Abstract
Species of the box jellyfish (Cubozoa) genus Alatina are notorious for their sting along the beaches of several localities of the Atlantic and Pacific. These species include Alatina alata on the Caribbean Island of Bonaire (the Netherlands), A. moseri in Hawaii, and A. mordens in Australia. Most cubozoans inhabit coastal waters, but Alatina is unusual in that specimens have also been collected in the open ocean at great depths. Alatina is notable in that populations form monthly aggregations for spermcast mating in conjunction with the lunar cycle. Nominal species are difficult to differentiate morphologically, and it has been unclear whether they are distinct or a single species with worldwide distribution. Here we report the results of a population genetic study, using nuclear and mitochondrial sequence data from four geographical localities. Our analyses revealed a general lack of geographic structure among Alatina populations, and slight though significant isolation by distance. These data corroborate morphological and behavioral similarities observed in the geographically disparate localities, and indicate the presence of a single, pan-tropically distributed species, Alatina alata. While repeated, human-mediated introductions of A. alata could explain the patterns we have observed, it seems more likely that genetic metapopulation cohesion is maintained via dispersal through the swimming medusa stage, and perhaps via dispersal of encysted planulae, which are described here for the first time in Alatina.
Introduction
Life-cycle and life-history characteristics have profound impacts on the basic biology of marine species, affecting geographic ranges and population dynamics. For instance, species possessing both benthic and planktonic life stages may be expected to display lower dispersal abilities and smaller geographic ranges than species lacking benthic stages (Gibbons et al., 2010). Many marine species possess pelagic larval stages that are often thought to be most responsible for dispersal (Bradbury et al., 2008; Bowen et al., 2013), but the mobility of all stages in a life cycle are relevant to determining species ranges (Johannesson, 1988). Species of the phylum Cnidaria exhibit a wide variety of life-cycle plasticity and dispersal capabilities (reviewed in Fautin, 2002). Jellyfish species (of the cnidarian subphylum Medusozoa) typically possess a benthic, sessile stage (polyp) that reproduces asexually by budding new polyps or medusae, the latter of which represent the sexually reproductive stage of the textbook medusozoan life cycle. Short-lived, ciliated larval forms known as planulae usually precede the benthic polyp stage. A sometimes overlooked life stage present in some medusozoans is the podocyst, a resting stage produced by polyps. Podocysts are essentially polyp-derived tissues encysted in a protective layer of perisarc, and are able to withstand extreme salinity changes and desiccation, sometimes for years (Dumont, 1994; Ikeda et al., 2011; Carrette et al., 2014).
For scyphozoan and cubozan species, the medusa stage is usually the life stage chosen for investigating global patterns of abundance, distribution, and diversity (Dawson et al., 2014; Lucas et al., 2014), because the polyp stage is often difficult to locate in the field. Recent focus has been on the sudden or periodic increases in biomass of scyphozoans that often have detrimental impacts on human activities; such increases are commonly known as “jellyfish blooms” (Condon et al., 2013; Dawson et al., 2014). Much debate exists surrounding the identification of the main factors driving jellyfish population dynamics, such as eutrophication, species introductions, and global climate change (for reviews of jellyfish blooms see Graham et al., 2001; Graham and Bayha, 2007; Purcell et al., 2007; Condon et al., 2013; Dawson et al., 2014; Lucas et al., 2014). While “blooms” of scyphozoans have been most heavily studied, the population dynamics of box jellyfish––in spite of their potentially large public health impacts due to their potent venoms and their generally coastal, shallow water distributions––remain poorly understood (Yoshimoto and Yanagihara, 2002; Bentlage et al., 2009; Gershwin et al., 2009). Periodic or seasonal population dynamics of box jellyfish have long been documented in, and associated with, tropical Australia (Barnes, 1966; Fenner, 1998; Fenner and Harrison, 2000), and an apparent increase in box jellyfish abundance on the Mediterranean Coast of Spain has been reported in recent years (Bordehore et al., 2011, 2015; Fontanet, 2014).
In several tropical to subtropical localities, species of the box jellyfish genus Alatina display monthly nearshore aggregations (Thomas et al., 2001; Chiaverano et al., 2013; Lewis et al., 2013; Carrette et al., 2014). Unlike typical jellyfish blooms, which consist of sudden, and often unpredictable, increases in single-species biomass linked to environmental conditions (Condon et al., 2013; Dawson et al., 2014), Alatina swarms are directly correlated with reproductive events related to the lunar cycle; mating aggregations occur 8 to 10 days after the full moon (Alatina moseri Mayer 1906 in Hawaii, A. mordens Gershwin 2005 in Australia, and A. alata (Reynaud, 1830) in Bonaire, the Netherlands). Though the animals are present in large numbers (hundreds to thousands of individuals) during these monthly reproductive swarms, the whereabouts of Alatina medusae in the interim is poorly known. Further, juveniles have been reported only on few occasions (Arneson, 1976; Arneson and Cutress, 1976; Lewis et al., 2013). Medusae of Alatina alata have been recorded swimming at depths greater than 540 m, and collected as deep as 1067 m (Morandini, 2003 as Carybdea alata; Lewis et al., 2013). Reports of box jellyfish at great depths are unusual; cubozoans are generally not thought to disperse across the open ocean (but see Bentlage et al., 2010). In addition, numerous Alatina specimens have been collected from surface waters in the open ocean and from depths as great as 2282 m (Lewis et al., 2013).
Among the eight nominal species of Alatina, A. alata is found in the eastern Atlantic and Caribbean, and displays what appears to be an identical life history to that of two Pacific species, A. moseri and A. mordens (Carrette et al., 2014). Bentlage et al. (2010) found that the two Pacific species share mtDNA (16S) haplotypes, and suggested that they belong to a widespread population from a single species, despite the large geographic distance separating them. In this contribution, we integrated molecular analyses and early development and morphological data to clarify the geographic distribution of Alatina from Bonaire, Hawaii, Saipan, and Australia. We show that Alatina does not follow the current paradigm, which restricts cnidarians with a bentho-pelagic life cycle to relatively narrow geographic ranges (Gibbons et al., 2010). Rather, we propose that they are members of a single, widespread, possibly circumtropical species known as Alatina alata.
Materials and Methods
Sampling and morphology
We sampled Alatina specimens from four localities (Fig. 1): Alatina alata from Karel’s Pier (Kralendijk, Bonaire, the Netherlands); Alatina mordens from Osprey Reef (Coral Sea, Queensland, Australia); Alatina moseri from Waikiki (Oahu, Hawaii), and Alatina sp. from Mañagaha Island (Saipan, Northern Mariana Islands). Medusae were collected individually by hand or by using dip nets at the sea surface from a boat, docks, or the beach. Specimens were preserved in 5%– 8% buffered formalin for morphological examination; a piece of tentacle was placed in 95% ethanol (EtOH) for DNA extractions. Other than two specimens from Saipan, which we tentatively identified as Alatina grandis (Agassiz & Mayer, 1902) (Bentlage et al., 2010) (see Results section), all specimens had the same general appearance in the field. The detailed morphology of Alatina samples was examined using the methods outlined in Bentlage and Lewis (2012) and Lewis et al. (2013). Bell height and bell width were measured, and the presence of taxon-defining morphological characters was confirmed by consulting the taxonomic keys provided in Gershwin (2005) and the recent redescription of Alatina alata (Lewis et al., 2013). A list of the museum specimens examined is provided in Appendix 1. Photography and videography were used to document the presence of A. alata in nearshore waters of Bonaire, the Netherlands (Fig. 2b), in connection with monthly spermcasting events (see Lewis et al., 2013 and this study). Male and female adults were placed in buckets of seawater and examined over a period of 24 hours. Embryos released from females (following internal fertilization) were photographed, and videos were taken to document developmental stages from blastulae to free-swimming planulae (following methods in Lewis et al., 2013).
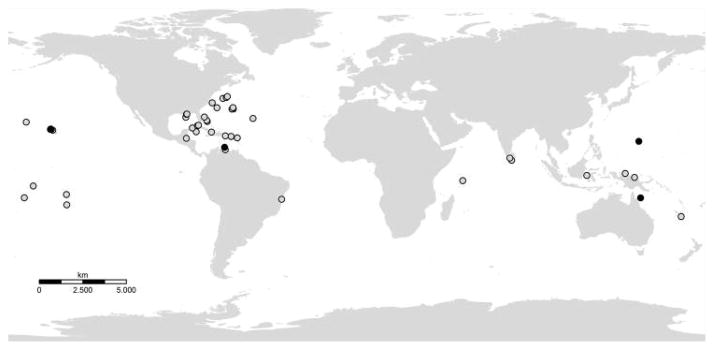
Figure 1.
Localities where Alatina medusae have been recorded in the literature or in museum collections (gray dots) and sampled for genetic analysis (black dots). Collection data and museum catalogue numbers, if applicable, are provided in Appendix 1.
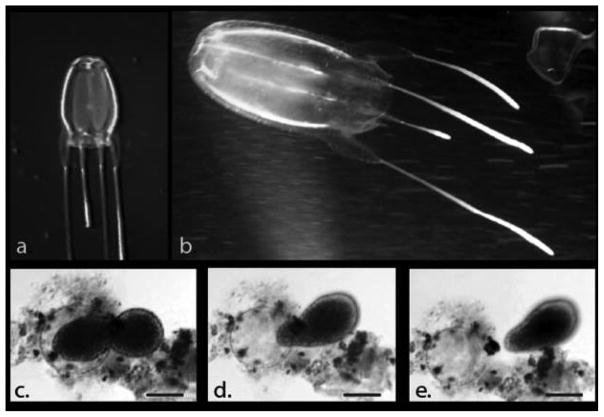
Figure 2.
Live Alatina alata medusae and encysted planula larva. (a) A. alata medusa recorded at a depth of 500–540 m, west off Gorda Cay, Bahamas, from the Johnson Sea Link I manned submersible (frame grab from video voucher USNM 1195809); (b) A. alata medusa next to diver 10–20 cm below the surface off Kralendijk, Bonaire, the Netherlands (Photograph courtesy of Jennifer Collins); (c–e) series of digital frame grabs taken from video footage of A. alata planula emerging from a cyst (scale bar = 50 μm).
