Summary
Stem cell antigen 1-positive (SCA1+) cells (SPCs) have been investigated in cell-based cardiac repair and pharmacological research, although improved cardiac function after injection has been variable and the mode of action remains unclear. Circadian (24-hr) rhythms are biorhythms regulated by molecular clocks that play an important role in (patho)physiology. Here, we describe (1) the presence of a molecular circadian clock in SPCs and (2) circadian rhythmicity in SPC function. We isolated SPCs from human fetal heart and found that these cells possess a molecular clock based on typical oscillations in core clock components BMAL1 and CRY1. Functional analyses revealed that circadian rhythmicity also governs SPC proliferation, stress tolerance, and growth factor release, with large differences between peaks and troughs. We conclude that SPCs contain a circadian molecular clock that controls crucial cellular functions. Taking circadian rhythms into account may improve reproducibility and outcome of research and therapies using SPCs.
Keywords: circadian rhythm, clock, stem cell, progenitor cell, SCA1, heart
Highlights
-
•
SCA1+ cells are a cell source used in pharmacology studies and cardiac repair
-
•
SCA1+ cells possess a molecular circadian (24-hr) clock
-
•
Proliferation, stress tolerance, and paracrine secretion follow a circadian pattern
-
•
Taking rhythmicity into account may improve studies using SCA1+ cells
SCA1+ cells (SPCs) have been used for pharmacological studies and cell-based cardiac repair. Van Laake and colleagues isolated human SPCs and demonstrated that these cells possess molecular circadian clocks. Functional analyses revealed circadian rhythmicity in SPC proliferation, stress tolerance, and growth factor release. Their findings could improve reproducibility, translation, and outcome of studies using SPCs and other progenitor-like cells.
Introduction
Stem cell antigen 1-positive cells (SCA1+ cells, SPCs) are self-renewing cells that have been isolated from fetal and adult heart (Dawn and Bolli, 2005, Zwetsloot et al., 2016). Together with C-KIT+ (Bearzi et al., 2007), and ISLET-1+ (Bu et al., 2009) cells, SPCs have been referred to as cardiac progenitor/stem cells, even though their in vivo cardiogenic differentiation potential is limited (Boland et al., 2014, Madonna et al., 2016, Santini et al., 2016). In vitro demethylation using compounds such as 5-azacytidine and transforming growth factor β to treat SPCs, however, does induce cardiomyocyte-like phenotypes with upregulation of cardiac genes and sarcomeric proteins and the development of functional action potentials (Goumans et al., 2007). SPCs have been investigated for their efficacy in cell-based cardiac repair. In pre-clinical and clinical studies, injection of these cardiac progenitor-like cells proved to be safe and has been described to lead to structural and functional cardiac improvements (Sanganalmath and Bolli, 2013, Smits et al., 2009a, Smits et al., 2009b).
Despite these promising results, challenges still remain. SCA1, C-KIT, and ISLET-1 cannot be used to distinguish a specific cell population (Noseda et al., 2015). As a result, studies show variability in cell-surface markers, proliferation, and differentiation, limiting the use of these cells in, for example, pharmacological studies (Takamiya et al., 2011). In addition to cell-surface marker, proliferation, and differentiation variability, pre-clinical transplantation studies face other drawbacks such as poor engraftment, cell survival, and in vivo differentiation (Madonna et al., 2016). Finally, there is an ongoing search for underlying mechanisms and ways to maximize beneficial effects, including novel effect modifiers.
Circadian (diurnal, 24-hr) rhythms are biorhythms with a period of approximately 24 hr. These rhythms are regulated by molecular clocks that consist of several oscillating proteins such as CLOCK, BMAL, PER, and CRY (Du Pre et al., 2014, Martino and Young, 2015). Many cardiovascular parameters, including heart rate, repolarization duration, and cardiac metabolism, display circadian rhythmicity (Durgan and Young, 2010). Circadian rhythms also play an important role in cardiovascular disease. The incidence of many cardiovascular diseases such as myocardial infarction and acute heart failure follows a 24-hr pattern. Second, disruption of 24-hr rhythms leads to an increase of cardiovascular disease. Lastly, outcome of disease and efficacy of therapy is influenced by this 24-hr rhythm (Durgan et al., 2010, Reiter et al., 2012, Vyas et al., 2012, Woon et al., 2007).
In short, circadian rhythms are an important factor in cardiovascular physiology and disease but have not yet been studied in SPCs, a potential cell source for cardiac cells that proved to be useful in pharmacological studies and pre-clinical cell-based cardiac repair. In the current study, we therefore analyzed the potential presence of circadian rhythms in SPC physiology. More specifically, we (1) sought to determine whether SPCs have functional molecular circadian clocks and (2) whether significant circadian oscillations determine SPC functions, such as proliferation, migration, stress tolerance, and paracrine factor secretion.
Results
Figure S1 shows the (simplified) experimental setup of the proliferation, migration, stress tolerance, and paracrine secretion experiments.
