ABSTRACT
Horizontal gene transfer plays a significant role in spreading antimicrobial resistance and virulence genes throughout the genus Staphylococcus, which includes species of clinical relevance to humans and animals. While phages and plasmids are the most well-studied agents of horizontal gene transfer in staphylococci, the contribution of integrative conjugative elements (ICEs) has been mostly overlooked. Experimental work demonstrating the activity of ICEs in staphylococci remained frozen for years after initial work in the 1980s that showed Tn916 was capable of transfer from Enterococcus to Staphylococcus. However, recent work has begun to thaw this field. To date, 2 families of ICEs have been identified among staphylococci – Tn916 that includes the Tn5801 subfamily, and ICE6013 that includes at least 7 subfamilies. Both Tn5801 and ICE6013 commonly occur in clinical strains of S. aureus. Tn5801 is the most studied of the Tn916 family elements in staphylococci and encodes tetracycline resistance and a protein that, when expressed in Escherichia coli, inhibits restriction barriers to incoming DNA. ICE6013 is among the shortest known ICEs, but it still includes many uncharacterized open reading frames. This element uses an IS30-like transposase as its recombinase, providing some versatility in integration sites. ICE6013 also conjugatively transfers among receptive S. aureus strains at relatively higher frequency than Tn5801. Continued study of these mobile genetic elements may reveal the full extent to which ICEs impact horizontal gene transfer and the evolution of staphylococci.
KEYWORDS: conjugation, horizontal gene transfer, ICE6013, integrative conjugative elements, Staphylococcus aureus, Tn916, Tn5801
Introduction
Mobile genetic elements (MGEs) often encode genes for antibiotic resistances and toxins and they may transfer within and between species of staphylococci, which includes major pathogens of humans and animals.1,2 The different classes of MGEs in staphylococci consist of genomic islands of various types, phages, plasmids, transposons, and integrative conjugative elements (ICEs).1-4 Methicillin-resistant S. aureus (MRSA) obtained its namesake resistance by acquisition of the mecA locus that resides on a genomic island called the staphylococcal cassette chromosome mec (SCCmec). SCCmec elements have been reported in multiple staphylococcal species and they may accumulate antibiotic resistance genes from insertion of other MGEs such as Tn554, Tn4001, pT181 and pUB110.5,6 The genomic islands νSaα, νSaβ and νSaγ, and numerous pathogenicity islands (SaPIs) often carry virulence factors.2,7 While much progress has been made in identifying staphylococcal MGEs, their characterization has been a slower process and the specific mechanisms for the horizontal gene transfer (HGT) of many of these elements is obscure.
New insights into classical agents of HGT in staphylococci
Bacteriophages are well-studied agents of HGT in staphylococci.8,9 Transduction of different phages and genetic elements between staphylococcal species and even to different gram-positive genera such as Listeria has been documented. For example, SaPIbov5 transfers via phage ϕ12 among several S. aureus strains and from S. aureus to Listeria monocytogenes at high frequency.10 SaPIbov1 transfers via phage ϕ187 specifically from S. aureus sequence type (ST) 395 strains to S. epidermidis, S. carnosus and L. monocytogenes, due to a common teichoic acid structure consisting of a glycerophosphate repeating unit that is externally attached to the cell wall and may act as the phage receptor.11,12 Other S. aureus strains have teichoic acid with ribitol-phosphate repeating units and are resistant to infection with ϕ187.11,12
While SaPIs have been demonstrated to make specific and exquisite use of helper phage for their mobilization,7 other DNA that is transferred by phage may use a mechanism that is indistinguishable from generalized transduction. For example, SCCmec elements from clonal complex (CC) 8 backgrounds can be transduced at low frequency by ϕ80α and ϕ29 into recipients that have homology to the region flanking the donor element's integration site.13 Such recombination is presumably recA-mediated and distinct from that mediated by the element's recombinases. Recent work has also shown the transduction of genomic island νSaβ by the linked prophage ϕSaBov, potentially by a population of phage carrying overlapping portions of the island.14 Transfer frequencies of the island's segments ranged from 4.3 × 10−4 to 5.0 × 10−8 CFU/PFU. Parts of the unlinked genomic islands νSaα and νSaγ have also been shown to transfer by ϕSaBov, which indicates that transduction may influence gene content variation in these islands.15 Finally, while much of the work on transduction in staphylococci has been done under in vitro conditions, a recent study has shown a high level of transfer of MGEs between S. aureus CC398 strains during colonization of gnotobiotic piglets.16 Although the mechanism of transfer was thought to be transduction by ϕ2 and ϕ6,16 it is important to point out that the S. aureus CC398 donor strain S0385 used in this study carries 4 integrative conjugative elements (described below) whose role in HGT cannot be discounted.
It has been estimated that ∼5% of S. aureus plasmids are conjugative, while the rest are non-conjugative due to their lack of conjugation transfer (tra) genes.17 Mobilization of the remaining 95% of S. aureus plasmids was thought to be rare.3 However, a recent review has summarized several mechanisms by which ∼92% of the non-conjugative plasmids might be mobilized.18 These mechanisms include the presence of mob genes, replicative relaxases, and oriT mimic sequences that interact in various ways with conjugative plasmids.19,20 The larger (> 14.5 kb) multidrug resistance plasmids often carry multiple mobilization determinants.19 These observations suggest that the contribution of conjugative elements to HGT in staphylococci may be much greater than previously thought.
