Summary
We performed genomic, epigenomic, transcriptomic and proteomic characterizations of uterine carcinosarcomas (UCSs). Cohort samples had extensive copy number alterations and highly recurrent somatic mutations. Frequent mutations were found in TP53, PTEN, PIK3CA, PPP2R1A, FBXW7 and KRAS, similar to endometrioid and serous uterine carcinomas. Transcriptome sequencing identified a strong epithelial-to-mesenchymal transition (EMT) gene signature in a subset of cases that was attributable to epigenetic alterations at microRNA promoters. The range of EMT scores in UCS was the largest amongst all tumor types studied via The Cancer Genome Atlas. UCSs shared proteomic features with gynecologic carcinomas and sarcomas with intermediate EMT features. Multiple somatic mutations and copy number alterations in genes that are therapeutic targets were identified.
Keywords: Uterine carcinosarcoma, endometrial cancer, the cancer genome atlas, translational science, gynecologic oncology, gynecologic cancer, EMT, epithelial to mesenchymal transition
Introduction
Uterine carcinosarcomas (UCSs) are rare, aggressive, biphasic neoplasms that are defined as tumors consisting of high-grade malignant epithelial and mesenchymal elements (Kurman et al., 2014). They account for less than 5% of all uterine malignancies. The mesenchymal component may contain cell types native to the uterus, which are termed “homologous”, or extra-uterine histologic subtypes, which are termed “heterologous”. Homologous components usually resemble high-grade undifferentiated sarcoma or fibrosarcoma, and heterologous components frequently appear to be rhabdomyosarcomas or chondrosarcomas. At the time of diagnosis, approximately one third of patients present with disease that has spread beyond the uterus (Bland et al., 2009; Kurman et al., 2014). Five-year progression-free survival for disease confined to the uterus at diagnosis is 40–75%, compared to 20–35% for disease with extra-uterine extension (Ferguson et al., 2007; Yamada et al., 2000). Reported risk factors for developing UCS include prior pelvic radiotherapy and exposure to tamoxifen (Brinton et al., 2013; Pothuri et al., 2006). Heterologous differentiation is a prognostic feature associated with a higher risk of tumor recurrence, particularly when present in early-stage disease (Ferguson et al., 2007; Lopez-Garcia and Palacios, 2010).
UCSs were thought to develop either from a collision between independent, geographically adjacent carcinomas and sarcomas (collision theory) or as a combination of cellular masses that underwent early divergence from a common precursor stem cell (combination theory) (McCluggage, 2002). Recent evidence suggests that most tumors are monoclonal in origin, with late divergence and metaplasia of the carcinoma into the sarcomatous components (conversion theory). Both components—carcinoma and sarcoma—express many similar immunohistochemical markers, share common somatic mutations and often have identical patterns of X-chromosome inactivation (Kounelis et al., 1998; Thompson et al., 1996; Wada et al., 1997). Additional studies using polymorphic microsatellite markers in both components demonstrated strongly shared patterns of loss of heterozygosity indicative of late patterns of divergent evolution (Abeln et al., 1997; Fujii et al., 2000). By resolving the clonal origins of UCSs, we will better understand patterns of metastases and approaches to therapeutic targeting.
Prior work that included 13 non-hypermutated UCS tumors found a high frequency of TP53 mutations and lower frequencies of FBXW7, PIK3CA and PPP2R1A mutations (Jones et al., 2014). Many of the mutations in chromatin-remodeling genes were found in hypermutated UCS samples or in carcinosarcoma tumors of ovarian origin. A recent publication of 24 non-hypermutated UCS tumor-normal exomes found lower frequencies of TP53 and FBXW7 mutations (Zhao et al., 2016). The purpose of this study was to conduct a comprehensive, multi-platform analysis of adjudicated UCS cases to identify key molecular insights into tumorigenesis.
Results
Samples and clinical data
Primary tumor samples and corresponding germline DNA were collected from 57 untreated patients with UCS who had a mean age of 70 years. Local Institutional Review Boards approved all tissue acquisition. Only cases that cleared rigorous pathology review were included. Three expert gynecological pathologists microscopically examined virtual slides prepared from hematoxylin and eosin–stained glass slides to ensure that tumors met WHO 2014 criteria and to exclude histologic mimics of UCS. Annotated morphologic data included carcinoma and sarcoma type, presence of heterologous differentiation and percentage distribution of carcinomatous and mesenchymal components. Clinical and pathological characteristics of the samples generally reflect a cross-section of individuals with UCS (Table S1). The median follow-up of the cohort was 25.7 months (range, 1–104 months); 64% of the patients had disease recurrence, and 58% died during follow-up. Comprehensive molecular analyses were performed at independent centers using six genomic, epigenomic or proteomic platforms (Supplemental Experimental Procedures).
Somatic alterations
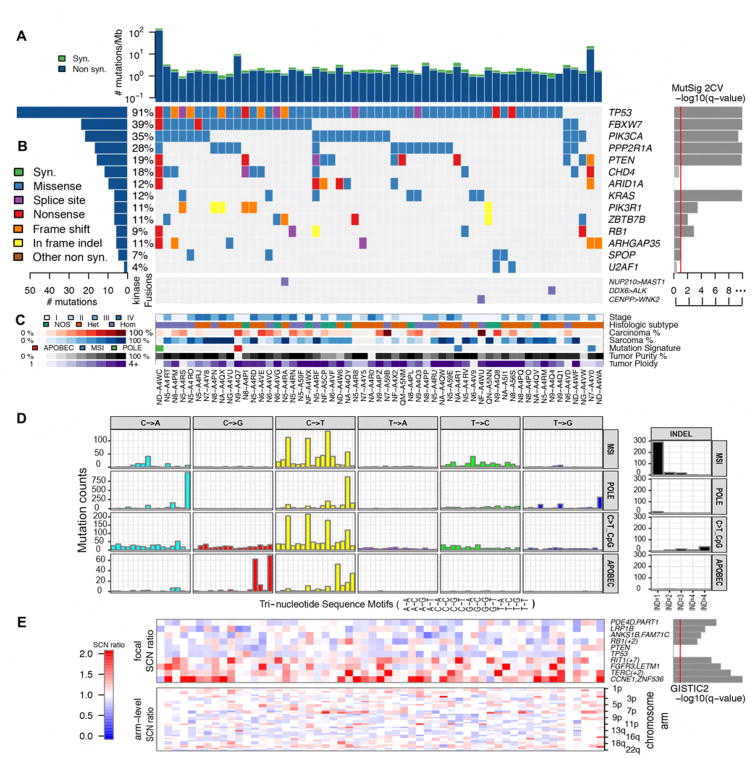
Exome sequencing of all 57 UCS tumors identified 9149 somatic mutations based on consensus calls from four mutation-detection algorithms (Table S2). From this set of mutations, we selected 492 sites for independent hybrid-capture validation, representing 161 genes from lists of significantly mutated genes in this study (below and Supplemental Experimental Procedures) as well as The Cancer Genome Atlas (TCGA) endometrioid endometrial cancer (UCEC) study. Of 417 sites with sufficient coverage to be 99% powered to validate mutations, 99.5% of these mutations were validated (376 of 378 single-nucleotide variants and all 35 indels). Similar to other gynecologic tumors, the median non-silent mutation density of this cohort was 1.59 mutations/Mb; however, a single tumor had an elevated mutation density of 120 mutations/Mb (Figures 1A and S1A), likely due to a mutation in the DNA polymerase gene POLE. Analysis of mutational signatures also identified an APOBEC signature with C>T and TC>AA/T substitutions in a single tumor that had elevated expression of several APOBEC3 genes (Figure S1B). Combined analysis using multiple versions of MutSig identified 14 significantly recurring mutations, many of which were also recurrent mutations in uterine endometrial cancer (Figure 1B and Table S3) (Cancer Genome Atlas Research et al., 2013; Lawrence et al., 2014; Lawrence et al., 2013). Most prominent were TP53 mutations, which were found in all but five tumors (91%). Approximately half of the UCS tumors had mutations in one or more of the PI3-kinase pathway genes—PIK3CA (35%), PTEN (19%) or PIK3R1 (11%). However, unlike previous studies of endometrial tumors where PTEN and TP53 mutations tended to be mutually exclusive and confined to one of the two common histologic subtypes (endometrioid or serous), eight (73%) of 11 tumors with PTEN mutations also had a TP53 mutation. Other significantly mutated genes shared with endometrioid or serous endometrial tumors included FBXW7 (28%), PPP2R1A (28%), CDH4 (18%), KRAS (12%), ARID1A (12%), ARHGAP35 (11%) and SPOP (7%). Genes that were significantly mutated in UCS but not in endometrial carcinomas include the tumor suppressor RB1 (11%) and the splicing factor U2AF1 (4%), which was twice mutated at a hotspot (p.S34F) also found in lung adenocarcinoma and acute myeloid leukemia tumors. The transcription factor ZBTB7B (11%) is a gene that was significantly mutated only in UCS. TP53 and KRAS hotspots are ubiquitous in many cancer types. The PIK3CA hotspots p.E545K and p.H1047R are the two most commonly reported mutated PIK3CA sites in this study and in cancer overall (COSMIC v78), both leading to a constitutively active enzyme. PTEN and ARHGAP35 mutations were more common in the few tumors without identified TP53 mutations. Through analysis of RNA sequencing data, three samples were found to have kinase-domain gene fusions (NUP210-MAST1, CENPP-WNK2 and DDX6-ALK) based on the consensus of two fusion-detection approaches (Supplemental Experimental Procedures). No recurring fusions leading to an in-frame protein were detected in this cohort. One sample was dominated by the APOBEC mutation signatures, whereas two samples showed microsatellite instability, and the sample with the highest mutation rate had a mutation in POLE (Figure 1C).
