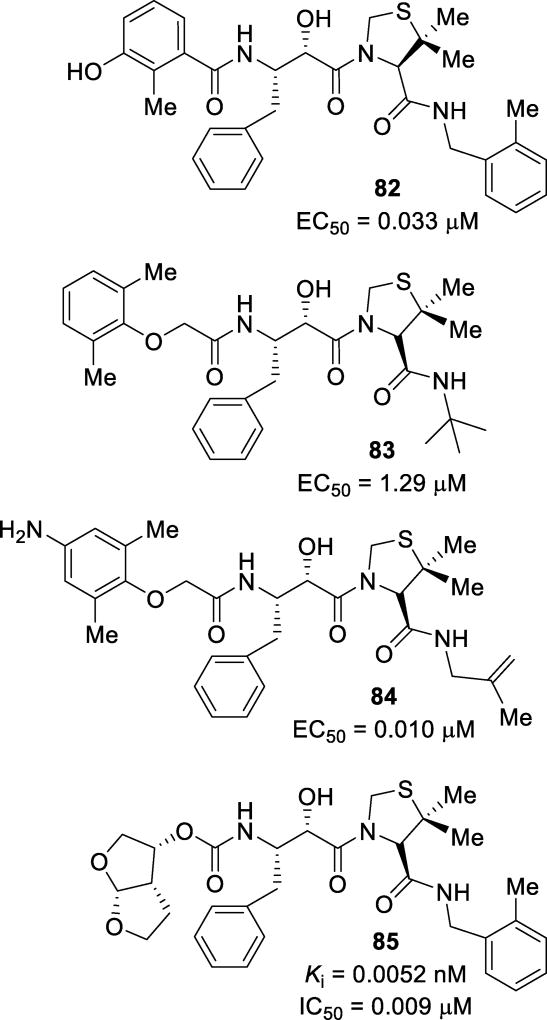
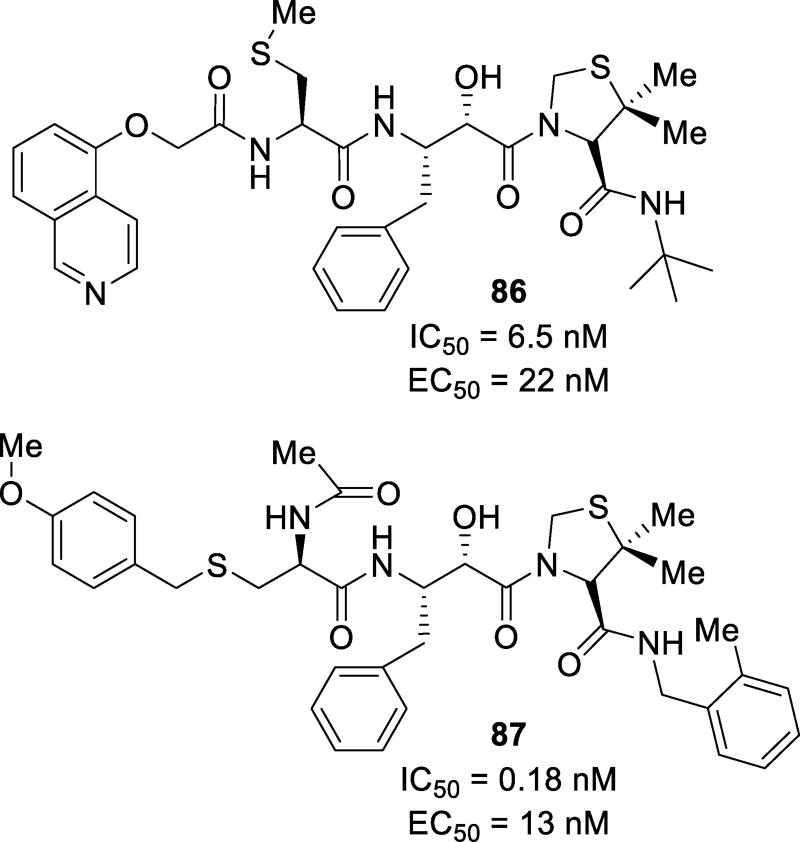
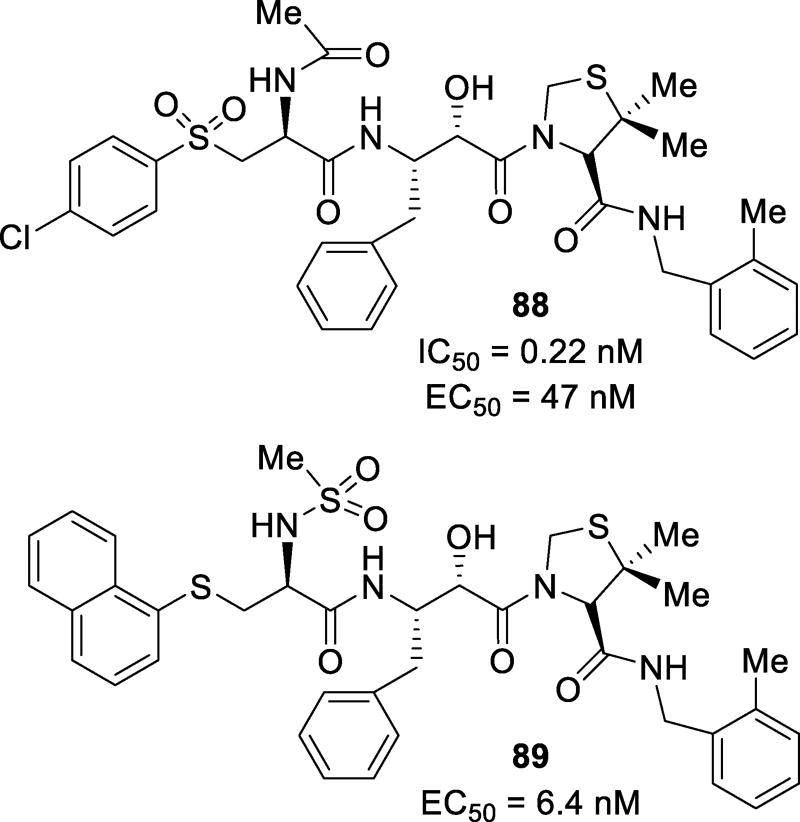
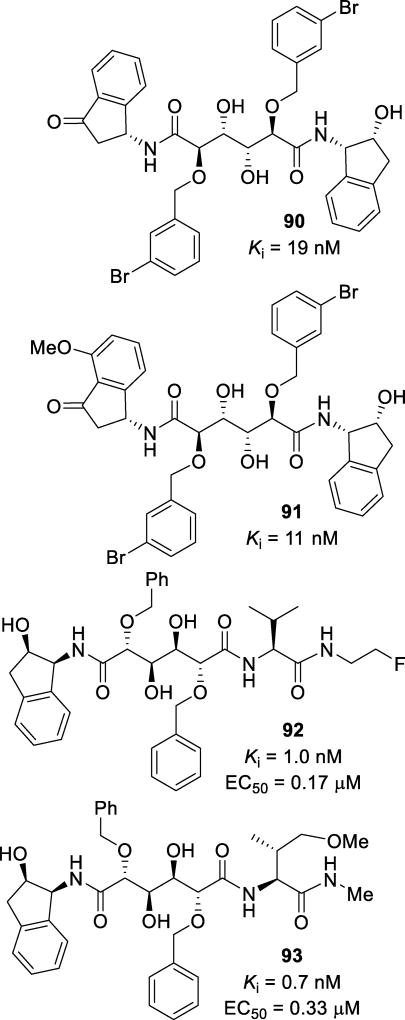
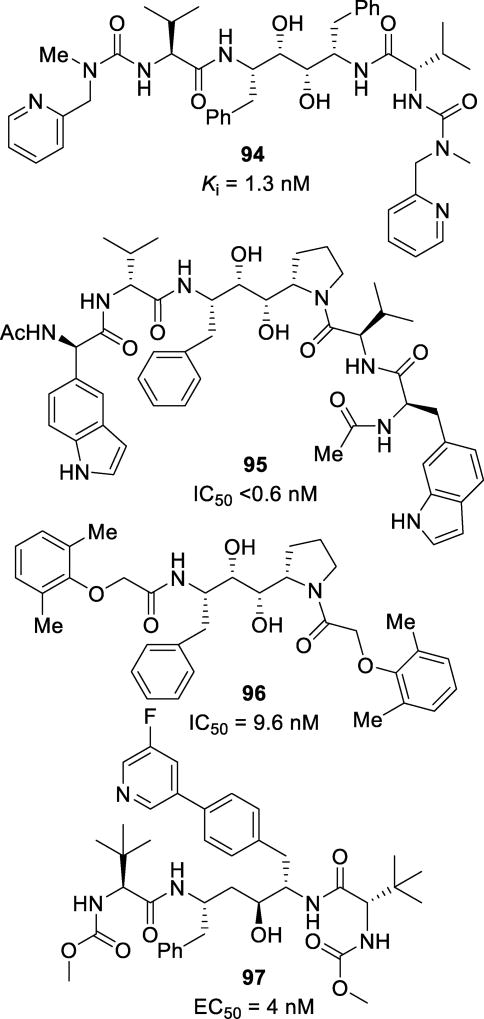
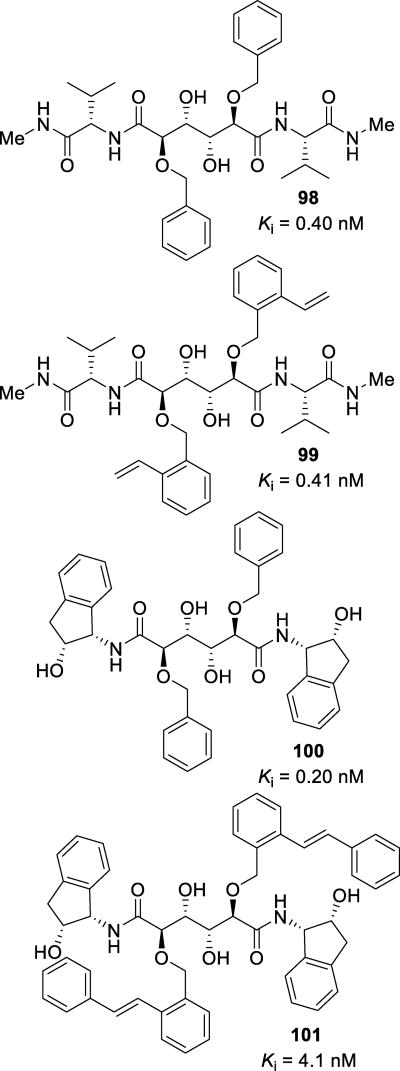
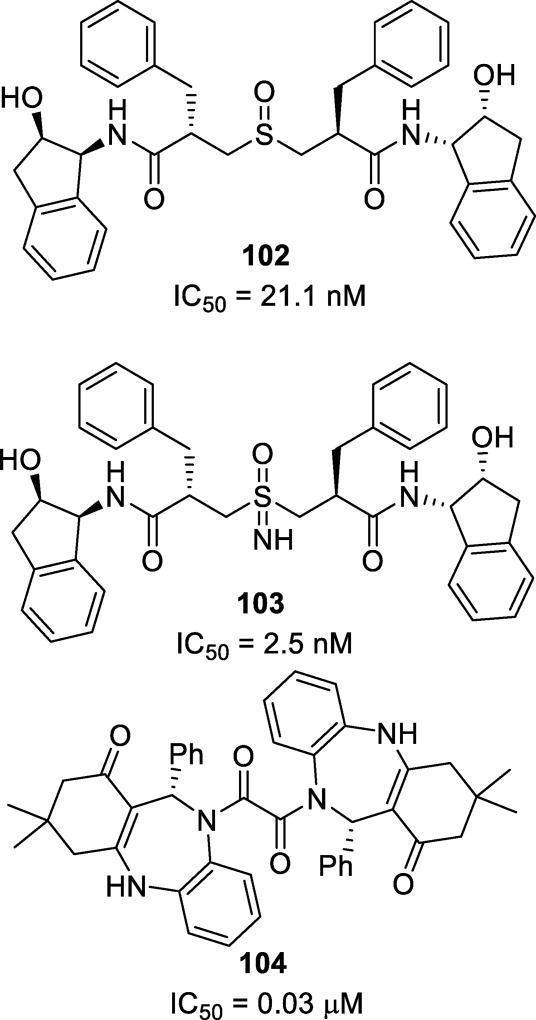
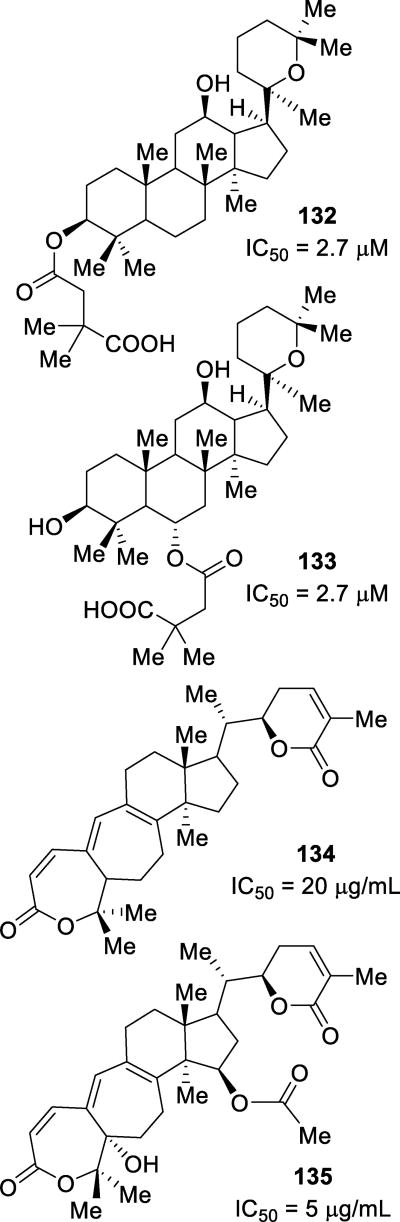
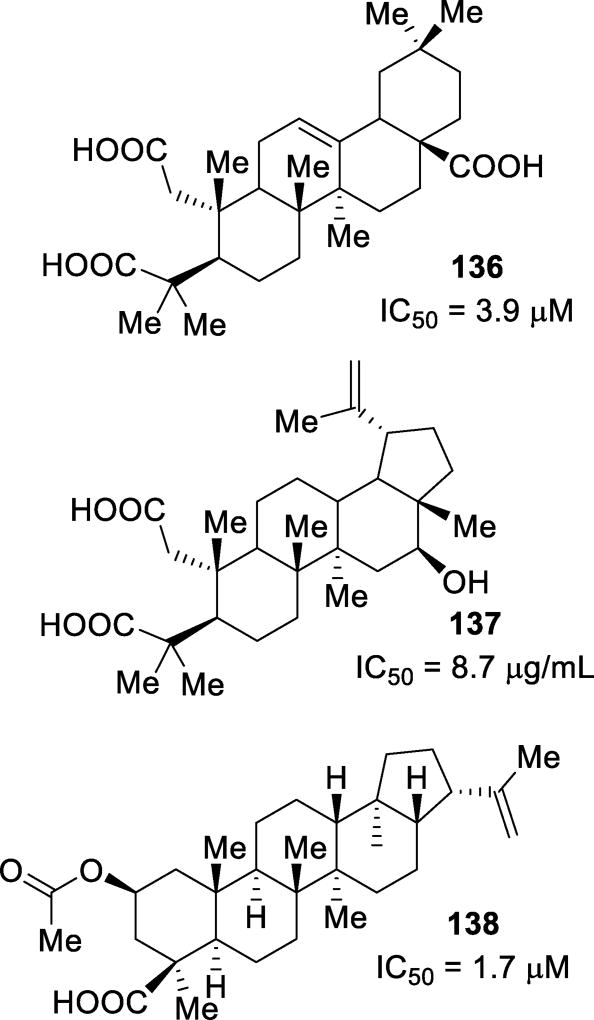
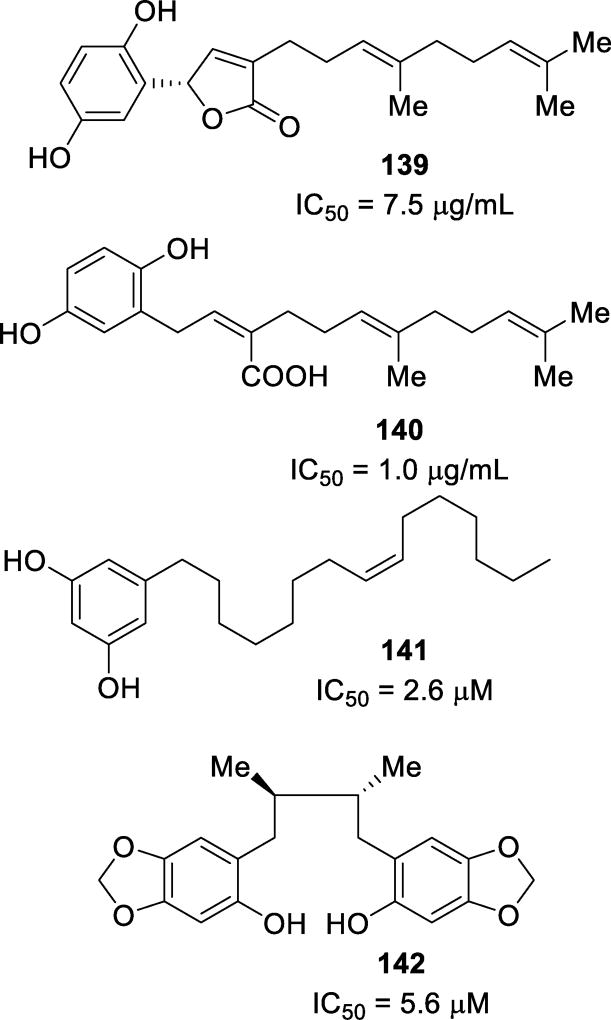
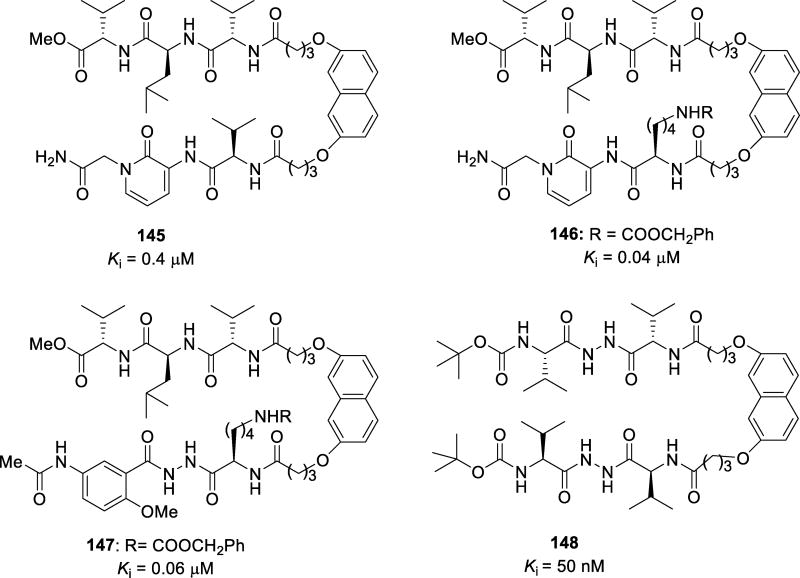
Abstract
HIV-1 protease inhibitors continue to play an important role in the treatment of HIV/AIDS, transforming this deadly ailment into a more manageable chronic infection. Over the years, intensive research led to a variety of approved protease inhibitors for the treatment of HIV/AIDS. In this review, we outline current drug design and medicinal chemistry efforts toward the development of new generation protease inhibitors beyond the currently approved drugs.
Graphical Table of Content
1. Introduction
It has been over three decades since human immunodeficiency virus (HIV), the causative agent for acquired immunodeficiency syndrome (AIDS), was identified.1, 2 Since the beginning of the global pandemic of HIV/AIDS in the early 1980’s, an estimated 78 million people have been infected with HIV and about 39 million people have died of AIDS-related causes according to the Joint United Nations Program on HIV/AIDS (UNAIDS). An estimated 37 million people worldwide are now living with HIV/AIDS.3, 4 These statistics are quite staggering by any measure. By the latter half of the 1980’s, advancements in the knowledge of HIV pathogenesis, biology, and pharmacology led to unprecedented efforts to translate basic findings into the development of novel antiviral drug therapies.5, 6 The progression and continuous evolution of antiretroviral therapy for HIV/AIDS treatment is quite unique in the history of medicine. Currently, there exists no treatment to eradicate the virus from an infected patient. However, the development of multiple therapeutic agents targeting various steps of the HIV life cycle helped transform HIV infection from an inevitably fatal disease into a manageable chronic ailment. This has resulted in dramatic improvement in HIV-related morbidity and mortality, particularly in developed countries where patients have access to potent antiretroviral drug combinations that allow sustained control of viral replication and combat drug-resistant virus.7, 8
The discovery of HIV as the causative agent and molecular events critical to HIV replication initially identified a number of important biochemical targets including reverse transcriptase (RT), protease (PR), and integrase (IN) for antiviral therapy development.9, 10 Nucleoside reverse transcriptase inhibitors were the first agents approved for the treatment of HIV infection by interfering with the transcription of double stranded viral RNA into DNA.11 Therapeutic inhibition of virally encoded HIV-1 protease was then specifically targeted since this enzyme plays a critical role in processing the gag and gag-pol gene product into essential viral proteins required for assembly of a new mature virus. An immense effort in the development of HIV-1 protease inhibitor drugs followed. The approval of several HIV-1 protease inhibitor drugs in the mid-1990’s and their combination with reverse transcriptase inhibitors marked the beginning of highly active antiretroviral therapy (HAART).12, 13 It became evident that combination chemotherapy was significantly more effective than dosing the drugs sequentially.14 The advent of HAART has resulted in dramatic improvement in HIV/AIDS treatment. Today, many different treatment regimens are known and new therapies with other targets including integrase inhibitors, viral attachment inhibitors, and membrane fusion inhibitors have been developed. Treatment regimens aim to be potent, convenient, well tolerated, and typically reduce HIV blood concentration to undetectable levels within a few weeks of treatment. Antiretroviral therapy (ART) regimes typically induce a robust and sustained increase of CD4 T-cell counts.7, 8
Despite major advances in HIV/AIDS therapies, there are significant drawbacks to current treatments. Drugs must be taken lifelong with unknown long-term side effects. Drug toxicity, drug-drug interactions, and evolution of different patterns of systemic complications involving heart, kidney, bone and other organs have emerged.6, 8 Since the central nervous system (CNS) is a major sanctuary for HIV-1 infection, HIV-1 associated neurocognitive disorders are increasing, possibly due to poor CNS penetration of current anti-HIV therapies.15, 16 Perhaps, the most alarming problem is the emergence of drug resistance, rendering current therapies ineffective within months in some cases. This has become a formidable challenge and may unravel the progress achieved toward HIV/AIDS management.17, 18 One of the greatest challenges that the World Health Organization faces today is that a large population of HIV infected patients are not diagnosed and treated until a late stage of the disease. This is due to limited diagnosis and ineffective treatment in areas like Africa and developing countries which contribute to nearly 70% of the global cases of HIV infection.4, 7 Some progress has been made in sub-Saharan Africa but significant challenges remain. This review will describe the progress made towards the development of novel next-generation protease inhibitors since the approval of darunavir, the most recent FDA-approved PI.19–21
2. HIV-1 Protease: Structure, Function, and Therapeutic Target
HIV-1 protease is responsible for the production of all viral enzymes and structural proteins necessary to produce mature, virulent virions. During replication, HIV infects T-cells via membrane fusion. Viral RNA then enters the cell and is turned into DNA via RT. The DNA enters the nucleus of the cell and is incorporated into the host cell’s DNA by IN. HIV then exploits the natural transcription and translation mechanism of the host cell to provide the viral polyprotein. The polypeptide is then hydrolyzed into mature proteins by PR. The viral RNA and proteins then accumulate at the cell surface and are released as infectious virions. Inhibition of PR represents intervention at a vital stage in the HIV life cycle.22
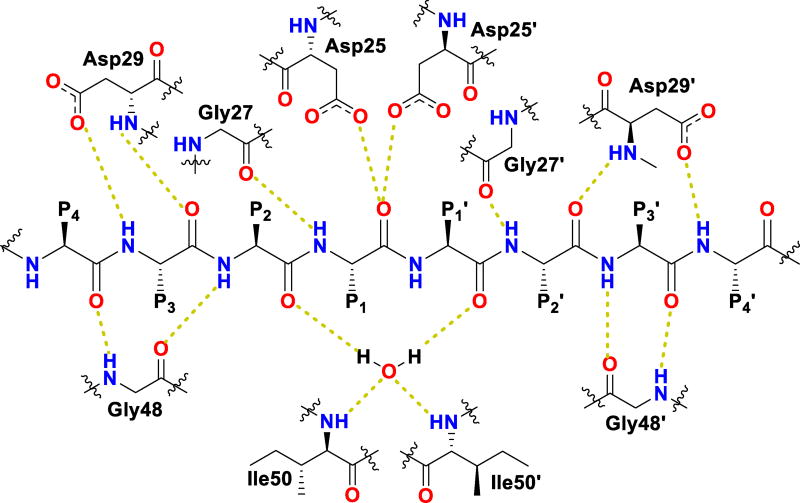
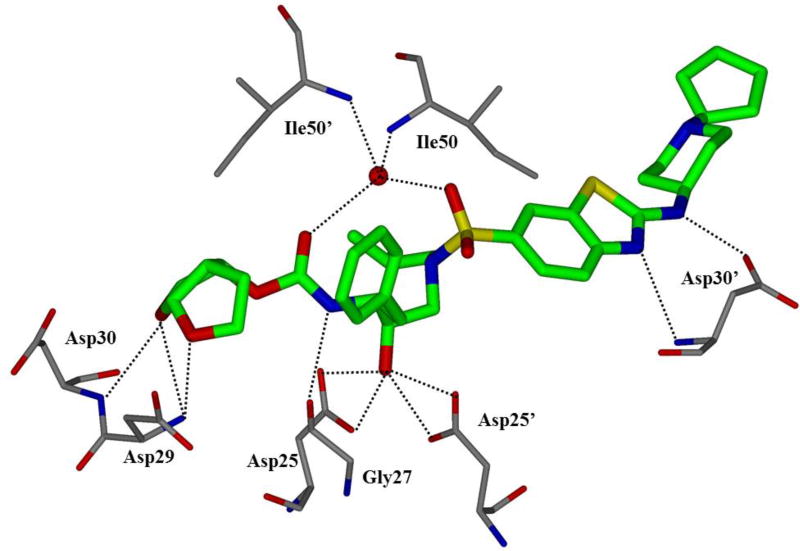
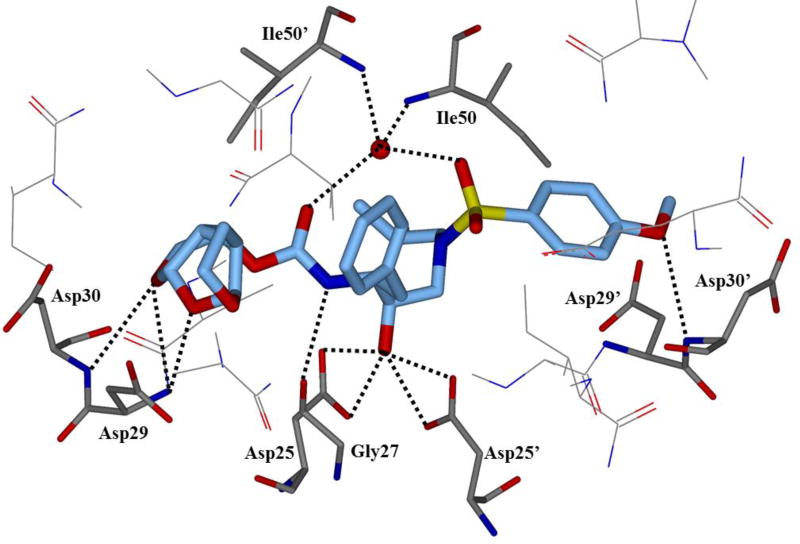
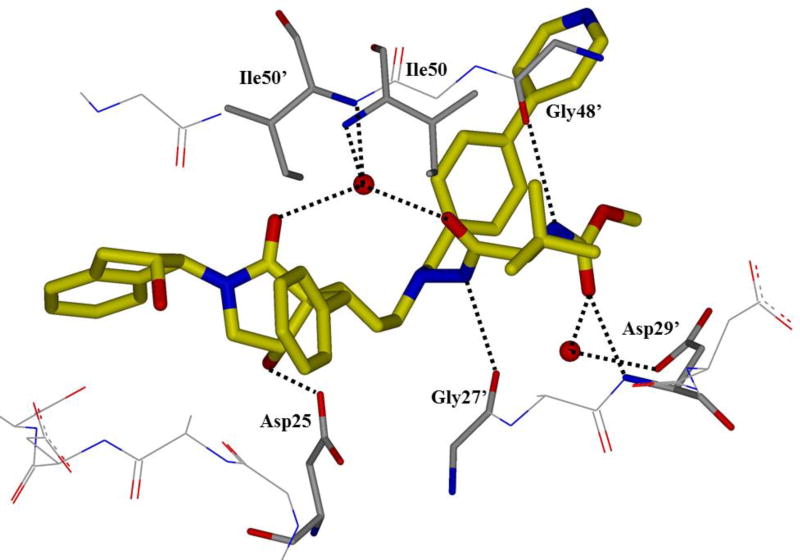
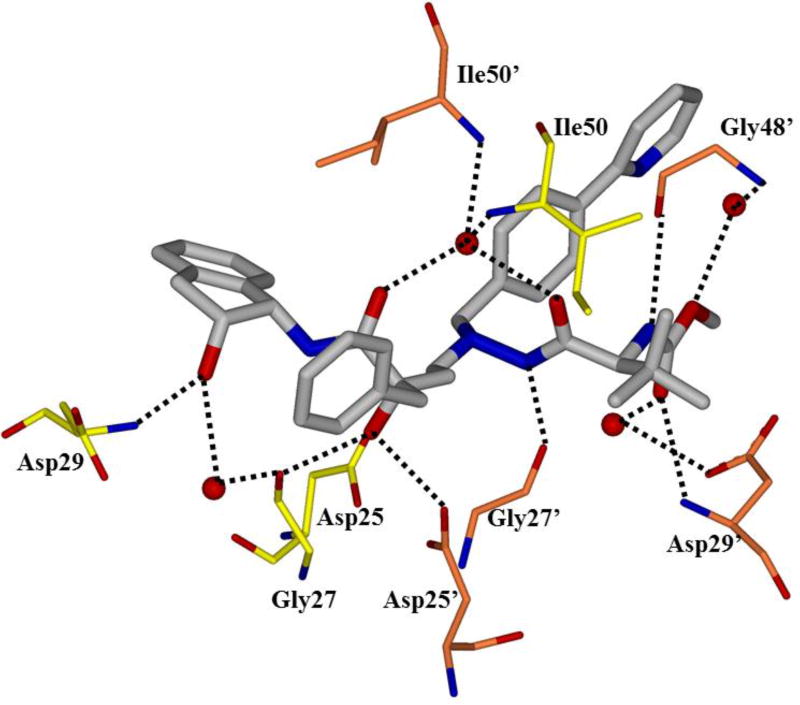
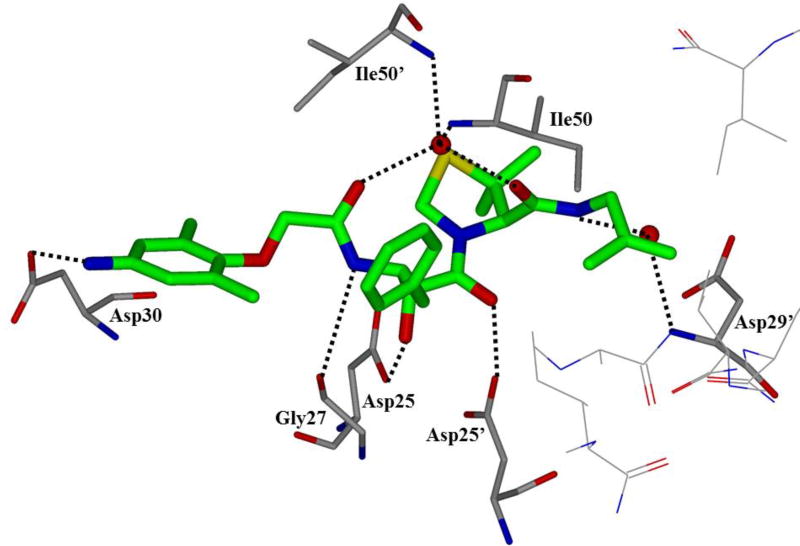
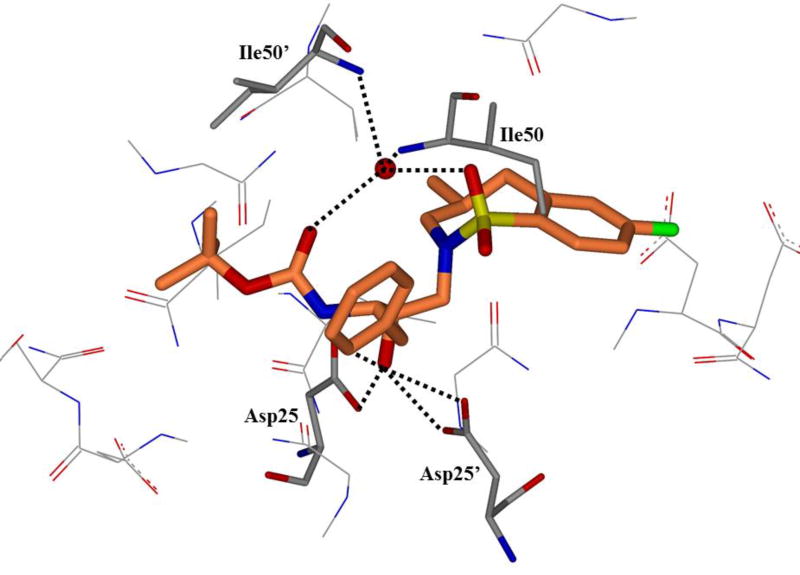
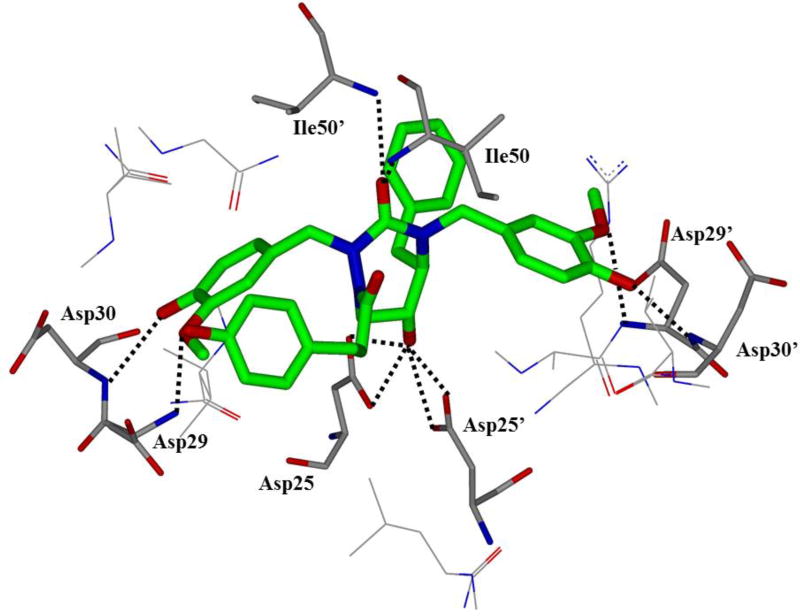
HIV protease catalyzes the hydrolysis of the Gag and Gag-Pol polyproteins at various cleavage sites which produce the structural proteins, such as the viral envelope glycoproteins, and the RT, IN, and PR enzymes for the new virion particles. X-ray crystallographic analysis provided insight into protease structure and function.23, 24 PR is a homodimer of two 99 amino acid subunits. Catalytic aspartic acid residues, Asp25 and Asp25’, one from each monomer, meet at the dimer interface and form the catalytic active site of the enzyme.22 Two flexible glycine-dense β-sheets form a flap region over the top of the active site. These flaps experience a conformational shift to close over the active site when the enzyme is bound to a substrate. Native substrate binds to the enzyme in an extended conformation with a minimum of seven amino acid residues interacting with the enzyme, indicated as P4 to P1 and P1’ to P4’ by standard nomenclature (Figure 1). Hydrolysis of the amide bond occurs between the P1 and P1’ residues. The enzyme is highly specific; each subsite has preference for which type of side chain can be accommodated. The S1 and S1’ subsites prefer hydrophobic residues, as do the S3 and S3’ subsites, while the S2 and S2’ subsites can accommodate both polar and hydrophobic side chains.25 Analysis of fifteen different peptidic, early inhibitors of HIV protease shed light onto the desired binding pattern of PR (Figure 1).26 Mean hydrogen bond lengths were 2.68 to 3.24 Å. Overall, hydrogen bonding distances were shortest between the structural water molecules and the inhibitor with an average distance of 2.86 Å while direct inhibitor-active site hydrogen bonds were slightly longer with an average length of 3.11 Å. It was shown that peptidic inhibitors and, by extension, substrates form stronger hydrogen bonds with the flexible flap regions consisting of residues 48–50 than with the more rigid portion of the active site consisting of residues 25–29.26
Figure 1.