ICEs, the overlooked agents of HGT in staphylococci
One possible source of conjugation machinery that is rarely considered in the staphylococcal literature consists of integrative conjugative elements (ICEs). In general, ICEs bring together the recombination features of phages and transposons with the conjugation features of plasmids. Most reported ICEs encode a phage-like tyrosine recombinase for integration/excision from a host genome, but examples exist of serine recombinases and DDE transposases serving these functions.21 ICEs are often recognizably modular in structure.22 The DNA transfer and replication functions of ICEs include a relaxase, and their mating pair functions include components of type 4-like secretion system (T4SS) such as a VirB4-like ATPase and a VirD4- or FtsK-like coupling protein.23,24 Some of the better studied ICEs in non-staphylococcal species are known to have additional plasmid-like functions, such as extrachromosomal replication and the ability to mobilize other elements that are not self-transmissible. For example, ICEBs1 and Tn916 from Bacillus subtilus are known to transiently replicate while excised from their host chromosome.25-27 Also, small, rolling-circle replication plasmids such as pC194 and pUB110 that were thought to be non-mobilizable can be mobilized by ICEBs1 or Tn916, respectively, potentially by different mechanisms.28,29
To investigate the relatedness between ICEs and conjugative plasmids, a phylogenetic analysis was performed on the VirB4 protein, which is the only protein conserved throughout the T4SS of ICEs and conjugative plasmids from multiple phyla.23 Results showed that VirB4 from ICEs or conjugative plasmids was monophyletic only at short evolutionary distances, which suggested that both types of elements have originated multiple times. However, challenges remain in identifying new ICEs, as each family may be very divergent from other elements and diverse in gene content.21 In staphylococci, only 3 families of conjugative plasmids (pSK41/pGO1, pWBG749, pWBG4) and 2 families of ICEs (Tn916, ICE6013) have been identified. The plasmids have been reviewed recently.19 Here, we review the 2 ICE families.
The Tn916 family of ICEs in staphylococci
Classification and diversity of the Tn5801 subfamily
Tn916 is a prototypical ICE of gram-positive bacteria and early reports showed its transfer from E. faecalis to S. aureus.30 Conjugation frequencies of Tn916 from E. faecalis to S. aureus in the absence of plasmids ranged between 6.0 × 10−9 to 7.5 × 10−10 transconjugants/donor.30 Conjugation of Tn916 was later documented between various gram-positive and gram-negative species at rates of 4.6 × 10−4 to 1.4 × 10−8 transconjugants/donor, proving the element to be extremely promiscuous.31 Based on recombinase detection, Tn916 is found in numerous staphylococcal species of human and animal origin including: S. aureus, S. epidermidis, S. hyicus, S. pseudintermedius, S. rostri, S. saprophyticus, S. simulans and S. xylosis.32-34 Tn916 uses a tyrosine recombinase (Int) and a recombination directionality factor (Xis) to insert into A-T rich sequences, though the recombinase differs in a couple of instances within the Tn916 family.22,35
The classification of genetic elements into the Tn916 family requires the general organization and a similar sequence identity to the founding Tn916 element from E. faecalis strain DS16, as well as the presence of both the conjugation and regulation modules.22 The tetM gene for tetracycline resistance is within the regulation module and located between the conjugation and recombination modules. The Tn5801 variant of Tn916 was originally identified in the vancomycin intermediately-resistant S. aureus strain Mu50 that was isolated in 1997.36 The average nucleotide identity (ANI) between the founding elements of Tn5801 and Tn916 is 75%, and large gaps in aligned sequences are accounted for by 4 orfs of unknown function encoded at the 3' end of Tn5801 that are not present in Tn916 (Fig. 1). Since most of the recent work on Tn916 family elements in staphylococci have focused on Tn5801, our discussion centers on that subfamily.
Figure 1.
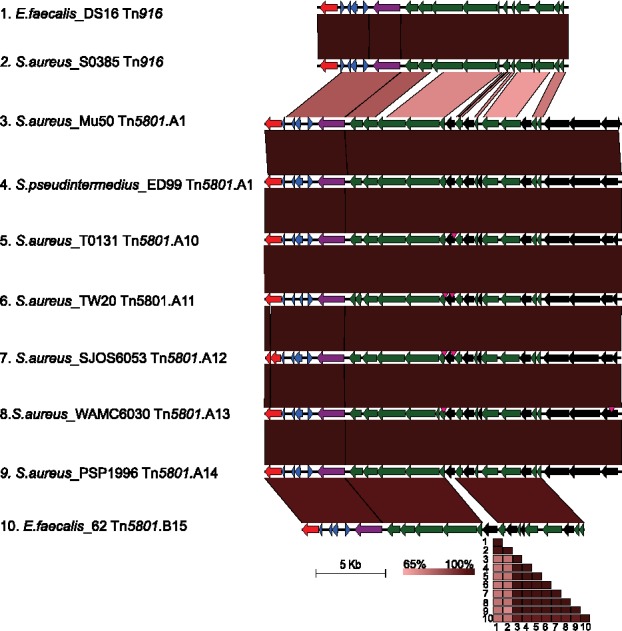
Comparison of Tn916 family elements from staphylococci and other species, as classified previously.39 Sequences are numbered and labeled by species, strain, and ICE identifier. Gene map shows orfs (open reading frames) predicted by Prodigal v1.2 as arrows and are colored according to the following: red for recombinase proteins, blue for regulation-related proteins, purple for tetM, and green for conjugation-related proteins. Black indicates orfs not present in the founding Tn916 element. Vertical pink arrowheads indicate insertions of transposons described elsewhere,39 which were removed to facilitate comparison. Percent nucleotide identity determined by BLASTn analysis is shown under sequences as red shading (EasyFig 2.2.2). A matrix of pairwise mean reciprocal ANI (average nucleotide identities) was computed using JSpecies v1.2.1 with default parameters, and colored according to the scale.
