Summary
Organogenesis and tissue development occur through sequential stepwise processes leading to increased lineage restriction and loss of pluripotency. An exception to this appears in the adult human breast, where rare variant epithelial cells exhibit pluripotency and multilineage differentiation potential when removed from the signals of their native microenvironment. This phenomenon provides a unique opportunity to study mechanisms that lead to cellular reprogramming and lineage plasticity in real time. Here, we show that primary human mammary epithelial cells (HMECs) lose expression of differentiated mammary epithelial markers in a manner dependent on paracrine factors and epigenetic regulation. Furthermore, we demonstrate that HMEC reprogramming is dependent on gene silencing by the DNA methyltransferase DNMT3A and loss of histone transcriptional marks following downregulation of the methyltransferase DOT1L. These results demonstrate that lineage commitment in adult tissues is context dependent and highlight the plasticity of somatic cells when removed from their native tissue microenvironment.
Keywords: mammary, DOT1L, lineage, vHMECs, DMNT3A
Graphical Abstract

Highlights
-
•
vHMECs arise through epigenetic modification of pre-existing human breast cells
-
•
DNA methylation by DNMT3a is required for vHMEC formation
-
•
Loss of DOT1L and active histone methylation marks accelerates vHMEC formation
-
•
Loss of mammary identity requires changes in both DNA and histone methylation
In this article, Kuperwasser and colleagues show that, upon removal from their native microenvironment, rare pre-existing human mammary epithelial cells undergo stochastic epigenetic reprogramming and lose mammary lineage identity. This dedifferentiation occurs through addition of repressive promoter methylation marks by DNMT3a and removal of active histone methylation marks through DOT1L loss, which together suppress mammary lineage commitment gene expression.
Introduction
Tissue development is orchestrated by stepwise progression of primitive multipotent progenitor cells giving rise to and differentiating into increasingly lineage-committed progenitors (Reya et al., 2001). During embryonic mammary gland development, the prospective mammary bud develops as a specialization of the uncommitted surface ectoderm (Macias and Hinck, 2012, Watson and Khaled, 2008). Microenvironmental signals, including crosstalk between the surface ectoderm and mesenchymal cells, are essential for mammary fate specification during this period; in the absence of specific growth factors and signals, mammary buds fail to organize or properly develop (Macias and Hinck, 2012). By birth, lineage restriction is established in the mammary epithelium and this highly specialized glandular structure, composed of bilayered epithelium of luminal and basal/myoepithelial (ME) cells, is organized into an extensive system of ducts and alveoli (Macias and Hinck, 2012, Van Keymeulen et al., 2011).
Despite restriction during development, mammary epithelial cells exhibit substantial plasticity in settings such as ex vivo culture, transplantation, wound healing, and tumorigenesis (Locke and Clark, 2012). The bulk population of primary human mammary epithelial cells (HMECs) isolated from breast tissue rapidly undergoes Rb-mediated senescence after 5–10 population doublings (Brenner et al., 1998, Foster and Galloway, 1996); however, rare proliferative clones, called variant HMECs (vHMECs), invariably emerge from primary cultures.
Apart from their proliferative potential, vHMECs display radically altered differentiation potential compared with HMECs (Garbe et al., 2009, Keller et al., 2012). In contrast to early-passage HMECs, vHMECs do not express many of the characteristic differentiation markers of mammary epithelial cells. Moreover, vHMECs display metaplastic differentiation potential, able to generate glandular structures or stratified squamous epithelium dependent on the 3D culture conditions. The latter structures exhibit full epidermal differentiation with expression of epidermal markers K10 and involucrin (Keller et al., 2012). Furthermore, when transformed, vHMECs generate aggressive, poorly differentiated metaplastic cancers with squamous, glandular, and papillary histologies, in contrast to early-passage HMECs which predominantly generate breast adenocarcinomas (Keller et al., 2012).
The origin of vHMECs is not known. vHMECs are present at a frequency of 1/100,000–1/250,000 and do not express the CDKN2a/p16 cell-cycle inhibitor due to promoter methylation (Foster and Galloway, 1996, Huschtscha et al., 1998, Hinshelwood et al., 2009). Therefore, it has been suggested that vHMECs represent a residual primitive population persisting in the adult breast (Bean et al., 2007, Holst et al., 2003, Roy et al., 2013), analogous to CDKN2a-negative cells identified in the esophagus (Wang et al., 2011). However, the appearance of vHMECs in culture as rare colonies arising after a long latency period is reminiscent of the nature and frequency of somatic cell reprogramming by defined factors typical in most induced pluripotent stem cell (iPSC) protocols (Takahashi and Yamanaka, 2006). Therefore, it is possible that vHMECs are epigenetically reprogrammed from lineage-committed precursors in a process resembling iPSCs. Supporting this, emergence of vHMECs is dependent on media composition. In serum-free media, vHMEC colonies arise 2–4 weeks after the bulk population of HMECs has undergone CDKN2a/p16-dependent growth arrest (Holst et al., 2003, Keller et al., 2012, Romanov et al., 2001). However, in media formulations that contain serum, vHMECs do not emerge (Stampfer et al., 1980), suggesting that microenvironmental factors may be important in controlling escape from growth arrest and/or dedifferentiation.
Interestingly, epigenetic modification of HMEC chromatin has been shown both locally and globally during progression to vHMECs. As vHMECs develop, DNA methylation occurs rapidly at CDKN2a and other targeted areas, similar to those methylated in breast cancer (Locke et al., 2015). As methylation of CDKN2a increases, global loss of H3K27 methylation occurs, suggesting that a coordinated epigenetic program could be responsible for HMEC dedifferentiation (Hinshelwood et al., 2009).
Here, we determined that the transition of HMEC to vHMEC is a model of epigenetic reprogramming, and identified specific mechanisms by which lineage-committed HMECs reprogram to a more primitive state. We conclude that reprogramming of lineage-committed HMECs requires gene silencing via coordinated regulation of both DNA and histone methylation.
Results
HMECs Lose Their Identity in the Absence of Native Microenvironmental Signals
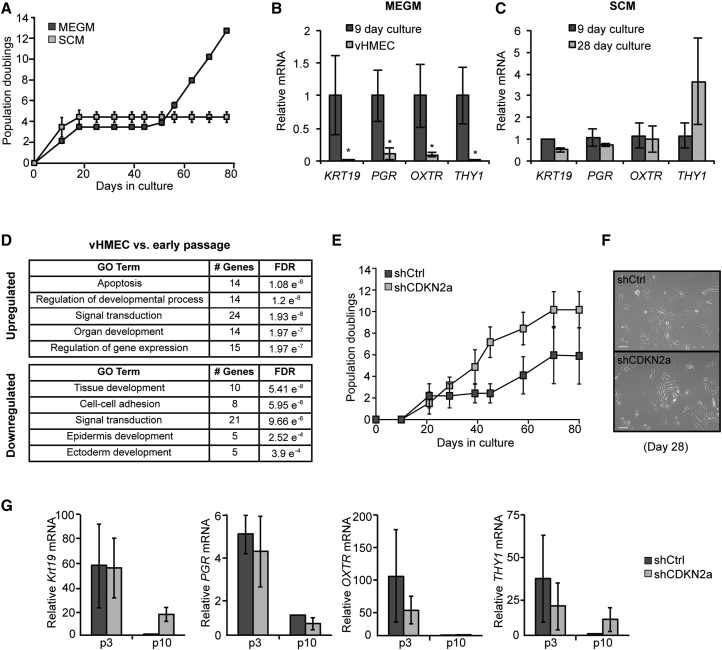
To characterize the phenotype of HMECs during culture, we derived HMECs from enzymatically dissociated reduction mammoplasty samples and analyzed these cells at different time points. In addition, to understand how media composition affects cellular differentiation and plasticity, we grew cells in either serum-free mammary epithelial growth medium (MEGM), which leads to vHMEC formation after 40–50 days, or in serum-containing medium (SCM), which leads to permanent cellular senescence (Stampfer and Bartley, 1985).
