The synthesis and crystal structure of 7-hydroxy-3-(2-methoxyphenyl)-2-trifluoromethyl-4H-chromen-4-one, C17H11F3O4, are reported. This isoflavone is used as a starting material in the preparation an array of potent and competitive FPR antagonists.
Keywords: crystal structure, isoflavone, chromone
Abstract
Herein, the synthesis and crystal structure of 7-hydroxy-3-(2-methoxyphenyl)-2-trifluoromethyl-4H-chromen-4-one, C17H11F3O4, are reported. This isoflavone is used as a starting material in the preparation an array of potent and competitive FPR antagonists. The pyran ring significantly deviates from planarity and the dihedral angle between the benzopyran mean plane and that of the exocyclic benzene ring is 88.18 (4)°. In the crystal, O—H⋯O hydrogen bonds connect the molecules into C(8) chains propagating in the [010] direction.
Chemical context
Isoflavones are a subclass of a larger chemical family, the flavonoids, being characterized by possessing a 3-phenylchromen-4-one (3-phenyl-1,4-benzopyrone) backbone instead of the 2-phenylchromen-4-one (3-phenyl-1,4-benzopyrone) structure of flavanones and flavones (Szeja et al., 2016 ▸). Dietary isoflavones are secondary metabolites that occur in plants of the Fabaceae family and as such are present in soy beans, soy foods and legumes. The health benefits of isoflavones have been linked to cholesterol-reducing, anti-inflammatory, chemotherapeutic and antioxidant properties (Jie et al., 2016 ▸). However, the best known property of isoflavones is related to their phytoestrogenic activity (Vitale et al., 2013 ▸). More recently, isoflavones of synthetic origin have been shown to be potent and competitive antagonists of formyl peptide receptors (FPRs), playing an important role in the regulation of inflammatory processes (Schepetkin et al., 2014 ▸).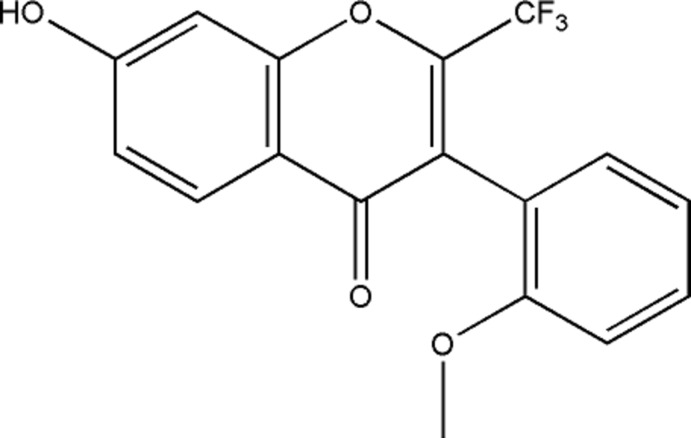
Herein we describe the synthesis and characterization of an isoflavone, 7-hydroxy-3-(2-methoxyphenyl)-2-trifluoromethyl-4H-chromen-4-one, 1, a precursor used in the preparation of relevant FPRs antagonists.
Structural commentary
The molecular structure of 1 is shown in Fig. 1 ▸ (left). This compound consists of a chromone core with several substituents, viz. a trifluoromethyl group at position 2, a 2-(methoxy)phenyl at position 3 and finally an hydroxy group at position 7 of the chromone ring.
Figure 1.
The molecular structure of 1 (left) and (right)the rotation of the exocyclic benzene relative to the benzopyran best plane [dihedral angle = 88.18 (4)°]. Displacement ellipsoids are drawn at the 70% probability level.
The pyran ring is not planar as the weighted average absolute torsion angle is 6.77 (7)°; for planarity this should be below 5.00° (Domenicano et al., 1975 ▸). In fact, as a Cremer & Pople puckering analysis shows, the pyran ring has a twist-boat pucker with puckering amplitude Q = 0.1085 (13) Å, θ = 90.0 (7)° and φ = 148.6 (7)°. The fused aromatic benzopyran ring system shows a slight distortion from planarity as a result of the puckering of the pyran ring. The dihedral angle between the pyran ring and the exocyclic benzene ring is 89.26 (6)°. The mean plane of the ten atoms of the benzopyran ring system was used to evaluate the degree of twisting of the 2-methoxyphenyl ring in relation to the chromone as depicted in Fig. 1 ▸ (right). The dihedral angle between the benzopyran mean plane and the exocyclic benzene ring is 88.18 (4)°; the major rotation is around the C3—C31 bond, which has sp 3 character, with a bond length of 1.4983 (17) Å. This conformation is to be expected and probably results from minimization of the steric hindrance between the 2-methoxy substituent of the exocyclic benzene ring with the voluminous –CF3 group and/or the oxo oxygen atom of the chromone ring.
Regarding the mean plane involving the benzopyran atoms, it is found that atoms O1 and C3 lie more than 0.1 Å out of it [the perpendicular vectors having values of 0.1039 (9) Å and −0.1398 (10) Å, respectively], showing again that the benzopyran ring itself does not show the typical planarity observed for similar chromone and coumarin structures (e.g. Gomes et al., 2016 ▸; Reis et al., 2013 ▸) in which the pyran and benzopyran ring systems are essentially planar. As can be seen in Table 1 ▸, the atoms of the pyran ring lie below the mean plane of the chromone benzene ring.
