Abstract
Giardia duodenalis is a common human and animal pathogen. It has been increasingly reported in wild and captive non-human primates (NHPs) in recent years. However, multilocus genotyping information for G. duodenalis infecting NHPs in southwestern China is limited. In the present study, the prevalence and multilocus genotypes (MLGs) of G. duodenalis in captive NHPs in southwestern China were determined. We examined 207 fecal samples from NHPs in Sichuan and Guizhou provinces, and 16 specimens were positive for G. duodenalis. The overall infection rate was 7.7%, and only assemblage B was identified. G. duodenalis was detect positive in northern white-cheeked gibbon (14/36, 38.9%), crab-eating macaque (1/60, 1.7%) and rhesus macaques (1/101, 0.9%). Multilocus sequence typing based on beta-giardin (bg), triose phosphate isomerase (tpi) and glutamate dehydrogenase (gdh) revealed nine different assemblage B MLGs (five known genotypes and four novel genotypes). Based on a phylogenetic analysis, one potentially zoonotic genotype of MLG SW7 was identified in a northern white-cheeked gibbon. A high degree of genetic diversity within assemblage B was observed in captive northern white-cheeked gibbons in Southwestern China, including a potentially zoonotic genotype, MLG SW7. To the best of our knowledge, this is the first report using a MLGs approach to identify G. duodenalis in captive NHPs in Southwestern China.
Introduction
Giardia duodenalis is the etiological agent of giardiasis, a gastrointestinal infection that is typically asymptomatic, but may also be severe in some individuals [1–3]. At present, there are eight distinct assemblages of G. duodenalis (A-H), assemblages A and B frequently infect humans and animals, assemblages C and D have been described in domestic and wild canines, assemblage E have been widely reported in ruminants but sporadically detected in NHPs and humans, assemblage F in cats, assemblage G in rodents and assemblage H in seals and gulls [4]. Assemblages A and B are considered zoonotic genotypes. In addition to humans, they are widely reported in non-human primates (NHPs) [4–6].
NHPs are valuable wildlife resources. Owing to their high genetic homology to humans, NHPs are important experimental models for clinical research and public health research. G. duodenalis have a monoxenous life cycle and can spread rapidly in captive NHPs [7]. Genetic polymorphism of G. duodenalis has been widely investigated in NHPs. Assemblages A, B and E are found in NHPs and assemblage B is dominant [5, 6]. Molecular analyses have revealed that assemblage A is further classified into three major subtypes (AI-AIII), but assemblage B includes many subtypes that have not been systematically categorized [4, 5].
However, little is known about genetic variation in G. duodenalis infecting NHPs based on multi-locus genotyping. Molecular analyses to date have typically focused on a single genetic locus [4, 8, 9]. Inconsistent genotyping results have sometimes been observed among different individual loci [4, 10]. To better understand the genetic heterogeneity and zoonotic potential of G. duodenalis, multi-locus genotyping (MLG) employing beta-giardin (bg), triose phosphate isomerase (tpi) and glutamate dehydrogenase (gdh) has been used for genotyping and subtyping G. duodenalis in humans and animals [11–13]. The aim of the present study was to characterize G. duodenalis in captive NHPs in Southwestern China. These findings improve our understanding of the genetic diversity and the transmission routes of G. duodenalis in NHPs.
Methods
Ethics statement
This study was reviewed and approved by the Institutional Animal Care and Use Committee of Sichuan Agricultural University under permit number DYY-S20156703. Prior to the collection of fecal specimens from NHPs, permission was obtained from owners.
Specimen collection
From March to May 2016 and September to November 2016, 207 fecal specimens from NHPs were collected from Sichuan and Guizhou provinces. Fresh fecal specimens were collected immediately after defecation on the ground and separately stored in 50-mL centrifuge tubes. The specimens were kept cool during transport and arrival at the Sichuan Agricultural University. Specifically, 101 samples were obtained from rhesus macaques from the National Experimental Macaque Reproduce Laboratory in Southwest China (n = 31), Chengdu Gaoxin rhesus macaque farm (n = 30), Chengdu zoo (n = 20) and Bifengxia zoo (n = 20). Thirty-six samples were from northern white-cheeked gibbons from zoos in Guiyang (n = 30), Chengdu (n = 2) and Bifengxia (n = 4). Nine samples were from Golden snub-nosed monkeys in the Chengdu zoo. Sixty samples were from crab-eating macaques in the National Experimental Macaque Reproduce Laboratory in Southwest China and the Chengdu Gaoxin rhesus macaque farm (Table 1). Samples were preserved in 2.5% potassium dichromate at 4°C in a refrigerator. All samples were processed within 24 h of collection.