As previously described, when cultured in MEGM, HMECs exhibited an initial proliferative arrest characterized by upregulation of CDKN2a expression and senescent morphology (Figure 1A). After 35–50 days, rare clones of small, refractive, proliferating cells overcame growth arrest and exhibited CDKN2a silencing; these cells possess extended proliferative capacity but are not immortalized. qPCR analysis revealed that, compared with early-passage HMECs, vHMECs had decreased expression of a panel of genes associated with mammary epithelial differentiation. These genes include basal/ME-specific OXTR and THY1, as well as luminal-specific PGR and KRT19 (Figure 1B). These data confirm that vHMECs exhibit an undifferentiated phenotype.
Figure 1.
HMECs Lose Lineage Commitment in the Absence of Stromal Cues
(A) Growth curve showing cumulative population doublings over time in primary HMECs grown in MEGM or SCM, n = 3.
(B and C) qPCR analysis of mammary lineage gene expression in HMECs grown in (B) MEGM or (C) SCM at different time points. mRNA levels are shown relative to the 9-day early culture, n = 3.
(D) Gene ontology (GO) analysis of the set of genes differentially expressed between vHMECs and early-passage HMECs.
(E) Growth curve showing cumulative population doublings over time in HMECs with knockdown of CDKN2a (shCDKN2a) or a firefly luciferase control (shCtrl), n = 3.
(F) Representative image of an shCDKN2a culture when shCtrl cells are undergoing growth arrest (day 28). shCtrl cells adopt a senescent morphology while shCDKN2a cells remain small, refractile, and highly proliferative. Scale bars, 100 μm.
(G) qPCR analysis of mammary lineage gene expression in shCtrl or shCDKN2a cultures at different time points, relative to shCtrl (p10), n = 4.
In all panels, error bars indicate the mean ± SEM and replicates are individual patient samples. ∗p < 0.05, Student's t test.
In contrast to MEGM, cells grown in SCM initially proliferated quickly, but exhibited growth arrest after 5–10 population doublings and never generated vHMECs (Figure 1A), behavior consistent with previous descriptions of HMECs grown in SCM (Stampfer and Bartley, 1985). Interestingly, unlike cells grown in MEGM, cells in SCM did not exhibit repression of mammary lineage differentiation genes even after several passages (Figure 1C). Therefore, the emergence and growth of vHMECs is dependent on media composition, which also affects loss of HMEC lineage identity in culture.
We next compared global gene expression profiles of early-passage HMECs, growth-arrested HMECs, or vHMECs using gene ontology (GO) analysis on the sets of differentially expressed genes between each group (Figure 1D; dataset GEO: GSE16058) (Garbe et al., 2009). Interestingly, GO terms related to epithelial/epidermal differentiation and development were highly enriched among genes differentially expressed between vHMECs and early-passage HMECs, consistent with an altered differentiation potential. Intriguingly, similar GO terms were enriched in a comparison of growth-arrested HMECs with the early-passage cultures, suggesting that loss of lineage commitment occurred in the bulk population of HMECs even before vHMECs arise (Figure S1A). In contrast, genes differentially expressed between vHMECs and growth-arrested HMECs were limited mainly to DNA replication- or proliferation-associated genes rather than differentiation-associated genes (Figure S1B). These data suggest that loss of lineage identity begins in HMECs prior to the emergence of vHMECs.
Since it has been reported that artificial silencing of CDKN2a in HMECs is sufficient to bypass growth arrest (Novak et al., 2009), we wondered whether loss of CDKN2a would also be sufficient to induce dedifferentiation of HMECs into a vHMEC-like state. Accordingly, we stably reduced CDKN2a in freshly dissociated HMECs using a lentiviral short hairpin RNA (shRNA) vector (shCDKN2a) (Figure S1C). As expected, HMECs infected with shCDKN2a continued to proliferate exponentially and did not experience the growth arrest seen in control cells (Figures 1E and 1F). To determine whether shCDKN2a cells lost mammary lineage identity, we measured the levels of mammary epithelial differentiation markers in early- and late-passage shCDKN2a cells compared with control cells. qPCR analysis revealed that transcripts for KRT19, PGR, OXTR, and THY1 were highly expressed in early-passage shCDKN2a cells at levels similar to those of early-passage control (shCtrl) cells, and decreased to similar levels as shCtrl cells in late passage (Figure 1G). Therefore, removing the CDKN2a senescence barrier does not enable HMECs to dedifferentiate more rapidly, but enables the bulk population of cells to bypass senescence.
Taken together, these data suggest that dedifferentiation and escape from growth arrest are distinct events during HMEC culture. The former occurs in the bulk population of HMECs in a manner dependent on media composition, while the latter occurs only in a small minority of cells (vHMECs).
vHMECs Arise from Cells that Undergo Spontaneous Epigenetic Reprogramming
The origin of vHMECs is not clear. Although it has been suggested that vHMECs are the outgrowth of a pre-existing rare population of CDKN2a-methylated cells that reside in the breast epithelium (Holst et al., 2003, Roy et al., 2013), our findings suggest that they may alternatively arise from dedifferentiation of lineage-committed cells that spontaneously silence the CDKN2a promoter. This would agree with the finding that HMECs can rapidly methylate and suppress CDKN2a during culture (Hinshelwood et al., 2009). Therefore, we next asked whether vHMECs arise spontaneously from epigenetic reprogramming of cells without CDKN2a silencing or whether they pre-exist in HMEC cultures prior to the growth arrest period.
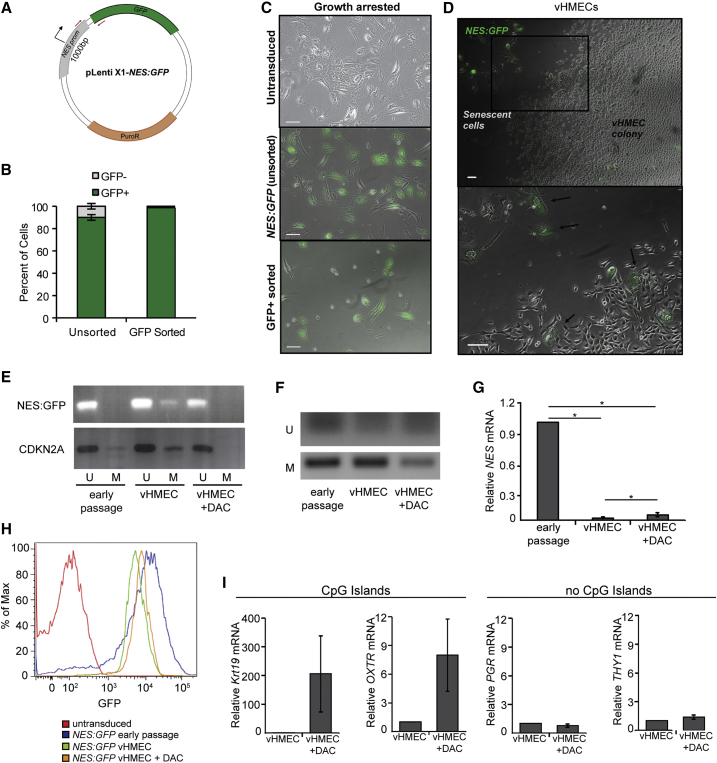
To assess this, we developed an assay to label HMECs that may potentially give rise to vHMECs. Our approach was to permanently mark early-passage cells with a promoter that invariably demonstrates CpG-island DNA methylation in vHMECs as a tool to trace epigenetic alterations during vHMEC progression. In addition to CDKN2a, vHMECs exhibit widespread differences in DNA methylation versus early-passage HMECs (Novak et al., 2009). We analyzed genes previously shown to be silenced in vHMECs and found that the nestin (NES) promoter was consistently methylated in vHMECs, but not in early-passage HMECs. NES is a neural stem cell gene that is also expressed in mammary epithelial stem/progenitor cells and is used in this study as a marker to track de novo DNA methylation (Cregan et al., 2007, Li et al., 2007).