Conserved binding mode of peptides in the HIV protease active site. Hydrogen bonds are shown as yellow dashed lines
3. First-Generation Protease Inhibitors and the Advent of HAART
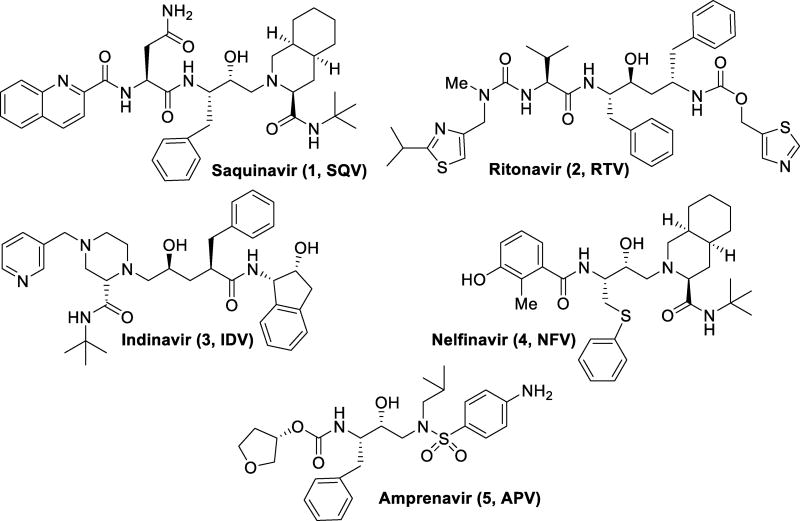
Initial therapies for HIV/AIDS patients consisted of nucleoside reverse transcriptase inhibitor monotherapy such as zidovudine or AZT. Introduction of protease inhibitors (PIs) to the market began with saquinavir in 1995 (1, Figure 2). The FDA approval of saquinavir marked the beginning of combination therapy for HIV/AIDS patients known as HAART. HAART therapy with a RT inhibitor greatly improved patient outcome by reducing viral loads, improving CD4 cell counts, and halting the progression to AIDS.25, 27, 28
Figure 2.
First-generation HIV protease inhibitors
First-generation inhibitors were based on hydroxyethylene and hydroxyethylamine isosteres.29 The central hydroxyl group mimics the transition state of the hydrolysis step by binding with the catalytic aspartic acid residues. Saquinavir (1, Figure 2) is a very potent inhibitor with a Ki of 0.12 nM.30 A crystal structure of saquinavir-bound HIV protease showed that the inhibitor bound in an extended conformation.31 The transition state hydroxyl group complexed between the catalytic aspartic acid residues Asp25 and Asp25’. The decahydroisoquinoline moiety fully occupies the S1’ pocket and makes contacts in the flap region. The (S,S,S) stereochemistry was found to be optimal for desired hydrophobic contacts in this area. The t-butyl amide moiety inserts into the S2’ pocket but does not make any polar contacts. The P1 phenyl moiety fills in the hydrophobic volume of the S1 pocket while the P2 carboxamide forms hydrogen bonds with the backbone amide moieties of Asp29 and Asp30. The quinadyl group fits tightly into the S3 subsite.31
Saquinavir marked the beginning of HAART therapy by the introduction of protease inhibitors into the clinic; however, many more first generation inhibitors have been developed since its approval. Ritonavir (2), developed by Abbott Laboratories, was approved by the FDA in 1996 and had a very potent Ki of 0.015 nM.32 However, ritonavir was later found to be a potent inhibitor of cytochrome P450 3A, a major metabolic enzyme for protease inhibitors.33 Due to this finding, ritonavir is used more frequently as a pharmacokinetic booster than a PI. Indinavir (3) was the next PI to be approved by the FDA. Developed by Merck, it was approved for use in 1996, only two weeks following the approval of ritonavir. Indinavir is very potent, with a Ki of 0.36 nM.34 Unfortunately, indinavir has a very short half-life (1.8h), requiring a multi-dose schedule to maintain low viral loads. Nelfinavir (4), approved in 1997, was developed by Agouron Pharmaceuticals in collaboration with Eli Lilly.35 With a Ki of 2 nM, nelfinavir features a very similar hydroxyethylamine isostere as was seen in saquinavir. Amprenavir (5), approved in 1999, also contains a hydroxyethylamine isostere with a Ki of 0.6 nM.36
4. Second-Generation Protease Inhibitors
The therapeutic efficacy of first generation PIs was limited. First, many of these inhibitors are highly peptidic in nature. This has resulted in high metabolic clearance, low half-life and largely poor oral bioavailability, requiring more frequent dosing. Furthermore, gastrointestinal distress including nausea, diarrhea, and abdominal pain, are common side effects for first-generation PIs. Most importantly, the emergence of drug-resistant strains of HIV has been a major problem for first-generation PIs. Research efforts aimed at addressing these issues led to the development of second-generation PIs.29
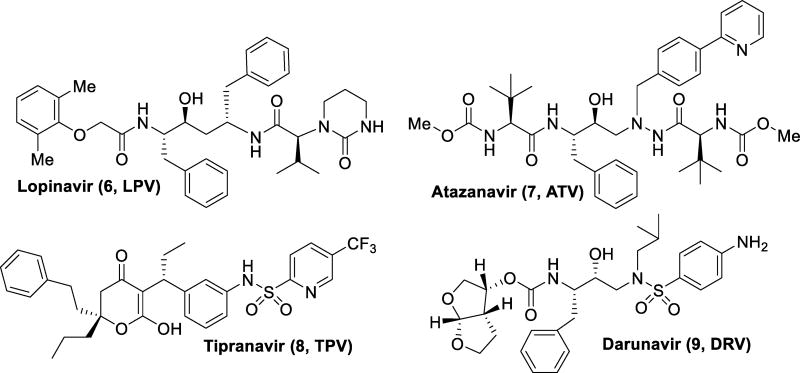
Lopinavir (6) was developed by Abbott to improve upon the properties of ritonavir. Unable to be dosed alone, lopinavir became the first PI available as a combination pill with ritonavir as a pharmacokinetic booster.37 Atazanavir (7, Figure 3) was approved in 2003, and it became the first protease inhibitor to be effectively dosed once daily. The balance of hydrophobicity and hydrophilicity was investigated to find a combination suitable for oral dosing with a longer half-life than previous protease inhibitors.38 This allowed for a lower pill burden and better patient compliance. Tipranavir (8), was approved in 2005 with extension to pediatric use in 2008. Due to its many structural differences, it maintains potency against some drug-resistant strains of HIV and appears to have a higher genetic barrier requiring many mutations to confer resistance.39 However, tipranavir has been rendered more useful in salvage therapy than as a first line treatment due to more severe side effects as compared to other protease inhibitors, such as intracranial hemorrhage, hepatitis, and diabetes mellitus. The most recently approved PI, darunavir (9), was approved by the FDA in 2006 for treatment experienced adult patients with extension to approval for treatment naïve and pediatric patients in 2008 and 2013 respectively.40–44 Darunavir maintained potency against multidrug-resistant strains of HIV with a high genetic barrier for the development of resistance in preclinical studies. However, multidrug-resistant HIV-1 variants have emerged from darunavir-experienced patients, showing the ability of the drug resistance issue to perpetuate.44–47
Figure 3.
Second-generation HIV protease inhibitors
5. Emergence of Drug-Resistant Variants
(a) Mechanisms of HIV-1 Protease Drug Resistance
HIV-1 protease is prone to resistance development in large part due to the process in which it is synthesized in vivo. Reverse transcriptase creates viral DNA from the viral RNA that gets incorporated into the host cell by the virus during infection. This process, unlike eukaryotic DNA synthesis, is extremely error prone due to its lack of proofreading mechanisms, causing HIV-1 to develop many polymorphic mutations.48, 49 The large error rate of this process increases the probability of genetic mutants to be introduced into the system, and this is influenced by the evolutionary pressures of PIs. As a consequence, the development of resistance is exacerbated.
HIV-1 protease mutations can be classified into two groups: primary and secondary mutations.50, 51 Primary mutations are changes in residues directly involved in substrate binding and manifest themselves in the active site of the enzyme. The residues considered to be involved in the active site are residues 25–32, 47–53, and 80–84. Changes in any other residues are classified as secondary mutations.52 Secondary mutations are located away from the active site and are usually compensatory mutations to relegate the detrimental effects of primary mutations on binding to the protease’s natural substrate.50, 53 There can also be a second group of secondary mutations considered which do not manifest in the protease itself but instead in the protein cleavage site on the Gag-Pol and Gag substrates.54 These changes in cleavage sites emerge to counteract the inefficiencies created by primary mutations on protease activity and are therefore considered secondary mutations.55
Primary mutations are usually observed to alter direct interactions between the protease and the PI but rarely involve residues that participate in active catalysis.56 These mutations are most often located in the substrate binding pockets, reducing the affinity for PI binding while retaining favorable interactions with the natural substrate and proteolytic activity.57 The effects of the active site double mutation V82F/I84V have been shown to change the site of the binding pocket to an unfavorable conformation for PIs. Reduction in van der Waals contacts because of this shape change leads to a significant reduction in binding enthalpy.56 Often, the protease requires additional mutations to compensate for deleterious effects of primary mutations on substrate processing, leading to a higher incidence of secondary mutations.58
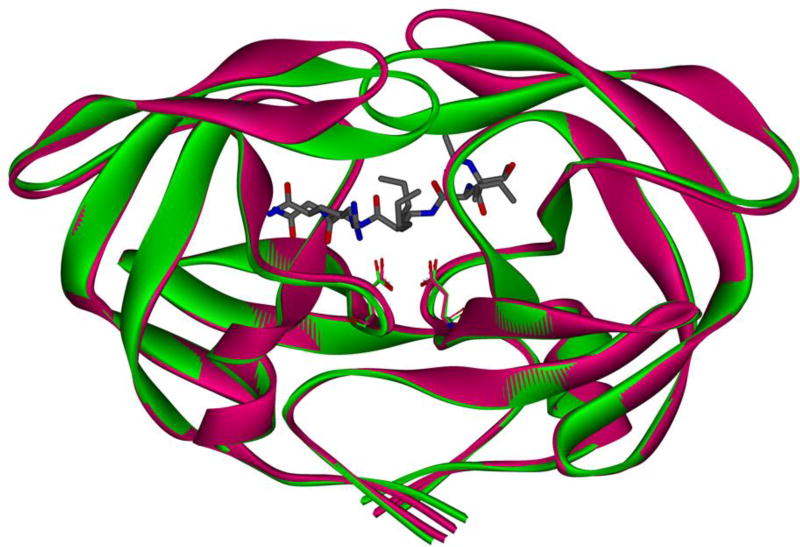
Of the 99 residues that compose a monomer of HIV-1 protease, mutations at 45 residues have been observed to confer drug resistance.59 Of these 45 mutations, only 11 have been identified as active site or primary mutations, leaving the majority of mutations classified as secondary mutations.59 Many secondary mutations are located at the dimerization interface and in the flap region.56 The flap region, composed of residues 39–57, is a main regulator for substrate and ligand binding.60 HIV protease exists in multiple conformations, including the free “flap-open” conformation and the ligand-bound “flap-closed” conformation (Figure 4). Mutations in this region tend to change the extent to which the flaps may be open or closed and thereby change the shape of and access to the binding pocket, consequently reducing the ability of the inhibitor to bind in the active site. A variant containing G48T/L89M mutations was shown to have an effect on the binding of saquinavir due to the flap being in a more open conformation. This reduced the van der Waals interactions between the PI and enzyme, reducing binding affinity.60
Figure 4.
HIV protease in both the flap open (magenta, PDB: 2PCO)61 and flap closed conformations (green, PDB: 2AOD)62 shown as ribbons. Catalytic residues Asp25 and Asp25’ of both enzymes are shown in sticks. Peptide inhibitor ace-Thr-Ile-Nle-r-Nle-Gln-Arg is shown in sticks, where r is the reduced peptide bond.
All currently available PIs have specific signature mutations associated with them.63, 64 As an example, the mutation I84V is commonly witnessed in patients treated with fosamprenavir. Viruses containing this mutation are also resistant to indinavir, atazanavir, tipranavir, and darunavir.13, 56, 63–65 This mutation changes a larger amino acid for a less bulky residue and reduces van der Waals contacts between the PI and the protease, thereby reducing affinity.64 Many PIs have been developed over the years with a very high affinity for the enzyme. The development of mutation-derived resistance has driven many in the medicinal community to develop strategies for designing inhibitors that interact with the protease in ways that create a high barrier to resistance. Swift response to PI introduction by mutations in the protease structure has been commonly observed and is the foremost concern in combating this disease. Nonetheless, there have been great strides in the battle against resistance by novel design techniques and a further understanding of the substrate binding mechanism.
(b) Inhibitor Design Strategy to Combat Drug-resistance
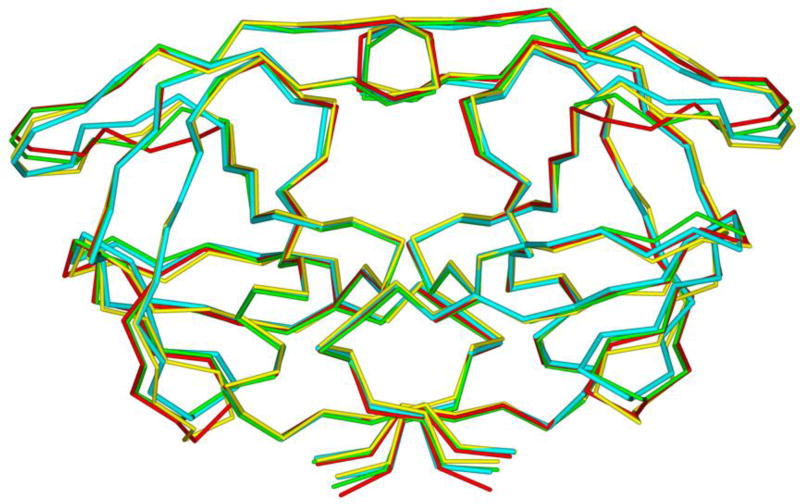
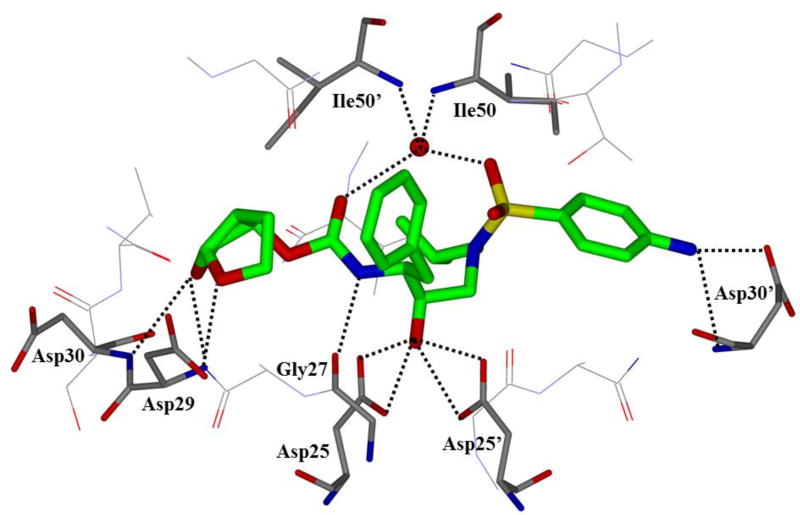
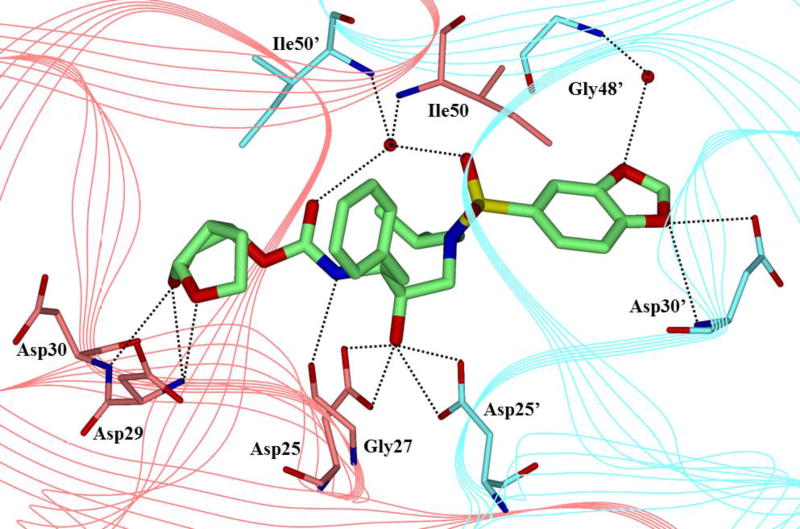
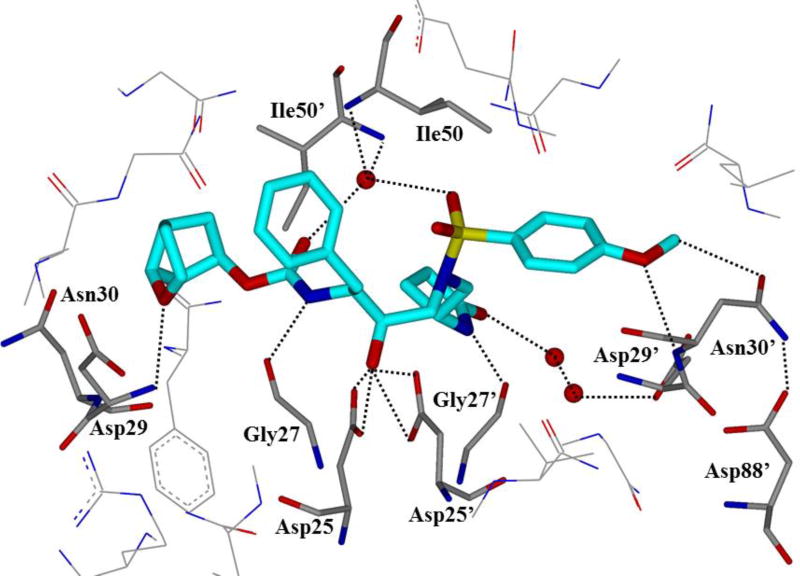
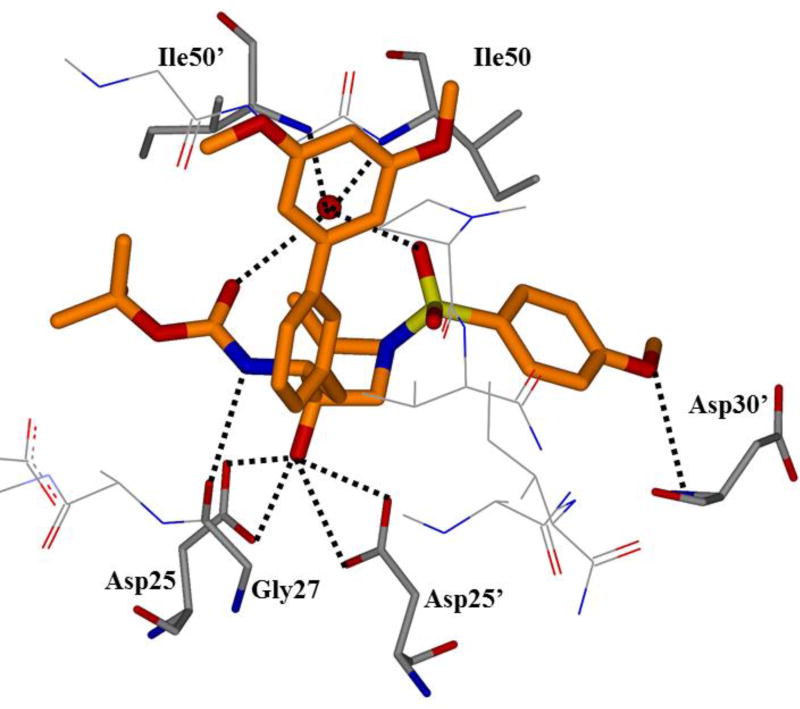
One of the major strategies utilized to combat protease drug resistance is the design of PIs by promoting hydrogen bonding interactions with the backbone atoms in the HIV-1 protease active site.42, 44 An overlay of the X-ray structures of inhibitor-bound wild-type HIV-1 protease and various mutant proteases (Figure 5) shows minimal distortion of active site backbone conformations.66, 67 As can be seen, the overall structural changes are minimal, even around the flexible flap region. Therefore, a catalytically viable mutant protease that maintains viral fitness shows only minimum distortion at the active site backbone conformation. Conceptually, this is intriguing since mutations that cause drug resistance cannot significantly alter protease active site backbone conformation and at the same time maintain protease functions. Based upon this molecular insight, the “backbone binding” molecular design strategy was developed.42, 68 It was speculated that an active site protease inhibitor that maximizes interactions in the active site, in particularly by making extensive interactions with protein backbone atoms from S2 and S2’ subsites of the wild-type enzyme, may likely retain these interactions in the active site of mutant proteases. A variety of protease inhibitors were then designed and synthesized, including darunavir, that showed extensive binding interactions with backbone atoms of the wild-type protease and maintained potent antiviral activity against panels of clinically relevant multidrug-resistant HIV-1 variants.42 Figure 6 depicts the X-ray crystal structure of darunavir(9)-bound HIV-1 protease, showing a network of hydrogen bonding interactions throughout the active site from S2 to S2’-subsites.
Figure 5.
Overlay of wild-type HIV-protease (cyan PDB: 2IEN) with three mutant proteases (red, PDB: 2FDD; green, PDB: 1SGU; yellow, PDB: 2HCO). Geometry of the backbone is conserved in the active site
Figure 6.
An X-ray crystal structure of darunavir-bound HIV protease (PDB: 2IEN)
The difficulty in combating resistance in HIV-1 protease is the tolerance for many mutations that lower affinity for PIs but maintain affinity for its natural substrate. Various cleavage sites in the natural substrates are vast and dissimilar. It has been speculated that the shape and volume of the substrate, with respect to the binding site, is of greater importance to binding affinity than the specific interactions with protease residues.69, 70 The observed loss of affinity for many PIs in the presence of the double mutant V82F/I84V can be rationalized by this hypothesis. The replacement of the isopropyl moiety of valine with a phenyl ring in phenylalanine and the loss of a methyl group from isoleucine to valine alters the shape of the active site. This results in loss of van der Waals interactions and gain in steric hindrances, leading to poor inhibitor binding.29
Resistance development is also thought to persist because of many PIs’ limited ability to maintain effective concentrations in the body and/or reach desired locations such as the CNS.71 The CNS and other regions of the body can act as viral reservoirs, which can result in increased virulence.72 To improve the effective pervasiveness of the PI, strategies have been employed to enhance their ability to cross blood brain barrier (BBB) by increasing the lipophilic character of inhibitors and introducing fluorine at various sites.71, 73
The majority of approved protease inhibitors, in general, show less than optimum pharmacokinetic properties. This is due to the fact that PIs are substrates for the isoenzyme CYP3A4, a subunit of the cytochrome P450 hepatic enzyme system, which is responsible for metabolic degradation of PIs.51 As a result, many approved PIs show low plasma trough levels and a short plasma half-life. Interestingly, many PIs act as both inhibitors and inducers of the isoenzymes. As it turns out, ritonavir (RTV) is a potent inhibitor of CYP3A4 and administration of a small dose of RTV with PIs significantly improves the PI’s pharmacokinetic parameters.74 PIs such as darunavir, fosamprenavir, and atazanavir can be administered as a once daily dose when given with a small dose of RTV.74 However, there are issues with RTV as a pharmacokinetic booster. When using RTV as a booster of other classes of antiviral compounds in HAART therapy which does not contain a PI, a subtherapeutic dose of RTV may accelerate emergence of HIV-1 variants resistant to PIs.75 Multidrug-resistant HIV-1 variants that are resistant to one PI are likely to show cross-resistance to other PIs. Concurrent RTV also possesses other side effects such as lipid disorders and undesired drug interactions. Recently, a new pharmacokinetic enhancer, cobicistat, has been developed which is stable and does not have anti-HIV activity.40, 76 Unlike RTV, it will not contribute toward the emergence of drug-resistant HIV-1 variants. Formulation of PIs with cobicistat has resulted in an all-in-one pill, making it convenient for patients by reducing pill burden. While this regimen has improved pharmacokinetic properties of currently approved PIs, the design of protease inhibitors that do not require a pharmacokinetic enhancer would be more ideal.
(5) Recent Progress Towards HIV-1 Protease Inhibitors
The second-generation PIs, particularly darunavir, show a number of important advantages. Darunavir (9, Figure 3) is a potent, non-peptidyl inhibitor of mature protease dimer.77, 78 In addition, it also potently inhibits dimerization of HIV-1 protease. Since dimerization of protease monomers is essential for the acquisition of catalytic proteolytic properties of HIV-1 protease, inhibition of protease dimerization represents a novel approach to halt HIV-1 progression. Except DRV and TPV, no other PIs exhibit dimerization inhibition properties. PIs with dual inhibitory properties may exhibit a high genetic barrier to resistance.
DRV set new standards in terms of its high genetic barrier to resistance as well as its dual mechanism of action.13, 25 As a result, the development of the next generation of PIs needs to focus on further improvement of drug-resistance profiles. For subsequent development of new PIs, molecular design efforts have focused on the design and synthesis of novel P2-ligands promoting enhanced backbone binding interaction to combat drug-resistance. Furthermore, efforts are underway to develop non-peptide PIs containing different structural scaffolds other than hydroxyethylsulfonamide isosteres. In this section, we outline development of various potent PIs following the approval of darunavir.
(a) Inhibitors Containing Bis-tetrahydrofuran as P2-Ligand
Many new inhibitors have retained the bis-tetrahydrofuran (bis-THF) moiety seen in darunavir while incorporating changes in other areas of the molecule to further explore the binding capacity of the enzyme. Most studies have focused on attempts to increase the hydrogen bonding capabilities of the P2’ ligand in hopes of increasing potency; however, the hydrophobic P1’ pocket has also been examined.
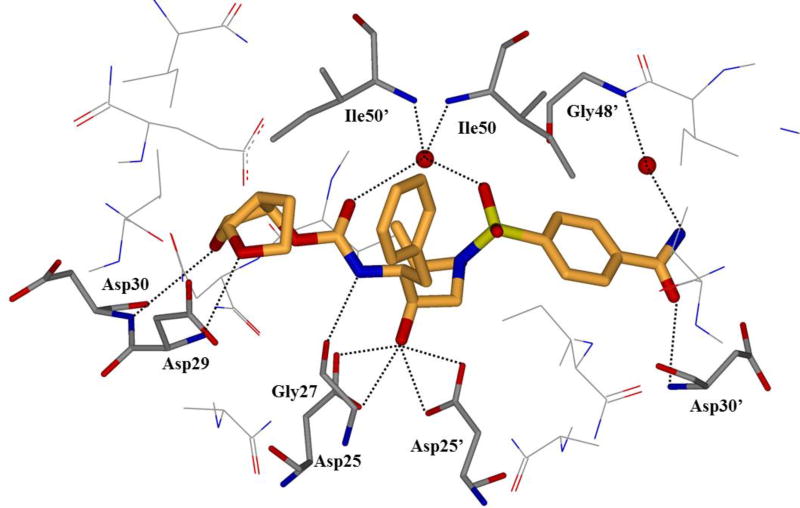
To probe the hydrogen bonding capacity of the P2’ region, compounds 11 and 12 (Figure 8), with a carboxylic acid and carboxamide moiety in the para position of the P2’ benzene ring, respectively, were designed and synthesized.79 It was envisioned that the acid moieties would be able to replace water-mediated hydrogen bonds with direct contacts, while maintaining the polar contact with Asp30’ observed with darunavir. These inhibitors were exceedingly potent in an enzymatic assay with 11 and 12 showing Ki values of 12.7 and 8.9 pM, respectively. The cellular assay exposed some issues with the enhanced polarity of 11, showing no significant affinity. Compound 12 maintained some activity with an EC50 value of 93 nM. The complete loss of activity for 11 in cells is presumably due to an issue with cellular uptake. The crystal structure of 12 with the wild-type protease enzyme was solved. The structure showed that the carboxamide moiety displaced one water molecule that was mediating a polar contact between darunavir and Gly48’ to form a direct hydrogen bond with the backbone amide NH of Gly48’.79 All other hydrogen bonding activities were maintained.
Figure 8.
Inhibitors with varying P2’ ligands
Since inhibitor 12 displayed promising affinity for the wild-type protease enzyme, it was tested against clinical isolates of mutant proteases.80 Inhibitor 12 displayed excellent retention of affinity against the mutant proteases with 0.8- and 2.6-fold changes. A crystal structure was solved for 12-bound PRA02 mutant protease to help elucidate the results of the cellular binding assay (Figure 9). The polar contact in the S2’ pocket was maintained through the P2’ amide of 12. This interaction was absent with darunavir, which may explain the observed difference in potency.80
Figure 9.
Crystal structure of 12 in the HIV-1 protease active site (PDB: 4I8Z)
An inhibitor containing a benzo-1,3-dioxole P2’ moiety was synthesized and evaluated.81, 82 Inhibitor 13 (Figure 8) was observed to have an impressive antiviral activity profile with an EC50 of 0.5 nM. When tested in the presence of a variety of laboratory selected HIV-1 variants, 13 maintained excellent activity against all PI-influenced mutant strains (5–20-fold change) except for the APV induced strain, where a 107-fold drop in affinity was observed, presumably due to the great structural similarities of amprenavir and 13.81 Similarly, inhibitor 13 maintained single-digit nanomolar EC50 values for all multidrug-resistant clinical isolates that were tested (Table 1). To determine the interactions of 13 with the enzyme, a crystal structure was solved of 13-bound wild-type protease (Figure 10). It was observed that 13 interacted with the wild-type protease in a similar manner to darunavir, with hydrogen bonding interactions to the backbone of residues Asp29, Asp30, Gly27, and Asp30’.82 In addition, inhibitor 13 formed a water-mediated hydrogen bond from the 1,3-benzodioxole P2’ substituent to the Gly48’ amide NH. Gly48’ is in the flexible flap region of the protease and strong interactions in this region could further stabilize the flap closed conformation of the enzyme.81
Table 1.
