Abstract
Circulating tumor cells (CTCs) are cancer cells that circulate in the blood stream after being naturally shed from original or metastatic tumors, and can lead to a new fatal metastasis. CTCs have become a hotspot research field during the last decade. Detection of CTCs, as a liquid biopsy of tumors, can be used for early diagnosis of cancers, earlier evaluation of cancer recurrence and chemotherapeutic efficacy, and choice of individual sensitive anti-cancer drugs. Therefore, CTC detection is a crucial tool to fight against cancer. Herein, we classify the currently reported CTC detection technologies, introduce some representative samples for each technology, conclude the advantages and limitations, and give a future prospective including challenges and opportunities of CTC detection.
Keywords: circulating tumor cells (CTCs), capture, enrichment, detection, release
1. Introduction
1.1 Circulating tumor cells
Circulating tumor cells (CTCs) are cancer cells that circulate in the blood stream after being naturally shed from original or metastatic tumors, and may lead to a new metastasis, which is the main reason of cancer-related death.1,2 With the development of new technologies of molecular imaging, genome sequencing, single cell analysis, metastasis model establishment and so on, the metastasis process has been demonstrated to be induced by the CTCs including invading distant tissues, settling in supportive niches and overtaking the host organs.3,4
CTCs were first described in 1869 by Prof. Ashworth in the blood of a cancer patient by thorough comparison of the CTC morphology to different tumor cells.5 Although it has been around 150 years since CTCs were discovered, little research was focused on CTCs before mid-1990s. That's because CTCs are a kind of extremely rare cells in the blood vessels (i.e. a few CTCs amongst 5 billion erythrocytes and 10 million leukocytes), and the detection of CTCs is technically challenging.6,7
With the advances of multidisciplinary studies including medicine, oncology, biology, materials science and chemistry, the CTC detection became promising and attracted more and more attention in the past two decades. Especially, in the last 10 years or so, CTCs have become a hotspot research field. There were 19,151 publications listed when searching the “circulating tumor cell” as key phrase in PubMed in Mar 2017. The number of new CTC publications each year has been increasing (Figure 1), and most of them focused on the detection technologies due to the importance of CTC detection, which is discussed below.
Figure 1.
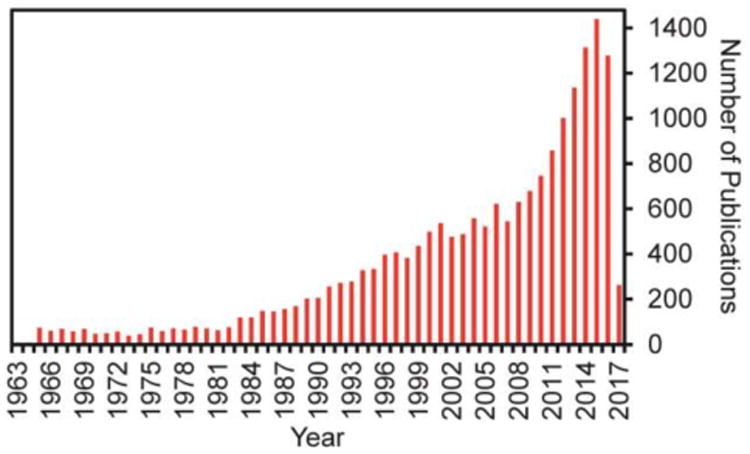
Publication numbers of each year with the key phrase of “circulating tumor cell” searched in PubMed in Dec 2016.
1.2 CTC detection
Invasive biopsy including a pathological analysis is usually required for making a definite diagnosis of cancer. However, the tumor biopsies are limited by sampling bias, sampling difficulty of deep tumors, and harm to patients. These problems do not exist for the detection of circulating tumor biomarkers (such as CTCs, circulating tumor-derived exosomes and circulating tumor nucleic acids) in the blood of patients, which can be considered as a liquid biopsy.8
The circulating tumor-derived exosomes are small membrane-bound cell fragments (sizes between 30 and 1000 nm diameter), and contain RNA populations, including microRNA. Therefore, determine whether the microRNAs contained within the cancer-derived exosomes could be used for tumor diagnosis. However, application of the circulating exosome detection has not yet been found in the clinical laboratory.8,9
Detection of the circulating tumor nucleic acids, which are released from the tumor cells in the body, is also feasible for tumor diagnosis. Although there are hundreds of articles on this topic, most have small study populations and only a few thousand patient samples in aggregate have been assayed for tumor nucleic acids.8 The studies are small and underpowered, they are often poorly controlled or uncontrolled, they examine one or a few markers, they use a variety of marginally validated methods, often with poor analytic sensitivities.8,10 Therefore, it's for sure that more assay validation studies are required before the detection technology of the circulating tumor nucleic acids can be brought into the clinic.
There are currently over 270 clinical trials that utilize CTCs as biomarkers based on the registration at ClinicalTrials.gov, and many studies confirm that CTCs could be used as prognostic markers for breast cancer (the survival time is longer when the CTC concentration is lower).11
CTCs are present in the peripheral blood of cancer patients even when the cancers are in the early stage. Therefore, CTC detection could be used for the early diagnosis of cancers. In addition, detection of CTC concentration in the blood can also be used for the earlier evaluation of cancer recurrence and chemotherapeutic efficacy.12-14 Furthermore, the phenotype identification and molecular analysis of CTCs are expected to provide much important information for the understanding of tumor metastasis and to play a key role in the personalized treatment of cancer patients (i.e. choice of individual sensitive anti-cancer drugs) in the future.15
In short, CTC detection is a crucial tool to fight against cancer and most of the new CTC publications focused on the detection technologies. There have been several published reviews that are CTC detection related.9,16,17 This tutorial review summarizes various CTC detection technologies.
The CellSearch® (Veridex, Raritan, NJ) is the first and only product on the market approved by U.S. food and drug administration (FDA) for detecting CTCs (in 2004). However, it has not been widely accepted by the medical community due at least to the following reasons: 1) the CellSearch system (equipment) is rather expensive (600,000∼800,000 USD); 2) the detection cost of each sample is high because antibodies are required to capture the CTCs in the blood; 3) the detection process requires a complicated enrichment step; 4) the detection time is long for each sample; 5) the purity of the captured CTCs is very low (< 0.5%);18 6) the CTCs cannot be isolated for phenotype identification and molecular analysis; 7) the sensitivity and selectivity are low (i.e. high false positive and false negative rates). Consequently, CellSearch will likely be obsolete in the near future due to these problems.
The CellCollector® (GILUPI GmbH, Potsdam, Germany) is a CE (French phrase “Conformité Européene” which literally means “European Conformity”) approved medical device, which is the first in vivo CTC isolation product worldwide. Its functional structured needle, which is inserted into the patient vein with 30 min of exposure for the CTC isolation, is flexible with 16 cm of length, and composed of stainless steel (0.5 mm of thickness), gold coating layer (2 μm of thickness) and hydrogel coating layer (2∼10 μm of thickness). On the hydrogel coating layer, the anti-EpCAM-antibodies are conjugated to identify and isolate the EpCAM-positive CTCs. Compared with the CellSearch®, the CellCollector® does not need expensive technical equipment, the sensitivity and selectivity are higher due to more blood in vivo, the captured CTCs can be isolated for phenotype identification and molecular analysis. However, the acceptance of invasive means by the patients is limited in contrast to the noninvasive one, the detection process also needs a complicated enrichment step, the detection time is also long for each sample (30 min), and the detection cost is also high as antibodies are required for CTC capture.
In order to overcome the limitations of CellSearch® and CellCollector®, many CTC detection technologies have been developed. In general, CTC detection may include 4 steps: 1) capture; 2) enrichment; 3) detection; 4) release. The capture step is known as the specific interaction (such as physical interaction and antibody/antigen interaction) between CTCs and materials (e.g. magnetic beads, microfluidic chips). This means that healthy blood cells do not interact with the materials. The enrichment step refers to isolation of CTCs from the blood. After enrichment, the CTCs could be detected by fluorescence (e.g. fluorescent microscope, fluorescent spectrophotometer and flow cytometry),18,19 surface-enhanced Raman scattering (SERS),20 or electrical impedance.21 The enriched CTCs can also be released for further phenotype identification and molecular analysis (e.g. mRNA profiling and cellular metabolism analysis). A technology “tree” classifying the current CTC detection technologies is shown in Scheme 1.
Scheme 1.
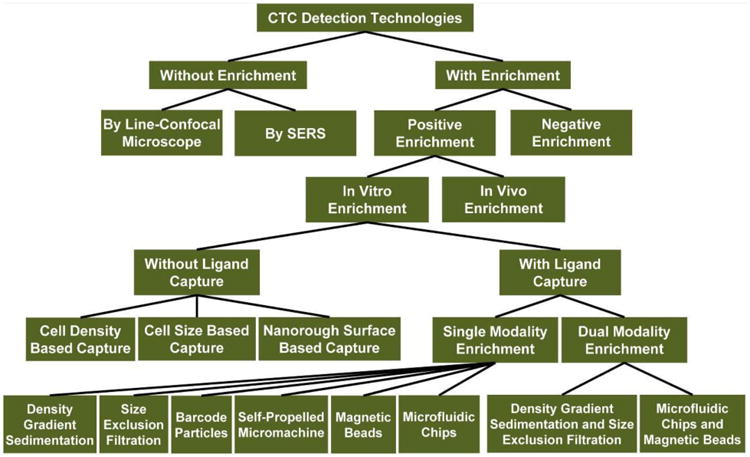
A technology “tree” showing the classification of current CTC detection technologies.
The important technical indicators for these CTC detection technologies include recovery rate, purity and limit of detection (LOD), which are summarized in Table 1. The recovery rate is defined as the number percentage of the enriched CTCs to the total CTCs in the blood, which is also named as capture efficiency or enrichment efficiency in some literatures. The purity refers to the number percentage of the enriched CTCs to the total number of cells in the enriched samples. The LOD is known as the limit of the CTC concentration in the blood that could be detected.
Table 1. Best results of recovery rate, purity and/or LOD of the current CTC detection technologies.
| CTC Detection Technologies a | Without Enrichment | Negative Enrichment | In vivo Enrichment | Without Ligand Capture | With Ligand Capture-Single Modality Enrichment | With Ligand Capture-Dual Modality Enrichment |
|---|---|---|---|---|---|---|
| Recovery Rate (%) b | - | ∼ 90 | ∼33 | 97 | 95 | >99 |
| Purity (%) c | - | 3.64 | - | < 10 | 84 | >92 |
| LOD (cells / mL) d | 1 | 1 | 0.12 | 2.2 | 1 | - |
| Refs | 23, 27 | 29, 30 | 32 | 35, 37 | 18, 19, 41 | 50 |
The currently reported CTC detection technologies as classified in Scheme 1.
