Abstract
Aminoacyl-tRNA synthetases (ARSs) are ubiquitously expressed enzymes that ligate amino acids onto tRNA molecules. Genes encoding ARSs have been implicated in myriad dominant and recessive disease phenotypes. Glycyl-tRNA synthetase (GARS) is a bi-functional ARS that charges tRNAGly in the cytoplasm and mitochondria. GARS variants have been associated with dominant Charcot-Marie-Tooth disease but have not been convincingly implicated in recessive phenotypes. Here we describe a patient from the NIH Undiagnosed Diseases Program with a multi-system, developmental phenotype. Whole-exome sequence analysis revealed that the patient is compound heterozygous for one frameshift (p.Glu83Ilefs*6) and one missense (p.Arg310Gln) GARS variant. Using in vitro and in vivo functional studies, we show that both GARS variants cause a loss-of-function effect: the frameshift variant results in depleted protein levels and the missense variant reduces GARS tRNA charging activity. In support of GARS variant pathogenicity, our patient shows striking phenotypic overlap with other patients having ARS-related recessive diseases, including features associated with variants in both cytoplasmic and mitochondrial ARSs; this observation is consistent with the essential function of GARS in both cellular locations. In summary, our clinical, genetic, and functional analyses expand the phenotypic spectrum associated with GARS variants.
Keywords: Glycyl-tRNA synthetase, GARS, aminoacyl-tRNA synthetase, growth retardation, developmental syndrome
INTRODUCTION
Aminoacyl-tRNA synthetases (ARS) are ubiquitously expressed, essential enzymes that charge tRNA molecules with cognate amino acids—the first step of cytoplasmic and mitochondrial protein synthesis. The human nuclear genome harbors 37 ARS genes including 17 that encode a cytoplasmic enzyme, 17 that encode a mitochondrial enzyme, and three that encode bi-functional enzymes that charge tRNA for protein synthesis in both cellular locations (Antonellis and Green, 2008). ARS loci and enzymes are named by the single-letter code for the amino acid that they charge, plus ‘ARS’ and a variable ‘2’ for enzymes that charge tRNA exclusively in the mitochondria. For example, AARS encodes cytoplasmic alanyl-tRNA synthetase and AARS2 encodes the mitochondrial form of alanyl-tRNA synthetase.
Glycyl-tRNA synthetase (GARS; MIM# 600287) encodes a bi-functional tRNA-charging enzyme that differentially localizes to the cytoplasm or mitochondria via two translation start sites that either include or exclude a mitochondrial targeting sequence (Antonellis and Green, 2008). GARS variants have been implicated in CMT2D, an autosomal dominant, axonal form of Charcot-Marie-Tooth disease (MIM# 600287) (Antonellis et al., 2003). CMT2D is characterized by an upper limb predominant peripheral neuropathy that affects motor and sensory function in the distal extremities (Sivakumar et al., 2005). One patient with a recessive mitochondrial phenotype was reported to be compound heterozygous for GARS missense variants: p.Arg596Gln and p.Ser635Leu (McMillan et al., 2014). However, certain limitations of the above study indicated that additional analyses were required to determine the role of GARS variants in recessive disease. First, the restriction of the phenotype to impaired mitochondrial function is surprising given the role of GARS in cytoplasmic protein translation and the severity of fly and mouse phenotypes associated with bi-allelic mutations in glycyl-tRNA synthetase upon severe depletion of GARS function (Chihara et al., 2007; Motley et al., 2011). Second, the authors of the above study did not provide functional data supporting the pathogenicity of the variants; enzyme kinetic analyses were not performed and, indeed, p.Ser635Leu was previously shown to encode a functional GARS enzyme (Nangle et al., 2007). Finally, p.Ser635Leu was previously shown to not be associated with dominant human disease (Griffin et al., 2014); however, the variant was not evaluated for a role in recessive phenotypes. Here we present clinical, genetic, and functional data that implicate compound heterozygosity for loss-of-function GARS variants in a recessive, early onset, multi-system, developmental phenotype. Our findings expand the phenotypic heterogeneity associated with GARS variants.
MATERIAL AND METHODS
Clinical evaluation and variant analysis
The proband was admitted to the National Institutes of Health (NIH) Undiagnosed Diseases Program (UDP) under the protocol 76-HG-0238, “Diagnosis and Treatment of Patients with Inborn Errors of Metabolism or Other Genetic Disorders,” approved by the National Human Genome Research Institute (NHGRI) Institutional Review Board (IRB). Clinical data were obtained after written informed consent from the proband’s parents. Genomic DNA was extracted from whole blood using the Gentra Puregene Blood Kit (Qiagen). Single-nucleotide polymorphism (SNP) analyses were performed using an Illumina Omni Express 12 (hg18) SNP array and the Genome Studio software program. Whole-exome sequencing was performed using the Illumina HiSeq2000 platform and the TrueSeq capture kit (Illumina) by the NIH Intramural Sequencing Center (NISC) as described (Bentley et al., 2008; Gnirke et al., 2009; Teer et al., 2010; Gahl et al., 2012). Sequence data were aligned to the human reference genome (hg19) using Novoalign (Novocraft Technologies). Variants were analyzed, sorted, and filtered with VarSifter and a graphical java tool to view, sort, and filter variants (Teer et al., 2012). Variants were filtered based on allele frequencies in the NIH-UDP (Gahl and Tifft, 2011; Gahl et al., 2012; Markello et al., 2012) cohort (<0.06). Variant annotation is based on NM_002047.3 and each variant was submitted to ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/): c.246_249del is accession number SCV000579486, and c.929G>A is accession number SCV000579487.
Protein isolation and western blot analysis
Primary fibroblasts from the affected individual and a control individual (i.e., bearing no GARS variants) were obtained and utilized for western blot analysis. Cells were cultured in T-75cm3 treated cell culture flasks (USA Scientific) and maintained at 37°C and 5% CO2 in complete medium [Dulbecco’s Modified Eagle Medium (DMEM; Invitrogen) with 10% fetal bovine serum (FBS; ThermoFisher Scientific), 1X L-glutamine (Corning), 1X Penicillin-Streptomycin (final concentration 50units/mL penicillin and 50μg/mL streptomycin; ThermoFisher Scientific)]. Cells were harvested at 90% confluence with 3mL .25% trypsin (ThermoFisher Scientific), quenched with 7mL complete cell medium, and collected at 2,000 rpm for 2 minutes. The resulting pellet was resuspended in 1mL cell lysis buffer [RIPA buffer (ThermoFisher Scientific) + 1X EDTA-free protease inhibitor (ThermoFisher Scientific)] and incubated at 4°C for 30 minutes with gentle shaking. Cell debris was collected at 4°C and 12,000 rpm for 5 minutes and the resulting supernatant was subjected to western blot analysis.