DNA extraction, polymerase chain reaction (PCR), and sequencing
DNA was extracted from tentacle tissue, using DNeasy Tissue kits (Qiagen Inc., Valencia, CA), following the manufacturer’s protocol for animal tissues. Cubozoans are unusual in that they have linear mitochondrial genomes consisting of eight chromosomes (Smith et al., 2012). From three separate mitochondrial chromosomes, we amplified and sequenced three markers: 1) a 1005-bp fragment from the region containing two open-reading frames, ORF314 and POLB (ORF-PolB) (see Smith et al., 2012); 2) a 558-bp fragment from the cytochrome c oxidase subunit I (COI); and 3) a ~545-bp fragment from the ribosomal RNA (16S). ORF314 and POLB are present in the mitochondrial genomes of several other medusozoan groups; ORF314 may have DNA-binding properties to help maintain the ends of mitochondrial telomeres, and POLB codes for a putative DNA polymerase beta (Kayal et al., 2012). From the nuclear genome, we obtained a ~558-bp fragment spanning the internal transcribed spacers (ITS), ITS1 and ITS2, of the ribosomal RNA operon, including 5.8S, as well as a 710-bp fragment of the large ribosomal RNA subunit (28S) spanning the expansion regions D1–D3 (Cannone et al., 2002). Polymerase chain reaction protocols followed standard procedures. Thermocycler profiles were conducted with initialization at 94–95 °C (3–5 min), followed by 36 – 40 cycles of denaturation at 94–95 °C (30 s), annealing at 52–54 °C (30 s), and extension at 72 °C (1–2 min). The final extension was further executed at 72 °C (5–10 min). Polymerase chain reaction products were purified by combining 3 μl of 0.75 units (U) of Exonuclease I and 0.5 U of Shrimp Alkaline Phosphatase (ExoSAP; USB Corp., Cleveland, OH) with 8 μl of PCR product, followed by incubation at 37 °C for 30 min and deactivation at 80 °C for 20 min. Cycle sequencing was accomplished using the same primers as those used in PCRs (Table 1). Cycle sequencing products with fluorescently labeled dideoxy terminators were visualized after clean up, using Sephadex columns on an Applied Biosystems (Thermo Fisher Scientific, Waltham, MA) 3130xl or 3730xl Genetic Analyzer at the Smithsonian’s Laboratory of Analytical Biology (LAB), of the National Museum of Natural History, Washington, D.C.
Table 1.
Primers used for sequencing
| Marker | Primer name | Primer | Reference |
|---|---|---|---|
| ORF-PolB | AM_ORF314-F1 | AGCGCTATGATTAGAGTATTTAAGG | This study |
| AM_ORF314-R1 | TCAATTCTAGTTTAGAGCTTCCTC | This study | |
| AM_polB-F1 | ATCCTGTACTAAGCCAAATCATC | This study | |
| AM_polB-R1 | ATATAATCGGTCGTTAGTCGGC | This study | |
| COI | med-cox1-F | ACNAAYCAYAAAGATATHGG | This study |
| med-cox1-R | TGGTGNGCYCANACNATRAANCC | This study | |
| 16S | med-rnl-F | GACTGTTTACCAAAGACATAGC | This study |
| med-rnl-R | AAGATAGAAACCTTCCTGTC | This study | |
| ITS | C2 | GAAAAGAACTTTGRARAGAGAGT | Chombard et al., 1997 |
| D2 | TCCGTGTTTCAAGACGGG | Chombard et al., 1997 | |
| 28S | 28S-F63 | AATAAGCGGAGGAAAAGAAAC | Medina et al., 2001 |
| 28S-R635 | GGTCCGTGTTTCAAGACGG | Medina et al., 2001 |
ORF-PolB primers were designed based on Kayal et al. (2012). COI and 16S were designed based on conserved regions among medusozoan cnidarians that overlap well with the commonly used Folmer et al. (1994) and Palumbi (1996) fragments, respectively.
Molecular genetic analyses
Sequences were assembled, trimmed, and aligned in Geneious ver. 6.1.8 (Kearse et al., 2012). The number of haplotypes and haplotype and nucleotide diversity were calculated in DnaSP ver. 5.10.1 (Librado and Rozas, 2009). Allelic states of nuclear sequences with more than one heterozygous site were estimated using PHASE 2.1, as implemented in DnaSP, with three runs, each a unique random-number seed, for each dataset. Each of these runs was conducted for 1000 iterations with 1000 burn-in iterations, and all runs returned consistent allele identities. The best-fit models of DNA sequence evolution for each alignment were determined using the Akaike information criterion (AIC) with a correction for finite sample sizes (AICc), as implemented in jModelTest 2.1.7 (Guindon and Gascuel, 2003; Darriba et al., 2012). To evaluate whether multiple species of Alatina were present in our sampling, we obtained all publicly available 16S sequences from non-Alatina cubozoans deposited in GenBank. These were then aligned to sequences generated for the Alatina specimens collected for this study (Appendix 2) using MAFFT with the E-INSi option (Katoh and Standley, 2013). To exclude regions of uncertain homology of 16S across Cubozoa, Gblocks ver. 0.91b (Castresana, 2000; Talavera and Castresana, 2007) was run with standard parameters, except that half the taxa were allowed to be gaps for any position. The maximum likelihood topology (ML) was inferred using PHYML (Guindon et al., 2010), assuming the best-fitting model for this dataset, TIM2+I+G. Node support was assessed by conducting ML searches using 1000 nonparametric bootstrap replicates in PHYML. The resulting alignments used herein, as well as the 16S phylogenetic tree, are available through Figshare (Collins et al., 2016).
Because many sequences of the five markers were obtained from non-overlapping sets of specimens (see Appendix 2), we did not combine markers for analyses. Arlequin ver. 3.5.1.2 (Excoffier et al., 2005) was used to perform an analysis of molecular variance (AMOVA) (Weir and Cockerham, 1984; Excoffier et al., 1992; Weir, 1996) and to estimate population differentiation using pairwise FSTs, both tested with 20,000 nonparametric permutations. AMOVA was used to estimate the proportion of variation explained among groups (FCT), among localities within groups (FSC), and among all localities (FST). To determine the possibility of a correlation between pairwise FSTs and geographic distance, we used the Isolation-by-Distance Web Service with a 10,000-permutations significance test (Jensen et al., 2005). Geographic distances were measured, using Google Earth, “as the crow flies” between known occurrence sites of Alatina worldwide (Fig. 1). For mitochondrial data, these previous analyses were performed with an analogue of Wright’s FST (ΦST), which incorporates the model of sequence evolution. The best-fitting model calculated for the alignments was not available in Arlequin; therefore, the TrN was chosen, as it was the model available with the highest best-fit score (lowest AICc). Haplotype networks were constructed in Network ver. 4.6.1.3 (Flux Technology Ltd., Suffolk, England), using the median-joining algorithm (Bandelt et al., 1999). Networks were post-processed using maximum parsimony calculations (Polzin and Daneshmand, 2003) to remove unessential median vectors in the network.
Results
Morphology
We compared the morphology of the specimens in the Alatina moseri (Mayer, 1906) syntype (Hawaii) series in the Smithsonian’s National Museum of Natural History collections (items are denoted by catalog reference code beginning USNM; USNM 21800, 22311, 29632, 42112) with the Alatina alata neotype (USNM 1195802; see Lewis et al., 2013). Museum specimens of A. moseri had gonads, and the three medusae ranged from 70 – 82 mm (bell height; BH) by 22–26 mm (bell width; BW). According to Mayer (1906), live material measured BH = 80 mm by BW = 47 mm. These measurements are consistent with those of the live, gonad-bearing A. alata neotype (70 mm × 40 mm) and additional nontype material from the Atlantic Ocean (see Lewis et al., 2013). Mayer (1906) described the 24 velarial canals of A. moseri medusae as unbranched. However, we examined the syntype series and found that while some velarial canals are simple, many are split into two or three short, secondary branches (similar to A. alata). Further, velarial lappets and corresponding warts characteristic of A. alata are also present, or at least partially visible, although some have sloughed off in the center, leaving just an outline of the wart. The lack of pit eyes noted by Gershwin (2005) is an artefact often seen in long-preserved box jellyfish material (see Bentlage et al., 2010; Lewis et al., 2013; Carrette et al., 2014). Although Gershwin (2005) argued that, in A. mordens, cirri are arranged in pairs rather than in bunches, we noted no differences with the gastric phacellae of the A. moseri syntype series and the A. alata neotype. Carrette et al. (2014) also found no morphological differences when comparing adult medusae of A. moseri with those of A. mordens from Osprey Reef, Australia. Furthermore, the cnidome (nematocyst composition) of adult A. alata medusae (see Arneson, 1976; Lewis et al., 2013) is indistinguishable from that of A. mordens and A. moseri (Gershwin, 2005, 2006; Yanagihara et al., 2002 as Carybdea alata) bearing euryteles (in tentacles) and isorhizas (in bell warts, and tentacle base) (Gershwin, 2005; Lewis et al., 2013). During early development, polyps of A. mordens, A. moseri (Carrette et al., 2014), and A. alata (Arneson and Cutress, 1976 as Carybdea alata) bear stenoteles and ovoid, heterotrichous, microbasic euryteles.
In the course of our collections, using pelagic, tethered drift SCUBA dives at night in the Philippine Sea off the west coast of Saipan, we obtained two exemplars of a more distinctive form of Alatina, which we tentatively identified as ripe females of Alatina grandis. Specimens of Carybdea grandis were collected off Fakarava and Anaa Island, in the Tuamotu Archipelago (formerly Paumotu Islands) and described by Agassiz and Mayer (1902). There were no subsequent reports for over a century (see Bentlage, 2010). Measuring 180 mm (bell height) by 46 mm (bell width), approximately twice the size of the average Alatina specimens in our study, these new exemplars share many characters with the original species description, but additional samples (with key morphological characters preserved) are needed to firmly establish its identity. Our phylogenetic analysis confirms our conclusion, that these large specimens are distinct from the remainder of the collected Alatina specimens (see Phylogenetics and population genetics below).
