Abstract
In diabetes, matrix metalloproteinase-9 (MMP-9) is activated, which damages mitochondria, resulting in accelerated capillary cell apoptosis. Regulation of MMP-9 is controlled by multiple transcription factors including nuclear factor-kB (NF-kB) and activator protein-1 (AP-1). Binding of these transcription factors, however, can be regulated by poly(ADP-ribose) polymerase-1 (PARP-1), which forms a strong initiation complex at the promoter region and facilitates multiple rounds of gene transcription. This complex formation with the transcription factors is regulated by posttranslational acetylation of PARP-1, and in diabetes, the deacetylating enzyme, Sirt1, is inhibited. Our aim was to understand the role of PARP-1 in transcriptional regulation of MMP-9 in the development of diabetic retinopathy. Using human retinal endothelial cells, the effect of PARP-1 inhibition (pharmacologically by PJ34, 1μM; or genetically by its siRNA) on MMP-9 expression was investigated. The effect of PARP-1 acetylation on its binding at the MMP-9 promoter, and with NF-kB/AP-1, was investigated in the cells transfected with Sirt1. In vitro results were validated in the retinal microvessels from diabetic mice either administered PJ34, or overexpressing Sirt1. Inhibition of PARP-1 ameliorated hyperglycemia-induced increase in the binding of NF-kB/AP-1 at the MMP-9 promoter, decreased MMP-9 expression and ameliorated mitochondrial damage. Overexpression of Sirt1 attenuated diabetes-induced increase in PARP-1 binding at MMP-9 promoter or with NF-kB/AP-1. Thus, PARP-1, via manipulating the binding of NF-kB/AP-1 at the MMP-9 promoter, regulates MMP-9 expression, which helps maintain mitochondrial homeostasis.
Keywords: Acetylation, Diabetic retinopathy, MMP-9, PARP-1, Transcriptional regulation
Graphical abstract
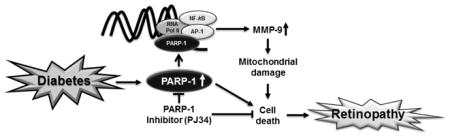
Increased retinal PARP-1 in diabetes increases its complex formation with the transcription factors at the MMP-9 promoter, subsequently increasing MMP-9 expression. Inhibition of PARP-1, inhibits, diabetes-induced increase in MMP-9, mitochondrial damage and capillary cell apoptosis.
Introduction
Diabetic retinopathy is one of the most serious ocular complications, and remains the leading cause of acquired blindness in working aged adults in developed countries [1, 2]. We have shown that diabetes activates matrix metalloproteinases -2 and -9 (MMP-2 and MMP-9) in the retina and its capillary cells. MMP-9 activation is associated with increased capillary cell apoptosis, and the enzyme remains active at duration of diabetes in rodents, which is associated with the development of histopathology characteristic of diabetic retinopathy. [3–6]. MMP-9 is secreted as a proenzyme, and on activation by proteolytic cleavage, it is capable of degrading extracellular matrix components. However, in diabetes, MMP-9 also accumulates inside the mitochondria and activated MMP-9 damages the mitochondrial membrane releasing cytochrome c into the cytosol, which activates the apoptotic machinery [4]. Diabetic MMP-9 knockout mice have normal retinal mitochondrial homeostasis, and are protected from histopathological characteristics observed in retinopathy [4].
Regulation of MMP-9 transcription is controlled by its promoter region, which has binding sites for a number of transcription factors including nuclear factor kappa B (NF-kB), activator protein 1 (AP-1) and specific protein 1 [7]. MMP-9 promoter has two AP-1 binding sites, a proximal site and a distal site [8, 9], and we have shown that in diabetes, expression of MMP-9 in the retina is closely associated with the binding of both NF-kB and AP-1 [10, 11]. Even though NF-kB and AP-1 transcription factors are regulated by different mechanisms, they can be activated simultaneously by the oxidative stress [12–14]. In response to stress stimulus, MMP-9 requires concomitant activation of NF-kB and AP-1. Activation of NF-kB and AP-1, however, may also require cofactors, and poly(ADP-ribose) polymerase-1 (PARP-1), a nuclear chromatin-associated protein, considered as one of the coactivators for both transcriptional factors [15–18]. The activity of PARP-1 and its interaction with NF-kB are increased in the retina of diabetic rats, and pharmacological inhibition of PARP-1 inhibits capillary cell apoptosis and the development of early lesions of diabetic retinopathy [19, 20].
Expression of genes can be regulated by posttranslational modification of associated transcription factors by regulating their DNA binding efficiency. Acetylation is one of the common modifications that regulate the activities of NF-kB and AP-1 [10, 11]; while acetylation is considered active, deacetylation results in the loss of their DNA binding activity. Sirtuin 1 (Sirt1), a class III histone deacetylase, plays an important role in the deacetylation of transcriptional factors. We have shown that in diabetes, Sirt1 is inhibited, and NF-kB and AP-1 are acetylated and their binding at the MMP-9 promoter is increased [10, 11]. PARP-1, in the presence of a histone acetyl transferase, p300, can act as a coactivator of NF-kB by promoting rapid formation of pre-initiation complexes at the transcription factor binding regions on the gene promoter [21]. In contrast, Sirt1 deacetylates PARP-1, blocks its activity and binding with other transcription factors [22]. However, the role of PARP-1 and its acetylation in the regulation of retinal MMP-9 transcription in diabetes remains elusive.
The aim of this study was to examine the role of PARP-1 in the regulation of transcription factor binding at the retinal MMP-9 promoter in diabetes. Using retinal endothelial cells, we have investigated the effect of high glucose on PARP-1 binding at the MMP-9 promoter, and how PARP-1 inhibition regulates the binding of transcription factors and expression of MMP-9. We have also examined the role of Sirt1-mediated PARP-1 binding with NF-kB, AP-1, and at MMP-9 promoter. The results are confirmed in the retinal microvessels from diabetic mice.
Methods
Retinal endothelial cells, isolated from non-diabetic human retina, with no documented ocular pathologies [23], were cultured in Dulbecco’s modified Eagle medium (DMEM)-F12 (HyClone, Waltham, MA, USA) containing 10% heat inactivated fetal bovine serum (Sigma-Aldrich St. Louis, MO, USA), 50μg/ml heparin (Sigma-Aldrich), 15μg/ml endothelial cell growth supplement (BD Bioscience, San Jose, CA, USA), 1% insulin transferrin selenium (Sigma-Aldrich), 1% Glutamax (Gibco-ThermoFisher Scientific, Waltham, MA) and antibiotic/antimycotic mixture (Sigma-Aldrich) as described previously [23]. Cells from 4th–7th passage (~80% confluency) were incubated in normal (5mM) or high (20mM) glucose for 4 days in DMEM-F12 containing 1% heat inactivated fetal bovine serum and low (5.0μg/ml) endothelial cell growth supplement in the presence or absence of PARP-1 inhibitor, PJ34 (1μM, PARP inhibitor VIII; Calbiochem/EMD Chemicals Inc., Gibbstown, NJ). A batch of cells from 4th–6th passage was transfected with PARP-1 siRNA (sc-29437, Santa Cruz Biotechnology, Santa Cruz, CA) or with Sirt1 plasmids (Sirt1 cDNA, RC227720; OriGene, Rockville, MD) using the methods described previously [10, 24–26]. After transfection, the cells were incubated in 5mM or 20mM glucose media for 4 additional days. Controls included cells transfected with a non-targeting scrambled siRNA (sc-37007, Santa Cruz Biotechnology) or reagent alone (transfection controls). Each incubation had osmotic controls, where the cells were incubated in 20mM mannitol instead of 20mM glucose.
