Abstract
Aminoacyl-transfer RNA (tRNA) synthetases ligate amino acids to specific tRNAs and are essential for protein synthesis. Although alanyl-tRNA synthetase (AARS) is a synthetase implicated in a wide range of neurological disorders from Charcot-Marie-Tooth (CMT) disease to infantile epileptic encephalopathy, there have been limited data on their pathogenesis. Here we report loss-of-function mutations in AARS in two siblings with progressive microcephaly with hypomyelination, intractable epilepsy and spasticity. Whole exome sequencing identified that the affected individuals were compound heterozygous for mutations in AARS gene, c.2067dupC (p.Tyr690Leufs*3) and c.2738G>A (p.Gly913Asp). A lymphoblastoid cell line developed from one of the affected individuals showed a strong reduction in AARS abundance. The mutations decrease aminoacylation efficiency by 70–90%. The p.Tyr690Leufs*3 mutation also abolished editing activity required for hydrolyzing misacylated tRNAs, thereby increasing errors during aminoacylation. Our study has extended potential mechanisms underlying AARS-related disorders to include destabilization of the protein, aminoacylation dysfunction, and defective editing activity.
Keywords: AARS, transfer RNA, microcephaly, hypomyelination, aminoacylation defect
Aminoacyl-transfer RNA synthetases (ARSs) have recently received increased attention because their dysfunctions are associated with many human disorders. ARSs catalyze the ligation of cognate amino acids to transfer RNAs, a critical step for maintaining the efficiency and fidelity of protein translation [Ibba and Soll 2000; Mascarenhas et al. 2008]. The human genome encodes 37 aminoacyl-tRNA synthetases that are present in the cytoplasm and mitochondria (18 and 17, respectively, and 2 bifunctional). To date, more than 20 ARSs have been found to be associated with human diseases [Antonellis and Green 2008; Yao and Fox 2013]. While mutations in mitochondrial ARSs lead to disorders affecting various organ systems, mutations in cytoplasmic ARSs have mainly been associated with autosomal dominant peripheral neuropathies [Antonellis et al. 2003; He et al. 2015]. Recently, a growing number of mutations in cytoplasmic ARSs genes, including QARS (MIM# 603727) [Zhang et al. 2014], DARS (MIM# 603084) [Taft et al. 2013; Novarino et al. 2014], MARS (MIM# 156560) [Novarino et al. 2014], KARS (MIM# 601421) [McMillan et al. 2015], AARS (MIM# 601065) [Simons et al. 2015], and RARS (MIM# 107820) [Wolf et al. 2014], have been identified in recessive disorders affecting the central nervous system. The shared phenotypes among these recessive conditions, including progressive microcephaly [Zhang et al. 2014; McMillan et al. 2015], hypomyelinating leukodystrophy [Taft et al. 2013; Wolf et al. 2014], and epileptic encephalopathy [Kodera et al. 2015], suggest a possible commonality in the ARSs-related molecular pathology underlying these neurodegenerative disorders.
Alanyl-tRNA synthetase (AARS) is an ARS that was initially implicated in autosomal dominant Charcot-Marie-Tooth (CMT) disease [Latour et al. 2010; McLaughlin et al. 2012; Bansagi et al. 2015; Motley et al. 2015]. Recently, biallelic mutations in AARS were identified in three individuals from two families, who presented with early-onset epileptic encephalopathy [Simons et al. 2015]. Similar to cases with mutations in other ARS genes, these affected individuals were reported to have microcephaly and hypomyelination of the cerebral white matter. However, the clinical spectrum of the AARS-related disorders has not been fully elucidated, and it remains unclear why AARS mutations cause distinct neurological disorders with different modes of inheritance.
Here we report the identification of novel biallelic AARS mutations in two siblings presenting with progressive microcephaly with hypomyelination and spastic paraplegia. We ascertained a non-consanguineous family of mixed European descent with two affected individuals presenting with congenital microcephaly and epilepsy (Fig. 1A). The parents (MC32505 and MC32506) and two siblings (MC32503 and MC32504) were healthy and did not have neurological symptoms, such as peripheral neuropathy. The female proband (MC32502) was born at full term via spontaneous vaginal delivery. Birth weight was 2.98 kg (−0.8 SD) and occipito-frontal circumference (OFC) at birth was 31.5 cm (−2.2 SDs). She had transient hypoglycemia at birth, developed progressive microcephaly and failure to thrive, and presented with infantile spasms at 3 months of age. Her electroencephalogram (EEG) showed discontinuous pattern consisting of bursts of high voltage sharp wave activity followed by spontaneous attenuation, which was reported as modified hypsarrhythmia. The infantile spasms transiently responded to adrenocorticotropic hormone (ACTH) therapy but then she had recurrent spasms and seizures, as well as occasional atypical absence seizures. At 7 years of age she developed recurrent tonic-clonic seizures with persistent slow spike and wave pattern and multifocal epileptiform discharges on EEG. The clinical evolution and multiple types of seizures with the typical EEG findings were consistent with Lennox-Gastaut syndrome.