To assess the ability of HMECs to undergo de novo methylation, we engineered a lentiviral vector coupling the NES promoter upstream of a GFP transgene, so that GFP expression is dependent on the methylation status of the exogenous NES promoter (NES:GFP, Figure 2A). Freshly dissociated HMECs from reduction mammoplasty tissues were transduced with NES:GFP, selected in puromycin, then purified to >99% homogeneity by fluorescence-activated cell sorting (FACS) (Figures 2B and S2A). Upon plating, early-passage NES:GFP cells remained GFP+ before and during senescence (Figure 2C).
Figure 2.
vHMECs Arise from HMECs Following Epigenetic Reprogramming
(A) Schematic of the construct used to GFP-label primary HMECs with an exogenous NES promoter; methylation-specific PCR primers are shown in red.
(B) Quantification of flow cytometry analysis of the percentage of GFP+ versus GFP− cells in HMEC cultures before and after FACS purification of GFP+ cells, n = 3.
(C) Representative images of untransduced, unsorted, and GFP+-sorted NES:GFP cultures at the point of growth arrest. The minority GFP− cells present in the unsorted cultures are absent from the FACS-purified cultures. Scale bars, 100 μm.
(D) Representative low-power (top) and high-power (bottom) images of a vHMEC colony derived from FACS-purified GFP+ cells showing GFP+ senescent cells (arrows) adjacent to GFP− vHMECs. Scale bars, 100 μm.
(E and F) Methylation-specific PCR of the (E) exogenous NES:GFP promoter, CDKN2a, or (F) endogenous NES promoter region, in early-passage HMECs, vHMECs, or in vHMECs treated with decitabine (DAC) from (E) NES:GFP or (F) untransduced cultures. U, unmethylated; M, methylated.
(G) qPCR analysis of NES expression in HMECs, vHMECs, and vHMECs treated with DAC, n = 3.
(H) Flow-cytometry analysis of high GFP expression in early-passage NES:GFP HMECs, decreased expression in NES:GFP vHMECs, and partial rescue by treatment of NES:GFP vHMECs with DAC.
(I) qPCR analysis of mammary lineage gene expression in early HMECs, vHMECs, and vHMECs treated with DAC, relative to vHMEC, n = 4.
In all panels, error bars indicated the mean ± SEM and replicates are individual patient samples. ∗p < 0.05, Student's t test.
Long-term culture of FACS-purified NES:GFP+ HMECs consistently led to the generation of vHMECs with reduced GFP expression, compared with nearby growth-arrested cells (Figure 2D). Furthermore, we performed methylation-specific PCR of the exogenous NES:GFP promoter using primers that spanned the promoter-transgene junction. This demonstrated that the exogenous NES:GFP promoter was methylated in vHMECs that arose from GFP+ cells, but was not methylated in early-passage GFP+ cultures (Figure 2E). As expected, the endogenous CDKN2a and NES promoters were also methylated in the vHMECs, but not in early-passage cells (Figures 2E and 2F). Importantly, promoter methylation was reversed and expression of both NES and GFP were rescued in the bulk population of cells upon treatment with the DNA methyltransferase inhibitor decitabine (DAC) (Figures 2E–2H). These findings suggest that vHMECs that exhibit NES silencing by DNA methylation can be derived from cells that do not exhibit NES methylation.
To determine how promoter methylation might affect lineage dedifferentiation of HMECs, we compared expression levels of mammary lineage differentiation genes in early-passage HMECs, vHMECs, and vHMECs treated with DAC. As a control, CDKN2a mRNA levels were rescued with DAC treatment, showing relief of DNA methylation at gene promoters (Figure S2B). Treatment of vHMECs with DAC led to re-expression of KRT19 and OXTR, but not PGR or THY1 (Figure 2I). Importantly, both KRT19 and OXTR have CpG islands in their gene promoters, while PGR and THY1 do not. This confirms that de novo DNA methylation is involved in HMEC reprogramming but that it likely coordinates with other epigenetic modifications to fully control dedifferentiation.
To further assess whether vHMECs originate either from rare pre-existing cells or from stochastic epigenetic reprogramming of lineage-committed cells, we used fluctuation analysis, an approach originally developed by Luria and Delbruck to study the emergence of phage-resistant clones in bacterial cultures (Luria, 1951). In fluctuation analysis, a mass culture of cells is split into many smaller bottlenecks and expanded, after which a selection pressure is applied to screen for the number of cells that survive. If the resistant clones arise stochastically during culture, their distribution amongst the bottlenecks will follow a Poisson distribution; however, if they are pre-existing the distribution will be non-Poisson, with some cultures containing no clones and some containing large amounts.
We applied fluctuation analysis to the HMEC system by dividing the mass HMEC culture into small bottlenecks of 5,000 cells each, and counting the number of vHMECs generated by each bottleneck. As a control, HMECs were also cultured en masse until the final passage before variant formation and plated at densities equal to those of the bottleneck cultures (Figure S2C).
Fluctuation analysis revealed that the bottlenecks did not follow a Poisson distribution (Figures S2D–S2F, chi-square p = 0.0034). A large number of bottlenecked cultures contained no vHMECs while others contained a large number of vHMECs (Figure S2F). These data are consistent with the existence of a rare pre-existing vHMEC population within breast tissue, as previously reported (Holst et al., 2003). However, it is inconsistent with previous data and our data that show de novo methylation of CDKN2a and NES occurs during vHMEC outgrowth (Figure 2; Hinshelwood et al., 2009).
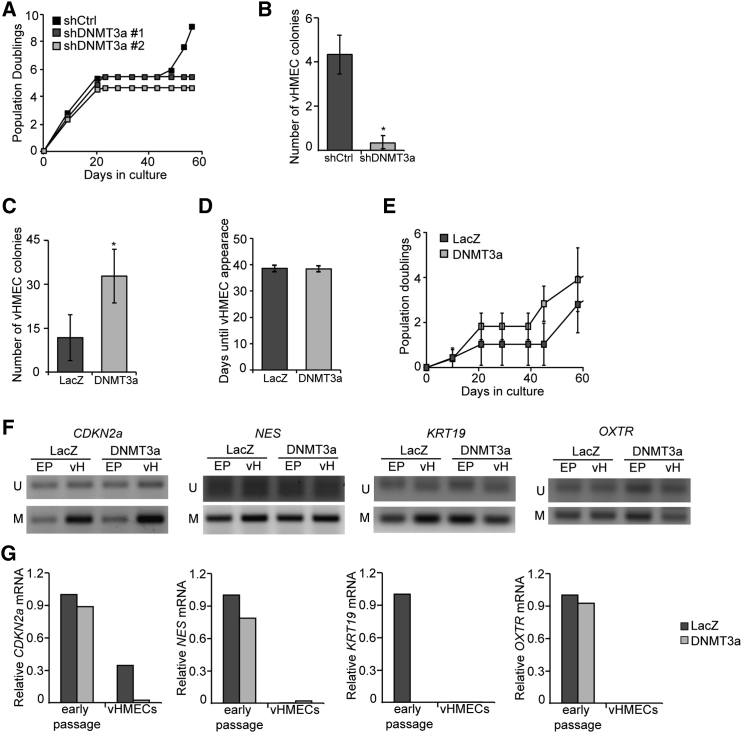
vHMEC Reprogramming Is Partially Controlled by DNMT3a
To reconcile this discrepancy, we reasoned that a rare subpopulation of HMECs residing in breast tissue is capable of giving rise to metaplastic vHMECs, but that this population undergoes epigenetic reprogramming through DNA methylation or other modifications. To test this, we manipulated the level of the de novo DNA methyltransferase, DNMT3a, in HMECs. Strikingly, when DNMT3a was silenced in early-passage HMECs, cells did not bypass senescence or form vHMEC colonies (Figures 3A, 3B, and S3A). Conversely, overexpression of DNMT3a markedly increased the number of vHMEC colonies without decreasing the time to colony formation or affecting cellular proliferation (Figures 3C–3E and S3B). Surprisingly, DNMT3a overexpression did not increase promoter methylation or expression of CDKN2a and NES, but greatly affected both promoter methylation and expression of the mammary lineage gene KRT19 (representative patient samples in Figures 3F, 3G, and S3C). OXTR was also methylated by DNMT3a; however, its expression was unchanged, indicating it may be regulated by additional epigenetic modifications (Figures 3F and 3G). Together, these data indicate that de novo methylation of specific gene promoter regions is a defining epigenetic event in the generation vHMECs from a rare population of HMECs and that loss of mammary lineage differentiation occurs through epigenetic modifications.
