Abstract
Protein kinases are an important class of enzymes and drug targets. New opportunities to discover medicines for neglected diseases can be leveraged by the extensive kinase tools and knowledge created in targeting human kinases. A valuable tool for kinase drug discovery is an enzyme assay that measures catalytic function. The functional assay can be used to identify inhibitors, estimate affinity, characterize molecular mechanisms of action (MMOAs) and evaluate selectivity. However, establishing an enzyme assay for a new kinases requires identification of a suitable substrate. Identification of a new kinase’s endogenous physiologic substrate and function can be extremely costly and time consuming. Fortunately, most kinases are promiscuous and will catalyze the phosphotransfer from ATP to alternative substrates with differing degrees of catalytic efficiency. In this manuscript we review strategies and successes in the identification of alternative substrates for kinases from organisms responsible for many of the neglected tropical diseases (NTDs) towards the goal of informing strategies to identify substrates for new kinases. Approaches for establishing a functional kinase assay include measuring auto-activation and use of generic substrates and peptides. The most commonly used generic substrates are casein, myelin basic protein, and histone. Sequence homology modeling can provide insights into the potential substrates and the requirement for activation. Empirical approaches that can identify substrates include screening of lysates (which may also help identify native substrates) and use of peptide arrays. All of these approaches have been used with a varying degree of success to identify alternative substrates.
Keywords: Alternative substrate, antiparasitic, assay development, neglected tropical disease, orphan kinase, protein kinase
INTRODUCTION
Protein kinases (PKs) catalyze the phosphorylation of proteins at serine, threonine and tyrosine residues. This post-translation modification is important to most physiological processes; there are hundreds of different protein kinases that catalyze specific phosphorylation reactions. Kinases are important targets for drug discovery with now over 30 medicines approved that inhibit protein kinases, primarily for cancer [1]. There is interest to exploit the success of targeting kinases for non-infectious diseases [2–5], as well as neglected tropical diseases (NTDs). The druggability of the kinomes of Plasmodia [6, 7], Kinetoplastids [8], and bacteria [9] have been reviewed elsewhere, and there are opportunities for kinases as drug targets in other protozoa [10], helminths [11] and ectoparasites [12, 13].
PKs are a large and pervasive group of enzymes. Over 1 % of all genes encode for kinases, and they can be grouped into families and subfamilies by their deduced primary structure. PKs which do not fall into these major groups are called “other protein kinases” (OPK) [14]. OPKs can have orthologues and consequent predictive behavior based on homology to known kinases, or they can have no known orthologs (often called ‘orphan protein kinases’).
Many essential kinases have been identified in infectious organisms with genetic techniques (e.g. RNAi knockdown), of which some have been pursued as drug targets. For example, the sequenced genomes of three human-infective trypanosomatid protozoa, Leishmania major, Trypanosoma brucei and Trypanosoma cruzi, have allowed the kinome for each parasite to be defined as 179, 156 and 171 eukaryotic protein kinases respectively [15]. In T. brucei, the causative agent of human African trypanosomiasis (also known as African sleeping sickness), RNAi screens have identified over 100 of these kinases to be essential [16, 17], of which only a few have reported activity assays and screening for inhibitors. Deduced from the genome, T. brucei has over 30 orphan kinases displaying no homology to known kinases [16, 17]. The malarial parasite, Plasmodium falciparum, has over 80 kinases with many members lacking clear orthologues in the human kinome and many having been shown to be essential by reverse genetic techniques [18]. Further, many kinases from P. falciparum exhibit a high degree of structural divergence from their host counterparts. A number of Plasmodium kinases have recently been shown by reverse genetics to be essential for various parts of the complex parasitic life cycle, and are thus genetically validated as potential targets [6, 19, 20]. There are over 70 members in the Cryptosporidium parvum kinome of which only a few have been investigated; 35 % of these are classified as other protein kinases, and 25 % of these have no known orthologues outside of Cryptosporidium [21]. The Apicomplexans, including Plasmodium, Toxo- plasma and Cryptosporidium, have two groups of kinases, the rhoptry kinases and FIKK kinases, unique to the clade [21]. Kinome of Entamoeba histolytica is predicted to have over 300 members, 112 of which are unclassified and 38 of which are classified as ‘other’ protein kinases [22].
A valuable tool for targeting kinases for drug discovery are enzyme assays. These can complement other approaches such as binding assays and cellular assays to provide new knowledge. They can be used for screening of compounds to identify new inhibitors and for evaluating specificity. Methodologies to determine physiological substrates have been developed, but can be difficult and expensive [23–25]. Developing assays for parasite kinases can identify new selective inhibitors may be useful probes to help determine the physiological function of the kinase [23] as well as valuable starting points for drug discovery.
The chemistries for the phosphotransfer reactions catalyzed by protein kinases are similar, magnesium/ATP transfers the γ-phosphate group to an activated hydroxyl group (Ser, Thr or Tyr) on the acceptor protein substrate through an SN2 displacement reaction. Specificity for the reaction is provided by the interactions between the kinase and the protein substrate. The similarity of the catalytic chemistries for all kinases has provided an approach that uses non-specific substrates to mimic the activity of the physiologic substrates in the phosphotransfer reactions. While the use of non-specific substrates provides little information as to the biological function of a kinase, they enable enzyme assays to be developed that identify specific inhibitors of the kinases. Subsequently, the inhibitors can be used as chemical biology tools to help reveal the physiological function of the kinase. In this paper we review the approaches and methods used to identify non-specific substrates with a focus to identify inhibitors of non-human kinases in organisms that cause neglected tropical diseases.
AUTOPHOSPHORYLATION
Autophosphorylation is the kinase-catalyzed phosphotransfer to a Ser, Thr or Tyr residue of the kinase itself. Many kinases require autophosphorylation for full or increased catalytic activity for phosphotransfer to other proteins. Autophosphorylation occurs on a kinase’s activation loop, in either a trans or cis (catalyzing phosphotransfer to another kinase molecule or to itself) fashion [26], and an autophosphorylated kinase may undergo a conformational change which better suits the kinase to accept exogenous substrates [27].
Several serine-threonine kinases have been screened using an autokinase assay. For example, a MAP kinase from Toxoplasma gondii, for which endogenous nor exogenous substrates have been identified, was recently shown to be inhibited SB505124, an inhibitor of transforming growth factor β type I receptors ALK4, ALK5 and ALK7 using the autokinase assay. Immunoprecipitated MAPK1 from T. gondii lysates was treated with SB505124. The inhibition of 32P incorporation into TgMAPK1 in lysates (from [γ-32P]ATP) by SB505124 revealed an apparent IC50 of 125 nM [28].
Another example of autokinase assay is with the C2-domain-containing protein kinase (C2PK) immunoprecipitated from E. histolytica. Activity was measured via radiolabeled phosphotransfer ([γ-32P]ATP [2.5 μM] for 1 h) to trans-autophosphorylate at Ser428. Biochemical analysis revealed a Vmax of 66 nmol/min/mg. Autokinase inhibition assays identified staurosporine as an inhibitor of C2PK from E. histolytica, with an IC50 of 150 nM [29].
Autophosphorylation assays generally require more enzyme, as the enzyme itself is also its substrate. Consequently, these assays can be costly. Nevertheless, it is important to consider autophosphorylation when developing new assays, as many kinases require autophosphorylation for full catalytic activity with exogenous and endogenous protein substrates. For instance, calcium-dependent protein kinase (CDPK) 3 from T. gondii required preincubation with ATP and CaCl2 to auto-activate, prior to activity measurements with exogenous substrate Syntide-2 and [β-32P] ATP. Purfalcamine, a 2,6,9-trisubstituted purine with antiplasmodial activity and inhibitory activity on a CDPK from T. gondii, was shown to inhibit CDPK3 (IC50 = 800 nM, [CDPK3] = 100 nM) activity.
Other recent examples of Ser/Thr protein kinases in which autophosphorylation assays are where used to test inhibitors include PK7 from P. falciparum in testing pyrazolopyrimidine compounds [30].
Putative tyrosine kinases from parasites have been shown to exhibit autokinase activity. A Wee1-like kinase from T. brucei was recently expressed, but no protein substrate has been identified. Assays with generic protein and peptide substrates were not phosphorylated, however [32P] was incorporated in the recombinant protein, and antiphosphotyrosine antibodies indicated that the phosphorylation site was a Tyr residue [31]. Tyrosine-kinase-like kinase from P. falciparum [32] and dual-specificity (Ser/Thr and Tyr) casein kinase from P. falciparum [33] have been shown to exhibit autophosphorylation, and it is believed that these activities regulate the kinases’ activity to other protein substrates.
