Summary
We serendipitously discovered that the marine bacterium Vibrio fischeri induces sexual reproduction in one of the closest living relatives of animals, the choanoflagellate Salpingoeca rosetta. Although bacteria influence everything from nutrition and metabolism to cell biology and development in eukaryotes, bacterial regulation of eukaryotic mating was unexpected. Here we show that a single V. fischeri protein, the previously uncharacterized EroS, fully recapitulates the aphrodisiac-like activity of live V. fischeri. EroS is a chondroitin lyase; although its substrate, chondroitin sulfate, was previously thought to be an animal synapomorphy, we demonstrate that S. rosetta produces chondroitin sulfate and thus extend the ancestry of this important glycosaminoglycan to the premetazoan era. Finally, we show that V. fischeri, purified EroS, and other bacterial chondroitin lyases induce S. rosetta mating at environmentally-relevant concentrations, suggesting that bacteria likely regulate choanoflagellate mating in nature.
Keywords: choanoflagellate, Vibrio, chondroitin sulfate, chondroitinase, swarming, mating, host-microbe
Graphical Abstract
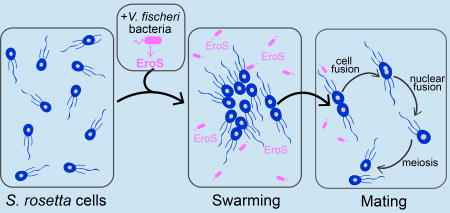
Introduction
Bacterial–eukaryotic interactions are ubiquitous, and the influences of bacteria on eukaryotes vary from subtle to profound. Yet, because eukaryotes are often associated with complex and unseen communities of bacteria, the breadth of eukaryotic biological processes regulated by bacteria and the underlying molecular dialogue often remain obscure. Nonetheless, studies of experimentally tractable host-microbe pairs have revealed a growing number of examples in which bacteria regulate eukaryotic cell biology and development, in some cases using molecular cues that mediate pathogenesis in other contexts (Koropatnick et al., 2004).
One of the closest living relatives of animals, the marine choanoflagellate S. rosetta, has emerged as an attractive model for uncovering bacterial cues that regulate eukaryotic development. Like all choanoflagellates, S. rosetta survives by eating bacteria (Dayel and King, 2014; Leadbeater, 2015). However, interactions between S. rosetta and bacteria extend far beyond those of predator and prey. In prior work, we demonstrated that the developmental switch triggering the formation of multicellular “rosettes” from a single founding cell (Fairclough et al., 2010) is regulated by specific lipids produced by the environmental bacterium Algoriphagus machipongonensis (Alegado et al., 2012; Cantley et al., 2016; Woznica et al., 2016). Rosette development is one of at least six different developmental switches in the sexual and asexual phases of S. rosetta’s dynamic life history (Dayel et al., 2011; Levin and King, 2013), but until now was the only choanoflagellate process known to be regulated by bacterial cues.
We report here on our serendipitous discovery that sexual reproduction in S. rosetta is regulated by a secreted cue from the marine bacterium Vibrio fischeri.
Results
S. rosetta forms mating swarms upon exposure to V. fischeri
V. fischeri is perhaps best understood as a model for bacterial quorum sensing and as a symbiont required for the induction of light organ development in the squid, Euprymna scolopes (Mcfall-Ngai, 2014). Although Vibrio spp. are known symbionts, commensals, and pathogens of animals (Thompson et al., 2006), V. fischeri does not induce rosette development (Alegado et al., 2012) and was not previously known to influence any aspect of S. rosetta biology. We were therefore surprised to observe that the addition of live V. fischeri bacteria to a culture of single-celled, motile S. rosetta induced cells to swarm into loose aggregates, each composed of between 2–50 cells (Figure 1A, B, Figure S1A, Movie 1). This dynamic and previously unobserved swarming behavior began as early as 15 minutes after induction with V. fischeri, with individual S. rosetta cells often moving between swarms that periodically broke apart or merged with other swarms. In its timescale, mechanism, and outcome, swarming was clearly unrelated to the Algoriphagus-induced developmental process by which a single S. rosetta cell divides repeatedly to form a rosette (Fairclough et al., 2010).
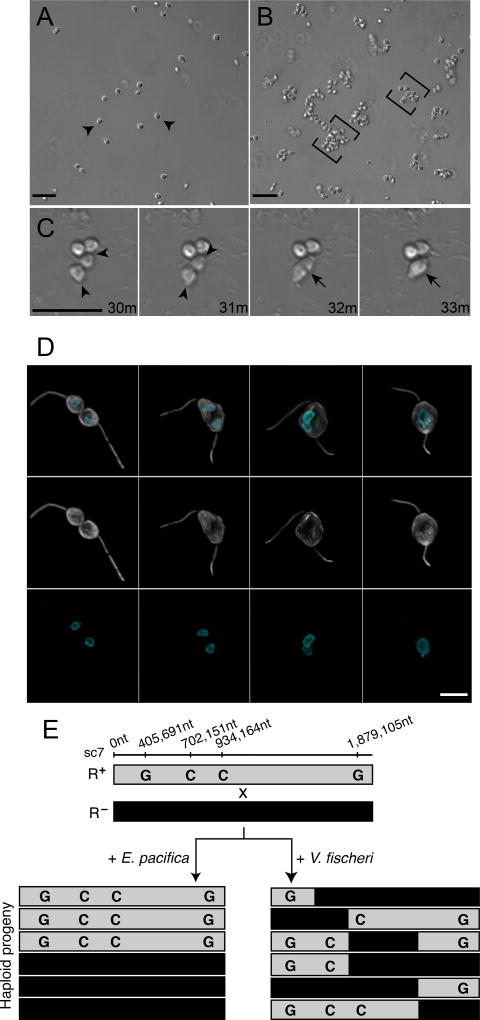
Figure 1. V. fischeri bacteria induce swarming and mating in the choanoflagellate, S. rosetta.
(A) In the absence of V. fischeri, motile S. rosetta cells (arrowheads) are evenly dispersed. (B) Within 30 minutes of exposure to V. fischeri, S. rosetta motile cells aggregate into large swarms (brackets). Scale bar = 20µm. (C) S. rosetta cells within a swarm pair and fuse. Prior to fusion, cells reposition themselves such that their basal membranes are adjacent and their apical flagella point away (31’; arrowheads mark apical pole of unfused cells). Cell fusion takes only minutes, and occurs along the basal membrane (32’; indicated by arrow), resulting in a single, elongated cell (33’; indicated by arrow). Scale bar = 20µm. (D) Stages of cell and nuclear fusion in S. rosetta mating pairs. Haploid mating pairs are oriented with their basal poles (opposite the flagellum) touching (D1), and cell fusion proceeds along the basal membrane, resulting in a binucleated cell with two flagella (D2). Nuclei then congress towards the midline (D3), where the nuclei undergo nuclear fusion, resulting in a diploid cell (D4). Anti-tubulin antibody (D1’-4’; white) highlights the cell body and flagellum, and Hoechst (D1’’-4’’; cyan) highlights the nucleus. Scale bar = 5µm (E) Evidence for meiotic recombination in S. rosetta following exposure to V. fischeri. Two haploid, genotypically distinct S. rosetta strains [R+(grey shading) and R- (black shading)] were mixed in the presence of either E. pacifica conditioned media (EPCM) or V. fischeri conditioned media (VFCM) for 16 hours. Haploid progeny were clonally isolated and genotyped at polymorphic markers across the genome (Supplemental Data). We show here genotyping results for four representative loci along supercontig 7 (sc7). All clones isolated from EPCM-treated cultures contained unrecombined parental genotypes, while haploid clones isolated from VFCM-treated cultures showed clear evidence of recombination. Top numbers show marker genomic positions along sc7.
Although swarming has not previously been reported in choanoflagellates and the biological significance of swarming in S. rosetta was not immediately obvious, swarming is associated with mating in diverse motile eukaryotes, including amoebae, ciliates, crustaceans, insects, fish, birds, and bats (Avery, 1984; Buskey, 1998; Downes, 1969; Giese, 1959; O'Day, 1979; Veith et al., 2004; Watson et al., 2003). Therefore, we hypothesized that swarming in S. rosetta might indicate mating. To investigate whether V. fischeri-induced swarming is a prelude to mating, we sought to determine whether the hallmarks of mating in microbial eukaryotes (cell fusion, nuclear fusion, and meiotic recombination (Bell, 1988; Dini and Nyberg, 1993; Levin and King, 2013)) occur in S. rosetta following treatment with V. fischeri.
Our lab previously found that S. rosetta cells mate at low frequencies (<2% of the population) after starvation for 11 days (Levin and King, 2013; Levin et al., 2014). In contrast, as early as 30 minutes after induction with live V. fischeri or conditioned medium isolated from a V. fischeri culture, S. rosetta cells formed swarms and then began to pair and fuse (Figure 1C and Movie 2). Once paired, cell fusion took as little as three minutes and all observed cell pairs were oriented with their basal poles (opposite the flagellum) touching. Paired cells subsequently fused along the basal pole, resulting in the formation of a binucleate cell harboring two flagella (Figure 1D). After cell fusion, the two nuclei congressed and fused, and one of the two parental flagella eventually retracted (Figure S1B), resulting in a diploid cell.
While cell fusion and nuclear fusion are consistent with mating, parasexual processes can occur in the absence of meiotic recombination (Goodenough and Heitman, 2014). Therefore, to test whether swarming was associated with the initiation of a true sexual cycle, we used V. fischeri to induce the formation of heterozygous diploids in S. rosetta cultures and then examined their offspring for evidence of meiosis and recombination (Figure 1E). To produce heterozygous diploid cells, two haploid S. rosetta strains (R+ and R−) containing previously characterized polymorphisms (Levin et al., 2014) were mixed either in the presence of V. fischeri conditioned medium (VFCM), or in conditioned medium from Echinicola pacifica (EPCM), a prey bacterium (Levin et al., 2014) that does not induce swarming, as a negative control (Figure 1E). After VFCM or EPCM exposure, 48 clones were isolated from each culture condition. Although we cannot directly measure the ploidy of live S. rosetta cells, heterozygous diploids can be readily identified by genotyping. While all clones (48/48) reared from the EPCM-treated culture contained un-recombined parental genotypes, 10/48 clones isolated following VFCM treatment were shown by genotyping to be heterozygous diploids. We surmised that the heterozygous diploids were the result of outcrossed mating, and found that further passaging of these clones yielded motile haploid progeny. 147 haploid progeny from three different heterozygous diploids were clonally isolated and genotyped at polymorphic sites across the genome, providing evidence for independent assortment and meiotic recombination (Figure 1E and Supplemental File 1). Taken together, these results demonstrate that S. rosetta swarms and mates in response to a compound produced by V. fischeri. Because the compound induces a switch from asexuality to sexuality, we hereafter refer to the molecular basis of the activity as an “aphrodisiac.”