Mice
Wild-type C57BL/6J and Sirt1 overexpressing (St, C57BL/6-Actbtm3.1 (Sirt1) Npa/J) mice, obtained from the Jackson Laboratory (Bar Harbor, ME), were made diabetic by streptozotocin injection (55 mg/kg BW) for 4 consecutive days. Mice with blood glucose >250 mg/dl after two days of the last injection were considered diabetic, and were maintained in the experiment for six months [10, 27]. Glycated hemoglobin values (GHb, measured using a kit from Helena Laboratories, Beaumont, TX) were similar in C57BL/6J and Sirt1 overexpressing diabetic mice (GHb >10%), and these values were significantly higher compared to their respective non-diabetic controls (GHb ~6%).
A group of C57BL/6J mice, soon after induction of diabetes, were administered PJ34 (15mgkg−1day−1, Intraperitoneal) for 4 weeks [28]. Age-matched normal mice were used as controls. The treatment of the animals conformed to the Association for Research in Vision and Ophthalmology Resolution on the Use of Animals in Research. Retinal microvessels were prepared by hypotonic shock method, as described previously [29, 30]. Retina (4–6 samples) was incubated at 37°C in 5–6 ml de-ionized water for 60 minutes in a shaking water bath. At the end of the incubation, nonvascular tissue was gently removed under the microscope, and the microvessel preparation was utilized for analyses.
Gene expression was quantified by SYBR green based qPCR using species and gene-specific primers (Table 1). Denaturation at 95 °C for 10 minutes was followed by 40 cycles of denaturation at 95 °C for 15 seconds, and annealing and extension at 60 °C for 60 seconds. This was followed by 95 °C for 15 seconds, 60 °C for 60 seconds, 95 °C for 15 seconds, and 60 °C for 15 seconds. Product specificity was confirmed by SYBR green single melting curve analysis and by agarose gel electrophoresis. The results were normalized to the expression of β-actin or 18S rRNA, and the relative fold change in the expression was calculated using the delta delta Ct method [10, 27].
Table 1.
Primer sequence
| Gene | Sequence |
|---|---|
| Human | |
| MMP-9 promoter, NF-kB binding site | 5′-GATTCAGCCTGCGGAAGACAGGG-3′ |
| 5′-CCAAACCCCTCCCCACACTCCA-3′ | |
| MMP-9 promoter, AP-1 proximal site | 5′-GAGTCAGCACTTGCCTGTCA-3′ |
| 5′-CTGCTGTTGTGGGGGCTTTA-3′ | |
| MMP-9 promoter, AP-1 distal site | 5′-CTTGCCTAGCAGAGCCCATT-3′ |
| 5′-TTTTTCCCTCCCTGACAGCC-3′ | |
| PARP-1 | 5′-GCTTCAGCCTCCTTGCTACA-3′ |
| 5′-TTCGCCACTTCATCCACTCC-3′ | |
| MMP-9 | 5′-CACTGTCCACCCCTCAGAGC-3′ |
| 5′-GCCACTTGTCGGCGATAAGG-3′ | |
| Cytb | 5′-TCACCAGACGCCTCAACCGC-3′ |
| 5′-GCCTCGCCCGATGTGTAGGA-3′ | |
| β-Actin | 5′-AGCCTCGCCTTTGCCGATCCG-3′ |
| 5′-TCTCTTGCTCTGGGCCTCGTCG-3′ | |
| Mouse | |
| MMP-9 promoter, NF-kB binding site | 5′-GCCCCATGGAATTCCCCAAA-3′ |
| 5′-CCGCCCCCTGATAGAGTCTT-3′ | |
| MMP-9 promoter, AP-1 proximal site | 5′-CAGGGCCTCGTCTTTCTTTC-3′ |
| 5′-CCATGGTTTGGTGTTGCTGTT-3′ | |
| MMP-9 promoter, AP-1 distal site | 5′-AGCGCCAGTTCTGTTAGCAT-3′ |
| 5′-TAGACGTCCACGAGTCTGGG-3′ | |
| MMP-9 | 5′-GGGGTTTGCCCCATGGAAT-3′ |
| 5′-GAGCCCATCCCCACACTGTA-3′ | |
| 18S | 5′-GCCCTGTAATTGGAATGAGTCCACTT-3′ |
| 5′-CTCCCCAAGATCCAACTACGAGCTTT-3′ | |
Nuclear localization of PARP-1 was investigated in retinal endothelial cells by immunofluorescence technique using PARP-1 primary antibody (Catalog No: sc-7150, Santa Cruz Biotechnology) and Alexa Fluor-488 conjugated anti-rabbit secondary antibody (green; Molecular Probes-Life Technologies, Grand Island, NE). Immuno-labelled cells were mounted using DAPI-containing (blue) Vectashield mounting medium (Vector Laboratories, Burlingame, CA, USA). The slides were imaged under a Zeiss ApoTome fluorescence microscope using 40X magnification (Carl Zeiss Inc., Chicago, IL) [25, 30, 31].
Chromatin immunoprecipitation (ChIP) was performed by immunoprecipitating 100–120μg crosslinked protein-DNA complex with antibodies against NF-kB (p65 subunit; ab7970, Abcam, Cambridge, MA), or AP-1 (c-Jun subunit; ab31419, Abcam), or PARP-1 (sc-7150, Santa Cruz Biotechnology). Phenol-chloroform-isoamyl isolated DNA was precipitated with ethanol and re-suspended in water, and the binding of NF-kB, AP-1 or PARP-1 at the MMP-9 promoter was quantified by SYBR green based qPCR. The controls included DNA from the input (internal control) and crosslinked sample precipitated with normal rabbit IgG (antibody control) [10, 11, 30].
PARP-1 acetylation and its interaction with NF-kB/AP-1 were performed by immunoprecipitating PARP-1, followed by incubation with Protein A/G Plus agarose beads (suspended in the lysis buffer). After washing the beads, the proteins were separated on a SDS-PAGE and were immunoblotted either with an antibody specific to acetyl-lysine (Catalog No: ab22550, Abcam) or with anti-NF-kB (p65 subunit) or AP-1 (c-Jun subunit) following standard procedures routinely used in the laboratory [5, 32].
Cell apoptosis was determined by Cell Death Detection ELISAPLUS kit from Roche Diagnostics (Indianapolis, IN) using monoclonal antibodies that can detect DNA and histones. Oligonucleosome formed in the cytoplasmic fraction was quantified by using monoclonal antibodies, followed by incubation with peroxidase conjugated anti-DNA and biotin-labeled anti-histone. The samples were then incubated with 2,2′-azino-di-[3-ethylbenzthiazoline sulfonate] diammonium salt, and the absorbance was measured at 405 and 490 nm [24, 25].
Statistical analysis was performed using Sigma Stat software. For multiple group comparison, one-way ANOVA followed by Student-Newman-Keuls test was used for data with normal distribution, while Kruskal-Wallis one-way analysis followed by Dunn’s test was performed for data that did not present normal distribution. Data are expressed as mean ± standard deviation, and a P < 0.05 was considered statistically significant.