Embryonic development
In this study, we documented that the early ontogeny of Bonaire Alatina alata embryos to the planula stage (Fig. 2, c–e) matched the findings of Arneson (1976) and Arneson and Cutress (1976) for the same species (as Carybdea alata) in Puerto Rico. The subsequent ontogenetic changes of A. alata from polyp to medusa stage (Arneson and Cutress, 1976) are also known to be identical in A. moseri from Hawaii and A. mordens from Australia (Carrette et al., 2014). Additionally, for all putative Alatina species, polyps can revert to a resting podocyst stage under adverse conditions (Carrette et al., 2014). During this study, while examining developing embryos in the lab approximately 24 h after their release into the aquarium water, we discovered multiple cysts (~150 μm in diameter), each containing a single planula bearing characteristic equatorial eye-spots, rotating on its longitudinal axis. During the course of our microscopic examination, one planula successfully bored through the outer, “shell-like” perisarc (within 5–50 min), emerging as swimming planula (Fig. 2, c–e). To our knowledge, this is the first documented occurrence of a planula hatching in a cnidarian. Whether this encysting stage is a form of diapause (i.e., a dormancy stage or delay in development in response to adverse environmental conditions) remains to be examined. Another open question is whether all or just some of maturing embryos develop a perisarc. The mechanisms involved in piercing the membrane were not investigated here, nor was the composition of the perisarc. Our findings resemble, to a certain extent, the “blastocysts” reported in Morbakka virulenta by Toshino et al. (2013), but in the case of M. virulenta, polyps (not zygotes) formed cysts that endured adverse conditions for many months, reemerging as polyps (not planulae).
Phylogenetics and population genetics
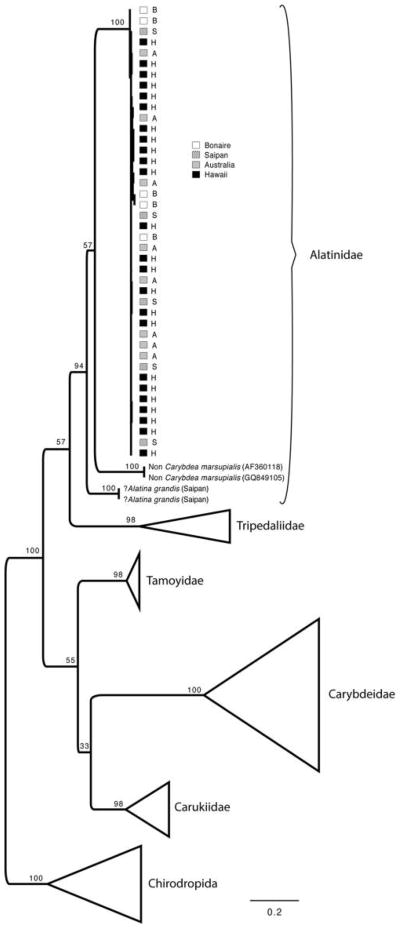
The 16S-ML phylogeny shows that specimens of Alatina alata from Bonaire, A. mordens from Australia, A. moseri from Hawaii, and Alatina sp. from Saipan fall into a single, well-supported clade with little differentiation among species and no apparent geographic structuring (Fig. 3). In contrast, the two large specimens that we tentatively identified as Alatina grandis were highly divergent from them (Fig. 3). While the placement of Alatina grandis is ambiguous due to low bootstrap support, it presents >20% sequence divergence from the remainder of Alatina spp., which in turn have <1.31% average sequence divergence in 16S (Table 2).
Figure 3.
Maximum-likelihood (ML) topology of the mitochondrial 16S of sampled Alatina specimens clustered within a cubozoan phylogeny, assuming the TIM2+I+G model of nucleotide evolution. ML nonpara-metric bootstrap support values are indicated for each node. Squares following legend indicate geographical origin of sampled specimens.
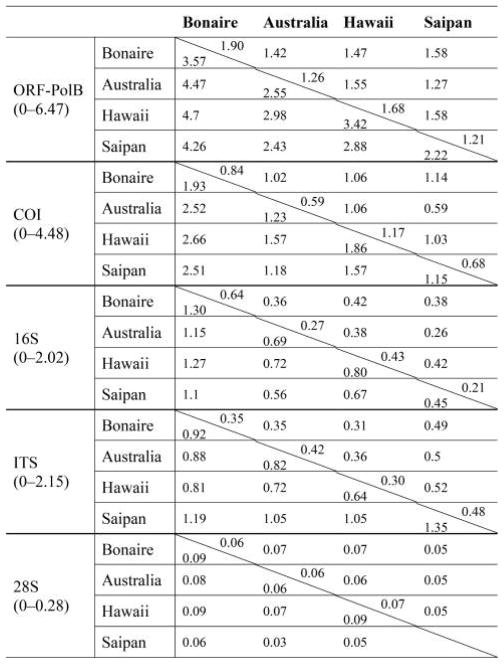
Table 2.
Percentage pairwise difference between sampling localities
Values below diagonal are the averages; values above diagonal are the standard deviations. Ranges below marker names are the minimum and maximum percentage pairwise differences recorded for the marker. Absent values indicate lack of sampling for average and standard deviation calculations.
In light of the 16S-based phylogenetic results, we removed A. grandis and all other cubozoans from further analysis, leaving just the representatives of Alatina alata from Bonaire, A. mordens from Australia, A. moseri from Hawaii, and Alatina sp. from Saipan, for which we realigned full-length sequences to preserve the largest amount of sequence information possible for further analysis. Table 3 provides the number of specimens sequenced per geographic region (N), number of haplotypes (Nh), haplotype diversity (h), and nucleotide diversity (π) for all markers. All mitochondrial markers and ITS had high overall haplotype diversity (h = 0.96 – 0.99; Table 3), with ORF-PolB and COI being the most diverse (0 – 6.47% and 0 – 4.48%, respectively; Table 2), 16S and ITS with intermediate divergence (0 –2.02% and 0 –2.15%, respectively; Table 2), and 28S the most conserved (0 – 0.28%; Table 2).
Table 3.
Molecular diversity indices
| Indices | Bonaire | Saipan | Australia | Hawaii | All | |
|---|---|---|---|---|---|---|
| ORF-PolB | N | 7 | 5 | 12 | 24 | 48 |
| Nh | 7 | 5 | 12 | 23 | 47 | |
| h | 1 ± 0.08 | 1 ± 0.13 | 1 ± 0.03 | 0.99 ± 0.01 | 0.99 ± 0 | |
| π | 0.03 ± 0.01 | 0.02 ± 0.01 | 0.02 ± 0 | 0.03 ± 0 | 0.03 ± 0 | |
| COI | N | 6 | 5 | 11 | 18 | 40 |
| Nh | 6 | 5 | 11 | 17 | 36 | |
| h | 1 ± 0.09 | 1 ± 0.13 | 1 ± 0.04 | 0.99 ± 0.02 | 0.99 ± 0.01 | |
| π | 0.02 ± 0 | 0.01 ± 0 | 0.01 ± 0 | 0.02 ± 0 | 0.02 ± 0 | |
| 16S | N | 5 | 5 | 8 | 24 | 42 |
| Nh | 4 | 5 | 8 | 15 | 26 | |
| h | 0.9 ± 0.16 | 1 ± 0.13 | 1 ± 0.06 | 0.94 ± 0.03 | 0.96 ± 0.02 | |
| π | 0.01 ± 0 | 0.01 ± 0 | 0.01 ± 0 | 0.01 ± 0 | 0.01 ± 0 | |
| ITS | N | 5 | 4 | 9 | 15 | 33 |
| Nh | 9 | 5 | 10 | 16 | 19 | |
| h | 0.98 ± 0.05 | 0.89 ± 0.09 | 0.92 ± 0.04 | 0.95 ± 0.02 | 0.97 ± 0.01 | |
| π | 0.01 ± 0 | 0.01 ± 0 | 0.01 ± 0 | 0.01 ± 0 | 0.01 ± 0 | |
| 28S | N | 7 | 1 | 13 | 23 | 44 |
| Nh | 3 | 1 | 4 | 8 | 9 | |
| h | 0.6 ± 0.08 | 0 ± 0 | 0.29 ± 0.11 | 0.52 ± 0.08 | 0.47 ± 0.06 | |
| π | 0.001 ± 0 | 0 ± 0 | 0.001 ± 0 | 0.001 ± 0 | 0 ± 0 |
Haplotype and nucleotide diversity are represented as mean ± standard deviation.
N, number of individuals sequenced; Nh, number of haplotypes or alleles; h, haplotype diversity; π, nucleotide diversity.
Overall ΦSTs and FSTs from the mitochondrial markers and nuclear ITS, respectively, indicated significant structure between sampled localities (ΦST = 0.086 – 0.17, FST = 0.088; Table 4). However, the majority of the overall variance was found within (>80%) rather than among geographic locations (Table 4). Pairwise comparisons across markers (measured by ΦST and FST; Table 5) showed that most of the Pacific sites were significantly different from Bonaire, but not significantly different from each other (samples were grouped by ocean basin: i.e., Pacific containing Saipan, Australia, and Hawaii vs. Atlantic containing Bonaire; see Materials and Methods). In this case, ΦST and FSTs were larger (ΦST = 0.25–0.35, FST = 0.11; Table 4), and variation within populations still explained the majority of the overall variance observed (>60%). ITS was the only marker to demonstrate significant structure within basins (FSC = 0.075; Table 4). We did not find significant evidence for isolation by distance among regions, other than for COI (Table 4).
Table 4.
AMOVA and Isolation by Distance (IBD) analyses
| ORF-PolB | COI | 16S | ITS | 28S | |
|---|---|---|---|---|---|
| All four sampled localities | |||||
| Among population variation | 14% | 17% | 9% | 9% | 3% |
| Within population variation | 86% | 83% | 91% | 91% | 97% |
| ΦST/FST | 0.137 | 0.170 | 0.086 | 0.088 | 0.032 |
| Atlantic vs. Pacific | |||||
| Among group variation | 31% | 35% | 27% | 4% | 12% |
| Within group variation | −1% | 0% | −2% | 7% | −2% |
| Within population variation | 70% | 65% | 75% | 89% | 90% |
| ΦST/FST | 0.302 | 0.354 | 0.249 | 0.110 | 0.099 |
| ΦSC/FSC | −0.010 | 0.001 | −0.031 | 0.075 | −0.027 |
| ΦCT/FCT | 0.309 | 0.353 | 0.272 | 0.038 | 0.122 |
| Isolation by distance (r) | 0.791 | 0.968 | 0.850 | 0.260 | 0.955 |
| P-value | 0.208 | 0.041 | 0.129 | 0.417 | 0.121 |
Analysis of molecular variance (AMOVA) is represented in the first part of the table with only one group encompassing the entire dataset including all four populations; then with two groups divided by ocean basin (i.e., Pacific containing Saipan, Australia, and Hawaii vs. Atlantic containing Bonaire). The mitochondrial AMOVA was calculated assuming the TrN model of nucleotide evolution. Molecular variance is divided into components of among groups (ΦCT and FCT), among localities within groups (ΦSC and FSC), and among all localities (ΦST and FST). Values in bold are significant (P < 0.05).
Table 5.