Bioactivity-guided fractionation revealed that the V. fischeri aphrodisiac is a protein
Automated image analysis of S. rosetta cells swarming in response to VFCM enabled a quantitative bioassay of mating induction (Figure 2A, B and Methods). As a baseline, we found that 30 minutes after treatment with VFCM, S. rosetta cells consistently formed swarms containing between 5–35 cells each, whereas cells did not form clusters in response to EPCM. Using this bioassay, we first tested whether V. fischeri cues involved in quorum sensing (e.g. homoserine lactones) (Lupp and Ruby, 2005; Lupp et al., 2003) or those required for its symbiosis with the squid Euprymna scolopes (e.g. lipopolysaccharide (LPS) and peptidoglycan (PGN)) (Koropatnick et al., 2004; Shibata et al., 2012) might contribute to swarming induction in S. rosetta. A set of five different V. fischeri mutant strains that are deficient in quorum sensing were all wild type for swarming induction (Lupp and Ruby, 2005; Lupp et al., 2003), as were seven mutants in polysaccharide export pathways required for symbiosis with E. scolopes (Shibata et al., 2012) (Table S1). Moreover, treatment of S. rosetta with purified quorum sensing molecules (Table S2) and V. fischeri outer membrane vesicles (OMVs) containing LPS and PGN (Beemelmanns et al., 2014) (Figure S2A) also failed to elicit mating, suggesting that the cue(s) required for S. rosetta mating induction likely differ from factors required either for quorum sensing or squid colonization.
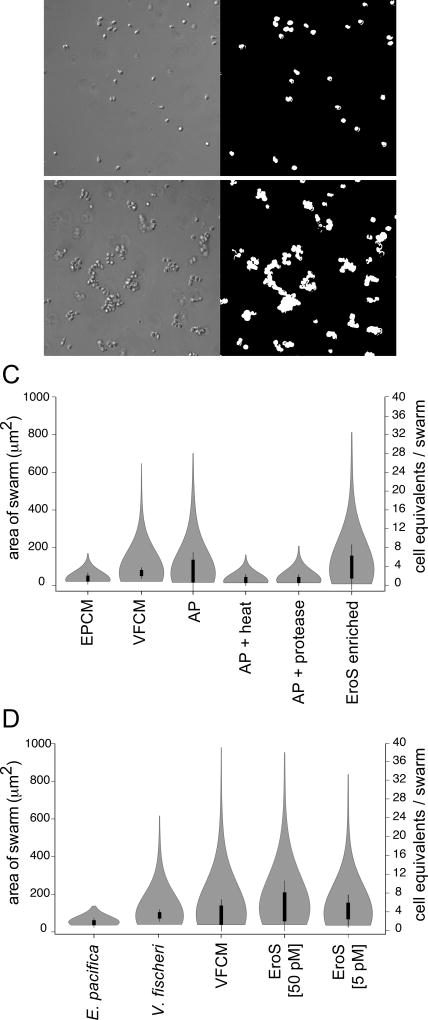
Figure 2. Bioactivity-guided isolation of the V. fischeri aphrodisiac.
(A, B) Automated image analysis allowed quantification of S. rosetta swarming in response to V. fischeri-derived activity. Pictured are S. rosetta cells 30 minutes after treatment with E. pacifica conditioned media (A) or V. fischeri conditioned media (B). By generating a binary mask (A’, B’) we could measure the area of each swarm and estimate the number of cells (“cell equivalents”) per swarm. (C) The V. fischeri aphrodisiac is a secreted protein. Swarming in S. rosetta is induced by V. fischeri culture supernatant (VFCM), as well as compounds in the ammonium sulfate precipitation of V. fischeri culture supernatant (AP), but not by AP exposed to heat (80°C for 10 minutes; AP + Heat) or proteases (AP + Protease), or by culture supernatant from the prey bacterium E. pacifica (EPCM). Proteins in the ammonium sulfate precipitation of V. fischeri culture supernatant were separated by size exclusion and anion exchange chromatography, and the aphrodisiac activity tracked with a ~90kD protein band (EroS enriched) that was revealed by mass spectrometry to be the V. fischeri EroS protein (also see Figure S2 and STAR Methods. Data are presented as violin box plots, showing the median cell number (white circle), interquartile range (thick line), and range excluding outliers (thin line). Surrounding the box plot is a kernel density trace, plotted symmetrically to show the swarm area frequency distribution. A minimum of 650 swarm areas were plotted for each condition. (D) EroS triggers mating at plausible environmental concentrations. Purified EroS induces swarming in S. rosetta at concentrations as low as 5 pM, and is sufficient to fully recapitulate the aphrodisiac activity of live V. fischeri bacteria and VFCM.
We next turned to an unbiased, activity-guided fractionation and found that the aphrodisiac activity was enriched in VFCM, including after depletion of OMVs (Figure S2A). The aphrodisiac produced by V. fischeri could be recovered from VFCM by ammonium sulfate precipitation, and the activity of the ammonium sulfate fraction was sensitive to both heat and protease treatment, suggesting that the activity might be proteinaceous (Figure 2C). We therefore separated all proteins precipitated from VFCM by size exclusion and anion exchange chromatography, and tested the protein fractions in the swarming bioassay (Figure 2A–C and Figure S2B; STAR Methods). The swarming activity tracked with a single ~90kD protein, which was revealed by mass spectrometry to be the uncharacterized V. fischeri protein VF_A0994, hereafter referred to as EroS for Extracellular regulator of Sex (Figure 2C and Supplemental File 2; GenPept Accession YP_206952). To test whether EroS was sufficient to induce swarming in S. rosetta, we heterologously expressed the eroS gene in E. coli and found that the purified protein recapitulated the swarm-inducing activity of live V. fischeri and VFCM (Figure 2D). Purified EroS protein was also sufficient to induce mating between two S. rosetta strains (R+ and R−; Table S3), demonstrating that a single protein secreted by V. fischeri is sufficient to induce both swarming and mating in S. rosetta.
EroS protein is a chondroitinase
To understand the mechanism by which V. fischeri induces choanoflagellate mating, we set out to determine the biochemical function of EroS. The EroS protein sequence contains a predicted glycosaminoglycan (GAG) lyase domain (supported by the detection of PFAM domains PF08124, PF02278, PF02884; (Finn et al., 2016)). GAGs are linear polysaccharides that are integral components of the animal extracellular matrix (ECM). GAG lyases depolymerize GAGs through an elimination mechanism that distinguishes them from hydrolases, and are produced by a subset of primarily pathogenic bacteria and fungi (Zhang et al., 2010), as well as by human commensals, including gut bacteria in the genus Bacteroides (Ahn et al., 2011; Hong et al., 2002). Through the alignment of the EroS protein sequence with multiple bacterial GAG lyases with solved structures, we found that EroS harbors conserved residues at sites required for catalytic activity (His-278 and Tyr-287; Figure 3A) (Han et al., 2014; Linhardt et al., 2006; Shaya et al., 2008; Weijun Huang et al., 2001).
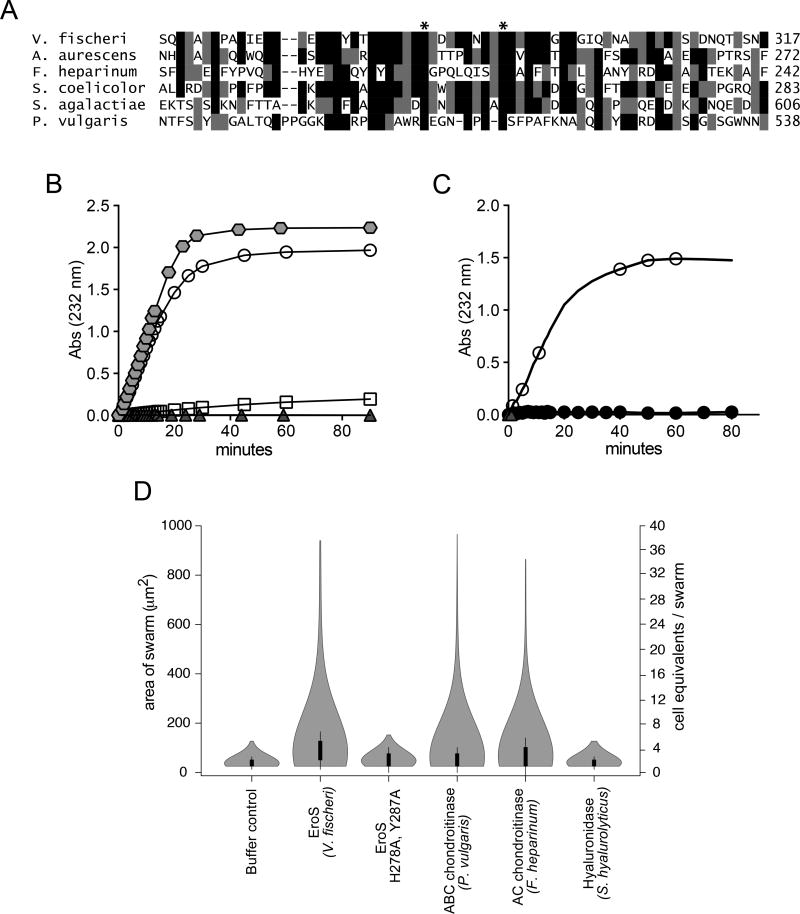
Figure 3. The V. fischeri aphrodisiac is a chondroitinase.