SPC Isolation
SPCs were derived from human fetal and adult hearts based on reaction to a SCA1 antibody, as described previously (Goumans et al., 2007). Although SCA1 itself is not present in the human heart, this technique has been shown previously to isolate a subset of proliferative and clonogenic cells with cardiogenic potential depending on the addition of exogenous factors as characterized extensively in previous work (Boland et al., 2014, de Boer et al., 2010, Madonna et al., 2016). SCA1+ cells from the human heart display a large degree of similarity with other progenitor-like cells previously referred to as cardiospheres and C-KIT+ cardiac stem cells (Gaetani et al., 2014).
The presence of early cardiac transcription factors Homeobox protein Nkx-2.5 (NKX2-5) and GATA4 in combination with the absence of cardiomyocyte markers β-myosin heavy chain (MYH7) confirmed their successful isolation (Table S1).
SPCs Have a Functional Molecular Clock
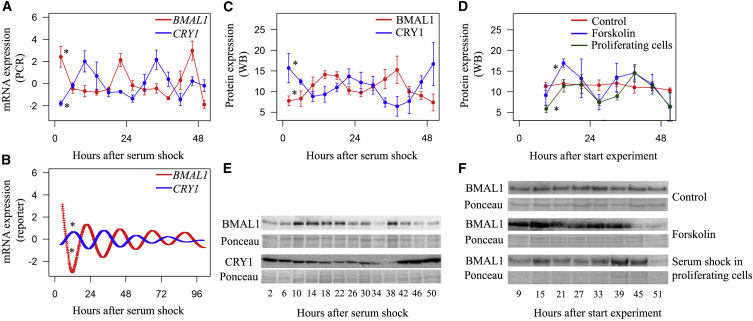
To analyze the presence of a molecular circadian clock in SPCs, we measured sequential mRNA and protein expression levels of core clock components BMAL1 and CRY1. After clock synchronization, BMAL1 and CRY1 mRNA levels oscillated in a 24-hr periodical pattern and were in counter phase with each other (Figure 1A: fold changes 2.25 and 2.28, peaks at 22.4 hr and 10.6 hr, respectively; Figure 1B: fold changes 1.79 and 1.40, peaks at 20.8 hr and 10.0 hr, respectively; p < 0.001 for all; n = 4), in concordance with predicted expression patterns in adult tissues (Young et al., 2001). This translated into corresponding 24-hr oscillations in protein levels of BMAL1 and CRY1 (Figures 1C and 1D: fold changes 1.26 and 1.34, peaks 13.8 hr and 22.7 hr, p < 0.001 and p = 0.014 for BMAL1 and CRY1, respectively; n = 4).
Figure 1.
Molecular Circadian Clockwork within SPCs
Twenty-four-hour oscillations in core clock components BMAL1 and CRY1 were present at the transcriptional and protein level.
(A) Forty-eight-hour analysis of mRNA using qRT-PCR (p < 0.001 for BMAL1 and CRY1; n = 4 independent experiments).
(B) One-hundred-hour analysis of mRNA using bioluminescence reporters (p < 0.001 for BMAL1 and CRY1).
(C and D) BMAL1 and CRY1 protein expression during 48 hr. Counter phased 24-hr rhythms were present (p < 0.001 and p = 0.014 for BMAL1 and CRY1, respectively; n = 4 independent experiments).
(E and F) BMAL1 protein expression during 48 hr. Twenty-four-hour rhythmicity was absent when SPCs were unsynchronized (p = 0.734; n = 4 independent experiments). Presence of rhythmicity was independent of synchronizer substance (forskolin p = 0.01; n = 4 independent experiments) and high/low proliferation rate (proliferating cells p = 0.002; n = 4 independent experiments).
Data are presented as mean ± SEM; ∗p < 0.05 for significant 24-hr oscillation. See also Figure S2.
To exclude the possibility that 24-hr fluctuations were caused by methodological artifacts, we confirmed that oscillations were absent when molecular clocks were not synchronized and were independent of the type of synchronizing substance (Figure 1D: fold change 1.38 and peak at 15.5 hr for synchronized-synchronized SPCs; p = 0.734 and 0.010 for circadian rhythmicity in unsynchronized and forskolin-synchronized SPCs, respectively; n = 4). In addition, proliferating SPCs had similar 24-hr fluctuations with respect to amplitude and phase as non-proliferating confluent SPCs (Figures 1E and 1F; fold change 1.38 and peak 17.3 hr, p = 0.002 for circadian rhythmicity; n = 4).
Next, we analyzed the presence of intracellular shuttling of BMAL1, one of the other characteristics of a functional molecular circadian clock (Darlington et al., 1998). Indeed, there was a 24-hr rhythm in the nuclear/cytoplasmic BMAL1 fraction in the first 24 hr after serum shock (Figure S2: fold change 1.36 and peak at 13.0 hr, p = 0.003; n = 3).
Because circadian rhythms are associated with aging, we repeated the clock component measurements in adult SPCs and found that similar to fetal cells, BMAL1 levels varied in a 24-hr periodical pattern (Figure S3: fold change 1.59 and 1.38, peak at 2.85 hr and 15.5 hr, p < 0.001 and 0.01 for reporter and western blot; n = 3).
Altogether, these observations demonstrate that the molecular circadian clockwork is present and active in SPCs.