Total protein concentration was measured using the Thermo Scientific Pierce BCA Protein Assay Kit (ThermoFisher Scientific) and 10μg protein/sample was analyzed unless otherwise specified. Protein samples were prepared in 1X SDS-sample buffer (ThermoFisher Scientific) and 1uL 2-mercaptoethanol (βME), denatured at 99°C for 5 minutes and separated on a pre-cast 4–20% tris-glycine protein gel (ThermoFisher Scientific) at 150 V for 2 hours at room temperature. Prior to protein transfer, a polyvinylidene difluoride (PVDF) membrane was pre-washed in 100% methanol and soaked in 1X transfer buffer (ThermoFisher Scientific) with extra thick blot paper (Bio Rad) and 3 mm Chromatography paper (Whatman PLC). Protein was transferred onto the membrane using a Transbot-SD Semi-Dry Transfer Cell (Bio Rad) at 23V for 1.5 hours at room temperature and subsequently blocked over night at 4°C in blocking solution [1 g dry milk powder, 50 mL 1X TBST (tris-buffered saline solution and tween 20, pH 7.5)]. The membrane was cut in half and incubated in blocking solution with either anti-GARS673 antibody (Antonellis et al., 2006) (at 1/2,000 dilution) or anti-actin antibody [at 1/5,000 dilution (Sigma Aldrich)] at room temperature for 1 hour. The membrane was washed three times in 1X TBST to remove unbound primary antibody and incubated in blocking solution with 10 ng/mL Donkey anti-Rabbit IgG HRP secondary antibody (ThermoFisher Scientific) at room temperature for 1 hour. The membrane was washed three times in 1X TBST to remove unbound secondary antibody and exposed using SuperSignal West Dura substrate and enhancer (ThermoFisher Scientific). Western blot analysis was repeated four independent times with independent fibroblast cell lysates and ImageJ (https://imagej.nih.gov/ij/) was used to quantify band intensity. The analyzed control and affected fibroblast cell lines were passage-matched (±3 passages).
Generation of GARS expression constructs
GARS expression constructs were generated using Gateway cloning technology (Invitrogen). The human GARS open-reading frame was amplified from human cDNA using primers designed with Gateway sequences attB1 (forward) and attB2 (reverse; primer sequences available on request). Entry clones were generated by recombining PCR-purified amplicons into the pDONR221 vector using BP clonase. After transformation into E. coli, DNA from individual entry clones was isolated and subjected to Sanger sequencing analysis to confirm the presence of the appropriate wild-type gene. Oligonucleotides containing sequences corresponding to the c.929G>A GARS variant (encoding p.Arg310Gln) were generated and mutagenesis was performed using the QuickChange II XL Site-Directed Mutagenesis Kit (Stratagene). After transformation into E. coli, DNA from individual clones was purified and sequenced to confirm the presence of each variant and the absence of any errors. Validated entry clones were purified and recombined into a Gateway-compatible pET-21a(+) vector for aminoacylation assays (Griffin et al., 2014) using LR clonase. DNA from the resulting expression constructs was purified and digested with BsrGI (New England Biosystems) to confirm the presence of the insert.
For yeast complementation assays, human GARS proteins were expressed from the pYY1 expression construct containing full-length, ΔMTS, or ΔMTSΔWHEP GARS, or no insert (empty); these constructs were a gift from Chin-I Chien and Chien-Chia Wang (National Central University, Taiwan) (Chien et al., 2014). We made pYY1 Gateway-compatible by digesting the empty pYY1 plasmid with SpeI (New England Biolabs), blunt-cloning in reading frame cassette A (Invitrogen), and subsequent transformation into ccdB resistant E. coli (Invitrogen) and purification of plasmid DNA. Sanger sequencing confirmed proper cassette orientation downstream of the ADH1 promoter. To generate human GARS expression constructs for yeast experiments, the full-length, ΔMTS, and ΔMTSΔWHEP GARS open-reading frames were amplified from the initial expression constructs with primers bearing Gateway flanking sequences. The resulting PCR products were gel-purified and BP-cloned into the pDONR221 entry vector (ThermoFisher Scientific). Sanger sequencing confirmed specificity and lack of PCR-induced errors. The p.Arg310Gln GARS variant was generated using the QuickChange II XL Site-Directed Mutagenesis Kit, a sequence validated wild-type full-length, ΔMTS, or ΔMTSΔWHEP GARS pDONR221 template, and variant-specific primers. The reaction was transformed into E. coli, plasmid DNA was purified from the individual colonies and then subjected to Sanger sequencing to confirm the presence of the variant and the absence of PCR-induced errors. A sequence-validated pDONR221 construct for wild-type or mutant full-length, ΔMTS, or ΔMTSΔWHEP GARS was LR-cloned into the Gateway compatible pYY1 construct. The resulting reactions were transformed into E. coli, purified, and digested with BsrG1 (New England Biolabs) to confirm the presence of the respective human GARS open-reading frame.
Aminoacylation assays
Wild-type and p.Arg310Gln GARS were expressed in E. coli with a C-terminal His tag and purified with nickel affinity resins (Novagen). The T7 transcript of human tRNAGly/UCU (UCU, anticodon) was prepared and purified as previously described (Hou et al., 1993), heat denatured at 85°C for 3 min, and annealed at 37°C for 20 min before use. Steady-state aminoacylation assays were monitored at 37°C in 50 mM HEPES (pH 7.5), 20 mM KCl, 10 mM MgCl2, 4 mM DTT, 2 mM ATP, and 50 mM 3H-glycine (Perkin Elmer) at a specific activity of 16,500 dpm/pmole. The reaction was initiated by mixing GARS enzyme (20–600 nM) with varying concentrations of tRNA (0.3–20 μM). Aliquots of a reaction mixture were spotted on filter paper, quenched by 5% trichloroacetic acid, washed, dried, and measured for radioactivity using a liquid scintillation counter (LS6000SC; Beckman Coulter Inc.). The amount of radioactivity retained on filter pads was corrected for quenching effects to determine the amount of synthesis of Gly-tRNAGly. Steady-state kinetics was determined by fitting the initial rate of aminoacylation as a function of tRNA concentration to the Michaelis-Menten equation (Schreier and Schimmel, 1972).
Yeast complementation assays
Yeast complementation assays were carried out using the haploid S. cerevisiae strain disrupted for the endogenous GRS1 and maintained viable via pRS316 bearing wild-type GRS1 (Turner et al., 2000). To assess the ability of wild-type and mutant human GARS alleles to support cellular growth, the haploid yeast strain was transformed with wild-type full-length, wild-type ΔMTS, or wild-type ΔMTSΔWHEP GARS, the corresponding p.Arg310Gln GARS expression constructs, or empty pYY1 (i.e., bearing no GARS insert) (Chien et al., 2014). Transformed yeast cells were selected for the presence of pYY1 by growth on solid media lacking leucine (pYY1 harbors the LEU2 gene) and uracil (pRS316 harbors the URA3 gene). Colonies were grown to saturation in 2 mL liquid medium lacking leucine and uracil (-leu-ura) at 30°C and shaking at 275 rpm for 48 hours. Each 2 mL saturated culture was spun down at 1,000 rpm and re-suspended in 100 uL UltraPure Rnase/DNase-free water. Undiluted cultures and dilutions of 1:10, 1:100, and 1:1,000 were spotted on 0.1% 5-FOA complete solid medium (Teknova, Hollister CA) to select for cells that spontaneously lost the URA3-bearing maintenance vector (Boeke et al., 1984). Yeast viability was visually assessed after 4 days of incubation at 30°C. Two colonies per transformation were assayed and each transformation was repeated twice.
