Abstract
Background
Immune‐mediated hemolytic anemia (IMHA) in dogs has a high risk of thrombosis and is associated with marked neutrophilia and necrosis. Cell death and release of neutrophil extracellular traps contribute to increased serum concentrations of cell‐free DNA, and in human autoimmune disease reduced DNase activity further increases cell‐free DNA. Free DNA in blood has prothrombotic properties and could contribute to hypercoagulability in IMHA.
Hypothesis
Cell‐free DNA is elevated and DNase activity reduced in dogs with IMHA compared to healthy dogs.
Animals
Dogs presenting to two referral hospitals with IMHA (n = 28) and healthy controls (n = 20).
Methods
Prospective observational study. Blood was collected and death and thrombotic events occurring in the first 14 days after hospitalization recorded. DNA was extracted from plasma with a commercial kit and quantified by PicoGreen fluorescence. DNase activity of serum was measured by radial diffusion assay.
Results
Cell‐free DNA was significantly higher in cases (median: 45 ng/mL, range: 10–2334 ng/mL) than controls (26 ng/mL, range 1–151 ng/mL, P = 0.0084). DNase activity was not different between cases and controls (P = 0.36). Four cases died and there were five suspected or confirmed thrombotic events. Cell‐free DNA concentration was associated with death (odds ratio for upper quartile versus lower 3 quartiles: 15; 95% confidence interval 1.62–201; P = 0.03) but not thrombosis (P = 0.57).
Conclusions and Clinical Importance
Cell‐free DNA is elevated in dogs with IMHA and likely reflects increased release rather than impaired degradation of DNA. Cell‐free DNA concentration is potentially associated with death and might be a prognostic indicator, but this requires confirmation in a larger population.
Keywords: Autoimmunity, Neutrophil extracellular trap, Prognosis, Thrombosis
Abbreviations
- DNA
deoxyribonucleic acid
- EDTA
ethylenediaminetetraacetic acid
- IMHA
immune‐mediated hemolytic anemia
- NET
neutrophil extracellular trap
- SLE
systemic lupus erythematosus
Dogs affected by immune‐mediated hemolytic anemia (IMHA) frequently die or are euthanized during, or shortly after, initial hospitalization.1, 2, 3 Thrombosis, necrosis, and neutrophilic inflammation are common postmortem findings.4 Both necrotic cells and activated neutrophils release DNA, either as a consequence of cell membrane breakdown or, in the case of neutrophils, through organized release of webs of DNA, histones, and extranuclear proteins known as neutrophil extracellular traps (NETs).5, 6, 7, 8 Cell‐free DNA released by these processes might contribute to thrombosis in dogs with IMHA. Cell‐free DNA might be further elevated in dogs with IMHA because of reduced DNA digestion by endogenous DNase 1, as occurs in human autoimmune diseases.9, 10
Cell‐free DNA has multiple prothrombotic properties, including activation of the contact pathway of coagulation11, 12 and inhibition of fibrinolysis.13, 14 Increased circulating DNA is associated with poor prognosis in humans with a variety of conditions including disseminated intravascular coagulopathy15 and sepsis,16 and blood concentrations of cell‐free DNA are correlated with disease severity in sick dogs.17 If cell‐free DNA is similarly associated with outcome in IMHA, the evidence that DNA has direct prothrombotic effects suggests it could provide a specific marker of thrombotic risk. As such, elevated cell‐free DNA might be capable of identifying those dogs with IMHA where the benefits of anticoagulant treatment outweigh the financial costs and risk of treatment‐associated hemorrhage. Furthermore, digestion of DNA by DNase improves outcome in murine models of thrombosis.18, 19, 20 DNase is already available as an inhalational treatment for humans with cystic fibrosis and the same product has been delivered systemically by intravenous and intraperitoneal routes in experimental animals.19 Cell‐free DNA might, therefore, provide a marker of thrombotic risk that could help to select dogs requiring aggressive antithrombotic treatment and provide a novel therapeutic target in dogs with IMHA.
This study tests the hypothesis that cell‐free DNA is increased in IMHA compared with healthy control dogs and seeks to determine if this is associated with reduced endogenous DNase activity. The study objectives were (1) to compare circulating cell‐free DNA concentration and serum DNase activity of IMHA cases and healthy controls; (2) to determine if DNase activity was negatively correlated with cell‐free DNA; (3) to determine if cell‐free DNA was positively correlated with D‐dimers, and (4) to provide a preliminary analysis of the association between cell‐free DNA and survival or suspected or confirmed thrombosis in the first 14 days after admission.
Methods
Ethical Permissions
Samples were collected with owner consent and ethical permission from the Iowa State and North Carolina State University institutional animal care and use committees (healthy controls: ISU 12‐13‐17687‐K; IMHA cases: NCSU 14.110‐O, ISU 1‐14‐7693K).
Inclusion Criteria for Clinical Cases
Clinical cases presenting to Iowa State University Lloyd Veterinary Medical Center or North Carolina State University Terry Center were prospectively enrolled between 1.2.14 and 8.26.14 respectively and 5.27.16, providing they met the following criteria: (1) anemia with one or more of (a) positive saline agglutination test, (b) positive Coombs test, or (c) ≥5 spherocytes per ×100 field; (2) no evidence of underlying disease; (3) no heparin treatment before sample collection and (4) at least 3 kg body weight or for dogs at North Carolina State, at least 7 kg if concurrently enrolled in another study to avoid collection of potentially excessive blood volume. Dogs which met these requirements but were not enrolled were recorded.
A saline agglutination test was considered positive if macro‐ or microscopic agglutination was present after adding four drops of saline to a single drop of EDTA blood. Coombs tests were performed at the clinical pathology laboratory of each institution, and considered positive if the titer was above the cut‐off established by that laboratory. Before experimental analysis of samples, all blood smears were examined for spherocytes by a single board‐certified clinical pathologist to confirm enrolled dogs met criteria for inclusion (UJ). Diagnostic investigations to confirm IMHA was primary were at the discretion of the attending clinician. Animals which had previously received heparin were excluded because of the potential for heparin to degrade NETs.21 Treatment after sample collection was at the discretion of the attending clinician. Diagnostic investigations and treatments after sampling were recorded from electronic medical records with further clarification from the attending clinician where necessary.
Inclusion Criteria for Healthy Controls
Healthy control dogs were pets of staff or students at Iowa State University, inclusion criteria were as follows: (1) aged between 1 and 9 years; (2) >5 kg body weight; (3) normal physical exam, hematology, biochemistry, and urinalysis, and (4) not receiving medication other than routine antiparasite treatment.
Sample Collection
Samples were collected by minimally traumatic venipuncture via 21‐gauge winged infusion sets1 inserted into a jugular, saphenous, or cephalic vein for all control dogs and either by the same method or via central sampling catheters for IMHA cases. Sampling via central lines was permitted only if flushed with saline rather than heparin, and a discard sample was collected to clear residual blood or flush fluid from the line before study sample collection. Blood was collected directly into sodium citrate2 (1:9 ratio, 3.8% sodium citrate) for measurement of D‐dimers and collected into a syringe then immediately transferred to K2EDTA3 for measurement of cell‐free DNA and a plain tube4 for the DNase assay. Within 20 minutes of collection, EDTA and citrate samples were centrifuged at 2500 × g for 20 minutes and serum samples for 1000 × g for 15 minutes. Plasma or serum was divided into aliquots and stored at −80°C until analysis.