GENERIC SUBSTRATES
Generic alternative kinase substrates are used to test activity in many instances where endogenous substrates are not known [34]. Isoforms of casein (α and β) were used as substrates as early as 1954 [35]. Mixtures of up to four isoforms of casein have been used as kinase substrates. Dephosphorylated casein also can serve as a kinase substrate, particularly with new kinases which align with casein kinases (see next section for homology strategies in substrate selection). Dephosphorylated casein is commercially available, or it can be prepared by phosphatase treatment of casein prior to incubation [36]. Other successful generic substrates for kinase assays include myelin basic protein (MBP) and isoforms of histone, as well as synthetic peptides like kemptide (sequence =LRRASLG), Syntide-2 (sequence =PLARTLSVAGLPGKK) [34], Crosstide (sequence =GRPRTSSFAEG) and CREBtide (sequence =KRREILSRRPSYR) [37].
In the next section, we review in more detail kinase family-specific successes in substrate selection for new kinases which align with known kinases. Several kinase families accept generic protein or peptide substrates. For example, there has been success with MBP as a substrate for new kinases which align with mitogen-activated protein kinases (MAPKs). Recent successes include MAPKs from Apicomplexan T. gondii [38], Amoebae E. histolytica [39], Kinetoplastids of Leishania spp. [40–44], T. cruzi [45] and T. brucei [Swinney DC, unpublished results], and helminths B. malayi [46] and E. multilocularis [47, 48]. Ashutosh and coworkers recently reported the specific activity of recombinant MAPK1 from Leishmania donovani to be 11.66 nmol ATP consumed/min/mg protein [43]. Likewise, new kinases aligning with kinases upstream in the MAPK signaling pathway from Apocomplexans (T. annulata) [49], Kinetoplastids (T. brucei) [Swinney DC, unpublished results] and Leishmania mexicana [50] and Schistosoma mansoni [51] can accept MBP as a protein substrate.
MBP has been used to show phosphotransfer activity in eukaryotic-like protein kinases (EPKs) from various bacteria [52–58]. Kimura and coworkers recently reported the cloning of 14 EPKs from Myxococcus xanthus. The selected EPKs had atypical motifs in their catalytic loops but contained all residues necessary for catalytic activity. Seven of the 14 showed activity in phosphorylating MBP, and four of those also showed autophosphorylation activity, with specific activities (200 μM ATP) ranging 1–13100 and 1–10 pmol/min/mg, respectively [56].
Specificity has been demonstrated with generic substrates. For instance, FIKK kinase (named for its conserved FIKK-motif in the Apicomplexan-specific group of kinases) from Plasmodium falciparum showed activity with human histone H1 and Xenopus laevis histone H3, but was not active with X. laevis histone H2 [59]. Two recently expressed NIMA-related serine/threonine kinases (Nek) from Giardia lamblia have been shown to phosphorylate recombinant human histone H1, while only one of the two Neks phosphorylated human histone H3 [60]. There are also examples of Nek accepting MBP instead of histone [61, 62].
Similarly, cdc2-related kinases (CRKs), a group of cyclin-dependent kinases, from protozoan parasites have catalytic activity using histone H1 [63–67]. Histone was incubated at 0.167 – 0.333 mg/mL [64–67]. A second isoform of CRK from E. tenella was recently cloned, and the recombinant enzyme was incubated with a fluorescent-tagged histone H1-derived peptide. Engels et al. validated EtCRK2 as a drug target using the cyclin-dependent kinase-specific inhibitor flavopiridol, (IC50 of 33 nM and Ki of 11 nM) which was similar to the human isoform (IC50 36 nM, Ki 19 nM). They also identified four chemically diverse compounds with Ki values in the low micromolar range [68].
Other generic peptide substrates were designed for specific kinase families. Kemptide (sequence = LRRASLG) is a synthetic oligomer derived from porcine liver pyruvate kinase which acts as a productive substrate for cyclic nucleoside mono- phosphate-dependent kinases (cyclic adenosine monophosphate (cAMP) -dependent kinases (PKAs) and cyclic guanosine monophosphate (cGMP) -dependent kinases (PKGs)) [69]. Another common peptide substrate for PKAs and PKGs is GRTGRRNSI, derived from PKI, the heat stable inhibitor protein of PKA [70]. New kinases, from protozoan parasites, which align with PKAs or PKGs have shown activity with Kemptide [71-77]. PKGs from apicomplexans were recently reported to have an apparent Km 28 μM for Kemptide (T. gondii PKG) [71] and 19 μM for biotinylated GRTGRRNSI (E. tenella PKG) [72]. Likewise, Crosstide [37] and Syntide-2 [78] were designed as generic substrates for protein kinase B (PKB) and calcium-dependent protein kinase (CDPK), respectively, and recently shown to be substrates for PKB [79] or CDPK [21, 80-83]. Typical peptide concentrations in kinase assays are about 200–400 μM for Kemptide [71–75] and 100–150 μM for Syntide-2 [82, 83]. Recently, four isoforms of CDPK from C. parvum were characterized (apparent Km values for Syntide-2 ranging from 156 μM to 426 μM and kcats ranging from 12 min-1 to 1370 min−1). Syntide-2 (500 μM) was used for an inhibitor screen [21]. For a recent inhibitor screen of CDPK3 from T. gondii, 1 mM Syntide-2 was chosen as a substrate concentration [81]. Biotinylated Syntide-2 was used in enzymatic assays of CDPK1 from Neospora caninum to discovery of seven inhibitors with IC50s in the low nanomolar range and one inhibitor with an IC50 of 513 nM [80].
Table 1 summarizes active generic substrates by kinase-type discussed in the text. Table 2 describes some recent successes with generic substrates in new kinases from protozoan parasites.
Table 1.
Canonical protein and peptide substrates for various types of kinases.
| Kinase Type | Kinase Abbreviation | Canonical Substrate |
|---|---|---|
| mitogen-activated protein kinase | MAPK | MBP |
| mitogen-activated protein kinase kinase | MAPKK | MBP |
| FIKK kinase | FIKK | Histone |
| NIMA-related kinase | Nek | MBP, Histone |
| cyclin-dependent kinase-related kinase | CRK | Histone |
| cAMP-dependent kinase | PKA | Kemptide |
| cGMP-dependent kinase | PKG | Kemptide |
| protein kinase B | PKB/AKT | Crosstide |
| calcium-dependent protien kinase | CDPK | Syntide-2 |
| glycogen synthase kinase | GSK | GSM, GSP-2, GS-1 |
| casein kinase 1 | CK1 | Casein, CKPep1, see text for others |
| casein kinase 2 | CK2 | Casein, CKPep2, see text for others |
Table 2.
Recent applications of generic substrates for assays of new protein kinases from parasitic protozoa.
| Organism | Kinase Assayeda | Successful Common Substratesb,c | Year | Reference |
|---|---|---|---|---|
| Plasmodium falciparum | CDPK4 | MBP, Syntide-2 | 2009 | [82] |
| Plasmodium falciparum | PK7 | MBP | 2005 | [84] |
| Plasmodium falciparum | eIK1 | α-cas., β-cas. | 2009 | [85] |
| Plasmodium falciparum | FIKK4.2 | MBP | 2014 | [86] |
| Plasmodium falciparum | FIKK8 | cas., MBP, Syntide-2 | 2015 | [87] |
| Plasmodium falciparum | SRPK1 | RSpep | 2010 | [88] |
| Plasmodium falciparum | CLK-1, CLK-2 | α/β cas., his. H1, MBP | 2011 | [89] |
| Plasmodium falciparum | CLK-1, CLK-2, CLK-3, CLK-4 | α/β cas., his. H1, MBP | 2014 | [90] |
| Plasmodium falciparum | PKB | Crosstide, his. IIAS | 2006 | [79] |
| Plasmodium falciparum | PKG | GRTGRRNSI, Kemptide, MCPD-4d, Glasstide | 2006 | [74] |
| Plasmodium falciparum | TKL2 | α-cas., β-cas., his. H1, MBP | 2013 | [32] |
| Plasmodium falciparum | TKL3 | α-cas., β-cas., his. H1, MBP | 2010 | [91] |
| Plasmodium falciparum | CDPK1 | β-cas., dephosphorylated cas., his. H1, his. H2, his. H3 | 2009 | [92] |
| Toxoplasma gondii | CDPKif3 | his. IIAS | 2009 | [93] |
| Toxoplasma gondii | FIKK | cas., MBP, Syntide-2 | 2015 | [87] |
| Giardia lamblia | Nek1, Nek2 | BSA, his. H1, his. H3 | 2012 | [60] |
| Giardia duodenalis | PKC | his. H1-IIIs | 2007 | [94] |
| Leishmania major | MPK7 | MBP | 2014 | [95] |
| Leishmania mexicana | PK4 | MBP | 2005 | [96] |
| Leishmania spp. | MPK10 | dephosphorylated cas., his. H1, MBP, Ets1 | 2014 | [42] |
| Trypanosoma brucei | TLK | cas., | unpublished results | |
| Trypanosoma brucei | AUK1 | MBP | 2013 | [97] |
| Trypanosoma.brucei | PK50, PK53 | α-cas., β-cas., his. H1, MBP | 2010 | [98] |
| Entamoeba histolytica | AK1 | actin, his. IIIs | 2014 | [99] |
Kinase abbreviations are taken from respective references
BSA, bovine serum albumin; cas., casein; his., histone; MBP, myelin basic protein; RSpep, Arginine-Serine repeat peptide (sequence = RSPSYGRSRSRSRSRSRSRSRSNSRSRSY)
casein and histone nomenclature are taken from respective references
MCPD-4 sequence = GKKRKRSRKES
HOMOLOGY AND KNOWN FUNCTIONAL SUBSTRATES OF HOMOLOGUES
Kinase sequence alignments and homology have been used to map conserved catalytic domains and deduce phylogeny across classes of protein kinases [14, 27]. While all known typical and atypical protein kinases have structural similarities [100], primary sequences of catalytic domains in phylogenetic clusters correlate to the biochemical properties of the protein kinase, including substrate specificity [14, 27]. Feature of substrate specificity have been deduced from these alignments. For example, Li and coworkers have described putative specificity-determining residues and suggested a role for hydrophobicity in substrate specificity [101].