In a survey of 205 tetracycline-resistant S.aureus isolates, Tn916 elements were found in isolates from both humans and animals, but Tn5801 elements were found only in isolates from humans.37 A more recent study identified Tn5801 in 26% (7/27) of veterinary tetracycline-resistant isolates of S. pseudintermedius, while 22% (6/27) had Tn916.38 Tn5801 can be classified into 2 or 3 groups based on gene content, overall nucleotide identity, and phylogenetic analysis. Using the gene content and nucleotide identity criteria, Group 1 contained Tn5801 elements from most S. aureus and S. pseudintermedius isolates.38 Group 2 contained elements from Streptococcus mitis strain B6 and E. faecium. Group 3 contained elements from most S. agalactiae isolates and E.faecalis strain 62,in addition to one Tn5801 element from S. pseudintermedius.38
Another recent study of Tn5801 diversity analyzed over 5,000 genomes of enterococci, staphylococci, and streptococci.39 The Tn5801 sequences clustered by phylogenetic analysis into 2 distinct groups: group A (subtypes A1-A14) and group B (subtypes B15-B23).39 These 2 groups differed in length from 25kb (group A) to 20kb (group B) due to the presence or absence of a 5kb fragment at the 3' end of the element (Fig. 1). Group A elements accounted for all (139/4130) of S. aureus and all (1/4) S. pseudintermedius isolates that were positive for Tn5801.39 Of these staphylococcal elements, 76% were subtype A1. In S. aureus, these Tn5801.A1 elements were represented mostly by strains of CC8 backgrounds. Other S. aureus elements belonged within 6 different group A subtypes (Fig. 1), which included 2 of the 3 previously reported groups.39 The ANI between Tn5801 and Tn916 elements is ∼75–77% (Fig. 1). The ANI between group A and group B Tn5801 elements is >96%, and >99% when comparing subtypes of group A (Fig. 1). Therefore, the Tn5801 elements that have described are all likely one subfamily of the Tn916 family.
Recombination and conjugation of Tn5801
Tn5801 encodes a tyrosine recombinase that shares only 22% amino acid identity with that of Tn916. The integration site of Tn5801 at the 3’ end of guaA (GMP synthetase) is consistent in all studies, denoting a strict site specificity.38-40 CW459tet(M), a truncated Tn916 family element in Clostridium perfringens, has an identical integrase and integration site as Tn5801.38 Tn5801 integration into secondary sites were found in enterococci transconjugants that contained more than one copy of the element, as analyzed by PFGE analysis.39 It has not been determined if the coupling sequences of excised Tn5801 are heterologous as occurs with Tn916. Also, it has not been determined if tetracycline induces excision and subsequent conjugation of Tn5801 as described for some elements of the Tn916 family,41,42 though the relevant regulatory region appears to be present in Tn5801.
Using different combinations of 14 donors and 2 recipients, conjugative transfer of Tn5801 was detected in S.aureus with 3 different donor strains at frequencies of 1 × 10−9 to 3 × 10−9 transconjugants/recipient.37 One Tn5801 element that was shown to conjugatively transfer was registered a new Tn number (Tn6014).37 Another study used 3 different S. agalactiae donors and S.aureus Mu50 as a donor in separate matings with either S. agalactiae, S. pyogenes, or S. aureus recipients, and was unable to show either the circularization of Tn5801 in the donors or its conjugative transfer.40 Overall, the conjugation frequencies of Tn5801 differ dramatically between species and strains, but are of relatively low frequency among S. aureus strains.
Potential biologic effects of hosting Tn5801
The acquisition of Tn5801 may have several consequences on its bacterial hosts including gain of tetracycline resistance, altered growth rates, and increased ability to acquire other mobile genetic elements. These effects have been scarcely studied in staphylococci. Tn5801 was present in ∼30% of 69 tetracycline-resistant human clinical isolates of S. agalactiae, which is noteworthy considering that the prevalence of tetracycline resistance in the species is close to 90%.40 The growth rate of several enterococcal transconjugants (Tn5801.B15 subtype) has been measured and compared with its parent recipient.39 Overall, the presence of Tn5801 increased growth rate by an average of 1.65%, but individual transconjugants could have increased or decreased growth rate depending on their specific donor and recipient pairing.
Tn916 (orf18) and Tn5801 (sav0405) encode an E. coli ArdA homolog that inhibits all 4 major classes of type 1 restriction/modification (RM) enzymes.43,44 This anti-restriction mechanism occurs by mimicking the DNA that contacts the restriction subunits (HsdR) of EcoKI while leaving the methyltransferase subunits (HsdM) still active.45 Interestingly, the Tn916 and Tn5801 homologs effectively prevented destruction of incoming DNA when expressed in E. coli.44,46 Thus, the homologs might provide defense against type I RM systems that are present in multiple backgrounds of S.aureus.47 This anti-restriction mechanism has not yet been studied in staphylococci, but would be expected to potentiate HGT of sequences that would normally be restricted.