Follow‐Up
Dogs were followed for 14 days after admission by clinical record review or contacting the owner or attending clinician. Survival to 14 days or date of death/euthanasia; suspected or confirmed thrombotic events; transfusion; and treatment with immunosuppressants, heparin, aspirin, clopidogrel, or intravenous immunoglobulin were recorded. Thrombosis was considered confirmed if at least one thrombus was documented grossly or microscopically postmortem, and suspected if the dog developed acute onset respiratory distress or neurologic signs consistent with a central nervous lesion or if a board‐certified radiologist considered thrombosis was the primary differential diagnosis for a finding on radiographs or ultrasound.
Sample Size Calculation
The study was initially designed principally to evaluate the association between cell‐free DNA and survival, with comparison between controls and cases as a secondary analysis. This would have required at least 80 IMHA cases. After 27 months, case recruitment was below targets and study design was revised to determine if analytes differed between cases and controls. To determine if case numbers were sufficient for this comparison, a sample size calculation based on reported cell‐free DNA concentrations and standard deviations for healthy humans and patients with systemic lupus22 was performed by G*Power v. 3.1.95.23 For cell‐free DNA, at least 12 cases and 12 controls were needed to detect a 49 ng/mL difference between the mean of cases and controls with an alpha of 0.05 and power of 80% assuming a standard deviation of 40 ng/mL.
Cell‐Free DNA
DNA was extracted from 200 μL EDTA‐plasma samples with a silica‐membrane‐based nucleic acid purification kit6 according to the manufacturer's instructions. Eluted DNA was quantified with the fluorescent double‐stranded DNA PicoGreen.7 Where necessary, samples were diluted to bring results into the linear range of the assay. All results are reported after correction for any dilutions.
DNase Activity
DNase activity was measured by a method modified from Macanovic and Lachmann.24 Briefly, 300 μL of a solution containing calf thymus DNA (final concentration 0.25 mg/mL); DNase reaction buffer (final concentrations 20 mM Tris–HCl; 1 mM CaCl2; 1 mM Mg Cl2); low melting point agarose8 (final concentration 1% weight by volume); and ethidium bromide9 (final concentration 1 μg/mL) were added to each well of a 24‐well plate10 and allowed to set. Serum (2 μL/well) was injected into the center of each test well and plates incubated for 19 hours at 37°C. Blank wells in which no solution was injected were included in each plate. Fluorescence of DNA‐bound ethidium bromide was imaged by ultraviolet transillumination. Controls and the first 25 cases were prepared on a single day, and camera settings were constant between each plate. The final three IMHA cases were analyzed as a separate experiment together with serum from a previously assayed control dog. Results were normalized to the previous result from the control dog by the formula [DNA lysis area for case*(average DNA lysis area for control, assay 2/average DNA lysis area for control, assay 1)]. Wells were set up in duplicate for each dog and the average result used in statistical analysis.
Images were analyzed in Image J.11 Histogram analysis was performed for a blank well on the plate of interest, and the threshold of the entire image adjusted so pixels equal to or brighter than the lowest gray value of the blank well were fully saturated, representing completely undigested DNA. The image was calibrated using known dimensions of the plate. Conversion to a binary image was then performed and the Analyze Particles plug‐in for Image J used to measure the area of the black circle at the center of the test well, which corresponds to the area of DNA lysis.
To determine the coefficient of variation, 10 duplicates were prepared with 1 U of DNase12 per well. To confirm the method could identify differences in DNase activity, a standard curve was prepared using 0.125, 0.25, 0.5, 0.75, 1, and 2 U of DNase per well. Attempts were made to assess effects of hemolysis and icterus using 0.5 U/μL DNase diluted in a 9 to 1 ratio with DNase‐free water, a commercial bilirubin control13 (final concentration 2.5 mg/dL) or hemoglobin (final concentration 0.18 g/dL or 0.35 g/dL) prepared from washed canine red cells by repeated freeze thaw cycles and osmotic lysis.
D‐Dimer Measurement
D‐dimers were measured in citrated plasma from cases using a quantitative immunoturbidimetric assay14 performed at Comparative Coagulation Section of the Animal Health Diagnostic Center, Cornell University.
Statistical Analysis
All analyses were performed by GraphPad Prism (v.7).15 Distribution was tested by the D'Agostino and Pearson normality test, and as one or both populations were non‐normal for cell‐free DNA and DNase activity, healthy controls and IMHA cases were compared by Mann–Whitney U‐tests. Spearman's correlation coefficient was calculated for correlation between cell‐free DNA and DNase activity and between cell‐free DNA and D‐dimer concentrations. Odds ratios and 95% confidence intervals were calculated for death and thrombosis by 14 days for IMHA cases in the upper quartile (n = 7) for cell‐free DNA concentration compared with those in the lower 3 quartiles (n = 21). Fisher's exact test was used to calculate p‐values for odds ratios. Receiver operator characteristic (ROC) curves were automatically constructed for cell‐free DNA as a predictor of death or thrombosis within 14 days, and the area under the curve compared to 0.5 (i.e. expected area if the ability of cell‐free DNA to predict outcome was no better than chance). For all analyses, P < 0.05 was considered significant.
Results
Study Population and Healthy Controls
Thirty dogs were initially recruited. One dog recruited based on spherocytosis was subsequently excluded because on blood‐smear exam, the cut‐off of five spherocytes per high power field was not reached. One other enrolled dog was not included because EDTA plasma was not collected. During the study period, 10 other potentially eligible dogs were not enrolled because caregivers did not consent to enrollment (n = 2); death before diagnostic investigations could be performed (n = 1) or lack of available personnel to process samples (n = 7).
Cases were of the following breeds: Australian Shepherd (n = 1); Beagle (n = 2); Bichon Frise (n = 1); Boston Terrier (n = 2); Cardigan Corgi (n = 1); Dachshund (n = 1); German Shorthaired Pointer (n = 1); Keeshond (n = 1); Labrador (n = 3); Maltese (n = 1); Mastiff breeds (n = 2); Miniature Schnauzer (n = 2); mix (n = 6); Pomeranian (n = 1); Rat Terrier (n = 1); Rottweiler (n = 1); Springer Spaniel (n = 1). For cell‐free DNA measurements, controls were of the following breeds: Border Collie (n = 1); Chesapeake Bay Retriever (n = 1); German Wirehair Pointer (n = 1); Golden Retriever (n = 1); Greyhound (n = 3); Labrador (n = 3); mix (n = 5); Pitbull Terrier (n = 3); Rhodesian Ridgeback (n = 1); Rough Collie (n = 1). Due to sample availability, for DNase activity, the Rhodesian Ridgeback control was replaced with an Australian Shepherd. Median age of cases was 8 years (range 0.3–14 years) and for controls was 4.5 years (range 1–9 years) for cell‐free DNA measurement and 4 (range 1–9 years) for DNase activity studies. Median body weight of cases was 15 kg (range 6–52 kg) and for controls was 28.5 kg (range 8–37 kg) for cell‐free DNA measurement and 30.5 kg (range 8–37 kg) for DNase activity measurement. Of the 28 cases, 14 were spayed females, 12 were neutered males, and two were intact males. For both cell‐free DNA measurement and DNase activity measurement, of the 20 controls, eight were spayed females and 12 were neutered males.
For the 28 included cases, median hematocrit at enrollment was 18% (range 8–28, reference interval 37–55%). All dogs with a hematocrit ≥25% had either received a transfusion (3/4) or long‐term immunosuppression (1/4) before enrollment. Five or more spherocytes per high power field were present in 18/28. Of the four dogs undergoing direct Coombs' testing, one was positive. Saline agglutination was positive for 24/28 and negative for 4/28. Of those dogs with negative saline agglutination tests, 1/4 had historically been positive and had been treated for 56 days before admission; 3/4 had five or more spherocytes per high power field and 1/4 had a positive direct Coomb's tests and spherocytes present but at a level below the threshold used for inclusion.