Results
Retinal endothelial cells
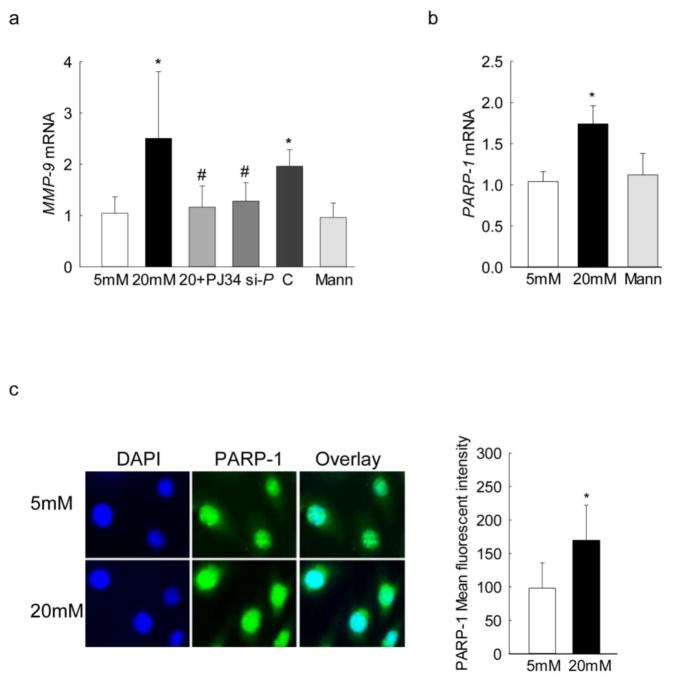
PARP-1 is an important co-factor in modulating the binding of transcription factors to regulate the subsequent gene expression [17, 33–35]; the role of PARP-1 in regulation of MMP-9 transcription was investigated using both pharmacological and genetic approaches. Inhibition of PARP-1 by PJ34 prevented glucose-induced increase in MMP-9 expression; the values obtained from the cells incubated in high glucose in the presence of PJ34 were significantly lower than without PJ34, and were not different from those obtained from cells in normal glucose (Figure 1a). Similarly, transfection of cells with PARP-1 siRNA, but not with scrambled siRNA, also ameliorated increase in MMP-9 seen under high glucose conditions (Figure 1a). In the same cell preparations, as expected, high glucose also increased PARP-1 expression by almost 2-fold compared to the cells in normal glucose (Figure 1b). Incubation of cells with 20mM mannitol, instead of 20mM glucose did not increase MMP-9 and PARP-1 expressions.
Figure 1.
Effect of PARP-1 on the glucose- induced MMP-9 expression: Gene transcripts of (a) MMP-9 and (b) PARP-1 were quantified in the cells incubated in normal or high glucose in the presence of PJ34 or PARP-1 siRNA or scrambled siRNA by qPCR. β-actin was used as the housekeeping gene. (c) Nuclear localization of PARP-1 was determined by immunofluorescence using Alexa-Flour 488 (green) conjugated secondary antibody. The cover slips were mounted using DAPI-containing mounting medium (blue). Each measurement was made in duplicate in 4–5 samples in each group, and the values are represented as mean ± SD. 5mM and 20mM = cells in 5mM or 20mM glucose; Mann = 20mM mannitol; 20+PJ34, si-P and C = cells incubated with PJ34, PARP-1 siRNA, or scrambled siRNA respectively, and incubated in 20mM glucose. *P<0.05 versus 5mM glucose and #P<0.05 versus 20mM glucose.
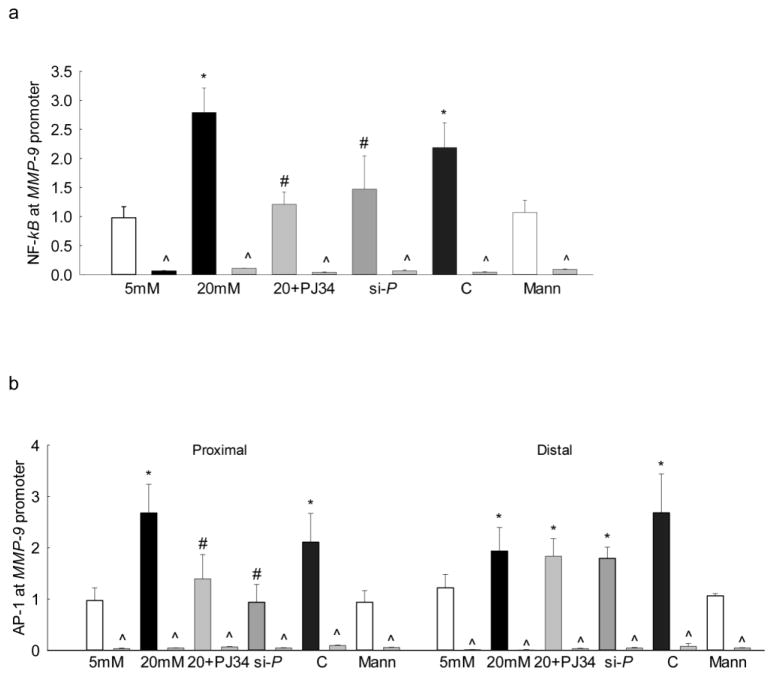
Since the binding of the transcription factors mainly occurs in the nucleus, the effect of high glucose on PARP-1 translocation into the nucleus was determined by immunofluorescence technique. As shown in figure 1c, nuclear localization of PARP-1 was significantly increased in cells exposed to high glucose compared to the cells in normal glucose. The accompanying histogram shows the mean fluorescent intensity of PARP-1 in the nucleus. Our recent work has shown that both NF-kB and AP-1 play important roles in the increased MMP-9 levels seen in hyperglycemic milieu [10, 11]. To evaluate the role of PARP-1 in the binding of these transcription factors at the MMP-9 promoter, the effect of regulation of PARP-1 on its binding with NF-kB and with AP-1, regions of the MMP-9 promoter was investigated. As expected, glucose increased binding of both NF-kB and AP-1 at the MMP-9 promoter (Figure 2a&b), and inhibition of PARP-1 with PJ34 or PARP-1 siRNA significantly ameliorated binding of these transcriptional factors at the proximal regions on the MMP-9 promoter (Figure 2a&b). However, cells transfected with scrambled siRNA had no effect on glucose-induced increased binding of either NF-kB or AP-1. The values obtained from IgG antibody control were less than 1% of the values obtained from the target genes.
Figure 2.
Regulation of PARP-1 and binding of NF-kB and AP-1 at the MMP-9 promoter: Endothelial cells incubated with PJ34 or PARP-1 siRNA, were analyzed for binding of (a) NF-kB, and (b) AP-1 at the proximal and the distal regions of MMP-9 promoter by ChIP technique. IgG was used as an antibody control (indicated as ^). Each measurement was made in duplicate in 3–4 samples/group. 5mM and 20mM=cells in 5mM or 20mM glucose; Mann = 20mM mannitol; 20+PJ34, si-P and C = cells incubated in 20mM glucose in the presence of PJ34, PARP-1 siRNA, and scrambled siRNA control respectively. *P<0.05 versus 5mM glucose and #P<0.05 versus 20mM glucose.
Since activated MMP-9 is implicated in mitochondrial damage and accelerated apoptosis [4], the effect of inhibition of PARP-1 on mitochondrial damage was investigated by quantifying the gene transcripts of mtDNA-encoded Cytochrome b (Cytb). Figure 3a shows that the inhibition of PARP-1 by PJ34 or its specific siRNA ameliorated glucose-induced decrease in Cytb expression. However, the values obtained from cells incubated in high glucose, with or without the scrambled siRNA, had similar decrease in Cytb expression. The values from the cells in 20mM mannitol were not different from those obtained from cells in 5mM glucose (Figure 3a). In the same cell preparation, PJ34 addition also ameliorated glucose-induced increase in cell apoptosis (Figure 3b).
