Recently refined methods for cfDNA quantification and detection of mutations have been developed. Results of clinical investigations of the predictive and prognostic value of total cfDNA and plasma mutations in mCRC suggest a strong prognostic value of baseline plasma levels of cfDNA in this setting. This article presents a systematic review and meta‐analysis of the prognostic value of total plasma cfDNA in patients with mCRC treated with chemotherapy and/or targeted agents.
Keywords: Cell‐free DNA plasma, Metastatic colorectal cancer, Prognosis, Meta‐analysis
Abstract
Background.
Circulating DNA can be detected and quantified in the blood of cancer patients and used for detection of tumor‐specific genetic alterations. The clinical utility has been intensively investigated for the past 10 years. The majority of reports focus on analyzing the clinical potential of tumor‐specific mutations, whereas the use of total cell‐free DNA (cfDNA) quantification is somehow controversial and sparsely described in the literature, but holds important clinical information in itself. The purpose of the present report was to present a systematic review and meta‐analysis of the prognostic value of total cfDNA in patients with metastatic colorectal cancer (mCRC) treated with chemotherapy. In addition, we report on the overall performance of cfDNA as source for KRAS mutation detection.
Materials and Methods.
A systematic literature search of PubMed and Embase was performed by two independent investigators. Eligibility criteria were (a) total cfDNA analysis, (b) mCRC, and (c) prognostic value during palliative treatment. The preferred reporting items for systematic reviews and meta‐analyses (PRISMA) guidelines were followed, and meta‐analysis applied on both aggregate data extraction and individual patients’ data.
Results.
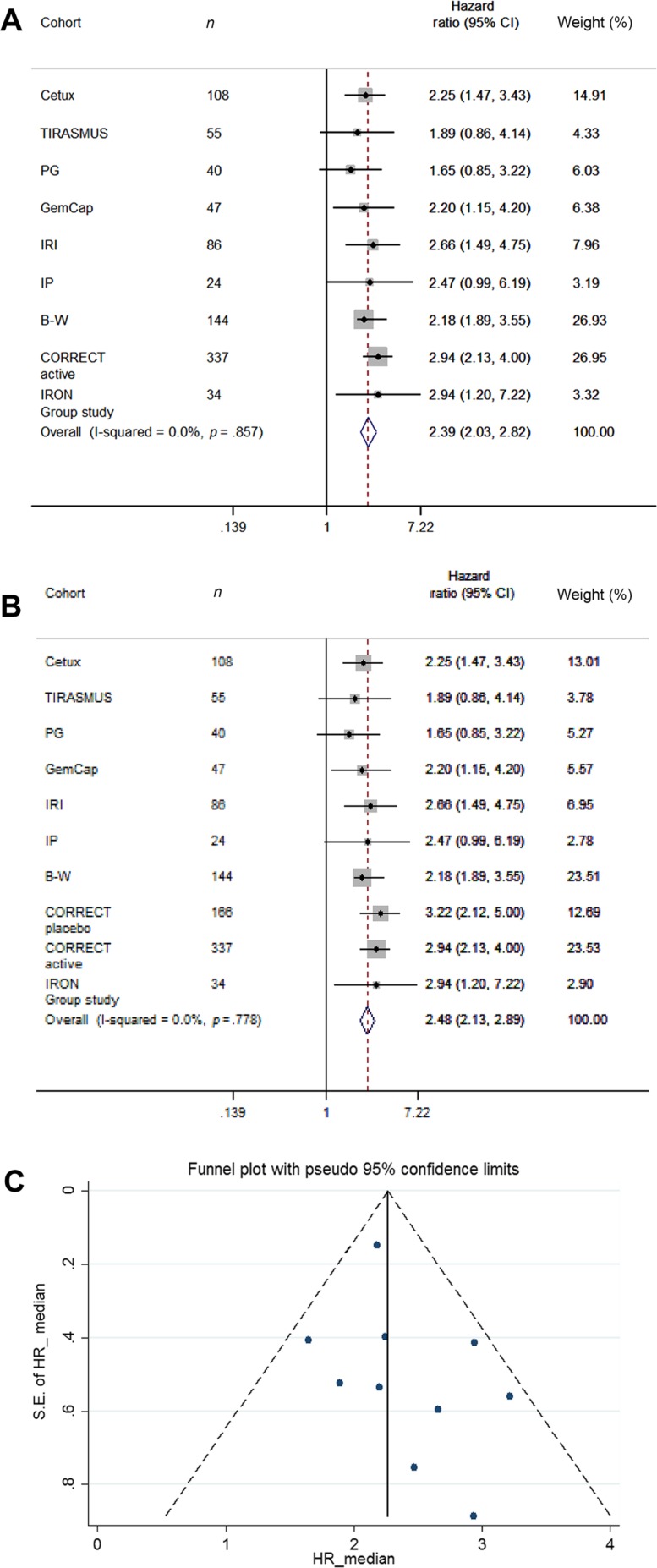
Ten eligible cohorts were identified, including a total of 1,076 patients. Seven studies used quantitative polymerase chain reaction methods, two BEAMing [beads, emulsification, amplification, and magnetics] technology, and one study digital droplet polymerase chain reaction. The baseline levels of cfDNA was similar in the presented studies, and all studies reported a clear prognostic value in favor of patients with lowest levels of baseline cfDNA. A meta‐analysis revealed a combined estimate of favorable overall survival hazard ratio (HR) in patients with levels below the median cfDNA (HR = 2.39, 95% confidence interval 2.03–2.82, p < .0001).