Median time from initial clinical signs to sample collection was 3 days (range 1–168). Samples were collected on day of admission for 18/28; the following day for 8/28; 3 days later for 1/28 and 4 days later for 1/28. Samples were collected from a peripheral vein in 22/28 IMHA cases and via central line sampling catheters for 6/28 IMHA cases. Based on available diagnostics, all dogs were considered affected by primary rather than secondary IMHA. Diagnostic investigations included abdominal radiographs and/or ultrasound 27/28; thoracic radiographs 20/28; cytology of one or more body system 5/28 (spleen 3/5; liver 1/5; lymph node 1/5; cutaneous masses 1/5, bone marrow 1/5); biochemistry 28/28; complete blood count 28/28; prothrombin time and activated partial thromboplastin time 18/28 and infectious disease testing 27/28 (PCR for Babesia, Bartonella, Ehrlichia, Rickettsia, and hemotropic Mycoplasma organisms16 10/27; point of care ELISA testing for Dirofilaria immitis antigen and antibodies against Borrelia burgdorferi (C6), Ehrlichia sp and Anaplasma sp17 26/27; quantitative ELISA for Borrelia burgdorferi antibodies (C6)18 1/27; Immunofluorescence antibody assays for Ehrlichia canis 16 , 18 12/27; Rickettsia rickettsia 16 , 18 12/27; Babesia canis 16 10/27, Babesia gibsoni 16 10/27 and Bartonella spp16 10/27; Microscopic agglutination testing for antibodies against Leptospira canicola, Leptospira ictero, Leptospira grippo, Leptospira Pomona, and Leptospira hardjo 19 3/27).
Before sample collection, 17/28 had received corticosteroids; 3/28 azathioprine; 3/28 cyclosporine; 16/28 one or more antibiotic; 10/28 intravenous fluid treatment; 7/28 blood product transfusion (2/7 whole blood, 5/7 packed red blood cells); 3/28 clopidogrel; 16/28 one or more other drug. In the 14 days after sample collection or the period between sample collection and death/euthanasia if <14 days, 26/28 received corticosteroids; 6/28 azathioprine; 11/28 cyclosporine; 3/28 mycophenolate mofetil; 4/28 intravenous human immunoglobulin, 24/28 clopidogrel; 13/28 unfractionated heparin; 2/28 aspirin; 13/28 at least one transfusion of packed red blood cells; 1/28 whole blood transfusion and 1/28 fresh frozen plasma transfusion.
Cell‐Free DNA
Median cell‐free DNA was significantly higher in dogs with IMHA (45 ng/mL, range 10–2334 ng/mL, n = 28) than healthy controls (26 ng/mL, range 1–151 ng/mL, n = 20, P = 0.0084) (Fig 1).
Figure 1.
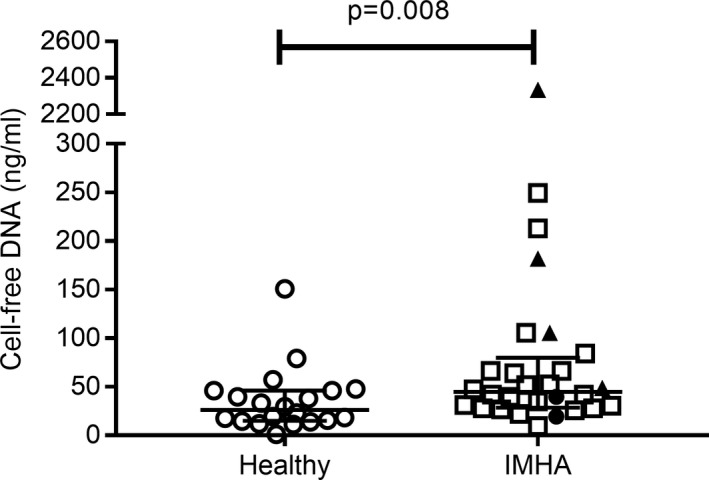
Cell‐free DNA in dogs with IMHA and in healthy dogs. Cases and controls were compared by Mann–Whitney U‐test (P < 0.05 significant; healthy n = 20; IMHA n = 28); error bars are median and interquartile range. Data points plotted as black triangles are from four dogs who did not survive to 14 days; black circles are from two dogs who survived to 14 days but had suspected (n = 1) or confirmed (n = 1) thrombosis; open squares represent dogs with IMHA that survived to 14 days and did not have suspected or confirmed thrombosis (n = 22).
DNase Activity
DNase activity was measured by area of lysis of an ethidium bromide‐containing DNA gel. Validation of the DNase assay produced a coefficient of variation of 6.5% based on 10 duplicates. R 2 for linear regression performed on a standard curve created using DNase concentrations from 0.125 U/well to 2 U/well was 0.94.
Area of lysis was not significantly different between cases (median 0.359 cm2, range 0.229–0.750 cm2) and healthy controls (median 0.365 cm2, range 0.157–0.456 cm2; P = 0.36) (Fig 2A). To determine if DNase results could have been affected by interference from hemoglobin of bilirubin, area of lysis was compared between gels to which water, hemoglobin, or bilirubin were added. Addition of 0.18 g/dL or 0.35 g/dL hemoglobin or 2.5 mg/dL bilirubin to gels increased areas of lysis by 140, 412, and 112% compared with water. These apparent interferences could have been due to presence of DNase within the biologically derived interferent sources or a genuine effect of hemoglobin or bilirubin. For this reason, area of lysis for IMHA cases with (median 0.348 cm2, range 0.229–0.724 cm2; n = 18) and without (median 0.406 cm2; range 0.294–0.750 cm2; n = 10) visible hemolysis and/or icterus was also compared by Mann–Whitney U‐tests and was not significantly different (P = 0.19; Fig 2B).
Figure 2.
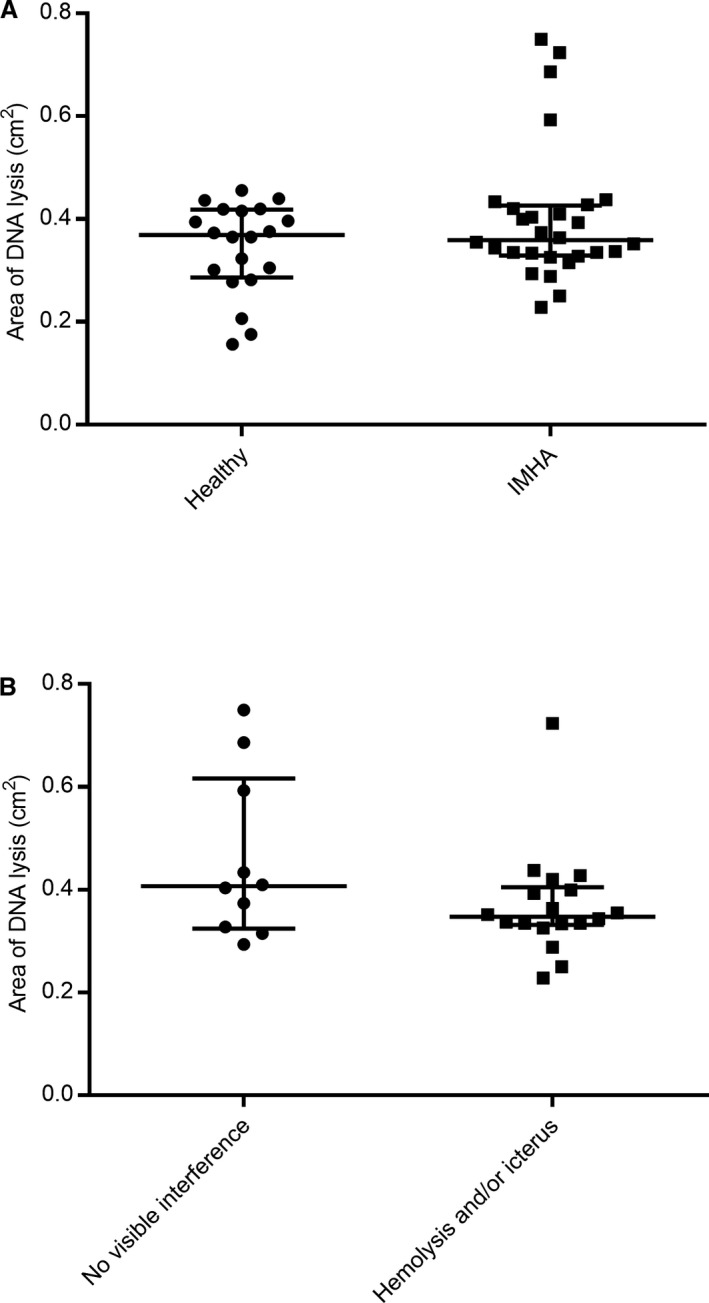
Serum DNase activity as measured by area of DNA lysis. (A) Area of lysis was not significantly different between IMHA cases (n = 28) and controls (n = 20) (P = 0.36). (B) Analysis of interference spikes suggested hemoglobin and bilirubin might both falsely increase area of DNA lysis, but both were biologically derived and might have spontaneous DNase activity making conclusions about their interference challenging. There was no significant difference between area of lysis for IMHA cases with (n = 18) or without (n = 10) visible hemolysis and/or icterus (P = 0.19). Comparisons were by Mann–Whitney U‐test (P < 0.05 significant); error bars are median and interquartile range.