(A) Alignment of the V. fischeri EroS amino acid sequence to diverse bacterial GAG lyases reveals that V. fischeri harbors conserved His and Tyr residues (indicated by *) at sites required for catalytic activity in characterized GAG lyases (Han et al., 2014; Linhardt et al., 2006). Amino acids with >50% conservation between sequences are shaded (black shading for identical amino acids and grey shading for similar amino acids. (B) Purified EroS degrades chondroitin sulfate and hyaluronan. EroS was incubated with purified chondroitin sulfate (open circle), hyaluronan (grey hexagon), dermatan sulfate (open square), and heparan sulfate (grey triangle), and GAG lyase activity of EroS was measured by monitoring the abundance of unsaturated oligosaccharide reaction products with an absorbance at 232nm. Chondroitin sulfate and hyaluronan oligosaccharides accumulated rapidly in the presence of EroS, indicating depolymerization, whereas heparan sulfate and dermatan sulfate were not depolymerized by EroS. (C) Alanine substitutions at two predicted catalytic residues in EroS (H278 and Y287) eliminated the protein’s ability to degrade chondroitin sulfate. The chondroitinase activity of either wild type EroS (open circle) or EroS-H278A,Y287A (filled circle) against purified chondroitin sulfate was measured by monitoring the abundance of unsaturated oligosaccharide products with an absorbance at 232nm. (D) The chondroitinase activity of EroS is necessary and sufficient for its function as an aphrodisiac. EroS-H278A,Y287A failed to induce swarming in S. rosetta. P. vulgaris ABC chondroitinase and F. heparinum AC chondroitinase were sufficient to induce swarming at levels similar to EroS, whereas S. hyalurolyticus hyaluronidase failed to induce swarming, indicating that chondroitinase activity is necessary and sufficient for aphrodisiac activity.
Sulfated GAGs are thought to be eumetazoan-specific innovations (DeAngelis, 2002a; Yamada et al., 2011), and are not known to exist in choanoflagellates. (Although some pathogenic bacteria evade host immune responses by producing extracellular GAGs, these molecules are produced by way of an independently evolved biosynthetic pathway and, unlike animal GAGs, are not modified by sulfation (DeAngelis, 2002b)). Moreover, GAGs are diverse and the substrate specificities of GAG lyases cannot be deduced from sequence alone (Zhang et al., 2010). Therefore, we next set out to answer three questions: (1) does EroS exhibit GAG-degrading activity, (2) is the enzymatic activity of EroS required for its ability to induce mating, and (3) what are its substrates in S. rosetta?
We found that purified EroS protein degrades GAG substrates in vitro, and is thus a bona fide GAG lyase. GAGs are classified based on their disaccharide units: heparan sulfate, chondroitin sulfate, dermatan sulfate, hyaluronic acid, and keratan sulfate (Zhang et al., 2010). EroS showed strong lyase activity toward purified chondroitin sulfate and hyaluronan, but not heparan sulfate or dermatan sulfate (Figure 3B and Figure S2C, D). We did not test keratan sulfate because it does not contain uronic acid and therefore cannot be degraded by GAG lyases (Garron and Cygler, 2010).
We next asked whether the enzymatic activity of EroS is important to its function as an aphrodisiac. Alanine substitution at conserved catalytic residues (His-278 and Tyr-287; Fig. 3A) required for chondroitin degradation by EroS (Fig. 3C), eliminated its ability to induce swarming in S. rosetta (Fig. 3D) (Linhardt et al., 2006). Moreover, well-characterized chondroitin lyases from other bacteria (ABC chondroitinase from Proteus vulgaris and AC chondroitinase from Flavobacterium heparinum) induced swarming and mating in S. rosetta at levels resembling those induced by EroS (Figure 3D and Table S3), indicating that the chondroitinase activity of EroS is both necessary and sufficient for its function as an aphrodisiac.
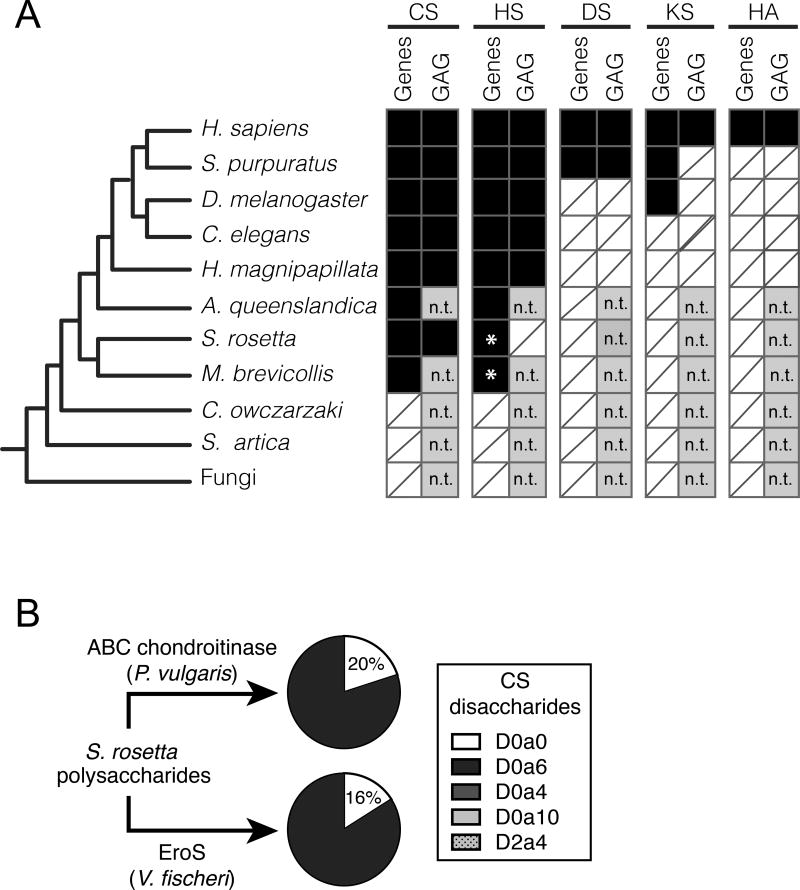
Although sulfated GAGs were previously thought to be restricted to animals, key heparan biosynthetic enzymes have been detected in the genome of the choanoflagellate Monosiga brevicollis (Ori et al., 2011), and we have further found that the S. rosetta genome encodes homologs of enzymes required for chondroitin biosynthesis (Figure 4A, Figure S3A, Supplemental File 3) (Fairclough et al., 2013; King et al., 2008). To test whether chondroitin is produced by S. rosetta, we treated polysaccharides isolated from motile S. rosetta cells with the broad specificity ABC chondroitinase from P. vulgaris. The P. vulgaris ABC chondroitinase liberated chondroitin disaccharides, demonstrating that S. rosetta indeed produces chondroitin sulfate (Figure 4B, Figure S3B,C). Finally, to test whether EroS can degrade S. rosetta chondroitin we treated S. rosetta polysaccharides with EroS and found that it released unsulfated chondroitin and chondroitin-6-sulfate disaccharides, indicating that S. rosetta chondroitin is a target of EroS (Figure 4B, Figure S3B,C).
Figure 4. S. rosetta produces chondroitin sulfate that can be degraded by EroS.
(A) Phylogenetic distribution of diverse GAGs [CS= chondroitin sulfate; HS= heparan sulfate; DS= dermatan sulfate; KS= keratan sulfate; HA= hyaluronan], and their biosynthetic genes. The presence (black box) and absence (white box with slash) of genes required for the biosynthesis of GAGs (“Gene”) and biochemical evidence for GAGs (“GAG”) in S. rosetta and select opisthokonts. *; Ori et al. (2011) identified putative homologs of a subset of HS biosynthetic enzymes in the M. brevicollis genome, and we detect homologs of the same limited set of HS biosynthetic enzymes in S. rosetta. Importantly, these enzymes are shared components of the chondroitin biosynthetic pathway, and digestion of S. rosetta polysaccharides with heparinases failed to liberate heparan sulfate disaccharides, suggesting S. rosetta does not produce heparan sulfate (also refer to Figure S3). n.t.; not tested (experiments have not been performed to biochemically profile GAGs). (B) S. rosetta produces chondroitin that can be degraded by EroS. Polysaccharides isolated from S. rosetta were treated with either P. vulgaris ABC chondroitinase, an enzyme that can degrade many modifications of chondroitin into its disaccharide units (CS disaccharides), or purified EroS. Both ABC chondroitinase and EroS yielded similar amounts of unsulfated chondroitin disaccharide (D0a0) and chondroitin-6-sulfate disaccharide (D0a6) degradation products, indicating that unsulfated chondroitin and chondroitin-6-sulfate are produced by S. rosetta. In contrast, we were unable to detect chondroitin-4-sulfate (D0a4), chondroitin-4,6-sulfate (D0a10), or chondroitin-2,4-sulfate (D2a4) following degradation of S. rosetta polysaccharides with either EroS or ABC chondroitinase.
Chondroitin sulfate in animal ECM can be found covalently linked to core proteins, thus forming proteoglycans. To determine whether chondroitin disaccharides released from proteoglycans play a role in stimulating mating, we tested various products of EroS digestion for aphrodisiac activity (Figure S2). Conditioned media from EroS-treated S. rosetta cells did not trigger swarming in naïve S. rosetta cells, nor did the digested products of commercial chondroitin sulfate treated with EroS. Moreover, swarming was not induced by any combination or concentration of unsulfated and 6-sulfated chondroitin disaccharides tested (Figure S2). These results lead us to hypothesize that the structural modification of S. rosetta proteoglycans by EroS, rather than the chondroitin disaccharide products of EroS digestion, are important for activating the swarming and mating pathway in S. rosetta.
Finally, because swarming has not been previously described in choanoflagellates, we investigated whether V. fischeri might induce swarming under plausible environmental conditions. We found that EroS is secreted constitutively by V. fischeri when grown under either high or low nutrient conditions (Table S4). Cultures of S. rosetta swarm in response to as little as 4×102 V. fischeri cells/mL – a density comparable to that of V. fischeri in oligotrophic oceans (from 6×102 cells/mL to >1×104 cells/mL during blooms (Jones et al., 2007)) – within 30 minutes of exposure (Figure S4). Moreover, EroS was sufficient to trigger robust swarming in S. rosetta at concentrations as low as 5 pM (Figure S4), making EroS as potent as the sex pheromones produced by volvocine algae (Kochert, 2012) and by marine invertebrates (Bartels-Hardege et al., 1996; Li et al., 2002). Together, these data suggest that V. fischeri or other chondroitinase-producing bacteria could plausibly trigger S. rosetta swarming and mating in natura.