Table 1. Prevalence of Giardia duodenalis in non-human primates.
| Common name (scientific name) | Area | No. tested | No. (%) of positive specimens |
|---|---|---|---|
| Rhesus macaque (Macaca mulatta) | National experimental Macaque Reproduce Laboratory in Southwest China | 31 | 0 (0) |
| Guiyang zoo | 20 | 1 (5) | |
| Bifengxia zoo | 20 | 0 (0) | |
| Chengdu Gaoxin rhesus macaque farm | 30 | 0 (0) | |
| Northern white-cheeked gibbon (Nomascus leucogenys) | Guiyang zoo | 30 | 14 (46.7) |
| Chengdu zoo | 2 | 0 (0) | |
| Bifengxia zoo | 4 | 0 (0) | |
| Golden snub-nosed mokey (Rhinopithecus roxellanae) | Chengdu zoo | 9 | 0 (0) |
| Crab-eating macaque (Macaca fascicularis) | Ya`an rhesus macaque base | 30 | 0 (0) |
| Chengdu Gaoxin rhesus macaque farm | 30 | 1 (3.3) |
DNA extraction and PCR amplification
Before extracting DNA, the fecal samples were washed with distilled water until potassium dichromate was removed. Genomic DNA was extracted using the PowerSoil® DNA Isolation Kit (MoBio, Carlsbad CA, USA) following the manufacturer`s instructions. DNA samples were stored in 100 μL of the kit's Solution Buffer at 20°C until use.
Each specimen was examined for G. duodenalis by nested PCR amplification of the beta-giardin (bg) gene [14], The bg-positive specimens were further characterized by PCR amplification of the tpi and gdh genes [11]. Secondary PCR products were visualized by staining with Golden View following 1% agarose gel electrophoresis.
Sequencing and phylogenetic analysis
The amplified products of the expected size were sequenced by Invitrogen (Shanghai, China). To determine the G. duodenalis assemblage, the sequences were aligned with sequences downloaded from the GenBank database based on a BLAST analysis (http://blast.ncbi.nlm.nih.gov) using ClustalX. For the phylogenetic analysis, sequences obtained in this study were used to construct a neighboring-joining tree using Mega 5 (http://www.megasoftware.net/). A total of 1000 replicates were used for the bootstrap analysis.
Statistical analysis
Differences in infection rates among NHPs and among animals in different areas were assessed using the chi-square test implemented in SPSS version 17.0 (SPSS Inc., Chicago, IL, USA). P < 0.05 was considered significant.
Results and discussion
In the bg-based PCR analysis of 207 specimens from 4 NHP species, 16 (7.7%) samples from 3 species were positive for G. duodenalis. All the positive specimens were successfully amplified and sequenced for the bg, tpi and gdh genes. Sequences were deposited in the GenBank database under the accession numbers KY696790-KY696821.
The infection rates ranged from 0% to 38.9% in the 4 species (Table 1). Specifically, 1 of 101 (0.9%) rhesus macaques and 1 of 60 (1.7%) crab-eating macaques were positive for G. duodenalis. Northern white-cheeked gibbons showed the highest infection rate (14/36, 38.9%). All golden snub-nosed monkeys (n = 9) were negative for G. duodenalis. The difference in infection rates among 4 species was significant (P<0.05). In China, six studies have examined G. duodenalis infection in NHPs in parks, zoos, farms and laboratories to date, and the overall infection rate was between 1.3% and 18.6% in these studies [7, 15–19]. The overall infection rate in our study (7.7%) was close to the total infection rate in Qianling Park in Guiyang (8.5%) [17], and was much lower than the total prevalence in zoos in China (18.6%) [15]. It was obviously higher than those reported in Guangxi (2.4%) [19], Qinling Mountain (2.0%) [16] and two other additional comprehensive parasite infection studies in China (2.2% and 1.3%) [7, 18]. Our results and those of previous studies indicate that G. duodenalis infection is common in wild and captive NHPs and has a wide geographic distribution in China.
