SUMMARY
The mucosal inflammatory response induced by Salmonella Typhimurium creates a favorable niche for this gut pathogen. Conventional wisdom holds that S. Typhimurium undergoes an incomplete TCA cycle in the anaerobic mammalian gut. One change during S. Typhimurium-induced inflammation is the production of oxidized compounds by infiltrating neutrophils. We show that inflammation-derived electron acceptors induce a complete, oxidative TCA cycle in S. Typhimurium, allowing the bacteria to compete with the microbiota for colonization. A complete TCA cycle facilitates utilization of the microbiota-derived fermentation end product succinate as a carbon source. S. Typhimurium succinate utilization genes contribute to efficient colonization in conventionally raised mice, but provide no growth advantage in germ-free mice. Mono-association of gnotobiotic mice with Bacteroides, a major succinate producer, restores succinate utilization in S. Typhimurium. Thus, oxidative central metabolism enables S. Typhimurium to utilize a variety of carbon sources, including microbiota-derived succinate.
eTOC BLURB
Spiga, et al. show that during colonization of the intestinal lumen, the enteric pathogen S. Typhimurium performs a complete TCA cycle. This oxidative central metabolism enables S. Typhimurium to utilize the microbiota-derived fermentation product succinate as a nutrient and to compete with the microbiota for colonization of the intestinal tract.

INTRODUCTION
The bacterial species dominating the microbiota in the large bowel are obligate anaerobic bacteria belonging to the phyla Bacteroidetes (class Bacteroidia) and Firmicutes (class Clostridia). With amino acids and simple sugars being absorbed in the small intestine, the primary carbon and energy sources for obligate anaerobic bacteria in the large intestine are complex glycans of dietary and host origin (reviewed in (Fischbach and Sonnenburg, 2011; Flint et al., 2012; Koropatkin et al., 2012; Martens et al., 2014)). Glycan degradation by the starch utilization system (sus) machinery has been studied extensively in obligate anaerobic commensal Bacteroides spp.. The genomes of sequenced Bacteroides strains are predicted to encode a large variety of distinct, sus-like systems that allow the utilization of a plethora of structurally unrelated glycans (Cuskin et al., 2015; El Kaoutari et al., 2013; Rogowski et al., 2015; Xu et al., 2003), with different Bacteroides strains exhibiting preference for distinct glycans (Pudlo et al., 2015). Similarly, commensal Bifidobacterium spp. and Clostridia produce extracellular glycoside hydrolases and other carbohydrate-active enzymes, allowing the fermentation of complex polysaccharides (Crost et al., 2013; El Kaoutari et al., 2013; Schell et al., 2002; Schwarz et al., 2004; Shimizu et al., 2002). Glycan degradation by Clostridia and Bacteroidia generates primary fermentation end products, which support the growth of syntrophic bacteria and archaea as minor constituents of the gut microbiota (Macy et al., 1975; Turton et al., 1983). Collectively, the ability to degrade a variety of complex polysaccharides directly correlates with the overall abundance of commensal gut microbes in the ecosystem (Eilam et al., 2014), indicating that polysaccharide utilization is a major determinant of the microbiota composition in the healthy gut.
Curiously, infection with enteric pathogens disturbs the normal gut microbiota structure, culminating in a bloom of the luminal pathogen population (Barman et al., 2008; Lupp et al., 2007; Stecher et al., 2007). Increased bacterial colonization of the intestinal tract enhances transmission success of the pathogen by the fecal oral route (Lawley et al., 2008; Rivera-Chavez et al., 2016). Proteobacteria express a very limited number of secreted glycoside hydrolases (El Kaoutari et al., 2013), suggesting that the metabolic pathways that allow gut colonization of pathogenic Enterobacteriaceae such as Salmonella enterica serotype Typhimurium (S. Tm; Phylum Proteobacteria, Family Enterobacteriaceae) must be different from the glycan-foraging mechanisms utilized by commensal Bacteroidia and Clostridia.
Several mechanisms have been shown to enhance fitness of S. Tm in the inflamed gut. Modifications of the outer membrane confer resistance to antimicrobial peptides and bile (Crawford et al., 2012; Gunn, 2008). To overcome nutritional immunity, siderophores facilitate the uptake of micronutrients such as iron and zinc (Liu et al., 2012; Raffatellu et al., 2009). Byproducts of reactive nitrogen and oxygen species generated in the wake of the host inflammatory are utilized as respiratory electron acceptors (Lopez et al., 2012; Winter et al., 2010). Furthermore, inflammation-associated changes in the colonocyte metabolism include leakage of molecular oxygen into the lumen, which supports oxygen respiration in S. Tm through high-affinity terminal oxidases (Rivera-Chavez et al., 2016). Utilization of microbiota-derived molecular hydrogen enhances initial colonization of the intestinal tract by S. Tm (Maier et al., 2013). While colonization of the inflamed gut by S. Tm has been studied extensively, the adaptation of the central metabolism to the nutritional environment of the inflamed intestine has not been elucidated. Here, we investigated the tricarboxylic acid (TCA) cycle as part of the central intermediary metabolism in the enteric pathogen S. Tm in an inbred mouse model of infection.
RESULTS
Contribution of TCA cycle enzymes to fitness of S. Tm in the streptomycin-treated mouse model
Current dogma holds that Enterobacteriaceae and many other bacteria do not express α-ketoglutarate dehydrogenase (SucAB), succinyl-CoA synthetase (SucCD), and succinate dehydrogenase (Sdh) under anaerobic conditions, resulting in a bifurcation of the TCA into a reductive and oxidative branch (Fig. 1A) (Amarasingham and Davis, 1965; Cronan and Laporte, 2013; Iuchi and Lin, 1988). In this arrangement, biosynthetic reactions of the TCA cycle can still occur, but acetyl-CoA cannot be oxidized. Surplus reduction equivalents are used to reduce the internal terminal electron acceptor fumarate. Based on these considerations, one would assume that the TCA cycle of S. Tm in the gut lumen is branched and α-ketoglutarate dehydrogenase, succinyl-CoA synthetase, and Sdh activity is dispensable (Fig. 1A). To test this idea, we generated a S. Tm mutant lacking the major subunit of Sdh (ΔsdhA) and determined fitness in a murine model of Salmonella-induced colitis (Fig. 1B – D; Fig. S1). Groups of streptomycin-treated C57BL/6 mice (Barthel et al., 2003) were intragastrically inoculated with an equal mixture of the S. Tm wild-type strain (AJB715), and an ΔsdhA mutant, or mock-treated. Four days after infection, the bacterial load for each strain was determined in the colonic and cecal content by plating on selective media and the ratio of wild-type strain bacteria to mutant bacteria (competitive index) calculated (Fig. 1C and D). Interestingly, the S. Tm wild-type strain was recovered in higher numbers than the ΔsdhA mutant, suggesting that Sdh activity was required for efficient colonization of the gut during infection.
Figure 1. Contribution of succinate dehydrogenase to colonization in the streptomycin-treated mouse model.
(A) Simplified model of the branched S. Tm TCA cycle under anaerobic conditions (blue arrows) and complete TCA in the inflamed gut. Frd, fumarate reductase; Sdh, succinate dehydrogenase; SucAB, α-ketoglutarate dehydrogenase; SucCD, succinyl-CoA synthetase. (B) Streptomycin-pretreated C57BL/6 mice were intragastrically inoculated with the S. Tm wild-type (WT) strain or a ΔinvA ΔspiB (ΔT3SS1/2) mutant. Representative images of hematoxylin and eosin-stained sections of the cecum and the colon. Scale bar = 500 μm. (C – D) Streptomycin-pretreated C57BL/6 mice were intragastrically inoculated with an equal mixture of the indicated S. Tm strains. The competitive index in the cecal (C) and colonic content (D) was determined four days after infection. (E – F) Mucin broth was inoculated with an equal mixture of the S. Tm wild-type strain (WT) and a ΔsucAB mutant (E), a ΔsucCD mutant (F) or a ΔsdhA mutant (G) and the competitive index determined after 16 h of anaerobic growth in the absence or presence of nitrate (40 mM) and tetrathionate (40 mM). Bars represent geometric means ± standard error. *, P < 0.05; **, P < 0.01; ***, P < 0.001. The number of animals per group (N) is indicated above each bar. See also Fig. S1.