The number percentage of the enriched CTCs to the total CTCs in the blood, which is also named as capture efficiency or enrichment efficiency in some literatures.
The number percentage of the enriched CTCs to the total cells in the enriched samples.
The limit of the CTC concentration in the blood that could be detected.
2. CTC detection technologies without enrichment
The CTC detection without an enrichment step is also called direct detection of CTCs. The reported technologies include: 1) line-confocal microscope; 2) SERS.
2.1 Direct detection by line-confocal microscope
Direct detection technology by line-confocal microscope is a fast and automated high-throughput screening method, which is developed based on the microfluidics and multicolor line-confocal detection techniques utilized in ensemble-decision aliquot ranking (eDAR).22
In the protocol of this direct detection technology, the cells in the blood are first simultaneously marked with multiple antibodies that are conjugated with different fluorescent agents. The blood is then pumped through a microfluidic channel, and interrogated by a line-confocal microscope. In this way, the number of CTCs could be automatically counted and reported in accordance with the fluorescent signals and labeling schemes.23 Figure S1a shows the schematic of direct CTC detection by a line-confocal microscope. The positive yellow signal for antiepithelial cell adhesion molecule (EpCAM), positive red signal for cytokeratin, and negative green signal for CD45 represent a CTC that is detected at 183 ms (Figure S1b). In Figure S1c, the top image of bright field and the following two fluorescent images show the blood in the microchannel. The MCF-7 cell in the fluorescent images, which is yellow due to the labeling of anti-EpCAM and red due to the labeling of anti-cytokeratin, shows the same location. However, it is not visible amongst plethora of blood cells in the bright field image, which is pointed out by a dashed white circle. The false positive rate is also evaluated for CTC counting (Figure S1d). For the analysis of healthy blood samples, 60% of the samples reported zero CTCs, and 1.2 CTCs on average were detected in 1.0 mL of the healthy blood samples. Figure S1e shows the recovery performance of the CTC counting. The healthy blood samples are spiked with MCF-7 cells, and then stained with PE-anti-EpCAM, Alexa-647-anti-cytokeratin, and FITC-anti-CD45. Figure S1f shows clinical results of the CTC analysis on 90 blood samples from 24 breast cancer patients by the line-confocal microscope (i.e. flow counting system) or CellSearch system. The line-confocal microscope method found a median of 90 CTCs per 7.5 mL of blood. However, the CellSearch system found a median of 0. Most of the events detected by the flow counting system are higher than the thresholds, and the events of false negative rate (∼ 40%) are much lower than that of the CellSearch system. This method is highly sensitive, with a LOD close to 1 cell/mL, but the throughput is suboptimal (20 min for 1 mL whole blood).23
The main advantages of the CTC detection by line-confocal microscope include: 1) the detection process is simple without an enrichment step; 2) the CTC counting is automated; 3) both avalanche photodiode (APD) signals and fluorescent images can be used to double confirm the CTC events; 4) false negative rate is much lower than that of the CellSearch system. The limitations of the line-confocal microscope methods are also obvious: 1) because the flow rate is limited in the microfluidic channels, the detection time is too long for each sample (analysis of 7.5 mL of blood requires about 3.5 h); 2) although the false negative rate is much lower than that of the CellSearch system, it is still very high (ca. 40 %); 3) the detection cost of each sample is high because several fluorescent dye-conjugated antibodies are required to label the cells in the blood; 4) the CTCs cannot be isolated for the following phenotype identification and molecular analysis.
2.2 Direct detection by SERS
SERS is another rapid analysis method of CTCs in the peripheral blood, which is developed based on the highly specific and sensitive SERS-active nanoparticles that have a targeting ligand to identify CTCs among healthy blood cells.
In the protocol of this direct detection technology, the peripheral blood samples are first added into a peripheral blood lymphocyte separation medium, and then centrifuged at room temperature. The low-density cell layers containing white blood cells (WBCs) and CTCs are transferred to new tubes, incubated with the SERS-active nanoparticles, and washed using phosphate buffered saline (PBS). The SERS spectra of the samples are finally analyzed by an instrument of Raman system.24,25
In this method, the design and construction of the SERS-active nanoparticles with strong SERS signal intensity and high affinity to CTCs rather than other healthy blood cells are the key issues. Wang et al. designed a kind of Raman reporter-encoded, PEG-stabilized, and EGF peptide-functionalized SERS nanoparticles.24 Gold nanoparticles (AuNPs) with a diameter of ∼60 nm were encoded with Raman reporter molecules (i.e. QSY21 quencher dye, Invitrogen Corporation, Carlsbad, CA), which were absorbed on the surface of AuNPs via electrostatic interaction. The QSY-encoded AuNPs (Au-QSY) were further functionalized with thiolated polyethylene glycol (HS-PEG) (85%) and HS-PEG-COOH (15%). The PEG protection layer was used to reduce nonspecific interaction with blood cells. The –COOH group was used for conjugation of EGF peptide as a targeting ligand to recognize and capture CTCs.24 It was found that the SERS signal intensity increased with increasing the number of spiked tumor cells. The LOD was in the range of 5 to 50 tumor cells in 1.0 mL of blood.24
However, in the design of the above-mentioned SERS-active nanoparticles,24 the PEG protection layer on the surface of AnNPs is hair-like and relatively thick. The thick layer of PEG reduces the intensity of SERS signal because the Raman reporter molecules are embedded inside the PEG layer. In order to overcome this problem, Wu et al. proposed another design of SERS-active nanoparticles for CTC detection (Figure 2a).25 4-Mercaptobenzoic acid (MBA) as a Raman reporter was first used to encode the AuNPs, and reductive bovine serum albumin (rBSA) was then used to stabilize the MBA-encoded AuNPs (AuNP-MBA) to reduce the non-specific interaction with blood cells. Finally, folic acid (FA) as a targeting ligand to capture CTCs was grafted to the surface of rBSA-stabilized AuNP-MBA (i.e. AuNP-MBA-rBSA) to prepare composite nanoparticle AuNP-MBA-rBSA-FA. Compared with the previous design,24 the rBSA layer on the nanoparticle surface like “lying hair” (Figure 2a) is far thinner than that of the PEG layer like “standing hair”, which results in stronger SERS signals. The rBSA layer is thin because many –SH groups in rBSA (broken up from disulfide bonds) can link to the surface of AuNPs via the formation of Au-S bonds.25
Figure 2.
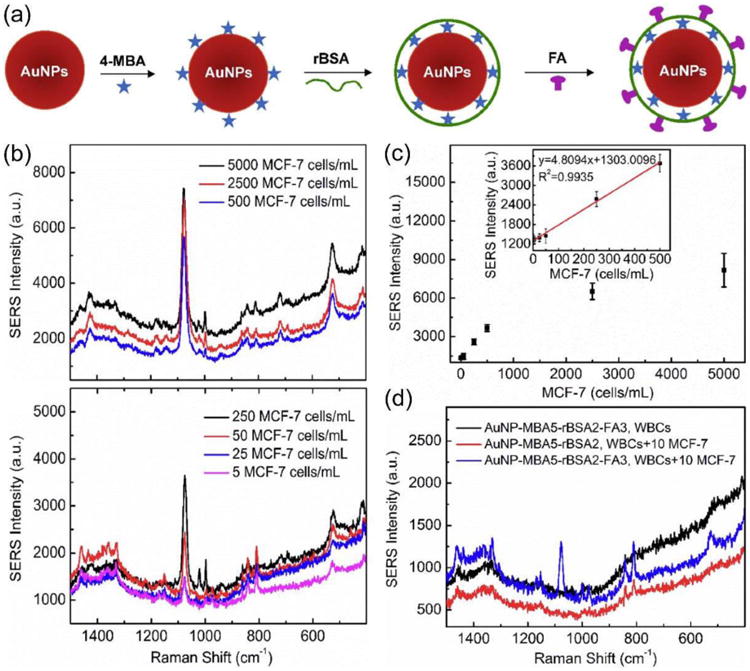
(a) Design of SERS-active nanoparticles for CTC detection. Gold nanoparticles (AuNPs) were loaded with 4-mercaptobenzoic acid (MBA) and stabilized by reductive bovine serum albumin (rBSA) followed by folic acid (FA) conjugation. (b) SERS signals of 2.0 mL rabbit blood spiked with different numbers of MCF-7 cells (range 5-5000 cells/mL). (c) Plot of SERS intensity versus the MCF-7 cell concentration in rabbit peripheral blood. (d) SERS signals with or without conjugation of targeting ligand FA in the rabbit blood (2 mL) spiked with or without 10 MCF-7 cells. Reproduced from reference 25 with permission from American Chemical Society, copyright 2015.
The SERS signals of the SERS-active nanoparticles in 2.0 mL of rabbit blood spiked with different numbers of MCF-7 cells in the range of 5-5000 cells/mL is shown in Figure 2b. A higher concentration of MCF-7 cells results in a stronger SERS intensity. Figure 2c shows the plot of the intensities of SERS signals versus the MCF-7 cell numbers in the peripheral blood of rabbit. A good linear relationship ranging from 5 to 500 cells/mL indicates that the cancer cells can be quantitatively analyzed by the designed SERS-active nanoparticles, and the LOD is 5 cells/mL.25 Figure 2d shows SERS signals of the SERS-active nanoparticles with or without conjugation of targeting ligand folic acid (FA) in the rabbit blood (2 mL) with or without the presence of 10 MCF-7 cells. The SERS peak at 1076 cm-1 is very clear in the case of FA-conjugated nanoparticles but not non-targeted nanoparticles, demonstrating folic acid receptor specificity of the SERS-active nanoparticles for cancer cell detection.25
Because the electromagnetic enhancement is the predominant contribution of the SERS signal, and the individual nanoparticle shape is an important feature to fine-tune the ability of electromagnetic enhancement,26 Wu et al. developed three types of SERS nanoparticles with similar particle size, similar modifications, but different shapes (spheres, rods and stars) to further enhance the sensitivity for CTC direct detection in the peripheral blood (Figure S2), and found that the gold nanostars were most effective with a LOD of as low as 1 cell/mL.27
The advantages of SERS detection mainly include: 1) similar to line-confocal microscopy, the detection process is simple and does not require an enrichment step; 2) many samples can be processed at the same time, leading to a short average detection time (several minutes) for each sample; 3) a relatively low-cost portable Raman spectrometer makes this technology easy to be popularized; 4) high sensitivity and specificity thus a low false positive rate and false negative rate. However, the limitations of this detection techniques are also obvious: 1) the CTC detection process is not easy to be automated; 2) the detection reagents are not cheap because antibodies are also required to capture the CTCs in the blood; 3) the CTCs cannot be isolated for the following phenotype identification and molecular analysis.
3. CTC detection technologies with enrichment
The low concentration of CTCs in blood can be enriched in two different ways. One is so-called negative enrichment, which aims to capture non-target cells (i.e. healthy blood cells) and elute target cells (i.e. CTCs). The other method is positive enrichment that captures CTCs and elute healthy blood cells.