Discussion
We have discovered that a secreted bacterial chondroitinase, EroS, induces mating in S. rosetta, one of the closest living relatives of animals. Through the study of this unexpected interkingdom interaction, we found that mating in S. rosetta is initiated in response to the degradation of chondroitin sulfate, a glycosaminoglycan previously thought to be restricted to animals.
The first hint that V. fischeri might induce mating came from the observation of S. rosetta swarms following exposure to the bacterium. By increasing local population density, swarming has previously been found to facilitate mating in diverse amoebae, flagellates, crustaceans, cnidarians, polychaetes, insects, fish, and birds (Avery, 1984; Buskey, 1998; Downes, 1969; Giese, 1959; Hamner and Dawson, 2009; Omori and Hamner, 1982; Sorensen and Wisenden, 2015; Watson et al., 2003). As in other organisms that swarm, the connection of swarming to mating may be critical, since their aquatic, pelagic lifestyle can make it challenging to find mates. Indeed, under the starvation conditions that trigger S. rosetta mating in the absence of swarming, mating takes >500X longer (~11 days) and occurs in a small fraction of the population (Levin et al., 2014).
Most previously characterized examples of coordinated mating behaviors are regulated by pheromonal cues. Conspecific swarming is initiated by diverse aggregation pheromones (for example, ester and isoprenoid pheromones in beetles (Kartika et al., 2015; Wertheim et al., 2005) and peptide pheromones in sea slugs (Painter et al., 2016) and polychaetes (Ram et al., 1999)), and free-spawning marine animals produce pheromones to synchronize gamete release and enhance fertilization success (Babcock et al., 2011). Biotic and abiotic cues from the environment can also help coordinate mating behavior in animals. For example, spawning in marine invertebrates is correlated with phytoplankton blooms, and some sea urchins, mussels, and polychaetes spawn after exposure to small molecules produced by environmental phytoplankton (Smith and Strehlow, 1983; Starr et al., 1990; 1992), although these cues remain structurally elusive. Just as phytoplankton blooms are hypothesized to signify a nutrient-rich environment for spawning, the presence of chondroitinase-producing bacteria may indicate an environmental condition, or the convergence of multiple environmental factors, that favor mating in S. rosetta. Although V. fischeri was the first bacterium observed to regulate mating in S. rosetta, we have since identified other bacteria that similarly induce swarming and mating (Table S1). Therefore, we predict that mating in S. rosetta might be regulated by diverse species of bacteria in nature, and hypothesize that swarming is a common occurrence within the natural life history of S. rosetta. Interestingly, the interaction between Vibrio bacteria and S. rosetta may enrich for conspecific mating, as no other tested choanoflagellate species swarmed or otherwise visibly responded following exposure to Vibrio spp. (Table S5).
Our discovery that V. fischeri produces a chondroitinase that induces mating also revealed that S. rosetta produces chondroitin sulfate, providing the first biochemical evidence for this important GAG in a non-animal and extending its evolutionary history to the premetazoan era. In an interesting parallel to the induction of S. rosetta mating by a chondroitinase, GAGs and sulfated polysaccharides mediate mating in diverse internally and externally fertilizing animals where they provide a protective and species-specific coating around oocytes (Miller and Ax, 1990). In the case of the mammalian oocyte, which is surrounded by the GAG hyaluronan, sperm secrete hyaluronidase to penetrate the hyaluronan-containing coating, whereas in sea urchins, sulfated polysaccharides coating the sea urchin oocyte ensure species-restricted sperm activation and binding (Mengerink and Vacquier, 2001). Of course, GAGs like hyaluronan and chondroitin sulfate are also essential components of the ECM in somatic cells of animals, where they contribute to a range of functions that include the maintenance of cell adhesion through interactions with ECM molecules, the integration of signals from the extracellular milieu, and the stabilization of collagen fibers. Future investigations may reveal whether chondroitin sulfate in S. rosetta functions to mediate species-specific cell recognition in the context of fertilization and may provide insight into the premetazoan roles of this important molecule. Moreover, it will be fascinating to explore whether bacteria influence mating in other aquatic organisms, for whom the triggers of mating are often obscure.
STAR Methods
Experimental Model and Subject Details
Culture media
Artificial seawater (ASW), cereal grass media (CG media), and Sea Water Complete media (SWC) were prepared as described previously (Levin and King, 2013; Woznica et al., 2016). Artificial sea water (ASW) was made by adding 32.9 g Tropic Marin sea salts (Wartenberg, Germany) to 1 L distilled water to a salinity of 32-27 parts per thousand. SWC media was made by adding 250 mg/L peptone, 150 mg/L yeast extract, 150 L/L glycerol in artificial sea water. CG media was made by infusing ASW with cereal grass pellets (Basic Science Supplies, Rochester NY).
Choanoflagellate husbandry
SrEpac (Levin and King, 2013) (S. rosetta grown in the presence of Echinicola pacifica bacteria, ATCC PRA-390) was propagated in 5% Sea Water Complete media (SWC diluted to 5% vol/vol in ASW) at 22°C. SrEpac was passaged 1:20 into 19mL fresh 5% SWC every other day to obtain stationary growth phase cultures (cells were grown in 25cm2 Corning cell culture flask). Prior to all induction bioassays, unless otherwise indicated, cells were diluted to approximately 1×105 cells/mL in ASW at the time of induction.
Method Details
Immunofluorescence microscopy
Stationary-phase cells were induced with V. fischeri conditioned media or E. pacifica conditioned media and fixed at intervals of 10 minutes, 30 minutes, 1 hour, 2 hours, and 4 hours after induction. After vortexing, cells were fixed for 5 min in 6% acetone followed by 10 minutes in 4% formaldehyde. Cells were then allowed to settle for 30 min onto poly-L-lysine coated coverslips (BD Bioscience). Cells were stained with E7 anti-tubulin antibody (1:200; Developmental Studies Hybridoma Bank), Alexa Fluor 488 anti-mouse (1:1000; Molecular Probes), and 0.01 mg/mL Hoechst 3342 (Thermo Fischer) before mounting in Prolong Gold antifade reagent (Molecular Probes). Cells were imaged at 63× using a Zeiss LSM 880 AxioExaminer with Airyscan.
Mating stages were assigned based on the following criteria: orientation of paired cells, fusion of cell bodies, localization and number nuclei, number of flagella. Cell fusion could be clearly distinguished from cell division for several reasons, including (1) fusing cells are paired basally, whereas recently divided sister cells are paired laterally, (2) flagella remain elongated during the fusion process, but are retracted throughout cell division and (3) DNA remains uncondensed throughout cell fusion, but is condensed during cell division.
Isolation of conditioned media (including VFCM and EPCM)
Vibrio fischeri ES114 (ATCC 700601) and all other Vibrio species (Table S2) were grown by shaking in 200mL 100% SWC media for 30 h at 20°C, and pelleted by centrifugation. E. pacifica was grown by shaking in 200mL 100% SWC for 30 h at 30°C, and pelleted by centrifugation. Cell-free supernatant was then vacuum filtered twice through a 0.22 µM filter (EMD Millipore Stericup) to obtain 100% CM. Concentrated conditioned media was obtained using 30kD and 50kD molecular weight cut off centrifugal filter units (Amicon).
Inducing mating and meiosis
S. rosetta strains
All crosses were performed between two S. rosetta strains with previously verified single nucleotide polymorphisms (SNPs), R- (previously referred to as Rosetteless) and R+ (previously referred to as Isolate B) (Levin et al., 2014). Prior to inducing mating, stationary phase cultures were obtained by passaging Rosetteless 1:20 into 19mLs fresh 5% SWC media every other day, and Isolate B 1:10 into 20mLs fresh 25% CG media every two days.
Inducing mating
Stationary phase R+ and R- cultures were counted and diluted to the same cell density (1×106 cells/mL). R+ and R- cultures were mixed in equal proportions, pelleted, and resuspended in fresh 25% CG media to obtain a final cell density of 1×106 cells/mL. Mating crosses were performed in 2 mL total volumes under the following induction conditions: 5% (V/V) E. pacifica conditioned media (EPCM), 5% (V/V) Vibrio fischeri conditioned media (VFCM), 0.5 nM VF_rGAG lyase, 0.0035 units Chondroitinase ABC (Sigma C3667), and 0.0035 units Chondroitinase AC (Sigma C2780). Cells were allowed to mate for 16 hours, after which the induced culture was pelleted and washed twice in 25% CG media to prevent further mating prior to limiting dilution.
Isolating diploids by limiting dilution
Mated cells were clonally isolated by limiting dilution into 96-well plates containing 25% CG media. For all crosses performed, the probability of clonal isolation at this step was between 0.85 and 0.92. Although we cannot directly measure the ploidy of live S. rosetta cells, the differentiation of planktonic motile cells into substrate-attached “thecate” cells correlates with the transition to diploidy (Levin et al., 2014). After five days of growth, isolates were phenotyped and then divided into two populations. For each isolate, one population was rapidly passaged to induce meiosis (see below), and the other population was used for DNA extraction. For DNA extraction, isolates were expanded into 1 mL of 5% CG media to prevent meiosis, and grown for three days in 24-well plates. DNA was extracted from each isolate using the following method: 500µL of cells were pelleted and resuspended in 20µL base solution (25mM NaOH, 2mM EDTA). Base solutions from the isolates were transferred to a PCR plate, boiled at 100°C fo r 20 min, and cooled at 4°C for 5 min. 20µL Tris solution (40mM Tris-HCl, pH 7.5) was then added to each sample. 1µL of this sample was used as the DNA template for genotyping reactions. To identify which isolates were the result of outcrossed mating, isolates were genotyped at two unlinked microsatellite markers that are polymorphic between the R+ and R-parental strain (Levin et al., 2014). All outcrossed diploids isolated were phenotypically thecate as opposed to motile planktonic. No thecate isolates were observed in control EPCM treated cultures.
Isolation of haploid meiotic progeny
Immediately after phenotyping, clones isolated by limiting dilution were passaged 1:10 into 1mL fresh 25% CG media to induce meiosis. Thecate clones that were outcrossed diploids typically gave rise to a clear mixture of haploid chains and rosettes after two days. Haploids were clonally isolated by limiting dilution into 96-well plates containing 25% CG media, and phenotyped after five days. Meiosis was confirmed either by 1) genotyping at two unlinked microsatellite markers, or 2) by genotyping at 38 markers using KASP technology (LGC Genomics, Beverly, MA) (Levin et al., 2014).