Cell Dissociation Disrupts Circadian Rhythmicity
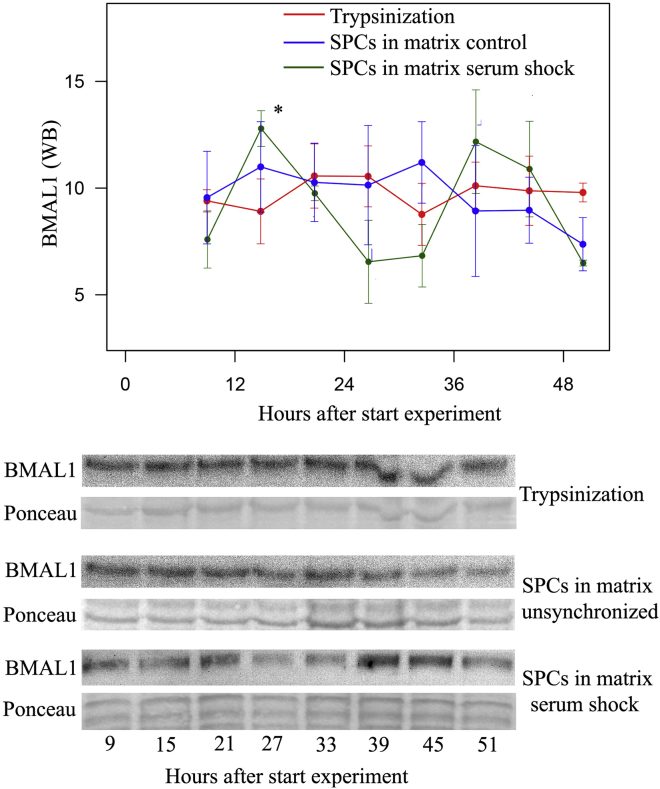
We analyzed whether cell dissociation using trypsinization or collagenase treatment of SPCs influences 24-hr rhythms of BMAL1. After SPCs were trypsinized or treated with collagenase, BMAL1 protein expression no longer oscillated, indicating that cell dissociation disrupts the synchrony of circadian rhythmicity in the cell dish (Figures 2 and S4A; p = 0.589 and p = 0.344, respectively, n = 3).
Figure 2.
Dissociation of SPCs Disrupts the 24-hr Rhythm in BMAL1 Protein Expression
p = 0.589; n = 3 independent experiments. SPCs were also loaded in collagen matrices that do not require cell dissociation. After synchronization, SPC in these matrices showed 24-hr oscillations in BMAL1 similar to SPCs plated in culture dishes (p = 0.014 and 0.734 for serum shock and unsynchronized cells in matrix, respectively; n = 3 independent experiments). Data are presented as mean ± SEM; ∗p < 0.05 for significant 24-hr oscillation.
This effect of dissociation could limit the use of circadian rhythmicity in SPCs. Synchronization and use of the most optimal time window for subsequent applications is problematic if SPCs have to be detached from the culture dish before injection. To circumvent this potential problem, we loaded SPCs in matrices, since matrices do not require dissociation before application. After loading of SPCs and synchronization, BMAL1 expression oscillated comparably with cells attached in a culture dish (Figure 2: fold change 1.38, peak at 15.5 hr, p = 0.014 for serum shock, p = 0.734 for unsynchronized cells in matrix; n = 3). This shows that by using extracellular matrix-based scaffolds, disruption of circadian rhythms by dissociation can be prevented.
Circadian Rhythms Are Present in Proliferation but Not Migration
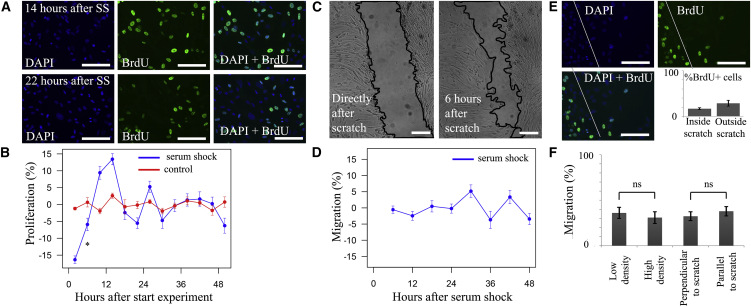
To explore the functional consequences of the molecular circadian clock, we analyzed circadian rhythmicity in SPC proliferation and migration. We found that bromodeoxyuridine (BrdU) incorporation, an indicator of proliferation, occurs in a clear circadian pattern with two peaks per 24 hr (Figures 3A and 3B: fold change 1.08 and peak at 15.5 hr for serum shock, p < 0.001 and p = 0.776 for serum shock and non-synchronized negative controls, respectively; n = 4). At its peaks, proliferation was 16.2% ± 6.9% higher compared with troughs (p = 0.015). We considered the possibility that clock synchronization also synchronizes cell cycle and that differences in BrdU incorporation were caused by changes in cell-cycle phase instead of proliferation. To analyze this possibility, we quantified the amount of cells during 48 hr of cell culture and found that SPC doubling time was 19.5 ± 0.8 hr (n = 4), making it an unlikely cause for the 12-hourly peaks in proliferation.
Figure 3.
Analysis of Circadian Rhythmicity in SPC Proliferation and Migration
(A) Two examples of BrdU staining showing BrdU incorporation 14–20 hr (47%) and 22–28 hr (31%) after serum shock.
