Abstract
Drought is a major abiotic stress that impairs growth and productivity of Italian ryegrass. Comparative analysis of drought responsive proteins will provide insight into molecular mechanism in Lolium multiflorum drought tolerance. Using the iTRAQ-based approach, proteomic changes in tolerant and susceptible lines were examined in response to drought condition. A total of 950 differentially accumulated proteins was found to be involved in carbohydrate metabolism, amino acid metabolism, biosynthesis of secondary metabolites, and signal transduction pathway, such as β-D-xylosidase, β-D-glucan glucohydrolase, glycerate dehydrogenase, Cobalamin-independent methionine synthase, glutamine synthetase 1a, Farnesyl pyrophosphate synthase, diacylglycerol, and inositol 1, 4, 5-trisphosphate, which might contributed to enhance drought tolerance or adaption in Lolium multiflorum. Interestingly, the two specific metabolic pathways, arachidonic acid and inositol phosphate metabolism including differentially accumulated proteins, were observed only in the tolerant lines. Cysteine protease cathepsin B, Cysteine proteinase, lipid transfer protein and Aquaporin were observed as drought-regulated proteins participating in hydrolysis and transmembrane transport. The activities of phospholipid hydroperoxide glutathione peroxidase, peroxiredoxin, dehydroascorbate reductase, peroxisomal ascorbate peroxidase and monodehydroascorbate reductase associated with alleviating the accumulation of reactive oxygen species in stress inducing environments. Our results showed that drought-responsive proteins were closely related to metabolic processes including signal transduction, antioxidant defenses, hydrolysis, and transmembrane transport.
Introduction
Drought is the largest abiotic stress factor leading to reduce the productivity of Italian ryegrass, especially at the seedling stage. The effect of drought stress on plants is usually characterized by reduced leaf water content, decreased cell growth, and inducted oxidative stress [1, 2]. The ability of plants to acclimate to such conditions through appropriate regulation is a key determinant of their survival [3]. Recent research has clearly demonstrated that stress responses rely on the functioning of complex gene networks. When plants regularly experience drought, extensive modification of gene accumulation occurs and results in alterations in protein synthesis (up- or down-regulation) [4]. These proteins react to stress by regulating metabolic homeostasis and detoxifying harmful elements such as reactive oxygen species (ROS) [5, 6]. Hence, identification of responsive proteins involved in drought tolerance is a major interest to plant scientists.
Early proteome analyses used two-dimensional gel electrophoresis for protein separation and tandem mass spectrometry for protein identification [7]. Out of a total of 455 proteins identified, 17 differentially accumulated proteins existed in L. multiflorum and F. arundinacea introgression lines [8]. With the rapid innovations in proteomics new methods have been developed for protein analysis. The iTRAQ (Isobaric tags for relative and absolute quantification) is a quantitative proteomic method for examining multiple samples in a single mass analysis, thereby enable sensitive assessment and quantification of protein levels [9, 10]. Based on the iTRAQ approach, by comparing tolerant and susceptible cultivars, many proteins were discovered that had the potential to enhance resistance in plants [11, 12].
Italian ryegrass (Lolium multiflorum L.) is one of most widespread cultivated cool-season forage grass in the world. Typically, it is grown in a mixture with other grass and legume species to improve pasture quality [13]. In southern China, L. multiflorum is most commonly served as an annual forage crop for feeding [14]. Although Italian ryegrass expresses some levels of drought tolerance, it still suffers a significant reduction of yield under drought conditions [15], and does not match F. arundinacea with respect to the potential of tolerance [16]. This potential, however, can be significantly improved in intergeneric L. multiflorum x F. arundinacea hybrids, and their introgression derivatives [8, 17].
However, there have been few reports on the regulatory mechanisms of drought tolerance at proteome level for Italian ryegrass. By using the iTRAQ-based method, two L. multiflorum lines, drought-tolerant “Abundant 10” and drought susceptible “Adrenalin 11” were used in the study to evaluate differentially accumulated proteins under drought stress. This study provides a novel proteomic data for further dissection the regulatory mechanisms of drought tolerance in Lolium multiflorum response to short-term drought.
Materials and methods
Plant materials and drought treatments
Two L. multiflorum lines, drought-tolerant “Abundant 10” and drought susceptible “Adrenalin 11” were used in this study [18]. Seeds were germinated on filter paper moistened with distilled water in an environment kept at 25°C.Seedlings were then transferred into plastic pots filled with the Hoagland’s nutrient solution and put into growth chambers with a 16/8 hour day-night cycle, a 25/18°C day-night temperature, and relative humidity of 60%.
One half of the 20-day-old seedlings of the two L. Multiflorum lines were grown in aerated hydroponics containing Hoagland’s nutrient solution at 25°C to be used as the control and the remaining seedlings were treated under drought stress condition, by lying on plastic trays and naturally air-drying for 2 hours at 25°C in the growth chamber. Ten individual plants for each L. Multiflorum line were used as biological replicate. We performed two biological replicates for each treatment in the experiment (S1 Fig). Therefore, 20 drought tolerant seedlings under control, 20 drought tolerant seedlings with drought stress treatment, 20 drought susceptible seedlings under control, and 20 drought susceptible seedlings with drought stress treatment, were immediately frozen in liquid nitrogen, and stored at -80°C until protein extraction.
Measurement of antioxidant activity
Three biological replicates were used for each treatment. Hydrogen peroxide content (H2O2), phospholipid hydroperoxide glutathione peroxidase (PHGPx), peroxiredoxin (Prx) and ascorbate peroxidase (APX) activities were assayed separately by hydrogen peroxide, PHGPx, Prx and APX assay kits (Comin Biotechnology Co., Ltd. Suzhou, China) according the manufacturer’s menu. Statistical analysis was performed with one-way ANOVA in SPSS 20.0 and all the data were average means of three independent experiments ± SDs.
Protein extraction
Total proteins of Italian ryegrass samples were extracted with Lysis Buffer 3 containing 1 mM PMSF and 2 mM EDTA, and suspended at 200 W for 15 min. Proteins were isolated by centrifuging at 30000×g for 15 min at 4°C, and were added 5× volume of chilled acetone and 10% (v/v) TCA at -20°C. After two rounds of centrifugation, the supernatant was carefully discarded and the precipitate was washed three times with cold acetone. The protein pellet was air-dried by lyophilization and dissolved in Lysis buffer (7 M urea, 2 M thiourea, 4% NP40, 20 mM Tris-HCl, pH 8.0–8.5). The protein pellet was suspended for 15 min and centrifuged at 4°C at 25000×g for 15 min, and the supernatant was collected. To reduce disulfide bonds in the proteins of the supernatant, 10 mM DTT was added and left at 56°C for 1 hr. Subsequently, 55 mM IAM was added to block the cysteines, so samples were kept in a darkened room for one hour. The supernatant of proteins were kept at -80°C.
iTRAQ labeling and SCX fractionation
Protein samples of 100 μg each was added to 2.5μg Trypsin (Promega, Madison, WI, USA) with a weight ratio of 40 protein: 1 trypsin and kept at 37°C for 4 hr. The peptides were dried using vacuum by Strata X and were reconstituted in 0.5 M TEAB based on the manufacture’s protocol for 8-plex iTRAQ reagent (Applied Biosystems). This included one unit of thawed and reconstituted iTRAQ reagent in 24 μL isopropanol. The peptides were labeled with isobaric tag, which were pooled and dried through vacuum.
SCX chromatography was performed using a LC-20AB HPLC Pump System (Shimadzu, Kyoto, Japan). The iTRAQ-labeled peptide mixtures were reconstituted with 2 mL buffer A (5% ACN, pH 9.8) and loaded onto a 5 um 4.6×250 mm Ultremex SCX column (Phenomenex, USA). The peptides were eluted at a flow rate of 1 mL/min with a gradient of buffers as following: 5% buffer B (95% ACN, pH 9.8) for 10 min, 5–35% buffer B for 40 min. 35–95% buffer B for 1 min. The system was then maintained in buffer B for 3 min before equilibrating with buffer B for 10 min. Elution was monitored by measuring the absorbance at 214 nm, and fractions were collected every 1 min. The eluted peptides were pooled into 20 fractions and desalted.
LC–ESI-MS/MS analysis using the Triple TOF 5600 System
Each fraction was re-suspended in buffer A (2% ACN, 0.1% FA) and centrifuged at 20000 g for 10 min, the final concentration of peptides was approximately 0.5 μg/μL on average. 10 μL supernatant was determined by a LC-20AD Nano-HPLC (Shimadzu, Kyoto, Japan) with an autosampler and the peptides were eluted onto analytical C18 column (inner diameter 75 μm and column length15 cm). The samples were loaded for 4 min, then gradient run from 5% buffer B (96% ACN, 0.1% FA) for 0–8 min, linear gradient to35% B for 8–43 min, keep at 60% B for 43–48 min, and return to 5% B for 55-65min.
Data acquisition was performed using a TripleTOF 5600 System (SCIEX, Framingham, MA, USA) fitted with a Nanospray III source (SCIEX, Framingham, MA,USA) and a pulled quartz tip as the emitter (New Objectives, Woburn, MA, USA). Data was acquired using an ion spray with 2.5 kV voltage and, curtain gas was set at 30 psi, nebulizer gas was set at 15 psi, and the interface heater temperature was 150°C. The MS was operated by a resolving power (RP) of 30,000FWHM for TOF/MS scans. Survey scans were obtained from 250 ms and up to 30 product ion scans (cut-off threshold was 120 counts per second (counts/s). Raw data files were transformed into MGF files using Proteome Discoverer software.