Table 1. Deviations (in Å) of the pyran ring atoms and attached atoms from the mean plane of the chromone benzene ring.
| Atom | O1 | C2 | C3′ | C4 | C21 | C31 | O4 |
|---|---|---|---|---|---|---|---|
| Distance | −0.0092 (18) | −0.243 (2) | −0.380 (2) | −0.155 (2) | −0.372 (3) | −0.805 (3) | −0.1267 (16) |
Supramolecular features
Details of the hydrogen-bonding interactions are given in Table 2 ▸. The O7—H7⋯O4( − x, −
− x, − + y, z) link forms a C(8) chain, which runs parallel to the b axis. This is formed by the action of the c-glide plane at y =
+ y, z) link forms a C(8) chain, which runs parallel to the b axis. This is formed by the action of the c-glide plane at y =  , Fig. 2 ▸.
, Fig. 2 ▸.
Table 2. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O7—H7⋯O4i | 0.87 (2) | 1.79 (2) | 2.6416 (13) | 166 (2) |
| C6—H6⋯O3ii | 0.95 | 2.59 | 3.4309 (16) | 148 |
| C36—H36⋯O7iii | 0.95 | 2.49 | 3.3886 (18) | 157 |
| C8—H8⋯Cg3iv | 0.95 | 2.93 | 3.5972 (14) | 128 |
| C321—H32b⋯Cg3v | 0.98 | 2.75 | 3.6348 (17) | 150 |
Symmetry codes: (i)  ; (ii)
; (ii)  ; (iii)
; (iii)  ; (iv)
; (iv)  ; (v)
; (v)  .
.
Figure 2.
Compound 1, the simple C8 chain formed by the O7—H7⋯O4i hydrogen bond. This chain extends along the b axis. Symmetry codes: (i) −x +  , y −
, y −  , z; (ii) −x +
, z; (ii) −x +  , y +
, y +  , z). Hydrogen atoms not involved in the hydrogen bonding have been omitted.
, z). Hydrogen atoms not involved in the hydrogen bonding have been omitted.
The molecules are linked into alternating pairs of dimers to form a ladder. The C36—H36⋯O7(1 − x, 1 − y, 1 − z) interaction forms an  (20) centrosymmetric dimer across the centre of symmetry at (
(20) centrosymmetric dimer across the centre of symmetry at ( ,
,  ,
,  ). The C6—H6⋯O3(−x, 1 − y, 1 − z) interaction forms an
). The C6—H6⋯O3(−x, 1 − y, 1 − z) interaction forms an  (18) centrosymmetric dimer across the centre of symmetry at (0,
(18) centrosymmetric dimer across the centre of symmetry at (0,  ,
,  ). Together, these interactions form the ladder, which lies in plane (011) and which runs parallel to the a axis, Fig. 3 ▸. There are also C—H⋯π interactions present (Table 2 ▸).
). Together, these interactions form the ladder, which lies in plane (011) and which runs parallel to the a axis, Fig. 3 ▸. There are also C—H⋯π interactions present (Table 2 ▸).
Figure 3.
Compound 1, view of the ladder of alternating linked  (18) and
(18) and  (20) structures formed by the interaction of centrosymmetrically related pairs of C6—H6⋯O3ii hydrogen bonds across the centre of symmetry at (0,
(20) structures formed by the interaction of centrosymmetrically related pairs of C6—H6⋯O3ii hydrogen bonds across the centre of symmetry at (0,  ,
,  ) and centrosymmetrically related pairs of C36—H36⋯O7iii hydrogen bonds across the centre of symmetry at (
) and centrosymmetrically related pairs of C36—H36⋯O7iii hydrogen bonds across the centre of symmetry at ( ,
,  ,
,  ). This chain extends by unit translation along the a axis. Hydrogen atoms not involved in the hydrogen bonding have been omitted.
). This chain extends by unit translation along the a axis. Hydrogen atoms not involved in the hydrogen bonding have been omitted.
Hirshfeld surfaces
The Hirshfeld surfaces and two-dimensional fingerprint (FP) plots (McKinnon et al., 2004 ▸) provide complementary information concerning the intermolecular interactions discussed above. They were generated using Crystal Explorer 3.1 (Wolff et al., 2012 ▸). The Hirshfeld surface mapped over d norm is scaled between −0.250 to 1.200. The electrostatic potential (ESP) was calculated with TONTO (Jayatilaka & Grimwood, 2003 ▸) as implemented in Crystal Explorer 3.
The contributions from various contacts, listed in Table 3 ▸, were selected by the partial analysis of the FP plots. Besides the H⋯H contacts the other most significant contacts are the H⋯F/F⋯H due to the fluorine atoms on the surface. The remaining high percentage contacts are H⋯O/O⋯H that include the relevant C—H⋯O and the O—H⋯O intermolecular interactions and also the H⋯C/C⋯H contacts including C—H⋯C contacts. The percentage of C⋯C contacts is 6.1% but they are too long to be considered as π–π stacking. The structure has four oxygen atoms, defining different functional groups, that may act as acceptors for hydrogen bonds: one oxo group, a methoxy group, a hydroxyl group and an alkoxy O atom, all of which participate in short atom–atom contacts with the exception of the chromone alkoxy O atom.