(B) Circadian biphasic rhythm in BrdU incorporation of SPCs after serum shock (period 12 hr; p < 0.001; n = 4 independent experiments) but not in unsynchronized cells (p = 0.776; n = 4 independent experiments).
(C) Example of scratch-wound migration assay picture taken directly and 6 hr after scratch.
(D) No rhythmicity was present in migration of SPCs into the scratch (p = 0.442, n = 4).
(E) Combination of proliferation and migration experiments showing that the majority (83.1% ± 2.0%) of cells present in the scratch after 6 hr did not proliferate. White line indicates border of the scratch.
(F) Percentage of growth back into scratch in conditions with lowest and highest cell density and in scratches perpendicular and parallel to cell orientation. No significant differences were found between groups (low density 36.1 ± 6.0 versus high density 31.0 ± 6.5, p = 0.59; perpendicular orientation 32.4 ± 4.9 versus parallel orientation 37.9 ± 5.2; p = 0.11; n = 12 locations for all).
Data are presented as mean ± SEM; ∗p < 0.05 for significant circadian oscillation. Scale bars, 100 μm.
SPC migration did not demonstrate any circadian fluctuations (Figures 3C and 3D: p = 0.442; n = 4). To rule out the possibility that concurrent proliferation clouded the results of our migration assay, we combined the migration assay with BrdU incorporation and found that 83.1% ± 2.0% of cells in the scratch were BrdU negative (Figure 3E; n = 12). This indicates that the majority of SPCs within the scratch originate from migration of cells rather than from proliferation of cells residing originally in the scratch border zone. In addition, we analyzed whether SPC density or orientation to the scratch (parallel or perpendicular to the orientation of the scratch) interfered, but these factors did not influence migration (Figure 3F: low density versus high density 36.1 ± 6.0 versus 31.0 ± 6.5, p = 0.59; perpendicular orientation versus parallel orientation 32.4 ± 4.9 versus 37.9 ± 5.2, p = 0.11; n = 12 for all).
Circadian Rhythmicity in Cell Death/Apoptosis
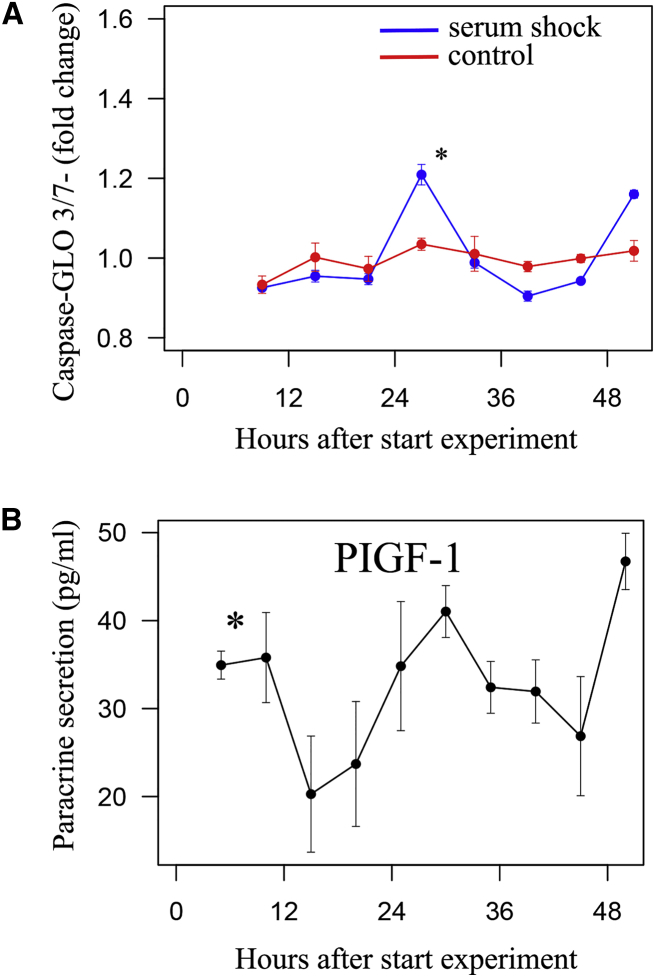
To further explore functional rhythmicity in SPCs, we analyzed rhythmicity in stress tolerance by exposing SPCs to doxorubicin, an anti-cancer drug well known for its cardiac toxicity (Zheng et al., 2013). We found a clear circadian rhythm in doxorubicin-induced apoptosis that was not present in unsynchronized control cells (Figure 4A: fold change 1.13 and peak at 27.2 hr, p < 0.001 for doxorubicin-treated SPCs, p = 0.396 for controls; n = 4). At its peaks, apoptosis as expressed by Caspase-Glo 3/7 was 27.4% ± 2.0% higher compared with the troughs (p < 0.001; n = 4). To test whether these findings were limited to the stressor doxorubicin, we repeated the experiment with or without tert-butyl hydroperoxide as an alternative, and found similar results (Figure S4B: fold change 1.06, peak at 26.9 hr, p < 0.001; n = 4).
Figure 4.