Pairwise ΦST and FST between sampling localities
| Bonaire | Australia | Hawaii | ||
|---|---|---|---|---|
| ORF-PolB | Bonaire | |||
| Australia | 0.33856 | |||
| Hawaii | 0.26632 | −0.00804 | ||
| Saipan | 0.30904 | 0.0088 | −0.01313 | |
|
| ||||
| COI | Bonaire | |||
| Australia | 0.40264 | |||
| Hawaii | 0.2944 | 0.00702 | ||
| Saipan | 0.37697 | −0.00964 | 0.00402 | |
|
| ||||
| 16S | Bonaire | |||
| Australia | 0.15865 | |||
| Hawaii | 0.24323 | −0.04435 | ||
| Saipan | 0.19975 | −0.05031 | 0.01688 | |
|
| ||||
| ITS | Bonaire | |||
| Australia | 0.0751 | |||
| Hawaii | 0.10827 | 0.02792 | ||
| Saipan | 0.15351 | 0.06368 | 0.18581 | |
|
| ||||
| 28S | Bonaire | |||
| Australia | 0.17493 | |||
| Hawaii | 0.05603 | −0.00225 | ||
| Saipan | 0.01712 | −0.31725 | −0.25806 | |
Values in bold are significant (P < 0.05). The Pairwise ΦSTs (mitochondrial) were calculated assuming the TrN model of nucleotide evolution.
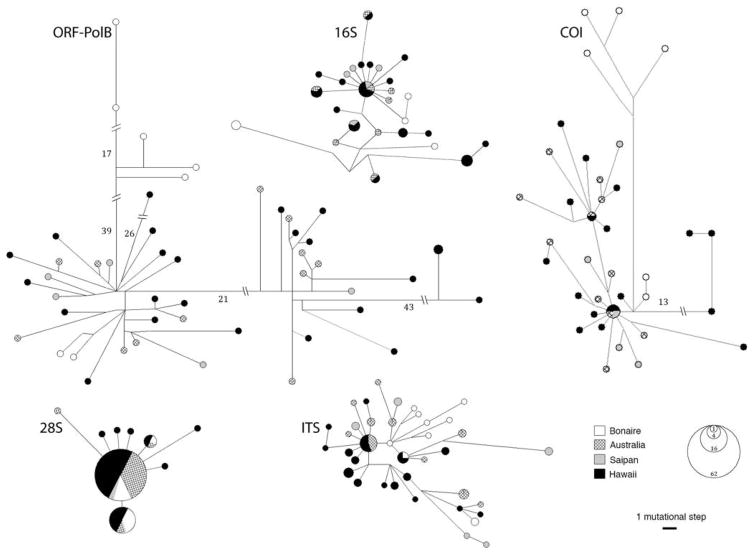
Haplotype networks (Fig. 4) show no well-defined geographic structure and each locality shares haplotypes with at least one other locality for one or more markers. Even though some specimens from Bonaire cluster together, others from this locality are more closely related to Pacific specimens. Nevertheless, pairwise ΦST and FST values involving Bonaire were mostly significant (at P < 0.05), with its lower range in the nuclear ITS (FST = 0.075–0.153) and upper range in the mitochondrial COI (ΦST = 0.294 – 0.403) (Table 5). Conversely, comparisons within the Pacific were primarily not significant, other than Hawaii–Saipan for ITS (FST = 0.186) (Table 5).
Figure 4.
Median-joining networks for mitochondrial (ORF-PolB, COI, and 16S) and nuclear (ITS and 28S) genes of Alatina specimens sampled. Each circle indicates one mitochondrial haplotype or nuclear allele, and symbols within indicate collection location (see key). The area of circles and symbols is proportional to its frequency in the dataset, according to circle size scale. Lines connecting haplotypes or alleles are proportional to the number of hypothesized mutational steps (see scale). Longer lines were reduced (indicated by parallel bars), and the number of mutational steps between closest nodes is indicated (not proportional to scale).
Revised systematics
Alatina alata diagnosis (from Lewis et al., 2013, table 1 and figs. 1– 6)
“Alatina with tall, narrow bell, flared at base, tapering into truncated pyramid at apex; 4 crescentric gastric phacellae at interradial corners of stomach; 3 simple to palmate branching velarial canals per octant, each with a velarial lappet bearing a row of 3 to 4 nematocyst warts; 4 long, wing-like (sensu Reynaud, 1830) pedalia, each with a pink tentacle. Cnidome: heterotrichous microbasic p– euryteles and small birhaploids in tentacles, and large isorhizas in nematocyst warts.” For more details on morphology and taxonomic history.
Neotype locality
Bonaire, the Netherlands (Atlantic Ocean).
Neotype specimen
National Museum of Natural History, Smithsonian Institution, Washington D.C.: USNM 1195802, 1 ind, female, BW 40 mm, BH 70 mm (live), BW 30 mm, BH 69 mm (8% formalin-preserved), 24 June 2011, Karel’s Pier, Kralendijk, Bonaire, the Netherlands, 12°09′06.37″ N, 68°16′06.37″ W; depth = surface.
Systematics
Phylum Cnidaria Verrill, 1865
Subphylum Medusozoa Peterson, 1979
Class Cubozoa Werner, 1973
Order Carybdeida Gegenbaur, 1857
Family Alatinidae Gershwin, 2005
Genus Alatina Gershwin, 2005
Species Alatina alata (Reynaud, 1830)
Synonymy list
Carybdea (medusa) alata
Reynaud, 1830 (in Lesson, 1830, pl. 33, fig. 1a)
La Marsupiale ailé
Lesson, 1837, p. 9, n. 26
Marsupialis alata
Lesson, 1843, p. 278
Charybdea alata
Haeckel, 1880, p. 441; 1940a, p. 5
Tamoya alata
Agassiz, 1862, p. 174
Carybdea alata
Mayer 1910, p. 508 –510; Mayer, 1915, p. 171; Bigelow, 1918, p. 400; 1938, pp. 144 –151, text–figs. 11–16; Kramp, 1961, p. 304; Arneson 1976, pp. 36, figs. 1, 2, table 1, 2, pls. I–V; Arneson and Cutress, 1976, pp. 227–236, table 1, pl. I A–G; Cutress, 1971, p. 19, pl. 1; Larson, 1976, pp. 242; Larson et al., 1991, p. 313, table 2; Thomas et al., 2001; Yoshimoto and Yanagihara, 2002; Humann and Deloach, 2002; Morandini, 2003, p. 15–17, fig. 2; Gershwin, 2005, pp. 501–523; Calder, 2009, pp. 12, 13, fig. 1; Bentlage, 2010, p. 52; Bentlage et al., 2010, p. 498; Bentlage and Lewis, 2012, p. 2602; Yanagihara and Shohet, 2012, pp. 1–2
Charybdea moseri
Mayer, 1906, pp. 1135–1136, pl. 1, fig. 2–2c; n. sp., description and illustrations; Bigelow, 1909, pp. 19–20, young stage of C. grandis; Bigelow, 1938, p. 144, junior synonym of C. alata; Chu and Cutress, 1954, p. 9, cause of dermatitis; Kramp, 1961, p. 304, in synonymy of C. alata
Carybdea moseri
Mayer, 1915, p. 171, probably young of C. alata var. grandis; Mayer, 1917, p. 189 [in part], fig. 3, only half-grown stage of C. alata
Carybdea alata var. moseri
Mayer, 1910, p. 512, probably a variety or young stage of C. grandis, probably identical with C. philippina; Light, 1914, p. 196 = Charybdea philippina [Semper 1860]; Mayer, 1915, p. 171, C. moseri is probably only a young of this medusa; Mayer, 1917, p. 189 [in part], fig. 3, only half-grown stage of C. alata; Stiasny, 1919, pp. 34, 37–38, fig. 5; Stiasny, 1940, pp. 5– 6; Bigelow, 1938, p. 144, in synonymy of C. alata
Alatina alata
Gershwin, 2005; Gershwin and Gibbons, 2009; Lewis et al., 2013; Yanagihara et al., 2016
Alatina moseri
Gershwin, 2005, 2006; Gershwin et al., 2009, 2013; Bentlage, 2010; Bentlage et al., 2010; Straehler-Pohl, 2011; Straehler-Pohl and Jarms, 2011; Kayal et al., 2012, 2013; Smith et al., 2012; Bentlage and Lewis, 2012; Kingsford et al., 2012; Yanagihara and Shohet, 2012; Chiaverano et al., 2013; Toshino et al., 2013, 2015; Carrette et al., 2014; Crow et al., 2015; Straehler-Pohl and Toshino, 2015
Alatina cf. moseri
Alatina mordens
Gershwin, 2005, 2006; Gershwin et al., 2009, 2013; Bentlage, 2010; Bentlage et al., 2010; Straehler-Pohl, 2011; Straehler-Pohl and Jarms, 2011; Bentlage and Lewis, 2012; Chiaverano et al., 2013; Toshino et al., 2013, 2015; Courtney and Seymour, 2013; Carrette et al., 2014; Crow et al., 2015
Alatina nr mordens
Alatina sp
Discussion
Historical background of the Alata species group
By the turn of the 19th century, 10 nominal species of the “alata” species group had been described for cubozoans from disparate geographic areas. These were eventually all united under the species name Carybdea alata (Bigelow, 1938; Kramp, 1961; Arneson, 1976). However, more recently Gershwin (2005) established the genus Alatina for all such cubomedusae, characterized by a tall and narrow bell, four crescentric, gastric phacellae, four “wing-like” pedalia, and three to four bifurcating to palmate velarial canals per octant. The taxonomic revision (Gershwin, 2005) resurrected five species of box jellyfish from the Indo-Pacific, previously synonymized under the name Carybdea alata, using the new genus-species combinations Alatina moseri, A. grandis, A. madraspatana, A. pyramis, and A. tetraptera. The revision also described two new Alatina species from Australia, A. mordens and A. rainensis, the former based mainly on medusa size, bell wart size, and an apparent reduced number of eyes per rhopalium (compared with other cubozoans). Lewis et al. (2013) established a neotype for the oldest species in the group, A. alata (Reynaud, 1830), bringing the number of currently recognized species to six. As body size is a factor of development influenced by environmental conditions, rhopalia eye spots are known to fade, and bell warts rub off following specimen preservation, doubts have been raised as to the validity of some of the nominal species, some of which have a single mention in the literature.