Protein identification and data analysis
The Mascot 2.3.02 search engine (Matrix Science, London, UK; version 2.3.02) was used to identify and quantify proteins. To identify proteins the following parameters were set: 1) a mass tolerance of 2 Da (ppm) was permitted for intact peptide masses, 2) the Peptides matching error was set at 0.05 Da, 3) Gln- > pyro-Glu (N-term Q), Oxidation (M), Deamidated (NQ) were set as potential variable modifications, 4) Carbamidomethyl (C), iTRAQ8plex (N-term), iTRAQ8plex (K) were set as fixed modifications, and 5) the charge states of the peptides were set to +2 and +3. An automatic decoy database search can be performed by choosing the decoy checkbox to produce a random sequence database and to test for raw spectra.
A 95% confidence interval was used to identify peptide using the Mascot probability analysis. Each protein was identified by at least one unique peptide and each protein should contain at least two unique spectra. The protein sequence database (NCBInr (http://www.ncbi.nlm.nih.gov), SwissProt (http://www.ebi.ac.uk/swissprot), and UniProt (http://www.uniprot.org) was used for the protein identifications. The quantitative protein ratios were measured and normalized in Mascot. A 2-fold change, statistical P-values < 0.05 and false discovery rate (FDR) < = 0.01 were used as criteria for identifying differentially accumulated proteins. Functional annotations of identified proteins were performed using Blast2GO against the Non-redundant protein database (NR). The KEGG database and the clusters of orthologous groups (COG) database were applied to classify the identified proteins. The data including the number of identified peptides, peptides masses, peptides sequence, and peptides scores were provided in S1 Table.
Western blotting
For each protein sample 10 ug was loaded on SDS PAGE gel (1.5 mm). Migration of proteins in the PAGE gel was conducted at 150 V until the blue band from the sample buffer run out of the gel. Protein-Marker IV was also loaded to determine the molecular weight of the proteins, Proteins were then transferred onto a Polyvinylidene fluoride (PVDF) membrane (Millipore, USA). The following antibodies were used in the western blot analysis: actin (ACT), ADP-glucose pyrophosphorylase (ADGP), β-amylase, isocitrate lyase (ICL), aquaporin, plasma membrane intrinistic protein 1–3 (PIP1), tonoplast intrinsic protein 1-1(TIP1), heat shock protein 90 (HSP90), dehydroascorbate reductase (DHAR1), and alpha-amylase (from Agrisera, Sweden). The PVDF membrane was probed with primary antibody and developed using enhanced chemilu-minescence detection (PerkinElmer, Waltham MA, USA). The blots were detected using the BeyoECL plus (P0018). The images were obtained with the ChemiDoc TM MP imaging system, and the quantifications were conducted with the software Image Lab TM V5.1.
Results
Changes in activities of antioxidative enzymes in drought treated L. multiflorum
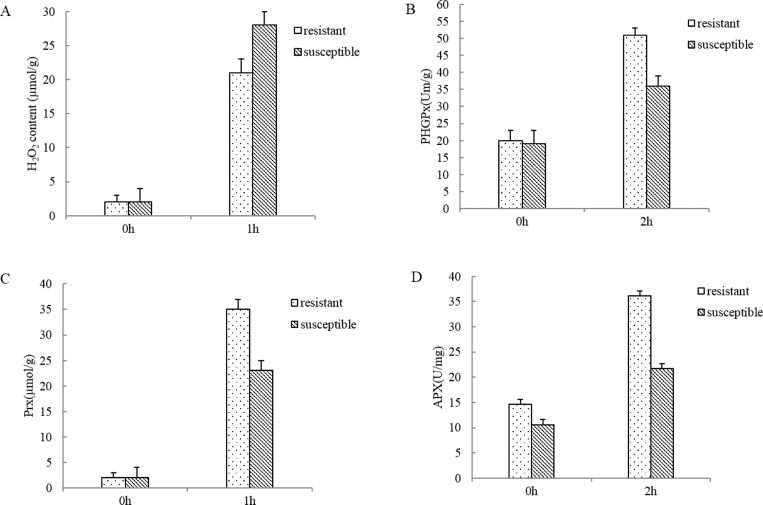
To investigate the effects of drought stress on the oxidative stress, the two L. multiflorum lines under control and drought treatment were tested for H2O2 content, enzyme activities of PHGPx, Prx and APX. Under drought stress, H2O2 content in both L. multiflorum lines showed dramatically increase compared with control seedlings (Fig 1A), especially in susceptible. A higher enzymatic activities of hospholipid PHGPx, Prx and APX were observed in the tolerant lines exposed to drought stress than under control (Fig 1B–1D). These results indicated that antioxidant enzymes contributed to alleviated oxidative stress- triggered ROS accumulation in tolerant lines during short-term drought stress treatment.
Fig 1. Changes in H2O2 content and enzyme activities of hospholipid hydroperoxide glutathione peroxidase (PHGPx), peroxiredoxin (Prx) and ascorbate peroxidase during drought stress in the two L. multiflorum lines.
The different letters above the columns indicate significant differences between different time points (P<0.05).
Differential accumulation analysis of L. multiflorum proteome under drought stress
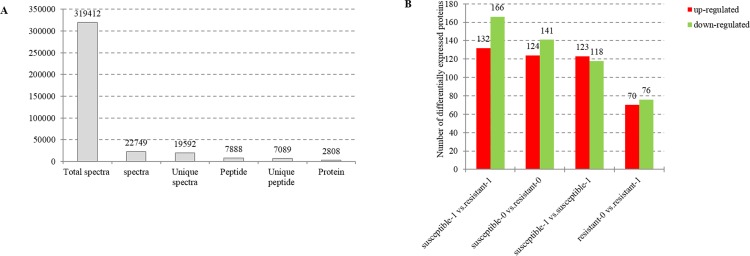
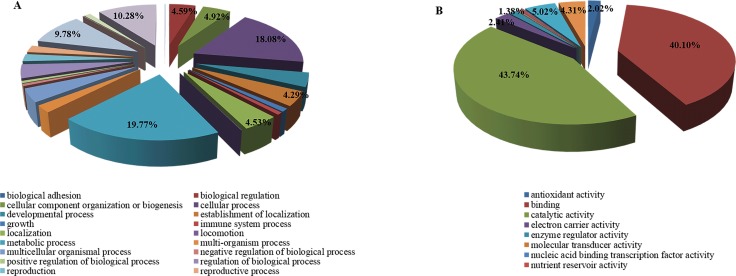
Drought stress-induced changes in the proteome of the two L. multiflorum lines were captured by analyzing quantitative information generated by iTRAQ-based quantitative analysis and LC-MS/MS method. A total of 7, 089 unique peptides matching to 2, 808 proteins were identified with a Mascot probability analysis (Fig 2A). A differentially accumulated analysis revealed that 449 proteins were up-regulated and 501 were down-regulated (Fig 2B). These proteins were categorized into biological process and molecular function based on blast2 GO program (Fig 3). The drought-regulated proteins were primarily related to metabolic process, cellular process, single-organism process and response to stimulus, which play a role in regulating the catalytic activity, structural molecule activity, transporter activity, electron carrier activity and antioxidant activity (Table 1).
Fig 2.
Statistics of total spectra, unique peptides and proteins in L. multiflorum proteome (A) and effects of drought stress on the accumulation of differentially accumulated proteins (B) of two L. multiflorum lines. The number 0 and 1 respectively reveal two Italian ryegrass lines subjected to well-water condition and naturally air drying for two hours. The red columns and green columns shows those proteins with significantly up-regulated or down-regulated accumulation.
Fig 3.
The percent of differentially accumulated proteins (DEPs) involved in biological process (A) and molecular function (B) in L. multiflorum lines.
Table 1. The detailed information of fifty-one drought-induced proteins identified from two Lolium multiflorum lines.