The ICE6013 family of ICEs in staphylococci
Classification and diversity of ICE6013 subfamilies
ICE6013 is the second family of ICEs discovered in staphylococci.48 The prototype element from S. aureus ST239 strain HDG2 encodes 15 orfs, 5 of which are homologous to genes on both ICEBs1 and Tn916/Tn5801. These ICE6013 orfs (orf6–8, orf12 and orf14) encode predicted conjugation-related proteins, the putative relaxase and helicase processivity factor.48 Phylogenetic analysis based on these homologs revealed ICE6013 to be more closely related to ICEBs1 than to Tn916/Tn5801.48 ICE6013 orf3, orf10, and orf11 encode putative membrane-associated proteins, and are homologous to genes found on ICEBs1 but not on Tn916/Tn5801. All ICE6013 homologs shared with ICEBs1 have <40% amino acid identity, and ICEBs1 has different regulation and recombination modules and other orfs not found on ICE6013. In addition, the gene for the putative coupling protein of ICE6013 (orf7) is in a different position compared with its homologs on ICEBs1 and Tn916/Tn5801.48
Currently, 7 subfamilies of ICE6013 have been identified, which are dispersed across 9 staphylococcal species: subfamily 1 (S. aureus, S. argenteus – previously S. aureus CC75, S. haemolyticus), subfamily 2 (S. agnetis), subfamily 3 (S. intermedius, S. pseudintermedius, S. schleiferi), subfamily 4 (S. saprophyticus), subfamilies 5 and 6 (S. aureus), subfamily 7 (S. microti).4 The ANI between ICE6013 subfamilies is ∼68–79%, while elements within the same subfamily share >98% ANI (Fig. 2). This range of nucleotide identities between and within ICE6013 subfamilies (Fig. 2) is similar to that observed between the Tn916 and Tn5801 subfamilies and within the Tn5801 subfamily, respectively, (Fig. 1) which indicates a consistency in the classification of these ICEs. In the literature, however, ICE6013 elements have been classified inconsistently as a transposon, a plasmid, and various ICEs,4 which illustrates the challenges in identifying and classifying ICEs.
Figure 2.
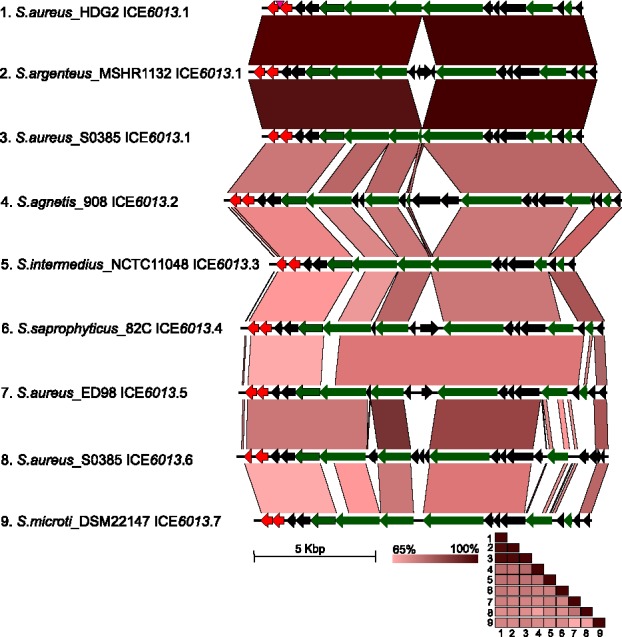
Comparison of ICE6013 family elements, as classified previously.4 Sequences are numbered by, species, strain, and subfamily. orfs (open reading frames) predicted by Prodigal v1.2 analysis are shown as arrows and are colored according to the following: red for IS30-like DDE transposase and green for conjugation-related proteins. Black indicates unknown function. Vertical pink arrowhead indicates insertion of Tn552 in strain HDG2,48 which was removed to facilitate comparison. Percent nucleotide identity determined by BLASTn analysis is shown under sequences as red shading (EasyFig 2.2.2). A matrix of pairwise mean reciprocal ANI (average nucleotide identities) was computed using JSpecies v1.2.1 with default parameters, and colored according to the scale.
ICE6013 is present in 44% (15/34) of different sequence types so far examined within S. aureus.48 In addition, all tested isolates of epidemic backgrounds ST239 and North American ST8-USA300 contained ICE6013.48,49 Several S. aureus strains harbor multiple ICE6013 copies and even multiple ICE families. For example, S. aureus ST239 strain TW20 carries both Tn5801 and ICE6013.50 S. aureus ST5 strain ED98 carries 3 different ICE6013 subfamilies. S. aureus ST398 strain S0385 carries Tn916 and 3 copies of ICE6013 spread between 2 subfamilies; the subfamily 1 elements are known as ICESa1A and ICESa1B and the subfamily 6 element is known as ICESa2.4,34 These findings suggest that neither the different families of staphylococcal ICEs nor the different subfamilies of ICE6013 strongly exclude each other within a given strain.
Before the discovery of ICE6013, the shortest known ICE was Tn916 at 18kb.51,52 The founding ICE6013 sequence in S. aureus ST239 strain HDG2 is 13.4kb, not including a Tn552 insertion that encodes the blaZ operon.48 The shortest example of ICE6013 is 12.7kb from S. intermedius strain NCTC11048, whereas the longest example is 16.5kb from S. agnetis strain 908.4 The S. agnetis element is predicted to encode 20 orfs, but no obvious antibiotic resistance genes are present on an ICE6013 element other than those provided by the Tn552 insertion in strain HDG2.Thus, ICE6013 may be the shortest known ICE and might provide information about the minimum requirements for a self-transmissible conjugative element within bacteria.
Recombination and conjugation of ICE6013
ICE6013 is the first family of ICEs identified to use an IS30-like DDE transposase (Tpase) as a recombinase. Previous studies on independent IS30 elements in E. coli showed that the Tpase has 2 insertion site preferences: a 24bp palindromic consensus sequence and sequences similar to its 26bp inverted repeats.53 IS30 has 2 helix-turn-helix motifs within the N-terminal DNA-binding domain of the Tpase. One motif was required for insertion into the consensus sequence, while the second motif was required for insertion into both types of insertion sites.53 The IS30-like Tpase from ICE6013 is encoded on 2 orfs, and is necessary for excision.4,48 This is contrary to most IS30-like elements that are encoded by one orf and not known to be associated with ICEs. The integration site of ICE6013 is a relatively arbitrary 3bp sequence that is duplicated upon integration and forms a coupling sequence for circular forms of the element upon excision. The imperfect inverted repeats that demarcate ICE6013 are 8bp. The integration site preference of ICE6013 was determined through a study of transconjugants to be a 15bp palindromic consensus sequence that surrounds the 3bp target site.4 At least 58 different integration sites of ICE6013 have been identified.4 Thus, there are some analogies in the site specificity of independent IS30 elements and ICE6013. The use of an IS30-like Tpase as ICE6013s recombinase clearly provides more versatility in integration sites than is the case with Tn5801s phage-like tyrosine recombinase.
