ABSTRACT
M2-polarized (alternatively activated) macrophages play an important role in the progression of hepatocellular carcinoma (HCC). Allograft inflammatory factor 1 (AIF1) is overexpressed in M2-polarized macrophages. This study explored the role of AIF1 in tumor-associated macrophages in HCC. Macrophages were stimulated with colony-stimulating factor 1 (CSF1) to characterize the regulatory pathway of AIF1 in macrophages. The chromatin immunoprecipitation and luciferase reporter gene assay were conducted to examine transcription factors associated with AIF1 expression. AIF1 was down or upregulated, and the effects on tumor progression were evaluated by using in vitro and in vivo co-culture systems. A cytokine array was performed to screen the downstream functional components of AIF1. Tumor tissue from 206 patients with HCC were used to explore the clinical significance of AIF1. AIF1 induced a M2-like phenotype of macrophages. By facilitating the binding of c-Jun to the promoter of AIF1, CSF1 secreted from hepatoma cells increased AIF1 expression through the CSF1R-MEK1/2-Erk1/2-c-Jun axis. AIF1 expressed in macrophages promoted the migration of hepatoma cells in co-culture system of RAW264.7 and Hepa1-6 and tumor growth in an animal model. The cytokine array showed that CXCL16 was increased in RAW264.7 cells with overexpressed AIF1, leading to enhanced tumor cell migration. In human HCC tissue, AIF1-positive macrophages in the adjacent microenvironment was associated with microvascular invasion and advanced TNM stages and with patients' overall and disease-free survival (p = 0.002 for both). AIF1 expression in macrophages plays a pivotal role in the interaction between macrophages and hepatoma cells.
KEYWORDS: AIF1, c-Jun, CSF1/CSF1R, CXCL16, hepatocellular carcinoma, tumor-associated macrophages
Introduction
Worldwide, hepatocellular carcinoma (HCC) is the sixth most common malignancy and the third leading cause of cancer-related death.1 Surgical resection is the primary treatment toward curative outcome for HCC patients,2 but the prognosis after surgical resection remains poor because of a high recurrence rate.3-6 For HCC, most tumor recurrences occur within the remaining liver tissue, usually adjacent to the surgical margin, implying that adjacent liver tissue may provide a favorable microenvironment for the growth of possible residual tumor cells.7, 8 The tumor microenvironment, which comprises tumor-associated immune cells, endothelial cells, fibroblasts, cytokines secreted by these cells, and the extracellular matrix, plays an important role in the progression of tumor cells.9 Tumor-associated macrophages (TAMs) are the most abundant immune cells within the tumor microenvironment, and they have been reported to be associated with poor prognosis in a wide variety of cancers. The number of infiltrated macrophages within the adjacent liver tissue was associated with poor prognosis after surgical resection of HCC.10 Although TAMs within the adjacent microenvironment appear to have a role in tumor progression,11, 12 the underlying mechanism is not fully understood. Based on various environmental stimulus, macrophages differentiate toward two different polarization statuses, termed M1 and M2. M1-polarized macrophages exhibit a high antigen-presenting capacity and participate in the antitumor immune response, while M2-polarized macrophages exhibit a low antigen-presenting capacity and facilitate tumor progression by secreting distinct protumor cytokines.13, 14 Notably, although M1-polarized macrophages show some antitumor effect, interleukin (IL)-1, IL-6, and other cytokines secreted by M1-polarized macrophages have been shown to be involved in a wide range of tumorigenic processes.15, 16 Determining the differences between M1- and M2-polarized macrophages may help elucidate the role of TAMs in HCC progression.
CSF2 or CSF1 and its receptor signaling independently regulate macrophages toward a M1 or M2 polarization status.17, 18 Gene Expression Omnibus data sets (GSE66805) comparing the gene expression profiling of human peripheral blood mononuclear cells (PBMCs) stimulated with colony-stimulating factor 2 (CSF2) (M1-polarized macrophages) and human PBMCs stimulated with CSF1 (M2-polarized macrophages) showed that allograft inflammatory factor 1 (AIF1) was differentially expressed in M1- and M2-polarized macrophages. The mRNA expression of AIF1 was significantly higher in M2 compared with M1 macrophages (p = 1.3 × 10−5). AIF1 is a 17-kDa IFNγ-induced calcium-binding protein.19 It is mainly expressed on macrophages and serves as a cytoskeleton-related protein, and it is associated with the migration and phagocytosis of macrophages.20, 21 Overexpression of AIF1 in the mouse macrophage cell line RAW264.7 resulted in increased secretion of IL-6, IL-10, and IL-12 upon lipopolysaccharide stimulation.22 However, whether AIF1 contributes to the M2 polarization of macrophages in an HCC microenvironment is unclear.
In the present study, we aimed to illustrate the regulatory mechanism of AIF1 in the HCC microenvironment and the role of AIF1 in the progression of HCC.
Results
AIF1 was exclusively expressed on macrophages within HCC microenvironment
Immunohistochemical staining was performed on human HCC tissue sections. AIF1-positive cells had a relatively smaller size and polymorphic branches. The expression of AIF1 was higher in the adjacent area than within tumor tissue, also than within normal liver tissue (Fig. 1A). Immunofluorescence confocal imaging showed that AIF1 was co-expressed with CD68, which is an accepted macrophage marker (Fig. 1B). In consistency with the immunohistochemical staining, the number of AIF1+CD68+ macrophages was higher in the adjacent area than within tumor tissue. Western blot analysis showed that AIF1 was exclusively expressed on different macrophage cell lines, including THP-1-derived macrophages, mouse bone marrow-derived macrophages (BMDMs) and RAW264.7, but not on various human and mouse hepatoma cell lines, human umbilical vein endothelial cells (HUVECs), and liver stellate cell lines (LX2) (Fig. 1C).
Figure 1.
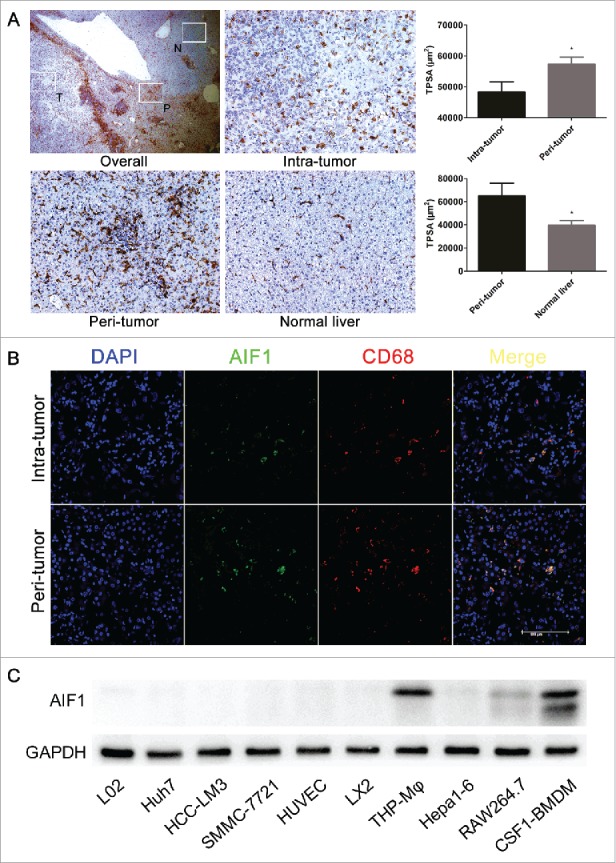
AIF1 was exclusively expressed in macrophages in HCC. (A) AIF1 positive cells were abundant in the adjacent peritumoral area of HCC ( × 25). The expression of AIF1 was significantly higher in adjacent liver tissue than within the tumor tissue (from 206 HCC patients), also than within normal liver tissue (from 20 HCC patients) ( × 200). (B) Immunofluorescence confocal analysis of HCC showed that AIF1 was co-expressed with CD68 within the intratumual and peritumoral microenvironment. The scale bar indicates a length of 100 μm. (C) Western blot showed that AIF1 was specifically expressed in various macrophage cell lines but not in liver cells, liver cancer cells, endothelial cells and astrocytes; *p < 0.05. TPSA, total positive staining area.