Figure 1. Landscape of somatic alterations in 57 uterine carcinosarcomas.
(A) Synonymous (syn.) and non-synonymous (non syn.) mutation density (mutations/Mb) across the cohort. (B) Gene-sample matrix of mutations in MutSig 2CV significant genes (Benjamini-Hochberg false-discovery rate q value < 0.1, left). Samples with kinase-domain fusions are shown below the mutations. Samples are ordered by number of mutations in significant genes. (C) Tumor stage, histologic subtype, carcinoma and sarcoma percentages, mutation signature type, purity and ploidy across the cohort. (D) Mutational signatures discovered in UCS cohort. A variant of non-negative matrix factorization (Bayesian NMF) to the mutation count matrix across samples (101 by 57). The vertical scale represents counts of mutations in each context bin labeled on the horizontal axis. Indel counts and bins are shown on the right. MSI, Micro-satellite instability. IND=1–4 refer to one, two, three, or four or more base indels. (E) Heatmap of focal somatic copy number (SCN) alterations (top) in genes with the highest GISTIC2 significance (right) for deletions (top 4), amplifications (lower 4), as well as for TP53 and PTEN, which were not significantly altered. The SCN ratio scale is corrected by purity and ploidy for each sample. Also shown is the arm-level SCN chromosome-sample heatmap across the cohort (bottom). See also Figures S1 and S2 and Tables S1 – S4.
Single nucleotide variants (SNVs) stratified by 96 tri-nucleotide base substitutions and indels grouped by the number of inserted or deleted bases identified four mutational signatures (Figure 1D). The APOBEC signature was characterized by C>T/G at TCW (W=A/T), the POLE signature had a predominance of C>A at TCT and C>T at TCG, the microsatellite instability (MSI) signature had a significant enrichment of one-base indels, and the spontaneous deamination signature was dominated by C>T at CpG di-nucleotides.
Somatic copy number alterations in 56 of 57 UCSs were determined using SNP 6.0 Arrays (Affymetrix). Most tumors were aneuploid with frequent arm- and focal-level alterations (Figures 1E and S2A). Analysis of ploidy and whole-genome doubling using the ABSOLUTE algorithm predicted that this cohort had a median ploidy of 3.3 and that 90% of the tumors had undergone at least one whole-genome-doubling event (Carter et al., 2012). This percentage is significantly higher than in serous ovarian tumors (54%, p < 0.001 Fisher’s exact test), the tumor type with the next highest frequency of genomic doubling in TCGA (Zack et al., 2013). GISTIC 2.0 analysis (Mermel et al., 2011) of significantly reoccurring copy number alterations identified 25 regions of focal amplifications and 37 regions of focal deletion (Table S4). Recurring focal amplifications include those containing the oncogenes TERC (3q26.2), FGFR3 (4p16.3), MYC (8q24.21), KAT6A (10q22.2), MDM2 (12q15), ERBB2 (17q12), CCNE1 (19q12) and BCL2L1 (20q11.21) (Figures 1E and S2B). A 1q22 amplification containing the recently discovered oncogene RIT1 (Berger et al., 2014), which was focally amplified in serous endometrial tumors, was also present. Recurring focal deletions contained the tumor suppressors PTPRD (9q23) and RB1 (13q12.2).
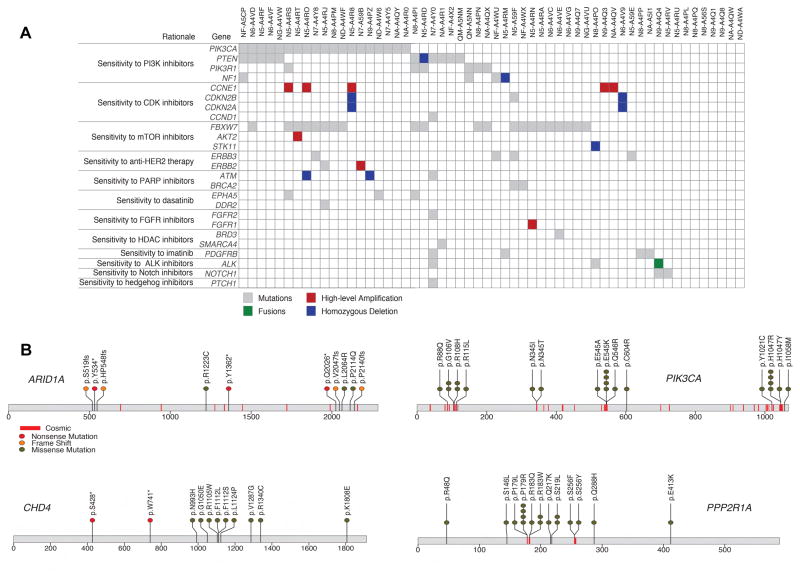
Analysis of clinically relevant somatic alterations was conducted using the PHIAL algorithm (Figures 2A) (Van Allen et al., 2014). Potentially actionable PI3-kinase-related alterations were found in 42.1% of patients. These alterations included somatic mutations in PIK3CA, PIK3R1 and PTEN, in addition to homozygous deletions of PTEN and NF1. Alterations were also identified in AKT/mTOR targets in 32% of patients, including frequent mutations in FBXW7, AKT2 and STK11. Together, 62% of tumors had one or more potentially clinically relevant alteration in the PI3K/AKT/mTOR pathway. Recurrent hotspot mutations were seen in PIK3CA and PPP2R1A (Figure 2B). Potentially clinically relevant alterations in cell cycle genes were found in 22.8% of patients, including somatic mutations in CCND1, CDKN2B; high-level amplification of CCNE1; and homozygous deletion of CDKN2A and CDKN2B. Potentially actionable alterations were also found in multiple cases for somatic mutations in ERBB2, ERBB3, BRCA2, and homozygous deletion of ATM. Mutations in chromatin remodeling genes including ARID1A and CHD4 were common and are becoming potentially actionable (Figure 2B). Single cases with FGFR2 and SMARCA4 mutations and an in-frame fusion with ALK were identified. Taken together, these data provide a rationale for the use of sequence-based diagnostics to direct patients with UCS to targeted therapy.
Figure 2. Clinically relevant alterations in uterine carcinosarcoma.
(A) Landscape of potentially clinically relevant mutations, copy number alterations and fusions in 56 UCS tumors, as identified by the PHIAL algorithm. Rows represent alterations in genes that lead to a given clinical rationale. Somatic mutations are shown in gray, homozygous deletions in blue, high-level amplifications in red (focal and more than two copies gained) and gene fusions in green. One POLE hypermutator case is excluded. (B) Mutation positions in amino acid coordinates for 4 of 14 significantly mutated genes. Each mutation is annotated by protein change (Gencode v18 transcripts). Also marked in red lines are sites of mutations from COSMIC v67 for which more than 5 experiments or samples had a mutation at the site.