Excision of ICE6013 ranged from 0.1 to 0.3% of a population regardless of the tested growth conditions.4 Frequencies of excision were significantly reduced upon disruption of a putative lipoprotein (orf3) and a putative helicase processivity factor (orf14) encoded by ICE6013.4 It is possible the lipoprotein acts as a pheromone, similar to the lipoprotein peptides that control the transfer of pCF10 conjugative plasmids in E. faecalis,54 though this remains to be tested. On the other hand, disruption of orf4 significantly increased excision frequencies of ICE6013. Orf4 is a putative membrane protein that contains a GlmU domain, which might be expected to synthesize a precursor (UDP-GlcNAc) used in several S. aureus macromolecules. The differing effects of orf3 and orf4 on excision suggest that these 2 orfs, along with one poly(A5) and 2 poly(A7) tracts in orf2, may comprise the regulation module for ICE6013 subfamily 1.
ICE6013 was shown to conjugatively transfer to an S. aureus recipient derived from strain RN4220 at a frequency of 1.16 × 10−7 ± 7.2 × 10−8 transconjugants/donor, which is on the order of 100x more efficient than reported for Tn5801.4,37 As observed with Tn5801, some mating pairs transfer ICE6013 much less efficiently or not at all.4,37,40,48 It has not been directly demonstrated that ICE6013 can replicate extrachromosomally, but the predominance of circular forms of the element in transconjugants after repeated passage strongly suggests the possibility.4 Insertional disruption of 5 of 6 genes for putative membrane-associated proteins disrupted conjugative transfer.4 In the S. aureus ST36 strain MRSA252, ICE6013 is integrated into a chromosomal site that flanks the strongest signal for homologous recombination in the species.55 This observation led to a test of the conjugative transfer of chromosomal segments flanking ICE6013, using an ST8-USA300 donor strain with ICE6013 integrated at a different site than in MRSA252, but no evidence for chromosomal transfer was obtained.4 It is not known whether ICE6013 of MRSA252 can mobilize flanking chromosomal DNA, or if the signal of recombination flanking its ICE6013 integration site indicates a region of reduced purifying selection that allows disruption by integrating MGEs or even positive selection driven by integrating MGEs at this site.
Future perspectives
It is interesting to note the common occurrence of Tn5801 and ICE6013 in the same clinically-relevant backgrounds of S. aureus. Unbiased strain sampling is needed to determine if there is a true association with the presence of ICEs in these backgrounds compared with other backgrounds. Both families of ICEs also occur in species that are associated with companion animals (S. aureus, S. pseudintermedius) and in S. aureus backgrounds that are associated with livestock animals (ST5 in poultry, ST398 in pigs). The possibility of these animal-associated species and strains acting as a reservoir of MGEs for human pathogenic strains has been proposed on several occasions.32,33,56,57 However, the reservoir of ICEs in staphylococci is essentially unexplored. Also, conjugation of ICEs between these human- and animal-associated species and strains has not yet been experimentally demonstrated.
The ability of ICE6013 to efficiently conjugate within S. aureus, and its presence in other staphylococci, suggests that the element may be a significant agent of HGT. Furthermore, the presence of ICE6013 in every tested isolate of the epidemic backgrounds S. aureus ST239 and North American ST8-USA300 suggests either a mechanism to prevent loss of the element, or some undiscovered advantage that the element confers upon its host. Mobilization of non-ICE DNA by Tn5801 and ICE6013 also has not yet been documented, but such an ability could have an impact on staphylococcal evolution.
ICE6013 may be minimal in size, but it has several orfs whose functions have yet to be elucidated. With the increased knowledge of gene families that signify the presence of ICEs, it is expected that additional ICEs will be identified from in silico studies of staphylococci. However, the most helpful knowledge will likely come through subsequent functional characterization of these elements. Finally, considering the high percentage of plasmids potentially mobilized by conjugative plasmids, in addition to the largely unexplored interactions between plasmids and ICEs, the paradigm that phages are the main mediators of HGT in staphylococci may eventually warrant a reevaluation.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was supported by National Institutes of Health grant GM080602
References
- [1].Lindsay J. Staphylococcus aureus genomics and the impact of horizontal gene transfer. Int J Med Microbiol. 2014;304(2):103-109. doi: 10.1016/j.ijmm.2013.11.010. PMID:24439196 [DOI] [PubMed] [Google Scholar]