Conclusion.
The total cfDNA levels are high in patients with mCRC and bear strong prognostic information, which should be tested prospectively by using a predefined cut‐off value based on normal values in healthy cohorts. Finally, the potential use of cfDNA for detection of tumor‐specific mutations was emphasized in a large individual patients’ data meta‐analysis.
Implications for Practice.
Reliable prognostic markers could help to guide patients and treating physicians regarding the relevance and choice of systemic therapy. Small fragments of circulating cell‐free DNA (cfDNA) can be measured in a simple blood sample. This report presents the first meta‐analysis of the prognostic value of total cfDNA measurement in patients with metastatic colorectal cancer. Data from 1,076 patients confirmed that patients with the lowest pre‐treatment levels of cfDNA had a significantly higher chance of longer survival than those with higher levels. Cell‐free DNA analysis can also be used for detection of tumor‐specific mutations, and hold potential as a valuable tool in colorectal cancer treatment.
Introduction
Palliative chemotherapy for metastatic colorectal cancer (mCRC) is offered to patients for the purpose of prolonging survival, achieving symptom control, and improving the quality of life. Unfortunately, tumor response and benefit from therapy is not achieved in all patients and, in addition, there is a high risk of unpleasant and potentially life threatening adverse effects [1]. Presently, we are unable to predict a positive effect of a specific drug, whereas a negative predictive value has been demonstrated for tumor‐specific mutations (e.g., KRAS, NRAS, BRAF) to the benefit of adding epidermal growth factor (EGFR) inhibitors to standard cytotoxic therapy in mCRC [2]. However, ground‐breaking improvement of the outcome for this group of patients is only possible if we identify reliable selection markers for optimal palliation in the individual patient. This implies both positive and negative predictive biomarkers and information on individual patient's prognosis.
Small fragments of cell‐free DNA (cfDNA) are present in the circulation, and the concentrations vary in patients with diverse non‐cancerous diseases and are increased in patients with cancer [3]. Several studies have indicated a potential diagnostic, prognostic, and predictive value of cfDNA for various cancer types [4], [5], and recent reports suggest that low levels of cfDNA are associated with a more favorable outcome than high levels [6]. The majority of studies have focused on the molecular characterization of the circulating tumor DNA and potential use for monitoring after surgery and systemic therapy, primarily EGFR inhibition [7], [8]. Only a few groups have directly analyzed the potential prognostic value of total cfDNA quantification in metastatic disease. Furthermore, there is no consensus regarding the optimal methods for DNA purification, quantification, and detection of mutations, and the same applies to laboratory validation and nomenclature. Consequently, due to the heterogeneity between study methods and endpoints, a statistical comparison has not yet been applicable.
Recently refined methods for cfDNA quantification and detection of mutations have been developed. This has allowed for clinical investigations of the predictive and prognostic value of total cfDNA and plasma mutations in mCRC [7], [8], [10], [11], [12], [13], [14], [15], [16], and results have suggested a strong prognostic value of baseline plasma levels of cfDNA in this setting. The purpose of the present report was to present a systematic review and meta‐analysis of the prognostic value of total plasma cfDNA in patients with mCRC treated with chemotherapy and/or targeted agents.
Materials and Methods
Literature Search and Selection of Patient Cohorts
A systematic search of literature was performed by two investigators (KLS and AKB) in September 2015. The databases PubMed and Embase were systematically searched using the search terms “cell free DNA,” “circulating DNA,” and “colorectal cancer.” Various alterations in spelling and abbreviations were applied due to the pronounced heterogeneity of this research field. First, title and abstracts were carefully screened for relevance and duplicates removed. Second, full text was retrieved and assessed for inclusion into analysis. Finally, all references were manually crosschecked for studies missed during the primary search. No limitations were applied in the search. When judged necessary, the authors of studies were contacted for clarification on data. The inclusion criteria for study cohorts were as follows:
Clinical study populations examined for the prognostic value of total cfDNA. Studies only reporting data on circulating tumor‐specific DNA were not eligible
Study cohorts evaluating patients on chemotherapeutic treatment for metastatic colorectal cancer
Presenting an objective statement for the prognosis on survival in relation to total cfDNA
Any disagreements were solved by consensus. Data extraction and synthesis were performed by one investigator (KLS), and validated in two different statistical programs. The literature search was guided by the recently published PRISMA statement [9].