Genotyping meiotic progeny
To confirm genome-wide recombination, haploid progeny isolated from the 5% VFCM-induced cross were genotyped at 38 markers (Levin et al., 2014). Briefly, three outcrossed diploids (named A2, A3, and H2) were rapidly passaged to induce meiosis, and clones from each outcrossed diploid were isolated by limiting dilution. The probability of clonal isolation at this step was 0.94 for A2, 0.93 for A3, and 0.91 for H2. A total of 147 haploid isolates from the three outcrossed diploids were phenotyped and expanded for subsequent DNA extraction and genotyping.
Quantifying mating swarms
Inductions were set up in 100 µL volumes in 96-well glass bottom plates (Ibidi 89626). Assays were imaged at 10X magnification using transmitted light (bright field) on the Zeiss Observer Z.1 platform using a Hamamatsu C11440 camera. An automated sequence was set up such that each sample was imaged at 4 distinct locations throughout the well.
Images were batch processed in ImageJ to ensure consistency. After applying the ‘Smooth’ command to reduce background bacterial signal, the ‘Find Edges’ command was applied to further highlight the phase-bright choanoflagellate cells. Images were then converted to black and white using the ‘Make Binary’ command, followed by the ‘Close’ command to fill in small holes.
Finally, images were analyzed using the ‘Analyze Particles’ command to calculate the area of each swarm (left Y axis; swarms correspond to the white space in Figure 2A’,B’). Cell equivalents / swarm (right Y axis) were calculated by dividing the area of a swarm by the area of an individual cell (the area of an individual cell was the averaged area of 100 single cells). Data are presented as violin box plots, showing the median cell number (white circle), interquartile range (thick line), and range excluding outliers (thin line). Surrounding the box plot is a kernel density trace, plotted symmetrically to show the swarm area frequency distribution. A minimum of 650 swarm areas are plotted for each condition.
Isolating the Vibrio fischeri mating induction factor
Preparation of >30kD-enriched VFCM
Eight 1 L cultures of V. fischeri ES114 were grown shaking for in 100% SWC for 24 h at 25°C. Cultures were pelleted at 16,000 × g, and the supernatants were concentrated to 120 mL using a tangential flow filtration device with a 30 kDa centramate filter (Pall #OS030T12). The supernatant was then further clarified by pelleting 39,000 × g.
Ammonium sulfate precipitation
45 mL of >30kD-enriched VFCM was treated with 5 mL of 1 M Tris-HCl (pH 7.6) and precipitated with increasing concentrations of ammonium sulfate to obtain precipitates with 40%, 45%, 50%, 55%, 60%, 65%, 70%, and 75% NH4SO4 saturation. Each precipitate was resuspended in 3 mL water, and tested in the swarming bioassay.
Size exclusion chromatography
Active ammonium sulfate precipitation fractions (fractions 50%–65%; Figure S2B) were combined and concentrated to 1 mL. 0.85 mL was injected on a HiPrep™ 16/60 Sephacryl™ S-200 High Resolution column (GE Healthcare Life Sciences #17-1166-01) using an AKTA Explorer FPLC instrument. Proteins were eluted with 30 mM Tris-HCl (pH 7.7, 4 °C) at 0.5 mL/min for 120 mL, and 2 mL fractions were collected. The majority of UV-absorbing material eluted between fractions 15 and 44. Adjacent SEC fractions were paired and tested in the swarming bioassay, as well as analyzed by PAGE. Fractions 25/26 and 27/28 exhibited robust swarm-inducing activity (Figure S2B).
Anion exchange chromatography
Active SEC fractions were combined and concentrated to 1 mL in Solvent A (20 mM L-histidine, pH 6.0) and injected at 2.5 mL/min into a HiPrep™ 16/10 Q XL column (GE Healthcare Life Sciences #17-5092-01). Proteins were eluted in 2 mL fractions over a 300 mL linear gradient (0–100%) of Solvent A to Solvent B (1 M NaCl in 20 mM L-histidine, pH 6.0). Of the 150 fractions collected, fractions 77–84 exhibited the most robust swarm-inducing activity (Figure S2B), and were further analyzed by PAGE.
Renaturing proteins after PAGE
PAGE analysis of active AEX fractions revealed that few proteins were present in the fractions that displayed robust swarm-inducing activity. Proteins from active AEX fractions were concentrated and mixed with 15 µg/mL β-lactoglobulin carrier protein, and run in adjacent lanes through a NuPAGE™ 4–12% Bis-Tris polyacrylamide gel. Evenly spaced bands were excised from one lane, and the remaining gel was stained with Coomassie blue R-250 and retained for mass spectrometry. The excised slices were crushed, and then extracted for 6 hours with 300 µL of elution buffer (50 mM Tris-HCl pH 7.7, 100 µM EDTA, 1 mM DDT, 150 mM NaCl, 0.1% sodium dodecyl sulfate [SDS], 100 µg/mL bovine serum albumin [BSA], pH 7.7) (Hager and Burgess, 1980). Proteins were precipitated with 1200 µL cold acetone, incubated on dry ice for 30 minutes, and pelleted at 16,000 × g for 10 minutes at 4 °C. Air-dried pellets were dissolved in 10 µL of solubilization buffer (50 mM Tris-HCl pH 7.7, 100 µM EDTA, 1 mM DDT, 150 mM NaCl, 20% glycerol, 6 M guanidine hydrochloride) for 20 minutes, and then diluted with 500 µL of dilution buffer (50 mM Tris-HCl pH 7.7, 100 µM EDTA, 1 mM DDT, 150 mM NaCl, 20% glycerol). Proteins further renatured for 3.5 h at room temperature, and were concentrated to 20 µL. Proteins excised from bands were then tested in the swarming bioassay.
Mass Spectrometry
Because only one excised protein band displayed bioactivity, the corresponding slice retained for mass spectrometry was subjected to trypsin digestion and LC-MS/MS at the Proteomics/Mass Spectrometry Laboratory at UC Berkeley. Complete mass spectrometry results are listed in Supplemental File 2.
Protein expression and purification
The eroS gene was amplified by PCR (Phusion® DNA polymerase, New England Biosciences) from V. fischeri ES114 genomic DNA (Forward primer: 5’-GCCTCTGTCGACGCAAAAAATACCCAAACACCAC; Reverse primer: 5’-AATTAAGCGGCCGCCGTCTTGAATTGTTACTTGGAAAGAATAAG). After digestion with SalI-HF and NotI-HF (New England Biolabs), the eroS gene was ligated in-frame into a pET6xHN-N vector (Clontech) for fusion to a His-tag and transformed into OneShot BL21(DE3) cells (Invitrogen) for expression. Transformed E. coli were grown at 37 °C, 200 rpm shaking in LB media supplemented with 100 µg/mL ampicillin. After growth to OD 1.0, the temperature was decreased to 16 °C, and protein expression was induced by addition of 1 mM IPTG. After 24 hours, cells were pelleted and lysed with xTractor™ buffer (Clontech), purified with HisTALON™ gravity columns (Clontech), and the His-tag was released by enterokinase cleavage (Millipore enterokinase cleave capture kit #69067).
A gBlock of the eroS gene sequence harboring H278A and Y287A mutations was purchased from Integrated DNA Technologies and ligated into the pET15b vector (Novagen) at NdeI and BamHI restriction sites. The mutant eroS gene was transformed into OneShot BL21 Star (DE3) cells (Invitrogen) for expression. Transformed E. coli were grown at 37 °C, 220 rpm shaking in LB media supplemented with 100 µg/mL ampicillin. After growth to OD 0.8, the temperature was decreased to 16 °C, and protein expression was induced by addition of 0.3 mM IPTG. After 24 hours, cells were lysed and protein was purified with HisPur Cobalt Resin (Thermo Scientific).
Amino acid sequence alignments
Amino acid sequences from V. fischeri (VF_A0994, GenBank: AAW88064.1) and characterized bacterial GAG lyases [A. aurescens (AC lyase, PDB: 1RWG_A), S. coelicolor (AC lyase, PDB: 2WDA_A), S. agalactiae (hyaluronate lyase, PDB: 1LXM_A), F. heparinum (AC lyase, PDB: 1CB8_A), and P. vulgaris (ABC chondroitinase, GenBank: ALL74069.1)] were aligned using Clustal Omega multiple sequence alignment (http://www.ebi.ac.uk/Tools/msa/clustalo/). Conserved amino acid residues were highlighted using BoxShade (http://www.ch.embnet.org/software/BOX_form.html).
EroS GAG lyase activity in vitro
The glycosaminoglycan cleavage activity of purified Eros was determined in vitro as previously described (Wang et al., 2015). Briefly, GAG standards [hyaluronic acid (Sigma #H5388), chondroitin sulfate (Sigma #C4384), dermatan sulfate (Sigma #C3788), and heparin (Sigma #H3393)] were dissolved to a concentration of 1 mg/mL in buffer solution (50 mM NaH2PO4/Na2HPO4, 0.5 M NaCl, pH 8.0). 1 mL of GAG standard was added to 5 µL of enzyme solution. The assays were performed in quartz cuvettes (1 cm pathlength) at 23 °C, and UV absorbance measurements (232 nm) were taken directly on the reacting mixture.
AQUA quantification of EroS
Absolute Quantification (AQUA) peptide (Gerber et al., 2003) was used to accurately quantify the concentration of purified EroS and EroS in Vibrio fischeri conditioned media. Briefly, V. fischeri was grown for 8 H in 100% SWC media and 5% SWC media. Purified EroS and concentrated Vibrio fischeri culture supernatants were loaded onto a PAGE gel. The gel was stained with Coomassie blue R-250, and bands containing VF_A0994 were excised and sent to the Taplin Mass Spectrometry facility (Harvard Medical School) for further analysis. Synthetic AQUA peptide (TQITDDTYQNFFD[KC13N15], Sigma-Aldrich) and trypsin were added to each excised band, and LC-MS/MS was performed on the digested peptides using a Thermo Scientific Orbitrap. The amount of EroS present in each gel slice was calculated by comparing MS2 peak intensities of the native peptide with the internal AQUA synthetic peptide standard.