RESULTS
Clinical and genetic analysis of patient UDP5316
A simplex pedigree was presented to the NIH Undiagnosed Diseases Program. The proband is a seven-year-old Caucasian female of non-consanguineous German and Polish ancestry with extreme growth retardation and multiple organ involvement as described below. She was a 1.52 kg infant born at 36 weeks gestation to a 27-year-old G1P0 mother by scheduled Caesarian section due to severe intrauterine growth retardation beginning at 29 weeks. There were no maternal exposures and maternal weight gain was only 5 pounds. APGAR scores were 4 and 9 at 1 and 4 minutes, respectively. Length, weight, and head circumference were all 50th percentile for 31 weeks gestation. She remained in the NICU for 2 weeks for poor weight gain.
TORCH titers (Group B streptococci, toxoplasma, CMV, HSV II) were negative. HSV I IgG was reported positive (greater than 5), indicating a previous immunological exposure but the patient did not show clinical signs of active infection and did not receive treatment; this might be due to an outbreak of cold sores as reported by the proband’s mother early in pregnancy. Hepatitis B, hepatitis C, Rubella, HIV, Herpes, and standard STD panel were negative. Ophthalmic exam was normal. An echocardiogram revealed peripheral pulmonic stenosis.
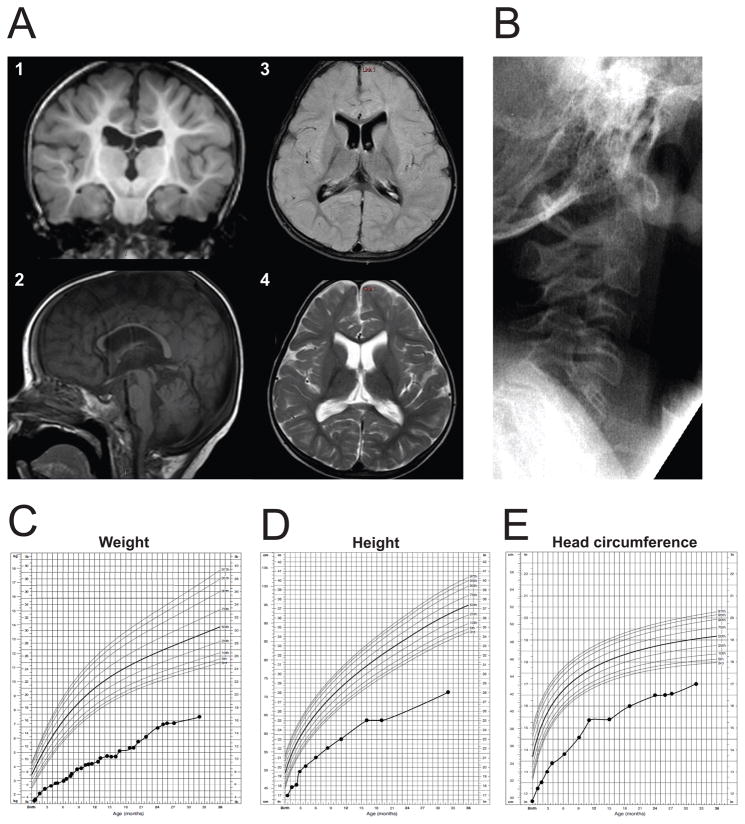
Despite adequate calories the child failed to thrive. X-rays at 7 months of age showed cephalofacial disproportion with a large calvaria, numerous wormian bones, and low-normal bone mineralization. Skull X-ray at 2 years, 9 months of age no longer showed the wormian bones, but the rest of the findings remained the same (Supp. Figure S1). Brain MRI at 29 months showed moderate-to-severe thinning of the corpus callosum, mild atrophy of the cerebellar vermis, and a small brain stem (Fig. 1A). There was decreased cerebral white matter volume consistent with delayed myelination (also observed at 11 months of age; data not shown), and mildly enlarged ventricles. Cervical X-ray revealed that C5 through C7 were fused (Fig. 1B). Cardiac evaluation at 4 years identified a moderately sized atrial septal defect and pulmonic stenosis. A sleep study demonstrated severe central and obstructive apnea. Recurrent ear and upper respiratory infections prompted myringotomy tube placement and prophylactic antibiotic treatments. She continues to suffer from atopic dermatitis and recurrent rhinitis. Developmentally, the child rolled at 4 months, sat at 16 months, commando crawled at 10 months, cruised at 22 months, and walked at 6 years. Fine motor skills include feeding herself with a pincer grasp at 5 years and holding a pencil at 5 years. Her verbal skills include 20–25 single words with poor intelligibility.
Figure 1.
Clinical features of patient UDP5316. (A) Representative MRI images for the proband at 29 months of age. Cerebral cortex appeared normal, but there was decreased cerebral white matter volume and delayed myelination (shown in 1, coronal image, T1-weighted), moderate-to-severe thinning of the corpus callosum, mild atrophy of the cerebellar vermis, small brain stem (shown in 2, sagittal image, T1-weighted), and mildly enlarged ventricles (axial images; 3, FLAIR; 4, T2-weighted). (B) Cervical spine X-ray shows fusion of C5–C7 vertebra. (C–E) Growth curves of the patient plotted in comparison with normal children 0–36 months of age; weight (C), height (D), and head circumference (E).
On initial NIH evaluation at 2 years 9 months her weight was 7.6 kgs, height was 72 cm, and head circumference was 43 cm. These measures are all below the 5th percentile for 33 months of age. The child was microcephalic with a patent anterior fontanelle and sparse, thin scalp hair. Facial features included a triangular face with broad forehead, prominent epicanthal folds, hypotelorism, smooth philtrum, and a high-arched palate. She was non-ambulatory and displayed upper motor neuron signs with hyperreflexia and spasticity of both upper and lower extremities but normal range of motion and hypermobility of her ankles. She did not have any signs of a lower motor neuron disorder or neuropathy and her sensation to temperature, light touch, and vibration were all normal.
Dilated ophthamologic evaluation revealed a normal macula and vessels but peripheral retinal changes including atrophy and depigmentation. Auditory evaluation showed bilateral sensorineural hearing loss.
