Abstract
Disruption of the laminar and columnar organization of the brain is implicated in several psychiatric disorders. Here, we show in utero gain-of-function of the psychiatric risk gene transcription factor 4 (TCF4) severely disrupts the columnar organization of medial prefrontal cortex (mPFC) in a transcription- and activity-dependent manner. This morphological phenotype was rescued by co-expression of TCF4 plus calmodulin in a calcium-dependent manner and by dampening neuronal excitability through co-expression of the inwardly rectifying potassium channel (Kir2.1). For the first time, we show that NMDA receptor-dependent Ca2+ transients are instructive to minicolumn organization because Crispr/Cas9-mediated mutation of NMDA receptors rescued TCF4-dependent morphological phenotypes. Furthermore, we demonstrate that the transcriptional regulation by the psychiatric risk gene TCF4 enhances NMDA receptor-dependent early network oscillations. Our novel findings indicate that TCF4-dependent transcription directs the proper formation of prefrontal cortical minicolumns by regulating the expression of genes involved in early spontaneous neuronal activity, and thus our results provides insights into potential pathophysiological mechanisms of TCF4 associated psychiatric disorders.
INTRODUCTION
Abnormal structural development of the cerebral cortex is implicated in a variety of psychiatric disorders including autism, dyslexia, epilepsy, Alzheimer disease, dementias, intellectual disability, and schizophrenia. These structural defects due to abnormal neuronal migration and/or morphological development can range from subtle, almost undetectable changes in the inter-neuronal spacing1–3, to large heterotopias and even double cortex 4. Structural defects have profound effects on the development and function of cortical circuits and can result in an imbalance of excitation and inhibition that is thought to underlie epilepsy and aspects of autism spectrum disorders 5. Therefore, understanding the cellular and molecular mechanisms that regulate cortical development is critical to identifying pathophysiology associated with these psychiatric disorders.
The mammalian cortex features arrays of radially oriented neuronal modules termed cortical minicolumns that are proposed to represent the smallest functional module of the neocortex 6. Minicolumns arise from pyramidal cell migration along radial glia fibers 7, and cells within a column are preferentially connected to each other8. Various aspects of radial migration are regulated by neuronal activity from a variety of sources including GABA, glutamate, and growth factors 9,10, and the rate of neuronal migration is shown to be regulated by voltage-gated Ca2+ channels and NMDARs 11. In addition, early neuronal activity is also critical for regulating the morphology of developing neurons 12. Therefore, neuronal excitability appears to guide the formation of the cortex at every step, from neurogenesis to neurite outgrowth, however a role for neuronal activity regulating minicolumn structure has not been described.
Transcription Factor 4 (TCF4; also known as E2-2, SEF2 or ITF2) is a basic helix-loop-helix (bHLH) transcription factor (TF) that is associated with a variety of neuropsychiatric disorders. Autosomal dominant mutation or deletion of TCF4 results in Pitt Hopkins syndrome (PTHS) and 18q deletion syndrome, two rare autism spectrum disorders 13–15. Common single nucleotide polymorphisms within introns of TCF4 are genome-wide significantly associated with schizophrenia 16, and some of these common genetic variants are associated with cognitive deficits in both schizophrenia patients and unaffected control carriers of the associated alleles 17,18. The direction of altered TCF4 expression in the brains of those with schizophrenia is unclear, although TCF4 expression was shown to be elevated in fibroblasts from schizophrenia patients 19. In addition, duplication of TCF4 is described in a patient with developmental delay 20, and partial duplication is observed in a patient with major depressive disorder21. A familial chromosomal translocation that disrupts expression of some TCF4 transcripts results in overexpression of the remaining isoforms and mild intellectual disability 22 and other familial mutations, resulting in deletion of some, but not all, TCF4 isoforms, cause mild intellectual disability with individuals being less severely affected than those with PTHS 23. In addition, TCF4 expression is upregulated in 2q23.1 deletion syndrome suggesting a common pathway with other genetic autism spectrum disorders 24. Taken together, it is clear that both gain and loss of TCF4 function results in abnormal brain development and increased risk of psychiatric disease.
TCF4 forms homo- or heterodimers with itself or other bHLH TFs25 and dimerization allows for recognition of E-box binding sites (motif: CANNTG). In vitro transcription by TCF4 is inhibited by calmodulin in a Ca2+-dependent manner and therefore TCF4 is potentially an activity-regulated TF 26, however evidence for activity-dependent regulation has never been observed in vivo. Expression of TCF4 is elevated during cortical development in rodents and humans, and while expression decreases after birth, TCF4 is expressed throughout the lifespan 27–29. Disruption of TCF4 expression in postnatal cortical neurons is shown to decrease neuronal excitability and, in mouse olfactory bulb granule cells, it restricts neurite branching and synapse formation 27,30. To begin to understand the biology of TCF4 during cortical development, when its expression is peaking in the cortex, we explored what effect TCF4 gain and loss of function would have on cortical formation in vivo. In utero electroporation (IUE) was used to deliver TCF4 constructs just prior to neurogenesis in the developing rat prefrontal cortex. We report here that TCF4 gain of function results in abnormal distribution of layer 2/3 pyramidal cells. We show this phenotype requires transcriptional regulation and is dependent on Ca2+-mediated neuronal activity through NMDA receptors. In addition, we demonstrate that TCF4 gain of function transcriptionally enhances spontaneous neuronal activity, thus providing the first evidence that the distribution of pyramidal cells into cortical minicolumns is activity-dependent. We propose these biological functions of TCF4 are critical for proper cortical development and likely underlie aspects of pathophysiology in TCF4-associated psychiatric disorders.
MATERIALS and METHODS
Animals and in utero electroporation
Pregnant Wistar rats were obtained from Charles River Laboratories, Inc. and maintained by SoBran on a 12-hour light cycle and fed ad libitum. In utero electroporation was performed as previously described 27,31. Electroporation was performed on embryonic day 16 (E16). The uterine horn was exposed via laparotomy under sterile conditions, and plasmid DNA (1.5 μg/μl) was injected into a single ventricle with a glass micropipette (Drummond). The head of each embryo was stabilized and three electrode pulses were delivered to target plasmid DNA to either the mPFC or somatosensory cortex (SCx) as noted (65V, 100ms). Following electroporation, the uterine horn was replaced in the abdominal cavity and the embryos were allowed to develop as normal. Pregnant dams were returned to the same housing conditions after recovery from surgery, and animals were euthanized as embryos or at postnatal ages as indicated. All the procedures were in accordance with the NIH Guide for the Care and Use of Laboratory Animals and approved by the institutional animal care and use committee.
Histological Analysis
For embryonic age points, rat dams were euthanized by CO2, embryos rapidly removed and their brains submerged in 4% paraformaldehyde in PBS for approximately 36 hours prior to coronal sectioning (50 μm) with a vibratome (Microm, HM650V). Neonatal animals (P1) and juvenile animals (p18-p24) were transcardially perfused under deep anesthesia with 4% paraformaldehyde in PBS. Brains were removed and post-fixed 24 hours in the same fixative solution, prior to coronal sectioning (50 μm) with a vibratome (Microm, HM650V). For all experiments, both male and female animals were used.
Microscopy and Column Analysis
Photomicrographs were taken with a Zeiss 700 confocal microscope equipped with a 20X Plan-Apochromat 0.8 NA objective at 1024 × 1024 pixel resolution at a scan speed of 7s or a Zeiss AxioImager 2 with Zeiss ApoTome2 module equipped with a 20X Plan-Apochromat 0.8 NA objective at 1-μm intervals. Acquisition parameters were consistent for all images. To control for inherent variability in transfection efficiency due to in utero electroporation, control animals were interwoven with experimental animals during surgeries, with multiple conditions in each dam. Due to the profound phenotype produced by TCF4 gain of function, the investigator was not blind to experimental condition. However, care was taken to minimize investigator bias by application of the following criteria: at least four images were obtained from each animal, and images were obtained only from matching coronal slices with uniform transfection through the entire cortical region of interest (mPFC or SCx). Additionally, multiple independent investigators performed image acquisition and analysis and consistent results were obtained. Images were reconstructed in ImageJ and analyzed for cellular distribution by generating small regions of interest perpendicular to the medial surface and calculating the pixel intensity within each bin. The coefficient of variance of these values was calculated, giving us a measure of cellular distribution, such that a high CV value is indicative of cellular aggregation while a low CV indicates an uniform distribution of cells (Supplementary Figure S1).
Plasmid constructs
Plasmid constructs were obtained and cloned into the pCAG-GFP vector 32 at the EcoRI and either NotI or BglII sites. Epitope tags, where indicated, were added at the N-terminus of the protein via PCR subcloning. Plasmids were kind gifts from the following: TCF4B, TCF4A, ΔAD2, R582P - Tonis Timmusk; CaM, CaM1,2,3,4 - John Adelman; Kir2.1, Kir2.1M pCAG-Kir2.1-T2A-tdTomato - Massimo Scanziani (Addgene plasmid # 60598, 60644); GCaMP6s - Douglas Kim (Addgene plasmid # 40753).
shRNA constructs
Hairpin sequences were designed to target the open reading frame of Tcf4, and control hairpin (shCon) was designed to target DsRed as previously described 27. Sense and anti-sense oligonucleotides were designed, annealed, and cloned into the BbsI and XbaI sites of the mU6pro vector, where expression of short hairpin RNA was under the control of mouse U6 promoter.