Resistance profile of inhibitor 13
| Virusa | EC50 values, µM (fold change)
|
||||
|---|---|---|---|---|---|
| 13 | APV | LPV | SQV | DRV | |
| HIV-1ERS104pre (WT) | 0.0005 | 0.029 | 0.007 | 0.008 | 0.0038 |
| HIV-1TM (MDR) | 0.0032 (6) | 0.30 (10) | 0.36 (51) | 0.18 (23) | 0.0043 (1) |
| HIV-1MM (MDR) | 0.0038 (8) | 0.48 (17) | 0.38 (54) | 0.14 (18) | 0.016 (4) |
| HIV-1JSL (MDR) | 0.006 (12) | 0.43 (15) | 0.70 (100) | 0.29 (36) | 0.027 (7) |
| HIV-1B (MDR) | 0.0039 (8) | 0.36 (12) | 0.30 (43) | 0.27 (34) | 0.04 (11) |
| HIV-1C (MDR) | 0.0027 (5) | 0.25 (9) | 0.31 (44) | 0.035 (4) | 0.009 (2) |
| HIV-1G (MDR) | 0.0034 (7) | 0.32 (11) | 0.16 (23) | 0.033 (4) | 0.007 (2) |
Amino acid substitutions in the protease-encoding region compared to type B: L63P in HIV-1ERS104pre; L10I, K14R, R41K, M46L, I54V, L63P, A71V, V82A, L90M, I93L in HIV-1TM; L10I, K43T, M46L, I54V, L63P, A71V, V82A, L90M, and Q92K in HIV-1MM; L10I, L24I, I33F, E35D, M36I, N37S, M46L, I54V, R57K, I62V, L63P, A71V, G73S, and V82A in HIV-1JSL; L10I, K14R, L33I, M36I, M46I, F53I, K55R, I62V, L63P, A71V, G73S, V82A, L90M, and I93L in HIV-1B; L10I, I15V, K20R, L24I, M36I, M46L, I54V, I62V, L63P, K70Q,V82A, and L89M in HIV-1C; and L10I, V11I, T12E, I15V, L19I, R41K, M46L, L63P, A71T, V82A, and L90M in HIV-1G
Figure 10.
X-ray crystal structure of 13-bound HIV protease (PDB: 2Z4O)
Further elaboration of P2’ ligands able to interact with extended residues in the S2’ pocket has been performed using inhibitors containing large heteroaromatic polycyclic P2’ ligands such as 14 (Figure 8).83 Inhibitor 14 was designed by focusing on discovering substituents which could extend far into the S2’ binding pocket of HIV-1 protease.84 This compound was evaluated against the wild-type protease and drug-resistant clinical isolates. Compound 14 displayed excellent affinity for the wild-type protease strains, with EC50 values ranging between 2.2 and 14 nM. This compound maintained its affinity (fold change <10) against clinical isolates containing less than 11 mutations. Some loss of potency was observed for isolates containing more than 11 mutations. When tested against laboratory-selected strains of PI-resistant recombinant clinical isolate HIV r13205 selected against 14 and darunavir, 14 was able to maintain potency against darunavir selected strains while maintain moderate potency against 14-selected strains (Table 2). To determine ligand-binding site interactions in the HIV-1 protease active site, a co-crystal structure of the inhibitor and enzyme was solved and examined (Figure 11). The interactions of 14 with the wild-type protease were comparable to that of darunavir, except for the interactions with Asp30’. Asp30’ forms two hydrogen bonding interactions with the aniline moiety of darunavir, but in 14, these two interactions are shared between the benzothiazole nitrogen and the nitrogen attached to this fragment. The piperidine ring may also form additional interactions with mutant proteases and this may explain the observed resistance profile. Compound 14 has been evaluated in Phase 2 clinical trials.85
Table 2.
Resistance profile of inhibitor 14
| Virusa | EC50 values, nM (fold change)
|
||
|---|---|---|---|
| 14 | DRV | LPV | |
| HIV-1r13205 | 26 | 22 | 28 |
| HIV-1T20760 (14-selected) | 65 (2.5) | 39 (1.8) | 112 (4.0) |
| HIV-1T21055 (14-selected) | 308 (12) | 122 (5.5) | 426 (15) |
| HIV-1T21217 (14-selected) | 312 (12) | 84 (3.8) | 198 (7.1) |
| HIV-1T13436 (DRV-selected) | 5.1 (0.20) | 49 (2.2) | 263 (9.4) |
| HIV-1T13572 (DRV-selected) | 1.5 (0.058) | 234 (11) | 511 (18) |
| HIV-1T13632 (DRV-selected) | 42 (1.6) | 624 (28) | 1597 (57) |
| HIV-1T13717 (DRV-selected) | 68 (2.6) | 670 (30) | 1900 (68) |
Figure 11.
X-ray crystal structure of 14 in the HIV protease active site (PDB: 3R4B)
To probe the S1’ site, inhibitors containing an N-alkoxy substituent at the P1’ position have been investigated.86 Cyclic, acyclic, and aromatic non-polar N-alkoxy moieties were incorporated at the P1’ position and evaluated for affinity against wild-type protease. The O-cyclopentyl (15; Ki <0.005 nM; IC50 = 7 nM) and O-cyclohexyl (16; Ki <0.005 nM; IC50 = 3 nM) derivatives proved to be the most potent (Figure 12).86 Unfortunately, potency against mutant enzyme was only moderate, with 26–28-fold change and 33–80-fold change in activity, respectively. The use of polar moieties at the P1’ position has also been investigated.87, 88 Inhibitor 17, containing a chiral methyl oxazolidinone as the P1’ ligand was synthesized.87 Incorporation of the oxazolidinone in the P1’ position was carried out to interact with the backbone of Gly27’ and Arg8 in the S1’ pocket. Interestingly, this inhibitor maintained full antiviral potency against a panel of multidrug-resistant HIV-1 variants.
Figure 12.
P1’ N-alkoxy derived inhibitors
(b) Inhibitors Containing Substituted Bis-tetrahydrofuran as P2-Ligand
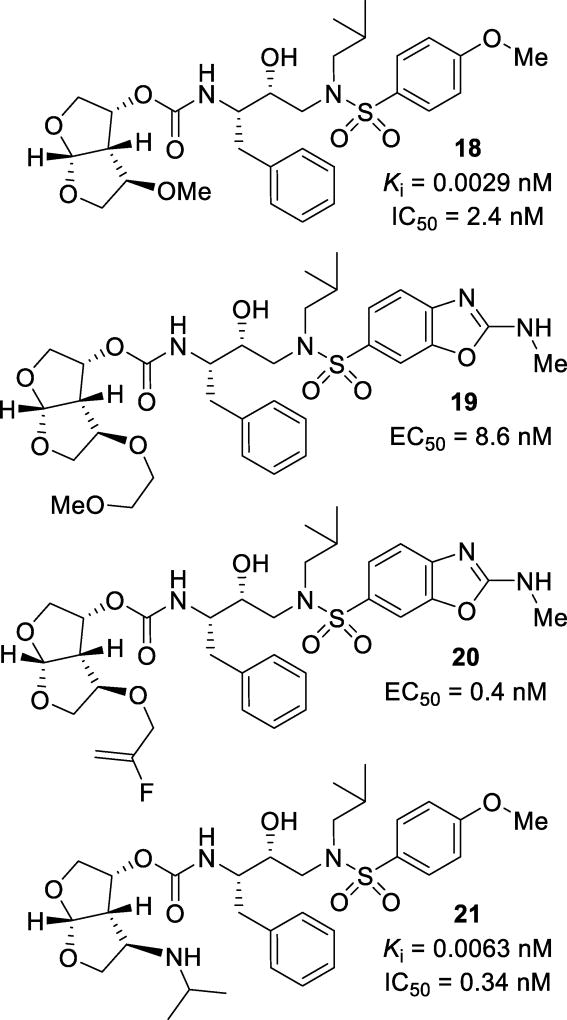
Novel substitutions of the bis-THF ligand have been a major focus to further improve ligand-binding site interactions. In particular, substituents have been specifically incorporated to interact with Gly48 in the flap region. Alkoxy substitutions at the C4 position of bis-THF were explored. A series of inhibitors containing simple alkoxy substituents resulted in inhibitor 18 (Figure 13) with a Ki of 0.0029 nM and an IC50 of 2.4 nM.89 It has been shown that the (R)-epimer at this position resulted in more potent inhibitors than the (S)-isomer. The X-ray structural studies of the inhibitor-HIV-1 protease complex showed that the oxygen of the methoxy substituent formed a water-mediated hydrogen bond with the amide nitrogen of Gly48 (Figure 14).
Figure 13.
Structure and activity of C4 substituted bis-THF inhibitors 18–21
Figure 14.
X-ray crystal structure of 18-bound HIV-protease (PDB: 3QAA)
As an extension, a series of compounds with longer alkoxyalkyl chains were designed and synthesized, resulting in inhibitor 19. This compound had an EC50 value of 8.6 nM.90 An energy-minimized model of this inhibitor showed binding similar to darunavir and a water mediated hydrogen bond to Gly48, similar to inhibitor 18. Further studies of these substitutions resulted in inhibitors with more structurally complex substitutions such as inhibitor 20, with an EC50 of 0.4 nM.91 The aim of this study was to obtain a direct hydrogen bond with Gly48. This compound was evaluated against three strains of multidrug-resistant HIV-1 variants and it showed 1–6 fold reduction in activity. The X-ray structural studies showed that the fluoroalkene had two conformations. This is due to the competing halogen bond interactions between the fluorine and both the Gly48 carbonyl and the Arg108 guanidinium moiety.
Incorporation of an N-alkyl amine at the C4 position of bis-THF was investigated. It was speculated that a hydrogen bond donor at this position may lead to the formation of a direct hydrogen bond with Gly48, rather than the water-mediated hydrogen bond that was observed in inhibitor 18. Compound 21, with an N-isopropyl C4 substitution, was a very potent inhibitor from this class of compounds. Inhibitor 21 displayed a Ki value of 6.3 pM and an antiviral IC50 of 0.34 nM.92 A number of these inhibitors were able to maintain good potency against laboratory selected multidrug-resistant HIV-1 variants, with EC50 values ranging from 0.021 µM to 0.26 µM. The X-ray structural studies revealed that the incorporation of an amine at this position enabled the inhibitor to form a direct hydrogen bond with the Gly48 carbonyl oxygen, while maintaining the water-mediated hydrogen bond with the backbone amide NH in the active site.
In an effort to improve lipophilicity as well as to interact with Gly48 backbone atoms, fluorine was introduced in the bis-THF P2 ligand. Inhibitors 22 and 23 (Figure 15) were also designed to improve blood-brain barrier penetration.73 Compound 22 showed a Ki of 0.0058 nM and an IC50 of 0.031 µM. Both inhibitors 22 and 23 displayed impressive selectivity indices of 12,333 and 21,875, respectively.71 When tested against clinical and laboratory variants, both 22 and 23 maintained comparable or better activity than darunavir with EC50 values ranging from 0.002 µM to 0.021 µM for clinical isolates (Table 3).71, 73 When tested against darunavir-resistant strains, 22 and 23 showed EC50 values of 0.034 and 0.030 µM, respectively, against the 10-pass virus and 0.043 and 0.020 µM, respectively, against the 20-pass virus. Darunavir showed an EC50 value of 0.174 µM against the 20-pass virus.71 The partition (logP) and distribution (logD) coefficient of these compounds were measured in an in vitro model of BBB penetration (Papp). Compound 23 was found to be the most lipophilic with a logP of −0.14 and a logD of −0.29. Compound 22 and darunavir have respective logP values of −0.83 and −0.63 and logD values of −1.01 and −1.03.71 A triple cell co-culture system of rat astrocytes, pericytes, and monkey endothelial cells was used to determine the Papp. The inhibitors were added to the lumenal interface. After 30 minutes, the concentration of the inhibitor in the ablumenal interface was detected via a spectrophotometer. Any compound with a Papp of more than 20 × 10−6 cm/s is considered to have efficient BBB penetration. Compound 22 had a Papp of 47.8 × 10−6 cm/s and compound 23 had a Papp of 61.8 × 10−6 cm/s. In the same assay, caffeine was used as a comparison and showed a Papp of 100 × 10−6 cm/s, showing that the fluorinated inhibitors have very efficient BBB penetration capabilities. Furthermore, these compounds were isolated and tested after the assay and it was observed that the inhibitors pass through the membrane without modification and retain their antiviral activity.71, 73
Figure 15.
Structures and activities of difluoro bis-THF and isosorbide derived inhibitors
Table 3.
Resistance profiles of inhibitors 22 and 23
| Virusa | EC50 values, µM (fold change)
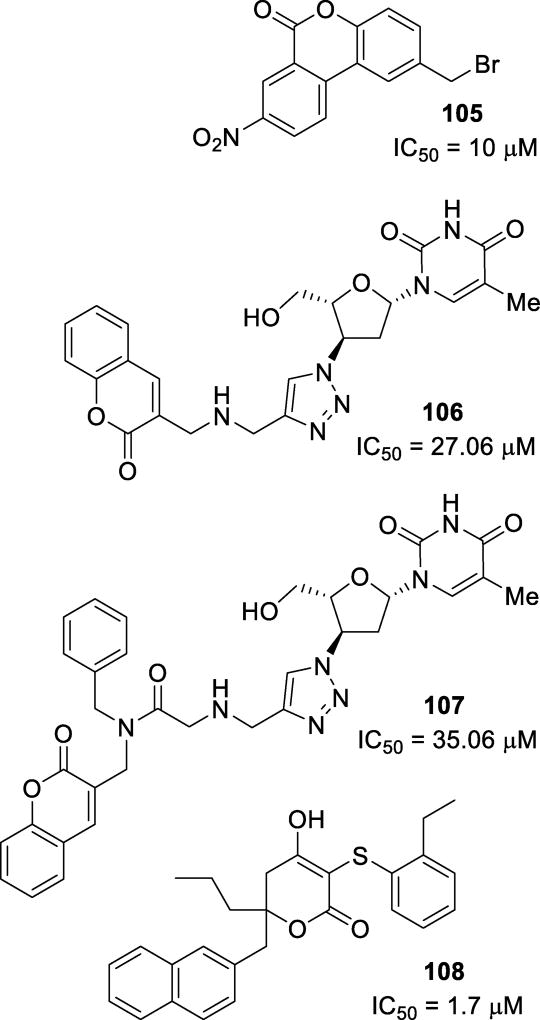
|
||||||
|---|---|---|---|---|---|---|---|
| SQV | LPV | ATV | APV | DRV | 22 | 23 | |
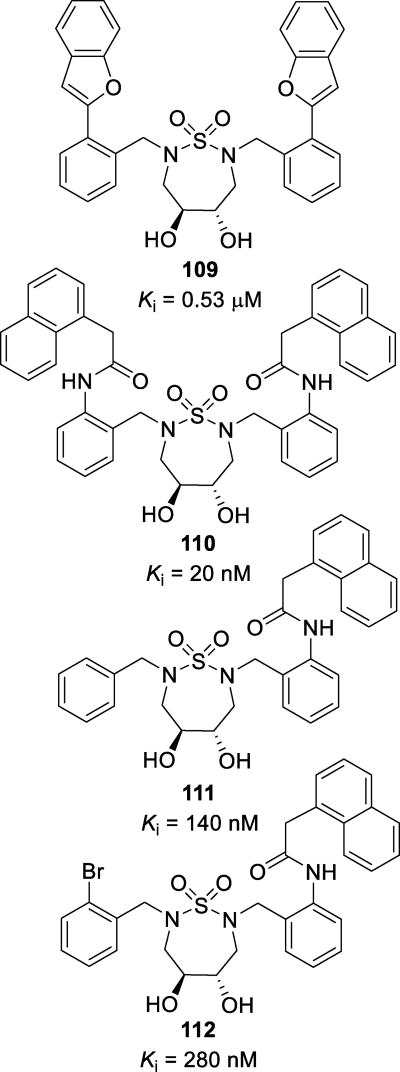
| HIV-1ERS104pre (WT) | 0.004 | 0.033 | 0.0021 | 0.0295 | 0.004 | 0.0027 | 0.0023 |
| HIV-1B (MDR) | 0.35 (90) | >1 (>33) | 0.45 (214) | 0.49 (15) | 0.021 (5) | 0.011 (3) | 0.014 (7) |
| HIV-1C (MDR) | 0.31 (78) | >1 (>33) | 0.43 (204) | 0.21 (7) | 0.005 (1) | 0.002 (1) | 0.002 (1) |
| HIV-1G (MDR) | 0.039 (10) | >1 (>33) | 0.042 (19) | 0.31 (11) | 0.014 (4) | 0.004 (2) | 0.004 (2) |
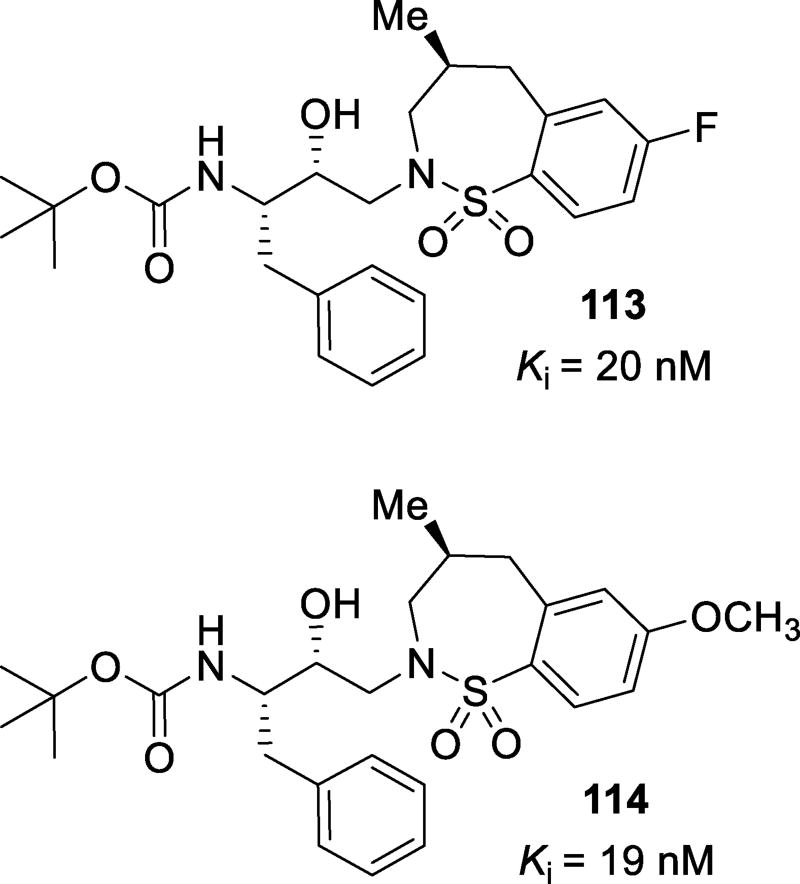
| HIV-1TM (MDR) | 0.10 (25) | >1 (>33) | 0.056 (24) | 0.328 (12) | 0.03 (9) | 0.004 (2) | 0.004 (2) |
| HIV-1JSL (MDR) | 0.53 (133) | >1 (>33) | >1 (>476) | 0.630 (22) | 0.025 (5) | 0.020 (7) | 0.021 (10) |
| HIV-1MM (MDR) | 0.11 (27 | >1 (>33) | 0.081 (38) | 0.27 (9) | 0.010 (3) | 0.003 (1) | 0.002 (1) |
Amino acid substitutions in the protease-encoding region compared to type B: L63P in HIV-1ERS104pre; L10I, K14R, L33I, M36I, M46I, F53I, K55R, I62V, L63P, A71V, G73S, V82A, L90M, and I93L in HIV-1B; L10I, I15V, K20R, L24I, M36I, M46L, I54V, I62V, L63P, K70Q,V82A, and L89M in HIV-1C; and L10I, V11I, T12E, I15V, L19I, R41K, M46L, L63P, A71T, V82A, and L90M in HIV-1G; L10I, K14R, R41K, M46L, I54V, L63P, A71V, V82A, L90M, I93L in HIV-1TM; L10I, L24I, I33F, E35D, M36I, N37S, M46L, I54V, R57K, I62V, L63P, A71V, G73S, and V82A in HIV-1JSL; L10I, K43T, M46L, I54V, L63P, A71V, V82A, L90M, and Q92K in HIV-1MM.
An isosorbide derived P2 ligand, as in compound 24 (Figure 15), was designed and synthesized. Inhibitor 24, with a methoxy substituent, showed an IC50 value of 0.05 nM.93 When docked in the X-ray crystal structure of wild-type HIV-1 protease, it was found that inhibitor 24 maintained a similar binding mode as darunavir. The top ring oxygen of the isosorbide P2 ligand formed hydrogen bonds with the amide NHs of Asp29 and Asp30, similar to the bis-THF moiety of darunavir. The C6 methoxy group did not make any polar interactions in the active site according to the docking model.93
(c) Inhibitors Containing Cyclopentyltetrahydrofuran as P2-Ligand
Various other bi- and tricyclic ether ring systems have been investigated as the P2 ligands. Cyclopentyltetrahydrofuran (Cp-THF) was designed to interact with residues in the S2 site similar to bis-THF ligand. Inhibitor 25 (Figure 16), with a Cp-THF P2 ligand, showed a Ki of 0.0045 nM and an IC50 of 1.8 nM.94 When screened against a panel of multidrug-resistant HIV-1 variants, 25 maintained potency with IC50 values ranging from 3–52 nM. Other FDA-approved protease inhibitors were much less active in this assay. An X-ray crystal structure of 25-bound HIV-1 protease showed that the THF oxygen of the P2 ligand formed a hydrogen bond with the amide NH of Asp29 (Figure 17).
Figure 16.
Structures and activities of Cp-THF containing inhibitors 25–27
Figure 17.
X-ray crystal structure of 25-bound protease (PDB: 2HB3)
Inhibitor 26 (Figure 16), with a dioxolane moiety in place of the THF of Cp-THF, was synthesized and evaluated. This inhibitor was potent with a Ki of 0.11 nM and an IC50 of 0.0038 µM.95 When tested against multidrug-resistant HIV-1 variants, inhibitor 26 maintained an excellent resistance profile with only a 1–19-fold reduction in potency. An X-ray crystal structure of a structurally related compound provided insight into the binding mode of 26. The oxygen of the dioxolane, which corresponds to the oxygen of the THF of Cp-THF, maintained its hydrogen bond with the amide NH of Asp29. The other oxygen formed a water-mediated hydrogen bond to Gly48.
It has been observed that the use of polar moieties instead of hydrophobic residues are also tolerated at the P1’ position.87, 88 Inhibitor 27 (Figure 16) containing chiral methyl-2-pyrrolidinone as the P1’ ligand was synthesized to form new backbone interactions in the active site.87 In particular, it was speculated that the incorporation of a polar substituent in the P1’ position could allow hydrogen bonding interactions with Gly27’ and Arg8 in the S1’ pocket. Inhibitor 27 displayed a very potent Ki of 0.099 nM and impressive IC50 of 0.026 nM. The crystal structure of 27 with the wild type HIV-protease showed two conformations of the pyrrolidinone. The structure revealed a strong hydrogen bonding interaction with the carbonyl of Gly27’ and a water mediated polar interaction with Arg8. The other conformation showed interactions with the backbone of residues Val82’ and Pro81’.87
The resistance profile of 27 was examined against multidrug-resistant HIV-1 variants (Table 4).84, 96, 94 Inhibitor 27 was shown to retain excellent potency against these viral strains.93 The V82A mutation of the protease has been shown to change the shape of the binding pocket by reducing hydrophobic interactions with the 80s loop of the protease with PIs.47, 97, 98 The dual binding mode of 27 is possibly due to the reduction of interactions with the 80s loop of the protease as it formed new interactions of the pyrrolidinone ring with Gly27’ and Arg8.96
Table 4.
Resistance profile of inhibitor 27
| Virusa | EC50 values, µM (fold change)
|
||||
|---|---|---|---|---|---|
| 27 | APV | LPV | SQV | DRV | |
| HIV-1ERS104pre (WT) | 0.027 | 0.025 | 0.032 | 0.009 | 0.0035 |
| HIV-1MOKW (WT) | 0.026 | 0.015 | 0.029 | 0.004 | 0.003 |
| HIV-1TM (MDR) | 0.026 (1) | 0.35 (14) | 0.23 (7) | 0.27 (30) | 0.004 (1) |
| HIV-1MM (MDR) | 0.041 (2) | 0.40 (16) | 0.62 (19) | 0.27 (30) | 0.017 (5) |
| HIV-1JSL (MDR) | 0.043 (2) | 0.66 (26) | 0.74 (23) | 0.32 (36) | 0.026 (7) |
| HIV-1A (MDR) | 0.014 (1) | 0.16 (6) | 0.32 (10) | 0.10 (11) | 0.003 (1) |
| HIV-1B (MDR) | 0.029 (1) | 0.31 (12) | 0.25 (8) | 0.30 (33) | 0.026 (7) |
| HIV-1C (MDR) | 0.027 (1) | 0.22 (9) | 0.46 (14) | 0.033 (4) | 0.007 (2) |
| HIV-1G (MDR) | 0.028 (1) | 0.23 (9) | 0.13 (4) | 0.027 (3) | 0.007 (2) |
Amino acid substitutions in the protease-encoding region compared to type B: L63P for HIV-1ERS104pre; L10I, K14R, R41K, M46L, I54V, L63P, A71V, V82A, L90M, I93L for HIV-1TM; L10I, K43T, M46L, I54V, L63P, A71V, V82A, L90M, Q92K for HIV-1MM; L10I, L24I, L33F, E35D, M36I, N37S, M46L, I54V, R57K, I62V, L63P, A71V, G73S, V82A for HIV-1JSL; L10I, I15V, E35D, N37E, K45R, I54V, L63P, A71V, V82T, L90M, I93L, C95F for HIV-1A; L10I, K14R, L33I, M36I, M46I, F53I, K55R, I62V, L63P, A71V, G73S, V82A, L90M, I93L for HIV-1B; L10I, I15V, K20R, L24I, M36I, M46L, I54V, I62V, L63P, K10O, V82A, L89M for HIV-1C; L10I, V11I, T12E, I15V, L19I, R41K, M46L, L63P, A71T, V82A, L90M for HIV-1G
Inhibitor 27 was evaluated for its affinity against a protease mutant bearing 20 mutations that is highly resistant to all clinical PIs.99, 100 The crystal structure of the mutant protease with 27 was determined (Figure 18). The pyrrolidinone ring which interacts with the S1’ appeared to be affected. The two binding conformations of the P1’ pyrrolidinone ring that was seen with the wild protease was not observed in the protease bearing the 20 mutations. This is due to the I54L mutation which displaces Pro81’ and makes interactions with this residue unfavorable. A L10F mutation displaces a structural water molecule with the phenylalanine side chain which previously mediated the interaction between Arg8 and the pyrrolidinone ring. Reduction in these interactions is thought to explain the loss of affinity for inhibitor 27.100
Figure 18.
X-ray structure of 27- bound to mutant HIV protease (PDB: 4J55)
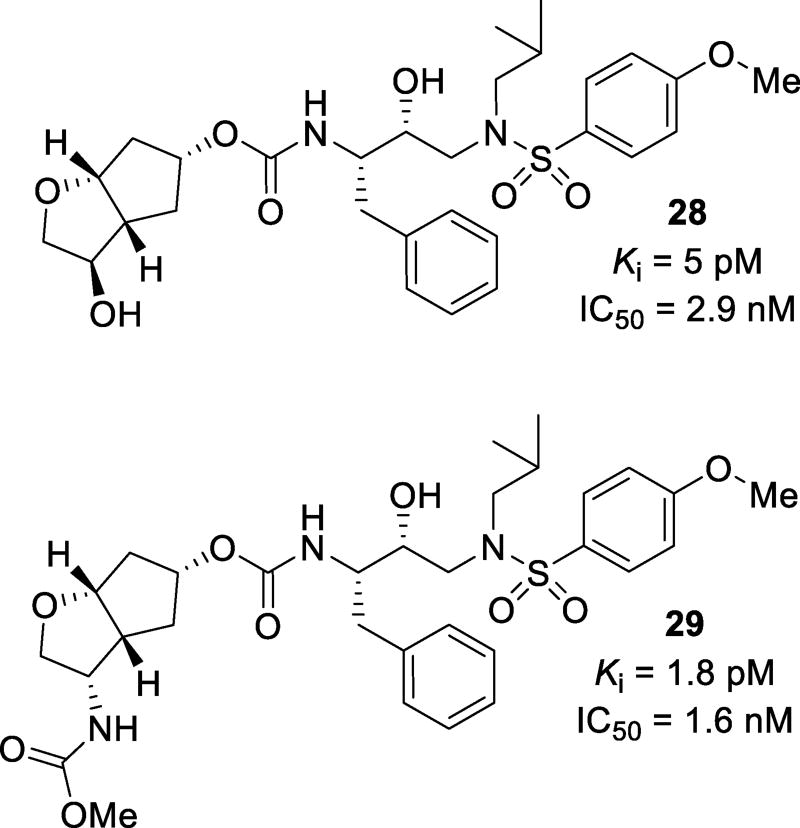
Substitutions on Cp-THF have been investigated. Inhibitors such as 28 (Figure 19) were designed to make a hydrogen bond with the Gly48 carbonyl in the S2 subsite. Compound 28 was very potent with a Ki of 5 pM and an IC50 of 2.9 nM.101 When tested against a panel of multidrug-resistant HIV-1 variants, 28 maintained potency with only a 1–10-fold reduction of potency. X-ray crystallographic analysis showed that the introduction of the C4-hydroxy group of 28 formed a water-mediated hydrogen bond with the carbonyl oxygen of Gly48. Other substituents including N-acyl, N-carbamate, and N-sulfonyl moieties at the C3 position were investigated. These substituents could function as both hydrogen bond donors and acceptors. Compound 29, with a methyl carbamate substituent, showed a Ki of 1.8 pM and an antiviral IC50 of 1.6 nM.102 When tested against a panel of multidrug-resistant HIV-1 variants, 29 maintained excellent potency with only a 1–3-fold reduction in activity (Table 5). Crystallographic analysis showed that the nitrogen of the carbamate formed a hydrogen bond with the carbonyl oxygen of Gly48 while the carbamate carbonyl interacts with Arg8’ via a water mediated hydrogen bond.
Figure 19.
C3 substituted Cp-THF containing inhibitors
Table 5.
Resistance profile of inhibitor 29
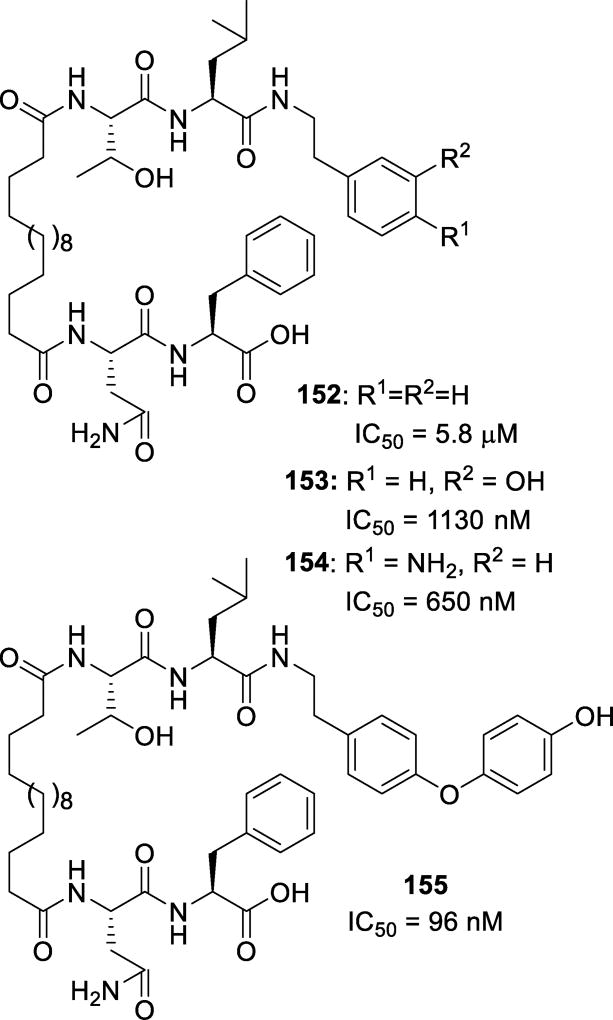
| Virusa | EC50 values, µM (fold change) |
|
|---|---|---|
| DRV | 29 | |
| HIV-1ERS104pre (WT) | 0.004 | 0.029 |
| HIV-1B (MDR) | 0.019 (5) | 0.075 (3) |
| HIV-1C (MDR) | 0.011 (3) | 0.030 (1) |
| HIV-1G (MDR) | 0.011 (3) | 0.039 (1) |
| HIV-1TM (MDR) | 0.028 (7) | 0.074 (3) |
Amino acid substitutions in the protease-encoding region compared to type B: L63P in HIV-1ERS104pre; L10I, K14R, L33I, M36I, M46I, F53I, K55R, I62V, L63P, A71V, G73S, V82A, L90M, and I93L in HIV-1B; L10I, I15V, K20R, L24I, M36I, M46L, I54V, I62V, L63P, K70Q,V82A, and L89M in HIV-1C; L10I, V11I, T12E, I15V, L19I, R41K, M46L, L63P, A71T, V82A, and L90M in HIV-1G; L10I, K14R, R41K, M46L, I54V, L63P, A71V, V82A, L90M, I93L in HIV-1TM.
