.1002/elps.200305614. [DOI] [PubMed] [Google Scholar]
- 10.Clark B.N., Gutstein H.B. The myth of automated, high-throughput two-dimensional gel analysis. Proteomics. 2008;8(6):1197–1203. doi: 10.1002/pmic.200700709. [DOI] [PubMed] [Google Scholar]
- 11.Corbett J.M., Dunn M.J., Posch A., Görg A. Positional reproducibility of protein spots in two-dimensional polyacrylamide gel electrophoresis using immobilised pH gradient isoelectric focusing in the first dimension: an interlaboratory comparison. Electrophoresis. 1994;15(8–9):1205–1211. doi: 10.1002/elps.11501501182. [DOI] [PubMed] [Google Scholar]
- 12.Doerrer N., Ladics G., McClain S., Herouet-Guicheney C., Poulsen L.K., Privalle L., Stagg N. Evaluating biological variation in non-transgenic crops: executive summary from the ILSI Health and Environmental Sciences Institute workshop, November 16–17, 2009, Paris, France. Regul. Toxicol. Pharmacol. 2010;58(3 Suppl.):S2–S7. doi: 10.1016/j.yrtph.2010.06.017. [DOI] [PubMed] [Google Scholar]
- 13.EFSA Guidance for risk assessment of food and feed from genetically modified plants – EFSA panel on genetically modified organisms (GMO) EFSA J. 2011;9:2150–2187. [Google Scholar]
- 14.Friry-Santini C., Rouquié D., Kennel P., Tinwell H., Benahmed M., Bars R. Correlation between protein accumulation profiles and conventional toxicological findings using a model antiandrogenic compound, flutamide. Toxicol. Sci. 2007;97(1):81–93. doi: 10.1093/toxsci/kfm022. [DOI] [PubMed] [Google Scholar]
- 15.Fuxius S., Eravci M., Broedel O., Weist S., Mansmann U., Eravci S., Baumgartner A. Technical strategies to reduce the amount of false significant results in quantitative proteomics. Proteomics. 2008;8(9):1780–1784. doi: 10.1002/pmic.200701074. [DOI] [PubMed] [Google Scholar]
- 16.Goldfarb M. Computer analysis of two-dimensional gels. J. Biomol. Tech. 2007;18(3):143–146. [PMC free article] [PubMed] [Google Scholar]
- 17.Gorg A., Weiss W., Dunn M.J. Current two-dimensional electrophoresis technology for proteomics. Proteomics. 2004;4(12):3665–3685. doi: 10.1002/pmic.200401031. [DOI] [PubMed] [Google Scholar]
- 18.Hajduch M., Ganapathy A., Stein J.W., Thelen J.J. A systematic proteomic study of seed filling in soybean. Establishment of high-resolution two-dimensional reference maps, expression profiles, and an interactive proteome database. Plant Physiol. 2005;137(4):1397–1419. doi: 10.1104/pp.104.056614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Houston N.L., Lee D.G., Stevenson S.E., Ladics G.S., Bannon G.A., McClain S., Privalle L., Stagg N., Herouet-Guicheney C., MacIntosh S.C., Thelen J.J. Quantitation of soybean allergens using tandem mass spectrometry. J. Proteome Res. 2011;10(2):763–773. doi: 10.1021/pr100913w. [DOI] [PubMed] [Google Scholar]
- 20.Hunt S.M., Thomas M.R., Sebastian L.T., Pedersen S.K., Harcourt R.L., Sloane A.J., Wilkins M.R. Optimal replication and the importance of experimental design for gel-based quantitative proteomics. J. Proteome Res. 2005;4(3):809–819. doi: 10.1021/pr049758y. [DOI] [PubMed] [Google Scholar]
- 21.Karp N.A., Lilley K.S. Design and analysis issues in quantitative proteomics studies. Proteomics. 2007;7(Suppl. 1):42–50. doi: 10.1002/pmic.200700683. [DOI] [PubMed] [Google Scholar]