Spearman's correlation coefficient (ρ) for cell‐free DNA and DNase activity was 0.38 (95% confidence interval −0.0006 to 0.67; P = 0.045) but visual inspection of the correlation plot revealed an obvious outlier (Fig 3A). Correlation was no longer significant after exclusion of this dog (Spearman's ρ 0.32, 95% confidence interval −0.08 to 0.63, P = 0.11) (Fig 3B).
Figure 3.
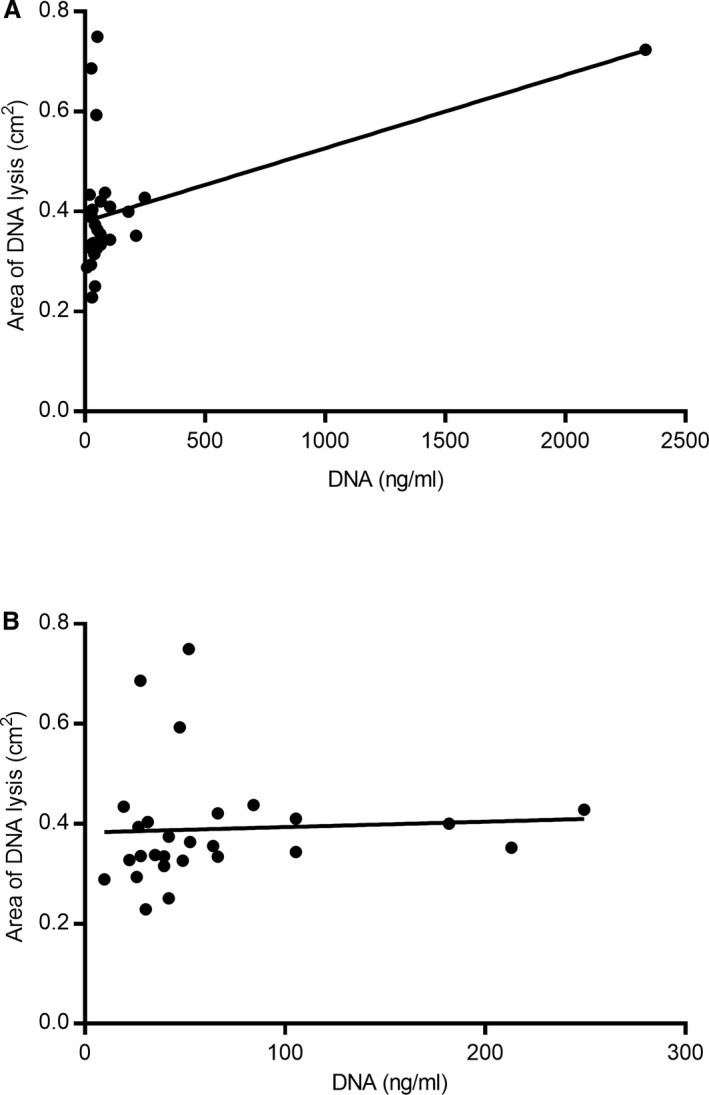
Correlation between cell‐free DNA concentration and area of DNA lysis. (A) Initial analysis suggested there was a positive correlation between cell‐free DNA concentration and DNase activity measured by area of lysis (Spearman's ρ 0.38; P = 0.045). However, this was largely the result of a single outlier. (B) Re‐analysis after exclusion of the outlier found no evidence of a correlation between the two parameters (P = 0.11).
Correlation between Cell‐Free DNA and D‐Dimer
D‐dimers were unmeasurable in four IMHA cases due to hemolytic interference, and in two dogs, insufficient plasma was available for analysis. For the remaining 22 dogs, median D‐dimer concentration was 760 ng/mL (range 120–5729 ng/mL). D‐dimers were not significantly correlated with cell‐free DNA (Spearman's ρ 0.17, P = 0.45).
Preliminary Analysis of Relationship between Cell‐Free DNA Concentration and Outcome
At day 14 postadmission, 25 dogs were alive, two dogs had died (day 3 and 6) and two had been euthanized (day 1 and 7). There were five suspected or confirmed thrombotic events. Necropsy was performed in three of four nonsurvivors and confirmed thrombosis in 3/3 (1/3 leptomeningeal vessels only; 1/3 jugular vein and multiple pulmonary thrombi; 1/3 pulmonary, splenic, hepatic, adrenal, and renal thrombi). The other nonsurvivor was euthanized due to financial constraints and permission for necropsy was not granted; thrombosis was not suspected based on physical exam, thoracic and abdominal radiographs and abdominal ultrasound. For survivors, in the first 14 days, thrombosis was suspected in two dogs based on acute onset neurologic signs. One of these dogs was euthanized after the 14‐day follow‐up period (day 16). Cerebral and renal arteriolar thrombi were confirmed at necropsy.
Of the seven dogs with cell‐free DNA in the upper quartile, 4/7 received unfractionated heparin and clopidogrel in the 14 days after sampling; 1/7 received unfractionated heparin only and 2/7 received no thromboprophylaxis. Of the 21 dogs with cell‐free DNA in the lower three quartiles, 8/21 received unfractionated heparin and clopidogrel; 2/21 received aspirin and clopidogrel; 10/21 received clopidogrel only, and one received no thromboprophylaxis. Of the dogs that died or were euthanized and had confirmed thrombosis, 2/3 received unfractionated heparin and clopidogrel and 1/3 received heparin only. The dog that was euthanized but in whom thrombosis was not suspected received no thromboprophylaxis. Of the two dogs that survived 14 days but where thrombosis was suspected or confirmed, one received clopidogrel and one received no thromboprophylaxis.
The odds ratio for death or euthanasia in dogs in the highest quartile for cell‐free DNA was 15 (95% confidence interval 1.62–201; P = 0.03), and on ROC curve analysis, the area under the curve was significantly different from 0.5 for prediction of death by 14 days (area 0.86, 95% confidence interval 0.69–1, P = 0.02) (Fig 4A). The odds ratio or suspected or confirmed thrombosis was 2.4 (95% confidence interval 0.34–14.3, P = 0.57), and the area under the ROC curve was not significantly different from 0.5 (area 0.57, 95% confidence interval 0.26–0.89, P = 0.61) (Fig 4B).