- [2].Malachowa N, DeLeo F. Mobile genetic elements of Staphylococcus aureus. Cell Mol Life Sci. 2010;67(18):3057-71. doi: 10.1007/s00018-010-0389-4. PMID:20668911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Shearer J, Wireman J, Hostetler J, Forberger H, Borman J, Gill J, Sanchez S, Mankin A, LaMarre J, Lindsay J, et al.. Major Families of Multiresistant Plasmids from Geographically and Epidemiologically Diverse Staphylococci. Genes Genomes Genetics. 2011;1(7):581-591. PMID:22384369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Sansevere E, Luo X, Park J, Yoon S, Seo K, Robinson D. Transposase-Mediated Excision, Conjugative Transfer, and Diversity of ICE6013 Elements in Staphylococcus aureus. J Bacteriol. 2017;199(8):e00629-16. doi: 10.1128/JB.00629-16. PMID:28138100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Zong Z, Peng C, Lü X. Diversity of SCCmec Elements in Methicillin-Resistant Coagulase-Negative Staphylococci Clinical Isolates. PloS One. 2011;6(5):e20191. doi: 10.1371/journal.pone.0020191. PMID:21637845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Liu J, Chen D, Peters B, Li L, Li B, Xu Z. Staphylococcal Chromosomal Cassettes mec (SCCmec): A mobile genetic element in methicillin-resistant Staphylococcus aureus. Microb Pathog. 2016;101:56-67. doi: 10.1016/j.micpath.2016.10.028. PMID:27836760. [DOI] [PubMed] [Google Scholar]
- [7].Martínez-Rubio R, Quiles-Puchalt N, Martí M, Humphrey S, Ram G, Smyth D, Chen J, Novick R, Penadés J. Phage-inducible islands in the Gram-positive cocci. ISME Journal. 2017;11(4):1029-1042. doi: 10.1038/ismej.2016.163. PMID:27959343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Deghorain M, Van Melderen L. The Staphylococci phages family: An overview. Viruses. 2012;4(12):3316-35. doi: 10.3390/v4123316. PMID:23342361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Xia G, Wolz C. Phages of Staphylococcus aureus and their impact on host evolution. Infect Genet Evol. 2014;21:593-601. doi: 10.1016/j.meegid.2013.04.022. PMID:23660485. [DOI] [PubMed] [Google Scholar]
- [10].Chen J, Carpena N, Quiles-Puchalt N, Ram G. Intra-and Inter-Generic Transfer of Pathogenicity Island-encoded Virulence Genes by cos phages. J ISME. 2015;9(5):1260-3. doi: 10.1038/ismej.2014.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Winstel V, Liang C, Sanchez-Carballo P, Steglich M, Munar M, Bröker B, Penadés J, Nübel U, Holst O, Dandekar T, et al.. Wall Teichoic Acid Structure Governs Horizontal Gene Transfer Between Major Bacterial Pathogens. Nat Commun. 2013;4:S92-S97. doi: 10.1038/ncomms3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Winstel V, Sanchez-Carballo P, Holst O, Xia G, Peschel A. Biosynthesis of the unique wall teichoic acid of Staphylococcus aureus lineage ST395. mBio. 2014;5(2):e00869. doi: 10.1128/mBio.00869-14. PMID:24713320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Scharn C, Tenover F, Goering R. Transduction of Staphylococcal Cassette Chromosome mec Elements between Strains of Staphylococcus aureus. Antimicrob Agents Chemother. 2013;57(11):5233-8. doi: 10.1128/AAC.01058-13. PMID:23939891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Moon B, Park J, Hwang S, Robinson D, Thomas J, Fitzgerald J, Park Y, Seo K. Phage-Mediated Horizontal Transfer of a Staphylococcus aureus Virulence-Associated Genomic Island. Sci Rep. 2015;5(1):9784. doi: 10.1038/srep09784. PMID:25891795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Moon B, Park J, Robinson D, Thomas J, Park Y, Thornton J, Seo K. Mobilization of Genomic Islands of Staphylococcus aureus by Temperate Bacteriophage. PLoS One. 2016;11(3):e0151409. doi: 10.1371/journal.pone.0151409. PMID:26953931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].McCarthy A, Loeffler A, Witney A, Gould K, Lloyd D, Lindsay J, McCarthy AJ, Loeffler A, Witney AA, Gould KA, et al.. Extensive Horizontal Gene Transfer during Staphylococcus aureus Co-colonization In Vivo. Genome Biol Evol. 2014;6(10):2697-2708. doi: 10.1093/gbe/evu214. PMID:25260585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].McCarthy A, Lindsay J. The distribution of plasmids that carry virulence and resistance genes in Staphylococcus aureus is lineage associated. BMC Microbiol. 2012;12(104). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Ramsay J, Firth N. Diverse Mobilization Strategies Facilitate Transfer of Non-Conjugative Mobile Genetic Elements. Curr Opin Microbiol. 2017;38:1-9. doi: 10.1016/j.mib.2017.03.003. PMID:28391142. [DOI] [PubMed] [Google Scholar]
- [19].Ramsay J, Kwong S, Murphy R, Yui Eto K, Price K, Nguyen Q, O'Brien F, Grubb W, Coombs G, Firth N. An Updated View of Plasmid Conjugation and Mobilization in Staphylococcus. Mob Genet Elements. 2016;6(4):e1208317. doi: 10.1080/2159256X.2016.1208317. PMID:27583185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].O'Brien F, Ramsay J, Monecke S, Coombs G, Robinson O, Htet Z, Alshaikh F, Grubb W. Staphylococcus aureus plasmids without mobilization genes are mobilized by a novel conjugative plasmid from community isolates. J Antimicrob Chemother. 2015;70(3):649-652. doi: 10.1093/jac/dku454. PMID:25411186. [DOI] [PubMed] [Google Scholar]
- [21].Cury J, Touchon M, Rocha E. Integrative and conjugative elements and their hosts: composition, distribution and organization. Nucleic Acids Res. 2017;46(GKX607):202-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Roberts A, Mullany P. A modular master on the move: The Tn916 Family of Mobile Genetic Elements. Trends Microbiol. 2009;17(6):251-8. doi: 10.1016/j.tim.2009.03.002. PMID:19464182. [DOI] [PubMed] [Google Scholar]
