Abstract
Circadian rhythm misalignment has been increasingly recognized to pose health risk for a wide range of diseases, particularly metabolic disorders. The liver maintains metabolic homeostasis and expresses many circadian genes, such as differentiated embryo chondrocyte-1 (DEC1) and small heterodimer partner (SHP). DEC1 is established to repress transcription through E-box elements, and SHP belongs to the superfamily of nuclear receptors and has multiple E-box elements in its promoter. Importantly, DEC1 and SHP are inversely oscillated. This study was performed to test the hypothesis that the SHP gene is a target gene of DEC1. Cotransfection demonstrated that DEC1 repressed the SHP promoter and attenuated the transactivation of the classic circadian activator complex of Clock/Bmal1. Site-directed mutagenesis, electrophoretic mobility shift assay and chromatin immunoprecipitation established that the repression was achieved through the E-box in the proximal promoter. Transfection of DEC1 suppressed the expression of SHP. In circadian-inducing cells, the epileptic agent valproate inversely altered the expression of DEC1 and SHP. Both DEC1 and SHP are involved in energy balance and valproate is known to induce hepatic steatosis. Our findings collectively establish that DEC1 participates in the negative loop of SHP oscillating expression with potential implications in metabolic homeostasis.
Keywords: DEC1, BHLHE40, SHP, NR0B2, Circadian rhythm
INTRODUCTION
Circadian rhythms are physiological changes that follow a 24-h cycle [1]. These changes ensure physiological processes to be coordinated with daily fluctuations of the environment [1–3]. Circadian rhythm misalignment has been increasingly recognized to pose health risk for a wide range of diseases including obesity, diabetes, cardiovascular diseases and stroke [4–7]. Mammals have central and peripheral circadian clocks [8]. The central clock is located in the suprachiasmatic nucleus (SCN) of the hypothalamus and is entrained by such environmental cues as the light-dark cycle [9, 10]. Through the retinohypothalamic tract, the SCN receives photic input signals and generates rhythms, which subsequently synchronize multiple peripheral clocks through neural and humoral signaling [9–12]. While each organ has its own clock, the liver clock is the most studied peripheral clocks, particularly related to food cues [13–16].
Circadian rhythms are controlled by a group of core clock genes such as Clock (circadian locomotor output cycles kaput) and Bmal1 (brain and muscle ARNT-like 1) [8, 17]. Clock and Bmal1 form a heterodimer and transactivate through E-Box elements the Per (Period-1 and 2) and Cry genes (Cryptochrome-1 and 2). Per and Cry proteins in turn interact with Clock/Bmal1 and attenuate their own transactivation [5, 8, 17]. In addition to Clock/Bmal1, we and other investigators have demonstrated that DEC transcription factors, differentiated embryo chondrocyte-1 and 2, are strong E-box binding proteins [18–20]. However, binding to E-box by DEC transcription factors, in contrast to the Clock/Bmal1 complex, leads to efficacious repression of the target genes including Per1 [8, 17, 19]. DEC1 and DEC2 are both central and peripheral oscillators [15]. In particular, DEC1 has been recognized to play fundamental roles in orchestrating metabolism and resetting the liver clock. Indeed, DEC1 is one of the most sensitive genes in responding to food cues. A 30 min feeding significantly induces the hepatic expression of DEC1 [15].
In addition to DEC1, there are several other well-characterized hepatic circadian genes such as the gene encoding small heterodimer partner gene (SHP) [5, 21, 22]. SHP has been established as a major regulator of diverse metabolic pathways, particularly in bile acid synthesis, lipid metabolism, glucose homeostasis and liver fibrogenesis [23]. Structurally, SHP belongs to the superfamily of nuclear receptors [23, 24]. However, it lacks the DNA binding domain, thus is referred to as an atypical nuclear receptor. SHP has been shown to interact with nuclear receptors and/or compete for co-factors (e.g., co-activators or co-repressor), delivering potent regulatory activities at the transcriptional level [23]. On the other hand, many nuclear receptors have been shown to support the induction of SHP in response to endobiotics such as bile acids [23]. In addition to nuclear receptors, the circadian complex of Clock/Bmal1 is a potent transactivator of the SHP gene [23]. The transactivation is achieved through the element CACGTG, a special type of E-box recognized by DEC transcription factors [19, 23]. In mice, the expression of DEC1 and SHP is inversely oscillating [25, 26].
The present study was performed to test the hypothesis that the SHP gene is a target of DEC1. Cotransfection experiments showed that DEC1 efficaciously repressed the SHP promoter and attenuated the transactivation of SHP by the Clock/Bmal1 complex. A set of molecular experiments including chromatin immunoprecipitation established that the repression was achieved through the E-box in the proximal promoter. Overexpression of DEC1 led to suppressed expression of SHP, and valproate inversely altered the oscillating expression of DEC1 and SHP. These findings collectively establish that DEC1 participates in the negative loop of the SHP oscillating expression and that the DEC1-SHP pathway is likely involved in metabolic homeostasis.
MATERIALS AND METHODS
Plasmid
Expression constructs of Bmal1 and Clock were gifts of Dr. Marina P Antoch of the Cleveland Clinic [27]. The Per1 promoter reporter (Per1-luc) was a gift of Dr. Joseph S. Takahashi of Northwestern University [28]. The SHP-2.2 Luc reporter was a gift of Dr. Hueng-Sik Choi of Keimyung University [29]. The 5’ deletion mutants of the SHP reporter were prepared by inserting a Nhe I-Hind III fragment into the pGL3 basic vector. These fragments were generated by PCR with primers as described in Table I. The SHP mutant reporter with a disrupted E-box (E1) was prepared with the same approach but the mutations were introduced in the forward primer (Table I). The DEC1 expression construct and its mutants (deletion or substitution) were described elsewhere [18, 30]. All constructs were subjected to sequencing analysis.
Table I.