AIF1 played a pivotal role in the proliferation, migration, and M2 polarization of macrophages
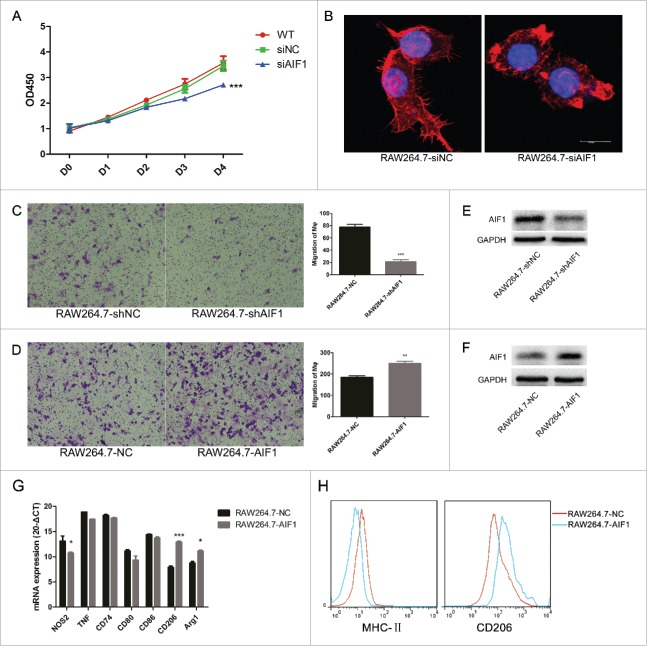
Inhibiting the expression of AIF1 using siRNAs impaired the proliferation of RAW264.7 cells by inducing the cell cycle arrest in the S phase (Fig. 2A, Fig. S1). Phalloidin-TRITC staining of RAW264.7 cells showed a disordered structure of the cytoskeleton after inhibition of the expression of AIF1 (Fig. 2B). Constitutively knocking down of AIF1 attenuated the migration of RAW264.7 cells upon CSF1 stimulation (Fig. 2C). In contrast, RAW264.7 cells overexpressing AIF1 were more attractant to CSF1 stimulation (Fig. 2D).
Figure 2.
AIF1 was associated with the proliferation, migration, and M2 polarization of macrophages. (A) Inhibiting AIF1 with siRNA transfection decreased the number of RAW264.7 cells. (B) Phalloidin-TRITC staining showed that inhibiting AIF1 with siRNA transfection resulted in malformation of the cytoskeleton. (C, D) RAW264.7 cells were less attractant to CSF1 stimulation (100 ng/mL) after constitutive knockdown of AIF1 and more attractant to CSF1 stimulation (100 ng/mL) after constitutive overexpression of AIF1. (E, F) Western blot confirmed AIF1 was effectively knocked down or overexpressed after infection with lentivirus containing shRNA targeting AIF1 or transfection with the AIF1 overexpression plasmid. (G) QRT-PCR showed that overexpression of AIF1 in RAW264.7 cells exhibited increased markers associated with M2 polarization and decreased those associated with M1 polarization. (H) Flow cytometric assay showed that overexpression of AIF1 in RAW264.7 cells showed increased CD206 and decreased MHC-II; *p < 0.05; ** p < 0.01; ***p < 0.001.
CSF1-induced BMDMs (CSF1-BMDMs), which were regarded as M2-like macrophages,17, 18 exhibited a higher migration capacity than CSF2-induced BMDMs (CSF2-BMDMs) (M1-like macrophage) (Fig. S2A). The expression of AIF1 was significantly higher in CSF1-BMDMs than in CSF2-BMDMs (Fig. S2B), so was in THP-1-derived M1 and M2 macrophages (Figs. S2C–F).
To investigate the effect of AIF1 on macrophage polarization, qRT-PCR was performed to evaluate the following markers associated with macrophage polarization: TNF, iNOS, CD80, CD86, CD74 (MHC-II-associated invariant chain), CD206, and Arg1. RAW264.7 cells overexpressing AIF1 exhibited a M2-polarized phenotype, which implicated AIF1 as having a role in M2 polarization (Fig. 2G). Flow cytometry of classic M2-associated marker CD206 and M1-associated marker MHC-II also showed that upregulation of AIF1 in RAW264.7 cells promoted an M2-like phenotype (Fig. 2H).
AIF1 expression in TAMs was induced by CSF1-CSF1R-c-Jun signaling pathway
Exogenous CSF1 induced the expression of AIF1 in RAW264.7 cells, which was blocked by the CSF1R inhibitor PLX3397 (Figs. 3A and B). The expression of AIF1 was also increased in TAMs. Twenty-four hours after being cultured in the CM of mouse Hepa1–6 and H22 cell lines, both RAW264.7 cells and BMDMs showed an increase of AIF1 expression (Fig. 3C, Fig. S3A). PLX3397 blocked the upregulation of AIF1 induced by the CM of Hepa1-6 and H22 cells (Fig. 3C). The expression of AIF1 in human THP-1-derived macrophages was also increased when cultured in the CM of HCCLM3, a human hepatoma cell line (Fig. S3B).
Figure 3.
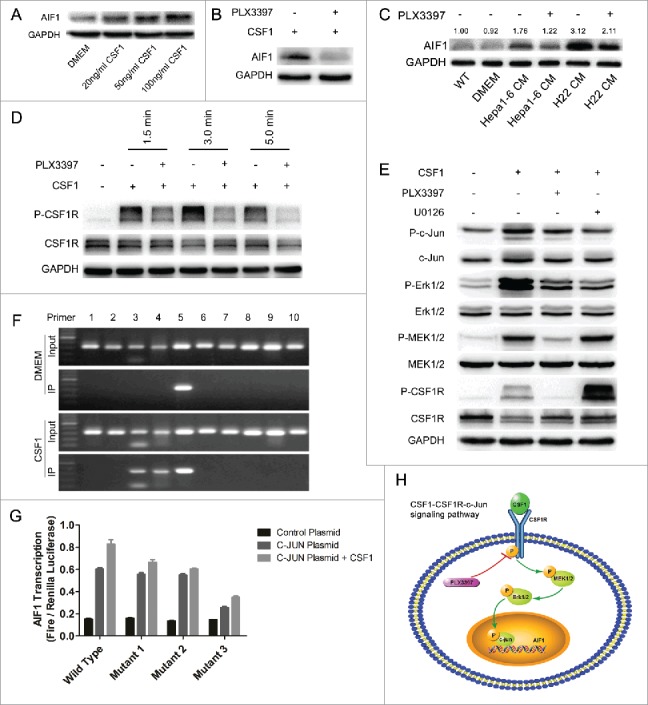
CSF1 secreted from hepatoma cells induced the expression of AIF1 in macrophages through the CSF1R-MEK1/2-Erk1/2-c-Jun signaling pathway. (A) CSF1 increased the expression of AIF1 in a dose-dependent manner. (B) The CSF1R inhibitor PLX3397 blocked the upregulation of AIF1 induced by CSF1. (C) The conditioned medium of Hepa1-6 and H22 cells increased the expression of AIF1 in macrophages, which was blocked by PLX3397. (D) Exposure to CSF1 induced the phosphorylation of CSF1R in RAW264.7 cells, which was blocked by PLX3397. (E) Thirty minutes after CSF1 stimulation, the phosphorylation of CSF1R, MEK1/2, Erk1/2, and c-Jun was increased, which was blocked by PLX3397. The MEK1/2 inhibitor U0126 decreased the phosphorylation of Erk1/2 and c-Jun.. (F) ChIP assay showed that c-Jun bound to the promoter region of AIF1. CSF1 increased the binding of c-Jun to the promoter region of AIF1. Sequence covered by primer 10 was near the start of the ORF. (G) Luciferase reporter gene assay showed that c-Jun promoted the expression of AIF1 by binding to the tgaattct sequence 1720bp–1728bp upstream of the ORF, which was accelerated by CSF1. Three mutated sequences were as follows: tgactttt (2156bp to 2164bp upstream of the ORF); tgatttaa (2037bp to 2045bp upstream of the ORF); tgaattct (1720bp to 1728bp upstream of the ORF). (H) An illustration showing the upstream pathway of AIF1 in the presence of CSF1; ***p < 0.001.