Statistical Analysis
The descriptive comparison of baseline plasma levels of cfDNA between the different cohorts was analyzed with a two‐sided t test and Wilcoxon rank sum test, and a receiver operating curve analysis was used to validate the performance of total cfDNA to discriminate between patients and controls. The meta‐analysis was performed on hazard ratios (HRs), which were calculated by log‐rank test for overall survival (OS) differences in patients with high and low cfDNA plasma concentrations. Calculations were based on the HRs from the original publications including 95% confidence interval (CI), and subsequent back calculation to Log(HR) and standard error (S.E) for overall estimates. The original datasets were used for recalculations in the seven Danish cohorts, using different cut‐offs for cfDNA, including the upper normal limit (UNL; based on the normal cohort as previously published [14]), the upper 75% quartiles, and median levels of cfDNA. Only HRs based on univariate analysis were used, because a multivariate analysis was not applicable in all trials due to low sample sizes. Log(HR) and S.E were entered in statistical software NCSS (NCSS, LLC, Kaysville, UT, https://www.ncss.com/) and analysis validated in comprehensive meta‐analysis (CMA; Biostat, Inc., Englewood, NJ, https://www.meta-analysis.com/), and STATA (StataCorp LLC, College Station, TX). Heterogeneity was assessed using chi‐square test and I2 statistics. An I2 value more than 50% was regarded as evidence of high heterogeneity, and results determined whether the random‐effects or fixed‐effect models should be used to pool the overall results. No subgroup analysis was planned. A funnel plot was generated to visually assess potential publication bias (i.e., an asymmetrical distribution). Results are presented with HRs, upper and lower limits, and p values, and illustrated in forest plots for the individual studies reporting both weighted and pooled effect. Cross‐tabulation was applied to test the concordance of KRAS status in plasma and tumor tissue, and presented as sensitivity, specificity, positive predictive (PPV), and negative predictive (NPV) values. All statistics were performed in the NCSS statistical software and p values <.05 were considered statistically significant.
Results
Search Results
Following the systematic search of literature, a total of 223 (PubMed 88, Embase 135) studies of potential interest were obtained. The majority of studies were excluded based on careful review of title and abstract and only 14 papers were necessarily retrieved in full text. In two cases, authors (Spindler and Sefrioui) supplemented the original paper with unpublished data. Following thorough assessment, a total of 10 patient cohorts were judged eligible for inclusion into the meta‐analysis. The reasons for categorizing the study population as not relevant were as follows: (a) not mCRC studied; (b) no objective statement of the prognostic value of cfDNA; and (c) studies with insufficient laboratory investigations (e.g., only investigating circulating tumor DNA). Furthermore, numerous review papers and doublets between the literature databases were excluded. A flowchart demonstrating the search is presented in supplemental online Figure 1.
Review of Eligible Studies
The identified 10 studies that have presented data on baseline cfDNA and prognosis in mCRC are listed in Table 1. There were no statistically significant differences between the baseline levels of cfDNA in the different cohorts but a significantly higher level in patients compared with healthy controls in the seven studies with available normal cohorts for control (Table 2; supplemental online Fig. 2).
Table 1. Studies presenting data on the prognostic value of total cfDNA in patients treated for metastatic colorectal cancer.

Colorectal cancer patients pooled with other cancers.
Abbreviations: B2M, beta2microglobulin; cfDNA, cell‐free DNA; LINE‐1, long interspersed nuclear element‐1; NA, not available; OS, overall survival; PCR, polymerase chain reaction; PFS, progression free survival; qRT‐PCR, quantitative real time polymerase chain reaction; UNL, upper normal limit; WT, wild type.
Table 2. The differences in baseline cfDNA levels between patients and healthy controls.

Seven samples excluded from final analysis due to positive test for contamination from normal lymphocytes.
Abbreviations: cfDNA, cell‐free DNA; NA, not available.