Sequences of oligonucleotides
| Oligonucleotide | Sequence |
|---|---|
| Native promoter reporters [numbered according to Kim et al., J. Biol. Chem. (2004), 279:28122-31] | |
| SHP-572 NheI | 5’-TCCTAGACTGGACAGTGGG-3’ |
| SHP-240 NheI | 5’-GTGAGCGGCAGGTGGCCCT-3’ |
| SHP-190 NheI | 5’-GTGATATCAGTGCCACGT-3’ |
| SHP-117 NheI | 5’-TGTCTGTGTGTTTTTTTCA-3’ |
| SHP-190m NheI | 5’-GTGATATCAGTGCAACGGGGGGTTCCCAATGCC-3’ |
| SHP+30 HindIII | 5’-GGTTAGGGATCTGCTCTC-3’ |
| EMSA | |
| SHP-187/162 (E1) | 5’-ATATCAGTGCCACGTGGGGTTCCCAA-3’ |
| SHP-242/217 (E2) | 5’-GAGTGAGCGGCAGGTGGCCCTGTGCC-3’ |
| SHP-280/255 (E4/5) | 5’- CTTGTTTATCCACTTGAGTCATCTGA-3’ |
| SHP-332/307 (E6) | 5’- GCTGATTGTGCACCTGGGGCCTTGGT-3’ |
| SHP-347/322 (E7) | 5’- CCAATGGGGACACCTGCTGATTGTGC-3’ |
| SHP-187/162m (E1) | 5’- ATATCAGTGCAACGGGGGGTTCCCAA-3’ |
| DEC2 E-box | 5’-TACGTTCCGCACGTGAGCTGGGTG-3’ |
| ChIP | |
| E1 element sense | 5’-GGCCCTGTGCCCTGCACCGGC-3’ |
| E1 element antisense | 5’-CTCATGGTTAGGGATCTGCTC-3’ |
| Other E-box element sense | 5’-CCTAGTCTTTTGTGCACACAA-3’ |
| Other E-box element antisense | 5’-TCACCTCAGTCAATGAAGTGG-3’ |
| Non-element sense | 5’-CAGAAATTCTTGTCACTGTTT-3’ |
| Non-element antisense | 5’-CACACCTCTTTCATTTGATTA-3’ |
Reporter activity and cotransfection
Cotransfection was performed, essentially as described previously [31, 32]. Unless otherwise specified, DEC1 stably transfected cells (293T) were plated in 24-well plates in DMEM media supplemented with 10% fetal bovine serum at a density of 1.6 × 105 cells per well. The cells were cultured in the absence or presence of tetracycline at various concentrations. The cells were then transfected by lipofection with Lipofectamine and Plus Reagents (Thermo Fisher Scientific, Waltham, MA). Generally, the transfection mixtures contained a reporter plasmid (50 ng) and the pRL-null Renilla plasmid (5 ng) unless otherwise specified. In some cases, vector plasmid was used to equalize the amount of plasmid DNA for each transfection. The transfected cells were cultured for additional 24 h, washed with PBS and resuspended in passive lysis buffer (Promega, Madison, WI). The lysed cells were subjected to 2 cycles of freezing/thawing. The reporter enzyme activities were assayed with a Dual-Luciferase Reporter Assay System (Promega). This system contained two substrates, which were used to determine the activity of two luciferases sequentially. The firefly luciferase activity, which represented the reporter gene activity, was initiated by mixing an aliquot of lysates (20 µl) with Luciferase Assay Reagent II. Then the firefly luminescence was quenched and the Renilla luminescence was simultaneously activated by adding Stop & Glo Reagent to the sample wells. The firefly luminescence signal was normalized based on the Renilla luminescence signal.
Electrophoretic mobility shift assay (EMSA)
DEC1 stably transfected cells with a tetracycline-inducible construct [33] were cultured in the presence or absence of tetracycline (0.1 µg/mL) for 24 h. Cells were harvested and nuclear extracts were prepared with a nuclear extraction kit (Active Motif, Carlsbad, CA). The EMSA experiment was performed with Lightshift Chemiluminescent EMSA Kit (Thermo Fisher Scientific, Waltham, MA) as described previously [34]. The sense strand for SHP-187/162 (E1) was synthesized as labeled or non-labeled form (for competition). Nuclear protein (5 µg) was mixed with binding buffer and then incubated with a double-stranded biotinylated probe (0.1 pmol) on ice for 20 min. In competition assays, nuclear extracts were first incubated with an unlabeled probe at a 50× excess for 30 min before addition of the labeled probe. For antibody-disruption assay, the nuclear extracts were first incubated with anti-DEC1 antibody on ice for 20 min and then with the labeled probe. As a positive control, the EMSA experiment was performed with a DEC2 E-box containing probe [18]. The protein-DNA complexes were resolved by non-denaturing polyacrylamide gel electrophoresis (6%) and transferred onto a Biodyne® nylon membrane. The biotinylated probe was detected with Chemiluminescent Nucleic Acid Detection Module Kit (Thermo Fisher Scientific, Waltham, MA). The chemiluminescent signal was captured by myECL Imager (Thermo Fisher Scientific, Waltham, MA).
Chromatin immunoprecipitation (ChIP)
ChIP experiment was performed with Active Motif ChIP-IT Express kit, essentially described previously [31, 34]. HepG2 Cells were transfected with DEC1 (Flag-tagged) by TransfeX (ATCC, Manassas, VA) for 24 h, washed and underwent cross-linking for 15 min by 1.0% formaldehyde at room temperature, and the cross-linking was terminated with glycine (final concentration of 125 mM). The soluble chromatins were prepared as described previously [31, 34]. Alternatively, DEC1 stably transfected cells were cultured in the presence of tetracycline (0.1 µg/mL) for 24 h and then processed as described for HepG2 cells. For ChIP experiment, chromatins were pre-cleared for 2 h at 4°C with protein G beads pre-treated with herring sperm DNA (0.2 mg/ml) and BSA (0.5 mg/ml). A fraction of the pre-cleared chromatins was stored at −80°C for later use as an input. The pre-cleared chromatins were incubated with anti-Flag antibody (HepG2) or anti-DEC1 (stably transfected cells) for overnight at 4°C. As a negative control, incubation was performed with pre-immune IgG. The antibody-bound chromatins and DNA input as well as IgG control chromatins were analyzed by PCR for the presence of the genomic fragments containing the E-box of interest, other E-boxes (not repressed by DEC1) with primers shown in Table I. The PCR was performed with Platinum Taq DNA polymerase for a total of 32 cycles at 94°C for 30 s, 58°C for 30 s and 68°C for 60 s. A 3-min initial denaturation was performed.
Suppressed expression of SHP by DEC1
HepG2 cells were seeded in 8-well chamber slides (2×104 cells/ well and cultured for 24 h. Cells were then transfected with Flag-DEC1 (500 ng) or the vector by TransfeX reagent. The transfected cells were cultured for another 24 h and subsequently washed with ice cold PBS and then fixed with 4% Paraformaldehyde for 10 min at pH 7.4. Cells were washed 3 times with PBS and permeabilization solution (0.1% Triton X-100) was added for 10 min. Chamber slides were incubated with 1% BSA (2 mg/mL) for 1 h to block nonspecific binding. The slides were incubated overnight with anti-SHP antibody (H-160, Santa Cruz Biotechnology, Dallas, TX) or anti-Flag antibody (M2, Sigma-Aldrich, St. Louis, MO) at a dilution of 1:200. The anti-SHP antibody was located by Alexa Fluor® 488 conjugated goat anti-rabbit IgG (green), whereas the anti-Flag antibody with Alexa Fluor® 555 conjugated goat anti-mouse IgG (red). Both secondary antibodies were purchased from Life Technologies (Carlsbad, CA). The slides were then mounted with ProLong Gold Antifade Mountant (Thermo Fisher). The mounting media contained 4',6-diamidino-2-phenylindole (DAPI) for staining nuclei (Blue). Cells were then imaged using confocal microscope. To provide more quantitative information, same experiments were performed in 6-well plates and the expression of SHP and DEC1 was determined by Western blotting.
Regulated expression of DEC1 and SHP by valproate in serum-shocked circadian induction
HepG2 cells were seeded in 6- or 24-well plates at a density of 6×105 or 1.5×105 cells/well in low glucose DMEM with 10% delipided FBS. When the confluency reached 75%, cells were shocked with media containing 50% horse serum for 2 h. Thereafter, the shocked cells were cultured in normal media or the same media containing valproate at 2 mM. Cells were harvested at 6 h interval. The expression of DEC1 and SHP was determined by RT-qPCR with Taqman probes. The TaqMan assay identification numbers were: DEC1: Hs00186419_m1; SHP: Hs00222677_m1; GAPDH, 4352934E; and RNA polymerase II, Hs00172187_m1.
Other analyses
The anti-DEC1 antibody against a peptide derived from the C-terminus was described elsewhere [33]. Protein concentration was determined with BCA assay (Pierce) with bovine serum albumin as the standard. Data are presented as mean ± SD of at least four separate experiments, except where results of blots are shown in which case a representative experiment is depicted in the figures. Statistical significance between 2 means with multiple groups was made according to two-way ANOVA followed by a DUNCAN’s multiple comparison test at P < 0.05 unless otherwise specified. Student t test was used for pairwise comparison with normally distributed data.