CRISPR-Cas9
The complete functional deletion of the GluN1 subunit of the NMDA receptor and GluA2 subunit of the AMPA receptor via CRISPR/Cas9 was previously shown in rat hippocampal neurons33. The validated oligos used for the specific sgRNA rat targets were (5′ to 3′): Grin1 forward CACCGaaccaggccaataagcgaca; Grin1 reverse AAACtgtcgcttattggcctggttC; Gria2 forward CACCGctaacagcatacagataggt; Gria2 reverse AAACacctatctgtatgctgttagC. The annealed oligonucleotides were cloned into the BbsI site of the pX458 vector34 (Addgene 48138) and sequenced for confirmation. A pX458 vector with no sgRNA guide (crEmpty) was used as a control.
Co-immunoprecipitation
Co-immunoprecipitation was performed as previously described 35. HEK 293 cells were grown in DMEM media supplemented with 10% FBS to 60% confluence in 6-well plates, and transfected pCAG-CaM or pCAG-CaM(1,2,3,4), pCAG-myc-TCF4, and pCAG-eGFP using the Lipofectamine 3000 reagent. After 48 hours, cells were washed, harvested in NP40 lysis buffer (50mM Tris-HCl pH 7.4, 137mM NaCl, 1% NP40) supplemented with Halt Protease/Phosphatase inhibitor cocktail, measure by Pierce BCA kit, and boiled in LDS/PAGE sample buffer. Protein A Dynabeads (Life Technologies) were prepared with anti-myc (AbCam, catalog number: ab9106) antibody per the manufacturers instructions. The bead-antibody complex was incubated with transfected cellular lysates overnight at 4°C on a rotating wheel. Washing and elution steps were followed per the manufacturer’s instructions. Immunoprecipitated protein (50μg) or input control protein lysates (50 μg) were resolved with Novex 4–12% gradient SDS/PAGE gel and then transferred onto a nitrocellulose membrane (Life Technologies), probed with appropriate antibodies (1:1000 anti-Myc (Abcam), 1:1000 anti-Calmodulin (Millipore, catalog number: 05-173)) and analyzed using 1:20000 LI-COR IR dye conjugated secondary antibodies.
Preparation of acute brain slices
Acute brain slices were performed as described previously 27,36, or with the following alterations described below. Male and female neonatal (P0-4) or juvenile (p18–24) rats were deeply anesthetized and transcardially perfused with ice-cold oxygenated (95% O2 and 5% CO2) dissecting buffer containing (in mM): 83 NaCl, 2.5 KCl, 1 NaH2PO4, 26.2 NaHCO3, 22 glucose, 72 sucrose, 0.5 CaCl2, and 3.3 MgCl2. The animal was decapitated and the brain was rapidly removed and immersed in ice-cold oxygenated dissection buffer. Coronal slices (300 μm) were cut using a vibratome (Microm HM650V), incubated in dissection buffer for 30–45 min at 34°C, and then stored at room temperature.
Calcium Imaging
Artificial cerebrospinal fluid (ACSF) was oxygenated (95% O2 and 5% CO2) and contained (in mM): 125 NaCl, 25 NaHCO3, 1.25 NaH2PO4, 3 KCl, 25 dextrose, 1 MgCl2, and 2 CaCl2, pH 7.3, unless otherwise noted. Slices were visualized using a CCD camera (QICAM, QImaging), and layer 2/3 pyramidal cells were visualized with epifluorescent illumination and a 20x Olympus XLUMPlanFl (0.95 numerical aperture) objective. Images were acquired at a frame rate of 2 frames per second with a resolution of 1392x1040 pixels for a total of 5 minutes. Time-lapse videos were stabilized with a python script using the Lucas-Kanade optical flow provided by OpenCV (2.4.13) to detect and correct for drift. Regions of interest (ROI) for each cell were detected from the mCherry fluorescence image using an adaptive thresholding with a 40 pixel square window then segmented with a watershed algorithm to differentiate adjacent cell bodies all with the R package EBImage 37, and regions with fewer than 500 pixels were dropped. The final list of ROIs was visually curated to discount false cells. Traces were taken from mean pixel intensities of each ROI over time, and normalized to the image background. Peaks were called from running max over 9 frame windows of the first derivative values above a 1.96 threshold. Peak regions lasting fewer than 5 frames were dropped as false peaks. The first and last 20 frames were drop to account for edge artifacts of the sliding window peak detection. The code used to process calcium imaging experiments is available by request from authors.
We modeled differential calcium activity across age and IUE transfection condition with the Poisson regression model: log(yi) ~ α + βCondi + γAgei + εi where yi is the number of calcium transients of sample i, β is the log effect ratio of activity by IUE transfection, and γ is the log effect ratio of activity by age. Differential activity by transfection condition (β ≠ 0) can formally be tested by constructing moderated T-statistics and corresponding p-values. We modeled the ages of the animals linearly (1 df). This Poisson modeling approach represents the standard ANOVA framework, but calculates the effects of the outcome and age directly as log incidence rates.
For some experiments, DL-AP5 (100μM) was added to the ACSF to block NMDA receptors. Baseline images were collected under control conditions and the manipulation was added to the bath after 200 frames had passed. Calcium peaks were counted in first and last 200 frames to allow for diffusion. We analyzed calcium activity effect due to pharmacological manipulation in each IUE condition using the Poisson regression model with a random intercept to account for repeated measures from each slice prep. We subsequently analyzed difference in activity due to pharmacology between IUE conditions using a regular regression model accounting for the same repeated measures.
Electrophysiology
For all experiments, as previously described 27,36, artificial cerebrospinal fluid (ACSF) was oxygenated (95% 02 and 5% CO2) and contained 125 mM NaCl, 25 mM NaHCO3, 1.25 mM NaH2PO4, 3 mM KCl, 25 mM dextrose, 1 mM MgCl2, and 2 mM CaCl2 (pH 7.3). Patch pipettes were fabricated from borosilicate glass (N51A, King Precision Glass) to a resistance of 2–5 MΩ. For current- and voltage-clamp measurements, pipettes were filled with 125 mM potassium gluconate, 10 mM HEPES, 4 mM Mg-ATP, 0.3 mM Na-GTP, 0.1 mM EGTA, and 10 mM phosophocreatine, adjusted to pH 7.3 with KOH. For generation of I/O curves, all cells were recorded from their inherent resting membrane potential. Current signals were recorded with either a Multiclamp 700A (Molecular Devices) or an Axopatch 200B amplifier (Molecular Devices) and were filtered at 2 kHz using a built in Bessel filter and digitized at 10 kHz. Voltage signals were filtered and 2 kHz and digitized at 10 kHz. Data were acquired using Axograph on a Dell PC. Data acquisition was terminated when series resistance was >15 MΩ.
Statistics
Statistics were calculated using GraphPad Prism Software (La Jolla, CA). For CV analysis, unpaired Student’s t-test or one-way analysis of variance tests were performed, followed by post hoc Bonferroni’s multiple comparisons were applicable. For electrophysiology experiments, two-way repeated measures analysis of variance were performed, followed by post hoc Sidak’s comparisons. Data are represented as mean±s.e.m., unless otherwise noted, and were uniformly distributed.
RESULTS
TCF4 gain of function severely alters the distribution of layer 2/3 pyramidal cells in a context-dependent manner
TCF4 expression peaks during neuronal migration and synaptogenesis in rodents and humans and indicates a critical period for TCF4 function 27–29. To begin to understand the role of TCF4 during this developmental peak we altered TCF4 expression using in utero electroporation (IUE). Electroporation of TCF4B, a full-length isoform, revealed a striking alteration in the distribution of transfected cells, resulting in the formation of compact cellular aggregates that were not observed following transfection with GFP alone (Figure 1a,d). The formation of these cellular aggregates became apparent on postnatal day 1 (PND 1), a developmental time point when the majority of neurons have completed migration, and cellular aggregates were not evident during neuronal migration (Supplemental Figure S2). Furthermore, we show that TCF4 loss-of-function using a previously validated shRNA construct (shTCF427) has no effect on pyramidal cell distribution when compared to a control shRNA construct (shCon, Figure 1b,e). These results indicate TCF4 gain of function, but not loss of function, significantly disrupts the distribution of pyramidal cells in the developing neocortex.
Figure 1. TCF4 gain-of-function results in abnormal columnar organization in the medial prefrontal cortex.