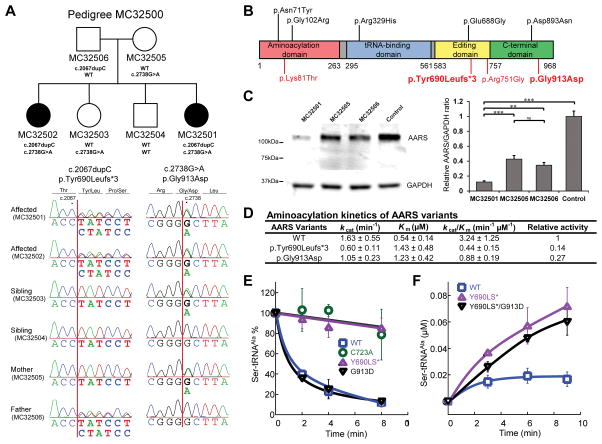
Figure 1. AARS mutations cause loss-of-fuction defects.
A: Identification of AARS mutations. Pedigree structure of the family studied is shown. Shaded symbols indicate affected individuals. Sanger sequencing confirmed the segregation of the identified variants in the affected individuals (MC32501 and MC32502). B: Schematic representation of the AARS mutations. Variants acting in dominant and recessive manners are shown in black and red, respectively. The variants in this study are in bold. The residue Tyr690 is in the editing domain and the residue Gly913 is in the C-terminal domain. C: Immunoblot analysis of AARS abundance in lymphoblastoid cell lines. AARS level in cells available from an affected individual (MC32501) showed a marked reduction of AARS, whereas cells from her parents (MC32505 and MC32506) showed a mild reduction. A representative image by using AARS antibody (A303-473A-T, Bethyl), with an epitope near the N-terminus, is shown. Quantification of AARS abundance was performed Relative AARS/GAPDH intensity ratio was normalized to the control. Values represent the average of three independent experiments. Error bars represent standard deviation. ***p<0.001, **p<0.01, ns; not significant. D: Aminoacylation kinetics of AARS protein variants. Relative acitivity is caliculated by ratio to WT activity. E: Hydrolytic editing activity of AARS. Deacylation of the incorrectly charged Ser-tRNAAla was monitored with wild type AARS (WT; blue square), p.Tyr690Leufs*3 (p.T690fs*; purple triangle), and p.Gly913Asp variants (p.G913D; black triangle). p.Cys723Ala (p.C723A; green circle) is served as a positive control of editing defect. F: Misacylation of AARS variants. Accumulation of misacylated Ser-tRNAAla was measured on wild type AARS (WT; blue square) and p.Tyr690Leufs*3 (p.T690fs*; purple triangle). Error bar shows the standard deviation. Values represent the average of at least three independent experiments.
At 3 months of age, her OFC was 37.5 cm (−1.9 SDs), her height was 58 cm (−0.5 SD), and her weight was 5.16 kg (−0.6 SD). She showed increased tone, with opisthotonic posturing when she was held. At 4 years 4 months of age, her OFC was 43 cm (−4.6 SDs), her height was 82.6 cm (−4.7 SDs), and her weight was 13.6 kg (−1.7 SDs). She had poor head control with increased reflexes in the upper and lower extremities bilaterally. On observation at 8 years 6 months of age, she displayed profound delay of growth and development. She was able to walk with a walker but did not have any speech. She had markedly increased tone in her lower extremities and showed no dysmorphic features.
Magnetic resonance imaging (MRI) at 2 years 5 months of age revealed reduced white matter volume with hypomyelination, suggesting underlying metabolic disorder (Supplmentary Fig. S1). The corpus callosum was fully formed but very thin. A series of tests including liver function, plasma amino acids, urine organic acids, lactic acid, pyruvate, acylcarnitine profile, lysosomal screen and ceruloplasmin was all normal.
