Summary
Influenza is a worldwide health and financial burden posing a significant risk to the immune-compromised, obese, diabetic, elderly, and pediatric populations. We identified increases in glucose metabolism in the lungs of pediatric patients infected with respiratory pathogens. Using quantitative mass spectrometry we found metabolic changes occurring after influenza infection in primary human respiratory cells, and validated infection-associated increases in c-Myc, glycolysis, and glutaminolysis. We confirmed these findings with a metabolic drug screen that identified the PI3K/mTOR inhibitor BEZ235 as a regulator of infectious virus production. BEZ235 treatment ablated the transient induction of c-Myc, restored PI3K/mTOR pathway homeostasis measured by 4E-BP1 and p85 phosphorylation, and reversed infection-induced changes in metabolism. Importantly, BEZ235 reduced infectious progeny but had no effect on the early stages of viral replication. BEZ235 significantly increased survival in mice while reducing viral titer. We show metabolic reprogramming of host cells by influenza virus exposes targets for therapeutic intervention.
Keywords: PET scan, proteomics, respiratory viral infection, metabolism, influenza, BEZ235
eTOC Blurb
Influenza infection is a significant burden on public health with increased morbidity and mortality in pediatric patients as well as diabetic, obese, and elderly populations. Smallwood et al. define the metabolic response in infected humans and primary human respiratory cells ex vivo and use this information for targeted drug discovery.
Introduction
Influenza A virus (IAV), is a segmented RNA virus that buds off the cell taking host proteins and lipids. IAV co-opts transcriptional and translational machinery to favor viral protein production, while undergoing antigenic drift and reducing vaccine efficacy. Additionally, in the absence of host targeted therapies, the current frontline therapy of neuraminidase inhibitors has been met with a rise in viral resistance (Li et al., 2015). These factors converge to reduce IAV antiviral therapeutic efficacy and necessitate annual vaccine formulations that are not always effective (Hue et al., 2002).
IAV replication predominantly occurs in respiratory epithelial cells causing local pathologic changes and initiating a systemic immune response that can contribute to poor outcomes. Immune cell responses to influenza are well documented while human respiratory epithelial cells remain less well-characterized (Legge and Braciale, 2003; Oshansky and Thomas, 2012; Sanders et al., 2013). Metabolic requirements in pathological processes have also been a topic of recent investigation, with a view towards therapeutic intervention. Increased glucose and glutamine metabolism links tumors and proliferating T cells, which share a need for ongoing macromolecular synthesis and replication (Marsin et al., 2002). Intriguingly these metabolic shifts also regulate effector T cell function (Chang et al., 2013; Pearce, 2010; Pearce et al., 2009). Despite the recent focus on immune metabolism, little is known about the metabolics of airway epithelia in response to infection. Human cytomegalovirus increases glycolysis, glutaminolysis, and lipid production by encoding viral proteins that direct metabolic change (Isler et al., 2005; Munger et al., 2008; Vastag et al., 2011; Yu et al., 2011a; Yu et al., 2011b). However, IAV is a much smaller RNA virus that mediates acute infections with no identified proteins directly regulating metabolism. In this study, we investigated the potential for influenza virus to alter host metabolism.
We performed a retrospective study of children diagnosed with respiratory viral infections and found elevated glucose uptake in their lungs. To determine if this effect was related to viral infection and to facilitate identification of molecular mechanisms coordinating the host response, we conducted quantitative proteomics in infected normal primary human bronchial epithelial (NHBE) cells from pediatric donors. We found a unique virus-associated metabolic program in NHBEs that was enriched for glycolysis and glutaminolysis, required viral replication, and was distinct from Toll-like receptor (TLR) stimulation. This finding led us to design a high-throughput metabolic drug screen, which identified putative cellular targets required for productive infection and some potential therapeutic compounds. One drug, BEZ235, reduced viral titers in vivo and significantly enhanced survival after sub-lethal infection, without altering early viral replication kinetics, but instead altering host cell metabolic phenotypes. Proteomic and network analysis facilitated discovery of several influenza-induced metabolic changes providing insights for targeted drug screening leading to the identification of promising therapeutic compounds.
Results
Metabolic increase in the lungs of respiratory virus–infected patients
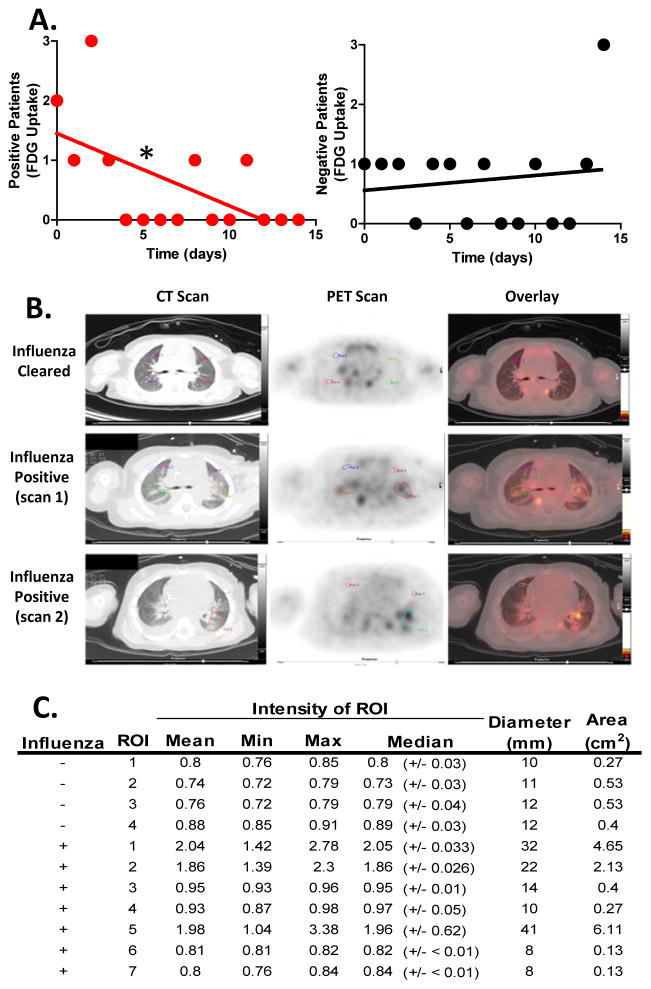
We performed a retrospective study of positron emission tomography (PET) scans from twenty pediatric cancer patients diagnosed with respiratory viral infections by PCR (in clinic), several of which showed increased glucose uptake in tumor free lungs (Table S1). A strong negative monotonic relationship (rs = −0.59, n = 15, p =0 .0198) between glucose uptake and respiratory viral infection was found in these patients by Spearman’s correlation (Figure 1A). 70% of patients scanned 0–3 days after diagnosis by PCR showed lung uptake of glucose (Table S1). Images from four infected patients are provided (Figure 1 and S1). One representative patient from this group (patient 10), a 4-year-old female with leukopenia from chemotherapy had a series of PET scans and was selected for in depth analysis (Figure 1B and C).
Figure 1. Glucose uptake in pediatric patients infected with respiratory viral infections.
(A–C) Patients with normal blood glucose were given 5.5 MBq/kg FDG intravenously after fasting ≥4 hours followed by 1 hr inactivity and image capture with transmission CT and PET obtained from the top of the skull to the feet. (A) We identified 20 respiratory virus-PCR positive pediatric patients retrospectively who received positron emission tomography (PET) scans. Linear regression compared the slopes of the PCR positive or negative patients over time to zero with the slope of the PCR positive line showing significant deviation from zero (P=0.0189). Significant relationship between FDG uptake and time of clinical PCR diagnosis with respiratory viral infection was found in these patients by Spearman’s correlation (rs = −0.59, number of XY pairs = 15, p =0 .0221). (B) Patient 10, a 4-year-old female tested PCR positive and underwent CT/PET scanning 3 days post infection diagnosis and 2 months later (Influenza Cleared). (C) Standardized uptake values of FDG were determined by placing regions-of-interest over areas of normal and abnormal uptake in the lungs for densitometry analysis. The mean, minimum, maximum and median densities are indicated with the ROI size (i.e. diameter and area). Where influenza negative ROI 1–4 are from the influenza cleared lungs 2 months post initial diagnosis, influenza positive 1–4 and 5–7 are from scans 1 and 2 respectively of patient 10 three days post PCR positive diagnosis for influenza A. 5 The Mann–Whitney U test was used to determine significance (i.e. p-values for mean = 0.037 and median = 0.03) with n=20. See also Figure S1 and Tables S1 & 2.