| Accession. | Protein name | 115/117 | 119/121 | GO Biological process |
|---|---|---|---|---|
| Tolerant (up) / Susceptible (up) | ||||
| Egi|92429455 | isocitrate lyase | 2.4 | 5.2 | |
| Cgi|39654150 | β-D-Glucan Glucohydrolase | 1.1 | 2.2 | |
| Pgi|262217337 | cathepsin B | 5.0 | 7.2 | /regulation of catalytic activity |
| Cgi|162462658 | α-amylase precursor | 1.5 | 2.5 | / |
| Cgi|269316344 | α-glucosidase | 1.0 | 1.4 | |
| Cgi|326503406 | predicted protein | 1.5 | 4.9 | |
| Cgi|1718236 | (1,4)-β-xylan endohydrolase | 3.7 | 3.7 | |
| Tgi|149392357 | Predicted protein | 1.0 | 1.5 | response to salt stress/glucosinolate biosynthetic process |
| Pgi|326533328 | predicted protein | 1.0 | 1.0 | protein refolding/chloroplast organization/embryo development |
| Pgi|262360187 | cysteine proteinase | 7.0 | 4.6 | |
| gi|393450 | β-amylase | 2.1 | 5.2 | |
| Cgi|326506982 | β-galactosidase | 2.0 | 1.7 | |
| Fgi|414881677 | LRR family protein | 4.1 | 2.8 | / kinase activity |
| Cgi|326489563 | α-L-arabinofuranosidase 2 | 2.3 | 6.5 | /L-arabinose metabolic process |
| gi|326497267 | Xylanase inhibitor protein 1 | 1.5 | 7.1 | |
| gi|413948511 | Phytepsin | 2.2 | 1.9 | /lipid metabolic process/ response to salt stress |
| Cgi|40363751 | putative β-xylosidase | 3.0 | 3.0 | |
| Egi|357126982 | cytochrome b5-like | 1.0 | 1.2 | electron transport chain |
| gi|56130862 | β-amylase | 1.7 | 4.3 | |
| gi|270311550 | α-amylase isoform | 3.2 | 7.3 | / / response to abscisic acid stimulus/ |
| gi|222618904 | Aspartic proteinase | 9.9 | 13.7 | / response to stimulus |
| Agi|326490063 | Serine carboxypeptidase | 1.9 | 2.2 | |
| gi|215398468 | globulin 3C | 2.7 | 7.7 | nutrient reservoir activity/binding |
| gi|115349894 | fasciclin-like protein | 1.6 | 2.0 | auxin polar transport/ regulation of cell size/root morphogenesis/plant-type cell wall organization |
| Agi|326533014 | Phospho-2-dehydro-3- deoxyheptonate aldolase 1 |
1.2 | 1.2 | shikimate biosynthetic process/chorismate biosynthetic process/aromatic amino acid family biosynthetic process |
| gi|149391177 | l-ascorbate peroxidase precursor | 1.0 | 1.0 | / /glucosinolate metabolic proces/ /response to oxidative stress |
| Tolerant (down) / Susceptible (down) | ||||
| Cgi|213536819 | tonoplast intrinsic protein | 1.0 | 0.9 | / water transport |
| gi|357121703 | uncharacterized protein LOC100836930 | 0.6 | 0.7 | electron carrier activity/cytochrome-c oxidase activity |
| Tgi|12651627 | 40S ribosomal protein S8 | 0.8 | 0.7 | |
| Pgi|86439735 | heat shock protein 90 | 0.9 | 0.8 | unfolded protein binding/ATP binding/protein folding/response to stress |
| gi|357134285 | putative mitochondrial 2-oxoglutarate | 0.9 | 0.7 | |
| gi|357111020 | Histone H1 | 0.8 | 0.3 | DNA binding/nucleosome assembly |
| gi|326500094 | Oxygen-evolving enhancer protein 3 | 0.6 | 0.9 | photosynthesis/cellular macromolecule metabolic process/ cellular component biogenesis/primary metabolic process |
| gi|357138855 | uncharacterized protein LOC100828601 | 0.6 | 0.9 | |
| Tgi|315113285 | 80s ribosomal protein L36 | 1.0 | 0.7 | |
| Tgi|326495694 | 60S ribosomal protein L | 0.3 | 0.8 | |
| Tgi|414873598 | 40S ribosomal protein S26 | 0.7 | 1.0 | |
| Pgi|326506180 | Peroxiredoxin Q | 0.5 | 0.6 | oxidation-reduction process |
| Ggi|375152034 | hsc70-interacting protein | 0.7 | 0.7 | response to cadmium ion |
| Tgi|357122371 | ribosome-recycling factor | 0.6 | 1.0 | pentose-phosphate shunt/ plastid translation/aromatic amino acid family biosynthetic process/dolichol biosynthetic process |
| Tgi|357134073 | proliferation-associated protein | 0.7 | 0.7 | proteolysis/cellular process |
| Egi|283896798 | phosphoenolpyruvate carboxylase | 0.9 | 0.8 | tricarboxylic acid cycle/oxaloacetate metabolic process/carbon fixation/ photosynthesis |
| Tolerant (down) / Susceptible (up) | ||||
| Cgi|161897650 | Probable aquaporin | 1.3 | 0.8 | / Water transport |
| Tolerant (up) / Susceptible (down) | ||||
| Egi|326502872 | Glycerate dehydrogenase | 1.1 | 0.8 | |
| Pgi|115444771 | peroxiredoxin 2 | 0.9 | 1.2 | |
| Pgi|375152308 | peroxiredoxin 5 | 0.8 | 1.3 | |
| Pgi|326504940 | predicted protein | 1.0 | 1.2 | Cysteine biosynthetic process |
| Cgi|357145851 | glucose-1-phosphate | 0.9 | 1.8 | |
| Pgi|375152246 | dehydroascorbate reductase | 0.9 | 1.3 | Response to cyclopentenone |
| gi|82780752 | lipid transfer protein | 0.7 | 1.8 | Lipid transport |
The 115/117 and 119/121 were the fold change of well-watered and drought-treated tolerant and susceptible plants, respectively. The COG category: c Carbohydrate transport and metabolism; E Energy production conversion; A Amino acid transport and metabolism; P Posttranslational modification, protein turnover, chaperones; T Translation, ribosomal structure and biogenesis; G General function prediction only.
Identification of proteins differentially accumulated in the two L. multiflorum Lines response to drought stress
A total of 51 drought-induced proteins were obtained from both tolerant and susceptible L. multiflorum proteomes (Table 1). Of them, 27 up-regulated and 16 down-regulated proteins having the same change trends were observed in the two L. multiflorum lines, of which up-regulated proteins associated with carbohydrate metabolism and proteolysis, while down-regulated proteins mostly participated in translation and transmembrane transport. Based on COG categories, the majority of shared proteins participated in regulation of carbohydrate transport and metabolism, post translational modification, protein turnover, translation, chaperones, ribosomal structure and biogenesis, and energy production and conversion. Comparison of the differentially accumulated proteins in the two lines identified eight responsive proteins having an opposite trend in the tolerant and susceptible lines, among them, three proteins as the important antioxidative enzymes, peroxiredoxin 2, peroxiredoxin 5 and dehydroascorbate reductase involved in oxidation-reduction processes were specifically identified in the tolerant lines. Other proteins, glycerate dehydrogenase (GDH), dehydroascorbate reductases (DHAR), glucose-1-phosphate (G1P) and lipid transfer protein (LTP), were also only up-regulated in the tolerant lines, but not susceptible line. The results not only revealed that drought tolerance of Italian ryegrass had a direct link to the specific proteins induced by drought stress, but also provided evidence that carbohydrate metabolism, oxidation-reduction processes, proteolysis, and transmembrane transport had a significant relationship with drought tolerance of Italian ryegrass.
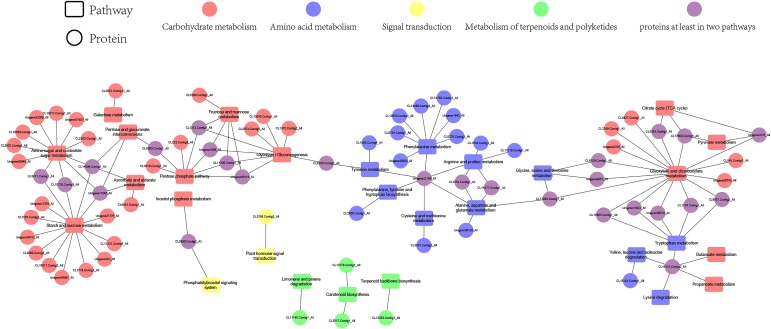
Network analysis for drought responsive proteins
Proteins in plant cells and subcellular fractions play interrelated roles together in the context of networks [19]. Within the differentially accumulated proteins (susceptible vs. tolerant), a total of 76 proteins were identified using the Cytoscape software that can be mapped onto an interaction network (Table 2; Fig 4). The left region showed that responsive proteins involved in carbohydrate metabolism-related pathways, e.g. fructose and mannose metabolism, glycolysis and gluconeogenesis, citrate cycle (TCA cycle), starch and sucrose metabolism, fructose and mannose metabolism, pentose and glucuronate interconversions, galactose metabolism, pointing to a potential importance of carbohydrates in modulating the homeostasis of the drought response of Italian ryegrass. The middle part revealed that drought-related proteins enriched in nine amino acid metabolic pathways including tyrosine metabolism, phenylalanine, tyrosine and tryptophan biosynthesis, cysteine and methionine metabolism, arginine and proline metabolism, alanine, aspartate and glutamate metabolism, glycine, serine and threonine metabolism, tryptophan metabolism, valine, leucine and isoleucine degradation. The amounts of special proteins involving both of the two pathways were observed in the right side. Interestingly, a predicted protein (CL16151.Contig2) existed in tryptophan metabolism, valine, leucine, lysine, and isoleucine degradation, and butanoate and propanoate metabolism. Furthermore, a special pathway with all the differentially accumulated protein, inositol phosphate metabolism, was only found in tolerant lines, and phosphoinositide-specific phospholipase C1 (PLC) showed obviously up-regulated in phosphatidylinositol signaling systems in tolerant lines subjected to drought condition.