Shortly after exogenous CSF1 stimulation, CSF1R phosphorylation was observed in both RAW264.7 cells and BMDMs (Fig. 3D, Fig. S3C). MEK1/2, Erk1/2, and c-Jun, which are downstream molecules of the CSF1R activation,23, 24 were phosphorylated or increased thereafter, outcomes that were blocked by the CSF1R inhibitor PLX3397 (Fig. 3E). The MEK1/2 inhibitor U0126 decreased the CSF1-induced phosphorylation of Erk1/2 and the downstream transcription factor c-Jun by inhibiting the activity of P-MEK1/2 (Fig. 3E). To show the interaction between c-Jun and AIF1 expression, a ChIP-RT-PCR assay was performed. Ten pairs of primers were designed to cover the full length of the promoter region of AIF1. RT-PCR showed that c-Jun bound to the promoter region of AIF1, sequence covered by primer 5 (Fig. 3F). Exogenous CSF1 stimulation increased the binding of c-Jun to the promoter region, sequence covered by primers 3, 4, and 5 (Fig. 3F). On the basis, a luciferase reporter gene assay was performed. C-Jun plasmid and the AIF1 promoter luciferase plasmids (wild type and three mutants targeting the preceding three possible binding sequences of c-Jun on the AIF1 promoter) were constructed. The results showed that c-Jun increased the expression of AIF1, which was accelerated by CSF1. The tgaattct sequence 1720bp to 1728bp upstream of the open reading frame (ORF) was the key binding site for c-Jun, mutated which resulted in significantly decreased AIF1 transcription (Fig. 3G). These results indicated that the CSF1-CSF1R-MEK1/2- Erk1/2-c-jun axis was a vital upstream regulator of AIF1 expression in macrophages (Fig. 3H).
AIF1 expression in macrophages-facilitated tumor progression by the secretion of CXCL16
AIF1 in RAW264.7 cells was downregulated with liposome-mediated siRNA transfection and lentivirus-mediated shRNA transfection or upregulated with lentivirus-mediated AIF1 overexpression plasmid transfection (Figs. 2E and F; Figs. S4A and B, 5A). In co-cultures of Hepa1-6 with RAW264.7 cells for 24 h, the RAW264.7 cells that overexpressed AIF1 increased the migration of Hepa1-6 (Fig. 4A). The migration of Hepa1-6 was attenuated when co-cultured with RAW264.7 cells with downregulated AIF1 expression (Fig. 4A, Figs. 5B and C). The proliferation of Hepa1-6 cells was impaired in the CM of RAW264.7 cells with downregulated AIF1 expression, compared with the CM of control RAW264.7 cells, by inducing cell cycle arrest in the G1 phase; the proliferation was accelerated in the CM of RAW264.7 cells overexpressing AIF1, with increased Hepa1-6 cells in the S and G2 phases (Fig. 4B, Fig. S6).
Figure 4.
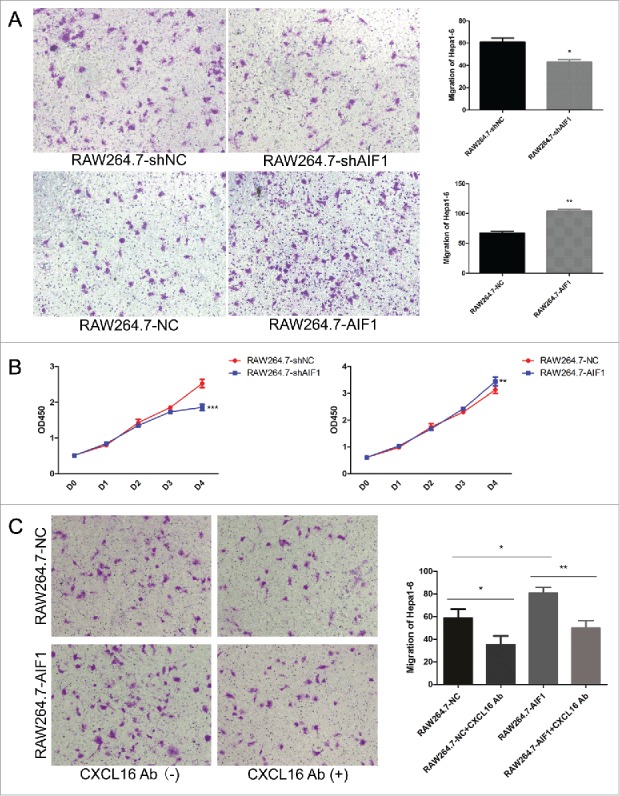
AIF1 enhanced the proliferation and migration of Hepa1-6 by the secretion of CXCL16. (A) In the Corning 3422 Boyden chambers, RAW264.7 cells in the lower chamber were co-cultured with Hepa1-6 cells in the upper chamber for 24 h. The migration of Hepa1-6 cells was attenuated with RAW264.7 cells downregulating AIF1 and enhanced with RAW264.7 cells overexpressing AIF1. (B) The conditioned medium of RAW264.7 cells downregulating AIF1 decreased the number of Hepa1-6 cells, and the conditioned medium of RAW264.7 cells overexpressing AIF1 increased the number of Hepa1-6 cells. (C) AIF1 promoted the migration of Hepa1-6 cells, which was blocked by the neutralizing antibody against CXCL16; *p < 0.05; **p < 0.01; ***p < 0.001.
Figure 5.
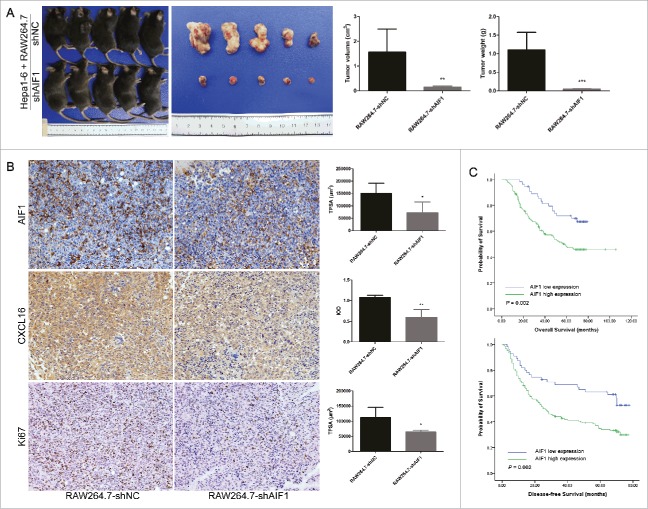
AIF1-positive macrophages promoted the progression of HCC. (A) RAW264.7-shNC/shAIF1 was co-injected with Hepa1-6 to establish a subcutaneous tumor model. Tumor sizes and weights were smaller after co-injection of RAW264.7 cells with downregulated AIF1 and Hepa1-6 cells as compared with that after co-injection of control RAW264.7 cells and Hepa1-6 cells. (B) A lower expression of AIF1, CXCL16 and Ki67 were detected in tumors derived from co-injection of RAW264.7 cells with downregulated AIF1 and Hepa1-6 cells, as compared with those from co-injection of control RAW264.7 cells and Hepa1-6 cells. (C) In human HCC tissue, high expression of AIF1 in the adjacent liver tissue was associated with poor overall survival (p = 0.002) and disease-free survival (p = 0.002) after surgical resection; *p < 0.05; **p < 0.01. TPSA, total positive staining area. IOD, integrated optical density.