Figure 3.
vHMEC Reprogramming Is Partially Controlled by DNMT3a
(A) Growth curve showing cumulative population doublings over time in cultures with stable depletion of DNMT3a (shDNMT3a) by two different lentiviral short hairpins versus shCtrl.
(B) Quantitative analysis of the number of vHMEC colonies formed by cultures lacking DNMT3a (shDNMT3a) versus the firefly luciferase control (shCtrl), n = 3.
(C) Quantitative analysis of the number of vHMEC colonies formed by DNMT3a-overexpressing cultures versus the LacZ control, n = 4.
(D) Latency of vHMEC colony formation, defined as the number of days required for vHMECs to appear, in LacZ versus DNMT3a overexpressing cultures, n = 4.
(E) Growth curve showing cumulative population doublings over time in cultures with LacZ or DNMT3a overexpression, n = 3.
(F) Methylation-specific PCR of gene promoters in early-passage HMECs (EP) versus vHMECs (vH) expressing LacZ or DNMT3a.
(G) Representative image of qPCR analysis of expression of DNA methylation-regulated genes in early HMECs versus vHMECs expressing LacZ or DNMT3a.
In all panels, error bars indicate the mean ± SEM and replicates are individual patient samples. ∗p < 0.05, Student's t test.
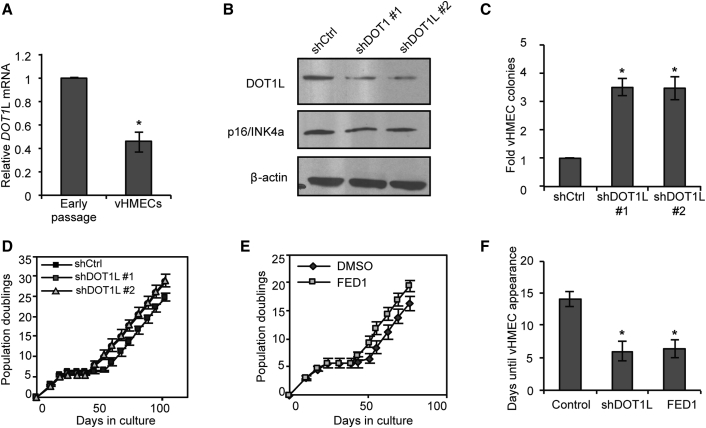
DOT1L Inhibition Enhances the Efficiency of vHMEC Cell Reprogramming
Induction and maintenance of pluripotency is dependent on multiple epigenetic processes including histone methylation, DNA methylation, and chromatin remodeling (Papp and Plath, 2013). To identify additional epigenetic processes that might play a role in imparting a primitive phenotype on lineage-committed HMECs, we profiled the expression of 184 chromatin-modifying factors in matched pre- and post-stasis HMECs using qPCR arrays. From this collection of chromatin modifiers, we identified four factors that exhibited differential expression in vHMECs compared with early-passage precursors after validation (DOT1L, KDM6B, SMYD3, and PRMT8) (Figure S4A).
Of these factors we focused on DOT1L, a histone lysine methyltransferase solely responsible for mono-, di-, and tri-methylation of Lys79 of histone H3 (Nguyen and Zhang, 2011). This chromatin mark is enriched in actively transcribed promoters and gene bodies and plays an important role in transcriptional elongation (Feng et al., 2002, Yao et al., 2011). Notably, DOT1L and H3K79 methylation were recently identified as an epigenetic barrier to iPSC reprogramming by maintaining expression of lineage-specific genes, and inhibition of DOT1L enhances the efficiency of iPSC reprogramming by blocking its anti-silencer effect on these genes (Onder et al., 2012). Given the similarities between iPSC reprogramming and vHMEC formation, we wondered whether DOT1L played a similar role in the generation of vHMECs. Since DOT1L is downregulated in vHMECs versus early-passage HMECs in a medium-dependent manner (Figures 4A and S4B), we wondered whether depletion of DOT1L would enhance the efficiency of vHMEC reprogramming. Interestingly, stable knockdown of DOT1L in freshly dissociated HMECs significantly increased the frequency of vHMEC colony formation (Figures 4B and 4C). Moreover, DOT1L knockdown significantly shortened the latency period between growth arrest and appearance of vHMEC colonies (Figures 4D and 4F). A similar effect was observed when cells were treated with FED1, a specific chemical inhibitor of DOT1L (Yao et al., 2011) (Figures 4E and 4F). Importantly, DOT1L depletion did not affect CDKN2a expression or growth rate in early-passage cultures, suggesting that shDOT1L cells do not just bypass stasis entirely.
Figure 4.
DOT1L Loss Enhances the Efficiency of Reprogramming
(A) qPCR of DOT1L mRNA levels in early-passage HMECs versus vHMECs, n = 5.
(B) Western blot of DOT1L and p16/INK4a protein levels following stable knockdown of DOT1L (shDOT1L) in early-passage HMECs.
(C) Number of vHMEC colonies generated by shDOT1L or shCtrl control cultures, shown relative to shCtrl, n = 3.
(D and E) Growth curves showing cumulative population doublings over time in cultures with (D) DOT1L depletion (shDOT1L) or (E) chemical inhibition with FED1, n = 3.
(F) Latency of vHMEC colony formation, defined as the number of days required after growth arrest for vHMECs to appear, in cells with DOT1L depletion (shDOT1L) or FED1 treatment, n = 3.
In all panels, error bars indicate the mean ± SEM and replicates are individual patient samples. ∗p < 0.05, Student's t test.
Histone Methylation by DOT1L Is Necessary to Maintain HMEC Differentiation
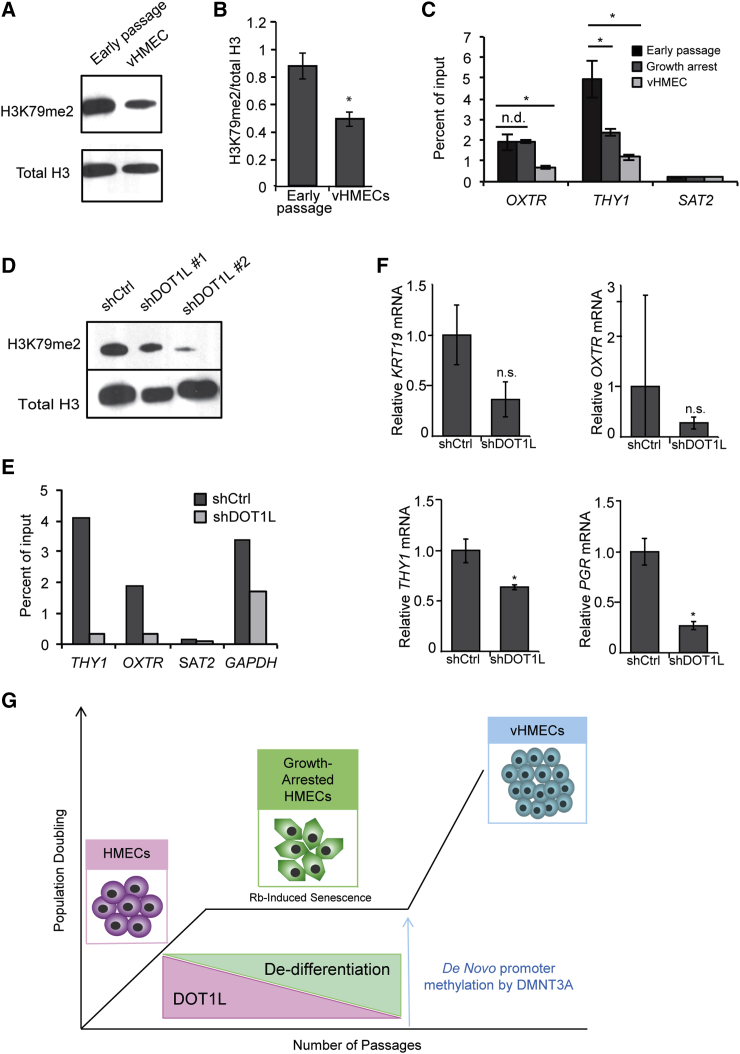
As in iPSC-reprogrammed fibroblasts, H3K79 may play an important role in preventing silencing of differentiation genes, and therefore in vHMEC reprogramming. As DOT1L is the sole known enzyme responsible for H3K79 methylation, a mark of actively transcribed chromatin (Feng et al., 2002), we wondered whether alterations in this mark contribute to the silencing of mammary-specific transcripts. We examined the total levels of H3K79me2 in vHMECs, as well as the levels of H3K79me2 at the promoters of genes silenced during vHMEC progression. Interestingly, we observed a decrease in global H3K79me2 levels in vHMECs compared with early-passage HMECs, correlating with the level of DOT1L (Figures 5A and 5B). In addition, chromatin immunoprecipitation (ChIP) assays of H3K79me2 at the promoters of THY1 and OXTR revealed decreased enrichment of H3K79me2 around the transcriptional start sites of mammary lineage genes in vHMECs compared with early-passage cells (Figure 5C). H3K79me2 levels were also moderately decreased in growth-arrested HMECs at the THY1 locus, though not at OXTR. Therefore, at both a global and local level, H3K79 methylation is progressively depleted as cells lose lineage commitment and become dedifferentiated.