The first report on the prognostic impact of total cfDNA in mCRC patients treated with chemotherapy was published by the Danish group in 2012 and revealed impaired survival with increasing levels of cfDNA in univariate analysis as well as multivariate validation [10]. These results have been repeatedly validated in six consecutive different Danish trials, comprising a total of 536 patients with mCRC treated with palliative chemotherapy, including 503 with available blood samples [11], [12], [13], [14], [15], [16]. Inclusion criteria for the clinical studies were largely similar. In summary, the majority of patients were male, in good performance status, and with indication for palliative second‐ to fourth‐line therapy. Treatment regimens varied, and are published as independent reports on the experimental phase II studies or translational studies on standard treatment regimes. These included cetuximab + irinotecan [10], [16], irinotecan + panitumumab [15], temsirolimus + irinotecan [11], gemcitabine + capecitabine [13], gemcitabine + pemetrexed [12], and irinotecan alone [14]. Baseline blood samples were drawn prior to start of chemotherapy in all patients, laboratory analysis was performed prospectively, and results were independently interpreted without knowledge of the clinical outcome data. A standard operating procedure for sampling ensured consistency in sample quality, and the concentration of cfDNA was determined by the same in‐house developed quantitative polymerase chain reaction method [10]. Different cut‐offs for survival analysis have been used, including quartiles and median levels of cfDNA, and finally, a predefined UNL (defined as mean+2 SD based on a large group of healthy controls) could be applied to the pooled analysis. Thus, these seven patient cohorts were homogeneous, marker analysis was performed by the same laboratory method, and data extraction was completed with the same effect size measures in all studies.
Recently, three other groups have presented similar data, confirming that higher levels of cfDNA were associated with poor prognosis [17], [18], [19]. Wong and colleagues treated 35 patients with chemotherapy refractory mCRC with regorafenib and investigated circulating DNA in pretreatment plasma samples. Data on total cfDNA quantification were available from 35 patients and analyzed as continuous variable in relation to progression‐free survival. An inverse correlation was reported. Total cfDNA was defined as the sum of mutated and non‐mutated DNA measured as long interspersed nuclear element‐1 (LINE‐1) sequence based assay [17]. Tabernero et al. reported on data from patients treated with regorafenib or placebo according to the large randomized CORRECT trial [18]. Baseline total cfDNA showed strong prognostic impact. In this report, the authors measured the total amount of human genomic DNA isolated from plasma samples using a modified version of human LINE‐1 quantitative real‐time (qRT)‐PCR, and DNA amount was reported in genome equivalents, with one genome equivalent being one haploid human genome weighing 3.3 pg. The median level was 10,653 genomes equivalences (range 1,114–804,450), in line with the broad range and median levels of all the Danish cohorts (Table 2). The clinical trial included 760 patients that were randomly assigned to receive regorafenib or placebo, 505 and 255 patients, respectively. Out of this, a total of 503 patients had mutational analysis done on plasma DNA via Beads, Emulsification, Amplification, and Magnetics (BEAMing) (337 on regorafenib and 166 on placebo), this constituting the single largest dataset to date. The median level of cfDNA was used for cut off in the log‐rank survival analysis. High levels of cfDNA were associated with poor outcome in both groups. Similarly, in a very recent study by the EquIpe de Recherche Onco‐Normande (IRON) group of 34 patients, the baseline cfDNA was measured by a Digital PCR method (QuantStudio 3D Digital PCR System, Thermo Fisher Scientific, Waltham, MA, https://www.thermofisher.com/us/en/home.html) targeting the KRAS gene (total cfDNA was defined as the sum of mutated and non‐mutated DNA) [19]. Patients were treated with different first‐ to fourth‐line chemotherapy regimens for mCRC. Baseline levels were similar to the remaining studies, although different methods were used, and data confirm the above‐mentioned observations of a poor prognosis in patients with the highest levels. Data were reported as log‐rank test of patients divided into four groups according to cfDNA quartiles and based on the 75% quartile cut‐off, similar to the pivotal Danish study. None of the three last studies included information of a predefined normal limit for external cut off.
In conclusion, ten studies were identified comprising 1,076 patients with mCRC, treated with various different first‐ to fourth‐line regimens. Three different methodological approaches were used, but an overall clear correlation between levels of cfDNA and survival was revealed. The applied cut‐offs for analysis also differed in the reports, and the Danish studies included evaluation of a predefined upper normal limit.
Meta‐Analysis of the Prognostic Effect of cfDNA
A meta‐analysis of prognostic effect calculated based on median levels and CI for the Danish studies, the CORRECT trial, and the IRON group study combined is presented in Figure 1. A meta‐analysis of all ten studies was not applicable due to the different cut‐offs used and lack of reported data in the latest study. A test for heterogeneity and funnel plots indicated symmetrical distribution of the studies, which included a total of 1,076 patients (910 who received active therapy). The combined estimate showed favorable HR in patients with levels below the median defined in the individual groups (HR = 2.39, 95% CI 2.03–2.82, p < .0001).
Figure 1.
Meta‐analysis presented as forest plot of HRs for overall survival based on dichotomisation by the median cell‐free DNA levels in the nine actively treated cohorts (A) and including the placebo group from the CORRECT trial (B). (C): Funnel plot of all available cohorts, including the placebo group from the CORRECT trial. Abbreviations: CI, confidence interval; HR, hazard ratio; S.E., standard error.