Table 3. Percentages for the most relevant atom–atom contacts in 1 .
| H⋯H’ | H⋯O/O⋯H | H⋯F/F⋯H | H⋯C/C⋯H | C⋯C | C⋯O/O⋯C | O⋯F/F⋯C | O⋯O | F⋯O/O⋯F | F⋯F |
|---|---|---|---|---|---|---|---|---|---|
| 22.1 | 18.3 | 25.1 | 18.3 | 6.1 | 2.3 | 0.2 | 3.0 | 0.8 | 3.4 |
The Hirshfeld surfaces mapped over d norm for 1 (see Fig. 4 ▸) show three sets of complementary red spot areas: one of those pairs consist of two intense red areas, circular in shape, that are located near the carbonyl oxygen atom O4 and near the hydroxyl substituent. This close contact accounts for the O4⋯H7—O7 hydrogen bond as indicated in Fig. 4 ▸. Another pair is consists of two light-red areas resulting from the overlap of two red spots near H36 and near the hydroxyl group; they suggest interactions between this hydrogen atom and O7 (that forms a C36—H36⋯O7 hydrogen contact) and with the carbon atom C7 of the chromone ring (Fig. 5 ▸ contains a detail of this contact and the corresponding FP plot area). Finally, there are two complementary very light-red spots of small diameter that suggest the existence of a close contact involving the oxygen atom O3 of the methoxy substituent with hydrogen atom H8 of the chromone ring (O3⋯H6—C6).
Figure 4.
Views of the Hirshfeld surface mapped over d norm for 1 and the corresponding FP plot. The highlighted red spots with large area on the top left image indicate O⋯H contact points involving the carbonyl oxygen atom of the chromone core and the hydrogen atom of the hydroxyl substituent while the pair of superposed light-red spots indicate C⋯H and O⋯H close contacts. The small red-spot areas on the concave and convex face of the right image are due to C⋯H close contacts. The bottom of the figure presents the FP plot for molecule 1 The light-blue area in the middle of the FP plot at d e/d i ∼ 1.9 Å shows a higher frequency of the pixels that are due to C⋯C contacts. The sharp spikes pointing to southwest are due to O⋯H contacts: the inner one on the right is related to O⋯H contacts and the short wings due to C⋯H close contacts (see Fig. 5 ▸ for details).
Figure 5.
Detail of the FP plot for 1 highlighting the H⋯C contacts (blue) and the corresponding areas in the Hirshfeld surface; The blue area covers the C⋯H/ H⋯C close contacts and displays two small wings as well as a pair of short spikes pointing to southwest ending at (d e/ d i)/(1.5/1.0) Å and vice versa that reflect the C36—H36⋯C7 contact area.
The FP plot for 1 is included in Fig. 4 ▸. The light-blue area in the middle of it at d e/d i approximately equal to 1.9 Å shows a higher frequency of the pixels that are due to C⋯C contacts. The sharp spikes pointing to the southwest are due to O⋯H contacts and the short wings due to C⋯H close contacts (see Fig. 5 ▸ for details).
In Fig. 6 ▸ the mapping of the molecular electrostatic potential (ESP) in the context of crystal packing is shown. As the Hirshfeld surface partitions of the crystal space give non-overlapping volumes associated with each molecule, these surfaces give a kind of ‘electrostatic complementarity’; red areas indicate negative electrostatic potential while blue areas indicate a positive one. The ESP mapped in the Hirshfeld surface for 1 reveals a red area of strongly negative electrostatic potential surrounding the carbonyl region of the chromone and light red areas surrounding the fluorine atoms of the –CF3 and as well on the areas covering the oxygen atoms of the hydroxyl and methoxy substituents showing the negative electrostatic potential. The blue region, strongly electropositive, is predominantly located on the hydrogen atom of the hydroxyl substituent and the light electropositive blue patch areas are also surrounding the H atoms of the methoxy substituent and well as H8 and H6 hydrogen atoms of the chromone. The remainder of the Hirshfeld surface is close to neutrality as seen by the grey regions. Thus, the figures highlight the electrostatic complementarity in the O4⋯H7—O7 contact as well as in the O3⋯H6—C6 contact.
Figure 6.
The electrostatic potential surfaces for 1 (ranging from −0.0920 to 0.2582 a.u.). The surfaces show the complementary of electropositive area (blue) near the hydrogen atom of the hydroxyl substituent and red electronegative area surrounding the vicinity of the lone pairs of the oxo oxygen atom O4.