Representative images from PET and CT scans of patient 10 shows healthy lung tissue (Figure 1B top row). In contrast, during influenza infection the lung CT scans show multiple nodular pulmonary densities and interstitial fluid densities (Figure 1B). The PET scan shows dark patches, with high signal in the dense walled areas as well as diffuse FDG uptake in the infected lungs (Figure 1B). Glucose uptake in the IAV–infected lungs was significantly higher (p3=30.03) than PCR negative virus cleared lungs from the follow-up scan 8 weeks later (Figure 1C). Prior to the PET scan, five days of chemotherapy (i.e. cyclophosphamide and topotecan) depleted the patient’s immune cells. Virtually all blood cell subtypes’ absolute values were low or absent (Table S2). White blood cell (WBC) counts, hemoglobin, hematocrit percent, and platelet counts were nearing critical levels with low WBC morphology and red blood cell mean corpuscular volume (Table S2). The absence of band cells was notable as one would expect increases in an infected patient. The low immune cell counts suggest the increased glucose uptake was not due to immune cell proliferation alone. Together, these data suggest that influenza virus induces increased glucose uptake in human lungs that is resolved temporally as the virus is cleared (Figure 1A–C, S1 and Table S1.).
Influenza infection induces significant global changes in metabolic proteins
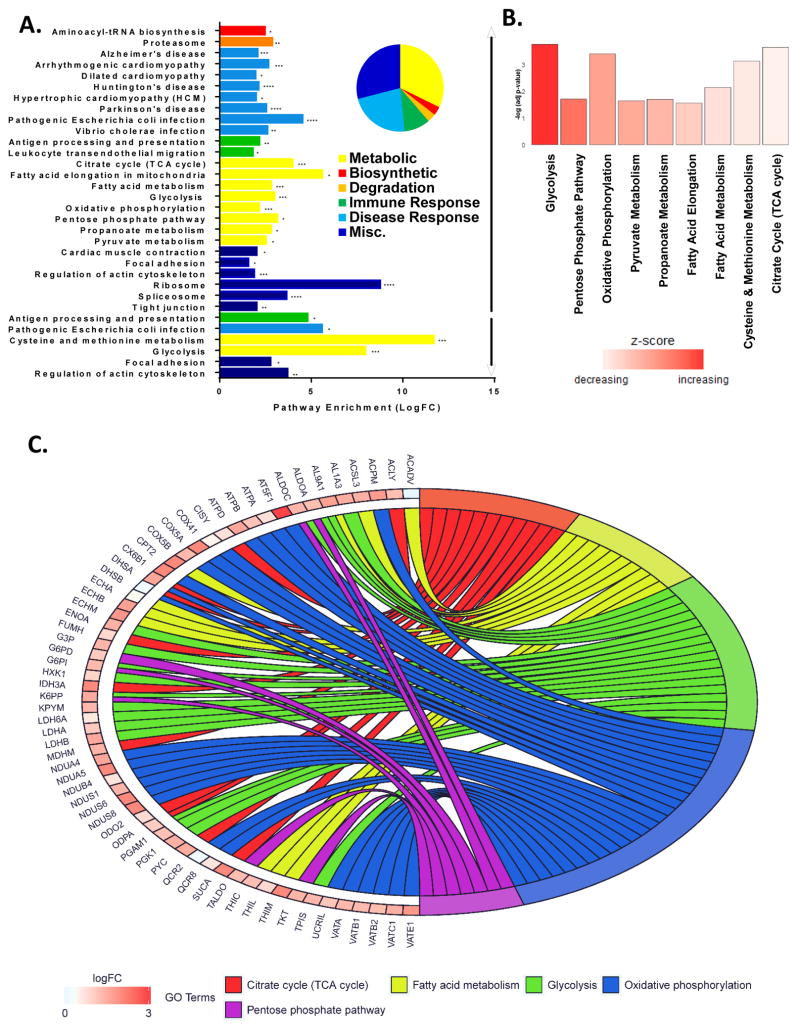
To identify molecular mechanisms coordinating this metabolic increase in infected patients’ lungs we conducted an unbiased global quantification of host proteins using mass spectrometry with iTRAQ labeling (Figure S2A). The responses in primary epithelial cells derived from the upper respiratory tract of 2 healthy young male donors were assessed, following 17 hours of in vitro infection with 1 MOI of a human H1N1 influenza virus pandemic strain (A/California/04/2009 (CA04)). The entire NHBE proteome (soluble and insoluble fractions +/− infection) is publicly available Proteomics Identifications (PRIDE) repository (Project accession PXD006265). FASP digestion and membrane separation vastly expanded proteomic coverage, mainly in the insoluble fraction (Figure S2B). We found infection-induced metabolic pathways dominate enrichment categories (Figure 2A). Confidently identified peptides with 2 fold or greater abundance changes were significantly enriched in glycolysis, TCA cycle, fatty acid metabolism, oxidative phosphorylation, and pentose phosphate pathway (Figure 2A and Table S3). After in silico combination of the soluble and insoluble proteomes fold changes among these pathways remained significant (Figure 2B). Metabolic pathway coverage was extensive, with specific protein fold changes indicated along with overall connectivity of the enriched categories in Figure 2C. These results are consistent with dramatically altered global metabolism following influenza infection.
Figure 2. Metabolic pathways dominate host-cell changes after influenza virus infection.
(A–C) NHBE from one donor were expanded for 10 days and monolayers were infected with CA04 (MOI 1 pfu for 17 hours) in two separate experiments. Lysates were collected on ice, homogenized in ammonium bicarbonate and separated into soluble and insoluble fractions), iTRAQ-labeled, FASP digested, desalted with C18 SPE, and analyzed with a Velos Orbitrap mass spectrometer. All confidently identified peptides with 2-fold or greater abundance changes were input into DAVID for enrichment analysis (using Human background). (A) Significantly enriched Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways are grouped by biological category (i.e. biosynthetic, degradative, disease and immune response, metabolic or misc.), and enumerated by fold enrichment. All significantly enriched KEGG pathways from the up regulated (↑) and down regulated (↓) soluble and insoluble peptides are colored by biological categories corresponding to the pie chart with fold enrichment significance is indicated with asterisk (* ≤0.05, ** ≤0.001, and *** ≤0.0001). (B) KEGG pathways within the metabolic category are graphed by the negative log of their respective p-values and bars are colored by their corresponding Z score. (C) Pathways of the metabolic biological category are colored as follows: Citric Acids Cycle (red), Fatty Acid Metabolism (yellow), Glycolysis (green), Oxidative Phosphorylation (blue), and Pentose Phosphate Pathway (purple). Proteins are labeled with their corresponding gene symbol, chords link each protein to its respective pathway(s) with the fold change (Log2) indicated by increasing red color intensity. See also Figure S2 and Table S3.
Influenza infection increases metabolic flux, distinct from TLR activation, inducing glucose and glutamine dependence to sustain host cell viability
To validate the virus-induced metabolic changes in the patient lungs and human proteome we monitored metabolite flux with the Seahorse XFe Analyzer (Agilent, Santa Clara, CA). Consistent with the PET scans and proteome, IAV infected NHBE cells had significant increases in metabolic flux (i.e. extracellular acidification rate (ECAR) from lactate production) and oxygen consumption rate (OCR), and proton production rate (PPR)) (Figure 3A–C). We also found glucose uptake markedly increased with IAV infection (Figure 3D). Previously Ritter et al found IAV infection elevated glycolysis in MDCK cells (Ritter et al., 2010). We too noted abnormally high glycolysis in MDCK irrespective of infection and distinct from NHBE (Data not shown). This makes good sense as elevated metabolism is the hallmark of transformed cells like MDCK. Thus in all independent assays performed on primary human epithelial cultures we find gross metabolic changes in keeping with our observation that respiratory infection increases glycolytic metabolism in children’s lungs.
Figure 3. Metabolic reprogramming by influenza confirmed and distinct from TLR stimulation.
(A–C) NHBE cells were grown and differentiated in 24-well Seahorse plates and left untreated (control), infected for 17 hours at MOI 1 with either viable virus or a β-propriolactone–inactivated virus (CA04 or BPL, respectively) or stimulated with TLR agonists lipopolysaccharide (LPS), polyinosinic polycytidylic acid (PolyIC), or Resiquimod (R848). After treatment, the extracellular acidification rate (ECAR) and oxygen consumption rate (OCR), and proton production rate (PPR) of NHBE cells were monitored for 2 hours. Error bars represent standard error of the mean of quadruplicate (control & CA04) or triplicate (BPL, LPS, PolyIC, & R848) samples, representative date from 3 independent experiments. Basal rate was established by measuring technical replicates measure 13 times/per well, to establish basal rate giving n=52 (control & CA04) or n=39 (BPL, LPS, PolyIC, & R848) for SEM.(D) NHBE cells were treated as in A–C and glucose uptake was monitored using a standard blood glucometer in 2 independent experiments, in quadruplicate with SEM. (E) NHBE cells were infected with CA04 for up to 17 hours at MOI 1, harvested at indicated time and lysates subjected to immunoblotting with a c-Myc protein standard from cMyc positive tumor lysate (“ctrl”) and probed for c-Myc and β-actin. (F–G) NHBE cells were grown and differentiated in 96-well Seahorse plates and left untreated (control), infected for 17 hours with CA04 viable virus at MOI 1, treated with cMyc inhibitor at 0.5 uM, 1.0 uM, and 2.0uM respectively or infected and treated. After treatment, the extracellular acidification rate (ECAR) and oxygen consumption rate (OCR) of NHBE cells were monitored for 2 hours. Error bars represent standard error of the mean of quintuplicate (control & CA04) and (cMyc 2.0uM & CA04+2.0uM) samples, representative plate from 5 independent experiments. Basal rate was established by measuring technical replicates measure 12 times per well, to establish basal rate giving n=60 (control, CA04, cMyc 2.0uM & CA04+2.0uM) for SEM. (H) BGEM was replaced with complete DMEM (red dotted line) or metabolite deplete DMEM for 2–4 hours followed by infection for 17 hours when viability was assessed using trypan blue exclusion. Two independent experiments were done with each control and virus in triplicate. (B–H) Error bars reflect the S.E.M. and significances are indicated as * p≤0.05, ** p≤0.001, *** p≤0.0001, and **** p<0.0001 from a Dunnett’s multiple comparison adjusted T test that followed ANOVA (<0.0001).