New kinases which align with the casein kinase (CK) family often are catalytically active, as mentioned above, with casein isoforms and dephosphorylated casein [36, 42, 102-112]. In addition CK-specific substrates have been reported. For instance, CK substrate CK-S (sequence = (RRKHAAIG(pS)AYSITA, where pS corresponds to phosphorylated serine) is a substrate for CK2 in L. donovani, [102]. Pep1 (sequence = RRKDLHDDEEDEAMSITA) and Pep2 (sequence = RRRADDSDDDDD) are selective substrates for CK1 and CK2, respectively, and not only have been used as substrates for new kinase activity, but have also been used to sub-classify CKs identified from parasites kinetoplastids [104-109, 111–113] and apicomplexans [33, 110, 114]. DDDEESITRR and KRRRAL(pS)VASLPGL [110] have also been used as CK1-specific peptide substrates, and RRREEE TEEE [110], RRREDEESDDEE, eIF2β-derived peptide MSGDEMIFDPTMSKKKKKKKKP [114] and RRASADDSDDEDL [115] have also been used as a CK2-specific peptide substrates. Typical Pep1 concentrations for activity assays are 100 – 800 μM [104-108, 113], while Pep2 concentrations usually range 40 -200 μM [33, 104–109, 112, 113]. Recombinant CK1.1 from T. cruzi had apparent Km for β-casein of 5.7 mg/mL and for Pep1 of 128 μM [108]. Recombinant CK1β from T. gondii had an apparent Km for partially dephosphorylated α-casein of 5 μM, for β-casein of 12 μM, for Pep1 of 15 μM and for phosphopeptide substrate KRRRAL(pS)VASLPGL of 79 μM [110]. CK2α from P. falciparum has an apparent Km for Pep2 of 137.5 μM [114].
Sequence alignment has been successfully employed for functional substrate discovery with homologues of kinases which accept eukaryotic translation initiation factor 2α and its orthologues as substrates. eIF2α kinases from Apicomplexa have been shown to phosphorylate generic substrates such as α- and β-casein [85], but also putative eIF2α isoforms from the parasite itself [85, 116] or from yeast [116, 117]. eIF2α from Kinetoplastids are phosphorylated at Thr169, as opposed to other eukaryotic eIF2α at Ser51; recombinant putative eIF2αs have been successfully used as in vitro substrates for their kinases in T. brucei [118], T. cruzi [119], and Leishmania infantum [120].
Homology of glycogen synthase kinase-3 (GSK3), showcased in Ref. [121], has informed assay development in homologues from parasites. GSK3 has several synthetic exogenous peptides which served as functional substrates, including CREBtide [122], GS-1 (sequence = YRRAAVPPS PSLSRHSSPHQ(pS)EDEEE) [123, 124], GSP-2 (sequence = YRRAAVPPSPSLSRHSSPHQ(pS) EDEEE) [124], and GSM (sequence = RRRPAS VPPSPSLSRHS(pS)HQRR) [125]. GS-1 has been used with recombinant homologues from P. falciparum [126] and L. donovani [127]. GSM was used as the substrate for the M. tuberculosis homologue [128]. In T. brucei, homologues of GSK3β have been assayed with GSP-2 [129–131] and GSM [132]. Biotinylated GSP-2 and untagged GSP-2 had apparent Km values of 2.4 μM [129] and 8.4 μM [131], respectively, and untagged GSM had an apparent Km of 23 μM [132].
LYSATES
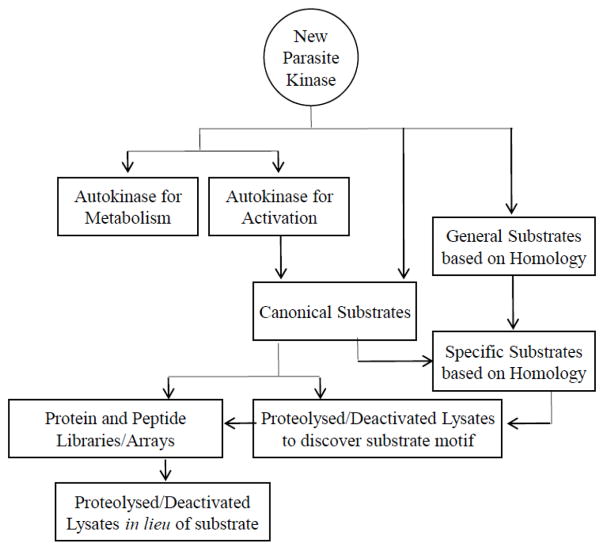
In the absence of a known peptide or protein substrate, and when generic substrate-strategies fail, lysates of the kinase’s organism can be used in lieu of substrate (Scheme 1). Cell lysates contain the native endogenous substrate, and have been used in kinase assays in lieu of purified active substrate [25]. It is worth noting that other kinases in the lysate must be inactivated. Treatments to inactivate the kinases from lysate include adding kinase inhibitors such as 5’-4-fluorosulphonylbenzoyladenosine [25], proteolytic digestion prior to kinase addition to the reaction [133, 134], and heat or acid denaturation. With some reaction mixtures, it may be necessary to treat trypic digests with phosphatases to dephosphorylate potential reaction sites from peptides, followed by heat to inactivate the activity of the phosphatase [134]. The use of lysates also provide an opportunity to identify physiological relevant substrates [25]. This can be an added value to justify pursuing this approach.
Scheme 1.
ROP18, a rhoptry kinase of T. gondii, has shown phosphorylation activity to both parasite cultures and human foreskin fibroblasts that had been heated for 30 min at 56 °C in order to inactivate endogenous kinases [135].
PEPTIDE LIBRARIES AND MICROARRAYS
For new kinases where orthologues are not known, in the absence of autokinase activity and activity from generic substrates, libraries of proteins and peptides can be screened for substrate specificity (Scheme 1). Commercial kits of randomly generated protein and peptide libraries, as well as kinase family-specific libraries are available [136]. Further, commercial microarrays, or chips, have been developed for substrate screening of kinases, which contain libraries of proteins or peptides on solid support. These microarrays often have their own assay protocol and detection and analysis methods [137] can be used with kinase specificity results to determine amino acid motifs preferentially accepted by a new kinase [136].
Libraries and library arrays are available in many different types, and often they are clustered by knowledge-based or overall random library sets. The random sets consist of arbitrarily generated peptides based on combinatorial methods, while knowledge-based libraries consist of peptides derived from naturally occurring proteins [136]. Though costly, new methods of peptide synthesis and array technology [136, 138] make these libraries more comprehensive. This method is particularly convenient for orphan kinases which do not have known orthologues.
ROP18 phosphorylates immunity-related p47 GTPases from the mammalian host, but the full function and substrate specificity of ROP18 are not fully understood to date. Lim and coworkers reported the cloning of the catalytic domain of ROP18 and, the use of a positional scanning peptide library array. The investigators used 50 μM cold ATP and approximately 1 μCi [γ-32P]-ATP in 8 h reactions and quantified the degree of phosphorylation in each library. They found TgROP18 to have a low peptide sequence selectivity, with a generalized motif of (X)-(X, not E)-(X)-(E)-(H)-(T)-(R/mixed, not P and not negatively charged)-(Ar)-(Ar)-(Ar), where X signifies any amino acid, the phosphoreceptor is underlined, and Ar denotes an aromatic amino acid, and they speculate the broad range of phosphoreceptor sequences suggests other specificity factors may exist to selectively allow TgROP18 to target its physiological substrates [139].
PfPK7, an atypical orphan plasmodial protein kinase, has been reported to autophosphorylate and phosphorylate the generic substrates MBP, histone H2A, and β-casein [84]. Peptide microarrays were used to further explore the substrate specificity of PfPK7 with peptide microarrays [30]. PfPK7 is one of many P. falciparum protein kinases which contains extensive insertions, reducing its identity to known kinases. Consequently, PfPK7 has no human orthologue, and is considered an orphan kinase. The two closest human homologues share 33 % and 26 % identity with PfPK7, and PfPK7 lacks activity related to these homologues. Using an arrayed peptide library, the sequence consensus for a phosphoreceptor of PfPK7 was found to be (r-R-R/K-K/R-S/T-P-K/R-K-R). The array-derived peptide specificity included a strong preference for positively charged amino acids lying 2, 3, and 4 residues on both sides of the Ser/Thr to be phosphorylated, in agreement with their structural observation of overall negative character in the active site cleft of PfPK7 [30].