RESULTS
Repression of the SHP promoter by DEC1
DEC1 and SHP are established to play critical roles in a wide range of biological activities including metabolic homeostasis [23, 35, 36]. Both DEC1 and SHP are circadian genes and their expression is inversely oscillated [25, 26]. We have shown that DEC1 is a sequence-specific transcription factor that acts on Sp1 site as well as a specific type of E-box: CACAGT [30, 37]. The SHP promoter and its immediate upstream sequence contain multiple E-boxes including a CACATG. We therefore hypothesized that DEC1 transcriptionally regulates the expression of SHP. To test this hypothesis, we first examined whether DEC1 represses the SHP promoter. Specifically, DEC1 stably transfected cells were cultured in the absence or presence of tetracycline to modulate the expression of DEC1 and transfected with an SHP promoter luciferase reporter. For comparison, a Per1 reporter was included. We and other investigators have demonstrated that Per1 is a circadian gene and negatively regulated by DEC1 [18, 30].
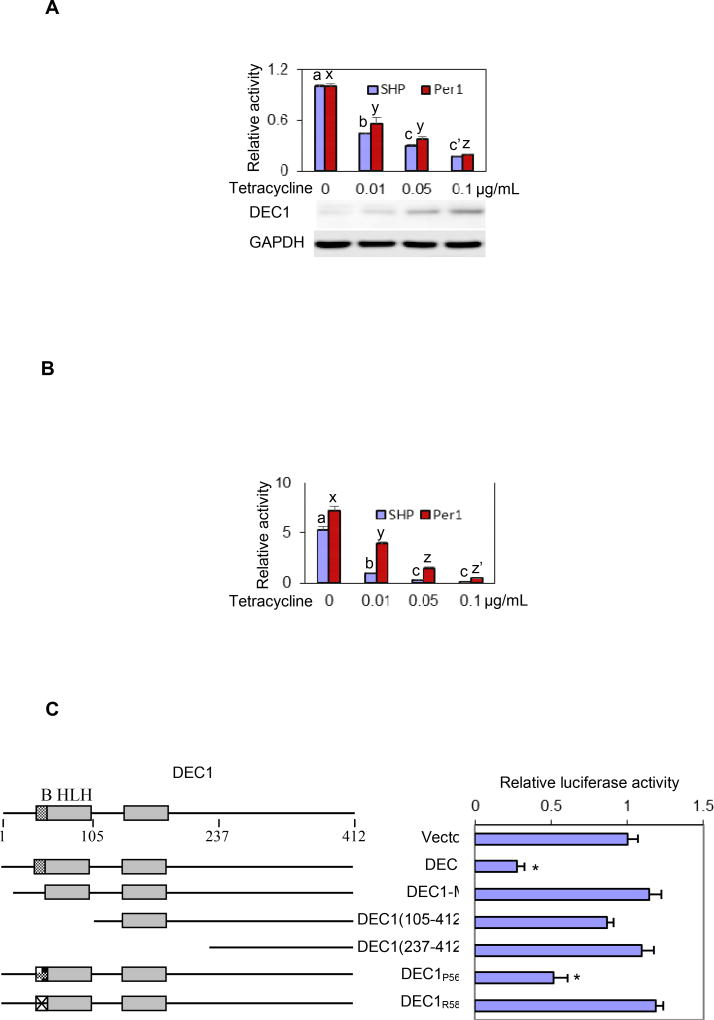
As shown in Fig. 1, DEC1 repressed both SHP and Per1 reporters and the repression occurred in a concentration-dependent manner. The repression was robust by as much as 90%. Nevertheless, the SHP reporter was repressed to a greater extent than the Per1 reporter, particularly when lower concentrations of tetracycline were used (Fig. 1A). As expected, Western blotting detected tetracycline-concentration dependent induction of DEC1 expression. We next tested whether DEC1 attenuates Clock/Bmal1-transactivation of the SHP and Per1 promoters, as the Clock/Bmal1 heterodimer has been shown to regulate both DEC1 and SHP in a circadian manner [23, 30]. As shown in Fig. 1B, Clock/Bmal1 strongly transactivated both the SHP and Per1 reporters with the Per1 reporter being transactivated to a greater extent (5 versus 7 fold) (Fig. 1B). However, the transactivation of the SHP reporter was attenuated to a much greater extent than that of the Per1 reporter by DEC1. For example, tetracycline at 0.01 µg/mL attenuated the Clock/Bmal1 transactivation of the SHP reporter by 80%. In contrast, the transactivation of the Per1 reporter was attenuated by 45% only. It should be noted that 293T cells transfected with different amounts of DEC1 expression construct produced similar repression and attenuation as DEC1 stably transfected. We next tested whether the repression of the SHP promoter requires DNA binding. As shown in Fig. 1C, no repression was detected with all constructs except DEC1 (wild-type) and DEC1P56A. We have previously shown that substitution of the residue proline-56 with an alanine remained the ability for DEC1 to bind to E-box and deliver repressive activity [18, 30]. In contrast, substitution of the residue arginine-58 with a proline no longer bound to E-box element. These results conclude that DEC1 is a transcriptional repressor of SHP.
Fig. 1. Regulation of SHP-luc and Per1-luc by Clock/Bmal1, DEC1 or in combination.
(A) Repression of SHP-Luc and Per1-Luc by DEC1 DEC1 stably transfected cells were cultured in the absence or presence of tetracycline at various concentrations (0.01 to 0.1 µg/mL) for 24 h and then transfected with a reporter construct (50 ng) and the pRL-null Renilla (5 ng). The transfected cells were cultured for 24 h (tetracycline remained the same), collected with PBS and resuspended in passive lysis buffer. The reporter activities were assayed with a Dual-Luciferase Reporter Assay System. The firefly luminescence signal was normalized based on the Renilla luminescence signal. The signal in the absence of tetracycline was recoded as 100%. Statistical significance (p<0.05) is denoted by a different letter of a, b, c and c’ or x, y and z. To specify tetracycline-induced expression of DEC1, lysates (1 µg) were analyzed by Western blotting with anti-DEC1 or anti-GAPDH antibody. (B) Attenuated Clock/Bmal1 activation by DEC1 DEC1 stably transfected cells were cultured in the absence or presence of tetracycline at various concentrations (0.01 to 0.1 µg/mL) for 24 h and then transfected with Clock/Bmal1 (100 ng each), a reporter construct (50 ng) and the pRL-null Renilla (5 ng). Cells were cultured for another 24 h and luciferase activities were determined. The signal was expressed relatively to that in the absence of DEC1. Statistical significance (p<0.05) is denoted by a different letter of a, b and c or x, y, z and z’. (C) Essentiality of DNA binding domain for DEC1 to repress SHP-Luc Cells (293T) were cultured in 24-well plates and transfected with DEC1 or a DEC1 mutant (100 ng), SHP-Luc (50 ng) and the pRL-null Renilla (5 ng). Vector plasmid was used to equalize the amount of plasmid DNA for each transfection. After a 24 h-incubation, cells were collected and analyzed for luciferase activities. Similarly, firefly luminescence signal was normalized based on the Renilla luminescence signal. The asterisk sign indicates statistical significance from vector control over DEC1 (p<0.01).