One explanation for the changes in NHBE metabolism is the activation of host cell response systems. We monitored metabolic flux after TLR agonist treatment to determine if the metabolic reprogramming is dependent on productive infection or is the result of activation of signaling cascades. Stimulation through TLR3, 4, or 7/8 by polyinosinic polycytidylic acid (PolyIC), lipopolysaccharide (LPS), or agonist R848 altered NHBE metabolism. However these alterations were significantly different from IAV (Figures 3A–C). PolyIC is structurally similar to IAV RNA, yet it produced a metabolic profile distinct from influenza infection (Figure 3A–C). Glucose uptake was significantly higher in infected cells compared to TLR treatment, another indication the metabolic reprogramming was not simply an attribute of pathogen recognition (Figure 3D).
To further dissect the molecular mechanisms underpinning these global changes we used β-propriolactone (BPL) inactivation to crosslink viral RNA. This permits entry while preventing replication allowing one to distinguish between cell-mediated responses to entry and metabolic reprogramming for viral replication (Jonges et al., 2010). The metabolic flux trend associated with BPL inactivated CA04 infection was similar to active virus, but was significantly lower in magnitude (Figure 3A and 3C). These data suggest that some of the metabolic effects observed in IAV-infected cells are likely initiated by cell-mediated responses to viral entry, but require viral replication to reinforce and amplify the metabolic changes.
c-Myc is a transcription factor and master regulator of cellular metabolism associated with cancer metabolism (Dang, 1999; Dang et al., 1999; Lin et al., 2009). Consistent with c-Myc mediated metabolic reprogramming found in other systems (i.e. cancer cells or T cells) we find a transient increase in c-Myc following influenza infection (Figure 3E). c-Myc inhibitor a well-described direct inhibitor of c-Myc, also known as 10058-F4 (Calbiochem, Billerica MA), was used to determine if blocking c-Myc could restore normal metabolic phenotype (Hammoudeh et al., 2009; Rahl et al., 2010; Wang et al., 2015; Yin et al., 2003). Indeed, while the inhibitor alone increased bioenergetics, in the context of IAV infection c-Myc inhibitor reduced both ECAR and OCR to near normal levels in a dose dependent manner that were significantly lower than infection alone (Figure 3F–G). c-Myc mediated metabolic changes in cancer often lead to a state of glucose and glutamine addiction, where cells become reliant on excesses of these substrates to survive. We tested viability of infected NHBE with glucose and glutamine restriction and found IAV infection significantly reduced viability in this context (Figure 3H). Influenza infection induces a transient increase in c-Myc that correlates with changes in metabolism that are modulated by c-Myc inhibition and may contribute to metabolite requirements that are critical for infected NHBE cell survival.
Targeted drug screen identifies compounds that modulate infection and expose integral pathways for potential therapeutic intervention
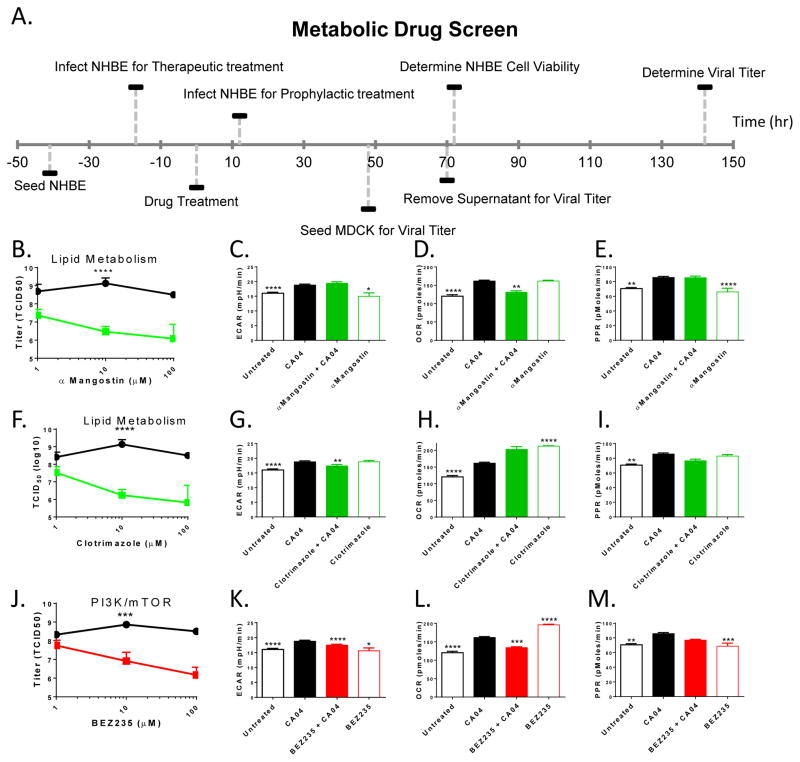
We screened 80 metabolic drugs with prophylactic and therapeutic treatment modalities (12 h prior to versus 17 h following infection) monitoring two endpoints: NHBE cell death and viral titer (Figure 4A). Table S4 provides a selected subset of the most active compounds. The NHBE viability curve data and corresponding titer data from one representative screen are in Supplementary File 1. Following the determination of NHBE cell viability in response to varying drug concentrations (10 point dose response), we titered IAV from each concentration of drug with both treatment modalities. The high throughput viral titer was done on MDCK cells with cytopathic effect (CPE) of the infection monitored using CellTiter-Glo viability assay. The area under the curve (AUC) for each titer was calculated and the average AUC was then subjected to K-means clustering to facilitate the selection of 23 drugs (Figure S3A).
Figure 4. Selective drugs reduce viral titer and restore metabolic flux.
(A) Timeline of drug treatment, infection, and titer. (B, F, J) Drugs acting on lipid metabolism (green), PI3K/mTOR (red), and vehicle controls (black). NHBE cells were seeded in BEGM medium rested and then infected at −17hr or +12 hr, drug treatment at 0 hr, and supernatant removed at 72 hours to recapitulate drug screen conditions. The viral laden supernatant removed from the drug dose response curves were then titrated to 8 serial dilutions on confluent MDCK cells and the infection was allowed to proceed, in the presence of TPCK trypsin, for 72 hours. Cytopathic effect was determined by staining remaining MDCK cells with Crystal Violet followed by 3 rinses with water. Each viral titer was performed in triplicate and calculated with the Reed Munch method. Vehicle was titered 8 times, α Mangostin was titered 7 times, clotrimazole was titered 3 times, and BEZ235 was titered 7 times (each in triplicate) corresponding to 3 separate experiments. Statistical differences in titer values between drug treatment were assessed using paired T test with P ≤ 0.0001 (***) and P < 0.0001 (****). (C–E, G–I, and K–M) NHBE cells were seeded in XF24 SEAHORSE cell culture plates, rested for 4–8 hours and treated with vehicle or 10nM BEZ235 (K–M), clotrimazole, (G–I) or α-Mangostin (C–E) in NMP/PEG (1:9), peanut oil, or DMSO/PBS (1:10), respectively, for 4 hours followed by infection at 1 MOI with CA04 where each drug treatment was in triplicate wells per plate, 1 plate per experiment in 2 separate experiments. 17 hours later plates were processed and read on Seahorse following manufacturer’s protocol. Basal levels established after 6 measurements, at approximately 1 hr. Error, error bars reflect the error of the mean calculated with n=6. Significance compared to CA04 treatment with Turkey’s multiple comparison adjusted T test that followed ANOVA (<0.0001). Adjusted P values from this test are as follows: * p ≤ 0.05, ** p ≤ 0.01, *** p ≤ 0.001, and **** p < 0.0001. See also Figure S3 and Table S4.