Calcium-dependent kinase 1 (CDPK1) from T. gondii is a member of a calcium-dependent signaling pathway that is necessary to the survival of the parasite. This signaling pathway is poorly understood and its kinases are not fully characterized. The substrate specificity of TgCDPK1 was further tested by positional-scanning peptide array. Lourido and coworkers found a strong preference for Ser over Thr, for Arg at the -3 position to the residue of phosphotransfer, and hydrophobic residues in the -5 position. The peptide-array-generated consensus was in agreement with the peptide motifs derived from sulfur-covalent linkages to T. gondii proteins in incubations of the lysate with a thio-ATP analog [140].
CRKs typically phosphorylate histone H1 in the presence of cyclins. Walker and coworkers recently reported leishmanial CRK3 was tested with histone H1. A substrate finder assay which contained a library of 61 potential serine/threonine phosphoreceptors was used to find a fluorescently labeled peptide for an inhibitor screen. Of the 61 peptides, 5 were sufficiently phosphorylated by leishmanial CRK3, including two histone H1-derived peptides, and a motif of x-S/T-P-x-R/K was common to all of them. GGGRSPGRRRRK was used for inhibitor screens with the CRK3:CYC6 kinase complex. The assay with the library-derived peptide had a Z’ score of 0.71, and potent inhibitors were identified for leishmanial CRK3 from two compound libraries [65].
FIKKs from P. falciparum and C. parvum were shown to have activity on canonical substrates (cf. Table 1), and their substrate specificities were further explored by means of a positional-screening peptide array. Osman and coworkers found a strong preference for basic residues, mainly arginine at positions three residues on either side of the peptide phosphorylation site, and a weaker preference for arginine at the -4 position. Knowing these preferences, an optimized substrate of sequence RRRAPSFYRK and three variants were tested, and the two isoforms had apparent kcats of about 20 min-1 to 30 min−1 (P. falciparum) or 70 min−1 to 120 min−1 (C. parvum) and Kms ranging from 7 μM to 150 μM [87].
CONCLUSION
Kinase enzyme assays are a powerful tool to help identify new inhibitors. The inhibitors can be used as tools for chemical biology to learn about the physiological function of the kinases as well as starting points for drug discovery optimization. Enzyme assays measure function which can complement other types of assays such as binding assays. However, the challenge is to identify substrates for new kinases with unknown function. In this manuscript we review different approaches to identify substrates for kinases with respect to developing assays suitable to screen for inhibitors.
There is no consensus way to identify the substrate. Methods including auto-activation and use of generic substrates and peptides are the most straight-forward. However, these may not provide robust activity, and, when that is the case, more significant quantities of protein will be required for the assays and screens. A concern with any of the assays is the potential need for the kinases to be activated. Sequence homology modeling can provide insights into the potential substrates and the requirement for activation. More empirical approaches that can provide substrates include screening lysates (which may also help identify native substrates) and peptide arrays. These later approaches are less biased but also more costly.
Our recommendation is to begin with the simplest approaches (auto-activation and generic substrates) and then work down the flow-chart shown in Scheme 1. This strategy has been used by numerous investigators to identify substrates for new kinases.
Acknowledgments
This work was funded by a grant from NIAID RO1AI103476.
LIST OF ABBREVIATIONS
- cAMP
Cyclic adenosine monophosphate
- cGMP
Cyclic guanosine monophosphate
- CRK
Cyclin-dependent kinase-related kinase
- eIF2α
Eukaryotic initiation factor 2α
- EPK
Eukaryotic-like protein kinase
- MAPK
Mitogen-activated protein kinase
- MBP
Myelin basic protein
- Nek
NIMA-related serine/threonine kinase
- PKA
cAMP-dependent kinase
- PKG
cGMP-dependent kinase
Footnotes
Send Orders for Reprints to reprints@benthamscience.ae
CONFLICT OF INTEREST
The authors confirm that this article content has no conflict of interest.
References
- 1.Fabbro D, Cowan-Jacob SW, Moebitz H. Ten things you should know about protein kinases: IUPHAR Review 14. Br J Pharmacol. 2015;172(11):2675–700. doi: 10.1111/bph.13096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sawyer TK, Wu JC, Sawyer JR, English JM. Protein kinase inhibitors: breakthrough medicines and the next generation. Expert Opin Investig Drugs. 2013;22(6):675–8. doi: 10.1517/13543784.2013.804509. [DOI] [PubMed] [Google Scholar]
- 3.Simmons DL. Targeting kinases: a new approach to treating inflammatory rheumatic diseases. Curr Opin Pharmacol. 2013;13(3):426–34. doi: 10.1016/j.coph.2013.02.008. [DOI] [PubMed] [Google Scholar]
- 4.Beurel E, Grieco SF, Jope RS. Glycogen synthase kinase-3 (GSK3): regulation, actions, and diseases. Pharmacol Ther. 2015;148:114–31. doi: 10.1016/j.pharmthera.2014.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaeberlein M. mTOR Inhibition: From Aging to Autism and Beyond. Scientifica (Cairo) 2013;2013:849186. doi: 10.1155/2013/849186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doerig C, Abdi A, Bland N, et al. Malaria: targeting parasite and host cell kinomes. Biochim Biophys Acta. 2010;1804(3):604–12. doi: 10.1016/j.bbapap.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 7.Lucet IS, Tobin A, Drewry D, Wilks AF, Doerig C. Plasmodium kinases as targets for new-generation antimalarials. Future Med Chem. 2012;4(18):2295–310. doi: 10.4155/fmc.12.183. [DOI] [PubMed] [Google Scholar]
- 8.Merritt C, Silva LE, Tanner AL, Stuart K, Pollastri MP. Kinases as druggable targets in trypanosomatid protozoan parasites. Chem Rev. 2014;114(22):11280–304. doi: 10.1021/cr500197d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schreiber M, Res I, Matter A. Protein kinases as antibacterial targets. Curr Opin Cell Biol. 2009;21(2):325–30. doi: 10.1016/j.ceb.2009.01.026. [DOI] [PubMed] [Google Scholar]
- 10.Artz JD, Wernimont AK, Lin LY, et al. Selective Inhibition of Parasite Protein Kinases. In: Doerig C, Spath G, Wiese M, editors. Protein Phosphorylation in Parasites Novel Targets for Antiparasitic Intervention. Weinheim, Germany: Wiley-VCH Verlag GmbH & Co. KGaA; 2013. [Google Scholar]
- 11.Beckmann S, Leutner S, Gouignard N, Dissous C, Grevelding CG. Protein kinases as potential targets for novel anti-schistosomal strategies. Curr Pharm Des. 2012;18(24):3579–94. [PubMed] [Google Scholar]
- 12.Logullo C, Witola WH, Andrade C, et al. Expression and activity of glycogen synthase kinase during vitellogenesis and embryogenesis of Rhipicephalus (Boophilus) microplus. Vet Parasitol. 2009;161(3–4):261–9. doi: 10.1016/j.vetpar.2009.01.029. [DOI] [PubMed] [Google Scholar]
- 13.Schuler AD, de Oliveira CJL, Coutinho ML, Vaz J, I da S. Characterization of Rhipicephalus microplus glycogen synthase kinase 3β protein by inhibition assays. Acta Sci Vet. 2014;42:1254–63. [Google Scholar]
- 14.Hanks SK, Hunter T. Protein kinases 6. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J. 1995;9(8):576–96. [PubMed] [Google Scholar]