Since the TCA cycle is an essential component for the intermediary metabolism, we were concerned that TCA cycle mutants were generally impaired for growth. We therefore sought to determine the fitness of TCA cycle mutants in the absence of inflammation. S. Tm employs two type three secretion systems to invade non-phagocytic epithelial cells and to mediate replication in the mucosa, respectively (Galan and Curtiss, 1989; Hensel et al., 1998). Mutants lacking both type three secretion systems (T3SS1/2; ΔinvA ΔspiB) do not induce overt inflammatory responses (Fig. 1B and Fig. S1A – C) and colonize the gut lumen to a limited extent (Coombes et al., 2005; Hapfelmeier et al., 2004; Hapfelmeier et al., 2005). The T3SS1/2 and the T3SS1/2 ΔsdhA mutants were recovered in similar numbers (Fig. 1C and D), indicating that Sdh activity is dispensable in the absence of inflammatory responses. These experiments raised the possibility that S. Tm uses a complete TCA cycle during infection.
Exogenous electron acceptors impact the S. Tm TCA cycle in vitro and in vivo
Addition of the alternative electron acceptor nitrate to anaerobic culture media can induce expression of TCA cycle enzymes in E. coli K-12, however this phenomenon only occurs in the absence of the regulatory protein ArcA (Perrenoud and Sauer, 2005; Prohl et al., 1998; Wimpenny and Cole, 1967). During mucosal inflammation, alternative electron acceptors such as nitrate and tetrathionate occur as byproducts of the oxidative burst (Winter et al., 2010). To test whether alternative electron acceptors could influence the TCA cycle in S. Tm, we cultured mixtures of the S. Tm wild-type strain and isogenic mutants deficient in TCA cycle enzymes in mucin broth under anaerobic conditions for 16 h. The wild-type strain outcompeted the ΔsucAB, ΔsucCD, and ΔsdhA mutant when nitrate or tetrathionate was added to the growth media while no growth advantage was apparent in the absence of exogenous electron acceptors (Fig. 1E – G). This outcome indicated that alternative electron acceptors are sufficient to alter operation of the TCA cycle in S. Tm.
Next, we investigated whether nitrate and tetrathionate increase transcription of sucA and sdhA during anaerobic growth in mucin broth. RNA was extracted after 16 h and sucA and sdhA mRNA levels were determined by RT-qPCR (Fig. 2A and B). Transcription of sucA and sdhA increased significantly in the presence of alternative electron acceptors. We then determined the transcriptome of S. Tm during colonization of the intestinal tract (Fig. S2). To this end, we colonized gnotobiotic mice with the S. Tm wild-type strain and performed RNAseq on RNA extracted from cecal content. A comparison to a published transcriptome database of S. Tm cultured in various in vitro conditions (Kroger et al., 2013) showed that the in vivo transcriptome formed a state distinct from all in vitro conditions in the database (Fig. S2A). To assess the overall metabolic state, we compared normalized mRNA levels of selected metabolic enzymes under aerobic and anaerobic culture conditions in vitro and in the mouse model (Fig. S2B – G). Transcription of general housekeeping enzymes such as glyceraldehyde-3-phosphate dehydrogenase (gapA), acetate kinase (ackA), and phosphate acetyltransferase (pta) was comparable in all three conditions (Fig. S2B). Consistent with the notion that nitrate and tetrathionate are inducers of the respective reductase operons (Hensel et al., 1999; Rabin and Stewart, 1993), transcription of the tetrathionate reductase, nitrate reductase-1, and the periplasmic nitrate reductase in the murine cecum was increased compared to standard aerobic and anaerobic in vitro conditions (Fig. S2C – E). Furthermore, transcription of sucAB, sucCD, and sdh in the murine model was found to be most similar to growth under aerobic in vitro conditions (Fig. S2F), which was suggestive of an overall oxidative central metabolism in S. Tm during gut colonization.
Figure 2. Effect of gut inflammation and electron acceptors on sucA and sdhA transcription.
(A and B). Relative transcription of sucA (A) and sdhA (B) in mucin broth supplemented with the indicated electron acceptors was determined by RT-qPCR. Transcription of target genes was normalized to 16S rRNA. (C – D) Streptomycin-pretreated C57BL/6 mice were intragastrically inoculated with the S. Tm wild-type strain, an ΔinvA ΔspiB (ΔT3SS1/2) mutant, or mock-treated (LB broth). (C and D) mRNA levels of Nos2 (black bars), Cxcl1 (gray bars), and Tnfa (white bars) in the colonic tissue was determined by RT-qPCR two days (C) and three days (D) post infection (p.i.). Transcription was normalized to Gapdh mRNA. (E and F) Bacterial RNA was extracted from the colon content. Relative transcription of sdhA (black bars) and sucA (white bars) normalized to S. Tm 16S rRNA was determined by RT-qPCR two (E) and three days (F) after infection. Bars represent geometric means ± standard error. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ns, not statistically significant. The number of animals per group (N) is indicated above each bar. See also Fig. S2.
We then explored whether inflammation alters expression of key TCA cycle enzymes in the streptomycin-treated mouse model (Fig. 2C – F). A marked increase in pro-inflammatory markers was observed three days after infection (Fig. 2D and Fig. S1C). Low levels of inflammation were noted early during infection with the wild-type strain (2 days after infection; Fig. 2C) and when mice were infected with a T3SS1/2-deficient mutant (Fig. 2C and 2D). Transcription of sdhA and sucA in the S. Tm wild-type strain and the T3SS1/2-deficient mutant were similar two days after infection (Fig. 2E). Concomitant with the onset of inflammation three days after infection, the wild-type strain exhibited a significant increase in sdhA and sucA mRNA levels compared to the ΔT3SS1/2 mutant (Fig. 2F), indicating that increased levels of mucosal inflammation correlates with increased expression of S. Tm TCA cycle enzymes.
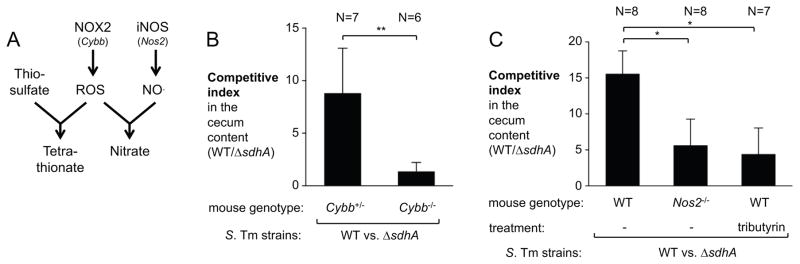
During Salmonella infection, neutrophils infiltrating the mucosa undergo an oxidative burst with the release of reactive oxygen (ROS) and nitrogen species (RNS) (Fig. 3A). NADPH oxidase 2 (NOX2, PHOX), comprised of the two chains CYBA and CYBB, catalyzes the first step in the generation of ROS. Inducible nitric oxide synthase (iNOS) is the sole source of nitric oxide. Oxidation of thiosulfate by ROS yields tetrathionate in the gut lumen (Winter et al., 2010) while peroxide can react with nitric oxide to form peroxynitrite, a reactive nitrogen species that decomposes to nitrate (Winter et al., 2013). To assess whether availability of tetrathionate and nitrate affects the S. Tm TCA cycle during infection, we repeated the competitive colonization assay in NOX2 (Cybb)- and iNOS (Nos2)-deficient mice. In heterozygous Cybb+/− littermate controls, the S. Tm wild-type strain outcompeted the sdhA mutant, while this competitive fitness advantage was abrogated in homozygous Cybb-deficient animals (Fig. 3B). The fitness advantage conferred by Sdh activity was significantly reduced in Nos2-deficient mice but not completely abrogated as in the Cybb-deficient mice suggesting that both tetrathionate and nitrate likely contribute to changes in the central metabolism of S. Tm in vivo (Fig. 3C). During S. Tm infection, butyrate-producing Clostridia spp. are diminished from the gut microbiota (Rivera-Chavez et al., 2016). Depletion of butyric acid, the main carbon source of intestinal epithelial cells, causes a perturbation in the epithelial metabolism and small amounts of oxygen become available at the mucosal interface. Thus, we considered the possibility that, in addition to nitrate and tetrathionate, molecular oxygen could influence the direction of the TCA cycle in the mammalian gut lumen. Supplementation with tributyrin, a source of butyrate that prevents oxygen leakage into the gut lumen (Rivera-Chavez et al., 2016), moderately reduced the competitive fitness of the wild-type strain over the sdhA mutant (Fig. 3C). Collectively, these data suggest that availability of exogenous electron acceptors, such as nitrate, tetrathionate, and oxygen, alter the central metabolism of S. Tm in a mouse model of infection.
Figure 3. Impact of inflammation-derived electron acceptors on the TCA cycle during infection.