3.1 Negative enrichment
In 2004, Lara et al. first developed a negative enrichment technology to enrich rare cells in blood.28 The enrichment procedures include: a red cell lysis step, immunomagnetically staining leukocytes with an anti-CD45 PE, anti-MACS sandwich, immunomagnetic sorting using a flow-through system (quadrupole magnetic cell sorter, QMS), and a final cell analysis step using either an automated cell counter, filtration, and visual counting or a cytospin analysis.28
In order to improve the efficiency of CTC negative enrichment, Hyun et al. developed a geometrically activated surface interaction (GASI) microfluidic chip using a herringbone shape to efficiently capture a large number of hematological cells rather than CTCs (Figure 3a).29 The herringbone structure was patterned on the channel to produce transverse flow that facilitates effective contact between antigens on the cells and antibodies on the channel surface (Figure 3 b,c). This was the first attempt to enrich CTCs from metastatic cancer patients using a negative enrichment microfluidic chip. The GASI chip successfully enriched CTCs and the same method may be useful for collecting other types of circulating rare cells whose phenotypes are poorly understood.29 For the conventional herringbone (HB) chip, the purity of the MCF-7 cells was less than 0.65% due to insufficient surface interaction between the blood cells and the CD45 antibody modified channel surface. For the GASI chip, the purity of the MCF-7 cells reached 3.64%. In addition, the number of isolated CTCs varied from 1 to 51 in 1 mL of blood.29
Figure 3.
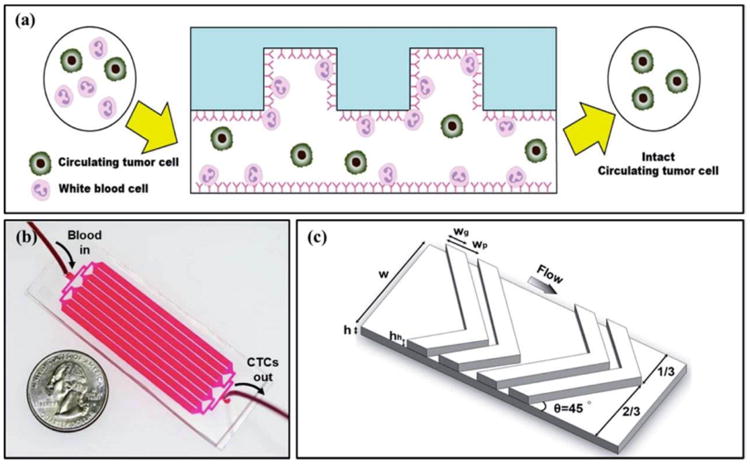
(a) Pictorial presentation of negative enrichment (side view). When the blood sample enters the inlet, the nontarget cells (i.e. leukocytes) are specifically captured on the CD45 antibody-coated channels, while the target cells (i.e. CTCs) are eluted through the outlet. (b) A photograph of the fabricated GASI chip. The GASI chip consists of an inlet, eight channels, and an outlet. Each channel is 2100 μm in width and 50 mm in length. (c) The herringbone structure and key parameters used in this study. Reproduced from reference 29 with permission from American Chemical Society, copyright 2013.
Sajay et al. developed another microfluidic platform that performs negative enrichment in two steps from 2 mL of whole blood in a total assay processing time of 60 min.30 This negative enrichment method employs upstream immunomagnetic depletion to remove CD45-positive WBCs followed by a microfabricated filter membrane to perform chemical-free RBC depletion and isolation of target cells. Experiments of spiking two types of cells, MCF-7 and NCIH1975, in the whole blood show an average of ∼ 90 % cell recovery over a range of spiked cell numbers.30
The advantages of the CTC detection technologies with negative enrichment include: 1) they do not rely on the biomarker expression of CTCs (e.g. EpCAM); 2) they can collect the CTCs in an intact form, which is desirable for downstream studies such as cellular and molecular analyses; 3) the negative enrichment process is easy to be automated. Some limitations of this strategy are also observed: 1) even a high capture rate of blood cells cannot lead to a high purity of CTCs (typical purity < 4%) because the CTCs are rare cells compared to the blood cells; 2) the identification of the obtained CTCs with a low purity needs further complicated analysis procedures; 3) large amount of antibodies are required to capture the blood cells using the negative enrichment technologies, which results in high cost.
3.2 Positive enrichment
As opposed to the negative enrichment strategy, positive enrichment captures CTCs and elutes healthy blood cells. The reported studies include two categories: 1) in vivo enrichment; 2) in vitro enrichment.
3.2.1 In vivo enrichment
In vivo positive enrichment of CTCs captures and enriches rare CTCs amongst large number of blood cells in vivo. For example, Galanzha et al. proposed a way to magnetically capture CTCs in the bloodstream of mice followed by rapid photoacoustic detection (Figure 4a).31 The magnetic nanoparticles (MNPs), which were functionalized to target a receptor commonly found in breast cancer cells (Figure 4b), bound and captured CTCs under a magnet. To improve detection sensitivity and specificity, gold-plated carbon nanotubes (GNTs) conjugated with folic acid (Figure 4c) were used as a second contrast agent for photoacoustic imaging and spectral measurement (Figure 4d). By integrating in vivo multiplex targeting, magnetic enrichment, signal amplification and multicolour recognition, this approach allows CTCs to be concentrated from a large volume of blood in the vessels of tumour-bearing mice, and this could have potential for the early diagnosis of cancer and the prevention of metastasis in humans.31
Figure 4.
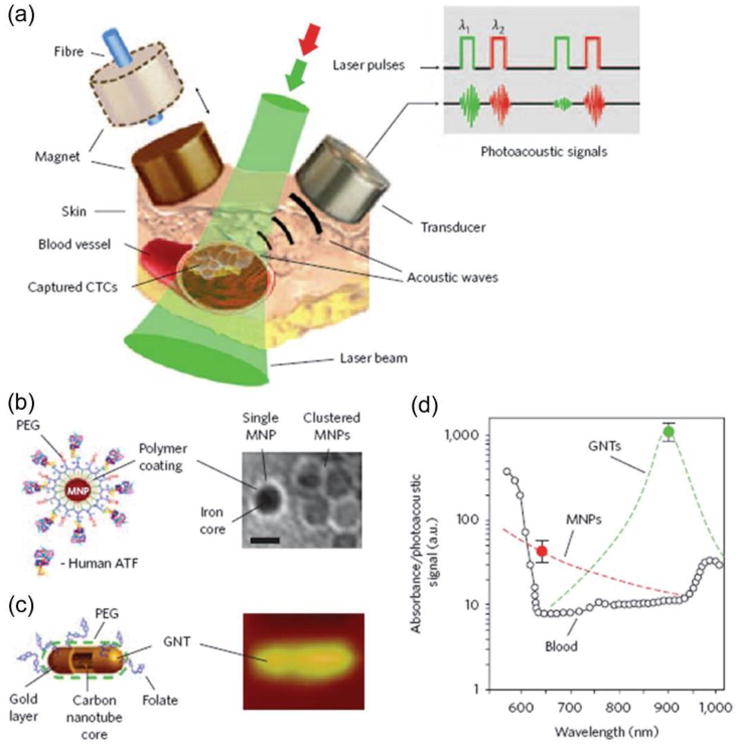
In vivo magnetic enrichment using two-color photoacoustic detection of CTCs. (a) Schematic showing the detection setup. (b) Schematic (left) and TEM image (right) of MNPs, each with a 10-nm core, a thin (∼2 nm) layer of amphiphilic triblock copolymers modified with short PEG chains and the amino-terminal fragment (ATF) of the urokinase plasminogen activator. Scale bar, 10 nm. (c) Schematic (left) and topographic atomic force microscopy image (right) of a GNT (12 nm × 98 nm) coated with PEG and folic acid. (d) Photoacoustic spectra of ∼70 μm veins in mouse ear (open circles). Reproduced from reference 31 with permission from Nature Publishing Group, copyright 2009.
However, this in vivo enrichment strategy is rather invasive. In order to overcome this problem, Zhang et al. developed a method of in vivo CTC enrichment based on transfusion with a vein indwelling needle (Figure 5).32 The vein indwelling needle was modified with an anti-EpCAM antibody, which could specifically recognize the EpCAM antigen on the surface of CTCs (Figure 5a). When this surface modified vein indwelling needle was applied for transfusion in the vein, the simultaneous capture of CTCs was performed. When the vein indwelling needle was drawn out, the captured CTCs were cleared from the vein and could be eluted for further microscope counting or component analysis (Figure 5b,c). The results showed that the vein indwelling needles could successfully capture the injected MCF-7 cells circulating in the veins of rabbits and wild type mice with a capture efficiency of 0.9 ∼ 33 %. In addition, 26 ± 3 cells and 24 ± 7 cells were captured from mice and rabbits, whose blood volumes were approximately 2 and 200 mL (i.e. the LOD is 0.12 cells/mL), respectively.32
Figure 5.
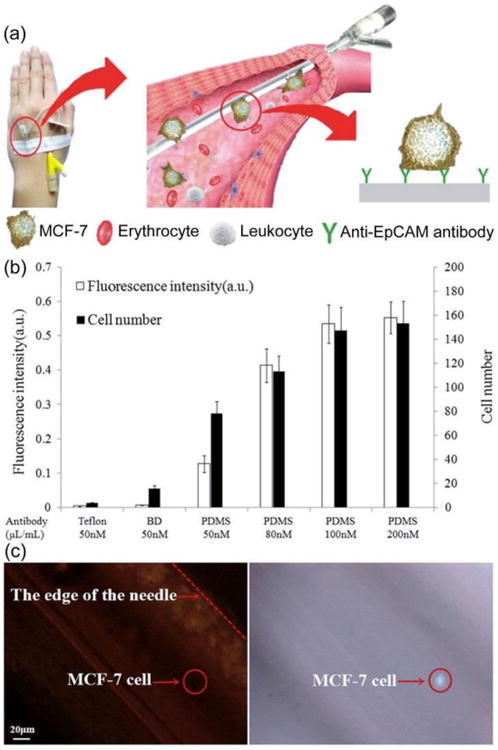
(a) Schematic of a method of in vivo CTC capture and enrichment based on transfusion with a vein indwelling needle. (b) Optimization of the antibody adsorbing on the needle via the fluorescence intensity and cell number of the CTCs captured using the nude or PDMS coated vein indwelling needles that are modified with various concentration of anti-EpCAM antibody. The error bars were estimated from three replicate measurements. (c) Microscope images of the DAPI stained MCF-7 cells captured on the needle. Reproduced from reference 32 with permission from American Chemical Society, copyright 2015.