Circadian Rhythmicity Was Present in Stress Tolerance and Four of Ten Paracrine Secretion Factors
(A) Apoptosis was measured after serum shock, subsequent time periods (as depicted on x axis), and 6 hr of doxorubicin exposure using a Caspase-Glo 3/7 assay. Apoptosis was 27.4% ± 2.0% higher 27 and 51 hr compared with 15 and 39 hr after serum shock (p < 0.001; n = 4 independent experiments). In non-synchronized cells, no circadian rhythm was present (p = 0.396; n = 4 independent experiments).
(B) Circadian rhythm in the excretion of paracrine factor PIGF-1 (placental growth factor).
Data are presented as mean ± SEM; ∗p < 0.05 for significant circadian oscillation.
Circadian Rhythm in Excretion of Growth Factors
Finally, we analyzed rhythmicity in SPC function by measuring the release of ten growth factors by SPCs during 48 hr. Four factors—β-nerve growth factor, human placental growth factor, stem cell factor, and vascular endothelial growth factor A (VEGFA)—showed a clear circadian rhythm, whereas in the remaining six factors (brain-derived neurotrophic factor, epidermal growth factor, basic fibroblast growth factor, leukemia inhibitory factor, hepatocyte growth factor, and VEGFD) no rhythmicity was found. The four oscillating factors have not been reported to influence circadian rhythmicity themselves. An example of a circadian rhythm in paracrine factor release is depicted in Figure 4B. A complete overview of all ten factors is given in Table S2. Peak release of the four oscillating factors occurred at the same time points (around 25 and 50 hr after synchronization). The amplitude of the four oscillating factors was 24.3% ± 1.2%, indicating that at its circadian peak, paracrine factor release was almost twice as large compared with the circadian trough.
Discussion
Our data demonstrate that Sca1-reactive cells, as an example of progenitor-like cells from the heart, possess a molecular circadian clock that is functional both at the transcriptional and protein level, resulting in circadian oscillations of core clock genes and proteins. In addition, we show that circadian rhythms are present in the important cell functions proliferation, stress tolerance, and paracrine factor secretion, but not in migration.
Taking circadian rhythmicity into account could improve the potential of progenitor-like cells in research and future therapeutic applications, and can potentially clarify some of the discrepancies that have been found between studies. In vitro experiments investigating proliferation, stress tolerance, and paracrine secretion will be more reliable and reproducible when circadian rhythmicity is included. For optimal testing, all different experimental conditions should be performed at the same molecular time, and preferably even at different time points. In our stress tolerance experiments, for example, we showed that the timing of adding a low dose of doxorubicin to SPC cultures is crucial: doxorubicin hardly increased apoptosis when applied 21 hr after the start of an experiment, whereas it did increase apoptosis significantly when added only 6 hr later, 27 hr after the start of the experiment.
In vivo experiments that apply transplantation of cultured stem- or progenitor-derived cells could also potentially benefit from knowledge about circadian rhythms: similar to in vitro experiments, reproducibility will likely be improved when all experiments are performed at the same (and preferably more) time frames. This may, for example, help translate results from nocturnal rodents to diurnal larger animals or patients, which are known to have opposite circadian rhythms. In addition, physiological circadian rhythmicity has the potential to improve SPC effects, with the advantage that it does not require genetic modifications, which often compromise translation from the experimental laboratory to the patient.
Circadian rhythms have previously been associated with cell proliferation and stress tolerance in vivo. Osteoblasts, adult stem cells responsible for bone formation, possess a molecular circadian clock which, when disrupted, inhibits proliferation (Fu et al., 2005). In epidermal stem cells, the phase of the molecular circadian clock determines whether stem cells are receptive to proliferative cues or stay in a dormant state (Janich et al., 2011). The link between stress tolerance and circadian rhythms was most prominently shown by studies investigating myocardial infarction. Several independent pre-clinical and clinical studies showed that infarct size and remaining left ventricular function depend on the time of day a myocardial infarction occurs (Durgan et al., 2010, Reiter et al., 2012). Reiter and coworkers quantified the potential impact of a circadian rhythm in infarct size for patients, with a difference of >7% in ejection fraction 1 year after the infarct between the best and worse time of day, even when taking possible confounders into account (Durgan et al., 2010, Reiter et al., 2012). Studies of Durgan et al., Xiao et al., and Bonney et al. provided mechanistic insight into this phenomenon by demonstrating the relation to the cardiomyocyte circadian clock and its output pathways, including possible paracrine excretion (Bonney et al., 2013, Durgan et al., 2010, Ho et al., 2016, Hsueh et al., 2014, Xiao et al., 2016).
The studies of Janich, Fu, Durgan, and others nicely demonstrated that genetic or environmental disruption of the molecular circadian clock in various stem cells and cardiac cells results in disrupted proliferation or stress tolerance (Durgan et al., 2010, Fu et al., 2005, Janich et al., 2011). Although our data are in line with these results, the setup of our experiments differs: we did not focus on (long-term) results of molecular clock disruption. Therefore, we cannot make any conclusions about the causative role of individual clock components. Instead we were interested in physiological circadian fluctuations in SPC functions, which had not been investigated previously.
In conclusion, to our knowledge this is the first time that circadian rhythmicity has been linked to a cell type with potential for cardiac in vitro research and cell-based cardiac repair. Our findings indicate that significant circadian rhythmicity is present in SPCs at both the molecular and functional level, and may influence the results of experiments and therapies using SPCs and other progenitor-like cells.