TCF4 was expressed in the developing cortex via in utero electroporation. Brains of the resultant pups were fixed at p21 and imaged using confocal microscopy. TCF4 overexpression (A, A1) resulted in abnormal distribution of layer 2/3 pyramidal cells in medial prefrontal cortex (mPFC) when compared to GFP controls (A2). Knockdown of TCF4 expression by shTCF4 did not alter neuronal distribution (B, B2) compared to shCon (B2). This morphological phenotype was specific to the mPFC because no cellular distribution defects were observed when TCF4B was expressed in somatosensory cortex (SCx; C, C1, C2). The distribution of cells was quantified by binning pixel intensity perpendicular to the surface and calculating the coefficient of variance (CV) across all bins, therefore a high CV value indicates a more clustered distribution. D) Expression of TCF4B resulted in a significant increase in the CV when compared to GFP-only controls in the mPFC (GFP n=4 0.30±0.036 vs. TCF4B n=5 0.57±0.036; p<0.01). E. TCF4 loss-of-function by shRNA does not significantly alter pyramidal neuron distribution in mPFC (shCon n=4 0.29±0.035; shTCF4 n=5 0.34±0.012; p=0.18). F. In SCx, coefficient of variance is unchanged between GFP control and TCF4B (GFP n=5 0.295±0.014 vs. TCF4B n=4 0.27±0.017; p=0.18). Scale bar 100μm, inset 50 μm.
The mammalian neocortex is regionalized such that subdivisions are distinguished by differences in architecture, axonal connections and function. This regionalization results from molecular gradients of morphogen expression across the embryonic ventricles that instruct specific gene expression 38. The ability of TCF4 to form homo- and heterodimers with itself and other bHLH TFs and its developmental and cell-specific expression patterns indicate the function of TCF4 is context-dependent 39. IUE can target specific regions of the cortex by directing the plasmid DNA to specific ventricle surfaces with focused electrical pulses 36,40. To determine if TCF4 regulates cellular distribution in a context-dependent manner we targeted TCF4 gain of function to specific ventricle surfaces that give rise to either the mPFC or somatosensory cortex (SCx). TCF4B gain of function directed at the SCx cortex had no effect on pyramidal cell distribution (Figure 1c,f). This result indicates TCF4 regulation of cortical development is context-dependent, such that the mPFC appears to be sensitive to TCF4 gain of function while the SCx is insensitive. This locational specificity further indicates TCF4-dependent disruption of cellular distribution is not the result of non-specific effects due to excessive overexpression, but rather indicates TCF4 gain of function is specifically exploiting the genetic program of mPFC but not SCx during development.
Altered pyramidal cell distribution requires transcriptional activation by TCF4
All TCF4 isoforms express common 3′ exons, which contain the bHLH domain and an activation domain (AD2, Figure 2a), that are essential for transcriptional regulation. To determine if regulation of pyramidal cell distribution by TCF4 requires transcription we first electroporated a PTHS patient-derived TCF4B construct that contains a point mutation in the bHLH domain (R582P) that renders TCF4 incapable of binding to DNA 41. Expression of R582P was ineffective at altering pyramidal cell distribution in the either the mPFC or SCx (Figure 2b,c, Supplemental Figure S3). Consistent with this result, expression of a TCF4B construct that lacks the AD2 domain (ΔAD2), which deletes the protein interaction domain necessary for binding of mediators of transcriptional activation to TCF4 42, also had no effect on pyramidal cell distribution in mPFC (Figure 2b,c). Together, these data indicate transcriptional regulation by TCF4B is required to disrupt the distribution of pyramidal cells.
Figure 2. TCF4-dependent regulation of cellular distribution requires transcription.
A) Diagram of three different TCF4 isoforms depicting the known protein motifs which include two activation domains (AD1 and AD2), a nuclear localization sequence (NLS) and the DNA-binding basic Helix-Loop-Helix domain (bHLH). B) TCF4B produces pyramidal cell aggregation in the mPFC, as does the short isoform TCF4A that lacks a NLS. A disease-causing point mutant, R582P, does not alter pyramidal cell distribution. Co-expression of TCF4B + R582P rescues TCF4B-dependent formation of abnormal minicolumn structures. Expression of TCF4B that lacks an AD2 (ΔAD2) does not disrupt pyramidal cell distribution. Abnormal column formation is not observed when the TCF4H isoform is overexpressed. Scale bar 50μm. C) Group analysis indicates that R582P (n=4, 0.33±0.036, p=0.0007), TCF4B + R582P (n=5, 0.38±0.025, p=0.021), ΔAD2 (n=6, 0.26±0.017; p<0.001), and TCF4H (n=8, 0.31±0.030, p<0.001) cause a significant reduction in CV compared to TCF4B (n=6, 0.57±0.061) while expression of TCF4A (n=9, 0.45±0.039, p>0.05; ANOVA p<0.0001) does not significantly reduce CV value when compared to TCF4B. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001
Previous work has demonstrated that in vitro co-expression of R582P with full-length TCF4 significantly inhibits transcription of a luciferase reporter construct, lending support for a dominant-negative model 41. Consistent with in vitro dimerization between mutant and full-length TCF4, in utero co-expression of TCF4B + R582P was effective at preventing the pyramidal cell aggregation phenotype (Figure 2b,c), thus indicating under these conditions, R582P acts in a dominant negative manner by forming homodimers with TCF4B and thereby prevents interaction with the genome and downstream disruption of pyramidal cell distribution.
The TCF4 gene is transcribed using numerous 5′ exon and results in a large diversity of TCF4 isoforms 42. Therefore, we also tested two other TCF4 isoforms that lack several 5′ exons and a nuclear localization signal (NLS) compared to TCF4B (Figure 2a,b,c). TCF4A and TCF4H differ between each other by only a single 5′ exon that is unique to each isoform and both isoforms were previously shown to be localized to both the nucleus and cytosol of transfected HEK293 cells 42. IUE of TCF4A, but not TCF4H, resulted in significant aggregation of pyramidal cells in the mPFC (Figure 2b,c) and this result indicates that a NLS signal is not required, but suggests the amino acids encoded by the unique 5′ exons between TCF4A and TCF4H must confer some functional differences. When expressed in the SCx, both TCF4A and TCF4H do not alter pyramidal cell distribution, suggesting that regional specificity is consistent among these isoforms (Supplemental Figure S3). Taken together, these results indicate that altered pyramidal cell distribution due to TCF4 gain of function requires transcriptional control in a regional- and isoform-specific manner.
TCF4-dependent regulation of pyramidal cell distribution is rescued by calmodulin in a Ca2+-dependent manner
Neuronal migration is dependent on Ca2+ signaling to regulate chemotaxis 9 and transcriptional control by TCF4 is regulated by calmodulin (CaM), a Ca2+ binding protein, such that in vitro transcription is inhibited upon binding of Ca2+/CaM to TCF4 43. To determine if Ca2+ signaling was converging on TCF4-dependent effects on pyramidal cell distribution in vivo, we co-expressed CaM with TCF4B. In agreement with CaM inhibiting TCF4 function, we failed to induce pyramidal cell aggregation when CaM was co-expressed with TCF4B (Figure 3a,b). Remarkably, a mutant version of CaM (CaM1,2,3,4), that contains a single point mutations in each of the four E-F hands and prevents Ca2+ binding 44, was ineffective at preventing TCF4B-dependent pyramidal cell aggregation (Figure 3a,b). Overexpression of CaM or CaM1,2,3,4 alone had no effect on pyramidal cell distribution (Figure 3a, b). To rule out the possibility that mutations in CaM1,2,3,4 preclude protein interactions with TCF4B, we performed co-immunoprecipitation experiments in the presence and absence of Ca2+. We coexpressed myc-TCF4B with either CaM or CaM1,2,3,4 and determined that both CaM and CaM1,2,3,4 are immunoprecipitated by an anti-myc antibody (Figure 3c). To determine if Ca2+ was necessary for binding of CaM or CaM1,2,3,4 to TCF4B in vitro, we altered the Ca2+ concentration in our protein lysates and performed additional co-immunoprecipitation experiments. We observed that both CaM and CaM1,2,3,4 were capable of binding to TCF4B regardless of Ca2+ concentration (Figure 3d). These results suggest that in vitro binding of TCF4B and CaM or CaM1,2,3,4 can occur independently of Ca2+ binding, but that in vivo CaM-dependent prevention of pyramidal cell aggregation by TCF4B gain of function requires Ca2+, because the Ca2+ insensitive CaM1,2,3,4 was ineffective at rescuing cellular aggregation. From these results, we propose that the distribution of pyramidal cells within the mPFC is regulated by TCF4 transcription in a Ca2+-dependent manner and therefore, we reason that that pyramidal cell distribution is regulated by neuronal activity.
Figure 3. Abnormal column formation is rescued by co-expression of calmodulin (CaM) and this rescue requires calcium binding to CaM.