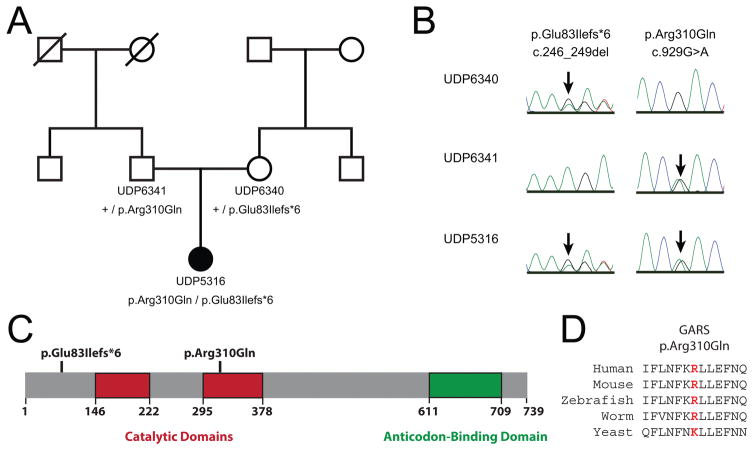
To identify candidate variants for the unexplained phenotype, whole-exome sequencing analysis was performed on genomic DNA isolated from the patient (UDP5316), her unaffected mother (UDP6340), and her unaffected father (UDP6341) (UDP; Fig. 2A). Using a Boolean sort with parents and proband as heterozygous in VarSifter, a mean allele frequency of <0.02 (1,000 genomes), and an allele frequency of 0 in the NIH-UDP cohort (Bentley et al., 2008; Gnirke et al., 2009; Teer et al., 2010; Gahl and Tifft, 2011; Gahl et al., 2012; Markello et al., 2012; Teer et al., 2012), we identified two variants in the gene encoding hexokinase 3 (HK3) and two variants in the gene encoding glycyl-tRNA synthetase (GARS). The HK3 variants were excluded from further analyses because one variant (c.1733A>G, NM_002115.2; rs61749651) is present in the exome aggregate (ExAC)/genome aggregate (gnomAD) database (Lek et al., 2016) at a relatively high frequency (3,862 alleles in 276,696 chromosomes including 54 homozygous genotypes). The GARS variants (NM_002047.3) include : (1) c.246_249del, which is predicted to cause an early frame-shift variant p.Glu83Ilefs*6; and (2) c.929G>A, which is predicted to cause a missense variant p.Arg310Gln (Table 1). The unaffected mother carries p.Glu83Ilefs*6 in the heterozygous state, the unaffected father carries p.Arg310Gln in the heterozygous state, and the affected proband is compound heterozygous for both GARS variants (Fig. 2A and 2B). Neither variant is present in the ExAC/gnomAD database (Table 1) (Lek et al., 2016). Modeling each of the identified GARS variants on the primary structure of the protein revealed that p.Glu83Ilefs*6 is predicted to ablate all key functional domains of the enzyme (both catalytic domains and the tRNA binding domain) and that p.Arg310Gln affects a residue in the catalytic core of the enzyme (Fig. 2C). Multiple-species conservation analysis revealed that p.Arg310Gln affects a residue that is conserved among diverse species including human, mouse, zebrafish, and worm (Fig. 2D).
Figure 2.
Identification of GARS variants in patient UDP5316. (A) The simplex pedigree is shown with squares representing males, circles representing females, and diagonal lines representing deceased individuals. Sample numbers and genotypes are indicated under each symbol for the mother, father, and proband (filled circle). The proband is the only affected individual in the pedigree. (B) Representative sequence chromatograms are shown for the identified variants (protein and cDNA annotations are provided at the top) in the indicated individuals (indicated along the left side). Arrows indicate the position of each variant. (C) GARS functional domains are indicated in red (catalytic domain) and green (anti-codon binding domain), and the position of each variant is shown across the top. Numbers along the bottom indicate amino acid positions. (D) The position of the p.Arg310Gln variant is shown along with flanking amino acid residues for multiple, evolutionarily diverse species. The position of the affected residue is shown in red for each species.
Table 1.
GARS variants identified in patient UDP5316
| Nucleotide changea | Amino-acid changeb | Detection in gnomADc |
|---|---|---|
| c.246_249del | p.Glu83Ilefs*6 | Not present |
| c.929G>A | p.Arg310Gln | Not present |
Human nucleotide positions correspond to GenBank Accession number NM_002047.3
Human amino-acid positions correspond to GenBank Accession number NP_002038.2 chttp://gnomad.broadinstitute.org
p.Glu83Ilefs*6 GARS reduces expression of the full-length GARS protein
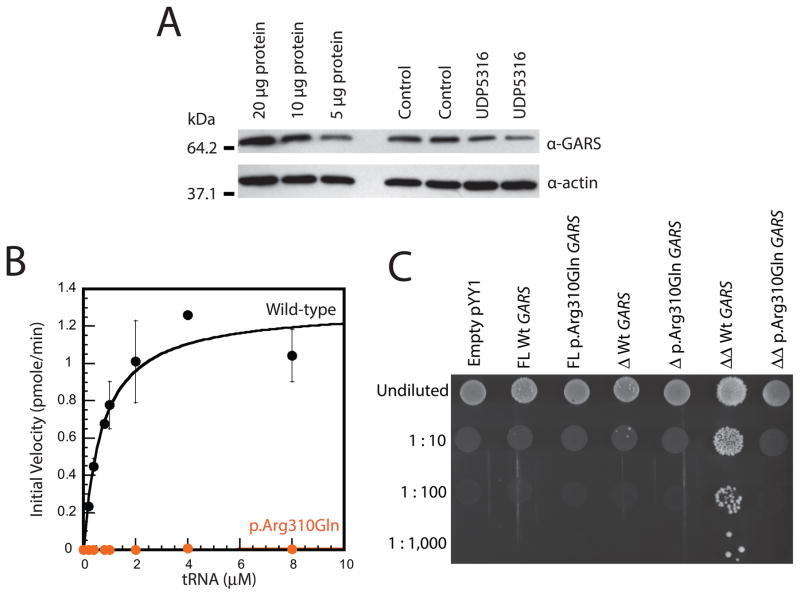
Because p.Gly83IllefsX6 GARS encodes a premature stop codon, the variant is predicted to ablate the expression of full-length GARS, which would result in a 50% decrease in full-length GARS protein levels in patient cells. To test this, we performed western blot analysis using whole-cell lysates from fibroblasts isolated from patient UDP5316 and from an unaffected control (i.e., a patient negative for GARS variants). Protein samples were electrophoresed, transferred onto a PVDF membrane, and incubated with an anti-GARS antibody(Antonellis et al., 2006). Our results revealed a ~50% reduction in full-length GARS protein levels in the patient fibroblast cells compared to the control cell line (Fig. 2A); a similar assessment of three additional control fibroblast samples confirmed these findings (Supp. Figure S2). To confirm the sensitivity of our western blot analysis, we analyzed 20μg, 10μg, and 5μg of total protein from the control cell lysate with the same antibody and were able to detect a 50% reduction in protein levels (Fig. 3A). We therefore conclude that p.Gly83IllefsX6 GARS ablates the expression of full-length GARS and posit that the remaining protein represents translation from the p.Arg310Gln GARS allele.