The younger sister (MC32501) had a similar clinical course with congenital microcephaly and global developmental delay. She was born via induced vaginal delivery at 38 weeks’ gestation. Ultrasound exam at about 36 weeks’ gestation revealed poor fetal growth. At birth, her weight was 1.96 kg (−2.8 SDs) and her OFC was 30.1 cm (−3.1 SDs). She had failure to thrive, microcephaly and generalized hypotonia at birth, and spent 6 days in the Neonatal Intensive Care Unit because of apnea and poor feeding. At 2 months of age she developed episodes of seizures that were described as head dropping. EEG showed multiple electrical seizures in the frontal and temporal regions. Possible decrements were noted at the time of the head drops, which are frequently associated with infantile spasms, although there were no high voltage discharges suggestive of hypsarrhythmia. At 6 years of age she presented multiple seizure patterns with tonic seizures and head drops. A follow-up EEG showed frequent slow-spike and wave discharges with bilateral epileptiform discharges (Supplementary Fig. S2), indicating encephalopathy of Lennox-Gastaut syndrome with infantile spasms as a clinical seizure manifestation.
MRI performed at 4 months of age showed relative hypomyelination with diminished white matter volume (Supplmentary Fig. S1). At 5 months of age, her OFC was 35.75 cm (−4.9 SDs), height was 63.5 cm (±0.0 SD), and weight was 4.72 kg (−2.6 SDs). She had bilateral hypertonia and spasticity with increased deep tendon reflexes in the upper and lower extremities, and acquisition of developmental milestones was delayed. Liver function tests were normal. Neither affected individual had nystagmus, tremors, or any congenital abnormalities of their extremities.
We performed whole-exome sequencing on the affected individuals (MC32501, MC32502). We screened forvariants that were protein altering and rare (allele frequency less than 0.1% in available population databases and less than 1% in our in-house exome database, consisting of 1466 samples from 512 families, mainly with autism and brain malformations), and fit the autosomal recessive mode of inheritance. Two genes, AARS and FLG (MIM# 135940), were found to have two variants shared by both siblings, for which they were heterozygous. Another gene, TAS2R31 (MIM# 612669), was found to have five variants shared by both siblings, for which they were heterozygous. FLG is associated with ichthyosis vulgaris, a condition not known to be present in this family, and TAS2R31 encodes a taste receptor. Therefore, these genes are unlikely to be causative of their neurological phenotype. The two variants in the AARS gene were c.2067dupC [p.Tyr690Leufs*3] and c.2738G>A [p.Gly913Asp] (positions of the variants are according to NM_001605.2 for cDNA and NP_001596.2 for protein), and these were confirmed by Sanger sequencing to segregate with the condition in the family (Fig. 1A). The frameshift variant c.2067dupC was inherited from the father (MC32506) and the missense variant c.2738G>A was inherited from the mother (MC32505). None of unaffected siblings (MC32503 and MC32504) were homozygous for AARS variants. The frameshift variant c.2067dupC is predicted to produce a truncated AARS (p.Tyr690Leufs*3). The missense variant c.2738G>A leads to a p.Gly913Asp change in AARS and it is predicted to be deleterious (Supplementary Fig. S3). Tyr690 is located in the editing domain of AARS [Beebe et al. 2003] and Gly913 is located in the C-terminal domain (Fig. 1B), which is crucial to its editing activity[Beebe et al. 2003]. Alignment of AARS orthologs shows that both residues are highly conserved (Supplementary Fig. S4).
We established a lymphoblastoid cell line from one of the affected individuals (MC32501) and both her parents (MC32505 and MC32506), and performed immunoblot analysis of AARS. The AARS protein level in the cells from the affected individual was markedly reduced compared to the parents and control (Figure 1C), suggesting that these mutations could impair AARS gene functions by reducing protein abundance. No truncated protein was identified in the affected individual (MC32501) or her father (MC32506), who carried the frameshift variant. We performed these experiments using two different AARS antibodies, with epitopes near the N-terminus and C-terminus, and all results were similar (Supplementary Fig. S5). The relative abundance of AARS protein in the affected individuals was estimated to be 3–12% of that in control individuals.