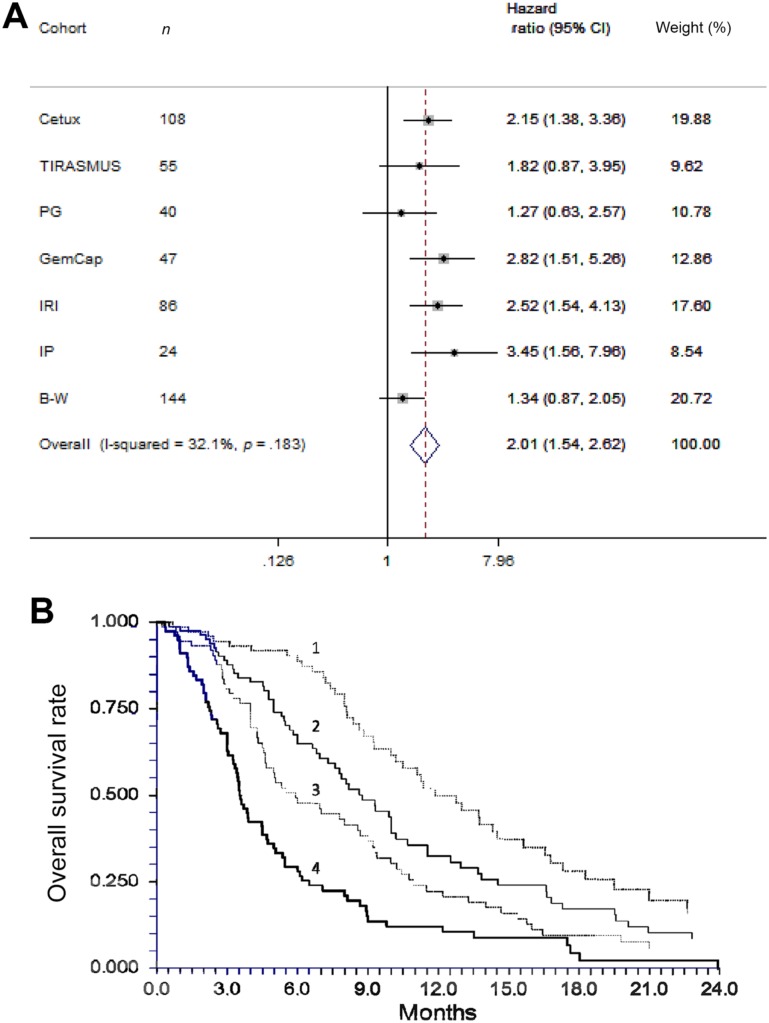
The potential clinical relevance of a prognostic biomarker is optimally analyzed if a reliable externally defined cut‐off level is applied. We were able to perform a meta‐analysis based on the seven Danish trials, using a previously defined UNL as cut‐off for survival analysis. The test for heterogeneity confirmed study homogeneity (I2 = 0.000, p = .88), and funnel plots shapes confirmed that the studies were symmetrically distributed. This allowed for using the random‐ and fixed‐effect models for the meta‐analysis and the forest plots to illustrate the results of the seven trials. All tests demonstrated a clear tendency for favorable HR in patients with low cfDNA levels, which was consistent with previous individual reports. Overall, a high cfDNA level (above the UNL) was significantly associated with a poor prognosis (pooled HR = 2.01, 95% CI 1.54–2.62, p < .0001; Fig. 2A). The meta‐analysis based on the individual data from the total cohort of 503 Danish patients is graphically illustrated in Figure 2B and confirms that increasing levels of cfDNA is associated with shorter survival. Different cut‐off can be applied with similar results, including the UNL, as also previously demonstrated in a pooled analysis of the four studies in chemotherapy refractory disease [6].
Figure 2.
Meta‐analysis on individual dataset presented as forest plot of hazard ratios based on dichotomisation by the upper normal limit from the normal cohort of 7,100 alleles per mL (A) in the seven Danish cohorts, and presented as Kaplan Meier Curves of the overall survival according to total cell‐free DNA levels in quartiles (B). In Figure 2B: 1, lower quartile; 2, second lowest quartile; 3, second highest quartile; 4, highest quartile.
Cell‐Free DNA as Source for KRAS Mutation Detection
There was a strong correlation between quantitative measures of total cfDNA and quantitative circulating tumor DNA concentrations reported from the Danish cohorts [10], [11], [12], [13], [14], [15], [16] and the study by Sefrioui et al. [19]. Similar data were not directly presented in the study by Wong et al. or the CORRECT trial, which analyzed a broader range of different mutations using different methods. The overall concordance of KRAS detection (in tumor and plasma) in the IRON group study [19] was reported to 85%, and 100% in the CORRECT trial [17]. In the study by Wong et al., 26 patients had available archival tumor tissue analyzed, and in four patients with tumor wild‐type status, KRAS mutations were detectable in the plasma [18]. As illustration, the comparison of KRAS mutational status in plasma and tumor tissue from the 498 patients with matching tumor and reliable cfDNA levels for mutation analysis from the Danish results are shown in supplemental online Table 1. The overall concordance between tumor and plasma mutational status was high (86%). The sensitivity, specificity, PPV, and NPV suggest an excellent performance of plasma KRAS testing.