Figure 5.
Histone Methylation by DOT1L Is Necessary for Maintaining HMEC Differentiation
(A and B) Representative western blot (A) and densitometric quantitation (B) of H3K79me2 levels in early-passage cultures versus vHMECs, n = 3.
(C) Chromatin immunoprecipitation (ChIP) of H3K79me2 levels in early-passage HMECs, growth-arrested cultures, or vHMECs (shown as the percent of input normalized to the total H3 level). SAT2 is a negative control, n = 3.
(D) Western blot of H3K79me2 levels in shDOT1L cultures versus shCtrl.
(E) ChIP analysis of H3K79me2 in shDOT1L versus shCtrl HMECs at early-passage (shown as the percent of input normalized to the total H3 level).
(F) qPCR analysis of mammary lineage gene expression in early-passage shDOT1L versus shCtrl HMECs analyzed 1 week after DOT1L depletion by shRNA, n = 3.
(G) Graphical depiction of the mechanism of HMEC reprogramming showing coordinated alterations in DNA and histone methylation to promote vHMEC formation.
In all panels, error bars indicate the mean ± SEM and replicates are individual patient samples. n.d., not determined; ∗p < 0.05, Student's t test.
We also examined whether the decrease in H3K79me2 could be induced by loss of DOT1L in early-passage HMECs. Within 7 days, DOT1L knockdown by shRNA led to decreased H3K79me2 levels in HMECs, both globally (Figure 5D) and locally at the THY1 and OXTR promoters (Figure 5E). Collectively, these data show that DOT1L is necessary to maintain di-methylation of H3K79 at promoters of lineage-related genes. Consistent with changes in H3K79me2 levels, there was reduced expression of lineage-committed genes upon knockdown of DOT1L (Figure 5F), suggesting that DOT1L depletion negatively regulates mammary epithelial differentiation. Interestingly, DOT1L depletion significantly reduced only THY1 and PGR, not KRT19 or OXTR. This is consistent with our DNA methylation results that both KRT19 and OXTR are also regulated by promoter methylation (Figure 2I), emphasizing that multiple epigenetic mechanisms collaborate to control dedifferentiation.
As DOT1L inhibition accelerated the dedifferentiation and outgrowth of vHMECs, we examined whether exogenous overexpression of DOT1L in early-passage HMECs might prevent or slow vHMEC reprogramming. However, despite robust overexpression of DOT1L and dramatically increased global levels of H3K79me2 (Figure S5A), the frequency of vHMEC outgrowth was unchanged (Figure S5B). Therefore, DOT1L is necessary but not sufficient to prevent epigenetic reprogramming, and other factors likely cooperate with DOT1L to maintain mammary lineage commitment in HMEC cultures (Figure 5G).
Discussion
The surprising plasticity of adult somatic cells has become an unanticipated theme of contemporary developmental biology, but the study of plasticity has remained largely descriptive and the precise mechanisms of dedifferentiation remain elusive. vHMECs are an interesting example of plasticity, as these cells arise from lineage-committed mammary epithelial cells that reprogram to a multipotent surface-ectoderm-like state capable of generating stratified epidermis (Keller et al., 2012). Hence, the intent of this study was to leverage the vHMEC model system to study loss of lineage commitment in real time, something that has been difficult to achieve with other systems.
Using this approach, we uncovered several important findings. First, our observations reinforce the importance of tissue architecture and stromal influences in the regulation of both cell growth and differentiation and underscore the highly plastic nature of somatic cells even without genetic mutations. When removed from instructive signals of the tissue microenvironment, lineage-committed mammary epithelial cells can spontaneously reprogram to a primitive state. Although the relevant microenvironmental factors remain to be determined, the requirement of specific signals for maintenance of lineage commitment is illustrated by the finding that HMECs grown in serum do not lose expression of lineage-specific genes. Presently it is unclear whether the signals that maintain lineage commitment of HMECs are secreted molecules, structural/mechanical cues from the tissue architecture, hetero-/homotypic interactions with other mammary epithelial cells in vivo, or some combination of the above. However, the fact that vHMECs did not form in SCM, nor did the cells dedifferentiate in this growth medium, suggests that hormonal and/or paracrine signaling mechanisms may be essential for maintaining lineage commitment.
Second, our data suggest that vHMECs originate from epigenetic reprogramming of non-vHMECs. vHMEC outgrowth involves de novo DNA methylation and gene silencing of both cell-cycle regulators (i.e., CDKN2a) and lineage-specific genes (NES, THY1, OXTR, PGR, and KRT19). Previously, it was unknown whether vHMECs arose from a rare subpopulation of breast epithelial cells that had already silenced these genes or from lineage-committed precursors by epigenetic reprograming. Here, we show by stable introduction of the NES promoter into pre-stasis cells that a de novo methylation event occurs during the transition from lineage-committed HMECs to vHMECs. Because vHMECs form only rarely in the absence of the de novo DNA methyltransferase, DNMT3a, it is very likely that vHMEC outgrowth is dependent on de novo DNA methylation of CDKN2a and other genes. As reported by others, and demonstrated here by silencing of CDKN2a by shRNA, CDKN2a silencing is the only event that is required for HMECs to escape proliferative arrest (Foster and Galloway, 1996). However, it has been shown that a large number of other genes are methylated and/or differentially expressed in vHMECs, suggesting that DNA methylation is not limited to the CDKN2a locus and that DNA methylation-independent mechanisms of gene silencing are also at play (Novak et al., 2009). Therefore, it is likely that epigenetic reprogramming of lineage-committed cells involves a coordinated process of DNA methylation and histone modification (Hinshelwood et al., 2009). Indeed, the four mammary lineage differentiation genes investigated in this study were downregulated through different mechanisms that involved a combination of both these processes. However, the temporal nature of reprogramming is not entirely clear from our experiments. While the transcriptional silencing of lineage-specific genes appears to be gradual over the period of tissue culture and likely is mediated by the loss of DOT1L, it is unclear whether all of the DNA methylation events occur as a single coordinated step or if HMECs gradually accumulate epigenetic modifications, such as CDKN2a methylation early, followed by other DNA methylation and histone alterations until escape from senescence as dedifferentiated variant colonies.