Figure 3.
The GARS variants identified in patient UDP5316 reduce gene function. (A) Western blot analyses were performed with total protein lysates from fibroblast cell lines from unaffected (Control) and affected (UDP5316) patients using an anti-GARS or anti-actin antibody, as indicated. Sample names are across the top and sizes (kDa) are indicated at the left. (B) Initial aminoacylation rates (pmole/min) of wild-type (black circles and line) and p.Arg310Gln GARS (orange circles and line) were plotted against tRNA concentrations and fit to the Michaelis-Menten equation. Error bars indicate standard deviations. (C) Representative cultures of the indicated yeast strains were plated on solid growth medium containing 5-FOA. Each strain was previously transfected with a vector containing no insert (Empty pYY1), or containing either wild-type (Wt) or mutant (p.Arg310Gln) GARS in open-reading frames to express full-length (FL) protein, a protein with the mitochondrial targeting sequence deleted (Δ), or a protein with the mitochondrial targeting sequence and the WHEP domain deleted (ΔΔ). Before inoculating on medium containing 5-FOA, each strain was diluted 1:10, 1:100, or 1:1,000 in water. Please note the minimal growth associated with the wild-type FL protein in the undiluted sample, the intermediate growth associated with the wild-type Δ protein in the undiluted and 1:10 diluted samples, and the robust growth associated with wild-type ΔΔ protein in all dilutions.
The p.Arg310Gln GARS variant reduces enzyme activity
The primary function of GARS is to charge tRNAGly molecules with glycine in a two-step aminoacylation reaction: (i) binding and activation of the amino acid with ATP and (ii) conjugation of the activated amino acid to the 3′-end of the appropriate tRNA (Delarue, 1995). The aminoacylation assay evaluates the completion of both steps and has been used to establish that CMT2D-associated variants disrupt GARS enzymatic activity (Nangle et al., 2007; Griffin et al., 2014). We assessed recombinant human GARS proteins (wild-type and p.Arg310Gln) for the ability to charge human cytoplasmic tRNAGly with tritium-labeled glycine as previously described (Griffin et al., 2014). Analysis of the initial rate of aminoacylation as a function of the tRNA substrate concentration showed that p.Arg310Gln GARS demonstrates less than 1% aminoacylation activity compared to wild-type GARS (Fig. 3B and Table 2) indicating that it is a hypomorphic allele.
Table 2.
Kinetic analysis of p.Arg310Gln GARS.
| Variant | kcat (s−1) | Km (μM) | kcat/Km (s−1 μM−1) | Ratio |
|---|---|---|---|---|
| Wild-type | 0.049 ± 0.014 | 0.74 ± 0.30 | 0.066 | 1 |
| p.Arg310Gln | 0.00040 ± 1.0×10−4 | 5.5 ± 2.2 | 7.2×10−5 | 1/920 |
The p.Arg310Gln GARS variant does not support yeast cell growth in complementation assays
Yeast complementation assays have been employed to reveal loss-of-function characteristics of CMT2D-associated GARS variants (Antonellis et al., 2006; Griffin et al., 2014). While previous efforts employed the yeast ortholog of GARS (GRS1) to assess human variants (Turner et al., 2000; Antonellis et al., 2006; Griffin et al., 2014), a recent study showed that three engineered isoforms of the human GARS protein were able to rescue deletion of GRS1: (1) full-length GARS allowed minimal cell growth; (2) GARS with the mitochondrial targeting sequence deleted (ΔMTS) allowed intermediate cell growth; and (3) GARS with both the MTS and WHEP domain deleted (ΔMTSΔWHEP) allowed robust cell growth (Chien et al., 2014) [the function of the WHEP domain has not been defined but it is not essential for enzyme activity (Chang et al., 2016)]. We therefore evaluated the effect of the p.Arg310Gln GARS variant in all three of the above GARS isoforms. Briefly, the wild-type or p.Arg310Gln GARS open-reading frames (full-length, ΔMTS, or ΔMTSΔWHEP) were cloned downstream of the yeast ADH1 promoter (Chien et al., 2014) and yeast complementation assays were performed using a haploid yeast strain deleted for the endogenous GRS1 locus (ΔGRS1); this strain carries wild-type GRS1 on a URA3-bearing maintenance vector to maintain viability (Turner et al., 2000). Wild-type or mutant GARS expression constructs, or one with no GARS insert (‘Empty’ in Fig. 3C), were introduced into the above strain and growth was subsequently evaluated on medium containing 0.1% 5-FOA, which selects for the spontaneous loss of the maintenance vector (Boeke et al., 1984). All of the wild-type human GARS expression constructs supported yeast growth, while the empty plasmid did not (Fig. 3C), consistent with human GARS supporting loss of the endogenous GRS1 locus and with GRS1 being an essential gene, respectively. In contrast, none of the p.Arg310Gln GARS expression constructs supported yeast cell growth, consistent with this being a loss-of-function allele (Fig. 3C). Combined, our functional studies indicate that p.Gly83IllefsX6 GARS represents a null allele and that p.Arg310Gln GARS represents a hypomorphic allele.
DISCUSSION
Twenty eight ARS loci encoding mitochondrial, cytoplasmic, and bi-functional ARS enzymes have been implicated in myriad recessive human diseases (Oprescu et al., 2017). GARS variants cause autosomal dominant, axonal Charcot-Marie-Tooth disease (CMT2D) (Antonellis et al., 2003) but have not been convincingly implicated in recessive phenotypes. Here we present clinical, genetic, and functional data that implicate GARS variants in a severe, multi-system, developmental phenotype. Our functional studies indicate that p.Gly83IllefsX6 GARS ablates the expression of full-length GARS and that p.Arg310Gln GARS represents a severely hypomorphic allele. It is important to note that aminoacylation and yeast complementation assays were previously employed to implicate loss-of-function variants at other ARS loci in recessive diseases similar to the phenotype described here (please see below): aminoacylation assays have been used to study disease-associated variants at 15 ARS loci and yeast complementation assays have been used to study disease-associated variants at eight ARS loci (Oprescu et al., 2017). Therefore, our informative and relevant functional analyses support the pathogenicity of p.Gly83IllefsX6 and p.Arg310Gln GARS in the phenotype reported here.