Mouse cytokine antibody array analysis of the CM from RAW264.7 cells (overexpressing or down-regulating AIF1) co-cultured with Hepa1-6 cells showed that AIF1 increased the secretion of CXCL16, which was among the most abundant cytokines in the supernatant of RAW264.7 cells (Fig. S7A). ELISA of the CM from RAW264.7 cells co-cultured with Hepa1-6 cells showed that the concentration of CXCL16 was increased in RAW264.7 cells overexpressing AIF1 and decreased in RAW264.7 cells downregulating AIF1 (Fig. S7B). Blocking CXCL16 using CXCL16-neutralizing antibody attenuated the migration of Hepa1-6 promoted by AIF1 (Fig. 4C).
Animal study showed that RAW264.7 cells with downregulated AIF1 decreased the growth of subcutaneous tumor, compared with control RAW264.7 cells (Fig. 5A). Accordingly, the expression of AIF1 was decreased in TAMs in the junctional area, and the expression of CXCL16 and proliferation-associated protein Ki67 were also significantly reduced within the junctional area as shown by immunohistochemical staining (Fig. 5B). The number of CXCL16-expressing macrophages was smaller in the junctional area of tumor co-implanted with RAW264.7-shAIF1 than RAW264.7-shNC (Fig. S8).
AIF1 expression on macrophages in the adjacent microenvironment was associated with poor prognosis after surgical resection of HCC
Immunohistochemical staining was performed to study the clinical significance of AIF1 expression in macrophages. AIF1 expression in the adjacent microenvironment was associated with the number of tumor nodules (p = 0.019), the presence of microvascular invasion (p = 0.005), and advanced TNM stage (p = 0.006) (Table 1), suggesting that AIF1 expression on macrophages may promote tumor cell dissemination. Furthermore, high infiltration of AIF1-positive cells in the adjacent liver tissue was associated with poor overall survival (p = 0.002) and disease-free survival (p = 0.002) after resection of HCC (Fig. 5C). Multivariate analysis of factors associated with the prognosis after resection of HCC showed that AIF1 was an independent prognostic factor of the overall survival (p = 0.039) and disease-free survival (p = 0.023) (Table 2).
Table 1.
Basic clinicopathological parameters of included patients.
| Peritumoral AIF1 expression |
|||
|---|---|---|---|
| Low | High | p value | |
| Age (year) | 52.51 ± 11.85 | 52.25 ± 9.99 | 0.873 |
| Gender | 0.133 | ||
| Male | 43 | 131 | |
| Female | 12 | 20 | |
| HBsAg | 0.170 | ||
| Negative | 11 | 19 | |
| Positive | 43 | 131 | |
| Concurrent cirrhosis | 0.304 | ||
| No | 15 | 31 | |
| Yes | 40 | 120 | |
| AFP (ng/mL) | |||
| ≤ 20 | 19 | 45 | 0.481 |
| >20 | 35 | 105 | |
| Tumor size (cm) | 4.58 ± 2.77 | 5.52 ± 3.90 | 0.059 |
| Number of tumor nodules | 0.019 | ||
| Solitary | 53 | 127 | |
| Multiple | 2 | 24 | |
| Tumor encapsulation | 0.743 | ||
| No | 28 | 72 | |
| Yes | 27 | 77 | |
| Microvascular invasion | 0.005 | ||
| No | 41 | 79 | |
| Yes | 14 | 71 | |
| Tumor differentiation | 0.132 | ||
| I - II | 43 | 104 | |
| III - IV | 11 | 47 | |
| TNM stage | 0.006 | ||
| I - II | 54 | 127 | |
| III - IV | 1 | 24 | |
Table 2.
Multivariate analysis of factors associated with overall and disease-free survivals of hepatocellular carcinoma after curative surgical resection.
| Overall survival |
Disease-free survival |
|||
|---|---|---|---|---|
| HR (95% CI) | p value | HR (95% CI) | p value | |
| Tumor size (cm) | ||||
| >5 vs. ≤ 5 | 1.74 (1.03–2.95) | 0.040 | 1.47 (0.91–2.37) | 0.113 |
| Tumor number | ||||
| Solitary vs. multiple | 0.77 (0.35–1.69) | 0.507 | 1.31 (0.69–2.45) | 0.409 |
| Vascular tumor thrombus | ||||
| Yes vs. no | 1.56 (0.96–2.54) | 0.075 | 1.35 (0.88–2.08) | 0.173 |
| TNM stage | ||||
| III - IV vs. I - II | 2.43 (1.05–5.58) | 0.037 | 1.66 (0.81–3.39) | 0.169 |
| GGT (U/L) | ||||
| >91 vs. ≤ 91 | 2.15 (1.31–3.53) | 0.002 | 1.72 (1.09–2.71) | 0.020 |
| AIF1 expression | ||||
| Low vs. high | 1.97 (1.03–3.74) | 0.039 | 1.92 (1.09–3.36) | 0.023 |
Discussion
We previously demonstrated that high expression of CSF1 in the adjacent tumor microenvironment was associated with poor prognosis after surgical resection of HCC.10, 25 Sorafenib treatment induces the expression of CSF1, which is accompanied by increased macrophage infiltration, partially reducing the antitumor effect of the drug.26 The present study showed that CSF1 induced the expression of AIF1 in macrophages through the CSF1R-MEK1/2-Erk1/2-c-Jun signaling pathway. AIF1 expression in TAMs increased the proliferation and migration of hepatoma cells in vitro, promoted the tumor growth in animal model, and was associated with poor prognosis after surgical resection of HCC.
AIF1 is a cytoskeleton-associated protein and increases the proliferation and migration of macrophages.27 AIF1 expression has been reported to be associated with poor prognosis in the context of glioma and breast cancer. AIF1-positive gliocytes were associated with poor prognosis in brain astrocytoma,28 and the expression of AIF1 in the epithelium of breast ductal carcinoma was associated with large tumor size and poor tumor differentiation.29 In vitro study showed that AIF1 promoted tumor growth via the NF-κB/cyclin D1 pathway.29 However, the relationship between AIF1 and HCC has not been previously reported. The present study showed that macrophages overexpressing AIF1 were more attractant to exogenous CSF1, which is abundant in the adjacent liver tissue of HCC. These data suggested that AIF1 is a vital functional component in the infiltration of peritumoral macrophages. The CSF1-activated CSF1R-MEK1/2-Erk1/2-c-Jun signaling pathway increased the accumulation of AIF1-positive macrophages in the adjacent microenvironment of HCC. Furthermore, the present study showed that AIF1 expression in macrophages in the adjacent microenvironment was associated with poor survival and more aggressive tumor features. We consistently found that macrophages overexpressing AIF1 promoted tumor migration and proliferation of tumor cells. Mouse cytokine antibody array analysis showed that macrophages overexpressing AIF1 were accompanied by increased secretion of CXCL16, which is reported to facilitate with the migration and invasion of HCC.30 The preceding results suggest that AIF1 promoted liver cancer progression by enhancing the M2 polarization and increasing CXCL16 secretion in macrophages.
The correlation between CSF1 and AIF1 has been reported before. Imai et al. found that CSF1 induced the activation of CSF1R-PI3K-PLCγ signaling pathway. AIF1, together with PLCγ further activated the RAC signaling pathway, which was associated with the rearrangement of the cytoskeleton, contributing to the proliferation and migration of macrophages,31 which is in consistency with the results of our study. However, the effect of CSF1 on AIF1 expression and its correlation with M2 polarization was not investigated, which was elucidated in detail in the present study. We revealed for the first time that CSF1 induced the expression of AIF1 through the activation of CSF1R-MEK1/2-Erk1/2-c-Jun signaling pathway.