(A) Simplified schematic outlining the contribution of NADPH oxidase 2 (NOX2) and inducible nitric oxide synthase (iNOS) to the generation of tetrathionate and nitrate via reactive oxygen species (ROS) and nitric oxide (NO). (B) Streptomycin-pretreated female Cybb+/− and Cybb−/− mice on the C57BL/6 background were intragastrically inoculated with the S. Tm wild-type strain and a ΔsdhA mutant. The competitive index in the cecal content was determined after four days. (C) Streptomycin-pretreated C57BL/6 wild-type mice and Nos2-deficient mice were inoculated with a mixture of the S. Tm wild-type strain and a ΔsdhA mutant. One group was treated with tributyrin as indicated. The competitive index in the cecal content was determined four days post infection. Bars represent geometric means ± standard error. *, P < 0.05; **, P < 0.01. The number of animals per group (N) is indicated above each bar.
A complete TCA cycle enhances S. Tm fitness in competition with the native microbiota
To determine whether a complete TCA cycle might enhance fitness of S. Tm in competition with the unperturbed, native gut microbiota, we used CBA mice. CBA mice develop neutrophilic inflammation of the large intestine after 7 to 10 days (Fig. S3A and B). The competitive fitness of the S. Tm wild-type strain and mutants lacking TCA cycle enzymes was determined 7 days after infection (Fig. 4A and B). The S. Tm wild-type strain outcompeted the ΔsucAB, ΔsucCD, and ΔsdhA mutants. The glyoxylate shunt, a pathway bypassing α-ketoglutarate dehydrogenase and succinyl-CoA synthetase, was dispensable for growth in the inflamed gut since a ΔaceAB mutant did not exhibit a fitness defect under the conditions tested (Fig. 4A and B). Sdh activity did not confer a fitness advantage in the absence of inflammation (ΔinvA ΔspiB mutant background) in the CBA mouse model. Complementation of the sdhA mutant by introducing the sdhA promoter and coding sequence into the neutral phoN recapitulated the phenotype of the wild-type strain (Fig. 4A and B). Similar observations were made using another S. Tm isolate, SL1344 (Fig. S3C).
Figure 4. Sdh, α-ketoglutarate dehydrogenase, and succinyl-CoA synthetase activity contribute to fitness of S. Tm in mice with a native microbiota.
(A – B) CBA mice were infected with an equal mixture of the indicated S. Tm strains by the intragastric route. Competitive index of the indicated strains in the cecal (A) and colonic lumen (B) 7 days after infection. WT, S. Tm wild-type strain; ΔT3SS1/2, ΔinvA ΔspiB mutant. Bars represent geometric means ± standard error. *, P < 0.05; ns, not statistically significant. (C – F) CBA mice were intragastrically infected with the S. Tm wild-type strain or a ΔsdhA mutant. (C) Representative images of Hematoxylin and Eosin-stained cecal tissue. Scale bar equals 100 μm. (D) Combined histopathology score of pathological lesions in the cecum. Each dot represents one animal. The lines represent the mean ± standard error. (E and F) S. Tm population levels in the cecum (E) and colon (F) content 7 days after infection. Bars represent geometric means ± standard error. *, P < 0.05. The number of animals per group (N) is indicated above each bar. See also Fig. S3.
To determine whether a complete TCA cycle contributed to growth of S. Tm in direct competition with the native microbiota, we infected groups of CBA animals intragastrically with either the S. Tm wild-type strain or the ΔsdhA mutant (single infection). Both strains induced host inflammation to a similar degree (Fig. 4C and D; Fig. S3). However, the sdhA-deficient mutant was recovered at significantly lower numbers than the wild-type strain in the cecum and colon content (29-fold and 80-fold, respectively) 7 days after infection (Fig. 4E and F). Collectively, the experiments support for the idea that switching from a branched to a full TCA cycle is a critical metabolic adaption for S. Tm to compete with the native microbiota for colonization of the intestinal tract.
Utilization of dicarboxylic acids by S. Tm
One potential consequence of the inflammation-associated oxidative central metabolism of S. Tm could be that poorly fermentable dicarboxylic acids, such as succinate, could serve as carbon sources by feeding directly into the TCA cycle. To test if uptake of di- and tri-carboxylic acids contributes to growth of S. Tm during colitis, we constructed mutants lacking the C4-dicarboxylate carriers DctA (succinate-proton symporter), DcuA (succinate-proton symporter), and DcuB (fumarate-succinate antiporter) (ΔdcuA ΔdcuB ΔdctA mutant; Δ3 mutant). In CBA mice, the S. Tm wild-type strain outcompeted the Δ3 mutant by a 5-fold (Fig. 5A). Similarly, the Δ3 mutant exhibited a gut colonization defect compared to the wild-type strain in single infection experiments (Fig. 5B and C), indicating that C4-dicarboxylate uptake enhances fitness of S. Tm in the lumen of the mammalian intestine.
Figure 5. Succinate uptake and utilization confer a fitness advantage in competition with the native gut microbiota.
(A) Groups of CBA mice were intragastrically infected with a mixture of the indicated S. Tm strains. Seven days after infection, the competitive index in the colon content was determined. Δ3, ΔdcuA ΔdcuB ΔdctA mutant. (B – E) As indicated, CBA mice were infected with the S. Tm wild-type strain (WT), the Δ3 mutant (ΔdcuA ΔdcuB ΔdctA mutant), or mock-treated. (B and C) S. Tm populations in the cecum (B) and colon content (C) 7 days after infection. (D and E) Succinate concentration in the cecum content (D) and colon content (E) as determined by GC/MS/MS. Bars represent geometric means ± standard error. *, P < 0.05; ns, not statistically significant. The number of animals per group (N) is indicated above each bar.
Microbiota-derived succinate supports growth of S. Tm during infection
Obligate anaerobic commensals, in particular Bacteroides spp., use a branched TCA cycle to support fumarate respiration. The end product of fumarate reduction, succinate, is secreted into the extracellular environment. In the human gut, luminal succinate levels are ranging from approximately 0.5 to 5 mM (Cummings et al., 1987; Meijer-Severs and van Santen, 1987; Rubinstein et al., 1969). In the CBA mouse model, the concentration of succinate was found to be about 0.4 mM in the cecum content and 3 mM in the colon content (Fig. 5D and E). No changes in extracellular succinate levels upon S. Tm infection were noted. Based on these findings, we hypothesized that Bacteroides-derived succinate could be utilized by S. Tm as a consequence of a complete TCA cycle. To investigate this idea, we cultured B. thetaiotaomicron in mucin broth for 3 days. Growth of B. thetaiotaomicron lead to the accumulation of succinate in the filter-sterilized culture supernatant (Fig. 6A). Next, we determined anaerobic growth of S. Tm using the B. thetaiotaomicron-fermented mucin as a growth media. In the absence of any exogenous electron acceptor, C4-dicarboxylate transporters did not provide a growth advantage. However, when nitrate was added to the media to switch from a branched to a complete TCA cycle, the wild-type strain outcompeted the ΔdcuA ΔdcuB ΔdctA mutant (Fig. 6B).
Figure 6. Microbiota-derived succinate enhances S. Tm growth during infection.
(A – B) Mucin broth was inoculated with B. thetaiotaomicron and incubated anaerobically for 3 days. (A) Concentration of succinate in the supernatant (B) The filter-sterilized B. thetaiotaomicron-digested mucin broth was inoculated with the indicated S. Tm strains and the competitive index after 16 h of anaerobic growth determined. Sodium nitrate (40 mM) was added as indicated. (C – E) Germ-free Swiss Webster mice and mice mono-associated with the B. thetaiotaomicron wild-type strain (wt) or a ΔfrdCAB (Δfrd) mutant were infected with an equal mixture of the S. Tm wild-type strain (WT) and the ΔdcuA ΔdcuB ΔdctA (Δ3) mutant or a mixture of a S. Tm ΔsdhA and a ΔsdhA ΔdcuA ΔdcuB ΔdctA (ΔsdhA Δ3) mutant. Samples were analyzed three days after infection. (C) Representative images of hematoxylin and eosin-stained sections of the cecum. Scale bar equals 200 μm. (D and E) Competitive index in the cecum (D) and colon content (E). Bars represent geometric means ± standard error. *, P < 0.05; **, P < 0.01; ***, P < 0.001. The number of animals per group (N) is indicated above each bar. See also Fig. S4–6.
C4-dicarboxylate transporters can facilitate the uptake of succinate and several other dicarboxylates, such as fumarate, aspartate, malate, and tartrate (reviewed in (Unden and Kleefeld, 2004)). In the absence of Sdh activity (ΔsdhA vs. ΔsdhA Δ3 mutant), the growth advantage conferred by C4-dicarboxylate transporters was abolished supporting the notion that the growth advantage conferred by these uptake systems was indeed due to succinate uptake. (Fig. 6B). Collectively, these experiments demonstrate that B. thetaiotaomicron-derived succinate can be utilized by S. Tm in the presence of exogenous electron acceptors in vitro.