Repression of the SHP promoter by DEC1 through the E-box in the proximal promoter
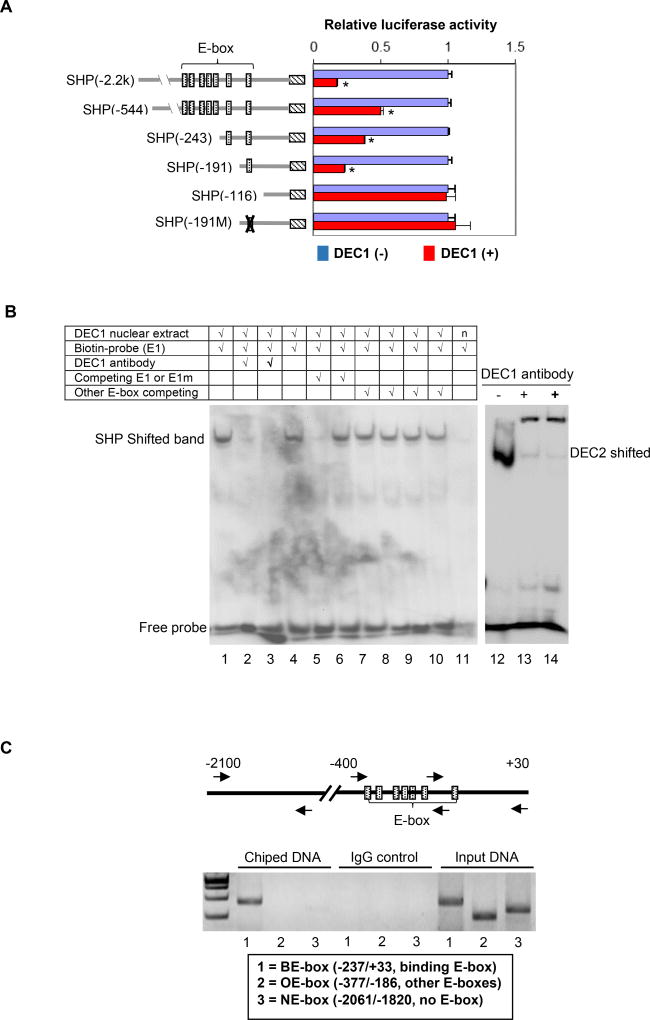
The proximal promoter of SHP is an E-box rich region and has as many as 7 E-box elements [29]. However, these elements differ slightly with 2 of them being CACCTG, and 1 of the following: CACTTG, CATCTG, CAGCTG, CAGGTG and CACGTG. To specify whether one or more of these elements support DEC1 repression, deletion and site-directed mutants of the SHP reporter were prepared and tested for the responsiveness to DEC1. Once again, DEC1 stably transfected cells were cultured in the presence or absence of tetracycline and then transfected with a reporter. As shown in Fig. 2A, all deletion SHP reporters were repressed except SHP-116Luc, suggesting that the E-box (E1: CACGTG) in the SHP-191Luc reporter supported the repression. To specify the role of this E-box in DEC1 repression, reporter SHP-191Luc was subjected to site-directed mutagenesis to selectively disrupt the E-box (CACGTG to AACGGG). As shown in Fig. 2A, disruption of this E-box completely attenuated DEC1-mediated repression of the SHP promoter.
Fig. 2. Identification and characterization of the E-box element in DEC1 binding.
(A) Identification of the E-box element in DEC1 binding DEC1 stably transfected cells were cultured in the absence or presence of tetracycline (0.1 µg/mL) to modulate the expression of DEC1 (tetracycline inducible) for 24 h. The cells were then transfected with a reporter construct (50 ng), the pRL-null Renilla (5 ng) and DEC1 (0–100 ng). The reporter constructs contained the 5’ sequence of the SHP promoter at different length or a sequence with a disrupted E-box element. The transfected cells were cultured, collected and analyzed for luciferase activity as described above. Once again, the firefly luminescence signal was normalized based on the Renilla luminescence signal. The signal in the absence of DEC1 was recoded as 100% for each reporter. The asterisk sign indicates statistical significance from vector control [labeled as DEC1(−)] over DEC1 (p<0.01). (B) EMSA analysis DEC1-stably transfected cells were cultured in the absence or presence of tetracycline (0.1 µg/ml) for 24 h, and the nuclear extracts were isolated. For EMSA, nuclear extracts (5 µg) were incubated with a biotinylated 187/162 (E1) probe for 20 min. In the competition assay, nuclear extracts were pre-incubated with the unlabeled element (50×) or oligonucleotides containing another E-box (not the same as the probe) for 30 min, and then incubated with the biotinylated probe. In the disruption assay, nuclear extracts were incubated first with an antibody against DEC1 on ice for 20 min and then with the biotinylated probe. Nuclear extracts (labeled as “n”) from cells cultured without tetracycline were used as a control (right lane of the left panel). For comparison, a probe derived from DEC2, known to interact with DEC1, was included. The protein-DNA complexes were electrophoretically resolved, transferred to a Biodyne® nylon membrane and located with streptavidin-conjugated horseradish peroxidase and chemiluminescent substrate. (C) ChIP analysis HepG2 cells were transfected with DEC1 (Flag-tagged) for 24 h, washed and underwent cross-linking for 15 min by 1% formaldehyde, and the cross-linking was terminated with 125 mM glycine. Alternatively DEC1 stably transfected cells were cultured in the presence of tetracycline (0.1 µg/ml) for 24 h and then processed as described for HepG2 cells. The soluble chromatins were prepared, pre-cleared with protein G beads and incubated with a Flag antibody (HepG2) or anti-DEC1 (stably transfected cells). As a control, the antibody was replaced with pre-immune IgG. The antibody-bound chromatins, DNA input (1/20 of the antibody-bound chromatins) as well as IgG-control were analyzed by PCR for the presence of the genomic fragment containing the E-box of interest, other E-boxes or no E-box. Both cells produced similar images and the image from HepG2 cells is shown).
We next tested whether this E-box interacted directly with DEC1. The DEC1-stable line was cultured in the presence or absence of tetracycline, and nuclear extracts were prepared. Double-stranded oligonucleotides harboring this E-box were synthesized and biotinylated. The labeled probe was incubated with the nuclear extracts and analyzed by EMSA. As shown in Fig. 2B, incubation with the extracts from the cells cultured in the presence of tetracycline yielded a shifted band. This band was not detected when incubation was performed with the extracts from the cell cultured without tetracycline (lane 11). The shifted band was competed completely by 50× un-biotinylated oligonucleotide (lane 5). A mutant of this oligonucleotide or oligonucleotides corresponding to other E-box elements showed no competitive activity (Fig. 2B, lanes 6–10). In addition, the shifted band disappeared by anti-DEC1 antibody (disrupted binding). As expected, a shifted band was detected with biotinylated oligonucleotide harboring an E-box derived from the DEC2 promoter [18]. In contrast to the disrupted binding with the SHP E-box (E1), the anti-DEC1 antibody caused the formation of a supershifted band (Right of Fig. 2B).
To determine whether DEC1 occupies the SHP promoter region that harbors this E-box (i.e., E1), ChIP experiment was performed. To gain specificity, primers were designed to amplify three fragments: the binding E-box (BE) fragment (E1 E-box); the other E-box (OE) fragment (other E-boxes but not binding) and the non E-box (NE) fragment (no E-box). As shown in Fig. 2C, chipped DNA showed the abundant presence of the BE-box fragment (labeled as lane 1) but not the other fragments. As expected, input DNA produced amplification of all three fragments (Right of Fig. 2C). It should be noted that pre-immune IgG was used as a control but did not enrich any fragments. The ChIP experiment was performed with HepG2 cells transfected with DEC1 (Flag-tagged) and DEC1 stably transfected cells. Same observations were made with both cell lines.
