Summary
HLA haplotype-homozygous (HLA-homo) induced pluripotent stem cells (iPSCs) are being prepared to be used for allogeneic transplantation of regenerated tissue into recipients carrying an identical haplotype in one of the alleles (HLA-hetero). However, it remains unaddressed whether natural killer (NK) cells respond to these regenerated cells. HLA-C allotypes, known to serve as major ligands for inhibitory receptors of NK cells, can be classified into group 1 (C1) and group 2 (C2), based on their binding specificities. We found that the T cells and vascular endothelial cells regenerated from HLA-homo-C1/C1 iPSCs were killed by specific NK cell subsets from a putative HLA-hetero-C1/C2 recipient. Such cytotoxicity was canceled when target cells were regenerated from iPSCs transduced with the C2 gene identical to the recipient. These results clarify that NK cells can kill regenerated cells by sensing the lack of HLA-C expression and further provide the basis for an approach to prevent such NK cell-mediated rejection responses.
Keywords: iPSCs, NK cells, KIR, KIR ligand, HLA-C, transplantation, regeneration, vascular endothelial cells, HLA haplotype-homozygote, missing-self recognition
Graphical Abstract
Highlights
-
•
Cells from HLA-homo iPSCs are killed by NK cells from an HLA-hetero C1/C2 individual
-
•
NK cells kill the regenerated cells by sensing the lack of KIR ligand expression
-
•
Cytotoxicity is cancelled when regenerated cells overexpress the missing KIR ligand
In this article, Kawamoto and colleagues show that NK cells derived from an HLA-hetero individual killed the cells regenerated from HLA-homo iPSCs in KIR ligand-mismatched cases, by sensing the lack of KIR ligand expression. Such cytotoxicity was cancelled when regenerated cells are enforced to express the missing KIR ligand, providing a novel approach to prevent NK cell-mediated rejection.
Introduction
Induced pluripotent stem cells (iPSCs) have received attention as a solution to overcome the limitations of using human embryonic stem cells (ESCs) in tissue regeneration (Takahashi et al., 2007). To cope with the high cost and the long duration required to develop autologous iPSCs, usage of pre-generated allogeneic iPSCs developed from human leukocyte antigen (HLA) haplotype-homozygous (HLA-homo) healthy individuals has been proposed. Banking of these iPSCs started as a national initiative in Japan in 2012, and iPSCs carrying the most frequent HLA haplotypes in the Japanese are now available for clinical use from the Center for iPS Cell Research and Application in Kyoto. Tissues regenerated from these iPSCs are assumed to trigger minimal T cell alloreactivity when transplanted into HLA-homo and HLA haplotype-heterozygous (HLA-hetero) recipients due to the lack of direct and indirect recognition of HLA (Nakatsuji et al., 2008, Okita et al., 2011, Taylor et al., 2012).
An issue remaining in the rejection of iPSC-derived tissue in the aforementioned homo-to-hetero transplantation setting is the allogeneic response by natural killer (NK) cells. Clinical studies are lacking because solid organ transplants in homo-to-hetero settings are often avoided to circumvent the risk of fatal graft-versus-host disease caused by contaminating T cells in the graft. We thus investigated whether an NK cell-based graft rejection response against iPSC-derived transplant tissue will occur in homo-to-hetero transplantation. Unique to NK cells is their ability to sense the lack of HLA class I expression on target cells: “missing self” (Joncker and Raulet, 2008, Kärre et al., 1986, Nakamura et al., 2013, Suzue et al., 2001, Velardi et al., 2009). Inhibitory receptors are responsible for sensing “missing self”; the killer cell immunoglobulin-like receptors (KIRs) and the NKG2A receptor. Heterogeneous receptor expression generates many NK cell subsets that differ in specificity toward HLA-A, -B, -C, and -E. Notably, only a fraction of these NK cell subsets in peripheral blood acquire the ability for missing-self response, a process termed “licensing” or “education,” whereby it is hypothesized that the cells that have encountered cognate receptor-HLA class I ligand interactions during cellular development become capable of mounting missing-self responses (Anfossi et al., 2006, Kim et al., 2005).
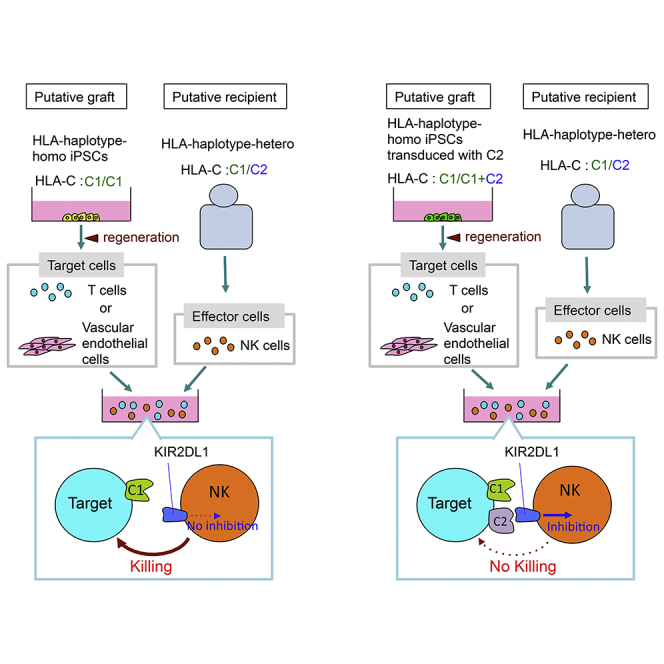
All HLA-C molecules are ligands for inhibitory KIRs (KIR2DL1, 2, 3), whereas only HLA-A, and -B allotypes that carry the Bw4 motif are recognized by KIR (KIR3DL1) (Moesta and Parham, 2012). The HLA-C allotypes are classified into two groups based on their sequence: “C1” and “C2” ligands (Moffett and Colucci, 2015). Based on their HLA-C genotypes, graft recipients are therefore divided broadly into C1/C1, C1/C2, and C2/C2 types. It is predicted that when regenerated cells from HLA-homo-C1/C1 or -C2/C2 iPSCs are transplanted into an HLA-hetero C1/C2 recipient, the recipient's licensed NK cells will respond against the lack of the KIR ligand (HLA-C2 or -C1, respectively) on the transplant and eventually kill the graft due to the lack of inhibitory signaling from KIR2DL1 or KIR2DL3, respectively, following the conventional mechanism of missing self unique to NK cells. Here, the missing-self response is thought to be a result of the lack of inhibitory signaling in the presence of activating receptor engagement.
To test whether iPSC-derived cells will undergo such NK cell-based killing, we first established iPSCs from an HLA-homo-C1/C1 healthy volunteer and produced T cells as well as vascular endothelial (VE) cells to be used as target cells in co-culture assays. This is because (1) we are planning to use iPSC-derived cytotoxic T cells for cell therapy against cancer (Maeda et al., 2016, Vizcardo et al., 2013), and (2) VE cells will likely become the first target of NK cells in peripheral circulation, as exemplified by NK cell-based vasculopathy in an allogeneic cardiac transplantation mouse model (Uehara et al., 2005). As effector cells of putative recipients, we used NK cells collected from HLA-hetero-C1/C2 healthy volunteers. We found that in this setting, NK cells exerted cytotoxicity against iPSC-derived cells, and that this is dependent on missing-self recognition.