Careful clinical comparisons between our patient and previously reported patients with ARS-associated recessive phenotypes (Oprescu et al., 2017) reveals remarkable overlap, further supporting the pathogenicity of the GARS variants reported here. For example, ARS genes have been associated with the following recessive phenotypes; (i) growth retardation in patients with AARS (MIM# 601065), IARS (MIM# 600709), MARS (MIM# 156560), MARS2 (MIM# 609728), QARS (MIM# 603727), VARS2 (MIM# 612802), and YARS (MIM# 603623) variants; (ii) microcephaly in patients with AARS, IARS, KARS (MIM# 601421), NARS2 (MIM# 612803), PARS2 (MIM# 612036), QARS, VARS (MIM# 192150), and VARS2 variants; (iii) thinning of the corpus callosum in patients with AARS2 (MIM# 612035), CARS2 (MIM# 612800), EARS2 (MIM# 612799), FARS2 (MIM# 611592), KARS, QARS, TARS2 (MIM# 612805), VARS, YARS, and YARS2 (MIM# 610957) variants; (iv) decreased white matter in patients with KARS and QARS variants; (v) brain stem involvement in patients with DARS2 (MIM# 610956), QARS, and RARS2 (MIM# 611524) variants; (vi) spasticity in patients with AARS, DARS, FARS2, IARS, MARS2, and RARS (MIM# 107820) variants; and (vii) sensorineural hearing loss in patients with CARS2, HARS2 (MIM# 600783), IARS, IARS2 (MIM# 612801), KARS, LARS2 (MIM# 604544), and MARS2 variants.
The recessive disorders caused by variants in the three bi-functional ARSs appear to share more clinical phenotypes compared to all ARS-associated disorders as a whole. Our patient shares many features with patients who are compound heterozygous for glutaminyl-tRNA synthetase (QARS) (Zhang et al., 2014) or lysyl-tRNA synthetase (KARS) variants (McLaughlin et al., 2010; Santos-Cortez et al., 2013; McMillan et al., 2015). Specifically, our patient shares growth retardation, microcephaly, thinning of the corpus callosum, decreased white matter, and brain stem involvement (all noted above), as well as large calvaria, cerebellar vermis atrophy, dysmorphic features, prominent epicanthal folds, hypotelorism, high-arched palate, delayed motor milestones, apnea, and sparse thin scalp hair with patients carrying loss-of-function QARS variants. Our patient also shares microcephaly, thinning of the corpus callosum, decreased white matter, and sensorineural hearing loss (all noted above), as well as dysmorphic features and delayed motor development with patients carrying loss-of-function KARS variants. These observations are significant since GARS, QARS, and KARS compose the three bi-functional ARSs that charge tRNA in the cytoplasm and mitochondria (Antonellis and Green, 2008). In summary, the clinical features of our patient compared to other patients with ARS variants and our functional studies showing that p.Gly83IllefsX6 and p.Arg310Gln dramatically impair GARS function both support the conclusion that compound heterozygosity for loss-of-function GARS variants results in the severe, multi-system, developmental phenotype reported here.
Loss-of-function missense variants in GARS have been associated with CMT2D, which is characterized by distal muscle weakness and sensory loss that is predominant in the upper extremities (Antonellis et al., 2003; Sivakumar et al., 2005). Interestingly, neither parent of the affected proband currently displays signs of CMT2D; EMG and nerve conduction studies performed on both parents were unremarkable at ages 30 (mother) and 32 (father); these ages are well-beyond the average age of onset for CMT2D, which is typically in the second decade of life (Sivakumar et al., 2005). With regard to the mother who carries the null allele (p.Gly83IllefsX6), this is perhaps not surprising since haploinsufficiency is not a likely mechanism of GARS-associated CMT2D; mice heterozygous for a null allele have no detectable phenotype while mice heterozygous for a missense variant present with a dominant neuropathy (Seburn et al., 2006; Motley et al., 2011). In contrast, p.Arg310Gln GARS shows features of CMT2D-associated variants (Griffin et al., 2014); it is a missense variant in the catalytic domain that affects tRNA charging capacity. Two possible explanations for this discrepancy are the reduced penetrance of GARS variants as seen for p.Glu71Gly and p.Leu129Pro GARS (Sivakumar et al., 2005) and the poorly defined mechanism of CMT2D (i.e., p.Arg310Gln GARS may be a loss-of-function missense variant that is not dominantly toxic to peripheral nerve axons). Addressing this discrepancy will require animal models and molecular studies to determine if, for example, p.Arg310Gln GARS inappropriately binds neuropilin-1, which has been observed for certain CMT2D-causing variants (He et al., 2015).
In conclusion, we present clinical, genetic, and functional data that associate GARS variants with a severe, multi-system, developmental phenotype. This report exemplifies the growing number of ARS loci and alleles implicated in recessive human diseases (Oprescu et al., 2017), expands the phenotypic heterogeneity associated with GARS variants, and underscores the importance of employing careful clinical comparisons and informative functional studies to support the pathogenicity of newly identified ARS variants.
Supplementary Material
Acknowledgments
The authors are indebted to the patient and their family for participating in this study. We would also like to thank Chin-I Chien and Chien-Chia Wang (National Central University, Taiwan) for the pYY1 expression construct and Laurie Griffin for assistance with developing GARS expression constructs.
Contract grant sponsors: This work was supported by the National Institute of General Medical Sciences (GM110184 and GM118647 to A.A.; GM108972 and GM114343 to Y.M.H). This study was also supported in part by the Intramural Research Program of the National Human Genome Research Institute and the NIH Common Fund, Office of the Director.
