Summary
Concomitant hepatocyte apoptosis and regeneration is a hallmark of chronic liver diseases (CLDs) predisposing to hepatocellular carcinoma (HCC). Here, we mechanistically link caspase-8-dependent apoptosis to HCC development via proliferation- and replication-associated DNA damage. Proliferation-associated replication stress, DNA damage, and genetic instability are detectable in CLDs before any neoplastic changes occur. Accumulated levels of hepatocyte apoptosis determine and predict subsequent hepatocarcinogenesis. Proliferation-associated DNA damage is sensed by a complex comprising caspase-8, FADD, c-FLIP, and a kinase-dependent function of RIPK1. This platform requires a non-apoptotic function of caspase-8, but no caspase-3 or caspase-8 cleavage. It may represent a DNA damage-sensing mechanism in hepatocytes that can act via JNK and subsequent phosphorylation of the histone variant H2AX.
Keywords: liver, hepatocellular carcinoma, DNA damage response, replication stress, apoptosis
Highlights
-
•
Hepatocyte apoptosis decisively determines and predicts HCC development
-
•
A non-apoptotic caspase-8/RIPK1/FADD/c-FLIP complex senses DNA damage
-
•
Caspase-8 deficiency is associated with impaired phosphorylation of H2AX
-
•
Low caspase-8 expression in HCC is associated with less aggressive behavior
Boege et al. identify persistent hepatocyte apoptosis as a determinant of hepatocellular carcinoma development. They show that caspase-8 not only executes hepatocyte apoptosis but also has a non-apoptotic role in proliferation-associated DNA damage response mediated by a caspase-8/RIPK1/FADD/c-FLIP complex.
Significance
We identified persistent hepatocyte apoptosis as a universally decisive determinant of HCC development in distinct mouse models and various human CLDs. Accordingly, levels of hepatocyte apoptosis and DNA damage predict the risk for liver cancer, the second leading cause of cancer-related death worldwide. Finding that caspase-8 not only executes hepatocyte apoptosis, but also has a non-apoptotic function in DNA damage response demonstrates its opposing functions. By orchestrating DNA damage response as part of the signaling platform, caspase-8 may protect against proliferation-associated genetic instability, and therefore early stages of hepatocarcinogenesis. Whereas once tumors are established, low caspase-8 expression is associated with less aggressive behavior of human HCC. Our data illustrate diverging mechanistic links of caspase-8 to cancer biology.
Introduction
Hepatocellular carcinoma (HCC) is the most common primary malignant liver tumor, the fifth most prevalent cancer, the second leading cause of cancer-related death and the fastest rising cancer worldwide (El-Serag and Kanwal, 2014). HCC arises on the background of chronic liver diseases (CLDs) such as chronic hepatitis B virus and hepatitis C virus (HCV) infections, alcohol, metabolically and dietary-induced fatty liver disease, and steatohepatitis, autoimmune, or chronic cholestatic diseases (Forner et al., 2012). Independent of the underlying etiology, all CLDs exhibit persistent hepatocyte damage. To maintain liver homeostasis and prevent the accumulation of mutations, damaged hepatocytes are eliminated by programmed cell death, regulated by key molecules, including caspase-8 and receptor-interacting protein kinase 1 (RIPK1) (Luedde et al., 2014). Hepatocyte-specific knock out of the anti-apoptotic Bcl2-family member myeloid cell leukemia 1 (Mcl-1) gene in mice (Mcl-1Δhep mice) recapitulates CLD pathophysiology including severe liver damage and regeneration early in life (Vick et al., 2009) and subsequent HCC development (Weber et al., 2010). Here we functionally and quantitatively examine the interplay between caspase-8-dependent hepatocyte apoptosis and regeneration-associated replication stress, genetic instability, and hepatocarcinogenesis. Moreover, we investigate a role for caspase-8, in conjunction with other regulators of cell death and inflammation, during DNA damage recognition within hepatocytes.
Results
CLDs Display High Levels of Hepatocyte Apoptosis, DNA Damage, Genetic Instability, and Risk for HCC
Hepatocyte apoptosis is an etiology-independent hallmark of human CLDs (Figures 1A, S1A, and S1B). Increased levels of apoptosis correlated with increased hepatocyte proliferation, reflecting regeneration, with significantly higher numbers of cells positive for the DNA damage marker γH2AX (Figures 1A, S1A, and S1C–S1F), and with higher expression of DNA damage-responsive (DDR) genes (Figures S1C and S1D). Liver tissues from CLD patients further displayed high levels of genetic instability at chromosomal common fragile sites (CFS) (Gao and Smith, 2014) as determined by TaqMan copy number assay (Figures 1B and S1G) and fragment length analysis (Figure 1C). Thus, our data suggest that genetic instability is established long before dysplastic changes are detectable. We next looked for an association between serum transaminase levels (Figure 1D) (as a surrogate marker for liver cell apoptosis) and subsequent HCC development. Elevated serum alanine and aspartate transaminase (ALT and AST) levels in CLDs were associated with subsequent HCC development: (1) retrospective analysis of patients with chronic HCV infection revealed that patients who developed HCC had significantly higher ALT and AST levels (p < 0.05) during a period of 6 years preceding HCC diagnosis compared with HCC-free individuals of the same cohort (matched for model of end-stage liver disease) score with similar albumin and bilirubin levels (Figures 1E, 1F, and S1H). (2) Retrospective analysis of liver transplant (LT) patients revealed that patients transplanted for HCC had significantly higher ALT and AST levels 1 year prior to LT compared with patients of the same cohort undergoing LT for non-HCC indications (p < 0.001; Figure S1I).
Figure 1.
DNA Damage and Genetic Instability CLDs Preceding Neoplastic Lesions and HCC
(A) Apoptosis (cl.Casp3), proliferation (Ki67), and DNA damage (γH2AX) in human CLDs of different etiology (viral hepatitis: hepatitis B virus [HBV] and [HCV], metabolic [NASH], and autoimmune [AIH] diseases). Arrowheads indicate cells with positive IHC staining. Scale bars, 100 μm.
(B) TaqMan copy number assay for allelic imbalances (AI). Each square represents one area of microdissected tissue, lines indicate different areas of the same liver (red, AI; black, no AI; NT, non-tumor CLD tissue).
(C) Fragment length analysis (loci DS31263 and DS31289) in CLD tissues. Arrowheads indicate changes in fragment length distribution.
(D) Serum ALT levels in CLDs (n = 4 HBV, n = 8 HCV, n = 4 NASH, and n = 4 AIH).
(E and F) Serum ALT levels in patients with HCC versus without HCC of the same cohort (n = 13 in both groups). (E) Time course 6 years prior to diagnosis and (F) mean of ALT values over time.
In (D), (E), and (F), data are presented as mean ± SEM. Statistical significance was calculated using Fisher's exact test (B), ANOVA with Bonferroni correction (D), or Student's t test (E and F). ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001. See also Figure S1.
Risk for HCC Development Correlates with Levels of Hepatocyte Apoptosis and DNA Damage Also in Mice
To functionally investigate the role of apoptosis for HCC development in vivo, we prospectively monitored Mcl-1Δhep mice for serum transaminase levels which, similarly to CLD patients, are characterized by chronically increased hepatocyte apoptosis and regeneration (Vick et al., 2009). Remarkably, the same Mcl-1Δhep mice that developed liver tumors at 1 year (50% of this cohort) also displayed higher serum ALT (and AST, data not shown) levels throughout life compared with Mcl-1Δhep mice without tumor development (Figures 2A–2C). Parallel to a reduced sensitivity toward tumor necrosis factor (TNF)-mediated apoptotic signaling with age (Figure S2A), ALT levels dropped and reached similar levels in both groups after 4 months. Nevertheless, differences were statistically significant at 2 and 4 months, i.e., the period of high levels of liver damage and regeneration. Livers of 2-month-old Mcl-1Δhep mice showed a moderate to high disease activity when applying a standard human scoring system (Batts and Ludwig, 1995) (Figure 2D). A statistically significant positive correlation between the percentage of cleaved caspase-3+ hepatocytes and serum ALT levels was also found (Figure 2E). Livers of 2-month-old Mcl-1Δhep mice revealed increased levels of proliferating hepatocytes and numbers of γH2AX+ hepatocytes (Figures 2F and 2G), which positively correlated with ALT levels (Figure 2H), strongly suggesting a link between hepatocyte apoptosis and DNA damage.
Figure 2.
Risk of HCC Development Correlates with Apoptosis and DNA Damage in Mcl-1Δhep Mice
(A) Livers from 12-month-old mice. Arrowheads indicate a tumor. Scale bar, 1 cm.
(B) Serum ALT levels throughout life time of wild-type mice, Mcl-1Δhep mice that developed HCC at 12 months (n = 12), and Mcl-1Δhep mice that did not.
(C) Serum ALT levels at 2 months (n = 8 animals per group).
(D) Hepatocyte death rates (n = 20).
(E) Correlation of ALT levels with hepatocytes apoptosis (n = 15).
(F) Hepatocyte mitosis (upper square and insert), apoptosis (lower square and insert), and signs of DNA damage (γH2AX, black arrow) in livers of Mcl-1Δhep mice. Scale bars, 50 μm.
(G) γH2AX+ hepatocytes per high-power field (HPF) in wild-type (n = 7) and Mcl-1Δhep mice (n = 12).
(H) Correlation of ALT levels with the number of γH2AX+ hepatocytes (n = 11).
(I) Pie chart displaying the percentage of genes at least 2-fold upregulated in Mcl-1Δhep mice and clustered according to KEGG pathway database analysis.
(J) Gene set enrichment analysis comparing all differentially regulated genes from Mcl-1Δhep mice with various gene sets. NES, normalized enrichment score.
In (B) and (C), data are presented as mean ± SEM. In (G), the bar indicates the mean. Statistical significance was calculated using Student's t test (B and G), ANOVA with Bonferroni correction (C). ∗p < 0.05. See also Figure S2.
Next, mRNA profiling unraveled genes differentially expressed in livers of 2-month-old wild-type, homozygous, and hemizygous Mcl-1Δhep mice, which were verified by qPCR (Figure S2B). KEGG pathway analysis revealed that genes upregulated by at least 2-fold were involved in diverse cellular functions including apoptosis, cell cycle, differentiation, metabolism, and DDR (Figure 2I). Gene set enrichment analysis revealed that livers of Mcl-1Δhep mice were not only significantly enriched for genes related to apoptosis and proliferation, but also to viral infection, wounding (not shown), alcoholic hepatitis, and, despite being non-neoplastic, also to HCC (Figure 2J). Collectively, our findings show that Mcl-1Δhep mice are appropriate for investigating HCC development on a CLD background.
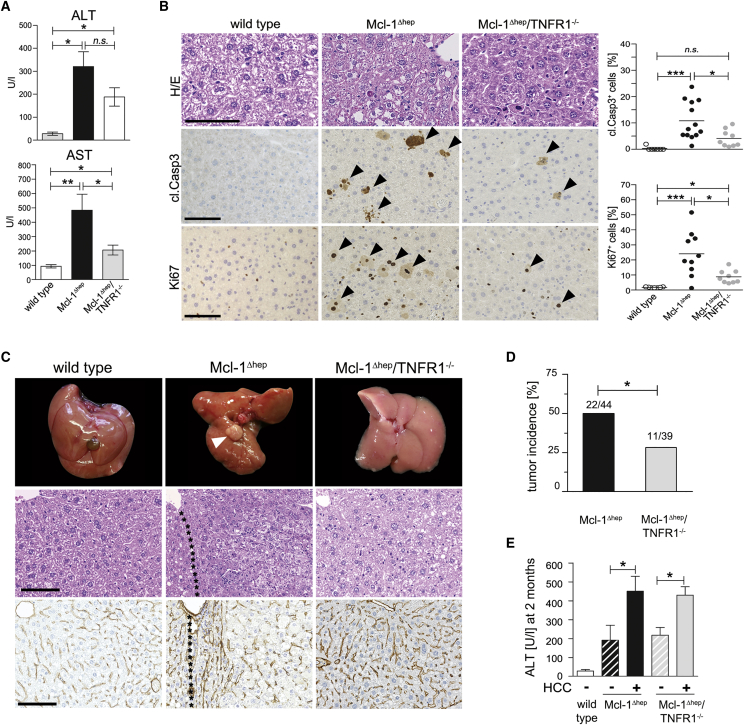
Next, we tested whether increased hepatocyte apoptosis through tumor necrosis factor receptor 1 (TNFR1) per se determined hepatocarcinogenesis in Mcl-1Δhep mice, rather than loss of a non-apoptotic function of Mcl-1. Mcl-1Δhep mice were crossed with TNFR1-deficient mice (Mcl-1Δhep/TNFR1−/− mice) to inhibit TNFR1-dependent apoptosis and downstream signaling via TNFR-caspase-8-BID/tBID-Mcl-1. Two-month-old Mcl-1Δhep/TNFR1−/− mice showed lower serum transaminase activity compared with age-matched Mcl-1Δhep mice (Figure 3A), and revealed significantly lower numbers of apoptotic and proliferating hepatocytes compared with Mcl-1Δhep mice (Figure 3B). This was paralleled by significantly increased hepatic mRNA levels of the death receptors Tnfr1 and Fas in Mcl-1Δhep mice compared with Mcl-1Δhep/TNFR1−/− mice, whereas Tnfr2, Trailr, and the ligands Tnfa, Fasl, and Trail (Figure S3A), and bilirubin and alkaline phosphatase showed no statistically significant difference (Figure S3B). Interestingly, livers of Mcl-1Δhep mice, but not Mcl-1Δhep/TNFR1−/− mice, displayed substantial caspase-8 cleavage (Figures S3C and S3D). Similar to LPS/DGal-challenged wild-type mice treated with the caspase-8 inhibitor zITED (Figure S3E), Mcl-1Δhep mice treated with zITED also displayed significantly decreased ALT levels (and AST, data not shown) and significantly less hepatocyte apoptosis (Figures S3F and S3G). In contrast, treating Mcl-1Δhep mice with the caspase-1 inhibitor, YVAD-CMK, used as an off-target control for zITED, did not affect ALT levels (Figure S3F). Thus, hepatocyte apoptosis in Mcl-1Δhep mice was caspase-8 dependent. Remarkably, Mcl-1Δhep/TNFR1−/− mice demonstrated a significantly reduced tumor incidence at 1 year compared with Mcl-1Δhep mice (28% versus 50%, p < 0.05; Figures 3C and 3D). In line with the data presented above, those Mcl-1Δhep/TNFR1−/− mice that developed liver tumors also displayed significantly higher transaminase levels at 2 months (Figure 3E).
Figure 3.
Reduced Apoptosis, Proliferation, and Tumor Development in Mcl-1Δhep/TNFR1−/− Mice
(A) AST and ALT levels from 2-month-old Mcl-1Δhep (n = 16), Mcl-1/TNFR1−/− (n = 10), and wild-type (n = 8) mice.
(B) Staining and quantification for H&E, cl.Casp3, and Ki67 in 2-month-old wild-type, Mcl-1Δhep/TNFR1−/−, and Mcl-1Δhep mice. Arrowheads indicate cells with positive IHC staining. Scale bars, 100 μm.
(C) Macroscopy, H&E, and collagen IV staining of livers at 12 months of age. The arrowhead indicates a tumor. Scale bars, 100 μm.
(D) Tumor development after 12 months in Mcl-1Δhep mice (n = 44) compared with Mcl-1Δhep/TNFR1−/− mice (n = 39).