Discussion
The number of reviews recently published illustrates the increasing interest for the potential use of cfDNA in patients with cancer. The majority of these publications have focused on a broad description of the potential utility in patients with colorectal cancer or cancer in general [20], [21], [22], [23], [24], [25]. The current systematic search confirmed significant heterogeneity between the identified studies, in terms of both different methodology and clinical approaches to this increasing research area. In addition, there were major variations in the use of nomenclature. Our primary search also revealed that the majority of published studies have focused on detection of tumor‐specific DNA, in terms of mutations or epigenetic alterations, especially concordance between tumor and blood, or the observed dynamics during therapy. The lack of large‐scale cohort analysis was emphasized. There is a high risk that unfocused broad studies will fail to bring this research area the important step past observational hypothesis‐generating data. A well‐defined clinical endpoint is consequently of utmost importance for a reliable assessment of a true clinical relevance.
Total cell‐free DNA quantification is a controversial topic, compared to tumor mutation detection. Circulating DNA is a mixture of DNA from normal cells as well as tumor cells, with only a fraction of the cfDNA originating from the tumor cells. Tumor DNA is detected and quantified in a broad range of different ways, but the most common approach to analyze tumor DNA is detection of mutations in the KRAS genes, which only account for approximately half of the patients. With improved methodology and addition of more tumor‐specific genetic alterations, the detection rate varies and is therefore difficult to use for direct comparison with other studies. In contrast, the total cfDNA can be analyzed in all patients, and a normal upper limit can be provided for standardization because it can be measured in healthy individuals. In addition, it is feasible, markedly cheaper, and less complicated to analyze than tumor‐specific DNA. The literature search revealed that the majority of studies have focused on tumor DNA analysis and not on the potential value of simple total DNA quantification; therefore, pooling of results from total cfDNA quantification and various tumor DNA analyses would imply a strong bias and limit conclusions of a subsequent meta‐analysis.
Having consistently observed a strong prognostic value in our line of Danish studies, we aimed to compare the findings with those of other groups. We consequently present the first systematic review of a specified clinical focus area regarding utility of cfDNA, and the first meta‐analysis in this setting. Ten studies were identified, and analysis showed a clear prognostic value in favor of patients with low levels of baseline cfDNA prior to treatment, despite the use of different laboratory approaches. The majority of data were based on patients with late‐line disease, but also a cohort of 86 patients in second‐line, and a small group of 13 patients in the first‐line settings, with similar results. These results illustrate the overseen potential of measuring the total cfDNA in itself in patients with advanced disease, and ongoing studies are validating the results in larger cohorts of first‐line chemotherapy.
The preferred gold standard for meta‐analysis is the Individual Patients Data (IPD) meta‐analysis, which has recently been added to the standard PRISMA statements—as specific requirements of reporting systematic reviews and meta‐analyses of IPD—the PRISMA IPD [9]. These were developed primarily to address optimal data synthesis in randomized trials. The predominant difference between this approach and aggregate data synthesis (extracted from the published reports or supplied by the investigators) is that the combined study results are generated from re‐analysis of the raw data from each study. This allows for more accurate data information and seeks to prevent publication bias and provide a better picture of the effect over time. This review includes both aggregate and IPD meta‐analysis, all of which showed consistent results, underlining the homogeneity of the study populations, methods, and endpoint extractions.
Subgroup analysis or meta‐regression was not performed because important biological information was only available from limited dataset. These would optimally have included measurements of carcinoembryonic antigen (CEA), lactate dehydrogenase (LDH), and other standard laboratory tests, and tumor burden and/or number of metastatic lesions. Of note, in two studies, the level of cfDNA was analyzed in relation to CEA, and no significant correlation was revealed [10], [16]. The same applied to standard laboratory parameters such as platelets and neutrophil counts. In a single study, a weak correlation to LDH was reported, which, however, failed in multivariate analysis [16]. Combining the information of cfDNA with tumor volume measures estimated by positron emission tomography ‐ computed tomography (PET‐CT) in a subset of patients has been done [16], but similar data were not available from the additional nine studies.
The potential clinical relevance of a prognostic marker is optimally analyzed if a reliable externally defined cut‐off level can be applied. Consequently, we performed an IPD meta‐analysis on the seven cohorts with available data and confirmed that a relevant cut point for prognostic estimation can be applied and a quantitative estimate provided. Homogeneity of the dataset and the IPD synthesis add major strength to the conclusions, but the true clinical relevance will only be validated if investigated in carefully designed prospective trials. Of note, another group has investigated the prognostic value of cfDNA based on an externally defined cut‐off [26]. Perkins et al. analyzed cfDNA in late‐stage patients with various different solid tumors, including 25 patients with CRC. Data showed poor prognosis in patients with levels above the upper value (detected in a cohort of 20 healthy individuals). However, data from the CRC patients could not be directly extracted, and the study was consequently not included in this meta‐analysis.