Experimental Procedures
SPC Isolation
SPCs were isolated and cultured as previously described (Goumans et al., 2007, Smits et al., 2009a, Smits et al., 2009b). In short, fetal hearts, obtained from elective abortion with prior informed consent and approval of the ethical committee of the University Medical Center Utrecht, were enzymatically dissociated. Magnetic cell sorting with an iron-labeled anti-SCA-1 antibody was performed to extract the SPCs from the dissociated cell suspension.
Synchronization of SPCs
Confluent SPCs were exposed to the synchronizers “serum shock” (SS, 50% culture medium/50% horse serum, Gibco), 10 μM forskolin (Sigma), or 100 nM dexamethasone (Sigma) at the start of the experiments to align circadian clocks (Balsalobre et al., 1998, Yagita and Okamura, 2000). Non-synchronized SPCs, with medium change more than 2 days before start of the experiment, served as controls.
qRT-PCR, Western Blotting, Immunofluorescence, and the Bioluminescent Reporter
Detailed techniques and protocols used for qRT-PCR, western blotting, immunofluorescence, and the bioluminescent reporter, including primer sequences and antibodies, are described in Supplemental Experimental Procedures and Table S3.
Cell Detachment, Matrix, Proliferation, Migration, Cell Death, and Cellular Secretion Assays
Specifics about the experiments performed can be found in Supplemental Experimental Procedures.
Statistical Analysis
Data are presented as mean ± SEM. Circadian rhythmicity was analyzed using Cosinor analysis in R (Refinetti et al., 2007). To compare differences between two groups, we used a two-tailed Student's t test. p Values of <0.05 were considered statistically significant.
Author Contributions
L.W.v.L. conceived the general concept of this work. L.W.v.L., B.C.d.P., D.A.M.F., P.D., and T.A.B.v.V. designed experiments and interpreted the data. B.C.d.P., E.J.D., D.A.M.F., P.D., S.C., and B.J.M.K. acquired and analyzed data. L.W.v.L., T.A.B.v.V., J.P.G.S., M.A.V., and P.A.D. supervised and interpreted the data. B.C.d.P., T.A.B.v.V., and L.W.v.L. drafted the manuscript. E.J.D. and D.A.M.F. contributed equally. All authors contributed to revising the manuscript and all authors approved the final version.
Acknowledgments
The authors would like to thank Professor A. Liu (University of Memphis) for sharing the lentiviral BMAL1-dLuc and CRY1-dLuc plasmids. Funding for this work was provided by the Netherlands Organization for Health Research and Development (ZonMW Veni 91612147), the Netherlands Heart Foundation (Dekker 2013T056), and the Alexandre Suerman program of the UMC Utrecht. We acknowledge the support from Innovation and the Netherlands CardioVascular Research Initiative (CVON): The Dutch Heart Foundation, the Dutch Federation of University Medical Centers, the Netherlands Organization for Health Research and Development, and the Royal Netherlands Academy of Science.
Published: August 10, 2017
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, four figures, and three tables and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2017.07.010.
Supporting Citations
The following references appear in the Supplemental Information: Liu et al., 2008, Liu et al., 2007, Ramanathan et al., 2012, van Mil et al., 2012.
Supplemental Information
References
- Balsalobre A., Damiola F., Schibler U. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell. 1998;93:929–937. doi: 10.1016/s0092-8674(00)81199-x. [DOI] [PubMed] [Google Scholar]
- Bearzi C., Rota M., Hosoda T., Tillmanns J., Nascimbene A., De Angelis A., Yasuzawa-Amano S., Trofimova I., Siggins R.W., Lecapitaine N. Human cardiac stem cells. Proc. Natl. Acad. Sci. USA. 2007;104:14068–14073. doi: 10.1073/pnas.0706760104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boland M.J., Nazor K.L., Loring J.F. Epigenetic regulation of pluripotency and differentiation. Circ. Res. 2014;115:311–324. doi: 10.1161/CIRCRESAHA.115.301517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonney S., Kominsky D., Brodsky K., Eltzschig H., Walker L., Eckle T. Cardiac Per2 functions as novel link between fatty acid metabolism and myocardial inflammation during ischemia and reperfusion injury of the heart. PLoS One. 2013;8:e71493. doi: 10.1371/journal.pone.0071493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu L., Jiang X., Martin-Puig S., Caron L., Zhu S., Shao Y., Roberts D.J., Huang P.L., Domian I.J., Chien K.R. Human ISL1 heart progenitors generate diverse multipotent cardiovascular cell lineages. Nature. 2009;460:113–117. doi: 10.1038/nature08191. [DOI] [PubMed] [Google Scholar]