There are several advantages of in vivo enrichment strategy in CTC detection: 1) compared with the limited volume of blood samples for in vitro methods, more flowing blood in vivo enables more CTCs of patients to be captured and enriched; 2) the relatively low requirement of sensitivity reduces the threshold of the technology; 3) the false negative rate is low. The caveats of in vivo enrichment strategy include: 1) the in vivo enrichment time is very long to ensure most of the body blood flows over the vein indwelling needle for CTC capture; 2) the technology is immature and no data of CTC purity has been reported; 3) the cost is high as antibodies are required for CTC capture.
3.2.2 In vitro enrichment
The in vitro enrichment captures and enriches rare CTCs in peripheral blood samples drawn from cancer patients. The capture can be realized with or without CTC-specific ligands prior to CTC enrichment (i.e. antibodies, antibody fragments, peptides, aptamers or small molecules).
3.2.2.1 Without ligand capture
In vitro CTC detection without specific ligand binding is sometimes called “physical capture”, which relies mainly on the physical properties of CTCs (cell density, cell size, and nanorough surface) to capture them.
In 2003, Baker et al. proposed a CTC enrichment technology via cell density based capture. Using fluorescently labeled breast cancer cells and flow cytometry, the treated peripheral blood by porous barrier density gradient centrifugation resulted in a 300-fold enrichment of breast cancer cells.33 In 2010, a CTC enrichment technology via cell size based capture (i.e. microcavity array system, MCA system) was reported by Hosokawa et al.34 The MCA system can specifically separate tumor cells from whole blood on the basis of differences in the size and deformability between tumor and hematologic cells. This device successfully detected approximately 97% of lung carcinoma NCI-H358 cells in 1 mL whole blood spiked with 10-100 NCI-H358 cells. On average, approximately 98% of recovered cells were viable.34 Subsequently, they further optimized the structure of MCA for enrichment of small-sized CTCs such as those found in the blood of small-cell lung cancer (SCLC) patients.35 They developed a rectangular MCA by electroforming to improve the number and purity of small tumor cells recovered from whole blood. The MCAs were made of nickel by electroforming as follows: a stainless steel plate was coated with SU-8 photoresists and exposed to UV light through a photomask to form the MCA pattern. By electroformation, nickel was built up in the bare areas of the stainless steel plate between the photoresists. Finally, the electroformed MCA was separated from the stainless steel plate. Each circular microcavity was fabricated with a diameter of 8-9 μm, and the rectangular microcavity was fabricated with a width of 5-9 μm and a length of 30 μm. The distance between each microcavity was 60 μm, and 10,000 (100 × 100) cavities were arranged in an 18 × 18 mm sheet. Under optimized conditions, ∼80% of SCLC (NCI-H69 and NCI-H82) cells spiked in 1 mL of whole blood were successfully recovered. The LODs by circular or rectangular MCA were 2.4 or 2.2 cells/mL, respectively.35
In 2013, Chen et al. reported a CTC enrichment technology via nanorough surface based capture.36 This method utilized the differential adhesion preference of cancer cells to nanorough surfaces over normal blood cells, and thus did not depend on their physical size or surface protein expression. They developed a microfluidic channel using polydimethylsiloxane (PDMS) integrated with smooth (Rq = 1 nm) or nanorough (Rq = 100 nm) glass substrates for direct measurements of adhesion strength of cancer cells and PBMCs. A low density of cancer cells (MCF-7, MBA-MB-231, and PC3 cells) or PBMCs was seeded uniformly inside the microfluidic channel for 12 h before they were exposed to constant directional fluid shear (0.1∼120 dyn cm-2) for 5 min. They quantified fractions of cancer cells, and PBMCs remained adherent on smooth and nanorough substrates after their treatments with this sustained 5-min directional fluid shear. The nanoroughness on glass surfaces was generated through reactive ion etching (Figure 6a,b). Figure 6c-f revealed significant enhancements of cancer cell capture yield by nanorough surfaces, and the yield improves with increasing nanoroughness, but is independent of anti-EpCAM antibody coating. Similar trend was observed on several other cancer cell lines (Figure 6g).36
Figure 6.
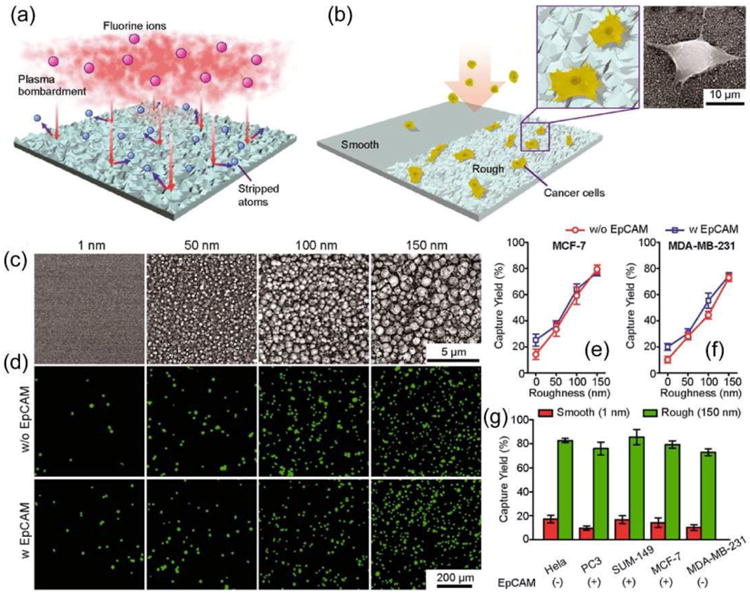
Intrinsic nanotopological sensing for capturing cancer cells. (a, b) Schematic of nanotopography generated by RIE on glass surfaces. Insets show a zoom-in (left) and SEM (right) images of cancer cells captured on nanorough glass surfaces. (c) SEM images of glass surfaces with their RMS nanoroughness indicated. (d-g) Cancer cells spiked in growth media at a concentration of 105 mL-1 captured on glass surfaces 1 h after cell seeding. Fluorescence images of MCF-7 cells (stained for nuclei, DAPI, green) captured on glass surfaces coated with (bottom) or without (top) EpCAM antibodies (d). Capture yields of MCF-7 (e), MDA-MB-231 (f), and other cancer cell lines (g) as a function of nanoroughness. Reproduced from reference 36 with permission from American Chemical Society, copyright 2013.
Furthermore, Wan et al. also developed an approach that combines high-throughput automated microscopy with a simple yet efficient approach for achieving a high level of tumor cell binding in standard tissue culture polystyrene (PS) well plates.37 A single 5 min high-power oxygen plasma treatment was used to create homogeneous nanoscale roughness on standard PS tissue culture plates and, in turn, drastically enhanced the binding of a range of tumor cells. After physical adsorption of a layer of poly-L-lysine, binding yields above 97% were obtained at 2 h for all tumor cell lines used in the study. However, the isolation purity from the physical methods is typically lower than that of biological methods with ligand capture.37
These reported CTC detection technologies without ligand capture, which rely on the physical properties of CTCs to capture them, have some advantages: 1) they do not rely on the biomarker expression of CTCs (e.g. EpCAM); 2) the enriched CTCs by these technologies are viable and intact; 3) the simple process of CTC enrichment without requiring biochemical modifications results in a short enrichment time and is amenable for downstream analyses; 4) the detection cost of each sample is not high because expensive ligands (e.g. antibodies, antibody fragments, peptides or aptamers) are not required to capture the CTCs in the blood. However, commercialization of these technologies can be a challenge as the purity of the enriched CTCs is low due to the low specificity. Further complicated analysis procedures are thus required to identify the obtained CTC samples with a low purity.
3.2.2.2 With ligand capture
There is a plethora of literature using ligand capture before CTC enrichment. The enrichment process this way can be either single modality enrichment or dual modality enrichment.
1) Single modality enrichment
Single modality enrichment means that only one of the following enrichment technologies with ligand capture is used for CTC detection: density gradient sedimentation, size exclusion filtration, barcode particles, self-propelled micromachine, magnetic beads, and microfluidic chips.
(1) Density gradient sedimentation
For the CTC enrichment technologies with ligand capture and density gradient sedimentation enrichment, ligands are conjugated onto the surface of microbeads with a high density to capture CTCs for effective separation of microbead-CTC complexes from healthy blood cells via selective sedimentation.
Yoo et al. enriched CTCs by using highly dense and transparent silica microbeads.38 The density of the transparent silica microbeads can be as high as 2.0 g/mL, which enables effective separation of microbead-CTC complexes from erythrocytes and leukocytes via selective sedimentation on a disc platform (i.e., selective sedimentation on a disc using silica microbeads, SSDS) (Figure 7a, b). Their optical transparency enables cellular and subcellular imaging of isolated CTCs (Figure 7c).38 The CTCs were defined as DAPI+/CD45-/CK+ and WBCs were defined as DAPI+ or DAPI+/CD45+ cells (Figure 7d, e). The SSDS method appeared to identify more CTCs than the established CellSearch system for most patients.
Figure 7.
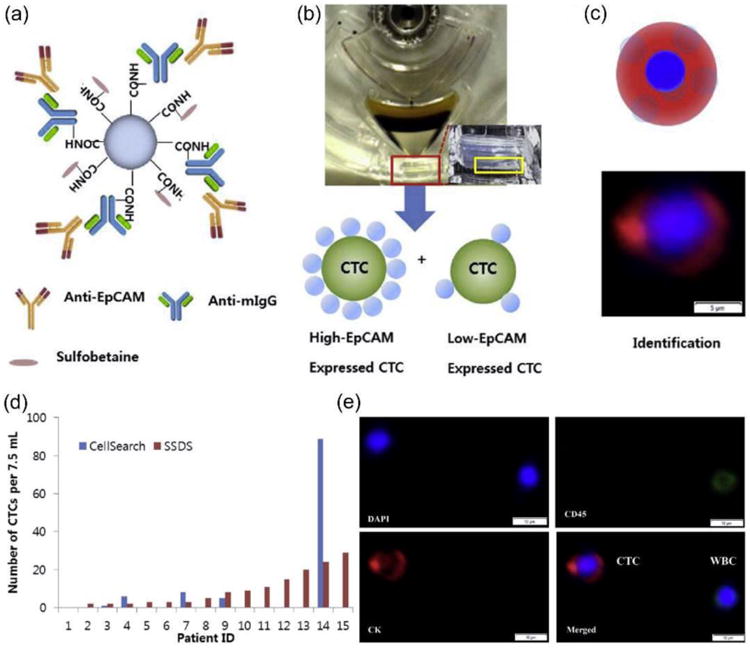
CTC enrichment based on highly dense and transparent silica microbeads. (a) Structure of silica microbeads prepared by the conjugation of the anti-EpCAM antibody with anti-mouse IgG coupled on the bead surface and blocking excess carboxylic groups with amine-functionalized sulfobetaine. (b) Isolation of CTCs by selective sedimentation on a disc using silica microbeads to capture CTCs forming CTC-microbead complexes. The high density (2.0 g/mL) of the silica microbeads enables the effective separation of CTC-microbead complexes, even when EpCAM expression is low, from erythrocytes and leukocytes via selective sedimentation on a disc platform. (c) Identification of CTCs isolated by silica microbeads via fluorescence staining and morphology. (d) The SSDS method isolated larger number of CTCs than the CellSearch System. (e) Stained image of isolated CTCs from a representative patient using the SSDS method. Reproduced from reference 38 with permission from Elsevier Ltd, copyright 2015.