Next, we investigated succinate uptake and utilization in gnotobiotic mice (genetically resistant Swiss Webster mice) and mice that had been mono-associated with B. thetaiotaomicron. Infection with S. Tm induced significant pathological changes in the cecal and colonic mucosa in both groups (Fig. 6C; Fig. S4 and S5A and B). Consistent with the idea that succinate is derived from the gut microbiota, C4-dicarboxylate uptake (Δ3 mutant) and Sdh activity (ΔsdhA mutant) were dispensable for intestinal colonization during infection of gnotobiotic mice (Fig. 6D and E; Fig. S5C). Importantly, mono-colonization with B. thetaiotaomicron was sufficient to reinstate the fitness advantage conferred by C4-dicarboxylate uptake (Fig. 6D and E; Fig. S5C). In the absence of succinate degradation (ΔsdhA vs. ΔsdhA Δ3), the effect of C4-dicarboxylate uptake on competitive fitness was minimal in the gnotobiotic mouse model as well as in conventionally-raised CBA mice (Fig. 5A; Fig. 6D and E). We also generated a B. thetaiotaomicron that is unable to generated succinate in vitro and in vivo (Fig. S6A and B) due to a lack of fumarate reductase activity (Δfrd). In contrast to the B. thetaiotaomicron wild-type strain, mono-association with the B. thetaiotaomicron Δfrd mutant was unable to restore the fitness advantage C4-dicarboxylate uptake in S. Tm (Fig. 6D and E). Metabolic profiling of large intestinal contents of mice mono-associated with the B. thetaiotaomicron wild-type strain and the isogenic Δfrd mutant revealed an absence of succinate in Δfrd colonized mice (Fig. S6B and C) and a compensatory increase in lactate levels (Fig. S6C). Lactate is not known to be transported by C4-dicarboxylate carriers and does not serve as a substrate for Sdh. Collectively, these experiments demonstrate that microbiota-derived succinate is taken up and is utilized by S. Tm during colonization of the inflamed intestinal tract.
DISCUSSION
The central metabolism of S. Tm has been analyzed extensively in vitro and in murine models of systemic infection. S. Tm mutants lacking key TCA cycle enzymes are defective for replication in tissue at systemic sites (Mercado-Lubo et al., 2008; Mercado-Lubo et al., 2009; Tchawa Yimga et al., 2006). TCA cycle reactions are predicted to have high metabolic conversion rates during growth in the spleen (Steeb et al., 2013). Furthermore, the central metabolism of S. Tm shapes host-microbe interactions during infection of macrophages. A complete, oxidative TCA cycle is of critical importance for S. Tm to avoid pyroptotic cell death in macrophages in cell culture (Wynosky-Dolfi et al., 2014). Activated macrophages restrict bacterial metabolism by releasing reactive nitrogen species to deactivate the lipoamide-dependent enzymes pyruvate and α-ketoglutarate dehydrogenase (Richardson et al., 2011). Infection of immune-competent individuals with non-typhoidal Salmonella serotypes results in a self-limiting gastroenteritis and no bacterial dissemination is observed. Curiously, the central metabolism of S. Tm during natural infection, i.e. during Salmonella-induced colitis, has not been investigated.
Strict anaerobic commensals have evolved to successfully compete for carbon and energy sources in the nutrient-limiting environment of the healthy large intestine. As a combined function of the metabolism of the bacterial community, all energetically valuable compounds are depleted, hampering intrusion of the ecosystem by enteric pathogens. To overcome this colonization resistance, S. Tm triggers an acute mucosal inflammatory response, which creates a niche in the lumen of the intestine that is suitable for the outgrowth of S. Tm over other commensals (Barman et al., 2008; Stecher et al., 2007). One prominent change in the gut environment during S. Tm infection is the production of oxidized compounds such as nitrate and tetrathionate as byproducts of the oxidative burst by infiltrating neutrophils (Lopez et al., 2012; Winter et al., 2010). Unlike obligate anaerobic commensals, S. Tm utilizes these oxidized compounds as electron acceptors for the electron transport chain. Here we show mucosal inflammation significantly alters the central metabolism of the S. Tm population residing in the gut lumen. Alternative electron acceptors, such as nitrate, change the direction of the TCA from a branched set of reactions to a full, oxidative TCA cycle in the mammalian intestine.
An oxidative central metabolism could enhance growth of S. Tm through several mechanisms. Carbon sources that are fully or partially degraded to acetyl-CoA could enter the TCA cycle and be oxidized to CO2 in vivo. In vitro, Salmonella is known to utilize a great variety of compounds as the sole carbon source during aerobic conditions (Gutnick et al., 1969). In the murine gut, only few carbon sources have been identified, such as ethanolamine, 1,2 propanediol, and fructose-asparagine (Ali et al., 2014; Faber et al., 2017; Thiennimitr et al., 2011). An oxidative TCA cycle may explain why utilization of these compounds in vivo strictly requires respiration. In this study, we demonstrate that C4-dicarboxylic acids may serve as nutrients for S. Tm during infection. Specifically, uptake and utilization of succinate, enhances S. Tm growth in the inflamed gut. Succinate is generated as a predominant fermentation end product of Bacteroides in the reductive branch of a split TCA cycle. During inflammation, this metabolism is mirrored by S. Tm as the set of reactions that make up the reductive branch is reversed. This metabolic adaptation to the inflamed gut allows the pathogen to utilize a microbiota-derived metabolic waste product as a nutrient.
Since the phenotype of the C4-dicarboyxlate uptake mutant only partially recapitulated the phenotype of the sdhA mutant, it is likely that the oxidative TCA cycle enhances S. Tm fitness by additional mechanisms. For example, in Proteus mirabilis, an oxidative TCA cycle is required to generate energy for swarming (Alteri et al., 2012). Flagella-mediated motility is required for S. Tm to efficiently colonize the inflamed gut lumen (Stecher et al., 2008). Furthermore, it is possible that other carbon sources could enter the oxidative TCA cycle, either as acetyl-CoA or through anaplerotic reactions. Of note, the phenotype of the C4-dicarboyxlate uptake mutant was not entirely abolished during infection in gnotobiotic mice, suggesting that dietary or host-derived C4-dicarboyxlates might be utilized by S. Tm as well. Collectively, an oxidative TCA cycle might give S. Tm flexibility in nutrient acquisition and energy generation during colonization of the gastrointestinal tract.
Apart from serving as a nutrient, succinate acts as a cue for some enteric pathogens. Expression of the Locus of Enterocyte Effacement (LEE), a major virulence factor of Enterohemorrhagic E. coli, is regulated in vivo by succinate (Curtis et al., 2014). Furthermore, oral antibiotic therapy is associated with blooms of C. difficile. In mouse models, an antibiotic-induced perturbation of the microbiota increases the local availability of succinate, which in return supports the expansion of C. difficile (Ferreyra et al., 2014). Our work shows that microbiota-derived succinate fuels S. Tm growth during natural infection without the need for antibiotic treatment, thus identifying a critical microbiota-pathogen interaction in the context of infection of the mammalian host.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Sebastian E. Winter (Sebastian.Winter@UTSouthwestern.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
C57BL/6, Cybb-deficient (on the C57BL/6 background), Nos2-deficient (on the C57BL/6 background) and CBA mice were originally obtained from the Jackson Laboratory and bred at UT Southwestern. Germ-free Swiss Webster mice were maintained in specific pathogen-free facilities at UT Southwestern Medical Center. Some of the germ-free mice were colonized with CBA mice microbiota to obtain the Swiss Webster mice used in this study. Conventional mice were housed in individually ventilated cages with ad libitum access to water and feed (Envigo Global 16% Protein Rodent Diet). The age at the begin of the experiment was 7–10 weeks for C57BL/6 WT, Cybb, and Nos2-deficient mice, 8–10 weeks for CBA mice, and 6–8 weeks for Swiss Webster mice. Unless indicated otherwise in the figure legend, both male and female mice were analyzed and no significant sex-specific differences were noted. Both sexes were equally represented in each experimental group.
Animals were randomly assigned into cages and treatment groups 3 days prior to experimentation. Unless stated otherwise, a minimum of 5 mice were used based on variability observed in previous experiments. At the end of the experiments, mice were humanely euthanized using carbon dioxide inhalation. Animals that had to be euthanized for humane reasons prior to reaching the predetermined time point were excluded from the analysis. All experiments involving mice were approved by the Institutional Animal Care and Use Committee at UT Southwestern Medical Center (APN# T-2013-0139, T2014-0061, T-2015-0031).