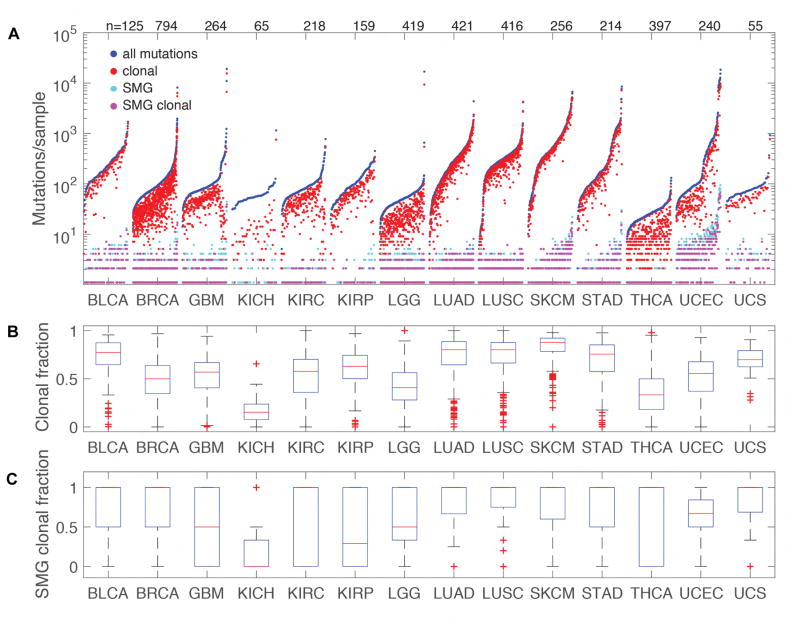
ABSOLUTE was used to predict the clonality of mutations, including an adjustment for cellularity (Figure 3A). Seventy-three percent of all mutations (Figure 3B) and 82% of those in significantly mutated genes (Figure 3C) were found to be clonal, which is similar to the frequencies seen in most other solid tumor types. The high clonal proportion of UCS mutations is consistent with mutations from bladder urothelial carcinoma, lung adenocarcinoma, lung squamous cell carcinoma, skin cutaneous melanoma, and stomach adenocarcinoma. This high degree of clonality in UCS is also consistent with the tumor being derived from a single cell of origin. A lower degree of clonality would be expected from the merger of two histologically distinct (collision) tumors, as well as from a tumor that diverged early from a common precursor (combination theory), resulting in greater differentiation between cell populations. These findings are consistent with the conversion theory of UCS metaplasia from an epithelial precursor.
Figure 3. Clonality of UCS tumors compared with other TCGA tumor types.
(A) Sorted mutation counts for each tumor separated by tumor type. All mutations are plotted in blue, clonal mutations in red, mutations in Significantly Mutated Genes (SMG) are light blue and clonal SMG mutations in magenta. All data was uniformly processed through the ABSOLUTE pipeline that includes adjustment for cellularity. SMGs for other tumor types were based on www.tumorportal.org (Lawrence et al., 2014). Labels across the top of the plot indicate the count of tumors included for each tumor type. (B) Boxplots showing the clonal proportion of mutations. Boxplot center lines (red) represent tumor medians, box limits (blue) are the inter-quartile range from 25% and 75%, whiskers (black) represent the extent of tumors out to 1.5 times the inter-quartile range, and red crosses are outliers beyond 1.5 times the inter-quartile range. (C) Boxplots showing the clonal proportion of SMGs. Components of the figure are as defined in panel (B). BLCA, bladder urothelial carcinoma; BRCA, breast invasive carcinoma; GBM, glioblastoma; KICH, kidney chromophobe; KIRC, kidney renal clear cell carcinoma; KIRP, kidney renal papillary cell carcinoma; LGG, low grade glioma; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; SKCM, skin cutaneous melanoma; STAD, stomach adenocarcinoma; THCA thyroid carcinoma; UCEC, uterine corpus endometrial carcinoma; UCS, uterine carcinosarcoma.
Based on prior work from TCGA and others (Cancer Genome Atlas Research et al., 2013; McConechy et al., 2012), we classified 55 of the 57 UCS samples as endometrioid-like or serous-like based on the presence of PTEN, ARID1A, PPP2R1A and/or TP53 somatic mutations (Table S3). Two samples were not classified because one was an ultramutated sample with a POLE mutation and one sample had no mutations in any of the classification genes. Of the 55 classified cases, 12 (22%) were endometrioid-like and 43 (78%) were serous-like. All 10 stage IV patients were classified as serous-like. Histologically, the endometrioid-like cases were more likely to have leiomyosarcoma components (p = 0.05) and the serous-like cases were more likely to have rhabdomyosarcoma components (p = 0.04, Table S1). There were no significant differences in progression-free or overall survival based on histologic subtype (data not shown).
Mesenchymal features of UCS
EMT is a process in which epithelial cells undergo loss of polarity and other changes that enable them to develop a mesenchymal phenotype. EMT is associated with greater metastatic potential (Radisky, 2005). Although different tumor types exhibit varying levels of EMT, studies have suggested that non-gynecologic carcinosarcomas (e.g., breast) might be examples of complete EMT (Sarrio et al., 2008). Using array-based gene expression profiling, an EMT phenotype has been suggested in a subset of UCS tumors (Chiyoda et al., 2012).
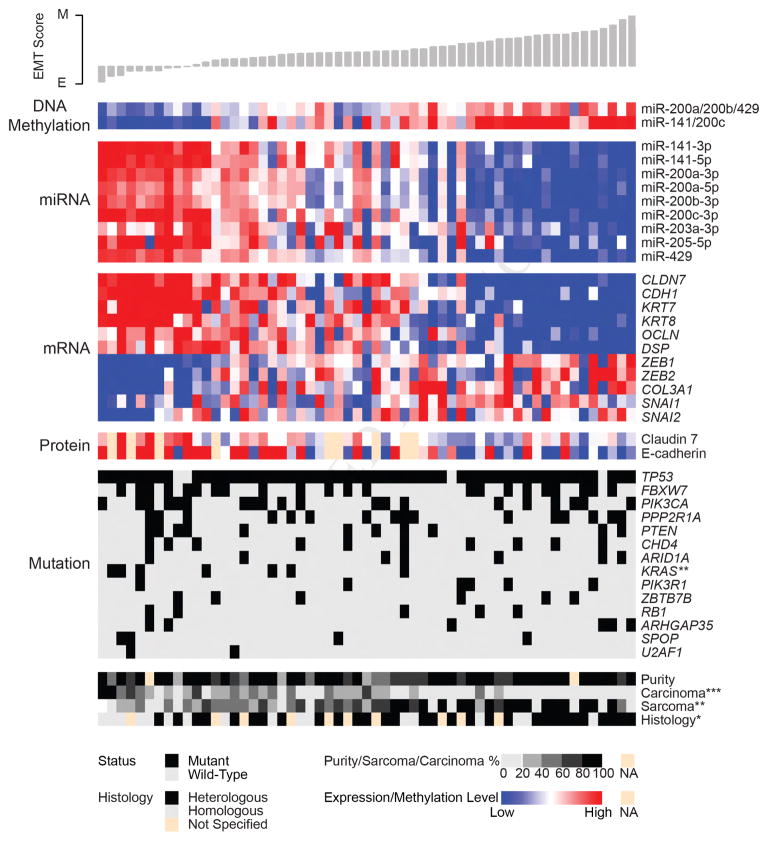
To quantify per-sample EMT levels across multiple tumor types and in our cohort, we calculated EMT scores based on a 76-gene signature (Byers et al., 2013). The EMT score provides a quantitative phenotypic measurement in which lower scores correspond to an epithelial phenotype and higher scores correspond to a mesenchymal phenotype. Based on EMT scores, high expression levels of many microRNAs (miRNAs), including all members of the miR-200 family, were significantly correlated with epithelial phenotype tumors (Figure 4). This result is noteworthy because the miR-200 family may have therapeutic relevance (Cortez et al., 2014). We also found a strong negative association between the expression of these miRNAs and their promoter methylation levels (Figure 4), which demonstrates epigenetic regulation of the miR-200 family and perhaps indicates a mechanism for the EMT in UCS. The significant association of EMT scores extends to expression of E-cadherin at the gene and protein levels, as well as to CDH1 regulators SNAI1/2 and ZEB1/2, which are direct miR-200 family targets. EMT scores were also higher in tumors with histologic findings that showed heterologous differentiation.
Figure 4. Evidence of EMT in uterine carcinosarcoma.
The heatmap shows methylation levels for miR-200 family members, expression of miR-200 family members, expression of select genes associated with EMT and protein expression of genes associated with EMT. Methylation and expression levels are median-centered by row and scaled by platform for visualization purposes. One DNA methylation probe from multiple highly correlated probes that map to the promoter of each miRNA cluster is shown (miR-200a/200b/429 = cg15822328, miR-141/200c = cg24702147). Samples (n = 57) appear in columns and are ordered according to EMT score, as shown by the histogram at the top. Annotation tracks at the bottom show mutation status for significantly mutated genes, as well as values of clinical variables of interest. Missing values (NA) for protein expression (n = 9) and tumor purity (n = 2) are indicated. Wilcoxon rank sum tests were used to assess the significance of associations between EMT scores and either gene mutations or histological type; Spearman correlation tests were used to assess the significance of associations between EMT scores and tumor purity, carcinoma percentage, and sarcoma percentage. Asterisks indicate statistically significant p values: *p < 0.05; **p < 0.01; ***p < 0.001. E, epithelial; M, mesenchymal; EMT, epithelial-to-mesenchymal transition. See also Figure S3 and Table S5.