The advantages of this CTC enrichment technology with ligand capture and density gradient sedimentation include: 1) compared with the FDA-approved CellSearch system, the sensitivity is improved and the CTC recovery from patient samples is increased; 2) the high density of the microbeads enables the effective separation of CTC-microbead complexes from blood, even when the EpCAM expression is low (i.e. the selectivity is good); 3) the CTC isolation disc platform based on density gradient sedimentation has achieved full automation resulting an easy operation and short enrichment time; 4) the CTC isolation platform based on the lab-on-a-disc has a large-volume capacity with 5 mL of blood chamber and 0.2 mL of CTC collection chamber.39 There are, however, also some limitations for this CTC enrichment technology as given below: 1) the usage of ligands (such as antibodies, antibody fragments, peptides, or aptamers) to capture CTCs in the blood significantly increases the detection cost, which is the common problem of the CTC enrichment technologies with ligand capture; 2) the microbeads may affect light scattering, quenching or eclipse which may interference with the fluorescence detection; 3) the microbeads may be internalized into the CTCs thus influencing the health of the living CTCs; 4) free microbeads mixed with the collected microbead-CTC complexes will interfere with the precise detection and analysis of CTCs.
(2) Size exclusion filtration
For size exclusion filtration enrichment, ligands are conjugated onto the surface of microfilters to isolate CTCs based on both size and surface receptor expression. Because the pore size of the polymer membrane filter has to be very small, the flow rate is very low resulting in low throughput and long enrichment time.
In order to overcome this problem, Kim et al. developed a high throughput CTC detection method that could separate heterogeneous population of CTCs with target markers in high efficiency.40 They designed a microchip filter device incorporating slit arrays and 3-dimensional flow that can separate heterogeneous population of CTCs. The density-based initial enrichment step, which took around 3 min, improved filter performance by not only enabling the removal of plasma and up to 80% of blood cells initially present in the blood sample but also reducing the total filtering time from 50 min to 10 min (Figure 8a).
Figure 8.
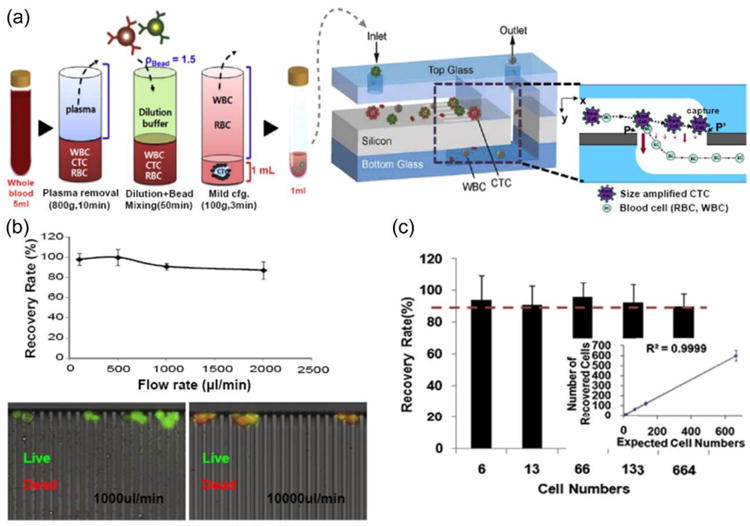
(a) Schematic illustration of CTC isolation principle using an automated platform based on antibody capture and size exclusion filtration. A density-based CTC enrichment method removes hematopoietic cells easily prior to introduction into the microchip filter device (left). Microchip filter device incorporates slit arrays and 3-dimensional flow with enlarged cross-sectional view around the slit (right). (b) Recovery rate as a function of input flow rate between 0.1 and 2 mL/min (top). Cell viability test after isolation of cancer cells, with green fluorescence indicating viable cells (bottom). (c) Recovery rate vs. input cell number. Reproduced from reference 40 with permission from Elsevier Ltd, copyright 2014.
Furthermore, by reducing the shear stress encountered by isolated cells in the microfilter, the “quality” of the isolated CTCs can be improved, as evidenced by more identifiable cell morphology in isolated cells. Furthermore, as melamine polymer beads are transparent, it is possible to observe CTCs inside the microchip filter by bright field and fluorescence microscopy. To assess the efficiency of the proposed device, they spiked MCF-7 cells in PBS at a concentration of 100 cells per mL and isolated the MCF-7 cells using the microchip filter. The recovery rate of spiked cells was maintained at above 90% for flow rates up to 1 mL/min (Figure 8b,c).40
Because accurate characterization of the enriched CTCs is important and removal of beads from the surface of CTCs prior to imaging is necessary, Lee et al. developed another approach for isolation and subsequent in situ protein-expression analysis of CTCs using detachable beads called RIA (reversible bead attachment for cell isolation and analysis) (Figure S3).
Based on the strategies of antibody capture and size exclusion filtration, Cheng et al. developed a new microchip embedded with a three-dimensional (3D) PDMS scaffold by a quadratic-sacrificing template method for high-efficiency capture of CTCs.41 The 3D scaffold microchip was functionalized with anti-EpCAM antibody, and gifted with a 3D interconnected macroporous structure, strong toughness, and excellent flexibility and transparency, enabling fast isolation and convenient observation of CTCs. The results showed that 1∼118 CTCs/mL were identified from 14 cancer patients' blood and 5 out of these cancer patients showed 1∼14 CTC clusters/mL. Therefore, the LOD of CTCs by using this method can be as low as 1 cell/mL.41
The CTC enrichment technologies with ligand capture and size exclusion filtration have some advantages including: 1) the CTC enrichment procedure is simple; 2) the CTC enrichment process is easy to be automated; 3) the sensitivity and selectivity towards CTCs are high due to dual capture by ligands and size exclusion filtration; 4) accurate in situ analysis of CTCs can be integrated with the enrichment process; 5) the size difference between CTCs and healthy blood cells can be amplified via conjugation microbeads with CTCs, and the pore size of the filters can thus be enlarged to increase the throughput (i.e. flow rate) of blood samples.
However, the common problem of the CTC enrichment technologies with ligand capture, i.e. the usage of ligands to capture CTCs in the blood increases the detection cost, is also one of the limitations of these technologies. Without coupling microbeads to CTCs, the size difference between CTCs and healthy blood cells is insignificant, and the flow rate is low due to the small pore size of the filters. To amplify the CTC size and filter pore size, microbeads can be conjugated onto the surface of CTCs. In that case, however, the microbeads may interfere with the precise detection and analysis of CTCs via fluorescence, and might be internalized into the CTCs influencing the health of the living CTCs.
(3) Barcode particles
Traditional barcode particles typically have two unfavorable characteristics: 1) they are small on the order of micrometers making them unsuitable as a substrate for cell capture and cell culturing; 2) when their surfaces are covered by cells, their encoding information would be obscured or incomprehensible.42 These characteristics, together with the debatable sensitivity, reliability, and specificity of the general surface morphology and biochemical modification, have limited the application of barcode particles in the capture and detection of CTCs.43
In 2014, Zheng et al. developed a new type of barcode particles with the desired capabilities.43 These barcode particles are able to capture, detect, and release multiple types of CTCs. The particles are spherical colloidal crystal clusters decorated with dendrimer-amplified aptamer probes. Their size can be adjusted to suit cell dimensions by their microfluidic-droplet templates. Encoding information in the particles is made possible by the characteristic reflection peaks originating from the photonic bandgap (PBG) structure of the colloidal crystals, and thus they are very stable. The encoding remains constant during multiple events of cell capture and cell culturing at the surface. In a typical experiment, the barcode particles are fabricated by the evaporation of droplet templates containing monodisperse silica nanoparticles. During this process, the nanoparticles form spherical assemblies with a regular arrangement (Figure S4), and when the droplet is dried, the barcode particles are obtained. It was found that when using the generation-5 (G5) poly(amidoamine) (PAMAM) dendrimer to decorate the barcode particles and immobilize the probe, the interaction of the barcode particles and the cells was very evident, and the corresponding capture efficiency was enhanced.43
The advantages of the barcode particle based CTC detection technology include: 1) it can simultaneously capture, detect and release multiple types of CTCs from a complex sample; 2) the etched surface of the barcode particles having a non-close-packed spherical array topography provides not only large surface area for probe immobilization and reaction, but also a nanopatterned platform for highly efficient bioreactions; 3) the specificity of CTC capture is high; 4) the released CTCs can be used for the following phenotype identification and molecular analysis. However, we have to bear in mind that in addition to the high detection cost due to the usage of ligands to capture CTCs, the sensitivity of CTC capture is low. Moreover, the release efficiency of the CTCs from the barcode particles is not high. The whole process of CTC capture, enrichment and detection is not easy to be automated resulting in a complicated operation process and a relatively long detection time.
(4) Self-propelled micromachine
Extending the scope of chemically powered nanomotors to physiological conditions is a key challenge since such nanomotors are commonly incompatible with the high ionic strength environment of biological fluids.44 In order to overcome the constraints to locomotion in physiological fluids, Balasubramanian et al. developed microrockets functionalized with an antibody specific for antigenic surface proteins expressed on CTCs, such as anti-carcinoembryonic antigen (anti-CEA) monoclonal antibody (mAb).44 Figure 9a illustrates the pickup and transport of CTCs by microrockets. Figure 9b, c illustrates the movement of the mAb-coated microrocket in PBS or human serum. These images show a long trail of microbubbles, catalytically generated on the inner platinum surface and released from the rear of the microtube. Such ejection of bubbles propels the microrocket in the diluted serum medium at a relatively high speed of ∼ 85 μm/s. The sandwiched ferromagnetic (Fe) layer of the microrocket offers convenient guidance of the microrocket through tuning of the external magnetic-field direction.44
Figure 9.