Effect of DEC1 on the expression of SHP
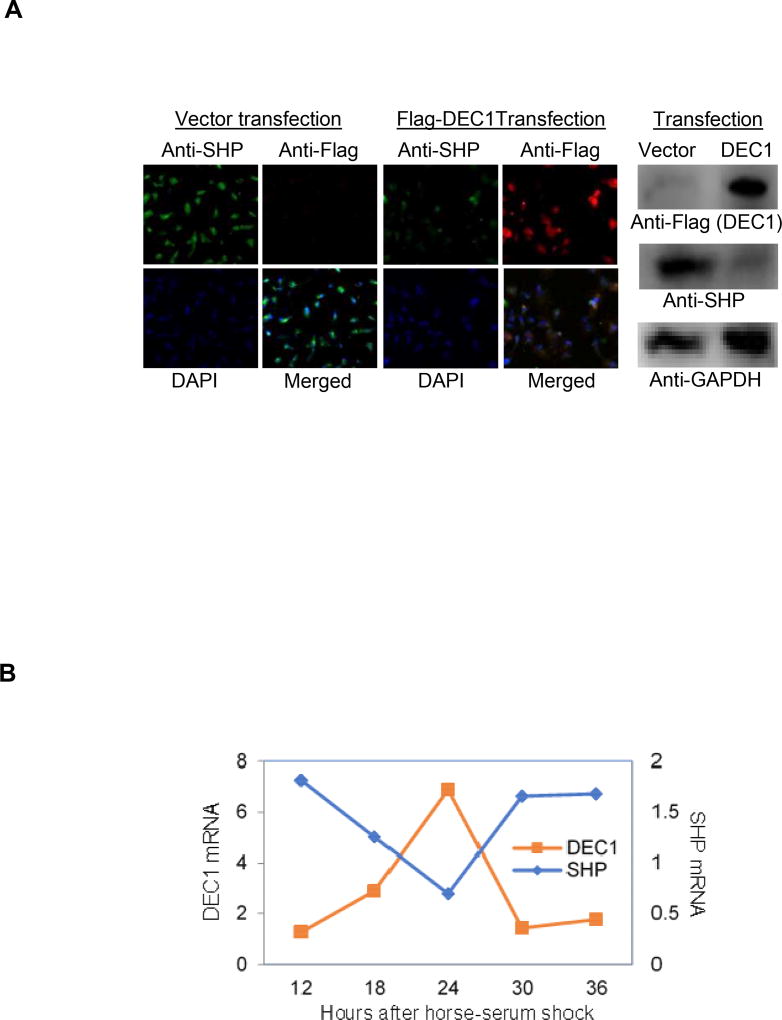
The reporter assay, EMSA and ChIP experiments established that DEC1 is a transcriptional repressor of SHP. Next we tested whether DEC1 suppresses the expression of endogenous SHP. To test this possibility, HepG2 cells were transfected with DEC1 (Flag-tagged) or the corresponding vector, and the expression of SHP was determined. Both Western blotting and immunocytochemistry were used to determine the changes of SHP expression. As expected, transfection of DEC1 increased immunodetection of DEC1. Importantly, transfection of DEC1 decreased the immunostaining for SHP (Right of Fig. 3A). The suppressed expression of SHP by DEC1 was confirmed by immunocytochemistry. Overall, transfection of DEC1 led to abundant Texas-Red staining (DEC1) accompanied by decreased FITC staining (green for SHP). Based on the overlay with DAPI staining (blue), both DEC1 and SHP are primarily present in the nuclei. It should be noted that the transfection was performed with TransfeX reagent, which delivered high transfection efficiency.
Fig. 3. Inverse expression between DEC1 and SHP.
(A) Suppressed expression of SHP by DEC1 HepG2 cells were seeded in 8-well chamber slides (2×104 cells/ well) and cultured for 24 h. Cells were then transfected with Flag-DEC1 (500 ng) or the vector by TransfeX reagent. The transfected cells were cultured for another 24 h and subsequently fixed with 4% paraformaldehyde for 10 min at pH 7.4. Cells were permeabilized with 0.1% Triton X-100 for 10 min. Chamber slides were incubated with 1% BSA (2mg/mL) for 1 h then incubated overnight with anti-SHP antibody or anti-Flag antibody. The anti-SHP antibody was located by Alexa Fluor® 488 conjugated goat anti-rabbit IgG (green), whereas the anti-Flag antibody with Alexa Fluor® 555 conjugated goat anti-mouse IgG (red). The slides were then mounted with ProLong Gold Antifade Mountant with DAPI (Blue). Cells were then imaged with confocal microscope. To provide more quantitative information, same experiments were performed in 6-well plates and cell lysates were prepared. Lysates (10 µg) were analyzed by Western blotting for the expression of SHP and DEC1 (Right panel). (B) Regulated expression of DEC1 and SHP by valproate in serum-shocked circadian induction HepG2 cells were seeded in 6- or 24-well plates at a density of 6×105 or 1.5×105 cells/well and cultured to reach 75% confluency. Cells were then shocked with media containing 50% horse serum for 2 h. Thereafter, the shocked cells were cultured in normal media or the same media containing valproate at 2 mM. Cells were harvested at a 6 h interval. The expression of DEC1 and SHP was determined by RT-qPCR with Taqman probes.
To complement the transfection study, we next tested whether SHP and DEC1 are inversely regulated for their oscillating expression by valproate, a widely used antiepileptic that was established to down-regulate SHP [38]. Importantly, valproate is a steatotic agent and both DEC1 and SHP are metabolic regulators. To mimic circadian rhythm, HepG2 cells were shocked by horse-serum and then treated with valproate. Cells were collected starting at 12 h after serum-shock and then at a 6 h interval. Total RNA was isolated and analyzed for the expression of SHP and DEC1. Both genes were expressed in a circadian manner and their expressions were inversely oscillated (Fig. 3B).
DISCUSSION
Normal circadian rhythms ensure physiological processes to be coordinated with daily changes of the environment [1–3]. Circadian rhythm misalignment has been increasingly recognized to pose health risk for a wide range of diseases [4–7]. The DEC1 and SHP genes are members of the liver clock and have been shown to play critical roles in metabolic homeostasis [5, 23, 35, 36]. In this study, we have shown that the SHP gene is a target of DEC1. DEC1 efficaciously repressed the SHP promoter and attenuated the transactivation of SHP by the Clock/Bmal1 complex. Site-directed mutagenesis, EMSA and ChIP identified that the E-box element in the SHP proximal promoter supported the repression. Overexpression of DEC1 led to decreased expression of SHP. In horse serum-shocked cells (induction of circadian rhythms), the steatotic drug valproate inversely altered the oscillating expression of DEC1 and SHP.
These findings establish that DEC1 participates in the negative loop of the SHP oscillating expression. It has been reported that the SHP gene is a target of the heterodimer Clock/Bmal1, a circadian activator complex. The Clock/Bmal1 and Per/Cry are well-established pairs that generate clock outputs by the transcriptional/translational feedback loop [5]. The Clock/Bmal1 dimer transactivates the Per and Cry genes through E-box elements, and the Per/Cry interacts with Clock/Bmal1 and attenuates the Clock/Bmal1 transactivation of Per/Cry. In this study, we have shown that DEC1 repressed the SHP promoter through the same E-box transactivated by the Clock/Bmal1 dimer, suggesting that DEC1 and Clock/Bmal1 form the transcriptional feedback loop for the oscillating expression of SHP (Figs. 1A, 1B and 2A). Consistent with the notion, the expression of DEC1 and SHP is inversely oscillated [25, 26].