Seven miRNA clusters were identified, suggesting that our cohort was molecularly heterogeneous (Figure S3A). We noted that miRNA cluster 1 had relatively high EMT scores and low miR-200b abundances (Figures S3B and S3C). We also noted that multiple members of the miR-200 family were associated with the EMT transcription factors ZEB1 and ZEB2 (Figure S3D and Table S5). Of 62 miRNAs previously associated with EMT, 14 were anticorrelated with the EMT score at a false discovery rate <0.05 (Figure S3E).
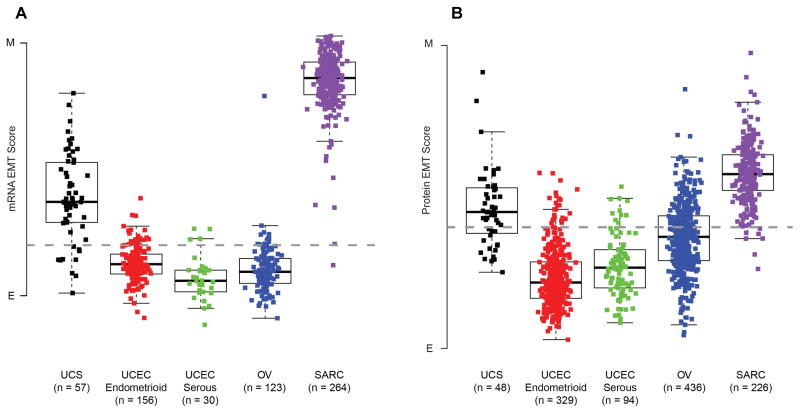
Although the EMT scores have significant positive and negative correlations with the percentages of sarcoma and carcinoma, respectively, no associations were observed with tumor purity estimates derived from ABSOLUTE (Carter et al., 2012). UCS has an EMT score that is intermediate between other gynecologic tumors and sarcomas (Figure 5A). Using a protein-derived EMT score also demonstrates that UCS is intermediate between other gynecologic tumors and sarcomas (Figure 5B). The expression of 131 genes identified as being correlated with EMT was more strongly correlated with the EMT scores in UCS than in other gynecologic cancers (Figure S4A). Furthermore, of 16 tumor types analyzed by TCGA, UCS had the most variable EMT scores (Figure S4B). There was no association between EMT scores and clinical outcome (data not shown). We calculated protein expression-based pathway scores for nine pathways, as described previously (Akbani et al., 2014), and correlated the nine pathway scores with the mRNA-based EMT pathway score. The nine pathways include apoptosis, cell cycle, DNA damage response, hormone_a, hormone_b, PI3K/AKT, RAS/MAPK, RTK, and TSC/mTOR. Two pathways, DNA damage response (R = 0.382, p = 0.007) and PI3K/AKT (R = 0.389, p = 0.006), were correlated with EMT (Table S6). Taken together, these findings show that, despite high tumor purity, UCS exhibits an epithelial-mesenchymal phenotypic diversity that distinguishes it from other tumors, which cannot be explained by variation in stromal content.
Figure 5. Expression and quantification of EMT across tumor types.
(A) The boxplot display shows differences of EMT scores within five groups of tumors: UCS, endometrioid UCEC, serous UCEC, OV, SARC (ANOVA p value < 0.001). Boxplot center lines represent tumor medians, box limits are the inter-quartile range from 25% and 75%, whiskers represent the extent of tumors out to 1.5 times the inter-quartile range. The dashed horizontal line corresponds to an EMT score of 0. (B) Boxplot display shows differences in protein-based EMT scores within five groups of tumors: UCS, endometrioid UCEC, serous UCEC, OV, SARC (ANOVA p value < 0.001). Components of the figure are as defined in panel (A). E, epithelial; M, mesenchymal; EMT, epithelial-to-mesenchymal transition, UCS, uterine carcinosarcoma; UCEC, uterine corpus endometrial carcinoma; OV, ovarian serous carcinoma; SARC, sarcoma. See also Figure S4.
Multi-platform integration
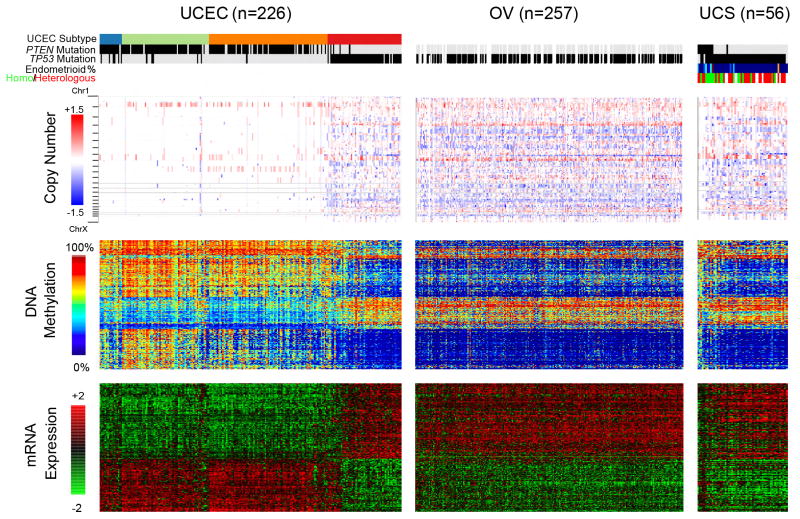
We used the Illumina Infinium HumanMethylation450 (HM450) BeadChips (Illumina, San Diego, CA, USA) to obtain DNA methylation profiles of the 57 UCS samples. An unsupervised cluster analysis of DNA methylation profiles grouped the tumors into three main clades according to their cancer-specific hypermethylation patterns (Figure S5A). Unsupervised consensus clustering of the 2500 most variably expressed genes from RNA sequencing suggested the presence of two expression subtypes (Table S7). One group of tumors from the DNA methylation clustering exhibited a hypermethylation pattern similar to that of endometrioid UCEC, whereas the others were much more similar to the serous subtype (Figure S5B). This finding prompted us to examine molecular characteristics distinguishing endometrioid versus serous histologic subtypes in UCEC across all genomic platforms (Figure 6). Although most UCS tumors resembled the serous/high somatic copy number alteration group in UCEC, a subgroup of UCS tumors appeared to have consistently more endometrioid features, including fewer overall somatic copy number alterations and greater DNA hypermethylation, as well as differential expression exemplified by lower S100A1 and HIF3A and higher TP73 mRNA abundance compared to the serous histologic subtypes (data not shown). This subset of tumors had frequent PTEN mutations, which were almost exclusive to the endometrioid subtype of UCEC, suggesting an independent molecular mechanism underlying the development of this histologic subtype. Tumors with PTEN mutations had frequent amplification of chromosome 8p (p = 0.001, Fisher’s exact test), whereas chromosome 8q was amplified in most tumors. In contrast to the serous-endometrioid dichotomy, the UCS tumors exhibited greater expression of mesenchymal genes than their carcinomatous counterparts (Figure S4A), which was proportional to the microscopically observed percentage of sarcomatous versus carcinomatous content of the tumor (Figure 4), highlighting a parallel molecular divergence between sarcoma and carcinoma in addition to the bifurcation to endometrioid and serous phenotypes. UCS tumors with heterologous differentiation had a higher EMT score than those with homologous differentiation, but did not show any other distinct molecular patterns (Figure 4).
Figure 6. Comparison of TCGA gynecological cancers.
Molecular features distinguishing serous versus endometrioid uterine carcinomas (UCEC) for DNA methylation and mRNA expression are shown for UCEC, OV and UCS samples (columns). In addition, all segments for DNA copy number are displayed, with blue and red indicating deletion and amplification respectively. Within each tumor group, the samples are ranked by increasing similarity to serous UCEC and decreasing similarity to endometrioid UCEC (as measured by a combined rank of difference in each tumor’s distance to the endometrioid UCEC centroid and serous UCEC centroid from four different platforms; see Supplemental Experimental Procedures). Top color bars denote molecular subtype (if UCEC: blue – POLE; green – MSI; orange – copy number low; red – copy number high), PTEN mutation, TP53 mutation (black – mutant; gray – wild-type; white - missing), and percent endometrioid content as estimated from slide review (for UCS: blue to red for 0% to 100%). One UCS sample failed DNA copy number analyses resulting in a total of 56 UCS samples for integrated analyses. See also Figures S5 and S6 and Table S7.