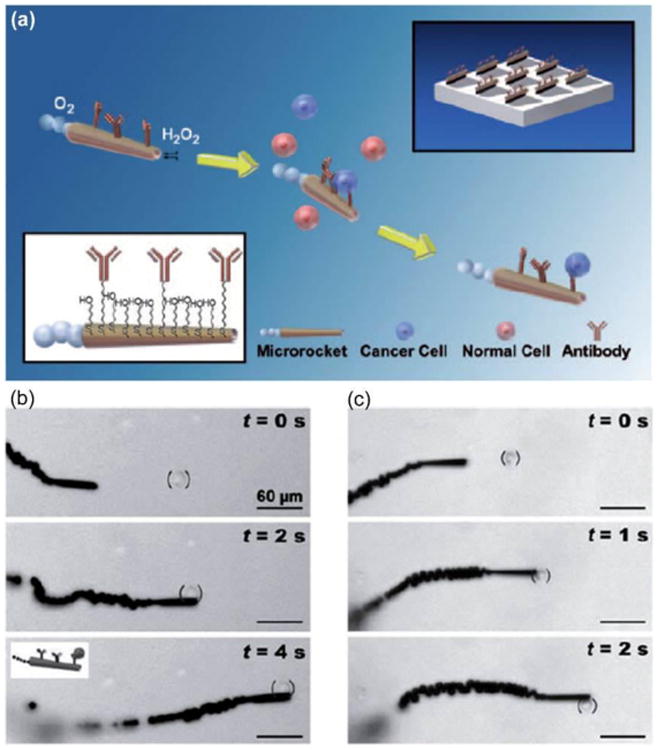
(a) Microrockets for capture and isolation of CTCs. The anti-CEA mAb-modified microrockets recognize the CEA surface antigens on the target CTCs, allowing selective pickup and transport. (b, c) Pickup and transport in PBS and diluted serum. Time-lapse images demonstrate the pickup and transport of a CEA+ pancreatic cancer cell by an anti-CEA mAb-modified microrocket in PBS (b) and human serum (c) at intervals of 2 and 1 s, respectively. Reproduced from reference 44 with permission from Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim, copyright 2011.
The self-propelled micromachine based CTC enrichment technology has two important advantages: 1) preprocessing of the biological samples is not necessary for the self-propelled micromachine based selective capture and transport of CTCs from biological fluids; 2) this technology can be extended to accumulate CTCs in a predefined “collection” area by detaching the captured CTCs. However, the motion direction and velocity of the micromachine are not easy to be controlled. This technology is still at the stage of concept verification. A full system for CTC detection based on this technology has yet to be developed.
(5) Magnetic beads
For the CTC enrichment with ligand capture and magnetic bead enrichment, ligands are conjugated onto the surface of magnetic beads to capture CTCs followed by effective separation of magnetic bead-CTC complexes from healthy blood cells via an external magnetic field. This is a relatively mature technique and the only commercialized CTC product approved by FDA (i.e. CellSearch system) belongs to this category.
In order to identify and detect the CTCs enriched using the magnetic beads by fluorescence-based instruments, fluorescent agents could be used to stain the magnetic beads. Xiong et al. developed biomimetic immuno-magnetosomes (IMSs) for the high-performance enrichment of CTCs (Figure 10a).18 Briefly, the magnetic nanoclusters (MNCs) were camouflaged with leukocyte membrane fragments (LMNC) through electrostatic interaction. The resulting magnetosome would be repelled when it encounters a leukocyte in the peripheral blood due to their homology; therefore, nonspecific leukocyte adsorption would be significantly suppressed, thus decreasing the corresponding background. Meanwhile, the leukocyte membrane was pre-engineered with azide (N3) via intrinsic biosynthesis and the metabolic incorporation of phospholipids to construct N3-LMNCs. Through mild and highly efficient click chemistry, the N3-LMNCs could be decorated with DBCO-Ab (dibenzocyclooctyne group modified antibody) at a controllable density with good fluidity (Figure 10b, c). The resulting IMSs were endowed with high CTC recognition efficiency. The capture efficiency was determined to be 79% for MACS beads but reached 95% for IMSs when they were exposed to 105 MCF-7 cells/mL in PBS medium. Around 90% of the rare CTCs could be captured from whole blood in 15 min with an undetectable leukocyte background, which demonstrates the great promise of the biomimetic IMSs for CTC enrichment.18
Figure 10.
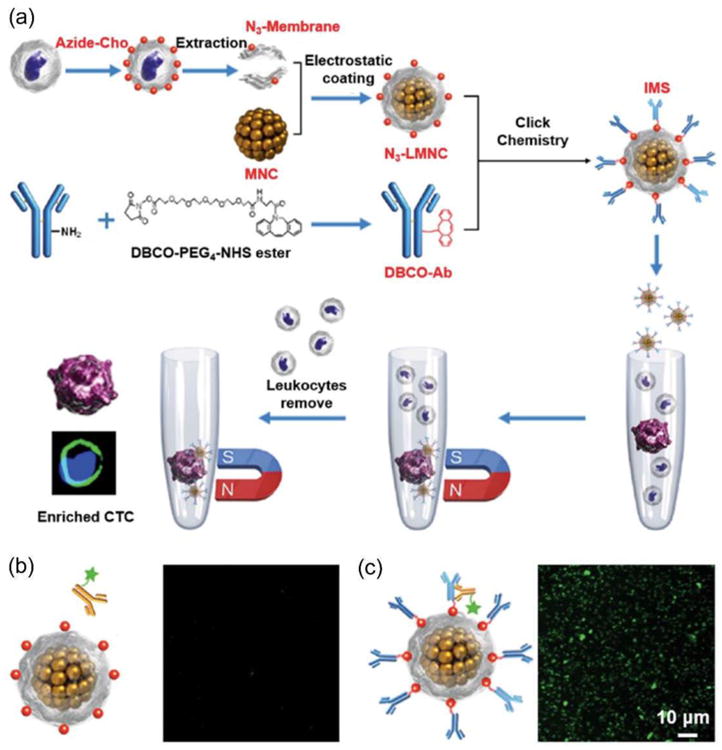
(a) Construction of immuno-magnetosomes (IMSs) and the procedure of CTC enrichment. The magnetic nanoclusters (MNCs) were camouflaged with leukocyte membrane fragments (LMNC) through electrostatic interaction. The leukocyte membrane is pre-engineered with azide (N3) via intrinsic biosynthesis and the metabolic incorporation of phospholipids to construct N3-LMNCs. The Ab is conjugated on the surface of N3-LMNCs through mild and highly efficient click chemistry. (b,c) CLSM images of N3-LMNCs (b) and IMSs (c) after reaction with fluorescent secondary antibody, demonstrating the success of DBCO-Abs conjugation to N3-LMNCs. Reproduced from reference 18 with permission from Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim, copyright 2016.
In addition, fluorescent agents could also be used to directly stain the CTCs enriched by the magnetic beads to identify and detect the CTCs. Lu et al. developed a biotin-triggered decomposable immunomagnetic system, in which peptide-tagged antibody designed by chemical conjugation was specifically immobilized on engineered protein coated magnetic beads.19 The interaction between peptide and engineered protein can be reversibly destroyed by biotin treatment, making capture and release of CTCs possible. Quantitative results showed that 70% of captured cells could be released by biotin addition, and 85% of released cells remained viable. In addition, the purity of separated SK-BR-3 cells was calculated to be 84 ± 3%.19 Wen et al. fabricated magnetic nanospheres (MNs) by a convenient and highly controllable layer-by-layer assembly method, and successfully captured rare tumor cells in whole blood with an efficiency of more than 94% via only a 5 min incubation (Figure S5).
In order to identify and detect the CTCs by SERS, SERS tags can be used together with the magnetic beads to bind to the CTCs. Figure S6a illustrates the concept in which magnetic beads, conjugated to anti-EpCAM, and the SERS tags, conjugated to an anti-her2 antibody (human epidermal growth factor receptor-2), bind to a tumor cell.20 The curve of CTC detection in buffer and whole blood (Figure S6b-d) demonstrates a high correlation between Raman signal intensity and cell concentration.20
The advantages of the CTC enrichment technologies with ligand capture and magnetic beads are given below: 1) the CTC enrichment process by an external magnetic field is very fast (only several minutes), and multiple samples could be treated simultaneously resulting in short average enrichment time of each sample; 2) it is the most widely studied and used technique for CTC enrichment, and the technology has led to commercialization and FDA approval of CellSearch system; 3) the magnetic field is not harmful to the CTCs; 4) the CTC capture efficiency by ligands and the CTC enrichment efficiency by magnetic microbeads are both high. However, there are unavoidable problems in this technique: 1) the use of expensive ligands to capture CTCs leads to a high detection cost; 2) the internalization of the microbeads may influence the CTC viability; 3) this technology relies on expression of CTC markers, such as EpCAM; 4) the whole process including CTC capture, enrichment and detection is not easy to be automated.
(6) Microfluidic chips
Microfluidics copes with the fluids' behavior and its accurate control at a small scale, typically sub-millimeter. Microfluidics emerged in the beginning of the 1980s and was then applied for the development of DNA chips, inkjet printheads, micro-propulsion, lab-on-a-chip technologies, and micro-thermal technologies. In recent years, the microfluidic chips are also developed for CTC detection in vitro because the blood samples drawn from patients are limited to a small scale. Regarding the CTC enrichment technologies with ligand capture and microfluidic chip enrichment, ligands are grafted to microfluidic chips to capture CTCs in the flowing blood resulting in isolation of CTCs from healthy blood cells (i.e. CTC enrichment).
There are many different kinds of ligand capture-based microfluidic chips, whose main difference is design of the microchips including the channel structures and substrate coating. For example, Nagrath et al. developed a unique microfluidic chip for CTC enrichment that consists of an array of microposts with functionalization of anti-EpCAM antibodies.45 Two essential parameters that determine the efficiency of cell capture on the CTC-chip are: (1) flow velocity, because it influences the duration of cell–micropost contact; and (2) shear force, which must be sufficiently low to ensure maximum cell-micropost attachment.45 Under precisely controlled laminar flow conditions, the microfluidic chip successfully identified CTCs in the peripheral blood of patients with cancer in 115 of 116 (99%) samples, with a range of 5∼1281 CTCs per mL and approximately 50% purity.45
Yoon et al. reported a graphene oxide microfluidic chip for sensitive capture of CTCs using functionalized graphene oxide nanosheets on a flat substrate.46 Graphene oxide nanosheets were adsorbed onto the patterned gold surface, then chemically functionalized with EpCAM antibodies. Graphene oxide nanosheets were non-covalently functionalized by phospholipid-polyethylene-glyco-amine (PL-PEG-NH2), and the hydrophobic lipid chains of PL-PEG-NH2 were strongly immobilized onto the graphene oxide surface. Tetrabutylammonium (TBA) hydroxide was added for intercalation and complete exfoliation of the graphene oxide. The TBA cations and the amino group of PL-PEG-NH2 interacted with the patterned gold surface by electrostatic attraction. N-γ-maleimidobutyryloxy succinimide ester (GMBS) was introduced, which has N-hydroxysuccinimide (NHS) esters that react with the amine groups of the graphene oxide-PEG to form amide bonds. The CTCs were then captured by subsequent NeutrAvidin and biotinylated EpCAM antibody interactions.46 The silicon substrate has 58,957 flower-shaped gold patterns with dimensions of 100 μm × 100 μm. CTCs were able to be captured at a low concentration of target cells (73 ± 32.4% at 3∼5 cells per mL blood).46
In addition, there are also many other microfluidic designs for the CTC detection technologies without enrichment (i.e. direct detection by line-confocal microscope),22 those with negative enrichment,29,30 and those with positive enrichment but without ligand capture.36 These designs have been described in the previous corresponding sections.