- [23].Guglielmini J, Quintais L, Garcillán-Barcia MP, de la Cruz F, Rocha EPC. The repertoire of ice in prokaryotes underscores the unity, diversity, and ubiquity of conjugation. PLoS Genet. 2011;7(8):e1002222. doi: 10.1371/journal.pgen.1002222. PMID:21876676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Peña A, Matilla I, Martín-Benito J, Valpuesta JM, Carrascosa JL, de la Cruz F, Cabezón E, Arechaga I. The Hexameric Structure of a Conjugative VirB4 protein ATPase Provides New Insights for a Functional and Phylogenetic Relationship with DNA Translocases. J Biol Chem. 2012;287(47):39925-32. doi: 10.1074/jbc.M112.413849. PMID:23035111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Thomas J, Lee C, Grossman A. A Conserved Helicase Processivity Factor is Needed for Conjugation and Replication of an Integrative and Conjugative Element. PLoS Genet. 2013, 9(1):e1003198. doi: 10.1371/journal.pgen.1003198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Wright L, Grossman A. Autonomous Replication of the Conjugative Transposon Tn916. J Bacteriol. 2016;198(24):3355-3366. doi: 10.1128/JB.00639-16. PMID:27698087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Lee C, Babic A, Grossman A. Autonomous plasmid-like replication of a conjugative transposon. Mol Microbiol. 2010;75(2):268-279. doi: 10.1111/j.1365-2958.2009.06985.x. PMID:19943900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Lee CA, Thomas J, Grossman AD. The Bacillus subtilis Conjugative Transposon ICEBs1 Mobilizes Plasmids Lacking Dedicated Mobilization Functions. J Bacteriol. 2012;194(12):3165-72. doi: 10.1128/JB.00301-12. PMID:22505685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Showsh S, Andrews R. Analysis of the Requirement for a pUB110 mob Region during Tn916-Dependent Mobilization. Plasmid. 1999;41(3):179-186. doi: 10.1006/plas.1999.1398. PMID:10366523. [DOI] [PubMed] [Google Scholar]
- [30].Jones J, Yost S, Pattee P. Transfer of the Conjugal Tetracycline Resistance Transposon Tn916 from Streptococcus faecalis to Staphylococcus aureus and Identification of Some Insertion Sites in the Staphylococcal Chromosome. J Bacteriol. 1987;169(5):2121-31. doi: 10.1128/jb.169.5.2121-2131.1987. PMID:3032908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Bertram J, Strätz M, Dürre P. Natural Transfer of Conjugative Transposon Tn916 Between Gram-Positive and Gram-Negative Bacteria. J Bacteriol. 1991;173(2):443-8. doi: 10.1128/jb.173.2.443-448.1991. PMID:1846142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Chajęcka-Wierzchowska W, Zadernowska A, Nalepa B. Coagulase-negative staphylococci (CoNS) Isolated from Ready-to-Eat Food of Animal Origin–Phenotypic and Genotypic Antibiotic Resistance. Food Microbiol. 2015;46:222-6. doi: 10.1016/j.fm.2014.08.001. PMID:25475289. [DOI] [PubMed] [Google Scholar]
- [33].Stegmann R, Perreten V. Antibiotic Resistance Profile of Staphylococcus rostri, a New Species Isolated from Healthy Pigs. Vet Microbiol. 2010;145(1):165-71. doi: 10.1016/j.vetmic.2010.03.015. PMID:20399039. [DOI] [PubMed] [Google Scholar]
- [34].Schijffelen M, Boel C, van Strijp J, Fluit A. Whole Genome Analysis of a Livestock-Associated Methicillin-Resistant Staphylococcus aureus ST398 Isolate from a Case of Human Endocarditis. BMC Genomics. 2010;11(1):376. doi: 10.1186/1471-2164-11-376. PMID:20546576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Roberts A, Mullany P. Tn916-like Genetic Elements: A Diverse Group of Modular Mobile Elements Conferring Antibiotic Resistance. FEMS Microbiol Rev. 2011;35(5):856-71. doi: 10.1111/j.1574-6976.2011.00283.x. PMID:21658082. [DOI] [PubMed] [Google Scholar]
- [36].Kuroda M, Ohta T, Uchiyama I, Baba T, Yuzawa H, Kobayashi I, Cui L, Oguchi A, Aoki KK, Nagai Y, et al.. Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet. 2001;357(9264):1225-1240. doi: 10.1016/S0140-6736(00)04403-2. PMID:11418146. [DOI] [PubMed] [Google Scholar]
- [37].de Vries L, Christensen H, Skov R, Aarestrup F, Agers Y. Diversity of the Tetracycline Resistance Gene tet(M) and Identification of Tn916-and Tn5801-like (Tn6014) Transposons in Staphylococcus aureus from Humans and Animals. J Antimicrob Chemother. 2009;64(3):490-500. doi: 10.1093/jac/dkp214. PMID:19531603. [DOI] [PubMed] [Google Scholar]
- [38].de Vries L Hasman H, Rabadán S, Jurado Rabadán S, Agersø Y. Sequence-Based Characterization of Tn5801-Like Genomic Islands in Tetracycline-Resistant Staphylococcus pseudintermedius and Other Gram-positive Bacteria from Humans and Animals. Front Microbiol. 2016;7(576):576. PMID:27199912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].León-Sampedro R, Novais C, Peixe L. Diversity and Evolution of the Tn5801-tet(M)-like Integrative and Conjugative Elements Among Enterococcus, Streptococcus, and Staphylococcus. Antimicrob Agents Chemother. 2016;60(3):1736-46. doi: 10.1128/AAC.01864-15. PMID:26729505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Mingoia M, Morici E, Tili E, Giovanetti E, Montanari MP, Varaldo PE. Characterization of Tn5801.Sag, a Variant of Staphylococcus aureus Tn916 Family Transposon Tn5801 that is Widespread in Clinical Isolates of Streptococcus agalactiae. Antimicrob Agents Chemother. 2013;57(9):4570-4. doi: 10.1128/AAC.00521-13. PMID:23817370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Showsh S, Andrews R. Tetracycline enhances Tn916-mediated conjugal transfer. Plasmid. 1992;28(3):213-224. doi: 10.1016/0147-619X(92)90053-D. PMID:1334267. [DOI] [PubMed] [Google Scholar]