The selected compounds were then subjected to traditional TCID50 titering. 8 compounds significantly reduced CPE with pretreatment; half of which block lipid metabolism: FABP4, Pioglitazone, αMangostin, and Clotrimazole (Figure S3B). 5 compounds showed a significant decrease in viral titer following therapeutic treatment, 1 of which targets glucose metabolism 2-deoxy-glucose (Figure S3C). The competitive inhibitor 2-Deoxy-D-glucose (2-DG) was included as an internal control to determine if the work flow could identify a known glycolytic inhibitor and to test our hypothesis that glycolysis is necessary IAV replication. Indeed, while 2-deoxy-glucose is not a viable therapeutic option (Raez et al., 2013), it demonstrates the efficacy of this workflow and bolsters our confidence in the drug screen.
Of the validated drug hits BEZ235, Clotrimazole, and αMangostin were selected for further study based on their differential effects on infected NHBE cell survival and consistent reduction in viral titers (Table S4, Figures 4B, F&J and S3B). BEZ235, a PI3K/mTOR dual inhibitor known to induce insulin resistance in vivo, significantly reduced viral titer (Figure 4J) (Smith et al., 2012). The other two drugs displaying similar profiles were Clotrimazole, a topical treatment for fungal infections that targets lipid metabolism, and αMangostin a known anti-viral effective against other RNA viruses (Chen et al., 1996; Choi et al., 2014; Shaneyfelt et al., 2006; Vlietinck et al., 1998). In keeping with our previous results with the first human donor, viral infection induced increases in metabolite flux (ECAR, OCR, and PPR) in a second NHBE donor (Figure 4C–E, G–I, K–M). Of the three drugs tested, BEZ235 was the only drug to reverse influenza induced metabolic flux to untreated levels (Figure 4C–E, G–I, K–M).
Mechanism of BEZ235
BEZ235 has been extensively characterized as a dual PI3K/mTOR inhibitor in transformed cell lines to facilitate its development as a cancer treatment. We sought to determine the activity of this inhibitor in primary NHBE. First we, performed label-free shotgun proteomics on NHBE, NHBE treated with BEZ235, and infected NHBE treated with BEZ235 (PRIDE Project accession PXD006263). Supplemental Figure S4 and Table S5 show the major pathways altered by BEZ235 treatment. In both infected and uninfected conditions, the strongest effects of BEZ treatment were on metabolic pathways. BEZ235 largely downregulated metabolic targets, consistent with its reported activity; in particular PI3K and mTOR pathways were significantly downregulated (Table S5). Few proteins were upregulated in response to treatment, these were also largely metabolic.
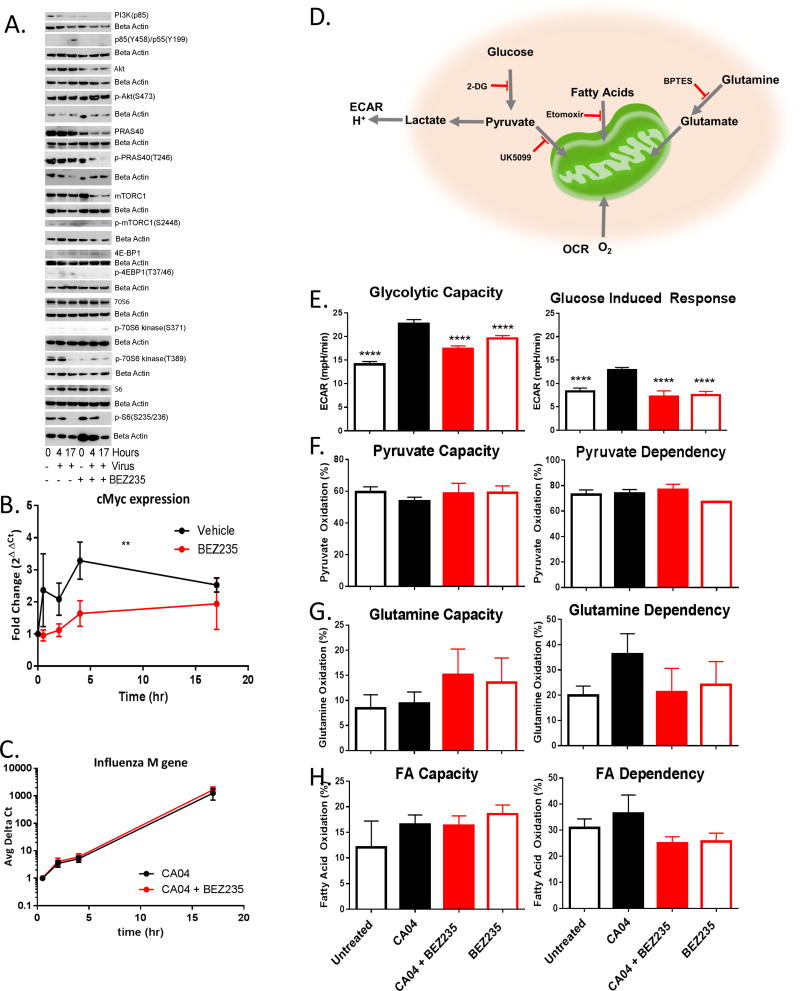
To validate the results of the proteomic measurements and establish the kinetics of activation of PI3K and mTOR pathways after IAV infection we tested a number of metabolic mediators by immunoblotting (Figure 5A). After infection 4E-BP1and PI3K (p85) are phosphorylated, indicating activation of the PI3K and mTOR pathways (Figure 5A). 70S6K(S371 & T389) and S6 (S235/236) are initially phosphorylated in uninfected cells while70S6K(T389) and S6 (S235/236) are desphosphorylated by 17 hours after infection (Figure 5A). Treatment with BEZ235 perturbs viral induction of the PI3K/mTOR pathways as is demonstrated by reduced PI3K and mTOR levels and phosphorylation products (p85(Y458)/p55(Y199) and mTORC1(S2448) respectively) compared to infection alone (Figure 5A, and consistent with the proteomic results, see Table S5). 70S6K(T389) and 4E-BP1 phosphorylation are abolished by BEZ235, consistent with mTORC1 inhibition (Figure 5A). BEZ235 treatment also leads to a time dependent decrease in protein and phosphorylation levels of the mTORC1 component and substrate PRAS40 (Figure 5A).
Figure 5. BEZ235 mechanism acquired via PI3K/mTOR pathway restriction on metabolism.
(A–H) NHBE cells were infected with CA04 for 17 hours at MOI 1 +/− BEZ235 1hr prior to infection. (A) NHBE were harvested at indicated times and lysates subjected to immunoblotting. (B–C) Two different donors’ cells in separate experiments (vehicle in duplicate, BEZ235 (−1hr) in triplicate per experiment per donor with 2 different donors) were harvested at indicated times and RNA purified and quantified with qPCR. The kinetic response of c-Myc (B) or viral M-gene (C) are graphed with significance determined by T test (P ≤ 0.05). (D) Schematic of metabolites and drug targets. (E–H) NHBE cells were grown and differentiated in Seahorse plates and left untreated (□ black open), infected (■ black solid), infected with 10nM BEZ235 (■ red solid), or 10nM BEZ235 alone (□ red open) and equilibrated in buffer free medium. Each stress test was performed following manufacture’s protocol at least 3 times in independent experiments in separate 96 well plates with each treatment in 4–12 replicate wells. Error bars reflecting standard error of the mean, with ANOVA (<0.0001) to distinguish differences among group means and treatment with post analysis by Dunnett’s multiple comparisons adjusted T test of significance in comparison to CA04 (**** = p<0.0001). (E) NHEB were equilibrated in non-buffered DMEM with glutamine (2 mM) for 1hr then ECAR kinetic reads taken in 6.22 min intervals with maximal rate measured after 3 reads followed by injection. Basal ECAR was determined then 10mM glucose was injected followed by 1μM oligomycin (to inhibit oxidative phosphorylation) with glycolytic capacity calculated by subtracting the basal ECAR from the maximal rate following both injections. Glucose induced response was calculated by subtracting the basal ECAR from the maximal ECAR after glucose injection. Untreated n=7, CA04 n=8, CA04+BEZ235 n=8, and BEZ235 n=8 for SEM. (F–H) NHBE bioenergetics studies were in supplemented medium with OCR kinetic reads taken in 6.22 min intervals with the first injection after 3 reads for basal OCR followed by 6 reads per inhibitor treatment. (F) Percent capacity to use pyruvate for respiration was calculated by dividing OCR from pyruvate alone by OCR with pyruvate, FA, and glutamine import inhibited with sequential injections of both 4 μM etomoxir and 3 μM BPTES followed by 2 μM UK5099 respectively. Percent dependence on pyruvate was calculated from the difference in basal OCR and pyruvate blocked OCR divided by basal OCR less OCR after all FA and glutamine import were blocked with sequential injections of 2 μM UK5099 followed by a combined of 4 μM etomoxir and 3 μM BPTES respectively. Untreated n=11, CA04 n=10, CA04+BEZ235 n=8, and BEZ235 n=9 for SEM. (G) Percent capacity to use glutamine for respiration was calculated by dividing OCR from glutamine alone by OCR with pyruvate, FA, and glutamine import inhibited with sequential injections of both 4 μM etomoxir and 2 μM UK5099 followed by 3 μM BPTES respectively. Percent dependence on glutamine was calculated from the difference in basal OCR and glutamine blocked OCR divided by basal OCR less OCR after all FA and pyruvate import were blocked with sequential injections of 3 μM BPTES followed by both 4 μM etomoxir and 2 μM UK5099 respectively. Untreated n=11, CA04 n=10, CA04+BEZ235 n=8, and BEZ235 n=9 for SEM. (H) Percent capacity to use fatty acids for respiration was calculated by dividing OCR from fatty acids alone by OCR with pyruvate, FA, and glutamine import inhibited with sequential injections of both 3 μM BPTES and 2 μM UK5099 followed by 4 μM etomoxir respectively. Percent dependence on fatty acids was calculated from the difference in basal OCR and fatty acid blocked OCR divided by basal OCR less OCR after all fatty acid, glutamine, and pyruvate import were blocked with sequential injections of 4 μM etomoxir followed by both 3 μM BPTES and 2 μM UK5099 respectively. Untreated n=11, CA04 n=12, CA04+BEZ235 n=11, and BEZ235 n=11 for SEM. See also Figure S4 and Table S5.