Bacterial culture
E. coli and S. Tm strains were routinely grown aerobically at 37°C in LB broth (10 g/l tryptone, 5 g/l yeast extr act, 10 g/l sodium chloride) or on LB agar plates (10 g/l tryptone, 5 g/l yeast extract, 10 g/l sodium chloride, 15 g/l agar). B. thetaiotaomicron strains were routinely cultured on blood agar plates (37 g/l brain heart infusion medium, 15 g/l agar, 50 ml sheep blood, 50 mg/l glutamine), Columbia blood plates (42 g/l Columbia agar, 5 % [v/v] defibrinated blood, 5 mg/l hemin, 0.02% [v/v] of a 0.5 % [v/v] Vitamin K1 solution in 95 % ethanol), or in modified TYG broth (10 g/l tryptone, 5 g/l yeast extract, 2 g/l glucose, 0.5 g/l cysteine 0.1 M potassium phosphate pH 7.2, 1 mg/l Vitamin K, 0.02 g/l magnesium sulfate heptahydrate, 0.4 g/l sodium bicarbonate, 0.08 g/l sodium chloride, 8 mg/l calcium chloride, 0.4 mg/l iron (II) sulfate, 1 mg/l resazurin, 40 μM histidine, 2.4 μg/l hematin) in an anaerobic chamber (Sheldon Manufacturing; 5 mol % H2, 5 % CO2, 90 % N2). When appropriate, agar plates and media were supplemented with 30 μg/ml chloramphenicol (Cm), 100 μg/ml carbenicillin (Carb), 50 μg/ml kanamycin (Kan), 50 μg/ml nalidixic acid (Nal), 50 μg/ml gentamycin (Gent), 25 μg/ml erythromycin (Erm), 25 μg/ml tetracycline (Tet) or 200 μg/ml 5-fluoro-2′-deoxyuridine (FUdR). For competitive growth assays, diluted samples were spread onto agar plates containing the chromogenic substrate 5-bromo-4-chloro-3-indolyl phosphate (40 mg/l) to detect acidic phosphatase (PhoN) activity. The competitive index was calculated by dividing the number of wild-type bacteria by the number of mutant bacteria in the output, divided by the same ratio obtained from the inoculum.
METHOD DETAILS
Construction of plasmids
To generate pSW284, pSW286 and pSW288, the upstream and downstream regions of S. Tm IR715 sucAB, sucCD and aceAB were amplified by PCR using the primer sets listed in the table below. Purified PCR products were digested with XbaI and ligated with T4 DNA ligase. The joint upstream and downstream regions were then amplified by PCR using the outside primers and cloned into pCR2.1. The DNA sequence of the cloned PCR product was verified by Sanger sequencing. The DNA fragment was subcloned into pRDH10 using BamHI and SalI restriction sites. To construct pSW306, pSW300, pSW301, and pSW299, the upstream regions of sdhA, dcuA dcuB, and dctA were PCR amplified from the IR715 chromosome. Purified PCR products and purified, SphI-digested pRDH10 were ligated using the Gibson cloning procedure in a three-part ligation. The DNA sequence of the upstream and downstream region was determined sequencing. For pSW328, an internal fragment of the phoN coding sequence was amplified by PCR and cloned into SacI-digested pGP704 in a Gibson cloning reaction. Subsequently, the promoter and coding sequence of the S. Tm sdh operon were amplified and cloned into the SphI restriction enzyme site of pSW327. To generate pMW5, regions upstream and downstream of frdCAB in B. thetaiotaomicron were PCR amplified and the products were inserted into the BamHI site of the suicide vector pExchange-tdk through Gibson assembly. The cloning strain for all suicide plasmids was DH5α λpir.
Generation of S. Tm mutants
Suicide plasmids were introduced into S. Tm by conjugation using S17-1 λpir as the donor strain. For pRDH10 and pGP704 derivatives, Cm and Carb, respectively were used to select for single cross over events. To select for a second crossover event, sucrose selection was performed (Lawes and Maloy, 1995). pSW306, pSW284, pSW286, pSW288 were used generate SW1397, SW1285, SW1286, SW1288 in the IR715 background and to generate MW156 in the SL1344 background, respectively. pSW328 was integrated into SW1397 to give rise to SW1410. Integration of pSW328 into the phoN gene was confirmed by PCR and lack of acidic phosphatase activity. SW1411 was generated by sequential introduction of pSW299, pSW300, and pSW301 into IR715 followed each time by sucrose selection. The phoN::KanR mutation was transduced by phage P22 HT int-105 (Schmieger, 1972) from AJB715 into SPN487 and SW1397 to generate SW1401 and SW1414, respectively. Similarly, the ΔsdhA::KanR from STM0734 was introduced into IR715, SPN487 and SW1411, thus generating SW1056, SW1203 and SW1413, respectively. To create MW231, the ΔinvA::TetR and ΔspiB::KanR mutations from SPN452 were transduced separately. Introduction of phoN::CmR and pSW306 into MW231 produced MW251 and MW256, respectively.
Construction of a B. thetaiotaomicron frd mutant
MW399, an isogenic derivative of the B. thetaiotaomicron strain ATCC 29148 Δtdk deficient for frdCAB, was generated via allelic exchange (Koropatkin et al., 2008). pMW5 was introduced into ATCC 29148 Δtdk via bacterial conjugation with the donor strain S17-1 λpir. The conjugation was carried out on blood plates under aerobic conditions at 37°C to enable the growth of the S17-1 λpir strain. All further steps were performed under anaerobic conditions to insure proper growth of the B. thetaiotaomicron strains. Exconjugates in which the suicide plasmid had integrated into the chromosome by single crossover were selected for on blood plates containing Gent and Erm. Then, the second homologous recombination event was selected for on blood plates containing FUdR. The second crossover event leads to an unmarked deletion of frdCAB, which was confirmed by PCR. The growth media was routinely supplemented with hemin.
Growth of Bacteroides thetaiotaomicron in mucin broth
Porcine mucin was sterilized by suspending 100 mg in 1 ml of 70 % ethanol for 24 h. The ethanol was removed by drying in a vacuum concentrator. Sterile mucin was dissolved in No-Carbon E medium (NCE) (3.94 g/l monopotassium phosphate, 5.9 g/l dipotassium phosphate, 4.68 g/l ammonium sodium hydrogen phosphate tetrahydrate, 2.46 g/l magnesium sulfate heptahydrate) at a final concentration of 0.5 % (w/v). Mucin broth was inoculated with a fresh colony of Bacteroides thetaiotaomicron and incubated under anaerobic conditions for 72 h at 37°C. Digested mucin broth was filter-sterilized (0.5 μm pore size).
Succinate concentration measurement in digested mucin
The succinate concentration in digested mucin was measured using a coupled enzymatic assay according to the recommendations of the manufacturer. Freshly prepared solutions of pure succinate were used as a standard curve. Some biological samples were spiked with known amounts of pure succinate as controls.
Anaerobic growth of S. Tm in mucin broth
Fresh hog mucin broth (0.5 % mucin in NCE media) or filter-sterilized supernatant of Bacteroides thetaiotaomicron-digested mucin was inoculated with equal mixtures of the Salmonella Typhimurium phoN mutant (wild-type strain; AJB715) and the ΔsdhA mutant (SW1397), the ΔsucAB mutant (SW1285), the ΔsucCD mutant (SW1286), and the ΔdcuA ΔdcuB ΔdctA mutant (SW1411) or the ΔsdhA phoN::KanR mutant (SW1414) and the ΔsdhA ΔdcuA ΔdcuB ΔdctA mutant (SW1413) at a final concentration of 1 × 103 CFU/ml for each strain. Sodium nitrate or sodium tetrathionate were added at a final concentration of 40 mM, as indicated. After 18 h of anaerobic growth at 37°C, bacterial numbers we re determined by spreading serial ten-fold dilutions on selective LB agar plates.
Animal models of S. Tm-induced colitis
Streptomycin-treated mouse model
Groups of 7–10 week old C57BL/6 mice received 20 mg per animal through the intragastric route. After 24 h, mice were intragastrically inoculated with 1 × 109 CFU for single strain infection experiments, 5 × 108 CFU of each strain for competitive infection experiments, or mock treated (LB broth). For the experiments shown in Fig. 1, groups of mice were infected with equal mixtures of AJB715 and SW1056 as well as SW1401 and SW1203. For the experiments shown in Fig. 2, groups of animals were infected with IR715 or SPN487. For the experiments shown in Fig. 3, groups of mice were infected with equal mixtures of AJB715 and SW1397. In some experiments, animals were orally treated with tributyrin (5 g/kg) 3 h post infection or mock-treated with PBS (Rivera-Chavez et al., 2016). Two, three, and four days after infection, samples for histopathology, flash frozen cecal and colonic tissue for RNA and protein extraction, and cecal and colonic luminal material (S. Tm colonization) were collected, as indicated. In some experiments, luminal content was flash frozen (liquid nitrogen) for bacterial gene expression analyses.