- [42].Doucet-Populaire F, Trieu-Cuot P, Andremont A, Courvalin P. Conjugal transfer of plasmid DNA from Enterococcus faecalis to Escherichia coli in digestive tracts of gnotobiotic mice. Antimicrob Agents Chemother. 1992;36(2):502-4. doi: 10.1128/AAC.36.2.502. PMID:1605622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Chen K, Reuter M, Sanghvi B, Roberts G, Cooper L, Tilling M, Blakely G, Dryden D. ArdA Proteins from Different Mobile Genetic Elements can Bind to the EcoKI Type I DNA Methyltransferase of E. coli K12. Biochim Biophys Acta – Proteins Proteomics. 2014;1844(3):505-511. doi: 10.1016/j.bbapap.2013.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Serfiotis-Mitsa D, Robert G, Cooper L, White J, Nutley M, Cooper A, Blakely G, Dryden D. The Orf18 Gene Product from Conjugative Transposon Tn916 is an ArdA Antirestriction Protein that Inhibits Type I DNA Restriction–Modification Systems. J Mol Biol. 2008;383(5):970-81. doi: 10.1016/j.jmb.2008.06.005. PMID:18838147. [DOI] [PubMed] [Google Scholar]
- [45].Thomas A, Brammar W, Wilkins B. Plasmid R16 ArdA Protein Preferentially Targets Restriction Activity of the Type I Restriction-Modification System EcoKI. J Bacteriol. 2003;185(6):2022-5. doi: 10.1128/JB.185.6.2022-2025.2003. PMID:12618468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].McMahon S, Robert SG, Johnson K, Cooper L, Liu H, White J, Carter L, Sanghvi B, Oke M, Walkinshaw M, et al.. Extensive DNA Mimicry by the ArdA Anti-Restriction Protein and its Role in the Spread of Antibiotic Resistance. Nucleic Acids Res. 2009;37(15):4887-97. doi: 10.1093/nar/gkp478. PMID:19506028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Waldron D, Lindsay J. Sau1: a Novel Lineage-Specific Type I Restriction-Modification System that Blocks Horizontal Gene Transfer into Staphylococcus aureus and between S. aureus Isolates of Different Lineages. J Bacteriol. 2006;188(15):5578-85. doi: 10.1128/JB.00418-06. PMID:16855248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Smyth D, Robinson D. Integrative and Sequence Characteristics of a Novel Genetic Element, ICE6013, in Staphylococcus aureus. J Bacteriol. 2009;191(19):5964-5975. doi: 10.1128/JB.00352-09. PMID:19648240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Jamrozy D, Harris S, Mohamed N, Peacock S, Tan C, Parkhill J, Anderson A, Holden M. Pan-Genomic Perspective on the Evolution of the Staphylococcus aureus USA300 Epidemic. Microb Genomics. 2016;2(5):1-14. doi: 10.1099/mgen.0.000058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Holden M, Lindsay J, Corton C, Quail M, Cockfield J, Pathak S, Batra R, Parkhill J, Bentley S, Edgeworth J. Genome Sequence of a Recently Emerged, Highly Transmissible, Multi-Antibiotic- and Antiseptic-Resistant Variant of Methicillin-Resistant Staphylococcus aureus, Sequence Type 239 (TW). J Bacteriol. 2010;192(3):888-892. doi: 10.1128/JB.01255-09. PMID:19948800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Johnson C, Grossman A. Integrative and Conjugative Elements (ICEs): What they do and how they work. Annu Rev Genet. 2015;49(1):577-601. doi: 10.1146/annurev-genet-112414-055018. PMID:26473380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Churchward G. Back to the future: the new ICE age. Mol Microbiol. 2008;70(3):554-556. doi: 10.1111/j.1365-2958.2008.06415.x. PMID:18761693. [DOI] [PubMed] [Google Scholar]
- [53].Nagy Z, Szabó M, Chandler M. Analysis of the N‐terminal DNA Binding Domain of the IS30 Transposase. Mol Microbiol. 2004;54:478-88. doi: 10.1111/j.1365-2958.2004.04279.x. PMID:15469518. [DOI] [PubMed] [Google Scholar]
- [54].Antiporta M, Dunny G. CcfA, the Genetic Determinant for the cCF10 Peptide Pheromone in Enterococcus faecalis OG1RF. J Bacteriol. 2002;184(4):1155-62. doi: 10.1128/jb.184.4.1155-1162.2002. PMID:11807076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Everitt R, Didelot X, Batty E, Miller R, Knox K, Young B, Bowden R, Auton A, Votintseva A, Larner-Svensson H, et al.. Mobile Elements Drive Recombination Hotspots in the Core Genome of Staphylococcus aureus. Nat Commun. 2014;5:3956. doi: 10.1038/ncomms4956. PMID:24853639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].McCarthy A, Harrison E, Stanczak-Mrozek K, Leggett B, Waller A, Holmes M, Lloyd D, Lindsay J, Loeffler A. Genomic Insights into the Rapid Emergence and Evolution of MDR in Staphylococcus pseudintermedius. J Antimicrob Chemother. 2015;70(4):997-1007. PMID:25527273. [DOI] [PubMed] [Google Scholar]
- [57].Otto M. Coagulase-negative staphylococci as reservoirs of genes facilitating MRSA infection. BioEssays. 2013;35(1):4-11. doi: 10.1002/bies.201200112. PMID:23165978. [DOI] [PMC free article] [PubMed] [Google Scholar]