In other countries, G. duodenalis infection in NHPs showed a similar trend to that observed in China. The overall infection rate of G. duodenalis in NHPs is between 2.2% and 47.0% [7, 20], indicating a wide range of infection rates. The prevalence in our present study was close to previous estimates in Italy (6.0%) [20] and Thailand (7.0%) [6], and it was lower than the infection rates reported in Uganda (11.1%) [21] and Croatia (50%) [22]. This result may be explained by differences among regions in climate, environmental management, NHPs species and animal exchange programs [5, 8, 20].
In this study, the infection rate for captive NHPs in Sichuan province was 0.6% (1/106), which is almost identical to that in a comprehensive parasite study performed in 2009–2015 in Sichuan (0.5%, 3/581) [18]. The infection rate in captive NHPs in Guizhou province was 30% (15/50), much higher than that of free-range NHPs in Guiyang (8.5%, 35/411) [17]. Additionally, 38.9% (14/36) of northern white-cheeked gibbons were positive for G. duodenalis, which was also higher than the infection rate in a previous study (14.3%, 2/14) [15]. These results suggest that captive northern white-cheeked gibbons are more prone to infection by G. duodenalis than wild animals. This might be explained by the single-host life cycle and the resilient infectious cysts of G. duodenalis [23]. Captive northern white-cheeked gibbons are closer to each other than free-range NHPs, and confined spaces result in the transmission of infectious cysts between NHPs. The high transmission between captive NHPs is consistent with those of previous studies in China [7, 15].
To date, assemblages A, B and E have been detected in NHP species in China. Assemblage A and B have both been found in captive and free-range NHPs, but assemblage E has only been found in captive NHPs [16]. In this study, only assemblage B was detected in 3 captive NHP species, consistent with a recent study in zoos in China [7]. In these previous studies, all specimens were obtained from captive NHPs inhabiting in zoos, farms or bases. The resilient infectious cysts of G. duodenalis may explain the low infection diversity of assemblages [4], and suggests that assemblage B is predominant in Sichuan province. Assemblage B was identified in rhesus macaques, northern white-cheeked gibbons and crab-eating macaques. According to a previous study in 2009–2015 [15], assemblage B was only identified in rhesus macaques and northern white-cheeked gibbons, but assemblages A and B were both identified in crab-eating macaques. This result may suggest that northern white-cheeked gibbons are more susceptible to assemblage B than to other assemblages, and assemblage A might be host-specific including few NHPs species.
No genetic variation was observed among the 16 gdh sequences. All of the 16 sequences were 100% similar to sequences of human isolates from Brazil (one strain was identical to EF507672 and 15 strains were identical to EF507682) [24]. The bg and tpi loci showed high levels of sequence polymorphism. 5 and 6 subtypes were identified in the 16 strains, including two new subtypes. At the bg locus, four known subtypes Bb-4 (KJ888977) [15], BIII (KF922976) [25], BIII-1 (EU637581) [26], and B1 (KM211793) [27] were found in 1, 7, 2 and 1 specimens, respectively. The novel subtype, named BIII-2, was found in 5 specimens in our study. At the tpi locus, five known subtypes B7 (JQ863259) [28], WB8 (KF679738) [7], EB5 (KT948110) [13], BIV (AB618783) [29] and WB6 (KJ888987) [15] were found in 1, 7, 1, 3 and 1 specimens, respectively. The novel subtypes, named B9, was found in 3 specimens (Table 2).
Table 2. Characterization of 16 specimens based on multi-locus sequences of bg, tpi and gdh genes.