The mismatch repair gene MLH1 was epigenetically silenced in two tumors that were independently identified by PCR as having high microsatellite instability (Figure S5A). These tumors, along with one tumor with a POLE mutation and another with an APOBEC mutation signature, had the highest mutation densities in the cohort (Figure 1A). No BRCA1 promoter methylation or BRCA1 somatic mutations were found in any tumors. This contrasts with serous ovarian tumors, in which 12% have epigenetic silencing and 3% have BRCA1 somatic mutations (Cancer Genome Atlas Research, 2011). We assessed microbes in the UCS cohort using whole exome sequencing and RNA-seq (Figure S6). None of the libraries were positive for infectious agents, either de novo or when we applied our viral integration pipeline.
Protein expression analysis
For 48 UCS samples with sufficient material available, we used reverse phase protein arrays (RPPA) (Hennessy et al., 2010) to generate protein expression data for 200 proteins, including 51 phosphoproteins and four post-translational modifications (Table S6). Unsupervised clustering of the RPPA data showed two clusters containing heterogeneously expressed proteins and lacking consistent expression patterns (Figure S7A). We therefore concluded that the UCS proteomics data did not reveal reliable unsupervised subtypes.
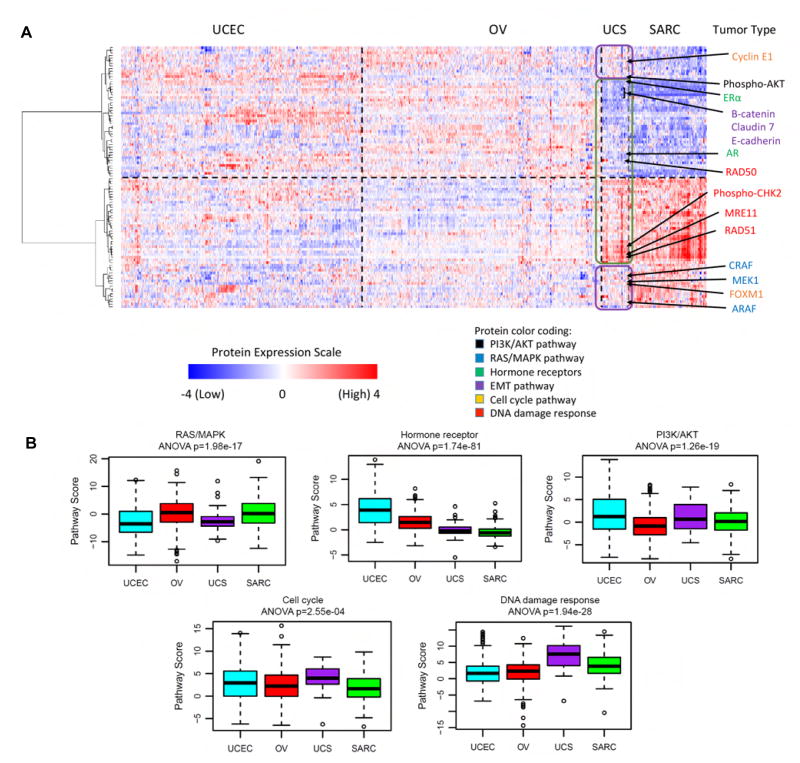
Due to the biphasic nature of UCS and relatively high EMT scores (Byers et al., 2013), we explored whether the proteomic profile of UCS more closely resembled gynecologic carcinomas (with low EMT scores) or sarcomas (with high EMT scores). For gynecologic carcinomas, we used TCGA protein expression data for endometrial (UCEC) and ovarian (OV) carcinomas and compared them against the TCGA sarcoma data (after removing leiomyosarcoma samples, which had very different expression profiles from the other sarcoma subtypes). To distinguish the gynecologic carcinomas from the sarcomas, we identified the top 100 out of 200 proteins that were most differentially expressed between them, independent of the UCS samples. Those 100 proteins were then used to cluster the two groups with the UCS samples included, supervised by tumor type. Pathway analysis was also performed, as described previously (Akbani et al., 2014), to determine which pathways in UCS were similar to the gynecologic carcinomas and which were similar to the sarcomas.
UCS tumors shared some protein expression patterns that were common to the gynecologic carcinomas and other patterns that were similar to the sarcomas without exclusively resembling either group (Figure 7A). For example, similar to endometrial carcinoma, but unlike the sarcomas, UCS tumors had elevated expression of phospho-AKT indicating high activity of the PI3K/AKT pathway and suggesting that UCS may be susceptible to PI3K inhibitor therapy. Also similar to endometrial carcinoma, UCS showed lower activity of the RAS/MAPK pathway compared to the sarcomas, with lower levels of MEK1, CRAF, and ARAF. By contrast and similar to the sarcomas, UCS tumors had surprisingly low levels of the hormone receptors ERα and AR compared to the gynecologic tumors, suggesting that UCS is unlikely to benefit from hormone therapy. In addition, Claudin 7, E-cadherin, and β-catenin levels were also low, indicating high EMT, as described previously. However, UCS showed higher activity of the cell cycle and DNA damage response pathways than either the gynecologic carcinomas or sarcomas, with elevated levels of Cyclin E1 and FOXM1 (cell cycle pathway), and phospho-CHK2, MRE11, RAD50 and RAD51 (DNA damage response pathway), delineating additional future targets for therapy (Figure 7B).
Figure 7. RPPA analysis demonstrating shared epithelial and mesenchymal features of UCS tumors.
(A) Supervised clustering heatmap showing the top 100 differentially expressed proteins between the gynecologic cancers (endometrial [UCEC] and ovarian [OV]) versus sarcomas [SARC] (without the leiomyosarcoma subtype). UCS was then added and the samples were clustered within each tumor type, but not across tumor types. The top half shows proteins that are not highly expressed in sarcomas, whereas the bottom half shows proteins that are highly expressed in sarcomas. The arrows point to proteins of interest color-coded to reflect pathway membership. UCS proteins outlined in purple have similar profiles to the gynecologic cancers and proteins outlined in green have profiles similar to sarcoma. (B) Box plots of pathway activities showing how UCS compares with UCEC, OV and SARC. Sample size of each tumor type is the same as panel (A). Boxplot center lines represent tumor medians, box limits are the inter-quartile range from 25% and 75%, whiskers represent the extent of tumors out to 1.5 times the inter-quartile range, and circles are outliers beyond 1.5 times the inter-quartile range. ANOVA-based p values are shown for each plot, indicating the statistical significance of the differences between the boxes. See also Figure S7 and Table S6.
The UCS samples did not exhibit a dichotomy in which some samples had features of gynecologic carcinomas but others had features of sarcomas; rather, they demonstrated features of both tumor cohorts simultaneously. We performed a similar analysis of the top 2,000 most differentially expressed genes using the mRNA data and those results also show that UCS shares some gene expression profiles with gynecologic carcinomas and others with sarcomas (Figure S7B), indicating good agreement between the observations from the proteomic and transcriptomic platforms. Whether these findings represent intratumoral heterogeneity versus heterogeneous features at the cellular level cannot be determined from these data.
Discussion
This integrated genomic and proteomic analysis of 57 UCSs provides insights into disease biology and offers therapeutic opportunities. Our analysis identified alterations in canonical pathways containing therapeutic targets that are currently in clinical trials. Half of the samples had a mutation in a PI3K pathway gene expected to result in activation of downstream elements. Additionally, more than three-quarters of the cases also had mutation in FBXW7, amplification of CCNE1 or RB1 loss, suggesting deregulated cell cycle function. ARID1A, which encodes a SWI/SNF epigenetic chromatin remodeler that has been implicated in many tumor types (Wu and Roberts, 2013; Wu et al., 2014), was also frequently altered. Taken together, these findings open possibilities for PI3K/AKT pathway, cell cycle and epigenetic therapies in stratified subpopulations of patients with UCS. A number of reports indicate that inactivation of chromatin remodeling genes such as ARID1A and CHD4 results in susceptibility to various targeted therapies including PARP and EZH2 inhibition (Bitler et al., 2015; Pan et al., 2012; Shen et al., 2015). A current clinical trial (NCT02059265) is testing the multi-target tyrosine kinase inhibitor dasatinib in clear cell ovarian and endometrial carcinomas with ARID1A loss (Miller et al., 2016). PI3K inhibition has been successful in advanced endometrial cancers with responses seen in patients with pathway alterations despite some limitations due to tolerability (Makker et al., 2016).
Although there was no clear evidence for traditional molecular subgroups, supervised comparisons with other gynecologic tumors across multiple platforms demonstrated that most UCS tumors share common features with high-grade serous ovarian and serous endometrial tumors. A minority of UCS tumors has molecular features consistent with an endometrioid subtype, including PTEN and KRAS mutations and corresponding gene expression profiles. These data support the theory that some UCS tumors develop from an endometrioid lineage, but most tumors likely de-differentiate from a serous precursor. De-differentiation from an epithelial precursor manifests through a mesenchymal phenotype, which might account for the aggressive nature of this tumor, and is highlighted by higher EMT scores in tumors that have heterologous differentiation. Individually, the UCS tumors have both epithelial and mesenchymal features that reflect their biphasic morphology. In addition to loss of cell adhesion to the basement membrane and apical-basal polarity, increased invasiveness and resistance to apoptosis are likely due to deregulated EMT control (Kalluri and Weinberg, 2009; Radisky, 2005).