Table 2. List of differentially accumulated proteins from the interaction network.
| Protein name | Description | NCBI accession number | Sources | |
|---|---|---|---|---|
| Starch and sucrose metabolism | ||||
| Unigene12376 | Starch-branching enzyme | gi|301090019 | Oryza sativa Indica Group | |
| CL7261.Contig1 | Β- glucosidase | gi|39654150 | Dictyostelium discoideum | |
| Unigene36731 | Cytosolic alpha-glucan phosphorylase | gi|229610905 | Hordeum vulgare | |
| CL2060.Contig1 | α-amylase precursor | gi|162462658 | Zea mays | |
| CL10671.Contig2 | β-glucosidase 4 | gi|359828768 | Triticum aestivum | |
| Unigene39957 | β-glucosidase 8 | gi|326515724 | Oryza sativa subsp. japonica | |
| CL1716.Contig1 | β-D-glucan exohydrolase | gi|1203832 | Hordeum vulgare | |
| Unigene342 | α-amylase | gi|270311550 | Dactylis glomerata | |
| CL11231.Contig4 | Sucrose synthase 1 | gi|326514918 | Hordeum vulgare | |
| Unigene31776 | β-amylase | gi|393450 | Secale cereale | |
| CL3871.Contig1 | Probable β-D-xylosidase 6 | gi|326491679 | Arabidopsis thaliana | |
| CL10726.Contig3 | ADP-glucose pyrophosphorylase | gi|52430025 | Triticum aestivum | |
| CL14998.Contig1 | PREDICTED: UDP-glucose 6-dehydrogenase-like | gi|357114933 | Brachypodium distachyon | |
| Unigene17245 | UDP-glucose 6-dehydrogenase | gi|108711177 | Oryza sativa Japonica Group | |
| Amino sugar and nucleotide sugar metabolism | ||||
| Unigene21923 | Chitinase 2 | gi|326492127 | Oryza sativa subsp. japonica | |
| Cl5263.Contig1 | Xylanase inhibitor protein 1 | |||
| Unigene22268 | PREDICTED: UDP-glucuronic acid decarboxylase 1-like | gi|357112854 | Brachypodium distachyon | |
| CL16558.Contig1 | Chloroplast stem-loop binding protein | gi|326531332 | Arabidopsis thaliana | |
| CL2607.Contig1 | Hypothetical protein OsJ_06514 | gi|222622743 | Oryza sativa Japonica Group | |
| CL3871.Contig1 | Probable β-D-xylosidase 6 | gi|326491679 | Arabidopsis thaliana | |
| CL10726.Contig3 | ADP-glucose pyrophosphorylase small subunit | gi|52430025 | Triticum aestivum | |
| CL14998.Contig1 | PREDICTED: UDP-glucose 6-dehydrogenase-like | gi|357114933 | Brachypodium distachyon | |
| Unigene17245 | UDP-glucose 6-dehydrogenase | gi|108711177 | Oryza sativa Japonica Group | |
| Unigene35646 | Endochitinase 3 | gi|326492127 | Arachis hypogaea | |
| CL13017.Contig1 | Xylanase inhibitor protein 1 | gi|326497267 | Oryza sativa subsp. japonica | |
| CL3871.Contig1 | Probable beta-D-xylosidase 6 | gi|326491679 | Arabidopsis thaliana | |
| CL10726.Contig3 | ADP-glucose pyrophosphorylase small subunit | gi|52430025 | Triticum aestivum | |
| CL14998.Contig1 | PREDICTED: UDP-glucose 6-dehydrogenase-like | gi|357114933 | Brachypodium distachyon | |
| Unigene17245 | UDP-glucose 6-dehydrogenase | gi|108711177 | Oryza sativa Japonica Group | |
| Ascorbate and aldarate metabolism | ||||
| CL6991.Contig1 | Chloroplast l-ascorbate peroxidase precursor | gi|149391177 | Oryza sativa Indica Group | |
| CL14998.Contig1 | PREDICTED: UDP-glucose 6-dehydrogenase-like | gi|357114933 | Brachypodium distachyon | |
| Unigene17245 | UDP-glucose 6-dehydrogenase | gi|108711177 | Oryza sativa Japonica Group | |
| Pentose and glucuronate interconversions | ||||
| CL14998.Contig1 | PREDICTED: UDP-glucose 6-dehydrogenase-like | gi|357114933 | Brachypodium distachyon | |
| Unigene17245 | UDP-glucose 6-dehydrogenase | gi|108711177 | Oryza sativa Japonica Group | |
| CL9560.Contig1 | Ribulose-phosphate 3-epimerase | gi|296040829 | Spartina alterniflora | |
| Galactose metabolism | ||||
| CL2001.Contig11 | β-galactosidase 9 | gi|357124049 | Oryza sativa subsp. japonica | |
| Pentose phosphate pathway | ||||
| CL3610.Contig1 | Glucose-6-phosphate dehydrogenase | gi|83267994 | Triticum dicoccoides | |
| CL9560.Contig1 | Ribulose-phosphate 3-epimerase | gi|296040829 | Spartina alterniflora | |
| CL223.Contig3 | PREDICTED: uncharacterized protein LOC100829451 | gi|357118665 | Brachypodium distachyon | |
| Unigene1889 | Fructose-bisphosphate aldolase 2 | gi|326499908 | Pisum sativum | |
| CL11236.Contig1 | Fructose-bisphosphate aldolase | gi|326493652 | Pisum sativum | |
| Unigene46118 | Fructose-1,6-bisphosphatase | gi|219362881 | Triticum aestivum | |
| Inositol phosphate metabolism/Phosphatidylinositol signaling system | ||||
| CL5690.Contig2 | Phosphoinositide-specific phospholipase C1 | gi|312618322| | Triticum aestivum | |
| Plant hormone signal transduction | ||||
| CL3788.Contig1 | Cytochrome P450 97B1 | gi|401831 | Hordeum vulgare | |
| Fructose and mannose metabolism | ||||
| Unigene1889 | Fructose-bisphosphate aldolase 2 | gi|326499908 | Pisum sativum | |
| CL11236.Contig1 | Fructose-bisphosphate aldolase | gi|326493652 | Pisum sativum | |
| CL11236.Contig1 | Fructose-bisphosphate aldolase | gi|326493652 | Pisum sativum | |
| Unigene46118 | Fructose-1,6-bisphosphatase | gi|219362881 | Triticum aestivum | |
| CL5390.Contig2 | Predicted protein | gi|326523467 | Hordeum vulgare | |
| Glycolysis/Gluconeogenesis | ||||
| Unigene1889 | Fructose-bisphosphate aldolase 2 | gi|326499908 | Pisum sativum | |
| CL11236.Contig1 | Fructose-bisphosphate aldolase | gi|326493652 | Pisum sativum | |
| Unigene46118 | Fructose-1,6-bisphosphatase | gi|219362881 | Triticum aestivum | |
| CL2073.Contig5 | Predicted protein | gi|326503406 | Hordeum vulgare | |
| CL13045.Contig1 | Cytosolic glyceraldehydes-3-phophate dehydrogenase | gi|168472723 | Lolium temulentum | |
| CL1372.Contig2 | 3-phosphoglycerate kinase | gi|226247069 | Leymus triticoides | |
| CL6250.Contig1 | Alcohol dehydrogenase class-3 | gi|357137596 | Oryza sativa subsp. japonica | |
| Tyrosine metabolism | ||||
| CL6250.Contig1 | Alcohol dehydrogenase class-3 | gi|357137596 | Oryza sativa subsp. japonica | |
| CL1800.Contig4 | Polyphenol oxidase | gi|46946548| | Triticum aestivum | |
| Unigene22199 | Aspartate aminotransferase | gi|165874483 | Oryza granulata | |
| Phenylalanine, tyrosine and tryptophan biosynthesis | ||||
| Unigene22199 | Aspartate aminotransferase | gi|165874483 | Oryza granulata | |
| CL2606.Contig1 | Phospho-2-dehydro-3-deoxyheptonate aldolase 1 | gi|357148189 | Brachypodium distachyon | |
| Cysteine and methionine metabolism | ||||
| Unigene22199 | Aspartate aminotransferase | gi|165874483 | Oryza granulata | |
| CL3501.Contig3 | Cobalamin-independent methionine synthase | gi|115589740 | Triticum monococcum | |
| Phenylalanine metabolism | ||||
| Unigene30853 | Peroxidase 1 | gi|326508456 | Oryza sativa subsp. japonica | |
| CL8724.Contig1 | Cationic peroxidase SPC4 | gi|357166838 | Sorghum bicolor | |
| CL10453.Contig2 | Peroxidase 1 | gi|326518626 | Zea mays | |
| CL14486.Contig3 | Peroxidase 12 | gi|5777628 | Arabidopsis thaliana | |
| CL12763.Contig1 | peroxidase POC1 | gi|8901180 | Oryza sativa Indica Group | |
| Unigene1942 | Peroxidase 2 | gi|125550742 | Zea mays | |
| Unigene22199 | Aspartate aminotransferase | gi|165874483 | Oryza granulata | |
| Arginine and proline metabolism | ||||