TAMs exhibit an M2-like phenotype, which is regulated by the interaction between macrophages and tumor cells.32 However, the mechanism by which HCC promotes an M2-polarized status among macrophages is not clearly understood. The CSF1-CSF1R signaling pathway was reported to be associated with the M2 polarization of macrophages.33 Blocking CSF1R showed an antitumor effect in glioma by decreasing the alternatively activated M2 markers.34 We speculated that AIF1 is a vital regulator downstream of the CSF1-CSF1R axis in the M2 polarization of macrophages. CSF1 induced the expression of AIF1, which resulted in the M2 polarization of macrophages with increased expression of markers associated with M2 polarization and reduced expression of markers associated with M1 polarization.13 However, the M2 polarization of macrophages is a complex process, which also involves multiple factor other than CSF1, such as CCL2. The CCL2-CCR2 axis was reported to regulate macrophage polarization, which is also regulated by the CSF1-CSF1R signaling pathway.35, 36
TAMs facilitate tumor angiogenesis, provide an immunosuppressive microenvironment, and promote the migration and invasion of tumor cells.32 The accumulation of TAMs in the adjacent liver tissue provides a suitable microenvironment for the seeding of tumor cells or circulating tumor cells.32 TAMs directly promote the migration and invasion of tumor cells by secreting a variety of cytokines or chemokines, including CCL18, CCL22, and EGF.11, 12, 37 CCL18 and CCL22 were reported to be increased in IL-4-activated macrophages,11, 12 but not in CSF1-activated macrophages. CSF1 secreted from cancer cells was reported to promote the expression of EGF by macrophages, which, in turn, promoted the invasiveness of cancer cells.37 Though in the present study, mouse cytokine antibody array showed that EGF secretion was slightly increased in RAW264.7 cells overexpressing AIF1 compared with the AIF1 knockdown, the quantity of EGF was generally low. CXCL16 was reported to be increased in cancer-associated fibroblasts compared with normal fibroblasts, which promoted the migration and invasion of HCC.30 Cho et al. compared the cytokine array of CM from macrophage/thyroid cancer co-cultures and CM from thyroid cancer culture alone and found that the CM of macrophage/thyroid cancer co-cultures contained a high level of CXCL16, which enhanced the migration of tumor cells.38 These findings were in line with our results and further confirmed that CXCL16 is an important downstream molecule of AIF1, which in the present study promoted hepatoma cell migration and further facilitated the progression of HCC. Consistent with the present study's findings, CXCL12 has also been reported to be secreted from M2-polarized macrophages and to promote the migration of cancer cells.39, 40
Our study has some limitations. First, the regulation of AIF1 expression in BMDMs, which are similar to tumor-infiltrating macrophages, was less effective. As we know, BMDMs are terminally differentiated cells, which are not susceptible to lentivirus- or liposome-mediated plasmid transfection. Whether AIF1 knockdown or overexpression in BMDMs was associated with macrophage polarization or tumor cell proliferation or migration is unclear. Second, since AIF1 expression in the adjacent liver tissue was associated with the prognosis of HCC, a premetastatic microenvironment with AIF1-transformed macrophages would be an ideal model to study the effect of peritumoral AIF1 on tumor progression. Third, the cytokines detected by the mouse cytokine antibody array were limited. Other cytokines, such as CCL18, CXCL12, or others, could potentially account for the increased migration of cancer cells.
This study has potential value for clinical practice. The AIF1 expression in the adjacent liver tissue provided an important prognostic indicator, which could guide proper postoperative adjuvant therapies. Since CSF1 induced M2 polarization through the CSF1R-c-Jun-AIF1 signaling pathway, blocking CSF1R with PLX3397 may reduce the M2 polarization34 and exert some antitumor effect, which has already been reported in the treatment of giant cell tumor.41 In vivo knockout of AIF1 may be possible in future for haling the M2 polarization of macrophages, which may reduce postoperative intrahepatic recurrence.
Materials and methods
Patients and follow-ups
From March 2004 to December 2006, 217 consecutive patients were diagnosed with HCC and received curative surgical resection in Zhongshan Hospital, Fudan University. Postoperative pathological examination confirmed the diagnosis of HCC, and no macroscopically tumor residual was observed. Eleven patients who died of factors related to surgery or were lost to follow-ups were excluded, leaving 206 patients to be included in our study. All patients were followed up every 2 to 3 mo after surgery. Informed consent was obtained from all patients, and this study was approved by the ethics committee of Zhongshan Hospital.
Cell lines
Mouse macrophage cell line RAW264.7 (Chinese Academy of Science), mouse hepatoma cell lines Hepa1-6 and H22 (Chinese Academy of Science), and human HCC cell line HCCLM3 (established and maintained in our institute) were cultured in Dulbecco's modified Eagle's medium (DMEM) (HyClone, Logan, UT, USA) containing 10% fetal bovine serum (FBS) (Invitrogen, Carlsbad, CA, USA) in a humidified atmosphere of 5% CO2 at 37°C. Human THP-1 cell lines (Chinese Academy of Science) were cultured in the RPMI Medium Modified (HyClone) containing 10% FBS (Invitrogen).
Mouse BMDMs were obtained by harvesting bone marrow cells from the long bones of C57BL/6 mice and culturing them in the presence of recombinant mouse CSF1 (20 ng/mL) or CSF2 (20 ng/mL) for 1 week according to the protocol provided by the David Hume Group – The Roslin Institute with slight modifications.42
Recombinant mouse CSF1 (51112-M08H) and CSF2 (51048-M07H) were purchased from Sino Biological (Beijing, China). The CSF1R inhibitor pexidartinib (PLX3397) (S7818) and MEK1/2 inhibitor U0126 were purchased from Selleck (Shanghai, China). The CXCL16 neutralizing antibody (MAB503) was purchased from R&D Systems, Inc. (Minneapolis, MN, USA).
Lentivirus infection and siRNA transfection
To inhibit AIF1 expression in macrophages, three small interfering RNAs (siRNAs) targeting mouse AIF1 were designed and constructed by Sigma Aldrich. The siRNA sequences (5′–3′) were as follows: CUGACUUUCUCAGAAUGAU, CUAGAGCUGAAGAGAUUAA, and GAUCUGCCAUCUUGAGAAU. The siRNAs were transfected into RAW264.7 using Lipofectamine 3000 transfection reagent (Invitrogen). The first siRNA sequence was used to construct recombinant lentivirus vectors containing small hairpin RNA (shRNA) targeting mouse AIF1 to constitutively inhibit the expression of AIF1 in macrophages. The vectors used for AIF1 knockdown were hU6-MCS-Ubiquitin-EGFP-IRES-puromycin (Genechem, Shanghai, China). To constitutively overexpress AIF1 in macrophages, we constructed recombinant lentivirus vectors containing the full-length sequence of AIF1. The vectors used for AIF1 overexpression were Ubi-MCS-3FLAG-CMV-EGFP (Genechem, Shanghai, China). RAW264.7 cells were then infected with lentivirus following the manufacturer's instructions, and Western blot was used to validate the efficiency of AIF1 knockdown and overexpression.
Cell proliferation and migration assay
Cell proliferation assay was performed using CCK-8 solution (Dojindo, Kumamoto, Japan) to detect the absorbance at a wavelength 450 nm. Cell migration assays were performed using Boyden chambers with an 8-μm pore size (Corning, Tewksbury, MA) to count the number of cells that migrated through the pore to the lower surface as described previously.43 Three independent experiments were performed in triplicate.
Tissue microarray and immunohistochemistry
Tissue microarrays (TMAs) were constructed according to a described previously protocol.10 In brief, tumor tissue and adjacent relatively noncancerous tissue within 10 mm of the tumor from 206 patients were sampled to construct three TMA sections (cooperation with Shanghai Biochip Company Ltd., Shanghai, China).
The immunohistochemistry protocol has been described elsewhere.44 Primary antibodies were rabbit monoclonal antibody combined with AIF1 (1:2000; Abcam, Cambridge, UK), rabbit polyclonal antibody combined with CXCL16 (1:50; Abcam) and rabbit monoclonal antibody combined with Ki67 (1:100; Abcam). The components of the Ultravision Quanto Detection system (Lab Vision Corporation, Fremont, CA) were applied for human tissue. The HRP-conjugated goat anti rabbit secondary antibody (1:200, Dako, Glostrup, Denmark) was used for mouse tissue. Reaction products were visualized by incubation with diaminobenzidine.