| Isolate | Host | Geographic source | subtype/host or source/GenBank accession number | MLG | ||
|---|---|---|---|---|---|---|
| bg | tpi | gdh | ||||
| YA053 | Rhesus macaque | National experimental Macaque Reproduce Laboratory in Southwest China | Bb-4/Lemur catta/ KJ888977 | B7/wastewater/JQ863259 | BIV/Human/EF507672 | SW1 |
| GY004 | Northern white-cheeked gibbon | Guiyang zoo | BIII/Human/KF922976 | WB8/rhesus macaque/KF679738 | BIV/Human/EF507682 | SW2 |
| GY006 | Northern white-cheeked gibbon | Guiyang zoo | BIII-2/ northern white-cheeked gibbon /KY696824# | WB8/rhesus macaque/KF679738 | BIV/Human/EF507682 | SW3* |
| GY007 | Northern white-cheeked gibbon | Guiyang zoo | BIII-2/ northern white-cheeked gibbon /KY696825# | WB8/rhesus macaque/KF679738 | BIV/Human/EF507682 | SW3* |
| GY013 | Northern white-cheeked gibbon | Guiyang zoo | BIII/Human/KF922976 | B9/ northern white-cheeked gibbon /KY696810# | BIV/Human/EF507682 | SW4* |
| GY014 | Northern white-cheeked gibbon | Guiyang zoo | BIII/Human/KF922976 | WB8/rhesus macaque/KF679738 | BIV/Human/EF507682 | SW2 |
| GY015 | Northern white-cheeked gibbon | Guiyang zoo | BIII-2/ northern white-cheeked gibbon /KY696828# | B9/ northern white-cheeked gibbon /KY696812# | BIV/Human/EF507682 | SW4* |
| GY019 | Northern white-cheeked gibbon | Guiyang zoo | BIII/Human/KF922976 | EB5/Human/KT948110 | BIV/Human/EF507682 | SW5 |
| GY020 | Northern white-cheeked gibbon | Guiyang zoo | BIII/Human/KF922976 | WB8/rhesus macaque/KF679738 | BIV/Human/EF507682 | SW2 |
| GY021 | Northern white-cheeked gibbon | Guiyang zoo | BIII/Human/KF922976 | WB8/rhesus macaque/KF679738 | BIV/Human/EF507682 | SW2 |
| GY025 | Northern white-cheeked gibbon | Guiyang zoo | BIII-2/ northern white-cheeked gibbon /KY696832# | BIV/Human/AB618783 | BIV/Human/EF507682 | SW6* |
| GY028 | Northern white-cheeked gibbon | Guiyang zoo | BIII-1/Barbary macaque/EU637581 | BIV/Human/AB618783 | BIV/Human/EF507682 | SW7 |
| GY031 | Northern white-cheeked gibbon | Guiyang zoo | BIII-1/Barbary macaque/EU637581 | BIV/Human/AB618783 | BIV/Human/EF507682 | SW7 |
| GY033 | Northern white-cheeked gibbon | Guiyang zoo | BIII/Human/KF922976 | WB8/rhesus macaque/KF679738 | BIV/Human/EF507682 | SW2 |
| GY034 | Northern white-cheeked gibbon | Guiyang zoo | BIII-2/ northern white-cheeked gibbon /KY696836# | B9/ northern white-cheeked gibbon /KY696820# | BIV/Human/EF507682 | SW8* |
| CD010 | Crab-eating macaque | Chengdu Gaoxin rhesus macaque farm | B1/rhesus macaque/KM211793 | WB6/Mandrill/KJ888987 | BIV/Human/EF507682 | SW9 |
Note:
“#”was the novel subtypes;
“*” was the novel MLGs
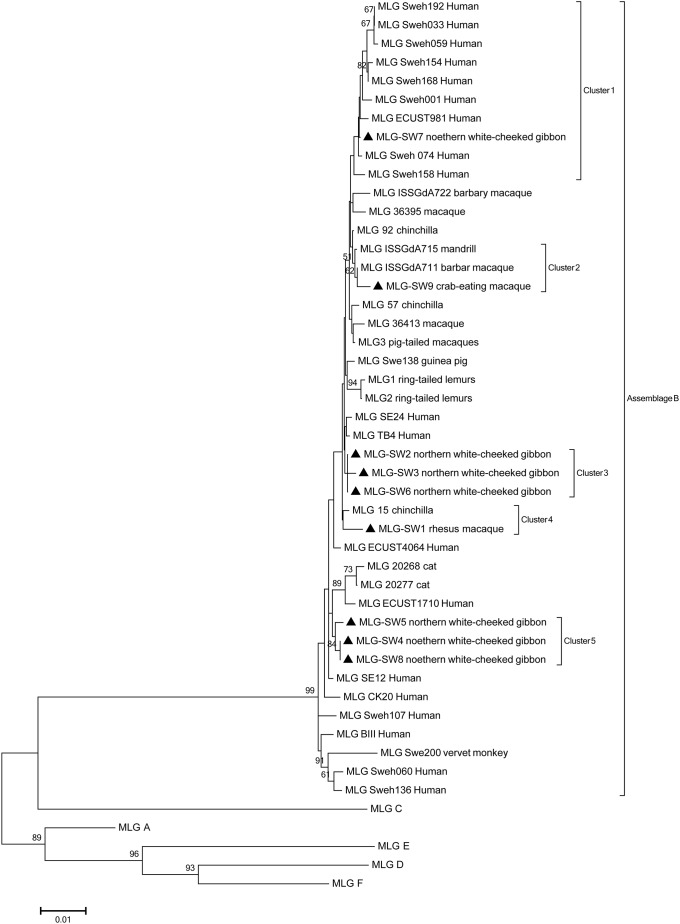
A total of 16 NHPs specimens (one from a rhesus macaque, one from a crab-eating macaque, and fourteen from northern white-cheeked gibbons) were classified as assemblage B and nine MLGs were identified among the 16 positive specimens. The subtype identities and geographical and host distributions of the nine MLGs are listed in Table 2. A phylogenetic analysis of the concatenated sequences of assemblage B revealed that nine MLGs in this study formed five clusters. MLG SW7, SW9 and SW1 were distributed in three separate clusters. SW 2, 3 and 6 isolated from northern white-cheeked gibbons formed cluster 3. Cluster 5 included SW4, 5 and 8, all of which were isolated from northern white-cheeked gibbons (Fig 1).