The presence of EMT in tumors has been a topic of intense debate (Li and Kang, 2016; Savagner, 2010), although carcinosarcomas are deemed by some as bona fide examples of EMT in vivo (Savagner, 2010). However, it has not been ruled out that transdifferentiation in the other direction exists; namely, it is possible that these tumors start out as sarcoma and then undergo epithelial differentiation (MET instead of EMT). The mutations that we discovered in this study were reminiscent of what was reported in the far more common endometrial carcinomas. In addition, molecular features including DNA methylation, copy number and gene expression recapitulate the endometrial versus serous dichotomy observed in endometrial carcinomas, which was in line with a previous report studying genetic mutation (McConechy et al., 2015). This paper also identified concordant mutation profiles in microdissected carcinoma and sarcomatous components of primary tumors as well as between primary and metastatic lesions. These data all suggest a carcinoma origin and retention of previous ancestral molecular alterations. However, our study was not designed to pinpoint the directionality of transition. A recent sequencing study of microdissected sarcoma and carcinoma components suggests that UCS begins as carcinoma followed by sarcomatous transformation (Zhao et al., 2016). In vivo cell lineage tracing would also help to establish this directionality.
In comparison to prior work where many of the mutations in chromatin-remodeling genes, including CHD4 and ARID1A, were found in hypermutated UCS samples or in carcinosarcoma tumors of ovarian origin (Jones et al., 2014), the present study identifies these mutations in more than one-third of non-hypermutated UCS samples. Despite initial treatment with surgery and/or chemotherapy, many UCS tumors recur, with limited salvage treatment options. The array of targets identified in this study provides compelling opportunities to develop molecularly driven clinical trials to individualize and improve cancer care for primary or recurrent UCSs. Combined with its rarity, the diverse molecular landscape of UCS tumors supports clinical trials in cooperative groups to develop independent treatment paradigms. These approaches have an opportunity to improve the outcome of women with this unique solid tumor.
Experimental procedures
Detailed description of each analysis presented in this study can be found within the Supplemental Experimental Procedures.
Biospecimen collection and clinical data
Biospecimens were collected at diagnosis after obtaining informed consent from patients with uterine carcinosarcoma. Local Institutional Review Board approval was obtained from Cedars-Sinai Medical Center, Duke University, ILSbio, LLC., Mayo Clinic, MD Anderson, Memorial Sloan Kettering Cancer Center, Roswell Park Cancer Institute, University of North Carolina, University of Oklahoma Health Sciences Center, University of Pittsburgh, and Washington University. Patients were selected only if their treatment plan required surgical resection and had received no prior treatment for their disease, including chemotherapy or radiotherapy. Each frozen primary tumor specimen had a companion normal tissue specimen.
Copy number analysis
DNA from each tumor or germline sample was hybridized to Affymetrix SNP 6.0 arrays. For each tumor, genome-wide copy number estimates were refined using tangent normalization, in which tumor signal intensities are divided by signal intensities from the linear combination of all normal samples that are most similar to the tumor. Significant focal copy number alterations were identified from segmented data using GISTIC 2.0. Allelic copy number, regions of homozygous deletions, whole genome doubling and purity and ploidy estimates were calculated using the ABSOLUTE algorithm.
Mutational Analysis
From each sample, 0.5–3 micrograms of DNA were used to prepare the sequencing library through shearing of the DNA followed by ligation of sequencing adaptors. Whole exome capture was performed using Agilent SureSelect Human All Exon protocol according to the manufacturers’ instructions. Basic alignment and sequence QC was done on the “Picard” and “Firehose” pipelines at the Broad Institute. Mutations from each calling center were combined into a single Mutation Annotation Format list (https://wiki.nci.nih.gov/display/TCGA/Mutation+Annotation+Format+%28MAF%29+Specification) and labeled according to the centers that detected each mutation with additional fields counting reference and alternate supporting allele counts from each center.
Each candidate mutation site was assessed in the matched RNA-seq tumor data to identify candidate mutations with independent confirmation from RNA. 35.6% of the candidate mutations occurred at expressed sites in the RNA-seq with sufficient coverage for >90% power to detect the mutation observed in the tumor DNA. The final list candidate mutations required that two or more centers made the call or that the mutation was supported by RNA-seq. Genes with a significant excess of the number of non-synonymous mutations relative to the estimated density of background mutations were identified using the MutSig algorithm (Lawrence et al., 2014; Lawrence et al., 2013).
Array-based DNA methylation assay
We used the Illumina Infinium HumanMethylation450 (HM450) BeadChips (Illumina, San Diego, CA) to obtain DNA methylation profiles of 57 uterine carcinosarcoma (UCS) samples. We performed bisulfite conversion on 1 μg of genomic DNA from each sample using the EZ-96 DNA Methylation Kit (Zymo Research, Irvine, CA) according to the manufacturer’s instructions. Bisulfite-converted DNA was whole genome amplified (WGA) and enzymatically fragmented prior to hybridization to the arrays.
RNA sequencing
One μg of total RNA was converted to mRNA libraries using the lllumina mRNA TruSeq kit (RS-122-2001 or RS-122-2002) following the manufacturer’s directions. Libraries were sequenced 48×7×48bp on the Illumina HiSeq 2000. FASTQ files were generated by CASAVA. RNA reads were aligned to the hg19 genome assembly. Gene expression measurements were obtained by replacing all RSEM values identically equal to zero with the smallest non-zero RSEM value, applying a log2 transformation, and median centering by gene. Using the median absolute deviation, the 2500 most variably expressed genes were identified. Consensus clustering was then performed.
miRNA Sequencing
We generated microRNA sequence (miRNA-seq) data for 57 tumor samples using 1 μg of total RNA (at 250 ng/μL) as input. We aligned reads to the GRCh37/hg19 reference human genome, and annotated miRNA read count abundance with miRBase v16. We used miRBase v20 to assign 5p and 3p mature strand (miR) names to MIMAT accession IDs. We identified groups of samples that had similar abundance profiles using unsupervised non-negative matrix factorization (NMF) consensus clustering.
Reverse-Phase protein array
Protein was extracted using RPPA lysis buffer from human tumors and RPPA was performed. Tumor lysates were manually serial diluted in two-fold of 5 dilutions with lysis buffer. Slides were probed with 200 validated primary antibodies followed by corresponding secondary antibodies. We performed median centering across all the antibodies for each sample to correct for sample loading differences.
Data Access
Data used to generate the analyses and all of the primary sequence files are deposited in the NCI’s Genomic Data Commons (https://gdc.cancer.gov/). The data can be explored via the cBio Cancer Genomics Portal (http://cbioportal.org).
Supplementary Material
Significance.
UCSs are fascinating tumors that contain morphologic components of both epithelial and mesenchymal cell types. They are thought to arise from a metaplastic conversion due to a strong EMT. Here we report an integrated genomic, epigenomic, transcriptomic and proteomic analysis of UCSs. We identify multiple somatic mutations and copy number alterations that offer expanded therapeutic options including potential use of PARP, EZH2, cell cycle and PI3K pathway inhibitors. We also dissect the interaction between the genomic and epigenomic regulation of EMT in UCSs highlighting the epigenetic alterations at microRNA promoters. The EMT features recognized in UCSs provide a mechanistic basis for the conversion of serous-like endometrial carcinoma precursors in a majority of cases.
Highlights.
Uterine carcinosarcomas (UCS) have frequent TP53 mutations
UCS demonstrate a strong and varied degree of epithelial-mesenchymal transition
MicroRNA expression in UCS is under epigenetic control
Multiple alterations are present in UCS in genes that are therapeutic targets
Acknowledgments
We wish to thank all patients and families who contributed to this study. This work was supported by the following grants from the US National Institutes of Health: U54 HG003273, U54 HG003067, U54 HG003079, U24 CA143799, U24 CA143835, U24 CA143840, U24 CA143843, U24 CA143845, U24 CA143848, U24 CA143858, U24 CA143866, U24 CA143867, U24 CA143882, U24 CA143883, U24 CA144025, and P30 CA016672. Andrew D. Cherniack and Matthew Meyerson declare research funding from Bayer AG. The remaining authors declare no competing financial interests.