It remains to be determined whether the Per/Cry proteins negatively regulate the expression of SHP. Nonetheless, it is likely that DEC1 exerts a dominant repression, particularly on the oscillating expression of SHP. In this study, we have shown that DEC1 repressed both the SHP promoter and the Per1 promoter with the SHP promoter being repressed to a greater extent (Fig. 1A), particularly when cells were cultured at lower concentrations of tetracycline, namely lower levels of DEC1. Importantly, tetracycline at 0.01 µg/mL attenuated the Clock/Bmal1 trans-activation of the SHP reporter by 80%. In contrast, the transactivation of the Per1 reporter was attenuated by 45% only (Fig. 1B). In both cases, DEC1 exerted repressive activity by binding to E-box CACGTG. The SHP E-box is flanked by GTGC (5’) and GGGT (3’), respectively, whereas the Per1 by TAGC and ACAG, respectively. It remains to be determined whether the differences in flanking sequences contribute to the differences in response to DEC1 repression. An early study demonstrated that the flanking sequences were important for interacting affinity with stra13, the mouse counterpart of DEC1 [26]. In this study, we have also shown that the antibody against DEC1 disrupted the shifted band with the SHP E-box (Fig. 2B). In contrast, the same antibody caused a supershift of the DEC2 E-box and the Per1 E-box [19, 30]. The SHP E-box is identical to those of DEC2 and Per1. However, it differs in the flanking sequences from both DEC1 and Per1 E-boxes.
The DEC1-SHP pathway likely plays critical roles in the synthesis of bile acids, particularly the circadian production of these endobiotics. While several pathways have been shown to regulate the synthesis of bile acids, the pathway mediated by SHP and F×R (farnesoid × receptor) has been extensively studied for the bile acid–activated regulatory cascade [5, 23]. This cascade is commonly referred as the bile acid negative feedback inhibition on the expression of the cytochrome P450 enzyme cholesterol 7α-hydroxylase (CYP7A1). CYP7A1 is the first and rate-limited enzyme in bile acid synthesis. Increased production of bile acids activates F×R, leading to the induction of SHP. Induction of SHP inactivates LRH-1 (liver receptor homolog-1) and HNF4α (hepatocyte nuclear factor 4α). In this study, we have shown that DEC1 downregulated SHP, thus counteracting the feedback inhibition. Interestingly, the transcription factor DEC2 (functionally related to DEC1) reportedly repressed the expression of rat CYP7A1 through an E-box: CACATG [39]. This E-box is conserved in human and mouse based on a BLAST search. It remains to be determined whether DEC1 binds to this E-box and causes repression. Nevertheless, we have reported that DEC1 negatively regulated the expression of DEC2 [19]. It is likely that DEC1 de-represses CYP7A1 by downregulating SHP and DEC2.
The DEC1-SHP pathway likely serves as an important mechanism for lipid metabolism. Although there are exceptions, SHP is generally considered to be lipogenic whereas DEC1 is anti-lipogenic. SHP reportedly augmented the transactivation by PPARγ (peroxisome proliferator-activated receptor-γ), leading to marked lipid accumulation in the liver [7]. Likewise, transgenic expression of SHP induced liver steatosis [41]. Consistent with these observations, SHP null mice were protected against diet-induced obesity. DEC1, on the other hand, has been shown to repress lipogenic genes such as fatty acid synthase and inhibit adipogenesis [42, 43]. Overexpression of DEC1 by viral transduction alleviated fatty liver phenotypes accompanied by suppressed expression of the lipogenic gene Srebp-1c (sterol regulatory element-binding protein -1c) [35]. Interestingly, SHP null mice supported higher induction of Srebp-1c in response to cholic acid treatment, suggesting that SHP is a repressor of Srebp-1c [44]. It is not clear whether the observed repression has a broad implication.
The DEC1-SHP pathway may have profound significance in carbohydrate homeostasis. Patients with type 2 diabetes had higher frequency of loss-of-function SHP mutants than those without type 2 diabetes (61.5 versus 28.1%) [45]. In addition, SHP mutation carriers had significantly higher fasting plasma insulin levels than non-carriers [45]. In mice, knockout of SHP developed hepatic insulin resistance [46], and the antidiabetic metformin ameliorated cytokine-induced hepatic insulin resistance by inducing SHP [47]. These observations suggest that SHP positively regulates glucose homeostasis. In contrast, high glucose and high insulin significantly induced DEC1 [48, 49]. The induction of DEC1 was inhibited by LY294002, a strong inhibitor of phosphoinositide 3-kinases. Importantly, DEC1 protein and the activity of AMPK (5’ AMP-activated protein kinase) showed an inverse circadian rhythm, and knockdown of DEC1 expression increased AMPK activity [50]. AMPK is known to regulate glucose homeostasis and prevent insulin resistance [51].
In summary, SHP belongs to the superfamily of nuclear receptors and has been established to exert a wide range of biological activities, particularly related to metabolic homeostasis. Many nuclear receptors and other transcription factors reportedly support the induction of SHP. On the other hand, the SHP gene is transactivated by the circadian complex of Clock/Bmal1. However, the negative loop of SHP oscillating expression remains unknown. In this study, we report that DEC1 repressed the SHP promoter and the repression was achieved through the E-box element in the proximal promoter region. We have also demonstrated that transfection of DEC1 led to decreased expression of SHP. In horse serum-shocked cells, the epileptic agent valproate (a steatotic drug) inversely altered the expression of DEC1 and SHP. Our findings collectively establish that DEC1 constitutes the negative loop of the SHP oscillating expression. Emerging evidence suggests that alterations on circadian systems are important risk factors for disease initiation and progression, and the expression of DEC1 and SHP is rapidly altered by many endobiotics and xenobiotics. Therefore, de-regulated expression of DEC1 and SHP genes likely alters normal circadian rhythms and contributes to the pathogenesis of many diseases, particularly metabolic disorders.
Abbreviation used
- AMPK
5’ AMP-activated protein kinase
- Bmal1
Brain and muscle ARNT-like 1
- Clock
circadian locomotor output cycles kaput
- Cry
Cryptochrome
- CYP7A1
Cytochrome P450 enzyme cholesterol 7α-hydroxylase
- DEC
Differentiated embryo chondrocyte
- DMEM
Dulbecco’s modified Eagle’s medium
- EMSA
Electrophoretic mobility shift assay
- F×R
Farnesoid × receptor
- Per
Period
- PCR
Polymerase chain reaction
- RT-PCR
Reverse transcription-polymerase chain reaction
- SCN
suprachiasmatic nucleus
- SHP
small heterodimer partner.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
This work was supported by National Institutes of Health Grants R01GM61988 and R01EB018748.