Database survey
A search made in the Cambridge Structural Database, (Groom et al., 2016 ▸), revealed the existence of seven polymorphic and pseudopolymorphic crystal structures of 7-hydroxy-3-phenyl-4H-chromen-4-one (Gong et al., 2016 ▸). In these structures, the pyran rings are essentially planar. The dihedral angles between the benzopyran ten-membered ring mean plane and the exocyclic benzene ring are given below. KUZJEW, 7-hydroxy-3-phenyl-4H-chromen-4-one: 2-methylpropan-2-ol solvate, dihedral angle= 48.28 (7)°. KUZJIA, 7-hydroxy-3-phenyl-4H-chromen-4-one, dihedral angle = 55.23 (8)°. KUZJIA01, 7-hydroxy-3-phenyl-4H-chromen-4-one, dihedral angle = 56.83 (7)°(molecule A), 48.27 (6)° (molecule B). KUZNIE, 7-hydroxy-3-phenyl-4H-chromen-4-one; dimethyl sulfoxide solvate, dihedral angle = 45.91 (7)°. KUZJUM, 7-hydroxy-3-phenyl-4H-chromen-4-one N,N-dimethylformamide solvate, dihedral angle = 41.70 (7)°. KUZKAT, 7-hydroxy-3-phenyl-4H-chromen-4-one propan-1-ol, solvate, dihedral angle = 45.18 (9)°. KUZKEX, 7-hydroxy-3-phenyl-4H-chromen-4-one butan-1-ol solvate, dihedral angle = 45.11 (11)°. In all solvated structures, the 7-OH hydroxyl group is involved in hydrogen bonding with the solvent. In the two KUZJIA(01) structures, –OH⋯O chains are formed as in 1.
Synthesis and crystallization
The title compound was obtained by a two-step synthesis (Balasubramanian & Nair, 2000 ▸; Eiffe et al., 2009 ▸). Resorcinol and 2-methoxyphenylacetic acid, in equimolar amounts, were suspended in boron trifluoride diethyl etherate (BF3·Et2O) and heated at 358 K, for 90 min. Then the mixture was poured into water and stirred until the formation of a solid and extracted with ethyl acetate. The combined organic phases were washed with water, dried over anhydrous sodium sulfate, filtered and evaporated. The product was recrystallized from ethyl acetate solution and used in the subsequent reaction to obtain the isoflavone.
1-(2,4-Dihydroxyphenyl)-2-(2-methoxyphenyl)ethan-1-one (1 mmol), trifluoroacetic anhydride (3 mmol) and trimethylamine (2 ml) were refluxed for 1 h. After cooling, water (15 ml) was added. The solution was acidified (pH 5) with 2 M HCl and stirred at room temperature for 2 h. After extraction with ethyl acetate, the combined organic phases were washed with water, dried over anhydrous sodium sulfate, filtered and evaporated. The isoflavone was recrystallized from ethyl acetate solution. Overall yield: 55%
1H NMR (DMSO-d 6): 3.69 (1H, s), 6.95 (1H, d, J = 2.20 Hz), 7.00 (1H, ddd, J = 0.97, 7.45, 7.45 Hz), 7.02 (1H, dd, J = 2.26, 8.82 Hz), 7.09 (1H, dd, J = 0.90, 8.38 Hz), 7.15 (1H, dd, J = 1.70, 7.46 Hz), 7.42 (1H, ddd, J = 1.73, 7.47, 8.31 Hz), 7.92 (1H, d, J = 8.76 Hz), 11.13 (1H, s).
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 4 ▸. The hydroxyl H atom, H7, was refined isotropically. All other H atoms were treated as riding atoms: C—H = 0.95–0.98 Å with U iso = 1.5U eq(C-methyl) and 1.2U eq(C) for other H atoms.
Table 4. Experimental details.
| Crystal data | |
| Chemical formula | C17H11F3O4 |
| M r | 336.27 |
| Crystal system, space group | Orthorhombic, P b c a |
| Temperature (K) | 100 |
| a, b, c (Å) | 7.9147 (5), 16.2171 (11), 22.5254 (16) |
| V (Å3) | 2891.2 (3) |
| Z | 8 |
| Radiation type | Mo Kα |
| μ (mm−1) | 0.14 |
| Crystal size (mm) | 0.21 × 0.07 × 0.01 |
| Data collection | |
| Diffractometer | Rigaku AFC12 |
| Absorption correction | Multi-scan (CrystalClear-SM Expert; Rigaku, 2012 ▸) |
| T min, T max | 0.775, 1.000 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 35487, 3314, 2777 |
| R int | 0.058 |
| (sin θ/λ)max (Å−1) | 0.649 |
| Refinement | |
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.036, 0.103, 1.02 |
| No. of reflections | 3314 |
| No. of parameters | 222 |
| H-atom treatment | H atoms treated by a mixture of independent and constrained refinement |
| Δρmax, Δρmin (e Å−3) | 0.34, −0.21 |
Supplementary Material
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S2056989017009896/hb7688sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989017009896/hb7688Isup2.hkl
Supporting information file. DOI: 10.1107/S2056989017009896/hb7688Isup3.cml
CCDC reference: 1453086
Additional supporting information: crystallographic information; 3D view; checkCIF report
Acknowledgments
The authors thank the staff at the National Crystallographic Service, University of Southampton, for the data collection, help and advice (Coles & Gale, 2012 ▸).