Results
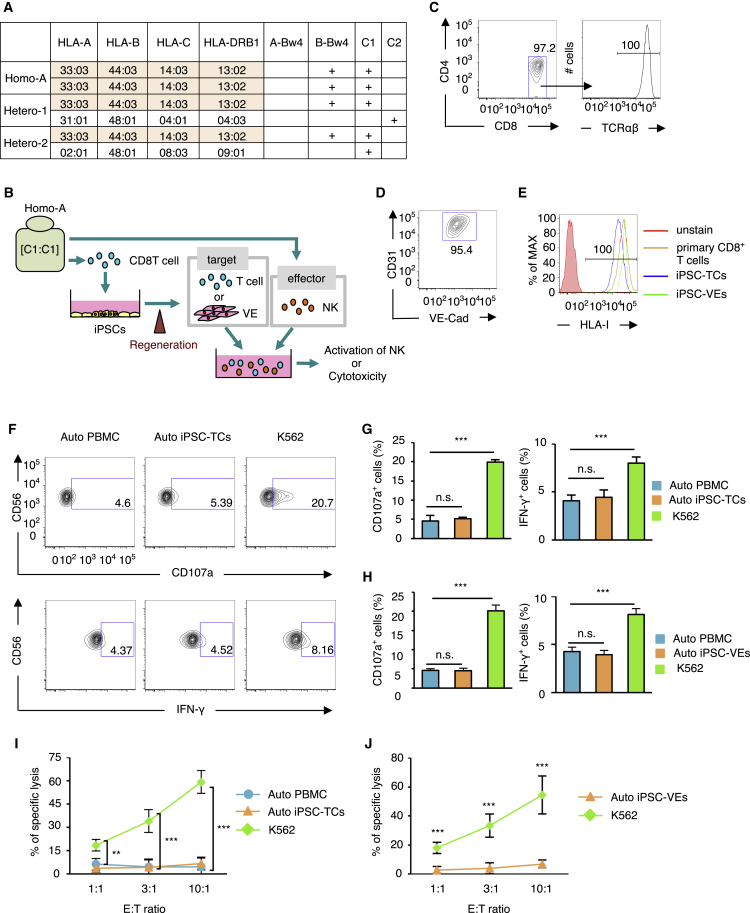
In the present study, we tested whether regenerated tissue/cells developed from HLA-homo iPSCs can be rejected by NK cells when transplanted into an HLA-hetero recipient. To this end, we selected three healthy volunteers, one putative donor and two putative recipients (Figure 1A). The first donor was homozygous for the second frequent HLA haplotype in the Japanese population (A∗3303, B∗4403, C∗1403, DR∗1302). As HLA-C∗1403 is group 1 HLA-C, this individual was designated HLA-homo-C1/C1, “Homo-A.” The other two individuals carried the same HLA haplotype on one allele as Homo-A; i.e., in a haploidentical setting, one individual bearing group 2 HLA-C on the other allele (HLA-hetero-C1/C2, “Hetero-1”), and the other individual carried different group 1 HLA-C on the other allele (HLA-hetero-C1/C1, “Hetero-2”). We did not include Bw4 ligand in this case, since this common HLA haplotype carries the Bw4 ligand (B∗4403). In the following experiments, we produced iPSCs from Homo-A and Hetero-1, and the pluripotency of established iPSCs (clone: Homo-A#1, Hetero-1#1, #4) were characterized (Table S1). Cells regenerated from those iPSC lines were used as target. As effector cells, NK cells in peripheral blood mononuclear cells (PBMCs) collected from Hetero-1 or Hetero-2 were used.
Figure 1.
NK Cells Do Not Respond to Cells Regenerated from Autologous iPSCs
(A) HLA typing and KIR ligand classification of the cells used to derive iPSCs (Homo-A) and the donors from which NK cells were isolated (Hetero-1, Hetero-2). The shared haplotype is depicted in orange.
(B) Schematic illustration of the experimental procedure. iPSCs were produced from CD8+ T cells isolated from Homo-A. T cells or vascular endothelial (VE) cells regenerated from the iPSCs were co-cultured with NK cells collected from donor Homo-A, and the activation status of NK cells or cytotoxicity against regenerated cells was assessed.
(C) Representative flow-cytometric profiles of the T cells (TCs) derived from Homo-A iPSCs.
(D) A flow-cytometric profile of iPSC-derived VE cells from Homo-A.
(E) Flow-cytometry-based profiling of HLA class I expression on primary CD8+ T cells, Homo-A iPSC-derived T cells (iPSC-TCs), and VE cells (iPSC-VEs).
(F–H) NK cell activation status (CD107a mobilization assay and intracellular staining for IFN-γ) was assessed on Homo-A derived NK cells after 12 hr of co-incubation with autologous (Auto) iPSC-derived cells, an HLA class I-deficient leukemic cell K562 (positive control), and Auto PBMCs (negative control). (F) Representative flow-cytometry analysis using the CD107a mobilization assay (top) and intracellular staining for IFN-γ (bottom). (G and H) Homo-A-derived NK cells did not respond to Auto iPSC-TCs (G) and Auto iPSC-VEs (H) in terms of cytotoxicity (left; CD107a) and cytokine production (right; IFN-γ) compared with the response against Auto PBMCs.
(I and J) Eighteen-hour 51Cr-release cytotoxicity assays using Homo-A NK cells against Auto iPSC-TCs (I) or Auto iPSC-VEs (J).
Results are presented as mean ± SD from three independent experiments. ∗∗p < 0.01, ∗∗∗p < 0.001, Student's t test. n.s., not significant.
NK Cells Are Tolerant to the T Cells and VE Cells Regenerated from Autologous iPSCs
We first tested whether NK cells respond to autologous (Auto) iPSC-derived cells. This experiment was designed as a human version of a previous report in mouse, whereby cardiomyocytes (CMs) regenerated from ESCs were not killed by NK cells from syngeneic mouse (Frenzel et al., 2009). iPSCs were produced from peripheral CD8 T cells collected from donor Homo-A (Figure 1B). From the iPSCs, T cells and VE cells were regenerated as target cells (designated iPSC-TCs or iPSC-VEs, respectively) using established methods (Maeda et al., 2016, Masumoto et al., 2014). The resulting iPSC-TCs were CD4−CD8+TCRαβ+ cells (Figure 1C), and iPSC-VEs were CD31+VE-Cadherin+ cells (Figure 1D). These cells expressed HLA class I at levels comparable with that of primary CD8 T cells (Figure 1E).
NK cell responses were then assessed in co-culture assays using Auto iPSC-TCs as target. Cytotoxicity and cytokine responses were measured using the CD107a mobilization assay and intracellular staining for interferon-γ (IFN-γ), respectively. As anticipated, the NK cells were hyporesponsive to Auto iPSC-TCs at levels similar to those of Auto PBMCs (Figures 1F and 1G) and of Auto iPSC-VEs as well (Figure 1H). The lack of NK cell responses against Auto iPSC-TCs and -VEs were validated further with a cytotoxicity assay; the NK cells did not exhibit cytotoxic activity against either Auto iPSC-TCs or -VEs (Figures 1I and 1J). Very similar results were obtained when target cells from Hetero-1-derived iPSCs and NK cells from Hetero-1 were used (Figure S1). These results indicate that human NK cells do not respond to cells “regenerated” from autologous iPSCs.
NK Cells Are Capable of Mounting a Missing-Self Response against Regenerated T Cells and VE Cells in a KIR-Ligand-Mismatched Setting
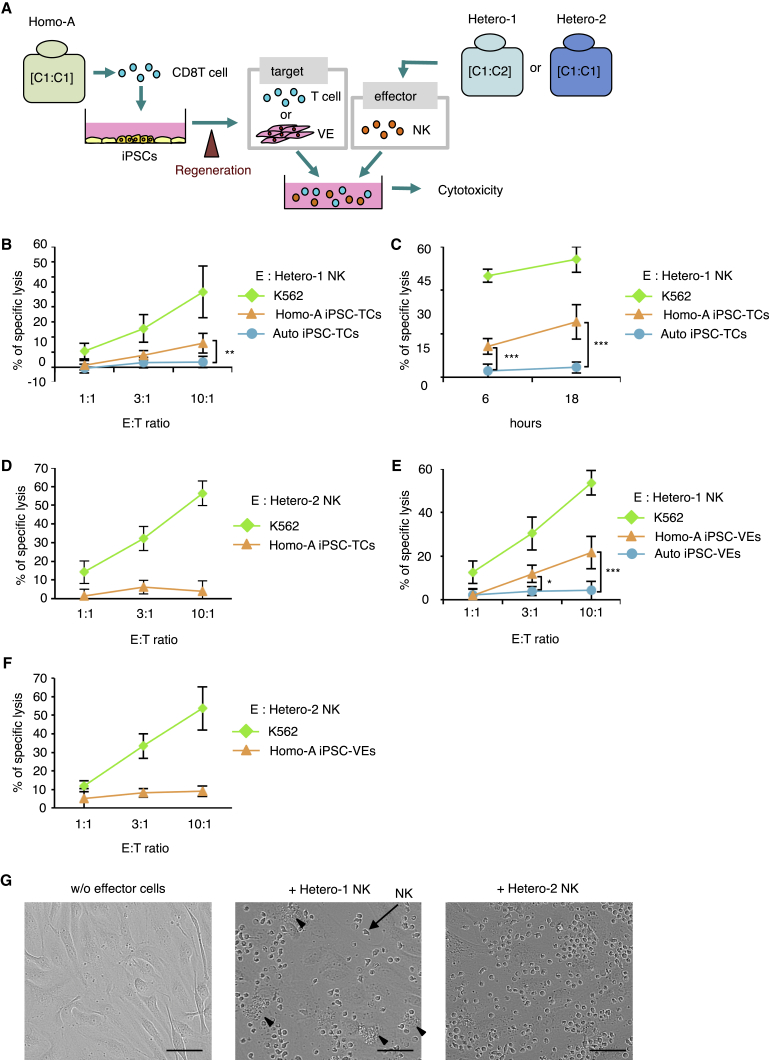
We next examined whether NK cells have the potential to reject regenerated, allogeneic cells when KIR ligands are mismatched in a putative “homo-to-hetero” setting. Again, T cells or VE cells were regenerated from iPSCs derived from donor Homo-A (Homo-A iPSC-TCs or Homo-A iPSC-VEs) (Figure 2A). NK cells from donors Hetero-1 (mismatched) or Hetero-2 (matched) were used as effector cells.