(d) Inhibitors Containing Tetrahydropyranyl-Tetrahydrofuran as P2-Ligand
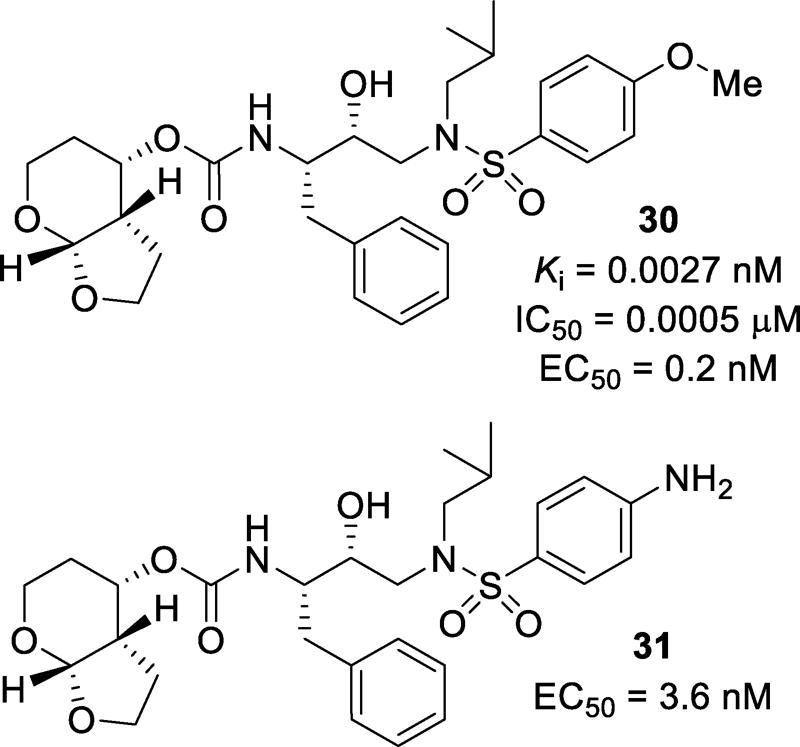
Other bicyclic scaffolds, such as the tetrahydropyranyl-tetrahydrofuran (Tp-THF) ligands in inhibitors 30 and 31 (Figure 20), were investigated. It was speculated that the addition of a methylene group into the top ring of bis-THF would promote stronger hydrogen bonding with the Asp30 amide NH. The methylene addition could also increase the dihedral angle and give more flexibility to the ring. Compound 30 was very potent with a Ki of 0.0027 nM and an IC50 of 0.0005 µM.103 Against a panel of MDR variants, inhibitor 30 only showed a 1–14-fold loss in potency. This may be due to more favorable van der Waals interactions of the pyran ring with the hydrophobic pocket created by Ile47, Val32, Ile84, Leu76, and Ile50’. Both 30 and 31 were further examined against both clinical isolates and selected laboratory strains. When tested against clinical isolates, both 30 and 31 maintained good potency with a 1–14-fold and 1–7-fold reduction of potency, respectively. It should be noted that while the fold-change of 30 was not high, the EC50 values obtained were substantially lower than darunavir when tested in the same assay, with values and 0.3–4.8 nM and 3.5–21.4 nM respectively.
Figure 20.
Structure and activity of inhibitors containing a Tp-THF P2 ligand
(e) Inhibitors Containing Tris-tetrahydrofuran as P2-Ligand
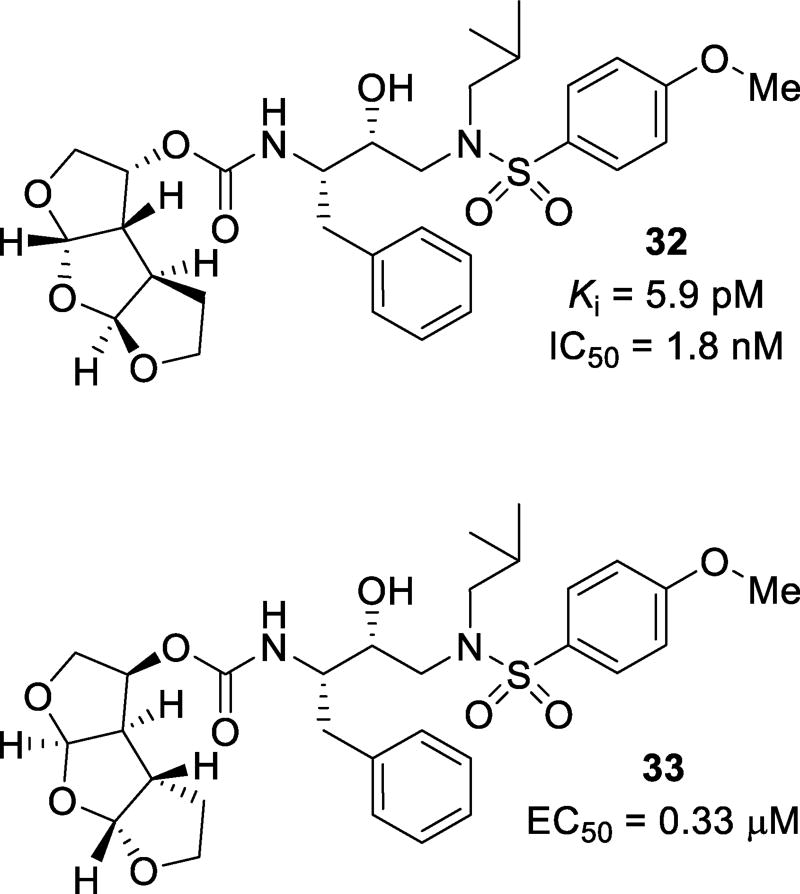
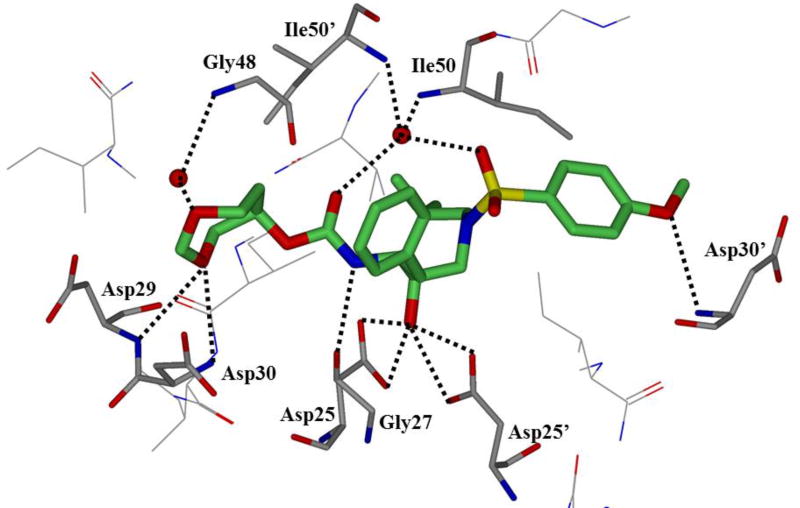
A tris-tetrahydrofuran (tris-THF) was designed and synthesized to interact with backbone atoms in the S2-site as well as to fill in the hydrophobic pocket in the S2-S3 region of the active site. Compounds 32 and 33 (Figure 21) were synthesized with varying stereochemistry on the tris-THF system. Inhibitor 32 with a syn-anti-syn structural motif was expected to maintain hydrogen bonding interactions with backbone atoms as well as fill the hydrophobic pocket of quinaldic acid amide of SQV. Inhibitor 32 was very potent with a Ki of 5.9 pM and an IC50 of 1.8 nM.104 Inhibitor 33, with a syn-syn-syn-structural motif, was less potent. Inhibitors 32 and 33 were screened against a panel of selected laboratory variants. Compound 32 only suffered a 6–7-fold loss in potency against ritonavir-, atazanavir-, and lopinavir-resistant strains. Two darunavir-resistant strains lowered the potency 9- and 50-fold (Table 6).105 The X-ray crystallographic studies revealed that the top two THF oxygens of the tris-THF ligand in 32-bound to Asp29 and Asp30 like the bis-THF ligand of darunavir (Figure 22). The third THF ring oxygen makes a water-mediated hydrogen bonding interactions with Arg8’. There is also an interaction between a methylene group CH of the THF ring and the carbonyl oxygen of Gly48.104, 105
Figure 21.
Structures and activities of tris-THF containing HIV protease inhibitors
Table 6.
Resistance profiles of inhibitors 32 and 33
| Virusa | EC50 values, nM (fold change) | |||||
|---|---|---|---|---|---|---|
|
|
||||||
| 32 | 33 | APV | ATV | LPV | DRV | |
| HIV-1ERS104pre (WT) | 0.6 | 347.4 | 33.8 | 2.7 | 31.4 | 3.9 |
| HIV-1B (MDR) | 3.4 (6) | 611.8 (2) | 459.4 (14) | 469.7 (174) | >1000 (>32) | 27.8 (7) |
| HIV-1C (MDR) | 0.8 (1) | 514.4 (1) | 356.1 (10) | 38.8 (14) | 436.5 (14) | 10.3 (3) |
| HIV-1G (MDR) | 2.6 (4) | 655.8 (2) | 462.6 (14) | 19.4 (7) | 181.3 (6) | 27.8 (7) |
| HIV-1TM (MDR) | 2.1 (4) | 530.0 (2) | 476.4 (14) | 74.5 (28) | 422.9 (13) | 30.0 (8) |
| HIV-1MM (MDR) | 2.5 (4) | 787.4 (2) | 338.9 (10) | 204.8 (76) | 622.5 (20) | 13.3 (3) |
| HIV-1JSL (MDR) | 2.5 (4) | >1000 (>3) | 436.3 (13) | 211.3 (78) | >1000 (>32) | 22.1 (6) |
| HIV-1DRVR10P | 5.6 (9) | >1000 (>3) | >1000 (>32) | 322.9 (77) | >1000 (>32) | 43.4 (11) |
| HIV-1DRVR20P | 30.0 (50) | >1000 (>3) | >1000 (>32) | >1000 (>370) | >1000 (>32) | 255.2 (64) |
Amino acid substitutions in the protease-encoding region compared to type B: L63P in HIV-1ERS104pre; L10I, K14R, L33I, M36I, M46I, F53I, K55R, I62V, L63P, A71V, G73S, V82A, L90M, and I93L in HIV-1B; L10I, I15V, K20R, L24I, M36I, M46L, I54V, I62V, L63P, K70Q,V82A, and L89M in HIV-1C; L10I, V11I, T12E, I15V, L19I, R41K, M46L, L63P, A71T, V82A, and L90M in HIV-1G; L10I, K14R, R41K, M46L, I54V, L63P, A71V, V82A, L90M, I93L in HIV-1TM; L10I, K43T, M46L, I54V, L63P, A71V, V82A, L90M, Q92K in HIV-1MM; L10I, L24I, L33F, E35D, M36I, N37S, M46L, I54V, R57K, I62V, L63P, A71V, G73S, V82A in HIV-1JSL; L10I, I15V, K20R, L24I, V32I, M36I, M46L, I54V, I62V, L63P, K70Q, V82A, L88M in HIV-1DRVR10P; L10I, I15V, K20R, L24I, V32I, M36I, M46L, L63P, A71T, V82A, L88M in HIV-1DRVR20P
Figure 22.
X-ray crystal structure of 32-protease co-crystal (PDB: 3OK9)
(f) Inhibitors Containing Tricyclic Derivatives as P2-Ligand
Carbocycles containing THF rings have also been investigated. Replacement of the middle THF ring of tris-THF with a cyclopentane ring resulted in inhibitors such as 34 (Figure 23), maintaining the same syn-anti-syn structural motif of 33. Inhibitor 34 had a Ki value of 1.39 nM.106 Replacement of the top THF ring of tris-THF with a cyclohexane ring resulted in inhibitors such as 35, with a Ki of 0.01 nM and an IC50 of 1.9 nM.107 These inhibitors maintained their potency against multidrug-resistant HIV variants with only 2–4-fold reduction of potency. The X-ray crystal structure of 35-bound wild type HIV-1 protease revealed a water-mediated hydrogen bond to Gly27 and Arg8’. Inhibitor 35 was evaluated against both multidrug-resistant clinical isolates as well as selected laboratory strains. It performed better than darunavir against all but one clinical isolate, with EC50 values ranging from 0.007 µM to 0.033 µM. In an in vitro BBB penetration assay, it displayed good lipophilicity and a Papp of 27.3 × 10−6 cm/s, indicating that this compound would likely have very good CNS penetration. Compound 35 has much greater hydrophobic contact with the active site of HIV protease than darunavir; however, it lacks in polar interactions.
Figure 23.
Structure and activity of carbocycle containing tricyclic P2 ligands
The use of cyclic polyethers with multiple sites available for hydrogen bonding activity resulted in the design of inhibitors such as 36 and 37 (Figure 24). It was envisioned that the use of a flexible ring, rather than the more rigid bis-THF motif, may be able to accommodate more variability in amino acid side chains and thus maintain potency across mutant viruses. Compound 36, with a 1,3-dioxepane P2 ligand, had a Ki of 26 pM and an IC50 of 4.9 nM, while a 1,3-dioxane P2 ligand, such as in 37 resulted in a Ki of 41 pM and an IC50 of 3.4 nM.108 Use of a symmetrical ligand, as in 37, was also successful in simplification of the inhibitor structure by elimination of a stereocenter. When tested against multidrug-resistant HIV-1 variants, 36 maintained good resistance profile with only a 1–25-fold change against the tested mutants (Table 7). The resistance profile of 37 was much more modest, with a 2–55-fold change against the tested mutants. The X- ray co-crystal structure of 36 was determined (Figure 25). The structure revealed that Asp29 and Asp30 formed hydrogen bonds directly to one of the oxygens of the dioxepane ligands, while the other oxygen formed a water-mediated hydrogen bond with the Gly48 backbone NH.
Figure 24.
HIV protease inhibitors containing monocyclic polyethers as the P2 ligand
Table 7.
Resistance profiles of inhibitors 36 and 37
| Virusa | IC50 values, nM (fold change) | |||
|---|---|---|---|---|
|
|
||||
| 36 | 37 | DRV | APV | |
| HIV-1ERS104pre (WT) | 20 | 6 | 3.5 | 33 |
| HIV-1TM (MDR) | 220 (11) | 64 (10) | 4 (1) | 290 (9) |
| HIV-1MM (MDR) | 250 (13) | 110 (18) | 17 (5) | 300 (9) |
| HIV-1JSL (MDR) | 500 (25) | 330 (55) | 26 (7) | 430 (13) |
| HIV-1B (MDR) | 340 (17) | 230 (38) | 26 (7) | 320 (10) |
| HIV-1C (MDR) | 210 (11) | 160 (27) | 7 (2) | 230 (7) |
| HIV-1G (MDR) | 360 (18) | 300 (50) | 7 (2) | 340 (10) |
| HIV-1A (MDR) | 20 (1) | 13 (2) | 3 (1) | 100 (3) |
Amino acid substitutions in the protease-encoding region compared to type B: L63P in HIV-1ERS104pre; L10I, K14R, R41K, M46L, I54V, L63P, A71V, V82A, L90M, I93L in HIV-1TM; L10I, K43T, M46L, I54V, L63P, A71V, V82A, L90M, Q92K in HIV-1MM; L10I, L24I, L33F, E35D, M36I, N37S, M46L, I54V, R57K, I62V, L63P, A71V, G73S, V82A in HIV-1JSL; L10I, K14R, L33I, M36I, M46I, F53I, K55R, I62V, L63P, A71V, G73S, V82A, L90M, and I93L in HIV-1B; L10I, I15V, K20R, L24I, M36I, M46L, I54V, I62V, L63P, K70Q,V82A, and L89M in HIV-1C; L10I, V11I, T12E, I15V, L19I, R41K, M46L, L63P, A71T, V82A, and L90M in HIV-1G; L10I, I15V, E35D, N37E, K45R, I54V, L63P, A71V, V82T, L90M, I93L, C95F in HIV-1A
Figure 25.
X-ray crystal structure of 36-bound protease (PDB: 3DJK)
(g) Inhibitors with Extended P1 Ligands
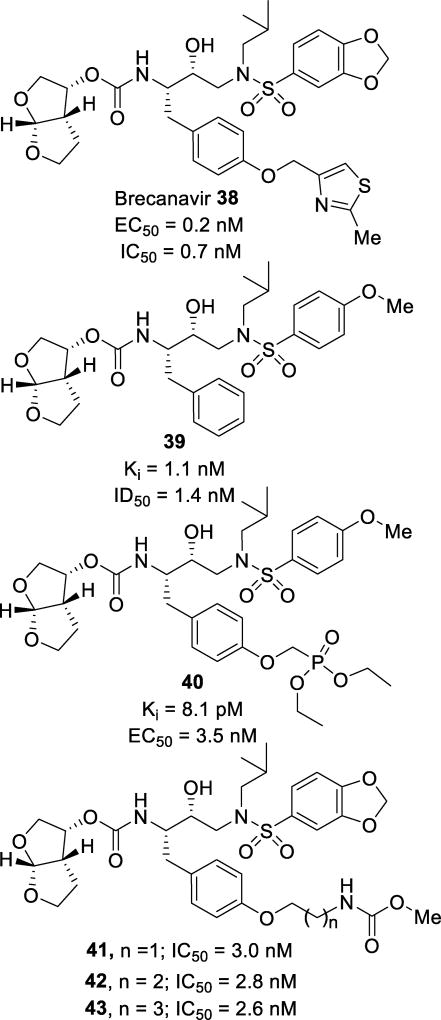
Brecanavir (38, Figure 26), was brought to clinical trials by GlaxoSmithKline but ultimately abandoned due to formulation issues.13 Researchers explored the P1 pocket via an extension of the phenyl group with a 2-methylthiazole ether. Further studies were focused on optimization of the P1 position by exploring both aryl and heteroaryl ethers.109 Inhibitor 38 displayed an impressive activity profile, with a wild-type IC50 of 0.7 nM and IC50 values of 4.8 nM and 1.1 nM against selected drug resistant variants (Table 8). When screened against a more extensive panel of ten multidrug-resistant HIV-1 mutants, 38 maintained potency with IC50 values ranging from 0.22 nM to 8.2 nM, a 1–12-fold change. When tested in the presence of 0% – 40% human serum, inhibitor 38 was seven-times more potent than darunavir.110
Figure 26.
Structures and activities of inhibitors 38–43 featuring phenyl ether P1 ligands
Table 8.
Selected resistance profile of inhibitor 38
| Virusa | IC50 values, nM (fold change) | |||||
|---|---|---|---|---|---|---|
|
|
||||||
| 38 | ATV | APV | SQV | IDV | DRV | |
| HIV-1HXB2 (WT) | 0.7 | 25 | 130 | 60 | 50 | 3.9 |
| HIV-1D545701 (MDR) | 4.8 (7) | 70 (3) | >1000 (>8) | 603 (10) | 440 (9) | 32 (8) |
| HIV-1EP13 (MDR) | 1.1 (2) | 93 (4) | 440 (3) | 80 (1.3) | 330 (7) | 6.8 (2) |
Amino acid substitutions in the protease-encoding region compared to type B: L10I, L19Q, K20R, E35D, M36I, S37N, M46I, I50V, I54V, I62V, L63P, A71V, V82A, L90M in HIV-1D545701; M46I, L63P, A71V, V82F, I84V in HIV-1EP13
Inhibitor 40 (Figure 26) explored the benefits of a phosphonate moiety off the P1 phenyl group.111 This inhibitor displayed an impressive Ki of 8.1 pM and an antiviral EC50 of 3.5 nM.112 The phosphonic acid derivative was also synthesized and tested. It was very potent in an enzymatic assay with a Ki of 13.3 pM; however, it did not display any cellular antiviral activity. This may be due to lower cellular permeability when incorporating a phosphonic acid moiety. When screened against darunavir-resistant viruses containing M46I/I50V or I84V/L90M mutations, 40 displayed only a 2.4- and 0.7-fold change, respectively. When examining the kinetic and thermodynamic parameters of binding, it was found that 40 bound to HIV protease with both favorable entropy and enthalpy. This is desired compared to inhibitor 39 (TMC-126), which binds with favorable enthalpy but unfavorable entropy.112–114 Examination of the X-ray crystal structure of the 40-HIV-1 protease complex showed that this inhibitor binds similar to inhibitor 39 (Figure 27). However, one ethyl group from the phosphonate moiety in the P1 ligand positions itself in a cleft created by Phe253 and Pro81. This displaces the waters solvating this portion of the enzyme, giving an entropic gain in the binding of the inhibitor.112 Exploration of the extended S1 pocket with long alkyl ether chains has been accomplished. Incorporation of a moiety capable of mediating potential hydrogen bonding opportunities resulted in inhibitors 41, 42, and 43 (Figure 26). The elongated alkyl chain, in addition to the carbamate moiety cap, helped to increase the cellular penetration and resulted in single digit nanomolar potencies against both wild-type and mutant HIV strains.115
Figure 27.
X-ray crystal structure of inhibitor 40-bound HIV-1 protease (PDB: 2I4W)
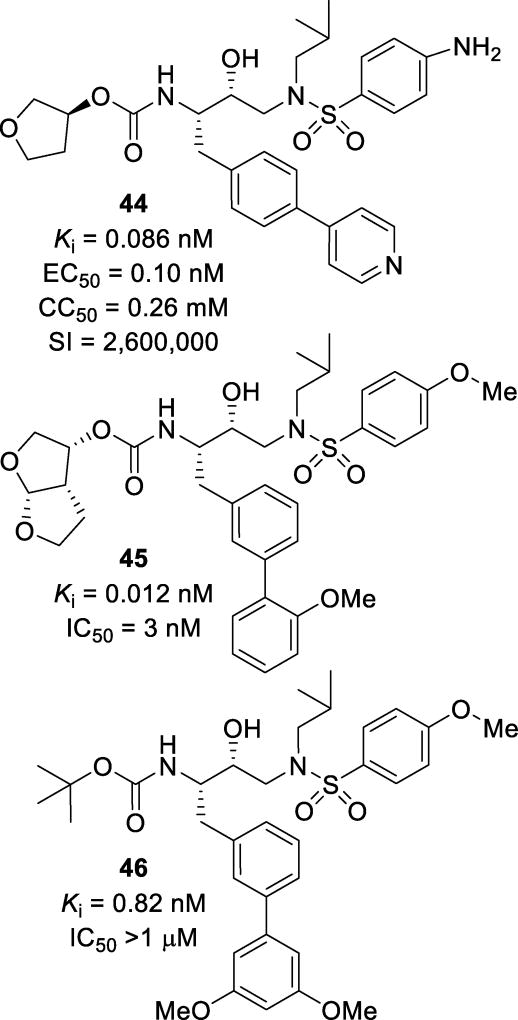
Other studies have explored a variety of biaryl motifs as the P1 ligand. Substitutions at the 4-position of the phenyl ring with aryl and heteroaryl moieties resulted in compounds such as inhibitor 44 (Figure 28) with a Ki of 0.086 nM. This inhibitor was well tolerated with a selectivity index of 2,600,000. Computational docking of this compound into the wild-type HIV protease showed that no new hydrogen bonds were made, but the S1 subsite was fully occupied as compared to amprenavir.116 Modifications at the 3-position with substituted phenyl moieties resulted in compounds such as inhibitor 45.117 These inhibitors were designed to fill the hydrophobic site of the S1 pocket in order to improve lipophilicity and CNS penetration. It was speculated that this extension would protrude towards the P2 ligand, filling in a small subsite between the S1 and S2 sites. This inhibitor had potent enzymatic and antiviral activity with a Ki of 0.012 nM and an IC50 of 3 nM. When tested against three different multidrug-resistant strains of HIV, this inhibitor maintained potency with only a 1–8-fold reduction in activity (Table 9). Elucidation of the crystal structure of related compound 46 showed that no additional polar interactions were made in the HIV active site (Figure 29). However, there were significant hydrophobic interactions with a pocket made from Leu23’, Arg8’, Pro81’, and Val182’. This was a different binding mode than originally predicted.117
Figure 28.
Inhibitors 44–46 with biaryl moieties
Table 9.
Resistance profile of inhibitor 45
| Virusa | EC50, µM (fold change) | ||
|---|---|---|---|
|
|
|||
| APV | DRV | 45 | |
| HIV-1ERS104pre (WT) | 0.037 | 0.0035 | 0.0048 |
| HIV-1B (MDR) | 0.044 (1.2) | 0.028 (8) | 0.036 (8) |
| HIV-1C (MDR) | 0.38 (10) | 0.019 (5) | 0.0029 (0.6) |
| HIV-1G (MDR) | 0.398 (11) | 0.023 (7) | 0.0047 (1) |
Amino acid substitutions in the protease-encoding region compared to type B: L63P in HIV-1ERS104pre; L10I, K14R, L33I, M36I, M46I, F53I, K55R, I62V, L63P, A71V, G73S, V82A, L90M, I93L in HIV-1B; L10I, I15V, K20R, L24I, M36I, M46L, I54V, I62V, L63P, K10O, V82A, L89M in HIV-1C; L10I, V11I, T12E, I15V, L19I, R41K, M46L, L63P, A71T, V82A, L90M in HIV-1G
Figure 29.
Binding mode of inhibitor 46 as determined by X-ray crystallography (PDB: 4ZLS)
(h) Benzamide-Based Inhibitors
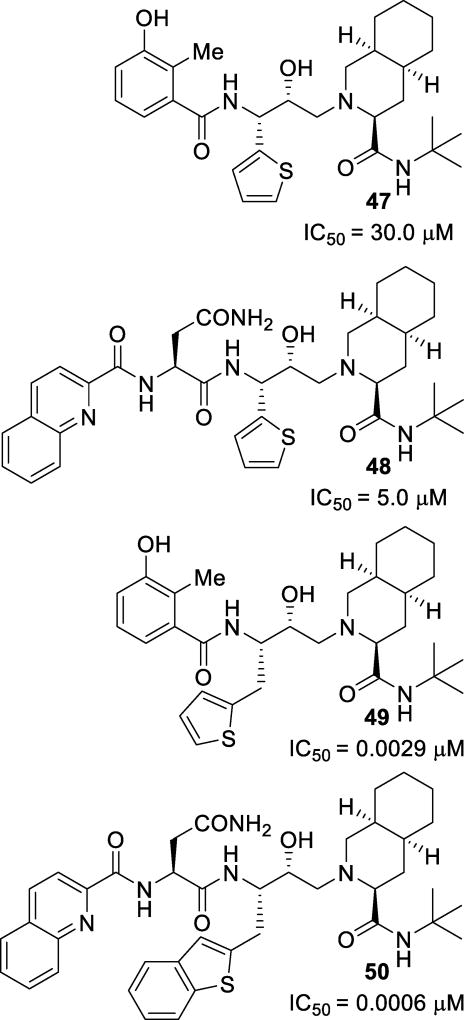
Replacement of the P1 phenyl rings of some PIs, particularly those of nelfinavir and saquinavir, with a thiophene ring and derivatives was investigated in an attempt to address drug resistance and pharmacokinetic liabilities. Initial investigations resulted in inhibitors 47 and 48 (Figure 30), which were not very active.118 A modeling study revealed that while most of the compound binds similarly to that of the parent compound, the P1 thiophene moiety cannot properly access the S1 pocket. Further expansion and derivatization of the series revealed inhibitors 49 and 50, with similar potencies as their parent compound.119 The incorporation of a one-carbon spacer allowed for greater flexibility and enabled these inhibitors to better access the binding site.
Figure 30.
Thiophene containing inhibitors 47–50
(i) Inhibitors Containing Proline Derivatives as P2-Ligand
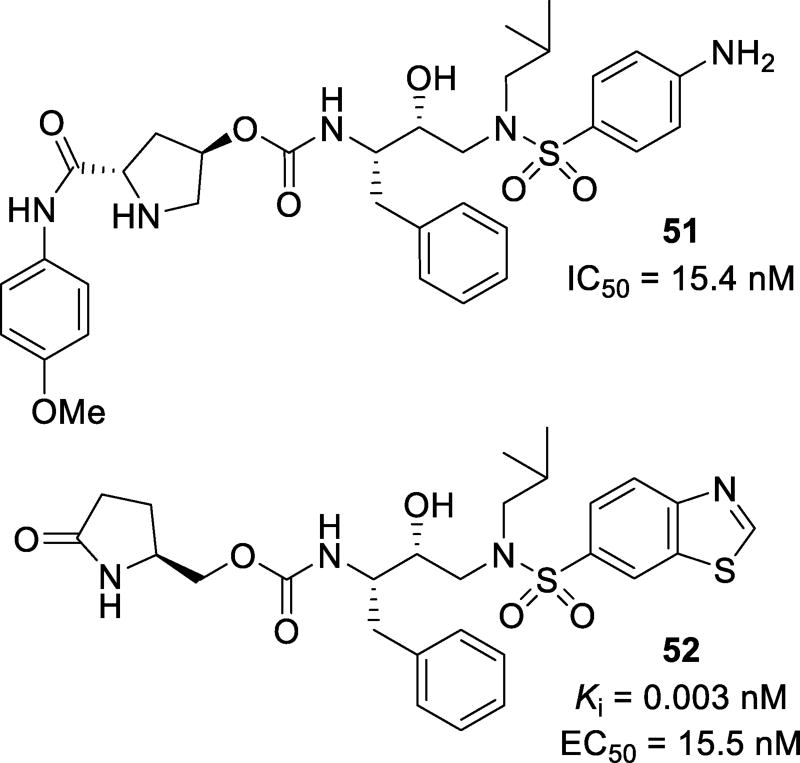
Proline derivatives have been incorporated in the design of potent inhibitors. Inhibitor 51 (Figure 31), containing a prolinamide as P2 ligand, was designed, synthesized, and evaluated. Compound 51 showed an antiviral IC50 of 15.4 nM. A docking study with this compound showed that inhibitors can make many hydrogen bonds in the S2 site, including a hydrogen bond between Arg8’ and the methoxy oxygen, Gly48 and the proline NH, and Asp30 with the amide NH.120 Compound 52 with a pyrrolidinone P2 ligand, was designed to exploit both structure-based design and the potential to promote hydrogen bonding in the S2 site similar to the bis-THF ligand. The use of a pyrrolidinone ligand reduced the structural complexity. Inhibitor 52 displayed a Ki of 0.003 nM and an EC50 of 15.5 nM.121 This inhibitor maintained potency against three tested multidrug-resistant strains, with Ki values ranging from 0.24 nM to 0.37 nM. The crystal structure for 52 was determined and showed that the pyrrolidinone carbonyl oxygen formed a hydrogen bond with the amide NH of Asp29 and a water-mediated hydrogen bond to Gly27. The pyrrolidinone NH formed a water-mediated hydrogen bond to the amide NH of Asp30 (Figure 32).
Figure 31.
Structure and activity of inhibitors 51 and 52
Figure 32.
X-ray structure of 52 in the HIV protease active site (PDB: 4DJR)
Pyrrolidinone based inhibitors have been designed in combination with hydroxyethylene isosteres.122 Inhibitors 53 and 54 (Figure 33) were synthesized and evaluated. They show better antiviral potency than amprenavir. Other pyrrolidinone containing inhibitors with a slightly different overall structure have been designed and evaluated. Inhibitors 55 and 56 achieved single-digit nanomolar potency.123 Both inhibitors were co-crystallized with protease containing the L63P, V82T, and I84V mutations. The β-hydroxy group of the pyrrolidinone ring in 55 forms tight hydrogen bonds with Asp25 and Asp25’. The carbonyl group of the pyrrolidinone and the carbonyl group adjacent to the hydrazide moieties formed water-mediated hydrogen bonds to Ile50 and Ile50’ in the protease flap region. The P1’ phenyl group forms hydrophobic contacts with Pro81 and an edge-face π-π interaction with Phe53’.123 Inhibitor 56 bound similarly (Figure 34).
Figure 33.
Pyrrolidinone containing inhibitors 53–56
Figure 34.