(E) Retrospective analysis of tumor development and correlation to ALT levels in the serum of 2-month-old mice (n = 11 Mcl-1Δhep/TNFR1−/− mice without HCC, n = 5 with HCC).
In (A), (B), (D), and (E), data are presented as mean ± SEM. Statistical significance was calculated using Student's t test (A and B), ANOVA with Bonferroni correction (E), or Fisher's exact test (D). ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; n.s., not significant. See also Figures S3 and S4.
Further analyses of the microenvironment of Mcl-1Δhep livers revealed: (1) no activation of canonical nuclear factor κB (NF-κB) signaling (Figure S3H), (2), no or only low levels of inflammasome activation as determined by cleaved caspase-1 and cleaved interleukin-1β (IL-1β) levels (Figure S3I and data not shown), and (3) a significant increase expression of several inflammatory cytokines IL6, IL33, and IFNγ (with reduced levels of IL6, IL33, and IFNγ in Mcl-1Δhep/TNFR1−/− livers; Figure S3J).
Collectively, these findings show that the association between high apoptotic activity of hepatocytes (in early disease stages) with subsequent liver cancer development previously described for CLD patients also exists in Mcl-1Δhep mice. Furthermore, they suggest that persistently increased hepatocyte apoptosis, resulting in regenerative proliferation and high DNA replication rate, determines hepatocarcinogenesis. This hypothesis is underpinned by stochastic considerations (Figure S4).
Reduced DNA Damage and Genetic Instability upon Ablation of TNFR1 and Caspase-8
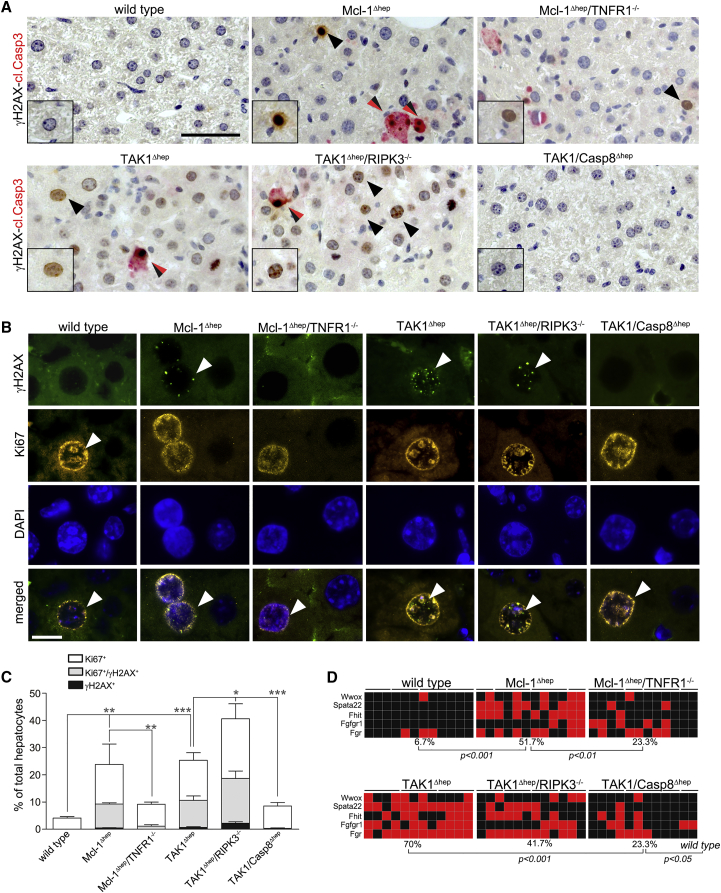
To identify the source of DNA damage and to determine the level of genetic instability in relation to hepatocyte apoptosis, we analyzed Mcl-1Δhep mice, and, to exclude Mcl-1-specific effects, TAK1Δhep mice characterized by increased hepatocyte apoptosis at 6 weeks and caspase-8-dependent HCC development at 35 weeks (100% incidence) (Bettermann et al., 2010). Co-staining for γH2AX and cleaved caspase-3 revealed that hepatocytes from 6- to 8-week-old Mcl-1Δhep mice as well as TAK1Δhep mice which were positive for γH2AX were mostly negative for cleaved caspase-3. Thus, γH2AX-positivity was unlikely to be a consequence of apoptosis of individual hepatocytes (Figure 4A). Immunofluorescence (IF) staining for γH2AX and Ki67 of livers from 6- to 8-week-old mice revealed virtually no γH2AX+ hepatocytes in wild-type livers, whereas Mcl-1Δhep and TAK1Δhep hepatocytes displayed the typical nuclear staining pattern and substantial γH2AX/Ki67 double positivity (∼10% and 20%, respectively; Figures 4B and 4C).
Figure 4.
Reduced DNA Damage And Genetic Instability in Mcl-1Δhep/TNFR1−/− and TAK1/Casp8Δhep Mice and Intercrossings
(A) Staining for γH2AX (black) and cleaved Casp3 (red), double-positive hepatocytes (black/red arrows). Scale bar, 50 μm.
(B) IF staining for γH2AX and Ki67 in wild-type, Mcl-1Δhep, and Mcl-1Δhep/TNFR1−/− mice, as well as TAK1Δhep, TAK1/Casp8Δhep, and TAK1Δhep/RIPK3−/− mice. Arrowheads indicate cells with positive IF staining. Scale bar, 10 μm.
(C) Quantification of Ki67+ and Ki67+/γH2AX+ hepatocytes (n = 4 mice per group, n = 5 for Mcl-1Δhep mice).
(D) Rate of AI in wild-type, Mcl-1Δhep, and Mcl-1Δhep/TNFR1−/− mice, TAK1Δhep, TAK1Δhep/RIPK3−/−, and TAK1/Casp8Δhep mice (TaqMan copy number assay, each square represents one area of microdissected liver tissue, lines indicate different areas of the same liver; red, AI; black, no AI). Mcl-1Δhep mice and intercrossings at 2 months; TAK1Δhep mice and intercrossings at 6 weeks of age.
In (C), data are presented as mean ± SEM. Statistical significance was calculated using ANOVA with Bonferroni correction (C), or Fisher's exact test (D). ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001. See also Figure S5.
To further investigate hepatocyte apoptosis, DDR, and genetic instability in relation to hepatocarcinogenesis, Mcl-1Δhep/TNFR1−/− mice and crossings of TAK1Δhep mice were analyzed: TAK1Δhep/RIPK3−/− mice (devoid of necroptosis, HCC-prone), and TAK1Δhep/Casp8Δhep mice (devoid of apoptosis, not HCC-prone; Table S1) (Vucur et al., 2013). The percentage of Ki67+/γH2AX+ hepatocytes was significantly reduced in intercrossings with reduced apoptosis (Mcl-1Δhep/TNFR1−/− < Mcl-1Δhep mice; and TAK1Δhep/Casp8Δhep < TAK1Δhep < TAK1Δhep/RIPK3−/− mice, respectively; Figures 4B and 4C). Similarly, intercrossings with increased HCC burden also displayed an increased percentage of Ki67+/γH2AX+ hepatocytes (Mcl-1Δhep > Mcl-1Δhep/TNFR1−/− mice; and TAK1Δhep/RIPK3−/− > TAK1Δhep/Casp8Δhep > TAK1Δhep mice, respectively; Figures 4B and 4C). The activation of DNA repair pathways in regenerative murine livers was further corroborated by western blot analysis and expression analysis of genes related to DNA replication, DDR, and DNA repair. Again, mRNA expression of DDR-related genes (and associated protein modifications) were reduced in parallel with hepatocyte apoptosis levels (Figures S5A–S5C). In contrast to wild-type mice, livers of Mcl-1Δhep, TAK1Δhep, and TAK1Δhep/RIPK3−/− mice showed widespread allelic imbalances (AI) at CFS, demonstrating genetic instability in hyper-apoptotic and hyper-proliferative mouse livers. Of note, although higher compared with wild-type mice, AI rates were much lower in Mcl-1Δhep/TNFR1−/− mice and TAK1Δhep/Casp8Δhep mice (Figure 4D).
Since almost all γH2AX+ hepatocytes were proliferating (Ki67+), replication stress (single-stranded DNA breaks accumulation and replication fork stalling) was considered the most likely source of DNA damage (Halazonetis et al., 2008). This was corroborated by fluorescence-activated cell-sorting analysis showing significantly less γH2AX+/RPA+ hepatocytes in low proliferating livers (Figure S5D). Treating Mcl-1Δhep and TAK1Δhep mice with the antioxidants, butylated hydroxyanisole or vitamin E, for 4 weeks revealed no evidence for reactive oxygen species being a major inducer of DNA damage in these mice (Figures S5E–S5H).
Hyper-proliferation-Associated Replicative Stress in Regenerating Livers Causes DNA Damage
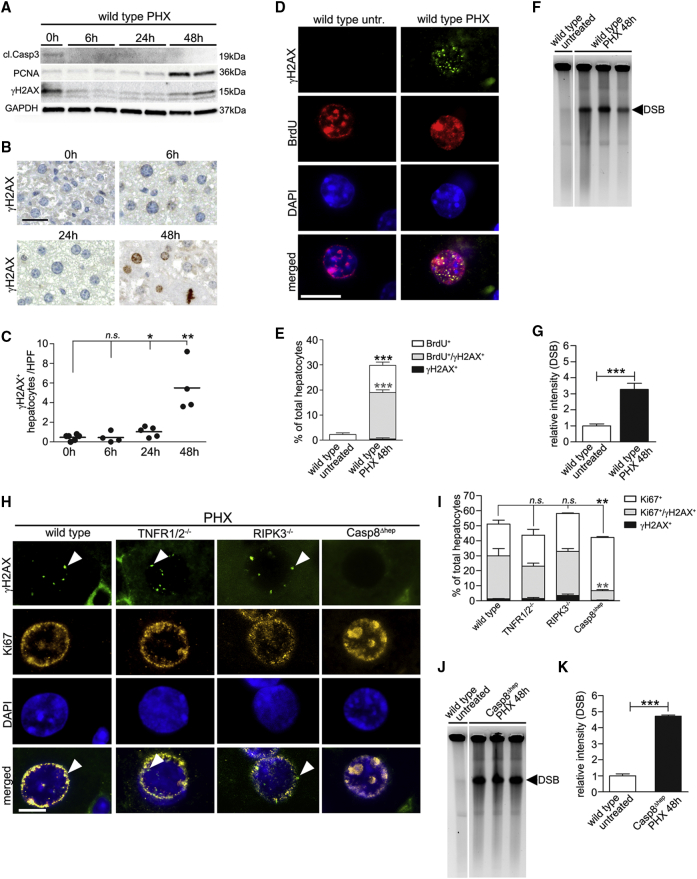
Next, to test whether proliferation by itself, i.e., independent of hepatocyte apoptosis, was sufficient to trigger DNA damage, we performed partial hepatectomy (PHX). Whereas low levels of baseline proliferation in wild-type mice were not associated with detectable levels of DNA damage, western blot analysis and immunohistochemistry for γH2AX peaked at 48 hr post-PHX, i.e., long after evidence for apoptosis (Speicher et al., 2014), and parallel to the proliferative activity (Figures 5A–5G). The correlation between hepatic hyper-proliferation and DNA damage was confirmed by γH2AX/bromodeoxyuridine (BrdU) double staining. Almost all γH2AX+ hepatocytes had incorporated BrdU, indicating that DNA damage occurred in replicating hepatocytes. To further investigate whether γH2AX in proliferating hepatocytes was marking DNA breakage, beside replication stress per se, pulse field gel electrophoresis (PFGE) was performed showing DNA double-strand breaks (DSB) in livers 48 hr after PHX (Figure 5F).
Figure 5.
Detection of Proliferation-Associated DNA Damage after PHX Is Impaired in Casp8Δhep Mice
(A–C) Western blot analysis of whole-liver lysates (A), immunostainings (B), and quantification of γH2AX+ hepatocytes 0, 6, 24, and 48 hr post-PHX (C). Scale bar, 50 μm.
(D and E) BrdU incorporation combined with γH2AX staining (n = 4). Scale bar, 10 μm.
(F and G) PFGE with densitometric quantification to visualize DNA DSB in livers of wild-type mice after PHX (n = 3).
(H and I) IF staining (H) and quantification of Ki67+/γH2AX+ hepatocytes in wild-type, TNFR1/2−/−, RIPK3−/−, and Casp8Δhep mice (I). Arrowheads indicate cells with positive IF staining. Scale bar, 10 μm.
(J and K) PFGE with densitometric quantification to visualize DNA DSB in livers of Casp8Δhep mice after PHX.
In (C), bar represents mean. In (E), (G), (I), and (K) data are presented as mean ± SEM. In (G), bar indicates the mean. Statistical significance was calculated using ANOVA with Bonferroni correction (C and I) or Student's t test (E, G, and K). ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; n.s., not significant. Irrelevant bands were omitted from gels (F and J). Areas in which lanes were omitted are indicated by white space between lanes. See also Figure S6.
To test if hyper-proliferation-associated replication stress also occurs in human livers, biopsies of patients after ALPPS (associating liver partition and portal vein ligation for staged hepatectomy) procedure were analyzed (Schadde et al., 2014). Biopsies taken post-liver partition and portal vein ligation from the patients' highly regenerating left lobe revealed significantly elevated numbers of Ki67+ hepatocytes and a substantial number of Ki67+/γH2AX+ hepatocytes. The latter were significantly lower prior to ALPPS, and in the non-regenerating right liver lobe that had been de-portalized (Figures S6A and S6B). Thus, replication stress due to increased proliferation also occurs in regenerating human livers.
Next, we aimed to investigate whether replication stress and the associated DNA DSB were determined mainly by hepatocyte proliferation, or also directly affected by TNFR1, caspase-8, or RIPK3. To this end, we analyzed livers of TNFR1/2−/−, RIPK3−/−, and Casp8Δhep mice 48 hr post-PHX. Whereas similar levels of Ki67+/γH2AX+ hepatocytes (between 25% and 35%) were detected in livers of wild-type, TNFR1/2−/−, and RIPK3−/− mice, unexpectedly <10% of hepatocytes from livers of Casp8Δhep mice were Ki67+/γH2AX+ (Figures 5H and 5I). Notably, at the same time PFGE clearly demonstrated DSB in Casp8Δhep mice (Figures 5J and 5K), suggesting that caspase-8 plays an important role in sensing or mediating DNA replication-associated damage in hyper-proliferating hepatocytes.