Figure 2.
NK Cells from HLA-Hetero C1/C2, but Not C1/C1, Exhibit Cytotoxic Activity against Regenerated T Cells and VE Cells Derived from HLA-Homo C1/C1 iPSCs
(A) Schematic illustration of the experimental procedure. iPSCs were produced from Homo-A, and subsequently T cells (Homo-A iPSC-TCs) or VE cells (Homo-A iPSC-VEs) were regenerated. Cytotoxicity against the regenerated cells was assessed in co-culture assays with NK cells isolated from Hetero-1 (C1/C2) or Hetero-2 (C1/C1).
(B–F) Cytotoxicity assays (B, 6 hr; C–F, 18 hr) of NK cells isolated from donors Hetero-1 or Hetero-2 against Homo-A iPSC-TCs and -VEs.
(G) Phase-contrast microscopy images of iPSC-VEs after co-culture with allogeneic NK cells display the presence of apoptotic iPSC-VEs (arrowheads). NK cells are indicated by arrows. The iPSC-VEs were plated on a 96-well plate and co-cultured with NK cells collected from Hetero-1 (middle) or Hetero-2 (right) at a 4:1 E/T ratio for 24 hr. Scale bars, 50 μm.
Results are presented as mean ± SD from three independent experiments. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, Student's t test.
NK cells from Hetero-1 displayed significant cytotoxicity against Homo-A iPSC-TCs at a 10:1 effector-to-target (E/T) ratio and at both the 6- and 18-hr time points (Figures 2B and 2C), whereas such specific lysis was absent with NK cells from donor Hetero-2 (Figure 2D). NK cells from Hetero-1 or Hetero-2 displayed the same pattern of response against VE cells regenerated from Homo-A-derived iPSCs (Homo-A iPSC-VEs) (Figures 2E and 2F). These results were further verified by phase-contrast microscopy whereby VE cell death was observed in co-culture with Hetero-1 NK cells but not with Hetero-2 NK cells (Figure 2G).
The conclusion is that the NK cells from an HLA-C1/C2 person elicit cytotoxicity against C1/C1 iPSC-derived cells, whereas NK cells from an HLA-C1/C1 person do not. These results support our prediction that NK cells have the capability to mount a rejection response against iPSC-derived grafts through missing-self recognition.
NK Cell Licensing Is a Key Factor in Cytotoxicity against Cells Regenerated from iPSCs
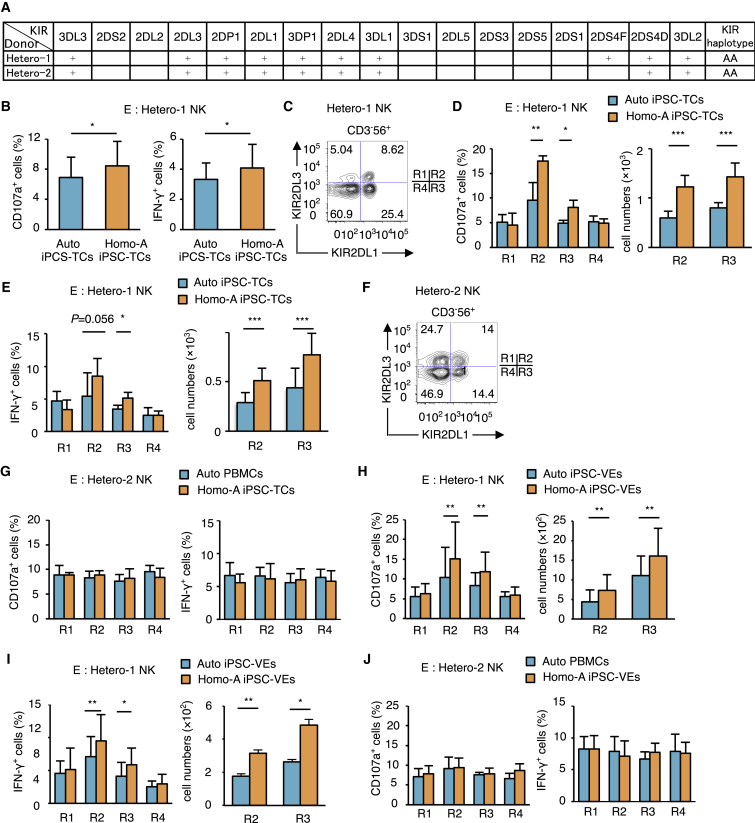
The results presented so far imply that NK cells are able to sense the absence of a KIR ligand in regenerated cells via the “missing-self recognition” mechanism. If this is the case, the NK cells that express inhibitory KIR for the missing ligand should be exclusively responsible for this killing. Taking KIR2DL1 and its ligand HLA-C2 as example, we approached this question by comparing the responses of four NK cell subsets expressing combinations of KIR2DL1 and KIR2DL3: KIR2DL3+2DL1−, KIR2DL3+2DL1+, KIR2DL3−2DL1+, and KIR2DL3−2DL1− (R1, R2, R3, and R4), respectively. The expectation is that NK cell subsets R2 and R3 that express KIR2DL1 will become exclusively activated upon encountering target cells of the C1/C1 genotype.
NK cells from donor Hetero-1 (homozygous for the group A KIR haplotype and thus carries the KIR2DL1 and KIR2DL3 genes but not the KIR2DL2 gene [Figure 3A]) (Yawata et al., 2002) were co-cultured with Homo-A iPSC-TCs. Consistent with the results shown in Figure 2B, we found a significant increase in CD107a+ cells and IFN-γ+ cells within the bulk NK cells (Figure 3B). When these NK cells were subdivided into R1–R4 subsets (Figure 3C), the R2 and R3 subsets both displayed allogeneic responses in terms of proportion and absolute number of CD107a+ cells and IFN-γ+ cells after co-culture with Homo-A iPSC-TCs (Figures 3D, 3E, S2A, and S2B). No significant increase of CD107a+ cells nor IFN-γ+ cells was seen in the other NK cell subsets (Figures 3D and 3E), indicating that sensing of missing self and licensing involving the KIR2DL1 receptor-ligand interaction was the primary mechanism inducing alloreactivity against the iPSC-derived cells. In addition, we co-cultured Hetero-2 NK cells (homozygous for the group A KIR haplotype) (Figures 3A and 3F) with Homo-A iPSC-TCs, where no KIR-ligand mismatch takes place, and investigated the proportion of CD107a+ cells and IFN-γ+ cells of NK cells in the R1–R4 subsets. No significant increase of CD107a+ cells nor IFN-γ+ cells was seen in any subset (Figure 3G), indicating that the NK cells expressing KIR2DL1 in this individual with the C1/C1 genotype had not been licensed to respond to the absence of C2, and were thus hyporesponsive to iPSC-derived cells carrying the C1/C1 type.
Figure 3.
KIR2DL1+ NK Cell Subsets Isolated from a C1/C2 Donor Respond to Regenerated C1/C1 T Cells or VE Cells
(A) The KIR genotypes for the two donors from which NK cells are isolated are shown. The full and deleted forms of KIR2DS4 are indicated by an F and D, respectively.
(B) NK cells isolated from a donor Hetero-1 were co-cultured for 12 hr with Homo-A iPSC-TCs and Auto iPSC-TCs.
(C) The variegated expression of KIR2DL1 and KIR2DL3 generates four distinct cell subsets (R1 to R4) within the CD3−CD56+ NK cells isolated from Hetero-1.
(D and E) Twelve-hour co-incubation assay by using Homo-A iPSC-TCs as target cells. CD107a+ (D) and IFN-γ+ (E) cell numbers are shown in right panels.
(F) The R1–R4 subsets within the NK cells isolated from donor Hetero-2, as defined by the expressed combinations of KIR2DL1 and KIR2DL3.
(G) Twelve-hour co-culture assay by using Homo-A iPSC-TCs as target cells. NK cells were isolated from donor Hetero-2.
(H–J) Twelve-hour co-culture assay by using Homo-A iPSC-VEs as target cells. NK cells were isolated from donor Hetero-1 (H and I) and Hetero-2 (J). CD107a+ (H) and IFN-γ+ (I) cell numbers are shown in right panels.
Results are presented as mean ± SD from three independent experiments. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, Student's t test.
This hypothesis was further supported when NK cells collected from Hetero-1 and Hetero-2 were co-cultured with Homo-A iPSC-VEs. The same R2 and R3 NK subsets of Hetero-1 were the primary responders against the target cells (Figures 3H and 3I) whereas the NK subsets of Hetero-2 did not respond (Figure 3J), indicating that the NK cells expressing KIR2DL1 in a C1/C2 heterozygote are exclusively activated when they encounter regenerated cells with the C1/C1 genotype. This infers that the results in Figures 2B, 2C, and 2E are based on recognition of missing self and NK cell licensing.