S. rosetta polysaccharide isolation and GAG disaccharide analysis
6 × 500 mL cultures of SrEpac were grown in 5% SWC until mid-stationary phase, and washed 3× to reduce bacterial load before being pelleted, flash frozen, and lyophilized. 125 mg of lyophilized S. rosetta sample was sent to the Complex Carbohydrate Research Center for GAG isolation, digestion, and SAX-HPLC.
Polysaccharides were isolated from the S. rosetta sample and digested with either EroS or chondroitinase ABC (Sigma C3667) for chondroitin disaccharide analysis, and heparinases I, II, and III (Dextra Laboratories) for heparan disaccharide analysis. Briefly, a ratio of 10 µL S. rosetta polysaccharides to 1uL of enzyme was incubated for 24 hours. Samples were heated to 100°C for 5 minutes to inactivate the enzyme, and centrifuged at 14,000 rpm for 30 minutes prior to SAX-HPLC.
SAX-HPLC was carried out on an Agilent system using a 4.6×250 mm Waters Spherisorb analytical column with 5µm particle size at 25°C. Detection was performed by post-column derivatization. Briefly, the eluent from the column was combined with a 1:1 mixture of 0.25 M NaOH and 1% 2-cyanoacetamide pumped at a flow rate of 0.5 mL/min from a post-column reactor. The eluent was heated to 130°C in a 10m reaction coil, then cooled in a 50-cm cooling coil and directed into a Shimadzu fluorescence detector (λex = 346 nm, λem = 410). Commercial standard disaccharides (Dextra Laboratories) were used for identification of each disaccharide based on elution time, as well as calibration.
Testing bioactivity of chondroitin disaccharides
Chondroitin disaccharides and chondroitin sulfate were tested for bioactivity by treating S. rosetta with unsulfated chondroitin disaccharides (Sigma C3920), chondroitin-6-sulfate disaccharides (Sigma C4170), unsulfated chondroitin + chondroitin-6-sulfate disaccharides, and chondroitin sulfate (from shark cartilage, Sigma C4384) at concentrations ranging from 0.0001M-0.1M. Cells were imaged and quantified after 30 minutes, 1 hour, and 3 hours.
Degradation products of chondroitin sulfate were generated by incubating 100 µg of chondroitin sulfate with either 50 pM EroS or 1 unit ABC chondroitinase (P. vulgaris) overnight. Enzymatic activity was killed by incubating samples at 80°C for 5 minutes. The resulting degradation products were tested for bioactivity at concentrations ranging from 0.0001M-0.1M. Cells were imaged and quantified after 30 minutes, 1 hour, and 3 hours.
Supplementary Material
(A) Stills of swarm formation after induction with V. fischeri bacteria. Arrowhead tracks the formation and movement of a single swarm over time. (B) Nuclear fusion in a mated pair of S. rosetta cells following treatment with V. fischeri. The final result of nuclear fusion is a diploid cell, harboring a single flagellum (B1-1’’). Hoechst (B1’; cyan) highlights the nucleus, and anti-tubulin antibody (B1’’; white) highlights the cell body and flagellum.
KASP genotyping of meiotic progeny isolated from V. fischeri- induced cross
Mass Spectrometry of bioactive V. fischeri protein
Putative homologs of GAG biosynthetic enzymes
(A) Swarming in S. rosetta is induced by large (>50kD), water-soluble factors present in V. fischeri conditioned media (VFCM). To identify the source of the aphrodisiac activity, proteins were precipitated from VFCM (AP 50%–65%) and separated by size exclusion (SEC) and anion exchange (AEX) chromatography. A protein band of ~90kD, later determined to be EroS (VF_A0994), was abundant in the bioactive SEC (SEC 25/26) and AEX (AEX 81/82) fractions. (B, C) EroS is a chondroitin AC lyase. (B) EroS degrades chondroitin sulfate AC, but not chondroitin sulfate B (dermatan sulfate) in vitro. EroS was incubated with purified chondroitin sulfate AC (open circle) and chondroitin sulfate B (dark grey square), and lyase activity of EroS was measured by monitoring the abundance of unsaturated oligosaccharide products with an absorbance at 232nm. Diamond represents a no enzyme control. (C) Chondroitinase ABC (P. vulgaris), a positive control for in vitro chondroitin degradation assays, rapidly depolymerizes both chondroitin sulfate AC (open circle) as well as chondroitin sulfate B (grey square). Diamond represents a no enzyme control. (E) Swarming in S. rosetta is not induced by chondroitin sulfate or chondroitin disaccharides. Neither commercial chondroitin disaccharides (D0a6 and D0a0), chondroitin disaccharides generated via the depolymerization of chondroitin sulfate by EroS, nor conditioned media isolated after EroS-treatment of S. rosetta cells are sufficient to induce swarming in S. rosetta.
(A) Orthologs of chondroitin sulfate (CS) synthesis are present in the genomes of choanoflagellates S. rosetta and M. brevicollis. Genes identified as “linker” synthesize the proteoglycan linker tetrasaccharide and are important for the biosynthesis of multiple types of GAG, whereas the genes identified as “CS synthesis” are specific to CS biosynthesis. The gene identified as “DS” is required for the formation of dermatan sulfate. All query sequences used were human orthologs. If multiple subject sequences were hits for a single query sequence, the ortholog with the highest Blastp score was chosen. Query and subject accession information is provided in Supplemental File 3. (B) S. rosetta produces chondroitin that can be degraded by ABC chondroitinase and EroS. Polysaccharides isolated from S. rosetta were treated with either ABC chondroitinase from P. vulgaris (center plot) or EroS (bottom plot). Degradation products from samples treated with ABC chondroitinase and EroS were separated by SAX-HPLC (X-axis indicates time, Y-axis indicates abundance) and compared to the following chondroitin disaccharide standards (top plot): (1) D0a0, unsulfated chondroitin; (2) D0a6, chondroitin-6-sulfate; (3) D0a4, chondroitin-4-sulfate; (4) D0a10, chondroitin-4,6-sulfate; (5) D2a4, chondroitin-2,4-sulfate. Unsulfated and 6-sulfated chondroitin disaccharides were present at similar abundance in both the ABC chondroitinase and EroS –treated samples, whereas all other chondroitin disaccharides were below the limit of detection. (C) Quantification of chondroitin disaccharide products produced by ABC chondroitinase (P. vulgaris) and EroS treatment of S. rosetta polysaccharides. Disaccharide abbreviations: D0a0=unsulfated chondroitin; D0a6=chondroitin-6-sulfate; D0a4= chondroitin-4-sulfate; D0a10=chondroitin-4,6-sulfate; D2a4=chondroitin-2,4-sulfate. (D) S. rosetta does not produce heparan sulfate. Polysaccharides isolated from S. rosetta (bottom plot) were treated with Heparinase I, Heparinase II, and Heparinase III (Dextra Laboratories) separated by SAX-HPLC (X-axis indicates time, Y-axis indicates abundance) and compared to the following heparan sulfate disaccharide standards (top plot): (1) D0A0; (2) D0S0; (3) D0A6; (4) D2A0; (5) D0S6; (6) D2S0; (7) D2A6; (8) D2S6. (E) No heparan disaccharides were present above the limit of detection in the S. rosetta polysaccharide sample.
(A) S. rosetta swarms in response to low numbers of V. fischeri bacteria in a cell density-dependent manner. S. rosetta at high cell densities (2.0×106 cells/mL) swarms in response to as few as one V. fischeri cell per 1000 S. rosetta cells within 30 minutes of exposure, whereas swarming in S. rosetta at lower cell densities (2.0×105 cells/mL) within a similar time frame requires at least one V. fischeri cell per 500 S. rosetta cells. (B) Picomolar concentrations of secreted (5% VFCM) and purified EroS are sufficient to induce swarming in S. rosetta.
The movie beings with a side-by-side comparison of S. rosetta treated with EPCM (control; left) and VFCM (right) one hour post induction. After the short transition (“Induction with V. fischeri”) is a time-lapse depicting early swarm formation after treatment with 5% VFCM. All footage is displayed at 35× real time.
Two cells within a four-cell swarm undergo cell fusion. Movie begins 30 minutes after addition of 5% VFCM. Cell fusion is displayed at 60X real time.
Highlights.
The bacterium Vibrio fischeri induces mating in the choanoflagellate S. rosetta.
The ‘aphrodisiac’ produced by V. fischeri is a chondroitinase that we name EroS.
The enzymatic activity of EroS is required for this function
Chondroitin sulfate, the EroS substrate, evolved before the origin of animals.
Bacteria generate a small molecule that stimulates swarming and sexual reproductive behavior in choanoflagellates, the closest living relatives of animals
Acknowledgments
This work was funded by the NIH (GM099533). We thank K. Visick for V. fischeri syp deletion mutants, N. Ruby for V. fischeri quorum sensing deletion mutants, and Y. Boucher for diverse species of Vibrio bacteria. We thank M. Abedin, D. Booth and B. Larson for helpful discussions and critical reading of the manuscript and B.L. Azpeitia for technical assistance. This work used the Vincent J. Coates Proteomics/Mass Spectrometry Laboratory at UC Berkeley, supported in part by the NIH (S10 RR025622), and the Complex Carbohydrate Research Center, supported by the NIH-funded Research Resource for Integrated Glycotechnology (P41GM103390). J.P.G. is supported by a Ruth L. Kirchstein Postdoctoral Fellowship from the NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
Conceptualization: A.W., J.P.G., J.C, and N.K.; Investigation: A.W., J.P.G., and R.E.H.; Writing- Original Draft: A.W. and N.K.; Writing- Review and Editing: A.W., N.K., J.P.G., and J.C.; Funding Acquisition: N.K. and J.C.
Supplemental Notes
Only 60 validated polymorphisms differentiate S. rosetta strains R+ and R− 21. Therefore, while we are able to detect independent assortment and recombination, because few markers sit within close proximity it was not possible to utilize genotyping data to accurately measure recombination frequency and construct linkage groups.