supplementary crystallographic information
Crystal data
| C17H11F3O4 | Dx = 1.545 Mg m−3 |
| Mr = 336.27 | Mo Kα radiation, λ = 0.71075 Å |
| Orthorhombic, Pbca | Cell parameters from 31907 reflections |
| a = 7.9147 (5) Å | θ = 2.5–27.5° |
| b = 16.2171 (11) Å | µ = 0.14 mm−1 |
| c = 22.5254 (16) Å | T = 100 K |
| V = 2891.2 (3) Å3 | Plate, colourless |
| Z = 8 | 0.21 × 0.07 × 0.01 mm |
| F(000) = 1376 |
Data collection
| Rigaku AFC12 diffractometer | 3314 independent reflections |
| Radiation source: Rotating Anode | 2777 reflections with I > 2σ(I) |
| Detector resolution: 28.5714 pixels mm-1 | Rint = 0.058 |
| profile data from ω–scans | θmax = 27.5°, θmin = 2.5° |
| Absorption correction: multi-scan (CrystalClear-SM Expert; Rigaku, 2012) | h = −10→10 |
| Tmin = 0.775, Tmax = 1.000 | k = −20→20 |
| 35487 measured reflections | l = −29→29 |
Refinement
| Refinement on F2 | 0 restraints |
| Least-squares matrix: full | Hydrogen site location: mixed |
| R[F2 > 2σ(F2)] = 0.036 | H atoms treated by a mixture of independent and constrained refinement |
| wR(F2) = 0.103 | w = 1/[σ2(Fo2) + (0.0543P)2 + 1.0823P] where P = (Fo2 + 2Fc2)/3 |
| S = 1.02 | (Δ/σ)max < 0.001 |
| 3314 reflections | Δρmax = 0.34 e Å−3 |
| 222 parameters | Δρmin = −0.21 e Å−3 |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| F21 | 0.37922 (12) | 0.49225 (7) | 0.27664 (4) | 0.0476 (3) | |
| F22 | 0.59157 (13) | 0.56872 (5) | 0.29861 (4) | 0.0459 (3) | |
| F23 | 0.59737 (12) | 0.43950 (5) | 0.31761 (4) | 0.0416 (2) | |
| O1 | 0.38627 (12) | 0.44652 (5) | 0.40256 (4) | 0.0248 (2) | |
| O3 | 0.13208 (12) | 0.65679 (5) | 0.33431 (4) | 0.0276 (2) | |
| O4 | 0.29964 (14) | 0.66084 (5) | 0.49162 (4) | 0.0323 (2) | |
| O7 | 0.08054 (13) | 0.29227 (6) | 0.54664 (4) | 0.0293 (2) | |
| H7 | 0.114 (3) | 0.2531 (13) | 0.5233 (9) | 0.055 (6)* | |
| C2 | 0.42085 (16) | 0.52119 (7) | 0.37796 (6) | 0.0235 (3) | |
| C3 | 0.38708 (16) | 0.59517 (7) | 0.40248 (6) | 0.0232 (3) | |
| C4 | 0.31834 (16) | 0.59616 (7) | 0.46352 (6) | 0.0243 (3) | |
| C5 | 0.17944 (17) | 0.50964 (7) | 0.54149 (6) | 0.0253 (3) | |
| H5 | 0.1607 | 0.5575 | 0.5649 | 0.030* | |
| C4A | 0.26922 (16) | 0.51638 (7) | 0.48767 (5) | 0.0230 (3) | |
| C6 | 0.11873 (17) | 0.43488 (7) | 0.56045 (6) | 0.0256 (3) | |
| H6 | 0.0579 | 0.4311 | 0.5967 | 0.031* | |
| C7 | 0.14709 (17) | 0.36333 (7) | 0.52579 (6) | 0.0248 (3) | |
| C8 | 0.24070 (17) | 0.36720 (7) | 0.47361 (6) | 0.0244 (3) | |
| H8 | 0.2647 | 0.3190 | 0.4512 | 0.029* | |
| C8A | 0.29805 (16) | 0.44429 (7) | 0.45536 (5) | 0.0224 (3) | |
| C21 | 0.49789 (19) | 0.50614 (7) | 0.31732 (6) | 0.0277 (3) | |
| C31 | 0.40573 (17) | 0.67677 (7) | 0.37198 (6) | 0.0244 (3) | |
| C32 | 0.26803 (16) | 0.70816 (7) | 0.33969 (5) | 0.0237 (3) | |
| C33 | 0.27695 (18) | 0.78732 (8) | 0.31499 (6) | 0.0274 (3) | |
| H33 | 0.1837 | 0.8090 | 0.2935 | 0.033* | |
| C34 | 0.42290 (19) | 0.83407 (8) | 0.32209 (6) | 0.0303 (3) | |
| H34 | 0.4285 | 0.8878 | 0.3055 | 0.036* | |
| C35 | 0.56005 (19) | 0.80327 (8) | 0.35305 (6) | 0.0314 (3) | |
| H35 | 0.6600 | 0.8352 | 0.3571 | 0.038* | |
| C36 | 0.55002 (18) | 0.72477 (8) | 0.37832 (6) | 0.0290 (3) | |
| H36 | 0.6433 | 0.7039 | 0.4002 | 0.035* | |
| C321 | −0.01472 (19) | 0.69009 (9) | 0.30555 (7) | 0.0343 (3) | |
| H32A | −0.1047 | 0.6486 | 0.3052 | 0.051* | |
