Summary
Mesoderm derived from human embryonic stem cells (hESCs) is a major source of the mesenchymal stem/stromal cells (MSCs) that can differentiate into osteoblasts and chondrocytes for tissue regeneration. While significant progress has been made in understanding of molecular mechanisms of hESC differentiation into mesodermal cells, little is known about epigenetic factors controlling hESC fate toward mesoderm and MSCs. Identifying potential epigenetic factors that control hESC differentiation will undoubtedly lead to advancements in regenerative medicine. Here, we conducted an epigenome-wide analysis of hESCs and MSCs and uncovered that EZH2 was enriched in hESCs and was downregulated significantly in MSCs. The specific EZH2 inhibitor GSK126 directed hESC differentiation toward mesoderm and generated more MSCs by reducing H3K27me3. Our results provide insights into epigenetic landscapes of hESCs and MSCs and suggest that inhibiting EZH2 promotes mesodermal differentiation of hESCs.
Keywords: mesoderm, mesenchymal stem cells, EZH2, human ESCs, osteoblast differentiation, epigenetics, histone methylation, cell fate
Highlights
-
•
Inhibiting EZH2 directs hESC differentiation to mesoderm and generates more MSCs
-
•
H3K27me3 levels decrease on specific gene clusters as hESCs differentiate to MSCs
-
•
EZH2 is downregulated as hESC differentiate to MSCs
In this article, Wang and colleagues show that EZH2 was enriched in hESCs and was downregulated significantly in MSCs. Using GSK126, a small-molecule inhibitor of EZH2, they found that reducing H3K27me3 epigenetically promotes mesodermal differentiation of hESCs and generated more MSCs with multipotency.
Introduction
Mesenchymal stem/stromal cells (MSCs) are attractive sources for a large number of cell-based therapies due to their self-renewal capacity and multi-lineage differentiation potential into bone, cartilage, and adipose tissues (Bianco et al., 2013, Alvarez et al., 2015). A major limitation of MSC-based therapy is that isolation of MSCs from bone marrow is an invasive process with potential adverse effects for the donor. Additionally, the proliferation and differentiation capacity of MSCs can vary between different donors with notable decreases as the donor patient age increases (Kern et al., 2006). Thus, while MSCs isolated from bone marrow exhibit a promising potential for bone regeneration, there are shortcomings that may limit their overall effectiveness.
MSCs derived from human embryonic stem cells (hESCs) represent an attractive alternative particularly for bone regeneration because they have higher proliferation and osteogenic potential (Barlow et al., 2008, Giuliani et al., 2011). Several groups have developed a variety of methods to derive MSCs from ESCs that are biologically and phenotypically similar to bone marrow MSCs (Brown et al., 2009, Deng et al., 2016). However, although there has been significant progress in developing protocols for hESC differentiation into MSCs, the differentiating or epigenetic programming of hESCs is a rather slow process that does not fulfill the requirements of tissue regeneration. Mesoderm derived from hESCs is a major source of multiple potent MSCs. Therefore, it is critical to identify potential inducing signals or epigenetic factors that might help to promote mesodermal differentiation of hESCs.
The highly conserved polycomb group (PcG) proteins are epigenetic modulators that carry out histone methylation to initiate and maintain the transcriptional repressive state of genes associated with differentiation to retain stem cell pluripotency in hESCs (Boyer et al., 2006, Deng et al., 2015). Mammalian PcG proteins are classified as two distinct complexes, the polycomb repressive complexes 1 and 2 (PRC1 and PRC2). While PRC1 recognizes the chromodomain of H3K27me3 to facilitate ubiquitination of H2AK119, PRC2 mediates the trimethylation of H3K27 at gene promoters. As a critical subunit of PRC2, EZH2 catalyzes the addition of methyl groups to H3K27 through the SET domain of its carboxy-terminal region and serves as a recruitment platform for DNA methyltransferases for gene silencing (Cao et al., 2002). Here, we performed epigenome-wide comparisons of the transcriptome and histone modifications between hESCs and human ESC-derived MSCs using publicly available RNA sequencing (RNA-seq) and chromatin immunoprecipitation sequencing (ChIP-seq) data. This epigenome-wide map revealed that EZH2 was enriched in hESCs and was downregulated significantly in MSCs. We found that reducing H3K27me3 epigenetically promoted mesodermal differentiation of hESCs and generated more MSCs with multipotency.