Phosphorylation of Histone H2AX Is Impaired in Caspase-8-Deficient Hepatocytes
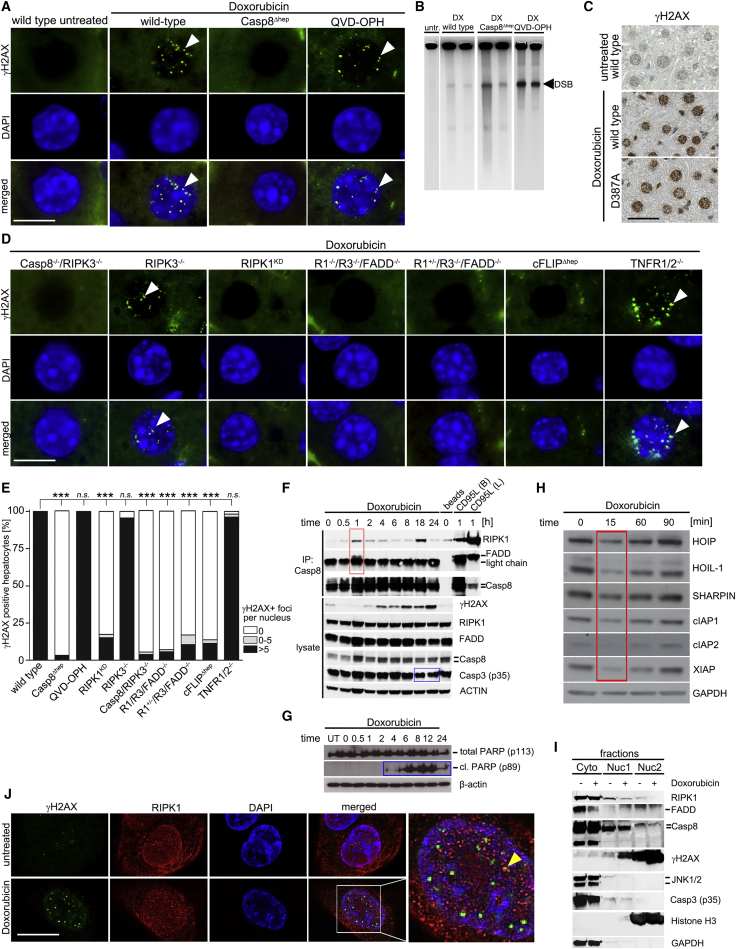
We then sought to determine whether caspase-8 also plays a role in mediating or sensing DNA DSB not related to hyper-proliferation. To this aim, wild-type and Casp8Δhep mice were treated with non-hepatotoxic doses of the genotoxic agent doxorubicin (Yang et al., 2014). Post-application (12 hr), wild-type mice displayed strong γH2AX reactivity in the liver (Figures 6A, 6B, S7A, and S7B) and other tissues (Figure S7C). Strikingly, although PFGE displayed DNA DSB in livers of Casp8Δhep mice (Figure 6B), Casp8Δhep hepatocytes were negative for γH2AX, whereas γH2AX+ nuclei were still detectable in Kupffer cells and several other caspase-8-proficient cell types (Figures 6A and S7C and data not shown). Post-doxorubicin treatment, no apoptotic hepatocytes, no cleavage of caspase-8 above baseline levels (Figures S7D–S7G), or no elevated transaminase levels were detectable (Figure S7H). Thus, H2AX phosphorylation under caspase-8 deficiency is impaired also following doxorubicin-induced DNA damage.
Figure 6.
Caspase-8, RIPK1, FADD, and c-FLIP Are Crucial for Phosphorylation of H2AX in Hepatocytes upon Doxorubicin Treatment
(A) IF for γH2AX in untreated wild-type mice and wild-type, Casp8Δhep, and QVD-OPH-treated wild-type mice following doxorubicin treatment. Arrow heads illustrate γH2AX+ foci in nuclei. Scale bar, 10 μm.
(B) PFGE on livers of doxorubicin-treated mice.
(C) γH2AX staining of doxorubicin-treated wild-type and caspase-8 D387-mutant mice. Scale bar, 50 μm.
(D) γH2AX IF staining 12 hr post-doxorubicin-induced DNA damage in hepatocytes of Casp8−/−/RIPK3−/− mice (n = 5), RIPK3−/− mice (n = 4), RIPK1KD mice (n = 9), RIPK1−/−RIPK3−/−FADD−/− (labeled as R1−/−R3−/−FADD−/−, n = 2), RIPK1+/−RIPK3−/−FADD−/− (labeled as R1+/−R3−/−FADD−/−, n = 2), c-FLIPΔhep (n = 6), and TNFR1/2−/− mice (n = 6). Arrowheads illustrate γH2AX+ foci in nuclei. Scale bar, 10 μm.
(E) Quantification of IF stainings (A and D).
(F) Immunoprecipitation with anti-caspase-8 antibody (upper panel) and immunoblotting of lysates (lower panel), 0–24 hr after doxorubicin (5 μM) treatment. Red box: RIPK1, FADD, and caspase-8 interaction at 1 hr; blue boxes: low-level activation of apoptosis starting at 4 hr post-treatment. (The signal visible in the t = 0 column, cl.PARP lane, does not originate from cl.PARP, but from a lower unspecific band.) Control cells treated for 1 hr with CD95L/FasL (B, beads; L, lysates).
(G) Immunoblotting of lysates, 0–24 hr after doxorubicin (5 μM) treatment looking at levels of total and cl.PARP, blue boxes (F and G): low-level activation of apoptosis starting at 4 hr post-treatment.
(H) Levels of LUBAC (HOIP, HOIL-1, and SHARPIN), cIAP1, cIAP2, and XIAP in U2OS cells at 15 min (red box) post-doxorubicin stimulation (5 μM).
(I) Subcellular fractionation of U2OS cells.
(J) RIPK1 and γH2AX IF staining in U2OS cells after doxorubicin treatment. The arrowhead indicates colocalizing signals. Scale bar, 10 μm.
Statistical significance was calculated using ANOVA with Bonferroni correction (E). ∗∗∗p < 0.001. Irrelevant bands were omitted from gels (B). Areas in which lanes were omitted are indicated by white space between lanes. See also Figure S7.
The Catalytic Activity of Caspase-8 Is Dispensable for H2AX Phosphorylation
Pre-treating wild-type mice with the pan-caspase inhibitor QVD-OPH did not abrogate H2AX phosphorylation after doxorubicin application (Figure 6A). In contrast, QVD-OPH strongly reduced liver damage in mice co-treated with LPS/DGal (Figures S7F–S7H). To exclude incomplete inhibition of caspase-8 activity using QVD-OPH, knockin mice expressing an uncleavable mutant of caspase-8 (D387A) were also treated with doxorubicin (Kang et al., 2008). Similar to wild-type mice, caspase-8 D387-mutant mice revealed γH2AX positivity in hepatocytes upon doxorubicin treatment (Figure 6C). Consistent with results from conditional caspase-8 knockout mice, we observed significantly reduced γH2AX positivity in Casp8−/−/RIPK3−/− livers 12 hr post-treatment (Figure 6D). In contrast, hepatocytes from RIPK3−/− littermates were positive for γH2AX, consistent with hepatocytes upon PHX of RIPK3−/− mice (Figure 5H). Collectively, these data show that full-length caspase-8, but not its cleaved form or catalytic activity, is required for H2AX phosphorylation.
H2AX Phosphorylation Is Impaired in Hepatocytes Deficient of c-FLIP, FADD, or RIPK1 Kinase Activity
We next investigated whether caspase-8-interacting proteins were involved in H2AX phosphorylation. Doxorubicin-induced H2AX phosphorylation was not affected in TNFR1/2−/− mice, indicating that hepatic H2AX phosphorylation activated by low levels of DNA DSB is not executed by TNFR1/2 signaling complexes. In contrast, c-FLIP-deficient hepatocytes lacked γH2AX positivity post-doxorubicin treatment (Figures 6D and 6E), pointing to a crucial role of the c-FLIP/Casp8 dimer in DDR. Pharmacological inhibition of RIPK1 by pre-treatment of wild-type mice with necrostatin-1 (Nec1) did not prevent DNA DSB formation, but prevented the appearance of γH2AX+ hepatocytes upon doxorubicin treatment (Figures S7A and S7B). This indicated a role of RIPK1 for H2AX phosphorylation in vivo. Since Nec1 blocks both RIPK1 assembly and RIPK1 kinase function, we analyzed knockin mice expressing a kinase-inactivated RIPK1 mutant (RIPK1KD mice) and observed significantly impaired H2AX phosphorylation, demonstrating that the kinase activity of RIPK1 is required for H2AX phosphorylation (Figure 6D). In addition, mice deficient for RIPK1, RIPK3, and FADD (R1−/−/R3−/−/FADD−/− mice), as well as RIPK1 (haplodeficient), RIPK3, and FADD (R1+/−/R3−/−/FADD−/− mice), also demonstrated impaired H2AX phosphorylation. Since RIPK3 was not involved in DDR and haploinsufficiency is not reported for RIPK1 (Dillon et al., 2014), the lack of γH2AX+ cells in R1+/−/R3−/−/FADD−/− mice was most likely due to the deletion of FADD. Of note, mice deficient for X-linked inhibitor of apoptosis protein (XIAP−/−) clearly showed H2AX phosphorylation upon doxorubicin treatment (Figure S7A). In summary, RIPK1KD, Casp8−/−/RIPK3−/−, R1−/−/R3−/−/FADD−/−, R1+/−/R3/FADD−/−, and c-FLIPΔhep mice all showed a significantly reduced percentage of γH2AX+ foci in hepatocyte nuclei.
Caspase-8 Functions within a Multi-Protein Complex to Orchestrate H2AX Phosphorylation
To test whether a caspase-8-containing protein complex forms in response to DNA DSB, U2OS cells were treated with doxorubicin, followed by time-course immunoprecipitation experiments with an anti-caspase-8 antibody. Starting 30 min post-treatment and peaking at 1 hr, RIPK1 was co-immunoprecipitated with caspase-8, FADD (Figure 6F, red box), and c-FLIP (Figure S7I, red box). In parallel, γH2AX positivity was detectable starting 1 hr post-doxorubicin treatment. Of note, complex formation 1 hr post-doxorubicin treatment was independent of any apoptotic activity, which was, however, observed between 4 hr (PARP cleavage) and 18–24 hr (caspase-3 cleavage) post-doxorubicin treatment (Figures 6F and 6G, blue boxes). Importantly, absolute quantification of caspase-8, FADD, c-FLIP, and RIPK1 in caspase-8 immunoprecipitates was performed 1 hr post-doxorubicin treatment by a mass spectrometry-based AQUA method (Schleich et al., 2015). This revealed a significant amount of FADD and RIPK1 in complex with caspase-8 in doxorubicin-treated cells (Figure S7J).
Since the linear ubiquitin chain assembly complex (LUBAC) plays a role in preventing cell-death-inducing complex formation in various cell types including hepatocytes (Lafont et al., 2017, Shimizu et al., 2017), it was considered a candidate signaling event. Indeed, LUBAC components HOIP, HOIL-1, and SHARPIN, as well as the inhibitor of apoptosis proteins (IAP) cIAP1, cIAP2, and XIAP, known to negatively regulate formation of the ripoptosome (Tenev et al., 2011), were transiently reduced 15 to 30 min post-doxorubicin treatment of U2OS cells (Figure 6H).
Next, we tested whether complex formation in response to DNA damage was paralleled by a change in subcellular localization. Western blot analysis after subcellular fractionation displayed a proportion of RIPK1, caspase-8, and c-FLIP in the nuclear fraction under steady-state conditions, but no enrichment upon doxorubicin-induced DNA damage (Figure 6I). Localization studies by IF staining and confocal imaging of U2OS cells, with and without doxorubicin, confirmed the induction of nuclear γH2AX positivity, while a minor fraction of RIPK1 was already detectable in the nucleus under steady state (Yoon et al., 2016). However, no increased nuclear signal was observed after doxorubicin treatment (Figure 6J).
JNK Is a Downstream Mediator of Caspase-8- and RIPK1-Dependent H2AX Phosphorylation in Hepatocytes
To identify candidate downstream signaling pathways of the DNA damage-sensing platform in hepatocytes, livers of doxorubicin-treated wild-type mice were analyzed for activation of ATM/ATR targets CHK1 and CHK2 (Figures 7A, S8A, and S8B and data not shown). Remarkably, no pCHK1+ or pCHK2+ hepatocytes were found 12 hr following doxorubicin-induced DNA damage, suggesting that the ATM and ATR kinase activity is rather low at that time, and pointing to DNA damage-transducing pathways other than ATM and ATR signaling. As control, LPS/DGal-induced cell death was associated with pCHK1+ and pCHK2+ apoptotic hepatocytes (Figure S8B). Furthermore, wild-type mice displayed pcJUN+ hepatocytes upon doxorubicin-induced DSB, indicative of activated c-JUN N-terminal kinase (JNK) signaling (Figures 7A and S8A). Of note, hepatocytes of Casp8Δhep, RIPKKD, Nec1-treated, c-FLIPΔhep, and Casp8−/−/RIPK3−/− mice all lacked substantial pcJUN staining after doxorubicin treatment, in contrast to QVD-OPH-treated wild-type, TNFR1/2−/−, RIPK3−/−, and XIAP−/−mice (Figures S8A and S8B). Co-IF staining for pJNK and γH2AX revealed that wild-type hepatocytes (independent of QVD-OPH pre-treatment) and TNFR1/2−/− hepatocytes had distinct nuclear pJNK signals following doxorubicin treatment, which partially co-localized with γH2AX signals, suggesting JNK as the responsible kinase for H2AX phosphorylation. Casp8Δhep, RIPK1KD, and c-FLIPΔhep hepatocytes were mostly devoid of pJNK signals (Figure 7B), suggesting that JNK signaling is downstream of the kinase function of RIPK1, and of caspase-8, FADD, and c-FLIP, and, as such, is involved in H2AX phosphorylation (Picco and Pages, 2013). Furthermore, mice lacking both JNKs in hepatocytes (JNK1/2Δhep mice) displayed DNA DSB by PFGE, but lacked γH2AX+ hepatocytes after doxorubicin treatment (Figures 7C and 7D). At the same time, PFGE displayed DNA DSB in livers of JNK1/2Δhep mice (Figure S8C).
Figure 7.
JNK Is a Downstream Mediator of Caspase-8-, c-FLIP-, and RIPK1-Dependent Phosphorylation of H2AX In Vivo and In Vitro
(A) Immunohistochemistry for pCHK1, pCHK2, and pcJUN in livers after doxorubicin treatment. Arrowheads indicate pcJUN-positive nuclei. Scale bar, 50 μm.
(B) γH2AX and pJNK co-stainings of livers 12 hr post-doxorubicin treatment. Merged: overlay of DAPI, γH2AX, and pJNK staining. Arrowheads indicate IF signals for γH2AX (green), pJNK (red), or overlapping signals of both (yellow). Scale bar, 10 μm.
(C and D) IF stainings (C) and quantification for γH2AX in wild-type and JNK1/2-deficient hepatocytes 12 hr post-doxorubicin treatment (D). Arrowheads indicate IF signals for γH2AX. Scale bar, 10 μm.
(E) Analysis of DDR signaling by western blotting of lysates from doxorubicin-treated caspase-8 knockdown cells, JNK inhibitor (SP600125) and ATM inhibitor (KU-55933) pre-treated control cells (U2OS). Red boxes: differences in γH2AX and pJNK activation post-doxorubicin treatment between control cells and lentiviral caspase-8 knockdown and JNK inhibitor treated cells. Statistical analysis was corrected for three tests using the Bonferroni method. See also Figure S8.