Alatina alata species complex
Bentlage et al. (2010) suggested that Alatina mordens from Australia and A. moseri from Hawaii likely represent a single species, and speculated about whether the two populations were connected at present, or were the result of human-mediated introductions. By increasing taxon and marker sampling from different ocean basins, we show that A. moseri and A. mordens from the Pacific, A. sp. from Saipan, as well as A. alata from its neotype locality (Bonaire, the Netherlands, Atlantic), are the same species. All nominal species sampled in this study (excluding A. grandis) form a single clade in our maximum likelihood (ML) analysis, with little divergence among geographic locations and lack of apparent geographic structuring (Fig. 3). Indeed, using several nuclear and mitochondrial markers we found that populations share haplotypes for all markers and lack a well-defined geographic clustering. All of the markers exhibit variability, particularly ORF-PolB and COI, but there are no clear divisions that would suggest the existence of multiple species among our samples (Fig. 4). Nevertheless, more comprehensive sampling of molecular data, especially of specimens from Bonaire and other localities in the Caribbean, could clarify population-level relationships among the distinct localities, to further investigate the possibility of a recent separation between populations in the different ocean basins in contrast to ongoing circumtropical gene flow.
Using a reverse taxonomic approach, we reevaluated the morphology of each of the nominal species and realized that they cannot be reliably delineated. Thus, in light of the genetic patterns presented and the uniformity in morphology, we conclude that the nominal species investigated herein all correspond to a single species, in spite of the large geographic distances among the populations sampled. Given that A. alata (Reynaud, 1830) is the oldest species described within the genus Alatina, A. moseri and A. mordens are to be considered junior synonyms of A. alata.
At this point, the status of the other nominal Alatina species (see Gershwin, 2005 for an overview) remains uncertain, as type material exists only for A. rainensis, which we have yet to examine or sample. However, we have tentatively identified the two large alatinid specimens from Saipan (USNM 1296954 and USNM 1296955) as A. grandis, a species that had been described from the Tuamotu Archipelago, French Polynesia, more than a century ago (Agassiz and Mayer, 1902), and whose type is badly damaged, making species identification difficult (Bentlage et al., 2010). Molecular data show that the specimens that we identified as A. grandis are distinct from A. alata within the family Alatinidae, and we expect that morphological examination of better-preserved samples will provide clear distinction between this species and A. alata.
Population structure and historical demography
Even though specimens from the different localities sampled share haplotypes in each of the markers analyzed, we detected some geographic structure, as measured by pairwise ΦST and FST. The Atlantic population shows the greatest separation from those of the Pacific localities. Nevertheless, even though Isolation by Distance (IBD) was significant and high for COI, it was not detected for any other marker, and no significant difference was revealed between the ocean basins in our AMOVA. Overall, there is no well-defined geographic structure, although some degree of divergence exists between Atlantic and Pacific specimens. This divergence could be due to lower rates of gene flow among distant localities or incomplete sorting of a large ancestral population.
Based on our findings, we suggest that Alatina alata is a single widespread species, found in several tropical and subtropical locations in the Pacific and Atlantic Oceans. Even though our findings are contrary to other studies of widespread marine invertebrates (Dawson and Jacobs, 2001; Goetze, 2011), similar results have been reported in fishes (Theisen et al., 2008; Lewallen, 2012). Numerous other cnidarian species with a bentho-pelagic life cycle are globally distributed, a phenomenon that is frequently attributed to repeated species introductions, often through commercial shipping activities (Bayha and Graham, 2013). The most likely means of cnidarian species introductions is via transport of polyps or cysts in ballast water or attachment to the hull of ships (Bayha and Graham, 2013). Under a scenario of species introductions, one would assume reduced haplotype diversities in the regions where it occurred due to the bottleneck created by the introduction of a few propagules into a locality. However, we observed very high haplotype diversities in all sampled localities and across markers. Thus, either there have been multiple separate introductions of Alatina alata from unknown source populations or A. alata is indeed capable of maintaining population cohesion across ocean basins. Of note is the observation of Smith et al. (2012), that Alatina alata (as A. moseri) mitochondrial haplotype diversity is nearly the highest of any metazoan species ever measured to date, which could be the result of extremely large, effective population size. What is clear from the literature is that A. alata has been present both in the Atlantic (Lewis et al., 2013) and at Hawaii for more than a century (Chiaverano et al., 2013). This indicates that putative introductions from one ocean basin to the other would have taken place prior to present-day commercial shipping activities. Unfortunately, the history of A. alata in other localities is not well documented, although large aggregations have been reported in Australia since 1999 (see Carrette et al., 2014, as A. mordens).
Do life-history characteristics favor dispersal abilities in Alatina alata?
Box jellyfish differ from the majority of scyphozoan jellyfish that possess a bentho-pelagic life cycle, which is defined by a sessile polyp stage that metamorphoses into one or more free-swimming medusae. In box jellyfish the sessile, asexually reproducing polyp (cubopolyp) generates either a single medusa through complete metamorphosis, or multiple medusae via metamorphosis coupled with transverse fission at the apical end of the polyp (Straehler-Pohl and Jarms, 2005). Conversely, in both scyphozoans and cubozoans sexual reproduction occurs exclusively during the adult medusa stage. In the case of A. alata, reproductive success is achieved by the formation of highly synchronized, monthly inshore spermcasting aggregations, occurring 8 –10 days after the full moon, during which males release sperm that is taken up by females for internal fertilization. These aggregations have been documented in several Atlantic and Pacific localities, including Bonaire, Hawaii, and Australia (Bentlage et al., 2010; Chiaverano et al., 2013; Lewis et al., 2013; Carrette et al., 2014). Reports of live A. alata medusae in the interim are rare, limited to a few documented cases in which a remotely operated vehicle (ROV) at ~100 m (USNM 1005621) and a manned submersible at ~540 m (USNM 1195809) were used (Lewis et al., 2013; Fig. 2a); therefore, A. alata is considered a deep-sea box jellyfish species (see Bentlage et al., 2010).
In the present study, we observed free-floating, encysted planulae (Fig. 2, c–e) with a morphology different from previously reported encysted life stages in box jellyfish, such as podocysts, which are sessile, encysted polyps (see Carrette et al., 2014). While the latter provides a good means of protection during bad conditions, and could potentially foul the hull of ships, the inherent ability of free-floating, encysted planulae to be immediately dispersed to the open ocean (by tides and currents) could ensure effective and broad distribution of planulae before their imminent settlement as polyps. That said, planula larvae themselves are short-lived in A. alata, from 2–3 days (Carrette et al., 2014), to 5–6 days (Arneson, 1976), while the lifespan of the encysted planulae newly described herein is unknown. Since the dispersal potential of larvae is generally highly correlated with the duration of its pelagic stage (Scheltema, 1971; Grantham et al., 2003), planulae are unlikely vectors for long-distance dispersal in A. alata.
Medusae of Alatina alata are strong swimmers (Chiaverano et al., 2013) with a potentially long lifespan (~1 year; see Arneson, 1976), and have been reported swimming at great depths and in the open ocean (see Lewis et al., 2013 and Fig. 1). This suggests that adults might contribute to dispersal, such as has been hypothesized for other marine organisms (see Kinlan et al., 2005). The deepwater habit of A. alata appears to be uncommon for box jellyfish, as most are reported, and predicted, to inhabit shallow, nearshore waters (Yoshimoto and Yanagihara, 2002; Bentlage et al., 2009; Gershwin et al., 2009). Recently, a Chironex box jellyfish was reported in waters at depths of around 50 m, which, although comparatively much shallower than the depths at which A. alata has been documented (Lewis et al., 2013), is much deeper than previously documented for Chironex (Keesing et al., 2016). Whether adult medusae, cysts, or a combination of the two provide a natural means for maintaining global population cohesion in A. alata is unclear at this point, although the deep-sea tendencies of A. alata medusae may partly explain how these widespread populations maintain genetic connectivity. Indeed, several deep-sea hydrozoan jellyfish appear to have distributions spanning ocean basins (Collins et al., 2008). In addition, jellyfish species living in the deep sea may also be globally distributed by natural means due to habitat homogeneity (Bentlage et al., 2013). However, these additional examples of widespread species distributions pertain specifically to cnidarians with holopelagic development, in which the benthic polyp stage has been lost.
Concluding Remarks
We conclude that Alatina alata is a single species, comprising multiple nominal species, with a wide geographical distribution. A. mordens and A. moseri should be regarded as junior synonyms of A. alata. Currently, it is impossible to determine with certainty whether the observed widespread distribution of A. alata is a result of natural dispersal mechanisms or repeated anthropogenic introductions occurring as long as 100 years ago. Future studies with increased locus sampling allowed by current sequencing technologies such as the ezRAD (Toonen et al., 2013), are needed to properly address this question. Further, expanding the geographic range to include Alatina samples from intermediate ocean basins, that is, the Indian and eastern Atlantic Oceans, should lead to a better understanding of the dispersal patterns and historical demography of this apparently cosmopolitan species.
Acknowledgments
Geoff Keel and the rest of the collections staff of the Department of Invertebrate Zoology at the Smithsonian NMNH are gratefully acknowledged for assistance in working with specimens. This work would not have been possible without the generous support of Rita Peachey and staff of CIEE Bonaire; citizen scientists Bud Gillan, Johan van Blerk, and Arjen van Dorsten; and the cooperation of the staff of STINAPA Bonaire. We also acknowledge Júlia Souza’s assistance, from the Marine Biodiversity Lab at UFSC, for phylogeographic analysis. Much of this work was performed using resources of the Laboratories of Analytical Biology at the Smithsonian NMNH. Saipan research was financially supported by the Brigham Young University–Hawaii Student Associateship Research Fund, Yamagata Foundation, and Biology Department Student Mentored Research Fund. Collection support by BYU–Hawaii students include Sheung Ting, Abigail Smith, Haley Sorenson-Pruitt, Tavaiilau Lueli, and Teylon Wilson.