Results
EZH2 Expression Is Decreased Following hESC Differentiation into MSCs
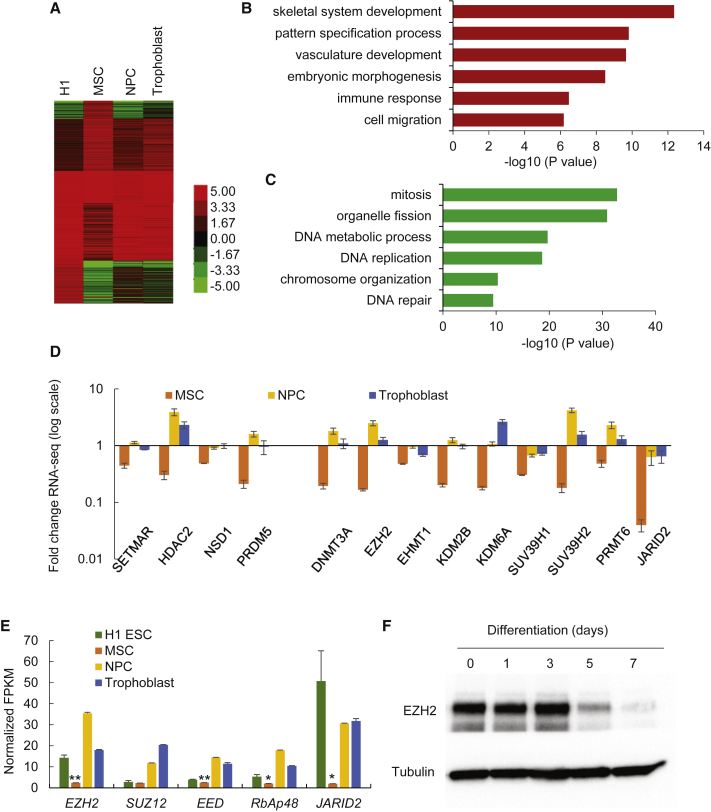
To generate the specific gene expression profile of MSCs following hESC differentiation, we used the RNA-seq data available from the NIH Roadmap Epigenomics project (Xie et al., 2013). The gene expression profiles of hESCs and three hESC-derived lineages showed gene clusters that were either upregulated or downregulated in MSCs compared with hESCs (Figure 1A). Gene ontology (GO) analysis indicated that upregulated genes are highly enriched in categories related to skeletal system development, embryonic morphogenesis, and cell migration (Figure 1B), whereas downregulated genes are enriched in categories related to mitosis, organelle fission, DNA replication and repair, and chromosome function (Figure 1C). As the importance of epigenetic regulation in ESC differentiation is well established, we next examined the expression of epigenetic modulators (Figure 1D). Among these, EZH2, JARID2, and DNMT3B were the three most downregulated genes in MSCs compared with hESCs. Specifically, these epigenetic modulators are involved in repressive function, with EZH2 and JARID2 being associated with the trimethylation of H3K27 and DNMT3B with the methylation of DNA (Chedin, 2011) (Figure 1D). qRT-PCR confirmed that JARID2, EZH2, SUV39H1, and KDM6A were downregulated in hESC-derived MSCs (Figures S1A and S1B). Since EZH2 and JARID2 are both components of PRC2, we investigated the gene expression of each individual PRC2 component. Comparative analysis of hESCs and MSCs revealed that all members of PRC2, EZH2, SUZ12, EED, RbAp48, and JARID2 are reduced in MSCs compared with hESCs (Figure 1E). While JARID2 exhibited the most significant decrease, its downregulation was observed in all three hESC-derived lineages (Figure 1E). Surprisingly, the downregulation of EZH2 was specific to MSCs, with its upregulation observed in the other hESC-derived lineages (Figure 1E). Consistently, protein expression of EZH2 decreased dramatically after 3 days of hESC differentiation (Figure 1F).
Figure 1.
Transcriptome Alterations in H1 hESC-Derived Lineages
(A) Heatmaps showing the expression levels of coding genes in H1 hESCs and H1 hESC-derived cell lineages.
(B and C) Gene ontology (GO) enrichment analysis for upregulated genes (B) and downregulated genes (C) in H1 hESC-derived MSCs. The GO terms include biological function. Bars represent −log10 of p values.
(D) Fold changes of FPKM (fragments per kilobase of transcript per million mapped reads) value for genes encoding epigenetic modifiers in H1 hESC-derived lineages comparing with H1 hESCs. The y axis shows the log scale. Data are shown as mean ± SD from 2 replicates of RNA-seq data.
(E) Average FPKM value of genes encoding core components of PRC2 complex in H1 hESCs and H1 hESC-derived lineages. Data are shown as mean ± SD from 2 replicates of RNA-seq data. ∗p < 0.05; ∗∗p < 0.01.
(F) Western blot analysis of EZH2 level during differentiation of H1 hESCs to MSC.
See also Figure S1.