- 15.Parsons M, Worthey EA, Ward PN, Mottram JC. Comparative analysis of the kinomes of three pathogenic trypanosomatids: Leishmania major, Trypanosoma brucei and Trypanosoma cruzi. BMC Genomics. 2005;6:127. doi: 10.1186/1471-2164-6-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alsford S, Turner DJ, Obado SO, et al. High-throughput phenotyping using parallel sequencing of RNA interference targets in the African trypanosome. Genome Res. 2011;21(6):915–24. doi: 10.1101/gr.115089.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jones NG, Thomas EB, Brown E, Dickens NJ, Hammarton TC, Mottram JC. Regulators of Trypanosoma brucei cell cycle progression and differentiation identified using a kinome-wide RNAi screen. PLoS pathogens. 2014;10(1):e1003886. doi: 10.1371/journal.ppat.1003886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doerig C, Billker O, Haystead T, Sharma P, Tobin AB, Waters NC. Protein kinases of malaria parasites: an update. Trends Parasitol. 2008;24(12):570–7. doi: 10.1016/j.pt.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 19.Ward P, Equinet L, Packer J, Doerig C. Protein kinases of the human malaria parasite Plasmodium falciparum: the kinome of a divergent eukaryote. BMC Genomics. 2004;5:79. doi: 10.1186/1471-2164-5-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Talevich E, Mirza A, Kannan N. Structural and evolutionary divergence of eukaryotic protein kinases in Apicomplexa. BMC Evol Biol. 2011;11:321. doi: 10.1186/1471-2148-11-321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Artz JD, Wernimont AK, Allali-Hassani A, et al. The Cryptosporidium parvum kinome. BMC Genomics. 2011;12:478. doi: 10.1186/1471-2164-12-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anamika K, Bhattacharya A, Srinivasan N. Analysis of the protein kinome of Entamoeba histolytica. Proteins. 2008;71(2):995–1006. doi: 10.1002/prot.21790. [DOI] [PubMed] [Google Scholar]
- 23.Sopko R, Andrews BJ. Linking the kinome and phosphorylome--a comprehensive review of approaches to find kinase targets. Mol Biosyst. 2008;4(9):920–33. doi: 10.1039/b801724g. [DOI] [PubMed] [Google Scholar]
- 24.Turk BE. Understanding and exploiting substrate recognition by protein kinases. Curr Opin Chem Biol. 2008;12(1):4–10. doi: 10.1016/j.cbpa.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Knight JD, Pawson T, Gingras AC. Profiling the kinome: current capabilities and future challenges. J Proteomics. 2013;81:43–55. doi: 10.1016/j.jprot.2012.10.015. [DOI] [PubMed] [Google Scholar]
- 26.Petsko GA, Ringe D. Protein Structure and Function. 1. London: New Science Press Ltd; 2003. [Google Scholar]
- 27.Hanks SK, Quinn AM, Hunter T. The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science. 1988;241(4861):42–52. doi: 10.1126/science.3291115. [DOI] [PubMed] [Google Scholar]
- 28.Brown KM, Suvorova E, Farrell A, et al. Forward genetic screening identifies a small molecule that blocks Toxoplasma gondii growth by inhibiting both host- and parasite-encoded kinases. PLoS Pathog. 2014;10(6):e1004180. doi: 10.1371/journal.ppat.1004180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Somlata, Kamanna S, Agrahari M, Babuta M, Bhattacharya S, Bhattacharya A. Autophosphorylation of Ser428 of EhC2PK plays a critical role in regulating erythrophagocytosis in the parasite Entamoeba histolytica. J Biol Chem. 2012;287(14):10844–52. doi: 10.1074/jbc.M111.308874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Merckx A, Echalier A, Langford K, et al. Structures of P.falciparum protein kinase 7 identify an activation motif and leads for inhibitor design. Structure. 2008;16(2):228–38. doi: 10.1016/j.str.2007.11.014. [DOI] [PubMed] [Google Scholar]
- 31.Boynak NY, Rojas F, D'Alessio C, et al. Identification of a Wee1-like kinase gene essential for procyclic Trypanosoma brucei survival. PLoS One. 2013;8(11):e79364. doi: 10.1371/journal.pone.0079364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abdi AI, Carvalho TG, Wilkes JM, Doerig C. A secreted Plasmodium falciparum kinase reveals a signature motif for classification of tyrosine kinase-like kinases. Microbiology. 2013;159(Pt 12):2533–47. doi: 10.1099/mic.0.070409-0. [DOI] [PubMed] [Google Scholar]
- 33.Graciotti M, Alam M, Solyakov L, et al. Malaria protein kinase CK2 (PfCK2) shows novel mechanisms of regulation. PLoS One. 2014;9(3):e85391. doi: 10.1371/journal.pone.0085391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peck SC. Analysis of protein phosphorylation: methods and strategies for studying kinases and substrates. Plant J. 2006;45(4):512–22. doi: 10.1111/j.1365-313X.2005.02613.x. [DOI] [PubMed] [Google Scholar]
- 35.Burnett G, Kennedy EP. The enzymatic phosphorylation of proteins. J Biol Chem. 1954;211(2):969–80. [PubMed] [Google Scholar]
- 36.Azevedo MF, Sanders PR, Krejany E, et al. Inhibition of Plasmodium falciparum CDPK1 by conditional expression of its J-domain demonstrates a key role in schizont development. Biochem J. 2013;452(3):433–41. doi: 10.1042/BJ20130124. [DOI] [PubMed] [Google Scholar]
- 37.Cross DA, Alessi DR, Cohen P, Andjelkovich M, Hemmings BA. Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature. 1995;378(6559):785–9. doi: 10.1038/378785a0. [DOI] [PubMed] [Google Scholar]
- 38.Brumlik MJ, Wei S, Finstad K, et al. Identification of a novel mitogen-activated protein kinase in Toxoplasma gondii. Int J Parasitol. 2004;34(11):1245–54. doi: 10.1016/j.ijpara.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 39.Ghosh AS, Ray D, Dutta S, Raha S. EhMAPK, the mitogen-activated protein kinase from Entamoeba histolytica is associated with cell survival. PLoS One. 2010;5(10):e13291. doi: 10.1371/journal.pone.0013291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Morales MA, Renaud O, Faigle W, Shorte SL, Spath GF. Over-expression of Leishmania major MAP kinases reveals stage-specific induction of phosphotransferase activity. Int J Parasitol. 2007;37(11):1187–99. doi: 10.1016/j.ijpara.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 41.Bengs F, Scholz A, Kuhn D, Wiese M. LmxMPK9, a mitogen-activated protein kinase homologue affects flagellar length in Leishmania mexicana. Mol Microbiol. 2005;55(5):1606–15. doi: 10.1111/j.1365-2958.2005.04498.x. [DOI] [PubMed] [Google Scholar]
- 42.Cayla M, Rachidi N, Leclercq O, et al. Transgenic analysis of the Leishmania MAP kinase MPK10 reveals an auto-inhibitory mechanism crucial for stage-regulated activity and parasite viability. PLoS Pathog. 2014;10(9):e1004347. doi: 10.1371/journal.ppat.1004347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ashutosh, Garg M, Sundar S, Duncan R, Nakhasi HL, Goyal N. Downregulation of mitogen-activated protein kinase 1 of Leishmania donovani field isolates is associated with antimony resistance. Antimicrob Agents Chemother. 2012;56(1):518–25. doi: 10.1128/AAC.00736-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Horjales S, Schmidt-Arras D, Limardo RR, et al. The crystal structure of the MAP kinase LmaMPK10 from Leishmania major reveals parasite-specific features and regulatory mechanisms. Structure. 2012;20(10):1649–60. doi: 10.1016/j.str.2012.07.005. [DOI] [PubMed] [Google Scholar]
- 45.Bao Y, Weiss LM, Ma YF, et al. Molecular cloning and characterization of mitogen-activated protein kinase 2 in Trypanosoma cruzi. Cell Cycle. 2010;9(14):2888–96. doi: 10.4161/cc.9.14.12372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Patel A, Chojnowski AN, Gaskill K, De Martini W, Goldberg RL, Siekierka JJ. The role of a Brugia malayi p38 MAP kinase ortholog (Bm-MPK1) in parasite anti-oxidative stress responses. Mol Biochem Parasitol. 2011;176(2):90–7. doi: 10.1016/j.molbiopara.2010.12.008. [DOI] [PubMed] [Google Scholar]
- 47.Spiliotis M, Konrad C, Gelmedin V, et al. Characterisation of EmMPK1, an ERK-like MAP kinase from Echinococcus multilocularis which is activated in response to human epidermal growth factor. Int J Parasitol. 2006;36(10–11):1097–112. doi: 10.1016/j.ijpara.2006.05.008. [DOI] [PubMed] [Google Scholar]
- 48.Gelmedin V, Caballero-Gamiz R, Brehm K. Characterization and inhibition of a p38-like mitogen-activated protein kinase (MAPK) from Echinococcus multilocularis: antiparasitic activities of p38 MAPK inhibitors. Biochem Pharmacol. 2008;76(9):1068–81. doi: 10.1016/j.bcp.2008.08.020. [DOI] [PubMed] [Google Scholar]
- 49.Ma M, Baumgartner M. Intracellular Theileria annulata promote invasive cell motility through kinase regulation of the host actin cytoskeleton. PLoS Pathog. 2014;10(3):e1004003. doi: 10.1371/journal.ppat.1004003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.John von Freyend S, Rosenqvist H, Fink A, et al. LmxMPK4, an essential mitogen-activated protein kinase of Leishmania mexicana is phosphorylated and activated by the STE7-like protein kinase LmxMKK5. Int J Parasitol. 2010;40(8):969–78. doi: 10.1016/j.ijpara.2010.02.004. [DOI] [PubMed] [Google Scholar]