We sought to determine if these mTOR/PI3K pathway changes and BEZ235-mediated restoration of metabolic flux correlated to changes in cMyc levels. BEZ235 treatment is known to block c-Myc induction via inhibition of p70S6K and eIF4B (Csibi et al., 2014; Duffy et al., 2014). Likewise we see that the transient increase in cMyc expression induced by influenza infection is ablated by BEZ235 treatment in both NHBE donors (Figure 5B). Akt and PI3K inhibitors can block viral entry in transformed cell lines (Hirata et al., 2014; Shin et al., 2007). Preventing viral entry could explain reduced viral titer, c-Myc expression, and altered PI3k/mTOR activity. We measured the production of viral M-gene levels in infected NHBE cells to determine the kinetics of viral entry, transcript production, and genome replication in BEZ235-treated cells. BEZ235 treatment had no effect on new viral transcript accumulation, indicating that viral entry, polymerase activity, and genome replication were intact (Figure 5C). Thus BEZ235’s replication blockade occurs after these initiating events.
Next, we examined how IAV infection and BEZ235 treatment modulate substrates fueling the infection by inhibiting glucose entry into glycolysis with 2-DG and pyruvate, palmitate, and glutamine entrance into the mitochondria with UK5099, etomoxir, or BPTES, respectively. Figure 5D summarizes the rates measured and inhibitors used in the Seahorse XF Glycolysis Stress Test and Seahorse XF Mito Fuel Flex Test (Agilent, Santa Clara CA). The glycolysis stress test begins with establishing basal ECAR levels followed by injection of 10mM Glucose to determine the increase in glycolysis at saturating substrate concentrations (Glucose Induced Response). Next mitochondrial ATP production is inhibited with 1 μM Oligomycin, to shift the cell’s energy production to glycolysis resulting in maximal ECAR (Glycolytic Capacity) followed by glycolysis inhibition by 50mM 2-DG treatment. We find the glycolytic capacity of NHBE was significantly increased by IAV infection (Figure 5E). Likewise, the addition of saturating glucose substrate resulted in a significant increase in ECAR in the IAV-infected cells (Figure 5E). BEZ235 treatment significantly reduced both the glycolytic capacity and glucose induced response, thereby abolishing the utilization of one major metabolic fuel needed in IAV infection (Figure 5E).
The Mito Fuel Flex Test determines cellular dependence on each metabolite to fuel mitochondrial respiration and the capacity for utilizing each substrate when the others are blocked. We found infection had very little impact on NHBE mitochondrial dependence on pyruvate import into the mitochondria or capacity for import when fatty acid oxidation and glutaminolysis were both inhibited (Figure 5F). As one would expect, in contrast to pyruvate, NHBE reliance on glutamine to fuel oxygen consumption was lower. However, the dependence of the NHBE on glutamine as a metabolic substrate doubles with infection (Figure 5G). Similar to glucose, we find BEZ235 limits glutamine oxidation of infected cells to normal levels (Figure 5G). Oxygen consumption from beta oxidation of fatty acids is relatively low, with infection having little impact (Figure 5H). Taken together we find that infection increases cellular dependence on glucose and glutamine to fuel metabolism and treatment with BEZ235 limits utilization of these fuels.
BEZ235 rescues mice from lethal influenza infection, reduces viral titer, and ameliorates respiratory distress
To test the efficacy of these drugs in vivo, 10- to 12-week-old female C57/BL6 mice were orally administered vehicle or drug daily for 1 week. Mice were infected intranasally with CA04 at an infectious dose of 103 EID50 after which respiratory parameters and weight losses were monitored for 15 days (Figure 6A). Clotrimazole and α-mangostin had little effect on the course of disease (Figure S5A and S5B). BEZ235 had no effect on weight loss (Figures 6B) and there were no signs of toxicity in blood chemistry analyses (data not shown). However, BEZ235 treated mice showed a profound improvement in survival following lethal infection with CA04 influenza virus (Figure 6C).
Figure 6. BEZ235 treatment protects mice from lethal influenza virus challenge.
(A) Daily drug treatment was started 48 hours before intranasal infection with CA04 and maintained for 7 days. Weight loss was checked daily, and respiratory parameters were monitored via whole-body plethysmography on days 0, 2, 4, 7, and 10. A subset of mice was euthanized on day 4, and lungs were harvested for viral titers. (B) Oral gavage of 200 μL of BEZ235 (25 mg/kg; red square) or vehicle (black circle) had no independent effect on weight loss. Error bars reflect standard error of the mean from 45 mice per treatment from 2 independent experiments. (C) BEZ235 treatment (red) dramatically improved survival. Error bars reflect standard error of the mean from 45 mice per treatment from 2 independent experiments (p<0.0001 by Mantel-Cox log rank test). (D) Lungs were harvested, homogenized, and seeded on MDCK cells for titer assays; hemagglutination activity was determined with chicken red blood cells. The viral titer was significantly lower in BEZ235–treated mice (red). The titer from 4 mice from each group was determined in triplicate per mouse (t-test p=0.0027). (E) Mice underwent whole-body plethysmography with an equilibration time of 30 minutes followed by 5 minutes of data collection. The tidal flow of air at the mid-point of expiration of four mice per treatment were monitored in 2 independent experiment groups, and t-testing was used to determine statistical differences among treatments, with p-values indicated by asterisks (**** p≤0.0001). See also Figure S5.
BEZ235 consistently reduced viral titer in vitro (Table S4 and Figures 4J and S3B). In keeping with these findings, BEZ235 significantly reduced lung titers in vivo 4 days after influenza infection (Figure 6D). Lung function is the primary pathologic impairment in influenza infection. Lung function (assessed by whole-body plethysmography (WBP)) was previously used to determine the efficacy of antivirals against influenza (Julander et al., 2011; Sanders et al., 2013). With WBP we find BEZ235 significantly reduced mid-expiratory tidal flow compared to vehicle treated animals (Figure 6E). A reduction in this parameter of lung function is notable as we have previously shown it strongly correlates with IAV infection survival outcomes (Julander et al., 2011; Sanders et al., 2013). In NHBE cells, BEZ235 reduces viral titer, restores metabolic function, restricts aberrant activation of the PI3K/mTOR pathway, and reduces c-Myc expression to normal levels while limiting glucose and glutamine utilization (Figure 4J–M and Figure 5). In vivo BEZ235 reduces influenza-induced mortality, decreases viral titer, augments respiratory function in infected adult mice, and is well tolerated (Figure 6C–E). Thus, influenza induced changes in host metabolism represent viable drug targets to ameliorate disease severity and infection induced mortality.
Discussion
Using pre-existing patient data combined with the human bronchial epithelium proteome, we identified a dramatic shift in metabolic pathway activity after influenza infection. The increase in glycolysis and glutaminolysis were validated with metabolic flux analyses and functional assays. This re-programming is consistent with the fact that each budding virus depletes cellular resources (lipids, proteins, and a small amount of nucleic acid) that need to be replaced to maintain cellular homeostasis. These changes in metabolism within infected primary cells can be seen as a fight for limited macromolecules fueling competing interests. Indeed, the substrates, energy, and products required for biomass in viral replication are also critical for the host response to infection. Glucose and glutamine are abundant substrates in the plasma that provide a nearly unlimited source of renewable energy for infected cells (Newsholme et al., 1985). Given the significant increases in glycolysis in NHBE cells as well as patients’ lungs, substrate availability does not appear to limit the metabolic program induced by viral infection. Influenza viral budding removes significant amounts of amino acids and fatty acids from the intracellular milieu, thereby preventing normal feedback inhibition by metabolic product buildup. Together, unlimited substrate availability (glutamine, glucose) and gross product export (lipids, proteins) enforce a “viral production” metabolic state. This state appears to be at least in part dependent on elevated levels of c-Myc, as c-Myc inhibition reverts the metabolic profile of infected cells to an uninfected profile.