Decreasing H3K27me3 on Specific Gene Clusters Associated with hESC Programming into MSCs
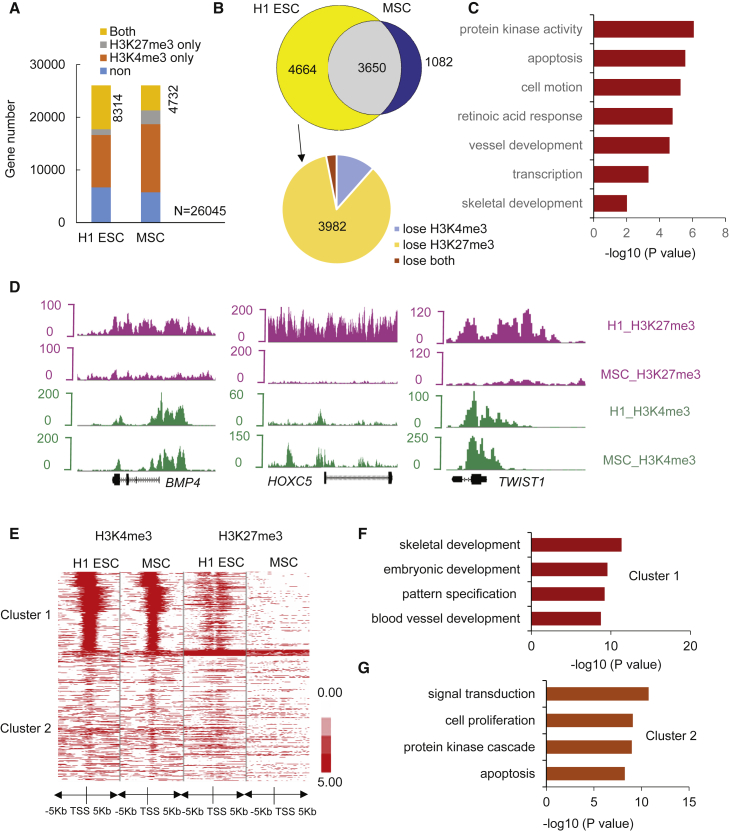
Both silencing H3K27me3 and activating H3K4me3 epigenetic marks, referred to as bivalent domains, occupy the EZH2-targeted genes in ESCs, working together to preserve ESC pluripotency and mediate rapid responses to differentiation signals (Bernstein et al., 2006). Therefore, to further examine the epigenetic basis of hESC differentiation into MSCs, we profiled H3K27me3 and H3K4me3 marks in hESCs and MSCs with the use of ChIP-seq data (Xie et al., 2013). Consequently, we identified 8,314 and 4,732 bivalent genes—those with binding sites for both H3K4me3 and H3K27me3—in H1 hESCs and MSCs, respectively (Figure 2A). Our data further indicated that, while there are 3,650 common bivalent genes in both H1 hESCs and MSCs, 4,664 bivalent genes disappear and 1,082 genes emerge solely in MSCs (Figure 2B, upper). We then selected developmental regulator genes that are bivalent in hESCs, but only have binding sites for H3K4me3—and are therefore transcriptionally active—in MSCs, identifying 3,982 genes (Figure 2B, lower). These genes are enriched for protein kinase activity, blood vessel development, and, notably, skeletal development (Figure 2C). Among them, we identified BMP4, HOXC5, and TWIST1 (Figure 2D), genes that have been known to play a role in MSC behavior, thereby demonstrating that histone profiles can independently predict key hESC to MSC differentiation regulators.
Figure 2.
Histone Marks in H1 hESC-Derived MSCs
(A) The classification of RefSeq genes in terms of enrichment of H3K4me3 and H3K27me3 in H1 hESCs and H1 hESC-derived MSCs. Both and H3K4me3 co-occupy with H3K27me3.
(B) Venn diagram showing bivalent domains change during hESC differentiation toward MSCs.
(C) GO enrichment analysis of genes that were developmental regulators (bivalent in ESCs) and were activated in MSCs (lose H3K27me3). Bars represent −log10 of p values.
(D) Representative peak tracks of H3K4me3 and H3K27me3 enrichment along BMP4, HOXC5, and TWIST1 genes in H1 hESCs and H1 hESC-derived MSCs.
(E) Profiles of H3K4me3 and H3K27me3 around TSS of genes that upregulated in MSCs. Red represents high intensity and white represents no signal. The profile plot shows the average reads at each relative position to TSS on the x axis with H3K4me3 and H3K27me3 coverage.
(F) GO enrichment analysis of cluster 1 genes. Bars represent −log10 of p values.
(G) GO enrichment analysis of cluster 2 genes. Bars represent −log10 of p values.
See also Figure S1.
We grouped the histone profiles of 3,087 RefSeq genes, whose expression levels were upregulated in MSCs. Clustering of the signal around the transcription start site (TSS) shows that H3K4me3 and H3K27me3 can be classified into two distinct clusters (Figure 2E). Cluster 1 heatmap showed high intensity and wide coverage (mean 3,200 bp) of H3K4me3 and H3K27me3 in H1 ESCs. Notably, H3K4me3 was also enriched around the TSS of MSCs, where H3K27me3 enrichment was absent. Cluster 2 showed no significant enrichment of either epigenetic signature within 5 kb of the TSS for both H1 ESCs and MSCs (Figure 2E). Interestingly, GO analysis using the functional annotation tool, DAVID Bioinformatics Resources 6.7, revealed that the co-presence of H3K4me3 and H3K27me3 at the TSS in hESCs, with little or no co-occupancy in MSCs, caused strong functional enrichment for genes involved in skeletal system development, embryonic development, and blood vessel development (Figure 2F). Genes enriched in cluster 2 were involved in signal transduction and cell proliferation (p < 10−8) (Figure 2G). We also examined the H3K4me3 and H3K27me3 enrichment peaks around the TSS of genes that were downregulated in MSCs and found that they can be classified into three distinct clusters (Figure S1C). Cluster 1 contained a wide distribution of H3K4me3 in H1 ESCs that co-occupied with H3K27me3. Cluster 2 showed enrichment of H3K4me3 in both H1 ESCs and MSCs, while cluster 3 showed no significant enrichment of either histone mark in both H1 ESCs and MSCs (Figure S1C). Importantly, cluster 1 displayed a combination of decreased H3K4me3 enrichment, and increased H3K27me3 enrichment was observed in downregulated genes. Among these genes, GO analysis indicated that those that were the most downregulated in MSCs compared with hESCs are involved in cell signaling, synaptic transmission, cell-cycle processes, chromosomal organization, and organelle fission (Figures S1D–S1F).