Footnotes
Author Contributions TCGA Research Network contributed collectively to this study. The National Cancer Institute and National Human Genome Research Institute project teams coordinated project activities. R.A. and D.A.L. both serve as corresponding authors as each made are responsible for all data, figures and text; have ensured that authorship is appropriately granted to contributors and worked together as a team to lead this TCGA project. D.A.L. serves as the lead contact. The contributions of other authors are as follows: Conceptualization, D.A.L; Methodology, A.C., H.S., V.W., C.S., R.B., R.A.S, R.R.B., R.E.Z., G.R., P.W.L., G.B.M., J.N.W., R.A.; Software, B.M., X.H., S.L., R.A.; Validation, V.W., C.S.; Formal Analysis, V.W., D.A.L., R.A., R.B., H.S.; Data Curation, A.C., H.S., V.W., C.S., B.M., R.B., X.H., S.L., R.A.;Writing – Original Draft, A.C., H.S., V.W., C.S., R.A., D.A.L.; Writing – Review & Editing, R.B., H.S., G.R., R.K., G.B.M., J.N.W., R.A., D.A.L.; Visualization, A.C., H.S., V.W., C.S., B.M., R.B., R.A.; Supervision, A.C., H.S., G.R., R.K., P.W.L., G.B.M., J.N.W., J.Z., R.A., D.A.L., Project Administration, J.Z., R.A., D.A.L.
Consortium
The members of The Cancer Genome Atlas Research Network Uterine Carcinosarcoma Working Group are: Rehan Akbani, Adrian Ally, J. Todd Auman, Miruna Balasundaram, Saianand Balu, Stephen B. Baylin, Rameen Beroukhim, Tom Bodenheimer, Faina Bogomolniy, Lori Boice, Moiz S. Bootwalla, Jay Bowen, Reanne Bowlby, Russell Broaddus, Denise Brooks, Rebecca Carlsen, Andrew D. Cherniack, Juok Cho, Eric Chuah, Sudha Chudamani, Kristian Cibulskis, Melissa Cline, Fanny Dao, Mutch David, John A. Demchok, Noreen Dhalla, Sean Dowdy, Ina Felau, Martin L. Ferguson, Scott Frazer, Jessica Frick, Stacey Gabriel, Julie M. Gastier-Foster, Nils Gehlenborg, Mark Gerken, Gad Getz, Manaswi Gupta, David Haussler, D. Neil Hayes, David I. Heiman, Julian Hess, Katherine A. Hoadley, Robert Hoffmann, Robert A. Holt, Alan P. Hoyle, Xin Hu, Mei Huang, Carolyn M. Hutter, Stuart R. Jefferys, Steven J.M. Jones, Corbin D. Jones, Rupa S. Kanchi, Cyriac Kandoth, Katayoon Kasaian, Sarah Kerr, Jaegil Kim, Phillip H. Lai, Peter W. Laird, Eric Lander, Michael S. Lawrence, Darlene Lee, Kristen M. Leraas, Ignaty Leshchiner, Douglas A. Levine, Tara M. Lichtenberg, Pei Lin, Shiyun Ling, Jia Liu, Wenbin Liu, Yuexin Liu, Laxmi Lolla, Yiling Lu, Yussanne Ma, Dennis T. Maglinte, Marco A. Marra, Michael Mayo, Shaowu Meng, Matthew Meyerson, Piotr A. Mieczkowski, Gordon B. Mills, Richard A. Moore, Lisle E. Mose, Andrew J. Mungall, Karen Mungall, Bradley A. Murray, Rashi Naresh, Michael S. Noble, Narciso Olvera, Joel S. Parker, Charles M. Perou, Amy H. Perou, Todd Pihl, Amie J. Radenbaugh, Nilsa C. Ramirez, W. Kimryn Rathmell, Jeffrey Roach, A. Gordon Robertson, Sara Sadeghi, Gordon Saksena, Helga B Salvesen, Jacqueline E. Schein, Steven E. Schumacher, Hui Shen, Margi Sheth, Yan Shi, Juliann Shih, Janae V. Simons, Payal Sipahimalani, Tara Skelly, Heidi J. Sofia, Matthew G. Soloway, Robert A. Soslow, Carrie Sougnez, Chip Stewart, Charlie Sun, Angela Tam, Donghui Tan, Roy Tarnuzzer, Nina Thiessen, Leigh B. Thorne, Kane Tse, Jill Tseng, David J. Van Den Berg, Umadevi Veluvolu, Roel G.W. Verhaak, Doug Voet, Amanda von Bismarck, Vonn Walter, Yunhu Wan, Zhining Wang, Chen Wang, John N. Weinstein, Daniel J. Weisenberger, Matthew D. Wilkerson, Boris Winterhoff, Lisa Wise, Tina Wong, Ye Wu, Liming Yang, Jean C. Zenklusen, Jiashan (Julia) Zhang, Hailei Zhang, Wei Zhang, Jing-chun Zhu, Erik Zmuda, Rosemary E. Zuna
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abeln EC, Smit VT, Wessels JW, de Leeuw WJ, Cornelisse CJ, Fleuren GJ. Molecular genetic evidence for the conversion hypothesis of the origin of malignant mixed mullerian tumours. The Journal of pathology. 1997;183:424–431. doi: 10.1002/(SICI)1096-9896(199712)183:4<424::AID-PATH949>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Akbani R, Ng PK, Werner HM, Shahmoradgoli M, Zhang F, Ju Z, Liu W, Yang JY, Yoshihara K, Li J, et al. A pan-cancer proteomic perspective on The Cancer Genome Atlas. Nature communications. 2014;5:3887. doi: 10.1038/ncomms4887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger AH, Imielinski M, Duke F, Wala J, Kaplan N, Shi GX, Andres DA, Meyerson M. Oncogenic RIT1 mutations in lung adenocarcinoma. Oncogene. 2014;33:4418–4423. doi: 10.1038/onc.2013.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitler BG, Aird KM, Garipov A, Li H, Amatangelo M, Kossenkov AV, Schultz DC, Liu Q, Shih Ie M, Conejo-Garcia JR, et al. Synthetic lethality by targeting EZH2 methyltransferase activity in ARID1A-mutated cancers. Nat Med. 2015;21:231–238. doi: 10.1038/nm.3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bland AE, Stone R, Heuser C, Shu J, Jazaeri A, Shutter J, Atkins K, Rice L. A clinical and biological comparison between malignant mixed mullerian tumors and grade 3 endometrioid endometrial carcinomas. International journal of gynecological cancer: official journal of the International Gynecological Cancer Society. 2009;19:261–265. doi: 10.1111/IGC.0b013e31819a1fa5. [DOI] [PubMed] [Google Scholar]
- Brinton LA, Felix AS, McMeekin DS, Creasman WT, Sherman ME, Mutch D, Cohn DE, Walker JL, Moore RG, Downs LS, et al. Etiologic heterogeneity in endometrial cancer: evidence from a Gynecologic Oncology Group trial. Gynecol Oncol. 2013;129:277–284. doi: 10.1016/j.ygyno.2013.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers LA, Diao L, Wang J, Saintigny P, Girard L, Peyton M, Shen L, Fan Y, Giri U, Tumula PK, et al. An epithelial-mesenchymal transition gene signature predicts resistance to EGFR and PI3K inhibitors and identifies Axl as a therapeutic target for overcoming EGFR inhibitor resistance. Clinical cancer research: an official journal of the American Association for Cancer Research. 2013;19:279–290. doi: 10.1158/1078-0432.CCR-12-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research N. Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research N. Kandoth C, Schultz N, Cherniack AD, Akbani R, Liu Y, Shen H, Robertson AG, Pashtan I, Shen R, et al. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497:67–73. doi: 10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter SL, Cibulskis K, Helman E, McKenna A, Shen H, Zack T, Laird PW, Onofrio RC, Winckler W, Weir BA, et al. Absolute quantification of somatic DNA alterations in human cancer. Nature biotechnology. 2012;30:413–421. doi: 10.1038/nbt.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiyoda T, Tsuda H, Tanaka H, Kataoka F, Nomura H, Nishimura S, Takano M, Susumu N, Saya H, Aoki D. Expression profiles of carcinosarcoma of the uterine corpus-are these similar to carcinoma or sarcoma? Genes, chromosomes & cancer. 2012;51:229–239. doi: 10.1002/gcc.20947. [DOI] [PubMed] [Google Scholar]
- Cortez MA, Valdecanas D, Zhang X, Zhan Y, Bhardwaj V, Calin GA, Komaki R, Giri DK, Quini CC, Wolfe T, et al. Therapeutic delivery of miR-200c enhances radiosensitivity in lung cancer. Molecular therapy: the journal of the American Society of Gene Therapy. 2014;22:1494–1503. doi: 10.1038/mt.2014.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson SE, Tornos C, Hummer A, Barakat RR, Soslow RA. Prognostic features of surgical stage I uterine carcinosarcoma. The American journal of surgical pathology. 2007;31:1653–1661. doi: 10.1097/PAS.0b013e3181161ba3. [DOI] [PubMed] [Google Scholar]
- Fujii H, Yoshida M, Gong ZX, Matsumoto T, Hamano Y, Fukunaga M, Hruban RH, Gabrielson E, Shirai T. Frequent genetic heterogeneity in the clonal evolution of gynecological carcinosarcoma and its influence on phenotypic diversity. Cancer research. 2000;60:114–120. [PubMed] [Google Scholar]