The major concern of this report is the risk of publication bias, which cannot be accounted for in this analysis, except for screening of relevant publications in our search. We systematically searched the publications for hidden data on total cfDNA in relation to clinical outcome. This especially applied to the major fraction of studies, which have analyzed the utility of tumor‐specific cfDNA. An overview of these studies is to be presented elsewhere. The most optimal way of addressing the issue is the initiation of an international cooperation for a prospective IPD meta‐analysis.
Conclusion
The present study indicates that the total cfDNA levels in plasma are high in patients with mCRC and bears strong prognostic information that should be tested prospectively by using a predefined cut‐off based on normal values in healthy cohorts. Finally, the potential use of plasma cfDNA for detection of tumor‐specific mutations has also been emphasized.
See http://www.TheOncologist.com for supplemental material available online.
Supplementary Material
Acknowledgments
The individual studies have previously received funding as acknowledged in the original publications. We thank the Novo Nordisk Foundation for funding of this report. We gratefully thank Ninna Aggerholm for help with the graphics.
Author Contributions
Conception/design: Karen‐Lise G. Spindler, Anders K. Boysen
Provision of study material or patients: Karen‐Lise G. Spindler, Niels Pallisgård, Julia S. Johansen, Josep Tabernero, Morten M. Sørensen, Benny V. Jensen, Torben F. Hansen, David Sefrioui, Rikke F. Andersen, Ivan Brandslund, Anders Jakobsen
Collection and/or assembly of data: Karen‐Lise G. Spindler, Anders K. Boysen, Niels Pallisgård, Julia S. Johansen, Josep Tabernero, Morten M. Sørensen, Benny V. Jensen, Torben F. Hansen, David Sefrioui, Rikke F. Andersen, Ivan Brandslund, Anders Jakobsen
Data analysis and interpretation: Karen‐Lise G. Spindler
Manuscript writing: Karen‐Lise G. Spindler, Anders K. Boysen, Niels Pallisgård, Julia S. Johansen, Josep Tabernero, Morten M. Sørensen, Benny V. Jensen, Torben F. Hansen, David Sefrioui, Rikke F. Andersen, Ivan Brandslund, Anders Jakobsen
Final approval of manuscript: Karen‐Lise G. Spindler, Anders K. Boysen, Niels Pallisgård, Julia S. Johansen, Josep Tabernero, Morten M. Sørensen, Benny V. Jensen, Torben F. Hansen, David Sefrioui, Rikke F. Andersen, Ivan Brandslund, Anders Jakobsen
Disclosures
Josep Tabernero: Amgen, Bayer, Boehringer Ingelheim, Celgene, Chugai, Lilly, Merck Sharp & Dohme, Merck Serono, Novartis, Pfizer, Roche (C/A). The other authors indicated no financial relationships.
(C/A) Consulting/advisory relationship; (RF) Research funding; (E) Employment; (ET) Expert testimony; (H) Honoraria received; (OI) Ownership interests; (IP) Intellectual property rights/inventor/patent holder; (SAB) Scientific advisory board
References
- 1. Cunningham D, Atkin W, Lenz HJ et al. Colorectal cancer. Lancet 2010;375:1030–1047. [DOI] [PubMed] [Google Scholar]
- 2. Therkildsen C, Bergmann TK, Henrichsen‐Schnack T et al. The predictive value of KRAS, NRAS, BRAF, PIK3CA and PTEN for anti‐EGFR treatment in metastatic colorectal cancer: A systematic review and meta‐analysis. Acta Oncol 2014;53:852–864. [DOI] [PubMed] [Google Scholar]
- 3. Jahr S, Hentze H, Englisch S et al. DNA fragments in the blood plasma of cancer patients: Quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res 2001;61:1659–1665. [PubMed] [Google Scholar]
- 4. Leon SA, Shapiro B, Sklaroff DM et al. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res 1997;37:646–650. [PubMed] [Google Scholar]
- 5. De Mattos‐Arruda L, Olmos D, Tabernero J. Prognostic and predictive roles for circulating biomarkers in gastrointestinal cancer. Future Oncol 2011;7:1385–1397. [DOI] [PubMed] [Google Scholar]
- 6. Spindler KL, Pallisgaard N, Andersen RF et al. Circulating free DNA as biomarker and source for mutation detection in metastatic colorectal cancer. PLoS One 2015;446:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Misale S, Yaeger R, Hobor S et al. Emergence of KRAS mutations and acquired resistance to anti‐EGFR therapy in colorectal cancer. Nature 2012;486:532–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yen LC, Yeh YS, Chen CW et al. Detection of KRAS oncogene in peripheral blood as a predictor of the response to cetuximab plus chemotherapy in patients with metastatic colorectal cancer. Clin Cancer Res 2009;15:4508–4513. [DOI] [PubMed] [Google Scholar]