Third, we implicate a new mechanism by which epigenetic reprogramming is controlled in breast tissue: H3K79 methylation by DOT1L. We found that DOT1L is necessary to maintain lineage commitment in HMECs, significantly accelerating vHMEC outgrowth when it is depleted. This acceleration of vHMEC reprogramming by DOT1L loss is correlated with reduced levels of H3K79me2 and can be mimicked by blocking the enzymatic activity of DOT1L by FED1. Therefore, it is likely that the erasure of active H3K79me2 marks over time is an important precursor to the silencing of lineage genes that accompanies vHMEC outgrowth. How H3K79me2 reduction leads to escape from growth arrest remains an unanswered question; however, it is likely through regulation of CDKN2a. One possibility is that the H3K79me2 mark may protect against DNA methylation of CDKN2a and other actively transcribed genes, a property that has been attributed to other active histone marks such as trimethyl-H3K4 (Li and Liu, 2004). While there is no evidence as yet of a specific link between H3K79me2 and DNA methylation, future studies in the vHMEC system should address whether H3K79me2 can prevent the recruitment of DNMT3a to genes that are methylated in vHMECs.
Lastly, how reprogramming relates to in vivo breast biology is currently unknown. Recent work by Tlsty's group demonstrated the existence of rare cells in human breast tissue that show CDKN2a methylation, lack THY1 expression, and demonstrate a pluripotent phenotype (Holst et al., 2003, Roy et al., 2013). However, while in vivo CDKN2a-negative cells of the breast share many features with vHMECs, they cannot be equated solely on the basis of similar immunophenotypes. Furthermore, even if they are equivalent, these cells may have different origins. Whether a vHMEC reprogramming-like event can occur in vivo therefore remains an open question. In addition, while an attractive hypothesis, it is not clear whether dedifferentiation or reprogramming of HMECs may play a role in the generation of metaplastic breast cancer. While CDKN2a DNA methylation is common in breast cancer, it is not specifically linked with poorly differentiated or metaplastic tumors (Herman et al., 1995, Hui et al., 2000). We previously reported that oncogenic transformation of primary CD10+ basal HMECs generates metaplastic tumors, similar to those generated by transformation of vHMECs, and these cells are also the source of adherent HMEC cultures that spontaneously generate vHMECs in culture (Keller et al., 2012). Therefore, it is possible that metaplastic breast cancers in xenografts and in human patients might result from dedifferentiation of CD10+ basal cells during oncogenic transformation, as previously suggested (Keller et al., 2012). Consistent with this notion, DOT1L levels are decreased in the metaplastic xenografts compared with other tumors (data not shown).
While there is more research needed to link vHMECs arising in cell culture with the in vivo equivalent in the mammary gland or breast cancer, this work sheds important light on the epigenetic basis of cellular plasticity, which could prove useful in understanding cellular reprogramming in other systems.
Experimental Procedures
Isolation of Primary HMECs
Reduction mammoplasty tissues were obtained from the Department of Pathology at Tufts Medical Center in full compliance with Institutional Research Board (IRB) guidelines. As the samples were de-identified prior to receipt, the requirement for written consent was waived by the IRB. Tissue processing to single cells was carried out as previously described (Keller et al., 2012). In short, tissue was minced into ∼1-mm pieces, resuspended in digestion medium (DMEM/F12 [Corning], 20 ng/mL human epidermal growth factor [hEGF], 10 μg/mL human insulin, 500 ng/mL hydrocortisone, 1.5 mg/mL collagenase, and 125 units/mL hyaluronidase [Sigma]) and incubated overnight at 37°C to release organoids. Stromal cells were depleted by plating on a tissue culture dish for 2 hr at 37°C. Following incubation, non-adherent organoids were harvested, washed with PBS and red blood cell lysis buffer (Sigma), further digested in 0.05% trypsin/EDTA, and filtered through 40-μm mesh prior to plating.
Cell Culture
To initiate primary HMEC cultures, 106 dissociated cells were plated on 10-cm tissue culture plates. Unless otherwise stated, all cultures were grown in MEGM (Lonza) with weekly subculture. SCM consisted of DMEM/F12 with 20 ng/mL recombinant hEGF, 10 μg/mL recombinant human insulin, and 500 ng/mL hydrocortisone (Sigma) with penicillin/streptomycin (Gibco). For methylation experiments, cells were treated with 500 nM decitabine (DAC) or DMSO for 72 hr, refreshing the drug daily.
For growth curves, cells were subcultured weekly with 0.05% trypsin-EDTA and 500,000 of the recovered cells were replated. At each passage, the total number of recovered cells was counted and the population doublings were computed as log2(number of recovered cells/number of plated cells). After growth arrest, the culture was maintained without passaging until vHMEC colonies were seen by microscopic inspection. Following vHMEC appearance, cultures were maintained with weekly subculture as described above. Quantitation of vHMEC colony formation was performed by daily microscopic inspection.
Fluctuation Analysis
For fluctuation analysis, freshly dissociated HMECs were plated either as a mass culture of 100,000 cells or as 20 bottleneck cultures of 5,000 cells each. Cells were passaged until proliferation slowed, at which point (1) 250,000 cells from each bottleneck were plated on a 10-cm dish and (2) the mass culture was also divided into 20 replicates of 250,000 cells each, to assess vHMEC colony formation. After 30 days, the number of vHMECs in each dish was visually determined by crystal violet staining and microscopy.
Retrovirus/Lentivirus Production and Generation of Stable Cell Lines
Packaging of replication-defective lentivirus and retrovirus was performed as previously described (Keller et al., 2012). To generate lentivirus, 293T cells were cotransfected with the vesicular stomatitis virus envelope (pCMV-VSV-G), the gag-pol genes (pCMV-Δ8.2Δvpr), and the appropriate lentiviral expression or shRNA construct in a 1:2:3 ratio, respectively. For generation of retrovirus, 293T cells were cotransfected with the pC10-A packaging plasmid with the appropriate retroviral expression or shRNA vector at a 1:1 ratio. Transfections were performed using Fugene 6 transfection reagent according to the manufacturer's protocol (Promega). Viral supernatants were harvested from the transfected cells 48 and 72 hr post transfection and filtered through a 0.45-μm syringe filter.
To generate stable lines of primary HMECs expressing exogenous transgenes or shRNAs, viral supernatant was applied to the cells 96 hr after initial dissociation and plating. Cells were infected with viral supernatants for 4 hr with 5 μg/mL protamine sulfate (Sigma), fed with normal growth medium, and a second infection was performed 24 hr later. Two days after transfection, cells were selected with 2 μg/mL puromycin (Sigma) or 10 μg/mL blasticidin (Life Technologies) and maintained in selective media until complete death of a mock-infected control plate, then MEGM for long-term growth.
Lentiviral/Retroviral Vectors
Retroviral DOT1L and DNMT3a shRNA clones, as well as DOT1L cDNA clones, were provided by Tamer S. Onder. The lentiviral CDKN2a and luciferase shRNAs for CDKN2a knockdown experiments were obtained from Addgene (#22264 and #20958).
To generate the NES:GFP construct, the proximal 1,000 base pairs upstream of the NES transcriptional start site was amplified by PCR from a bacterial artificial chromosome template and cloned into the CSCG vector upstream of the GFP coding sequence. The resulting NES:GFP cassette was then PCR amplified from the CSCG template with flanking attB sites added during PCR. The attB-tailed NES:GFP cassette was first cloned into pDONR221, then transferred to the promoterless pLenti X1-puro backbone (Addgene, #17297) using the Gateway BP Clonase II enzyme kit (Life Technologies) according to the manufacturer's instructions.
qRT-PCR
For all qPCR experiments, total RNA was extracted from cells using the RNeasy Mini Kit (Qiagen). cDNA was generated from 1 μg of RNA with the iScript cDNA synthesis kit (Bio-Rad) according to the manufacturer's instructions. qPCR was performed with 50 ng of cDNA using SYBR Green Supermix (Bio-Rad) with a primer concentration of 250 nM each (forward and reverse). Three-step amplification and quantitation was performed on a CFX96 Real-time Thermal Cycler (Bio-Rad) using the following program: initial denaturation at 95°C for 3 min, followed by 40 cycles of 95°C denaturation for 30 s, annealing at 60°C 30 s, and extension at 72°C for 30 s. Threshold cycle (Ct) values were converted to relative gene expression values using the 2−ΔΔCt method. Primer sequences are provided in Supplemental Experimental Procedures.
qPCR Arrays
For analysis of chromatin-modifying factors, we used the Epigenetic Chromatin Modification Enzymes PCR Array (Qiagen/SABiosciences, PAHS-085Z). For these arrays, we used the associated RT2 First Strand cDNA kit (Qiagen #330401) and the RT2 qPCR Mastermix (Qiagen #330522) for cDNA synthesis and qPCR, respectively, according to the manufacturer's instructions. Gene expression values were normalized to the mean of five housekeeping genes present on each array. Hits were validated by qPCR.