Given that AARS is the only enzyme responsible for alanyl-tRNA aminoacylation reaction, we speculate that a low but functional level of AARS abundance is essential for survival in these affected individuals. Low levels of mischarged transfer alanyl-transfer RNAs due to a defective tRNA synthetase could lead to an intracellular accumulation of misfolded proteins in neurons (Lee et al., Nature 2006). To examine whether the residual p.Tyr690Leufs*3 and p.Gly913Asp mutants compromise the aminoacylation activity of AARS, we next overexpressed and purified recombinant variants carrying each mutation. The truncation caused by p.Tyr690Leufs*3 was confirmed by the observation that the recombinant variant migrated at a smaller molecular weight than the wild-type AARS on SDS-PAGE (Supplemenary Fig. S6). The aminoacylation activities of both variants were then tested using 14C-labeled alanine with various concentrations of tRNA. The results showed that the p.Tyr690Leufs*3 mutation caused a 86% decrease in catalytic efficiency (kcat/Km) (Fig. 1D). The p.Gly913Asp mutation led to a 73% decrease in catalytic efficiency. The loss of aminoacylation activity was due to increased Km and decreased kcat.
tRNAAla can be mischarged with serine and glycine by AARS to yield Ser-tRNAAla and Gly-tRNAAla, respectively [Tsui and Fersht 1981]. To maintain the fidelity of aminoacylation, AARS possesses an editing domain that is capable of hydrolyzing the mischarged tRNAAla [Beebe et al. 2003; Guo et al. 2009]. As p.Tyr690 is located within the editing domain, we subsequently investigated whether p.Tyr690Leufs*3 impairs this editing activity. An in vitro editing activity assay was carried out using [3H] Ser-tRNAAla as a substrate. AARS p.Cys723Ala, a mutant that has been previously shown to have defective editing activity in mice [Liu et al. 2014a], was included as a positive control. In the presence of AARS p.Tyr690Leufs*3, the deacylation of [3H] Ser-tRNAAla was diminished to a level comparable to the AARS p.Cys723Ala mutant (Fig. 1E). In contrast, the AARS p.Gly913Asp mutant maintained unaffected deacylation activity compared to the wild-type AARS. This result indicates that the p.Tyr690Leufs*3 mutation drastically decreases the editing activity, whereas the p.Gly913Asp mutation does not impact the editing activity of AARS. Decreased editing activity leads to the production and release of misacylated tRNAs. To further assess whether the defective editing of the p.Tyr690Leufs*3 mutant increases the level of misacylation, we measured production of [3H] Ser-tRNAAla by wild-type AARS and the p.Tyr690Leufs*3 AARS mutant. Compared with the wild-type, more than 3-fold higher accumulation of [3H] Ser-tRNAAla was observed in the p.Tyr690Leufs*3 mutant (Fig. 1F), confirming that misacylation resulted from the editing deficiency.
In this study, we demonstrate that mutations in AARS are associated with autosomal recessive progressive microcephaly with hypomyelination, intractable epilepsy and spasticity. We found that the mutations could potentially lead to the loss of function of AARS in multiple ways, including a significant reduction of AARS protein abundance, impairment of aminoacylation activity and impairment of evolutionarily conserved editing function, which maintains fidelity during aminoacylation and overall protein synthesis. Multiple mutations in AARS have been implicated in autosomal dominant Charcot-Marie-Tooth disease type 2N (CMT2N), characterized by peripheral neuropathies due to axonal pathology [Latour et al. 2010; McLaughlin et al. 2012; Bansagi et al. 2015] (Fig. 1B). Individuals with CMT2N typically demonstrate distal lower limb-predominant sensorimotor neuropathy with decreased action potentials indicating primary axonal damage [England et al. 2009], though several cases with mild associated myelopathy have been reported [Motley et al. 2015]. Recently, biallelic mutations in AARS were reported in three individuals with severe epileptic encephalopathy [Simons et al. 2015]. The affected individuals reported herein represent additional cases of AARS mutations with a phenotype distinct from autosomal dominant CMT2N. Similar to the previously reported autosomal recessive cases, the affected individuals demonstrated congenital and progressive microcephaly, central hypomyelination and severe early-onset epilepsy, including infantile spasms and Lennox-Gastaut syndrome. In contrast to the reported cases, the affected individuals showed retained or exaggerated deep tendon reflexes, suggesting that peripheral neuropathy might not be a major phenotype in these individuals, at least in young ages. Infantile syndromic liver failure has been reported in association with leucyl- and methionyl-tRNA synthetase mutations, but the affected individuals herein did not have abnormalities in their liver function tests. Thus, AARS mutations appear to lead to two distinct phenotypes, a peripheral nervous system disorder that is a dominant trait and a central nervous system condition that is a recessive trait.