| H32B | 0.0137 | 0.7053 | 0.2647 | 0.051* | |
| H32C | −0.0533 | 0.7391 | 0.3271 | 0.051* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| F21 | 0.0446 (6) | 0.0703 (7) | 0.0280 (5) | 0.0018 (5) | −0.0036 (4) | −0.0067 (4) |
| F22 | 0.0623 (6) | 0.0276 (4) | 0.0478 (5) | −0.0090 (4) | 0.0258 (5) | −0.0027 (4) |
| F23 | 0.0549 (6) | 0.0325 (4) | 0.0373 (5) | 0.0188 (4) | 0.0129 (4) | 0.0034 (3) |
| O1 | 0.0320 (5) | 0.0160 (4) | 0.0265 (5) | 0.0010 (3) | 0.0023 (4) | −0.0003 (3) |
| O3 | 0.0260 (5) | 0.0259 (4) | 0.0308 (5) | 0.0002 (4) | −0.0032 (4) | 0.0043 (4) |
| O4 | 0.0432 (6) | 0.0178 (4) | 0.0357 (5) | −0.0001 (4) | 0.0037 (4) | −0.0044 (4) |
| O7 | 0.0375 (6) | 0.0178 (4) | 0.0327 (5) | −0.0016 (4) | 0.0040 (4) | 0.0008 (4) |
| C2 | 0.0248 (6) | 0.0188 (6) | 0.0269 (6) | 0.0001 (5) | −0.0016 (5) | 0.0017 (4) |
| C3 | 0.0228 (6) | 0.0189 (5) | 0.0280 (6) | 0.0008 (4) | −0.0030 (5) | 0.0005 (5) |
| C4 | 0.0251 (6) | 0.0184 (5) | 0.0294 (6) | 0.0017 (5) | −0.0031 (5) | −0.0006 (5) |
| C5 | 0.0307 (7) | 0.0191 (5) | 0.0262 (6) | 0.0022 (5) | −0.0019 (5) | −0.0027 (5) |
| C4A | 0.0257 (6) | 0.0174 (5) | 0.0258 (6) | 0.0016 (5) | −0.0030 (5) | −0.0012 (4) |
| C6 | 0.0299 (7) | 0.0223 (6) | 0.0245 (6) | 0.0027 (5) | 0.0006 (5) | −0.0003 (5) |
| C7 | 0.0270 (7) | 0.0188 (5) | 0.0286 (6) | 0.0008 (5) | −0.0037 (5) | 0.0018 (5) |
| C8 | 0.0296 (7) | 0.0161 (5) | 0.0275 (6) | 0.0012 (5) | −0.0022 (5) | −0.0018 (4) |
| C8A | 0.0246 (6) | 0.0197 (6) | 0.0231 (6) | 0.0016 (5) | −0.0017 (5) | −0.0007 (4) |
| C21 | 0.0336 (7) | 0.0202 (6) | 0.0294 (6) | 0.0015 (5) | 0.0020 (5) | 0.0004 (5) |
| C31 | 0.0296 (7) | 0.0176 (5) | 0.0258 (6) | 0.0016 (5) | 0.0017 (5) | 0.0001 (4) |
| C32 | 0.0276 (6) | 0.0203 (5) | 0.0231 (6) | 0.0016 (5) | 0.0025 (5) | −0.0006 (4) |
| C33 | 0.0339 (7) | 0.0223 (6) | 0.0259 (6) | 0.0055 (5) | 0.0028 (5) | 0.0019 (5) |
| C34 | 0.0426 (8) | 0.0196 (6) | 0.0287 (7) | 0.0007 (5) | 0.0081 (6) | 0.0019 (5) |
| C35 | 0.0361 (7) | 0.0220 (6) | 0.0361 (7) | −0.0056 (5) | 0.0043 (6) | −0.0028 (5) |
| C36 | 0.0301 (7) | 0.0222 (6) | 0.0346 (7) | 0.0000 (5) | −0.0015 (5) | −0.0007 (5) |
| C321 | 0.0292 (7) | 0.0375 (7) | 0.0362 (7) | 0.0034 (6) | −0.0058 (6) | 0.0056 (6) |
Geometric parameters (Å, º)
| F21—C21 | 1.3314 (17) | C6—C7 | 1.4164 (17) |
| F22—C21 | 1.3256 (15) | C6—H6 | 0.9500 |
| F23—C21 | 1.3371 (15) | C7—C8 | 1.3908 (19) |
| O1—C2 | 1.3594 (15) | C8—C8A | 1.3921 (17) |
| O1—C8A | 1.3796 (15) | C8—H8 | 0.9500 |
| O3—C32 | 1.3662 (15) | C31—C36 | 1.3895 (19) |
| O3—C321 | 1.4356 (16) | C31—C32 | 1.4057 (18) |
| O4—C4 | 1.2340 (15) | C32—C33 | 1.4008 (17) |
| O7—C7 | 1.3514 (15) | C33—C34 | 1.391 (2) |
| O7—H7 | 0.87 (2) | C33—H33 | 0.9500 |
| C2—C3 | 1.3475 (17) | C34—C35 | 1.384 (2) |
| C2—C21 | 1.5158 (18) | C34—H34 | 0.9500 |
| C3—C4 | 1.4788 (18) | C35—C36 | 1.3967 (18) |
| C3—C31 | 1.4983 (17) | C35—H35 | 0.9500 |
| C4—C4A | 1.4564 (16) | C36—H36 | 0.9500 |
| C5—C6 | 1.3723 (17) | C321—H32A | 0.9800 |
| C5—C4A | 1.4094 (18) | C321—H32B | 0.9800 |
| C5—H5 | 0.9500 | C321—H32C | 0.9800 |
| C4A—C8A | 1.3960 (16) | ||
| C2—O1—C8A | 118.46 (9) | F22—C21—F21 | 107.78 (11) |
| C32—O3—C321 | 116.62 (10) | F22—C21—F23 | 106.92 (12) |
| C7—O7—H7 | 107.1 (13) | F21—C21—F23 | 106.39 (11) |
| C3—C2—O1 | 125.88 (12) | F22—C21—C2 | 112.84 (10) |