- Darlington T.K., Wager-Smith K., Ceriani M.F., Staknis D., Gekakis N., Steeves T.D., Weitz C.J., Takahashi J.S., Kay S.A. Closing the circadian loop: CLOCK-induced transcription of its own inhibitors per and tim. Science. 1998;280:1599–1603. doi: 10.1126/science.280.5369.1599. [DOI] [PubMed] [Google Scholar]
- Dawn B., Bolli R. Cardiac progenitor cells: the revolution continues. Circ. Res. 2005;97:1080–1082. doi: 10.1161/01.RES.0000195610.71671.c2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer T.P., van Veen T.A., Jonsson M.K., Kok B.G., Metz C.H., Sluijter J.P., Doevendans P.A., de Bakker J.M., Goumans M.J., van der Heyden M.A. Human cardiomyocyte progenitor cell-derived cardiomyocytes display a maturated electrical phenotype. J. Mol. Cell. Cardiol. 2010;48:254–260. doi: 10.1016/j.yjmcc.2009.05.004. [DOI] [PubMed] [Google Scholar]
- Du Pre B.C., Van Veen T.A., Young M.E., Vos M.A., Doevendans P.A., Van Laake L.W. Circadian rhythms in cell maturation. Physiology. 2014;29:72–83. doi: 10.1152/physiol.00036.2013. [DOI] [PubMed] [Google Scholar]
- Durgan D.J., Young M.E. The cardiomyocyte circadian clock: emerging roles in health and disease. Circ. Res. 2010;106:647–658. doi: 10.1161/CIRCRESAHA.109.209957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durgan D.J., Pulinilkunnil T., Villegas-Montoya C., Garvey M.E., Frangogiannis N.G., Michael L.H., Chow C.W., Dyck J.R., Young M.E. Short communication: ischemia/reperfusion tolerance is time-of-day-dependent: mediation by the cardiomyocyte circadian clock. Circ. Res. 2010;106:546–550. doi: 10.1161/CIRCRESAHA.109.209346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu L., Patel M.S., Bradley A., Wagner E.F., Karsenty G. The molecular clock mediates leptin-regulated bone formation. Cell. 2005;122:803–815. doi: 10.1016/j.cell.2005.06.028. [DOI] [PubMed] [Google Scholar]
- Gaetani R., Feyen D.A., Doevendans P.A., Gremmels H., Forte E., Fledderus J.O., Ramjankhan F.Z., Messina E., Sussman M.A., Giacomello A. Different types of cultured human adult cardiac progenitor cells have a high degree of transcriptome similarity. J. Cell. Mol. Med. 2014;18:2147–2151. doi: 10.1111/jcmm.12458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goumans M.J., de Boer T.P., Smits A.M., van Laake L.W., van Vliet P., Metz C.H., Korfage T.H., Kats K.P., Hochstenbach R., Pasterkamp G. TGF-beta1 induces efficient differentiation of human cardiomyocyte progenitor cells into functional cardiomyocytes in vitro. Stem Cell Res. 2007;1:138–149. doi: 10.1016/j.scr.2008.02.003. [DOI] [PubMed] [Google Scholar]
- Ho Y.S., Tsai W.H., Lin F.C., Huang W.P., Lin L.C., Wu S.M., Liu Y.R., Chen W.P. Cardioprotective actions of TGFbetaRI inhibition through stimulating autocrine/paracrine of survivin and inhibiting wnt in cardiac progenitors. Stem Cells. 2016;34:445–455. doi: 10.1002/stem.2216. [DOI] [PubMed] [Google Scholar]
- Hsueh Y.C., Wu J.M., Yu C.K., Wu K.K., Hsieh P.C. Prostaglandin E(2) promotes post-infarction cardiomyocyte replenishment by endogenous stem cells. EMBO Mol. Med. 2014;6:496–503. doi: 10.1002/emmm.201303687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janich P., Pascual G., Merlos-Suarez A., Batlle E., Ripperger J., Albrecht U., Cheng H.Y., Obrietan K., Di Croce L., Benitah S.A. The circadian molecular clock creates epidermal stem cell heterogeneity. Nature. 2011;480:209–214. doi: 10.1038/nature10649. [DOI] [PubMed] [Google Scholar]
- Liu A.C., Welsh D.K., Ko C.H., Tran H.G., Zhang E.E., Priest A.A., Buhr E.D., Singer O., Meeker K., Verma I.M. Intercellular coupling confers robustness against mutations in the SCN circadian clock network. Cell. 2007;129:605–616. doi: 10.1016/j.cell.2007.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu A.C., Tran H.G., Zhang E.E., Priest A.A., Welsh D.K., Kay S.A. Redundant function of REV-ERBalpha and beta and non-essential role for Bmal1 cycling in transcriptional regulation of intracellular circadian rhythms. PLoS Genet. 2008;4:e1000023. doi: 10.1371/journal.pgen.1000023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madonna R., Van Laake L.W., Davidson S.M., Engel F.B., Hausenloy D.J., Lecour S., Leor J., Perrino C., Schulz R., Ytrehus K. Position paper of the European Society of Cardiology Working Group cellular biology of the heart: cell-based therapies for myocardial repair and regeneration in ischemic heart disease and heart failure. Eur. Heart J. 2016;37:1789–1798. doi: 10.1093/eurheartj/ehw113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martino T.A., Young M.E. Influence of the cardiomyocyte circadian clock on cardiac physiology and pathophysiology. J. Biol. Rhythms. 2015;30:183–205. doi: 10.1177/0748730415575246. [DOI] [PubMed] [Google Scholar]