A) TCF4B was expressed alone, co-expressed with CaM or with a mutant calmodulin that is unable to bind Ca2+ (CaM1,2,3,4); CaM (n=6) or CaM1,2,3,4 (n=4) were also expressed alone as controls. Scale bar 50μm. B) Cortical column formation was significantly reduced in the TCF4B + CaM condition (TCF4B n=6, 0.51±0.049 vs. TCF4B + CaM n=5, 0.31±0.012, p=0.028), the CaM condition (n=6, 0.18±0.019, p=0.001), or the CaM1,2,3,4 condition (n=4, 0.25±0.057, p=0.004) but not in the TCF4B + CaM1,2,3,4 condition (TCF4B + CaM1,2,3,4 n=5, 0.56±0.071; p>0.05; ANOVA p<0.0001). C) Co-immunoprecipitation experiments demonstrate that myc-TCF4B can immunoprecipitate both CaM (left) and CaM1,2,3,4 (right). D) Co-immunoprecipitation is observed regardless of the amount of Ca2+ present during the immunoprecipitation reaction.
Pyramidal cell distribution within the mPFC is dependent on neuronal activity and TCF4
Neuronal activity is known to be present in every step during cortical development including proliferation, neuronal migration, neurite outgrowth, and synaptogenesis 10,45, however a role for activity regulating cellular distribution has not been described. To determine if neuronal activity is required for TCF4B-induced neuronal aggregation, we altered activity by expressing a recombinant inward rectifying potassium channel (Kir2.1). IUE of Kir2.1 was previously shown to impair the morphological maturation of layer 2/3 pyramidal cells by lowering the resting membrane potential and thereby reducing neuronal excitability 12. Co-expression of TCF4B + Kir2.1 resulted in prevention of pyramidal cellular distribution compared to co-expression of TCF4B + Kir2.1M, an inactive version of Kir2.1 (Figure 4a,b). IUE of either Kir2.1 or Kir2.1M alone had no effect on cellular distribution (Figure 4a,b). These results indicate that neuronal activity in addition to TCF4 gain of function is required to disrupt pyramidal cell distribution in the mPFC.
Figure 4. TCF4-induced abnormal column formation is activity-and NMDA receptor-dependent.
A) An inwardly rectifying potassium channel (Kir2.1) rescues TCF4B-dependent disruption of pyramidal cell distribution. A mutant Kir2.1 channel (Kir2.1M) that is expressed on the membrane but does not pass current was ineffective at rescue. Expression of either channel alone has no effect on cellular distribution. Scale bar 50μm. B) Group analysis showing that co-expression of TCF4B + Kir2.1 prevents abnormal column formation (TCF4B n=6, 0.46±0.065 vs. TCF4B + Kir2.1 n=5, 0.23±0.022; p=0.012; ANOVA p=0.007). TCF4B + Kir2.1M did not prevent column formation (TCF4B + Kir2.1M n=6, 0.42±0.051, p>0.05), while either Kir2.1 (n=4, 0.24±0.020; p=0.032) or Kir2.1M (n=5, 0.28±0.046, p=0.07) alone had no effect on cellular distribution. (** P<0.01; * P<0.05; # P<0.10). C) TCF4B-dependent cellular aggregation was observed when TCF4B was co-expressed with a Crispr/Cas9 vector lacking a guide RNA that, when expressed alone, did not result in cellular aggregation. (TCF4B + crEmpty, n=5 0.73±0.11 vs. crEmpty, n=5, 0.27±0.036; p<0.001; ANOVA p<0.0001). Co-expression TCF4B with a Crispr/Cas9 construct that targets Grin1 rescues cellular aggregation, while co-expression of a Crispr/Cas9 that targets the AMPA receptor subunit Gria2 failed at rescue of cellular aggregation. Scale bar 50μm. D) Cellular distribution analysis indicates TCF4B + crGrin1 significantly rescued pyramidal cell aggregation compared to TCF4B + crEmpty (TCF4B + crGrin1 n=5, 0.36±0.024, p<0.0001), and TCF4B + crGria2 did not rescue TCF4B-dependent cellular aggregation (TCF4B + crGria2 n=4, 0.55±0.009, p>0.05) Expression of crGrin1 (n=5, 0.29±0.032, p<0.001) or crGria2 (n=5, 0.33±0.033, p=0.002) alone has no effect on pyramidal cell distribution (** P<0.01; ****p<0.0001).
TCF4-dependent disruption of pyramidal cell distribution requires NMDA receptor activity
Once migrating neurons reach the cortical plate, functional NMDA receptors are expressed 46 and their activation coincides with cessation of neuronal migration 47. In addition, NMDAR-mediated Ca2+ currents are known to regulate dendrite outgrowth, dendrite complexity, and synapse formation 45,48. Given our prior result that expression of CaM can prevent TC4B-dependent disruption of pyramidal cell distribution in a Ca2+-dependent manner, we hypothesize that NMDAR-mediated glutamatergic transmission may be a source of neuronal activity and Ca2+ that is required for regulating pyramidal cell distribution. To test this hypothesis, we suppressed NMDA receptor function using Crispr/Cas9 mutation. Genetic mutation of Grin1 or Gria2 with Crispr/Cas9 was shown to be effective at blocking glutamatergic transmission 33 and is compatible with IUE delivery 27,49. Co-expression of TCF4B plus a Crispr/Cas9 vector that lacks a guide RNA (crEmpty) resulted in cellular aggregation (Figure 4c,d) and this phenotype was rescued by co-expressing TCF4B plus a Crispr/Cas9 that targets Grin1 (crGrin1). This effect was specific to NMDA receptors because a Crispr/Cas9 that targets AMPA receptors (crGria2) was ineffective at rescue (Figure 4c,d). Furthermore, Crispr-mediated mutation by expression of either crGrin1 or crGria2 alone had no effect on pyramidal cell distribution (Fig 4c,d). These data indicate Ca2+ influx via NMDA receptor activation appears to be a sufficient source of the Ca2+ required for TCF4’s effect on cellular distribution, and further indicates that the spatial orientation of neurons within a minicolumn is regulated in an activity- and TCF4-dependent manner.
Does TCF4B respond inappropriately to endogenous neuronal activity or does TCF4B gain of function transcriptionally enhance neuronal activity?
We have shown that pyramidal cell distribution is sensitive to TCF4 gain of function and this effect requires TCF4B-dependent transcription and spontaneous neuronal activity. From these results we reasoned that there are two potential mechanisms for how pyramidal cell distribution is regulated by TCF4B during corticogenesis. One hypothesis is that elevated TCF4B protein levels are inappropriately responding to spontaneous early network activity and this leads to aberrant transcription of genes that regulate pyramidal cell distribution. In this case, we would expect to observe a similar amount of spontaneous Ca2+ transients between TCF4B and control conditions. Alternatively, TCF4B gain of function could be directly increasing network activity through aberrant transcription of genes that regulate neuronal activity and pyramidal cell distribution. In this case, we should observe increased activity in TCF4B condition compared to control. To differentiate between these two cellular mechanisms we performed calcium imaging in acute slices prepared from neonatal (P0-4) animals that were IUE transfected with the genetically encoded calcium indicator GCaMP6s 50 (Figure 5a). This developmental time window coincides with the first significant appearance of pyramidal cell aggregation phenotype (Supplemental Figure S2). Remarkably, we observed that TCF4B gain of function significantly increased the number of Ca2+ transients compared to control cells expressing GCaMP6s + mCherry (Figure 5b, c). At this developmental stage in the cerebral cortex, network oscillations are known to be partially due to NMDA receptor activation 45. We thus asked if the calcium transients we observed were due to Ca2+ influx through NMDAR channels. Consistent with rescue of pyramidal cell aggregation by co-expression of TCF4B + crGrin1 (Figure 4c,d), spontaneous Ca2+ transients were significantly reduced in the presence of the NMDA receptor antagonist DL-AP5 (Figure 5e), thus confirming previous observations 45 that these early spontaneous network oscillations are partially NMDA receptor-dependent. Furthermore, we compared the level of DL-AP5 inhibition between the TCF4B and control condition and observed a greater magnitude of inhibition was produced in the TCF4B condition, suggesting that TCF4 gain of function leads to a greater amount of NMDA receptor activity. These experiments indicate that TCF4B-dependent transcription regulates the level of spontaneous neuronal activity in the developing neocortex and potentially enhances NMDA receptor function.