References
- Antonellis A, Ellsworth RE, Sambuughin N, Puls I, Abel A, Lee-Lin S-Q, Jordanova A, Kremensky I, Christodoulou K, Middleton LT, Sivakumar K, Ionasescu V, Funalot B, Vance JM, Goldfarb LG, Fischbeck KH, Green ED. Glycyl tRNA synthetase mutations in Charcot-Marie-Tooth disease type 2D and distal spinal muscular atrophy type V. 2003;72:1293–1299. doi: 10.1086/375039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonellis A, Green ED. The role of aminoacyl-tRNA synthetases in genetic diseases. Annual review of genomics and human genetics. 2008;9:87–107. doi: 10.1146/annurev.genom.9.081307.164204. [DOI] [PubMed] [Google Scholar]
- Antonellis A, Lee-Lin S-Q, Wasterlain A, Leo P, Quezado M, Goldfarb LG, Myung K, Burgess S, Fischbeck KH, Green ED. Functional analyses of glycyl-tRNA synthetase mutations suggest a key role for tRNA-charging enzymes in peripheral axons. J Neurosci. 2006;26:10397–10406. doi: 10.1523/JNEUROSCI.1671-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, Hall KP, Evers DJ, Barnes CL, Bignell HR, Boutell JM, Bryant J, Carter RJ, Keira Cheetham R, Cox AJ, Ellis DJ, Flatbush MR, Gormley NA, Humphray SJ, Irving LJ, Karbelashvili MS, Kirk SM, Li H, Liu X, Maisinger KS, Murray LJ, Obradovic B, Ost T, Parkinson ML, Pratt MR, Rasolonjatovo IM, Reed MT, Rigatti R, Rodighiero C, Ross MT, Sabot A, Sankar SV, Scally A, Schroth GP, Smith ME, Smith VP, Spiridou A, Torrance PE, Tzonev SS, Vermaas EH, Walter K, Wu X, Zhang L, Alam MD, Anastasi C, Aniebo IC, Bailey DM, Bancarz IR, Banerjee S, Barbour SG, Baybayan PA, Benoit VA, Benson KF, Bevis C, Black PJ, Boodhun A, Brennan JS, Bridgham JA, Brown RC, Brown AA, Buermann DH, Bundu AA, Burrows JC, Carter NP, Castillo N, Chiara E, Catenazzi M, Chang S, Neil Cooley R, Crake NR, Dada OO, Diakoumakos KD, Dominguez-Fernandez B, Earnshaw DJ, Egbujor UC, Elmore DW, Etchin SS, Ewan MR, Fedurco M, Fraser LJ, Fuentes Fajardo KV, Scott Furey W, George D, Gietzen KJ, Goddard CP, Golda GS, Granieri PA, Green DE, Gustafson DL, Hansen NF, Harnish K, Haudenschild CD, Heyer NI, Hims MM, Ho JT, Horgan AM, Hoschler K, Hurwitz S, Ivanov DV, Johnson MQ, James T, Huw Jones TA, Kang GD, Kerelska TH, Kersey AD, Khrebtukova I, Kindwall AP, Kingsbury Z, Kokko-Gonzales PI, Kumar A, Laurent MA, Lawley CT, Lee SE, Lee X, Liao AK, Loch JA, Lok M, Luo S, Mammen RM, Martin JW, McCauley PG, McNitt P, Mehta P, Moon KW, Mullens JW, Newington T, Ning Z, Ling NgB, Novo SM, O’Neill MJ, Osborne MA, Osnowski A, Ostadan O, Paraschos LL, Pickering L, Pike AC, Pike AC, Chris Pinkard D, Pliskin DP, Podhasky J, Quijano VJ, Raczy C, Rae VH, Rawlings SR, Chiva Rodriguez A, Roe PM, Rogers J, Rogert Bacigalupo MC, Romanov N, Romieu A, Roth RK, Rourke NJ, Ruediger ST, Rusman E, Sanches-Kuiper RM, Schenker MR, Seoane JM, Shaw RJ, Shiver MK, Short SW, Sizto NL, Sluis JP, Smith MA, Ernest Sohna Sohna J, Spence EJ, Stevens K, Sutton N, Szajkowski L, Tregidgo CL, Turcatti G, Vandevondele S, Verhovsky Y, Virk SM, Wakelin S, Walcott GC, Wang J, Worsley GJ, Yan J, Yau L, Zuerlein M, Rogers J, Mullikin JC, Hurles ME, McCooke NJ, West JS, Oaks FL, Lundberg PL, Klenerman D, Durbin R, Smith AJ. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke JD, LaCroute F, Fink GR. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet. 1984;197:345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- Chang C-Y, Chien C-I, Chang C-P, Lin B-C, Wang C-C. A WHEP Domain Regulates the Dynamic Structure and Activity of Caenorhabditis elegansGlycyl-tRNA Synthetase. J Biol Chem. 2016;291:16567–16575. doi: 10.1074/jbc.M116.730812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien C-I, Chen Y-W, Wu Y-H, Chang C-Y, Wang T-L, Wang C-C. Functional substitution of a eukaryotic glycyl-tRNA synthetase with an evolutionarily unrelated bacterial cognate enzyme. PLoS ONE. 2014;9:e94659. doi: 10.1371/journal.pone.0094659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chihara T, Luginbuhl D, Luo L. Cytoplasmic and mitochondrial protein translation in axonal and dendritic terminal arborization. Nat Neurosci. 2007;10:828–837. doi: 10.1038/nn1910. [DOI] [PubMed] [Google Scholar]
- Delarue M. Aminoacyl-tRNA synthetases. Curr Opin Struct Biol. 1995;5:48–55. doi: 10.1016/0959-440x(95)80008-o. [DOI] [PubMed] [Google Scholar]
- Gahl WA, Markello TC, Toro C, Fajardo KF, Sincan M, Gill F, Carlson-Donohoe H, Gropman A, Pierson TM, Golas G, Wolfe L, Groden C, Godfrey R, Nehrebecky M, Wahl C, Landis DM, Yang S, Madeo A, Mullikin JC, Boerkoel CF, Tifft CJ, Adams D. The National Institutes of Health Undiagnosed Diseases Program: insights into rare diseases. Genet Med. 2012;14:51–59. doi: 10.1038/gim.0b013e318232a005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gahl WA, Tifft CJ. The NIH Undiagnosed Diseases Program: lessons learned. JAMA. 2011;305:1904–1905. doi: 10.1001/jama.2011.613. [DOI] [PubMed] [Google Scholar]
- Gnirke A, Melnikov A, Maguire J, Rogov P, LeProust EM, Brockman W, Fennell T, Giannoukos G, Fisher S, Russ C, Gabriel S, Jaffe DB, Lander ES, Nusbaum C. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat Biotechnol. 2009;27:182–189. doi: 10.1038/nbt.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin LB, Sakaguchi R, McGuigan D, Gonzalez MA, Searby C, Züchner S, Hou Y-M, Antonellis A. Impaired Function is a Common Feature of Neuropathy-Associated Glycyl-tRNA Synthetase Mutations. Hum Mutat. 2014;35:1363–1371. doi: 10.1002/humu.22681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W, Bai G, Zhou H, Wei N, White NM, Lauer J, Liu H, Shi Y, Dumitru CD, Lettieri K, Shubayev V, Jordanova A, Guergueltcheva V, Griffin PR, Burgess RW, Pfaff SL, Yang XL. CMT2D neuropathy is linked to the neomorphic binding activity of glycyl-tRNA synthetase. Nature. 