- Noseda M., Harada M., McSweeney S., Leja T., Belian E., Stuckey D.J., Abreu Paiva M.S., Habib J., Macaulay I., de Smith A.J. PDGFRalpha demarcates the cardiogenic clonogenic Sca1+ stem/progenitor cell in adult murine myocardium. Nat. Commun. 2015;6:6930. doi: 10.1038/ncomms7930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanathan C., Khan S.K., Kathale N.D., Xu H., Liu A.C. Monitoring cell-autonomous circadian clock rhythms of gene expression using luciferase bioluminescence reporters. J. Vis. Exp. 2012;67:4234. doi: 10.3791/4234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Refinetti R., Lissen G.C., Halberg F. Procedures for numerical analysis of circadian rhythms. Biol. Rhythm Res. 2007;38:275–325. doi: 10.1080/09291010600903692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter R., Swingen C., Moore L., Henry T.D., Traverse J.H. Circadian dependence of infarct size and left ventricular function after ST elevation myocardial infarction. Circ. Res. 2012;110:105–110. doi: 10.1161/CIRCRESAHA.111.254284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanganalmath S.K., Bolli R. Cell therapy for heart failure: a comprehensive overview of experimental and clinical studies, current challenges, and future directions. Circ. Res. 2013;113:810–834. doi: 10.1161/CIRCRESAHA.113.300219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santini M.P., Forte E., Harvey R.P., Kovacic J.C. Developmental origin and lineage plasticity of endogenous cardiac stem cells. Development. 2016;143:1242–1258. doi: 10.1242/dev.111591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits A.M., van Vliet P., Metz C.H., Korfage T., Sluijter J.P., Doevendans P.A., Goumans M.J. Human cardiomyocyte progenitor cells differentiate into functional mature cardiomyocytes: an in vitro model for studying human cardiac physiology and pathophysiology. Nat. Protoc. 2009;4:232–243. doi: 10.1038/nprot.2008.229. [DOI] [PubMed] [Google Scholar]
- Smits A.M., van Laake L.W., den Ouden K., Schreurs C., Szuhai K., van Echteld C.J., Mummery C.L., Doevendans P.A., Goumans M.J. Human cardiomyocyte progenitor cell transplantation preserves long-term function of the infarcted mouse myocardium. Cardiovasc. Res. 2009;83:527–535. doi: 10.1093/cvr/cvp146. [DOI] [PubMed] [Google Scholar]
- Takamiya M., Haider K.H., Ashraf M. Identification and characterization of a novel multipotent sub-population of Sca-1(+) cardiac progenitor cells for myocardial regeneration. PLoS One. 2011;6:e25265. doi: 10.1371/journal.pone.0025265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Mil A., Grundmann S., Goumans M.J., Lei Z., Oerlemans M.I., Jaksani S., Doevendans P.A., Sluijter J.P. MicroRNA-214 inhibits angiogenesis by targeting Quaking and reducing angiogenic growth factor release. Cardiovasc. Res. 2012;93:655–665. doi: 10.1093/cvr/cvs003. [DOI] [PubMed] [Google Scholar]
- Vyas M.V., Garg A.X., Iansavichus A.V., Costella J., Donner A., Laugsand L.E., Janszky I., Mrkobrada M., Parraga G., Hackam D.G. Shift work and vascular events: systematic review and meta-analysis. BMJ. 2012;345:e4800. doi: 10.1136/bmj.e4800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woon P.Y., Kaisaki P.J., Braganca J., Bihoreau M.T., Levy J.C., Farrall M., Gauguier D. Aryl hydrocarbon receptor nuclear translocator-like (BMAL1) is associated with susceptibility to hypertension and type 2 diabetes. Proc. Natl. Acad. Sci. USA. 2007;104:14412–14417. doi: 10.1073/pnas.0703247104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J., Pan Y., Li X.H., Yang X.Y., Feng Y.L., Tan H.H., Jiang L., Feng J., Yu X.Y. Cardiac progenitor cell-derived exosomes prevent cardiomyocytes apoptosis through exosomal miR-21 by targeting PDCD4. Cell Death Dis. 2016;7:e2277. doi: 10.1038/cddis.2016.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagita K., Okamura H. Forskolin induces circadian gene expression of rPer1, rPer2 and dbp in mammalian rat-1 fibroblasts. FEBS Lett. 2000;465:79–82. doi: 10.1016/s0014-5793(99)01724-x. [DOI] [PubMed] [Google Scholar]
- Young M.E., Razeghi P., Taegtmeyer H. Clock genes in the heart: characterization and attenuation with hypertrophy. Circ. Res. 2001;88:1142–1150. doi: 10.1161/hh1101.091190. [DOI] [PubMed] [Google Scholar]
- Zheng W., Lu Y.B., Liang S.T., Zhang Q.J., Xu J., She Z.G., Zhang Z.Q., Yang R.F., Mao B.B., Xu Z. SIRT1 mediates the protective function of Nkx2.5 during stress in cardiomyocytes. Basic Res. Cardiol. 2013;108:364. doi: 10.1007/s00395-013-0364-y. [DOI] [PubMed] [Google Scholar]
- Zwetsloot P.P., Vegh A.M., Jansen Of Lorkeers S.J., van Hout G.P., Currie G.L., Sena E.S., Gremmels H., Buikema J.W., Goumans M.J., Macleod M.R. Cardiac stem cell treatment in myocardial infarction: a systematic review and meta-analysis of preclinical studies. Circ. Res. 2016;118:1223–1232. doi: 10.1161/CIRCRESAHA.115.307676. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.