- Hennessy BT, Lu Y, Gonzalez-Angulo AM, Carey MS, Myhre S, Ju Z, Davies MA, Liu W, Coombes K, Meric-Bernstam F, et al. A Technical Assessment of the Utility of Reverse Phase Protein Arrays for the Study of the Functional Proteome in Non-microdissected Human Breast Cancers. Clinical proteomics. 2010;6:129–151. doi: 10.1007/s12014-010-9055-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S, Stransky N, McCord CL, Cerami E, Lagowski J, Kelly D, Angiuoli SV, Sausen M, Kann L, Shukla M, et al. Genomic analyses of gynaecologic carcinosarcomas reveal frequent mutations in chromatin remodelling genes. Nature communications. 2014;5:5006. doi: 10.1038/ncomms6006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. The Journal of clinical investigation. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kounelis S, Jones MW, Papadaki H, Bakker A, Swalsky P, Finkelstein SD. Carcinosarcomas (malignant mixed mullerian tumors) of the female genital tract: comparative molecular analysis of epithelial and mesenchymal components. Human pathology. 1998;29:82–87. doi: 10.1016/s0046-8177(98)90394-x. [DOI] [PubMed] [Google Scholar]
- Kurman RJ International Agency for Research on Cancer., and World Health Organization. WHO classification of tumours of female reproductive organs. 4. Lyon: International Agency for Research on Cancer; 2014. [Google Scholar]
- Lawrence MS, Stojanov P, Mermel CH, Robinson JT, Garraway LA, Golub TR, Meyerson M, Gabriel SB, Lander ES, Getz G. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, Carter SL, Stewart C, Mermel CH, Roberts SA, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499:214–218. doi: 10.1038/nature12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Kang Y. Probing the Fifty Shades of EMT in Metastasis. Trends Cancer. 2016;2:65–67. doi: 10.1016/j.trecan.2016.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Garcia MA, Palacios J. Pathologic and molecular features of uterine carcinosarcomas. Seminars in diagnostic pathology. 2010;27:274–286. doi: 10.1053/j.semdp.2010.09.005. [DOI] [PubMed] [Google Scholar]
- Makker V, Recio FO, Ma L, Matulonis UA, Lauchle JO, Parmar H, Gilbert HN, Ware JA, Zhu R, Lu S, et al. A multicenter, single-arm, open-label, phase 2 study of apitolisib (GDC-0980) for the treatment of recurrent or persistent endometrial carcinoma (MAGGIE study) Cancer. 2016 doi: 10.1002/cncr.30286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCluggage WG. Malignant biphasic uterine tumours: carcinosarcomas or metaplastic carcinomas? Journal of clinical pathology. 2002;55:321–325. doi: 10.1136/jcp.55.5.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConechy MK, Ding J, Cheang MC, Wiegand KC, Senz J, Tone AA, Yang W, Prentice LM, Tse K, Zeng T, et al. Use of mutation profiles to refine the classification of endometrial carcinomas. The Journal of pathology. 2012;228:20–30. doi: 10.1002/path.4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConechy MK, Hoang LN, Chui MH, Senz J, Yang W, Rozenberg N, Mackenzie R, McAlpine JN, Huntsman DG, Clarke BA, et al. In-depth molecular profiling of the biphasic components of uterine carcinosarcomas. J Pathol Clin Res. 2015;1:173–185. doi: 10.1002/cjp2.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12:R41. doi: 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller RE, Brough R, Bajrami I, Williamson CT, McDade S, Campbell J, Kigozi A, Rafiq R, Pemberton H, Natrajan R, et al. Synthetic Lethal Targeting of ARID1A-Mutant Ovarian Clear Cell Tumors with Dasatinib. Mol Cancer Ther. 2016;15:1472–1484. doi: 10.1158/1535-7163.MCT-15-0554. [DOI] [PubMed] [Google Scholar]
- Pan MR, Hsieh HJ, Dai H, Hung WC, Li K, Peng G, Lin SY. Chromodomain helicase DNA-binding protein 4 (CHD4) regulates homologous recombination DNA repair, and its deficiency sensitizes cells to poly(ADP-ribose) polymerase (PARP) inhibitor treatment. J Biol Chem. 2012;287:6764–6772. doi: 10.1074/jbc.M111.287037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pothuri B, Ramondetta L, Eifel P, Deavers MT, Wilton A, Alektiar K, Barakat R, Soslow RA. Radiation-associated endometrial cancers are prognostically unfavorable tumors: a clinicopathologic comparison with 527 sporadic endometrial cancers. Gynecol Oncol. 2006;103:948–951. doi: 10.1016/j.ygyno.2006.05.039. [DOI] [PubMed] [Google Scholar]
- Radisky DC. Epithelial-mesenchymal transition. Journal of cell science. 2005;118:4325–4326. doi: 10.1242/jcs.02552. [DOI] [PubMed] [Google Scholar]
- Sarrio D, Rodriguez-Pinilla SM, Hardisson D, Cano A, Moreno-Bueno G, Palacios J. Epithelial-mesenchymal transition in breast cancer relates to the basal-like phenotype. Cancer research. 2008;68:989–997. doi: 10.1158/0008-5472.CAN-07-2017. [DOI] [PubMed] [Google Scholar]
- Savagner P. The epithelial-mesenchymal transition (EMT) phenomenon. Ann Oncol. 2010;21(Suppl 7):vii89–92. doi: 10.1093/annonc/mdq292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J, Peng Y, Wei L, Zhang W, Yang L, Lan L, Kapoor P, Ju Z, Mo Q, Shih Ie M, et al. ARID1A Deficiency Impairs the DNA Damage Checkpoint nd Sensitizes Cells to PARP Inhibitors. Cancer discovery. 2015;5:752–767. doi: 10.1158/2159-8290.CD-14-0849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson L, Chang B, Barsky SH. Monoclonal origins of malignant mixed tumors (carcinosarcomas). Evidence for a divergent histogenesis. The American journal of surgical pathology. 1996;20:277–285. doi: 10.1097/00000478-199603000-00003. [DOI] [PubMed] [Google Scholar]
- Van Allen EM, Wagle N, Sucker A, Treacy DJ, Johannessen CM, Goetz EM, Place CS, Taylor-Weiner A, Whittaker S, Kryukov GV, et al. The genetic landscape of clinical resistance to RAF inhibition in metastatic melanoma. Cancer discovery. 2014;4:94–109. doi: 10.1158/2159-8290.CD-13-0617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada H, Enomoto T, Fujita M, Yoshino K, Nakashima R, Kurachi H, Haba T, Wakasa K, Shroyer KR, Tsujimoto M, et al. Molecular evidence that most but not all carcinosarcomas of the uterus are combination tumors. Cancer research. 1997;57:5379–5385. [PubMed] [Google Scholar]
- Wu JN, Roberts CW. ARID1A mutations in cancer: another epigenetic tumor suppressor? Cancer discovery. 2013;3:35–43. doi: 10.1158/2159-8290.CD-12-0361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu RC, Wang TL, Shih Ie M. The emerging roles of ARID1A in tumor suppression. Cancer Biol Ther. 2014;15:655–664. doi: 10.4161/cbt.28411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada SD, Burger RA, Brewster WR, Anton D, Kohler MF, Monk BJ. Pathologic variables and adjuvant therapy as predictors of recurrence and survival for patients with surgically evaluated carcinosarcoma of the uterus. Cancer. 2000;88:2782–2786. doi: 10.1002/1097-0142(20000615)88:12<2782::aid-cncr17>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- Zack TI, Schumacher SE, Carter SL, Cherniack AD, Saksena G, Tabak B, Lawrence MS, Zhang CZ, Wala J, Mermel CH, et al. Pan-cancer patterns of somatic copy number alteration. Nature genetics. 2013;45:1134–1140. doi: 10.1038/ng.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Bellone S, Lopez S, Thakral D, Schwab C, English DP, Black J, Cocco E, Choi J, Zammataro L, et al. Mutational landscape of uterine and ovarian carcinosarcomas implicates histone genes in epithelial-mesenchymal transition. Proc Natl Acad Sci U S A. 2016;113:12238–12243. doi: 10.1073/pnas.1614120113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.