X-ray crystal structure of the 56-mutant protease co-crystal (L63P, V82T, and I84V PDB: 4A6B)
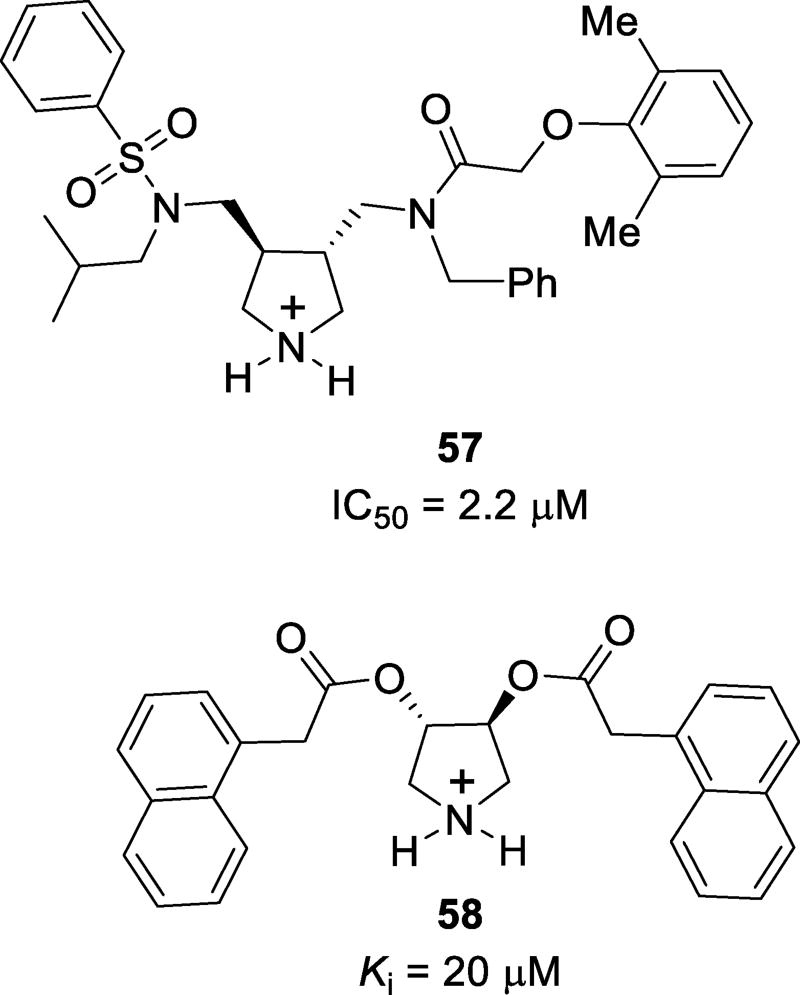
Inhibitors featuring a pyrrolidinium moiety have been synthesized and evaluated. These inhibitors were designed to incorporate an endocyclic amine with a protonated nitrogen atom which may bind the catalytic aspartic acids. Also, the two carbonyl moieties are expected to bind to the structural flap water molecule, and the side chains are expected to interact favorably in the extended binding site.124 Inhibitor 57 (Figure 35) was the most potent of the series with an IC50 value of 2.2 µM as a racemic mixture. The X-ray crystal structure of the 57-protease complex was determined and the structure revealed that the (R,R) isomer bound to the enzyme over the (S,S) isomer, as predicted by docking studies. However, the protonated amine of the pyrrolidine was able to bind the catalytic residues Asp25 and Asp25’. The sulfone and carbonyl did not bind the structural water molecule in the flap region. Instead, the sulfone oxygen forms a direct hydrogen bond with Ile50, displacing the water molecule and leaving the amide carbonyl of the inhibitor unbound. Interestingly, substituents on the inhibitor did not occupy the expected subsites. Instead, an overall rotated orientation was observed resulting in mismatched occupancy of the subsites.124 Further investigation of pyrrolidinium-based inhibitors includes compound 58.125 This series of compounds was designed to target the open-flap conformation of the enzyme. Compound 58 achieved modest enzyme inhibition with a Ki value of 20 µM. This compound was co-crystallized with a mutant protease containing an I84V mutation. Two binding modes were shown as α and β. In the α binding mode, 58 forms hydrogen bonds with the Asp25 and Asp25’ catalytic residues. These are the only polar contacts observed. In the β binding mode, the ligand binds to the flap region of the protease. Extensive van der Waals contacts are observed with many amino acids in the flap region and water mediated hydrogen bonds to Gly51 and Gly51’ from the amine of the pyrrolidinium ring are also observed.125
Figure 35.
Structures of pyrrolidinium-based inhibitors
(j) Oxazolidinone-Based P2 Ligands
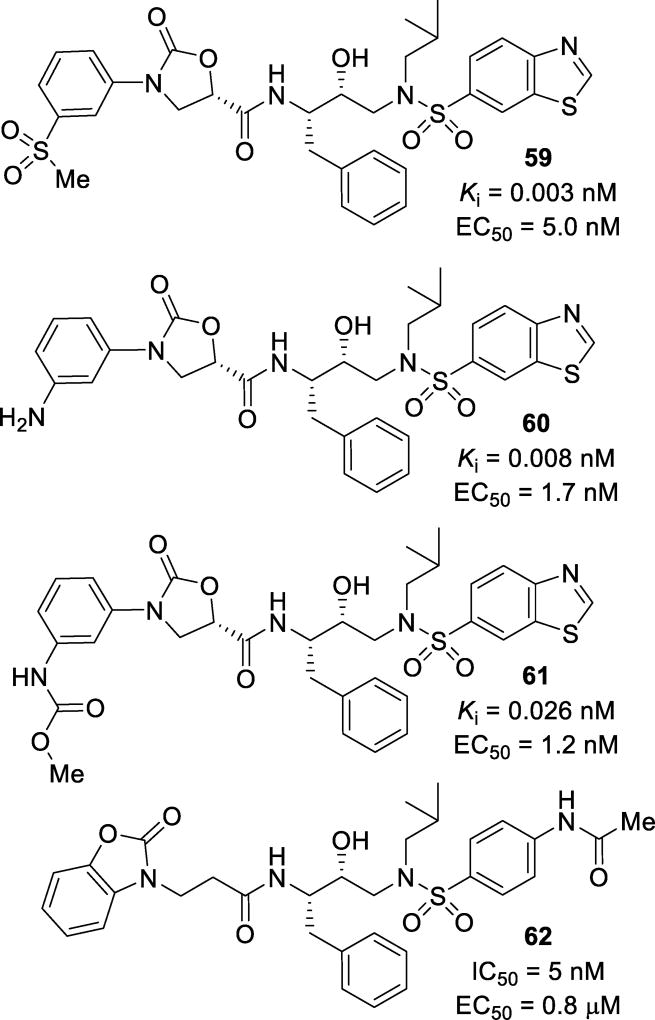
In further attempts to address drug resistance, N-aryl oxazolidinone derived P2 ligands have been explored. Inhibitors 59, 60, and 61 (Figure 36) demonstrated potent antiviral activity against wild-type virus with Ki values of 0.003 nM, 0.008 nM, and 0.026 nM, respectively, and EC50 values of 5.0 nM, 1.7 nM, and 1.2 nM, respectively. Remarkably, each of these inhibitors maintained potency against mutant virus and had better activity than lopinavir against each virus.126 A structurally related compound 62, with a fused ring benzoxazolidinone ligand, has been investigated. In these studies, substituted phenyl- and benzo-heterocycle derivatives were investigated for their ability to accept hydrogen bonding. Compound 62 proved to be the most potent of the series with an IC50 of 5 nM and an EC50 of 0.8 µM. Elongation of the chain between the benzo-heterocycle and the amide carbonyl resulted in decreased potency.127
Figure 36.
Inhibitors with oxazolidinone-derived P2 ligands
(k) Urea and Triazole Derived Inhibitors
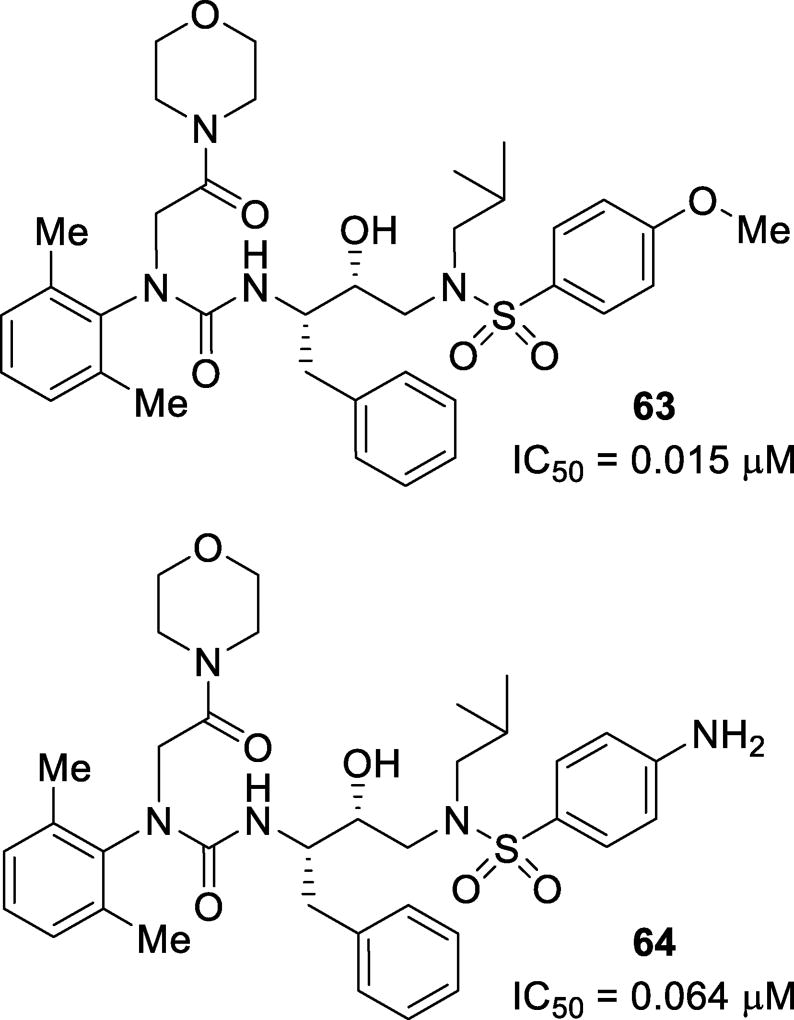
Inhibitors featuring a urea moiety as the P2 ligand have been designed and synthesized. These inhibitors were designed to mimic the P2 ligand of nelfinavir while adding an acetamide moiety to form strong hydrogen bonds in the S2 site. Inhibitors 63 and 64 (Figure 37) showed decent inhibitory activity with IC50 values of 0.015 µM and 0.064 µM, respectively.128 Click chemistry has been utilized to form triazoles which can serve as a peptide surrogate in the P2 position. These inhibitors can be synthesized in situ to form potent inhibitors of HIV protease.129 Inhibitor 65 (Figure 38), with a N-Boc benzylamine moiety, and 66, with an aminoindanol moiety, were both very potent, showing Ki values of 4 nM and 1.7 nM and IC50 values of 13 nM and 6 nM, respectively. The X-ray crystallographic studies revealed that the triazole moiety forms hydrogen bonds in the active site such as C5 of the triazole with Gly27 and a water-mediated hydrogen bond to nitrogen 2 of the triazole.130
Figure 37.
Inhibitors with tertiary amine P2 ligands
Figure 38.
Inhibitors synthesized via click chemistry containing triazole P2 fragments
(l) Atazanavir-Based Compounds
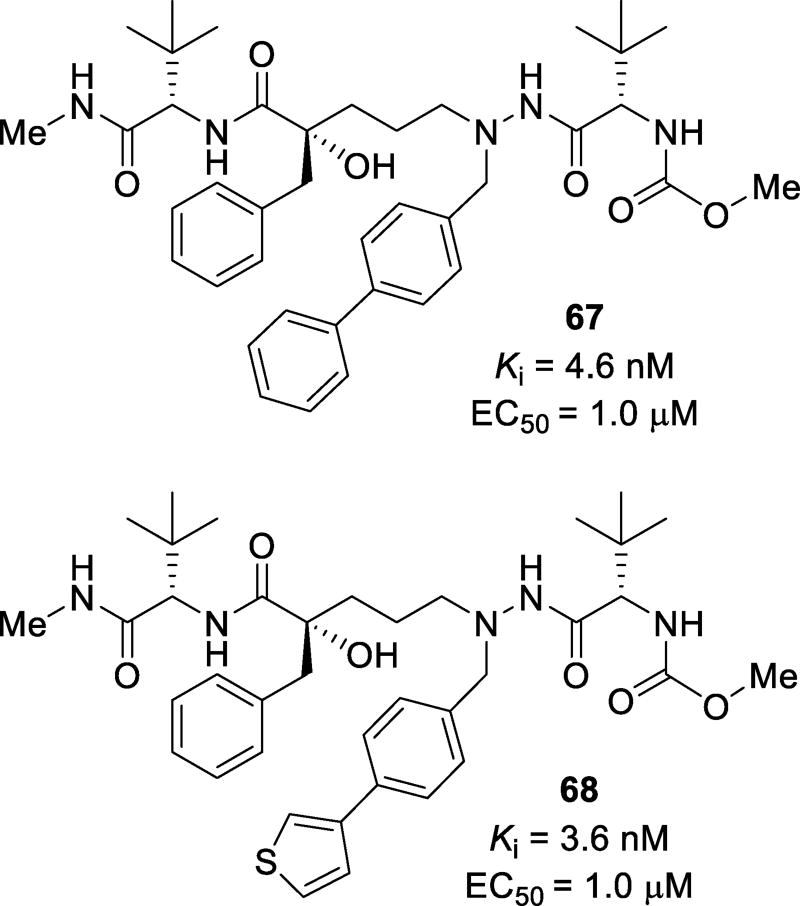
A series of inhibitors based on a modified substructure of atazanavir have been synthesized and evaluated.131 A tertiary chiral alcohol was investigated as the transition state mimetic. Modified derivatives were examined as the P1’ ligand. The pyridyl benzyl moiety of atazanavir was replaced with a series of heteroaromatic functional groups, while the chain length between the tertiary alcohol and the hydrazide functional groups were varied between one and three carbons.131 Inhibitors containing the chiral tertiary alcohol were less potent against wild-type protease. The elongation of the hydrocarbon chain resulted in inhibitors with a three-carbon linker as the most potent compounds. Inhibitor 67 (Figure 39), with a para-biphenyl moiety, displayed a Ki of 4.6 nM and an EC50 of 1 µM, while inhibitor 68, with a 3-thiophene substitution, displayed a Ki of 3.6 nM and an EC50 of 1 µM. The X-ray crystallographic analysis of the protease-inhibitor complex showed that the tertiary hydroxyl group was unable to adequately bind to the catalytic aspartic acid residues Asp25 and Asp25’. This may explain the loss of potency of inhibitors 67 and 68 compared to inhibitors with the atazanavir isostere. Furthermore, the three carbon chain positioned the aromatic P1’ ligand into the S1’ pocket facilitating π-π and hydrophobic interactions with Phe53 and Phe153 and forcing the S1’ pocket to accommodate larger residues. This was observed when the carbon chain length was three and the P1’ substituents were 4-phenyl phenylalanine and 4-(3-thiophene)phenyl alanine as described by the co-crystal structure.131
Figure 39.
Structure of atazanavir based inhibitors with biaryl P1’ motifs
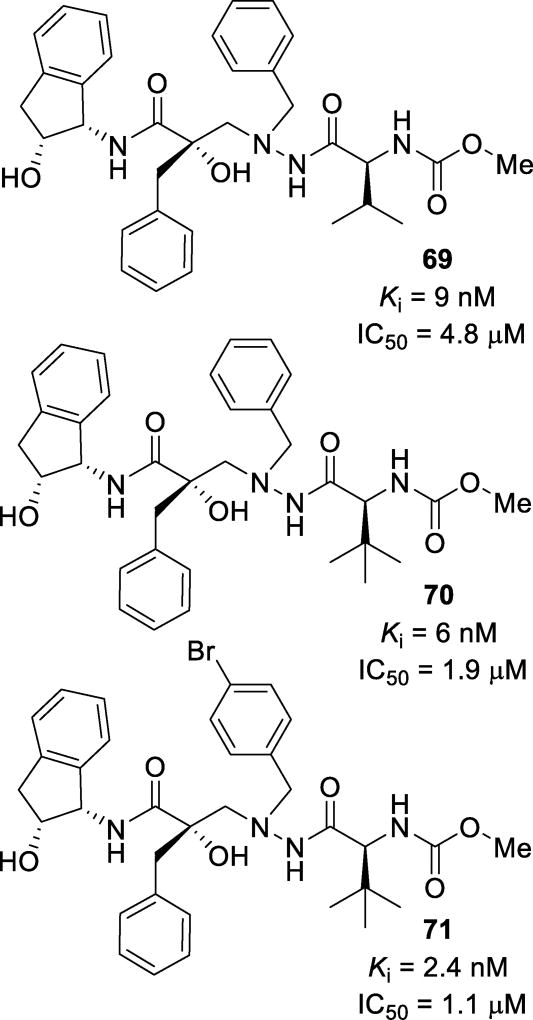
Further derivatization resulted in inhibitors 69, 70, and 71 (Figure 40), which displayed single-digit nanomolar Ki values.132 Inhibitors 70 and 71 were tested against multidrug-resistant HIV variants. Both inhibitors either maintained potency (70; IC50 = 2.0 µM and 1.6 µM) or showed enhanced potency (71; IC50 = 0.7 and 0.8 µM) against two strains of resistant HIV. Inhibitors 70 and 71 showed good cellular permeability in a Caco-2 assay, Papp = 35 and 42 × 10−6 cm/s, respectively. Unfortunately, metabolic hydroxylation of the benzylic position of the aminoindanol moiety led to fast intrinsic clearance of these compounds. A crystal structure of the HIV protease-69 complex was determined (Figure 41). Instead of the tertiary hydroxyl group positioning itself tightly between the catalytic aspartic acids, as seen with darunavir, it makes a longer hydrogen bond with Asp25’. The β-nitrogen of the hydrazido group makes a hydrogen bond with Asp25. The P3’ carbamate makes hydrogen bonds with Asp29’, Gly48’, and Gly49’.132
Figure 40.
Tertiary hydroxyl inhibitors 69–71
Figure 41.
X-ray crystal structure of 69-bound HIV protease (PDB: 2BQV)
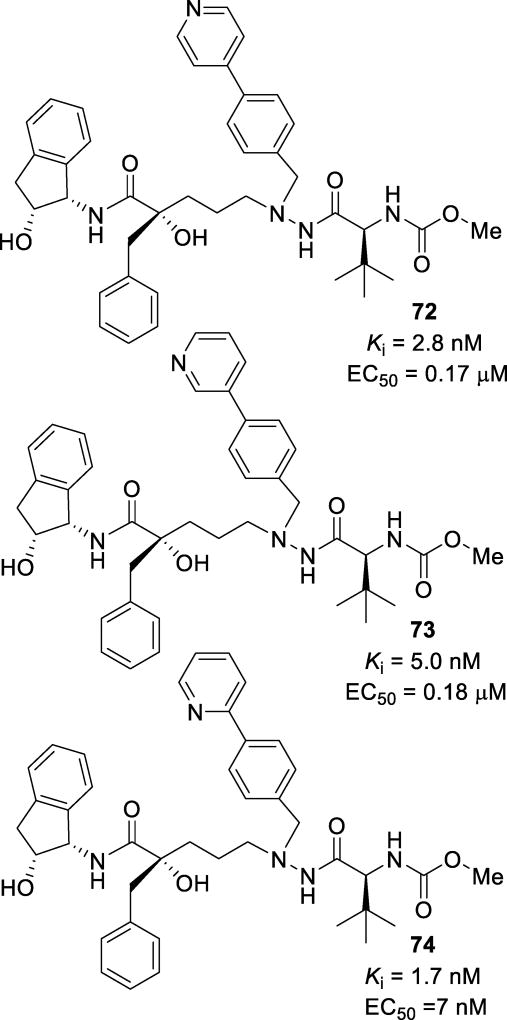
Further studies into this motif included extension of the benzyl amine of the α-nitrogen of the hydrazido moiety into biaryl systems. Inhibitor 72 (Figure 42), with a 4-pyridyl extension of the aryl system, displayed good activity with a Ki of 2.8 nM and an EC50 of 0.17 µM.133 The 3-pyridyl derivative 73 showed similar activity, with a Ki of 5 nM and an EC50 of 0.18 µM.134 Inhibitor 73 showed excellent cellular permeability with a Caco-2 Papp of 33 × 10−6 cm/s. The X-ray crystallographic studies revealed that this inhibitor interacted with the enzyme through a series of hydrogen bonds. The tertiary alcohol hydrogen bonded to Asp25 and the hydrazide nitrogens bound to Gly27. The structure showed a hydrogen bond between the pyridine nitrogen and Arg8’ as well as conserved water-mediated hydrogen bonds with the NHs of Ile50 and Ile50’ and the carbonyls of the inhibitor. Inhibitor 74, the 2-pyridyl derivative, was tested against a panel of multidrug-resistant HIV-1 variants and was found to maintain EC50 values ranging from 0.006 µM to 0.130 µM (Table 10).135 The X-ray structural studies showed that it maintained a similar binding mode as previously observed, including the binding of the tertiary alcohol to the catalytic Asp25, water-mediated hydrogen bonding to Ile50 and Ile50’ via the carbonyls, and an indanol hydroxyl group forming a hydrogen bond to Asp29 (Figure 43).
Figure 42.
Inhibitors 72–74 featuring a biaryl motif
Table 10.
Resistance profile of inhibitor 74
| Entry | Mutant Protease | EC50 (µM) of 74 |
|---|---|---|
| 1 | wild-type | 0.007 |
| 2 | G48V, L90M | 0.008 |
| 3 | A71V, I84V, L90M | 0.007 |
| 4 | V32I, M46I, A71V, V82A | 0.006 |
| 5 | V32I, M46I, V82A | 0.024 |
| 6 | M46I, V82F, I84V | 0.13 |
Figure 43.
X-ray crystal structure of the 74-protease co-crystal (PDB: 2WKZ)
(m) Macrocyclic Inhibitors
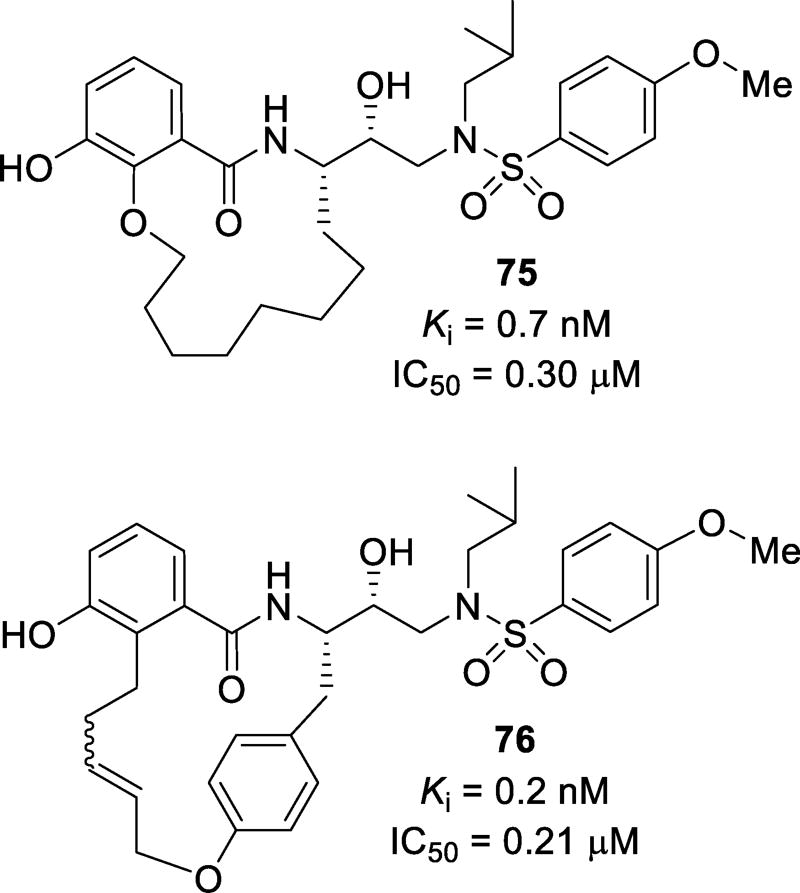
Macrocyclic inhibitor design spanning the S1 to S2 sites have been explored with and without involving the P1 phenyl group. A 14-membered macrocycle 75 (Figure 44), with 3-hydroxybenzamide as the P2 ligand, was designed and synthesized. This compound showed a Ki of 0.7 nM and IC50 of 0.30 µM.136 An energy-minimized model revealed that the hydroxyl group of the phenolic moiety forms hydrogen bonds with the amide NH of Asp29, while the amide carbonyl forms a water-mediated hydrogen bond with the structural water molecule that forms hydrogen bonds with amide NHs of Ile50 and Ile50’. When the P1 phenyl group is incorporated, a 16-membered macrocycle is preferred. Inhibitor 76 displayed a Ki of 0.2 nM and an antiviral IC50 value of 0.21 µM.137 An energy-minimized model of inhibitor 76 in the HIV-1 protease active site show that the phenolic hydroxyl group of the P2 ligand forms a hydrogen bond with amide NHs of Asp29 and Asp30 like the bis-THF portion of darunavir. The flexible macrocycle fills the S1 site.
Figure 44.
Macrocyclic inhibitors 75 and 76 spanning the S2 to S1 subsites
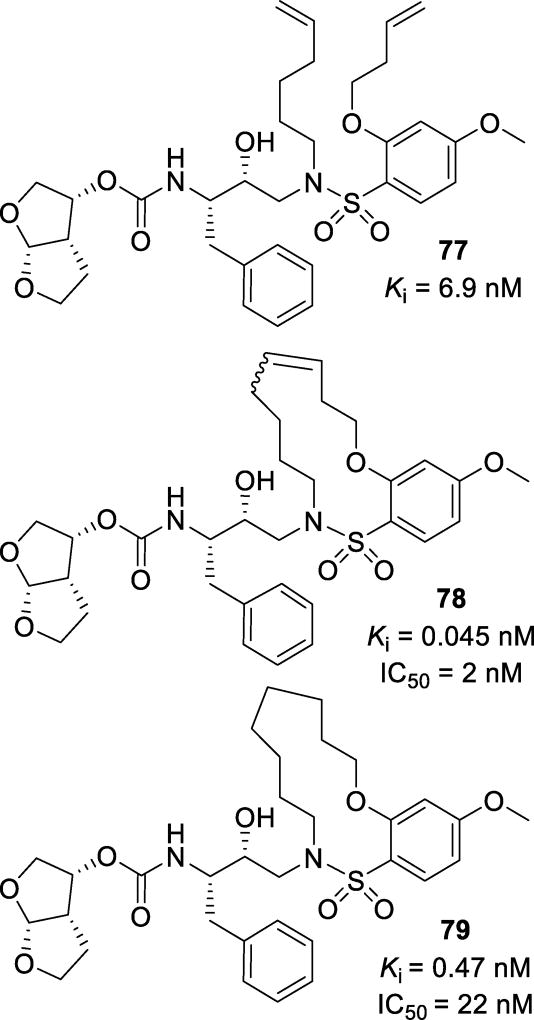
A series of macrocyclic structures have been designed as the P1’-P2’ ligands.138, 139 Various 9–15 membered ring macrocycles were synthesized and evaluated. To compare the effect of the cyclization on inhibitor activity, the pre-cyclized linear analogs (77, Figure 45) were also evaluated.138 The macrocyclic analogs (78 and 79) were more potent than their linear analogs. Unsaturated inhibitor 78 was formed as an E:Z ratio of 3:1; the mixture showed a Ki of 0.045 nM and an IC50 of 2 nM. Once saturated, inhibitor 79 displayed a Ki of 0.47 nM and an IC50 of 22 nM. The X-ray crystal structure showed that the cyclic P1’ structure fits into the S1’ binding pocket in a zig-zag conformation, allowing for optimal hydrophobic contacts and dipole interactions between the ligand and the protease (Figure 46).138
Figure 45.
Macrocyclic inhibitors spanning the P1’ and P2’ subsites
Figure 46.
X-ray crystal structure of 78-bound HIV protease (PDB: 3I6O)
(n) Non-Sulfonamide Inhibitors
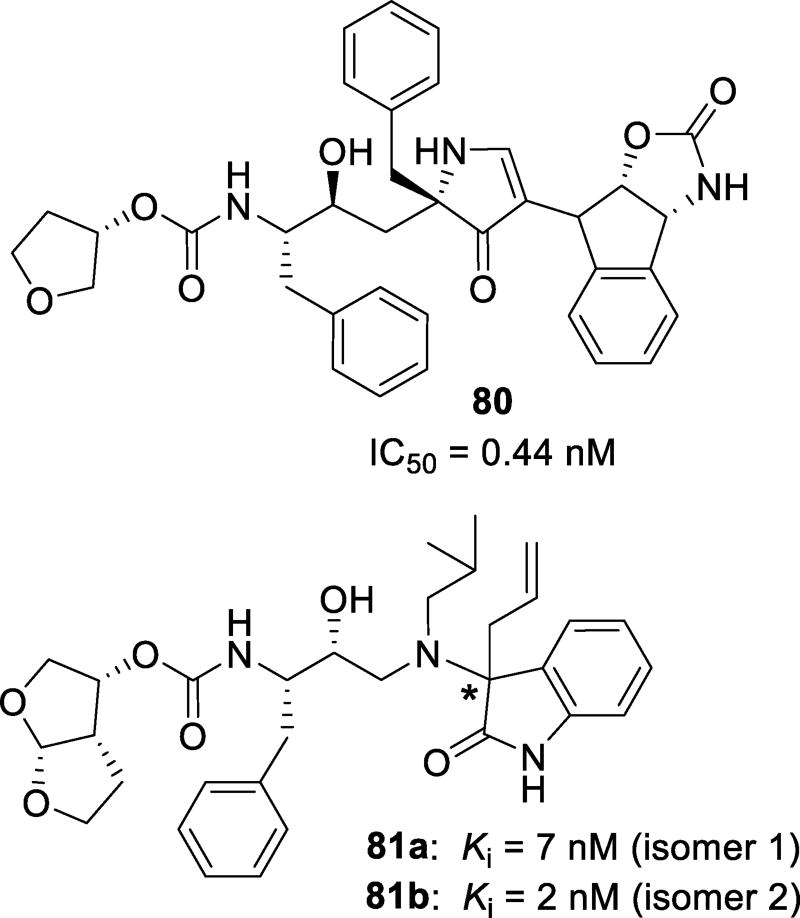
A 3,5-disubstituted pyrrolidinone scaffold was previously developed as a successful β-sheet/strand mimic. This template showed great potential for acting as a β-turn mimic.140 Since many aspartic acid proteases, including HIV-1 protease, bind their targets in an extended β-strand conformation, it was speculated that appropriately substituted pyrrolidinones can be developed as a PI scaffold.141 A series of PIs were designed based on the structure of amprenavir with a 3,5-disubstituted pyrrolidinone as the P2’ substituent. The most potent inhibitor, 80 (Figure 47), contained a chiral indane oxazolidinone skeleton. This compound exhibited a sub-nanomolar IC50 value. An X-ray crystal structure of this inhibitor complexed with wild-type protease revealed the displacement of a water molecule in the S2’ binding pocket of the enzyme led to an entropic gain.141
Figure 47.
Non-sulfonamide inhibitors 80, 81a, and 81b
A series of inhibitors with oxyindole as the P2’ ligands was designed in an attempt to maximize the water-mediated hydrogen bonding interactions with amide NHs of Ile50 and Ile50’.142 The two most potent compounds were diastereomers 81a and 81b (Figure 47). The absolute stereochemistry of these isomers was not determined. The enzyme did not show a preference for either isomer. Both derivatives 81a and 81b were nearly equipotent, showing Ki values of 7 and 2 nM. Six and seven membered spirocyclic oxyindole derivatives were also synthesized; however, they were less potent compared to their linear analogs.142
(o) Allophenylnorstatine-Containing Inhibitors
A series of PIs was designed by incorporating the allophenylnorstatine isostere and (R)-5,5-dimethyl-1,3-thiazolidine-4-carboxamide as the P1’ substituent.143 Inhibitors 82 and 83 (Figure 48) showed EC50 values of 0.033 µM and 1.29 µM, respectively.144 Compound 84, containing a β-methyl allyl derivative as the P2’ moiety, showed significant affinity for the wild-type protease (EC50 = 0.010 µM). However, inhibitor solubility was moderate. An X-ray structure of the inhibitor-protease complex revealed important ligand-binding site interactions (Figure 49). The β-methyl allyl group formed hydrophobic contacts with residues Ala28, Ile84, Ile50’, Val32 and Ile 47. This allyl moiety was thought to compensate for the interactions of the phenyl group found in 82.144
Figure 48.
Structure and activity of inhibitors 82–85
Figure 49.
X-ray crystal structure of 84-bound HIV protease (PDB: 3A2O)
The (R)-5,5-dimethylthiazolidine-4-carboxamide derivatives were further modified to improve antiviral properties.145 A number of compounds based on the 82 substructure were designed and synthesized. Among these compounds, inhibitor 85 (Figure 48), containing bis-THF as the P2 ligand, showed an enzymatic Ki of 0.0052 nM and antiviral IC50 of 0.009 µM. When tested against multidrug-resistant HIV-1 variants, this inhibitor showed a 1–8-fold decrease in potency.145 The X-ray structural studies of inhibitor 85 with HIV-1 protease revealed the enzyme-inhibitor interactions in the S1’ and S2’ binding pockets are similar to inhibitor 82. The potency enhancing effect of the bis-THF ligand along with the extensive hydrophobic contacts in the S1’ and S2’ pockets may be responsible for the potency and resistance profile of inhibitor 85.145
Further exploration of P2-ligands provided inhibitor 86 (Figure 50) which showed an EC50 value of 22 nM against wild-type virus.146 Inhibitor 87, with an acetyl P2 substituent and a p-methoxybenzyl-thiomethyl group as the P3 ligand, displayed an enzymatic IC50 of 0.18 nM and an antiviral EC50 of 13 nM. The resistance profile of this compound was evaluated with PI-resistant virus. Compound 87 retained good activity against multidrug-resistant HIV-1 variants (Table 11).143
Figure 50.
Allophenylnorstatine inhibitors 86 and 87
Table 11.
Resistance profile of inhibitor 87
| Virusa | EC50, nM (fold change) | ||
|---|---|---|---|
|
|
|||
| 87 | RTV | NFV | |
| HIV-1NL-432 | 17 | 80 | 25 |
| HIV-1RTVR | 18 (1.1) | 220 (2.8) | 28 (1.1) |
| HIV-1NFVR | 2.4 (0.14) | 16 (0.20) | 308 (12) |
| HIV-1RTV,NFVR | 14 (0.82) | 162 (2.0) | 67 (2.7) |
Amino acid substitutions in the protease-encoding region: N37S, G57R, I84V in HIV-1RTVR; D30N, N37S, G57R in HIV-1NFVR; L19V, V32I, M46L, G57R, L63P, I85V in HIV-1RTV,NFVR
A small library of thirty-two inhibitors containing the allophenylnorstatine core structure was synthesized and evaluated in enzymatic and cellular assays.147 Among the derivatives, compounds 88 and 89 (Figure 51) displayed the best activity. Resistance profiles of these inhibitors showed excellent properties against RTV- and NFV-resistant HIV-1 variants (Table 12). Inhibitor 89 displayed excellent antiviral activity showing an EC50 value of 6.4 nM and retained good antiviral activity against resistant strains. These compounds were evaluated in the presence of fetal calf serum and α1-acid glycoprotein to mimic blood. Both inhibitors showed reduced antiviral activity.147
Figure 51.