JNK Is a Downstream Mediator of Caspase-8- and RIPK1-dependent DDR Also in Cell Types Other than Hepatocytes
To determine whether caspase-8 and JNK were involved in H2AX phosphorylation in non-hepatocytic cells, caspase-8 was knocked down in U2OS cells. As expected, shCASP8 lentivirus-transfected U2OS cells were less sensitive to TNF-α-mediated apoptosis compared with shCTRL cells (Figure S8D). As observed in vivo, shCTRL cells also displayed a clear increase in γH2AX 30 min post-doxorubicin administration and a strong pJNK signal, but no obvious ATM or ATR activation (Figure 7E). Strikingly, upon doxorubicin treatment, knock down of caspase-8 substantially decreased γH2AX, pJNK, and pcJUN in shCASP8 cells compared with shCTRL cells (Figures 7E and S8E). Pre-treatment of shCTRL cells with a JNK inhibitor (JNKi), but not with an ATM inhibitor (ATMi), abolished c-JUN activation and decreased H2AX phosphorylation similar to shCASP8 cells (Figure 7E, red boxes). Combining ATMi and JNKi reduced γH2AX signals in shCASP8 cells, and led to minimal activation of ATR (Figure S8E). To exclude that data were cell line-specific, HepG2 cells were analyzed, revealing a caspase-8- and JNK-dependent γH2AX increase 30 min post-doxorubicin administration (Figure S8F, red boxes).
Next, we analyzed potential downstream targets of the caspase-8-containing complex regulating H2AX phosphorylation. Of note, IF staining for p53-binding protein 1 (53BP1), an important regulator of the cellular response to DNA DSB (Panier and Boulton, 2014), revealed that, in response to doxorubicin treatment, 53BP1 nuclear positivity was absent in shCASP8 cells, similar to γH2AX 30 min following doxorubicin treatment (Figure S8G). This indicated impaired recruitment of 53BP1 to sites of DNA DSB under caspase-8 deficiency. Aiming to identify further signaling pathways involved in caspase-8-dependent H2AX phosphorylation, we analyzed MAPK and phosphatidylinositol 3-kinase signaling pathways. We found reduced activation of p38 and ERK1/2 under steady-state conditions in shCASP8 cells. At the same time total levels of ERK1/2 and also AKT2 were increased (Figure S8H, red boxes). Doxorubicin treatment induced activation of ERK1/2 and AKT2, whereas p38 activation was impaired in shCASP8 cells. Thus, caspase-8 interferes with or controls MAPK signaling under steady-state and doxorubicin-challenged conditions. Looking for an interaction between JNK and γH2AX in human livers, we found mostly overlapping signals for pJNK and γH2AX in liver tissues of patients with chronic low-level, CLD-related liver regeneration and acute high-level liver regeneration after ALPPS, indicating a role of JNK in mediating DDR (Figure 8A).
Figure 8.
Evidence for JNK-Dependent DDR in Human Regenerating Livers and Caspase-8 in Human HCC
(A) γH2AX and pJNK co-stainings demonstrating JNK-dependent phosphorylation of H2AX in liver tissue of CLD patients or the left lobe of patients after (right) portal vein ligation and liver transection (PVL/LT). Arrowheads indicate IF signals for γH2AX (green), pJNK (red), or overlapping signals of both (yellow). Scale bar, 10 μm.
(B) Overall survival of HCC patients depending on HCC caspase-8 expression level (<mean+1SD; n = 307 patients; >mean+1SD; n = 51 patients, log rank test, statistical analysis was corrected for three tests using the Bonferroni method. The Cancer Genome Atlas [TCGA] cohort).
(C) Overall survival of HCC patients depending on HCC caspase-8 methylation status (n = 358 patients, TCGA cohort, log rank test).
Finally, we analyzed publically accessible databases to address whether caspase-8 expression affects HCC biology. Although different datasets yielded variable results, analysis of the largest, most stringent cohort (n = 358 patients) from The Cancer Genome Atlas data portal, validated by the Universal exPression Codes method, revealed that HCC with low caspase-8 expression levels were associated with a better overall survival compared with HCC with high caspase-8 expression (Figure 8B). Moreover, high caspase-8 expression correlated with high PCNA and Ki67 expression (PCNA versus CASP8: p = 2 × 10−22, Ki67 versus CASP8: p = 3 × 10−25; data not shown), indicating high proliferative activity. Finally, HCC with methylation of the caspase-8 gene exhibited a better overall survival compared with caspase-8-unmethylated HCC (Figure 8C). These findings suggest that low caspase-8 expression is associated with a less aggressive behavior of HCC.
Discussion
Hepatocyte apoptosis, a hallmark of CLDs, plays opposing roles in liver homeostasis: on the one hand, it constitutes a hepato-protective mechanism by eliminating damaged hepatocytes. On the other hand, chronically increased hepatocyte apoptosis is harmful.
Here, we show that persistently increased levels of hepatocyte apoptosis tightly correlate with subsequent HCC development: (1) CLD patients who developed HCC had higher preceding transaminase levels compared with case-control matched pairs. (2) Mcl-1Δhep mice and Mcl-1Δhep/TNFR1−/− mice that developed liver tumors had higher levels of transaminase activity early in life compared with littermates without tumors. (3) Genetically reducing apoptosis in Mcl-1Δhep mice, by additional deletion of TNFR1 (here) or BAK (Hikita et al., 2012), decreased tumorigenesis similar to TAK1Δhep mice with additional caspase-8 (apoptosis) but not RIPK3 (necroptosis) deficiency (Vucur et al., 2013). Collectively, these data identify chronically increased hepatocyte apoptosis as a major risk for subsequent HCC development.
By demonstrating (1) increased γH2AX+ hepatocytes, indicative of DDR, in both hyper-apoptotic, hyper-regenerating livers of CLD patients and CLD mouse models, and (2) a significant level of AI at CFS in these same livers, we link chronically increased hepatocyte apoptosis with HCC development. Liver regeneration results in DNA replication stress, making hepatocyte proliferation a genotoxic stimulus inducing DNA damage and genetic instability. DNA replication is a major genotoxic stress due to the risk of nucleotide misincorporation, the intrinsic fragility of replicating chromosomes, and the abundance of repetitive and unusual DNA structures in particular at CFS. Genome stability pathways address these challenges and minimize replication-associated risks, but require extra time in cell cycle progression and are often limited in hyper-proliferative states (Halazonetis et al., 2008).
Our findings in human CLD and murine CLD models, underpinned by stochastic considerations (Figure S4), argue that persistently increased hepatocyte apoptosis resulting in regenerative proliferation and high DNA replication rate (independent of etiology) is a decisive determinant of hepatocarcinogenesis. This is in line with a recent report on the carcinogenic effect of replication errors stochastically occurring in highly proliferative stem cells (Tomasetti and Vogelstein, 2015). This concept also explains most HCC epidemiological data, i.e., that CLD patients are at risk to develop HCC, and that the risk increases with disease activity and duration.
Dissecting the role of caspase-8 for hepatocyte apoptosis, we discovered a non-apoptotic function of caspase-8 in H2AX phosphorylation. Firstly, by performing PHX in C57BL/6 and Casp8Δhep mice, we show that caspase-8 is needed for an efficient DDR to replication stress. Secondly, we found that doxorubicin-induced H2AX phosphorylation in mice was not deficient in livers pretreated with the pan-caspase inhibitor (QVD-OPH), or livers of caspase-8 D387A mutant mice (Kang et al., 2008), but in mice that were full knockout for caspase-8. Thus, DDR was not dependent on caspase-8 catalytic activity, but rather on a non-apoptotic, e.g., a scaffold function of caspase-8. Aiming to identify caspase-8-interacting molecules, we discovered a signaling platform comprising also RIPK1, FADD, and c-FLIP. These molecules are also central in a complex which assembles independently of death receptor activation, referred to as the ripoptosome (Tenev et al., 2011). It acts cooperatively in different combinations: (1) to control cell fate upon genotoxic stress in concert with NEMO (Biton and Ashkenazi, 2011), (2) together with RIPK3 to control the non-canonical inflammasome activation (Kang et al., 2013), (3) together with RIPK1 to control TNF-α expression NF-κB independently via JNK (Christofferson et al., 2012), and together with caspase-3 to suppress necrosis (Brown et al., 2015). Moreover, (4) a different complex, the PIDDosome (Tinel and Tschopp, 2004), is activated by ATM and executes apoptosis in response to DNA damage (Ando et al., 2012). Finally, (5) it was demonstrated recently that a structural (rather than enzymatic) function of these signaling complex is central for the production of chemotactic cytokines (Hartwig et al., 2017, Henry and Martin, 2017). Although not providing a direct proof of their regulatory function, the downregulation of LUBAC components and IAPs occurring in temporal association with the formation of the complex discovered here is remarkable. It is reminiscent of the formation of the above-mentioned related complexes. Collectively, findings from all these studies suggest that a defined set of molecules constitutes a dynamic and temporary signaling platform. This platform integrates various inputs (e.g., genotoxic stress, inflammatory signals) resulting in different outputs (e.g., cell death, cytokine production, DDR), thus efficiently coordinating cell fate. We consider the complex found in this study to represent one possibility of these responses, most likely to DNA DSB. Although we found no obvious nuclear localization after induction of DNA damage, our observations do not preclude that under steady state a minor nuclear fraction of the above described components remains functionally activatable to form a complex, as described elsewhere (Yoon et al., 2016).
We observed (1) impaired pJNK and pcJUN response to doxorubicin in caspase-8-deficient cells, (2) impaired H2AX phosphorylation after pharmacological inhibition of JNK in various cell lines (U2OS, HepG2) in response to doxorubicin, and (3) impaired H2AX phosphorylation in doxorubicin-treated livers of JNK1/2Δhep mice, all implicating a possible involvement of JNK signaling. However, performing several in vivo, ex vivo, and in vitro experiments to investigate the downstream signaling modes of this pathway did not give conclusive results. The co-localization of pJNK and γH2AX in human and murine hepatocyte nuclei suggested a link between pJNK and H2AX phosphorylation, in line with previous reports (Picco and Pages, 2013). Moreover, activation of the ATM/ATR and JNK signaling in human and murine CLDs suggested that all pathways might be present. In contrast, doxorubicin-induced DNA DSB appeared to preferably activate the JNK signaling pathway, whereas the ATM/ATR signaling pathway was activated less in a model- and time-dependent fashion. This inter-experimental variation made it challenging to clearly demonstrate whether or not the ATM/ATR signaling was generally activated by the caspase-8/RIPK1/c-Flip/FADD pathway. Due to the different kinetics in in vivo and also in distinct in vitro model systems, a uniform pattern of ATM/ATR activation was not identifiable. Interestingly, combined use of ATM and JNK inhibitors led to the strongest and most consistent effects in suppressing H2AX phosphorylation upon doxorubicin in vitro. Thus, we conclude that (1) doxorubicin might not necessarily reflect the overall in vivo signaling behavior of the complex (e.g., kinetics, signaling candidates), and (2) JNK signaling might be only one of several possible downstream mediators of this complex. Based on our findings, we cannot exclude that phosphatidylinositol 3-kinase-related kinases contribute to the DNA damage signaling pathway discovered in this study. Further studies are needed to identify all important downstream signaling mediators of caspase-8-dependent DDR.
Showing that the caspase-8-containing complex triggers H2AX phosphorylation suggests that it controls DNA integrity and thus potentially prevents malignant transformation. If this holds true, loss of caspase-8 can be expected to be genotoxic and generate an environment of genetic instability. In line with these findings, caspase-8 deficiency has been shown to facilitate cellular transformation independently of its killing function (Krelin et al., 2008). Loss of caspase-8 expression by either mutations or epigenetic silencing has been reported in murine and human HCC (Liedtke et al., 2005, Soung et al., 2005). Therefore, it is conceivable that loss of caspase-8 in one and the same cell not only confers apoptosis resistance (a hallmark of cancer), but also promotes replication errors, and thus contributes to cancer development. Based on our observations, caspase-8 deficiency is thus expected to predispose to mutations in proliferating non-neoplastic hepatocytes, whereas at the same time it should confer a fitness disadvantage to neoplastic hepatocytes. In line with the latter, data mining using distinct, already published HCC cohorts revealed that low caspase-8 expression in HCC is associated with a less aggressive behavior, reflected by a less proliferative phenotype and a better overall survival. This is reminiscent to the biology of mismatch repair (MMR) deficiency in the colorectum. MMR deficiency on the one hand predisposes to replication errors and cancer development, and on the other hand it results in hyper-mutated tumors with a better prognosis compared with MMR-proficient carcinomas (Gryfe et al., 2000).
Given the here described role of caspase-8 in DDR, it is at first glance counter-intuitive that deletion of caspase-8 rescued HCC development in TAK1Δhep mice (Vucur et al., 2013). However, DNA damage in a hyper-apoptotic environment such as in CLD patients, Mcl-1Δhep, or TAK1Δhep mice, is provoked by constantly enhanced regeneration causing replication stress. Taking into account that caspase-8 deficiency in TAK1Δhep mice abolished apoptosis and nearly normalized proliferative levels (Figure 4C; Vucur et al., 2013), it is obvious that TAK1/Casp8Δhep mice are not tumor prone.