Appendix 1. Alatina alata records from museum collections or the literature
| Species | Described as | Locality | Latitude | Longitude | Depth (m) | Reference (museum collection code no. or literature) |
|---|---|---|---|---|---|---|
| Alatina alata | Charybdea alata | Samoa | −13.83 | −171.76 | 16 | Stiasny, 1940 |
| Alatina alata | Alatina moseri | Hawaii | 25.12 | −170.83 | – | USNM 22311 |
| Alatina alata | Charybdea alata | South Pacific Ocean | −7.77 | −167.17 | 3800 | Stiasny, 1940 |
| Alatina alata | Carybdea moseri | Hawaii | 21.38 | −158.32 | – | USNM 22309 |
| Alatina alata | Alatina alata | Hawaii | 21.59 | −158.11 | – | USNM 1245460 |
| Alatina alata | Alatina moseri | Hawaii | 21.28 | −157.84 | – | USNM 1124426–1124451, 1245923, 1245925, 1155723, 42112, 51962 |
| Alatina alata | Alatina moseri | Hawaii | 20.94 | −157.08 | – | USNM 21800, 29632 |
| Alatina alata | Alatina moseri | South Pacific Ocean | −12.18 | −150.00 | 860 | USNM 1195808 |
| Alatina alata | Carybdea alata | French Polynesia | −17.48 | −149.84 | 0 | FLMNH 8380 |
| Alatina alata | Alatina alata | Mississippi (Gulf of Mexico) | 27.64 | −88.35 | 0 | USNM 1131245 |
| Alatina alata | Carybdea alata | Belize | 16.79 | −88.08 | – | USNM 58207–58211 |
| Alatina alata | Alatina alata | Mississippi (Gulf of Mexico) | 29.16 | −88.02 | 98–133 | USNM 1131246 |
| Alatina alata | Alatina alata | Mississippi (Gulf of Mexico) | 29.32 | −87.76 | 96.5–108.7 | USNM 1005621 |
| Alatina alata | Charybdea alata | Cuba | 22.10 | −84.97 | – | Stiasny, 1940 |
| Alatina alata | Charybdea alata | Cuba | 20.13 | −82.98 | – | Stiasny, 1940 |
| Alatina alata | Charybdea alata | Cuba | 23.22 | −82.35 | – | Stiasny, 1940 |
| Alatina alata | Carybdea alata | Cuba | 23.53 | −81.80 | – | USNM 41920 |
| Alatina alata | Carybdea alata | Bahamas | 27.77 | −78.77 | – | USNM 41921 |
| Alatina alata | Carybdea alata | Bahamas | 26.06 | −77.55 | 457–610 | USNM 1195809 |
| Alatina alata | Carybdea alata | Bahamas | 25.45 | −77.27 | – | USNM 41919 |
| Alatina alata | Carybdea alata | Cuba | 20.00 | −75.12 | – | USNM 94780 |
| Alatina alata | Carybdea alata | North Carolina | 35.05 | −74.68 | 204–228 | USNM 53694 |
| Alatina alata | Carybdea alata | South Carolina | 32.55 | −72.23 | 0–100 | USNM 42017 |
| Alatina alata | Carybdea alata | Northwest Atlantic | 37.35 | −69.17 | 0–90 | USNM 56737 |
| Alatina alata | Alatina alata | Bonaire | 12.19 | −68.30 | – | USNM 1248604, 1248677 |
| Alatina alata | Alatina alata | Bonaire | 12.15 | −68.28 | – | USNM 1195801–1195907, 1205447–1205450, 1156074, 1156075 |
| Alatina alata | Carybdea alata | Venezuela | 10.90 | −67.97 | – | USNM 53659 |
| Alatina alata | Carybdea alata | Puerto Rico | 18.07 | −67.88 | – | USNM 54398, 54472 |
| Alatina alata | Carybdea alata | Northwest Atlantic | 37.83 | −67.42 | 0–150 | USNM 56735 |
| Alatina alata | Carybdea alata | Northwest Atlantic | 38.31 | −66.86 | 0–50 | USNM 56736 |
| Alatina alata | Charybdea alata | Virgin Islands | 17.72 | −64.93 | – | Stiasny, 1940 |
| Alatina alata | Charybdea alata | Virgin Islands | 17.75 | −64.92 | 950 | Stiasny, 1940 |
| Alatina alata | Carybdea alata | Bermuda | 31.88 | −64.45 | 327–335 | USNM 58691 |
| Alatina alata | Carybdea alata | Bermuda | 31.93 | −64.42 | 55 | USNM 58692, 58316 |
| Alatina alata | Carybdea alata | Bermuda | 32.58 | −63.97 | 550–675 | USNM 58655 |
| Alatina alata | Carybdea alata | Bermuda | 31.92 | −63.95 | 0–300 | USNM 54367 |
| Alatina alata | Carybdea alata | Bermuda | 31.93 | −63.77 | 350 | USNM 54366 |
| Alatina alata | Carybdea alata | Antigua and Barbuda | 17.00 | −61.76 | – | USNM 54385 |
| Alatina alata | Charybdea alata | North Atlantic Ocean | 27.03 | −53.65 | – | Stiasny, 1940 |
| Alatina alata | Carybdea alata | Brazil | −14.62 | −38.83 | 1067 | Morandini, 2003 |
| Alatina alata | Charybdea alata | Seychelles | −5.02 | 54.77 | 1880 | Stiasny, 1940 |
| Alatina alata | Charybdea alata | Sri Lanka | 6.60 | 79.10 | 2530 | Stiasny, 1940 |
| Alatina alata | Charybdea alata | Sri Lanka | 5.47 | 80.00 | 4000 | Stiasny, 1940 |
| Alatina alata | Carybdea alata | Indonesia | −2.33 | 118.83 | – | USNM 42094 |
| Alatina alata | Charybdea alata | Papua | −1.33 | 138.70 | 3450 | Stiasny, 1940 |
| Alatina alata | Alatina alata | Papua New Guinea | −3.38 | 143.53 | 1–2 | USNM 1296950, 1296951 |
| Alatina alata | Alatina alata | Saipan | 15.24 | 145.71 | – | USNM 1296956–1296960 |
| Alatina alata | Alatina mordens | Australia | −13.90 | 146.63 | – | USNM 1124410–1124425 |
| Alatina alata | Charybdea alata | New Caledonia | −23.53 | 167.60 | 1060 | Stiasny, 1940 |
(USNM, collection catalog coding of the National Museum of Natural History, Smithsonian Institution, Washington, D.C.; FLMNH, Florida Museum of Natural History, Gainesville, FL). Latitude and longitude are presented in decimal degrees.
Appendix 2. Sequences from Alatina alata and Alatina grandis specimens collected in this study and from other cubozoans available in GenBank
Literature Cited
- Agassiz AE, Mayer AG. Reports on the scientific results of the expedition to the tropical Pacific in charge of Alexander Agassiz, by the U.S. Fish Commission steamer “Albatross,” from August, 1899, to March, 1900, Commander Jefferson F. Moser, U. S. N., Commanding. III. Medusa. Mem Mus Comp Zool Harv Coll. 1902;26:139–176. [Google Scholar]
- Agassiz L. Contributions to the Natural History of the United States of America. IV. Little, Brown and Co; Boston: 1862. [[2016, Oct. 13]]. [Online]. Available: http://www.biodiversitylibrary.org/item/54510#page/9/mode/1up. [Google Scholar]
- Arneson AC. Master’s thesis. University of Puerto Rico; Mayaguez: 1976. Life history of Carybdea alata Reynaud, 1830 (Cubomedusae) [Google Scholar]
- Arneson AC, Cutress CE. Life history of Carybdea alata Reynaud, 1830 (Cubomedusae) In: Mackie GO, editor. Coelenterate Ecology and Behaviour. Springer; New York: 1976. pp. 227–236. [Google Scholar]
- Bandelt HJ, Forster P, Röhl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- Barnes JH. Studies on three venomous cubomedusae. Symp Zool Soc Lond. 1966;16:307–332. [Google Scholar]
- Bayha KM, Graham WM. Nonindigenous marine jellyfish: invasiveness, invasibility, and impacts. In: Pitt KA, Lucas CH, editors. Jellyfish Blooms. Springer; New York: 2013. pp. 45–77. [Google Scholar]
- Bentlage B. Carybdea alata auct. (Cubozoa): rediscovery of the Alatina grandis type. Zootaxa. 2010;2713:52–54. [Google Scholar]
- Bentlage B, Lewis C. An illustrated key and synopsis of the families and genera of carybdeid box jellyfishes (Cnidaria: Cubozoa: Carybdeida), with emphasis on the “Irukandji family” (Carukiidae) J Nat Hist. 2012;46:2595–2620. [Google Scholar]
- Bentlage B, Peterson AT, Cartwright P. Inferring distributions of chirodropid box-jellyfishes (Cnidaria: Cubozoa) in geographic and ecological space using ecological niche modeling. Mar Ecol Prog Ser. 2009;384:121–133. [Google Scholar]
- Bentlage B, Cartwright P, Yanagihara AA, Lewis C, Richards GS, Collins AG. Evolution of box jellyfish (Cnidaria: Cubozoa), a group of highly toxic invertebrates. Proc R Soc B Biol Sci. 2010;277:493–501. doi: 10.1098/rspb.2009.1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentlage B, Peterson AT, Barve N, Cartwright P. Plumbing the depths: extending ecological niche modelling and species distribution modelling in three dimensions. Global Ecol Biogeogr. 2013;22:952–961. [Google Scholar]
- Bigelow HB. Reports of the scientific results of the Expedition to the Eastern Tropical Pacific, in charge of Alexander Agassiz, by the U. S. Fish Commission Steamer “Albatross,” from October, 1904, to March, 1905, Lieut. Commander L. M. Garrett, U.S.N., commanding. XVI. The Medusae. Mem Mus Comp Zool. 1909;37:1–243. [Google Scholar]
- Bigelow HB. Some Medusae and Siphonophora from the western Atlantic. Bull Mus Comp Zool Harv Coll. 1918;62:363–442. [Google Scholar]
- Bigelow HB. Plankton of Bermuda Oceanographic Expeditions. VIII. Medusae taken during the years 1929 and 1930. Zoologica. 1938;23:99–189. [Google Scholar]
- Bordehore C, V, Fuentes L, Atienza D, Barberá C, Fernandez-Jover D, Roig M, Acevedo-Dudley MJ, Canepa AJ, Gili JM. Detection of an unusual presence of the cubozoan Carybdea marsupialis at shallow beaches located near Denia, Spain (southwestern Mediterranean) Mar Biodivers Rec. 2011;4:e69. [Google Scholar]
- Bordehore C, V, Fuentes L, Segarra JG, Acevedo M, Canepa A, Raventós J. Use of an inverse method for time series to estimate the dynamics of and management strategies for the box jellyfish Carybdea marsupialis . PLoS One. 2015;10:e0137272. doi: 10.1371/journal.pone.0137272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen BW, Rocha LA, Toonen RJ, Karl SA the ToBo Laboratory. The origins of tropical marine biodiversity. Trends Ecol Evol. 2013;28:359–366. doi: 10.1016/j.tree.2013.01.018. [DOI] [PubMed] [Google Scholar]
- Bradbury IR, Laurel B, Snelgrove PVR, Bentzen P, Campana SE. Global patterns in marine dispersal estimates: the influence of geography, taxonomic category and life history. Proc R Soc B Biol Sci. 2008;275:1803–1809. doi: 10.1098/rspb.2008.0216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calder DR. Cubozoan and scyphozoan jellyfishes of the Carolinian Biogeographic Province, southeastern USA. R Ont Mus Contrib Sci. 2009;3:1–58. [Google Scholar]