Allophenylnorstatine containing inhibitors 88 and 89
Table 12.
Resistance profiles of inhibitors 88 and 89
| Virusa | EC50, nM (fold change) | |||
|---|---|---|---|---|
|
|
||||
| 88 | 89 | RTV | NFV | |
| HIV-1NL-432 | 88 | 9.3 | 80 | 25 |
| HIV-1RTVR | 44 (0.50) | 17 (1.8) | 220 (2.8) | 28 (1.1) |
| HIV-1NFVR | 9.6 (0.11) | 0.67 (0.07) | 16 (0.2) | 308 (12) |
| HIV-1RTV,NFVR | 19 (0.22) | 13 (1.4) | 162 (2.0) | 67 (2.7) |
Amino acid substitutions in the protease-encoding region: N37S, G57R, I84V in HIV-1RTVR; D30N, N37S, G57R in HIV-1NFVR; L19V, V32I, M46L, G57R, L63P, I85V in HIV-1RTV,NFVR
(p) Diol and Pseudo-symmetric Inhibitors
Exploiting the symmetry of HIV protease, a number of new symmetric inhibitors have been designed and synthesized. As PR is a homodimer, it is conceivable that an identical binding pattern in both the prime and non-prime side of the enzyme could be accommodated. Pseudo-symmetric inhibitors featuring indanol ligands were shown to provide potent inhibitors. Compound 90 (Figure 52) was very potent with an enzymatic Ki of 19 nM.
Figure 52.
Indanol containing inhibitors 90–93
Compound 91 also showed good enzyme affinity with a Ki of 11 nM.148 Inhibitors 92 and 93 were designed with a similar isosteric core along with a P2 aminoindanol moiety and other varying P2’ ligands.149 Inhibitor 92 showed good potency with a Ki of 1.0 nM and an EC50 of 0.17 µM. Docking studies of inhibitor 92 did not show any polar interactions in the S2’ subsite as initially speculated. Incorporation of a more polar residue as a P2’ ligand resulted in compound 93. While this inhibitor was more potent in an enzymatic assay (Ki = 0.7 nM), it was less potent in an antiviral assay (EC50 = 0.33 µM). Inhibitor 93 has been shown to maintain good potency against multidrug-resistant HIV-1 variants with an EC50 of 0.30 µM. Docking studies, however, revealed that the methoxy substituent was not able to make any polar interactions in the S2’ subsite.149
Pseudo-symmetric inhibitor 94 (Figure 53), containing a Phe-Phe dihydroxyethylene isostere and valine derivatives as the P2 and P2’ ligands, was designed. This compound exhibited a Ki value of 1.3 nM.150 Incorporation of a phenylalanine-proline dihydroxyethylene isostere as the transition state mimic has also been investigated.151 Inhibitors 95 and 96 showed good enzymatic activity with IC50 values of <0.6 and 9.6 nM, respectively. Other pseudo-C2-symmetric inhibitors have been investigated, such as inhibitor 97.152 Featuring a biaryl P1’ moiety, inhibitor 97 showed an EC50 of 4 nM and very good resistance profile, with only 2- and 38-fold change in potency against two mutant patient isolates. When dosed with a subtherapeutic does of RTV in rats, no appreciable metabolism of inhibitor 97 was observed.152
Figure 53.
Pseudo-symmetric inhibitors 94–97
Further replacement of the phenylalanine side chain with a benzyloxy moiety provided a new symmetric dipeptide isostere. A series of inhibitors were designed and synthesized with arylmethoxy P1 and P1’ side chains. Inhibitors 98–101 (Figure 54) showed good potency against the enzyme with nanomolar to subnanomolar Ki values.153 Docking studies of stilbene derivative 101 showed that one of the hydroxyl groups was positioned between catalytic Asp25 and Asp25’. This inhibitor bound in three main conformations. One positioned the terminal phenyl rings between Phe53/53’ and Pro81/81’. In the second position, the benzyloxy group has hydrophobic interactions with Val82/82’. The third conformation finds the extended aryl system in a groove formed by Arg8/8’ and Leu10/10’.
Figure 54.
Symmetric inhibitors 98–101
Other symmetric inhibitors incorporating sulfoxides and sulfoximines as the transition state mimic to bind the catalytic aspartic acids were reported. Inhibitor 102 (Figure 55), with a sulfoxide functionality, showed potent inhibitor activity with an IC50 of 21.1 nM.154 It was envisioned that the incorporation of a sulfoximine rather than a sulfoxide may better act as a transition state mimic. Inhibitor 103 proved to be much more potent than the sulfoxide derivative 102, with an IC50 of 2.5 nM. Docking studies of this compound in the HIV-1 protease active site revealed that the inhibitor formed extensive hydrogen bonding interactions in the active site.155 The amide NHs formed hydrogen bonds with the carbonyl of Gly27/27’. The hydroxyl groups of the aminoindanol moiety made hydrogen bonds with the carboxylate oxygens of Asp29/29’. As expected, the sulfoximine is within hydrogen bonding distance to the catalytic Asp25/25’.155 Symmetric inhibitor 104, based on a benzodiazepine scaffold, showed promise with an IC50 of 0.03 µM.156
Figure 55.
Other symmetric HIV protease inhibitors
(q) Non-Transition State Inhibitors
A series of inhibitors were designed that do not utilize a transition-state mimic to bind the catalytic aspartic acids. Inhibitor 105 (Figure 56) displayed an HIV-1 protease inhibitory IC50 of 10 µM.157 Chromanone derivatives 106 and 107 were designed to serve as dual HIV-1 protease and HIV-1 reverse transcriptase inhibitors.158, 159 A part of the inhibitors resembles AZT, the first FDA approved treatment for HIV, to target reverse transcriptase. Inhibitor 106 inhibited protease with an IC50 of 27.06 µM and reverse transcriptase with an IC50 of 5.59 µM. It was speculated that incorporation of the N-benzyl in 107 would improve HIV-1 protease inhibitory activity; however, it resulted in decreased PR activity and improved RT inhibition with IC50 values of 35.06 and 3.40 µM, respectively. Docking studies showed that both 106 and 107 fit into the active site of PR similarly, forming hydrogen bonds at both the AZT and coumarin parts of the inhibitors. Inhibitor 108 with a dihydropyranone core had an IC50 of 1.7 µM.160 Presumably, the enol hydroxyl group binds the catalytic Asp25 and Asp25’ similar to TPV.
Figure 56.
Coumarin and pyrone inhibitors 105–108
(r) Cyclic Sulfamide Inhibitors
C2 symmetric cyclic sulfamide-based inhibitors were designed based upon the success of cyclic urea-derived HIV-1 protease inhibitors.161 A variety of biaryl structural motifs were investigated. Inhibitor 109 (Figure 57) contained 2-benzofuran substituents and showed a Ki of 0.53 µM against the wild-type HIV-1 protease. Based on computational modeling, it was speculated that the oxygen of the benzofurans may interact favorably with the NH of the backbone of Gly48 and better fill the S3 pocket.161 Other cyclic sulfamide based inhibitors 110 and 111 were synthesized incorporating 2-naphthalene acetamide substituents on the benzyl side chain.162 The symmetric inhibitor 110 was much more potent than nonsymmetric inhibitor 111 with Ki values of 20 nM and 140 nM, respectively. Other substituents were investigated and, in general, the symmetric inhibitors were significantly more potent than their asymmetric analogs. Incorporation of an ortho-bromo substituent on the N-benzyl side chain of 111 resulted in inhibitor 112. This compound displayed a Ki of 280 nM, a 14-fold loss of activity compared to its symmetric analog, 110.163
Figure 57.
Cyclic sulfamide inhibitors 109–112
Cyclic sulfonamide-derived HIV-1 protease inhibitors were designed by restricting the conformation of acyclic derivatives.164 Inhibitors 113 (Figure 58), with a para-fluorosulfonamide P2 ligand, and 114, with a para-methoxysulfonamide P2 ligand, were found to be equipotent with Ki values of 20 nM and 19 nM respectively. The X-ray crystallographic structure of 113 with mutant HIV-1 protease was determined (Figure 59). The hydroxyl group of this inhibitor makes the expected hydrogen bonds with Asp25 and Asp25’. The key conserved structural water molecule formed a tetra-coordinated hydrogen bonding interaction with Ile50 and Ile50’, the sulfonamide oxygen, and the carbamate carbonyl oxygen. The methyl group on the sulfonamide ring forms hydrophobic interactions with Leu23, Val82, and Ile84.164
Figure 58.
Cyclic sulfonamide inhibitors 113 and 114
Figure 59.
X-ray crystal structure of 113-bound mutant protease (Q7K, L33I, L63I PDB: 3TH9)
(s) Lysinol Containing Inhibitors
A series of compounds incorporating the structures of lysine, ornithine, and arginine have been synthesized in an attempt to inhibit protease activity.165 Compounds 115, 116, and 117 (Figure 60) were very potent, showing IC50 values of 5.0, 3.9, and 2.1 nM, respectively. It was observed that the alkyl chain length is very important. Variation of even one carbon leads to compounds that either displayed a significant drop in affinity or a complete loss of activity.165 Further modification of the lysine-derived structures resulted in inhibitors 118 and 119, with Ki values of 40 and 20 pM, respectively.166 These compounds were investigated in cellular assays against two viral strains (Table 13). Both compounds maintained good antiviral activity against all strains evaluated in this study.166
Figure 60.
Lysine-derived inhibitors 115–119
Table 13.
Resistance profile of inhibitors 118 and 119
| Virusa | EC50, nM (fold change) | |
|---|---|---|
|
|
||
| 118 | 119 | |
| HIV-1NL4-3 | 16 | 20 |
| HIV-14596 | 38 (2.3) | 13 (0.65) |
| HIV-1SaqR | 19 (1.1) | 4 (0.2) |
Amino acid substitutions in the protease-encoding region: L10R, M46I, L63P, V82T, in HIV-14596; I48V, L90M in HIV-1SaqR
Further modification of lysine derivatives was carried out by incorporation of chiral alkyl substituents adjacent to the lysine amine. Compound 120 (Figure 61), with an S-methyl group at the ε position, resulted in a very potent inhibitor with an IC50 of 7 pM. Replacement of the methyl group with an ethyl group, while retaining the stereochemical configuration, resulted in 121 with an IC50 of 16 pM.167 Initial pharmacokinetic studies showed good oral bioavailability, showing an AUC of 0.32 and 1.05 µM·h, respectively, when dosed at 4 mpk in beagle dogs. Inhibitor 121 showed a half-life of 7.4 hours. In an attempt to further improve the pharmacokinetic properties of these compounds, this series was further elaborated to investigate both the N-alkyl and ε-alkyl substituents. Inhibitors 122 and 123, featuring an N-propyl substituent, while maintaining the methyl or, in the case of 123, trifluoromethyl, at the ε-position, were synthesized.168 Both of these inhibitors lost potency as compared to 120, displaying IC50 values of 28 and 22 pM, respectively. However, these compounds displayed a better pharmacokinetic profile, particularly in dogs (Table 14).168
Figure 61.
Lysinol derived inhibitors 120–123
Table 14.
Pharmacokinetic data for inhibitors 121, 122, and 123
| Compound | Rat PK | Dog PK | ||||
|---|---|---|---|---|---|---|
|
|
||||||
| Cla (ml/min·kg) |
AUCb (µM·h) |
t1/2a (h) |
Clc (ml/min·kg) |
AUCd (µM·h) |
t1/2c (h) |
|
| 121 | 48.5 | 1.17 | 2.13 | 10.1 | 1.05 | 7.4 |
| 122 | 30 | 1.35 | 12.4 | 5.3 | 2.48 | 9.1 |
| 123 | 17.4 | 0.56 | 3.38 | 4.7 | 3.62 | 5.2 |
Cl, clearance; dosed at 2 mpk IV;
AUC, area under the curve; dosed at 10 mpk PO;
Cl, clearance; dosed at 1 mpk IV;
AUC, area under the curve; dosed at 4 mpk PO.
(t) Cyclic Urea-derived Inhibitors
Based upon the tetrahydropyrimidinone P2 scaffold of lopinavir, a series of inhibitors was designed and synthesized to incorporate an imidizolidinone moiety as the P2 ligand. Compound 124 (Figure 62) contains a thiazole extension off the imidizolidinone ring. Compound 124 displayed an enzymatic EC50 of 0.005 µM.169 In order to address solubility issues, the thiazole moiety was optimized. Inhibitor 125, with a 3-pyridyl aryl group and an exocyclic alkene on the imidizolidinone, proved to be a potent inhibitor against both wild-type and mutant HIV proteases, showing EC50 values of 1 nM against both enzymes. The cyclic urea analogs were found to have better aqueous solubility than their imide counterparts. Inhibitor 125 displayed better overall exposure when in vivo pharmacokinetic studies were done in rats with ritonavir as a pharmacokinetic booster.170 Inhibitor 126, with a dione derivative, was very potent, displaying an EC50 of 1 nM. When tested against two patient isolates, inhibitor 126 showed EC50 values of 8.4 nM and 11.3 nM, respectively.171 A cyclic urea derivative with a biaryl P1’ ligand has been designed and synthesized to address high metabolic clearance and subsequent side effects. Inhibitor 127 was potent in a cellular assay against wild-type HIV with an EC50 of 3 nM. When tested against a panel of thirteen mutant strains of HIV, only a 1–33-fold loss in potency was observed.172
Figure 62.
Structures and activities of imidizolidinone-derived inhibitors 124–127
Cyclic lactams and azaureas have also been investigated as potential protease inhibitors. A series of inhibitors containing a seven-membered lactam was investigated.173 Inhibitor 128 (Figure 63), containing an epoxide at the scissile site, was more potent than the corresponding cis-diol derivative 129.174 Inhibitors 130 and 131 were very potent displaying IC50 values of 61 nM and 4 nM, respectively. The X-ray crystal structure of 130-bound HIV-1 protease was determined (Figure 64). The 3-methoxy and 4-hydroxy substituents of the aromatic ring formed hydrogen bonds with Asp29, Asp30, Asp29’, and Asp30’. The carbonyl of the urea formed direct hydrogen bonding interactions with the backbone NH of the Ile50 and Ile50’. The acyl side chain forms many favorable hydrophobic interactions with Leu23’, Val82’, and Pro81’. The phenyl ring nestles between Gly49’, Val82, and Ile84 and shows good van der Waals interactions. The hydroxyl group binds to the catalytic Asp25 and Asp25’.174 Inhibitor 131 was evaluated against drug-resistant HIV strains (Table 15). It suffered a 14- and 16-fold loss of potency against these strains; however, it showed 7- and 16-fold better activity than lopinavir. Preliminary pharmacokinetic studies showed that these compounds had very poor bioavailibilities ranging between 0–23%, even when co-dosed with ritonavir as a pharmacokinetic booster.174
Figure 63.
Lactam and azaurea containing inhibitors 128–131
Figure 64.
X-ray crystal structure of 130 in the active site of HIV-protease (PDB: 2A4F)
Table 15.
Resistance profile of 131. NT=not tested
| Virusa | EC50, µM (fold change) | ||
|---|---|---|---|
|
|
|||
| 131 | RTV | LPV | |
| HIV-1WT | 0.004 | 0.022 | 0.016 |
| HIV-1INDR | 0.054 (13.5) | NT | 0.385 (24.1) |
| HIV-1A17 | 0.065 (16.3) | 1.24 (56.4) | 1.06 (66.3) |
Amino acid substitutions in the protease-encoding region L10R, M46I, L63P, V82T, I84V for HIV-1INDR; L10F, V32I, M46I, I47V, Q58E, I84V for HIV-1A17
(u) Natural Product-based inhibitors
Nature continues to provide structurally intriguing protease inhibitor leads which may be further optimized to novel inhibitors. Semi-synthetic dammarane-type triterpene derivatives were examined against HIV-1 protease. Compounds 132 and 133 (Figure 65) showed mild HIV-1 protease inhibitory activity with both compounds showing an IC50 of 2.7 µM.175 Schisanlactone A (134) was isolated from the evergreen shrub Kadsura longipedunculata.176 Structurally related compound 135 was isolated from Ganoderma colossum, a Vietnamese mushroom. These compounds displayed IC50 values of 20 and 5 µg/mL, respectively.177 Other terpene derivatives, both natural and semi-synthetic, have been evaluated as protease inhibitors. Semi-synthetic triterpene derivative 136 (Figure 66) was derived from oleanolic acid and it showed moderate anti-HIV-1 PR activity with an IC50 of 3.9 µM.178 A similar triterpenoid, 137, was isolated from Stauntonia obovatifolia. It showed modest activity with an IC50 of 8.7 µg/mL.179 2-O-acetyldryopteric acid, 138, which was isolated from the rhizome of Dryopteris crossrhizoma, showed similar activity with an IC50 value of 1.7 µM.180
Figure 65.
Structures and activities of natural products 132–135
Figure 66.
Natural products 136–138 and their activity against HIV-1 protease
Ganomycin I and ganomycin B (139 and 140, Figure 67) were also isolated from Ganoderma colossum. They showed similar anti-HIV-1 activity with IC50 values of 7.5 and 1.0 µg/mL, respectively.181 A structurally related molecule, bilobol C (141), was isolated from the sarcotestas of Ginkgo biloba. It showed moderate activity against HIV-1 protease with an IC50 of 2.6 µM.182 Symmetric saururin B (142) was isolated from the Korean medicinal plant Saururus chinensis.183 It inhibited HIV-1 protease with an IC50 of 5.6 µM.
Figure 67.
Structure and activity of natural products 139–142
(v) Pentacycloundecane-Containing Inhibitors
In an attempt to enhance activity and slow metabolic degradation of inhibitors, caged structures, in particular, pentacycloundecanes (PCU), have been investigated as PIs. PCU was introduced into the natural substrate in place of phenylalanine. The most potent compound of the series is derivative 143 (Figure 68) which showed an IC50 of 78 nM.184 Molecular docking studies revealed that the PCU hydroxyl group formed hydrogen bonds with Asp25 and Asp25’, and the rest of the inhibitor showed binding interactions similar to its substrate.
Figure 68.
PCU containing inhibitors 143 and 144
Inhibitors containing a PCU moiety were further modified to improve enzyme affinity.185 The most potent compound in this series was compound 144 which displayed an IC50 of 75 nM. It has been hypothesized that the long flexible peptides can exist in many conformations and that rigidifying these structures can greatly improve binding affinity. The binding affinity of both the PCU diol and PCU lactam are similarly active for the protease, though they are structurally different.184, 185
(w) Dimerization Inhibitors
With growing resistance to active site inhibitors, other mechanisms of inhibition have been explored. As HIV-1 protease is only proteolytically active as a dimer, it is feasible that the inhibition of this protein-protein interaction will stop the reproduction of the virus. Four antiparallel β sheets of the C- and N-terminus of each monomer create the dimer interface, consisting of residues 1–5 and 95–99.186, 187 Ile3, Leu5, Thr96, and Leu97 are particularly important for dimerization. It has been shown that mutations of these residues disrupts the dimerization process. Thr26, Asp29, and Arg87 are also important and mutations of these residues result in destabilized dimers prone to dissociation.188 Protease dimerization occurs through a two-step process. First, initial weak intermolecular interactions in the active site brings the two monomer units in proximity, creating a loosely associated dimer. Second, subsequent interactions at the dimerization interface associates the dimer and creates the mature enzyme.77 As a secondary mechanism of action, darunavir and structurally-related inhibitors block the dimerization of HIV protease by binding to monomers, but does not dissociate mature dimerized protease.188, 78 Other inhibitors have been designed and synthesized specifically to target the dimerization of HIV protease.
A class of dimerization inhibitors termed “molecular tongs” have been designed to intercalate into the monomer dimerization interface and prevent dimerization. Inhibitor 145 (Figure 69) incorporates a pyrimidinone moiety in an attempt to reduce the peptidic character and create more proteolytically stable inhibitors. Compound 145 showed a dimerization inhibition Ki of 0.4 µM; however, its poor water solubility limits its clinical viability.189 Polar substituents were incorporated into this scaffold to address this issue. Compounds 146 and 147 incorporate a carbamate moiety into this scaffold. This increased the anti-dimerization activity of these compounds (Ki = 0.04 µM and 0.06 µM, respectively) and improved their aqueous solubility.190 Further investigations included incorporation of a carbonyl hydrazide motif to further address solubility concerns which resulted in more stable inhibitors. Inhibitor 148, with pseudopeptidic arms containing a hydrazine bridge, proved to be a very potent derivative with a Ki of 50 nM.191
Figure 69.
Dimerization inhibitors of HIV protease
Other dimerization inhibitors are composed of two peptide chains connected by alkyl linkers. Inhibitor 149 (Figure 70) was potent with a Ki of 85 nM. However, the peptidic features of this inhibitor may leave it susceptible to proteolytic cleavage. It was envisioned that a peptoid containing inhibitor may be able to make similar interactions with the dimerization interface while circumventing potential peptidase degradation. Inhibitor 150, the fully peptoid derivative of 149, only displayed a Ki of 801 nM. However, optimization of the linker length, side chain length, and side chain functionality resulted in inhibitor 151 which showed an improved Ki of 135 nM.192
Figure 70.
Peptoid-based dimerization inhibitors
Inhibitors such as these can also be modulated between allosteric competitive inhibitors and dimerization inhibitors. Inhibitor 152 (Figure 71) was found to be a weak competitive allosteric inhibitor with an IC50 of 5.8 µM. Substitution on one of the benzene rings showed that this activity can be changed from allosteric to dimerization depending on the location of the substituent. Inhibitor 153, containing a meta-hydroxy derivative, displayed an IC50 value of 1130 nM. It showed competitive inhibition. Inhibitor 154, with a para-amine substituent, showed dimerization inhibition with an IC50 of 650 nM. Phenyl ether derivative 155, also exhibited potent dimerization inhibitory activity with an IC50 value of 96 nM.193
Figure 71.
Inhibitors with allosteric and dimerization inhibitory activity
Future Directions
Due to the sustained advances of antiretroviral therapies in the last three decades, HIV/AIDS can now be perceived as a chronic illness. HIV-1 protease inhibitors continue to play an important role in today’s combination therapy with reverse transcriptase inhibitors and other antiviral drugs. HIV/AIDS patients have to maintain lifelong adherence to antiretroviral therapy and are confronted with managing this chronic disease for many years. Therefore, long-term HIV care requires more efficient therapy with less drug-related side effects and toxicities. In this context, the design and development of new and robust protease inhibitors with broad spectrum activity against multidrug-resistant HIV-1 variants is essential. The first-generation protease inhibitors demonstrated effectiveness of combination therapy. However, rapid emergence of resistance and cross-resistance brought new challenges in HIV management. Second-generation HIV-1 protease inhibitors, particularly darunavir, were developed to address these concerns. However, there is no doubt that drug-resistance will remain a challenging issue to the scientific community. In addition, new therapy must demonstrate superiority over current treatment with convenient dosing regimens. These stringent requirements have resulted in reduced investment in new protease inhibitors by the pharmaceutical industry; yet, the overall outlook for new protease inhibitors is not so grim.
As outlined in this review, many new classes of potent HIV-1 protease inhibitors with innovative ligands and functionalities have been developed. A number of protease inhibitors have novel structural features with favorable resistance profiles, showing clinical potential. Protease inhibitor TMC310911 (14), with structural features similar to darunavir, is in advanced clinical development. A few other new inhibitors with promising results are also in preclinical development, bringing hope for improved long-term treatment of HIV/AIDS. HIV-1 protease is a validated biochemical target and novel transition-state binding inhibitors with innovative ligands and scaffolds have emerged, providing potent inhibitors with excellent antiviral activity against multidrug-resistant HIV-1 variants. Further structural evolution will continue and more effective novel inhibitors will emerge. Inhibitor design targeting the protein backbone will continue to play an important role in the design of inhibitors to combat drug resistance. High resolution inhibitor-protease structures, offering molecular insight into their interactions, will further inspire innovative design and medicinal chemistry efforts leading to new classes of protease inhibitors.
Figure 7.
Structures of ritonavir and cobicistat
Acknowledgments
Financial support of this research by the National Institutes of Health (GM53386) and Purdue University is gratefully acknowledged. We would like to thank our colleagues Mr. Luke Kassekert, Ms. Anne Veitschegger and Mr. Anindya Sarkar (Purdue University) for helpful discussions.
Abbreviations Used
- AIDS
acquired immunodeficiency syndrome
- APV
Amprenavir
- ATV
Atazanavir
- bis-THF
bis-tetrahydrofuran
- CNS
central nervous system
- CYP3A4
Cytochrome P450-3A4
- DRV
Darunavir
- HAART
highly active antiretroviral therapy
- HIV-1
human immunodeficiency virus type 1
- IDV
Indinavir; isothermal titration calorimetry
- LPV
Lopinavir
- MDR
multidrug-resistant
- NFV
Nelfinavir
- PCU
pentacycloundecane
- PDB
Protein Data Bank
- PI
protease inhibitor
- PR
protease
- RTV
Ritonavir
- SQV
Saquinavir
- TPV
Tipranavir
- Tris-THF
tris-tetrahydrofuran
Biographies
Arun K. Ghosh received his B.S. and M.S. in chemistry from University of Calcutta and Indian Institute of Technology, Kanpur, India, respectively. He obtained his Ph.D. in chemistry from University of Pittsburgh (1985) and then pursued postdoctoral research with Professor E. J. Corey at Harvard University (1985–1988). He was a research fellow at Merck Research Laboratories prior to joining the faculty at University of Illinois at Chicago as an assistant Professor in 1994. In 2005, he moved to Purdue University with a joint appointment in the department of Chemistry and the department of Medicinal Chemistry. In 2009, he was named the Ian P. Rothwell Distinguished Professor of Chemistry and Medicinal Chemistry. His research interests are in the areas of synthetic organic, bioorganic, and medicinal chemistry.
Heather L. Osswald received her B.S. in chemistry from Missouri State University in Springfield, MO. Upon graduation in 2012, she came to Purdue University to join the laboratory of Professor Ghosh to pursue a Ph.D. in organic and medicinal chemistry. Her current research focuses on the structure-based drug design of small molecule inhibitors, including inhibitors of HIV-1 protease and BACE1 for HIV/AIDS and Alzheimer’s disease, respectively. She maintains independent interests in neurological disorders, including neurodegeneration and mental illness, and diseases of the developing world.
Gary Prato received his bachelor’s degree in chemistry with a concentration in biochemistry from Monmouth University in West Long Branch, NJ. After graduating in 2014, he began his pursuit towards a Ph.D. in medicinal chemistry at Purdue University in West Lafayette, IN, under the supervision of Professor Ghosh. His current research involves the structure-based design of aspartic acid protease inhibitors for the treatment of HIV/AIDS and Alzheimer’s disease.