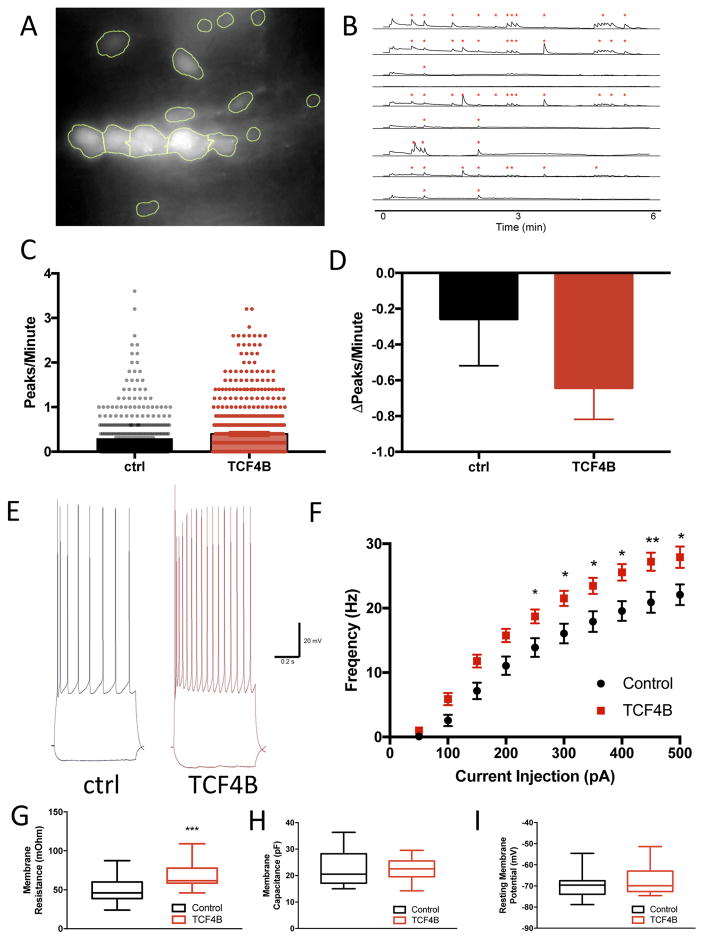
Figure 5. TCF4B gain of function significantly enhances spontaneous neuronal activity.
A) Example image of an abnormal minicolumn structure in pyramidal neurons expressing TCF4B + mCherry + GCaMP6s. A custom R script was written to identify individual cells (ROI) using the mCherry channel. B) Example traces of spontaneous Ca2+ events that were detected (*). Pixel intensity was normalized to background for each image, and peaks were called using a custom R script. C) Total activity was calculated by number of peaks per cell per minute, and was significantly increased in the TCF4B condition compared to control (TCF4B n=743 cells, 0.3950±0.0202 vs. Ctrl n=450, 0.3020±0.0232; t-test; P<0.001). D) Ca2+ transients were inhibited by bath application of the NMDA receptor antagonist DL-AP5 (100μM). A Poisson mix effect model was used to discern the difference in activity by DL-AP5 with each transfection condition. Addition of DL-AP5 inhibited activity both transfection conditions (TCF4B n=201, −0.2140±0.0294; ctrl n=91, −0.0857±0.0248; p<4.06e-06). TCF4B transfect cells were more affected by AP5 compared to ctrl cells (post-hoc t-test; P<0.00655). E). Sample current-clamp traces recorded from GFP- or TCF4B-expressing neurons showing increased action potential output in response to current injection (−50 pA, +400 pA). F) Summary data shows that the frequency of action potentials increases in response to increasing current injection. Post hoc analysis indicated that TCF4B cells (n=27) produced significantly more spikes than GFP control cells (n=19) between 300 and 500 pA of current injection. G) TCF4B cells show in an increase in membrane resistance compared to control (control n=19, 49.94±4.223: TCF4B n=27, 68.67±2.829; p=0.004). H) Membrane capacitance is unchanged between TCF4B and control cells (control n=19, 22.96±1.565: TCF4B n=27, 21.89±0.877; p=0.525). I) Resting membrane potential is unchanged between TCF4B and control cells (control n=19, −69.94±1.387: TCF4B n=27, −67.74±1.178; p=0.238).
We previously showed that reduced expression of TCF4 in this population of neurons results in decreased intrinsic excitability in the form of enhanced spike frequency adaptation 27. Given that TCF4B gain of function increased the frequency of spontaneous Ca2+ transients in developing neurons we reasoned it might also increase intrinsic excitability in more mature cortical neurons. Therefore, we prepared acute slices and recorded from layer 2/3 pyramidal neurons at p18–24 expressing either TCF4B and GFP or GFP alone (control). We observed an increase in intrinsic excitability, such that TCF4B neurons fired more action potentials than control cells in response to a given current injection (Figure 5e, f) and this effect was likely due to the significant increase in membrane resistance (Figure 5g–i), which results in neurons that are more sensitive to electrical and/or synaptic input thereby increasing their intrinsic excitability.
Overall, these results suggest TCF4 regulates expression of genes associated with neuronal excitability as TCF4 gain of function produces a general increase in neuronal excitability as measured by spontaneous Ca2+ transients and whole-cell electrophysiology. When considering these data together with our morphological rescue due to reduction of spontaneous neuronal activity by Kir2.1 expression or Crispr-mediate mutation of NMDA receptors it suggests that spontaneous neuronal activity appears to be critical to the distribution of layer 2/3 pyramidal cells in the mPFC. Importantly, these results further indicate that the psychiatric risk gene TCF4 is an upstream regulator of gene expression and neuronal activity that appears to be instructive to the columnar organization of the prefrontal cortex.
DISCUSSION
We provide a series of experiments that identify a cellular function of TCF4 during cortical development that has implications for TCF4-associated psychiatric risk. Using IUE, we manipulated the expression of TCF4 during its developmental peak in expression 27, and observed that TCF4 gain of function, but not loss of function, resulted in profound defects in the distribution of principal cells of the mPFC. The cortical minicolumn is considered to be the smallest processing unit in the brain and defects in its structure are linked to several psychiatric disorders including autism, schizophrenia, dyslexia and dementia 6. Our results suggest TCF4 plays a role in the formation of cortical minicolumns in the mPFC and we show that this function is sensitive to both neuronal activity and Ca2+, two critical signals that are necessary for cortical development 51. Furthermore, we show that TCF4B gain of function significantly enhances early spontaneous neuronal activity and this increased activity regulates the distribution of pyramidal cells in the mPFC. Therefore, we suggest TCF4 is an activity-dependent transcription factor that links instructional activity signals and transcriptional regulation.
TCF4 can form homodimers with itself or heterodimers with other bHLH TFs and this confers specificity to transcriptional regulation in a context-dependent manner52. We provide evidence that TCF4 gain of function is context-dependent as we show specific TCF4 isoforms (TCF4A and TCF4B, but not TCF4H) severely disrupt the distribution of pyramidal cells in the mPFC but show no effect on the cellular distribution in the SCx (Figure 2 and Supplemental Figure S3). These results indicate TCF4’s effect on pyramidal cell distribution is context-dependent. Furthermore, both the isoform specificity and the regionalized effect of TCF4 indicates cellular aggregation due to TCF4 does not result from a non-specific effect of TCF4 overexpression, because if this were the case we would expect to observe a similar phenotypes across the entire cortex and by overexpression of every TCF4 isoform.
Exactly how this isoform- and regional-specificity is occurring is not clear from our data and therefore will require further experimentation. TCF4A and TCF4H both lack a NLS and only differ by a unique 5′ exon that encodes for 23 amino acids. In transfected HEK293 cells, both isoforms are localized to the cytoplasm and nucleus and TCF4H was shown to be relatively more effective at in vitro transcription compared to TCF4A 42. Therefore, it appears that the differential effect on pyramidal cell aggregation between TCF4A and TCF4H is related to their unique N-terminal amino acid sequences. As for regional-specificity, TCF4 is relatively equally expressed across the entire cortex 53, and therefore differences in regional expression of TCF4 is not a likely explanation. In general, regional specification of the cortex is dependent on morphogenetic gradients 38 and one potential explanation could be that TCF4 gain of function is inappropriately tapping into a morphogenetic gradient that is specific to the frontal cortex. One example is Id2 (Inhibitor of DNA binding 2), which is specifically expressed at high levels in layer 2/3 cell of the frontal cortex and is thought to regulate regionalization 54,55. ID2 functions to inhibit bHLH transcription factors, including TCF4 56, in a dominant-negative manner by forming heterodimers and thus preventing DNA binding. TCF4 gain of function in the frontal cortex could result in excess binding of TCF4 with ID2 and consequently alter downstream transcription that leads to increased neuronal excitability and pyramidal cell aggregation.
The observed defects in cellular distribution required transcriptional regulation by TCF4 are supported by three independent experiments. First, we show that a PTHS patient-specific point mutation in the bHLH domain (R582P), that prevents TCF4 interaction with DNA 41 and produces a functional dominant-negative protein in vivo, was ineffective at altering cellular distribution (Figure 2). Secondly, deletion of the AD2 domain (ΔAD2), which is necessary for recruitment of transcriptional machinery57, also was ineffective at producing cellular aggregation (Figure 2). Lastly, we show that CaM, a Ca2+ binding protein known to inhibit TCF4 transcription in vitro 26, was capable of preventing cellular aggregation when co-expressed with TCF4B. Together, these results clearly indicated TCF4 gain of function in the mPFC produces pyramidal cell aggregation by directly binding to the genome and regulating transcription.