Fig 1. Phylogenetic relationship of Giardia duodenalis assemblage B MLGs inferred by the neighbor-joining analysis of concatenated bg, tpi and gdh sequences.
Bootstrap values greater than 50% from 1000 replicates are shown. Concatenated sequences from this study are marked by filed roundness.
Phylogenetic analyses showed that MLG SW7 belonged to a zoonotic group. Given the zoonotic potential of this subtype, epidemiological and source tracking investigations as well as strict surveillance in captive NHPs in southwestern China are needed. MLG SW9 was closely related to the sequences obtained from NHPs in other studies [11]. MLG SW1 was similar to the sequences isolated from chinchillas, suggesting the potential for transmission of G. duodenalis between animals [16]. Other MLGs formed two separate clusters. In this study, most MLGs (7 MLGs) were found in northern white-cheeked gibbons suggesting greater genetic heterogeneity in G. duodenalis from this species [15].
Conclusion
The results of the present study confirm previous findings that assemblage B is dominant in northern white-cheeked gibbons. We first used a MLGs approach to identify G. duodenalis in captive NHPs in Southwestern China. One genotype of the potentially zoonotic assemblage B of MLG SW7 strain was identified in a northern white-cheeked gibbon. This suggests that the zoonotic transmission of Giardia might occur between the northern white-cheeked gibbon and humans. Additionally, high degree genetic diversity of assemblage B MLGs (7 MLGs) was detected in captive northern white-cheeked gibbons in Southwestern China. Additional MLGs studies of captive NHPs are needed, to better characterize genetic diversity and the routes of transmission of G. duodenalis between NHPs and humans or other animals.
Acknowledgments
We thank Xuehan Liu and Lei Deng for giving advice on sample collection. We sincerely thank Tao Wang from University of Melbourne for comment on the draft manuscript.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by the National Key Research and Development Program of China (2016YFD0501009); the National Natural Science Foundation of China (31272620); and the Chengdu Giant Panda Breeding Research Foundation (CPF2015-4). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Thompson SC. Giardia lamblia in children and the child care setting: a review of the literature. J Paediatr Child Health. 1994: 30(3):202–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ehsan AM, Geurden T, Casaert S, Parvin SM, Islam TM, Ahmed UM, et al. Assessment of zoonotic transmission of Giardia and Cryptosporidium between cattle and humans in rural villages in Bangladesh. PloS one. 2015: 10(2):e0118239 doi: 10.1371/journal.pone.0118239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jenkins EJ, Castrodale LJ, de Rosemond SJ, Dixon BR, Elmore SA, Gesy KM, et al. Tradition and transition: parasitic zoonoses of people and animals in Alaska, northern Canada, and Greenland. Adv Parasitol. 2013: 82:33–204. doi: 10.1016/B978-0-12-407706-5.00002-2 . [DOI] [PubMed] [Google Scholar]
- 4.Feng Y, Xiao L. Zoonotic potential and molecular epidemiology of Giardia species and giardiasis. Clin Microbiol Rev. 2011: 24(1):110–140. doi: 10.1128/CMR.00033-10 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ryan U, Caccio SM. Zoonotic potential of Giardia. Int J Parasitol. 2013: 43(12–13):943–956. doi: 10.1016/j.ijpara.2013.06.001 . [DOI] [PubMed] [Google Scholar]