In summary, we identified a role of caspase-8 in sensing DNA damage, and have mechanistically linked increased hepatocyte apoptosis with subsequent HCC development.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-RIP3 (phospho S227) antibody | Abcam | Cat# ab209384 |
| Anti-c-Jun Rabbit polyclonal Antibody | Abcam | Cat# ab31367; RRID: AB_731606 |
| Purified Mouse Anti-RIP Antibody (38/RIP) (Immunofluorescent imaging) | BD Biosciences | Cat# 610459; RRID: AB_397832 |
| Phospho Histone H2A.X (ser139) (20E3) Rabbit mAb | Cell Signaling Technologies | Cat# 9718; RRID: AB_2118009 |
| RIP (D94C12) XP Rabbit Mab | Cell Signaling Technology | Cat# 3493; RRID: AB_2305314 |
| Anti-Caspase-3 antibody | Cell Signaling Technology | Cat# 9662; RRID: AB_331439 |
| Phospho-p53 (Ser15) (D4S1H) Rabbit mAb | Cell Signaling Technology | Cat# 12571 |
| GAPDH (D16H11) XP Rabbit mAb | Cell Signaling Technology | Cat# 5174; RRID: AB_10622025 |
| PCNA (PC10) Mouse mAb | Cell Signaling Technology | Cat# 2586; RRID: AB_2160343 |
| Phospho-ATM (Ser1981) (D6H9) Rabbit mAb | Cell Signaling Technology | Cat# 5883; RRID: AB_10835213 |
| Phospho-Chk1 (Ser345) (133D3) Rabbit mAb | Cell Signaling Technology | Cat# 2348; RRID: AB_331212 |
| Phospho-SAPK/JNK (Thr183/Tyr185) Antibody | Cell Signaling Technology | Cat# 9251; RRID: AB_331659 |
| Phospho-Chk2 (Thr68) Antibody | Cell Signaling Technology | Cat# 2661; RRID: AB_331479 |
| Phospho-ATR (Ser428) Antibody | Cell Signaling Technology | Cat# 2853; RRID: AB_2290281 |
| Phospho-BRCA1 (Ser1524) Antibody | Cell Signaling Technology | Cat# 9009; RRID: AB_491003 |
| Cleaved Caspase-3 (Asp175) (5A1E) Rabbit mAb | Cell Signaling Technology | Cat# 9664; RRID: AB_2070042 |
| Cleaved Caspase-8 (Asp387) (D5B2) XP Rabbit mAb | Cell Signaling Technology | Cat# 8592 |
| RIP (D94C12) XP Rabbit mAb | Cell Signaling Technology | Cat# 3493; RRID: AB_2305314 |
| Cleaved Caspase-1 (Asp296) Antibody | Cell Signaling Technology | Cat# 67314 |
| Phospho-p38 MAPK (Thr180/Tyr182) (D3F9) XP Rabbit mAb | Cell Signaling Technology | Cat# 4511; RRID: AB_2139682 |
| p38 MAPK (D13E1) XP Rabbit mAb | Cell Signaling Technology | Cat# 8690; RRID: AB_10999090 |
| c-IAP1 Rabbit polyclonal Antibody | Cell Signaling Technology | Cat# 4952; RRID: AB_2063660 |
| c-IAP2 (58C7) Rabbit mAb | Cell Signaling Technology | Cat# 3130; RRID: AB_10693298 |
| XIAP Rabbit polyclonal Antibody | Cell Signaling Technology | Cat# 2042; RRID: AB_2214870 |
| PARP Rabbit polyclonal Antibody | Cell Signaling Technology | Cat# 9542; RRID: AB_2160739 |
| Caspase-8 Rabbit polyclonal Antibody | Cell Signaling Technology | Cat# 4927; RRID: AB_2068301 |
| Alexa Fluor 488 Goat anti-Rat IgG (Immunofluorescent imaging) | Life Technologies | Cat# A11006; RRID: AB_141373 |
| Alexa Fluor 546 Goat anti-Rabbit (Immunofluorescent imaging) | Life Technologies | Cat# A11010; RRID: AB_143156 |
| Goat anti-Mouse IgG Alexa Fluor 488 (Immunofluorescent imaging) | Life Technologies | Cat# A11029; RRID: AB_138404 |
| NA19L Anti-replication Protein A (Ab-3) Mouse mAb (RPA34-20) | Merck (Calbiochem) | Cat# NA19L; RRID: AB_565123 |
| Ki-67 (SP6) Rabbit mAb | Neomarkers / Lab vision Corporation | Cat# RM9106; RRID: AB_2335745 |
| Cleaved caspase-8 Rabbit polyclonal Antibody | Novus Biologicals | Cat# NB100-56116; RRID: AB_837874 |
| Chk1 [p Ser317] Rabbit polyclonal Antibody | Novus Biologicals | Cat# NB100-92499; RRID: AB_1216466 |
| p-Chk2 [p Thr68] Rabbit polyclonal Antibody | Novus Biologicals | Cat# NB100-92502; RRID: AB_1216474 |
| gamma H2AX [p Ser139] Rabbit polyclonal Antibody (Immunofluorescent imaging) | Novus Biologicals | Cat# NB100-2280; RRID: AB_10000580 |
| gamma H2AX (p Ser139) Rabbit polyclonal Antibody | Novus Biologicals | Cat# NB100-384; RRID: AB_350295 |
| SHARPIN Rabbit polyclonal Antibody | Proteintech | Cat# 14626-1-AP; RRID: AB_2187734 |
| Caspase-8 p18 Antibody (H-134) | Santa Cruz Biotechnology | Cat# sc-7890; RRID: AB_2068330 |
| p-c-Jun Goat polyclonal Antibody (Ser 63/73) | Santa Cruz Biotechnology | Cat# sc-16312; RRID: AB_2129883 |
| p-Akt1/2/3 Rabbit polyclonal Antibody (Ser 473) | Santa Cruz Biotechnology | Cat# sc-7985-R; RRID: AB_667741 |
| Akt1/2 Goat polyclonal Antibody (N-19) | Santa Cruz Biotechnology | Cat# sc-1619; RRID: AB_671713 |
| 53BP1 Rabbit polyclonal Antibody (H-300) (Immunofluorescent imaging) | Santa Cruz Biotechnology | Cat# sc-22760; RRID: AB_2256326 |
| Human HOIP/RNF31 Antibody | R&D Systems | Cat# AF8039 |
| Anti-BrdU antibody, Mouse Monoclonal (clone BU-33) | Sigma-Aldrich | Cat# B8434; RRID: AB_476811 |
| Anti-actin N terminal antibody | Sigma Aldrich | Cat# A2103; RRID: AB_476694 |
| Anti-ATM Mouse mAb | Sigma-Aldrich | Cat# A1106; RRID: AB_796190 |
| C15 (anti-caspase 8) | Prof. Peter H Krammer (DKFZ, Heidelberg) | N/A |
| 1C4 (anti-FADD) | Prof. Peter H Krammer (DKFZ, Heidelberg) | N/A |
| Human HOIL-1 Antibody | Prepared in house | Haas et al., 2009 |
| Bacterial and Virus Strains | ||
| lentiviral particles for caspase-8 | Santa Cruz | Cat# sc-29930-V |
| lentiviral particles for control | Santa Cruz | Cat# sc-108080 |
| Biological Samples | ||
| Liver tissue from mice after Vitamin E diet | This paper | N/A |
| Liver tissue from mice after BHA diet | This paper | N/A |
| Liver tissue from mice after two-third partial hepatectomy | Speicher et al., 2014 | N/A |
| Liver Tissue from mice after LPS/D-Gal treatment | This paper | N/A |
| Liver tissue from mice after Doxorubicin treatment | This paper | N/A |
| Liver tissue from various mutant mice and intercrossings | Vick et al., 2009, Vucur et al., 2013, Olayioye et al., 2005, Kang et al., 2008, Das et al., 2011, Dillon et al., 2014, this paper | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Doxorubicin | Sigma-Aldrich | Cat# D1515 |
| DMSO | Sigma-Aldrich | Cat# 276855 |
| Q-VD-OPH | Sigma-Aldrich | Cat# SML0063 |
| Necrostatin1 | Sigma-Aldrich | Cat# N9037 |
| D-(+)-Galactosamine | Sigma-Aldrich | Cat# G0500 |
| Lipopolysaccharide | Sigma-Aldrich | Cat# F3665 |
| Buprenorphine | MSD Sharp & Dohme GmbH | NDC 12496-0757-5 |
| DAPI | Life Technologies |
Cat# D1306 |
| Puromycin (CAS 53-79-2) | Santa Cruz | Cat# sc-205821 |
| Caspase 8 inhibitor (Z-IETD-FMK) | Selleckchem | Cat# S7314 |
| ATM kinase inhibitor Ku-55933 | Selleckchem | Cat# S1092 |
| JNK inhibitor SP600125 | Selleckchem | Cat# S1460 |
| Caspase 1 inhibitor (YVAD-CMK) | Merck (Calbiochem) | Cat# 400012 |
| Critical Commercial Assays | ||
| LIVE/DEAD Fixable Dead Cell Stain Kit | Invitrogen | Cat# L23102 |
| RNeasy Mini Kit | Qiagen | Cat# 74106 |
| Quantitect Reverse Transcription Kit |
Qiagen | Cat# 205313 |
| Fast Start SYBR Green Master Rox | Roche | Cat# 04913850001 |
| TaqMan™ Copy Number Assays | Thermo Fisher | Cat# 4400291 |
| Human Wwox | Thermo Fisher | Cat# 4400291 |
| Human Spata22 | Thermo Fisher | Cat# 4400291 |
| Human Fhit | Thermo Fisher | Cat# 4400291 |
| Human Fgfr1 | Thermo Fisher | Cat# 4400291 |
| Human Fgr | Thermo Fisher | Cat# 4400291 |
| Murine Wwox | Thermo Fisher | Cat# 4400291 |
| Murine Spata22 | Thermo Fisher | Cat# 4400291 |
| Murine Fhit | Thermo Fisher | Cat# 4400291 |
| Murine Fgfr1 | Thermo Fisher | Cat# 4400291 |
| Murine Fgr | Thermo Fisher | Cat# 4400291 |
| Mouse DNA Microarray 4x 44K |
Agilent | Cat# G4122F |
| Deposited Data | ||
| Mouse RNA expression data | This paper | The accession number for the data reported in this paper is: GSE75730 |
| Datasets for Gene Set Enrichment Analysis (GSEA) | Molecular Signatures Database | http://www.broadinstitute.org |
| Clinical and RNA sequencing data from human HCC | The Cancer Genome Atlas (TCGA) | https://tcga-data.nci.nih.gov/ |
| Methylation data from human HCC | TCGA, via cBioPortal | http://www.cbioportal.org/ |
| Experimental Models: Cell Lines | ||
| U2OS | Massimo Lopes | N/A |
| p19-/- MEFs | Emmanuel Dejardin | N/A |
| Experimental Models: Organisms/Strains | ||
| JNK1/2flox/flox | Roger J. Davis | N/A |
| Mcl-1flox/flox | Joseph T. Opferman | N/A |
| Tak1Δhep | Tom Luedde | N/A |
| Casp8Δhep | Tom Luedde | N/A |
| Tak1/Casp8Δhep | Tom Luedde | N/A |
| Tak1Δhep/RIPK3-/- | Tom Luedde | N/A |
| Xiap−/− | Philip Jost | N/A |
| caspase 8 D387 | David Wallach | N/A |
| cFLIPΔhep | Jörn M Schattenberg | |
| TNFR1-/- | Mathias Heikenwälder | N/A |
| TNFR1/2-/- | Jackson | JAX: 003243 |
| Ripk1-/-/Ripk3−/−/Casp8−/− | Douglas Green | N/A |
| Ripk1-/-/Ripk3−/−FADD−/− | Douglas Green | N/A |
| Ripk3−/−/Casp8−/− | Douglas Green | N/A |
| Ripk3−/− | Douglas Green/ Tom Luedde | N/A |
| RIPK1KD | Douglas Green | N/A |
| Oligonucleotides | ||
| Murine Mcl-1 Fwd TCAAAGATGGCGTAACAAACTGG Rev CCCGTTTCGTCCTTACAAGAAC |
This paper | N/A |
| Murine Tnf-α Fwd CATCTTCTCAAAATTCGAGTGACAA Rev TGG GAGTAGACAAGGTACAACCC |
This paper | N/A |
| Murine Trail Fwd CGGGCAGATCACTACACCC Rev TGTTACTGGAACAAAGACAGCC |
This paper | N/A |
| Murine TrailR Fwd AGTAGTGCTGCTGATTGGAG Rev CCTGTTTTCTGAGTCTTGCC |
This paper | N/A |
| Murine Fas Fwd TGCACCCTGACCCAGAATAC Rev GCCAGGAGAATCGCAGTAGAA |
This paper | N/A |
| Murine FasL Fwd GCAAATAGCCAACCCCAGTACAC Rev GCCACCTTTCTTATACTTCACTCCAG |
This paper | N/A |
| Murine Tnfr1 Fwd CACCGTGACAATCCCCTGTAA Rev TTTGCAAGCGGAGGAGGTAG |
This paper | N/A |
| Murine Tnfr2 Fwd ACAAAGTACCAAGGGTGGCA Rev GGGCTTCTTTTTCCTCTGCAC |
This paper | N/A |
| Murine IL-6 Fwd TAGTCCTTCCTACCCCAATTTCC Rev TTGGTCCTTAGCCACTCCTTC |
This paper | N/A |
| Murine IL-1α Fwd CGA AGC TCT CCG TAC ATT CC Rev TAA GGA CGG GAG GGA GAA AG |
This paper | N/A |
| Murine IL-1β Fwd TAA GGA CGG GAG GGA GAA AG Rev GAT CCA CAC TCT CCA GCT GCA |
This paper | N/A |
| Murine IL-18 Fwd GAC TCT TGC GTC AAC TTC AAG G Rev CAG GCT GTC TTT TGT CAA CGA |
This paper | N/A |
| Murine Ifn-γ Fwd GCA TCC AAA AGA GTG TGG AG Rev GCA GGC AGG ACA ACC ATT AC |
This paper | N/A |
| Murine Gadd45a Fwd AGC ACG CAA AAG GTC ACA TTG Rev GGG AAA GCA CTG CAC GAA CT |
This paper | N/A |
| Murine Actin Fwd GTGGGCCGCCCTAGGCACCA Rev CTCTTTGATGTCACGCACGATTTC |
This paper | N/A |
| Murine GAPDH Fwd CCACCCCAGCAAGGAGACT Rev GAAATTGTGAGGGAGATGCT |
This paper | N/A |
| Murine Rad51 Fwd CGGGAGTTGGTGGGTTATCC Rev CCGGCACATCTTGGTTTATTTGT |
This paper | N/A |
| Murine Exo1 Fwd ATGGGGATTCAAGGGTTACTTCA Rev AGCCAACAGTAGGTATCCACAG |
This paper | N/A |
| Murine Ddit3 Fwd CTCGCTCTCCAGATTCCAGTC Rev CTTCATGCGTTGCTTCCCA |
This paper | N/A |
| Murine PolE2 Fwd TCCTCGAACATGATCGAACGA Rev ACGTGGAATATCAAAAGCTCCAA |
This paper | N/A |
| Murine PolQ Fwd GCTTGGTCACGTCTTGGAAG Rev GGGCAAAATAAACAACGCTTTCT |
This paper | N/A |
| Murine Ddb1 Fwd ATGTCGTACAACTACGTCGTAAC Rev CTGAAGTAAAGTGTCCGGTCAC |
This paper | N/A |
| Murine Chek2 Fwd CTGAAGTAAAGTGTCCGGTCAC Rev CACCACCCGGTCAAATAGTTC |
This paper | N/A |
| Murine Lig1 Fwd CCAGCTCATAGTCCCCTCTGA Rev GTCTTGGCACCTCTAGCAGG |
This paper | N/A |
| Human Actin Fwd ATGGCCCTGTGCCTTAGTAG Rev GGTCTCAAACATGATCTGGG |
This paper | N/A |
| Human GAPDH Fwd CCT GGT CAC CAG GGC TGC Rev CCG TTC TCA GCC TTG ACG G |
This paper | N/A |
| Human Rad51 Fwd TTTGGTGAGTTTCCCGCTGTC Rev AACTTCTTTGCTAAGCTCGGAG |
This paper | N/A |
| Human Exo1 Fwd CCTCGTGGCTCCCTATGAAG Rev AGGAGATCCGAGTCCTCTGTAA |
This paper | N/A |
| Human Ddit3 Fwd GGAAACAGAGTGGTCATTCCC Rev CTGCTTGAGCCGTTCATTCTC |
This paper | N/A |
| Human PolE2 Fwd TGAGAAGCAACCCTTGTCATC Rev TCATCAACAGACTGACTGCATTC |
This paper | N/A |
| Human PolQ Fwd ACCTCTCCATCAAGGCATTTCT Rev GCAAAAGTTCCAGCAGATACC |
This paper | N/A |
| Human Ddb1 Fwd ACCGGACACTTTACTTCGGC Rev TCGGCGGTGACCACATAGA |
This paper | N/A |
| Human Chek2 Fwd TGAGAACCTTATGTGGAACCCC Rev ACAGCACGGTTATACCCAGC |
This paper | N/A |
| Human Lig1 Fwd ACAGTTCCCCATCAGGGATTC Rev CTCTGTGAGGCTTTCTTTCGG |
This paper | N/A |
| Human Gpnmb Fwd AAGTGAAAGATGTGTACGTGGTAACAG Rev TCGGATGAATTTCGATCGTTCT |
This paper | N/A |
| Human Tinag Fwd CGAAAGCTTCAGACACATGC Rev TTTCTTTCTGCCCTTGTGCT |
This paper | N/A |
| Human Plk1 Fwd GCTTAATGACGAGTTCTTTACTTC Rev TCGAAAACCTTGGTGGAATG |
This paper | N/A |
| Human Bcl2a1b Fwd ACGACAGCAAATTGCCCCGGAT Rev AAGCCATTTTCCCAGCCTCCGT |
This paper | N/A |
| Human Tpx2 Fwd CGAAAGCATCCTTCATCTCC Rev TCCTTGGGACAGGTTGAAAG |
This paper | N/A |
| Human CD44 Fwd CCGCTATGTCCAGAAAGGA Rev CTGTCTGTGCTGTCGGTGAT |
This paper | N/A |
| Human Glypican3 Fwd CCTTTGAAATTGTTGTTCGCCA Rev CCTGGGTTCATTAGCTGGGTA |
This paper | N/A |
| Human Epcam Fwd AATCGTCAATGCCAGTGTAC Rev TCTCATCGCAGTCAGGATCATAA |
This paper | N/A |
| Human Afp Fwd AACTATTGGCCTGTGGCGAG Rev TCATCCACCACCAAGCTG |
This paper | N/A |
| D3S1263-Fwd (FAM labeled) CTG TTG ACC CAT TGA TAC CC |
Thermo Fisher | N/A |
|
D3S1263-Rev (HEX labeled) TAA AAT CAC AGC AGG GGT TC |
Thermo Fisher | N/A |
|
D3S1289-Fwd (HEX labeled) AAA GCA ACT TGT AAG AGA GCA |
Thermo Fisher | N/A |
| D3S1289-Rev (FAM labeled) CTC CTA GAT ATA ATC ACT GGC A |
Thermo Fisher | N/A |
| Software and Algorithms | ||
| Copy Caller Software | Life Technologies | https://www.thermofisher.com |
| FlowJo sofware | TreeStar | https://www.flowjo.com |
| Summit software v4.3 | Beckman Coulter | https://www.beckman.com |
| GeneMapper software | Applied Biosystems | https://www.thermofisher.com |
| NDP Viewer v1.2.36 | NDP View | https://www.hamamatsu.com |
| Tissue IA image 2.0 | Leica Biosystems | www.leicabiosystems.com |
| GeneSpring GX | Agilent | http://www.genomics.agilent.com |
| GESA Molecular Signatures Database | http://www.broadinstitute.org | |
| Xcalibur software 2.2 | Thermo Fisher Scientific | https://www.thermofisher.com |
| R statistical programming language 3.2.2 | R Foundation for Statistical Computing | https://www.R-project.org/ |
| cgdsr R package (R-Based API for Accessing the MSKCC Cancer Genomics Data Server (CGDS)) | CRAN repository | https://CRAN.R-project.org/package=cgdsr |
| SCAN.UPC R package | Bioconductor project | https://bioconductor.org/packages/SCAN.UPC |
| Other | ||
| ABI 3130XL Genetic Analyzer | Applied Biosystems | https://www.thermofisher.com |
| Stella 3200 imaging system | Raytest | https://www.raytest.com/ |
| Nano Zoomer C9600 Virtual Slide Light microscope scanner | Hamamatsu | https://www.hamamatsu.com |
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Achim Weber (achim.weber@usz.ch).