Figure 3.
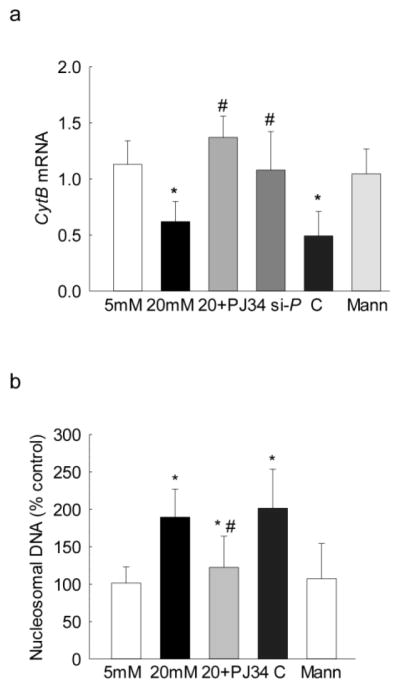
Regulation of PARP-1 and its effect on mtDNA damage and cell apoptosis: Effect of PARP-1 regulation on (a) mtDNA-encoded Cytb expression was measured by qPCR using β-actin as the housekeeping gene, and (b) apoptosis by an ELISA kit for histone-associated-DNA-fragments in the cells incubated with PJ34 or PARP-1 siRNA, or scrambled siRNA control. The values are represented as mean ± SD from 3–4 samples/group. 5mM and 20mM=cells in 5mM or 20mM glucose; Mann = 20mM mannitol; 20+PJ34, si-P and C = cells incubated in 20mM glucose in the presence of PJ34, PARP-1 siRNA and scrambled siRNA control, respectively. *P<0.05 versus 5mM glucose and #P<0.05 versus 20mM glucose.
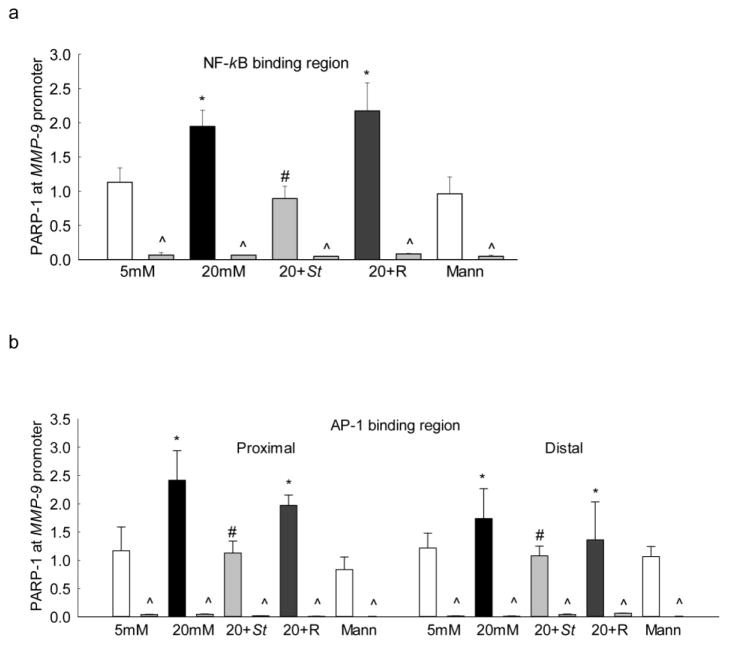
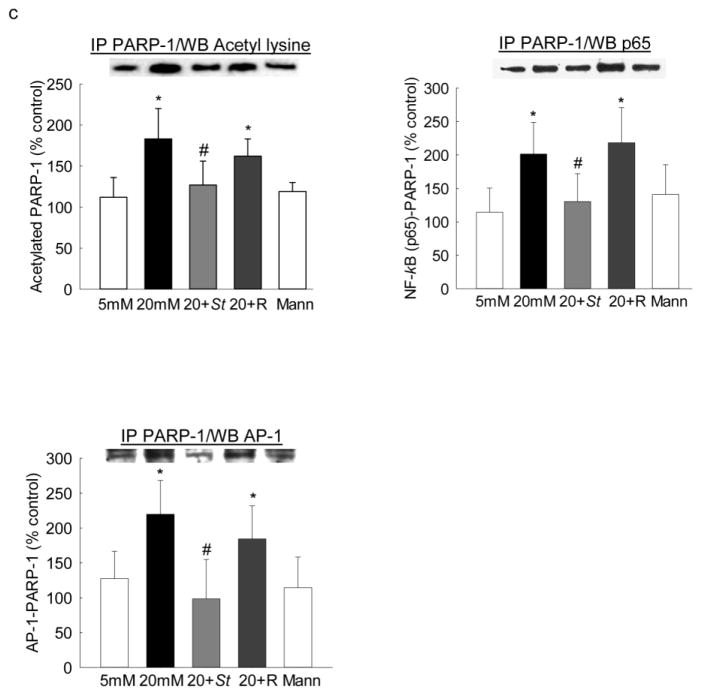
To further confirm the role of PARP-1 in MMP-9 regulation, effect of high glucose on PARP-1 bindings at both NF-kB and AP-1 regions of the MMP-9 promoter were determined. Incubation of cells in high glucose increased PARP-1 binding at the NF-kB region by ~80% compared to the cells in normal glucose. Similarly, PARP-1 binding was also increased by ~2 fold at the both proximal and the distal AP-1 binding regions of the MMP-9 promoter (Figures 4a&b). In addition to increased binding of PARP-1 at the MMP-9 promoter, high glucose also increased its binding with NF-kB and with AP-1 by ~2 folds (Figures 4c).
Figure 4.
Effect of Sirt1 regulation on PARP-1 binding: Endothelial cells transfected with Sirt1 cDNA and incubated in 20mM glucose, were analyzed for the binding of PARP-1 at (a) NF-kB and (b) AP-1 binding regions of the MMP-9 promoter by ChIP technique. IgG was used as an antibody control (indicated as ^). (c) PARP-1 acetylation and its effect on the binding of NF-kB/AP-1 with PARP-1 were determined by immunoprecipitating total proteins using PARP-1 antibody, followed by western blotting for NF-kB/AP-1. The values are represented as mean ± SD from 3–4 samples/group. 5mM and 20mM = cells in 5mM or 20mM glucose; Mann = 20mM mannitol; 20+St and 20+R = cells transfected with Sirt1 plasmids or transfection reagent alone respectively, and incubated in 20mM glucose. *P<0.05 versus 5mM glucose and #P<0.05 versus 20mM glucose.
PARP-1 regulates transcription by forming complex with other transcription factors, and this is regulated by its posttranslational modifications [17]. The role of acetylation of PARP-1 in forming complex with the transcription factors was investigated in the cells overexpressing Sirt1. As shown in figure 4, Sirt1 overexpression inhibited glucose-induced increase in PARP-1 binding at both NF-kB and AP-1 regions of the MMP-9 promoter. The values obtained from Sirt1 transfected cells incubated in high glucose were not significantly different from the cells incubated with normal glucose. In contrast, transfection of cells with reagent alone had no effect on glucose-induced increase in PARP-1 binding. In the same cells, Sirt1 overexpression prevented increased PARP-1 acetylation and its subsequent binding with NF-kB and AP-1 transcription factors (Figure 4).