The difference in manifestation of dominant and recessive AARS-related phenotypes could be explained in part by the residual aminoacylation activities of the enzyme. The mutations identified in this study, p.Tyr690Leufs*3 and p.Gly913Asp, cause an aminoacylation defect but retain 10–30% residual activity compared to wild type (7.4-fold and 3.7-fold decrease in kcat/Km respectively). This is similar to the previously described missense AARS mutations, p.Lys81Thr and p.Arg751Gly [Simons et al. 2015], which possess aminoacylation activity of 10–20% of the wild type (2-fold and 10-fold decrease in kcat/Km respectively). On the other hand, the AARS mutations identified in CMT2N, typified by p.Arg329His and p.Asn71Tyr, result in almost no residual aminoacylation activity [McLaughlin et al. 2012]. In these cases, the wild-type AARS allele in the same cell is fully functional and ~50% of overall cellular aminoacylation activity is expected to remain. This suggests a possible genotype-phenotype correlation, where a total residual aminoacylation activity of AARS from both alleles defines the clinical phenotype ranging from CMT2N to progressive microcephaly. It is notable that the parents of the affected siblings described herein (MC32505 and MC32506) are both healthy, with no symptoms suggestive of peripheral neuropathies, even though they are heterozygous for an AARS mutation, possibly implicating another threshold of residual aminoacylation activity, above which individuals are asymptomatic. This genotype-phenotype model is in accord with the recently proposed idea that common molecular pathways link CMT neuropathies and hereditary spastic paraplegia [Timmerman et al. 2013; Liu et al. 2014b]. Alternatively, AARS mutations might cause CMT in heterozygous individuals through impairment of non-canonical functions outside protein synthesis [Guo and Schimmel 2013; Yao and Fox 2013; He et al. 2015].
In addition to aminoacylation activity, AARS possesses an editing activity. Here we show that the p.Tyr690Leufs*3 mutation affects the editing activity and causes serine misacylation. To our knowledge, editing deficiency caused by mutations in human aminoacyl-tRNA synthetases has not been experimentally demonstrated prior to our study. In mice, a biallelic missense mutation in the editing domain of Aars (p.Ala734Glu) has been shown to diminish the editing activity by 40–50%, which leads to accumulation of misfolded proteins resulting in neurodegeneration [Lee, et al., 2006]. Another Aars mutation, p.Cys723Ala, causes a more severe editing deficiency, leading to cardiomyopathy in mice when placed in trans with p.Ala734Glu [Liu, et al., 2014a]. However, the affected individuals reported herein, with the editing-deficient p.Tyr690Leufs*3 mutation, presented with a very similar phenotype to the individuals with previously reported AARS mutations with no editing defects, except for the retained deep tendon reflexes. Therefore, the contribution of editing deficiency to the disease phenotype in humans remains unclear. Even though the recombinant p.Tyr690Leufs*3 AARS was stable, we found a very low level of the AARS protein in the lymphoblast cell line of the affected individual. Thus, we cannot rule out the possibility that the p.Tyr690Leufs*3 mutant may not be present at a high enough level to exert editing defects in vivo.
In summary, we have identified loss-of-function mutations in the AARS gene as the cause of a human recessive disorder affecting the central nervous system and characterized by progressive microcephaly with hypomyelination and spasticity. We present functional data indicating that both mutations impair stability and aminoacylation activity of AARS, and one mutation, p.Tyr690Leufs*3, disrupts its editing activity. Further investigation at the cellular and organism level would help further delineate the roles of the AARS gene in human neurological development and function.
Supplementary Material
Table 1.