Experimental Model and Subject Details
Human Material
Snap-frozen and formalin-fixed, paraffin-embedded (FFPE) human liver tissue samples were retrieved from the archives and the biobank of the Department of Pathology and Molecular Pathology, University Hospital Zurich, for morphological and molecular analyses. The study was and approved by the local ethics committee (“Kantonale Ethikkommission Zürich”, application numbers StV26/2005 and KEK-ZH-Nr. 2013-0382).
Mice
All animal experiments conformed to the relvant regulatory standards and were approved by the Swiss Veterinary Office (134/2014, 217/2012, 63/2011 Zurich). Animals were maintained under pathogen-free conditions and experiments were performed in accordance to the guidelines of the Swiss Animal Protection Law, Veterinary Office, Canton Zurich. Generation of mice with hepatocyte-specific Mcl-1 knock-out (homozygous: AlbCretg/+/Mcl-1flox/flox (Mcl-1Δhep), heterozygous AlbCretg/+/Mcl-1flox/wt) (Vick et al., 2009), with hepatocyte-specific c-Flip knockout and Tak1Δhep, Casp8Δhep, Tak1/Casp8Δhep and Tak1Δhep/RIPK3-/- mice (Vucur et al., 2013), Xiap−/− mice (Olayioye et al., 2005), and caspase 8 D387-mutant mice (Kang et al., 2008), was as described (see also Table S1). TNFR1-/- and TNFR1/2-/- mice were purchased from Jackson Laboratories and TNFR1-/- mice intercrossed to Mcl-1Δhep mice and bred in JNK1/2Δhep mice were generated by crossing with JNK1/2loxP/loxP mice (Das et al., 2011). Alb-Cre mice were bred in house (Haybaeck et al., 2009). Ripk1-/-/Ripk3−/−/Casp8−/−, Ripk3−/−/Casp8−/− and Ripk3−/− mice were previously described (Dillon et al., 2014).
Cell Lines and Drug Treatments
U2OS and HepG2 were grown in DMEM containing 10% FBS and 1% penicillin/streptomycin. Cells were transfected with lentiviral particles for caspase-8 (Santa Cruz, sc-29930-V) or control particles (Santa Cruz, sc-108080) according to the manufacturer's protocol and cells stably expressing the shRNA were isolated by puromycin selection (Santa Cruz). Cells were treated as indicated with Doxorubicin (Sigma) and for inhibition experiments, cells were pretreated for 4h with 10μM of the ATM inhibitor KU-55933 (Selleckchem) or pretreated with the JNK inhibitor (SP600125, Selleckchem) at 25μM and Doxorubicin added and cells incubated for indicated time. Cells 2h post irradiation with 10Gy were used as controls.
Method Details
Human Cohort Studies
For evaluation of liver function tests as potential predictors of HCC development, HCV patients with confirmed diagnosis of HCC and HCV patients without HCC were selected from the patient database as matched pairs according to MELD score for the given time point before the HCC diagnosis. The MELD score was chosen as the current international standard for assessment of severity of liver disease e.g. in liver transplant organ allocation and is based on laboratory values bilirubin, creatinine and INR. HCV patients who underwent liver transplantation (Swiss Hepato-Pancreato-Biliary Center, University-Hospital Zurich) due to liver tumors were chosen for the transplantation study, and compared to HCV patients which underwent liver transplantation, but did not develop liver tumors.
Mouse Strains and Intercrossings
Live damage of mice at the indicated age and tumor incidence analyzed at 12 months of age for Mcl-1Δhep and Mcl-1Δhep/TNFR1-/- mice and 33-35 weeks of age for TAK1Δhep mice and intercrossings. For overview, please also see Table S1.
Measurement of Serum Parameters
The analysis of aminotransferases (ALT/AST) and bilirubin was performed with mouse serum on a Roche Modular System (Roche Diagnostics) with a commercially available automated colorimetric system at the Institute of Clinical Chemistry, University Hospital Zurich, using a Hitachi P-Modul (Roche).
RNA Isolation from Liver Tissue
Total RNA from snap-frozen human liver biopsies or mouse livers was isolated using RNeasy Mini Kit (Qiagen) according to the manufacturer's protocol. The quantity and quality of the RNA was determined spectroscopically using a Nanodrop (Thermo Scientific).
Real-time PCR
Total RNA (1 μg) was reversely transcribed into cDNA using Quantitect Reverse Transcription Kit (Qiagen) according to the manufacturer’s protocol. For mRNA expression analysis real-time PCR was performed (reactions in duplicates) using Fast Start SYBR Green Master Rox (Roche). Real-time PCR was performed on an ABI PRISM 7900 HT and VIIA7 Fast Real-Time PCR System (AB). Data were generated and analyzed using SDS 2.4 and RQ manager 1.2 software. mRNA expression levels were normalized to the housekeeping genes Hprt for human samples, and Gapdh for murine samples.
DNA Extraction
Genomic DNA was isolated from 2 μm sections of murine or human FFPE slides by scrapings and tissue digested with Proteinase K overnight. After Proteinase K inactivation for 10 min at 95°C DNA concentration was determined spectroscopically using a Nanodrop (Thermo Scientific) and appropriate genomic DNA was directly used for PCR reactions in duplicates.
Taqman Copy Number Analysis
Taqman copy number analysis was carried out as multiplex PCR in duplicates with 20 ng DNA per reaction and Ttert as internal reference according to the manufacturer’s protocol. Wwox, Spata22, Fhit were selected as described markers from common fragile sites in humans and Fgfr1 and Fgr were selected as genes of interest in previously published areas of genetic instability in Tak1Δhep-/- mice (Bettermann et al., 2010). Data analysis was performed using Copy Caller Software (Life Technologies).
Flow Cytometry for DNA Damage
Primary murine hepatocytes were isolated by the two-step collagenase perfusion method, purified by Percoll gradient and finally collected in RPMI 1640 medium for flow cytometry procedures. Next, hepatcytes were fixed and permeabilized, followed by incubation with antibodies against γH2AX (#9718; Cell Signaling Technology) and RPA (NA19L, Calbiochem) and suitable secondary antibodies. DNA was stained with 1 μg/ml DAPI. Samples were measured on a Cyan ADP flow cytometer (Beckman Coulter) and analyzed with Summit software v4.3 (Beckman Coulter).
Partial Hepatectomy (PHX)
Eight- to twelve-week-old male mice with indicated genotype received food and water ad libitum before surgery. Mice were anaesthetized by inhalation of isoflurane (2%). PHX was performed between 8 and 12 a.m..Three liver lobes, including the previously emptied gall bladder, were removed. After surgery, mice were injected with buprenorphine for analgesia (Temgesic; Essex Chemie AG, Luzern, Switzerland; 0.1 mg/kg of body weight). Mice were euthanized by CO2 inhalation, and the remaining liver was harvested at different time points after PHX for further analysis (Speicher et al., 2014).
BrdU Assay
Proliferating cells were identified by 5-bromo-2′-deoxyuridine (BrdU) labelling. For this purpose, BrdU (Sigma, Buchs, Switzerland; 250 μg/g body weight) was injected i.p. prior to PHX. Two hours later, PHX was performed and mice were sacrificed at indicated time points. Detection of BrdU-positive cells was performed by immunofluorescence stainings using a peroxidase-coupled antibody against BrdU (1:30; Roche, Switzerland) (Speicher et al., 2014).
Immunoprecipitation
Cells were lysed in 1 ml lysis buffer (20 mM Tris HCl, pH 7.4, 137 mM NaCl, 2 mM EDTA, 10% glycerine, 1% Triton X-100, 1 mM PMSF, Protease Inhibitor mix (Roche)) for 30 min on ice. Afterwards, the samples were centrifuged at 14.000 rpm for 15 min at 4°C. 50 μl supernatant was used as lysate control. The remaining supernatant was immunoprecipitated by mixing with 30 μl protein A-Sepharose and 2 μg of C15 antibodies. Immunoprecipitations were performed for at least 2h at 4°C and washed four times with PBS. Samples were subjected to SDS PAGE (Biorad) and transferred to Hybond nitrocellulose membrane using the Western Blot system (Biorad). Membranes were blocked with 5% nonfat dry milk in PBS-T (PBS + 0.05% Tween 20) for 1 h, washed with PBS-T 3x for 10 min and incubated with the primary antibody in PBS/Tween for 1h at room temperature. C15 (anti-caspase 8) and 1C4 (anti-FADD) antibodies were a kind gift of Prof. Peter H. Krammer (DKFZ, Heidelberg). The following antibodies were used: anti-RIPK1 (D94C12), Cell Signaling, anti-Casp3 (9662), Cell Signaling and anti-Actin (A2103), Sigma.
Fragment Length Analysis for Allelic Imbalance
For analysis of AI the markers D3S1263 and D3S1289 at known common fragile sites (Gorgoulis et al., 2005) were selected. Four distinct regions (non-inflamed) of interest per liver-needle biopsy were identified by pathologists and gDNA isolated from 2 μm unstained consecutive FFPE sections. PCR products were separated by capillary electrophoresis using the ABI 3130XL Genetic Analyzer (Applied Biosystems) and results were analyzed with the help of GeneMapper software (Applied Biosystems). AI was identified by calculating the fluorescence ratios of heterozygous (informative) markers for each biopsy. The following primers were used: D3S1263-Fwd CTG TTG ACC CAT TGA TAC CC (FAM labeled), D3S1263-Rev TAA AAT CAC AGC AGG GGT TC, D3S1289-Fwd AAA GCA ACT TGT AAG AGA GCA (HEX labeled), D3S1289-Rev CTC CTA GAT ATA ATC ACT GGC A.
Pulse Field Gel Electrophoresis (PFGE)
PFGE was performed as published previously (Neelsen et al., 2013). Briefly, snap-frozen liver tissue was directly put into 4% formaldehyde without thawing and incubated for 10 min at 37°C. Tissue was mechanically dissociated (gentleMACS Dissociator, Miltenyi Biotec), filtered through a 70 μm cell strainer (Falcon) and 2.5x105 cell were embedded in a 0.8% agarose plus, digested in lysis buffer (100 mM EDTA, 1% (wt/vol) sodium lauryl sarcosyne, 0.2% (wt/vol) sodium deoxycholate, and 1 mg/ml proteinase K) at 37°C for 48 h, and washed in 10 mM Tris-HCl, pH 8.0, and 100 mM EDTA. Electrophoresis was performed at 14°C in 0.9% (wt/vol) Pulsed Field Certified Agarose (Bio-Rad Laboratories) containing Tris-borate/EDTA buffer in a CHEF DR III apparatus (9 h, 120°, 5.5 V/cm, 30-18 s switch time; 6 h, 117°, 4.5 V/cm, 18-9 s switch time; 6 h, 112°, 4 V/cm, 9-5 s switch time; Bio-Rad Laboratories). The gel was stained with ethidium bromide and imaged on an Alpha Innotech Imager.
Immunoblot Analysis
Snap-frozen liver tissue was dissociated (gentle MACS Dissociator, Miltenyi Biotec) and homogenates (10%) were prepared in RIPA buffer (50 mM Tris; 1% NP40; 0.25% Deoxycholic acid sodium salt; 150 mM NaCl; 1 mM EGTA) containing Halt Protease and Phosphatase Inhibitor Cocktail (Thermo Scientific). Quantification with a BCA protein assay kit (Thermo Scientific) according to the manufacturer’s manual was followed by denaturation of 80 μg protein in Laemmli buffer containing 5% β-mercaptoethanol and separated by gel electrophoresis (Mini Protean Gels, Bio Rad) and blotted by semi-dry blotting (Trans-Blot Turbo Transfer, Bio Rad) onto nitrocellulose membranes (Bio Rad) and stained with Ponceau Red. Membranes were blocked in 5% milk/PBS-T for at least 1 hr at RT. Primary antibodies against γH2AX, p-p53, GAPDH, PCNA, pATM, pCHK1, pJNK1/2, pCHK2, pATR, pBRCA1, cleaved-Casp1, cleaved-Casp3, cleaved-Casp8, RIPK1, RIPK3, total-JNK1/2, p-p38, p38, cIAP1, cIAP2, XIAP (all Cell Signaling Technology), total-Casp8, p-cJUN, pAKT1/2/3, total AKT (SantaCruz), total-ATM (Sigma), total-cJUN (BD Bioscience), HOIP (R&D), SHARPIN (Proteintech), HOIL-1 (Walczak lab) and Casp8 C15 (provided by Dr. P. Krammer, Heidelberg), were incubated at 4°C overnight under shaking conditions. Incubation with the secondary antibody (HRP-anti rabbit IgG, 1:5000; Promega) was performed under shaking conditions for 1 hr. Detection was achieved with Clarity Western ECL Substrate (Bio Rad) using Stella 3200 imaging system Raytest.