- Cannone JJ, Subramanian S, Schnare MN, Collett JR, D’Souza LM, Du Y, Feng B, Lin N, Madabusi LV, Müller KM, et al. The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics. 2002;3:2. doi: 10.1186/1471-2105-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrette T, Straehler-Pohl I, Seymour J. Early life history of Alatina cf. moseri populations from Australia and Hawaii with implications for taxonomy (Cubozoa: Carybdeida, Alatinidae) PLoS One. 2014;9:e84377. doi: 10.1371/journal.pone.0084377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- Chiaverano LM, Holland BS, Crow GL, Blair L, Yanagihara AA. Long-term fluctuations in circalunar beach aggregations of the box jellyfish Alatina moseri in Hawaii, with links to environmental variability. PLoS One. 2013;8:e77039. doi: 10.1371/journal.pone.0077039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chombard C, Boury-Esnault N, Tillier A, Vacelet J. Polyphyly of “Sclerosponges” (Porifera, Demospongiae) supported by 28S ribosomal sequences. Biol Bull. 1997;193:359–367. doi: 10.2307/1542938. [DOI] [PubMed] [Google Scholar]
- Chu GW, Cutress CE. Human dermatitis caused by marine organisms in Hawaii. Proc Hawaii Acad Sci. 1954;9:1953–54. [Google Scholar]
- Collins AG, Bentlage B, Lindner A, Lindsay D, Haddock SHD, Jarms G, Norenburg JL, Jankowski T, Cartwright P. Phylogenetics of Trachylina (Cnidaria: Hydrozoa) with new insights on the evolution of some problematical taxa. J Mar Biol Assoc UK. 2008;88:1673–1685. [Google Scholar]
- Collins AG, Lawley J, Bentlage B, Kayal E, Lewis CA, Yanagihara A, Goodwill R, Hurwitz K. Genetic data for Alatina alata, a box jellyfish with a circumtropical distribution. Figshare; 2016. [[2016, Feb. 3]]. [Online]. Available: https://figshare.com/articles/Genetic_Data_for_Alatina_alata_a_box_jellyfish_with_a_circumtropical_distribution/3410104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condon RH, Duarte CM, Pitt KA, Robinson KL, Lucas CH, Sutherland KR, Mianzan HW, Bogeberg M, Purcell JE, Decker MB, et al. Recurrent jellyfish blooms are a consequence of global oscillations. Proc Natl Acad Sci USA. 2013;110:1000–1005. doi: 10.1073/pnas.1210920110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtney R, Seymour J. Seasonality in polyps of a tropical cubozoan: Alatina nr mordens. PLoS One. 2013;8:e69369. doi: 10.1371/journal.pone.0069369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow GL, Chiaverano LM, Crites J, Khramov MA, Holland BS. Box jellyfish (Cubozoa: Carybdeida) in Hawaiian waters, and the first record of Tripedalia cystophora in Hawai‘i. Bishop Mus Bull Zool. 2015;108:93–108. [Google Scholar]
- Cutress CE. Investigation of the Biology and Control of Noxious Coelenterates Occurring in the Coastal Waters of Puerto Rico. Department of Agriculture, Commonwealth of Puerto Rico; Mayaguez, P.R: 1971. Second Annual Report (January 1 to December 31, 1970). Jellyfish Project JF 2– 6. [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 2012;9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson MN, Jacobs DK. Molecular evidence for cryptic species of Aurelia aurita (Cnidaria, Scyphozoa) Biol Bull. 2001;200:92–96. doi: 10.2307/1543089. [DOI] [PubMed] [Google Scholar]
- Dawson MN, Cieciel K, Decker MB, Hays GC, Lucas CH, Pitt KA. Population-level perspectives on global change: genetic and demographic analyses indicate various scales, timing, and causes of scyphozoan jellyfish blooms. Biol Invasions. 2014;17:851–867. [Google Scholar]
- Dumont HJ. The distribution and ecology of the fresh- and brackish-water medusae of the world. Hydrobiologia. 1994;272:1–12. [Google Scholar]
- Excoffier L, Smouse PE, Quattro JM. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics. 1992;131:479–491. doi: 10.1093/genetics/131.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Laval G, Schneider S. Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol Bioinform Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Fautin DG. Reproduction of Cnidaria. Can J Zool. 2002;80:1735–1754. [Google Scholar]
- Fenner PJ. Dangers in the ocean: the traveler and marine envenomation. I. Jellyfish. J Travel Med. 1998;5:135–141. doi: 10.1111/j.1708-8305.1998.tb00487.x. [DOI] [PubMed] [Google Scholar]
- Fenner PJ, Harrison SL. Irukandji and Chironex fleckeri jellyfish envenomation in tropical Australia. Wilderness Environ Med. 2000;11:233–240. doi: 10.1580/1080-6032(2000)011[0233:iacfje]2.3.co;2. [DOI] [PubMed] [Google Scholar]
- Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol. 1994;3:294–299. [PubMed] [Google Scholar]
- Fontanet CB. PhD thesis. University of Alicante; Spain: 2014. Studies on the ecology of Carybdea marsupialis (Cubozoa) and jellyfish sting risk management. [Google Scholar]
- Gegenbaur C. Versuch eines Systemes der Medusen, mit Beschreibung neuer oder wenig gekannter Formen; zugleich ein Beitrag zur Kenntniss der Fauna des Mittelmeeres. Z Wissen Zool. 1857;8:202–73. plates vii–x. [Google Scholar]
- Gershwin L. Nematocysts of the Cubozoa. Zootaxa. 2006;1232:1–57. [Google Scholar]
- Gershwin LA. Carybdea alata auct. and Manokia stiasnyi, reclassification to a new family with description of a new genus and two new species. Mem Qld Mus. 2005;51:501–523. [Google Scholar]
- Gershwin L-A, Gibbons MJ. Carybdea branchi, sp. nov., a new box jellyfish (Cnidaria: Cubozoa) from South Africa. Zootaxa. 2009;2088:41–50. [Google Scholar]
- Gershwin LA, De Nardi M, Winkel KD, Fenner PJ. Marine stingers: review of an under-recognized global coastal management issue. Coast Manage. 2009;38:22–41. [Google Scholar]
- Gershwin L-A, Richardson AJ, Winkel KD, Fenner PJ, Lippmann J, Hore R, Avila-Soria G, Brewer D, Kloser RJ, Steven A, Condie S. Biology and ecology of Irukandji jellyfish (Cnidaria: Cubozoa) Adv Mar Biol. 2013;66:1–85. doi: 10.1016/B978-0-12-408096-6.00001-8. [DOI] [PubMed] [Google Scholar]
- Gibbons MJ, Janson LA, Ismail A, Samaai T. Life cycle strategy, species richness and distribution in marine Hydrozoa (Cnidaria: Medusozoa) J Biogeogr. 2010;37:441–448. [Google Scholar]
- Goetze E. Population differentiation in the open sea: insights from the pelagic copepod Pleuromamma xiphias. Integr Comp Biol. 2011;51:580–597. doi: 10.1093/icb/icr104. [DOI] [PubMed] [Google Scholar]
- Graham WM, Bayha KM. Biological invasions by marine jellyfish. In: Nentwig W, editor. Biological Invasions, Ecological Studies. Vol. 193. Springer; Berlin: 2007. pp. 239–255. [Google Scholar]
- Graham WM, Pagès F, Hamner WM. A physical context for gelatinous zooplankton aggregations: a review. Hydrobiologia. 2001;451:199–212. [Google Scholar]
- Grantham BA, Eckert GL, Shanks AL. Dispersal potential of marine invertebrates in diverse habitats. Ecol Appl. 2003;13:108–116. [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- Haeckel E. System der Acraspeden – Zweite Hälfte des Systems der Medusen. Denkschriften der Medizinisch–Naturwissenschaftlichen Gesellschaft zu Jena; Germany: 1880. [Google Scholar]
- Humann P, Deloach N. Reef Creature Identification: Florida, Caribbean, Bahamas. 2. New World Publications; Jacksonville, FL: 2002. p. 420. [Google Scholar]
- Ikeda H, Ohtsu K, Uye SI. Fine structure, histochemistry, and morphogenesis during excystment of the podocysts of the giant jellyfish Nemopilema nomurai (Scyphozoa, Rhizostomeae) Biol Bull. 2011;221:248–60. doi: 10.1086/BBLv221n3p248. [DOI] [PubMed] [Google Scholar]
- Jensen JL, Bohonak AJ, Kelley ST. Isolation by distance, web service. BMC Genetics. 2005;6 doi: 10.1186/1471-2156-6-13. 13. v.3.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johannesson K. The paradox of Rockall: why is a brooding gastropod (Littorina saxatilis) more widespread than one having a planktonic larval dispersal stage (L. littorea)? Mar Biol. 1988;99:507–513. [Google Scholar]
- Katoh K, Standley DM. MAFFT Multiple Sequence Alignment Software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayal E, Bentlage B, Collins AG, Kayal M, Pirro S, Lavrov DV. Evolution of linear mitochondrial genomes in medusozoan cnidarians. Gen Biol Evol. 2012;4:1–12. doi: 10.1093/gbe/evr123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayal E, Roure B, Philippe H, Collins AG, Lavrov DV. Cnidarian phylogenetic relationships as revealed by mitogenomics. BMC Evol Biol. 2013;13:5. doi: 10.1186/1471-2148-13-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keesing JK, Strzelecki J, Stowar M, Wakeford M, Miller KJ, Gershwin LA, Liu D. Abundant box jellyfish, Chironex sp. (Cnidaria: Cubozoa: Chirodropidae), discovered at depths of over 50 m on western Australian coastal reefs. Sci Rep. 2016;6:22290. doi: 10.1038/srep22290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsford MJ, Seymour JE, O’Callaghan MD. Abundance patterns of Cubozoans on and near the Great Barrier Reef. Hydrobiologia. 2012;690:257–268. [Google Scholar]
- Kinlan BP, Gaines SD, Lester SE. Propagule dispersal and the scales of marine community process. Divers Distrib. 2005;11:139–148. [Google Scholar]