Inhibition of EZH2 Directs hESC Differentiation toward Mesoderm and Generates More MSCs
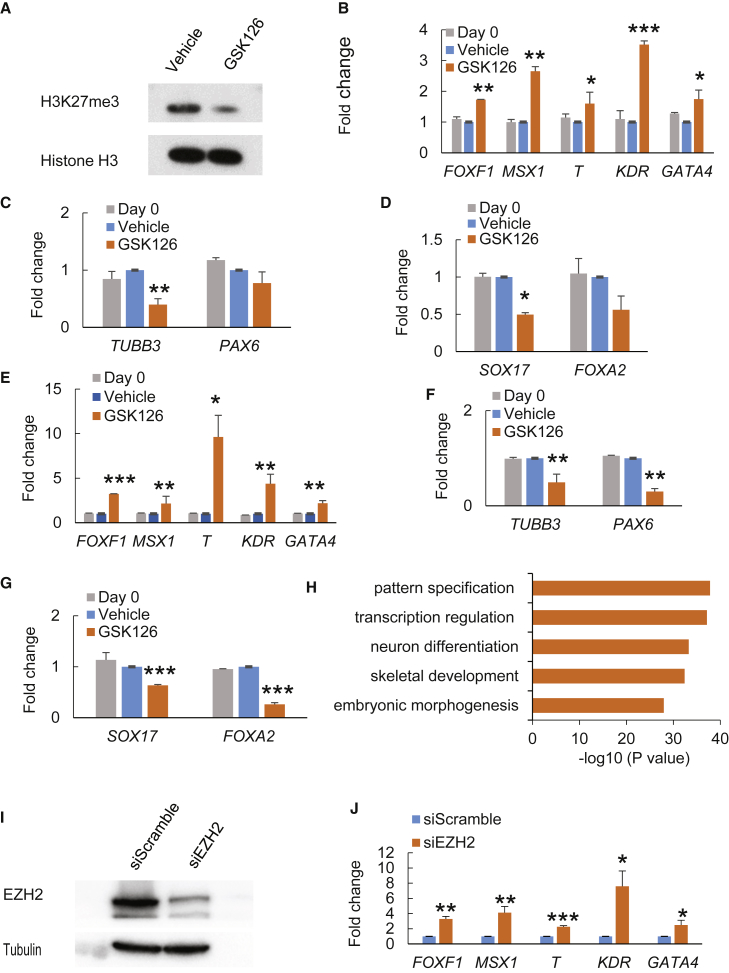
Next, we sought to understand the role of EZH2 in the differentiation of hESCs into MSCs. We treated hESCs with GSK126, a highly selective and potent inhibitor of EZH2. Western blot analysis showed that GSK126 potently inhibited EZH2's histone methyltransferase activity in hESCs, as demonstrated by an evident reduction in H3K27me3 (Figure 3A). Alkaline phosphatase (ALP) staining showed that GSK126 treatment dramatically inhibited ALP activities in both H1 and H9 hESCs (Figure S2). We assessed the effects of EZH2 inactivation on the expression pattern of markers for the three germ layers. qRT-PCR revealed that multiple mesodermal markers, including KDR, FOXF1, MSX1, T, and GATA4, were significantly elevated upon GSK126 treatment (Figures 3B–3D). On the contrary, GSK126 significantly inhibited the expression of the ectodermal mark gene TUBB3 and the endodermal gene SOX17 and had a minimal effect on the expression of the ectodermal marker gene PAX6 and endodermal marker gene FOXA2. Flow cytometry via fluorescence-activated cell sorting (FACS) staining confirmed that GSK126 treatment promoted KDR expression while it did not affect the expression of PAX6 and FOXA2 (Figure S2B). Interestingly, a similar pattern was seen in H9 hESCs, but with even more pronounced changes (Figures 3E–3G and S2C). GO enrichment analysis of EZH2 ChIP-seq data in H1 hESCs revealed that EZH2 is enriched at the promoters of genes involved in developmental processes such as skeletal development and pattern specification (Figure 3H). To rule out the non-specific effect of GSK126, we knocked down EZH2 in hESCs using small interfering RNA (siRNA) (Figure 3I). Consistently, siRNA-mediated EZH2 depletion also led to enhanced expression of mesodermal markers (Figure 3J).
Figure 3.
Effect of GSK126 Treatment on Differentiation of H1 hESCs and H9 hESCs
(A) Western blot analysis of H3K27me3 level after 10 μM GSK126 treatment (upper panel); western blot of histone H3 serves as loading control (lower panel).
(B–D) qRT-PCR analysis of mesodermal genes (FOXF1, MSX1, T, KDR, GATA4) (B), ectodermal genes (TUBB3, PAX6) (C), and endodermal genes (SOX17, FOXA2) (D) for H1 cells treated with DMSO or GSK126.
(E–G) qRT-PCR analysis of mesodermal genes (FOXF1, MSX1, T, KDR, GATA4) (E), ectodermal genes (TUBB3, PAX6) (F), and endodermal genes (SOX17, FOXA2) (G) for H9 cells treated with DMSO or GSK126.