Figure 4.
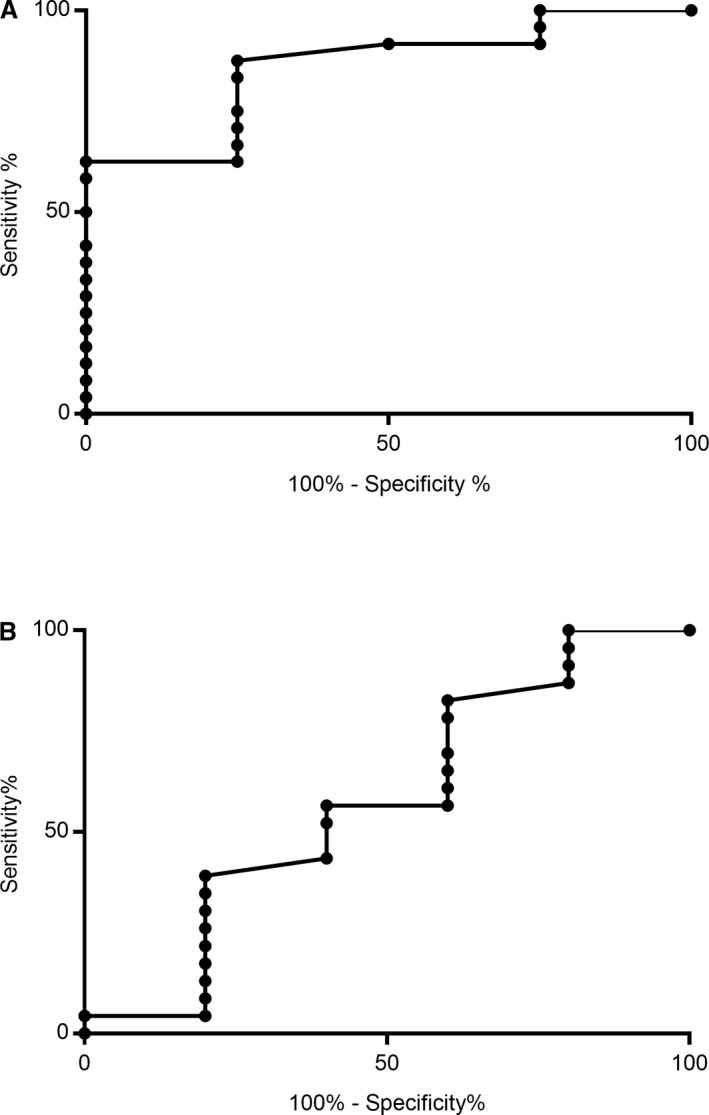
ROC curve analysis for cell‐free DNA as a predictive marker for death (A) or thrombosis (B). Area under the ROC curve was significantly greater than 0.5 for the ability of cell‐free DNA to predict death/euthanasia by day 14 postadmission (four died/euthanized; 24 survived; P = 0.02); but was not significantly different from 0.5 for prediction of confirmed or suspected thrombotic events (five affected; 23 unaffected; P = 0.61).
Discussion
Results of this study indicate that cell‐free DNA concentration was increased in dogs with dogs with IMHA compared with healthy controls. DNase activity was not decreased in dogs with IMHA, nor was it negatively correlated with cell‐free DNA concentration. This suggests increased cell‐free DNA in dogs with IMHA is the result of increased production rather than impaired degradation. The analysis of the ability of cell‐free DNA to predict death or thrombosis should be viewed as pilot data as the number of negative outcomes was low. With this caveat, elevated cell‐free DNA was associated with increased odds of death in the first 14 days, and as the area under the ROC curve was significantly greater than 0.5, it might have value as a prognostic indicator in dogs with IMHA.
Potential sources of cell‐free DNA include necrosis,5 apoptosis,5 and release of extracellular traps by leukocytes, particularly neutrophils.25, 26, 27 Necrosis is well documented at necropsy in dogs with IMHA, with moderate to severe necrosis present in one or more organ in 82% of dogs.4 Elevated serum cardiac troponin I concentration in 74% of IMHA dogs suggests that necrotic myocardial tissue damage is common even in surviving dogs.28 Apoptosis is also likely to contribute to cell‐free DNA as it is induced both by effects of the disease itself, such as oxidative stress29, 30 and by treatment31 Sixty‐one percent of dogs enrolled in the current study received glucocorticoids, a stimulus for lymphocyte apoptosis, before sample collection.31 Twenty‐five percent also received blood products before enrollment. Blood units, particularly nonleukoreduced units, do contain cell‐free DNA but it is not known if the concentration is sufficient to alter circulating cell‐free DNA.32
Although NETs trap bacteria, they are often increased in sterile inflammatory conditions. In humans, circulating markers associated with NETs are increased in many diseases including rheumatoid arthritis,33 systemic lupus erythematosus (SLE),22 transfusion‐related acute lung injury,26 cancer,34 and in intensive care patients.35 Similarly, NET formation is a plausible source of cell‐free DNA in dogs with IMHA. Marked neutrophilia is a common feature of canine IMHA,2, 36, 37 sometimes to the point of a leukemoid response,38 and neutrophilic tissue inflammation is frequent in postmortem specimens.4 Canine neutrophils release NETs in vitro 39 and many of the stimuli known to induce NET formation in humans and experimental animals, including free heme/hemin;40, 41 immune complexes,42 and hypoxia,43 are likely often elevated in IMHA.44, 45, 46, 47
This study also investigated the possibility that impaired clearance contributes to increased cell‐free DNA. Reduced DNA degradation by endogenous DNase enzymes is reported in human SLE10, 48, 49 and antiphospholipid syndrome9 and is associated with disease severity.10, 50, 51 DNA degradation was not significantly decreased in IMHA cases compared with controls. Correlation between cell‐free DNA concentration and DNA lysis was also absent after exclusion of an outlier, or positive if this dog was included. It therefore appears that impaired DNA degradation does not contribute to elevated cell‐free DNA in dogs with IMHA.
It should, however, be noted that we could not completely rule out the possibility that interference from hemolysis or icterus falsely elevated the apparent DNase activity of IMHA sera. We measured DNase activity using a previously published ethidium bromide radial diffusion assay adapted to allow automated quantification of lysis area.24 Addition of hemoglobin and bilirubin did increase the area of lysis. Absorption peaks for hemoglobin are at 415, 540, and 570 nm51 and for bilirubin the peak is between 415 and 440 nm.53 Therefore, hemoglobin in particular might be falsely increasing lysis area by absorption of light emitted by ethidium bromide at close to its emission peak of approximately 600 nm. However, both hemolysis and icterus controls were derived from biological specimens and so might contain DNase 1 activity. Statistical comparison of results for dogs with or without visible hemolysis or icterus did not reveal a significant difference in DNA lysis. Furthermore, serum was diluted 150 to 1 for the DNase assay, so end bilirubin and icterus concentrations would have been low. Therefore, on balance we consider systematic alterations in DNase activity due to hemolysis or icterus unlikely. Effects of lipemia were not investigated as it was not visible in any control or case samples.