CBA colitis model
Groups of 8–10 week old CBA mice were intragastrically infected with 1 × 109 CFU for single strain infection experiments, 5 × 108 CFU of each strain for competitive infection experiments, or mock treated (LB broth). After 7 days, samples were collected as described above. For the experiments shown in Fig. 4A and B, mice were infected with AJB715 and SW1285, AJB715 and SW1286, AJB715 and SW1288, AJB715 and SW1056, SW1401 and SW1203, SW1410 and SW1397, respectively. For the experiments shown in Fig. 4C–F, groups of animals were inoculated with IR715 and SW1397, respectively. For Fig. 5A–C, groups of animals were infected with AJB715 and SW1397, AJB715 and SW1411, or SW1410 and SW1413. For Fig. 5C–E, groups of animals were infected with IR715, SW1411, or mock-treated.
Germ-free and conventionally-raised Swiss Webster mice
6–8 week old germ-free mice were intragastrically inoculated with 3 × 109 CFU of Bacteroides thetaiotaomicron. After 3 days, mono-associated and germ-free control animals were infected with an equal mixture of AJB715 and SW1411, SW1410 and SW1413, or AJB715 and SW1397 as described above. Furthermore, cecal microbiota from one CBA donor mouse was orally transferred to a germ-free Swiss Webster breeder pair, which was then maintained under conventional housing conditions. Offspring from this breeder pair was used for the experiment shown in Fig. S5C.
Quantification of inflammatory markers by RT-qPCR
Relative mRNA levels of Nos2, Cxcl1, and Tnfa was determined by RT-qPCR as described previously (Winter et al., 2009). Briefly, tissue was homogenized in a Mini beadbeater (Biospec Products) and RNA was extracted using the TRI reagent method. cDNA was generated by TaqMan reverse transcription reagents. Real-time PCR was performed using SYBR Green qPCR master mix. Data was acquired in a QuantStudio 6 Flex instrument (Life Technologies) and analyzed using the comparative Ct method. Target gene transcription of each sample was normalized to Gapdh mRNA levels.
Bacterial gene expression
To determine bacterial gene expression in vitro, mucin broth was inoculated with 1 × 103 CFU/ml of S. Tm and anaerobically cultured at 37°C for 16 h. Sodium nitrate and tetra thionate was added at a concentration of 40 mM, as indicated. RNA was extracted using the Aurum Total RNA Mini Kit. To investigate bacterial gene expression in the intestinal content, flash frozen cecal material in TRI reagent was homogenized for 1 min in a bead beater (BioSpec) and RNA isolated using the TRI reagent method. RT-PCR and qPCR were performed as described above. Gene expression was normalized to S. Tm 16S rRNA levels. A mock-RT-PCR, lacking reverse transcriptase, was performed for each sample and gene of interest to control for DNA contamination.
Histopathology
Cecal and colonic tissue was fixed in phosphate-buffered formalin for 48 h and embedded in paraffin. Sections (5 μm) were stained with hematoxylin and eosin. Stained sections were blinded and evaluated by a veterinary pathologist according to the criteria listed in the supplementary material. The contrast for the images was uniformly (linear) adjusted using Photoshop CS6.
iNOS expression in intestinal tissue by Western Blot
After murine colonic tissue homogenization with a Mini-BeadBeater (BioSpec Products), colonic proteins were extracted with TRI Reagent according to the manufacturer’s specifications (Molecular Research Center). Precipitated proteins were resuspended in a 1 % w/v sodium dodecyl sulfate (SDS) and 10 mM β-mercaptoethanol solution. Protein concentration of each sample was calculated based on the absorbance at 280 nm measured with an Epoch Microplate Spectrophotometer (BioTek Instruments). Samples were boiled for 1 min and then 10 μg of each sample were resolved by 10% SDS-PAGE. Proteins were transferred to polyvinylidene fluoride (PVDF) membranes (Millipore Immobilon-P) by wet transfer (Bio-Rad Laboratories). Membranes were blocked in 3 % non-fat dry milk and 0.1 % Tween 20 in phosphate-buffered saline (pH 7.4) solution. To detect tubulin and inducible nitric oxide synthase (iNOS) expression, membranes were incubated overnight with primary antibodies. Horseradish peroxidase-conjugated antibodies were used as secondary antibodies. A G:Box imaging system (Syngene) was used to detect the secondary antibodies after 1 min incubation with Immobilon Western Substrate. Images were processed with Photoshop CS6 (Adobe) to uniformly adjust brightness levels.
Metabolite profiling and quantification of succinate
Colon and cecal content from mice was collected in sterile PBS. Samples were agitated for 2 min and debris and bacterial cells were removed by centrifugation at 6,000 g for 15 min at 4°C. For in vitro bacterial cultures, 2 ml of culture were centrifuged at 20,000 g at 4°C. The supernatant was removed and su ccinic-2,2,3,3,-d4 acid was added as an internal control. Samples were dried using a SpeedVac concentrator (Eppendorf) for 2 h and stored at −80°C. External standards and biological samples were derivatized as follows: After adding 0.1 ml of water-free pyridine, each sample was sonicated for 1 min and incubated for 20 min at 80°C. Then 0.1 ml of N-tert-Butyldimethylsilyl-N-methyltrifluoroacetamide with 1 % tert-Butyldimethylchlorosilane was added to the samples and incubated at 80°C for 1 h. After centrifugation at 20,000 g, derivatized samples were transferred to autosampler vials for gas chromatography-mass spectrometry (GC-MS) analysis (Shimadzu, TQ8040). The injection temperature was 250°C and the injection split ratio was set to 1:50 with an injection volume of 1 μl. The oven temperature started at 50°C for 2 min, increasing to 100°C at 2 0°C per min and to 330°C at 40°C per min with a final hold at this temperature for 3 min. Flow rate of the helium carrier gas was kept constant at a linear velocity of 50 cm/s. The column used was a 30 m × 0.25 mm × 0.25 μm Rtx-5Sil MS (Shimadzu). The interface temperature was 300°C. The electron impact ion sour ce temperature was 200°C, with 70 V ionization voltage and 150 μA current. For qualitative experiments, Q3 scans (range of 50–550 m/z, 1000 m/z per second) were performed. The retention time for succinate and deuterated succinate was 10.877 and 10.868 minutes respectively. Multiple reaction monitoring mode was used to quantitatively measure succinate and deuterated succinate, target ion m/z 289>147 reference ion m/z 331>189, and target ion m/z 293>147 reference ion m/z 335>189, respectively.
Transcriptional profile of S. Tm in the large intestine
Gnotobiotic Swiss Webster mice were orally inoculated with 1 × 105 CFU of AJB715 for 48 h as described above. Cecal content was collected and stored in RNALater at −80°C until further processing. Total RNA was extrac ted and cleaned using RNeasy PowerMicrobiome kit and RNeasy Purification kit (Qiagen, MD) according to the recommendations of the manufacturer. RNAseq library was constructed using TruSeq Stranded Total RNA Library Prep kit (Illumina, CA) and the resulting cDNA library was analyzed using TapeStation 4200 (Agilent, CA). Single-end 150 bp sequencing was conducted on an Illumina NextSeq system (Illumina, CA). RNAseq reads were trimmed and decontaminated using BBmap software suite. Reads that failed to align to mouse genome were mapped to the S. Tm LT2 genome using Bowtie2 (Langmead and Salzberg, 2012). Number of reads of each gene was determined using the Subread package (Liao et al., 2014). Relative expression of genes was determined, grouped into indicated pathways, and compared to previously published data (Kroger et al., 2013). The Bray-Curtis distances between samples was calculated using R Vegan packages implemented through Qiime (Caporaso et al., 2010) and visualized via Emperor (Vazquez-Baeza et al., 2013). The RNAseq dataset was deposited at the European Nucleotide Archive under accession number PRJEB21324.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analysis
Statistical data analysis was performed using Graphpad PRISM. Succinate concentrations, fold changes in mRNA transcription, competitive indices, and abundance of Salmonella (CFU/g) underwent logarithmic transformation prior to descriptive and inferential statistical analysis. All transformed data was normally distributed, as determined by D’Agostino-Pearson normality test for large group sizes and Shapiro-Wilk for smaller sizes. The statistical significance of differences between groups was determined using the parametric Student’s t-test applied to the log-transformed, normally distributed data. Cumulative histopathology scores did not follow a normal distribution and thus were analyzed using the non-parametric Mann-Whitney U test. Unless indicated otherwise, *, P < 0.05; **, P < 0.01; ***, P < 0.001; and ns, not statistically significant. Unless indicated otherwise in the figure legend, bars represent the geometric mean +/− standard error. When displaying aggregate data from mouse experiments, the number of animals per group (N) is indicated above each graph or in the figure legend. Animals that had to be euthanized for humane reasons prior to reaching the predetermined time point were excluded from the analysis.