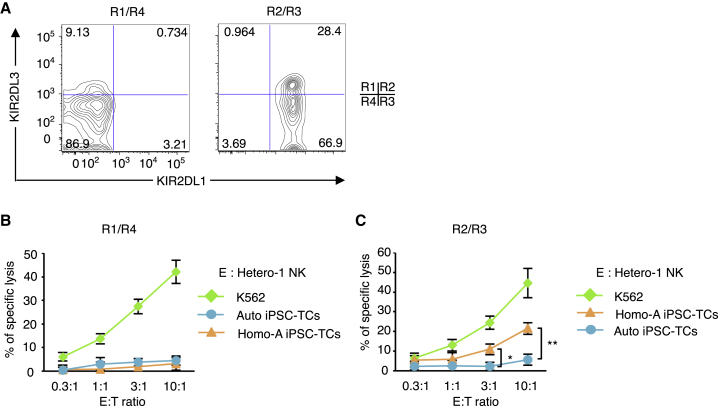
As a confirmation, we performed a cytotoxicity assay using KIR2DL1− cells (R1 + R4 subsets) or KIR2DL1+ cells (R2 + R3 subsets) isolated by magnetic beads (Figure 4A, purity >95%) as effector cells, and Homo-A iPSC-TCs and Auto iPSC-TCs as targets. In agreement with the previous results, the R1 + R4 cells were hyporesponsive to both types of target cells but were able to kill K562 cells (Figure 4B). In contrast, the R2 + R3 cells showed significant cytotoxicity against Homo-A iPSC-TCs but not against Auto iPSC-TCs (Figure 4C). Thus in this reductionist experiment, it is the NK cell subsets expressing inhibitory KIR for the group 2 HLA-C ligand that exert cytotoxicity against the regenerated C1/C1 cells.
Figure 4.
Cytotoxic Responses against Regenerated C1/C1 T Cells Is Limited to the KIR2DL1+ NK Cell Subsets Isolated from a C1/C2 Donor
(A) Flow-cytometric profiles of KIR2DL1 and KIR2DL3 in NK cells isolated from donor Hetero-1 after sorting by magnetic beads. KIR2DL1− cells (R1/R4) were enriched by negative selection (depletion of KIR2DL1+ cells), whereas KIR2DL1+ (R2/R3) cells were enriched by positive selection.
(B and C) Six-hour cytotoxicity assays of NK cells isolated from donor Hetero-1 against Homo-A iPSC-TCs. Results are presented as mean ± SD from three independent experiments. ∗p < 0.05, ∗∗p < 0.01, Student's t test.
Overexpression of a Missing KIR Ligand Enables NK Cell Tolerance against Cells Regenerated from Allogeneic iPSCs
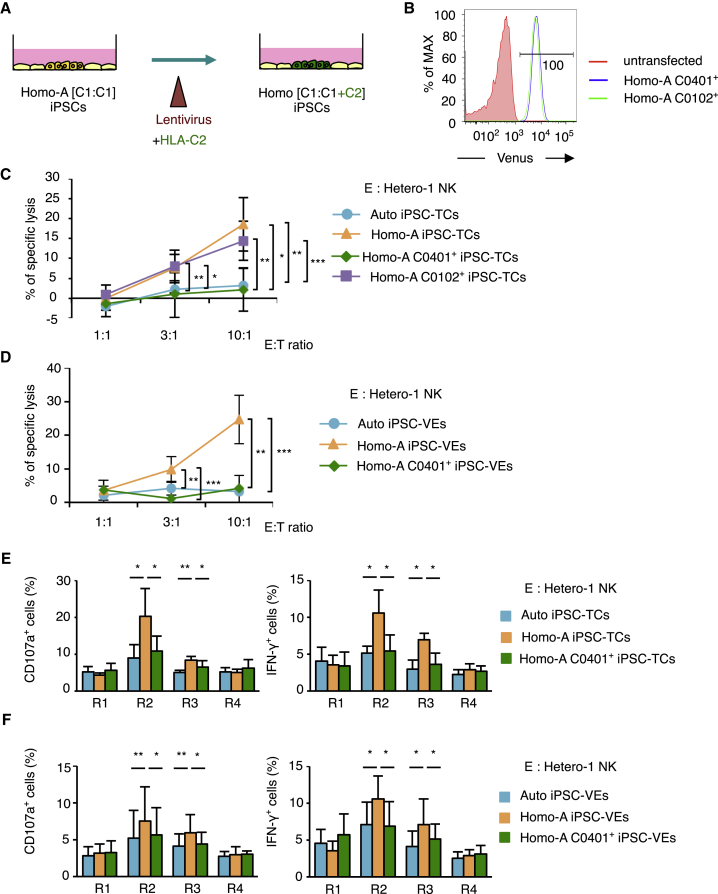
We then proceeded to investigate whether the alloreactivity that could lead to a graft rejection response can be antagonized by overexpression of the C2 ligand on target cells. For this purpose, and to alleviate an alloreactive T cell response at the same time, we transduced Homo-A iPSCs with a group 2 HLA-C allotype that is identical to the recipient.
Thus, we lentivirally transduced Homo-A iPSCs with the HLA-C∗04:01 (“C0401”) gene together with the Venus gene as a reporter (Figure 5A). As a negative control, we transduced Homo-A iPSCs with a group 1 HLA-C allotype, HLA-C∗01:02 (“C0102”). The resulting cells (designated Homo-A C0401+ iPSCs and Homo-A C0102+ iPSCs) were found to express Venus (Figure 5B). We then regenerated T cells from these iPSCs (designated Homo-A C0401+ iPSC-TCs and Homo-A C0102+ iPSC-TCs) and co-cultured them with NK cells isolated from Hetero-1. Here we observed that the ectopic expression of the C2 ligand on Homo-A iPSC-TCs efficiently blocked the cytotoxicity of NK cells, as specific lysis did not increase against Homo-A C0401+ iPSC-TCs (Figure 5C). Furthermore, the cytotoxicity against Homo-A C0102+ iPSC-TCs was at a level comparable with that against Homo-A iPSC-TCs (Figure 5C), indicating that the blockade of cytotoxicity against Homo-A C0401+ iPSC-TCs was unlikely to be simply due to the overexpression of the HLA-C gene. The experiments using regenerated VE cells as target cells resulted in the same observation (Figure 5D).
Figure 5.
Ectopic Expression of the HLA-C2 Molecule in T Cells and VE Cells Regenerated from iPSCs Suppresses NK Cell Alloreactivity
(A) Schematic illustration of the experimental procedure. Homo-A-derived C1/C1 iPSCs were transduced to express group 2 HLA-C, using a lentiviral construct encoding UbC-HLA-C∗04:01-IRES-Venus (Homo-A C0401+ iPSCs). As a control, Homo-A iPSCs transduced with a group 1 HLA-C gene was also produced (Homo-A C0102+ iPSCs). T cells and VE cells were regenerated from the Homo-A C0401+ iPSCs and used as target cells.
(B) Flow-cytometric analysis indicated a homogeneous high expression of Venus in Homo-A C0401+ iPSCs and Homo-A C0102+ iPSCs.
(C and D) Results from 6-hr cytotoxicity assays using NK cells isolated from donor Hetero-1 against T cells (C) and VE cells (D) regenerated from Homo-A C0401+ iPSCs.
(E and F) Twelve-hour co-culture assay by using Homo-A C0401+ iPSC-TCs (E) and -VEs (F) as target cells.
Results are presented as mean ± SD from three independent experiments. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, Student's t test.
The specificity of blockade by HLA-C allotypes was confirmed by comparing the suppression of NK cell subsets defined by the expression of KIR2DL3 versus KIR2DL1. The upregulation of CD107a and increased intracellular IFN-γ production observed in the aforementioned experiments against Homo-A C0401+ iPSC-TCs or Homo-A C0401+ iPSC-VEs was reduced to the level of the autologous control (Figures 5E and 5F). These results are in line with those from a previous study in which NK cells expressing inhibitory KIR for self HLA ligands are silenced by target cells enforced to express corresponding KIR ligands (Yu et al., 2007).
Collectively, these results provide compelling evidence that the cytotoxic activity of NK cells against iPSC-derived regenerated cells depends on recognition of the missing self. Moreover, the method for overexpression of HLA has potential for clinical application to circumvent NK cell-mediated immune responses in KIR-ligand-mismatched transplantation.