Identified from a large scale screening of metabolic compounds, we found that BEZ235 improved clinical outcomes of infected animals and reduced viral titers in vitro and in vivo. While BEZ235 alters glucose metabolism in vivo, it is primarily thought to act via inhibition of PI-3 kinase isoforms and is currently in clinical trials in combination with antitumor drugs (Smith et al., 2012). Our proteomic data suggests BEZ235 largely affects metabolism, indeed the PI3K-AKT-mTOR pathway was heavily modulated after treatment. Small interfering RNA (siRNA)-mediated knockdown of PI3K-AKT-mTOR host gene expression previously identified several genes in this pathway as potential therapeutic targets and for influenza infection (Murray et al., 2012). However, in spite of in vitro antiviral activity in this study the mTOR inhibitor everolimus had no effect in vivo (Murray et al., 2012). Hirata and coworkers found using a peptide inhibitor of Akt kinase they could suppress entry and replication of the mouse adapted PR8 virus into the A549 cell line, in contrast to BEZ235 which we found had no effect on entry and genome replication in human primary epithelial cells (Hirata et al., 2014) (Fig. 5C).
A transient dysregulation of glucose metabolism induced by BEZ235, consistent with the alterations in metabolite flux, glucose uptake, and blood amylase levels, may limit viral replication. Indeed, in vitro we see a reversion of global cellular metabolism to normal levels and reduced glutamine dependence induced by infection provided infected cells are treated with BEZ235. This reversion to normal metabolism occurred despite high levels of viral genome replication. These data suggest that the primary metabolic disruption by influenza virus is independent of genome replication; the significant depletion of lipid membranes would seem to be a likely candidate. Future studies will address how late into the course of influenza infection treatment with BEZ235 remains effective. Also, a thorough analysis of the drug’s effects on components of the adaptive immune system would be useful to determine if treatment might limit memory recall responses from B cells and T cells.
Experimental Procedures
Additional details of all experimental procedures and statistical analysis can be found in the Supplemental Experimental Procedures section.
PET scans of an influenza virus–infected patient
The Institutional Review Board of St. Jude Children’s Research Hospital approved the study, which was a retrospective analysis of data acquired for diagnostic purposes on consented patients. PCR tests were performed in clinical labs at St Jude. Blood glucose levels were tested and if found to be normal the patients were given intravenous injections of 5.5 MBq/kg fluorodeoxyglucose (FDG; maximum, 444 MBq) after at least 4 hours of fasting. The patients were kept in a quiet, dark room after injection and were told to lie down and relax with their arms at their sides. Approximately 1 h later, attenuation correction and lesion localization were acquired by using a GE Discovery LS PET/CT system (GE Medical Systems, Waukesha, WI) to capture transmission CT and PET images. The following CT parameters were: tube rotation, 0.8 s; slice thickness, 0.5 cm; table speed, 1.5 cm/rotation; pitch, 1.5:1; 120 kV; 90 mA; with dose modulation. PET images were obtained from the top of the skull to the feet for 5 minutes per bed position in 2-dimensional mode. Anesthesia-sedation was used if needed. Scans after July 2011 were acquired on a GE Discovery 690 PET/CT system. Those images were acquired in 3-dimensional mode for 3–5 min per bed position. Vendor-supplied software was used for reconstruction. Standardized uptake values were determined by placing regions-of-interest over areas of normal and abnormal uptake in the lungs.
Institutional Permissions
The Institutional Review Board of St. Jude Children’s Research Hospital approved the study, which was a retrospective analysis of data acquired for diagnostic purposes on consented patients. All animal procedures were approved by the St. Jude Institutional Animal Care and Use Committee (Protocol number: 098), following the guidelines established by the Institute of Laboratory Animal Resources, approved by the Governing Board of the U.S. National Research Council.
Gender and age
PET scans were performed on twenty children, eight female and twelve male. NHBE were from young male donors (ages 3, 10, and 17). Mice were female C57/BL6 mice aged 6 to 8 weeks were used in infection studies due to the maturity of immune response at this developmental stage.
Proteomics sample preparation, mass spectrometry, and data processing
NHBE cells (Lonza, Walkersville, MD) were infected for 17 hours with CA04 (MOI 1) followed by lysis, homogenization, and fractionation by centrifugation. NHBE cells were desalted by C18 solid-phase extraction (SPE) before isobaric labeling (SUPELCO, Bellefonte, PA). Digested samples were then processed according to the manufacturer’s directions for iTRAQ 4-plex labeling (ABSciex, Redwood City, CA). Before fractionating via high-pH reverse-phase fractionation with concatenated pooling, samples were desalted by C18 SPE (SUPELCO). All samples were processed in a custom liquid chromatography system using reversed-phase C18 columns. The iTRAQ samples were analyzed by using a Velos Orbitrap mass spectrometer (Thermo Scientific, San Jose, CA), and 18O-labeled samples were analyzed by using an LTQ-Orbitrap mass spectrometer (Thermo Scientific). Both systems were equipped with custom ion funnel–based atmospheric pressure ionization sources and electrospray ionization interfaces.
Raw files were compared with a concatenated NCBI Mus musculus database and contaminant database by using SEQUEST v.27 (rev. 12). The resulting sequence identifications were rescored by using MS-GF and filtered to a 1% false-discovery rate by using the target-decoy approach and MS-GF–derived spectral probabilities. Reporter-ion intensities were quantified by using the MASIC tool (Monroe et al., 2008). Missing reporter ion channel results were excluded from analysis. Redundant peptide identification reporter ions were summed across fractions, and median central tendency normalization was used to account for channel bias and then log2-transformed. Soluble and insoluble fractions were analyzed separately by using DanteR software. The analysis of variance model included treatment and peptide effects. The remaining treatment effect and p-value were calculated by using Student’s t-test. Benjamin-Hochberg multiple-testing error correction was applied to the results of this hypothesis testing, and proteins having a p-value ≤0.05 were considered to be statistically significant.
Metabolic Fuel Flux Assays
Glycolytic Stress and Mito Fuel Flex tests were performed on XFe 96 Bioanalyzer (Agilent Santa Clara, CA). At 16 hours post infection all assays were performed following manufacture’s protocols. Briefly, the Mito Fuel Flex Test inhibits import of three major metabolic substrates (pyruvate, fatty acids, and/or glutamine) with mitochondrial pyruvate carrier inhibitor UK5099 (2 μM), carnitine palmitoyl transferase 1A inhibitor etomoxir (4 μM), or glutaminase inhibitor BPTES (3 μM). This test determines cellular dependence on each of metabolite to fuel mitochondrial metabolism by inhibiting the individual substrate import or the capacity for utilizing that substrate when the others are blocked. Baseline OCR was monitored for 18 minutes followed by sequential inhibitor injections (i.e. “Treatment 1” or “Treatment 2” below) with OCR readings for an hour following each treatment. The inhibitor treatments and calculations are as follows:
| Metabolite Test | Treatment 1 | Treatment 2 |
|---|---|---|
| Pyruvate Dependence | UK5099 | Etomoxir + BPTES |
| Pyruvate Capacity | Etomoxir + BPTES | UK5099 |
| Glutamine Dependence | BPTES | Etomoxir + UK5099 |
| Glutamine Capacity | Etomoxir + UK5099 | BPTES |
| Fatty Acid Dependence | Etomoxir | BPTES + UK5099 |
| Fatty Acid Capacity | BPTES + UK5099 | Etomoxir |
High-throughput drug screening
The drug library included 80 compounds FDA-approved drugs and tool compounds directed at well-known metabolic targets that were screened in NHBE cells. The dose-response of each drug was determined (4 nM to 10 μM) for its use as a prophylactic treatment given 8 h before CA04 infection (MOI3=31) and its use as a therapeutic treatment given 17 h after infection. After 72 h, the supernatant was removed for viral titer, and cell viability was determined by performing CellTiter-Glo luminescent cell viability assays (Promega, Fitchburg, WI) measuring ATP production. Raw data were log10-transformed, and the percentage of inhibition was estimated in relation to that of previously and empirically determined controls (DMSO3=3negative control [0% cell death]; cycloheximide3=3positive control [100% cell death]). Virus-laden supernatant was added to confluent MDCK cells and titrated to 8 serial dilutions or 3 serial dilutions for uninfected controls (3×384-well plates per drug were set up in triplicate for each of 3 conditions). After 72 h, the cytopathic effect was determined by using the CellTiter-Glo viability assay. Data processing for the titer (171× 384-well plates) was done by determining the area under the curve for each titer. From these data, 23 compounds were selected for titer validation by traditional TCID50 assays.