Methodologic issues are also an important consideration for cell‐free DNA isolation. We followed recommendations for sample handling,54, 55, 56, 57 except delay between plasma collection and DNA extraction did exceed the recommended interval of 9 months for two dogs (both IMHA cases).57 The difference between cell‐free DNA in cases and controls remained significant after excluding these dogs (Table S1). It has been suggested that direct measurement of DNA in plasma is preferable in canine samples because there is some loss of DNA during extraction.17 Direct measurement of cell‐free DNA in plasma by PicoGreen fluorescence is a simple assay which correlates well with qPCR.58 However, during preliminary studies, we found that direct measurement of DNA in plasma was not reliable in IMHA due to marked interference from hemolysis and icterus with the PicoGreen assay (Table S2). It is also notable that although PicoGreen has been used in human studies without prior isolation of DNA,22, 59 the product's datasheet reports interferences from both albumin and IgG.60
The time involved in DNA isolation and quantification would be acceptable for clinical use, and the equipment required is readily available. The association between cell‐free DNA and survival is therefore intriguing, but it should be emphasized that the number of dogs that died or were euthanized is low. Not only does this mean that the confidence intervals for area under ROC curves and odds ratios are wide, but there is also a risk of a false positive result.61 The possibility of a spurious association between cell‐free DNA and survival is suggested by the lack of a similar relationship between cell‐free DNA and thrombosis or D‐dimers.
On the other hand, accurate diagnosis of thrombosis in dogs is notoriously difficult and the specificity of D‐dimers for thrombosis is limited, making death or euthanasia a more robust measure of prognostic ability.62, 63 Furthermore, this is not the first report of a relationship between cell death and prognosis in dogs: cell‐free DNA has a reported negative predictive value of 97% for death in a population of dogs with various diseases,17 and high cell‐free DNA was a negative prognostic indicator in canine lymphoid neoplasia.64 The data presented here could provide a starting point for a large scale evaluation of cell‐free DNA as a prognostic indicator in IMHA.
The biological plausibility of a relationship between cell‐free DNA and survival in IMHA provides further support for such a study. This is in part because DNA provides an indirect measure of cell death. It will therefore be important in future studies of its prognostic ability to compare performance with simpler measures of hypoxia and tissue damage such as lactate.44 However, DNA itself also has direct deleterious effects. Most notably for IMHA, a disease with a high rate of thrombosis, DNA has been shown to have prothrombotic effects on the human and canine coagulation system.12, 13, 14, 65 The negative charge of DNA provides a surface for contact pathway activation,11, 12 and the DNA backbone of neutrophil extracellular traps locally concentrates tissue factor and a potential activator of tissue factor, protein disulfide isomerase.18 DNA is also antifibrinolytic, partly because it forms inhibitory complexes with fibrin and plasmin.13, 14
This link between cell‐free DNA and hypercoagulability suggests that cell‐free DNA could also be a therapeutic target in IMHA. In experimental models of venous thrombosis, DNase treatment improves outcome.18, 19 Additionally, in a human proof‐of‐concept trial, SLE flare‐ups were reduced by metformin, an inhibitor of mitochondrial neutrophil extracellular trap release.66 Although the current study does show overlap in cell‐free DNA concentrations between IMHA survivors, nonsurvivors and healthy dogs, one nonsurvivor had a marked increase. Therefore, evaluation of therapies that reduce DNA release or accelerate its breakdown might be warranted, at least in a subset of IMHA cases.
It should be emphasised that in the current study, treatment was not standardized except for the requirement that IMHA cases could not receive heparin before study enrollment. The study was not designed to detect the effect of medications on outcome, so to avoid misleading conclusions due to low‐power, statistical analysis of the relationships between cell‐free DNA, treatment and outcome have not been performed. However, it is noteworthy that 5/7 (71%) dogs with cell‐free DNA concentrations in the upper quartile received unfractionated heparin compared with only 8/21 (38%) dogs with cell‐free DNA concentrations in the lower three quartiles. It is possible the more aggressive thromboprophylaxis received by dogs with high cell‐free DNA might have led to underestimation of the strength of the relationship between cell‐free DNA and thrombotic risk. Similarly, variation in immunosuppressive protocols could be a confounding factor. In future studies of the prognostic value of cell‐free DNA or of the utility of anti‐DNA therapies, it will therefore be important to standardize treatment.
In this discussion, we have highlighted several limitations of the study, the most important of which is the low number of enrolled dogs with IMHA that died or were euthanized. There are other factors which might have influenced our results. Firstly, effects of breed, age, or sex on circulating cell‐free DNA have not been reported for dogs, but it is possible that factors other than disease status might alter the concentration of cell‐free DNA. Secondly, seven dogs were excluded due to lack of available staff for sample processing.
In conclusion, this study demonstrates that cell‐free DNA is increased in dogs with IMHA compared with healthy controls and this is likely due to increased production not reduced degradation. Preliminary data suggest that cell‐free DNA might have value in predicting death or euthanasia, but this requires confirmation in a larger independent cohort of dogs with IMHA.
Supporting information
Table S1. Effect of removing samples stored for >9 months.
Table S2. Validation of PicoGreen for measurement of cell‐free DNA
Acknowledgments
We thank the owners and dogs who participated in the study; the clinicians who recruited cases and technicians who assisted with sample collection. The study was partially funded by a Research Incentive Grant from the Department of Veterinary Clinical Sciences, College of Veterinary Medicine, Iowa State University.
Conflict of Interest Declaration
Authors declare no conflict of interest.
Off‐label Antimicrobial Declaration
Authors declare no off‐label use of antimicrobials.
Cases were recruited at Iowa State and North Carolina State University. Laboratory investigations were performed at Iowa State University.
The study was partially funded by a Research Incentive Grant from the Department of Veterinary Clinical Sciences, College of Veterinary Medicine, Iowa State University.
This study was presented at the 2017 ACVIM Forum, National Harbor, Maryland, and the data forms part of Unity Jeffery's PhD thesis.