The immunohistochemical images were recorded with a computerized image system composed of a Leica CCD camera DFC420 connected to a Leica DM IRE2 microscope (Leica Microsystems Imaging Solutions Ltd, Cambridge, UK). At high-power magnification (×200), images of three representative fields were captured by the Leica QWin Plus v3 software. The parameters were constant for each captured image. The total positive staining area of AIF1 or Ki67, and the integrated optical density of CXCL16 were calculated by Image-Pro Plus v6.2 software (Media Cybernetics Inc., Bethesda, MD). A uniform setting was applied, and the total positive staining area or the integrated optical density was recorded for each slice.
Western blot
Western blot procedures have been described elsewhere.43 Primary antibodies used are listed as follows: AIF1/Iba1 (#ab178847, Abcam), c-Jun (#9165, Cell Signaling Technologies, Massachusetts, USA), phospho c-Jun (#3270, Cell Signaling Technologies), Erk1/2 (#4695, Cell Signaling Technologies), phospho Erk1/2 (#4370, Cell Signaling Technologies), MEK1/2 (#8787, Cell Signaling Technologies), phospho MEK1/2 (#9154, Cell Signaling Technologies), and GAPDH (#5174, Cell Signaling Technologies). Peroxidase-conjugated goat anti-rabbit second antibody (KC-RB-035) were purchased from Kangchen Biotech (Shanghai, China).
Real-time quantitative PCR
RNA isolation and qRT-PCR procedures have been described previously.43 Reverse transcription reagents (RR820) and PCR reagents (RR047) were purchased from TaKaRa Biotechnology (Dalian, China), and primers were synthesized by Sangon Biotech (Shanghai, China). Primer sequences used are listed in Table S1.
Immunofluorescence staining and confocal microscopy
Procedures for immunofluorescence staining and confocal microscopy were described in our previous study.43 Primary rabbit anti-Iba1 antibody (10904-1-AP, Proteintech, Chicago, USA), mouse anti-CD68 antibody (ab955, Abcam), rabbit anti-CXCL16 antibody (abs122925, Absin, Shanghai, China) and rat anti-CD68 (ab53444, Abcam) antibody were used. The mouse, rabbit and rat IgGs were used as isotype controls to rule out non-specific binding of primary antibodies. Alexa Fluor 546-conjugated anti-rabbit IgG (1:250, Invitrogen) and Alexa Fluor 488-conjugated anti-mouse IgG (1:250, Invitrogen) were used as secondary antibodies. Phalloidin-TRITC (P1951, Sigma Aldrich) was used to label the cytoskeleton of macrophages. The immunofluorescence staining was observed under the confocal fluorescent microscope (×400 magnification, Olympus).
Flow cytometry
Single-cell suspensions were prepared. Antibodies for cell surface staining were APC-conjugated anti-MHC-II (130-108-004, Miltenyi Biotec, Bergisch Gladbach, Germany) and Alexa Fluor 488-conjugated anti-mannose receptor (ab195191, Abcam). FACS data were acquired using a FACSCalibur (BD Biosciences, Franklin Lakes, NJ, USA), which was provided by Shu-Hui Sun of the Public Technology Platform, School of Basic Medical Sciences, Fudan University. CELLQuest software (BD Biosciences) and FlowJo (Tree Star, Inc., Ashland, OR) were used to examine the expression of CD206 and MHC-II on 10,000 cells.
For the analysis of cell-cycle distribution, the PI/RNase Staining Buffer (BD PharMingen) was used according to the manufacturer's protocol before flow cytometry. In brief, cells were pelleted, centrifuged and resuspended in 75% ethanol at −20℃ overnight. Then, 106 cells were centrifuged and resuspended in 0.5 mL of PtdIns/RNase Staining Buffer at room temperature for 15 min. CELLQuest and FlowJo were used to analyze the cell-cycle distribution in the G1, S and G2 phases in 10,000 cells.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) against c-Jun was conducted in RAW264.7 cells stimulated with exogenous CSF1 using the Pierce Agarose ChIP Kit (#26156, Thermo). The ChIP procedure is described elsewhere45 and was used with minor modifications. For immunoprecipitation of c-Jun in RAW264.7 cells, rabbit polyclonal c-Jun antibody (ab31419, Abcam) was used. Normal rabbit IgG (#2729P, Cell Signaling Technologies) was used as the negative control. Primer sequences for detection of c-Jun target promoter by RT-PCR are listed in Table S2.
Luciferase reporter gene assay
The Luciferase reporter gene assay was performed using Dual Luciferase Assay Kits purchased from Promega (Madison, WI) according to the manufacturer's protocol. The GL3b-AIF1 promoter luciferase plasmids (wild and mutant types) and c-Jun plasmid were constructed by Genechem. The 293T cells were co-transfected with AIF1 promoter luciferase plasmid and c-Jun plasmid with or without the stimulation of CSF1 with different combinations. The Firefly luciferase activity normalized to Renilla luciferase activity was calculated for each sample.
Collection of the conditioned medium (CM)
Tumor cells were cultured in the FBS-free DMEM for 24 h. The macrophages were co-cultured with tumor cells in Boyden chambers (Corning) in DMEM containing 1% FBS for 24 h. The CM was centrifuged for 20 min at 3,000 rpm, and the resultant pellet was collected for further study.
Mouse cytokine antibody array
The quantities of secreted cytokines in the CM were compared between RAW264.7 cells with AIF1 knockdown and RAW264.7 cells with AIF1 overexpression using C-Series Mouse Cytokine Antibody Array C6 for the semiquantitative detection of 97 mouse proteins (RayBiotech, Norcross, GA, USA) as directed by the manufacturer. The concentration of CXCL16 in the CM was confirmed by ELISA (Boster, Wuhan, China).
Animal model
RAW264.7-shNC/shAIF1 cells and Hepa1-6 cells were mixed at a ratio of 1 : 4 and a total of 5 × 106 cells were subcutaneously injected to the right flank of C57BL/6 mice. The longest diameter (a), shortest diameter (b) and mass of tumors were measured for each group after 3 weeks and the volume of tumors was equaled as a × b × b / 2 (cm3). Immunohistochemical staining was performed to confirm the knockdown of AIF1 in TAMs and assess the expression of CXCL16 and Ki67 within tumor. The animal experiments were approved by the animal care committee of Zhongshan Hospital.
Statistical analysis
Data were analyzed with SPSS 18.0. All quantitative data were recorded as the means ± SD and compared with the two-tailed Student's t-test. Qualitative data were recorded as relativity and compared with the chi-square test. Kaplan-Meier analysis and log-rank test were used to compare the differences in patients' overall and disease-free survival. p values < 0.05 were considered statistically significant.
Supplementary Material
Acknowledgments
We thank Ke Qiao form Key Laboratory of Medical Molecular virology, Ministry of Education and Public Health, School of Basic Medical Sciences, Fudan University for her excellent technical expertise in microscopic imaging techniques.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by grants from the National Natural Science Foundation of China (81372655, 81472224, 81672326), the Leading Investigator Program of Shanghai municipal government (17XD1401100) and the National Key Basic Research Program (973 project) (2015CB554005) from the Ministry of Science and Technology of China.