DATA AND SOFTWARE AVAILABILITY
The accession number for the RNAseq data reported in this paper is PRJEB21324 at the European Nucleotide Archive.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-rabbit alpha/beta-tubulin | Cell Signaling Technology | Cat# 2148S |
| Anti-mouse iNOS | Becton Dickinson | Cat# 610431 |
| Anti-rabbit peroxidase-conjugated | Bio-Rad Laboratories | Cat# 170-6515 |
| Anti-mouse peroxidase-conjugated | Bio-Rad Laboratories | Cat# 170-6516 |
| Bacterial and Virus Strains | ||
| E. coli, DH5α λpir, F− endA1 hsdR17 (r−m+) supE44 thi-1 recA1 gyrA relA1 Δ(lacZYA-argF)U189 Φ80lacZΔM15 λpir | Pal et al., 2005 | DH5α λpir |
| E. coli, S17-1 λpir, zxx::RP4 2-(TetR::Mu) (KanR::Tn7) λpir recA1 thi pro hsdR (r−m+) | Simon et al., 1983 | S17-1 λpir |
| S. Typhimurium, IR715 ATCC14028 NalR | Stojiljkovic et al., 1995 | IR715 |
| S. Typhimurium, SL1344 StrepR | Hoiseth and Stocker, 1981 | SL1344 |
| S. Typhimurium, IR715 phoN::KanR | Kingsley et al., 2003 | AJB715 |
| S. Typhimurium, IR715 ΔinvA::TetR ΔspiB::KanR | Raffatellu et al., 2009 | SPN452 |
| S. Typhimurium, IR715 ΔinvA(−9 to +2057) ΔspiB(+25 to +1209) | Rivera-Chavez et al., 2013 | SPN487 |
| S. Typhimurium, ATCC14028 ΔsdhA::KanR | Porwollik et al., 2014 | STM0734 |
| S. Typhimurium, SL1344 ΔsdhA | This study | MW156 |
| S. Typhimurium, SL1344 ΔinvA::TetR ΔspiB::KanR | This study | MW231 |
| S. Typhimurium, SL1344 ΔinvA::TetR ΔspiB::KanR phoN::CmR | This study | MW251 |
| S. Typhimurium, SL1344 ΔinvA::TetR ΔspiB::KanR ΔsdhA | This study | MW256 |
| S. Typhimurium, SL1344 phoN::CmR | Winter et al., 2014 | SW759 |
| S. Typhimurium, IR715 ΔsdhA::KanR | This study | SW1056 |
| S. Typhimurium, IR715 ΔinvA ΔspiB ΔsdhA::KanR | This study | SW1203 |
| S. Typhimurium, IR715 ΔsucAB | This study | SW1285 |
| S. Typhimurium, IR715 ΔsucCD | This study | SW1286 |
| S. Typhimurium, IR715 ΔaceAB | This study | SW1288 |
| S. Typhimurium, IR715 ΔsdhA | This study | SW1397 |
| S. Typhimurium, IR715 ΔinvA ΔspiB phoN::KanR | This study | SW1401 |
| S. Typhimurium, IR715 ΔsdhA phoN::sdhA | This study | SW1410 |
| S. Typhimurium, IR715 ΔdcuA ΔdcuB ΔdctA | This study | SW1411 (Δ3) |
| S. Typhimurium, IR715 ΔdcuA ΔdcuB ΔdctA ΔsdhA::KanR | This study | SW1413 |
| S. Typhimurium, IR715 ΔsdhA phoN::KanR | This study | SW1414 |
| B. thetaiotaomicron, VPI 5482 Δtdk | Koropatkin et al., 2008 | VPI 5482 Δtdk |
| B. thetaiotaomicron, Δtdk ΔfrdCAB | This study | MW399 |
| Biological Samples | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| Gibson Assembly Master Mix | NEB | Cat# E2611L |
| LB Broth, Miller (Luria Bertani) | Becton Dickinson | Cat# 244520 |
| LB Agar, Miller (Luria Bertani) | Becton Dickinson | Cat# 244620 |
| Bacto Brain Heart Infusion | Becton Dickinson | Cat# 237500 |
| Sheep Blood | Hemostat | Cat# DSB1 |
| Columbia Agar | Sigma | Cat# 27688 |
| Bacto Tryptone | Becton Dickinson | Cat# 211705 |
| Sodium Nitrate | Sigma | Cat# S5506 |
| Sodium Tetrathionate | Sigma | Cat# P2926 |
| 5-fluoro-2′-deoxyuridine (FUdR) | ARK Pharm | Cat# AK-24802-1 |
| Mucin from porcine stomach, Type II | Sigma-Aldrich | Cat# M2378 |
| TRI Reagent | Molecular Research | Cat# TR118 |
| TaqMan Reverse Transcription Reagents | Life Technologies | Cat# N8080234 |
| SYBR Green qPCR Master Mix | Life Technologies | Cat# 4309155 |
| Succinic-2,2,3,3,-d4 acid | CDN Isotopes | Cat# D-197 |
| Pyridine anhydrous | Sigma | Cat# 270970 |
| MTBSTFA (with 1% t-BDMCS) | Sigma | Cat# M-108 |
| Critical Commercial Assays | ||
| Succinic acid enzymatic assay | Megazyme | Cat# K-SUCC |
| Aurum Total RNA Mini Kit | BioRad | Cat# 7326820 |
| PowerMicrobiome RNA Isolation Kit | MoBio | Cat# 26000-50 |
| RNeasy Purification kit | Qiagen | Cat# 74204 |
| TruSeq Stranded Total RNA Library Prep kit | Illumina | Cat# RS-122-2201 |
| Tributyrin | Sigma | Cat# W222305 |
| Immobilon Western Chemiluminescent HRP Substrate | Millipore | Cat# WBKLS0500 |
| Deposited Data | ||
| Salmonella in vivo RNAseq dataset | European Nucletide Archive | PRJEB21324 |
| Salmonella transcriptomic compendium from Kroger et al., 2013 (http://bioinf.gen.tcd.ie/cgi-bin/salcom.pl?_HL) | GEO database | GSE49829 |
| Experimental Models: Cell Lines | ||
| Experimental Models: Organisms/Strains | ||
| C57BL/6 WT | Jackson Laboratory | Cat# 000664 |
| C57BL/6 Cybb−/− | Jackson Laboratory | Cat# 002365 |
| C57BL/6 Nos2−/− | Jackson Laboratory | Cat# 002609 |
| CBA/J | Jackson Laboratory | Cat# 000656 |
| Swiss Webster, ex Germ-Free colonized with CBA/J microbiota | This study | N/A |
| Germ-free Swiss-Webster | Hooper Lab | N/A |
| Oligonucleotides | ||
| Primer used in this study, see Table S1 | This paper | N/A |
| Recombinant DNA | ||
| Plasmid: pExchange-tdk, ori(R6K) mobRP4 tdk CarbR ErmR | Koropatkin et al., 2008 | pExchange-tdk |
| Plasmid: pGP704, ori(R6K) mobRP4 CarbR | Miller and Mekalanos, 1988 | pGP704 |
| Plasmid: pRDH10, ori(R6K) mobRP4 sacRB CmR TetR | Kingsley et al., 1999 | pRDH10 |
| Plasmid: Up- and downstream region of sucAB in pRDH10 | This study | pSW284 |
| Plasmid: Up- and downstream region of sucCD in pRDH10 | This study | pSW286 |
| Plasmid: Up- and downstream region of aceAB in pRDH10 | This study | pSW288 |
| Plasmid: Up- and downstream region of sdhA in pRDH10 | This study | pSW306 |
| Plasmid: Internal fragment of the phoN coding sequence cloned into pGP704 | This study | pSW327 |
| Plasmid: sdhA promoter and coding sequence cloned into pSW327 | This study | pSW328 |
| Plasmid: Up- and downstream region of dctA in pRDH10 | This study | pSW299 |
| Plasmid: Up- and downstream region of dcuA in pRDH10 | This study | pSW300 |
| Plasmid: Up- and downstream region of dcuB in pRDH10 | This study | pSW301 |
| Plasmid: Up- and downstream region of frdCAB in pExchange-tdk | This study | pMW5 |
| Software and Algorithms | ||
| Prism 7 | GrapPad Software | https://www.graphpad.com/scientific-software/prism/ |
| TapeStation 4200 | Agilent, CA | |
| BBmap software suite | http://jgi.doe.gov/data-and-tools/bbtools/ | |
| Bowtie2 | Langmead and Salzberg, 2012 | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| R Vegan | https://cran.r-project.org/web/packages/vegan/index.html | |
| Qiime | Caporaso et al., 2010 | http://qiime.org |
| Emperor | Vazquez-Baeza et al., 2013 | https://biocore.github.io/emperor/ |
| GCMS Real Time Analysis | Shimadzu, TQ8040 | N/A |
| Photoshop CS6 | Adobe Photoshop | N/A |
| G:Box Imaging System | Syngene | N/A |
| Other | ||
Supplementary Material
HIGHLIGHTS.