Histology and Immuno Stainings
Sections (2 μm) of livers (fixed in 4% paraformaldehyde and paraffin-embedded) were stained with Hematoxylin/Eosin or various antibodies. Incubation in Ventana buffer and staining was performed on a NEXES immunohistochemistry robot (Ventana Instruments) using an IVIEW DAB Detection Kit (Ventana) or on a Bond MAX (Leica). Immunostainings were performed as described before (Wolf et al., 2014) with antibodies against the following proteins: Ki67, 1:200 dilution (SP6, NeoMarkers / Lab Vision Corporation); γH2AX, 1:300 dilution (Novus Biologicals); p-cJUN, 1:100 (Abcam); cleaved-Caspase8, 1:500; p-CHK1, 1:50 and p-CHK2, 1:500 (Novus Biologicals). For virtual microscopy and archiving, histological and immunohistochemical images were digitalized using a Nano Zoomer C9600 Virtual Slide Light microscope scanner by Hamamatsu using NDP, View Software, version 1.2.36. Alternatively, for quantification of stainings, slides were scanned using a SCN 400 slide scanner (Leica) and analyzed using Tissue IA image analysis software (Slidepath, Leica).
RNA Microarray
An Agilent one-color microarray-based gene expression analysis (mouse DNA Microarray 4x44K) was performed according to the manufacturer’s protocol. Two- and twelve-months-old mice were analyzed. For 12 months, HCC and corresponding non-tumor tissue (n=3) from the same animal (n=5) as well as livers from Cre-negative littermates as controls (n=3) were analyzed. For 2 months, Mcl-1Δhep (AlbCretg/+Mcl-1flox/flox), hemizygous Mcl-1Δhep (AlbCretg/+/Mcl-1flox/wt) and Cre-negative controls were analyzed. Gene expression was quantified using Agilent Feature Extraction Software Version 9.5.3.1. Gene Ontology microarray data analysis: Lists of significantly differentially expressed genes were investigated in respect to enrichment of Gene Ontology categories using the Gene Ontology Browser as implemented in GeneSpring 7.3. Fisher’s exact test was used to show whether more genes belonging to a Gene Ontology category are found in the list under investigation than in a randomized gene list of the same size.
Gene Set Enrichment Analysis (GSEA)
Gene sets from the biological process gene ontology for GSEA analysis http://www.broadinstitute.org were downloaded from the Molecular Signatures Database or integrated manually into the GSEA for human HCV-induced hepatitis gene expression sets (Sarasin-Filipowicz et al., 2008) or alcohol-induced hepatitis (Affo et al., 2013). GSEA tests whether genes sets were overrepresented in microarray expression data were performed with standard settings.
Immunofluorescence Stainings and Confocal Microscopy
U2OS cells were seeded on cover slips and grown to 50-80% confluence for 24 h. Cells were incubated with medium or 1 μM doxorubicin for 30 min and fixed with 4% fomaldehyde for 1 h. Cells were washed with PBS and Tris buffer (100 mM Tris, pH=7.4, 50 mM NaCl), permeabilized with 0.5% Triton X-100 in PBS for 10 min, incubated in blocking solution (PBS, pH=7.4, 1% BSA, 2% FCS) for 20 min, incubated with primary antibody in blocking solution for 12 h at 4°C, washed twice with PBS, incubated with secondary antibody in blocking solution for 3 h, washed with PBS, incubated with 1 μg/ml DAPI (life technologies) for 5-15 min, washed 4 times with PBS and mounted using Vectashield (Vector Laboratories). Primary antibodies and concentrations used for immunofluorescence were γH2AX (Abcam, ab26350, 1:200), γH2AX (Novus Biologicals, NB100-2280, 1:200), RIP1 (BD Biosciences, 610459, 1:50), Caspase 8-C15 and 53BP1 (Santa-Cruz, sc-22760, 1:50). Secondary, Alexa-488 or Alexa-546 labelled antibodies were all from life technologies and used at a 1:500 dilution. Imaging was performed on a Leica SP8 confocal microscope equipped with 405, 488, 552, and 638 nm diode lasers and a 63x oil objective (HC PL APO CS2 / 1.40 oil) using LAS software (Leica Microsystems, Wetzlar Germany) and processed using ImageJ.
Subcellular Fractionation
Cells grown on dishes were washed twice with ice-cold PBS and carefully scraped in PBS, centrifuged at 1000 x g for 2 min at 4°C and resuspended in 300 μl buffer A (20 mM Tris/HCl pH 7.9, 10 mM NaCl, 1.5 mM MgCl2, 10 % Glycerol, 0.5 mM DTT, 0.5 mM AEBSF, 1 mM Na3VO4, 1 mM Na2MoO4, 10 mM NaF, 10 mM K2HPO4, 20 mM glycerol-2-phosphate, phosphatase inhibitor tablet (Roche)). After incubation on ice for 10 min, swelling of cells was monitored by microscopy using 0.4% trypan blue staining. For disruption of the cytoplasm membrane cells were treated with 0.125% NP-40 and incubated on ice for 5 min. After centrifugation at 2000 x g for 10 min at 4°C the supernatant was kept as cytosolic fraction (additional centrifugation at 13000 x g for 10 min at 4°C freed the lysate from cell debris). Nuclear fractions were resuspended in 100 μl buffer C (20 mM Tris/HCl pH 7.9, 420 mM NaCl, 1.5 mM MgCl2, 10% gycerol, 0.5 mM DTT, 0.2 mM EDTA, 0.5 mM AEBSF, 1 mM Na3VO4, 1 mM Na2MoO4, 10 mM NaF, 10 mM K2HPO4, 20 mM glycerol-2-phosphate, phosphatase inhibitor tablet (Roche)). After incubation for 15 min on ice with occasional vortexing N1 fractions were collected by centrifugation at 13000 x g for 10 min at 4°C. Pellets were resuspended in 50 μl buffer E (20 mM Tris/HCl pH 7.9, 150 mM NaCl, 1.5 mM MgCl2, 10% glycerol, 0.5 mM AEBSF, 1 mM Na3VO4, 1 mM Na2MoO4, 10 mM NaF, 20 mM glycerol-2-phosphate, phosphatase inhibitor tablet (Roche), 1 μl Benzonase Nuclease (25 U/μl, Novagen) and 2% SDS). After incubation for 30 min at 4°C with shaking N2 fractions were collected by centrifugation at 13000 x g for 10 min at 4°C.
AQUA-Mass Spectrometry
Immunoprecipitates (sepharose beads) were suspended in 50 mM NH4HCO3 and cysteins were ß-thiomethylated by dithiothreitol reduction (1 mM DTT, 56°C, 45 min) and subsequent S-Methyl methanethiosulphonate (MMTS) treatment (5 mM MMTS, 30 min). Tryptic digestion was performed by addition of 0.5 μg Trypsin (Trypsin Gold, Promega) and incubated at 37°C for 24 h. AQUA peptides for caspase 8, FADD, c-Flip and RIPK1 were spiked into the digestion solution in an absolute amount of 50 fmol of each peptide as described previously (Schleich et al., 2015). Sequences of AQUA peptides could be received upon request. After digestion, the supernatant was collected and dried down in a vacuum centrifuge. The peptides were redissolved in 5 μl 0.1 % trifluoroacetic acid (TFA) and purified on ZIP-TIP, C18-nanocolumns (Millipore, Billerica, USA). Peptides were eluted in 7 μl 70% (v/v) acetonitrile (ACN) and subsequently dried in a vacuum centrifuge. Dried samples were dissolved in 10 μl 2% ACN/0.1% TFA and separated on a 75 μm I.D., 25 cm PepMap C18-column (Dionex, Sunnyvale, USA) applying a gradient from 2% to 45% ACN in 0.1% formic acid over 120 min at 300 nl/min using an Ultimate 3000 Nano-HPLC (Thermo Scientific, San Jose, USA). Mass spectrometry was performed on a hybrid dual-pressure linear ion trap/orbitrap mass spectrometer (LTQ Orbitrap Velos Pro, Thermo Scientific, San Jose, USA) in exclusive orbitrap full MS mode (FTMS; resolution 60,000; m/z range 400-2000). Instrument control, data acquisition and peak integration were performed using the Xcalibur software 2.2 (Thermo Fisher Scientific). Extracted ion chromatograms derived from Orbitrap mass scans from each AQUA/target peptide pair were generated and the peak areas of the light and heavy peptide were obtained by manual integration, respectively. The heavy to light ratio of each AQUA peptide pair was calculated and the resulting absolute amount of the endogenous (light) peptide was determined.
Analysis of Data from Human Hepatocellular Carcinoma
Data mining was performed to assess the relevance of caspase 8 in HCC. All analyses were performed using the R statistical programming environment, version 3.2.2. Publicly available data from the Cancer Genome Atlas (TCGA) project were used. Clinical data and Level 3 RNA sequencing data were downloaded from the TCGA website (https://tcga-data.nci.nih.gov/ – files nationwidechildrens.org_LIHC.bio.Level_2.0.54.0 and unc.edu_LIHC.IlluminaHiSeq_RNASeqV2.Level_3.1.14.0, respectively). Methylation data were downloaded via cBioPortal (http://www.cbioportal.org/) using the cgdsr R package on 11th November 2015. Patients with fibrolamellar hepatocellular carcinoma and combined hepatocellular and cholangiocarcinoma were excluded. The UPC (Universal exPression Codes) method, as implemented by the SCAN.UPC R/Bioconductor package (Piccolo et al., 2013), was used to evaluate whether genes within the TCGA dataset should be considered to be expressed in individual patients (UPC values > 0.5 indicating expression of the gene). Count data from RNA sequencing were transformed using the formula log2 (x + 1) to better approximate a normal distribution. Methylation data from cBioPortal were dichotomized using a percentage of methylation (β) cutoff of 0.3 (< 0.3 unmethylated, > 0.3 methylated). The log rank test was used to assess association with survival. For RNA expression, the median, the mean + 1 standard deviation (shown in the final figure), and the mean - 1 standard deviation were initially used as cutoffs to separate patients into two groups according to the level of CASP8 expression; the p value shown in the final figure was corrected for 3 tests using the Bonferroni method. Spearman’s rank correlation coefficient was used to assess the correlation between genes.
Quantification and Statistical Analysis
Statistical Analysis
Statistical analysis was performed using GraphPad Prism software (version 5.0) or SPSS. All data are presented as mean ± SEM and were analyzed by ANOVA with Bonferroni correction. Analysis of two samples was performed with Student t test, statistics for HCC incidence were calculated using Fisher’s Exact test. Statistical significance is indicated as follows: ∗∗∗∗p < 0.0001; ∗∗∗p < 0.001; ∗∗p < 0.01; ∗p < 0.05; n.s. not significant.
Modelling of Mutation Rates Depending on Proliferative Activity
A rough calculation of the replication error rates depending on the replication rate in wild type mice and Mcl-1Δhep mice with low transaminase levels and corresponding low proliferative activity or Mcl-1Δhep mice with high transaminase levels and corresponding high proliferative activity. Modelling was based on proliferation rates determined by Ki67+ hepatocytes at 2 months of age revealing that Mcl-1Δhep mice have about 10- to 25-fold higher Ki67 rates (see Figure 2). Calculation is based on the following assumptions: A wild type mouse liver weighting 2 g consists of ∼2x108 hepatocytes, and assuming a proliferation index of 0.2 (Eldridge and Goldsworthy, 1996), 4x105 proliferating hepatocytes. Further, assuming an replication error rate of 10-8 per cell per generation (Tago et al., 2005), and 24 h cycle duration (Alexiades and Cepko, 1996), taking Poisson distribution as basis, the expected number of replications errors after 1 year of is 1.46 for wild type mice, 14.6 for Mcl-1Δhep mice with low hepatocyte proliferative activity, and 36.8 for Mcl-1Δhep mice with high hepatocyte proliferative activity.
Data and Software Availability
Gene expression microarray data are deposited in the GEO database under accession number GSE75730.
Author Contributions
Y.B., M.H., and A.W. designed the study, co-ordinated experiments, wrote the manuscript with input from all co-authors. Y.B., M.M., M.E.H., M.V., F.B., C.K., R.M., J.J., L.L., S.Z., R.B., N.H., H.S.B., S.W., and L.B., developed and analyzed the described mouse models and performed in vitro and in vivo experiments. Y.B., M.E.H., A.L., F.B., D.S., H.M., M.D.V., M.H., and A.W. conducted morphological analyses. Y.B., M.M., K.B., M.E.H., Y.S., A.K.A., F.B., C.R., T.K., M.J.W., T.S., S.P.A., R.M., K.M., E.D., M.N., M.L., H.W., L.H., A.B., and I.L. contributed to in vitro molecular signaling studies. Y.B. and R.M. performed in vitro allelic imbalance analysis. Y.B., H.R., L.F., and K.U. performed in silico analyses. Y.B., F.B., J.M., B.M., P.A.C., and A.W. identified, collected, and provided human tissue and data. J.M.S., R.J.D., S.A.L., P.J., R.W., C.D., D.W., L.Z., D.R.G., and T.L. provided genetically modified cell lines and mice for in vivo studies.
Acknowledgments
We thank M. Bawohl, V. Schüppel, J. Tracy, R. Hillermann, D. Kull, O. Seelbach, M. Storz, P. Tzscheetzsch, A. Fitsche, P. Schraml, S. Dettwiler, J.F. Glaus Garzon, T.B. Kang, M. Egger, J. Schmitt, K. Weber, S.M. Kwon, and X.W. Wang for their excellent support, B. Seifert for statistical guidance, M. Bertrand for helpful discussion, and T. O'Connor for critical reading. This study was supported by Krebsliga Schweiz (Oncosuisse), Promedica Stiftung, Stiftung zur Krebsbekämpfung, Zürich, Swiss National Research Foundation (SNF; project 310030_146940/1) (to A.W.), Helmholtz Association, “Stiftung Experimentelle Biomedizin” (Hofschneider Foundation), European Research Council (ERC Consolidator Grant, HepatoMetaboPath), Graduierten Kolleg (GRK) (482) and Sonderforschungsbereiche (SFB) (36, 179 and 209), the European Union's Horizon 2020 research and innovation program (no. 667273) (to M.H.), Mildred-Scheel Endowed Professorship, German Cancer Aid (Deutsche Krebshilfe, project 110043), German Research Foundation (SFB-TRR57/P06) (to T.L.), SNF (grant 310030_132884) (to S.W.), DFG (FOR2314 and SFB685), Gottfried Wilhelm Leibniz Program (to L.Z.), DFG (DFG-LA 2386) (to I.L.), NIH grant DK107220 (to R.D.), and Hartmann Müller Stiftung and PhD program of the Cancer Network Zurich (to Y.B.). M.D.V. is a Helmholtz Young Investigator (Helmholtz Association).
Published: September 11, 2017
Footnotes
Supplemental Information includes eight figures and one table and can be found with this article online at http://dx.doi.org/10.1016/j.ccell.2017.08.010.
Contributor Information
Mathias Heikenwalder, Email: m.heikenwaelder@dkfz.de.
Achim Weber, Email: achim.weber@usz.ch.