NK Cells Respond to VE Cells Regenerated from an iPSC Line Homozygous for the Most Frequent HLA Haplotype in Japanese
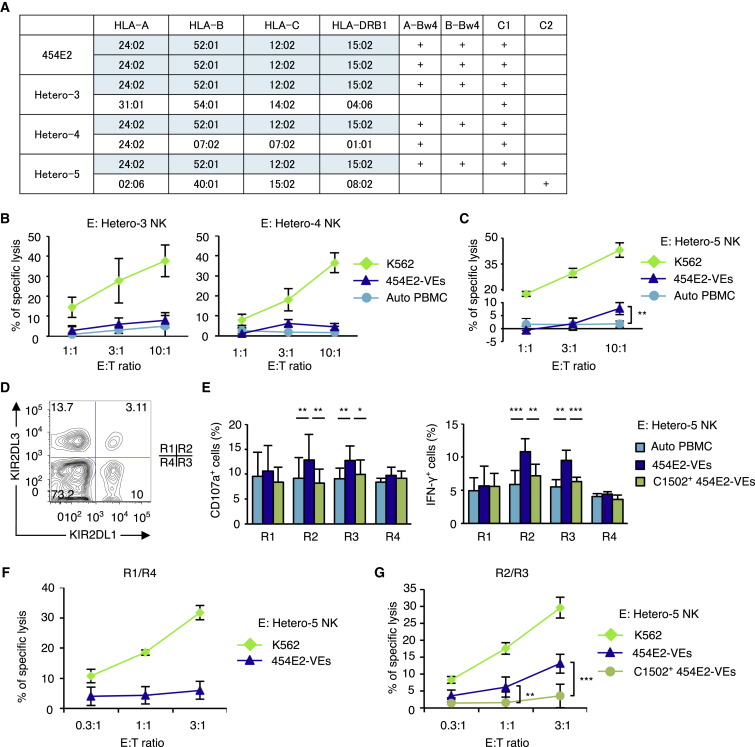
To confirm our hypothesis in other HLA haplotypes, we investigated NK cell alloreactivity in the co-culture assays, using as target cells the iPSC line homozygous for the most frequent HLA haplotype in the Japanese population, namely 454E2 (Figure 6A) (Okita et al., 2011). NK cells were isolated from three healthy volunteers bearing the same HLA haplotype on one chromosome, hereafter designated “Hetero-3,” “Hetero-4,” and “Hetero-5.” Donors Hetero-3 and Hetero-4 were of C1/C1 genotype, while Hetero-5 was of C1/C2 genotype. Donors Hetero-3, Hetero-4, and Hetero-5 were KIR group A homozygotes (Figure S3A).
Figure 6.
KIR-Ligand-Mismatched NK Cells Respond to VE Cells Regenerated from iPSCs Homozygous for the Most Frequent HLA Haplotype in Japanese
(A) HLA haplotypes and KIR ligands of an iPSC line (454E2) and those of three blood donors. The 454E2 line is homozygous for the most frequent HLA haplotype in Japanese (shaded in blue), and the blood donors are all heterozygous for this haplotype. Note that donor Hetero-5 is the only source of NK cells that carries the group 2 HLA-C ligand, so the KIR2DL1+ NK cells are licensed only in this donor within the panel.
(B and C) Eighteen-hour cytotoxicity assay of NK cells collected from (B) Hetero-3 (C1/C1), Hetero-4 (C1/C1), and (C) Hetero-5 (C1/C2).
(D) The R1–R4 subsets within the NK cells isolated from donor Hetero-5, as defined by the heterogeneous expression of KIR2DL1 and KIR2DL3.
(E) Twelve-hour co-culture assay using NK cells isolated from Hetero-5.
(F and G) Eighteen-hour cytotoxicity assays of NK cells isolated from Hetero-5 against 454E2-VEs. The KIR2DL1-positive and -negative NK cell subsets were isolated from Hetero-5 using magnetic beads.
Results are presented as mean ± SD from three independent experiments. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, Student's t test.
Target cells were prepared by regenerating VE cells from 454E2 cells (454E2-VEs). NK cells were isolated from donors Hetero-3 or Hetero-4, and their responses quantified in co-culture assays. As anticipated from the previous experiments, NK cells exhibited little cytotoxic activity against 454E2-VEs (Figure 6B), regardless of whether they were KIR2DL1-positive or -negative (Figures S3B–S3D). Slightly increased levels of specific lysis were observed when NK cells from Hetero-5 were used as effector cells (Figure 6C), likely due to the licensing through the extra KIR ligand (group 2 HLA-C).
We then performed C2-overexpression experiments by transducing 454E2 cells with HLA-C∗15:02 (C1502). Similarly, VE cells expressing C∗15:02 (C1502+ 454E2-VE cells) were regenerated from 454E2, and co-culture assays were conducted using NK cells collected from Hetero-5. NK cell subsets were defined by the expressed combinations of KIR2DL1 and KIR2DL3 (Figure 6D), and responses were compared between subsets to enable assessment of HLA-specific NK response mechanisms. In the positive control (Figure 6E, dark blue bars), the results agreed with those from the previous experiments that indicated cytotoxicity and proinflammatory cytokine responses exclusively in the R2 and R3 NK cells. This exclusive response against 454E2-VEs by the R2 and R3 NK cells from donor Hetero-5 was suppressed when C1502+ 454E2-VEs were used as target cells (Figure 6E, green bars), demonstrating the efficacy of our KIR-ligand-transduction approach in inducing NK cell tolerance against tissue regenerated from iPSCs.
Finally, we enriched KIR2DL1− or KIR2DL1+ NK cells from donor Hetero-5 and used these cells in cytotoxic assays, targeting 454E2-VEs. As anticipated, KIR2DL1− NK cells did not exert specific lysis against 454E2-VEs (Figure 6F). In contrast, KIR2DL1+ NK cells exhibited cytotoxic activity against 454E2-VEs; however, this response was suppressed when C1502+ 454E2-VEs were used as target cells (Figure 6G), indicating that KIR2DL1+ NK cells are exclusively responsible for the observed cytotoxicity of NK cells against 454E2-VEs, and that such killing is based on missing-self recognition.
Discussion
Generating iPSCs for individual persons with various HLA backgrounds is now readily achieved, and it has been assumed that allogeneic immune responses can be circumvented in transplant using autologous iPSC-derived tissue. However, the cost and the long duration for iPSC development are obstacles, and thus the clinical solution is to bank and utilize HLA-homozygous iPSCs for tissue regeneration with the goal of haploidentical allogeneic transplantation.
The present study demonstrates that NK cells can respond to KIR-ligand-mismatched iPSC-derived regenerated cells in vitro, and that the mechanisms are through the missing-self recognition response distinct to NK cells. Therefore, careful consideration should be given to NK cell alloreactivity in the case of KIR-ligand-mismatched transplantations of iPSC-derived cells. This is because transplantation in this context is planned in the near future in Japan, and clinical trials will soon be conducted globally. We further propose a method that has potential to prevent possible rejection responses by NK cells, whereby an HLA class I gene is ectopically expressed in cells regenerated from iPSCs to cancel the missing-self NK cell responses.
In this study, we first approached the question as to whether NK cells have the potential to reject iPSC-derived “hematopoietic” cells. This is of particular relevance in utilizing regenerated T cells in cell therapy (Maeda et al., 2016, Themeli et al., 2013), which will be applied in allogeneic settings in the near future. Through our experiments, it is clear that iPSC-derived T cells are killed by NK cells in a KIR-ligand mismatch situation.
Our second question centered on whether allogeneic iPSC-derived tissues/organs other than hematopoietic cells could be attacked by NK cells. NK cells play a role in graft rejection of solid organs in mouse and human (Harmon et al., 2016, Villard, 2011). In a murine parent-to-F1 hybrid cardiac transplantation model, NK cells exerted graft rejection in collaboration with CD4+ T cells (Uehara et al., 2005). In human, biopsy samples of cardiac allografts showed infiltration of NK cells that was likely involved in allograft rejection (Sorrentino et al., 2006). Some other studies have shown that KIR-ligand mismatch is associated with allograft rejection in renal (van Bergen et al., 2011) and liver transplant (Legaz et al., 2013).
In this context, in the present study we have demonstrated that NK cells can kill iPSC-derived VE cells. This result is important, since VE cells will be initially exposed to NK cells when transplanted. Indeed, it was previously demonstrated in the aforementioned model of murine parent-to-F1 cardiac transplantation that NK cells initially cause vasculopathy in the allograft, which eventually leads to rejection of the graft (Uehara et al., 2005). In regenerative medicine, some studies are trying to produce 3-dimensional structures containing vasculature (Takebe et al., 2013). The potential for NK cell alloreactivity against VE cells would become a serious issue when such approaches are realized.
Another consideration affecting the fate of the iPSC-derived graft is the environment into which the regenerated tissue is transplanted. In transplant of iPSC-derived CMs (Kawamura et al., 2016, Shiba et al., 2016) and retinal pigmentation membrane (RPM) (Sugita et al., 2016) in monkey models, immune reactions were dramatically reduced in the homo-to-hetero setting as compared with major histocompatibility complex total-mismatch cases. These findings formed the basis for the current strategy in regenerative medicine to utilize homo-to-hetero transplantation. However, whereas the regenerated CM transplants required administration of an immune-suppressive agent the regenerated RPM did not, inferring that the microenvironment of the retina, known as one of immune-privileged sites, is more tolerant of allogeneic grafts.