Methylation-Specific PCR
For methylation analysis of gene promoters, genomic DNA was extracted from the cells using the PureLink Genomic DNA Mini Kit (Life Technologies). Cytosine deamination was performed using the MethylCode bisulfite conversion kit (Life Technologies) according to the manufacturer's instructions. The bisulfite-converted genomic DNA was then used as the template for a genomic PCR reaction using primers specific either for the methylated (unconverted) or unmethylated (C → U converted) CpG islands near the transcriptional start sites of NES, NES:GFP, Krt19, OXTR, and CDKN2a. The PCR products were analyzed by gel electrophoresis in 2% agarose. Primer sequences are provided in Supplemental Experimental Procedures.
Western Blotting
To evaluate total levels of H3K79me2 and H3, we performed western blot analysis on acid-extracted nuclear lysates. Cell membranes were lysed by suspension in PBS/0.05% Triton X-100, 2 mM PMSF, and 0.02% sodium azide at a density of 107/mL for 10 min on ice. Nuclei were pelleted by centrifugation at 2,000 rpm for 10 min at 4°C, washed in PBS/PMSF/NaN3, resuspended in 0.2 N HCl, acid-extracted overnight at 4°C with rotation, and centrifuged at 2,000 rpm for 10 min, and the acid-insoluble pellet was discarded. For all other western blotting, protein lysates were prepared by resuspension of harvested cell pellets in RIPA buffer (50 mM Tris, 150 mM NaCl, 2 mM EDTA, 1% NP-40, and 0.1% SDS [pH 7.4]) on ice for 20 min with occasional agitation, followed by passage five times through a 20-gauge needle.
Protein samples were subjected to SDS-PAGE and transferred to either 0.45 μm or 0.2 μm nitrocellulose (Bio-Rad) for 1 hr at 100 V using the Criterion system (Bio-Rad) in 25 mM Tris, 200 mM glycine buffer with 20% methanol. Following transfer, membranes were blocked in 2% milk in Tris-buffered saline (TBS; 50 mM Tris, 150 mM NaCl) with 0.1% Tween 20 (TBST) for 1 hr at room temperature, followed by overnight incubation in the blocking buffer and primary antibody at 4°C. Membranes were washed with TBST, incubated with horseradish peroxidase-conjugated secondary antibodies for 1 hr at room temperature, washed in TBST, and developed with either West Pico or West Dura ECL substrates (Pierce) according to the manufacturer's protocol. Antibody information is provided in Supplemental Experimental Procedures.
Flow Cytometry
For analysis of GFP positivity of NES:GFP cultures, cells were trypsinized, washed twice with FACS buffer (2% FBS/PBS), resuspended at 106 cells/mL, and either sorted for GFP+ cells or analyzed on a flow cytometry to determine levels of GFP expression.
Chromatin Immunoprecipitation
ChIP was performed using the MagnaChIP A kit (Millipore) according to the manufacturer's recommended protocol. In brief, two 15-cm of near-confluent HMECs were crosslinked with 1% formaldehyde for 10 min at room temperature, followed by quenching with glycine for 5 min at room temperature. The crosslinked chromatin was sheared by sonication in a Misonix 3000 tip sonicator (power level 3.0, five cycles of 20 s “on,” 60 s “off”). 10% of the sheared crosslinked chromatin was used in each immunoprecipitation.
Antibody incubation and elution steps were performed using protein A magnetic beads according to manufacturer's instructions. Immunoprecipitated chromatin was quantified by genomic qPCR, with the following parameters: initial denaturation for 3 min at 95°C, then 40 cycles of denaturation at 95°C for 10 s, annealing at 60°C for 30 s, and extension at 72°C for 30 s. Cycle thresholds were converted to linear quantities using the 2−ΔCt formula and expressed as a percentage of the original input. The antibodies and primers used for ChIP are provided in Supplemental Experimental Procedures.
Gene Ontology Analysis
For transcriptomic GO analysis of HMECs during culture, the dataset GEO: GSE16058 was downloaded from the GEO website (Garbe et al., 2009).
A list of differentially expressed genes between early-passage HMECs, growth-arrested HMECs, and vHMECs was generated using the GEO2R web tool, and the top 200 genes with a false discovery rate (FDR) < 0.05 were used for GO analysis. Enrichment of GO terms was performed by hypergeometric overlap using the Broad Institute's online tool against the GO Biological Process gene sets. The enriched gene sets were selected from the top 20 most highly enriched GO terms.
Author Contributions
Conceptualization, C.K.; Methodology, J.L.B., A.S., M.S., A.W.-C., W.Z., P.J.K., J.M., J.B., T.O., and C.K.; Investigation, J.L.B., A.S., M.S., A.W.-C., W.Z., P.J.K., J.M., J.B., T.O., and C.K.; Acquisition of data, J.L.B., A.S., M.S., A.W.-C., W.Z., P.J.K., J.M., J.B., T.O., and C.K.; Analysis and interpretation of data, J.L.B., A.S., M.S., A.W.-C., W.Z., P.J.K., J.M., J.B., T.O., and C.K.; Writing, J.L.B., A.S., and C.K; Funding Acquisition, C.K.
Acknowledgments
We acknowledge Lisa Arendt and Ravi Subramanian for technical and scientific assistance and advice. We thank Dr. Stephen Naber for assistance with the procurement of human reduction mammoplasty tissue samples and Kayla Gross for careful reading and editing of the manuscript.
This work was supported by funding from the Raymond & Beverly Sackler Convergence Laboratory and grants from ArtBeCAUSE, the Breast Cancer Research Foundation, and the NIH (NICDH HD073035 and NCI CA170851). J.L.B. is supported by award K12GM074869 from the NIGMS.
Published: August 3, 2017
Footnotes
Supplemental Information includes Supplemental Experimental Procedures and five figures and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2017.06.019.