| CL1034.Contig1 | Acetylornithine aminotransferase | gi|326510053 | Alnus glutinosa | |
| CL17218.Contig1 | PREDICTED: acetylornithine deacetylase-like | gi|357137096 | Brachypodium distachyon | |
| Unigene22199 | Aspartate aminotransferase | gi|165874483 | Oryza granulata | |
| CL2354.Contig2 | delta-1-pyrroline-5-carboxylate dehydrogenase | gi|73913053 | Triticum aestivum | |
| CL14377.Contig1 | GdhA protein | gi|129920003 | Triticum durum | |
| Alanine, aspartate and glutamate metabolism | ||||
| CL2354.Contig2 | delta-1-pyrroline-5-carboxylate dehydrogenase | gi|73913053 | Triticum aestivum | |
| CL14377.Contig1 | GdhA protein | gi|129920003 | Triticum durum | |
| Unigene36136 | glutamine-dependent asparagine synthetase | gi|53680379 | Triticum aestivum | |
| Unigene22199 | Aspartate aminotransferase | gi|165874483 | Oryza granulata | |
| CL17063.Contig1 | Glutamate—glyoxylate aminotransferase 1 | gi|357111762 | Arabidopsis thaliana | |
| Glycine, serine and threonine metabolism | ||||
| CL17063.Contig1 | Glutamate—glyoxylate aminotransferase 1 | gi|357111762 | Arabidopsis thaliana | |
| Unigene8857 | Serine hydroxymethyltransferase | gi|375152224 | Lolium perenne | |
| CL6530.Contig1 | Glycerate dehydrogenase | gi|326502872 | Cucumis sativus | |
| Glyoxylate and dicarboxylate metabolism | ||||
| Unigene8857 | serine hydroxymethyltransferase | gi|375152224 | Lolium perenne | |
| CL6530.Contig1 | Glycerate dehydrogenase | gi|326502872 | Cucumis sativus | |
| Unigene2226 | Glycine cleavage system H protein, | gi|326505670 | Oryza sativa subsp. japonica | |
| CL2359.Contig1 | Isocitrate lyase | gi|92429455 | Lolium perenne | |
| CL5827.Contig1 | Peroxisomal (S)-2-hydroxy-acid oxidase GLO1 | gi|357112622 | Oryza sativa subsp. indica | |
| CL5363.Contig1 | Predicted protein | gi|326523589 | Hordeum vulgare subsp. vulgare | |
| CL17600.Contig5 | Malate dehydrogenase | gi|13517921 | Lolium perenne | |
| CL14863.Contig1 | Catalase | gi|90264977 | Festuca arundinacea | |
| Unigene13422 | Catalase isozyme 2 | gi|326516518 | Hordeum vulgare | |
| Unigene36570 | Catalase | gi|90264977 | Festuca arundinacea | |
| CL7784.Contig1 | Catalase-1 | gi|2493543 | Triticum aestivum | |
| CL9937.Contig2 | Catalase isozyme 1 | gi|238802280 | Hordeum vulgare | |
| Unigene26743 | Peroxisomal (S)-2-hydroxy-acid oxidase GLO5 | gi|357111705 | Oryza sativa subsp. japonica | |
| CL545.Contig1 | 60S ribosomal protein L18a | gi|357132550 | Oryza sativa subsp. japonica | |
| Unigene7931 | Malate dehydrogenase | gi|357147942 | Arabidopsis thaliana | |
| Pyruvate metabolism | ||||
| CL17600.Contig5 | Malate dehydrogenase | gi|13517921 | Lolium perenne | |
| Unigene7931 | Malate dehydrogenase | gi|357147942 | Arabidopsis thaliana | |
| Citrate cycle (TCA cycle) | ||||
| CL5363.Contig1 | Predicted protein | gi|326523589 | Hordeum vulgare subsp. vulgare | |
| CL17600.Contig5 | Malate dehydrogenase | gi|13517921 | Lolium perenne | |
| Unigene7931 | Malate dehydrogenase | gi|357147942 | Arabidopsis thaliana | |
| Tryptophan metabolism | ||||
| CL14863.Contig1 | Catalase | gi|90264977 | Festuca arundinacea | |
| Unigene13422 | Catalase isozyme 2 | gi|326516518 | Hordeum vulgare | |
| Unigene36570 | catalase | gi|90264977 | Festuca arundinacea | |
| CL7784.Contig1 | Catalase-1 | gi|2493543 | Triticum aestivum | |
| CL9937.Contig2 | Catalase isozyme 1 | gi|238802280 | Hordeum vulgare | |
| CL16151.Contig2 | Predicted protein | gi|326528605 | Hordeum vulgare | |
| Valine, leucine and isoleucine degradation | ||||
| CL16151.Contig2 | Predicted protein | gi|326528605 | Hordeum vulgare | |
| CL18181.Contig1 | Predicted protein | gi|326513882 | Hordeum vulgare subsp. vulgare | |
| Lysine degradation/Propanoate metabolism/Butanoate metabolism | ||||
| CL16151.Contig2 | Predicted protein | gi|326528605 | Hordeum vulgare | |
| Limonene and pinene degradation | ||||
| CL11140.Contig1 | Indole-2-monooxygenase | gi|13661750 | Zea mays | |
| Carotenoid biosynthesis | ||||
| CL3517.Contig1 | GDSL esterase/lipase | gi|326516774 | Arabidopsis thaliana | |
| CL13778.Contig1 | Cytochrome P450 97B1 | gi|194699820 | Pisum sativum | |
| Terpenoid backbone biosynthesis | ||||
| CL12582.Contig1 | Farnesyl pyrophosphate synthase | gi|326490760 | Zea mays | |
Fig 4. Interaction network of differentially accumulated proteins.
Confirmation of protein abundance changes by Western blotting
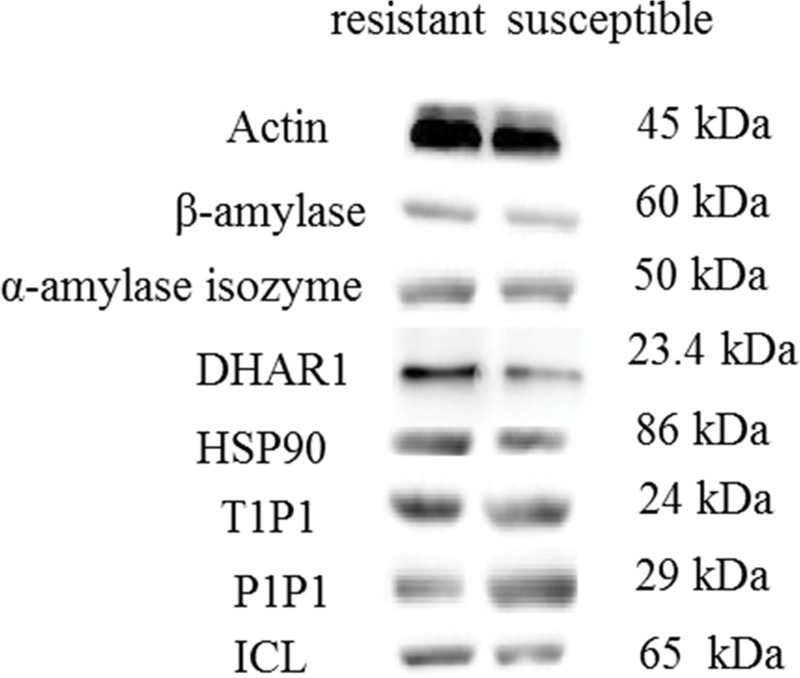
We confirmed the accumulation of seven proteins shared in the tolerant and susceptible lines by Western blotting. As shown in Fig 5, ICL (gi|92429455), α-amylase isozyme (gi|270311550), β-amylase (gi|56130862), HSP90(gi|86439735) and TIP1 (gi|213536819), showed a similar accumulation level as iTRAQ results in Table 1. Compared with drought susceptible lines, DHAR1 (gi|375152246) showed higher accumulation in tolerant lines, whereas the opposite result was observed in aquaporin, PIP1 (gi|161897650).
Fig 5. Validation of accumulation levels among differentially accumulated proteins of two L. multiflorum lines using Western blotting.
Discussion
Drought-responsive proteins related to tolerance to oxidative stress
The overproduction of reactive oxygen species (ROS) occurs when plants suffer from drought stress [20]. To prevent damage caused by ROS, plants can synthesize antioxidants, such as ascorbate, glutathione, and flavonoids, and enhance antioxidative enzymes [21]. Tolerance to abiotic stresses is involved with the ROS scavenging capacity in plants [22]. Phospholipid PHGPx is an antioxidant enzyme that directly reduces the phospholipid hydroperoxides in bio-membranes and protects cells from oxidative damage [23]. In our study, PHGPx was involved in the metabolism of arachidonic acid, which reduced the oxidative damage in the drought tolerant lines. Prx and catalase (CAT) can reduce the accumulation of ROS in plants [24, 25]. Consistent with the results of previous studies, the activity of Prx exhibited an increase trend in drought tolerant line. Dehydroascorbate reductase (DHAR) helps to enhance plant tolerance to various abiotic stresses [26]. Actually, the protective role of DHAR was found and confirmed by developing transgenic tobacco plants with cytosolic DHAR gene [27]. For Italian ryegrass, it can be speculated that DHAR has a potent protective role in defending oxidative stress. pAPX play a key role in protecting plants against oxidative stress and thus conferred abiotic stress tolerance [28]. The up-regulation of peroxisomal ascorbate peroxidase (pAPX) was observed in this study, indicating that the enzyme might reduce cell damage caused by oxidative stress in Lolium multiflorum. Similarly, monodehydroascorbate reductase (MDHAR) was a key enzyme in the ascorbate-glutathione cycle and served as an important antioxidant [29]. In our results, MDHAR was down-regulated in ascorbate and aldarate metabolism, but the exact mechanism is still not clear and needs further study.