(H) GO enrichment analysis of EZH2 enrichment within 1 kb of TSS for RefSeq genes in H1 hESCs.
(I) Western blot analysis of EZH2 protein level after EZH2 siRNA knockdown in H1 hESCs.
(J) qRT-PCR analysis of mesodermal genes (FOXF1, MSX1, T, KDR, GATA4) after EZH2 siRNA knockdown.
Data are shown as mean ± SD; n = 3 independent experiments. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001 by Student's t test. See also Figure S2.
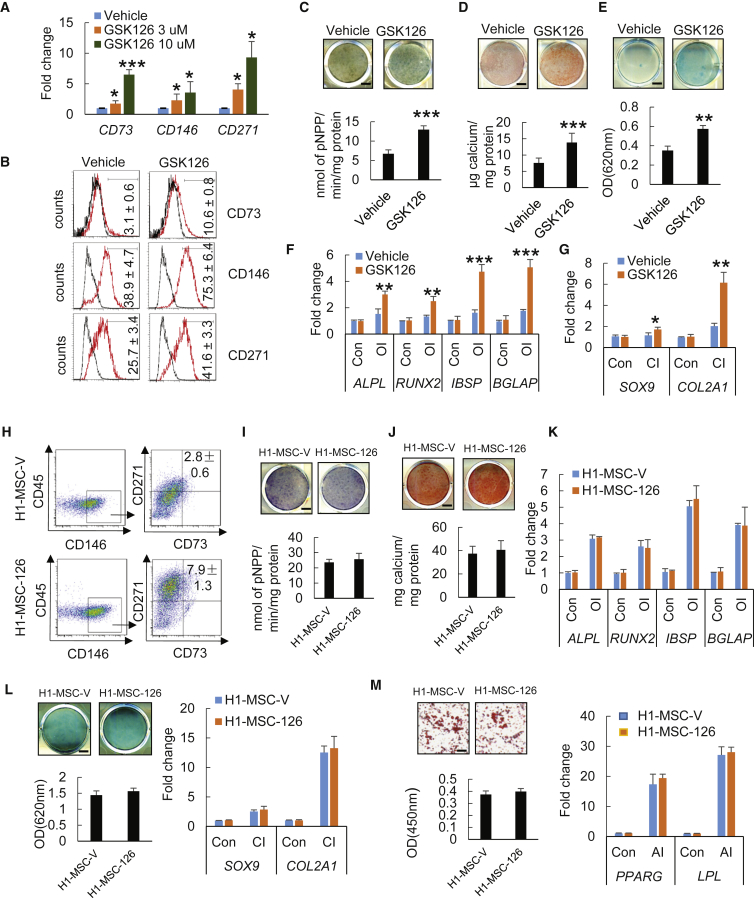
Since the mesoderm is a major source of the mesenchymal precursors that enable the formation of skeletal and connective tissues (Vodyanik et al., 2010), we further explored lineage specification to MSCs by examining MSC surface markers. qRT-PCR analysis indicated that the mRNA levels of CD73, CD146, and CD271 were significantly upregulated following 3 days of GSK126 treatment in H1 hESCs (Figure 4A) and H9 ESCs (Figure S3A) in a dose-dependent manner. Such enhanced expression of MSC markers was further confirmed by flow-cytometry analysis (Figures 4B and S2C).
Figure 4.
Effect of GSK126 Treatment on Mesenchymal Lineage Commitment of H1 hESCs
(A) qRT-PCR gene expression analysis of well-known MSC surface markers (CD73, CD146, and CD271).
(B) Flow-cytometry analysis for CD73, CD146, and CD271 expression of cells treated with DMSO or GSK126.
(C) ALP staining and quantitative ALP activity assay after 14 days of osteogenic induction (OI) for DMSO- or GSK126-treated cells. Scale bar, 300 μm.
(D) ARS staining and quantification after 14 days of OI for DMSO- or GSK126-treated cells. Scale bar, 440 μm.
(E) Alcian blue staining and quantification after 21 days of chondrogenic induction (CI) for DMSO- or GSK126-treated cells. Scale bar, 440 μm.
(F and G) qRT-PCR gene expression analysis of osteogenic markers (ALPL, RUNX2, IBSP, and BGLAP) (F) and chondrogenic markers (SOX9 and COL2a1) (G) after lineage-specific differentiation in H1 cells treated with or without GSK126.
(H) Proportions of CD90+CD146+CD271+CD45– H1-MSC-V and H1-MSC-126 are compared.
(I) ALP staining and quantitative ALP activity assay of H1-MSC-V and H1-MSC-126 after 14 days of OI. Scale bar, 440 μm.
(J) ARS staining and quantification of H1-MSC-V and H1-MSC-126 after 14 days of OI. Scale bar, 440 μm.
(K) qRT-PCR gene expression analysis of osteogenic markers (ALPL, RUNX2, IBSP, and BGLAP) in H1-MSC-V and H1-MSC-126 after 14 days of OI.
(L) Alcian blue staining and quantification (left) and qRT-PCR gene expression analysis of chondrogenic markers (SOX9 and COL2a1) (right) of H1-MSC-V and H1-MSC-126 after 21 days of CI. Scale bar, 440 μm.