The ability of CaM to prevent TCF4-dependent cellular aggregation appears to be Ca2+-dependent as co-expression of TCF4B with a mutant CaM (CaM1,2,3,4) 44 was ineffective at rescue. We were able to biochemically separate Ca2+ binding to CaM from CaM binding to TCF4, as both CaM and the EF-hand mutant CaM1,2,3,4 were immunoprecipitated by recombinant myc-TCF4 (Figure 3c). This result indicates that in vitro, CaM and CaM1,2,3,4 can bind to TCF4B without Ca2+, but that prevention of cellular aggregation in vivo by CaM requires the ability of CaM to bind Ca2+. The current model suggests that Ca2+/CaM complexes regulate bHLH transcription by preventing dimerization or blocking contacts between bHLH dimers and nucleotide sequences 43. Our results suggest that blockade of TCF4-dependent transcription by Ca2+/CaM complexes may require Ca2+-dependent binding of additional unknown proteins or that Ca2+ binding to TCF4/CaM complexes leads to a specific conformational change that is necessary for inhibition of transcription. Further biochemical analysis will be necessary to understand the exact mechanism of Ca2+/CaM effects on TCF4-dependent transcription, but in general, our results taken together clearly indicate TCF4 transcriptional activity is regulating pyramidal cell distribution in a Ca2+-dependent manner.
The Ca2+/CaM dependence of our TCF4 phenotype led us to investigate whether neuronal activity in the form of Ca2+ signaling was involved in abnormal pyramidal cell distribution due to TCF4B-dependent transcription. We showed that dampening neuronal excitability by expression of Kir2.1 resulted in rescue of cellular aggregation. Previously, Kir2.1 expression was shown to hyperpolarize pyramidal neurons and reduce dendritic elaboration 12. To further validate a causal role for neuronal activity converging on TCF4 gain of function to disrupt pyramidal cell distribution, we suppressed NMDA receptor function by expressing a Crispr/Cas9 construct that specifically targets Grin1, which encodes for GluN1, a constitutive subunit of NMDA receptors 58 59. Consistent with a role for Ca2+ signaling to regulate TCF4 transcription, crGrin1 was effective at rescuing TCF4-dependent cellular aggregation. This effect was specific to glutamate transmission through NMDA receptors because Crispr/Cas9 mutation of Gria2, which encodes GluA2 subunit of AMPA receptors, had no effect on TCF4B dependent aggregation of pyramidal cells (Figure 4). We predict these two molecular manipulations produce rescue by reducing Ca2+ influx. Expression of Kir2.1 will hyperpolarize the membrane and reduce Ca2+ influx through voltage-gated Ca2+ channels while also favoring Mg2+ block of NMDA receptors and crGrin1 will directly inhibit Ca2+ influx through NMDA receptors. Together, these experiments are the first to show that a convergence of TCF4 transcription and neuronal activity is regulating the distribution of pyramidal neurons, which is critically important for the proper formation of prefrontal circuitry.
The dosage of TCF4 expression appears to regulate the excitability of prefrontal neurons and suggests TCF4 may be an activity-dependent transcription factor. We previously reported that suppression of TCF4 decreases intrinsic excitability through direct transcriptional regulation of two ion channels 27 and here we show TCF4 gain of function increases the frequency of early network oscillations and increases intrinsic excitability in mature neurons (Figure 5). Furthermore it appears that TCF4B gain of function may directly regulate NMDA receptor activity because the NMDA receptor antagonist DL-AP5 lead to a greater amount of activity blockade in the TCF4B condition compared to control (Figure 5). Together these results indicate TCF4 expression appears to be positively correlated with the level of neuronal activity and because TCF4 can be directly regulated by the Ca2+ binding protein CaM, we speculate that it is a bone fide activity-dependent transcription factor that can both sense neuronal activity and modify neuronal activity through transcription.
Although TCF4 dosage is correlated well with neuronal activity we didn’t observe a perfect correlation with TCF4 expression and its effect on pyramidal cell distribution, as shRNA knockdown of TCF4 did not appear to alter pyramidal cell distribution (Figure 1). We think this lack of correlation likely reflects our inability to quantify widening of cortical minicolumns, as there is no known morphological marker of the boundary of each cortical minicolumn. Furthermore, the exact mechanism for how TCF4 gain of function leads to pyramidal cell aggregation is not completely understood. Our data clearly indicates that TCF4-dependent transcription leads to increased neuronal activity and that a convergence of TCF4-dependent transcription and neuronal activity is necessary to produce the phenotype, but exactly what genes are being regulated by TCF4 during this developmental time period is not fully understood. Our results suggest one potential candidate is Grin1 and the NMDA receptors that are encoded by this gene (Figure 4,5), but clearly many other genes associated with other biological functions such as neurite motility, migration, cell adhesion, microtubule and actin biology are all likely involved. A candidate gene approach and validation by phenotype rescue can begin to decipher the underlying molecular mechanism for this TCF4-dependent phenotype.
TCF4 is a pleiotropic gene and has clinical association with many psychiatric disorders. For some of these disorders, such as PTHS, a clear deficit in TCF4 function is causal. However, examples exist where increased TCF4 expression is associated with psychiatric risk. For instance, duplication of TCF4 and increased expression of other specific TCF4 isoforms are associated with intellectual disability 20. In addition, TCF4 expression is enhanced in the fibroblasts of patients diagnosed with Schizophrenia 19. Endogenous TCF4 expression is elevated during corticogenesis, suggesting that it plays an important role in regulating cortical development. In Drosophila, both increase and decrease in gene expression of the TCF4 homolog Daughterless affects the morphology of postmitotic neurons and synaptic transmission at the neuromuscular junction, and similar morphological changes were observed when expression levels of TCF4 were altered in mature mouse olfactory bulb neurons 30. We previously showed that TCF4 regulates intrinsic excitability of pyramidal neurons in the mPFC by altering the expression of two ion channels in the central nervous system 27 and together with the results shown here, we suggest TCF4 is strongly associated with transcriptional regulation of neuronal activity and morphology during all phases of development. Taken together, this evidence suggests that the dosage of TCF4 is critical to proper neurodevelopment and understanding its effects in models of both deficiency and gain of function are relevant to gaining insights into etiology of psychiatric disorders. The convergence of Ca2+ signaling and TCF4-dependent transcription implies TCF4 likely regulates a variety of cellular functions including neuronal and circuit development, and synaptic plasticity, all of which are critical to building the brain and implicated as pathophysiological in psychiatric disorders.
Supplementary Material
A) Representative images of pyramidal cell distribution in the mPFC following transfection with TCF4B (left) or R582P (right). Small bins with a diameter of 10 pixels were generated perpendicular to the medial (for mPFC) or pial (for SCx) surface of the brain. Total pixel intensity within each bin was obtained. B) Graphical representation of pixel intensity versus bin number. The coefficient of variance (CV) of this distribution was calculated, providing a measure of cellular distribution. A high CV value indicates cellular aggregation, while a low CV value indicates uniform cellular distribution.
A) Example images showing the progression of neuronal migration and the formation of abnormal cortical columns over development. B) The appearance of abnormal cortical structures due to TCF4B expression is not observed until after migration is complete (E19: TCF4B n=5 0.20±0.031, R582P n=5 0.30±0.029, GFP n=5 0.28±0.033; p>0.05 (TCF4B vs. GFP); p>0.05 (R582P vs. GFP); E20: TCF4B n=4 0.22±0.048, R582P n=5 0.27±0.031, GFP n=4 0.21±−.057; p>0.05 (TCF4B vs. GFP); p>0.05 (R582P vs. GFP); P1: TCF4B n=8 0.45±0.056, R582P n=7 0.21±0.029, GFP n=8 0.22±0.030; p<0.001 (TCF4B vs. GFP); p>0.05 (R582P vs. GFP); P21: TCF4B n=5 0.57±0.061, R582P n=4 0.29±0.047, GFP n=4 0.30±0.036; p=0.004 (TCF4B vs. GFP); p>0.05 (R582P vs. GFP); two-factor ANOVA, * P<0.05; *** P<0.001).
A) Representative images of pyramidal cell distribution following transfection of TCF4B, R582P, TCF4A, and TCF4H. B) Group data showing that pyramidal cell distribution is significantly disrupted only in the mPFC by TCF4B (mPFC n=5 0.57±0.061 vs. SCx n=4 0.27±0.17; p<0.001) and TCF4A (mPFC n=9 0.45±0.039 vs. SCx n=5 0.32±0.015; p=0.038). Expression of R582P (mPFC n=4 0.33±0.036 vs. SCx n=4 0.29±0.021; p<0.05) or TCF4H (mPFC n=8 0.31±0.030 vs. SCx n=4 0.35±0.006; p<0.05) does not alter pyramidal cell distribution in either cortical region.
Acknowledgments
We are grateful for the vision and generosity of the Lieber and Maltz families, who made this work possible. This work was supported by the Lieber Institute, NIH (R56MH104593), NIH (R01MH110487), NARSAD Young Investigator Award, and Pitt-Hopkins Research Foundation Award to B.J.M.
Footnotes
AUTHOR CONTRIBUTION
SCP contributed to all aspects of histological and calcium imaging experiments, data analysis and manuscript preparation. GRH and MDR performed electrophysiological experiments. GRH, RAG, and MDR performed histological experiments. NEC performed co-IP experiments. RAG, MNC, BM, AB cloned and validated plasmid constructs. BHP, AEJ performed statistical analysis of calcium imaging experiments. BJM contributed to all aspects of experiments, electrophysiology, experimental design, writing, and data analysis.