Metabolism-related proteins contributed to enhanced drought tolerance
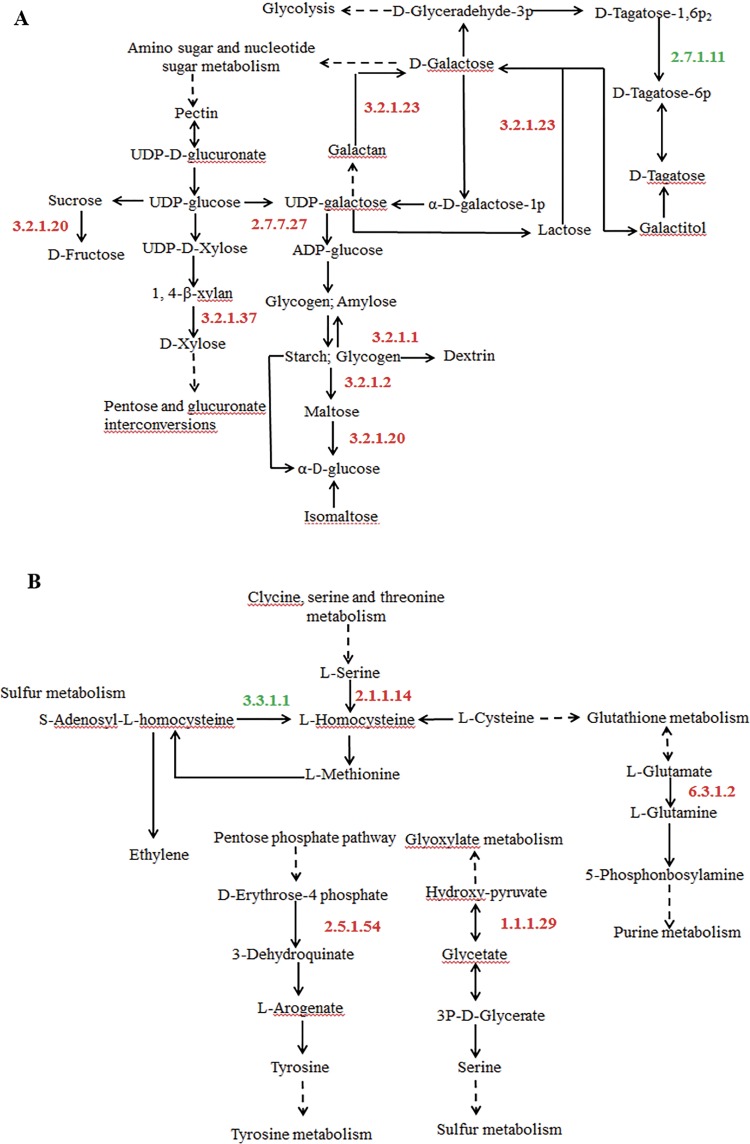
The levels of the readily metabolizable carbohydrates significantly increased in plants under drought stress to maintain metabolic homeostasis [30]. In this study, changes in the accumulation levels of a number of carbohydrate metabolism-associated proteins in drought tolerant lines were observed (Fig 6A). The α-glucosidase (EC 3.2.1.20) played role in catalyzing the liberation of α-D-glucose from the non-reducing end of polysaccharides [31], especially the Type I of α-glucosidase can rapidly hydrolyzes sucrose [32]. The up-regulation of regulators, such as α-glucosidase (EC 3.2.1.20) and β-galactosidase (EC 3.2.1.23), were possibly stimulated to accumulate abundant galactose in control of drought stress in Italian ryegrass. Conversely, drought stress induced a pronounced increase in the activity of enzymes that hydrolyzed starch and sucrose according to Keller et al (1993). Glucose-1-phosphate (EC 2.7.7.27), β-amylase (EC 3.2.1.2), α-amylase (EC 3.2.1.1), α-glucosidase, and putative β-D-xylosidase (EC 3.2.1.37), showed significant up-regulation and were linked with the hydrolysis of starch and sucrose under drought condition, suggesting that the drought-tolerant lines invested more carbohydrate into immediate defense against drought stress than drought susceptible lines. A putative β-D-xylosidase gene (AtBXL1) has been reported to be involved in secondary cell wall xylan synthesis [33]. Hemicelluloses were usually grouped into xylans and β-glucans [34]. The increase of the concentrations of β-D-glucan glucohydrolase (GGH) led to decrease of cell wall β-glucan concentrations in Hordeum vulgare [35]. In our study, down-regulated GGH not only catalyzed the hydrolytic removal of β-D-glucosyl residues [36], but also associated with enhancing drought tolerance by stimulating the degradation of β-glucans in abiotic stress.
Fig 6.
Drought stress induced proteins accumulation associating with carbohydrate metabolism (A) and amino acid metabolism (B) in tolerant L. multiflorum lines.
Plant growth inhibition due to drought stress was accompanied by increased amino acid concentrations [37]. Our analysis revealed that drought-responsive proteins as regulators participated in amino acid metabolism in tolerant lines of Italian ryegrass (Fig 6B). Cobalamin-independent methionine synthase (MetE) (EC 2.1.1.14), glycerate dehydrogenase (GDH) (EC 1.1.1.29) and Phospho-2-dehydro-3-deoxyheptonate aldolase 1 (EC 2.5.1.54) were found to be up-regulated in tolerant lines, except for unnamed protein product (EC 3.3.1.1). Cobalamin-independent methionine synthase wass involved in methionine synthesis and connected sulfur and carbon metabolic networks [38]. The activity of MetE could modulate methionine biosynthesis in E.coli under oxidative stress conditions [39]. We observed the up-regulation of MetE specifically in tolerant lines, suggesting that this enzyme might has a positive regulatory role in methionine biosynthesis to mitigate oxidative stress. Glycerate dehydrogenase is one of the precursors of glycine betaine, which was accumulated in both plants and animals in response to drought stress [40]. Results in this study might explain that up-regulated GDH was associated with drought tolerance in Italian ryegrass. The salt-inducible cDNA had high homology to phospho-2-dehydro-3-deoxyheptonate aldolase 1 in salt-tolerant rice [41]. Our results provided evidence that phospho-2-dehydro-3-deoxyheptonate aldolase 1 as regulator contributed to drought tolerance of Italian ryegrass. It was found in previous research that proline analogues and galactose levels were higher in drought resistant varieties of potato and barley [11, 42].The higher amounts of proline involved in osmoprotection and stress signaling under drought [43]. Similar to results from other studies, we also noticed that the glutamine synthetase 1a (GS1a; EC 6.3.1.2) was up-regulated indicating that it participated in proline and glutamine synthesis to enhance drought tolerance in the tolerant line, and no change was observed in the drought susceptible lines [44–46]. A wide range of experiments showed that plants subjected to drought stress indeed expressed highconcentrations of secondary metabolites [47]. Terpenoids constitute a large and structurally diverse group of chemicals, playing diverse functional roles in plants as hormones, electron carriers and structural component of membranes [48]. Farnesyl pyrophosphate synthase (FPS) not only played a vital role in terpenoid metabolism, but also functioned as a key regulatory enzyme to control the sterol biosynthetic pathway [49]. In our study, FPS as key regulators was significantly accumulated and involved in Terpenoid backbone biosynthesis.
Inositol phosphates, diacylglycerol (DAG) and inositol 1, 4, 5-trisphosphate (IP3) assecond messengers play an important role in signal transduction pathway. Actually, the PI-PLC could mediate the production of the DAG and IP3 [50]. In our study, PI-PLC as up-regulator played role in both inositol phosphate metabolism and phosphatidylinositol signaling system in tolerant lines, indicating that PI-PLC contributed to drought-mediated abiotic stress tolerance in Italian ryegrass. Moreover, DAG as a structural lipid changed its abundance during drought condition in grasses from Lolium-Festuca complex was observed by Perlikowski et al. (2014) [8].
Hydrolysis proteins and transport proteins respond to drought stress
The increase of amino acid concentration was caused by proteolysis in the advanced stages of drought [51]. Our data indicated that cathepsin B was up-regulated in the vacuole of L. multiflorum under drought stress, and might relate to the degradation of proteins. Recently, cysteine protease cathepsin B (TbCatB) was described as being involved in host protein degradation in Trypanosoma brucei [52]. Cysteine proteinase (CysP) showed to respond to environmental stress, such as cold or water deficiency [52] and its accumulation was observed in the leaves of tomato plants submitted to drought-stress [53]. In agreement with all the previous researches, our results indicated that CysP as an up-regulator controled protein degradation, which allowed L. multiflorum to adapt to drought stress. Aspartic proteinase (APs) are a large family of proteolytic enzymes, which are found in almost every plant [54]. APs were participated in some biological processes, e.g., stress responses, and programmed cell death [55]. We also observed the up-regulated accumulation of APs in the drought tolerant line of Italian ryegrass subjected to drought stress. This result suggested that aspartic proteinases might be important for improving drought tolerance.
Lipid transfer proteins (LTP) are small, basic, soluble proteins and are involved in stress response processes [56]. Furthermore, LTPs may repair stress-induced damage in membranes [57], and also may be responsible for increasing wax deposition [58]. In our study, LTP accumulation had dramatically up-regulated in the tolerant line, which might relate to enhance drought tolerance. Aquaporin (AQP) is plasma membrane water-transporting protein that facilitates water movement across cell membranes against osmotic gradients [59]. AQP transports water and other small molecules through biological membranes, which is vital for plants to tolerate drought [60]. According to our results, the drought tolerant line might have a lower water evaporation and higher water transport than the drought susceptible lines in drought conditions.
Predicted (hypothetical) proteins functionally relevant to drought tolerance
The functions of hypothetical proteins are still unknown, which is a challenge not only to genome annotation but also to in depth biological interpretation [61]. Hypothetical proteins were reported previously in cereal crops under abiotic stress [12, 62]. Similarly, some predicted proteins are considered as key regulators of molecular mechanisms, with an important function in stress conditions. Kaneko et al. (1997) [63] found 3,189 predicted proteins as salt-responsive proteins in the Synechocystis genome (a cyanobacterium). Yang et al (2015) [64] detected 7% predicted protein that may improve drought tolerance and maintain photosynthetic activity in Purpurea seedlings. In our study, predicted proteins (CL16151.Contig2) played a vital role in metabolic processes of amino acids, propanoate, and butanoate. Five predicted proteins (gi|326533328, gi|149392357, gi|326503406, gi|357121703, and gi|357138855) (Table 1), were significantly accumulated in response to drought stress in two L. multiflorum lines. Based on these results, predicted proteins are valuable though with unknown functions in understanding the effects caused by complex metabolic processes.