- 22.Koller A., Wätzig H. Precision and variance components in quantitative gel electrophoresis. Electrophoresis. 2005;26(12):2470–2475. doi: 10.1002/elps.200500024. [DOI] [PubMed] [Google Scholar]
- 23.Lehesranta S.J., Davies H.V., Shepherd L.V., Nunan N., McNicol J.W., Auriola S., Koistinen K.M., Suomalainen S., Kokko H.I., Kärenlampi S.O. Comparison of tuber proteomes of potato varieties, landraces, and genetically modified lines. Plant Physiol. 2005;138(3):1690–1699. doi: 10.1104/pp.105.060152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lopez M.F., Patton W.F. Reproducibility of polypeptide spot positions in two-dimensional gels run using carrier ampholytes in the isoelectric focusing dimension. Electrophoresis. 1997;18(3–4):338–343. doi: 10.1002/elps.1150180307. [DOI] [PubMed] [Google Scholar]
- 25.Mahon P., Dupree P. Quantitative and reproducible two-dimensional gel analysis using Phoretix 2D Full. Electrophoresis. 2001;22(10):2075–2085. doi: 10.1002/1522-2683(200106)22:10<2075::AID-ELPS2075>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 26.Millioni R., Sbrignadello S., Tura A., Iori E., Murphy E., Tessari P. The inter- and intra-operator variability in manual spot segmentation and its effect on spot quantitation in two-dimensional electrophoresis analysis. Electrophoresis. 2010;31(10):1739–1742. doi: 10.1002/elps.200900674. [DOI] [PubMed] [Google Scholar]
- 27.Millioni R., Puricelli L., Sbrignadello S., Iori E., Murphy E., Tessari P. Operator- and software-related post-experimental variability and source of error in 2-DE analysis. Amino Acids. 2012;42(5):1583–1590. doi: 10.1007/s00726-011-0873-7. [DOI] [PubMed] [Google Scholar]
- 28.Molloy M.P., Brzezinski E.E., Hang J., McDowell M.T., Van Bogelen R.A. Overcoming technical variation and biological variation in quantitative proteomics. Proteomics. 2003;3(10):1912–1919. doi: 10.1002/pmic.200300534. [DOI] [PubMed] [Google Scholar]
- 29.Natarajan S.S., Xu C., Bae H., Caperna T.J., Garrett W.M. Characterization of storage proteins in wild (Glycine soja) and cultivated (Glycine max) soybean seeds using proteomic analysis. J. Agric. Food Chem. 2006;54(8):3114–3120. doi: 10.1021/jf052954k. [DOI] [PubMed] [Google Scholar]
- 30.Natarajan S., Xu C., Bae H., Bailey B.A., Cregan P., Caperna T.J., Garrett W.M., Luthria D. Proteomic and genetic analysis of glycinin subunits of sixteen soybean genotypes. Plant Physiol. Biochem. 2007;45(6–7):436–444. doi: 10.1016/j.plaphy.2007.03.031. [DOI] [PubMed] [Google Scholar]
- 31.Natarajan S.S., Xu C., Cregan P., Caperna T.J., Garrett W.M., Luthria D. Utility of proteomics techniques for assessing protein expression. Regul. Toxicol. Pharmacol. 2009;54(Suppl. 3):S32–S36. doi: 10.1016/j.yrtph.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 32.Norbeck A.D., Callister S.J., Monroe M.E., Jaitly N., Elias D.A., Lipton M.S., Smith R.D. Proteomic approaches to bacterial differentiation. J. Microbiol. Methods. 2006;67(3):473–486. doi: 10.1016/j.mimet.2006.04.024. [DOI] [PubMed] [Google Scholar]
- 33.Olsson I., Larsson K., Palmgren R., Bjellqvist B. Organic disulfides as a means to generate streak-free two-dimensional maps with narrow range basic immobilized pH gradient strips as first dimension. Proteomics. 2002;2:1630–1632. doi: 10.1002/1615-9861(200211)2:11<1630::AID-PROT1630>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 34.Ramagli L.S., Rodriguez L.V. Quantitation of microgram amounts of protein in two-dimensional polyacrylamide gel electrophoresis sample buffer. Electrophoresis. 1985;6:559–563. [Google Scholar]