References
- 1.Barré-Sinoussi F, Chermann JC, Rey F, Nugeyre MT, Chamaret S, Gruest J, Dauguet C, Axlerblin C, Vézinet-Brun F, Rouzioux C, Rozenbaum W, Montagnier L. Isolation of a T-lymphotropic retrovirus from a patient at risk for acquired immune-deficiency syndrome (AIDS) Science. 1983;220:868–871. doi: 10.1126/science.6189183. [DOI] [PubMed] [Google Scholar]
- 2.Gallo RC, Sarin PS, Gelmann EP, Robert-Guroff M, Richardson E, Kalyanaraman VS, Mann D, Sidhu GD, Stahl RE, Zolla-Pazner S, Leibowitch J, Popovic M. Isolation of human T-cell leukemia virus in acquired immune deficiency syndrome (AIDS) Science. 1983;220:865–867. doi: 10.1126/science.6601823. [DOI] [PubMed] [Google Scholar]
- 3.UNAIDS. The Gap Report. Joint United Nations Programme on HIV/AIDS; Geneva: 2014. [Google Scholar]
- 4. [accessed February 4];WHO Global Health Observatory (GHO) data. http://www.who.int/gho/hiv/en/
- 5.Mehellou Y, De Clercq E. Twenty-six years of anti-HIV drug discovery: where do we stand and where do we go? J. Med. Chem. 2010;53:521–538. doi: 10.1021/jm900492g. [DOI] [PubMed] [Google Scholar]
- 6.Palmisano L, Vella S. A brief history of antiretroviral therapy of HIV infection: success and challenges. Ann. Ist Super Sanita. 2011;47:44–48. doi: 10.4415/ANN_11_01_10. [DOI] [PubMed] [Google Scholar]
- 7.Piot P, Abdool Karim SS, Hecht R, Legido-Quigley H, Buse K, Stover J, Resch S, Ryckman T, Møgedal S, Dybul M, Goosby E, Watts C, Kilonzo N, McManus J, Sidibé M. Defeating AIDS—advancing global health. The Lancet. 2015;386:171–218. doi: 10.1016/S0140-6736(15)60658-4. [DOI] [PubMed] [Google Scholar]
- 8.Maartens G, Celum C, Lewin SR. HIV infection: epidemiology, pathogenesis, treatment, and prevention. The Lancet. 2014;384:258–271. doi: 10.1016/S0140-6736(14)60164-1. [DOI] [PubMed] [Google Scholar]
- 9.Graves MC, Lim JJ, Heimer EP, Kramer RA. An 11-kDa form of human immunodeficiency virus protease expressed in Escherichia coli is sufficient for enzymatic activity. Proc. Nat. Acad. Sci. U. S. A. 1988;85:2449–2453. doi: 10.1073/pnas.85.8.2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farmerie WG, Loeb DD, Casavant NC, Hutchison CA, 3rd, Edgell MH, Swanstrom R. Expression and processing of the AIDS virus reverse transcriptase in Escherichia coli. Science. 1987;236:305–308. doi: 10.1126/science.2436298. [DOI] [PubMed] [Google Scholar]
- 11.Mitsuya H, Erickson J. Discovery and development of antiretroviral therapeutics for HIV infection. In: Merigan TC, Bartlett JG, Bolognesi D, editors. Textbook of AIDS Medicine. 2. Lippincott, Williams, & Wilkins; Baltimore, MD, USA: 1999. pp. 751–780. [Google Scholar]
- 12.Mitsuya H, Maeda K, Das D, Ghosh AK. Development of protease inhibitors and the fight with drug-resistant HIV-1 variants. In: Jeang K-T, editor. HIV-1: Molecular Biology and Pathogenesis: Clinical Applictions. 2. Vol. 56. Elsevier Inc; 2007. pp. 169–197. [DOI] [PubMed] [Google Scholar]
- 13.Ghosh AK, Chapsal BD. Second-generation approved HIV protease inhibitors for the treatment of HIV/AIDS. In: Ghosh AK, editor. Aspartic Acid Proteases as Therapeutic Targets. Vol. 45. Wiley-VCH; Weinheim, Germany: 2010. pp. 169–204. [Google Scholar]
- 14.Yarchoan R, Lietzau JA, Nguyen BY, Brawley OW, Pluda JM, Saville MW, Wyvill KM, Steinberg SM, Agbaria R, Mitsuya H, Broder S. A randomized pilot study of alternating or simultaneous zidovudine and didanosine therapy in patients with symptomatic human immunodeficiency virus infection. J. Infect. Dis. 1994;169:9–17. doi: 10.1093/infdis/169.1.9. [DOI] [PubMed] [Google Scholar]
- 15.McArthur JC, Brew BJ, Nath A. Neurological complications of HIV infection. Lancet Neurol. 2005;4:543–555. doi: 10.1016/S1474-4422(05)70165-4. [DOI] [PubMed] [Google Scholar]
- 16.Kramer-Hämmerle S, Rothenaigner I, Wolff H, Bell JE, Brack-Werner R. Cells of the central nervous system as targets and reservoirs of the human immunodeficiency virus. Virus Res. 2005;111:194–213. doi: 10.1016/j.virusres.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 17.Hué S, Gifford RJ, Dunn D, Fernhill E, Pillay D. Demonstration of sustained drug-resistant human immunodeficiency virus type 1 lineages circulating among treatment-naïve individuals. J. Virol. 2009;83:2645–2654. doi: 10.1128/JVI.01556-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Conway B. HAART in treatment-experienced patients in the 21st century: the audacity of hope. Fut. Virol. 2009;4:39–41. [Google Scholar]
- 19.Randolph J, DeGoey D. Peptidomimetic inhibitors of HIV protease. Curr. Top. Med. Chem. 2004;4:1079–1095. doi: 10.2174/1568026043388330. [DOI] [PubMed] [Google Scholar]
- 20.Zhan P, Pannecouque C, De Clercq E, Liu X. Anti-HIV drug discovery and development: current innovations and future trends. J. Med. Chem. [Online early access] doi: 10.1021/acs.jmedchem.5b00497. Published Online: October 28, 2015. [DOI] [PubMed] [Google Scholar]
- 21.Lv Z, Chu Y, Wang Y. HIV protease inhibitors: a review of molecular selectivity and toxicity. HIV/AIDS. 2015;7:95–104. doi: 10.2147/HIV.S79956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brik A, Wong CH. HIV-1 protease: mechanism and drug discovery. Org. Biomol. Chem. 2003;1:5–14. doi: 10.1039/b208248a. [DOI] [PubMed] [Google Scholar]
- 23.Wlodawer A, Miller M, Jaskólski M, Sathyanarayana B, Baldwin E, Weber I, Selk L, Clawson L, Schneider J, Kent S. Conserved folding in retroviral proteases: crystal structure of a synthetic HIV-1 protease. Science. 1989;245:616–621. doi: 10.1126/science.2548279. [DOI] [PubMed] [Google Scholar]
- 24.Lapatto R, Blundell T, Hemmings A, Overington J, Wilderspin A, Wood S, Merson JR, Whittle PJ, Danley DE, Geoghegan KF, et al. X-ray analysis of HIV-1 proteinase at 2.7 A resolution confirms structural homology among retroviral enzymes. Nature. 1989;342:299–302. doi: 10.1038/342299a0. [DOI] [PubMed] [Google Scholar]
- 25.Ghosh AK, Chapsal BD. Design of the anti-HIV protease inhibitor darunavir. In: Ganellin R, Roberts SM, Jefferies R, editors. Introduction to Biological and Small Molecule Drug Research and Development. Elsevier Ltd; 2013. pp. 355–385. [Google Scholar]
- 26.Gustchina A, Sansom C, Prevost M, Richelle J, Wodak SY, Wlodawer A, Weber IT. Energy calculations and analysis of HIV-1 protease-inhibitor crystal structures. Protein Eng. 1994;7:309–317. doi: 10.1093/protein/7.3.309. [DOI] [PubMed] [Google Scholar]
- 27.Hammer SM, Squires KE, Hughes MD, Grimes JM, Demeter LM, Currier JS, Eron JJ, Jr, Feinberg JE, Balfour HH, Jr, Deyton LR, Chodakewitz JA, Fischl MA. A controlled trial of two nucleoside analogues plus indinavir in persons with human immunodeficiency virus infection and CD4 cell counts of 200 per cubic millimeter or less. AIDS Clinical Trials Group 320 Study Team. N. Engl. J. Med. 1997;337:725–733. doi: 10.1056/NEJM199709113371101. [DOI] [PubMed] [Google Scholar]
- 28.Gulick RM, Mellors JW, Havlir D, Eron JJ, Gonzalez C, McMahon D, Richman DD, Valentine FT, Jonas L, Meibohm A, Emini EA, Chodakewitz JA. Treatment with indinavir, zidovudine, and lamivudine in adults with human immunodeficiency virus infection and prior antiretroviral therapy. N. Engl. J. Med. 1997;337:734–739. doi: 10.1056/NEJM199709113371102. [DOI] [PubMed] [Google Scholar]
- 29.Ghosh AK, Anderson DD, Mitsuya H. The FDA Approved HIV-1 Protease Inhibitors for Treatment of HIV/AIDS. In: Abraham DJ, Rotella DP, editors. Burger's Medicinal Chemistry and Drug Discovery. 7. Vol. 7. John Wiley & Sons, Inc; 2010. pp. 1–74. [Google Scholar]
- 30.Roberts NA, Martin JA, Kinchington D, Broadhurst AV, Craig JC, Duncan IB, Galpin SA, Handa BK, Kay J, Kröhn A, Lambert RW, Merrett JH, Mills JS, Parkes KE, Redshaw S, Ritchie AJ, Taylor DL, Thomas GJ, Machin PJ. Rational design of peptide-based HIV proteinase inhibitors. Science. 1990;248:358–361. doi: 10.1126/science.2183354. [DOI] [PubMed] [Google Scholar]
- 31.Krohn A, Redshaw S, Ritchie JC, Graves BJ, Hatada MH. Novel binding mode of highly potent HIV-proteinase inhibitors incorporating the (R)-hydroxyethylamine isostere. J. Med. Chem. 1991;34:3340–3342. doi: 10.1021/jm00115a028. [DOI] [PubMed] [Google Scholar]
- 32.Kempf DJ, Marsh KC, Denissen JF, McDonald E, Vasavanonda S, Flentge CA, Green BE, Fino L, Park CH, Kong XP, Wideburg NE, Saldivar A, Ruiz L, Kati WM, Sham HL, Robins T, Stewart KD, Hsu A, Plattner JJ, Leonard JM, Norbeck DW. ABT-538 is a potent inhibitor of human immunodeficiency virus protease and has high oral bioavailability in humans. Proc. Nat. Acad. Sci.U. S. A. 1995;92:2484–2488. doi: 10.1073/pnas.92.7.2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kumar GN, Rodrigues AD, Buko AM, Denissen JF. Cytochrome P450-mediated metabolism of the HIV-1 protease inhibitor ritonavir (ABT-538) in human liver microsomes. J. Pharmacol. Exp. Ther. 1996;277:423–431. [PubMed] [Google Scholar]
- 34.Vacca JP, Dorsey BD, Schleif WA, Levin RB, Mcdaniel SL, Darke PL, Zugay J, Quintero JC, Blahy OM, Roth E, Sardana VV, Schlabach AJ, Graham PI, Condra JH, Gotlib L, Holloway MK, Lin J, Chen IW, Vastag K, Ostovic D, Anderson PS, Emini EA, Huff JR. L-735,524 - an Orally Bioavailable Human-Immunodeficiency-Virus Type-1 Protease Inhibitor. Proc. Nat. Acad. Sci. U. S. A. 1994;91:4096–4100. doi: 10.1073/pnas.91.9.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaldor SW, Kalish VJ, Davies JF, 2nd, Shetty BV, Fritz JE, Appelt K, Burgess JA, Campanale KM, Chirgadze NY, Clawson DK, Dressman BA, Hatch SD, Khalil DA, Kosa MB, Lubbehusen PP, Muesing MA, Patick AK, Reich SH, Su KS, Tatlock JH. Viracept (nelfinavir mesylate, AG1343): a potent, orally bioavailable inhibitor of HIV-1 protease. J. Med. Chem. 1997;40:3979–3985. doi: 10.1021/jm9704098. [DOI] [PubMed] [Google Scholar]
- 36.Kim EE, Baker CT, Dwyer MD, Murcko MA, Rao BG, Tung RD, Navia MA. Crystal structure of HIV-1 protease in complex with VX-478, a potent and orally bioavailable inhibitor of the enzyme. J. Am. Chem. Soc. 1995;117:1181–1182. [Google Scholar]
- 37.Sham HL, Kempf DJ, Molla A, Marsh KC, Kumar GN, Chen CM, Kati W, Stewart K, Lal R, Hsu A, Betebenner D, Korneyeva M, Vasavanonda S, McDonald E, Saldivar A, Wideburg N, Chen X, Niu P, Park C, Jayanti V, Grabowski B, Granneman GR, Sun E, Japour AJ, Leonard JM, Plattner JJ, Norbeck DW. ABT-378, a highly potent inhibitor of the human immunodeficiency virus protease. Antimicrob. Agents Chemother. 1998;42:3218–3224. doi: 10.1128/aac.42.12.3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bold G, Fässler A, Capraro HG, Cozens R, Klimkait T, Lazdins J, Mestan J, Poncioni B, Rösel J, Stover D, Tintelnot-Blomley M, Acemoglu F, Beck W, Boss E, Eschbach M, Hürlimann T, Masso E, Roussel S, Ucci-Stoll K, Wyss D, Lang M. New aza-dipeptide analogues as potent and orally absorbed HIV-1 protease inhibitors: candidates for clinical development. J. Med. Chem. 1998;41:3387–3401. doi: 10.1021/jm970873c. [DOI] [PubMed] [Google Scholar]
- 39.Doyon L, Tremblay S, Bourgon L, Wardrop E, Cordingley MG. Selection and characterization of HIV-1 showing reduced susceptibility to the non-peptidic protease inhibitor tipranavir. Antiviral Res. 2005;68:27–35. doi: 10.1016/j.antiviral.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 40.Deeks ED. Darunavir: a review of its use in the management of HIV-1 infection. Drugs. 2014;74:99–125. doi: 10.1007/s40265-013-0159-3. [DOI] [PubMed] [Google Scholar]
- 41.De Meyer S, Azijn H, Surleraux D, Jochmans D, Tahri A, Pauwels R, Wigerinck P, de Béthune MP. TMC114, a novel human immunodeficiency virus type 1 protease inhibitor active against protease inhibitor-resistant viruses, including a broad range of clinical isolates. Antimicrob. Agents Chemother. 2005;49:2314–2321. doi: 10.1128/AAC.49.6.2314-2321.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ghosh AK, Anderson DD, Weber IT, Mitsuya H. Enhancing protein backbone binding--a fruitful concept for combating drug-resistant HIV. Angew. Chem. Int. Ed. Engl. 2012;51:1778–1802. doi: 10.1002/anie.201102762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ghosh AK, Dawson ZL, Mitsuya H. Darunavir, a conceptually new HIV-1 protease inhibitor for the treatment of drug-resistant HIV. Bioorg. Med. Chem. 2007;15:7576–7580. doi: 10.1016/j.bmc.2007.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Koh Y, Nakata H, Maeda K, Ogata H, Bilcer G, Devasamudram T, Kincaid JF, Boross P, Wang YF, Tie Y, Volarath P, Gaddis L, Harrison RW, Weber IT, Ghosh AK, Mitsuya H. Novel bis-tetrahydrofuranylurethane-containing nonpeptidic protease inhibitor (PI) UIC-94017 (TMC114) with potent activity against multi-PI-resistant human immunodeficiency virus in vitro. Antimicrob. Agents Chemother. 2003;47:3123–3129. doi: 10.1128/AAC.47.10.3123-3129.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Koh Y, Amano M, Towata T, Danish M, Leshchenko-Yashchuk S, Das D, Nakayama M, Tojo Y, Ghosh AK, Mitsuya H. In vitro selection of highly darunavir-resistant and replication-competent HIV-1 variants by using a mixture of clinical HIV-1 isolates resistant to multiple conventional protease inhibitors. J Virol. 2010;84:11961–9. doi: 10.1128/JVI.00967-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ghosh AK. Harnessing nature's insight: design of aspartyl protease inhibitors from treatment of drug-resistant HIV to Alzheimer's disease. J. Med. Chem. 2009;52:2163–2176. doi: 10.1021/jm900064c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tie Y, Boross PI, Wang YF, Gaddis L, Hussain AK, Leshchenko S, Ghosh AK, Louis JM, Harrison RW, Weber IT. High resolution crystal structures of HIV-1 protease with a potent non-peptide inhibitor (UIC-94017) active against multi-drug-resistant clinical strains. J. Mol. Biol. 2004;338:341–352. doi: 10.1016/j.jmb.2004.02.052. [DOI] [PubMed] [Google Scholar]
- 48.Kozal MJ, Shah N, Shen N, Yang R, Fucini R, Merigan TC, Richman DD, Morris D, Hubbell E, Chee M, Gingeras TR. Extensive polymorphisms observed in HIV-1 clade B protease gene using high–density oligonucleotide arrays. Nat. Med. 1996;2:753–759. doi: 10.1038/nm0796-753. [DOI] [PubMed] [Google Scholar]
- 49.De Conto V, Braz AS, Perahia D, Scott LP. Recovery of the wild type atomic flexibility in the HIV-1 protease double mutants. J. Mol. Graph. Model. 2015;59:107–116. doi: 10.1016/j.jmgm.2015.04.006. [DOI] [PubMed] [Google Scholar]
- 50.Mitsuya H, Maeda K, Das D, Ghosh AK. Development of protease inhibitors and the fight with drug-resistant HIV-1 variants. Adv. Pharmacol. 2008;56:169–197. doi: 10.1016/S1054-3589(07)56006-0. [DOI] [PubMed] [Google Scholar]
- 51.Wensing AM, van Maarseveen NM, Nijhuis M. Fifteen years of HIV protease inhibitors: raising the barrier to resistance. Antiviral Res. 2010;85:59–74. doi: 10.1016/j.antiviral.2009.10.003. [DOI] [PubMed] [Google Scholar]
- 52.Gulnik SV, Suvorov LI, Liu B, Yu B, Anderson B, Mitsuya H, Erickson JW. Kinetic characterization and cross-resistance patterns of HIV-1 protease mutants selected under drug pressure. Biochemistry. 1995;34:9282–9287. doi: 10.1021/bi00029a002. [DOI] [PubMed] [Google Scholar]
- 53.Colman PM. New antivirals and drug resistance. Annu. Rev. Biochem. 2009;78:95–118. doi: 10.1146/annurev.biochem.78.082207.084029. [DOI] [PubMed] [Google Scholar]
- 54.Perno CF, Cozzi-Lepri A, Balotta C, Forbici F, Violin M, Bertoli A, Facchi G, Pezzotti P, Cadeo G, Tositti G, Pasquinucci S, Pauluzzi S, Scalzini A, Salassa B, Vincenti A, Phillips AN, Dianzani F, Appice A, Angarano G, Monno L, Ippolito G, Moroni M, d' Arminio Monforte A. Secondary mutations in the protease region of human immunodeficiency virus and virologic failure in drug-naive patients treated with protease inhibitor-based therapy. J. Infect. Dis. 2001;184:983–991. doi: 10.1086/323604. [DOI] [PubMed] [Google Scholar]
- 55.Condra JH, Holder DJ, Schleif WA, Blahy OM, Danovich RM, Gabryelski LJ, Graham DJ, Laird D, Quintero JC, Rhodes A, Robbins HL, Roth E, Shivaprakash M, Yang T, Chodakewitz JA, Deutsch PJ, Leavitt RY, Massari FE, Mellors JW, Squires KE, Steigbigel RT, Teppler H, Emini EA. Genetic correlates of in vivo viral resistance to indinavir, a human immunodeficiency virus type 1 protease inhibitor. J. Virol. 1996;70:8270–8276. doi: 10.1128/jvi.70.12.8270-8276.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Todd MJ, Luque I, Velázquez-Campoy A, Freire E. Thermodynamic basis of resistance to HIV-1 protease inhibition: calorimetric analysis of the V82F/I84V active site resistant mutant. Biochemistry. 2000;39:11876–11883. doi: 10.1021/bi001013s. [DOI] [PubMed] [Google Scholar]
- 57.Kozísek M, Bray J, Rezácová P, Sasková K, Brynda J, Pokorná J, Mammano F, Rulísek L, Konvalinka J. Molecular analysis of the HIV-1 resistance development: enzymatic activities, crystal structures, and thermodynamics of nelfinavir-resistant HIV protease mutants. J. Mol. Biol. 2007;374:1005–1016. doi: 10.1016/j.jmb.2007.09.083. [DOI] [PubMed] [Google Scholar]
- 58.Ohtaka H, Schön A, Freire E. Multidrug resistance to HIV-1 protease inhibition requires cooperative coupling between distal mutations. Biochemistry. 2003;42:13659–13666. doi: 10.1021/bi0350405. [DOI] [PubMed] [Google Scholar]
- 59.Turner D, Schapiro JM, Brenner BG, Wainberg MA. The influence of protease inhibitor resistance profiles on selection of HIV therapy in treatment-naive patients. Antivir. Ther. 2004;9:301–314. [PubMed] [Google Scholar]
- 60.Goldfarb NE, Ohanessian M, Biswas S, McGee TD, Jr, Mahon BP, Ostrov DA, Garcia J, Tang Y, McKenna R, Roitberg A, Dunn BM. Defective hydrophobic sliding mechanism and active site expansion in HIV-1 protease drug resistant variant Gly48Thr/Leu89Met: mechanisms for the loss of saquinavir binding potency. Biochemistry. 2015;54:422–433. doi: 10.1021/bi501088e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Heaslet H, Rosenfeld R, Giffin M, Lin YC, Tam K, Torbett BE, Elder JH, McRee DE, Stout CD. Conformational flexibility in the flap domains of ligand-free HIV protease. Acta Crystallogr. D Biol. Crystallogr. 2007;63:866–875. doi: 10.1107/S0907444907029125. [DOI] [PubMed] [Google Scholar]
- 62.Tie Y, Boross PI, Wang YF, Gaddis L, Liu F, Chen X, Tozser J, Harrison RW, Weber IT. Molecular basis for substrate recognition and drug resistance from 1.1 to 1.6 angstroms resolution crystal structures of HIV-1 protease mutants with substrate analogs. FEBS J. 2005;272:5265–5277. doi: 10.1111/j.1742-4658.2005.04923.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Virgil SC. First-generation HIV-1 protease inhibitors for the treatment of HIV/AIDS. In: Ghosh AK, editor. Aspartic Acid Proteases as Therapeutic Targets. Vol. 45. Wiley-VCH; Weinheim, Germany: 2010. pp. 139–168. [Google Scholar]
- 64.Weber IT, Agniswamy J. HIV-1 protease: structural perspectives on drug resistance. Viruses. 2009;1:1110–1136. doi: 10.3390/v1031110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mitsuya H, Ghosh AK. Development of HIV-1 protease inhibitors, antiretroviral resistance, and current challenges of HIV/AIDS management. In: Ghosh AK, editor. Aspartic Acid Proteases as Therapeutic Targets. Vol. 45. Wiley-VCH; Weinheim, Germany: 2010. pp. 245–262. [Google Scholar]
- 66.Hong L, Zhang XC, Hartsuck JA, Tang J. Crystal structure of an in vivo HIV-1 protease mutant in complex with saquinavir: insights into the mechanisms of drug resistance. Protein Sci. 2000;9:1898–1904. doi: 10.1110/ps.9.10.1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Laco GS, Schalk-Hihi C, Lubkowski J, Morris G, Zdanov A, Olson A, Elder JH, Wlodawer A, Gustchina A. Crystal structures of the inactive D30N mutant of feline immunodeficiency virus protease complexed with a substrate and an inhibitor. Biochemistry. 1997;36:10696–10708. doi: 10.1021/bi9707436. [DOI] [PubMed] [Google Scholar]
- 68.Ghosh AK, Chapsal BD, Weber IT, Mitsuya H. Design of HIV protease inhibitors targeting protein backbone: an effective strategy for combating drug resistance. Acc. Chem. Res. 2008;41:78–86. doi: 10.1021/ar7001232. [DOI] [PubMed] [Google Scholar]
- 69.Nalam MN, Ali A, Altman MD, Reddy GS, Chellappan S, Kairys V, Ozen A, Cao H, Gilson MK, Tidor B, Rana TM, Schiffer CA. Evaluating the substrate-envelope hypothesis: structural analysis of novel HIV-1 protease inhibitors designed to be robust against drug resistance. J. Virol. 2010;84:5368–5378. doi: 10.1128/JVI.02531-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Prabu-Jeyabalan M, Nalivaika EA, King NM, Schiffer CA. Viability of a drug-resistant human immunodeficiency virus type 1 protease variant: structural insights for better antiviral therapy. J. Virol. 2003;77:1306–1315. doi: 10.1128/JVI.77.2.1306-1315.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Salcedo Gómez PM, Amano M, Yashchuk S, Mizuno A, Das D, Ghosh AK, Mitsuya H. GRL-04810 and GRL-05010, difluoride-containing nonpeptidic HIV-1 protease inhibitors (PIs) that inhibit the replication of multi-PI-resistant HIV-1 in vitro and possess favorable lipophilicity that may allow blood-brain barrier penetration. Antimicrob. Agents Chemother. 2013;57:6110–6121. doi: 10.1128/AAC.01420-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kaul M, Garden GA, Lipton SA. Pathways to neuronal injury and apoptosis in HIV-associated dementia. Nature. 2001;410:988–994. doi: 10.1038/35073667. [DOI] [PubMed] [Google Scholar]
- 73.Ghosh AK, Yashchuk S, Mizuno A, Chakraborty N, Agniswamy J, Wang YF, Aoki M, Gomez PM, Amano M, Weber IT, Mitsuya H. Design of gem-difluoro-bis-tetrahydrofuran as P2 ligand for HIV-1 protease inhibitors to improve brain penetration: synthesis, X-ray studies, and biological evaluation. ChemMedChem. 2015;10:107–115. doi: 10.1002/cmdc.201402358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gerber JG. Using pharmacokinetics to optimize antiretroviral drug-drug interactions in the treatment of human immunodeficiency virus infection. Clin. Infect. Dis. 2000;30(Suppl 2):S123–S129. doi: 10.1086/313857. [DOI] [PubMed] [Google Scholar]
- 75.Xu L, Liu H, Murray BP, Callebaut C, Lee MS, Hong A, Strickley RG, Tsai LK, Stray KM, Wang Y, Rhodes GR, Desai MC. Cobicistat (GS-9350): a potent and selective inhibitor of human CYP3A as a novel pharmacoenhancer. ACS Med. Chem. Lett. 2010;1:209–213. doi: 10.1021/ml1000257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.McDonald CK, Martorell C, Ramgopal M, Laplante F, Fisher M, Post FA, Liu Y, Curley J, Abram ME, Custodio J, Graham H, Rhee MS, Szwarcberg J. Cobicistat-boosted protease inhibitors in HIV-infected patients with mild to moderate renal impairment. HIV Clin. Trials. 2014;15:269–273. doi: 10.1310/hct1506-269. [DOI] [PubMed] [Google Scholar]
- 77.Hayashi H, Takamune N, Nirasawa T, Aoki M, Morishita Y, Das D, Koh Y, Ghosh AK, Misumi S, Mitsuya H. Dimerization of HIV-1 protease occurs through two steps relating to the mechanism of protease dimerization inhibition by darunavir. Proc. Nat. Acad. Sci. U. S. A. 2014;111:12234–12239. doi: 10.1073/pnas.1400027111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Koh Y, Aoki M, Danish ML, Aoki-Ogata H, Amano M, Das D, Shafer RW, Ghosh AK, Mitsuya H. Loss of protease dimerization inhibition activity of darunavir is associated with the acquisition of resistance to darunavir by HIV-1. J. Virol. 2011;85:10079–10089. doi: 10.1128/JVI.05121-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yedidi RS, Maeda K, Fyvie WS, Steffey M, Davis DA, Palmer I, Aoki M, Kaufman JD, Stahl SJ, Garimella H, Das D, Wingfield PT, Ghosh AK, Mitsuya H. P2' benzene carboxylic acid moiety is associated with decrease in cellular uptake: evaluation of novel nonpeptidic HIV-1 protease inhibitors containing P2 bis-tetrahydrofuran moiety. Antimicrob. Agents Chemother. 2013;57:4920–4927. doi: 10.1128/AAC.00868-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yedidi RS, Garimella H, Aoki M, Aoki-Ogata H, Desai DV, Chang SB, Davis DA, Fyvie WS, Kaufman JD, Smith DW, Das D, Wingfield PT, Maeda K, Ghosh AK, Mitsuya H. A conserved hydrogen-bonding network of P2 bis-tetrahydrofuran-containing HIV-1 protease inhibitors (PIs) with a protease active-site amino acid backbone aids in their activity against PI-resistant HIV. Antimicrob. Agents Chemother. 2014;58:3679–3688. doi: 10.1128/AAC.00107-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Amano M, Koh Y, Das D, Li J, Leschenko S, Wang YF, Boross PI, Weber IT, Ghosh AK, Mitsuya H. A novel bis-tetrahydrofuranylurethane-containing nonpeptidic protease inhibitor (PI), GRL-98065, is potent against multiple-PI-resistant human immunodeficiency virus in vitro. Antimicrob. Agents Chemother. 2007;51:2143–2155. doi: 10.1128/AAC.01413-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang YF, Tie Y, Boross PI, Tozser J, Ghosh AK, Harrison RW, Weber IT. Potent new antiviral compound shows similar inhibition and structural interactions with drug resistant mutants and wild type HIV-1 protease. J. Med. Chem. 2007;50:4509–4515. doi: 10.1021/jm070482q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dierynck I, Van Marck H, Van Ginderen M, Jonckers TH, Nalam MN, Schiffer CA, Raoof A, Kraus G, Picchio G. TMC310911, a novel human immunodeficiency virus type 1 protease inhibitor, shows in vitro an improved resistance profile and higher genetic barrier to resistance compared with current protease inhibitors. Antimicrob. Agents Chemother. 2011;55:5723–5731. doi: 10.1128/AAC.00748-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Surleraux DL, de Kock HA, Verschueren WG, Pille GM, Maes LJ, Peeters A, Vendeville S, De Meyer S, Azijn H, Pauwels R, de Bethune MP, King NM, Prabu-Jeyabalan M, Schiffer CA, Wigerinck PB. Design of HIV-1 protease inhibitors active on multidrug-resistant virus. J. Med. Chem. 2005;48:1965–1973. doi: 10.1021/jm049454n. [DOI] [PubMed] [Google Scholar]
- 85.Stellbrink HJ, Arastéh K, Schürmann D, Stephan C, Dierynck I, Smyej I, Hoetelmans RM, Truyers C, Meyvisch P, Jacquemyn B, Marien K, Simmen K, Verloes R. Antiviral activity, pharmacokinetics, and safety of the HIV-1 protease inhibitor TMC310911, coadministered with ritonavir, in treatment-naive HIV-1-infected patients. J. Acquir. Immune Defic. Syndr. 2014;65:283–289. doi: 10.1097/QAI.0000000000000003. [DOI] [PubMed] [Google Scholar]
- 86.Sherrill RG, Furfine ES, Hazen RJ, Miller JF, Reynolds DJ, Sammond DM, Spaltenstein A, Wheelan P, Wright LL. Synthesis and antiviral activities of novel N-alkoxy-arylsulfonamide-based HIV protease inhibitors. Bioorg. Med. Chem. Lett. 2005;15:3560–3564. doi: 10.1016/j.bmcl.2005.05.101. [DOI] [PubMed] [Google Scholar]
- 87.Ghosh AK, Leshchenko-Yashchuk S, Anderson DD, Baldridge A, Noetzel M, Miller HB, Tie Y, Wang YF, Koh Y, Weber IT, Mitsuya H. Design of HIV-1 protease inhibitors with pyrrolidinones and oxazolidinones as novel P1'-ligands to enhance backbone-binding interactions with protease: synthesis, biological evaluation, and protein-ligand X-ray studies. J. Med. Chem. 2009;52:3902–3914. doi: 10.1021/jm900303m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Koh Y, Das D, Leschenko S, Nakata H, Ogata-Aoki H, Amano M, Nakayama M, Ghosh AK, Mitsuya H. GRL-02031, a novel nonpeptidic protease inhibitor (PI) containing a stereochemically defined fused cyclopentanyltetrahydrofuran potent against multi-PI-resistant human immunodeficiency virus type 1 in vitro. Antimicrob. Agents Chemother. 2009;53:997–1006. doi: 10.1128/AAC.00689-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ghosh AK, Martyr CD, Steffey M, Wang Y-F, Agniswamy J, Amano M, Weber IT, Mitsuya H. Design, synthesis, and X-ray structure of substituted bis-tetrahydrofuran (Bis-THF)-derived potent HIV-1 protease inhibitors. ACS Med. Chem. Lett. 2011;2:298–302. doi: 10.1021/ml100289m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hohlfeld K, Tomassi C, Wegner JK, Kesteleyn B, Linclau B. Disubstituted bis-THF moieties as new P2 ligands in nonpeptidal HIV-1 protease inhibitors. ACS Med. Chem. Lett. 2011;2:461–465. doi: 10.1021/ml2000356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hohlfeld K, Wegner JK, Kesteleyn B, Linclau B, Unge J. Disubstituted bis-THF moieties as new P2 ligands in nonpeptidal HIV-1 protease inhibitors (II) J. Med. Chem. 2015;58:4029–4038. doi: 10.1021/acs.jmedchem.5b00358. [DOI] [PubMed] [Google Scholar]
- 92.Ghosh AK, Martyr CD, Osswald HL, Sheri VR, Kassekert LA, Chen S, Agniswamy J, Wang YF, Hayashi H, Aoki M, Weber IT, Mitsuya H. Design of HIV-1 protease inhibitors with amino-bis-tetrahydrofuran derivatives as P2-ligands to enhance backbone-binding interactions: synthesis, biological evaluation, and protein-ligand X-ray studies. J. Med. Chem. 2015;58:6994–7006. doi: 10.1021/acs.jmedchem.5b00900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Qiu X, Zhao GD, Tang LQ, Liu ZP. Design and synthesis of highly potent HIV-1 protease inhibitors with novel isosorbide-derived P2 ligands. Bioorg. Med. Chem. Lett. 2014;24:2465–2468. doi: 10.1016/j.bmcl.2014.04.008. [DOI] [PubMed] [Google Scholar]
- 94.Ghosh AK, Sridhar PR, Leshchenko S, Hussain AK, Li J, Kovalevsky AY, Walters DE, Wedekind JE, Grum-Tokars V, Das D, Koh Y, Maeda K, Gatanaga H, Weber IT, Mitsuya H. Structure-based design of novel HIV-1 protease inhibitors to combat drug resistance. J. Med. Chem. 2006;49:5252–5261. doi: 10.1021/jm060561m. [DOI] [PubMed] [Google Scholar]
- 95.Ghosh AK, Gemma S, Takayama J, Baldridge A, Leshchenko-Yashchuk S, Miller HB, Wang YF, Kovalevsky AY, Koh Y, Weber IT, Mitsuya H. Potent HIV-1 protease inhibitors incorporating meso-bicyclic urethanes as P2-ligands: structure-based design, synthesis, biological evaluation and protein-ligand X-ray studies. Org. Biomol. Chem. 2008;6:3703–3713. doi: 10.1039/b809178a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chang YC, Yu X, Zhang Y, Tie Y, Wang YF, Yashchuk S, Ghosh AK, Harrison RW, Weber IT. Potent antiviral HIV-1 protease inhibitor GRL-02031 adapts to the structures of drug resistant mutants with its P1'-pyrrolidinone ring. J. Med. Chem. 2012;55:3387–3397. doi: 10.1021/jm300072d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mahalingam B, Wang YF, Boross PI, Tozser J, Louis JM, Harrison RW, Weber IT. Crystal structures of HIV protease V82A and L90M mutants reveal changes in the indinavir-binding site. Eur. J. Biochem. 2004;271:1516–1524. doi: 10.1111/j.1432-1033.2004.04060.x. [DOI] [PubMed] [Google Scholar]