Clinical summary of affected individuals
| Study | Current study | Simons et al., 2015 | |||
|---|---|---|---|---|---|
| MC32501 | MC32502 | LD_0115.0A | LD_0115.B | LD_0857.0 | |
| Protein variants | p.Tyr690Leufs*3, p.Gly913Asp | p.Lys81Thr, p.Arg751Gly | p.Arg751Gly, homozygous | ||
| Gender | Female | Female | Female | Female | Male |
| Age at examination | 5 months | 4 years 4 months | 7 years 6 months | 3 years 8 months | 1 years 4 months |
| Head circumference at birth | 30.1cm (−3.12 SDs) | 31.5 cm (−2.17 SDs) | N/A (congenital microcephaly) | N/A (congenital microcephaly) | N/A (congenital microcephaly) |
| Weight at birth | 1.96 kg (−2.75 SDs) | 2.98 kg (−0.80 SD) | N/A | N/A | N/A |
| Intrauterine growth retardation | Yes | Yes | Yes | Yes | Yes |
| Head circumference at examination | 35.75 cm (−4.90 SDs) | 43 cm (−4.59 SDs) | 44 cm (−6.14 SDs) | 41.5 cm (−4.84 SDs) | 38 cm (−7.29 SDs) |
| Weight at examination | 4.72 kg (−2.64 SDs) | 13.6 kg (−1.67 SDs) | N/A | N/A | N/A |
| Cognition | Fixating on objects | Tracking objects, no speech | No tracking or fixation | Searching eye movement with no visual tracking | Poor intraction |
| Motor examination | Increased tone and spasticity | Increased tone and spasticity | Dystonia and spasticity | Dystonia and spasticity | Dystonia and tremulousness |
| Deep tendon reflexes | 1+ bilaterally in the upper and lower extremities, no ankle clonus | 2+ bilaterally in the upper and lower extremities, positive ankle clonus | Not elicited | Not elicited | Not elicited |
| Remarks | 6 days of NICU stay due to apnea and poor feeding | Transient hypoglycemia at birth | Severe GI reflux, vertical tali | Severe GI reflux, vertical tali | Poor feeding, vertical tali |
| Epilepsy/Seizure | |||||
| Onset | 2 months | 3.5 months | 2 months | 3 months | 5 months |
| Type of seizures | Head dropping | Infantile spasms giving way to LGS | Myoclonic tonic- clonic epilepsy | Myoclonic epilepsy | Myoclonic epilepsy |
| EEG findings | Multifocal epileptiform discharges | Modified hypsarrhythmia, multifocal discharges with infrequent electrodecrements | Multifocal, posterior- predominant discharges | Multifocal, posterior- predominant discharges | Multifocal, psterior- predominant discharges with background suppression |
| MRI findings | |||||
| Age at MRI | 4 months | 3 years 4 months | 1 year 2 months | 1 year 5 months | 1 year 2 months |
| Myelination | Hypomyelination | Hypomyelination | Hypomyelination | Hypomyelination | Hypomyelination |
| White matter volume | Moderately diminished | Moderately diminished | Markedly diminished | Markedly diminished | Markedly diminished |
| Corpus callosum | Thin | Thin | Very thin | Very thin | Very thin |
| Cellebellum/brain stem | Normal | Normal | Mild volume loss | Marked volume loss | Hypoplastic |
EEG: electroencephalography, GI: Gastrointestinal LGS: Lennox Gastaut syndrome, MRI: Magnetic resornace imaging, N/A: not available, NICU: Neonatal intensive care unit, SD: Standard deviation
Acknowledgments
Grant Sponsor
The study was supported by grants from the Manton Center for Orphan Disease Research, NINDS (R01NS035129), NIH/Fogarty International Center (R21TW008223), NIGMS (R01GM115431 to J.L.), and Dubai Harvard Foundation for Medical Research. TN was supported by fellowship grants from the Japan Foundation for Pediatric Research and the Japan Society for the Promotion of Science. GHM was also supported by grants from F. Hoffmann-La Roche Ltd., Qatar National Research Fund, and Boston Children’s Hospital Faculty Career Development Award.
The study was supported by grants from the Manton Center for Orphan Disease Research, NINDS (R01NS035129), NIH/Fogarty International Center (R21TW008223), NIGMS (R01GM115431 to J.L.), and Dubai Harvard Foundation for Medical Research. TN was supported by fellowship grants from the Japan Foundation for Pediatric Research and the Japan Society for the Promotion of Science. GHM was also supported by grants from F. Hoffmann-La Roche Ltd., Qatar National Research Fund, and Boston Children’s Hospital Faculty Career Development Award. We thank the family for their participation in this research, Dr. Gail I. Schuman for her clinical assessments.
Footnotes
Disclosure statement: The authors declare no conflicts of interest.