- 9. Stewart LA, Clarke M, Rovers M et al. Preferred Reporting Items for Systematic Review and Meta‐Analyses of individual participant data: The PRISMA‐IPD Statement. JAMA 2015;313:1657–1665. [DOI] [PubMed] [Google Scholar]
- 10. Spindler KL, Pallisgaard N, Vogelius I et al. Quantitative cell free DNA, KRAS and BRAF mutations in plasma from patients with metastatic colorectal cancer during treatment with cetuximab and irinotecan. Clin Cancer Res 2012;18:1177–1185. [DOI] [PubMed] [Google Scholar]
- 11. Spindler KL, Sorensen MM, Pallisgaard N et al. Phase II trial of temsirolimus alone and in combination with irinotecan for KRAS mutant metastatic colorectal cancer: Outcome and results of KRAS mutational analysis in plasma. Acta Oncol 2013;52:963–970. [DOI] [PubMed] [Google Scholar]
- 12. Spindler KL, Pallisgaard N, Andersen RF et al. Pemetrexed and gemcitabine for chemotherapy refractory colorectal cancer—Results of a phase II and translational research study. J Cancer Ther 2013;4:44–50. [Google Scholar]
- 13. Spindler KL, Pallisgaard N, Andersen RF et al. Gemcitabine and capecitabine for heavily pre‐treated metastatic colorectal cancer patients—A phase II and translational research study. Anticancer Res 2014;34:845–850. [PubMed] [Google Scholar]
- 14. Spindler KL, Appelt AL, Pallisgaard N et al. Cell‐free DNA in healthy individuals, noncancerous disease and strong prognostic value in colorectal cancer. Int J Cancer 2014;135:2984–2991. [DOI] [PubMed] [Google Scholar]
- 15. Hansen TF, Andersen RF, Pallisgaard N et al. A three weekly schedule of irinotecan and panitumumab as third‐line treatment for patients with wild type KRAS metastatic colorectal cancer: Results from a phase II trial. Clin Colorectal Cancer 2014;3:135–145. [Google Scholar]
- 16. Spindler KL, Pallisgaard N, Appelt AL et al. Clinical utility of KRAS status in circulating plasma DNA compared to archival tumor tissue from patients with metastatic colorectal cancer treated with anti‐EGFR therapy. Eur J Cancer 2015;51:2678–2685. [DOI] [PubMed] [Google Scholar]
- 17. Wong AL, Lim JS, Sinha A et al. Tumour pharmacodynamics and circulating cell free DNA in patients with refractory colorectal carcinoma treated with regorafenib. J Transl Med 2015;13:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tabernero J, Lenz HJ, Siena S et al. Analysis of circulating DNA and protein biomarkers to predict the clinical activity of regorafenib and assess prognosis in patients with metastatic colorectal cancer: A retrospective, exploratory analysis of the CORRECT trial. Lancet Oncol 2015;16:937–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sefrioui D, Sarfan‐Vasseur N, Beaussire L et al. Clinical value of chip‐based digital‐PCR platform for the detection of circulating DNA in metastatic colorectal cancer. Dig Liver Dis 2015;47:884–890. [DOI] [PubMed] [Google Scholar]
- 20. Karampini E, McCaughan F. Circulating DNA in solid organ cancers‐Analysis and clinical application. QJM 2016. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Aarthy R, Mani S, Velusami S et al. Role of Circulating Cell-Free DNA in Cancers. Mol Diagn Ther 2015;19:339–350. [DOI] [PubMed] [Google Scholar]
- 22. Kato S, Janku F. Cell‐free DNA as a novel marker in cancer therapy. Biomark Med 2015;9:703–712. [DOI] [PubMed] [Google Scholar]
- 23. Francis G, Stein S. Circulating cell‐free tumour DNA in the management of cancer. Int J Mol Sci 2015;9:14122–14142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lim SH, Becker TM, Chua W et al. Circulating tumour cells and circulating free nucleic acid as prognostic and predictive biomarkers in colorectal cancer. Cancer Lett 2014;28:24–33. [DOI] [PubMed] [Google Scholar]
- 25. Kin C, Kidess E, Poultsides GA et al. Colorectal cancer diagnostics: Biomarkers, cell‐free DNA, circulating tumor cells and defining heterogeneous populations by single‐cell analysis. Expert Rev Mol Diagn 2013;13:581–599. [DOI] [PubMed] [Google Scholar]
- 26. Perkins G, Yap TA, Pope L et al. Multi‐purpose utility of circulating plasma DNA testing in patients with advanced cancers. PLoS One 2012;7:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.