(M) Oil red O staining and quantification (left) and qRT-PCR gene expression analysis of adipogenic markers (PPARG and LPL) (right) of H1-MSC-V and H1-MSC-126 after 21 days of adipogenic induction (AI). Scale bar, 440 μm.
Data are shown as mean ± SD; n = 3 independent experiments. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001 by Student's t test. See also Figures S3 and S4.
Since the inhibition of EZH2 by GSK126 appeared to generate more mesenchymal precursors from hESCs, we examined whether these differentiated cells from hESCs could be induced to differentiate into osteoblasts, chondrocytes, or adipocytes. We first treated H1 hESCs with either GSK126 or a vehicle control for 3 days, then transitioned them to a monolayer and cultured them in appropriate differentiation media. When these cells were induced to undergo osteogenic differentiation with osteogenic induction (OI) medium for 14 days, both ALP activity and the capacity to form mineralized nodules were significantly elevated in hESCs treated with GSK126 compared with the vehicle control (Figures 4C and 4D). Consistently, real-time RT-PCR showed elevated expression levels of osteogenic markers including ALPL, RUNX2, IBSP, and BGLAP (Figure 4F). Furthermore, we evaluated the chondrogenic capacity of differentiated hESCs with or without GSK126 treatment. Following treatment with chondrogenic induction (CI) medium for 21 days, GSK126-treated hESCs showed enhanced chondrogenic potential demonstrated by the presence of increased glycosaminoglycans and increased expression of chondrogenic markers SOX9 and COL2A1 (Figure 4E). Comparable results were also observed in GSK126-treated H9 hESCs compared with vehicle treatment (Figures S3C–S3G). Interestingly, we were unable to induce adipogenic differentiation in these unsorted cells (data not shown).
Next, we sought to determine whether the increased osteogenic potentials following GSK126 treatment was due to an enhanced potency of individual mesenchymal precursors or a generation of more MSCs from hESCs. In general, MSCs positively express multiple cell-surface markers, including CD51, CD73, CD90, CD146, and CD271, and are negative for CD34 and CD45. To accomplish this, we treated H1 hESCs with either GSK126 or a vehicle control for 3 days. Subsequently, cells were trypsinized to generate a single-cell suspension for further differentiation for 5 days and then sorted by flow cytometry. Our preliminary analysis found that more than 95% of the cells were positive for CD51 and CD90 (Figures S4A and S4C) and negative for CD34 (Figures S4B and S4D). Therefore, we utilized the combination of markers CD73+CD146+CD271+CD45– to isolate MSCs from differentiating hESCs excluding CD51, CD90, and CD34. FACS analysis indicated that, whereas 2.8% MSCs (H1-MSC-V) from differentiating hESCs without GSK126 treatment were isolated, GSK126 treatment generated an approximately 3-fold greater number of MSCs (H1-MSC-126) (Figure 4H). Since GSK126 treatment yielded more MSCs, it was important to demonstrate whether accelerating MSC fates by an epigenetic modifier did not impair the multiple potentials of MSCs from the aspect of regenerative medicine. The isolated H1-MSC-126 and H1-MSC-V cells by FACS were compared for their terminal differentiation capacity to develop into osteoblasts, chondrocytes, and adipocytes in the absence of GSK126. Importantly, ALP or Alizarin red staining revealed that H1-MSC-126 had same strong osteogenic potentials as H1-MSC-V (Figures 4I and 4J). Real-time RT-PCR showed that the expression of the osteogenic markers in H1-MSC-126, including ALPL, RUNX2, IBSP, and BGLAP, was induced at levels similar to those in H1-MSC-V upon OI treatment (Figure 4K). Following CI, both H1-MSC-126 and H1-MSC-V equivalently differentiated into chondrocytes (Figure 4L). Unlikely unsorted cells, both H1-MSC-126 and H1-MSC-V could differentiate into adipocytes as determined by oil red O staining. Real-time RT-PCR also confirmed that the adipogenic markers PPARG and LPL were similarly induced in both H1-MSC-126 and H1-MSC-V (Figure 4M). To further confirm our results, we also isolated MSCs from H9 hESCs treated with or without GSK126 using CD73+CD146+CD271+CD45− markers. Consistently, we found that the inhibition of EZH2 by GSK126 also significantly increased the proportion of MSCs from H9 hESCs by 3-fold (Figure S3H). MSCs derived from GSK126-treated H9 hESCs maintained osteogenic, chondrogenic, and adipogenic potentials in vitro (Figures S3I–S3M).
Discussion
In this study, we identified an epigenetic mechanism that facilitates controlled differentiation of pluripotent hESCs into mesodermal lineage. Through a comparative analysis of the transcriptome and epigenetic signatures between hESCs and MSCs, we found that PRC2 components were significantly downregulated in MSCs compared with hESCs. Further analysis revealed that among the most downregulated of PRC2 components in MSCs was EZH2. EZH2 serves as an essential force in the epigenetic landscape of embryonic development by mediating the silencing of a diverse group of developmental genes. Indeed, obliterating EZH2's function in replenishing H3K27me3 repressive signatures disturbed the equilibrium in the epigenetic landscape, leading to differentiation. Surprisingly, however, we observed a discerning rise in the expression of mesoderm and MSC markers and a concurrent decline in endoderm and ectoderm markers, indicating EZH2's distinguishing role in restricting commitment to mesodermal lineage. Importantly, we showed that the inhibition of EZH2 by the small-molecule inhibitor GSK126 potently produced higher volumes of mesodermal progenitors from hESCs, positioning EZH2 as a vital epigenetic modifier that regulates hESC differentiation into mesodermal lineage.