Diabetic mice
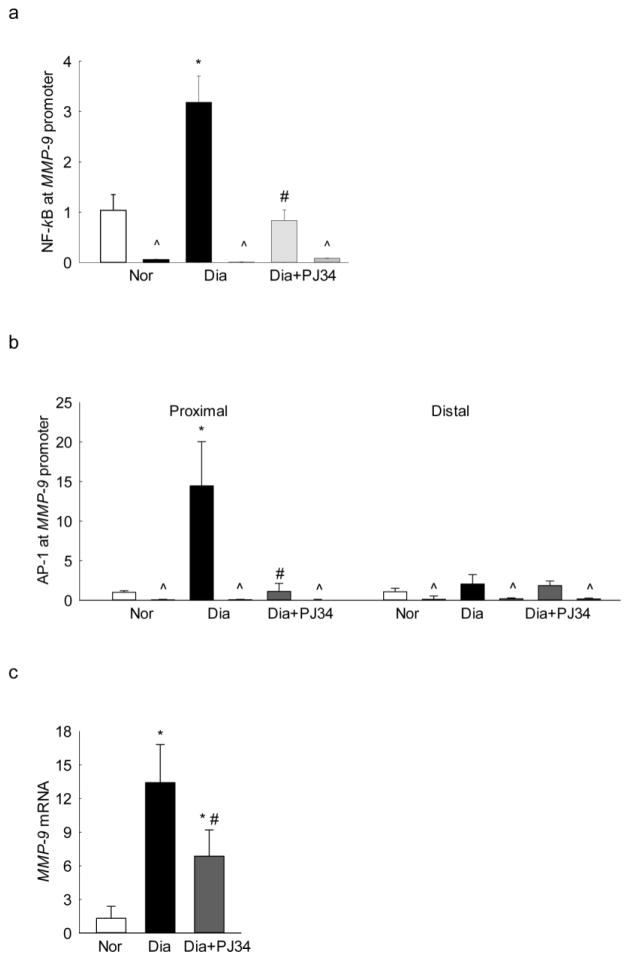
To confirm the role of PARP-1 in the regulation of MMP-9 in an in vivo model, retinal microvessels from streptozotocin-induced diabetic mice were analyzed. Consistent with the results from isolated cells, administration of PJ34 prevented diabetes-induced increase in NF-kB binding at the MMP-9 promoter; the values obtained from diabetic mice receiving PJ34 were not different from those obtained from nondiabetic mice without any treatment (Figure 5a). In the same microvessels preparation, IgG control values were <1% of the values obtained from the target gene. Similarly, PJ34 also inhibited diabetes-induced increased AP-1 binding at the proximal region of the MMP-9 promoter (Figure 5b), however, AP-1 binding at the distal region though slightly higher in diabetes, was not affected. In the same retinal microvessels, diabetes-induced increase in MMP-9 gene transcripts was also ameliorated (Figure 5c). Interestingly, in our in vivo model, AP-1 binding at the MMP-9 promoter and MMP-9 expression were significantly higher compared to the cells incubated in high glucose; the possibility that these could be due to the complexity of diabetes and/or of the retinal structure, cannot be ruled out.
Figure 5.
Effect of PARP-1 on diabetes-induced NF-kB/AP-1 binding at MMP-9 promoter in retinal microvessels: Binding of (a) NF-kB, and (b) AP-1 at MMP-9 promoter was measured in retinal microvessels using NF-kB and AP-1 antibodies. IgG was used as an antibody control (indicated as ^). (c) Retinal microvasculature from diabetic mice receiving PJ34 was quantified for MMP-9 expression by qPCR using 18S as the housekeeping gene. The values are represented as mean ± SD from 4–6 mice/group. Nor and Dia = C57BL/6J mice normal and diabetic respectively, and Dia+PJ34 = C57BL/6J diabetic mice administered with PJ34. *P<0.05 compared to Nor and #P<0.05 compared to diabetes.
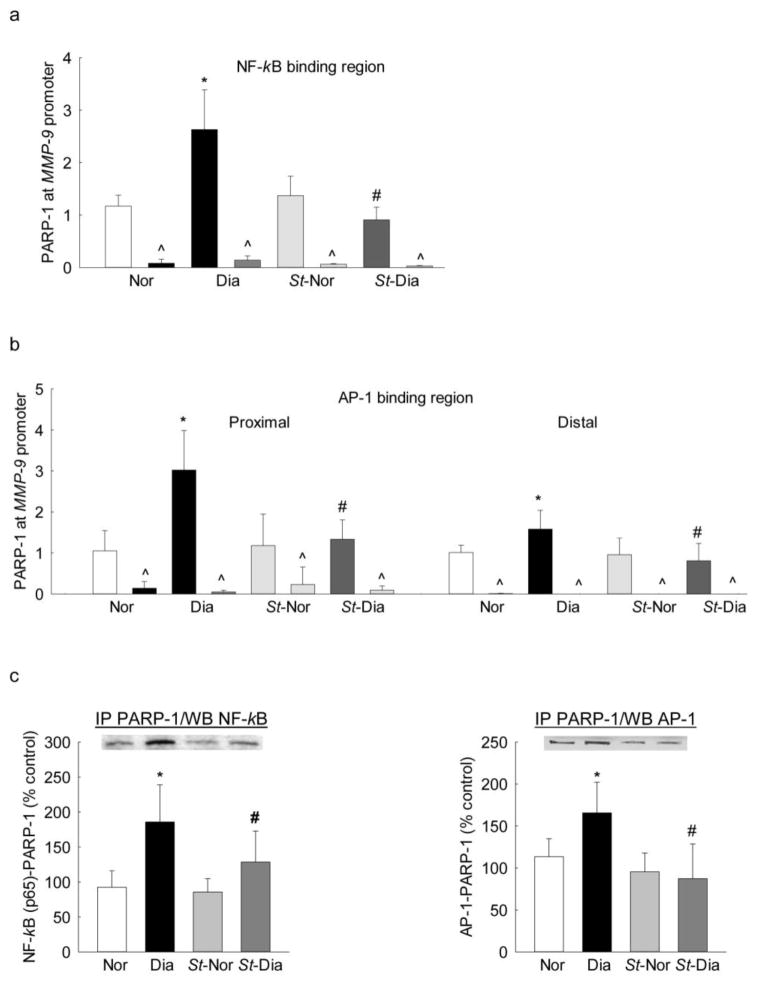
To investigate the role of posttranslational modification in PARP-1 mediated increased transcription factor binding at the MMP-9 promoter, retinal microvessels from diabetic mice overexpressing Sirt1 were analyzed. As shown in figure 6a, diabetes increased PARP-1 binding at NF-kB region of the MMP-9 promoter, which was ameliorated by Sirt1 overexpression; the values obtained from Sirt1 overexpressing diabetic mice were not different from those obtained from nondiabetic Sirt1 overexpressing or wildtype control mice. Consistent with NF-kB binding, PARP-1 binding at both the distal and proximal regions of the AP-1 binding sites of the MMP-9 promoter was also significantly decreased in diabetic mice overexpressing Sirt1 compared to those from age-matched wild type diabetic mice (Figures 6b). In the same Sirt1 overexpressing diabetic mice, increase in the binding of both NF-kB and of AP-1 with PARP-1 was also prevented; and the values obtained from wildtype non-diabetic mice and Sirt1 overexpressing mice (non-diabetic or diabetic) were not different from each other (Figures 6c).
Figure 6.