- 6.Sricharern W, Inpankaew T, Keawmongkol S, Supanam J, Stich RW, Jittapalapong S. Molecular detection and prevalence of Giardia duodenalis and Cryptosporidium spp. among long-tailed macaques (Macaca fascicularis) in Thailand. Infect Genet Evol. 2016: 40:310–314. doi: 10.1016/j.meegid.2016.02.004 . [DOI] [PubMed] [Google Scholar]
- 7.Karim MR, Zhang S, Jian F, Li J, Zhou C, Zhang L, et al. Multilocus typing of Cryptosporidium spp. and Giardia duodenalis from non-human primates in China. Int J Parasitol. 2014: 44(13):1039–1047. doi: 10.1016/j.ijpara.2014.07.006 . [DOI] [PubMed] [Google Scholar]
- 8.Levecke B, Geldhof P, Claerebout E, Dorny P, Vercammen F, Cacciò SM, et al. Molecular characterisation of Giardia duodenalis in captive non-human primates reveals mixed assemblage A and B infections and novel polymorphisms. Int J Parasitol. 2009: 39(14):1595–1601. doi: 10.1016/j.ijpara.2009.05.013 . [DOI] [PubMed] [Google Scholar]
- 9.Takumi K, Swart A, Mank T, Lasek-Nesselquist E, Lebbad M, Cacciò SM, et al. Population-based analyses of Giardia duodenalis is consistent with the clonal assemblage structure. Parasit Vectors. 2012: 5(1):168 doi: 10.1186/1756-3305-5-168 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lebbad M, Petersson I, Karlsson L, Botero-Kleiven S, Andersson JO, Svenungsson B, et al. Multilocus genotyping of human Giardia isolates suggests limited zoonotic transmission and association between assemblage B and flatulence in children. PLoS Negl Trop Dis. 2011: 5(8):e1262 doi: 10.1371/journal.pntd.0001262 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cacciò SM, Beck R, Lalle M, Marinculic A, Pozio E. Multilocus genotyping of Giardia duodenalis reveals striking differences between assemblages A and B. Int J Parasitol. 2008, 38(13):1523 doi: 10.1016/j.ijpara.2008.04.008 . [DOI] [PubMed] [Google Scholar]
- 12.Huey CS, Mahdy MA, Al-Mekhlafi HM, Nasr NA, Lim YA, Mahmud R, et al. Multilocus genotyping of Giardia duodenalis in Malaysia. Infect Genet Evol. 2013: 17:269–276. doi: 10.1016/j.meegid.2013.04.013 . [DOI] [PubMed] [Google Scholar]
- 13.Wegayehu T, Karim MR, Li J, Adamu H, Erko B, Zhang L, et al. Multilocus genotyping of Giardia duodenalis isolates from children in Oromia Special Zone, central Ethiopia. BMC Microbiol. 2016: 16:89 doi: 10.1186/s12866-016-0706-7 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li J, Qi M, Chang Y, Wang R, Li T, Dong H, et al. Molecular Characterization of Cryptosporidium spp., Giardia duodenalis, and Enterocytozoon bieneusi in Captive Wildlife at Zhengzhou Zoo, China. J Eukaryot Microbiol. 2015: 62(6):833–839. doi: 10.1111/jeu.12269 . [DOI] [PubMed] [Google Scholar]
- 15.Karim MR, Wang R, Yu F, Li T, Dong H, Li D, Zhang L et al. Multi-locus analysis of Giardia duodenalis from nonhuman primates kept in zoos in China: Geographical segregation and host-adaptation of assemblage B isolates. Infect Genet Evol. 2015: 30(30):82–88. doi: 10.1016/j.meegid.2014.12.013 . [DOI] [PubMed] [Google Scholar]
- 16.Du SZ, Zhao GH, Shao JF, Fang YQ, Tian GR, Zhang LX, et al. Cryptosporidium spp., Giardia duodenalis, and Enterocytozoon bieneusi in Captive Non-Human Primates in Qinling Mountains. Korean J Parasitol. 2015: 53(4):395–402. doi: 10.3347/kjp.2015.53.4.395 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ye J, Xiao L, Ma J, Guo M, Liu L, Feng Y. Anthroponotic enteric parasites in monkeys in public park, China. Emerg Infect Dis. 2012: 18(10):1640–1643. doi: 10.3201/eid1810.120653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li J, Dong H, Wang R, Yu F, Wu Y, Chang Y, et al. An investigation of parasitic infections and review of molecular characterization of the intestinal protozoa in nonhuman primates in China from 2009 to 2015. Int J Parasitol Parasites Wildl. 