- Kramp PL. Synopsis of the medusae of the world. J Mar Biol Assoc UK. 1961;40:7–382. [Google Scholar]
- Larson RJ. Marine Flora and Fauna of the Northeastern United States. U.S. Dept. of Commerce, National Oceanic and Atmospheric Administration, National Marine Fisheries Service; Seattle: 1976. Cnidaria: Scyphozoa (003) [Google Scholar]
- Larson RJ, Mills CE, Harbison GR. Western Atlantic midwater hydrozoan and scyphozoan medusae: in situ studies using manned submersibles. Hydrobiologia. 1991;216:311–317. [Google Scholar]
- Lesson RP. In: Centurie Zoologique, ou, Choix d’Animaux Rares, Nouveaux ou Imparfaitement Connus: enrichi de planches in-édites, dessinées d’après nature par M. Prêtre, gravées et coloriées avec le plus grand soin. Chez FG, editor. Levrault; Paris: 1830. p. 244. [Google Scholar]
- Lesson RP. Prodrome d’une Monographie des Méduses. Extr. d’une Histoire manuscrite des méduses, en 3 vol. 4e avec 200 planches coloriées, ouvrage entièrement terminé; Rochefort, France. 1837. p. 200. [Google Scholar]
- Lesson RP. Acalèphes. Librairie Encyclopédique de Roret; Paris: 1843. Historie Naturelle des Zoophytes; p. 663. [Google Scholar]
- Lewallen EA. PhD Thesis. University of Toronto; Canada: 2012. Evolution and Ecology of Flyingfishes (Teleostei: Exocoetidae) [Google Scholar]
- Lewis C, Bentlage B, Yanagihara A, Gillan W, Van Blerk J, Keil DP, Bely AE, Collins AG. Redescription of Alatina alata (Reynaud, 1830) (Cnidaria: Cubozoa) from Bonaire, Dutch Caribbean. Zootaxa. 2013;3737:473–487. doi: 10.11646/zootaxa.3737.4.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- Light SF. Some Philippine Scyphomedusae, including two new genera, five new species, and one new variety. Philipp J Sci. 1914;9:195–231. [Google Scholar]
- Lucas CH, Jones DOB, Hollyhead CJ, Condon RH, Duarte CM, Graham WM, Robinson KL, Pitt KA, Schildhauer M, Regetz J. Gelatinous zooplankton biomass in the global oceans: geographic variation and environmental drivers. Glob Ecol Biogeogr. 2014;23:701–714. [Google Scholar]
- Mayer AG. Medusae of the Hawaiian Islands collected by the steamer Albatross in 1902. Bull US Fish Comm. 1906;23:1131–1143. [Google Scholar]
- Mayer AG. Medusae of the World, Vol. III, The Scyphomedusae. Carnegie Inst Washington Publ. 1910;109:499–735. [Google Scholar]
- Mayer AG. Papers from the Department of Marine Biology. VIII. Carnegie Inst. of Washington; Washington, D.C: 1915. Medusae of the Philippines and of Torres Straits. Being a report on the scyphomedusae collected by the U.S. Fisheries Bureau steamer ‘Albatross’ in the Philippine Islands and Malay Archipelago, 1907–1910, and upon the medusae collected by the expedition of the Carnegie Institution of Washington to Torres Straits, Australia in 1913; pp. 157–202. [Google Scholar]
- Mayer AG. Report upon the scyphomedusae collected by the United States Bureau of Fisheries steamer “Albatross” in the Philippine Islands and Malay Archipelago. Bull US Nat Mus. 1917;100:175–233. [Google Scholar]
- Medina M, Collins AG, Silberman JD, Sogin ML. Evaluating hypothesis of basal animal phylogeny using complete sequences of large and small subunit rRNA. Proc Natl Acad Sci USA. 2001;98:9707–9712. doi: 10.1073/pnas.171316998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morandini AC. Deep-sea medusae (Cnidaria: Cubozoa, Hydrozoa and Scyphozoa) from the coast of Bahia (western South Atlantic, Brazil) Mitt hamb zool Mus Inst. 2003;100:13–25. [Google Scholar]
- Palumbi SR. Nucleic acids II: the polymerase chain reaction. In: Hillis DM, Moritz C, Mable BK, editors. Molecular Systematics. Sinauer; Sunderland, MA: 1996. pp. 205–247. [Google Scholar]
- Petersen KW. Development of coloniality in Hydrozoa. In: Larwood G, Rosen BR, editors. Biology and Systematics of Colonial Organisms. Academic Press; London: 1979. pp. 105–139. [Google Scholar]
- Polzin T, Daneshmand SV. On Steiner trees and minimum spanning trees in hypergraphs. Oper Res Lett. 2003;31:12–20. [Google Scholar]
- Purcell JE, Uye SI, Lo WT. Anthropogenic causes of jellyfish blooms and their direct consequences for humans: a review. Mar Ecol Prog Ser. 2007;350:153–174. [Google Scholar]
- Reynaud M. Carybdea alata n. sp. In: Lesson RP, editor. Centurie Zoologique. Levrault; Paris: 1830. p. 95. Pl. 33. [Google Scholar]
- Scheltema RS. Larval dispersal as a means of genetic exchange between geographically seperated populations of shallow-water benthic marine gastropods. Biol Bull. 1971;140:284–322. [Google Scholar]
- Smith DR, Kayal E, Yanagihara AA, Collins AG, Pirro S, Keeling PJ. First complete mitochondrial genome sequence from a box jellyfish reveals a highly fragmented linear architecture and insights into telomere evolution. Genome Biol Evol. 2012;4:52–58. doi: 10.1093/gbe/evr127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiasny G. Die Scyphomedusen-Sammlung des Naturhistorischen Reichsmuseums in Leiden: Die Carybdeiden (Cubomedusen) Zool Meded. 1919;5:33–58. [Google Scholar]
- Stiasny G. Die Scyphomedusen. Dana Rep. 1940;18:1–28. pls. 1–2. [Google Scholar]
- Straehler-Pohl I. Biology of the box jellyfish Carybdea sivickisi at Akajima. Midoriishi. 2011;22:8–13. (in Japanese) [Google Scholar]
- Straehler-Pohl I, Jarms G. Life cycle of Carybdea marsupialis Linnaeus, 1758 (Cubozoa, Carybdeidae) reveals metamorphosis to be a modified strobilation. Mar Biol. 2005;147:1271–1277. [Google Scholar]
- Straehler-Pohl I, Jarms G. Morphology and life cycle of Carybdea morandinii, sp. nov. (Cnidaria), a cubozoan with zooxan-thellae and peculiar polyp anatomy. Zootaxa. 2011;2755:36–56. [Google Scholar]
- Straehler-Pohl I, Toshino S. Carybdea morandinii: New investigations on its life cycle reveal its true genus: Carybdea morandinii Straehler-Pohl & Jarms, 2011 becomes Alatina morandinii (Straehler-Pohl & Jarms, 2011) Plankton Benthos Res. 2015;10:167–177. [Google Scholar]
- Talavera G, Castresana J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 2007;56:564–577. doi: 10.1080/10635150701472164. [DOI] [PubMed] [Google Scholar]
- Theisen TC, Bowen BW, Lanier W, Baldwin JD. High connectivity on a global scale in the pelagic wahoo, Acanthocybium solandri (tuna family Scombridae) Mol Ecol. 2008;17:4233–4247. doi: 10.1111/j.1365-294x.2008.03913.x. [DOI] [PubMed] [Google Scholar]
- Thomas CS, Scott SA, Galanis DJ, Goto RS. Box jellyfish (Carybdea alata) in Waikiki: their influx cycle plus the analgesic effect of hot and cold packs on their stings to swimmers at the beach: a randomized, placebo-controlled, clinical trial. Hawaii Med J. 2001;60:100–107. [PubMed] [Google Scholar]
- Toonen RJ, Puritz JB, Forsman ZH, Whitney JL, Fernandez-Silva I, Andrews KR, Bird CE. ezRAD: a simplified method for genomic genotyping in non-model organisms. [[2016, Feb. 3]];PeerJ. 2013 1:e203. doi: 10.7717/peerj.203. [Online]. Available: https:/doi.org/10.7717/peerJ.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toshino S, Miyake H, Ohtsuka S, Okuizumi K, Adachi A, Hamatsu Y, Urata M, Nakaguchi K, Yamaguchi S. Development and polyp formation of the giant box jellyfish Morbakka virulenta (Kishinouye, 1910) (Cnidaria: Cubozoa) collected from the Seto Inland Sea, western Japan. Plankton Benthos Res. 2013;8:1–8. [Google Scholar]
- Toshino S, Miyake H, Shibata H. Meteorona kishinouyei, a new family, genus and species (Cnidaria, Cubozoa, Chirodropida) from Japanese waters. ZooKeys. 2015;503:1–21. doi: 10.3897/zookeys.503.9047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verrill AE. Synopsis of the polyps and corals of the North Pacific exploring expedition under Commodore C. Ringgold and Captain John Rodgers, U.S.N. Commun Essex Inst. 1865;4:145–152. [Google Scholar]
- Weir BS. Genetic Data Analysis II: Methods for Discrete Population Genetic Data. Sinauer; Sunderland, MA: 1996. [DOI] [PubMed] [Google Scholar]
- Weir BS, Cockerham CC. Estimating F-Statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- Werner B. New investigations on systematics and evolution of the class Scyphozoa and the phylum Cnidaria. Publ Seto Mar Biol Lab. 1973;20:35–61. [Google Scholar]
- Yanagihara AA, Shohet RV. Cubozoan venom-induced cardiovascular collapse is caused by hyperkalemia and prevented by zinc gluconate in mice. PLoS One. 2012;2012:e51368. doi: 10.1371/journal.pone.0051368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagihara AA, Kuroiwa JM, Oliver LM, Chung JJ, Kunkel DD. Ultrastructure of a novel eurytele nematocyst of Carybdea alata Reynaud (Cubozoa, Cnidaria) Cell Tissue Res. 2002;308:307–318. doi: 10.1007/s00441-002-0545-8. [DOI] [PubMed] [Google Scholar]
- Yanagihara AA, Wilcox C, King R, Hurwitz K, Castelfranco AM. Experimental assays to assess the efficacy of vinegar and other topical first-aid approaches on Cubozoan (Alatina alata) tentacle firing and venom toxicity. Toxins. 2016;8:19. doi: 10.3390/toxins8010019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimoto CM, Yanagihara AA. Cnidarian (coelenterate) application envenomations in Hawai’i improve following heat application. Trans R Soc Trop Med Hyg. 2002;96:300–303. doi: 10.1016/s0035-9203(02)90105-7. [DOI] [PubMed] [Google Scholar]