Diabetes-induced PARP-1 binding at MMP-9 promoter in retinal microvessels, and its regulation by Sirt1: DNA isolated from crosslinked retinal microvessels from Sirt1 overexpressing mice was analyzed for the binding of PARP-1 at (a) NF-kB and (b) AP-1 binding regions of the MMP-9 promoter by ChIP technique. IgG was used as an antibody control (indicated as ^). (c) Effect of acetylation on the binding of NF-kB and of AP-1 with PARP-1 was determined by immunoprecipitating total proteins using PARP-1 antibody, followed by western blotting for NF-kB (p65) and of AP-1 (c-Jun). The values are represented as mean ± SD from 4–5 samples/group. Nor and Dia = C57BL/6J mice normal and diabetic respectively, and St-Nor and St-Dia = Sirt1 overexpressing mice normal and diabetic respectively. *P<0.05 compared to Nor and #P<0.05 compared to diabetes.
Discussion
In diabetes, the expression of retinal MMP-9 is increased and mitochondria are damaged. We have shown that, MMP-9 is transported into the mitochondria with the help of cytosolic heat shock (Hsp70/Hsp60) and mitochondrial membrane transporter proteins [4]. Once inside the mitochondria, MMP-9 damages mitochondrial membrane to help release of cytochrome c and initiate capillary cell apoptosis, a phenomenon, which precedes the development of diabetic retinopathy [36–39]. Expression of MMP-9 is regulated by multiple transcription factors, and in the development of diabetic retinopathy, increase in the binding of NF-kB and AP-1 transcription factors at the MMP-9 promoter induces its expression [10, 11]. Binding of these transcription factors are delimited by other co-factors including, PARP-1, and the activity of PARP-1 is increased in diabetes [19, 20]. Here, using in vivo and in vitro models, we show that in the hyperglycemic milieu, the binding of PARP-1 at NF-kB/AP-1 transcription factor binding regions of the MMP-9 promoter is increased, and inhibition of PARP-1 consequently inhibits this binding and ameliorates MMP-9 expression. Additionally, the study also highlights that due to decreased Sirt1 in diabetes, PARP-1 is acetylated, and this further facilitates the binding of the transcription factors at the MMP-9 promoter. Overexpression of Sirt1 prevents PARP-1 acetylation and its subsequent binding with the transcription factors. Thus, this study marks an important observation that the activation of transcription factor binding at MMP-9 is mediated through the active participation of PARP-1.
Cellular presence of PARP-1 is critically important for maintaining chromatin dynamics and transcriptional regulation of genes; PARP-1 deficiency alters transcription of p53 targets in mouse embryonic fibroblasts [40] and regulates heat shock factor 1 activity and its response in murine fibroblasts [41]. Efficiency of transcriptional regulation of genes is governed by the potency of the transcription factor binding and the formation of strong initiation complexes at the gene promoter. PARP-1 has an important function during assembly of the pre-initiation complex, where it facilitates the regulation of transcription factor binding [17, 33–35]. It efficiently interacts with NF-kB and AP-1 [15–18], and plays a pathophysiological role in the number of inflammatory disorders [33]. Additionally, several gene-specific and genomic studies have also demonstrates that PARP-1 efficiently binds to the promoter region of many genes and regulates their expression [42]. In diabetes, PARP-1 expression and activity are increased in the retina, and inhibition of PARP-1 activity inhibits development of diabetic complications including neuropathy, nephropathy and retinopathy [20, 43–45]. Here, we show that the inhibition of PARP-1 ameliorates diabetes-induced increase in MMP-9, suggesting an important role of PARP-1 in regulation of MMP-9 transcription. In a recent study, we have shown that the binding of NF-kB and AP-1 is increased at retinal MMP-9 promoter in diabetes, and here our result shows that PARP-1 binding at the NF-kB and AP-1 binding regions on the MMP-9 promoter is also significantly increased in the retinal vasculature. The integral role of PARP-1 in the regulation of transcription factor binding and development of diabetic retinopathy was further confirmed by manipulating the levels of PARP-1 pharmacologically or genetically in in vitro and in vivo model systems. Incubation of retinal endothelial cells with PJ34 or PARP-1 siRNA, or administration of PJ34 in vivo significantly inhibited the binding of NF-kB and AP-1 at the MMP-9 promoter, suggesting an important role of PARP-1 in promoting transcription initiation complex formation at the MMP-9 promoter. This is in agreement with a previous observation where PARP-1 deletion is shown to contribute to a defective activation of transcription factors critical in tumor development [46, 47]
PARP-1 is shown to be required for specific NF-kB transcriptional activation in vivo; NF-kB and PARP-1 form a stable immunoprecipitable nuclear complex and this interaction is required for NF-kB-DNA binding [34, 48]. Here, our study show that PARP-1 physically interacts with both NF-kB, and AP-1, and their binding is increased at the MMP-9 promoter. These results imply that in diabetes, PARP-1 assists the binding of the transcription factors at the MMP-9 promoter resulting in its increased expression.
Acetylation regulates the transcription process by changing the protein-DNA interaction activity, and acetylation of PARP-1 is required for its NF-kB coactivator activity [17, 33]. We show that in diabetes, PARP-1 acetylation is not only important for its binding with NF-kB, but also regulates its binding with AP-1. In support, others have shown that acetylation of PARP-1 increases its activity and its deacetylation by Sirt1, suppresses PARP-1-mediated gene activation [17, 22, 49]. In addition, increase in PARP-1 acetylation is also shown in hearts of Sirt1 knockout mice further, demonstrating that the activity of PARP-1 is regulated by Sirt1, [22]. The activity of Sirt1 is decreased in the retina and its microvasculature in diabetes, and regulation of Sirt1 by its pharmacological activator attenuates glucose-induced activation of MMP-9 [4, 5, 10]. Here our results demonstrate a strong correlation between Sirt1 regulation and PARP-1 acetylation along with its binding at the MMP-9 promoter and with transcription factors; overexpression of Sirt1 reduces PARP-1 binding at the MMP-9 promoter, and with NF-kB/AP-1 transcription factors, which ultimately decreases MMP-9 expression. Thus, Sirt1 appears to have a direct effect on PARP-1 binding, which, in turn, affects NF-kB/AP-1 binding at the MMP-9 promoter.
In summary, using in vivo and in vitro models of diabetic retinopathy, we have demonstrated a novel mechanism for transcriptional regulation of retinal MMP-9. We show that PARP-1 has an important role in binding of the transcription factors at the MMP-9 promoter and its transcription. Activation of PARP-1 in diabetes increases transcription factor binding, and acetylation of PARP-1 appears to be an important posttranslational modification for its regulation and the formation of transcription factor complex. Due to inhibition of Sirt1 in diabetes, PARP-1 remains acetylated, and this enables their binding at the MMP-9 promoter, resulting in increased binding of the transcription factors. Taken together, present study suggests that PARP-1 is an essential positive cofactor of NF-kB and AP-1 in the regulation of retinal MMP-9 expression in diabetes. These results also provide a fresh insight into the mechanism associated with the reduced function of Sirt1 during pathological processes in diabetic retinopathy.
Highlights.
PARP-1 augments NF-kB/AP-1 binding at MMP-9 promoter and induces its expression.
Sirt1 overexpression reduces PARP-1 and NF-kB/AP-1 complex formation.
PARP-1 inhibition prevents NF-kB/AP-1 binding at MMP-9 promoter.
Acknowledgments
The authors thank Mangayarkarasi Thandampallayam Ajjeya for her help with the maintenance of the animal colony. The work is supported in part by grants to RAK from the National Institutes of Health (R01-EY014370, R01-EY017313 and R01-EY022230), Thomas Foundation, and from Research to Prevent Blindness to the Ophthalmology department.