Conclusions
The current study revealed how drought-related proteins involved in regulatory system to adapt to abiotic stress in Italian ryegrass. Some differentially accumulated proteins, such as AtBXL1, GGH, GDH, MetE, GS1a, FPS, DAG, and PI-PLC, were found to be involved in carbohydrate metabolism, amino acid metabolism, biosynthesis of secondary metabolites, and signal transduction pathway, which might contributed to enhance drought tolerance or adaptation in Lolium multiflorum. The two specific metabolic processes, arachidonic acid and inositol phosphate metabolism, were differentially accumulated only in the tolerant lines. CatB, CysP, LTP and AQP were observed as drought-regulated proteins participating in hydrolysis and transmembrane transport. The activities of PHGPx, Prx, CAT, DHAR, pAPX and MDHAR associated with alleviate the accumulation of reactive oxygen species in stress inducing environments. In addition, a significant number of predicted proteins might play a vital of role in the modulation of metabolic pathways.
Supporting information
(TIF)
(XLSX)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This research work was funded by the earmarked fund for Modern Agro-industry Technology Research System (No. CARS-35-05), the National Basic Research Program (973 program) in China (No. 2014CB138705) and the National Natural Science Foundation of China (NSFC 31372363). XZ is the funder who had role in study design and decision to publish.
References
- 1.Jaleel CA, Manivannan P, Wahid A, Farooq M, Al-Juburi HJ, Somasundaram R, et al. Drought stress in plants: a review on morphological characteristics and pigments composition. Int J Agric Biol. 2009;11(1):100–5. [Google Scholar]
- 2.Noctor G, Mhamdi A, Foyer CH. The roles of reactive oxygen metabolism in drought: not so cut and dried. Plant physiology. 2014;164(4):1636–48. doi: 10.1104/pp.113.233478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Claeys H, Inzé D. The agony of choice: how plants balance growth and survival under water-limiting conditions. Plant Physiology. 2013;162(4):1768–79. doi: 10.1104/pp.113.220921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caruso G, Cavaliere C, Foglia P, Gubbiotti R, Samperi R, Laganà A. Analysis of drought responsive proteins in wheat (Triticum durum) by 2D-PAGE and MALDI-TOF mass spectrometry. Plant Science. 2009;177(6):570–6. [Google Scholar]
- 5.Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K. AP2/ERF family transcription factors in plant abiotic stress responses. Biochimica et Biophysica Acta (BBA)-Gene Regulatory Mechanisms. 2012;1819(2):86–96. [DOI] [PubMed] [Google Scholar]
- 6.Kissoudis C, Kalloniati C, Flemetakis E, Madesis P, Labrou NE, Tsaftaris A, et al. Maintenance of metabolic homeostasis and induction of cytoprotectants and secondary metabolites in alachlor-treated GmGSTU4-overexpressing tobacco plants, as resolved by metabolomics. Plant Biotechnology Reports. 2015;9(5):287–96. [Google Scholar]
- 7.Jiang Y, Guo L, Xie L-Q, Zhang Y-Y, Liu X-H, Zhang Y, et al. Proteome profiling of mitotic clonal expansion during 3T3-L1 adipocyte differentiation using iTRAQ-2DLC-MS/MS. Journal of proteome research. 2014;13(3):1307–14. doi: 10.1021/pr401292p [DOI] [PubMed] [Google Scholar]
- 8.Perlikowski D, Kosmala A, Rapacz M, Kościelniak J, Pawłowicz I, Zwierzykowski Z. Influence of short‐term drought conditions and subsequent re‐watering on the physiology and proteome of Lolium multiflorum/Festuca arundinacea introgression forms, with contrasting levels of tolerance to long‐term drought. Plant Biology. 2014;16(2):385–94. doi: 10.1111/plb.12074 [DOI] [PubMed] [Google Scholar]
- 9.Chen S, Harmon AC. Advances in plant proteomics. Proteomics. 2006;6(20):5504– doi: 10.1002/pmic.200600143 [DOI] [PubMed] [Google Scholar]
- 10.Wiese S, Reidegeld KA, Meyer HE, Warscheid B. Protein labeling by iTRAQ: a new tool for quantitative mass spectrometry in proteome research. Proteomics. 2007;7(3):340–50. doi: 10.1002/pmic.200600422 [DOI] [PubMed] [Google Scholar]
- 11.Wendelboe‐Nelson C, Morris PC. Proteins linked to drought tolerance revealed by DIGE analysis of drought resistant and susceptible barley varieties. Proteomics. 2012;12(22):3374–85. doi: 10.1002/pmic.201200154 [DOI] [PubMed] [Google Scholar]
- 12.Ji K, Wang Y, Sun W, Lou Q, Mei H, Shen S, et al. Drought-responsive mechanisms in rice genotypes with contrasting drought tolerance during reproductive stage. Journal of plant physiology. 2012;169(4):336–44. doi: 10.1016/j.jplph.2011.10.010 [DOI] [PubMed] [Google Scholar]
- 13.Han K, Alison M, Pitman W, McCormick M. Contribution of Field Pea to Winter Forage Production and Nutritive Value in the South-Central United States. Crop Science. 2013;53(1):315–21. [Google Scholar]
- 14.ZHANG L, LIU D-y, SHAO T. The feeding value and utilization prospect of Lolium multiflorum [J]. Pratacultural Science. 2008;4. [Google Scholar]
- 15.Franca A, Loi A, Davies W. Selection of annual ryegrass for adaptation to semi-arid conditions. European journal of agronomy. 1998;9(2):71–8. [Google Scholar]
- 16.Kosmala A., Perlikowski D, Pawłowicz I, and Rapacz M. Changes in the chloroplast proteome following water deficit and subsequent watering in a high-and a low-drought-tolerant genotype of Festuca arundinacea. Journal of experimental botany. 2012; 63(17), 6161–6172. doi: 10.1093/jxb/ers265 [DOI] [PubMed] [Google Scholar]
- 17.Humphreys M.W. and Thomas H. Improved drought resistance in introgression lines derived from Lolium multiflorum × Festuca arundinacea hybrids. Plant Breeding. 1993; 11:155–161. [Google Scholar]
- 18.Pan L, Zhang X, Wang J, Xiao M, Zhou M, Huang LK, et al. Transcriptional Profiles of Drought-Related Genes in Modulating Metabolic Processes and Antioxidant Defenses in Lolium multiflorum. Frontiers in Plant Science. 2016;7(170). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang L, Jiang T, Fountain JC, Scully BT, Lee RD, Kemerait RC, et al. Protein profiles reveal diverse responsive signaling pathways in kernels of two maize inbred lines with contrasting drought sensitivity. International journal of molecular sciences. 2014;15(10):18892–918. doi: 10.3390/ijms151018892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smirnoff N. The role of active oxygen in the response of plants to water deficit and desiccation. New Phytologist. 1993;125(1):27–58. [DOI] [PubMed] [Google Scholar]
- 21.Egert M, Tevini M. Influence of drought on some physiological parameters symptomatic for oxidative stress in leaves of chives (Allium schoenoprasum). Environmental and Experimental Botany. 2002;48(1):43–9. [Google Scholar]
- 22.You J, Zong W, Hu H, Li X, Xiao J, Xiong L. A STRESS-RESPONSIVE NAC1-regulated protein phosphatase gene rice protein phosphatase18 modulates drought and oxidative stress tolerance through abscisic acid-independent reactive oxygen species scavenging in rice. Plant physiology. 2014;166(4):2100–14. doi: 10.1104/pp.114.251116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sakamoto T, Maebayashi K, Nakagawa Y, Imai H. Deletion of the four phospholipid hydroperoxide glutathione peroxidase genes accelerates aging in Caenorhabditis elegans. Genes to Cells. 2014;19(10):778–92. doi: 10.1111/gtc.12175 [DOI] [PubMed] [Google Scholar]
- 24.Rhee SG, Woo HA, Kil IS, Bae SH. Peroxiredoxin functions as a peroxidase and a regulator and sensor of local peroxides. Journal of Biological Chemistry. 2012;287(7):4403–10. doi: 10.1074/jbc.R111.283432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nazıroğlu M. Molecular role of catalase on oxidative stress-induced Ca2+ signaling and TRP cation channel activation in nervous system. Journal of Receptors and Signal Transduction. 2012;32(3):134–41. doi: 10.3109/10799893.2012.672994 [DOI] [PubMed] [Google Scholar]
- 26.Eltayeb AE, Kawano N, Badawi GH, Kaminaka H, Sanekata T, Shibahara T, et al. Overexpression of monodehydroascorbate reductase in transgenic tobacco confers enhanced tolerance to ozone, salt and polyethylene glycol stresses. Planta. 2007;225(5):1255–64. doi: 10.1007/s00425-006-0417-7 [DOI] [PubMed] [Google Scholar]
- 27.Eltayeb AE, Kawano N, Badawi GH, Kaminaka H, Sanekata T, Morishima I, et al. Enhanced tolerance to ozone and drought stresses in transgenic tobacco overexpressing dehydroascorbate reductase in cytosol. Physiologia Plantarum. 2006;127(1):57–65. [Google Scholar]
- 28.Singh N, Mishra A, Jha B. Over-expression of the peroxisomal ascorbate peroxidase (SbpAPX) gene cloned from halophyte Salicornia brachiata confers salt and drought stress tolerance in transgenic tobacco. Marine biotechnology. 2014;16(3):321– doi: 10.1007/s10126-013-9548-6 [DOI] [PubMed] [Google Scholar]