2015;526:710–714. doi: 10.1038/nature15510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou YM, Westhof E, Giegé R. An unusual RNA tertiary interaction has a role for the specific aminoacylation of a transfer RNA. Proc Natl Acad Sci USA. 1993;90:6776–6780. doi: 10.1073/pnas.90.14.6776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lek M, Karczewski KJ, Minikel EV, Samocha KE, Banks E, Fennell T, O’Donnell-Luria AH, Ware JS, Hill AJ, Cummings BB, Tukiainen T, Birnbaum DP, Kosmicki JA, Duncan LE, Estrada K, Zhao F, Zou J, Pierce-Hoffman E, Berghout J, Cooper DN, Deflaux N, DePristo M, Do R, Flannick J, Fromer M, Gauthier L, Goldstein J, Gupta N, Howrigan D, Kiezun A, Kurki MI, Moonshine AL, Natarajan P, Orozco L, Peloso GM, Poplin R, Rivas MA, Ruano-Rubio V, Rose SA, Ruderfer DM, Shakir K, Stenson PD, Stevens C, Thomas BP, Tiao G, Tusie-Luna MT, Weisburd B, Won HH, Yu D, Altshuler DM, Ardissino D, Boehnke M, Danesh J, Donnelly S, Elosua R, Florez JC, Gabriel SB, Getz G, Glatt SJ, Hultman CM, Kathiresan S, Laakso M, McCarroll S, McCarthy MI, McGovern D, McPherson R, Neale BM, Palotie A, Purcell SM, Saleheen D, Scharf JM, Sklar P, Sullivan PF, Tuomilehto J, Tsuang MT, Watkins HC, Wilson JG, Daly MJ, MacArthur DG. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markello TC, Han T, Carlson-Donohoe H, Ahaghotu C, Harper U, Jones M, Chandrasekharappa S, Anikster Y, Adams DR, Gahl WA, Boerkoel CF NISC Comparative Sequencing Program. Recombination mapping using Boolean logic and high-density SNP genotyping for exome sequence filtering. Mol Genet Metab. 2012;105:382–389. doi: 10.1016/j.ymgme.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin HM, Sakaguchi R, Liu C, Igarashi T, Pehlivan D, Chu K, Iyer R, Cruz P, Cherukuri PF, Hansen NF, Mullikin JC, Biesecker LG, Wilson TE, Ionasescu V, Nicholson G, Searby C, Talbot K, Vance JM, Züchner S, Szigeti K, Lupski JR, Hou YM, Green ED, Antonellis A NISC Comparative Sequencing Program. Compound heterozygosity for loss-of-function lysyl-tRNA synthetase mutations in a patient with peripheral neuropathy. 2010;87:560–566. doi: 10.1016/j.ajhg.2010.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan HJ, Humphreys P, Smith A, Schwartzentruber J, Chakraborty P, Bulman DE, Beaulieu CL, Majewski J, Boycott KM, Geraghty MT FORGE Canada Consortium. Congenital Visual Impairment and Progressive Microcephaly Due to Lysyl–Transfer Ribonucleic Acid (RNA) Synthetase ( KARS) Mutations. Journal of Child Neurology. 2015;30:1037–1043. doi: 10.1177/0883073814553272. [DOI] [PubMed] [Google Scholar]
- McMillan HJ, Schwartzentruber J, Smith A, Lee S, Chakraborty P, Bulman DE, Beaulieu CL, Majewski J, Boycott KM, Geraghty MT. Compound heterozygous mutations in glycyl-tRNA synthetase are a proposed cause of systemic mitochondrial disease. BMC Med Genet. 2014;15:36. doi: 10.1186/1471-2350-15-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motley WW, Seburn KL, Nawaz MH, Miers KE, Cheng J, Antonellis A, Green ED, Talbot K, Yang X-L, Fischbeck KH, Burgess RW. Charcot-Marie-Tooth-linked mutant GARS is toxic to peripheral neurons independent of wild-type GARS levels. 2011;7:e1002399. doi: 10.1371/journal.pgen.1002399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nangle LA, Zhang W, Xie W, Yang X-L, Schimmel P. Charcot-Marie-Tooth disease-associated mutant tRNA synthetases linked to altered dimer interface and neurite distribution defect. Proc Natl Acad Sci USA. 2007;104:11239–11244. doi: 10.1073/pnas.0705055104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oprescu SN, Griffin LB, Beg AA, Antonellis A. Predicting the pathogenicity of aminoacyl-tRNA synthetase mutations. Methods. 2017;113:139–151. doi: 10.1016/j.ymeth.2016.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Cortez RL, Lee K, Azeem Z, Antonellis PJ, Pollock LM, Khan S, Irfanullah, Andrade-Elizondo PB, Chiu I, Adams MD, Basit S, Smith JD, Nickerson DA, McDermott BM, Jr, Ahmad W, Leal SM University of Washington Center for Mendelian Genomics. Mutations in KARS, encoding lysyl-tRNA synthetase, cause autosomal-recessive nonsyndromic hearing impairment DFNB89. Am J Hum Genet. 2013;93:132–140. doi: 10.1016/j.ajhg.2013.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreier AA, Schimmel PR. Transfer ribonucleic acid synthetase catalyzed deacylation of aminoacyl transfer ribonucleic acid in the absence of adenosine monophosphate and pyrophosphate. Biochemistry. 1972;11:1582–1589. doi: 10.1021/bi00759a006. [DOI] [PubMed] [Google Scholar]
- Seburn KL, Nangle LA, Cox GA, Schimmel P, Burgess RW. An active dominant mutation of glycyl-tRNA synthetase causes neuropathy in a Charcot-Marie-Tooth 2D mouse model. Neuron. 2006;51:715–726. doi: 10.1016/j.neuron.2006.08.027. [DOI] [PubMed] [Google Scholar]
- Sivakumar K, Kyriakides T, Puls I, Nicholson GA, Funalot B, Antonellis A, Sambuughin N, Christodoulou K, Beggs JL, Zamba-Papanicolaou E, Ionasescu V, Dalakas MC, Green ED, Fischbeck KH, Goldfarb LG. Phenotypic spectrum of disorders associated with glycyl-tRNA synthetase mutations. Brain. 2005;128:2304–2314. doi: 10.1093/brain/awh590. [DOI] [PubMed] [Google Scholar]
- Teer JK, Bonnycastle LL, Chines PS, Hansen NF, Aoyama N, Swift AJ, Abaan HO, Albert TJ, Margulies EH, Green ED, Collins FS, Mullikin JC, Biesecker LG NISC Comparative Sequencing Program. Systematic comparison of three genomic enrichment methods for massively parallel DNA sequencing. Genome Res. 2010;20:1420–1431. doi: 10.1101/gr.106716.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teer JK, Green ED, Mullikin JC, Biesecker LG. VarSifter: visualizing and analyzing exome-scale sequence variation data on a desktop computer. Bioinformatics. 2012;28:599–600. doi: 10.1093/bioinformatics/btr711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner RJ, Lovato M, Schimmel P. One of two genes encoding glycyl-tRNA synthetase in Saccharomyces cerevisiae provides mitochondrial and cytoplasmic functions. J Biol Chem. 2000;275:27681–27688. doi: 10.1074/jbc.M003416200. [DOI] [PubMed] [Google Scholar]
- Zhang X, Ling J, Barcia G, Jing L, Wu J, Barry BJ, Mochida GH, Hill RS, Weimer JM, Stein Q, Poduri A, Partlow JN, Ville D, Dulac O, Yu TW, Lam AT, Servattalab S, Rodriguez J, Boddaert N, Munnich A, Colleaux L, Zon LI, Söll D, Walsh CA, Nabbout R. Mutations in QARS, encoding glutaminyl-tRNA synthetase, cause progressive microcephaly, cerebral-cerebellar atrophy, and intractable seizures. 2014;94:547–558. doi: 10.1016/j.ajhg.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.