Footnotes
Conflict of interest
No financial interests for Manish Mishra or Renu A. Kowluru.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Brownlee M. The pathobiology of diabetic complications: a unifying mechanism. Diabetes. 2005;54:1615–1625. doi: 10.2337/diabetes.54.6.1615. [DOI] [PubMed] [Google Scholar]
- 2.Kowluru RA, Chan PS. Oxidative stress and diabetic retinopathy. Exp Diabetes Res. 2007;2007:43603. doi: 10.1155/2007/43603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kowluru RA. Role of matrix metalloproteinase-9 in the development of diabetic retinopathy and its regulation by H-Ras. Invest Ophthalmol Vis Sci. 2010;51:4320–4326. doi: 10.1167/iovs.09-4851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kowluru RA, Mohammad G, dos Santos JM, Zhong Q. Abrogation of MMP-9 gene protects against the development of retinopathy in diabetic mice by preventing mitochondrial damage. Diabetes. 2011;60:3023–3033. doi: 10.2337/db11-0816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kowluru RA, Santos JM, Zhong Q. Sirt1, a negative regulator of matrix metalloproteinase-9 in diabetic retinopathy. Invest Ophthalmol Vis Sci. 2014;55:5653–5660. doi: 10.1167/iovs.14-14383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Santos JM, Tewari S, Lin JY, Kowluru RA. Interrelationship between activation of matrix metalloproteinases and mitochondrial dysfunction in the development of diabetic retinopathy. Biochem Biophys Res Commun. 2013;438:760–764. doi: 10.1016/j.bbrc.2013.07.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Robert I, Aussems M, Keutgens A, Zhang X, Hennuy B, Viatour P, Vanstraelen G, Merville MP, Chapelle JP, de Leval L, Lambert F, Dejardin E, Gothot A, Chariot A. Metalloproteinase-9 gene induction by a truncated oncogenic NF-kappaB2 protein involves the recruitment of MLL1 and MLL2 H3K4 histone methyltransferase complexes. Oncogene. 2009;28:1626–1628. doi: 10.1038/onc.2009.6. [DOI] [PubMed] [Google Scholar]
- 8.Chakraborti S, Mandal M, Das S, Mandal A, Chakraborti T. Regulation of matrix metalloproteinases: an overview. Mol Cell Biochem. 2003;253:285–303. doi: 10.1023/a:1026028303196. [DOI] [PubMed] [Google Scholar]
- 9.Farina AR, Mackay AR. Gelatinase B/MMP-9 in Tumour Pathogenesis and Progression. Cancers (Basel) 2014;6:240–296. doi: 10.3390/cancers6010240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mishra M, Flaga J, Kowluru RA. Molecular mechanism of transcriptional regulation of matrix metalloproteinase-9 in diabetic retinopathy. J Cell Physiol. 2016;231:1709–1718. doi: 10.1002/jcp.25268. [DOI] [PubMed] [Google Scholar]
- 11.Zhong Q, Kowluru RA. Regulation of matrix metalloproteinase-9 by epigenetic modifications and the development of diabetic retinopathy. Diabetes. 2013;62:2559–2568. doi: 10.2337/db12-1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abate C, Patel L, Rauscher FJ, 3rd, Curran T. Redox regulation of fos and jun DNA-binding activity in vitro. Science. 1990;249:1157–1161. doi: 10.1126/science.2118682. [DOI] [PubMed] [Google Scholar]
- 13.Karin M, Takahashi T, Kapahi P, Delhase M, Chen Y, Makris C, Rothwarf D, Baud V, Natoli G, Guido F, Li N. Oxidative stress and gene expression: the AP-1 and NF-kappaB connections. Biofactors. 2001;15:87–89. doi: 10.1002/biof.5520150207. [DOI] [PubMed] [Google Scholar]
- 14.von Knethen A, Callsen D, Brune B. NF-kappaB and AP-1 activation by nitric oxide attenuated apoptotic cell death in RAW 264.7 macrophages. Mol Biol Cell. 1999;10:361–372. doi: 10.1091/mbc.10.2.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aguilar-Quesada R, Munoz-Gamez JA, Martin-Oliva D, Peralta-Leal A, Quiles-Perez R, Rodriguez-Vargas JM, Ruiz de Almodovar M, Conde C, Ruiz-Extremera A, Oliver FJ. Modulation of transcription by PARP-1: consequences in carcinogenesis and inflammation. Curr Med Chem. 2007;14:1179–1187. doi: 10.2174/092986707780597998. [DOI] [PubMed] [Google Scholar]
- 16.Andreone TL, O’Connor M, Denenberg A, Hake PW, Zingarelli B. Poly(ADP-ribose) polymerase-1 regulates activation of activator protein-1 in murine fibroblasts. J Immunol. 2003;170:2113–2120. doi: 10.4049/jimmunol.170.4.2113. [DOI] [PubMed] [Google Scholar]
- 17.Hassa PO, Haenni SS, Buerki C, Meier NI, Lane WS, Owen H, Gersbach M, Imhof R, Hottiger MO. Acetylation of poly(ADP-ribose) polymerase-1 by p300/CREB-binding protein regulates coactivation of NF-kappaB-dependent transcription. J Biol Chem. 2005;280:40450–40464. doi: 10.1074/jbc.M507553200. [DOI] [PubMed] [Google Scholar]
- 18.Zerfaoui M, Errami Y, Naura AS, Suzuki Y, Kim H, Ju J, Liu T, Hans CP, Kim JG, Abd Elmageed ZY, Koochekpour S, Catling A, Boulares AH. Poly(ADP-ribose) polymerase-1 is a determining factor in Crm1-mediated nuclear export and retention of p65 NF-kappa B upon TLR4 stimulation. J Immunol. 2010;185:1894–1902. doi: 10.4049/jimmunol.1000646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanwar M, Kowluru R. Role of glyceraldehyde 3-phosphate dehydrogenase in the development and progression of diabetic retinopathy. Diabetes. 2009;58:227–234. doi: 10.2337/db08-1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zheng L, Szabo C, Kern TS. Poly(ADP-ribose) polymerase is involved in the development of diabetic retinopathy via regulation of nuclear factor-kappaB. Diabetes. 2004;53:2960–2967. doi: 10.2337/diabetes.53.11.2960. [DOI] [PubMed] [Google Scholar]
- 21.Chen LF, Williams SA, Mu Y, Nakano H, Duerr JM, Buckbinder L, Greene WC. NF-kappaB RelA phosphorylation regulates RelA acetylation. Mol Cell Biol. 2005;25:7966–7975. doi: 10.1128/MCB.25.18.7966-7975.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rajamohan SB, Pillai VB, Gupta M, Sundaresan NR, Birukov KG, Samant S, Hottiger MO, Gupta MP. SIRT1 promotes cell survival under stress by deacetylation-dependent deactivation of poly(ADP-ribose) polymerase 1. Mol Cell Biol. 2009;29:4116–4129. doi: 10.1128/MCB.00121-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen W, Jump DB, Grant MB, Esselman WJ, Busik JV. Dyslipidemia, but not hyperglycemia, induces inflammatory adhesion molecules in human retinal vascular endothelial cells. Invest Ophthalmol Vis Sci. 2003;44:5016–5022. doi: 10.1167/iovs.03-0418. [DOI] [PubMed] [Google Scholar]