The enriched CTCs on the microfluidic chips can also be released for further phenotype identification and molecular analysis. For example, Yoon et al. developed a microfluidic chip with thermal-sensitive polymer-controlled CTC release.47 Lv et al. designed a near-infrared (NIR) light-responsive substrate for highly efficient immune-capture and biocompatible site-release of CTCs.48
The microfluidic chip enrichment strategy has several advantages: 1) the peripheral blood of patients can be directly applied on the microfluidic chips without pre-dilution, pre-labelling, pre-fixation or other processing steps; 2) the cost of the microfluidic chips is not high; 3) the sensitivity is not bad (∼70 % of CTCs can be captured at 3∼5 cells per mL blood); 4) the captured and enriched CTCs by the microfluidic chips can be released for the following phenotype identification and molecular analysis. However, there are still some problems that limit the commercialization of the microfluidic chips for CTC detection: 1) the capability to deal with large volumes (milliliters) of whole blood samples is limited; 2) the slow flow rate of the microfluid results in a long time of the CTC enrichment; 3) the shear force must be sufficiently low to ensure maximum cell-substrate attachment.
2) Dual modality enrichment
In order to improve the CTC enrichment efficiency from blood samples, two of the above-mentioned six enrichment technologies can be combined. The combined enrichment technologies are termed as dual modality enrichment, such as density gradient sedimentation plus size exclusion filtration,49 and microfluidic chips plus magnetic beads.50
Park et al. developed a sensitive, selective, and fast dual modality enrichment method for the isolation and detection of CTCs.49 The assay platform consists of three steps: (i) capturing CTCs with anti-EpCAM conjugated microbeads, (ii) removal of unwanted hematologic cells (e.g., leukocytes, erythrocytes, etc.) by selective sedimentation of CTCs within a density gradient medium, and (iii) simple microfiltration to collect these cells. The results showed a near perfect recovery rate (~99%) for MCF-7 cells.49
Kim et al. reported a single-cell isolation technology for CTCs based on a microfluidic chip (SIM-Chip) and immunomagnetic nanobeads.50 The SIM-Chip comprises a lateral magnetophoretic microseparator and a microdispenser as a two-step cascade platform. First, CTCs were enriched from whole blood by the lateral magnetophoretic microseparator based on immunomagnetic nanobeads. Next, the enriched CTCs were electrically identified by single-cell impedance cytometer and isolated as single cells using the microshooter. The average volume of the droplets was 4.5 μL, which faithfully matches the simulation result using the equivalent circuit mode. Single-cell isolation throughput of the microdispenser was 700 ms, as required for shooting the buffer and positioning of the 96-well plate. The single-cell isolation experiment on the SIM-Chip using blood samples spiked with 50 MCF7 cells showed that the average proportion of droplets containing a single MCF7 cell was 82.4 % (i.e. isolation efficiency of the SIM-Chip). The average number of total MCF7 cells isolated by the SIM-Chip was 49, and the number of contaminated WBCs were 4.0. This result indicate that the purity of MCF7 cells among the isolated nucleated cells, including MCF7 cells and WBCs, was 92.45%. The recovery rate of the SIM-Chip was measured for various numbers (from 10 to 90) of MCF7 cells spiked into 200 μL of whole blood. The results showed that the recovery rate was consistently around 99.78% for various numbers of spiked MCF7 cells. This high recovery rate demonstrates that neither the lateral magnetophoretic microseparator nor the microdispenser loses MCF7 cells.50
The advantages of the dual modality enrichment methods include: 1) the enrichment efficiency is very high (> 99%); 2) the purity of the enriched CTCs is very high (> 92 %). However, this technology is immature and not yet widely investigated, even no LOD value is presented in the literature. It can be foreseen that the detection sensitivity could be significantly enhanced after systematic investigation of the enrichment technology combinations (i.e. combining two or three of the above-mentioned single modality enrichment technologies).
4. Conclusions and future prospective
The important technical indicators for the currently reported CTC detection technologies (Scheme 1) include recovery rate, purity and LOD (Table 1). The LOD of the CTC detection technologies without enrichment (i.e. direct detection) could be very low (1 cell/mL) due to the supersensitive detection method (e.g. SERS). The recovery rate and purity are not applicable to this method because CTCs are detected directly without isolation. The recovery rate and LOD of the CTC detection technologies with negative enrichment are both reasonably good, but the purity is rather low (< 4 %) because they aim to capture huge amount of healthy blood cells and elute rare CTCs. Regarding the CTC detection technologies with in vivo enrichment, the LOD is impressive (as low as 0.12 cells/mL) but the recovery rate (∼ 33%) is not so high due to the large volume of the samples (i.e. the whole body blood). All the technical indicators of the CTC detection technologies with ligand capture are significantly better than those without ligand capture, but ligand capture relies on the biomarker expression of CTCs. In order to further improve the technical aspects of the CTC detection technologies with ligand capture, some research teams tried to combine two or more enrichment technologies with ligand capture. In this way, both the recovery rate and purity are enhanced.
Although CTCs have become a hot pursuit and many new CTC detection technologies have been reported, translation of these technologies from laboratory to clinical practice is non-trivial. CellSearch system was approved by FDA for CTC based cancer diagnosis more than a decade ago, it is still the only CTC product approved by FDA until now. In this review, we discussed all the existing CTC detection technologies and analyzed the pros and cons of each technology. Without enrichment, CTCs cannot be isolated for the subsequent phenotype identification and molecular analysis. With negative enrichment, the purity of the enriched cells is low. With in vivo enrichment, the detection time is long as the whole body blood need to flow over the vein indwelling needle for CTC capture. Without ligand capture, the CTC purity of the enriched cells is also low. With ligand capture and density gradient sedimentation, the free microbeads will likely interfere with the collected microbead-CTC complexes. With ligand capture and size exclusion filtration, the low flow rate results in low throughput and long enrichment time. With ligand capture and barcode particles, automation is not currently possible and the whole process of CTC capture, enrichment and detection is time-consuming. With ligand capture and self-propelled micromachine, the motion direction and velocity of the micromachine are not easily controllable. With ligand capture and magnetic beads, the CTCs without expression (or with very low expression) of the biomarker (e.g. EpCAM) cannot be captured. With ligand capture and microfluidic chips, the slow flow rate of the microfluid results in a long time of the CTC enrichment.
Opportunities often come with challenges. Combination of several complimenting CTC detection technologies might allow to overcome some of the above-mentioned challenges. For example, the direct detection by SERS (i.e. CTC detection technology without enrichment) could be combined to that with negative enrichment. That means the blood samples can be pre-processed to remove most of the blood cells prior to SERS detection. In that case, the LOD of CTCs can be further improved theoretically. In addition, in order to enhance the CTC purity enriched by the technologies with negative enrichment, the post-processing samples can be subjected to the technologies with ligand capture. Furthermore, combination of physical capture (i.e. without ligand capture) and ligand capture, the CTC purity can be substantially improved and the dependence on the CTC biomarker expression can be reduced. Similar with the technologies with dual modality enrichment, more than two of the CTC enrichment technologies might be combined (i.e. multiple modality enrichment technology) to obtain high technical indicators, such as recovery rate, purity and/or LOD. Finally, in addition to the current CTC analysis methods by fluorescence, SERS, electrical impedance and mRNA profiling, there are also ample opportunities to analyze CTCs systematically using proteomics and genomics in the future.
Supplementary Material
Key learning points.
Circulating tumor cells (CTCs) are cancer cells that circulate in the blood stream after being naturally shed from original or metastatic tumors, and CTC detection plays a key role in early diagnosis of cancers, earlier evaluation of cancer recurrence and chemotherapeutic efficacy, and choice of individual sensitive anti-cancer drugs.
Limitations of CellSearch® system that is the first and only product on the market approved by FDA for detecting of CTCs.
Classification, important processing steps (capture, enrichment, detection and release) and essential technical indicators (recovery rate, purity and limit of detection) of the currently reported CTC detection technologies.
Advantages and limitations of the currently reported CTC detection technologies.
Future perspective including challenges and opportunities of the CTC detection.
Acknowledgments
This work is supported in part, by the Intramural Research Program (IRP), National Institute of Biomedical Imaging and Bioengineering (NIBIB), National Institutes of Health (NIH) (Grant No. ZIA EB000073), Youth Innovation Promotion Association of Chinese Academy of Sciences (2016269), Public Welfare Technology Application Research Project of Zhejiang Province (2017C33129), the National Key Research & Development Program (2016YFC1400600), National Natural Science Foundation of China (Grant Nos. 61571278 and 21305148), Bureau of Science and Technology of Ningbo Municipality City (Grant Nos. 2015B11002), and NSFC-Guangdong Province Joint Project on National Supercomputer Centre in Guangzhou (NSCC-GZ) (A. Wu).