Whereas the aforementioned monkey homo-to-hetero transplantation experiments have provided important information, from the viewpoint of the present study criticism could be raised: KIR-ligand match or mismatch was not taken into consideration in experimental design and interpretation of data in these studies, which could have influenced the outcomes. Further studies are required to assess the involvement of KIR-ligand match/mismatch in animal models.
As a practical clinical question, we estimated the frequency at which KIR-ligand mismatch may occur at the population level, if recipients are selected without taking into consideration the status of KIR-ligand matching, using the data of haplotype frequencies in the Japanese as a test case (Ikeda et al., 2015). Because the allotype frequencies of C1 versus C2 in Japanese is 92.7:7.3, most of the HLA-homo iPSCs (if generated at random) will be of the C1/C1 type. Indeed, among the HLA-homo iPSC lines carrying the frequently observed top-10 HLA haplotypes, only one (no. 6) is of the C2/C2 type (Table S3). In the case of selecting recipients for HLA-homo-C1/C1 iPSC-derived tissues, the frequency of C1/C2 recipients within HLA-hetero recipients is predicted to be 7.3%. On the other hand, when we use HLA-homo-C2/C2 iPSC-derived tissues, KIR-ligand mismatch will occur at 92.7%. Thus, in general the frequency of KIR-ligand mismatch for HLA-C is rather rare in homo-to-hetero transplantations in Japanese persons. In contrast, the allotype frequency of C2 is much higher in other populations, and the chances of a C1 or C2 mismatch are thus considerably higher. For example, in the Polish population the ratio of C1 to C2 is approximately 6:4 (Nowak et al., 2010). If a regenerated iPSC-derived tissue is to be transplanted in this population, the chances of a C1/C1 to C1/C2 mismatch is ∼40%, and a C2/C2 to C1/C2 transplant will occur at ∼60% frequency.
Ligand frequencies of HLA alleles containing the Bw4 motif in Japanese (Tables S2 and S3) reveal a different picture. Among the HLA-A allele types, A∗24:02, the most frequent HLA-A allele in the Japanese population, generally categorized in the A-Bw4 ligand group, was excluded from this consideration, because KIR3DL1 has been shown to bind HLA-A allotypes with the Bw4 motif, but the signaling does not seem to be strong enough to induce licensing of KIR3DL1+ cells (Yawata et al., 2008). HLA-B Bw4 ligand frequency is 31% (Table S4), with the dominant presence of KIR3DL1 (99.2%) (Yawata et al., 2006). Consequently, KIR-ligand mismatch will occur at ∼30.8% frequency when an HLA-homo graft lacking a Bw4 ligand is transplanted into an HLA-hetero recipient. It should be noted that, among the HLA-homo iPSC lines carrying the top-10 frequent HLA haplotypes, seven of them lack the HLA-B Bw4 ligand (nos. 3, 4, 5, 6, 8, 9, and 10) (Table S3) (Ikeda et al., 2015). Therefore, KIR-ligand mismatch regarding the Bw4 motif will take place at considerably high frequency in the iPSC-stock project in the near future. Since ∼40% of HLA haplotypes encode an HLA-B Bw4 ligand in most populations globally (Norman et al., 2007), a KIR3DL1-HLA-Bw4 mismatch will likely occur at similar frequencies elsewhere as well.
The A haplotype is one that carries most of the KIR known to be involved in licensing (KIR2DL1, KIR2DL3, KIR3DL1), and the receptors encoded by this haplotype recognize the full range of KIR ligands (groups 1 and 2 HLA-C, and Bw4). Therefore, donors homozygous for this haplotype are suitable in analyzing the effects of NK cell licensing and recognition of missing self, and elucidating the rejection mechanism of cells regenerated from iPSCs. In this study, the five donors from whom NK cells were derived were all homozygotes for the group A haplotype (Figures 3A and S3A). The Japanese population is highly suitable for the scope of such mechanistic studies, as the group A KIR haplotype is the predominant haplotype in the population (Yawata et al., 2002).
The contributions from activating KIR in rejecting iPSC-derived cells are as yet unclear. In our study, KIR2DS4 was the only “activating KIR gene” present in our NK donors (Hetero-1 to -5). The KIR2DS4 gene has subtypes; KIR2DS4F carries the full length of KIR2DS4 gene, while KIR2DS4D is a variant of the deleted form encoding a soluble (membrane-unbound) form of the molecule (Maxwell et al., 2002). Although subtype variations can be seen in the five NK donors (Figures 3A and S3A), we thought that the contribution of KIR2DS4 in NK response to the regenerated cells is minimal, if any, since NK cells from Hetero-3 and Hetero-4 carrying the KIR2DS4F gene did not create a response while those from Hetero-5 carrying KIR2DS4D did.
In addition, recent studies have suggested that the strength of licensing and inhibitory signaling varies among the alleles of KIR and the HLA allotypes (Yawata et al., 2008). For example, variations in KIR3DL1/HLA-Bw4 subtypes have been shown to affect clinical outcomes, such as in antibody-based immunotherapy against neuroblastoma (Forlenza et al., 2016) or in hematopoietic stem cell transplantation against leukemia (Boudreau et al., 2017). Such subtype differences will also need to be considered in the field of regenerative medicine.
In summary, our present study demonstrates that human NK cells have the potential to reject iPSC-derived tissue in a homo-to-hetero transplantation setting when KIR ligands are mismatched. This has special relevance in the transplantation of tissues derived from HLA-homozygous iPSCs from cell banks. Nonetheless, we feel that transplantation across a KIR-ligand mismatch need not be contraindicated. This is because in homo-to-hetero transplantation cases, immune-suppressive drugs will be more or less administrated to recipients to suppress the inevitable immune reaction caused by minor histocompatibility mismatch. It is likely that such drugs will have the additional effect of suppressing NK cell-mediated responses. However, in KIR-ligand “matched” cases, it may be possible to reduce the use of immune-suppressive agents. This would benefit recipients, since immune-suppressive agents are known to increase the frequency of post-transplant malignancy and infectious diseases. In this context, we can also propose that agents with NK cell-suppressive action, such as mycophenolate mofetil, may be preferable in KIR-ligand-mismatched cases (Harmon et al., 2016, Ohata et al., 2011). Finally, we propose that KIR-ligand matching should be assessed in the coming homo-to-hetero transplantations using HLA-homo iPSCs, and that KIR-ligand-mismatched transplantation cases will require intensified follow-up examinations.
Experimental Procedures
Study Approval
This study was approved by the institutional review board of the Graduate School of Medicine, Kyoto University (approval number: G1020) and abided by the tenets of the Declaration of Helsinki. All specimens from healthy individuals were collected after written informed consent was obtained.
Human Subjects
Blood samples were obtained from healthy, unrelated volunteer donors after obtaining informed consent for sample procurement as approved by Kyoto University Hospital. PBMCs were isolated using Ficoll-Paque PLUS (GE Healthcare). CD8+ T cells were isolated by positive selection using magnetic microbeads (Miltenyi Biotec). NK cells were purified by positive selection of CD56+ cells after negative selection of CD3+, CD4+, CD8+, CD14+, and CD19+ cells with microbeads (Miltenyi Biotec). The post-sort purity of NK cells (CD56+CD3−) was >95% for all experiments.
Plasmid Constructs
cDNA clones of HLA-C∗04:01:01, HLA-C∗15:02:01, and HLA-C∗01:02 cDNA genes were provided by the RIKEN BRC through the National Bio-Resource Project of the MEXT, Japan, cloned by methods described previously (Akatsuka et al., 2002).
HLA Typing and KIR Next-Generation Sequencing Analysis
HLA and KIR typing were performed at the HLA Foundation Laboratory (Kyoto, Japan). HLA typing was performed with PCR-rSSOP using WAKFlow (Wakunaga Pharmaceutical). Next-generation sequencing-based KIR typing used PCR-based amplification of KIR and sequencing using Illumina MiSeq technology as described by Nelson et al. (2015). In brief, the laboratory steps consist of consecutive PCR reactions with barcoding for individual sample tracking. Robust assays for each of the target loci for all KIR loci were developed. The depth of genotyping was extended to all KIR loci to include exons 3, 4, 5, and 9. The analytical tools to define haplotypes and genotypes were developed by Scisco Genetics (Seattle, WA). The HLA-C allele typing results were classified according to the current C1, C2 classification.
Establishment of iPSCs
iPSCs from peripheral blood CD8+ T cells were established by the method of Maeda et al. (2016) with slight modifications (see Supplemental Experimental Procedures).