| C3—C2—C21 | 126.33 (11) | F21—C21—C2 | 111.33 (12) |
| O1—C2—C21 | 107.75 (10) | F23—C21—C2 | 111.26 (10) |
| C2—C3—C4 | 117.63 (11) | C36—C31—C32 | 119.18 (11) |
| C2—C3—C31 | 125.38 (12) | C36—C31—C3 | 121.91 (12) |
| C4—C3—C31 | 116.93 (10) | C32—C31—C3 | 118.73 (11) |
| O4—C4—C4A | 122.10 (12) | O3—C32—C33 | 124.28 (12) |
| O4—C4—C3 | 122.03 (11) | O3—C32—C31 | 115.83 (10) |
| C4A—C4—C3 | 115.84 (10) | C33—C32—C31 | 119.89 (12) |
| C6—C5—C4A | 120.84 (11) | C34—C33—C32 | 119.71 (13) |
| C6—C5—H5 | 119.6 | C34—C33—H33 | 120.1 |
| C4A—C5—H5 | 119.6 | C32—C33—H33 | 120.1 |
| C8A—C4A—C5 | 117.78 (11) | C35—C34—C33 | 120.85 (12) |
| C8A—C4A—C4 | 120.36 (11) | C35—C34—H34 | 119.6 |
| C5—C4A—C4 | 121.66 (11) | C33—C34—H34 | 119.6 |
| C5—C6—C7 | 119.78 (12) | C34—C35—C36 | 119.34 (13) |
| C5—C6—H6 | 120.1 | C34—C35—H35 | 120.3 |
| C7—C6—H6 | 120.1 | C36—C35—H35 | 120.3 |
| O7—C7—C8 | 122.67 (11) | C31—C36—C35 | 121.02 (13) |
| O7—C7—C6 | 116.44 (12) | C31—C36—H36 | 119.5 |
| C8—C7—C6 | 120.88 (11) | C35—C36—H36 | 119.5 |
| C7—C8—C8A | 117.63 (11) | O3—C321—H32A | 109.5 |
| C7—C8—H8 | 121.2 | O3—C321—H32B | 109.5 |
| C8A—C8—H8 | 121.2 | H32A—C321—H32B | 109.5 |
| O1—C8A—C8 | 116.31 (10) | O3—C321—H32C | 109.5 |
| O1—C8A—C4A | 120.69 (10) | H32A—C321—H32C | 109.5 |
| C8—C8A—C4A | 123.01 (12) | H32B—C321—H32C | 109.5 |
| C8A—O1—C2—C3 | 4.95 (19) | C4—C4A—C8A—O1 | 5.63 (18) |
| C8A—O1—C2—C21 | −172.99 (10) | C5—C4A—C8A—C8 | 0.76 (19) |
| O1—C2—C3—C4 | 5.0 (2) | C4—C4A—C8A—C8 | −174.13 (12) |
| C21—C2—C3—C4 | −177.39 (12) | C3—C2—C21—F22 | 25.4 (2) |
| O1—C2—C3—C31 | −171.91 (12) | O1—C2—C21—F22 | −156.64 (11) |
| C21—C2—C3—C31 | 5.7 (2) | C3—C2—C21—F21 | −95.91 (15) |
| C2—C3—C4—O4 | 172.60 (12) | O1—C2—C21—F21 | 82.03 (13) |
| C31—C3—C4—O4 | −10.19 (19) | C3—C2—C21—F23 | 145.61 (13) |
| C2—C3—C4—C4A | −9.24 (17) | O1—C2—C21—F23 | −36.46 (15) |
| C31—C3—C4—C4A | 167.97 (11) | C2—C3—C31—C36 | −95.96 (16) |
| C6—C5—C4A—C8A | −1.73 (19) | C4—C3—C31—C36 | 87.07 (16) |
| C6—C5—C4A—C4 | 173.08 (12) | C2—C3—C31—C32 | 88.86 (16) |
| O4—C4—C4A—C8A | −177.67 (12) | C4—C3—C31—C32 | −88.10 (14) |
| C3—C4—C4A—C8A | 4.17 (18) | C321—O3—C32—C33 | −5.40 (18) |
| O4—C4—C4A—C5 | 7.6 (2) | C321—O3—C32—C31 | 175.24 (11) |
| C3—C4—C4A—C5 | −170.51 (12) | C36—C31—C32—O3 | 178.68 (11) |
| C4A—C5—C6—C7 | 0.3 (2) | C3—C31—C32—O3 | −6.01 (17) |
| C5—C6—C7—O7 | −178.66 (12) | C36—C31—C32—C33 | −0.71 (18) |
| C5—C6—C7—C8 | 2.2 (2) | C3—C31—C32—C33 | 174.60 (11) |
| O7—C7—C8—C8A | 177.81 (12) | O3—C32—C33—C34 | −178.71 (12) |
| C6—C7—C8—C8A | −3.06 (19) | C31—C32—C33—C34 | 0.63 (18) |
| C2—O1—C8A—C8 | 169.42 (11) | C32—C33—C34—C35 | 0.3 (2) |
| C2—O1—C8A—C4A | −10.35 (17) | C33—C34—C35—C36 | −1.2 (2) |
| C7—C8—C8A—O1 | −178.16 (11) | C32—C31—C36—C35 | −0.2 (2) |
| C7—C8—C8A—C4A | 1.6 (2) | C3—C31—C36—C35 | −175.30 (12) |
| C5—C4A—C8A—O1 | −179.48 (11) | C34—C35—C36—C31 | 1.1 (2) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| O7—H7···O4i | 0.87 (2) | 1.79 (2) | 2.6416 (13) | 166 (2) |
| C6—H6···O3ii | 0.95 | 2.59 | 3.4309 (16) | 148 |
| C36—H36···O7iii | 0.95 | 2.49 | 3.3886 (18) | 157 |
| C8—H8···Cg3iv | 0.95 | 2.93 | 3.5972 (14) | 128 |
| C321—H32b···Cg3v | 0.98 | 2.75 | 3.6348 (17) | 150 |
Symmetry codes: (i) −x+1/2, y−1/2, z; (ii) −x, −y+1, −z+1; (iii) −x+1, −y+1, −z+1; (iv) x, −y−3/2, z−1/2; (v) x−3/2, y, −z−1/2.