Because MSCs demonstrate high osteogenic potential with greater consistency and higher proliferation rate compared with MSCs from bone marrow, they may be better suited for regenerative medicine. Here, we identified the central epigenetic mechanism that directs hESCs to mesodermal linage as the incapacitation of EZH2 remodeled the epigenetic landscape in favor of MSC derivation. These results underscore a new duty of EZH2 in governing mesodermal commitment, beyond its established roles in ESC identity preservation and terminal differentiation. Mechanistically, when an inhibitor such as GSK126 binds to EZH2, its histone methyltransferase activity on H3K27 is disabled and repressive H3K27me3 marks are no longer sufficiently replenished, leading to disinhibition of target genes. As increasing the efficiency of MSC production is an important clinical goal, modulation of EZH2 will have profound implications for the future of regenerative medicine. While drugs targeting epigenetic modifiers are currently being investigated for therapeutic use in the treatment of cancer, autoimmune diseases, and neurological disorders, we provide strong evidence that epigenetic modifiers can also be utilized for enriched acquisition of MSCs with osteogenic potential from hESCs.
Experimental Procedures
Cell Culture and MSC Differentiation
The protocol for the hESCs study was approved by UCLA Embryonic stem cells research oversight committee (IRB: 10-001711-CR-00001). H1 and H9 hESCs were obtained from the UCLA Broad Stem Cell Research Center. hESCs were cultured on a mitotically inactivated mouse embryonic feeder (MEF) layer, as previously described (Thomson et al., 1998), and cells were maintained for 35–45 passages. hESC colonies were detached by type IV collagenase (1 mg/mL), plated onto Matrigel-coated tissue culture dishes, and grown in mTeSR1 medium (catalog #85850, STEMCELL Technologies; Yu et al., 2011, Xie et al., 2013). Subsequently, GSK126 (10 μM; CAS #1346574-57-9, Cayman Chemical) was added to the medium for 3 days every day. After 3 days of treatment, the derived cells were trypsinized to generate a single-cell suspension for further differentiation for 5 days and then sorted by flow cytometry. Osteogenic, chondrogenic, and adipogenic differentiation was induced as described previously (Chang et al., 2009, Deng et al., 2016).
qRT-PCR
Total RNA was isolated using the RNeasy Micro kit (Qiagen). First-strand cDNA was synthesized using random hexamers and reverse transcriptase according to the manufacturer's protocol (Invitrogen). RT-PCR was performed using the QuantiTect SYBR Green PCR kit (Qiagen) and the Icycler iQ Multi-color Real-time PCR Detection System (Bio-Rad). The primers for GAPDH are 5′-GGA GCG AGA TCC CTC CAA AAT-3′ (forward), 5′-GGC TGT TGT CAT ACT TCT CAT GG-3′ (reverse). The primers for PAX6 are 5′-TGG GCA GGT ATT ACG AGA CTG-3′ (forward), 5′-ACT CCC GCT TAT ACT GGG CTA-3′ (reverse). The primers for PDGFR-α are 5′-TAT GTG CCA GAC CCA GAT GT-3′ (forward), 5′-GGA GTC TCG GGA TCA GTT GT-3′ (reverse). The primers for CD73 are 5′-TTA CAC AGG CAA TCC ACC TTC-3′ (forward), 5′-TTA CAC AGG CAA TCC ACC TTC-3′ (reverse). The primers for CD146 are 5′-CTG CTG AGT GAA CCA CAG GA-3′ (forward), 5′-CAC CTG GCC TGT CTC TTC TC-3′ (reverse).
Flow Cytometry and Fluorescence-Activated Cell Sorting
Cells were collected and washed twice with FACS buffer (PBS, 10 mM EDTA, and 2% fetal bovine serum) and suspended at a maximum concentration of 2 × 105 cells per 100 μL. Cells were incubated with antibodies for 30 min on ice in the dark, washed, and suspended in PBS. Samples were analyzed on a BD LSR II analyzer or sorted on a BD FACSAria III. Cell gating was based on comparison with isotype-negative controls and single-stained controls. Cells were sorted into serum-free DMEM for gene expression analysis or into complete medium for cell culture. Antibodies used (all from BioLegend) included PE-CD34 (catalog #343606), PerCP-Cy5.5-CD45 (#368506), PE-CD51 (#327910), APC-CD73 (#344006), FITC-CD90 (#328108), FITC-CD146 (#361012), and PE-CD271 (#345106).
External Data Source, ChIP-Seq Data Analysis, and RNA-Seq Data Analysis
Raw data for histone H3K4me3 and H3K27me3 ChIP-seq and raw data for RNA-seq of hESCs and hESC-derived lineages were downloaded from the NCBI epigenome roadmap (http://www.ncbi.nlm.nih.gov/geo/roadmap/epigenomics/). Data analysis was performed as described in Supplemental Experimental Procedures.