Lentivirus Preparation and Infection
Each cDNA clone of HLA-C allele was subcloned into the pENTR/D-TOPO vector and further subcloned into an expression vector, CS-UbC-RfA-IRES2-Venus (a gifted from Atsushi Miyawaki, RIKEN) using pENTR Directional TOPO Cloning Kits (Thermo Fisher Scientific). Lentiviruses were collected 48–72 hr after transfection of Lenti-X 293T cells (Clontech) with appropriate amounts of lentiviral vectors, pRSV-Rev, pMDLg/pRRE, and pMD2.G (Addgene) using ViaFect (Promega). iPSCs (5 × 104) were infected by centrifugation and seeded onto mouse embryonic fibroblasts with iPSC medium. Venus-expressing iPSC colonies were picked up manually and maintained in iPSC medium.
Differentiation of iPSCs to CD8 Single-Positive T Cells
The following iPSC clones were differentiated into iPSC-TCs: Homo-A#1 and Hetero-1#1. CD8 single-positive T cells derived from iPSCs were differentiated using the method described by Maeda et al. (2016) with slight modifications (see Supplemental Experimental Procedures).
Differentiation of iPSCs to Vascular Endothelial Cells
The following iPSC clones were differentiated into iPSC-VEs: Homo-A#1, Hetero-1#4, and 454E2. VE cells derived from iPSCs were differentiated using the method of Masumoto et al. (2014) with some modifications (see Supplemental Experimental Procedures).
In Vitro Cytotoxicity Assay
Frozen NK cells were thawed and cultured for 12 hr with RPMI with 10% human serum. NK cells were then used as effector cells for 51Cr release assays against iPSC-TCs and iPSC-VEs. A total of 4,000 51Cr-labeled cells were used as target per well. In some experiments KIR2DL1+ or KIR2DL1− cells were isolated using anti-KIR2DL1 monoclonal antibody conjugated with allophycocyanin (APC) and anti-APC microbeads (Miltenyi Biotec) after thaw, and were cultured for 12 hr. After 6 or 18 hr of co-culture, culture supernatant was applied to Picoplate (PerkinElmer) and analyzed by TopCount NXT (PerkinElmer). The percentage of specific lysis was calculated as follows: specific lysis (%) = (sample lysis with NK [%] − basal lysis without NK [%])/(100 − basal lysis without NK [%]).
CD107a Mobilization and Intracellular Cytokine Assays
Freshly isolated NK cells were co-cultured with target at an 1:1 E/T ratio for 12 hr in the presence of BD GolgiStop (BD Biosciences) and Brefeldin A (Sigma). For measurement of degranulation by NK cells, anti-CD107a was added in a 1:200 dilution to the co-cultures. Cells were permeabilized with a BD Cytofix/Cytoperm Kit (BD Biosciences). Flow cytometry was performed on a FACSCanto II instrument.
Author Contributions
H.I. designed the experiments, performed the major experiments, interpreted the results, and wrote the manuscript. S.N. and T.M. provided technical help with the experiments. M.M. helped in designing the experiments. Y.M., H. Kojima, H.T., and H.S. conducted KIR typing analysis. N.Y., M.Y., H.T., and H.S. gave conceptual advice. N.Y. and M.Y. wrote the manuscript. K.M. and H. Kawamoto conceived and designed the study. H. Kawamoto interpreted the results and wrote the manuscript.
Acknowledgments
We thank Dr. Kouetsu Ogasawara, Nobuyoshi Arima, Keiko Iwaisako, and Masataka Asagiri for helpful discussions, Eri Satoh, Toshika Senba, and all laboratory members for technical support and kind help, Dr. Atsushi Miyawaki for providing plasmid vectors, Dr. Mahito Nakanishi and Manami Ohtaka for providing Sendai virus and Yamanaka factors, RegCell Co., Ltd. and ASTEM for grant funding, and the Kyoto University Radioisotope Research Center for use of facilities.
Published: August 31, 2017
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, three figures, and four tables and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2017.07.020.
Supplemental Information
References
- Akatsuka Y., Goldberg T.A., Kondo E., Martin E.G., Obata Y., Morishima Y., Takahashi T., Hansen J.A. Efficient cloning and expression of HLAclass I cDNA in human B-lymphoblastoid cell lines. Tissue Antigens. 2002;59:502–511. doi: 10.1034/j.1399-0039.2002.590607.x. [DOI] [PubMed] [Google Scholar]
- Anfossi N., André P., Guia S., Falk C.S., Roetynck S., Stewart C.A., Breso V., Frassati C., Reviron D., Middleton D. Human NK cell education by inhibitory receptors for MHC class I. Immunity. 2006;25:331–342. doi: 10.1016/j.immuni.2006.06.013. [DOI] [PubMed] [Google Scholar]
- Boudreau J.E., Giglio F., Gooley T.A. KIR3DL1/HL AB subtypes govern acute myelogenous leukemia relapse after hematopoietic cell transplantation. J. Clin. Oncol. 2017;35:2268–2278. doi: 10.1200/JCO.2016.70.7059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forlenza C.J., Boudreau J.E., Zheng J., Le Luduec J.-B.T., Chamberlain E., Heller G., Cheung N.-K.V., Hsu K.C. KIR3DL1Allelic polymorphism and HLA-B epitopes modulate response to anti-GD2 monoclonal antibody in patients with neuroblastoma. J. Clin. Oncol. 2016;34:2443–2451. doi: 10.1200/JCO.2015.64.9558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenzel L.P., Abdullah Z., Kriegeskorte A.K., Dieterich R., Lange N., Busch D.H., Krönke M., Utermöhlen O., Hescheler J., Šarić T. Role of natural-killer group 2 member D ligands and intercellular adhesion molecule 1 in natural killer cell-mediated lysis of murine embryonic stem cells and embryonic stem cell-derived cardiomyocytes. Stem Cells. 2009;27:307–316. doi: 10.1634/stemcells.2008-0528. [DOI] [PubMed] [Google Scholar]
- Harmon C., Sanchez Fueyo A., O'Farrelly C., Houlihan D.D. Natural killer cells and liver transplantation: orchestrators of rejection or tolerance? Am. J. Transplant. 2016;16:751–757. doi: 10.1111/ajt.13565. [DOI] [PubMed] [Google Scholar]
- Ikeda N., Kojima H., Nishikawa M., Hayashi K., Futagami T., Tsujino T., Kusunoki Y., Fujii N., Suegami S., Miyazaki Y. Determination of HLA-A, -C, -B, -DRB1 allele and haplotype frequency in Japanese population based on family study. Tissue Antigens. 2015;85:252–259. doi: 10.1111/tan.12536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joncker N.T., Raulet D.H. Regulation of NK cell responsiveness to achieve self-tolerance and maximal responses to diseased target cells. Immunol. Rev. 2008;224:85–97. doi: 10.1111/j.1600-065X.2008.00658.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura T., Miyagawa S., Fukushima S., Maeda A., Kashiyama N., Kawamura A., Miki K., Okita K., Yoshida Y., Shiina T. Cardiomyocytes derived from MHC-homozygous induced pluripotent stem cells exhibit reduced allogeneic immunogenicity in MHC-matched non-human primates. Stem Cell Rep. 2016;6:312–320. doi: 10.1016/j.stemcr.2016.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kärre K., Ljunggren H.-G., Piontek G., Kiessling R. Selective rejection of H|-2-deficient lymphoma variants suggests alternative immune defence strategy. Nature. 1986;319:675–678. doi: 10.1038/319675a0. [DOI] [PubMed] [Google Scholar]
- Kim S., Poursine-Laurent J., Truscott S.M., Lybarger L., Song Y.-J., Yang L., French A.R., Sunwoo J.B., Lemieux S., Hansen T.H., Yokoyama W.M. Licensing of natural killer cells by host major histocompatibility complex class I molecules. Nature. 2005;436:709–713. doi: 10.1038/nature03847. [DOI] [PubMed] [Google Scholar]
- Legaz I., López-Álvarez M.R., Campillo J.A., Moya-Quiles M.R., Bolarín J.M., la Peña de J., Salgado G., Gimeno L., García-Alonso A.M., Muro M. KIR gene mismatching and KIR/C ligands in liver transplantation. Transplantation. 2013;95:1037–1044. doi: 10.1097/TP.0b013e318286486c. [DOI] [PubMed] [Google Scholar]
- Maeda T., Nagano S., Ichise H., Kataoka K., Yamada D., Ogawa S., Koseki H., Kitawaki T., Kadowaki N., Takaori-Kondo A. Regeneration of CD8αβ T cells from T-cell-derived iPSC imparts potent tumor antigen-specific cytotoxicity. Cancer Res. 2016;76:6839–6850. doi: 10.1158/0008-5472.CAN-16-1149. [DOI] [PubMed] [Google Scholar]