CONFLICT OF INTEREST
The authors declare no conflict of interest.
References
- 1.Casanova MF, Buxhoeveden DP, Switala AE, Roy E. Minicolumnar pathology in autism. Neurology. 2002;58:428–432. doi: 10.1212/wnl.58.3.428. [DOI] [PubMed] [Google Scholar]
- 2.Casanova MF, Buxhoeveden DP, Cohen M, Switala AE, Roy EL. Minicolumnar pathology in dyslexia. Ann Neurol. 2002;52:108–110. doi: 10.1002/ana.10226. [DOI] [PubMed] [Google Scholar]
- 3.Casanova MF, de Zeeuw L, Switala A, Kreczmanski P, Korr H, Ulfig N, et al. Mean cell spacing abnormalities in the neocortex of patients with schizophrenia. Psychiatry Research. 2005;133:1–12. doi: 10.1016/j.psychres.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 4.Gleeson JG. Classical lissencephaly and double cortex (subcortical band heterotopia): LIS1 and doublecortin. Curr Opin Neurol. 2000;13:121–125. doi: 10.1097/00019052-200004000-00002. [DOI] [PubMed] [Google Scholar]
- 5.Belmonte MK. Autism and Abnormal Development of Brain Connectivity. Journal of Neuroscience. 2004;24:9228–9231. doi: 10.1523/JNEUROSCI.3340-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mountcastle V. The columnar organization of the neocortex. Brain. 1997;120:701–722. doi: 10.1093/brain/120.4.701. [DOI] [PubMed] [Google Scholar]
- 7.Noctor SC, Flint AC, Weissman TA, Dammerman RS, Kriegstein AR. Neurons derived from radial glial cells establish radial units in neocortex. Nature. 2001;409:714–720. doi: 10.1038/35055553. [DOI] [PubMed] [Google Scholar]
- 8.Yu Y-C, Bultje RS, Wang X, Shi S-H. Specific synapses develop preferentially among sister excitatory neurons in the neocortex. Nature. 2009;458:501–504. doi: 10.1038/nature07722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luhmann HJ, Fukuda A, Kilb W. Control of cortical neuronal migration by glutamate and GABA. Frontiers in Cellular Neuroscience. 2015;9:83. doi: 10.3389/fncel.2015.00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spitzer NC. Electrical activity in early neuronal development. Nature. 2006;444:707–712. doi: 10.1038/nature05300. [DOI] [PubMed] [Google Scholar]
- 11.Komuro H, Rakic P. Orchestration of neuronal migration by activity of ion channels, neurotransmitter receptors, and intracellular Ca2+ fluctuations. J Neurobiol. 1998;37:110–130. [PubMed] [Google Scholar]
- 12.Cancedda L, Fiumelli H, Chen K, Poo M-M. Excitatory GABA action is essential for morphological maturation of cortical neurons in vivo. J Neurosci. 2007;27:5224–5235. doi: 10.1523/JNEUROSCI.5169-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brockschmidt A, Todt U, Ryu S, Hoischen A, Landwehr C, Birnbaum S, et al. Severe mental retardation with breathing abnormalities (Pitt-Hopkins syndrome) is caused by haploinsufficiency of the neuronal bHLH transcription factor TCF4. Hum Mol Genet. 2007;16:1488–1494. doi: 10.1093/hmg/ddm099. [DOI] [PubMed] [Google Scholar]
- 14.Amiel J, Rio M, Pontual L, de Redon R, Malan V, Boddaert N, et al. Mutations in TCF4, Encoding a Class I Basic Helix-Loop-Helix Transcription Factor, Are Responsible for Pitt-Hopkins Syndrome, a Severe Epileptic Encephalopathy Associated with Autonomic Dysfunction. The American Journal of Human Genetics. 2007;80:988–993. doi: 10.1086/515582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zweier C, Peippo MM, Hoyer J, Sousa S, Bottani A, Clayton-Smith J, et al. Haploinsufficiency of TCF4 Causes Syndromal Mental Retardation with Intermittent Hyperventilation (Pitt-Hopkins Syndrome) The American Journal of Human Genetics. 2007;80:994–1001. doi: 10.1086/515583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ripke S, O’Dushlaine C, Chambert K, Moran JL, Kähler AK, Akterin S, et al. Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat Genet. 2013;45:1150–1159. doi: 10.1038/ng.2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhu X, Gu H, Liu Z, Xu Z, Chen X, Sun X, et al. Associations between TCF4 gene polymorphism and cognitive functions in schizophrenia patients and healthy controls. Neuropsychopharmacology. 2013;38:683–689. doi: 10.1038/npp.2012.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Quednow BB, Ettinger U, Mössner R, Rujescu D, Giegling I, Collier DA, et al. The schizophrenia risk allele C of the TCF4 rs9960767 polymorphism disrupts sensorimotor gating in schizophrenia spectrum and healthy volunteers. J Neurosci. 2011;31:6684–6691. doi: 10.1523/JNEUROSCI.0526-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cattane N, Minelli A, Milanesi E, Maj C, Bignotti S, Bortolomasi M, et al. Altered gene expression in schizophrenia: findings from transcriptional signatures in fibroblasts and blood. PLoS ONE. 2015;10:e0116686. doi: 10.1371/journal.pone.0116686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Talkowski ME, Rosenfeld JA, Blumenthal I, Pillalamarri V, Chiang C, Heilbut A, et al. Sequencing chromosomal abnormalities reveals neurodevelopmental loci that confer risk across diagnostic boundaries. Cell. 2012;149:525–537. doi: 10.1016/j.cell.2012.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ye T, Lipska BK, Tao R, Hyde TM, Wang L, Li C, et al. Analysis of copy number variations in brain DNA from patients with schizophrenia and other psychiatric disorders. Biol Psychiatry. 2012;72:651–654. doi: 10.1016/j.biopsych.2012.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maduro V, Pusey BN, Cherukuri PF, Atkins P, Souich du C, Rupps R, et al. Complex translocation disrupting TCF4 and altering TCF4 isoform expression segregates as mild autosomal dominant intellectual disability. Orphanet J Rare Dis. 2016;11:62. doi: 10.1186/s13023-016-0439-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kalscheuer VM, Feenstra I, Van Ravenswaaij-Arts CMA, Smeets DFCM, Menzel C, Ullmann R, et al. Disruption of the TCF4 gene in a girl with mental retardation but without the classical Pitt-Hopkins syndrome. Am J Med Genet A. 2008;146A:2053–2059. doi: 10.1002/ajmg.a.32419. [DOI] [PubMed] [Google Scholar]
- 24.Mullegama SV, Alaimo JT, Chen L, Elsea SH. Phenotypic and molecular convergence of 2q23. 1 deletion syndrome with other neurodevelopmental syndromes associated with autism spectrum disorder. Int J Mol Sci. 2015;16:7627–7643. doi: 10.3390/ijms16047627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Henthorn P, Kiledjian M, Kadesch T. Two distinct transcription factors that bind the immunoglobulin enhancer microE5/kappa 2 motif. Science. 1990;247:467–470. doi: 10.1126/science.2105528. [DOI] [PubMed] [Google Scholar]
- 26.Saarikettu J, Sveshnikova N, Grundström T. Calcium/calmodulin inhibition of transcriptional activity of E-proteins by prevention of their binding to DNA. J Biol Chem. 2004;279:41004–41011. doi: 10.1074/jbc.M408120200. [DOI] [PubMed] [Google Scholar]
- 27.Rannals MD, Hamersky GR, Page SC, Campbell MN, Briley A, Gallo RA, et al. Psychiatric Risk Gene Transcription Factor 4 Regulates Intrinsic Excitability of Prefrontal Neurons via Repression of SCN10a and KCNQ1. Neuron. 2016;90:43–55. doi: 10.1016/j.neuron.2016.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Colantuoni C, Lipska BK, Ye T, Hyde TM, Tao R, Leek JT, et al. Temporal dynamics and genetic control of transcription in the human prefrontal cortex. Nature. 2011;478:519–523. doi: 10.1038/nature10524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Pontual L, Mathieu Y, Golzio C, Rio M, Malan V, Boddaert N, et al. Mutational, functional, and expression studies of the TCF4 gene in Pitt-Hopkins syndrome. Hum Mutat. 2009;30:669–676. doi: 10.1002/humu.20935. [DOI] [PubMed] [Google Scholar]
- 30.D’Rozario M, Zhang T, Waddell EA, Zhang Y, Sahin C, Sharoni M, et al. Type I bHLH Proteins Daughterless and Tcf4 Restrict Neurite Branching and Synapse Formation by Repressing Neurexin in Postmitotic Neurons. Cell Rep. 2016;15:386–397. doi: 10.1016/j.celrep.2016.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maher BJ, LoTurco JJ. Controlled Genetic Manipulations. Humana Press; Totowa, NJ: 2011. In Utero Electroporation for Cellular Transgenesis in the Developing Mammalian Forebrain; pp. 113–128. [Google Scholar]