- 24.Mishra M, Kowluru RA. Retinal mitochondrial DNA mismatch repair in the development of diabetic retinopathy, and its continued progression after termination of hyperglycemia. Invest Ophthalmol Vis Sci. 2014;55:6960–6967. doi: 10.1167/iovs.14-15020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mohammad G, Kowluru RA. Matrix metalloproteinase-2 in the development of diabetic retinopathy and mitochondrial dysfunction. Lab Invest. 2010;90:1365–1372. doi: 10.1038/labinvest.2010.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tewari S, Santos JM, Kowluru RA. Damaged mitochondrial DNA replication system and the development of diabetic retinopathy. Antioxid Redox Signal. 2012;17:492–504. doi: 10.1089/ars.2011.4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kowluru RA, Shan Y, Mishra M. Dynamic DNA methylation of matrix metalloproteinase-9 in the development of diabetic retinopathy. Lab Invest. 2016;96:1040–1049. doi: 10.1038/labinvest.2016.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rodriguez MI, Peralta-Leal A, O’Valle F, Rodriguez-Vargas JM, Gonzalez-Flores A, Majuelos-Melguizo J, Lopez L, Serrano S, de Herreros AG, Rodriguez-Manzaneque JC, Fernandez R, Del Moral RG, de Almodovar JM, Oliver FJ. PARP-1 regulates metastatic melanoma through modulation of vimentin-induced malignant transformation. PLoS Genet. 2013;9:e1003531. doi: 10.1371/journal.pgen.1003531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kanwar M, Kowluru RA. Diabetes regulates small molecular weight G-protein, H-Ras, in the microvasculature of the retina: Implication in the development of retinopathy. Microvasc Res. 2008;76:189–193. doi: 10.1016/j.mvr.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mishra M, Kowluru RA. Epigenetic modification of mitochondrial DNA in the development of diabetic retinopathy. Invest Ophthalmol Vis Sci. 2015;56:5133–5142. doi: 10.1167/iovs.15-16937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhong Q, Mishra M, Kowluru RA. Transcription factor Nrf2-mediated antioxidant defense system in the development of diabetic retinopathy. Invest Ophthalmol Vis Sci. 2013;54:3941–3948. doi: 10.1167/iovs.13-11598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Santos JM, Mishra M, Kowluru RA. Posttranslational modification of mitochondrial transcription factor A in impaired mitochondria biogenesis: implications in diabetic retinopathy and metabolic memory phenomenon. Exp Eye Res. 2014;121:168–177. doi: 10.1016/j.exer.2014.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hassa PO, Hottiger MO. The functional role of poly(ADP-ribose)polymerase 1 as novel coactivator of NF-kappaB in inflammatory disorders. Cell Mol Life Sci. 2002;59:1534–1553. doi: 10.1007/s00018-002-8527-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hassa PO, Buerki C, Lombardi C, Imhof R, Hottiger MO. Transcriptional coactivation of nuclear factor-kappaB-dependent gene expression by p300 is regulated by poly(ADP)-ribose polymerase-1. J Biol Chem. 2003;278:45145–45153. doi: 10.1074/jbc.M307957200. [DOI] [PubMed] [Google Scholar]
- 35.Kraus WL, Lis JT. PARP goes transcription. Cell. 2003;113:677–683. doi: 10.1016/s0092-8674(03)00433-1. [DOI] [PubMed] [Google Scholar]
- 36.Kern TS, Tang J, Mizutani M, Kowluru R, Nagraj R, Lorenzi M. Response of capillary cell death to aminoguanidine predicts the development of retinopathy: Comparison of diabetes and galactosemia. Invest Ophthalmol Vis Sci. 2000;41:3972–3978. [PubMed] [Google Scholar]
- 37.Kowluru RA, Abbas SN. Diabetes-induced mitochondrial dysfunction in the retina. Invest Ophthalmol Vis Sci. 2003;44:5327–5334. doi: 10.1167/iovs.03-0353. [DOI] [PubMed] [Google Scholar]
- 38.Kowluru RA, Kanwar M. Oxidative stress and the development of diabetic retinopathy: Contributory role of matrix metalloproteinase-2. Free Radic Biol Med. 2009;46:1677–1685. doi: 10.1016/j.freeradbiomed.2009.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mizutani M, Kern TS, Lorenzi M. Accelerated death of retinal microvascular cells in human and experimental diabetic retinopathy. J Clin Invest. 1996;97:2883–2890. doi: 10.1172/JCI118746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Valenzuela MT, Guerrero R, Nunez MI, Ruiz De Almodovar JM, Sarker M, de Murcia G, Oliver FJ. PARP-1 modifies the effectiveness of p53-mediated DNA damage response. Oncogene. 2002;21:1108–1116. doi: 10.1038/sj.onc.1205169. [DOI] [PubMed] [Google Scholar]
- 41.Fossati S, Formentini L, Wang ZQ, Moroni F, Chiarugi A. Poly(ADP-ribosyl)ation regulates heat shock factor-1 activity and the heat shock response in murine fibroblasts. Biochem Cell Biol. 2006;84:703–712. doi: 10.1139/o06-083. [DOI] [PubMed] [Google Scholar]
- 42.Krishnakumar R, Kraus WL. PARP-1 regulates chromatin structure and transcription through a KDM5B-dependent pathway. Mol Cell. 2010;39:736–749. doi: 10.1016/j.molcel.2010.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kassan M, Choi SK, Galan M, Bishop A, Umezawa K, Trebak M, Belmadani S, Matrougui K. Enhanced NF-kappaB activity impairs vascular function through PARP-1-, SP-1-, and COX-2-dependent mechanisms in type 2 diabetes. Diabetes. 2013;62:2078–2087. doi: 10.2337/db12-1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Obrosova IG, Li F, Abatan OI, Forsell MA, Komjati K, Pacher P, Szabo C, Stevens MJ. Role of poly(ADP-ribose) polymerase activation in diabetic neuropathy. Diabetes. 2004;53:711–720. doi: 10.2337/diabetes.53.3.711. [DOI] [PubMed] [Google Scholar]
- 45.Szabo C, Biser A, Benko R, Bottinger E, Susztak K. Poly(ADP-ribose) polymerase inhibitors ameliorate nephropathy of type 2 diabetic Leprdb/db mice. Diabetes. 2006;55:3004–3012. doi: 10.2337/db06-0147. [DOI] [PubMed] [Google Scholar]
- 46.Abu El-Asrar AM, Mohammad G, Nawaz MI, Siddiquei MM, Van den Eynde K, Mousa A, De Hertogh G, Opdenakker G. Relationship between vitreous levels of matrix metalloproteinases and vascular endothelial growth factor in proliferative diabetic retinopathy. PloS one. 2013;8:e85857. doi: 10.1371/journal.pone.0085857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martin-Oliva D, Aguilar-Quesada R, O’Valle F, Munoz-Gamez JA, Martinez-Romero R, Garcia Del Moral R, Ruiz de Almodovar JM, Villuendas R, Piris MA, Oliver FJ. Inhibition of poly(ADP-ribose) polymerase modulates tumor-related gene expression, including hypoxia-inducible factor-1 activation, during skin carcinogenesis. Cancer Res. 2006;66:5744–5756. doi: 10.1158/0008-5472.CAN-05-3050. [DOI] [PubMed] [Google Scholar]
- 48.Hassa PO, Covic M, Hasan S, Imhof R, Hottiger MO. The enzymatic and DNA binding activity of PARP-1 are not required for NF-kappa B coactivator function. J Biol Chem. 2001;276:45588–45597. doi: 10.1074/jbc.M106528200. [DOI] [PubMed] [Google Scholar]
- 49.Luna A, Aladjem MI, Kohn KW. SIRT1/PARP1 crosstalk: connecting DNA damage and metabolism. Genome integr. 2013;4:6. doi: 10.1186/2041-9414-4-6. [DOI] [PMC free article] [PubMed] [Google Scholar]