- 51.Yan Y, Tulasne D, Browaeys E, et al. Molecular cloning and characterisation of SmSLK, a novel Ste20-like kinase in Schistosoma mansoni. Int J Parasitol. 2007;37(14):1539–50. doi: 10.1016/j.ijpara.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 52.Shakir SM, Bryant KM, Larabee JL, et al. Regulatory interactions of a virulence-associated serine/threonine phosphatase-kinase pair in Bacillus anthracis. J Bacteriol. 2010;192(2):400–9. doi: 10.1128/JB.01221-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lima A, Duran R, Schujman GE, et al. Serine/threonine protein kinase PrkA of the human pathogen Listeria monocytogenes: biochemical characterization and identification of interacting partners through proteomic approaches. J Proteomics. 2011;74(9):1720–34. doi: 10.1016/j.jprot.2011.03.005. [DOI] [PubMed] [Google Scholar]
- 54.Yuk JM, Shin DM, Yang CS, et al. Role of apoptosis-regulating signal kinase 1 in innate immune responses by Mycobacterium bovis bacillus Calmette-Guerin. Immunol Cell Biol. 2009;87(1):100–7. doi: 10.1038/icb.2008.74. [DOI] [PubMed] [Google Scholar]
- 55.Nagarajan SN, Upadhyay S, Chawla Y, et al. Protein kinase A (PknA) of Mycobacterium tuberculosis is independently activated and is critical for growth in vitro and survival of the pathogen in the host. J Biol Chem. 2015;290(15):9626–45. doi: 10.1074/jbc.M114.611822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kimura Y, Urata M, Okamoto R. Characterizing activities of eukaryotic-like protein kinases with atypical catalytic loop motifs from Myxococcus xanthus. J Biosci Bioeng. 2015;119(5):511–4. doi: 10.1016/j.jbiosc.2014.10.002. [DOI] [PubMed] [Google Scholar]
- 57.Hu X, Prehna G, Stebbins CE. Targeting plague virulence factors: a combined machine learning method and multiple conformational virtual screening for the discovery of Yersinia protein kinase A inhibitors. J Med Chem. 2007;50(17):3980–3. doi: 10.1021/jm070645a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Theeya N, Ta A, Das S, et al. An inducible and secreted eukaryote-like serine/threonine kinase of Salmonella enterica serovar Typhi promotes intracellular survival and pathogenesis. Infect Immun. 2015;83(2):522–33. doi: 10.1128/IAI.02521-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brandt GS, Bailey S. Dematin, a human erythrocyte cytoskeletal protein, is a substrate for a recombinant FIKK kinase from Plasmodium falciparum. Mol Biochem Parasitol. 2013;191(1):20–3. doi: 10.1016/j.molbiopara.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 60.Smith AJ, Lauwaet T, Davids BJ, Gillin FD. Giardia lamblia Nek1 and Nek2 kinases affect mitosis and excystation. Int J Parasitol. 2012;42(4):411–9. doi: 10.1016/j.ijpara.2012.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lye YM, Chan M, Sim TS. Pfnek3: an atypical activator of a MAP kinase in Plasmodium falciparum. FEBS Lett. 2006;580(26):6083–92. doi: 10.1016/j.febslet.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 62.Desoubzdanne D, Marcourt L, Raux R, et al. Alisiaquinones and alisiaquinol, dual inhibitors of Plasmodium falciparum enzyme targets from a New Caledonian deep water sponge. J Nat Prod. 2008;71(7):1189–92. doi: 10.1021/np8000909. [DOI] [PubMed] [Google Scholar]
- 63.Kinnaird JH, Bumstead JM, Mann DJ, et al. EtCRK2, a cyclin-dependent kinase gene expressed during the sexual and asexual phases of the Eimeria tenella life cycle. Int J Parasitol. 2004;34(6):683–92. doi: 10.1016/j.ijpara.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 64.Grant KM, Dunion MH, Yardley V, et al. Inhibitors of Leishmania mexicana CRK3 cyclin-dependent kinase: chemical library screen and antileishmanial activity. Antimicrob Agents Chemother. 2004;48(8):3033–42. doi: 10.1128/AAC.48.8.3033-3042.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Walker RG, Thomson G, Malone K, et al. High throughput screens yield small molecule inhibitors of Leishmania CRK3:CYC6 cyclin-dependent kinase. PLoS Negl Trop Dis. 2011;5(4):e1033. doi: 10.1371/journal.pntd.0001033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dorin-Semblat D, Carvalho TG, Nivez MP, et al. An atypical cyclin-dependent kinase controls Plasmodium falciparum proliferation rate. Kinome. 2013;1:4–16. [Google Scholar]
- 67.Maity AK, Goswami A, Saha P. Identification of substrates of an S-phase cell cycle kinase from Leishmania donovani. FEBS Lett. 2011;585(17):2635–9. doi: 10.1016/j.febslet.2011.06.017. [DOI] [PubMed] [Google Scholar]
- 68.Engels K, Beyer C, Suarez Fernandez ML, et al. Inhibition of Eimeria tenella CDK-related kinase 2: From target identification to lead compounds. ChemMedChem. 2010;5(8):1259–71. doi: 10.1002/cmdc.201000157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kemp BE, Graves DJ, Benjamini E, Krebs EG. Role of multiple basic residues in determining the substrate specificity of cyclic AMP-dependent protein kinase. J Biol Chem. 1977;252(14):4888–94. [PubMed] [Google Scholar]
- 70.Mitchell RD, Glass DB, Wong CW, Angelos KL, Walsh DA. Heat-stable inhibitor protein derived peptide substrate analogs: phosphorylation by cAMP-dependent and cGMP-dependent protein kinases. Biochemistry. 1995;34(2):528–34. doi: 10.1021/bi00002a018. [DOI] [PubMed] [Google Scholar]
- 71.Donald RG, Allocco J, Singh SB, et al. Toxoplasma gondii cyclic GMP-dependent kinase: chemotherapeutic targeting of an essential parasite protein kinase. Eukaryot Cell. 2002;1(3):317–28. doi: 10.1128/EC.1.3.317-328.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gurnett AM, Liberator PA, Dulski PM, et al. Purification and molecular characterization of cGMP-dependent protein kinase from Apicomplexan parasites. A novel chemotherapeutic target. J Biol Chem. 2002;277(18):15913–22. doi: 10.1074/jbc.M108393200. [DOI] [PubMed] [Google Scholar]
- 73.Gibson C, Schanen B, Chakrabarti D, Chakrabarti R. Functional characterisation of the regulatory subunit of cyclic AMP-dependent protein kinase A homologue of Giardia lamblia: Differential expression of the regulatory and catalytic subunits during encystation. Int J Parasitol. 2006;36(7):791–9. doi: 10.1016/j.ijpara.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 74.Diaz CA, Allocco J, Powles MA, et al. Characterization of Plasmodium falciparum cGMP-dependent protein kinase (PfPKG): antiparasitic activity of a PKG inhibitor. Mol Biochem Parasitol. 2006;146(1):78–88. doi: 10.1016/j.molbiopara.2005.10.020. [DOI] [PubMed] [Google Scholar]
- 75.Bhattacharya A, Biswas A, Das PK. Identification of a protein kinase A regulatory subunit from Leishmania having importance in metacyclogenesis through induction of autophagy. Mol Microbiol. 2012;83(3):548–64. doi: 10.1111/j.1365-2958.2011.07950.x. [DOI] [PubMed] [Google Scholar]
- 76.Malki-Feldman L, Jaffe CL. Leishmania major: effect of protein kinase A and phosphodiesterase activity on infectivity and proliferation of promastigotes. Exp Parasitol. 2009;123(1):39–44. doi: 10.1016/j.exppara.2009.05.010. [DOI] [PubMed] [Google Scholar]
- 77.Donald RG, Liberator PA. Molecular characterization of a coccidian parasite cGMP dependent protein kinase. Mol Biochem Parasitol. 2002;120(2):165–75. doi: 10.1016/s0166-6851(01)00451-0. [DOI] [PubMed] [Google Scholar]
- 78.Hashimoto Y, Soderling TR. Calcium . calmodulin-dependent protein kinase II and calcium. phospholipid-dependent protein kinase activities in rat tissues assayed with a synthetic peptide. Arch Biochem Biophys. 1987;252(2):418–25. doi: 10.1016/0003-9861(87)90048-8. [DOI] [PubMed] [Google Scholar]
- 79.Vaid A, Sharma P. PfPKB, a protein kinase B-like enzyme from Plasmodium falciparum: II. Identification of calcium/calmodulin as its upstream activator and dissection of a novel signaling pathway. J Biol Chem. 2006;281(37):27126–33. doi: 10.1074/jbc.M601914200. [DOI] [PubMed] [Google Scholar]