References
- 1.Global Burden of Disease Cancer C, Fitzmaurice C, Dicker D, Pain A, Hamavid H, Moradi-Lakeh M, MacIntyre MF, Allen C, Hansen G, Woodbrook R et al.. The Global Burden of Cancer 2013. JAMA Oncol 2015; 1:505-27; PMID:26181261; https://doi.org/ 10.1001/jamaoncol.2015.0735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bruix J, Sherman M, American Association for the Study of Liver D . Management of hepatocellular carcinoma: An update. Hepatology 2011; 53:1020-2; PMID:21374666; https://doi.org/ 10.1002/hep.24199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roayaie S, Obeidat K, Sposito C, Mariani L, Bhoori S, Pellegrinelli A, Labow D, Llovet JM, Schwartz M, Mazzaferro V. Resection of hepatocellular cancer </ = 2 cm: Results from two Western centers. Hepatology 2013; 57:1426-35; PMID:22576353; https://doi.org/ 10.1002/hep.25832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheung TT, Poon RT, Yuen WK, Chok KS, Jenkins CR, Chan SC, Fan ST, Lo CM. Long-term survival analysis of pure laparoscopic versus open hepatectomy for hepatocellular carcinoma in patients with cirrhosis: A single-center experience. Ann Surg 2013; 257:506-11; PMID:23299521; https://doi.org/ 10.1097/SLA.0b013e31827b947a [DOI] [PubMed] [Google Scholar]
- 5.Shrager B, Jibara G, Schwartz M, Roayaie S. Resection of hepatocellular carcinoma without cirrhosis. Ann Surg 2012; 255:1135-43; PMID:22258064; https://doi.org/ 10.1097/SLA.0b013e31823e70a3 [DOI] [PubMed] [Google Scholar]
- 6.Hasegawa K, Kokudo N, Imamura H, Matsuyama Y, Aoki T, Minagawa M, Sano K, Sugawara Y, Takayama T, Makuuchi M. Prognostic impact of anatomic resection for hepatocellular carcinoma. Ann Surg 2005; 242:252-9; PMID:16041216; https://doi.org/ 10.1097/01.sla.0000171307.37401.db [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shi M, Zhang CQ, Zhang YQ, Liang XM, Li JQ. Micrometastases of solitary hepatocellular carcinoma and appropriate resection margin. World J Surg 2004; 28:376-81; PMID:15022021; https://doi.org/ 10.1007/s00268-003-7308-x [DOI] [PubMed] [Google Scholar]
- 8.Shi M, Guo RP, Lin XJ, Zhang YQ, Chen MS, Zhang CQ, Lau WY, Li JQ. Partial hepatectomy with wide versus narrow resection margin for solitary hepatocellular carcinoma: A prospective randomized trial. Ann Surg 2007; 245:36-43; PMID:17197963; https://doi.org/ 10.1097/01.sla.0000231758.07868.71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang JD, Nakamura I, Roberts LR. The tumor microenvironment in hepatocellular carcinoma: Current status and therapeutic targets. Semin Cancer Biol 2011; 21:35-43; PMID:20946957; https://doi.org/ 10.1016/j.semcancer.2010.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu XD, Zhang JB, Zhuang PY, Zhu HG, Zhang W, Xiong YQ, Wu WZ, Wang L, Tang ZY, Sun HC. High expression of macrophage colony-stimulating factor in peritumoral liver tissue is associated with poor survival after curative resection of hepatocellular carcinoma. J Clin Oncol 2008; 26:2707-16; PMID:18509183; https://doi.org/ 10.1200/JCO.2007.15.6521 [DOI] [PubMed] [Google Scholar]
- 11.Chen J, Yao Y, Gong C, Yu F, Su S, Chen J, Liu B, Deng H, Wang F, Lin L et al.. CCL18 from tumor-associated macrophages promotes breast cancer metastasis via PITPNM3. Cancer Cell 2011; 19:541-55; PMID:21481794; https://doi.org/ 10.1016/j.ccr.2011.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yeung OW, Lo CM, Ling CC, Qi X, Geng W, Li CX, Ng KT, Forbes SJ, Guan XY, Poon RT et al.. Alternatively activated (M2) macrophages promote tumour growth and invasiveness in hepatocellular carcinoma. J Hepatol 2015; 62:607-16; PMID:25450711; https://doi.org/ 10.1016/j.jhep.2014.10.029 [DOI] [PubMed] [Google Scholar]
- 13.Murray PJ, Allen JE, Biswas SK, Fisher EA, Gilroy DW, Goerdt S, Gordon S, Hamilton JA, Ivashkiv LB, Lawrence T et al.. Macrophage activation and polarization: Nomenclature and experimental guidelines. Immunity 2014; 41:14-20; PMID:25035950; https://doi.org/ 10.1016/j.immuni.2014.06.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Capece D, Fischietti M, Verzella D, Gaggiano A, Cicciarelli G, Tessitore A, Zazzeroni F, Alesse E. The inflammatory microenvironment in hepatocellular carcinoma: A pivotal role for tumor-associated macrophages. Biomed Res Int 2013; 2013:187204; PMID:23533994; https://doi.org/ 10.1155/2013/187204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell 2010; 140:883-99; PMID:20303878; https://doi.org/ 10.1016/j.cell.2010.01.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lanaya H, Natarajan A, Komposch K, Li L, Amberg N, Chen L, Wculek SK, Hammer M, Zenz R, Peck-Radosavljevic M et al.. EGFR has a tumour-promoting role in liver macrophages during hepatocellular carcinoma formation. Nat Cell Biol 2014; 16:972-81, 1-7; PMID:25173978; https://doi.org/ 10.1038/ncb3031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Van Overmeire E, Stijlemans B, Heymann F, Keirsse J, Morias Y, Elkrim Y, Brys L, Abels C, Lahmar Q, Ergen C et al.. M-CSF and GM-CSF receptor signaling differentially regulate monocyte maturation and macrophage polarization in the tumor microenvironment. Cancer Res 2016; 76:35-42; PMID:26573801; https://doi.org/ 10.1158/0008-5472.CAN-15-0869 [DOI] [PubMed] [Google Scholar]
- 18.Joshi S, Singh AR, Zulcic M, Bao L, Messer K, Ideker T, Dutkowski J, Durden DL. Rac2 controls tumor growth, metastasis and M1-M2 macrophage differentiation in vivo. PloS One 2014; 9:e95893; PMID:24770346; https://doi.org/ 10.1371/journal.pone.0095893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Utans U, Arceci RJ, Yamashita Y, Russell ME. Cloning and characterization of allograft inflammatory factor-1: A novel macrophage factor identified in rat cardiac allografts with chronic rejection. J Clin Invest 1995; 95:2954-62; PMID:7769138; https://doi.org/ 10.1172/JCI118003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mishima T, Iwabuchi K, Fujii S, Tanaka SY, Ogura H, Watano-Miyata K, Ishimori N, Andoh Y, Nakai Y, Iwabuchi C et al.. Allograft inflammatory factor-1 augments macrophage phagocytotic activity and accelerates the progression of atherosclerosis in ApoE-/- mice. Int J Mol Med 2008; 21:181-7; PMID:18204784; https://doi.org/ 10.3892/ijmm.21.2.181 [DOI] [PubMed] [Google Scholar]
- 21.Wang J, Zhao Y, Wang W, Du Z, Yan D, Li C, Chen Z. Daintain/AIF-1 plays roles in coronary heart disease via affecting the blood composition and promoting macrophage uptake and foam cell formation. Cell Physiol Biochem 2013; 32:121-6; PMID:23867161; https://doi.org/ 10.1159/000350130 [DOI] [PubMed] [Google Scholar]
- 22.Watano K, Iwabuchi K, Fujii S, Ishimori N, Mitsuhashi S, Ato M, Kitabatake A, Onoé K. Allograft inflammatory factor-1 augments production of interleukin-6, -10 and -12 by a mouse macrophage line. Immunology 2001; 104:307-16; PMID:11722645; https://doi.org/ 10.1046/j.1365-2567.2001.01301.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morandi A, Barbetti V, Riverso M, Dello Sbarba P, Rovida E. The colony-stimulating factor-1 (CSF-1) receptor sustains ERK1/2 activation and proliferation in breast cancer cell lines. PloS One 2011; 6:e27450; PMID:22096574; https://doi.org/ 10.1371/journal.pone.0027450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hamilton JA. CSF-1 signal transduction. J Leukocyte Biol 1997; 62:145-55; PMID:9261328. [DOI] [PubMed] [Google Scholar]