Author Contributions
Y.Y., C.H., and C.Y.W. conceived of this study. Y.Y. and P.D. performed most experiments and analyzed data. B.Y., J.M.S., C.H., and T.A. assisted P.D. for cell differentiation studies and flow cytometry. Y.Y. performed bioinformatics. B.Y. and C.H. helped to analyze the data. Y.Y., C.H., and C.Y.W. wrote the paper.
Acknowledgments
This work was supported by the Shapiro Family Charitable Funds. Flow cytometry was performed in the UCLA Flow Cytometry Core Facility that is supported by NIH awards P30CA016042 and 5P30AI028697.
Published: August 17, 2017
Footnotes
Supplemental Information includes Supplemental Experimental Procedures and four figures and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2017.07.016.
Contributor Information
Christine Hong, Email: chong@dentistry.ucla.edu.
Cun-Yu Wang, Email: cwang@dentistry.ucla.edu.
Supplemental Information
References
- Alvarez R., Lee H.L., Wang C.Y., Hong C. Characterization of the osteogenic potential of mesenchymal stem cells from human periodontal ligament based on cell surface markers. Int. J. Oral Sci. 2015;7:213–219. doi: 10.1038/ijos.2015.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow S., Brooke G., Chatterjee K., Price G., Pelekanos R., Rossetti T., Doody M., Venter D., Pain S., Gilshenan K. Comparison of human placenta- and bone marrow-derived multipotent mesenchymal stem cells. Stem Cells Dev. 2008;17:1095–1107. doi: 10.1089/scd.2007.0154. [DOI] [PubMed] [Google Scholar]
- Bernstein B.E., Mikkelsen T.S., Xie X., Kamal M., Huebert D.J., Cuff J., Fry B., Meissner A., Wernig M., Plath K. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- Bianco P., Cao X., Frenette P.S., Mao J.J., Robey P.G., Simmons P.J., Wang C.Y. The meaning, the sense and the significance: translating the science of mesenchymal stem cells into medicine. Nat. Med. 2013;19:35–42. doi: 10.1038/nm.3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer L.A., Plath K., Zeitlinger J., Brambrink T., Medeiros L.A., Lee T.I., Levine S.S., Wernig M., Tajonar A., Ray M.K. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Brown S.E., Tong W., Krebsbach P.H. The derivation of mesenchymal stem cells from human embryonic stem cells. Cells Tissues Organs. 2009;189:256–260. doi: 10.1159/000151746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R., Wang L., Wang H., Xia L., Erdjument-Bromage H., Tempst P., Jones R.S., Zhang Y. Role of histone H3 lysine 27 methylation in polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- Chang J., Wang Z., Tang E., Fan Z., McCauley L., Franceschi R., Guan K., Krebsbach P.H., Wang C.Y. Inhibition of osteoblastic bone formation by nuclear factor-kappaB. Nat. Med. 2009;15:682–689. doi: 10.1038/nm.1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chedin F. The DNMT3 family of mammalian de novo DNA methyltransferases. Prog. Mol. Biol. Transl. Sci. 2011;101:255–285. doi: 10.1016/B978-0-12-387685-0.00007-X. [DOI] [PubMed] [Google Scholar]
- Deng P., Chen Q.M., Hong C., Wang C.Y. Histone methyltransferases and demethylases: regulators in balancing osteogenic and adipogenic differentiation of mesenchymal stem cells. Int. J. Oral Sci. 2015;7:197–204. doi: 10.1038/ijos.2015.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng P., Zhou C., Alvarez R., Hong C., Wang C.Y. Inhibition of IKK/NF-κB signaling enhances differentiation of mesenchymal stromal cells from human embryonic stem cells. Stem Cell Reports. 2016;6:456–465. doi: 10.1016/j.stemcr.2016.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuliani M., Fleury M., Vernochet A., Ketroussi F., Clay D., Azzarone B., Lataillade J.J., Durrbach A. Long-lasting inhibitory effects of fetal liver mesenchymal stem cells on T-lymphocyte proliferation. PLoS One. 2011;6:e19988. doi: 10.1371/journal.pone.0019988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kern S., Eichler H., Stoeve J., Klüter H., Bieback K. Comparative analysis of mesenchymal stem cells from bone marrow, umbilical cord blood, or adipose tissue. Stem Cells. 2006;24:1294–1301. doi: 10.1634/stemcells.2005-0342. [DOI] [PubMed] [Google Scholar]
- Thomson J.A., Itskovitz-Eldor J., Shapiro S.S., Waknitz M.A., Swiergiel J.J., Marshall V.S., Jones J.M. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Vodyanik M.A., Yu J., Zhang X., Tian S., Stewart R., Thomson J.A., Slukvin I.I. A mesoderm-derived precursor for mesenchymal stem and endothelial cells. Cell Stem Cell. 2010;7:718–729. doi: 10.1016/j.stem.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W., Schultz M.D., Lister R., Hou Z., Rajagopal N., Ray P., Whitaker J.W., Tian S., Hawkins R.D., Leung D. Epigenomic analysis of multilineage differentiation of human embryonic stem cells. Cell. 2013;153:1134–1148. doi: 10.1016/j.cell.2013.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu P., Pan G., Yu J., Thomson J.A. FGF2 sustains NANOG and switches the outcome of BMP4-induced human embryonic stem cell differentiation. Cell Stem Cell. 2011;8:326–334. doi: 10.1016/j.stem.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.