Footnotes
Surflo winged infusion set, Terumo, Tokyo, Japan
Sodium citrate dehydrate, Sigma‐Aldrich Corp, St. Louis, MO
Plastic whole blood tube with spray‐coated K2EDTA (3 mL), BD Vacutainer, Franklin Lakes, NJ
Serum tube, no additive, BD Vacutainer, Franklin Lakes, NJ
G*Power version 3, Universität Düsseldorf, Germany
QIAamp DNA Blood Mini kit, Qiagen, Valencia, CA
Quant‐iT PicoGreen, Thermofisher scientific, Waltham MA
DNase‐free low melting point agarose, Sigma‐Aldrich Corp, St. Louis, MO
Ethidium Bromide 0.625 mg/mL solution, Thermofisher scientific, Waltham, MA
Corning Inc., Corning, NY
National institutes of health, Bethesda, MD
Pulmozyme (dornase alfa), Genentech, San Francisco, CA
MAS Bilirubin (level 3); Thermoscientific, Waltham, MA
Hemosil D‐Dimer, Instrumentation Laboratories, Orangeburg, NY
GraphPad Software Inc., La Jolla, CA
Canine Comprehensive Panel, Vector Borne Disease Diagnostic Lab, Raleigh, NC
SNAP 4Dx Plus, IDEXX, Westbrook, ME
Tick Profile with Lyme Quant C6 Test, IDEXX, Westbrook, ME
Lepto 5 MAT, Iowa State Veterinary Diagnostic Laboratory, Ames, IA
References
- 1. Weinkle TK, Center SA, Randolph JF, et al. Evaluation of prognostic factors, survival rates, and treatment protocols for immune‐mediated hemolytic anemia in dogs: 151 cases (1993–2002). J Am Vet Med Assoc 2005;226:1869–1880. [DOI] [PubMed] [Google Scholar]
- 2. McAlees TJ. Immune‐mediated haemolytic anaemia in 110 dogs in Victoria, Australia. Aust Vet J 2010;88:25–28. [DOI] [PubMed] [Google Scholar]
- 3. Goggs R, Dennis SG, Di Bella A, et al. Predicting outcome in dogs with primary immune‐mediated hemolytic anemia: Results of a multicenter case registry. J Vet Intern Med 2015;29:1603–1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. McManus PM, Craig LE. Correlation between leukocytosis and necropsy findings in dogs with immune‐mediated hemolytic anemia: 34 cases (1994–1999). J Am Vet Med Assoc 2001;218:1308–1313. [DOI] [PubMed] [Google Scholar]
- 5. Jahr S, Hentze H, Englisch S, et al. DNA fragments in the blood plasma of cancer patients: Quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res 2001;15:1659–1665. [PubMed] [Google Scholar]
- 6. Brinkmann V, Reichard U, Goosmann C, et al. Neutrophil extracellular traps kill bacteria. Science 2004;303:1532–1535. [DOI] [PubMed] [Google Scholar]
- 7. Pilsczek FH, Salina D, Poon KK, et al. A novel mechanism of rapid nuclear neutrophil extracellular trap formation in response to Staphylococcus aureus . J Immunol 2010;185:7413–7425. [DOI] [PubMed] [Google Scholar]
- 8. Yousefi S, Mihalache C, Kozlowski E, et al. Viable neutrophils release mitochondrial DNA to form neutrophil extracellular traps. Cell Death Differ 2009;16:1438–1444. [DOI] [PubMed] [Google Scholar]
- 9. Leffler J, Stojanovich L, Shoenfeld Y, et al. Degradation of neutrophil extracellular traps is decreased in patients with antiphospholipid syndrome. Clin Exp Rheumatol 2014;32:66–70. [PubMed] [Google Scholar]
- 10. Leffler J, Ciacma K, Gullstrand B, et al. A subset of patients with systemic lupus erythematosus fails to degrade DNA from multiple clinically relevant sources. Arthritis Res Ther 2015;17:205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gould TJ, Vu TT, Swystun LL, et al. Neutrophil extracellular traps promote thrombin generation through platelet‐dependent and platelet‐independent mechanisms. Arterioscler Thromb Vasc Biol 2014;34:1977–1984. [DOI] [PubMed] [Google Scholar]
- 12. Oehmcke S, Mörgelin M, Herwald H. Activation of the human contact system on neutrophil extracellular traps. J Innate Immun 2009;1:225–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gould TJ, Vu TT, Stafford AR, et al. Cell‐free DNA modulates clot structure and impairs fibrinolysis in sepsis. Arterioscler Thromb Vasc Biol 2015;35:2544–2553. [DOI] [PubMed] [Google Scholar]
- 14. Varjú I, Longstaff C, Szabó L, et al. DNA, histones and neutrophil extracellular traps exert anti‐fibrinolytic effects in a plasma environment. Thromb Haemost 2015;113:1289–1298. [DOI] [PubMed] [Google Scholar]
- 15. Kim JE, Lee N, Gu JY, et al. Circulating levels of DNA‐histone complex and dsDNA are independent prognostic factors of disseminated intravascular coagulation. Thromb Res 2015;135:1064–1069. [DOI] [PubMed] [Google Scholar]
- 16. Dwivedi DJ, Toltl LJ, Swystun LL, et al., Prognostic utility and characterization of cell‐free DNA in patients with severe sepsis. Crit Care 2012;16:R151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Burnett DL, Cave NJ, Geye KR, Bridges JP. Investigation of cell‐free DNA in canine plasma and its relation to disease. Vet Q 2016;36:122–129. [DOI] [PubMed] [Google Scholar]
- 18. von Brühl ML, Stark K, Steinhart A, et al. Monocytes, neutrophils, and platelets cooperate to initiate and propagate venous thrombosis in mice in vivo. J Exp Med 2012;209:819–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Brill A, Fuchs TA, Savchenko AS, et al. Neutrophil extracellular traps promote deep vein thrombosis in mice. J Thromb Haemost 2012;10:136–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. De Meyer SF, Suidan GL, Fuchs TA, et al. Extracellular chromatin is an important mediator of ischemic stroke in mice. Arterioscler Thromb Vasc Biol 2012;32:1884–1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kolaczkowska E, Jenne CN, Surewaard BG, et al. Molecular mechanisms of NET formation and degradation revealed by intravital imaging in the liver vasculature. Nat Commun 2015;6:6673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang S, Lu X, Shu X, et al. Elevated plasma cfDNA may be associated with active lupus nephritis and partially attributed to abnormal regulation of neutrophil extracellular traps (NETs) in patients with systemic lupus erythematosus. Intern Med 2014;53:2763–2771. [DOI] [PubMed] [Google Scholar]
- 23. Faul F, Erdfelder E, Lang A‐G, Buchner A. G*Power 3: A flexible statistical power analysis program for the social, behavioral, and biomedical sciences. Behav Res Methods 2007;39:175–191. [DOI] [PubMed] [Google Scholar]
- 24. Macanovic M, Lachmann PJ. Measurement of deoxyribonuclease I (DNase) in the serum and urine of systemic lupus erythematosus (SLE)‐prone NZB/NZW mice by a new radial enzyme diffusion assay. Clin Exp Immunol 1997;108:220–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sur Chowdhury C, Hahn S, Hasler P, et al. Elevated levels of total cell‐free DNA in maternal serum samples arise from the generation of neutrophil extracellular traps. Fetal Diagn Ther 2016;40:263–267. [DOI] [PubMed] [Google Scholar]
- 26. Caudrillier A, Kessenbrock K, Gilliss BM, et al. Platelets induce neutrophil extracellular traps in transfusion‐related acute lung injury. J Clin Invest 2012;122:2661–2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Surmiak M, Hubalewska‐Mazgaj M, Wawrzycka‐Adamczyk K, et al. Neutrophil‐related and serum biomarkers in granulomatosis with polyangiitis support extracellular traps mechanism of the disease. Clin Exp Rheumatol 2016;34:S98–S104. [PubMed] [Google Scholar]
- 28. Gow DJ, Gow AG, Bell R, et al. Serum cardiac troponin I in dogs with primary immune‐mediated haemolytic anaemia. J Small Anim Pract 2011;52:259–264. [DOI] [PubMed] [Google Scholar]
- 29. Kannan K, Jain SK. Oxidative stress and apoptosis. Pathophysiology 2000;7:153–163. [DOI] [PubMed] [Google Scholar]
- 30. Pesillo SA, Freeman LM, Rush JE. Assessment of lipid peroxidation and serum vitamin E concentration in dogs with immune‐mediated hemolytic anemia. Am J Vet Res 2004;65:1621–1624. [DOI] [PubMed] [Google Scholar]
- 31. Ammersbach MA, Kruth SA, Sears W, Bienzle D. The effect of glucocorticoids on canine lymphocyte marker expression and apoptosis. J Vet Intern Med 2006;20:1166–1171. [DOI] [PubMed] [Google Scholar]