Funding Statement
This work was funded by Portuguese Foundation for Science and Technology (FCT) grant UID/Multi/04546/2013. FEDER/COMPETE grants UID/QUI/UI0081/2013 and POCI-01-0145-FEDER-006980.
References
- Balasubramanian, S. & Nair, M. G. (2000). Synth. Commun. 30, 469–484.
- Coles, S. J. & Gale, P. A. (2012). Chem. Sci. 3, 683–689.
- Domenicano, A., Vaciago, A. & Coulson, C. A. (1975). Acta Cryst. B31, 221–234.
- Eiffe, E., Heaton, A., Walker, C. & Husband, A. (2009). US Patent WO 2009003229 A1.
- Gomes, L. R., Low, J. N., Fonseca, A., Matos, M. J. & Borges, F. (2016). Acta Cryst. E72, 1121–1125. [DOI] [PMC free article] [PubMed]
- Gong, N., Zhang, G., Jin, G., Du, G. & Lu, Y. (2016). J. Pharm. Sci. 105, 1387–1397. [DOI] [PubMed]
- Groom, C. R., Bruno, I. J., Lightfoot, M. P. & Ward, S. C. (2016). Acta Cryst. B72, 171–179. [DOI] [PMC free article] [PubMed]
- Hübschle, C. B., Sheldrick, G. M. & Dittrich, B. (2011). J. Appl. Cryst. 44, 1281–1284. [DOI] [PMC free article] [PubMed]
- Jayatilaka, D. & Grimwood, D. J. (2003). ICCS, 4, 142–151.
- Jie Yu, J., Bi, X. J., Yu, B. & Chen, D. (2016). Nutrients, 8, 361–365. [DOI] [PMC free article] [PubMed]
- Macrae, C. F., Edgington, P. R., McCabe, P., Pidcock, E., Shields, G. P., Taylor, R., Towler, M. & van de Streek, J. (2006). J. Appl. Cryst. 39, 453–457.
- McArdle, P., Gilligan, K., Cunningham, D., Dark, R. & Mahon, M. (2004). CrystEngComm, 6, 303–309.
- McKinnon, J. J., Spackman, M. A. & Mitchell, A. S. (2004). Acta Cryst. B60, 627–668. [DOI] [PubMed]
- Reis, J., Gaspar, A., Borges, F., Gomes, L. R. & Low, J. N. (2013). Acta Cryst. C69, 1527–1533. [DOI] [PubMed]
- Rigaku (2012). CrystalClear-SM Expert. Rigaku Corporation, Tokyo, Japan.
- Schepetkin, I. A., Kirpotina, L. N., Khlebnikov, A. I., Cheng, N., Ye, R. D. & Quinn, M. T. (2014). Biochem. Pharmacol. 92, 627–641. [DOI] [PMC free article] [PubMed]
- Sheldrick, G. M. (2015a). Acta Cryst. A71, 3–8.
- Sheldrick, G. M. (2015b). Acta Cryst. C71, 3–8.
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
- Szeja, W., Grynkiewicz, G. & Rusin, A. (2016). Curr. Org. Chem. 21, 218–235. [DOI] [PMC free article] [PubMed]
- Vitale, D. C., Piazza, C., Melilli, B., Drago, F. & Salomone, S. (2013). Eur. J. Drug Metab. Pharmacokinet. 38, 15–25. [DOI] [PubMed]
- Wolff, S. K., Grimwood, D. J., McKinnon, J. J., Turner, M. J., Jayatilaka, D. & Spackman, M. A. (2012). Crystal Explorer. The University of Western Australia.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I, global. DOI: 10.1107/S2056989017009896/hb7688sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989017009896/hb7688Isup2.hkl
Supporting information file. DOI: 10.1107/S2056989017009896/hb7688Isup3.cml
CCDC reference: 1453086
Additional supporting information: crystallographic information; 3D view; checkCIF report