In vivo drug tests
All animal procedures were approved by the St. Jude Institutional Animal Care and Use Committee. Female C57/BL6 mice aged 6 to 8 weeks were given either 0.2 mL oral gavage with BEZ235 at 25 mg/kg or 0.5 mL intraperitoneal injections of either 120 mg/kg clotrimazole or 8 mg/kg α-mangostin, with appropriate vehicle controls performed in littermates (Supplementary Methods). Mice were dosed daily for 7 days beginning 2 days before infection and then monitored for signs of infection and weight loss daily for 15 days (no significant difference by t-test). On days 0, 2, 4, 7, and 10, after 30 minutes of chamber equilibration time, respiratory parameters were monitored for 5–10 minutes by performing whole-body plethysmography (Buxco, Wilmington, NC). On day 4, a subset of mice was euthanized for arterial blood and lung collection for blood chemistry and complete blood count or titer. Lungs were homogenized and subjected to TCID50 assays with hemagglutination titer readout, with significance determined by t-testing (p<0.003). Moribund mice were euthanized, and survival percentage differences were calculated, with significance determined by Mantel-Cox log rank testing (p<0.0001). 45 mice were included for each condition, more than doubling the total events needed (21) to satisfy the following criteria: α (two-tailed) = 0.05, β = 0.20, q1 = 0.692, q0 = 0.308 with relative hazard ratio 3.776. Mice were excluded from an isolated catastrophic cage failure altering survival patterns (e.g. cage flooding); no randomization or blinding was used.
Statistics
Proteome
Redundant peptide identification reporter ions were summed across fractions similar to a weighted average. Median central tendency normalization was applied to account for channel bias associated with sample handling. The results were then Log2 transformed. The processed results from the soluble and insoluble fraction were analyzed separately using the DanteR software suite. The ANOVA model applied consists of a treatment and peptide effect. This removes the variance associated with each peptide for a given protein having differing ionization efficiencies or due to the position on the elution profile when a peptide is sent for fragmentation. The remaining treatment effect and p-value can then be calculated using a student’s t-test. The Benjamin-Hochberg multiple-testing error correction was applied to the results of this hypothesis testing and proteins having a p-value of 0.05 or less were considered significant. With the following predetermined exclusion criteria: Peptide to spectrum matches were filtered at a 1% false discovery rate using target-decoy approach and MS-GF derived spectral probabilities as the scoring metric. All peptides identified with missing reporter ion channel results were removed from the analysis.
Enrichment analysis
Peptide identifiers were input into DAVID for enrichment analysis, with significantly enriched KEGG pathways indicated (i.e. p-value ≤ 0.01), grouped by similarity (i.e. biosynthetic, degradative, disease and immune response, metabolic or misc.), and enumerated by fold enrichment. Fold enrichment significance is indicated with asterisk. Fold changes from the proteins identified in the glycolysis/gluconeogenesis and TCA cycle enriched KEGG pathways are graphed with identification significance in the inset. Group Enrichment score ranks the biological significance of gene groups based on overall EASE scores of all enriched annotation terms. Significance is indicated as * p≤0.05, ** p≤0.001, and *** p≤0.0001. With the following predetermined exclusion criteria: Peptides with < 2 fold abundance changes were removed.
Basal metabolic rates, glucose, viability assay, TCID50, glycolysis stress test, mitochondrial flex fuel, and lung titer
ANOVA was used to distinguish differences among group means and treatment with post analysis by Dunnett’s multiple comparisons adjusted T test of significance in comparison to CA04. Asterisks represent p values following standard convention in all figures as follows: p<0.05 (*), p< 0.01 (**), p<0.001(***), and p< 0.0001 (****). With the following predetermined exclusion criteria (bioanalyzer): 1) injection failure 2) bubbles in well 3) negative values.
qPCR and plethysmography
ANOVA analysis was used with asterisks represent p values following standard convention in all figures as follows: p = 0.0069 (**) and p < 0.0001 (****).
Supplementary Material
Highlights.
Increased glucose metabolism in pediatric patients with respiratory infection
Proteomic and functional analyses identified metabolic changes induced by influenza
Metabolic drug screen on primary human epithelial cells identified therapeutic targets
PI3K/mTOR modulator rescued mice from a lethal influenza infection
Acknowledgments
We would like to thank and acknowledge Asli Goktug, Jaeki Min, Josiah Becker, and Lavanya Bezavada for their technical assistance in reading the high-throughput plates, programming, running the Biomek, viral tittering, and running the XFe96 Bioanalyzer assays respectively as well as David Kakhniashvili for help with the unlabeled proteomes.
Funding: This work was supported by AI091938, HHSN266200700005C, HHSN272201400006C, a V Foundation Grant and Cancer Center Grant CA-21765 (MFR) and the American Lebanese Syrian Associated Charities (ALSAC). Proteomic portions of this research were supported by the NIH National Center for Research Resources (Grant RR018522) and the Department of Energy’s W.R. Wiley Environmental Molecular Science Laboratory at the Pacific Northwest National Laboratory under contract DE-AC05-76RL0 1830.
Footnotes
Data and Software Availability: The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD006265 (10.6019/PXD006265) and PXD006263.
Author Contributions: H.S.S. performed analysis of PET scans and CBC, prepared proteomic samples, analyzed proteomes (i.e. performed gene ontology, KEGG enrichment analysis and protein interaction networks), performed and analyzed BIOLOG metabolic array, prepared samples, analyzed, and ran XF24 and XFe96 Bioanalyzer assays, performed glucose uptake experiments, performed metabolite restriction assays, developed NHBE screening protocol and titer conditions, performed high throughput titer, analyzed titer data, performed TCID50, mTOR/PI3K immunoblots, designed and conducted in vivo mouse studies, and wrote the manuscript; S.D. drugged mice, infected mice, weighed mice, titered lungs, and contributed intellectually to in vivo experimental design; M.M. performed western blot for c-Myc, seeded cells for drug screen and titer, as well as drugged and read NHBE plates; S.R. prepared label- free proteomic samples, analyzed proteomes (i.e. performed gene ontology, KEGG enrichment analysis and protein interaction networks), assisted in c-Myc XFe96 Bioanalyzer assays, B.L.S. analyzed PET scans and coordinated clinical experiments; A.S. selected bioactive metabolic compounds, developed analysis software and analyzed high-throughput screening data; E.M.Z. prepared and ran samples for M.S.; R.B. assisted in mouse experiments; A.O. performed enrichment analysis in R; S.M. performed Seahorse experiments; M.F.R. interpreting data and editing manuscript; D.R.G. interpreting data and editing manuscript; L.P.T contributed to proteome experimental design, coordinated M.S. experiments, and intellectually contributed to proteomic analysis; and P.G.T. interpreting data, experiment coordination, and substantially contributed to editing and writing the manuscript.