- 32. Fuchs TA, Alvarez JJ, Martinod K, et al. Neutrophils release extracellular DNA traps during storage of red blood cell units. Transfusion 2013;53:3210–3216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sur Chowdhury C, Giaglis S, Walker UA, et al. Enhanced neutrophil extracellular trap generation in rheumatoid arthritis: Analysis of underlying signal transduction pathways and potential diagnostic utility. Arthritis Res Ther 2014;16:R122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Thålin C, Demers M, Blomgren B, et al. NETosis promotes cancer‐associated arterial microthrombosis presenting as ischemic stroke with troponin elevation. Thromb Res 2016;139:56–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hirose T, Hamaguchi S, Matsumoto N, et al. Presence of neutrophil extracellular traps and citrullinated histone H3 in the bloodstream of critically ill patients. PLoS ONE 2014;9:e111755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Scott‐Moncrieff JC, Treadwell NG, McCullough SM, Brooks MB. Hemostatic abnormalities in dogs with primary immune‐mediated hemolytic anemia. J Am Anim Hosp Assoc 2001;37:220–227. [DOI] [PubMed] [Google Scholar]
- 37. Piek CJ, Brinkhof B, Teske E, et al. High intravascular tissue factor expression in dogs with idiopathic immune‐mediated haemolytic anaemia. Vet Immunol Immunopathol 2011;144:346–354. [DOI] [PubMed] [Google Scholar]
- 38. Reimer ME, Troy GC, Warnick LD. Immune‐mediated hemolytic anemia: 70 cases (1988–1996). J Am Anim Hosp Assoc 1999;35:384–391. [DOI] [PubMed] [Google Scholar]
- 39. Jeffery U, Kimura K, Gray R, et al. Dogs cast NETs too: Canine neutrophil extracellular traps in health and immune‐mediated hemolytic anemia. Vet Immunol Immunopathol 2015;168:262–268. [DOI] [PubMed] [Google Scholar]
- 40. Chen G, Zhang D, Fuchs TA, et al. Heme‐induced neutrophil extracellular traps contribute to the pathogenesis of sickle cell disease. Blood 2014;123:3818–3827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kono M, Saigo K, Takagi Y, et al. Heme‐related molecules induce rapid production of neutrophil extracellular traps. Transfusion 2014;54:2811–2819. [DOI] [PubMed] [Google Scholar]
- 42. Behnen M, Leschczyk C, Möller S, et al. Immobilized immune complexes induce neutrophil extracellular trap release by human neutrophil granulocytes via FcγRIIIB and Mac‐1. J Immunol 2014;193:1954–1965. [DOI] [PubMed] [Google Scholar]
- 43. McInturff AM, Cody MJ, Elliott EA, et al. Mammalian target of rapamycin regulates neutrophil extracellular trap formation via induction of hypoxia‐inducible factor 1α. Blood 2012;120:3118–3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Holahan ML, Brown AJ, Drobatz KJ. The association of blood lactate concentration with outcome in dogs with idiopathic immune‐mediated hemolytic anemia: 173 cases (2003–2006). J Vet Emerg Crit Care (San Antonio) 2010;20:413–420. [DOI] [PubMed] [Google Scholar]
- 45. Harkin KR, Hicks JA, Wilkerson MJ. Erythrocyte‐bound immunoglobulin isotypes in dogs with immune‐mediated hemolytic anemia: 54 cases (2001–2010). J Am Vet Med Assoc 2012;241:227–232. [DOI] [PubMed] [Google Scholar]
- 46. Lobetti R. Changes in the serum urea: Creatinine ratio in dogs with babesiosis, haemolytic anaemia, and experimental haemoglobinaemia. Vet J 2012;191:253–256. [DOI] [PubMed] [Google Scholar]
- 47. Caviezel LL, Raj K, Giger U. Comparison of 4 direct Coombs' test methods with polyclonal antiglobulins in anemic and nonanemic dogs for in‐clinic or laboratory use. J Vet Intern Med 2014;28:583–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Villanueva E, Yalavarthi S, Berthier CC, et al. Netting neutrophils induce endothelial damage, infiltrate tissues, and expose immunostimulatory molecules in systemic lupus erythematosus. J Immunol 2011;187:538–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Chitrabamrung S, Rubin RL, Tan EM. Serum deoxyribonuclease I and clinical activity in systemic lupus erythematosus. Rheumatol Int 1981;1:55–60. [DOI] [PubMed] [Google Scholar]
- 50. Leffler J, Gullstrand B, Jönsen A, et al. Degradation of neutrophil extracellular traps co‐varies with disease activity in patients with systemic lupus erythematosus. Arthritis Res Ther 2013;15:R84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hakkim A, Fürnrohr BG, Amann K, et al. Impairment of neutrophil extracellular trap degradation is associated with lupus nephritis. Proc Natl Acad Sci USA 2010;107:9813–9818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lippi G, Blanckaert N, Bonini P, et al. Haemolysis: An overview of the leading cause of unsuitable specimens in clinical laboratories. Clin Chem Lab Med 2008;46:764–772. [DOI] [PubMed] [Google Scholar]
- 53. Lee KS, Gartner LM. Spectrophotometric characteristics of bilirubin. Pediatr Res 1976;10:782–788. [DOI] [PubMed] [Google Scholar]
- 54. Taback B, O'Day SJ, Hoon DS. Quantification of circulating DNA in the plasma and serum of cancer patients. Ann N Y Acad Sci 2004;1022:17–24. [DOI] [PubMed] [Google Scholar]
- 55. Chiu RW, Poon LL, Lau TK, et al. Effects of blood‐processing protocols on fetal and total DNA quantification in maternal plasma. Clin Chem 2001;47:1607–1613. [PubMed] [Google Scholar]
- 56. Xue X, Teare MD, Holen I, et al. Optimizing the yield and utility of circulating cell‐free DNA from plasma and serum. Clin Chim Acta 2009;404:100–104. [DOI] [PubMed] [Google Scholar]
- 57. El Messaoudi S, Rolet F, Mouliere F, Thierry AR. Circulating cell free DNA: Preanalytical considerations. Clin Chim Acta 2013;424:222–230. [DOI] [PubMed] [Google Scholar]
- 58. Ramachandran K, Speer CG, Fiddy S, et al. Free circulating DNA as a biomarker of prostate cancer: Comparison of quantitation methods. Anticancer Res 2013;33:4521–4529. [PubMed] [Google Scholar]
- 59. Wang H, Sha LL, Ma TT, et al. Circulating level of neutrophil extracellular traps is not a useful biomarker for assessing disease activity in antineutrophil cytoplasmic antibody‐associated vasculitis. PLoS ONE 2016;11:e0148197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Quant‐iT PicoGreen ds DNA reagent and kits . ThermoFisher Scientific. https://tools.thermofisher.com/content/sfs/manuals/mp07581.pdf. Accessed: 6‐3‐2015.
- 61. Button KS, Ioannidis JP, Mokrysz C, et al. Power failure: Why small sample size undermines the reliability of neuroscience. Nat Rev Neurosci 2013;14:365–376. [DOI] [PubMed] [Google Scholar]
- 62. Epstein SE, Hopper K, Mellema MS, Johnson LR. Diagnostic utility of D‐dimer concentrations in dogs with pulmonary embolism. J Vet Intern Med 2013;27:1646–1649. [DOI] [PubMed] [Google Scholar]
- 63. Goggs R, Chan DL, Benigni L, et al. Comparison of computed tomography pulmonary angiography and point‐of‐care tests for pulmonary thromboembolism diagnosis in dogs. J Small Anim Pract 2014;55:190–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Schaefer DM, Forman MA, Kisseberth WC, et al. Quantification of plasma DNA as a prognostic indicator in canine lymphoid neoplasia. Vet Comp Oncol 2007;5:145–155. [DOI] [PubMed] [Google Scholar]
- 65. Jeffery U, LeVine DN. Canine neutrophil extracellular traps enhance clot formation and delay lysis. Vet Pathol 2017; https://doi.org/10.1177/0300985817699860. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 66. Wang H, Li T, Chen S, et al. Neutrophil extracellular trap mitochondrial DNA and its autoantibody in systemic lupus erythematosus and a proof‐of‐concept trial of metformin. Arthritis Rheumatol 2015;67:3190–3200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Effect of removing samples stored for >9 months.
Table S2. Validation of PicoGreen for measurement of cell‐free DNA