- 80.Ojo KK, Reid MC, Kallur Siddaramaiah L, et al. Neospora caninum calcium-dependent protein kinase 1 is an effective drug target for neosporosis therapy. PLoS One. 2014;9(3):e92929. doi: 10.1371/journal.pone.0092929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Garrison E, Treeck M, Ehret E, et al. A forward genetic screen reveals that calcium-dependent protein kinase 3 regulates egress in Toxoplasma. PLoS Pathog. 2012;8(11):e1003049. doi: 10.1371/journal.ppat.1003049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ranjan R, Ahmed A, Gourinath S, Sharma P. Dissection of mechanisms involved in the regulation of Plasmodium falciparum calcium-dependent protein kinase 4. J Biol Chem. 2009;284(22):15267–76. doi: 10.1074/jbc.M900656200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ahmed A, Gaadhe K, Sharma GP, et al. Novel insights into the regulation of malarial calcium-dependent protein kinase 1. FASEB J. 2012;26(8):3212–21. doi: 10.1096/fj.12-203877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dorin D, Semblat JP, Poullet P, et al. PfPK7, an atypical MEK-related protein kinase, reflects the absence of classical three-component MAPK pathways in the human malaria parasite Plasmodium falciparum. Mol Microbiol. 2005;55(1):184–96. doi: 10.1111/j.1365-2958.2004.04393.x. [DOI] [PubMed] [Google Scholar]
- 85.Fennell C, Babbitt S, Russo I, et al. PfeIK1, a eukaryotic initiation factor 2alpha kinase of the human malaria parasite Plasmodium falciparum, regulates stress-response to amino-acid starvation. Malar J. 2009;8:99. doi: 10.1186/1475-2875-8-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kats LM, Fernandez KM, Glenister FK, et al. An exported kinase (FIKK4. 2) that mediates virulence-associated changes in Plasmodium falciparum-infected red blood cells. Int J Parasitol. 2014;44(5):319–28. doi: 10.1016/j.ijpara.2014.01.003. [DOI] [PubMed] [Google Scholar]
- 87.Osman KT, Lou HJ, Qiu W, et al. Biochemical characterization of FIKK8 - A unique protein kinase from the malaria parasite Plasmodium falciparum and other apicomplexans. Mol Biochem Parasitol. 2015;201(2):85–9. doi: 10.1016/j.molbiopara.2015.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Dixit A, Singh PK, Sharma GP, Malhotra P, Sharma P. PfSRPK1, a novel splicing-related kinase from Plasmodium falciparum. J Biol Chem. 2010;285(49):38315–23. doi: 10.1074/jbc.M110.119255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Agarwal S, Kern S, Halbert J, et al. Two nucleus-localized CDK-like kinases with crucial roles for malaria parasite erythrocytic replication are involved in phosphorylation of splicing factor. J Cell Biochem. 2011;112(5):1295–310. doi: 10.1002/jcb.23034. [DOI] [PubMed] [Google Scholar]
- 90.Kern S, Agarwal S, Huber K, et al. Inhibition of the SR protein-phosphorylating CLK kinases of Plasmodium falciparum impairs blood stage replication and malaria transmission. PLoS One. 2014;9(9):e105732. doi: 10.1371/journal.pone.0105732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Abdi A, Eschenlauer S, Reininger L, Doerig C. SAM domain-dependent activity of PfTKL3, an essential tyrosine kinase-like kinase of the human malaria parasite Plasmodium falciparum. Cell Mol Life Sci. 2010;67(19):3355–69. doi: 10.1007/s00018-010-0434-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lemercier G, Fernandez-Montalvan A, Shaw JP, et al. Identification and characterization of novel small molecules as potent inhibitors of the plasmodial calcium-dependent protein kinase 1. Biochemistry. 2009;48(27):6379–89. doi: 10.1021/bi9005122. [DOI] [PubMed] [Google Scholar]
- 93.Sugi T, Kato K, Kobayashi K, et al. Molecular analyses of Toxoplasma gondii calmodulin-like domain protein kinase isoform 3. Parasitol Int. 2009;58(4):416–23. doi: 10.1016/j.parint.2009.08.005. [DOI] [PubMed] [Google Scholar]
- 94.Bazan-Tejeda ML, Arguello-Garcia R, Bermudez-Cruz RM, Robles-Flores M, Ortega-Pierres G. Protein kinase C isoforms from Giardia duodenalis: identification and functional characterization of a beta-like molecule during encystment. Arch Microbiol. 2007;187(1):55–66. doi: 10.1007/s00203-006-0174-9. [DOI] [PubMed] [Google Scholar]
- 95.Norris-Mullins B, VanderKolk K, Vacchina P, Joyce MV, Morales MA. LmaPA2G4, a homolog of human Ebp1, is an essential gene and inhibits cell proliferation in L. major. PLoS Negl Trop Dis. 2014;8(1):e2646. doi: 10.1371/journal.pntd.0002646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kuhn D, Wiese M. LmxPK4, a mitogen-activated protein kinase kinase homologue of Leishmania mexicana with a potential role in parasite differentiation. Mol Microbiol. 2005;56(5):1169–82. doi: 10.1111/j.1365-2958.2005.04614.x. [DOI] [PubMed] [Google Scholar]
- 97.Ochiana SO, Pandarinath V, Wang Z, et al. The human Aurora kinase inhibitor danusertib is a lead compound for anti-trypanosomal drug discovery via target repurposing. Eur J Med Chem. 2013;62:777–84. doi: 10.1016/j.ejmech.2012.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ma J, Benz C, Grimaldi R, et al. Nuclear DBF-2-related kinases are essential regulators of cytokinesis in bloodstream stage Trypanosoma brucei. J Biol Chem. 2010;285(20):15356–68. doi: 10.1074/jbc.M109.074591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mansuri MS, Bhattacharya S, Bhattacharya A. A novel alpha kinase EhAK1 phosphorylates actin and regulates phagocytosis in Entamoeba histolytica. PLoS Pathog. 2014;10(10):e1004411. doi: 10.1371/journal.ppat.1004411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cheek S, Ginalski K, Zhang H, Grishin NV. A comprehensive update of the sequence and structure classification of kinases. BMC Struct Biol. 2005;5:6. doi: 10.1186/1472-6807-5-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Li L, Shakhnovich EI, Mirny LA. Amino acids determining enzyme-substrate specificity in prokaryotic and eukaryotic protein kinases. Proc Natl Acad Sci U S A. 2003;100(8):4463–8. doi: 10.1073/pnas.0737647100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Rachidi N, Taly JF, Durieu E, et al. Pharmacological assessment defines Leishmania donovani casein kinase 1 as a drug target and reveals important functions in parasite viability and intracellular infection. Antimicrob Agents Chemother. 2014;58(3):1501–15. doi: 10.1128/AAC.02022-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Silva-Neto MA, Carneiro AB, Vieira DP, Mesquita RD, Lopes AH. Platelet-activating factor (PAF) activates casein kinase 2 in the protozoan parasite Herpetomonas muscarum muscarum. Biochem Biophys Res Commun. 2002;293(5):1358–63. doi: 10.1016/S0006-291X(02)00395-9. [DOI] [PubMed] [Google Scholar]
- 104.Justiniano I, Noris-Suarez K, De Lima A, Contreras V, Bubis J. An unusual casein kinase 1 from Trypanosoma cruzi epimastigotes. Bio Chem Comp. 2014;2:1. [Google Scholar]
- 105.Calabokis M, Kurz L, Gonzatti MI, Bubis J. Protein kinase CK1 from Trypanosoma cruzi. J Protein Chem. 2003;22(6):591–9. doi: 10.1023/b:jopc.0000005509.60532.af. [DOI] [PubMed] [Google Scholar]
- 106.Galan-Caridad JM, Calabokis M, Uzcanga G, Aponte F, Bubis J. Identification of casein kinase 1, casein kinase 2, and cAMP-dependent protein kinase-like activities in Trypanosoma evansi. Mem Inst Oswaldo Cruz. 2004;99(8):845–54. doi: 10.1590/s0074-02762004000800011. [DOI] [PubMed] [Google Scholar]
- 107.Uzcanga G, Galan-Caridad JM, Noris Suarez K, Bubis J. Divalent cation hinder the solubilization of a tubulin kinase activity from Trypanosoma cruzi epimastigotes. Biol Res. 2003;36(3–4):367–79. doi: 10.4067/s0716-97602003000300008. [DOI] [PubMed] [Google Scholar]
- 108.Calabokis M, Kurz L, Wilkesman J, et al. Biochemical and enzymatic characterization of a partially purified casein kinase-1 like activity from Trypanosoma cruzi. Parasitol Int. 2002;51(1):25–39. doi: 10.1016/s1383-5769(01)00104-0. [DOI] [PubMed] [Google Scholar]
- 109.Zylbersztejn A, de Morais C, Lima A, et al. CK2 secreted by Leishmania braziliensis mediates microphage association invasion: A comparative study between virulent and avirulent promastigotes. BioMed Res Int. 2015;2015:11. doi: 10.1155/2015/167323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Donald RG, Zhong T, Meijer L, Liberator PA. Characterization of two T.gondii CK1 isoforms. Mol Biochem Parasitol. 2005;141(1):15–27. doi: 10.1016/j.molbiopara.2005.01.011. [DOI] [PubMed] [Google Scholar]