- 98.Tie Y, Kovalevsky AY, Boross P, Wang YF, Ghosh AK, Tozser J, Harrison RW, Weber IT. Atomic resolution crystal structures of HIV-1 protease and mutants V82A and I84V with saquinavir. Proteins. 2007;67:232–242. doi: 10.1002/prot.21304. [DOI] [PubMed] [Google Scholar]
- 99.Zhang Y, Chang YC, Louis JM, Wang YF, Harrison RW, Weber IT. Structures of darunavir-resistant HIV-1 protease mutant reveal atypical binding of darunavir to wide open flaps. ACS Chem. Biol. 2014;9:1351–1358. doi: 10.1021/cb4008875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Agniswamy J, Shen CH, Wang YF, Ghosh AK, Rao KV, Xu CX, Sayer JM, Louis JM, Weber IT. Extreme multidrug resistant HIV-1 protease with 20 mutations is resistant to novel protease inhibitors with P1'-pyrrolidinone or P2-tris-tetrahydrofuran. J. Med. Chem. 2013;56:4017–4027. doi: 10.1021/jm400231v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ghosh AK, Chapsal BD, Parham GL, Steffey M, Agniswamy J, Wang YF, Amano M, Weber IT, Mitsuya H. Design of HIV-1 protease inhibitors with C3-substituted hexahydrocyclopentafuranyl urethanes as P2-ligands: synthesis, biological evaluation, and protein-ligand X-ray crystal structure. J. Med. Chem. 2011;54:5890–5901. doi: 10.1021/jm200649p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ghosh AK, Chapsal BD, Steffey M, Agniswamy J, Wang YF, Amano M, Weber IT, Mitsuya H. Substituent effects on P2-cyclopentyltetrahydrofuranyl urethanes: design, synthesis, and X-ray studies of potent HIV-1 protease inhibitors. Bioorg. Med. Chem. Lett. 2012;22:2308–2311. doi: 10.1016/j.bmcl.2012.01.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ghosh AK, Chapsal BD, Baldridge A, Steffey MP, Walters DE, Koh Y, Amano M, Mitsuya H. Design and synthesis of potent HIV-1 protease inhibitors incorporating hexahydrofuropyranol-derived high affinity P(2) ligands: structure-activity studies and biological evaluation. J. Med. Chem. 2011;54:622–634. doi: 10.1021/jm1012787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ghosh AK, Xu CX, Rao KV, Baldridge A, Agniswamy J, Wang YF, Weber IT, Aoki M, Miguel SG, Amano M, Mitsuya H. Probing multidrug-resistance and protein-ligand interactions with oxatricyclic designed ligands in HIV-1 protease inhibitors. ChemMedChem. 2010;5:1850–1854. doi: 10.1002/cmdc.201000318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Amano M, Tojo Y, Salcedo-Gómez PM, Campbell JR, Das D, Aoki M, Xu CX, Rao KV, Ghosh AK, Mitsuya H. GRL-0519, a novel oxatricyclic ligand-containing nonpeptidic HIV-1 protease inhibitor (PI), potently suppresses replication of a wide spectrum of multi-PI-resistant HIV-1 variants in vitro. Antimicrob. Agents Chemother. 2013;57:2036–2046. doi: 10.1128/AAC.02189-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ghosh AK, Xu CX, Osswald HL. Enantioselective synthesis of dioxatriquinane structural motifs for HIV-1 protease inhibitors using a cascade radical cyclization. Tetrahedron Lett. 2015;56:3314–3317. doi: 10.1016/j.tetlet.2015.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ghosh AK, Parham GL, Martyr CD, Nyalapatla PR, Osswald HL, Agniswamy J, Wang YF, Amano M, Weber IT, Mitsuya H. Highly potent HIV-1 protease inhibitors with novel tricyclic P2 ligands: design, synthesis, and protein-ligand X-ray studies. J. Med. Chem. 2013;56:6792–6802. doi: 10.1021/jm400768f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ghosh AK, Gemma S, Baldridge A, Wang YF, Kovalevsky AY, Koh Y, Weber IT, Mitsuya H. Flexible cyclic ethers/polyethers as novel P2-ligands for HIV-1 protease inhibitors: design, synthesis, biological evaluation, and protein-ligand X-ray studies. J. Med. Chem. 2008;51:6021–6033. doi: 10.1021/jm8004543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Miller JF, Andrews CW, Brieger M, Furfine ES, Hale MR, Hanlon MH, Hazen RJ, Kaldor I, McLean EW, Reynolds D, Sammond DM, Spaltenstein A, Tung R, Turner EM, Xu RX, Sherrill RG. Ultra-potent P1 modified arylsulfonamide HIV protease inhibitors: the discovery of GW0385. Bioorg. Med. Chem. Lett. 2006;16:1788–1794. doi: 10.1016/j.bmcl.2006.01.035. [DOI] [PubMed] [Google Scholar]
- 110.Hazen R, Harvey R, Ferris R, Craig C, Yates P, Griffin P, Miller J, Kaldor I, Ray J, Samano V, Furfine E, Spaltenstein A, Hale M, Tung R, St Clair M, Hanlon M, Boone L. In vitro antiviral activity of the novel, tyrosyl-based human immunodeficiency virus (HIV) type 1 protease inhibitor brecanavir (GW640385) in combination with other antiretrovirals and against a panel of protease inhibitor-resistant HIV. Antimicrob. Agents Chemother. 2007;51:3147–3154. doi: 10.1128/AAC.00401-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.He G-X, Yang Z-Y, Williams M, Callebaut C, Cihlar T, Murray BP, Yang C, Mitchell ML, Liu H, Wang J, Arimilli M, Eisenberg E, Stray KM, Tsai LK, Hatada M, Chen X, Chen JM, Wang Y, Lee MS, Strickley RG, Iwata Q, Zheng X, Kim CU, Swaminathan S, Desai MC, Lee WA, Xu L. Discovery of GS-8374, a potent human immunodeficiency virus type 1 protease inhibitor with a superior resistance profile. MedChemComm. 2011;2:1093–1098. [Google Scholar]
- 112.Callebaut C, Stray K, Tsai L, Williams M, Yang ZY, Cannizzaro C, Leavitt SA, Liu X, Wang K, Murray BP, Mulato A, Hatada M, Priskich T, Parkin N, Swaminathan S, Lee W, He GX, Xu L, Cihlar T. In vitro characterization of GS-8374, a novel phosphonate-containing inhibitor of HIV-1 protease with a favorable resistance profile. Antimicrob. Agents Chemother. 2011;55:1366–1376. doi: 10.1128/AAC.01183-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ghosh AK, Kincaid JF, Cho W, Walters DE, Krishnan K, Hussain KA, Koo Y, Cho H, Rudall C, Holland L, Buthod J. Potent HIV protease inhibitors incorporating high-affinity P2-ligands and (R)-(hydroxyethylamino)sulfonamide isostere. Bioorg. Med. Chem. Lett. 1998;8:687–690. doi: 10.1016/s0960-894x(98)00098-5. [DOI] [PubMed] [Google Scholar]
- 114.Yoshimura K, Kato R, Kavlick MF, Nguyen A, Maroun V, Maeda K, Hussain KA, Ghosh AK, Gulnik SV, Erickson JW, Mitsuya H. A potent human immunodeficiency virus type 1 protease inhibitor, UIC-94003 (TMC-126), and selection of a novel (A28S) mutation in the protease active site. J. Virol. 2002;76:1349–1358. doi: 10.1128/JVI.76.3.1349-1358.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Miller JF, Brieger M, Furfine ES, Hazen RJ, Kaldor I, Reynolds D, Sherrill RG, Spaltenstein A. Novel P1 chain-extended HIV protease inhibitors possessing potent anti-HIV activity and remarkable inverse antiviral resistance profiles. Bioorg. Med. Chem. Lett. 2005;15:3496–3500. doi: 10.1016/j.bmcl.2005.05.129. [DOI] [PubMed] [Google Scholar]
- 116.Yan J, Huang N, Li S, Yang LM, Xing W, Zheng YT, Hu Y. Synthesis and biological evaluation of novel amprenavir-based P1-substituted bi-aryl derivatives as ultra-potent HIV-1 protease inhibitors. Bioorg. Med. Chem. Lett. 2012;22:1976–1979. doi: 10.1016/j.bmcl.2012.01.037. [DOI] [PubMed] [Google Scholar]
- 117.Ghosh AK, Yu X, Osswald HL, Agniswamy J, Wang YF, Amano M, Weber IT, Mitsuya H. Structure-based design of potent HIV-1 protease inhibitors with modified P1-biphenyl ligands: synthesis, biological evaluation, and enzyme-inhibitor X-ray structural studies. J. Med. Chem. 2015;58:5334–5343. doi: 10.1021/acs.jmedchem.5b00676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Bonini C, Chiummiento L, Bonis MD, Funicello M, Lupattelli P, Suanno G, Berti F, Campaner P. Synthesis, biological activity and modelling studies of two novel anti HIV PR inhibitors with a thiophene containing hydroxyethylamino core. Tetrahedron. 2005;61:6580–6589. [Google Scholar]
- 119.Bonini C, Chiummiento L, De Bonis M, Di Blasio N, Funicello M, Lupattelli P, Pandolfo R, Tramutola F, Berti F. Synthesis of new thienyl ring containing HIV-1 protease inhibitors: promising preliminary pharmacological evaluation against recombinant HIV-1 proteases. J. Med. Chem. 2010;53:1451–1457. doi: 10.1021/jm900846f. [DOI] [PubMed] [Google Scholar]
- 120.Gao BL, Zhang CM, Yin YZ, Tang LQ, Liu ZP. Design and synthesis of potent HIV-1 protease inhibitors incorporating hydroxyprolinamides as novel P2 ligands. Bioorg. Med. Chem. Lett. 2011;21:3730–3733. doi: 10.1016/j.bmcl.2011.04.070. [DOI] [PubMed] [Google Scholar]
- 121.Parai MK, Huggins DJ, Cao H, Nalam MN, Ali A, Schiffer CA, Tidor B, Rana TM. Design, synthesis, and biological and structural evaluations of novel HIV-1 protease inhibitors to combat drug resistance. J. Med. Chem. 2012;55:6328–6341. doi: 10.1021/jm300238h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sherrill RG, Andrews CW, Bock WJ, Davis-Ward RG, Furfine ES, Hazen RJ, Rutkowske RD, Spaltenstein A, Wright LL. Optimization of pyrrolidinone based HIV protease inhibitors. Bioorg. Med. Chem. Lett. 2005;15:81–84. doi: 10.1016/j.bmcl.2004.10.029. [DOI] [PubMed] [Google Scholar]
- 123.Wu X, Ohrngren P, Joshi AA, Trejos A, Persson M, Arvela RK, Wallberg H, Vrang L, Rosenquist A, Samuelsson BB, Unge J, Larhed M. Synthesis, X-ray analysis, and biological evaluation of a new class of stereopure lactam-based HIV-1 protease inhibitors. J. Med. Chem. 2012;55:2724–2736. doi: 10.1021/jm201620t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Specker E, Böttcher J, Brass S, Heine A, Lilie H, Schoop A, Müller G, Griebenow N, Klebe G. Unexpected novel binding mode of pyrrolidine-based aspartyl protease inhibitors: design, synthesis and crystal structure in complex with HIV protease. ChemMedChem. 2006;1:106–117. doi: 10.1002/cmdc.200500008. [DOI] [PubMed] [Google Scholar]
- 125.Böttcher J, Blum A, Dörr S, Heine A, Diederich WE, Klebe G. Targeting the open-flap conformation of HIV-1 protease with pyrrolidine-based inhibitors. ChemMedChem. 2008;3:1337–1344. doi: 10.1002/cmdc.200800113. [DOI] [PubMed] [Google Scholar]
- 126.Ali A, Reddy GS, Nalam MN, Anjum SG, Cao H, Schiffer CA, Rana TM. Structure-based design, synthesis, and structure-activity relationship studies of HIV-1 protease inhibitors incorporating phenyloxazolidinones. J. Med. Chem. 2010;53:7699–7708. doi: 10.1021/jm1008743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.He M, Zhang H, Yao X, Eckart M, Zuo E, Yang M. Design, biologic evaluation, and SAR of novel pseudo-peptide incorporating benzheterocycles as HIV-1 protease inhibitors. Chem. Biol. Drug Des. 2010;76:174–180. doi: 10.1111/j.1747-0285.2010.00995.x. [DOI] [PubMed] [Google Scholar]
- 128.Yang ZH, Bai XG, Zhou L, Wang JX, Liu HT, Wang YC. Synthesis and biological evaluation of novel HIV-1 protease inhibitors using tertiary amine as P2-ligands. Bioorg. Med. Chem. Lett. 2015;25:1880–1883. doi: 10.1016/j.bmcl.2015.03.047. [DOI] [PubMed] [Google Scholar]
- 129.Whiting M, Muldoon J, Lin YC, Silverman SM, Lindstrom W, Olson AJ, Kolb HC, Finn MG, Sharpless KB, Elder JH, Fokin VV. Inhibitors of HIV-1 protease by using in situ click chemistry. Angew. Chem. Int. Ed. Engl. 2006;45:1435–1439. doi: 10.1002/anie.200502161. [DOI] [PubMed] [Google Scholar]
- 130.Brik A, Alexandratos J, Lin YC, Elder JH, Olson AJ, Wlodawer A, Goodsell DS, Wong CH. 1,2,3-triazole as a peptide surrogate in the rapid synthesis of HIV-1 protease inhibitors. ChemBioChem. 2005;6:1167–1169. doi: 10.1002/cbic.200500101. [DOI] [PubMed] [Google Scholar]
- 131.Öhrngren P, Wu X, Persson M, Ekegren JK, Wallberg H, Vrang L, Rosenquist Å, Samuelsson B, Unge T, Larhed M. HIV-1 protease inhibitors with a tertiary alcohol containing transition-state mimic and various P2 and P1' substituents. MedChemComm. 2011;2:701–709. [Google Scholar]
- 132.Ekegren JK, Unge T, Safa MZ, Wallberg H, Samuelsson B, Hallberg A. A new class of HIV-1 protease inhibitors containing a tertiary alcohol in the transition-state mimicking scaffold. J. Med. Chem. 2005;48:8098–8102. doi: 10.1021/jm050790t. [DOI] [PubMed] [Google Scholar]
- 133.Wu X, Ohrngren P, Ekegren JK, Unge J, Unge T, Wallberg H, Samuelsson B, Hallberg A, Larhed M. Two-carbon-elongated HIV-1 protease inhibitors with a tertiary-alcohol-containing transition-state mimic. J. Med. Chem. 2008;51:1053–1057. doi: 10.1021/jm070680h. [DOI] [PubMed] [Google Scholar]
- 134.Ekegren JK, Ginman N, Johansson A, Wallberg H, Larhed M, Samuelsson B, Unge T, Hallberg A. Microwave-accelerated synthesis of P1'-extended HIV-1 protease inhibitors encompassing a tertiary alcohol in the transition-state mimicking scaffold. J. Med. Chem. 2006;49:1828–1832. doi: 10.1021/jm051239z. [DOI] [PubMed] [Google Scholar]
- 135.Mahalingam AK, Axelsson L, Ekegren JK, Wannberg J, Kihlström J, Unge T, Wallberg H, Samuelsson B, Larhed M, Hallberg A. HIV-1 protease inhibitors with a transition-state mimic comprising a tertiary alcohol: improved antiviral activity in cells. J. Med. Chem. 2010;53:607–615. doi: 10.1021/jm901165g. [DOI] [PubMed] [Google Scholar]
- 136.Ghosh AK, Swanson LM, Cho H, Leshchenko S, Hussain KA, Kay S, Walters DE, Koh Y, Mitsuya H. Structure-based design: synthesis and biological evaluation of a series of novel cycloamide-derived HIV-1 protease inhibitors. J. Med. Chem. 2005;48:3576–3585. doi: 10.1021/jm050019i. [DOI] [PubMed] [Google Scholar]
- 137.Ghosh AK, Schiltz GE, Rusere LN, Osswald HL, Walters DE, Amano M, Mitsuya H. Design and synthesis of potent macrocyclic HIV-1 protease inhibitors involving P1-P2 ligands. Org. Biomol. Chem. 2014;12:6842–6854. doi: 10.1039/c4ob00738g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ghosh AK, Kulkarni S, Anderson DD, Hong L, Baldridge A, Wang YF, Chumanevich AA, Kovalevsky AY, Tojo Y, Amano M, Koh Y, Tang J, Weber IT, Mitsuya H. Design, synthesis, protein-ligand X-ray structure, and biological evaluation of a series of novel macrocyclic human immunodeficiency virus-1 protease inhibitors to combat drug resistance. J. Med. Chem. 2009;52:7689–7705. doi: 10.1021/jm900695w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Tojo Y, Koh Y, Amano M, Aoki M, Das D, Kulkarni S, Anderson DD, Ghosh AK, Mitsuya H. Novel protease inhibitors (PIs) containing macrocyclic components and 3(R),3a(S),6a(R)-bis-tetrahydrofuranylurethane that are potent against multi-PI-resistant HIV-1 variants in vitro. Antimicrob. Agents Chemother. 2010;54:3460–3470. doi: 10.1128/AAC.01766-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Smith AB, Wang W, Sprengeler PA, Hirschmann R. Design, synthesis, and solution structure of a pyrrolinone-based β-turn peptidomimetic. J. Am. Chem. Soc. 2000;122:11037–11038. [Google Scholar]
- 141.Smith AB, 3rd, Charnley AK, Harada H, Beiger JJ, Cantin LD, Kenesky CS, Hirschmann R, Munshi S, Olsen DB, Stahlhut MW, Schleif WA, Kuo LC. Design, synthesis, and biological evaluation of monopyrrolinone-based HIV-1 protease inhibitors possessing augmented P2' side chains. Bioorg. Med. Chem. Lett. 2006;16:859–863. doi: 10.1016/j.bmcl.2005.11.011. [DOI] [PubMed] [Google Scholar]
- 142.Ghosh AK, Schiltz G, Perali RS, Leshchenko S, Kay S, Walters DE, Koh Y, Maeda K, Mitsuya H. Design and synthesis of novel HIV-1 protease inhibitors incorporating oxyindoles as the P2'-ligands. Bioorg. Med. Chem. Lett. 2006;16:1869–1873. doi: 10.1016/j.bmcl.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 143.Ami E, Nakahara K, Sato A, Nguyen JT, Hidaka K, Hamada Y, Nakatani S, Kimura T, Hayashi Y, Kiso Y. Synthesis and antiviral property of allophenylnorstatine-based HIV protease inhibitors incorporating D-cysteine derivatives as P2/P3 moieties. Bioorg. Med. Chem. Lett. 2007;17:4213–4217. doi: 10.1016/j.bmcl.2007.05.039. [DOI] [PubMed] [Google Scholar]
- 144.Hidaka K, Kimura T, Abdel-Rahman HM, Nguyen JT, McDaniel KF, Kohlbrenner WE, Molla A, Adachi M, Tamada T, Kuroki R, Katsuki N, Tanaka Y, Matsumoto H, Wang J, Hayashi Y, Kempf DJ, Kiso Y. Small-sized human immunodeficiency virus type-1 protease inhibitors containing allophenylnorstatine to explore the S2' pocket. J. Med. Chem. 2009;52:7604–7617. doi: 10.1021/jm9005115. [DOI] [PubMed] [Google Scholar]
- 145.Ghosh AK, Gemma S, Simoni E, Baldridge A, Walters DE, Ide K, Tojo Y, Koh Y, Amano M, Mitsuya H. Synthesis and biological evaluation of novel allophenylnorstatine-based HIV-1 protease inhibitors incorporating high affinity P2-ligands. Bioorg. Med. Chem. Lett. 2010;20:1241–1246. doi: 10.1016/j.bmcl.2009.11.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Mimoto T, Imai J, Kisanuki S, Enomoto H, Hattori N, Akaji K, Kiso Y. Kynostatin (KNI)-227 and −272, highly potent anti-HIV agents: conformationally constrained tripeptide inhibitors of HIV protease containing allophenylnorstatine. Chem. Pharm. Bull. 1992;40:2251–2253. doi: 10.1248/cpb.40.2251. [DOI] [PubMed] [Google Scholar]
- 147.Nakatani S, Hidaka K, Ami E, Nakahara K, Sato A, Nguyen JT, Hamada Y, Hori Y, Ohnishi N, Nagai A, Kimura T, Hayashi Y, Kiso Y. Combination of non-natural D-amino acid derivatives and allophenylnorstatine-dimethylthioproline scaffold in HIV protease inhibitors have high efficacy in mutant HIV. J. Med. Chem. 2008;51:2992–3004. doi: 10.1021/jm701555p. [DOI] [PubMed] [Google Scholar]
- 148.Arefalk A, Wannberg J, Larhed M, Hallberg A. Stereoselective synthesis of 3-aminoindan-1-ones and subsequent incorporation into HIV-1 protease inhibitors. J. Org. Chem. 2006;71:1265–1268. doi: 10.1021/jo0521504. [DOI] [PubMed] [Google Scholar]
- 149.Adrian Meredith J, Wallberg H, Vrang L, Oscarson S, Parkes K, Hallberg A, Samuelsson B. Design and synthesis of novel P2 substituents in diol-based HIV protease inhibitors. Eur. J. Med. Chem. 2010;45:160–170. doi: 10.1016/j.ejmech.2009.09.038. [DOI] [PubMed] [Google Scholar]
- 150.Clemente JC, Robbins A, Graña P, Paleo MR, Correa JF, Villaverde MC, Sardina FJ, Govindasamy L, Agbandje-McKenna M, McKenna R, Dunn BM, Sussman F. Design, synthesis, evaluation, and crystallographic-based structural studies of HIV-1 protease inhibitors with reduced response to the V82A mutation. J. Med. Chem. 2008;51:852–860. doi: 10.1021/jm701170f. [DOI] [PubMed] [Google Scholar]
- 151.Benedetti F, Berti F, Budal S, Campaner P, Dinon F, Tossi A, Argirova R, Genova P, Atanassov V, Hinkov A. Synthesis and biological activity of potent HIV-1 protease inhibitors based on Phe-Pro dihydroxyethylene isosteres. J. Med. Chem. 2012;55:3900–3910. doi: 10.1021/jm3001136. [DOI] [PubMed] [Google Scholar]
- 152.DeGoey DA, Grampovnik DJ, Chen HJ, Flosi WJ, Klein LL, Dekhtyar T, Stoll V, Mamo M, Molla A, Kempf DJ. P1-substituted symmetry-based human immunodeficiency virus protease inhibitors with potent antiviral activity against drug-resistant viruses. J. Med. Chem. 2011;54:7094–7104. doi: 10.1021/jm201109t. [DOI] [PubMed] [Google Scholar]
- 153.Wannberg J, Sabnis YA, Vrang L, Samuelsson B, Karlén A, Hallberg A, Larhed M. A new structural theme in C2-symmetric HIV-1 protease inhibitors: ortho-substituted P1/P1' side chains. Bioorg. Med. Chem. 2006;14:5303–5315. doi: 10.1016/j.bmc.2006.03.045. [DOI] [PubMed] [Google Scholar]
- 154.Lu D, Vince R. Discovery of potent HIV-1 protease inhibitors incorporating sulfoximine functionality. Bioorg. Med. Chem. Lett. 2007;17:5614–5619. doi: 10.1016/j.bmcl.2007.07.095. [DOI] [PubMed] [Google Scholar]
- 155.Lu D, Sham YY, Vince R. Design, asymmetric synthesis, and evaluation of pseudosymmetric sulfoximine inhibitors against HIV-1 protease. Bioorg. Med. Chem. 2010;18:2037–2048. doi: 10.1016/j.bmc.2010.01.020. [DOI] [PubMed] [Google Scholar]
- 156.Schimer J, Cígler P, Veselý J, Grantz Šašková K, Lepšík M, Brynda J, Řezáčová P, Kožíšek M, Císařová I, Oberwinkler H, Kraeusslich HG, Konvalinka J. Structure-aided design of novel inhibitors of HIV protease based on a benzodiazepine scaffold. J. Med. Chem. 2012;55:10130–10135. doi: 10.1021/jm301249q. [DOI] [PubMed] [Google Scholar]
- 157.Garino C, Bihel F, Pietrancosta N, Laras Y, Quéléver G, Woo I, Klein P, Bain J, Boucher JL, Kraus JL. New 2-bromomethyl-8-substituted-benzo[c]chromen-6-ones. Synthesis and biological properties. Bioorg. Med. Chem. Lett. 2005;15:135–138. doi: 10.1016/j.bmcl.2004.10.018. [DOI] [PubMed] [Google Scholar]
- 158.Olomola TO, Klein R, Mautsa N, Sayed Y, Kaye PT. Synthesis and evaluation of coumarin derivatives as potential dual-action HIV-1 protease and reverse transcriptase inhibitors. Bioorg. Med. Chem. 2013;21:1964–1971. doi: 10.1016/j.bmc.2013.01.025. [DOI] [PubMed] [Google Scholar]
- 159.Olomola TO, Klein R, Lobb KA, Sayed Y, Kaye PT. Towards the synthesis of coumarin derivatives as potential dual-action HIV-1 protease and reverse transcriptase inhibitors. Tetrahedron Lett. 2010;51:6325–6328. [Google Scholar]
- 160.Sun CL, Pang RF, Zhang H, Yang M. Design, synthesis, and biological evaluation of novel 4-hydroxypyrone derivatives as HIV-1 protease inhibitors. Bioorg. Med. Chem. Lett. 2005;15:3257–3262. doi: 10.1016/j.bmcl.2005.04.057. [DOI] [PubMed] [Google Scholar]
- 161.Ax A, Schaal W, Vrang L, Samuelsson B, Hallberg A, Karlén A. Cyclic sulfamide HIV-1 protease inhibitors, with sidechains spanning from P2/P2' to P1/P1'. Bioorg. Med. Chem. 2005;13:755–764. doi: 10.1016/j.bmc.2004.10.042. [DOI] [PubMed] [Google Scholar]
- 162.Gold H, Ax A, Vrang L, Samuelsson B, Karlén A, Hallberga A, Larhed M. Fast and selective synthesis of novel cyclic sulfamide HIV-1 protease inhibitors under controlled microwave heating. Tetrahedron. 2006;62:4671–4675. [Google Scholar]
- 163.Ax A, Joshi AA, Orrling KM, Vrang L, Samuelsson B, Hallberg A, Karlén A, Larhed M. Synthesis of a small library of non-symmetric cyclic sulfamide HIV-1 protease inhibitors. Tetrahedron. 2010;66:4049–4056. [Google Scholar]
- 164.Ganguly AK, Alluri SS, Caroccia D, Biswas D, Wang CH, Kang E, Zhang Y, McPhail AT, Carroll SS, Burlein C, Munshi V, Orth P, Strickland C. Design, synthesis, and X-ray crystallographic analysis of a novel class of HIV-1 protease inhibitors. J. Med. Chem. 2011;54:7176–7183. doi: 10.1021/jm200778q. [DOI] [PubMed] [Google Scholar]
- 165.Bouzide A, Sauvé G, Yelle J. Lysine derivatives as potent HIV protease inhibitors. Discovery, synthesis and structure-activity relationship studies. Bioorg. Med. Chem. Lett. 2005;15:1509–1513. doi: 10.1016/j.bmcl.2004.12.068. [DOI] [PubMed] [Google Scholar]
- 166.Stranix BR, Lavallée JF, Sévigny G, Yelle J, Perron V, LeBerre N, Herbart D, Wu JJ. Lysine sulfonamides as novel HIV-protease inhibitors: Nepsilon-acyl aromatic alpha-amino acids. Bioorg. Med. Chem. Lett. 2006;16:3459–3462. doi: 10.1016/j.bmcl.2006.04.011. [DOI] [PubMed] [Google Scholar]
- 167.Jones KL, Holloway MK, Su HP, Carroll SS, Burlein C, Touch S, DiStefano DJ, Sanchez RI, Williams TM, Vacca JP, Coburn CA. Epsilon substituted lysinol derivatives as HIV-1 protease inhibitors. Bioorg. Med. Chem. Lett. 2010;20:4065–4068. doi: 10.1016/j.bmcl.2010.05.082. [DOI] [PubMed] [Google Scholar]
- 168.Rajapakse HA, Walji AM, Moore KP, Zhu H, Mitra AW, Gregro AR, Tinney E, Burlein C, Touch S, Paton BL, Carroll SS, DiStefano DJ, Lai MT, Grobler JA, Sanchez RI, Williams TM, Vacca JP, Nantermet PG. Strategies towards improving the pharmacokinetic profile of epsilon-substituted lysinol-derived HIV protease inhibitors. ChemMedChem. 2011;6:253–257. doi: 10.1002/cmdc.201000395. [DOI] [PubMed] [Google Scholar]
- 169.Yeung CM, Klein LL, Flentge CA, Randolph JT, Zhao C, Sun M, Dekhtyar T, Stoll VS, Kempf DJ. Oximinoarylsulfonamides as potent HIV protease inhibitors. Bioorg. Med. Chem. Lett. 2005;15:2275–2278. doi: 10.1016/j.bmcl.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 170.Randolph JT, Huang PP, Flosi WJ, DeGoey D, Klein LL, Yeung CM, Flentge C, Sun M, Zhao C, Dekhtyar T, Mo H, Colletti L, Kati W, Marsh KC, Molla A, Kempf DJ. Synthesis, antiviral activity, and pharmacokinetic evaluation of P3 pyridylmethyl analogs of oximinoarylsulfonyl HIV-1 protease inhibitors. Bioorg. Med. Chem. 2006;14:4035–4046. doi: 10.1016/j.bmc.2006.02.013. [DOI] [PubMed] [Google Scholar]
- 171.Flosi WJ, DeGoey DA, Grampovnik DJ, Chen HJ, Klein LL, Dekhtyar T, Masse S, Marsh KC, Mo HM, Kempf D. Discovery of imidazolidine-2,4-dione-linked HIV protease inhibitors with activity against lopinavir-resistant mutant HIV. Bioorg. Med. Chem. 2006;14:6695–6712. doi: 10.1016/j.bmc.2006.05.063. [DOI] [PubMed] [Google Scholar]
- 172.Dekhtyar T, Ng TI, Lu L, Masse S, DeGoey DA, Flosi WJ, Grampovnik DJ, Klein LL, Kempf DJ, Molla A. Characterization of a novel human immunodeficiency virus type 1 protease inhibitor, A-790742. Antimicrob. Agents Chemother. 2008;52:1337–1344. doi: 10.1128/AAC.01132-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Zaman S, Campaner P, Abell AD. Synthesis of amino acid derived seven-membered lactams by RCM and their evaluation against HIV protease. Bioorg. Med. Chem. 2006;14:8323–8331. doi: 10.1016/j.bmc.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 174.Zhao C, Sham HL, Sun M, Stoll VS, Stewart KD, Lin S, Mo H, Vasavanonda S, Saldivar A, Park C, McDonald EJ, Marsh KC, Klein LL, Kempf DJ, Norbeck DW. Synthesis and activity of N-acyl azacyclic urea HIV-1 protease inhibitors with high potency against multiple drug resistant viral strains. Bioorg. Med. Chem. Lett. 2005;15:5499–5503. doi: 10.1016/j.bmcl.2005.08.093. [DOI] [PubMed] [Google Scholar]
- 175.Wei Y, Ma CM, Hattori M. Synthesis of dammarane-type triterpene derivatives and their ability to inhibit HIV and HCV proteases. Bioorg. Med. Chem. 2009;17:3003–3010. doi: 10.1016/j.bmc.2009.03.019. [DOI] [PubMed] [Google Scholar]
- 176.Sun QZ, Chen DF, Ding PL, Ma CM, Kakuda H, Nakamura N, Hattori M. Three new lignans, longipedunins A-C, from Kadsura longipedunculata and their inhibitory activity against HIV-1 protease. Chem. Pharm. Bull. 2006;54:129–132. doi: 10.1248/cpb.54.129. [DOI] [PubMed] [Google Scholar]
- 177.El Dine RS, El Halawany AM, Ma CM, Hattori M. Anti-HIV-1 protease activity of lanostane triterpenes from the vietnamese mushroom Ganoderma colossum. J. Nat. Prod. 2008;71:1022–1026. doi: 10.1021/np8001139. [DOI] [PubMed] [Google Scholar]
- 178.Wei Y, Ma CM, Hattori M. Synthesis and evaluation of A-seco type triterpenoids for anti-HIV-1protease activity. Eur. J. Med. Chem. 2009;44:4112–4120. doi: 10.1016/j.ejmech.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 179.Wei Y, Ma CM, Chen DY, Hattori M. Anti-HIV-1 protease triterpenoids from Stauntonia obovatifoliola Hayata subsp. intermedia. Phytochemistry. 2008;69:1875–1879. doi: 10.1016/j.phytochem.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 180.Lee JS, Maiyashiro H, Nakamura N, Hattori M. Two new triterpenes from the Rhizome of Dryopteris crassirhizoma, and inhibitory activities of its constituents on human immunodeficiency virus-1 protease. Chem. Pharm. Bull. 2008;56:711–714. doi: 10.1248/cpb.56.711. [DOI] [PubMed] [Google Scholar]
- 181.El Dine RS, El Halawany AM, Ma CM, Hattori M. Inhibition of the dimerization and active site of HIV-1 protease by secondary metabolites from the Vietnamese mushroom Ganoderma colossum. J. Nat. Prod. 2009;72:2019–2023. doi: 10.1021/np900279u. [DOI] [PubMed] [Google Scholar]
- 182.Lee JS, Hattori M, Kim J. Inhibition of HIV-1 protease and RNase H of HIV-1 reverse transcriptase activities by long chain phenols from the sarcotestas of Ginkgo biloba. Planta Med. 2008;74:532–534. doi: 10.1055/s-2008-1074497. [DOI] [PubMed] [Google Scholar]
- 183.Lee J, Huh MS, Kim YC, Hattori M, Otake T. Lignan, sesquilignans and dilignans, novel HIV-1 protease and cytopathic effect inhibitors purified from the rhizomes of Saururus chinensis. Antiviral Res. 2010;85:425–428. doi: 10.1016/j.antiviral.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 184.Makatini MM, Petzold K, Sriharsha SN, Soliman ME, Honarparvar B, Arvidsson PI, Sayed Y, Govender P, Maguire GE, Kruger HG, Govender T. Pentacycloundecane-based inhibitors of wild-type C-South African HIV-protease. Bioorg. Med. Chem. Lett. 2011;21:2274–2277. doi: 10.1016/j.bmcl.2011.02.105. [DOI] [PubMed] [Google Scholar]
- 185.Makatini MM, Petzold K, Arvidsson PI, Honarparvar B, Govender T, Maguire GE, Parboosing R, Sayed Y, Soliman ME, Kruger HG. Synthesis, screening and computational investigation of pentacycloundecane-peptoids as potent CSA-HIV PR inhibitors. Eur. J. Med. Chem. 2012;57:459–467. doi: 10.1016/j.ejmech.2012.06.019. [DOI] [PubMed] [Google Scholar]
- 186.Todd MJ, Semo N, Freire E. The structural stability of the HIV-1 protease. J. Mol. Biol. 1998;283:475–488. doi: 10.1006/jmbi.1998.2090. [DOI] [PubMed] [Google Scholar]
- 187.Davis DA, Brown CA, Singer KE, Wang V, Kaufman J, Stahl SJ, Wingfield P, Maeda K, Harada S, Yoshimura K, Kosalaraksa P, Mitsuya H, Yarchoan R. Inhibition of HIV-1 replication by a peptide dimerization inhibitor of HIV-1 protease. Antiviral Res. 2006;72:89–99. doi: 10.1016/j.antiviral.2006.03.015. [DOI] [PubMed] [Google Scholar]
- 188.Koh Y, Matsumi S, Das D, Amano M, Davis DA, Li J, Leschenko S, Baldridge A, Shioda T, Yarchoan R, Ghosh AK, Mitsuya H. Potent inhibition of HIV-1 replication by novel non-peptidyl small molecule inhibitors of protease dimerization. J. Biol. Chem. 2007;282:28709–28720. doi: 10.1074/jbc.M703938200. [DOI] [PubMed] [Google Scholar]
- 189.Bannwarth L, Kessler A, Pèthe S, Collinet B, Merabet N, Boggetto N, Sicsic S, Reboud-Ravaux M, Ongeri S. Molecular tongs containing amino acid mimetic fragments: new inhibitors of wild-type and mutated HIV-1 protease dimerization. J. Med. Chem. 2006;49:4657–4664. doi: 10.1021/jm060576k. [DOI] [PubMed] [Google Scholar]
- 190.Vidu A, Dufau L, Bannwarth L, Soulier JL, Sicsic S, Piarulli U, Reboud-Ravaux M, Ongeri S. Toward the first nonpeptidic molecular tong inhibitor of wild-type and mutated HIV-1 protease dimerization. ChemMedChem. 2010;5:1899–1906. doi: 10.1002/cmdc.201000308. [DOI] [PubMed] [Google Scholar]
- 191.Dufau L, Marques Ressurreição AS, Fanelli R, Kihal N, Vidu A, Milcent T, Soulier JL, Rodrigo J, Desvergne A, Leblanc K, Bernadat G, Crousse B, Reboud-Ravaux M, Ongeri S. Carbonylhydrazide-based molecular tongs inhibit wild-type and mutated HIV-1 protease dimerization. J. Med. Chem. 2012;55:6762–6775. doi: 10.1021/jm300181j. [DOI] [PubMed] [Google Scholar]
- 192.Lee SG, Chmielewski J. Cross-linked peptoid-based dimerization inhibitors of HIV-1 protease. ChemBioChem. 2010;11:1513–1516. doi: 10.1002/cbic.201000248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Bowman MJ, Byrne S, Chmielewski J. Switching between allosteric and dimerization inhibition of HIV-1 protease. Chem. Biol. 2005;12:439–444. doi: 10.1016/j.chembiol.2005.02.004. [DOI] [PubMed] [Google Scholar]