2017: 6(1):8–15. doi: 10.1016/j.ijppaw.2016.12.003 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ye J, Xiao L, Li J, Huang W, Amer SE, Guo Y, et al. Occurrence of human-pathogenic Enterocytozoon bieneusi, Giardia duodenalis and Cryptosporidium genotypes in laboratory macaques in Guangxi, China. Parasitol Int. 2013: 63(1):132–137. doi: 10.1016/j.parint.2013.10.007 . [DOI] [PubMed] [Google Scholar]
- 20.Berrilli F, Prisco C, Friedrich KG, Di Cerbo P, Di Cave D, De Liberato C. Giardia duodenalis assemblages and Entamoeba species infecting non-human primates in an Italian zoological garden: zoonotic potential and management traits. Parasit Vectors. 2011: 4:199 doi: 10.1186/1756-3305-4-199 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnston AR, Gillespie TR, Rwego IB, Mclachlan TL, Kent AD, Goldberg TL. Molecular epidemiology of cross-species Giardia duodenalis transmission in Western Uganda. PLoS Negl Trop Dis. 2012: 6(4):e1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beck R, Sprong H, Bata I, Lucinger S, Pozio E, Caccio SM. Prevalence and molecular typing of Giardia spp. in captive mammals at the zoo of Zagreb, Croatia. Vet Parasitol. 2011: 175(1–2):40–46. doi: 10.1016/j.vetpar.2010.09.026 . [DOI] [PubMed] [Google Scholar]
- 23.Lee MF, Cadogan P, Eytle S, Copeland S, Walochnik J, Lindo JF. Molecular epidemiology and multilocus sequence analysis of potentially zoonotic Giardia spp. from humans and dogs in Jamaica. Parasitol Res. 2016: 116(1):1–6. [DOI] [PubMed] [Google Scholar]
- 24.Souza SL, Gennari SM, Richtzenhain LJ, Pena HF, Funada MR, Cortez A, et al. Molecular identification of Giardia duodenalis isolates from humans, dogs, cats and cattle from the state of Sao Paulo, Brazil, by sequence analysis of fragments of glutamate dehydrogenase (gdh) coding gene. Vet Parasitol. 2007: 149(3–4):258–264. doi: 10.1016/j.vetpar.2007.08.019 . [DOI] [PubMed] [Google Scholar]
- 25.Durigan M, Abreu AG, Zucchi MI, Franco RM, de Souza AP. Genetic diversity of Giardia duodenalis: multilocus genotyping reveals zoonotic potential between clinical and environmental sources in a metropolitan region of Brazil. PloS one. 2014: 9(12):e115489 doi: 10.1371/journal.pone.0115489 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Caccio SM, Beck R, Lalle M, Marinculic A, Pozio E. Multilocus genotyping of Giardia duodenalis reveals striking differences between assemblages A and B. Int J Parasitol. 2008: 38(13):1523–1531. doi: 10.1016/j.ijpara.2008.04.008 . [DOI] [PubMed] [Google Scholar]
- 27.Li W, Zhong Z, Qu Y, Liu X, Xie N, Deng j, et al. Genetic identification of new subtype B11 of Giardia from a monkey. Chinese Vet Sci. 2015: (2):156–160. [Google Scholar]
- 28.Li N, Xiao L, Wang L, Zhao S, Zhao X, Duan L, et al. Molecular surveillance of Cryptosporidium spp., Giardia duodenalis, and Enterocytozoon bieneusi by genotyping and subtyping parasites in wastewater. PLoS Nefl Trop Dis. 2012: 6(9):e1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Abe N, Teramoto I. Molecular evidence for person-to-person transmission of a novel subtype in Giardia duodenalis assemblage B at the rehabilitation institution for developmentally disabled people. Parasitol Res. 2012: 110(2):1025–1028. doi: 10.1007/s00436-011-2564-4 . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.