References
- Ahn MY, Shin KH, Kim DH, Jung E-A, Toida T, Linhardt RJ, Kim YS. Characterization of a Bacteroides species from human intestine that degrades glycosaminoglycans. Can. J. Microbiol. 2011;44:423–429. doi: 10.1139/cjm-44-5-423. [DOI] [PubMed] [Google Scholar]
- Alegado RA, Brown LW, Cao S, Dermenjian RK, Zuzow R, Fairclough SR, Clardy J, King N. A bacterial sulfonolipid triggers multicellular development in the closest living relatives of animals. Elife. 2012;1:e00013. doi: 10.7554/eLife.00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avery MI. Lekking in birds: choice, competition and reproductive constraints. Ibis. 1984;126:177–187. [Google Scholar]
- Babcock R, Mundy C, Keesing J, Oliver J. Predictable and unpredictable spawning events: in situ behavioural data from free-spawning coral reef invertebrates. Invertebrate Reproduction & Development. 2011;22:213–227. [Google Scholar]
- Bartels-Hardege HD, Hardege JD, Zeeck E, Müller C, Wu BL, Zhu MY. Sex pheromones in marine polychaetes: a biologically active volatile compound from the coelomic fluid of female Nereis (Neanthes) japonica. Journal of Experimental Marine Biology and Ecology. 1996;201:275–284. [Google Scholar]
- Beemelmanns C, Woznica A, Alegado RA, Cantley AM, King N, Clardy J. Synthesis of the rosette-inducing factor RIF-1 and analogs. J. Am. Chem. Soc. 2014;136:10210–10213. doi: 10.1021/ja5046692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell G. Sex and death in protozoa: the history of an obsession. Cambridge University Press; 1988. [Google Scholar]
- Buskey EJ. Components of mating behavior in planktonic copepods. Journal of Marine Systems. 1998;15:13–21. [Google Scholar]
- Cantley AM, Woznica A, Beemelmanns C, King N, Clardy J. Isolation and Synthesis of a Bacterially Produced Inhibitor of Rosette Development in Choanoflagellates. J. Am. Chem. Soc. 2016;138:4326–4329. doi: 10.1021/jacs.6b01190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayel MJ, King N. Prey capture and phagocytosis in the choanoflagellate Salpingoeca rosetta. PloS One. 2014;9:e95577. doi: 10.1371/journal.pone.0095577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayel MJ, Alegado RA, Fairclough SR, Levin TC, Nichols SA, McDonald K, King N. Cell differentiation and morphogenesis in the colony-forming choanoflagellate Salpingoeca rosetta. Developmental Biology. 2011;357:73–82. doi: 10.1016/j.ydbio.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeAngelis PL. Evolution of glycosaminoglycans and their glycosyltransferases: implications for the extracellular matrices of animals and the capsules of pathogenic bacteria. Anat Rec. 2002a;268:317–326. doi: 10.1002/ar.10163. [DOI] [PubMed] [Google Scholar]
- DeAngelis PL. Microbial glycosaminoglycan glycosyltransferases. Glycobiology. 2002b;12:9R–16R. doi: 10.1093/glycob/12.1.9r. [DOI] [PubMed] [Google Scholar]
- Dini F, Nyberg D. Advances in Microbial Ecology. Boston, MA: Springer US; 1993. Sex in Ciliates; pp. 85–153. [Google Scholar]
- Downes JA. The swarming and mating flight of Diptera. Annual Review of Entomology. 1969;14:271–298. [Google Scholar]
- Fairclough SR, Chen Z, Kramer E, Zeng Q, Young S, Robertson HM, Begovic E, Richter DJ, Russ C, Westbrook MJ, et al. Premetazoan genome evolution and the regulation of cell differentiation in the choanoflagellate Salpingoeca rosetta. Genome Biol. 2013;14:R15. doi: 10.1186/gb-2013-14-2-r15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairclough SR, Dayel MJ, King N. Multicellular development in a choanoflagellate. Curr Biol. 2010;20:R875–R876. doi: 10.1016/j.cub.2010.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, Potter SC, Punta M, Qureshi M, Sangrador-Vegas A, et al. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2016;44:D279–D285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garron M-L, Cygler M. Structural and mechanistic classification of uronic acid-containing polysaccharide lyases. Glycobiology. 2010;20:1547–1573. doi: 10.1093/glycob/cwq122. [DOI] [PubMed] [Google Scholar]
- Gerber SA, Rush J, Stemman O, Kirschner MW, Gygi SP. Absolute quantification of proteins and phosphoproteins from cell lysates by tandem MS. Proceedings of the National Academy of Sciences. 2003;100:6940–6945. doi: 10.1073/pnas.0832254100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giese AC. Comparative physiology: annual reproductive cycles of marine invertebrates. Annu Rev Physiol. 1959;21:547–576. doi: 10.1146/annurev.ph.21.030159.002555. [DOI] [PubMed] [Google Scholar]
- Goodenough U, Heitman J. Origins of eukaryotic sexual reproduction. Cold Spring Harbor Perspectives in Biology. 2014;6:a016154–a016154. doi: 10.1101/cshperspect.a016154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hager DA, Burgess RR. Elution of proteins from sodium dodecyl sulfate-polyacrylamide gels, removal of sodium dodecyl sulfate, and renaturation of enzymatic activity: Results with sigma subunit of Escherichia coli RNA polymerase, wheat germ DNA topoisomerase, and other enzymes. Anal. Biochem. 1980;109:76–86. doi: 10.1016/0003-2697(80)90013-5. [DOI] [PubMed] [Google Scholar]
- Hamner WM, Dawson MN. A review and synthesis on the systematics and evolution of jellyfish blooms: advantageous aggregations and adaptive assemblages. Hydrobiologia. 2009;616:161–191. [Google Scholar]
- Han W, Wang W, Zhao M, Sugahara K, Li F. A novel eliminase from a marine bacterium that degrades hyaluronan and chondroitin sulfate. J Biol Chem. 2014;289:27886–27898. doi: 10.1074/jbc.M114.590752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SW, Kim BT, Shin HY, Kim WS, Lee KS, Kim YS, Kim DH. Purification and characterization of novel chondroitin ABC and AC lyases from Bacteroides stercoris HJ 15, a human intestinal anaerobic bacterium. The FEBS Journal. 2002;269:2934–2940. doi: 10.1046/j.1432-1033.2002.02967.x. [DOI] [PubMed] [Google Scholar]
- Jones BW, Maruyama A, Ouverney CC, Nishiguchi MK. Spatial and Temporal Distribution of the Vibrionaceae in Coastal Waters of Hawaii, Australia, and France. Microb Ecol. 2007;54:314–323. doi: 10.1007/s00248-006-9204-z. [DOI] [PubMed] [Google Scholar]
- Kartika T, Shimizu N, Yoshimura T. Identification of Esters as Novel Aggregation Pheromone Components Produced by the Male Powder-Post Beetle, Lyctus africanus Lesne (Coleoptera: Lyctinae) PloS One. 2015;10:e0141799. doi: 10.1371/journal.pone.0141799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King N, Westbrook MJ, Young SL, Kuo A, Abedin M, Chapman J, Fairclough S, Hellsten U, Isogai Y, Letunic I, et al. The genome of the choanoflagellate Monosiga brevicollis and the origin of metazoans. Nature. 2008;451:783–788. doi: 10.1038/nature06617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochert G. Sexual Pheromones in Volvox Development. Sexual Interactions in Eukaryotic Microbes. 2012:73–93. [Google Scholar]
- Koropatnick TA, Engle JT, Apicella MA, Stabb EV, Goldman WE, McFall-Ngai MJ. Microbial factor-mediated development in a host-bacterial mutualism. Science. 2004;306:1186–1188. doi: 10.1126/science.1102218. [DOI] [PubMed] [Google Scholar]
- Leadbeater BSC. The Choanoflagellates: Evolution, Ecology, and Biology. Cambridge University Press; 2015. [Google Scholar]
- Levin TC, King N. Evidence for sex and recombination in the choanoflagellate Salpingoeca rosetta. Current Biology. 2013;23:2176–2180. doi: 10.1016/j.cub.2013.08.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin TC, Greaney AJ, Wetzel L, King N. The Rosetteless gene controls development in the choanoflagellate S. rosetta. Elife. 2014:3. doi: 10.7554/eLife.04070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Scott AP, Siefkes MJ, Yan H, Liu Q, Yun S-S, Gage DA. Bile acid secreted by male sea lamprey that acts as a sex pheromone. Science. 2002;296:138–141. doi: 10.1126/science.1067797. [DOI] [PubMed] [Google Scholar]
- Linhardt RJ, Avci FY, Toida T, Kim YS, Cygler M. Chondroitin Sulfate: Structure, Role and Pharmacological Activity. Elsevier; 2006. CS Lyases: structure, activity, and applications in analysis and the treatment of diseases; pp. 187–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupp C, Ruby EG. Vibrio fischeri uses two quorum-sensing systems for the regulation of early and late colonization factors. Journal of Bacteriology. 2005;187:3620–3629. doi: 10.1128/JB.187.11.3620-3629.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupp C, Urbanowski M, Greenberg EP, Ruby EG. The Vibrio fischeri quorum-sensing systems ain and lux sequentially induce luminescence gene expression and are important for persistence in the squid host. Molecular Microbiology. 2003;50:319–331. doi: 10.1046/j.1365-2958.2003.t01-1-03585.x. [DOI] [PubMed] [Google Scholar]
- Mcfall-Ngai M. Divining the Essence of Symbiosis: Insights from the Squid-Vibrio Model. PLoS Biol. 2014;12:e1001783. doi: 10.1371/journal.pbio.1001783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengerink KJ, Vacquier VD. Glycobiology of sperm-egg interactions in deuterostomes. Glycobiology. 2001;11:37R–43R. doi: 10.1093/glycob/11.4.37r. [DOI] [PubMed] [Google Scholar]
- Miller DJ, Ax RL. Carbohydrates and fertilization in animals. Mol. Reprod. Dev. 1990;26:184–198. doi: 10.1002/mrd.1080260213. [DOI] [PubMed] [Google Scholar]
- O'Day DH. Aggregation during sexual development in Dictyostelium discoideum. Can. J. Microbiol. 1979;25:1416–1426. doi: 10.1139/m79-221. [DOI] [PubMed] [Google Scholar]
- Omori M, Hamner WM. Patchy distribution of zooplankton: Behavior, population assessment and sampling problems. Marine Biology. 1982;72:193–200. [Google Scholar]