Supplemental Information
References
- Bean G.R., Ibarra Drendall C., Goldenberg V.K., Baker J.C., Jr., Troch M.M., Paisie C., Wilke L.G., Yee L., Marcom P.K., Kimler B.F. Hypermethylation of the breast cancer-associated gene 1 promoter does not predict cytologic atypia or correlate with surrogate end points of breast cancer risk. Cancer Epidemiol. Biomarkers Prev. 2007;16:50–56. doi: 10.1158/1055-9965.EPI-06-0598. [DOI] [PubMed] [Google Scholar]
- Brenner A.J., Stampfer M.R., Aldaz C.M. Increased CDKN2a expression with first senescence arrest in human mammary epithelial cells and extended growth capacity with CDKN2a inactivation. Oncogene. 1998;17:199–205. doi: 10.1038/sj.onc.1201919. [DOI] [PubMed] [Google Scholar]
- Cregan M.D., Fan Y., Appelbee A., Brown M.L., Klopcic B., Koppen J., Mitoulas L.R., Piper K.M., Choolani M.A., Chong Y.S. Identification of nestin-positive putative mammary stem cells in human breastmilk. Cell Tissue Res. 2007;329:129–136. doi: 10.1007/s00441-007-0390-x. [DOI] [PubMed] [Google Scholar]
- Feng Q., Wang H., Ng H.H., Erdjument-Bromage H., Tempst P., Struhl K., Zhang Y. Methylation of H3-lysine 79 is mediated by a new family of HMTases without a SET domain. Curr. Biol. 2002;12:1052–1058. doi: 10.1016/s0960-9822(02)00901-6. [DOI] [PubMed] [Google Scholar]
- Foster S.A., Galloway D.A. Human papillomavirus type 16 E7 alleviates a proliferation block in early passage human mammary epithelial cells. Oncogene. 1996;12:1773–1779. [PubMed] [Google Scholar]
- Garbe J.C., Bhattacharya S., Merchant B., Bassett E., Swisshelm K., Feiler H.S., Wyrobek A.J., Stampfer M.R. Molecular distinctions between stasis and telomere attrition senescence barriers shown by long-term culture of normal human mammary epithelial cells. Cancer Res. 2009;69:7557–7568. doi: 10.1158/0008-5472.CAN-09-0270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman J.G., Merlo A., Mao L., Lapidus R.G., Issa J.P., Davidson N.E., Sidransky D., Baylin S.B. Inactivation of the CDKN2/CDKN2a/MTS1 gene is frequently associated with aberrant DNA methylation in all common human cancers. Cancer Res. 1995;55:4525–4530. [PubMed] [Google Scholar]
- Hinshelwood R.A., Melki J.R., Huschtscha L.I., Paul C., Song J.Z., Stirzaker C., Reddel R.R., Clark S.J. Aberrant de novo methylation of the CDKN2aINK4A CpG island is initiated post gene silencing in association with chromatin remodeling and mimics nucleosome positioning. Hum. Mol. Genet. 2009;18:3098–3109. doi: 10.1093/hmg/ddp251. [DOI] [PubMed] [Google Scholar]
- Holst C.R., Nuovo G.J., Esteller M., Chew K., Baylin S.B., Herman J.G., Tlsty T.D. Methylation of CDKN2a(INK4a) promoters occurs in vivo in histologically normal human mammary epithelia. Cancer Res. 2003;63:1596–1601. [PubMed] [Google Scholar]
- Hui R., Macmillan R.D., Kenny F.S., Musgrove E.A., Blamey R.W., Nicholson R.I., Robertson J.F., Sutherland R.L. INK4a gene expression and methylation in primary breast cancer: overexpression of CDKN2aINK4a messenger RNA is a marker of poor prognosis. Clin. Cancer Res. 2000;6:2777–2787. [PubMed] [Google Scholar]
- Huschtscha L.I., Noble J.R., Neumann A.A., Moy E.L., Barry P., Melki J.R., Clark S.J., Reddel R.R. Loss of CDKN2aINK4 expression by methylation is associated with lifespan extension of human mammary epithelial cells. Cancer Res. 1998;58:3508–3512. [PubMed] [Google Scholar]
- Keller P.J., Arendt L.M., Skibinski A., Logvinenko T., Klebba I., Dong S., Smith A.E., Prat A., Perou C.M., Gilmore H. Defining the cellular precursors to human breast cancer. Proc. Natl. Acad. Sci. USA. 2012;109:2772–2777. doi: 10.1073/pnas.1017626108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Cherukuri P., Li N., Cowling V., Spinella M., Cole M., Godwin A.K., Wells W., DiRenzo J. Nestin is expressed in the basal/myoepithelial layer of the mammary gland and is a selective marker of basal epithelial breast tumors. Cancer Res. 2007;67:501–510. doi: 10.1158/0008-5472.CAN-05-4571. [DOI] [PubMed] [Google Scholar]
- Li J.X., Liu H.L. The relationship of DNA methylation and histone methylation. Yi Chuan. 2004;26:267–270. [PubMed] [Google Scholar]
- Locke W.J., Clark S.J. Epigenome remodelling in breast cancer: insights from an early in vitro model of carcinogenesis. Breast Cancer Res. 2012;14:215. doi: 10.1186/bcr3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke W.J., Zotenko E., Stirzaker C., Robinson M.D., Hinshelwood R.A., Stone A., Reddel R.R., Huschtscha L.I., Clark S.J. Coordinated epigenetic remodeling of transcriptional networks occurs during early breast carcinogenesis. Clin. Epigenetics. 2015;7:52. doi: 10.1186/s13148-015-0086-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luria S.E. The frequency distribution of spontaneous bacteriophage mutants as evidence for the exponential rate of phage reproduction. Cold Spring Harb. Symp. Quant. Biol. 1951;16:463–470. doi: 10.1101/sqb.1951.016.01.033. [DOI] [PubMed] [Google Scholar]
- Macias H., Hinck L. Mammary gland development. Wiley Interdiscip. Rev. Dev. Biol. 2012;1:533–557. doi: 10.1002/wdev.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen A.T., Zhang Y. The diverse functions of Dot1 and H3K79 methylation. Genes Dev. 2011;25:1345–1358. doi: 10.1101/gad.2057811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak P., Jensen T.J., Garbe J.C., Stampfer M.R., Futscher B.W. Stepwise DNA methylation changes are linked to escape from defined proliferation barriers and mammary epithelial cell immortalization. Cancer Res. 2009;69:5251–5258. doi: 10.1158/0008-5472.CAN-08-4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onder T.T., Kara N., Cherry A., Sinha A.U., Zhu N., Bernt K.M., Cahan P., Marcarci B.O., Unternaehrer J., Gupta P.B. Chromatin-modifying enzymes as modulators of reprogramming. Nature. 2012;483:598–602. doi: 10.1038/nature10953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papp B., Plath K. Epigenetics of reprogramming to induced pluripotency. Cell. 2013;152:1324–1343. doi: 10.1016/j.cell.2013.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reya T., Morrison S.J., Clarke M.F., Weissman I.L. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- Romanov S.R., Kozakiewicz B.K., Holst C.R., Stampfer M.R., Haupt L.M., Tlsty T.D. Normal human mammary epithelial cells spontaneously escape senescence and acquire genomic changes. Nature. 2001;409:633–637. doi: 10.1038/35054579. [DOI] [PubMed] [Google Scholar]
- Roy S., Gascard P., Dumont N., Zhao J., Pan D., Petrie S., Margeta M., Tlsty T.D. Rare somatic cells from human breast tissue exhibit extensive lineage plasticity. Proc. Natl. Acad. Sci. USA. 2013;110:4598–4603. doi: 10.1073/pnas.1218682110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stampfer M., Hallowes R.C., Hackett A.J. Growth of normal human mammary cells in culture. In Vitro. 1980;16:415–425. doi: 10.1007/BF02618365. [DOI] [PubMed] [Google Scholar]
- Stampfer M.R., Bartley J.C. Induction of transformation and continuous cell lines from normal human mammary epithelial cells after exposure to benzo[a]pyrene. Proc. Natl. Acad. Sci. USA. 1985;82:2394–2398. doi: 10.1073/pnas.82.8.2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K., Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Van Keymeulen A., Rocha A.S., Ousset M., Beck B., Bouvencourt G., Rock J., Sharma N., Dekoninck S., Blanpain C. Distinct stem cells contribute to mammary gland development and maintenance. Nature. 2011;479:189–193. doi: 10.1038/nature10573. [DOI] [PubMed] [Google Scholar]
- Wang W.C., Wu T.T., Chandan V.S., Lohse C.M., Zhang L. Ki-67 and ProExC are useful immunohistochemical markers in esophageal squamous intraepithelial neoplasia. Hum. Pathol. 2011;42:1430–1437. doi: 10.1016/j.humpath.2010.12.009. [DOI] [PubMed] [Google Scholar]
- Watson C.J., Khaled W.T. Mammary development in the embryo and adult: a journey of morphogenesis and commitment. Development. 2008;135:995–1003. doi: 10.1242/dev.005439. [DOI] [PubMed] [Google Scholar]
- Yao Y., Chen P., Diao J., Cheng G., Deng L., Anglin J.L., Prasad B.V., Song Y. Selective inhibitors of histone methyltransferase DOT1L: design, synthesis, and crystallographic studies. J. Am. Chem. Soc. 2011;133:16746–16749. doi: 10.1021/ja206312b. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.