- 32.Matsuda T, Cepko CL. Controlled expression of transgenes introduced by in vivo electroporation. Proc Natl Acad Sci USA. 2007;104:1027–1032. doi: 10.1073/pnas.0610155104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Incontro S, Asensio CS, Edwards RH, Nicoll RA. Efficient, complete deletion of synaptic proteins using CRISPR. Neuron. 2014;83:1051–1057. doi: 10.1016/j.neuron.2014.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ran FA, Hsu PD, Lin C-Y, Gootenberg JS, Konermann S, Trevino AE, et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Calcaterra NE, Hoeppner DJ, Wei H, Jaffe AE, Maher BJ, Barrow JC. Schizophrenia-Associated hERG channel Kv11.1–3. 1 Exhibits a Unique Trafficking Deficit that is Rescued Through Proteasome Inhibition for High Throughput Screening. Sci Rep. 2016;6:19976. doi: 10.1038/srep19976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maher BJ, LoTurco JJ. Disrupted-in-schizophrenia (DISC1) functions presynaptically at glutamatergic synapses. PLoS ONE. 2012;7:e34053. doi: 10.1371/journal.pone.0034053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pau G, Fuchs F, Sklyar O, Boutros M, Huber W. EBImage--an R package for image processing with applications to cellular phenotypes. Bioinformatics. 2010;26:979–981. doi: 10.1093/bioinformatics/btq046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rakic P, Ayoub AE, Breunig JJ, Dominguez MH. Decision by division: making cortical maps. Trends in Neurosciences. 2009;32:291–301. doi: 10.1016/j.tins.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Powell LM, Jarman AP. Context dependence of proneural bHLH proteins. Current Opinion in Genetics & Development. 2008;18:411–417. doi: 10.1016/j.gde.2008.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saito A, Taniguchi Y, Rannals MD, Merfeld EB, Ballinger MD, Koga M, et al. Early postnatal GABAA receptor modulation reverses deficits in neuronal maturation in a conditional neurodevelopmental mouse model of DISC1. Molecular Psychiatry. 2016 doi: 10.1038/mp.2015.203.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sepp M, Pruunsild P, Timmusk T. Pitt-Hopkins syndrome-associated mutations in TCF4 lead to variable impairment of the transcription factor function ranging from hypomorphic to dominant-negative effects. Hum Mol Genet. 2012;21:2873–2888. doi: 10.1093/hmg/dds112. [DOI] [PubMed] [Google Scholar]
- 42.Sepp M, Kannike K, Eesmaa A, Urb M, Timmusk T. Functional Diversity of Human Basic Helix-Loop-Helix Transcription Factor TCF4 Isoforms Generated by Alternative 5′ Exon Usage and Splicing. PLoS ONE. 2011;6:e22138. doi: 10.1371/journal.pone.0022138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Corneliussen B, Holm M, Waltersson Y, Onions J, Hallberg B, Thornell A, et al. Calcium/calmodulin inhibition of basic-helix-loop-helix transcription factor domains. Nature. 1994;368:760–764. doi: 10.1038/368760a0. [DOI] [PubMed] [Google Scholar]
- 44.Lee W-S, Ngo-Anh TJ, Bruening-Wright A, Maylie J, Adelman JP. Small conductance Ca2+-activated K+ channels and calmodulin: cell surface expression and gating. J Biol Chem. 2003;278:25940–25946. doi: 10.1074/jbc.M302091200. [DOI] [PubMed] [Google Scholar]
- 45.Allène C, Cattani A, Ackman JB, Bonifazi P, Aniksztejn L, Ben-Ari Y, et al. Sequential generation of two distinct synapse-driven network patterns in developing neocortex. J Neurosci. 2008;28:12851–12863. doi: 10.1523/JNEUROSCI.3733-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.LoTurco JJ, Blanton MG, Kriegstein AR. Initial expression and endogenous activation of NMDA channels in early neocortical development. J Neurosci. 1991;11:792–799. doi: 10.1523/JNEUROSCI.11-03-00792.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Behar TN, Scott CA, Greene CL, Wen X, Smith SV, Maric D, et al. Glutamate acting at NMDA receptors stimulates embryonic cortical neuronal migration. J Neurosci. 1999;19:4449–4461. doi: 10.1523/JNEUROSCI.19-11-04449.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Corlew R, Bosma MM, Moody WJ. Spontaneous, synchronous electrical activity in neonatal mouse cortical neurones. J Physiol (Lond) 2004;560:377–390. doi: 10.1113/jphysiol.2004.071621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen F, Maher BJ, LoTurco JJ. piggyBac transposon-mediated cellular transgenesis in mammalian forebrain by in utero electroporation. Cold Spring Harb Protoc. 2014;2014:741–749. doi: 10.1101/pdb.prot073650. [DOI] [PubMed] [Google Scholar]
- 50.Chen T-W, Wardill TJ, Sun Y, Pulver SR, Renninger SL, Baohan A, et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature. 2013;499:295–300. doi: 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen Y, Ghosh A. Regulation of dendritic development by neuronal activity. J Neurobiol. 2005;64:4–10. doi: 10.1002/neu.20150. [DOI] [PubMed] [Google Scholar]
- 52.Massari ME, Murre C. Helix-Loop-Helix Proteins: Regulators of Transcription in Eucaryotic Organisms. Molecular and Cellular Biology. 2000;20:429–440. doi: 10.1128/mcb.20.2.429-440.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2006;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- 54.Bishop KM, Rubenstein JLR, O’Leary DDM. Distinct actions of Emx1, Emx2, and Pax6 in regulating the specification of areas in the developing neocortex. Journal of Neuroscience. 2002;22:7627–7638. doi: 10.1523/JNEUROSCI.22-17-07627.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Miyashita-Lin EM, Hevner R, Wassarman KM, Martinez S, Rubenstein JL. Early neocortical regionalization in the absence of thalamic innervation. Science (New York, NY) 1999;285:906–909. doi: 10.1126/science.285.5429.906. [DOI] [PubMed] [Google Scholar]
- 56.Langlands K, Yin X, Anand G, Prochownik EV. Differential interactions of Id proteins with basic-helix-loop-helix transcription factors. J Biol Chem. 1997;272:19785–19793. doi: 10.1074/jbc.272.32.19785. [DOI] [PubMed] [Google Scholar]
- 57.Tovar KR, Tovar KR, McGinley MJ, McGinley MJ, Westbrook GL, Westbrook GL. Triheteromeric NMDA Receptors at Hippocampal Synapses. Journal of Neuroscience. 2013;33:9150–9160. doi: 10.1523/JNEUROSCI.0829-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cull-Candy S, Brickley S, Farrant M. NMDA receptor subunits: diversity, development and disease. Current Opinion in Neurobiology. 2001;11:327–335. doi: 10.1016/s0959-4388(00)00215-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A) Representative images of pyramidal cell distribution in the mPFC following transfection with TCF4B (left) or R582P (right). Small bins with a diameter of 10 pixels were generated perpendicular to the medial (for mPFC) or pial (for SCx) surface of the brain. Total pixel intensity within each bin was obtained. B) Graphical representation of pixel intensity versus bin number. The coefficient of variance (CV) of this distribution was calculated, providing a measure of cellular distribution. A high CV value indicates cellular aggregation, while a low CV value indicates uniform cellular distribution.
A) Example images showing the progression of neuronal migration and the formation of abnormal cortical columns over development. B) The appearance of abnormal cortical structures due to TCF4B expression is not observed until after migration is complete (E19: TCF4B n=5 0.20±0.031, R582P n=5 0.30±0.029, GFP n=5 0.28±0.033; p>0.05 (TCF4B vs. GFP); p>0.05 (R582P vs. GFP); E20: TCF4B n=4 0.22±0.048, R582P n=5 0.27±0.031, GFP n=4 0.21±−.057; p>0.05 (TCF4B vs. GFP); p>0.05 (R582P vs. GFP); P1: TCF4B n=8 0.45±0.056, R582P n=7 0.21±0.029, GFP n=8 0.22±0.030; p<0.001 (TCF4B vs. GFP); p>0.05 (R582P vs. GFP); P21: TCF4B n=5 0.57±0.061, R582P n=4 0.29±0.047, GFP n=4 0.30±0.036; p=0.004 (TCF4B vs. GFP); p>0.05 (R582P vs. GFP); two-factor ANOVA, * P<0.05; *** P<0.001).
A) Representative images of pyramidal cell distribution following transfection of TCF4B, R582P, TCF4A, and TCF4H. B) Group data showing that pyramidal cell distribution is significantly disrupted only in the mPFC by TCF4B (mPFC n=5 0.57±0.061 vs. SCx n=4 0.27±0.17; p<0.001) and TCF4A (mPFC n=9 0.45±0.039 vs. SCx n=5 0.32±0.015; p=0.038). Expression of R582P (mPFC n=4 0.33±0.036 vs. SCx n=4 0.29±0.021; p<0.05) or TCF4H (mPFC n=8 0.31±0.030 vs. SCx n=4 0.35±0.006; p<0.05) does not alter pyramidal cell distribution in either cortical region.