Supplemental Information
References
- Affo S., Dominguez M., Lozano J.J., Sancho-Bru P., Rodrigo-Torres D., Morales-Ibanez O., Moreno M., Millan C., Loaeza-del-Castillo A., Altamirano J. Transcriptome analysis identifies TNF superfamily receptors as potential therapeutic targets in alcoholic hepatitis. Gut. 2013;62:452–460. doi: 10.1136/gutjnl-2011-301146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexiades M.R., Cepko C. Quantitative analysis of proliferation and cell cycle length during development of the rat retina. Dev. Dyn. 1996;205:293–307. doi: 10.1002/(SICI)1097-0177(199603)205:3<293::AID-AJA9>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- Ando K., Kernan J.L., Liu P.H., Sanda T., Logette E., Tschopp J., Look A.T., Wang J., Bouchier-Hayes L., Sidi S. PIDD death-domain phosphorylation by ATM controls prodeath versus prosurvival PIDDosome signaling. Mol. Cell. 2012;47:681–693. doi: 10.1016/j.molcel.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batts K.P., Ludwig J. Chronic hepatitis. An update on terminology and reporting. Am. J. Surg. Pathol. 1995;19:1409–1417. doi: 10.1097/00000478-199512000-00007. [DOI] [PubMed] [Google Scholar]
- Bettermann K., Vucur M., Haybaeck J., Koppe C., Janssen J., Heymann F., Weber A., Weiskirchen R., Liedtke C., Gassler N. TAK1 suppresses a NEMO-dependent but NF-kappaB-independent pathway to liver cancer. Cancer Cell. 2010;17:481–496. doi: 10.1016/j.ccr.2010.03.021. [DOI] [PubMed] [Google Scholar]
- Biton S., Ashkenazi A. NEMO and RIP1 control cell fate in response to extensive DNA damage via TNF-alpha feedforward signaling. Cell. 2011;145:92–103. doi: 10.1016/j.cell.2011.02.023. [DOI] [PubMed] [Google Scholar]
- Brown M.F., Leibowitz B.J., Chen D., He K., Zou F., Sobol R.W., Beer-Stolz D., Zhang L., Yu J. Loss of caspase-3 sensitizes colon cancer cells to genotoxic stress via RIP1-dependent necrosis. Cell Death Dis. 2015;6:e1729. doi: 10.1038/cddis.2015.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christofferson D.E., Li Y., Hitomi J., Zhou W., Upperman C., Zhu H., Gerber S.A., Gygi S., Yuan J. A novel role for RIP1 kinase in mediating TNFalpha production. Cell Death Dis. 2012;3:e320. doi: 10.1038/cddis.2012.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das M., Garlick D.S., Greiner D.L., Davis R.J. The role of JNK in the development of hepatocellular carcinoma. Genes Dev. 2011;25:634–645. doi: 10.1101/gad.1989311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon C.P., Weinlich R., Rodriguez D.A., Cripps J.G., Quarato G., Gurung P., Verbist K.C., Brewer T.L., Llambi F., Gong Y.N. RIPK1 blocks early postnatal lethality mediated by caspase-8 and RIPK3. Cell. 2014;157:1189–1202. doi: 10.1016/j.cell.2014.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Serag H.B., Kanwal F. Epidemiology of hepatocellular carcinoma in the United States: where are we? Where do we go? Hepatology. 2014;60:1767–1775. doi: 10.1002/hep.27222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eldridge S.R., Goldsworthy S.M. Cell proliferation rates in common cancer target tissues of B6C3F1 mice and F344 rats: effects of age, gender, and choice of marker. Fundam. Appl. Toxicol. 1996;32:159–167. doi: 10.1006/faat.1996.0119. [DOI] [PubMed] [Google Scholar]
- Forner A., Llovet J.M., Bruix J. Hepatocellular carcinoma. Lancet. 2012;379:1245–1255. doi: 10.1016/S0140-6736(11)61347-0. [DOI] [PubMed] [Google Scholar]
- Gao G., Smith D.I. Very large common fragile site genes and their potential role in cancer development. Cell Mol. Life Sci. 2014;71:4601–4615. doi: 10.1007/s00018-014-1753-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorgoulis V.G., Vassiliou L.V., Karakaidos P., Zacharatos P., Kotsinas A., Liloglou T., Venere M., Ditullio R.A., Jr., Kastrinakis N.G., Levy B. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434:907–913. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- Gryfe R., Kim H., Hsieh E.T., Aronson M.D., Holowaty E.J., Bull S.B., Redston M., Gallinger S. Tumor microsatellite instability and clinical outcome in young patients with colorectal cancer. N. Engl. J. Med. 2000;342:69–77. doi: 10.1056/NEJM200001133420201. [DOI] [PubMed] [Google Scholar]
- Halazonetis T.D., Gorgoulis V.G., Bartek J. An oncogene-induced DNA damage model for cancer development. Science. 2008;319:1352–1355. doi: 10.1126/science.1140735. [DOI] [PubMed] [Google Scholar]
- Hartwig T., Montinaro A., von Karstedt S., Sevko A., Surinova S., Chakravarthy A., Taraborrelli L., Draber P., Lafont E., Arce Vargas F. The TRAIL-induced cancer secretome promotes a tumor-supportive immune microenvironment via CCR2. Mol. Cell. 2017;65:730–742.e5. doi: 10.1016/j.molcel.2017.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas T.L., Emmerich C.H., Gerlach B., Schmukle A.C., Cordier S.M., Rieser E., Feltham R., Vince J., Warnken U., Wenger T. Recruitment of the linear ubiquitin chain assembly complex stabilizes the TNF-R1 signaling complex and is required for TNF-mediated gene induction. Mol. Cell. 2009;36:831–844. doi: 10.1016/j.molcel.2009.10.013. [DOI] [PubMed] [Google Scholar]
- Haybaeck J., Zeller N., Wolf M.J., Weber A., Wagner U., Kurrer M.O., Bremer J., Iezzi G., Graf R., Clavien P.A. A lymphotoxin-driven pathway to hepatocellular carcinoma. Cancer Cell. 2009;16:295–308. doi: 10.1016/j.ccr.2009.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry C.M., Martin S.J. Caspase-8 acts in a non-enzymatic role as a scaffold for assembly of a pro-inflammatory “FADDosome” complex upon TRAIL stimulation. Mol. Cell. 2017;65:715–729.e5. doi: 10.1016/j.molcel.2017.01.022. [DOI] [PubMed] [Google Scholar]
- Hikita H., Kodama T., Shimizu S., Li W., Shigekawa M., Tanaka S., Hosui A., Miyagi T., Tatsumi T., Kanto T. Bak deficiency inhibits liver carcinogenesis: a causal link between apoptosis and carcinogenesis. J. Hepatol. 2012;57:92–100. doi: 10.1016/j.jhep.2012.01.027. [DOI] [PubMed] [Google Scholar]
- Kang T.B., Oh G.S., Scandella E., Bolinger B., Ludewig B., Kovalenko A., Wallach D. Mutation of a self-processing site in caspase-8 compromises its apoptotic but not its nonapoptotic functions in bacterial artificial chromosome-transgenic mice. J. Immunol. 2008;181:2522–2532. doi: 10.4049/jimmunol.181.4.2522. [DOI] [PubMed] [Google Scholar]
- Kang T.B., Yang S.H., Toth B., Kovalenko A., Wallach D. Caspase-8 blocks kinase RIPK3-mediated activation of the NLRP3 inflammasome. Immunity. 2013;38:27–40. doi: 10.1016/j.immuni.2012.09.015. [DOI] [PubMed] [Google Scholar]
- Krelin Y., Zhang L., Kang T.B., Appel E., Kovalenko A., Wallach D. Caspase-8 deficiency facilitates cellular transformation in vitro. Cell Death Differ. 2008;15:1350–1355. doi: 10.1038/cdd.2008.88. [DOI] [PubMed] [Google Scholar]
- Lafont E., Kantari-Mimoun C., Draber P., De Miguel D., Hartwig T., Reichert M., Kupka S., Shimizu Y., Taraborrelli L., Spit M. The linear ubiquitin chain assembly complex regulates TRAIL-induced gene activation and cell death. EMBO J. 2017;36:1147–1166. doi: 10.15252/embj.201695699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liedtke C., Zschemisch N.H., Cohrs A., Roskams T., Borlak J., Manns M.P., Trautwein C. Silencing of caspase-8 in murine hepatocellular carcinomas is mediated via methylation of an essential promoter element. Gastroenterology. 2005;129:1602–1615. doi: 10.1053/j.gastro.2005.08.007. [DOI] [PubMed] [Google Scholar]
- Luedde T., Kaplowitz N., Schwabe R.F. Cell death and cell death responses in liver disease: mechanisms and clinical relevance. Gastroenterology. 2014;147:765–783.e4. doi: 10.1053/j.gastro.2014.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neelsen K.J., Zanini I.M., Mijic S., Herrador R., Zellweger R., Ray Chaudhuri A., Creavin K.D., Blow J.J., Lopes M. Deregulated origin licensing leads to chromosomal breaks by rereplication of a gapped DNA template. Genes Dev. 2013;27:2537–2542. doi: 10.1101/gad.226373.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olayioye M.A., Kaufmann H., Pakusch M., Vaux D.L., Lindeman G.J., Visvader J.E. XIAP-deficiency leads to delayed lobuloalveolar development in the mammary gland. Cell Death Differ. 2005;12:87–90. doi: 10.1038/sj.cdd.4401524. [DOI] [PubMed] [Google Scholar]
- Panier S., Boulton S.J. Double-strand break repair: 53BP1 comes into focus. Nat. Rev. Mol. Cell Biol. 2014;15:7–18. doi: 10.1038/nrm3719. [DOI] [PubMed] [Google Scholar]
- Picco V., Pages G. Linking JNK activity to the DNA damage response. Genes Cancer. 2013;4:360–368. doi: 10.1177/1947601913486347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piccolo S.R., Withers M.R., Francis O.E., Bild A.H., Johnson W.E. Multiplatform single-sample estimates of transcriptional activation. Proc. Natl. Acad. Sci. USA. 2013;110:17778–17783. doi: 10.1073/pnas.1305823110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarasin-Filipowicz M., Oakeley E.J., Duong F.H., Christen V., Terracciano L., Filipowicz W., Heim M.H. Interferon signaling and treatment outcome in chronic hepatitis C. Proc. Natl. Acad. Sci. USA. 2008;105:7034–7039. doi: 10.1073/pnas.0707882105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schadde E., Ardiles V., Robles-Campos R., Malago M., Machado M., Hernandez-Alejandro R., Soubrane O., Schnitzbauer A.A., Raptis D., Tschuor C. Early survival and safety of ALPPS: first report of the international ALPPS registry. Ann. Surg. 2014;260:829–836. doi: 10.1097/SLA.0000000000000947. discussion 836–828. [DOI] [PubMed] [Google Scholar]
- Schleich K., Buchbinder J.H., Pietkiewicz S., Kahne T., Warnken U., Ozturk S., Schnolzer M., Naumann M., Krammer P.H., Lavrik I.N. Molecular architecture of the DED chains at the DISC: regulation of procaspase-8 activation by short DED proteins c-FLIP and procaspase-8 prodomain. Cell Death Differ. 2015;23:681–694. doi: 10.1038/cdd.2015.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu Y., Peltzer N., Sevko A., Lafont E., Sarr A., Draberova H., Walczak H. The linear ubiquitin chain assembly complex acts as a liver tumor suppressor and inhibits hepatocyte apoptosis and hepatitis. Hepatology. 2017;65:1963–1978. doi: 10.1002/hep.29074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soung Y.H., Lee J.W., Kim S.Y., Sung Y.J., Park W.S., Nam S.W., Kim S.H., Lee J.Y., Yoo N.J., Lee S.H. Caspase-8 gene is frequently inactivated by the frameshift somatic mutation 1225_1226delTG in hepatocellular carcinomas. Oncogene. 2005;24:141–147. doi: 10.1038/sj.onc.1208244. [DOI] [PubMed] [Google Scholar]
- Speicher T., Siegenthaler B., Bogorad R.L., Ruppert R., Petzold T., Padrissa-Altes S., Bachofner M., Anderson D.G., Koteliansky V., Fassler R., Werner S. Knockdown and knockout of beta1-integrin in hepatocytes impairs liver regeneration through inhibition of growth factor signalling. Nat. Commun. 2014;5:3862. doi: 10.1038/ncomms4862. [DOI] [PubMed] [Google Scholar]
- Tago Y., Imai M., Ihara M., Atofuji H., Nagata Y., Yamamoto K. Escherichia coli mutator (Delta)polA is defective in base mismatch correction: the nature of in vivo DNA replication errors. J. Mol. Biol. 2005;351:299–308. doi: 10.1016/j.jmb.2005.06.014. [DOI] [PubMed] [Google Scholar]
- Tenev T., Bianchi K., Darding M., Broemer M., Langlais C., Wallberg F., Zachariou A., Lopez J., MacFarlane M., Cain K., Meier P. The Ripoptosome, a signaling platform that assembles in response to genotoxic stress and loss of IAPs. Mol. Cell. 2011;43:432–448. doi: 10.1016/j.molcel.2011.06.006. [DOI] [PubMed] [Google Scholar]
- Tinel A., Tschopp J. The PIDDosome, a protein complex implicated in activation of caspase-2 in response to genotoxic stress. Science. 2004;304:843–846. doi: 10.1126/science.1095432. [DOI] [PubMed] [Google Scholar]
- Tomasetti C., Vogelstein B. Cancer etiology. Variation in cancer risk among tissues can be explained by the number of stem cell divisions. Science. 2015;347:78–81. doi: 10.1126/science.1260825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vick B., Weber A., Urbanik T., Maass T., Teufel A., Krammer P.H., Opferman J.T., Schuchmann M., Galle P.R., Schulze-Bergkamen H. Knockout of myeloid cell leukemia-1 induces liver damage and increases apoptosis susceptibility of murine hepatocytes. Hepatology. 2009;49:627–636. doi: 10.1002/hep.22664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vucur M., Reisinger F., Gautheron J., Janssen J., Roderburg C., Cardenas D.V., Kreggenwinkel K., Koppe C., Hammerich L., Hakem R. RIP3 inhibits inflammatory hepatocarcinogenesis but promotes cholestasis by controlling caspase-8- and JNK-dependent compensatory cell proliferation. Cell Rep. 2013;4:776–790. doi: 10.1016/j.celrep.2013.07.035. [DOI] [PubMed] [Google Scholar]
- Weber A., Boger R., Vick B., Urbanik T., Haybaeck J., Zoller S., Teufel A., Krammer P.H., Opferman J.T., Galle P.R. Hepatocyte-specific deletion of the antiapoptotic protein myeloid cell leukemia-1 triggers proliferation and hepatocarcinogenesis in mice. Hepatology. 2010;51:1226–1236. doi: 10.1002/hep.23479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf M.J., Adili A., Piotrowitz K., Abdullah Z., Boege Y., Stemmer K., Ringelhan M., Simonavicius N., Egger M., Wohlleber D. Metabolic activation of intrahepatic CD8(+) T cells and NKT cells causes nonalcoholic steatohepatitis and liver cancer via cross-talk with hepatocytes. Cancer Cell. 2014;26:549–564. doi: 10.1016/j.ccell.2014.09.003. [DOI] [PubMed] [Google Scholar]
- Yang F., Teves S.S., Kemp C.J., Henikoff S. Doxorubicin, DNA torsion, and chromatin dynamics. Biochim. Biophys. Acta. 2014;1845:84–89. doi: 10.1016/j.bbcan.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon S., Bogdanov K., Kovalenko A., Wallach D. Necroptosis is preceded by nuclear translocation of the signaling proteins that induce it. Cell Death Differ. 2016;23:253–260. doi: 10.1038/cdd.2015.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.