References
- 1.Adams AA, Okagbare PI, Feng J, Hupert ML, Patterson D, Gottert J, McCarley RL, Nikitopoulos D, Murphy MC, Soper SA. J Am Chem Soc. 2008;130:8633–8641. doi: 10.1021/ja8015022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sun N, Li X, Wang Z, Zhang R, Wang J, Wang K, Pei R. ACS Appl Mater Interfaces. 2016;8:12638–12643. doi: 10.1021/acsami.6b02178. [DOI] [PubMed] [Google Scholar]
- 3.Massague J, Obenauf AC. Nature. 2016;529:298–306. doi: 10.1038/nature17038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weigelt B, Peterse JL, van't Veer LJ. Nat Rev Cancer. 2015;5:591–602. doi: 10.1038/nrc1670. [DOI] [PubMed] [Google Scholar]
- 5.Ashworth TR. Aust Med J. 1869;14:146–147. [Google Scholar]
- 6.Baccelli I, Schneeweiss A, Riethdorf S, Stenzinger A, Schillert A, Vogel V, Klein C, Saini M, Bauerle T, Wallwiener M, Holland-Letz T, Hofner T, Sprick M, Scharpff M, Marme F, Sinn HP, Pantel K, Weichert W, Trumpp A. Nat Biotechnol. 2013;31:539–544. doi: 10.1038/nbt.2576. [DOI] [PubMed] [Google Scholar]
- 7.Nellore BPV, Kanchanapally R, Pramanik A, Sinha SS, Chavva SR, Hamme A, Ray PC. Bioconjugate Chem. 2015;26:235–242. doi: 10.1021/bc500503e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gold B, Cankovic M, Furtado LV, Meier F, Gocke CD. J Mol Diagn. 2015;17:209–224. doi: 10.1016/j.jmoldx.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taylor DD, Gercel-Taylor C. Gynecol Oncol. 2008;110:13–21. doi: 10.1016/j.ygyno.2008.04.033. [DOI] [PubMed] [Google Scholar]
- 10.Fleischhacker M, Schmidt B. Biochim Biophys Acta. 2007;1775:181–232. doi: 10.1016/j.bbcan.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 11.Alix-Panabieres C, Pantel K. Nat Rev Cancer. 2014;14:623–631. doi: 10.1038/nrc3820. [DOI] [PubMed] [Google Scholar]
- 12.Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L, Smith MR, Kwak EL, Digumarthy S, Muzikansky A, Ryan P, Balis UJ, Tompkins RG, Haber DA, Toner M. Nature. 2007;450:1235–1239. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galanzha EI, Shashkov EV, Kelly T, Kim JW, Yang L, Zharov VP. Nat Nanotechnol. 2009;4:855–860. doi: 10.1038/nnano.2009.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stott SL, Hsu CH, Tsukrov DI, Yu M, Miyamoto DT, Waltman BA, Rothenberg SM, Shah AM, Smas ME, Korir GK, Floyd FP, Jr, Gilman AJ, Lord JB, Winokur D, Springer S, Irimia D, Nagrath S, Sequist LV, Lee RJ, Isselbacher KJ, Maheswaran S, Haber DA, Toner M. Proc Nat Acad Sci U S A. 2010;107:18392–18397. doi: 10.1073/pnas.1012539107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nima ZA, Mahmood M, Xu Y, Mustafa T, Watanabe F, Nedosekin DA, Juratli MA, Fahmi T, Galanzha EI, Nolan JP, Basnakian AG, Zharov VP, Biris AS. Sci Rep. 2014;4:4752. doi: 10.1038/srep04752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin M, Chen JF, Lu YT, Zhang Y, Song J, Hou S, Ke Z, Tseng HR. Acc Chem Res. 2014;47:2941–2950. doi: 10.1021/ar5001617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Green BJ, Safaei TS, Mepham A, Labib M, Mohamadi RM, Kelley SO. Angew Chem Int Ed. 2016;55:1252–1265. doi: 10.1002/anie.201505100. [DOI] [PubMed] [Google Scholar]
- 18.Xiong K, Wei W, Jin Y, Wang S, Zhao D, Wang S, Gao X, Qiao C, Yue H, Ma G, Xie HY. Adv Mater. 2016;28:7929–7935. doi: 10.1002/adma.201601643. [DOI] [PubMed] [Google Scholar]
- 19.Lu NN, Xie M, Wang J, Lv SW, Yi JS, Dong WG, Huang WH. ACS Appl Mater Interfaces. 2015;7:8817–8826. doi: 10.1021/acsami.5b01397. [DOI] [PubMed] [Google Scholar]
- 20.Sha MY, Xu H, Natan MJ, Cromer R. J Am Chem Soc. 2008;130:17214–17215. doi: 10.1021/ja804494m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Han SI, Han KH. Anal Chem. 2015;87:10585–10592. doi: 10.1021/acs.analchem.5b03147. [DOI] [PubMed] [Google Scholar]
- 22.Schiro PG, Zhao M, Kuo JS, Koehler KM, Sabath DE, Chiu DT. Angew Chem Int Ed. 2012;51:4618–4622. doi: 10.1002/anie.201108695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao M, Schiro PG, Kuo JS, Koehler KM, Sabath DE, Popov V, Feng Q, Chiu DT. Anal Chem. 2013;85:2465–2471. doi: 10.1021/ac400193b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang X, Qian X, Beitler JJ, Chen ZG, Khuri FR, Lewis MM, Shin HJC, Nie S, Shin DM. Cancer Res. 2011;71:1526–1532. doi: 10.1158/0008-5472.CAN-10-3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu X, Luo L, Yang S, Ma X, Li Y, Dong C, Tian Y, Zhang L, Shen Z, Wu A. ACS Appl Mater Interfaces. 2015;7:9965–9971. doi: 10.1021/acsami.5b02276. [DOI] [PubMed] [Google Scholar]
- 26.Lu XM, Rycenga M, Skrabalak SE, Wiley B, Xia YN. Annu Rev Phys Chem. 2009;60:167–192. doi: 10.1146/annurev.physchem.040808.090434. [DOI] [PubMed] [Google Scholar]
- 27.Wu X, Xia Y, Huang Y, Li J, Ruan H, Luo L, Yang S, Shen Z, Wu A. ACS Appl Mater Interfaces. 2016;8:19928–19938. doi: 10.1021/acsami.6b07205. [DOI] [PubMed] [Google Scholar]
- 28.Lara O, Tong X, Zborowski M, Chalmers JJ. Exp Hematol. 2004;32:891–904. doi: 10.1016/j.exphem.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 29.Hyun KA, Lee TY, Jung HI. Anal Chem. 2013;85:4439–4445. doi: 10.1021/ac3037766. [DOI] [PubMed] [Google Scholar]
- 30.Sajay BNG, Chang CP, Ahmad H, Khuntontong P, Wong CC, Wang Z, Puiu PD, Soo R, Rahman ARA. Biomed Microdevices. 2014;16:537–548. doi: 10.1007/s10544-014-9856-2. [DOI] [PubMed] [Google Scholar]
- 31.Galanzha EI, Shashkov EV, Kelly T, Kim JW, Yang L, Zharov VP. Nat Nanotechnol. 2009;4:855–860. doi: 10.1038/nnano.2009.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang H, Jia Z, Wu C, Zang L, Yang G, Chen Z, Tang B. ACS Appl Mater Interfaces. 2015;7:20477–20484. doi: 10.1021/acsami.5b06874. [DOI] [PubMed] [Google Scholar]
- 33.Baker MK, Mikhitarian K, Osta W, Callahan K, Hoda R, Brescia F, Kneuper-Hall R, Mitas M, Cole DJ, Gillanders WE. Clin Cancer Res. 2003;9:4865–4871. [PubMed] [Google Scholar]
- 34.Hosokawa M, Hayata T, Fukuda Y, Arakaki A, Yoshino T, Tanaka T, Matsunaga T. Anal Chem. 2010;82:6629–6635. doi: 10.1021/ac101222x. [DOI] [PubMed] [Google Scholar]
- 35.Hosokawa M, Yoshikawa T, Negishi R, Yoshino T, Koh Y, Kenmotsu H, Naito T, Takahashi T, Yamamoto N, Kikuhara Y, Kanbara H, Tanaka T, Yamaguchi K, Matsunaga T. Anal Chem. 2013;85:5692–5698. doi: 10.1021/ac400167x. [DOI] [PubMed] [Google Scholar]
- 36.Chen W, Weng S, Zhang F, Allen S, Li X, Bao L, Lam RHW, Macoska JA, Merajver SD, Fu J. ACS Nano. 2013;7:566–575. doi: 10.1021/nn304719q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wan Y, Winter M, Delalat B, Hardingham JE, Grover PK, Wrin J, Voelcker NH, Price TJ, Thierry B. ACS Appl Mater Interfaces. 2014;6:20828–20836. doi: 10.1021/am505201s. [DOI] [PubMed] [Google Scholar]
- 38.Yoo CE, Moon HS, Kim YJ, Park JM, Park D, Han KY, Park K, Sun JM, Park WY. Biomaterials. 2016;75:271–278. doi: 10.1016/j.biomaterials.2015.10.033. [DOI] [PubMed] [Google Scholar]
- 39.Park JM, Kim MS, Moon HS, Yoo CE, Park D, Kim YJ, Han KY, Lee JY, Oh JH, Kim SS, Park WY, Lee WY, Huh N. Anal Chem. 2014;86:3735–3742. doi: 10.1021/ac403456t. [DOI] [PubMed] [Google Scholar]
- 40.Kim YJ, Koo GB, Lee JY, Moon HS, Kim DG, Lee DG, Lee JY, Oh JH, Park JM, Kim MS, Woo HG, Kim SI, Kang P, Choi W, Sim TS, Park WY, Lee JG, Kim YS. Biomaterials. 2014;35:7501–7510. doi: 10.1016/j.biomaterials.2014.05.039. [DOI] [PubMed] [Google Scholar]
- 41.Cheng SB, Xie M, Xu JQ, Wang J, Lv SW, Guo S, Shu Y, Wang M, Dong WG, Huang WH. Anal Chem. 2016;88:6773–6780. doi: 10.1021/acs.analchem.6b01130. [DOI] [PubMed] [Google Scholar]
- 42.Liu X, Wang S. Chem Soc Rev. 2014;43:2385–2401. doi: 10.1039/c3cs60419e. [DOI] [PubMed] [Google Scholar]
- 43.Zheng F, Cheng Y, Wang J, Lu J, Zhang B, Zhao Y, Gu Z. Adv Mater. 2014;26:7333–7338. doi: 10.1002/adma.201403530. [DOI] [PubMed] [Google Scholar]
- 44.Balasubramanian S, Kagan D, Hu CMJ, Campuzano S, Lobo-Castanon MJ, Lim N, Kang DY, Zimmerman M, Zhang L, Wang J. Angew Chem Int Ed. 2011;50:4161–4164. doi: 10.1002/anie.201100115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nagrath S, Sequist LV, Maheswaran S, Bell DW, Irimia D, Ulkus L, Smith MR, Kwak EL, Digumarthy S, Muzikansky A, Ryan P, Balis UJ, Tompkins RG, Haber DA, Toner M. Nature. 2007;450:1235–1239. doi: 10.1038/nature06385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yoon HJ, Kim TH, Zhang Z, Azizi E, Pham TM, Paoletti C, Lin J, Ramnath N, Wicha MS, Hayes DF, Simeone DM, Nagrath S. Nat Nanotechnol. 2013;8:735–741. doi: 10.1038/nnano.2013.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yoon HJ, Shanker A, Wang Y, Kozminsky M, Jin Q, Palanisamy N, Burness ML, Azizi E, Simeone DM, Wicha MS, Kim J, Nagrath S. Adv Mater. 2016;28:4891–4897. doi: 10.1002/adma.201600658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lv SW, Liu Y, Xie M, Wang J, Yan XW, Li Z, Dong WG, Huang WH. ACS Nano. 2016;10:6201–6210. doi: 10.1021/acsnano.6b02208. [DOI] [PubMed] [Google Scholar]
- 49.Park JM, Lee JY, Lee JG, Jeong H, Oh JM, Kim YJ, Park D, Kim MS, Lee HJ, Oh JH, Lee SS, Lee WY, Huh N. Anal Chem. 2012;84:7400–7407. doi: 10.1021/ac3011704. [DOI] [PubMed] [Google Scholar]
- 50.Kim J, Cho H, Han SI, Han KH. Anal Chem. 2016;88:4857–4863. doi: 10.1021/acs.analchem.6b00570. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