- Masumoto H., Ikuno T., Takeda M., Fukushima H., Marui A., Katayama S., Shimizu T., Ikeda T., Okano T., Sakata R., Yamashita J.K. Human iPS cell-engineered cardiac tissue sheets with cardiomyocytes and vascular cells for cardiac regeneration. Sci. Rep. 2014;4:6716. doi: 10.1038/srep06716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell L.D., Wallace A., Middleton D., Curran M.D. A common KIR2DS4 deletion variant in the human that predicts a soluble KIR molecule analogous to the KIR1D molecule observed in the rhesus monkey. Tissue Antigens. 2002;60:254–258. doi: 10.1034/j.1399-0039.2002.600307.x. [DOI] [PubMed] [Google Scholar]
- Moffett A., Colucci F. Co-evolution of NK receptors and HLA ligands in humans is driven by reproduction. Immunol. Rev. 2015;267:283–297. doi: 10.1111/imr.12323. [DOI] [PubMed] [Google Scholar]
- Moesta A.K., Parham P. Diverse functionality among human NK cell receptors for the C1 epitope of HLA-C: KIR2DS2, KIR2DL2, and KIR2DL3. Front. Immunol. 2012;3:336. doi: 10.3389/fimmu.2012.00336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K., Nakayama M., Kawano M., Ishii T., Harigae H., Ogasawara K. NK-cell fratricide: dynamic crosstalk between NK and cancer cells. Oncoimmunology. 2013;2:e26529. doi: 10.4161/onci.26529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsuji N., Nakajima F., Tokunaga K. HLA-haplotype banking and iPS cells. Nat. Biotechnol. 2008;26:739–740. doi: 10.1038/nbt0708-739. [DOI] [PubMed] [Google Scholar]
- Nelson W.C., Pyo C.-W., Vogan D., Wang R., Pyon Y.-S., Hennessey C., Smith A., Pereira S., Ishitani A., Geraghty D.E. An integrated genotyping approach for HLA and other complex genetic systems. Hum. Immunol. 2015;76:928–938. doi: 10.1016/j.humimm.2015.05.001. [DOI] [PubMed] [Google Scholar]
- Norman P.J., Abi-Rached L., Gendzekhadze K., Korbel D., Gleimer M., Rowley D., Bruno D., Carrington C.V.F., Chandanayingyong D., Chang Y.-H. Unusual selection on the KIR3DL1/S1 natural killer cell receptor in Africans. Nat. Genet. 2007;39:1092–1099. doi: 10.1038/ng2111. [DOI] [PubMed] [Google Scholar]
- Nowak I., Majorczyk E., Wiśniewski A., Pawlik A., Magott-Procelewska M., Passowicz-Muszyńska E., Malejczyk J., Płoski R., Giebel S., Barcz E. Does the KIR2DS5 gene protect from some human diseases? PLoS One. 2010;5:e12381. doi: 10.1371/journal.pone.0012381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohata K., Espinoza J.L., Lu X., Kondo Y., Nakao S. Mycophenolic acid inhibits natural killer cell proliferation and cytotoxic function: a possible disadvantage of including mycophenolate mofetil in the graft-versus-host disease prophylaxis regimen. Biol. Blood Marrow Transpl. 2011;17:205–213. doi: 10.1016/j.bbmt.2010.08.014. [DOI] [PubMed] [Google Scholar]
- Okita K., Matsumura Y., Sato Y., Okada A., Morizane A., Okamoto S., Hong H., Nakagawa M., Tanabe K., Tezuka K.-I. A more efficient method to generate integration-free human iPS cells. Nat. Meth. 2011;8:409–412. doi: 10.1038/nmeth.1591. [DOI] [PubMed] [Google Scholar]
- Shiba Y., Gomibuchi T., Seto T., Wada Y., Ichimura H., Tanaka Y., Ogasawara T., Okada K., Shiba N., Sakamoto K. Allogeneic transplantation of iPS cell-derived cardiomyocytes regenerates primate hearts. Nature. 2016;538:388–391. doi: 10.1038/nature19815. [DOI] [PubMed] [Google Scholar]
- Sorrentino C., Scarinci A., D'Antuono T., Piccirilli M., Di Nicola M., Pasquale M., Di Iorio C., Di Carlo E. Endomyocardial infiltration by B and NK cells foreshadows the recurrence of cardiac allograft rejection. J. Pathol. 2006;209:400–410. doi: 10.1002/path.1980. [DOI] [PubMed] [Google Scholar]
- Sugita S., Iwasaki Y., Makabe K., Kamao H., Mandai M., Shiina T., Ogasawara K., Hirami Y., Kurimoto Y., Takahashi M. Successful transplantation of retinal pigment epithelial cells from MHC homozygote iPSCs in MHC-matched models. Stem Cell Rep. 2016;7:635–648. doi: 10.1016/j.stemcr.2016.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzue K., Reinherz E.L., Koyasu S. Critical role of NK but not NKT cells in acute rejection of parental bone marrow cells in F1 hybrid mice. Eur. J. Immunol. 2001;31:3147–3152. doi: 10.1002/1521-4141(200111)31:11<3147::aid-immu3147>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Takebe T., Sekine K., Enomura M., Koike H., Kimura M., Ogaeri T., Zhang R.-R., Ueno Y., Zheng Y.-W., Koike N. Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature. 2013;499:481–484. doi: 10.1038/nature12271. [DOI] [PubMed] [Google Scholar]
- Taylor C.J., Peacock S., Chaudhry A.N., Bradley J.A., Bolton E.M. Generating an iPSC bank for HLA-matched tissue transplantation based on known donor and recipient HLA Types. Cell Stem Cell. 2012;11:147–152. doi: 10.1016/j.stem.2012.07.014. [DOI] [PubMed] [Google Scholar]
- Themeli M., Kloss C.C., Ciriello G., Fedorov V.D., Perna F., Gonen M., Sadelain M. Generation of tumor-targeted human T lymphocytes from induced pluripotent stem cells for cancer therapy. Nat. Biotechnol. 2013;31:928–933. doi: 10.1038/nbt.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara S., Chase C.M., Kitchens W.H., Rose H.S., Colvin R.B., Russell P.S., Madsen J.C. NK cells can trigger allograft vasculopathy: the role of hybrid resistance in solid organ allografts. J. Immunol. 2005;175:3424–3430. doi: 10.4049/jimmunol.175.5.3424. [DOI] [PubMed] [Google Scholar]
- van Bergen J., Thompson A., Haasnoot G.W., Roodnat J.I., de Fijter J.W., Claas F.H.J., Koning F., Doxiadis I.I.N. KIR-ligand mismatches are associated with reduced long-term graft survival in HLA-compatible kidney transplantation. Am. J. Transplant. 2011;11:1959–1964. doi: 10.1111/j.1600-6143.2011.03621.x. [DOI] [PubMed] [Google Scholar]
- Velardi A., Ruggeri L., Mancusi A., Aversa F., Christiansen F.T. Natural killer cell allorecognition of missing self in allogeneic hematopoietic transplantation: a tool for immunotherapy of leukemia. Curr. Opin. Immunol. 2009;21:525–530. doi: 10.1016/j.coi.2009.07.015. [DOI] [PubMed] [Google Scholar]
- Villard J. The role of natural killer cells in human solid organ and tissue transplantation. J. Innate Immun. 2011;3:395–402. doi: 10.1159/000324400. [DOI] [PubMed] [Google Scholar]
- Vizcardo R., Masuda K., Yamada D., Ikawa T., Shimizu K., Fujii S.-I., Koseki H., Kawamoto H. Regeneration of human tumor antigen-specific T cells from iPSCs derived from mature CD8+ T cells. Cell Stem Cell. 2013;12:31–36. doi: 10.1016/j.stem.2012.12.006. [DOI] [PubMed] [Google Scholar]
- Yawata M., Yawata N., Draghi M., Little A.-M., Partheniou F., Parham P. Roles for HLA and KIR polymorphisms in natural killer cell repertoire selection and modulation of effector function. J. Exp. Med. 2006;203:633–645. doi: 10.1084/jem.20051884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yawata M., Yawata N., Draghi M., Partheniou F., Little A.-M., Parham P. MHC class I-specific inhibitory receptors and their ligands structure diverse human NK-cell repertoires toward a balance of missing self-response. Blood. 2008;112:2369–2380. doi: 10.1182/blood-2008-03-143727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yawata M., Yawata N., McQueen K.L., Cheng N.W., Guethlein L.A., Rajalingam R., Shilling H.G., Parham P. Predominance of group AKIR haplotypes in Japanese associated with diverse NK cell repertoires of KIR expression. Immunogenetics. 2002;54:543–550. doi: 10.1007/s00251-002-0497-x. [DOI] [PubMed] [Google Scholar]
- Yu J., Heller G., Chewning J., Kim S. Hierarchy of the human natural killer cell response is determined by class and quantity of inhibitory receptors for self-HLA-B and HLA-C ligands. J. Immunol. 2007;179:5977–5989. doi: 10.4049/jimmunol.179.9.5977. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.