References
- Antonellis A, Ellsworth RE, Sambuughin N, Puls I, Abel A, Lee-Lin SQ, Jordanova A, Kremensky I, Christodoulou K, Middleton LT, Sivakumar K, Ionasescu V, et al. Glycyl tRNA synthetase mutations in Charcot-Marie-Tooth disease type 2D and distal spinal muscular atrophy type V. Am J Hum Genet. 2003;72(5):1293–9. doi: 10.1086/375039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonellis A, Green ED. The role of aminoacyl-tRNA synthetases in genetic diseases. Annu Rev Genomics Hum Genet. 2008;9:87–107. doi: 10.1146/annurev.genom.9.081307.164204. [DOI] [PubMed] [Google Scholar]
- Bansagi B, Antoniadi T, Burton-Jones S, Murphy SM, McHugh J, Alexander M, Wells R, Davies J, Hilton-Jones D, Lochmuller H, Chinnery P, Horvath R. Genotype/phenotype correlations in AARS-related neuropathy in a cohort of patients from the United Kingdom and Ireland. J Neurol. 2015;262(8):1899–908. doi: 10.1007/s00415-015-7778-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beebe K, Ribas De Pouplana L, Schimmel P. Elucidation of tRNA-dependent editing by a class II tRNA synthetase and significance for cell viability. EMBO J. 2003;22(3):668–75. doi: 10.1093/emboj/cdg065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- England JD, Gronseth GS, Franklin G, Carter GT, Kinsella LJ, Cohen JA, Asbury AK, Szigeti K, Lupski JR, Latov N, Lewis RA, Low PA, et al. Practice Parameter: evaluation of distal symmetric polyneuropathy: role of laboratory and genetic testing (an evidence-based review). Report of the American Academy of Neurology, American Association of Neuromuscular and Electrodiagnostic Medicine, and American Academy of Physical Medicine and Rehabilitation. Neurology. 2009;72(2):185–92. doi: 10.1212/01.wnl.0000336370.51010.a1. [DOI] [PubMed] [Google Scholar]
- Guo M, Chong YE, Shapiro R, Beebe K, Yang XL, Schimmel P. Paradox of mistranslation of serine for alanine caused by AlaRS recognition dilemma. Nature. 2009;462(7274):808–12. doi: 10.1038/nature08612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M, Schimmel P. Essential nontranslational functions of tRNA synthetases. Nat Chem Biol. 2013;9(3):145–53. doi: 10.1038/nchembio.1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He W, Bai G, Zhou H, Wei N, White NM, Lauer J, Liu H, Shi Y, Dumitru CD, Lettieri K, Shubayev V, Jordanova A, et al. CMT2D neuropathy is linked to the neomorphic binding activity of glycyl-tRNA synthetase. Nature. 2015;526(7575):710–4. doi: 10.1038/nature15510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibba M, Soll D. Aminoacyl-tRNA synthesis. Annu Rev Biochem. 2000;69:617–50. doi: 10.1146/annurev.biochem.69.1.617. [DOI] [PubMed] [Google Scholar]
- Kodera H, Osaka H, Iai M, Aida N, Yamashita A, Tsurusaki Y, Nakashima M, Miyake N, Saitsu H, Matsumoto N. Mutations in the glutaminyl-tRNA synthetase gene cause early-onset epileptic encephalopathy. J Hum Genet. 2015;60(2):97–101. doi: 10.1038/jhg.2014.103. [DOI] [PubMed] [Google Scholar]
- Latour P, Thauvin-Robinet C, Baudelet-Mery C, Soichot P, Cusin V, Faivre L, Locatelli MC, Mayencon M, Sarcey A, Broussolle E, Camu W, David A, et al. A major determinant for binding and aminoacylation of tRNA(Ala) in cytoplasmic Alanyl-tRNA synthetase is mutated in dominant axonal Charcot-Marie-Tooth disease. Am J Hum Genet. 2010;86(1):77–82. doi: 10.1016/j.ajhg.2009.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Satz JS, Vo MN, Nangle LA, Schimmel P, Ackerman SL. Deficiencies in tRNA synthetase editing activity cause cardioproteinopathy. Proc Natl Acad Sci U S A. 2014a;111(49):17570–5. doi: 10.1073/pnas.1420196111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YT, Laura M, Hersheson J, Horga A, Jaunmuktane Z, Brandner S, Pittman A, Hughes D, Polke JM, Sweeney MG, Proukakis C, Janssen JC, et al. Extended phenotypic spectrum of KIF5A mutations: From spastic paraplegia to axonal neuropathy. Neurology. 2014b;83(7):612–9. doi: 10.1212/WNL.0000000000000691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascarenhas AP, An S, Rosen AE, Martinis SA, Musier-Forsyth K. Fidelity Mechanisms of the Aminoacyl-tRNA Synthetases. In: RajBhandary UL, Köhrer C, editors. Protein Engineering. New York: Springer-Verlag; 2008. pp. 153–200. [Google Scholar]