References
- 1.Millius A, Ueda HR. Systems Biology-Derived Discoveries of Intrinsic Clocks. Front Neurol. 2017;8:25. doi: 10.3389/fneur.2017.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Riede SJ, van der Vinne V, Hut RA. The flexible clock: predictive and reactive homeostasis, energy balance and the circadian regulation of sleep-wake timing. J Exp Biol. 2017;220:738–749. doi: 10.1242/jeb.130757. [DOI] [PubMed] [Google Scholar]
- 3.Dominoni DM, Borniger JC, Nelson RJ. Light at night, clocks and health: from humans to wild organisms. Biol Lett. 2016;12:20160015. doi: 10.1098/rsbl.2016.0015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Touitou Y, Reinberg A, Touitou D. Association between light at night, melatonin secretion, sleep deprivation, and the internal clock: Health impacts and mechanisms of circadian disruption. Life Sci. 2017;173:94–106. doi: 10.1016/j.lfs.2017.02.008. [DOI] [PubMed] [Google Scholar]
- 5.Ferrell JM, Chiang JY. Circadian rhythms in liver metabolism and disease. Acta Pharm Sin B. 2015;5:113–122. doi: 10.1016/j.apsb.2015.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Potter GD, Skene DJ, Arendt J, Cade JE, Grant PJ, Hardie LJ. Circadian rhythm and sleep disruption: causes, metabolic consequences, and countermeasures. Endocr Rev. 2016;37:584–608. doi: 10.1210/er.2016-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kunze KN, Hanlon EC, Prachand VN, Brady MJ. Peripheral circadian misalignment: contributor to systemic insulin resistance and potential intervention to improve bariatric surgical outcomes. Am J Physiol Regul Integr Comp Physiol. 2016;311:R558–563. doi: 10.1152/ajpregu.00175.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Korenčič A, Košir R, Bordyugov G, Lehmann R, Rozman D, Herzel H. Timing of circadian genes in mammalian tissues. Sci Rep. 2014;4:5782. doi: 10.1038/srep05782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakamura TJ, Takasu NN, Nakamura W. The suprachiasmatic nucleus: age-related decline in biological rhythms. J Physiol Sci. 2016;66:367–374. doi: 10.1007/s12576-016-0439-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kalsbeek A, Palm IF, La Fleur SE, Scheer FA, Perreau-Lenz S, Ruiter M, Kreier F, Cailotto C, Buijs RM. SCN outputs and the hypothalamic balance of life. J Biol Rhythms. 2006;21:458–469. doi: 10.1177/0748730406293854. [DOI] [PubMed] [Google Scholar]
- 11.Besharse JC, McMahon DG. The retina and other light-sensitive ocular clocks. J Biol Rhythms. 2016;31:223–243. doi: 10.1177/0748730416642657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tan E, Scott EM. Circadian rhythms, insulin action, and glucose homeostasis. Curr Opin Clin Nutr Metab Care. 2014;17:343–348. doi: 10.1097/MCO.0000000000000061. [DOI] [PubMed] [Google Scholar]
- 13.Hirao A, Nagahama H, Tsuboi T, Hirao M, Tahara Y, Shibata S. Combination of starvation interval and food volume determines the phase of liver circadian rhythm in Per2::Luc knock-in mice under two meals per day feeding. Am J Physiol Gastrointest Liver Physiol. 2010;299:G1045–1053. doi: 10.1152/ajpgi.00330.2010. [DOI] [PubMed] [Google Scholar]
- 14.Wu T, Yao C, Huang L, Mao Y, Zhang W, Jiang J, Fu Z. Nutrients and circadian rhythms in mammals. J Nutr Sci Vitaminol (Tokyo) 2015;61(Suppl):S89–91. doi: 10.3177/jnsv.61.S89. [DOI] [PubMed] [Google Scholar]
- 15.Wu T, Ni Y, Kato H, Fu Z. Feeding-induced rapid resetting of the hepatic circadian clock is associated with acute induction of Per2 and Dec1 transcription in rats. Chronobiol Int. 2010;27:1–18. doi: 10.3109/07420520903398625. [DOI] [PubMed] [Google Scholar]
- 16.Oike H, Nagai K, Fukushima T, Ishida N, Kobori M. Feeding cues and injected nutrients induce acute expression of multiple clock genes in the mouse liver. PLoS One. 2011;6:e23709. doi: 10.1371/journal.pone.0023709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Challet E, Caldelas I, Graff C, Pevet P. Synchronization of molecular clock by light- and food-related cues in mammals. Biol Chem. 2003;384:711–719. doi: 10.1515/BC.2003.079. [DOI] [PubMed] [Google Scholar]
- 18.Honma S, Kawamoto T, Takagi Y, Fujimoto K, Sato F, Noshiro M, Kato Y, Honma K. DEC1 and DEC2 are regulators of the mammalian molecular clock. Nature. 2002;419:841–844. doi: 10.1038/nature01123. [DOI] [PubMed] [Google Scholar]
- 19.Li Y, Zhang H, Xie M, Ge S, Hu M, Yang D, Wan Y, Yan B. DEC1 negatively regulates the expression of DEC2 through the E-box in the proximal promoter. J. Biol. Chem. 2003;278:16899–16907. doi: 10.1074/jbc.M300596200. [DOI] [PubMed] [Google Scholar]
- 20.St-Pierre B, Flock G, Zacksenhaus E, Egan SE SE. Stra13 homodimers repress transcription through class B E-box elements. J Biol Chem. 2002;277:46544–46551. doi: 10.1074/jbc.M111652200. [DOI] [PubMed] [Google Scholar]
- 21.Wang L, Liangpunsakul S. Circadian clock control of hepatic lipid metabolism: role of small heterodimer partner (Shp) J Investig Med. 2016;64:1158–1161. doi: 10.1136/jim-2016-000194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu N, Kim KH, Zhou Y, Lee JM, Kettner NM, Mamrosh JJ, Choi S, Fu L, Moore DD. Small Heterodimer Partner (NR0B2) Coordinates nutrient signaling and the circadian clock in mice. Mol Endocrinol. 2016;30:988–995. doi: 10.1210/me.2015-1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Y, Hagedorn CH, Wang L. Role of nuclear receptor SHP in metabolism and cancer. Biochim Biophys Acta. 2011;1812:893–908. doi: 10.1016/j.bbadis.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chiang JY. Bile acids: regulation of synthesis. J Lipid Res. 2009;50:1955–1966. doi: 10.1194/jlr.R900010-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oiwa A, Kakizawa T, Miyamoto T, Yamashita K, Jiang W, Takeda T, Suzuki S, Hashizume K. Synergistic regulation of the mouse orphan nuclear receptor SHP gene promoter by CLOCK-BMAL1 and LRH-1. Biochem Biophys Res Commun. 2007;353:895–901. doi: 10.1016/j.bbrc.2006.12.131. [DOI] [PubMed] [Google Scholar]
- 26.Grechez-Cassiau A, Panda S, Lacoche S, Teboul M, Azmi S, Laudet V, Hogenesch JB, Taneja R, Delaunay F. The transcriptional repressor STRA13 regulates a subset of peripheral circadian outputs. J Biol Chem. 2004;279:1141–1150. doi: 10.1074/jbc.M305369200. [DOI] [PubMed] [Google Scholar]
- 27.Kondratov RV, Chernov MV, Kondratova AA, Gorbacheva VY, Gudkov AV, Antoch MP. BMAL1-dependent circadian oscillation of nuclear CLOCK: posttranslational events induced by dimerization of transcriptional activators of the mammalian clock system. Genes Dev. 2003;17:1921–1932. doi: 10.1101/gad.1099503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wilsbacher LD, Yamazaki S, Herzog ED, Song EJ, Radcliff LA, Abe M, Block G, Spitznagel E, Menaker M, Takahash JS. Photic and circadian expression of luciferase in mPeriod1-luc transgenic mice in vivo. Proc Natl Acad Sci USA. 2002;99:489–499. doi: 10.1073/pnas.012248599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim HL, Kim JK, Kim JY, Park SK, Seo JH, Kim JB, Lee IK, Kim KS, Choi HS. Differential regulation of human and mouse orphan nuclear receptor small heterodimer partner promoter by sterol regulatory element binding protein-1. J Biol Chem. 2004;279:28122–28131. doi: 10.1074/jbc.M313302200. [DOI] [PubMed] [Google Scholar]