- Ori A, Wilkinson MC, Fernig DG. A systems biology approach for the investigation of the heparin/heparan sulfate interactome. J Biol Chem. 2011;286:19892–19904. doi: 10.1074/jbc.M111.228114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Painter SD, Clough B, Garden RW, Sweedler JV, Nagle GT. Characterization of Aplysia Attractin, the First Water-borne Peptide Pheromone in Invertebrates. The Biological Bulletin. 2016;194:120–131. doi: 10.2307/1543042. [DOI] [PubMed] [Google Scholar]
- Ram JL, Müller CT, Beckmann M, Hardege JD. The spawning pheromone cysteine-glutathione disulfide (“nereithione”) arouses a multicomponent nuptial behavior and electrophysiological activity in Nereis succinea males. Faseb J. 1999;13:945–952. doi: 10.1096/fasebj.13.8.945. [DOI] [PubMed] [Google Scholar]
- Shaya D, Hahn B-S, Bjerkan TM, Kim WS, Park NY, Sim J-S, Kim YS, Cygler M. Composite active site of chondroitin lyase ABC accepting both epimers of uronic acid. Glycobiology. 2008;18:270–277. doi: 10.1093/glycob/cwn002. [DOI] [PubMed] [Google Scholar]
- Shibata S, Yip ES, Quirke KP, Ondrey JM, Visick KL. Roles of the structural symbiosis polysaccharide (syp) genes in host colonization, biofilm formation, and polysaccharide biosynthesis in Vibrio fischeri. Journal of Bacteriology. 2012;194:6736–6747. doi: 10.1128/JB.00707-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JR, Strehlow DR. Algal-induced spawning in the marine mussel Mytilus californianus 4. International Journal of Invertebrate Reproduction. 1983;6:129–133. [Google Scholar]
- Sorensen PW, Wisenden BD. Fish Pheromones and Related Cues. John Wiley & Sons; 2015. [Google Scholar]
- Starr M, Himmelman JH, Therriault J-C. Direct Coupling of Marine Invertebrate Spawning with Phytoplankton Blooms. Science. 1990;247:1071–1074. doi: 10.1126/science.247.4946.1071. [DOI] [PubMed] [Google Scholar]
- Starr M, Himmelman JH, Therriault J-C. Isolation and properties of a substance from the diatom Phaeodactylum tricornutum which induces spawning in the sea urchin Strongylocentrotus droebachiensis. Marine Ecology Progress Series. 1992;79:275–287. [Google Scholar]
- Thompson FL, Austin B, Swings J. The biology of vibrios. Washington (D.C.): ASM Press; 2006. [Google Scholar]
- Veith M, Beer N, Kiefer A, Johannesen J, Seitz A. The role of swarming sites for maintaining gene flow in the brown long-eared bat. Heredity (Edinb) 2004;93:342–349. doi: 10.1038/sj.hdy.6800509. [DOI] [PubMed] [Google Scholar]
- Wang W, Han W, Cai X, Zheng X, Sugahara K, Li F. Cloning and characterization of a novel chondroitin sulfate/dermatan sulfate 4-O-endosulfatase from a marine bacterium. J Biol Chem. 2015;290:7823–7832. doi: 10.1074/jbc.M114.629154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson GJ, Bentley MG, Gaudron SM, Hardege JD. The role of chemical signals in the spawning induction of polychaete worms and other marine invertebrates. Journal of Experimental Marine Biology and Ecology. 2003;294:169–187. [Google Scholar]
- Huang Weijun, Boju Lorena, Tkalec Lydia, Su Hongsheng, Yang Hyun-Ok, Gunay Nur Sibel, Linhardt Robert J, Kim Yeong Shik, Allan Matte A, Cygler Miroslaw. Active site of chondroitin AC lyase revealed by the structure of enzyme–oligosaccharide complexes and mutagenesis. Biochemistry. 2001;40:2359–2372. doi: 10.1021/bi0024254. [DOI] [PubMed] [Google Scholar]
- Wertheim B, van Baalen E-JA, Dicke M, Vet LEM. Pheromone-mediated aggregation in nonsocial arthropods: an evolutionary ecological perspective. Annual Review of Entomology. 2005;50:321–346. doi: 10.1146/annurev.ento.49.061802.123329. [DOI] [PubMed] [Google Scholar]
- Woznica A, Cantley AM, Beemelmanns C, Freinkman E, Clardy J, King N. Bacterial lipids activate, synergize, and inhibit a developmental switch in choanoflagellates. Proc Natl Acad Sci USA. 2016;113:7894–7899. doi: 10.1073/pnas.1605015113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada S, Sugahara K, Özbek S. Evolution of glycosaminoglycans. Communicative & Integrative Biology. 2011;4:150–158. doi: 10.4161/cib.4.2.14547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Zhang Z, Linhardt RJ. Handbook of Glycomics. Elsevier; 2010. Glycosaminoglycans; pp. 59–80. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Stills of swarm formation after induction with V. fischeri bacteria. Arrowhead tracks the formation and movement of a single swarm over time. (B) Nuclear fusion in a mated pair of S. rosetta cells following treatment with V. fischeri. The final result of nuclear fusion is a diploid cell, harboring a single flagellum (B1-1’’). Hoechst (B1’; cyan) highlights the nucleus, and anti-tubulin antibody (B1’’; white) highlights the cell body and flagellum.
KASP genotyping of meiotic progeny isolated from V. fischeri- induced cross
Mass Spectrometry of bioactive V. fischeri protein
Putative homologs of GAG biosynthetic enzymes
(A) Swarming in S. rosetta is induced by large (>50kD), water-soluble factors present in V. fischeri conditioned media (VFCM). To identify the source of the aphrodisiac activity, proteins were precipitated from VFCM (AP 50%–65%) and separated by size exclusion (SEC) and anion exchange (AEX) chromatography. A protein band of ~90kD, later determined to be EroS (VF_A0994), was abundant in the bioactive SEC (SEC 25/26) and AEX (AEX 81/82) fractions. (B, C) EroS is a chondroitin AC lyase. (B) EroS degrades chondroitin sulfate AC, but not chondroitin sulfate B (dermatan sulfate) in vitro. EroS was incubated with purified chondroitin sulfate AC (open circle) and chondroitin sulfate B (dark grey square), and lyase activity of EroS was measured by monitoring the abundance of unsaturated oligosaccharide products with an absorbance at 232nm. Diamond represents a no enzyme control. (C) Chondroitinase ABC (P. vulgaris), a positive control for in vitro chondroitin degradation assays, rapidly depolymerizes both chondroitin sulfate AC (open circle) as well as chondroitin sulfate B (grey square). Diamond represents a no enzyme control. (E) Swarming in S. rosetta is not induced by chondroitin sulfate or chondroitin disaccharides. Neither commercial chondroitin disaccharides (D0a6 and D0a0), chondroitin disaccharides generated via the depolymerization of chondroitin sulfate by EroS, nor conditioned media isolated after EroS-treatment of S. rosetta cells are sufficient to induce swarming in S. rosetta.
(A) Orthologs of chondroitin sulfate (CS) synthesis are present in the genomes of choanoflagellates S. rosetta and M. brevicollis. Genes identified as “linker” synthesize the proteoglycan linker tetrasaccharide and are important for the biosynthesis of multiple types of GAG, whereas the genes identified as “CS synthesis” are specific to CS biosynthesis. The gene identified as “DS” is required for the formation of dermatan sulfate. All query sequences used were human orthologs. If multiple subject sequences were hits for a single query sequence, the ortholog with the highest Blastp score was chosen. Query and subject accession information is provided in Supplemental File 3. (B) S. rosetta produces chondroitin that can be degraded by ABC chondroitinase and EroS. Polysaccharides isolated from S. rosetta were treated with either ABC chondroitinase from P. vulgaris (center plot) or EroS (bottom plot). Degradation products from samples treated with ABC chondroitinase and EroS were separated by SAX-HPLC (X-axis indicates time, Y-axis indicates abundance) and compared to the following chondroitin disaccharide standards (top plot): (1) D0a0, unsulfated chondroitin; (2) D0a6, chondroitin-6-sulfate; (3) D0a4, chondroitin-4-sulfate; (4) D0a10, chondroitin-4,6-sulfate; (5) D2a4, chondroitin-2,4-sulfate. Unsulfated and 6-sulfated chondroitin disaccharides were present at similar abundance in both the ABC chondroitinase and EroS –treated samples, whereas all other chondroitin disaccharides were below the limit of detection. (C) Quantification of chondroitin disaccharide products produced by ABC chondroitinase (P. vulgaris) and EroS treatment of S. rosetta polysaccharides. Disaccharide abbreviations: D0a0=unsulfated chondroitin; D0a6=chondroitin-6-sulfate; D0a4= chondroitin-4-sulfate; D0a10=chondroitin-4,6-sulfate; D2a4=chondroitin-2,4-sulfate. (D) S. rosetta does not produce heparan sulfate. Polysaccharides isolated from S. rosetta (bottom plot) were treated with Heparinase I, Heparinase II, and Heparinase III (Dextra Laboratories) separated by SAX-HPLC (X-axis indicates time, Y-axis indicates abundance) and compared to the following heparan sulfate disaccharide standards (top plot): (1) D0A0; (2) D0S0; (3) D0A6; (4) D2A0; (5) D0S6; (6) D2S0; (7) D2A6; (8) D2S6. (E) No heparan disaccharides were present above the limit of detection in the S. rosetta polysaccharide sample.
(A) S. rosetta swarms in response to low numbers of V. fischeri bacteria in a cell density-dependent manner. S. rosetta at high cell densities (2.0×106 cells/mL) swarms in response to as few as one V. fischeri cell per 1000 S. rosetta cells within 30 minutes of exposure, whereas swarming in S. rosetta at lower cell densities (2.0×105 cells/mL) within a similar time frame requires at least one V. fischeri cell per 500 S. rosetta cells. (B) Picomolar concentrations of secreted (5% VFCM) and purified EroS are sufficient to induce swarming in S. rosetta.
The movie beings with a side-by-side comparison of S. rosetta treated with EPCM (control; left) and VFCM (right) one hour post induction. After the short transition (“Induction with V. fischeri”) is a time-lapse depicting early swarm formation after treatment with 5% VFCM. All footage is displayed at 35× real time.
Two cells within a four-cell swarm undergo cell fusion. Movie begins 30 minutes after addition of 5% VFCM. Cell fusion is displayed at 60X real time.