- 29.Eltelib H, Fujikawa Y, Esaka M. Overexpression of the acerola (Malpighia glabra) monodehydroascorbate reductase gene in transgenic tobacco plants results in increased ascorbate levels and enhanced tolerance to salt stress. South African Journal of Botany. 2012;78:295–301. [Google Scholar]
- 30.Merchant A, Tausz M, Arndt SK, Adams MA. Cyclitols and carbohydrates in leaves and roots of 13 Eucalyptus species suggest contrasting physiological responses to water deficit. Plant, Cell & Environment. 2006;29(11):2017–29. [DOI] [PubMed] [Google Scholar]
- 31.Zhou M-L, Zhang Q, Zhou M, Sun Z-M, Zhu X-M, Shao J-R, et al. Genome-wide identification of genes involved in raffinose metabolism in Maize. Glycobiology. 2012;22(12):1775–85. doi: 10.1093/glycob/cws121 [DOI] [PubMed] [Google Scholar]
- 32.Nakai H, Ito T, Hayashi M, Kamiya K, Yamamoto T, Matsubara K, et al. Multiple forms of alpha-glucosidase in rice seeds (Oryza sativa L., var Nipponbare). Biochimie. 2007;89(1):49–62. doi: 10.1016/j.biochi.2006.09.014 [DOI] [PubMed] [Google Scholar]
- 33.Arsovski AA, Popma TM, Haughn GW, Carpita NC, McCann MC, Western TL. AtBXL1 encodes a bifunctional β-d-xylosidase/α-l-arabinofuranosidase required for pectic arabinan modification in Arabidopsis mucilage secretory cells. Plant Physiology. 2009;150(3):1219–34. doi: 10.1104/pp.109.138388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoch G. Cell wall hemicelluloses as mobile carbon stores in non‐reproductive plant tissues. Functional Ecology. 2007;21(5):823–34. [Google Scholar]
- 35.Roulin S, Buchala AJ, Fincher GB. Induction of (1→ 3, 1→ 4)-β-D-glucan hydrolases in leaves of dark-incubated barley seedlings. Planta. 2002;215(1):51–9. doi: 10.1007/s00425-001-0721-1 [DOI] [PubMed] [Google Scholar]
- 36.Rouyi C, Baiya S, Lee S-K, Mahong B, Jeon J-S, Ketudat-Cairns JR, et al. Recombinant expression and characterization of the cytoplasmic rice β-glucosidase Os1BGlu4. PloS one. 2014;9(5):e96712 doi: 10.1371/journal.pone.0096712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schubert S, Serraj RE, Mengel K. EFFECT OF DROUGHT STRESS ON GROWTH, SUGAR CONCENTRATIONS AND AMINO ACID ACCUMULATION IN N-2-FIXING ALFALFA (MEDICAGO SATIVA). Journal of Plant Physiology. 1995;146(4):541–6. [Google Scholar]
- 38.Ravanel S, Block MA, Rippert P, Jabrin S, Curien G, Rébeillé F, et al. Methionine metabolism in plants chloroplasts are autonomous for de novo methionine synthesis and can import s-adenosylmethionine from the cytosol. Journal of Biological Chemistry. 2004;279(21):22548–57. doi: 10.1074/jbc.M313250200 [DOI] [PubMed] [Google Scholar]
- 39.Hondorp ER, Matthews RG. Oxidation of cysteine 645 of cobalamin-independent methionine synthase causes a methionine limitation in Escherichia coli. Journal of bacteriology. 2009;191(10):3407–10. doi: 10.1128/JB.01722-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mitton JB, Grant MC, Yoshino AM. Variation in allozymes and stomatal size in pinyon (Pinus edulis, Pinaceae), associated with soil moisture. American Journal of Botany. 1998;85(9):1262–5. [PubMed] [Google Scholar]
- 41.Shiozaki N, Yamada M, Yoshiba Y. Analysis of salt-stress-inducible ESTs isolated by PCR-subtraction in salt-tolerant rice. Theoretical and applied genetics. 2005;110(7):1177–86. doi: 10.1007/s00122-005-1931-x [DOI] [PubMed] [Google Scholar]
- 42.Kirby EG, Molina-Rueda JJ. Drought Stress Tolerance Genes and Methods of Use Thereof to Modulate Drought Resistance in Plants. Google Patents; 2014.
- 43.Perlikowski D, Kierszniowska S, Sawikowska A, Krajewski P, Rapacz M, Eckhardt Ä, et al. Remodeling of Leaf Cellular Glycerolipid Composition under Drought and Re-hydration Conditions in Grasses from the Lolium-Festuca Complex. Frontiers in Plant Science. 2016;7(54):1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bates L, Waldren R, Teare I. Rapid determination of free proline for water-stress studies. Plant and soil. 1973;39(1):205–7. [Google Scholar]
- 45.Szabados L, Savoure A. Proline: a multifunctional amino acid. Trends in plant science. 2010;15(2):89–97. doi: 10.1016/j.tplants.2009.11.009 [DOI] [PubMed] [Google Scholar]
- 46.Veeranagamallaiah G, Chandraobulreddy P, Jyothsnakumari G, Sudhakar C. Glutamine synthetase expression and pyrroline-5-carboxylate reductase activity influence proline accumulation in two cultivars of foxtail millet (Setaria italica L.) with differential salt sensitivity. Environmental and Experimental Botany. 2007;60(2):239– [Google Scholar]
- 47.Selmar D, Kleinwächter M. Influencing the product quality by deliberately applying drought stress during the cultivation of medicinal plants. Industrial Crops and Products. 2013;42:558–66. [Google Scholar]
- 48.Paulick MG, Bertozzi CR. The Glycosylphosphatidylinositol Anchor: A Complex Membrane-Anchoring Structure for Proteins†. Biochemistry. 2008;47(27):6991–7000. doi: 10.1021/bi8006324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao Y-J, Chen X, Zhang M, Su P, Liu Y-J, Tong Y-R, et al. Molecular cloning and characterisation of farnesyl pyrophosphate synthase from Tripterygium wilfordii. PloS one. 2015;10(5):e0125415 doi: 10.1371/journal.pone.0125415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xiong L, Schumaker KS, Zhu J-K. Cell signaling during cold, drought, and salt stress. The plant cell. 2002;14(suppl 1):S165–S83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fukutoku Y, Yamada Y. Sources of proline‐nitrogen in water‐stressed soybean (Glycine max). II. Fate of 15N‐labelled protein. Physiologia plantarum. 1984;61(4):622–8. [Google Scholar]
- 52.Redecke L, Nass K, Deponte DP, White TA, Rehders D, Barty A, et al. Natively inhibited Trypanosoma brucei cathepsin B structure determined by using an X-ray laser. Science. 2014;339(6116):227–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Harrak H, Azelmat S, Baker EN, Tabaeizadeh Z. Isolation and characterization of a gene encoding a drought-induced cysteine protease in tomato (Lycopersicon esculentum). Genome. 2001;44(3):368–74. [DOI] [PubMed] [Google Scholar]
- 54.Singh R, Tiwari J K, Sharma V, et al. Role of Pathogen related protein families in defence mechanism with potential role in applied biotechnology. International Journal of Advanced Research, 2014; 2(8): 210–226. [Google Scholar]
- 55.Guo R, Xu X, Carole B, Li X, Gao M, Zheng Y, et al. Genome-wide identification, evolutionary and expression analysis of the aspartic protease gene superfamily in grape. BMC genomics. 2013;14(1):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Abid G, Muhovski Y, Mingeot D, Watillon B, Toussaint A, Mergeai G, et al. Identification and characterization of drought stress responsive genes in faba bean (Vicia faba L.) by suppression subtractive hybridization. Plant Cell, Tissue and Organ Culture (PCTOC). 2015;121(2):367–79. [Google Scholar]
- 57.Seki M, Narusaka M, Ishida J, Nanjo T, Fujita M, Oono Y, et al. Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high‐salinity stresses using a full‐length cDNA microarray. The Plant Journal. 2002;31(3):279–92. [DOI] [PubMed] [Google Scholar]
- 58.Cameron KD, Teece MA, Smart LB. Increased accumulation of cuticular wax and expression of lipid transfer protein in response to periodic drying events in leaves of tree tobacco. Plant physiology. 2006;140(1):176–83. doi: 10.1104/pp.105.069724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Papadopoulos MC, Verkman AS. Aquaporin water channels in the nervous system. Nature Reviews Neuroscience. 2013;14(4):265–77. doi: 10.1038/nrn3468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhou S, Hu W, Deng X, Ma Z, Chen L, Huang C, et al. Overexpression of the wheat aquaporin gene, TaAQP7, enhances drought tolerance in transgenic tobacco. PLoS One. 2012;7(12):e52439 doi: 10.1371/journal.pone.0052439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Qiao J, Shao M, Chen L, Wang J, Wu G, Tian X, et al. Systematic characterization of hypothetical proteins in Synechocystis sp. PCC 6803 reveals proteins functionally relevant to stress responses. Gene. 2013;512(1):6–15. doi: 10.1016/j.gene.2012.10.004 [DOI] [PubMed] [Google Scholar]
- 62.Budak H, Akpinar BA, Unver T, Turktas M. Proteome changes in wild and modern wheat leaves upon drought stress by two-dimensional electrophoresis and nanoLC-ESI–MS/MS. Plant molecular biology. 2013;83(1–2):89–103. doi: 10.1007/s11103-013-0024-5 [DOI] [PubMed] [Google Scholar]
- 63.Kaneko T, Tabata S. Complete genome structure of the unicellular cyanobacterium Synechocystis sp. PCC6803. Plant and Cell Physiology. 1997;38(11):1171–6. [DOI] [PubMed] [Google Scholar]
- 64.Yang Y, Dong C, Yang S, Li X, Sun X, Yang Y. Physiological and proteomic adaptation of the alpine grass Stipa purpurea to a drought gradient. Plos One. 2015;10(2):e0117475 doi: 10.1371/journal.pone.0117475 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.