- 30.Li Y, Song X, Ma Y, Liu J, Yang D, Yan B. DNA binding, but not interaction with Bmal, is responsible for DEC1-mediated transcription regulation of the circadian gene Per1. Biochem J. 2004;382:895–904. doi: 10.1042/BJ20040592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xiao D, Yang D, Guo L, Lu W, Charpentier M, Yan B. Regulation of carboxylesterase-2 expression by p53 family proteins and enhanced anticancer activities among 5-fluorouracil, irinotecan and doxazolidine prodrug. Brit J Pharmacol. 2013;168:1989–1999. doi: 10.1111/bph.12125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vachirayonsti T, Yan B. MicroRNA-30c-1 suppresses the pregnane × receptor by targeting the 3’-untranslated region and alters the expression of its target gene cytochrome P450 3A4. BBA-Gene Regulatory Mechanisms. 2016;1859:1238–1244. doi: 10.1016/j.bbagrm.2016.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Y, Zhang H, Xie M, Hu M, Ge S, Yang D, Wan Y, Yan B. DEC1/STRA13/SHARP2 is abundantly expressed in colon carcinoma, antagonizes serum deprivation induced apoptosis and selectively inhibits the activation of procaspases. Biochem J. 2002;367:413–422. doi: 10.1042/BJ20020514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vachirayonsti T, Yang D, Yan B. Suppression of the pregnane × receptor during endoplasmic reticulum stress is achieved by down-regulating hepatocyte nuclear factor-4α and up-regulating liver-enriched inhibitory protein. Toxicol Sci. 2015;144:382–392. doi: 10.1093/toxsci/kfv008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shen L, Cui A, Xue Y, Cui Y, Dong X, Gao Y, Yang H, Fang F, Chang Y. Hepatic differentiated embryo-chondrocyte-expressed gene 1 (Dec1) inhibits sterol regulatory element-binding protein-1c (Srebp-1c) expression and alleviates fatty liver phenotype. J Biol Chem. 2014;289:23332–23342. doi: 10.1074/jbc.M113.526343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shen M, Yoshida E, Yan W, Kawamoto T, Suardita K, Koyano Y, Fujimoto K, Noshiro M, Kato Y. Basic helix-loop-helix protein DEC1 promotes chondrocyte differentiation at the early and terminal stages. J Biol Chem. 2002;277:50112–50120. doi: 10.1074/jbc.M206771200. [DOI] [PubMed] [Google Scholar]
- 37.Li Y, Xie M, Yang J, Yang D, Deng R, Wan Y, Yan B. The expression of antiapoptotic protein survivin is transcriptionally upregulated by DEC1 primarily through multiple Sp1 binding sites in the proximal promoter. Oncogene. 2006;25:3296–3306. doi: 10.1038/sj.onc.1209363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Benet M, Guzmán C, Pisonero-Vaquero S, García-Mediavilla MV, Sánchez-Campos S, Martínez-Chantar ML, Donato MT, Castell JV, Jover R. Repression of the nuclear receptor small heterodimer partner by steatotic drugs and in advanced nonalcoholic fatty liver disease. Mol Pharmacol. 2015;87:582–594. doi: 10.1124/mol.114.096313. [DOI] [PubMed] [Google Scholar]
- 39.Noshiro M, Kawamoto T, Furukawa M, Fujimoto K, Yoshida Y, Sasabe E, Tsutsumi S, Hamada T, Honma S, Honma K, Kato Y. Rhythmic expression of DEC1 and DEC2 in peripheral tissues: DEC2 is a potent suppressor for hepatic cytochrome P450s opposing DBP. Genes Cells. 2004;9:317–329. doi: 10.1111/j.1356-9597.2004.00722.x. [DOI] [PubMed] [Google Scholar]
- 40.Nishizawa H, Yamagata K, Shimomura I, Takahashi M, Kuriyama H, Kishida K, Hotta K, Nagaretani H, Maeda N, Matsuda M, Kihara S, Nakamura T, Nishigori H, Tomura H, Moore DD, Takeda J, Funahashi T, Matsuzawa Y. Small heterodimer partner, an orphan nuclear receptor, augments peroxisome proliferator-activated receptor gamma transactivation. J Biol Chem. 2002;277:1586–1592. doi: 10.1074/jbc.M104301200. [DOI] [PubMed] [Google Scholar]
- 41.Boulias K, Katrakili N, Bamberg K, Underhill P, Greenfield A, Talianidis I. Regulation of hepatic metabolic pathways by the orphan nuclear receptor SHP. EMBO J. 2005;24:2624–2633. doi: 10.1038/sj.emboj.7600728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ozaki N, Noshiro M, Kawamoto T, Nakashima A, Honda K, Fukuzaki-Dohi U, Honma S, Fujimoto K, Tanimoto K, Tanne K, Kato Y Y. Regulation of basic helix-loop-helix transcription factors Dec1 and Dec2 by RORα and their roles in adipogenesis. Genes Cells. 2012;17:109–21. doi: 10.1111/j.1365-2443.2011.01574.x. [DOI] [PubMed] [Google Scholar]
- 43.Iizuka K, Horikawa Y. Regulation of lipogenesis via BHLHB2/DEC1 and ChREBP feedback looping. Biochem Biophys Res Commun. 2008;374:95–100. doi: 10.1016/j.bbrc.2008.06.101. [DOI] [PubMed] [Google Scholar]
- 44.Wang L, Lee YK, Bundman D, Han Y, Thevananther S, Kim CS, Chua SS, Wei P, Heyman RA, Karin M, Moore DD DD. Redundant pathways for negative feedback regulation of bile acid production. Dev Cell. 2002;2:721–731. doi: 10.1016/s1534-5807(02)00187-9. [DOI] [PubMed] [Google Scholar]
- 45.Yang Z, Wu S, Zheng T, Lu H, Ma X, Jia W, Hu R R. Gender-dependent penetrance of small heterodimer partner (SHP) gene deficiency in overweight/obese Chinese pedigrees. J Int Med Res. 2010;38:142–149. doi: 10.1177/147323001003800116. [DOI] [PubMed] [Google Scholar]
- 46.Park YJ, Kim SC, Kim J, Anakk S, Lee JM, Tseng HT, Yechoor V, Park J, Choi JS, Jang HC, Lee KU, Novak CM, Moore DD, Lee YK. Dissociation of diabetes and obesity in mice lacking orphan nuclear receptor small heterodimer partner. J Lipid Res. 2011;52:2234–2244. doi: 10.1194/jlr.M016048. (2011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim YD, Kim YH, Cho YM, Kim DK, Ahn SW, Lee JM, Chanda D, Shong M, Lee CH, Choi HS. Metformin ameliorates IL-6-induced hepatic insulin resistance via induction of orphan nuclear receptor small heterodimer partner (SHP) in mouse models. Diabetologia. 2012;55:1482–1494. doi: 10.1007/s00125-012-2494-4. [DOI] [PubMed] [Google Scholar]
- 48.Chen R, Wang Y, Ning R, Hu J, Liu W, Xiong J, Wu L, Liu J, Hu G, Yang J. Decreased carboxylesterases expression and hydrolytic activity in type 2 diabetic mice through Akt/mTOR/HIF-1α/Stra13 pathway. Xenobiotica. 2015;45:782–793. doi: 10.3109/00498254.2015.1020353. [DOI] [PubMed] [Google Scholar]
- 49.Yamada K, Kawata H, Shou Z, Mizutani T, Noguchi T, Miyamoto K K. Insulin induces the expression of the SHARP-2/Stra13/DEC1 gene via a phosphoinositide 3-kinase pathway. J Biol Chem. 2003;278:30719–30724. doi: 10.1074/jbc.M301597200. [DOI] [PubMed] [Google Scholar]
- 50.Sato F, Muragaki Y, Zhang Y. DEC1 negatively regulates AMPK activity via LKB1. Biochem Biophys Res Commun. 2015;467:711–716. doi: 10.1016/j.bbrc.2015.10.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nandipati KC, Subramanian S, Agrawal DK. Protein kinases: mechanisms and downstream targets in inflammation-mediated obesity and insulin resistance. Mol Cell Biochem. 2017;426:27–45. doi: 10.1007/s11010-016-2878-8. [DOI] [PMC free article] [PubMed] [Google Scholar]