- 111.Dutra PM, Vieira DP, Meyer-Fernandes JR, Silva-Neto MA, Lopes AH. Stimulation of Leishmania tropica protein kinase CK2 activities by platelet-activating factor (PAF) Acta Trop. 2009;111(3):247–54. doi: 10.1016/j.actatropica.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 112.De Lima AR, Medina R, Uzcanga GL, et al. Tight binding between a pool of the heterodimeric alpha/beta tubulin and a protein kinase CK2 in Trypanosoma cruzi epimastigotes. Parasitology. 2006;132(Pt 4):511–23. doi: 10.1017/S0031182005009352. [DOI] [PubMed] [Google Scholar]
- 113.Spadafora C, Repetto Y, Torres C, et al. Two casein kinase 1 isoforms are differentially expressed in Trypanosoma cruzi. Mol Biochem Parasitol. 2002;124(1–2):23–36. doi: 10.1016/s0166-6851(02)00156-1. [DOI] [PubMed] [Google Scholar]
- 114.Holland Z, Prudent R, Reiser JB, Cochet C, Doerig C. Functional analysis of protein kinase CK2 of the human malaria parasite Plasmodium falciparum. Eukaryot Cell. 2009;8(3):388–97. doi: 10.1128/EC.00334-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Foucher AL, Rachidi N, Gharbi S, et al. Apoptotic marker expression in the absence of cell death in staurosporine-treated Leishmania donovani. Antimicrob Agents Chemother. 2013;57(3):1252–61. doi: 10.1128/AAC.01983-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zhang M, Mishra S, Sakthivel R, et al. PK4, a eukaryotic initiation factor 2alpha(eIF2alpha) kinase, is essential for the development of the erythrocytic cycle of Plasmodium. Proc Natl Acad Sci U S A. 2012;109(10):3956–61. doi: 10.1073/pnas.1121567109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sullivan WJ, Jr, Narasimhan J, Bhatti MM, Wek RC. Parasite-specific eIF2 (eukaryotic initiation factor-2) kinase required for stress-induced translation control. Biochem J. 2004;380(Pt 2):523–31. doi: 10.1042/BJ20040262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Moraes MC, Jesus TC, Hashimoto NN, et al. Novel membrane-bound eIF2alpha kinase in the flagellar pocket of Trypanosoma brucei. Eukaryot Cell. 2007;6(11):1979–91. doi: 10.1128/EC.00249-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.da Silva Augusto L, Moretti NS, Ramos TC, et al. A membrane-bound eIF2 alpha kinase located in endosomes is regulated by heme and controls differentiation and ROS levels in Trypanosoma cruzi. PLoS Pathog. 2015;11(2):e1004618. doi: 10.1371/journal.ppat.1004618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Chow C, Cloutier S, Dumas C, Chou MN, Papadopoulou B. Promastigote to amastigote differentiation of Leishmania is markedly delayed in the absence of PERK eIF2alpha kinase-dependent eIF2alpha phosphorylation. Cell Microbiol. 2011;13(7):1059–77. doi: 10.1111/j.1462-5822.2011.01602.x. [DOI] [PubMed] [Google Scholar]
- 121.Osolodkin DI, Zakharevich NV, Palyulin VA, Danilenko VN, Zefirov NS. Bioinformatic analysis of glycogen synthase kinase 3: human versus parasite kinases. Parasitology. 2011;138(6):725–35. doi: 10.1017/S0031182011000151. [DOI] [PubMed] [Google Scholar]
- 122.Wang QM, Roach PJ, Fiol CJ. Use of a synthetic peptide as a selective substrate for glycogen synthase kinase 3. Anal Biochem. 1994;220(2):397–402. doi: 10.1006/abio.1994.1356. [DOI] [PubMed] [Google Scholar]
- 123.Woodgett JR. Use of peptide substrates for affinity purification of protein-serine kinases. Anal Biochem. 1989;180(2):237–41. doi: 10.1016/0003-2697(89)90423-5. [DOI] [PubMed] [Google Scholar]
- 124.Sutherland C, Leighton IA, Cohen P. Inactivation of glycogen synthase kinase-3 beta by phosphorylation: new kinase connections in insulin and growth-factor signalling. Biochem J. 1993;296(Pt 1):15–9. doi: 10.1042/bj2960015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ryves WJ, Fryer L, Dale T, Harwood AJ. An assay for glycogen synthase kinase 3 (GSK-3) for use in crude cell extracts. Anal Biochem. 1998;264(1):124–7. doi: 10.1006/abio.1998.2832. [DOI] [PubMed] [Google Scholar]
- 126.Droucheau E, Primot A, Thomas V, et al. Plasmodium falciparum glycogen synthase kinase-3: molecular model, expression, intracellular localisation and selective inhibitors. Biochim Biophys Acta. 2004;1697(1–2):181–96. doi: 10.1016/j.bbapap.2003.11.023. [DOI] [PubMed] [Google Scholar]
- 127.Xingi E, Smirlis D, Myrianthopoulos V, et al. 6-Br-5methylindirubin-3'oxime (5-Me-6-BIO) targeting the leishmanial glycogen synthase kinase-3 (GSK-3) short form affects cell-cycle progression and induces apoptosis-like death: exploitation of GSK-3 for treating leishmaniasis. Int J Parasitol. 2009;39(12):1289–303. doi: 10.1016/j.ijpara.2009.04.005. [DOI] [PubMed] [Google Scholar]
- 128.Chan MM, Cheung BK, Li JC, Chan LL, Lau AS. A role for glycogen synthase kinase-3 in antagonizing mycobacterial immune evasion by negatively regulating IL-10 induction. J Leukoc Biol. 2009;86(2):283–91. doi: 10.1189/jlb.0708442. [DOI] [PubMed] [Google Scholar]
- 129.Ojo KK, Gillespie JR, Riechers AJ, et al. Glycogen synthase kinase 3 is a potential drug target for African trypanosomiasis therapy. Antimicrob Agents Chemother. 2008;52(10):3710–7. doi: 10.1128/AAC.00364-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Urich R, Grimaldi R, Luksch T, Frearson JA, Brenk R, Wyatt PG. The design and synthesis of potent and selective inhibitors of Trypanosoma brucei glycogen synthase kinase 3 for the treatment of human african trypanosomiasis. J Med Chem. 2014;57(18):7536–49. doi: 10.1021/jm500239b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Woodland A, Grimaldi R, Luksch T, et al. From on-target to off-target activity: identification and optimisation of Trypanosoma brucei GSK3 inhibitors and their characterisation as anti-Trypanosoma brucei drug discovery lead molecules. ChemMedChem. 2013;8(7):1127–37. doi: 10.1002/cmdc.201300072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Swinney ZT, Haubrich BA, Xia S, et al. A four-point screening method for assessing molecular mechanism of action (MMOA) identifies tideglusib as a time-dependent inhibitor of Trypanosoma brucei GSK3beta. doi: 10.1371/journal.pntd.0004506. Manuscript submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hennrich ML, Marino F, Groenewold V, Kops GJ, Mohammed S, Heck AJ. Universal quantitative kinase assay based on diagonal SCX chromatography and stable isotope dimethyl labeling provides high-definition kinase consensus motifs for PKA and human Mps1. J Proteome Res. 2013;12(5):2214–24. doi: 10.1021/pr400074f. [DOI] [PubMed] [Google Scholar]
- 134.Wang C, Ye M, Bian Y, et al. Determination of CK2 specificity and substrates by proteome-derived peptide libraries. J Proteome Res. 2013;12(8):3813–21. doi: 10.1021/pr4002965. [DOI] [PubMed] [Google Scholar]
- 135.El Hajj H, Lebrun M, Arold ST, Vial H, Labesse G, Dubremetz JF. ROP18 is a rhoptry kinase controlling the intracellular proliferation of Toxoplasma gondii. PLoS Pathog. 2007;3(2):e14. doi: 10.1371/journal.ppat.0030014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Schutkowski M, Reineke U, Reimer U. Peptide arrays for kinase profiling. Chembiochem. 2005;6(3):513–21. doi: 10.1002/cbic.200400314. [DOI] [PubMed] [Google Scholar]
- 137.Kim M, Shin DS, Kim J, Lee YS. Substrate screening of protein kinases: detection methods and combinatorial peptide libraries. Biopolymers. 2010;94(6):753–62. doi: 10.1002/bip.21506. [DOI] [PubMed] [Google Scholar]
- 138.Sun H, Chen GY, Yao SQ. Recent advances in microarray technologies for proteomics. Chem Biol. 2013;20(5):685–99. doi: 10.1016/j.chembiol.2013.04.009. [DOI] [PubMed] [Google Scholar]
- 139.Lim D, Gold DA, Julien L, et al. Structure of the Toxoplasma gondii ROP18 kinase domain reveals a second ligand binding pocket required for acute virulence. J Biol Chem. 2013;288(48):34968–80. doi: 10.1074/jbc.M113.523266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Lourido S, Jeschke GR, Turk BE, Sibley LD. Exploiting the unique ATP-binding pocket of toxoplasma calcium–dependent protein kinase 1 to identify its substrates. ACS Chem Biol. 2013;8(6):1155–62. doi: 10.1021/cb400115y. [DOI] [PMC free article] [PubMed] [Google Scholar]