- McLaughlin HM, Sakaguchi R, Giblin W, Program NCS, Wilson TE, Biesecker L, Lupski JR, Talbot K, Vance JM, Zuchner S, Lee YC, Kennerson M, et al. A recurrent loss-of-function alanyl-tRNA synthetase (AARS) mutation in patients with Charcot-Marie-Tooth disease type 2N (CMT2N) Hum Mutat. 2012;33(1):244–53. doi: 10.1002/humu.21635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan HJ, Humphreys P, Smith A, Schwartzentruber J, Chakraborty P, Bulman DE, Beaulieu CL, Majewski J, Boycott KM, Geraghty MT Consortium FC. Congenital Visual Impairment and Progressive Microcephaly Due to Lysyl-Transfer Ribonucleic Acid (RNA) Synthetase (KARS) Mutations: The Expanding Phenotype of Aminoacyl-Transfer RNA Synthetase Mutations in Human Disease. J Child Neurol. 2015;30(8):1037–43. doi: 10.1177/0883073814553272. [DOI] [PubMed] [Google Scholar]
- Motley WW, Griffin LB, Mademan I, Baets J, De Vriendt E, De Jonghe P, Antonellis A, Jordanova A, Scherer SS. A novel AARS mutation in a family with dominant myeloneuropathy. Neurology. 2015;84(20):2040–7. doi: 10.1212/WNL.0000000000001583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novarino G, Fenstermaker AG, Zaki MS, Hofree M, Silhavy JL, Heiberg AD, Abdellateef M, Rosti B, Scott E, Mansour L, Masri A, Kayserili H, et al. Exome sequencing links corticospinal motor neuron disease to common neurodegenerative disorders. Science. 2014;343(6170):506–11. doi: 10.1126/science.1247363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons C, Griffin LB, Helman G, Golas G, Pizzino A, Bloom M, Murphy JL, Crawford J, Evans SH, Topper S, Whitehead MT, Schreiber JM, et al. Loss-of-function alanyl-tRNA synthetase mutations cause an autosomal-recessive early-onset epileptic encephalopathy with persistent myelination defect. Am J Hum Genet. 2015;96(4):675–81. doi: 10.1016/j.ajhg.2015.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taft RJ, Vanderver A, Leventer RJ, Damiani SA, Simons C, Grimmond SM, Miller D, Schmidt J, Lockhart PJ, Pope K, Ru K, Crawford J, et al. Mutations in DARS cause hypomyelination with brain stem and spinal cord involvement and leg spasticity. Am J Hum Genet. 2013;92(5):774–80. doi: 10.1016/j.ajhg.2013.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmerman V, Clowes VE, Reid E. Overlapping molecular pathological themes link Charcot-Marie-Tooth neuropathies and hereditary spastic paraplegias. Exp Neurol. 2013;246:14–25. doi: 10.1016/j.expneurol.2012.01.010. [DOI] [PubMed] [Google Scholar]
- Tsui WC, Fersht AR. Probing the principles of amino acid selection using the alanyl-tRNA synthetase from Escherichia coli. Nucleic Acids Res. 1981;9(18):4627–37. doi: 10.1093/nar/9.18.4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf NI, Salomons GS, Rodenburg RJ, Pouwels PJ, Schieving JH, Derks TG, Fock JM, Rump P, van Beek DM, van der Knaap MS, Waisfisz Q. Mutations in RARS cause hypomyelination. Ann Neurol. 2014;76(1):134–9. doi: 10.1002/ana.24167. [DOI] [PubMed] [Google Scholar]
- Yao P, Fox PL. Aminoacyl-tRNA synthetases in medicine and disease. EMBO Mol Med. 2013;5(3):332–43. doi: 10.1002/emmm.201100626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Ling J, Barcia G, Jing L, Wu J, Barry BJ, Mochida GH, Hill RS, Weimer JM, Stein Q, Poduri A, Partlow JN, et al. Mutations in QARS, Encoding Glutaminyl-tRNA Synthetase, Cause Progressive Microcephaly, Cerebral-Cerebellar Atrophy, and Intractable Seizures. Am J Hum Genet. 2014;94(4):547–58. doi: 10.1016/j.ajhg.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.