- 25.Jia JB, Wang WQ, Sun HC, Zhu XD, Liu L, Zhuang PY, Zhang JB, Zhang W, Xu HX, Kong LQ et al.. High expression of macrophage colony-stimulating factor-1 receptor in peritumoral liver tissue is associated with poor outcome in hepatocellular carcinoma after curative resection. Oncologist 2010; 15:732-43; PMID:20551429; https://doi.org/ 10.1634/theoncologist.2009-0170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang W, Zhu XD, Sun HC, Xiong YQ, Zhuang PY, Xu HX, Kong LQ, Wang L, Wu WZ, Tang ZY. Depletion of tumor-associated macrophages enhances the effect of sorafenib in metastatic liver cancer models by antimetastatic and antiangiogenic effects. Clin Cancer Res 2010; 16:3420-30; PMID:20570927; https://doi.org/ 10.1158/1078-0432.CCR-09-2904 [DOI] [PubMed] [Google Scholar]
- 27.Tian Y, Kelemen SE, Autieri MV. Inhibition of AIF-1 expression by constitutive siRNA expression reduces macrophage migration, proliferation, and signal transduction initiated by atherogenic stimuli. Am J Physiol Cell Physiol 2006; 290:C1083-91; PMID:16291819; https://doi.org/ 10.1152/ajpcell.00381.2005 [DOI] [PubMed] [Google Scholar]
- 28.Deininger MH, Seid K, Engel S, Meyermann R, Schluesener HJ. Allograft inflammatory factor-1 defines a distinct subset of infiltrating macrophages/microglial cells in rat and human gliomas. Acta Neuropathologica 2000; 100:673-80; PMID:11078219; https://doi.org/ 10.1007/s004010000233 [DOI] [PubMed] [Google Scholar]
- 29.Liu S, Tan WY, Chen QR, Chen XP, Fu K, Zhao YY, Chen ZW. Daintain/AIF-1 promotes breast cancer proliferation via activation of the NF-kappaB/cyclin D1 pathway and facilitates tumor growth. Cancer Sci 2008; 99:952-7; PMID:18341653; https://doi.org/ 10.1111/j.1349-7006.2008.00787.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu J, Chen S, Wang W, Ning BF, Chen F, Shen W, Ding J, Chen W, Xie WF, Zhang X. Cancer-associated fibroblasts promote hepatocellular carcinoma metastasis through chemokine-activated hedgehog and TGF-beta pathways. Cancer Lett 2016; 379:49-59; PMID:27216982; https://doi.org/ 10.1016/j.canlet.2016.05.022 [DOI] [PubMed] [Google Scholar]
- 31.Imai Y, Kohsaka S. Intracellular signaling in M-CSF-induced microglia activation: Role of Iba1. Glia 2002; 40:164-74; PMID:12379904; https://doi.org/ 10.1002/glia.10149 [DOI] [PubMed] [Google Scholar]
- 32.Chanmee T, Ontong P, Konno K, Itano N. Tumor-associated macrophages as major players in the tumor microenvironment. Cancers 2014; 6:1670-90; PMID:25125485; https://doi.org/ 10.3390/cancers6031670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martinez FO, Gordon S, Locati M, Mantovani A. Transcriptional profiling of the human monocyte-to-macrophage differentiation and polarization: New molecules and patterns of gene expression. J Immunol 2006; 177:7303-11; PMID:17082649; https://doi.org/ 10.4049/jimmunol.177.10.7303 [DOI] [PubMed] [Google Scholar]
- 34.Pyonteck SM, Akkari L, Schuhmacher AJ, Bowman RL, Sevenich L, Quail DF, Olson OC, Quick ML, Huse JT, Teijeiro V et al.. CSF-1R inhibition alters macrophage polarization and blocks glioma progression. Nat Med 2013; 19:1264-72; PMID:24056773; https://doi.org/ 10.1038/nm.3337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li X, Yao W, Yuan Y, Chen P, Li B, Li J, Chu R, Song H, Xie D, Jiang X et al.. Targeting of tumourinfiltrating macrophages via CCL2/CCR2 signalling as a therapeutic strategy against hepatocellular carcinoma. Gut 2017; 66:157–67; PMID:26452628; https://doi.org/ 10.1136/gutjnl-2015-310514 [DOI] [PubMed] [Google Scholar]
- 36.Sierra-Filardi E, Nieto C, Dominguez-Soto A, Barroso R, Sanchez-Mateos P, Puig-Kroger A, López-Bravo M, Joven J, Ardavín C, Rodríguez-Fernández JL et al.. CCL2 shapes macrophage polarization by GM-CSF and M-CSF: Identification of CCL2/CCR2-dependent gene expression profile. J Immunol 2014; 192:3858-67; PMID:24639350; https://doi.org/ 10.4049/jimmunol.1302821 [DOI] [PubMed] [Google Scholar]
- 37.Goswami S, Sahai E, Wyckoff JB, Cammer M, Cox D, Pixley FJ, Stanley ER, Segall JE, Condeelis JS. Macrophages promote the invasion of breast carcinoma cells via a colony-stimulating factor-1/epidermal growth factor paracrine loop. Cancer Res 2005; 65:5278-83; PMID:15958574; https://doi.org/ 10.1158/0008-5472.CAN-04-1853 [DOI] [PubMed] [Google Scholar]
- 38.Cho SW, Kim YA, Sun HJ, Kim YA, Oh BC, Yi KH, Park DJ, Park YJ. CXCL16 signaling mediated macrophage effects on tumor invasion of papillary thyroid carcinoma. Endocr Relat Cancer 2016; 23:113-24; PMID:26559672; https://doi.org/ 10.1530/ERC-15-0196 [DOI] [PubMed] [Google Scholar]
- 39.Qiao J, Liu Z, Yang C, Gu L, Deng D. SRF promotes gastric cancer metastasis through stromal fibroblasts in an SDF1-CXCR4-dependent manner. Oncotarget 2016; 7:46088-99; PMID:27323859; https://doi.org/ 10.18632/oncotarget.10024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park JY, Sung JY, Lee J, Park YK, Kim YW, Kim GY, Won KY, Lim SJ. Polarized CD163+ tumor-associated macrophages are associated with increased angiogenesis and CXCL12 expression in gastric cancer. Clin Res Hepatol Gastroenterol 2016; 40:357-65; PMID:26508473; https://doi.org/ 10.1016/j.clinre.2015.09.005 [DOI] [PubMed] [Google Scholar]
- 41.Tap WD, Wainberg ZA, Anthony SP, Ibrahim PN, Zhang C, Healey JH, Chmielowski B, Staddon AP, Cohn AL, Shapiro GI et al.. Structure-guided blockade of CSF1R kinase in tenosynovial giant-cell tumor. N Engl J Med 2015; 373:428-37; PMID:26222558; https://doi.org/ 10.1056/NEJMoa1411366 [DOI] [PubMed] [Google Scholar]
- 42.Robert C, Lu X, Law A, Freeman TC, Hume DA. Macrophages.com: An on-line community resource for innate immunity research. Immunobiology 2011; 216:1203-11; PMID:21924793; https://doi.org/ 10.1016/j.imbio.2011.07.025 [DOI] [PubMed] [Google Scholar]
- 43.Ao JY, Chai ZT, Zhang YY, Zhu XD, Kong LQ, Zhang N, Ye BG, Cai H, Gao DM, Sun HC. Robo1 promotes angiogenesis in hepatocellular carcinoma through the Rho family of guanosine triphosphatases' signaling pathway. Tumour Biol 2015; 36:8413-24; PMID:26022159; https://doi.org/ 10.1007/s13277-015-3601-1 [DOI] [PubMed] [Google Scholar]
- 44.Qian YB, Zhang JB, Wu WZ, Fang HB, Jia WD, Zhuang PY, Zhang BH, Pan Q, Xu Y, Wang L et al.. P48 is a predictive marker for outcome of postoperative interferon-alpha treatment in patients with hepatitis B virus infection-related hepatocellular carcinoma. Cancer 2006; 107:1562-9; PMID:16948122; https://doi.org/ 10.1002/cncr.22206 [DOI] [PubMed] [Google Scholar]
- 45.Suzuki K, Juelich T, Lim H, Ishida T, Watanebe T, Cooper DA, Rao S, Kelleher AD. Closed chromatin architecture is induced by an RNA duplex targeting the HIV-1 promoter region. J Biol Chem 2008; 283:23353-63; PMID:18519571; https://doi.org/ 10.1074/jbc.M709651200 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.