Competing interests: The authors of this manuscript declare they have no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Chang CH, Curtis JD, Maggi LB, Jr, Faubert B, Villarino AV, O’Sullivan D, Huang SC, van der Windt GJ, Blagih J, Qiu J, et al. Posttranscriptional control of T cell effector function by aerobic glycolysis. Cell. 2013;153:1239–1251. doi: 10.1016/j.cell.2013.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen SX, Wan M, Loh BN. Active constituents against HIV-1 protease from Garcinia mangostana. Planta medica. 1996;62:381–382. doi: 10.1055/s-2006-957916. [DOI] [PubMed] [Google Scholar]
- Choi M, Kim YM, Lee S, Chin YW, Lee C. Mangosteen xanthones suppress hepatitis C virus genome replication. Virus genes. 2014;49:208–222. doi: 10.1007/s11262-014-1098-0. [DOI] [PubMed] [Google Scholar]
- Csibi A, Lee G, Yoon SO, Tong H, Ilter D, Elia I, Fendt SM, Roberts TM, Blenis J. The mTORC1/S6K1 pathway regulates glutamine metabolism through the eIF4B-dependent control of c-Myc translation. Current biology: CB. 2014;24:2274–2280. doi: 10.1016/j.cub.2014.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Dang CV. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Molecular and cellular biology. 1999;19:1–11. doi: 10.1128/mcb.19.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang CV, Resar LM, Emison E, Kim S, Li Q, Prescott JE, Wonsey D, Zeller K. Function of the c-Myc oncogenic transcription factor. Experimental cell research. 1999;253:63–77. doi: 10.1006/excr.1999.4686. [DOI] [PubMed] [Google Scholar]
- Duffy DJ, Krstic A, Schwarzl T, Higgins DG, Kolch W. GSK3 inhibitors regulate MYCN mRNA levels and reduce neuroblastoma cell viability through multiple mechanisms, including p53 and Wnt signaling. Molecular cancer therapeutics. 2014;13:454–467. doi: 10.1158/1535-7163.MCT-13-0560-T. [DOI] [PubMed] [Google Scholar]
- Hammoudeh DI, Follis AV, Prochownik EV, Metallo SJ. Multiple independent binding sites for small-molecule inhibitors on the oncoprotein c-Myc. Journal of the American Chemical Society. 2009;131:7390–7401. doi: 10.1021/ja900616b. [DOI] [PubMed] [Google Scholar]
- Hirata N, Suizu F, Matsuda-Lennikov M, Edamura T, Bala J, Noguchi M. Inhibition of Akt kinase activity suppresses entry and replication of influenza virus. Biochemical and biophysical research communications. 2014;450:891–898. doi: 10.1016/j.bbrc.2014.06.077. [DOI] [PubMed] [Google Scholar]
- Hue L, Beauloye C, Marsin AS, Bertrand L, Horman S, Rider MH. Insulin and ischemia stimulate glycolysis by acting on the same targets through different and opposing signaling pathways. Journal of molecular and cellular cardiology. 2002;34:1091–1097. doi: 10.1006/jmcc.2002.2063. [DOI] [PubMed] [Google Scholar]
- Isler JA, Maguire TG, Alwine JC. Production of infectious human cytomegalovirus virions is inhibited by drugs that disrupt calcium homeostasis in the endoplasmic reticulum. Journal of virology. 2005;79:15388–15397. doi: 10.1128/JVI.79.24.15388-15397.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonges M, Liu WM, van der Vries E, Jacobi R, Pronk I, Boog C, Koopmans M, Meijer A, Soethout E. Influenza virus inactivation for studies of antigenicity and phenotypic neuraminidase inhibitor resistance profiling. Journal of clinical microbiology. 2010;48:928–940. doi: 10.1128/JCM.02045-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julander JG, Hagloch J, Latimer S, Motter N, Dagley A, Barnard DL, Smee DF, Morrey JD. Use of plethysmography in assessing the efficacy of antivirals in a mouse model of pandemic influenza A virus. Antiviral research. 2011;92:228–236. doi: 10.1016/j.antiviral.2011.08.011. [DOI] [PubMed] [Google Scholar]
- Legge KL, Braciale TJ. Accelerated migration of respiratory dendritic cells to the regional lymph nodes is limited to the early phase of pulmonary infection. Immunity. 2003;18:265–277. doi: 10.1016/s1074-7613(03)00023-2. [DOI] [PubMed] [Google Scholar]
- Li TC, Chan MC, Lee N. Clinical Implications of Antiviral Resistance in Influenza. Viruses. 2015;7:4929–4944. doi: 10.3390/v7092850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CH, Lin C, Tanaka H, Fero ML, Eisenman RN. Gene regulation and epigenetic remodeling in murine embryonic stem cells by c-Myc. PloS one. 2009;4:e7839. doi: 10.1371/journal.pone.0007839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsin AS, Bouzin C, Bertrand L, Hue L. The stimulation of glycolysis by hypoxia in activated monocytes is mediated by AMP-activated protein kinase and inducible 6-phosphofructo-2-kinase. The Journal of biological chemistry. 2002;277:30778–30783. doi: 10.1074/jbc.M205213200. [DOI] [PubMed] [Google Scholar]
- Monroe ME, Shaw JL, Daly DS, Adkins JN, Smith RD. MASIC: a software program for fast quantitation and flexible visualization of chromatographic profiles from detected LC-MS(/MS) features. Computational biology and chemistry. 2008;32:215–217. doi: 10.1016/j.compbiolchem.2008.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munger J, Bennett BD, Parikh A, Feng XJ, McArdle J, Rabitz HA, Shenk T, Rabinowitz JD. Systems-level metabolic flux profiling identifies fatty acid synthesis as a target for antiviral therapy. Nature biotechnology. 2008;26:1179–1186. doi: 10.1038/nbt.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray JL, McDonald NJ, Sheng J, Shaw MW, Hodge TW, Rubin DH, O’Brien WA, Smee DF. Inhibition of influenza A virus replication by antagonism of a PI3K-AKT-mTOR pathway member identified by gene-trap insertional mutagenesis. Antiviral chemistry & chemotherapy. 2012;22:205–215. doi: 10.3851/IMP2080. [DOI] [PubMed] [Google Scholar]
- Newsholme EA, Crabtree B, Ardawi MS. The role of high rates of glycolysis and glutamine utilization in rapidly dividing cells. Bioscience reports. 1985;5:393–400. doi: 10.1007/BF01116556. [DOI] [PubMed] [Google Scholar]
- Oshansky CM, Thomas PG. The human side of influenza. Journal of leukocyte biology. 2012;92:83–96. doi: 10.1189/jlb.1011506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce EL. Metabolism in T cell activation and differentiation. Current opinion in immunology. 2010;22:314–320. doi: 10.1016/j.coi.2010.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce EL, Walsh MC, Cejas PJ, Harms GM, Shen H, Wang LS, Jones RG, Choi Y. Enhancing CD8 T-cell memory by modulating fatty acid metabolism. Nature. 2009;460:103–107. doi: 10.1038/nature08097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raez LE, Papadopoulos K, Ricart AD, Chiorean EG, Dipaola RS, Stein MN, Rocha Lima CM, Schlesselman JJ, Tolba K, Langmuir VK, et al. A phase I dose-escalation trial of 2-deoxy-D-glucose alone or combined with docetaxel in patients with advanced solid tumors. Cancer chemotherapy and pharmacology. 2013;71:523–530. doi: 10.1007/s00280-012-2045-1. [DOI] [PubMed] [Google Scholar]
- Rahl PB, Lin CY, Seila AC, Flynn RA, McCuine S, Burge CB, Sharp PA, Young RA. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritter JB, Wahl AS, Freund S, Genzel Y, Reichl U. Metabolic effects of influenza virus infection in cultured animal cells: Intra- and extracellular metabolite profiling. BMC systems biology. 2010;4:61. doi: 10.1186/1752-0509-4-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders CJ, Vogel P, McClaren JL, Bajracharya R, Doherty PC, Thomas PG. Compromised respiratory function in lethal influenza infection is characterized by the depletion of type I alveolar epithelial cells beyond threshold levels. American journal of physiology Lung cellular and molecular physiology. 2013;304:L481–488. doi: 10.1152/ajplung.00343.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaneyfelt ME, Burke AD, Graff JW, Jutila MA, Hardy ME. Natural products that reduce rotavirus infectivity identified by a cell-based moderate-throughput screening assay. Virology journal. 2006;3:68. doi: 10.1186/1743-422X-3-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin YK, Liu Q, Tikoo SK, Babiuk LA, Zhou Y. Effect of the phosphatidylinositol 3-kinase/Akt pathway on influenza A virus propagation. The Journal of general virology. 2007;88:942–950. doi: 10.1099/vir.0.82483-0. [DOI] [PubMed] [Google Scholar]
- Smith GC, Ong WK, Rewcastle GW, Kendall JD, Han W, Shepherd PR. Effects of acutely inhibiting PI3K isoforms and mTOR on regulation of glucose metabolism in vivo. The Biochemical journal. 2012;442:161–169. doi: 10.1042/BJ20111913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vastag L, Koyuncu E, Grady SL, Shenk TE, Rabinowitz JD. Divergent effects of human cytomegalovirus and herpes simplex virus-1 on cellular metabolism. PLoS pathogens. 2011;7:e1002124. doi: 10.1371/journal.ppat.1002124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlietinck AJ, De Bruyne T, Apers S, Pieters LA. Plant-derived leading compounds for chemotherapy of human immunodeficiency virus (HIV) infection. Planta medica. 1998;64:97–109. doi: 10.1055/s-2006-957384. [DOI] [PubMed] [Google Scholar]
- Wang H, Sharma L, Lu J, Finch P, Fletcher S, Prochownik EV. Structurally diverse c-Myc inhibitors share a common mechanism of action involving ATP depletion. Oncotarget. 2015;6:15857–15870. doi: 10.18632/oncotarget.4327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin X, Giap C, Lazo JS, Prochownik EV. Low molecular weight inhibitors of Myc-Max interaction and function. Oncogene. 2003;22:6151–6159. doi: 10.1038/sj.onc.1206641. [DOI] [PubMed] [Google Scholar]
- Yu Y, Clippinger AJ, Alwine JC. Viral effects on metabolism: changes in glucose and glutamine utilization during human cytomegalovirus infection. Trends in microbiology. 2011a;19:360–367. doi: 10.1016/j.tim.2011.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Clippinger AJ, Pierciey FJ, Jr, Alwine JC. Viruses and metabolism: alterations of glucose and glutamine metabolism mediated by human cytomegalovirus. Advances in virus research. 2011b;80:49–67. doi: 10.1016/B978-0-12-385987-7.00003-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.