- 35.Righetti P.G., Campostrini N., Pascali J., Hamdan M., Astner H. Quantitative proteomics: a review of different methodologies. Eur. J. Mass. Spectrom. (Chichester, Eng.) 2004;10(3):335–348. doi: 10.1255/ejms.600. [DOI] [PubMed] [Google Scholar]
- 36.Rouquie D., Capt A., Eby W.H., Sekar V., Herouet-Guicheney C. Investigation of endogenous soybean food allergens by using a 2-dimensional gel electrophoresis approach. Regul. Toxicol. Pharmacol. 2010;58:S47–S53. doi: 10.1016/j.yrtph.2010.09.013. [DOI] [PubMed] [Google Scholar]
- 37.Ruebelt M.C., Leimgruber N.K., Lipp M., Reynolds T.L., Nemeth M.A., Astwood J.D., Engel K.H., Jany K.D. Application of two-dimensional gel electrophoresis to interrogate alterations in the proteome of genetically modified crops. 1. Assessing analytical validation. J. Agric. Food Chem. 2006;54(6):2154–2161. doi: 10.1021/jf0523566. [DOI] [PubMed] [Google Scholar]
- 38.Schlags W., Walther M., Masree M., Kratzel M., Noe C.R., Lachmann B. Towards validating a method for two-dimensional electrophoresis/silver staining. Electrophoresis. 2005;26(12):2461–2469. doi: 10.1002/elps.200410347. [DOI] [PubMed] [Google Scholar]
- 39.Schröder S., Zhang H., Yeun E.S., Jänsch L., Zabel C., Wätzig H. Quantitative gel electrophoresis: sources of variation. J. Proteome Res. 2008;7(3):1226–1234. doi: 10.1021/pr700589s. [DOI] [PubMed] [Google Scholar]
- 40.Stevenson S.E., Woods C.A., Hong B., Kong X., Thelen J.J., Ladics G.S. Environmental effects on allergen levels in commercially grown non-genetically modified soybeans: assessing variation across North America. Front. Plant Sci. 2012;3(196):1–13. doi: 10.3389/fpls.2012.00196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Terry D.E., Desiderio D.M. Between-gel reproducibility of the human cerebrospinal fluid proteome. Proteomics. 2003;3(10):1962–1979. doi: 10.1002/pmic.200300463. [DOI] [PubMed] [Google Scholar]
- 42.Teshima R., Nakamura R., Satoh R., Nakamura R. 2D-DIGE analysis of rice proteins from different cultivars. Regul. Toxicol. Pharmacol. 2010;58:S30–S35. doi: 10.1016/j.yrtph.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 43.Voss T., Haberl P. Observations on the reproducibility and matching efficiency of two-dimensional electrophoresis gels: consequences for comprehensive data analysis. Electrophoresis. 2000;21(16):3345–3350. doi: 10.1002/1522-2683(20001001)21:16<3345::AID-ELPS3345>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 44.Wheelock A.M., Buckpitt A.R. Software-induced variance in two-dimensional gel electrophoresis image analysis. Electrophoresis. 2005;26(23):4508–4520. doi: 10.1002/elps.200500253. [DOI] [PubMed] [Google Scholar]
- 45.Yaklich R.W. beta-Conglycinin and glycinin in high-protein soybean seeds. J. Agric. Food Chem. 2001;49(2):729–735. doi: 10.1021/jf001110s. [DOI] [PubMed] [Google Scholar]
- 46.Zhan X., Desiderio D.M. Differences in the spatial and quantitative reproducibility between two second-dimensional gel electrophoresis systems. Electrophoresis. 2003;24(11):1834–1846. doi: 10.1002/elps.200305389. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.