During gut colonization, S. Typhimurium performs a complete, oxidative TCA cycle
Host-derived alternative electron acceptors impact S. Tm central metabolism
Uptake of C4-dicarboxylates enhances S. Tm fitness in the inflamed intestine
Utilization of microbiota-derived succinate supports growth of S. Tm in the gut lumen
Acknowledgments
Work in SEW’s lab was funded by the NIH (AI118807, AI103248, AI128151) and The Welch Foundation (I-1858). Work in LVH’s lab was funded by the NIH (DK070855), the Welch Foundation (I-1874) and the Howard Hughes Medical Institute. JK is supported by the Cancer Prevention and Research Institute of Texas (grant RP150596). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the funding agencies. We would like to thank Drs. David Hendrixson, Julie Pfeiffer, and Vanessa Sperandio for helpful discussion and Madeline Smoot for technical assistance.
Footnotes
AUTHOR CONTRIBUTIONS
SEW, LVH, DC, HAP, DPB, and RLS designed and conceived the study; LS, MGW, WZ, ERH, CCG, CLB, and SEW performed all experiments. RLS and TFC performed the histopathology analysis. WZ, DPB, and JK generated and analyzed the RNAseq data. All authors contributed to data analysis and writing the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ali MM, Newsom DL, Gonzalez JF, Sabag-Daigle A, Stahl C, Steidley B, Dubena J, Dyszel JL, Smith JN, Dieye Y, et al. Fructose-asparagine is a primary nutrient during growth of Salmonella in the inflamed intestine. PLoS pathogens. 2014;10:e1004209. doi: 10.1371/journal.ppat.1004209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alteri CJ, Himpsl SD, Engstrom MD, Mobley HL. Anaerobic respiration using a complete oxidative TCA cycle drives multicellular swarming in Proteus mirabilis. mBio. 2012;3 doi: 10.1128/mBio.00365-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amarasingham CR, Davis BD. Regulation of alpha-ketoglutarate dehydrogenase formation in Escherichia coli. The Journal of biological chemistry. 1965;240:3664–3668. [PubMed] [Google Scholar]
- Barman M, Unold D, Shifley K, Amir E, Hung K, Bos N, Salzman N. Enteric salmonellosis disrupts the microbial ecology of the murine gastrointestinal tract. Infection and immunity. 2008;76:907–915. doi: 10.1128/IAI.01432-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthel M, Hapfelmeier S, Quintanilla-Martinez L, Kremer M, Rohde M, Hogardt M, Pfeffer K, Russmann H, Hardt WD. Pretreatment of mice with streptomycin provides a Salmonella enterica serovar Typhimurium colitis model that allows analysis of both pathogen and host. Infection and immunity. 2003;71:2839–2858. doi: 10.1128/IAI.71.5.2839-2858.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coombes BK, Coburn BA, Potter AA, Gomis S, Mirakhur K, Li Y, Finlay BB. Analysis of the contribution of Salmonella pathogenicity islands 1 and 2 to enteric disease progression using a novel bovine ileal loop model and a murine model of infectious enterocolitis. Infection and immunity. 2005;73:7161–7169. doi: 10.1128/IAI.73.11.7161-7169.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford RW, Keestra AM, Winter SE, Xavier MN, Tsolis RM, Tolstikov V, Baumler AJ. Very long O-antigen chains enhance fitness during Salmonella-induced colitis by increasing bile resistance. PLoS pathogens. 2012;8:e1002918. doi: 10.1371/journal.ppat.1002918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronan JE, Laporte D. Tricarboxylic Acid Cycle and Glyoxylate Bypass. EcoSal Plus. 2013 doi: 10.1128/ecosalplus.3.5.2. [DOI] [PubMed] [Google Scholar]
- Crost EH, Tailford LE, Le Gall G, Fons M, Henrissat B, Juge N. Utilisation of mucin glycans by the human gut symbiont Ruminococcus gnavus is strain-dependent. PLoS One. 2013;8:e76341. doi: 10.1371/journal.pone.0076341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings JH, Pomare EW, Branch WJ, Naylor CP, Macfarlane GT. Short chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut. 1987;28:1221–1227. doi: 10.1136/gut.28.10.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis MM, Hu Z, Klimko C, Narayanan S, Deberardinis R, Sperandio V. The gut commensal Bacteroides thetaiotaomicron exacerbates enteric infection through modification of the metabolic landscape. Cell host & microbe. 2014;16:759–769. doi: 10.1016/j.chom.2014.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuskin F, Lowe EC, Temple MJ, Zhu Y, Cameron EA, Pudlo NA, Porter NT, Urs K, Thompson AJ, Cartmell A, et al. Human gut Bacteroidetes can utilize yeast mannan through a selfish mechanism. Nature. 2015;517:165–169. doi: 10.1038/nature13995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eilam O, Zarecki R, Oberhardt M, Ursell LK, Kupiec M, Knight R, Gophna U, Ruppin E. Glycan degradation (GlyDeR) analysis predicts mammalian gut microbiota abundance and host diet-specific adaptations. mBio. 2014;5 doi: 10.1128/mBio.01526-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Kaoutari A, Armougom F, Gordon JI, Raoult D, Henrissat B. The abundance and variety of carbohydrate-active enzymes in the human gut microbiota. Nature reviews Microbiology. 2013;11:497–504. doi: 10.1038/nrmicro3050. [DOI] [PubMed] [Google Scholar]
- Faber F, Thiennimitr P, Spiga L, Byndloss MX, Litvak Y, Lawhon S, Andrews-Polymenis HL, Winter SE, Baumler AJ. Respiration of Microbiota-Derived 1,2-propanediol Drives Salmonella Expansion during Colitis. PLoS pathogens. 2017;13:e1006129. doi: 10.1371/journal.ppat.1006129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreyra JA, Wu KJ, Hryckowian AJ, Bouley DM, Weimer BC, Sonnenburg JL. Gut microbiota-produced succinate promotes C. difficile infection after antibiotic treatment or motility disturbance. Cell host & microbe. 2014;16:770–777. doi: 10.1016/j.chom.2014.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischbach MA, Sonnenburg JL. Eating for two: how metabolism establishes interspecies interactions in the gut. Cell host & microbe. 2011;10:336–347. doi: 10.1016/j.chom.2011.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint HJ, Scott KP, Duncan SH, Louis P, Forano E. Microbial degradation of complex carbohydrates in the gut. Gut Microbes. 2012;3:289–306. doi: 10.4161/gmic.19897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galan JE, Curtiss R., 3rd Cloning and molecular characterization of genes whose products allow Salmonella typhimurium to penetrate tissue culture cells. Proceedings of the National Academy of Sciences of the United States of America. 1989;86:6383–6387. doi: 10.1073/pnas.86.16.6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godinez I, Haneda T, Raffatellu M, George MD, Paixao TA, Rolan HG, Santos RL, Dandekar S, Tsolis RM, Baumler AJ. T cells help to amplify inflammatory responses induced by Salmonella enterica serotype Typhimurium in the intestinal mucosa. Infect Immun. 2008;76:2008–2017. doi: 10.1128/IAI.01691-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunn JS. The Salmonella PmrAB regulon: lipopolysaccharide modifications, antimicrobial peptide resistance and more. Trends Microbiol. 2008;16:284–290. doi: 10.1016/j.tim.2008.03.007. [DOI] [PubMed] [Google Scholar]
- Gutnick D, Calvo JM, Klopotowski T, Ames BN. Compounds which serve as the sole source of carbon or nitrogen for Salmonella typhimurium LT-2. Journal of bacteriology. 1969;100:215–219. doi: 10.1128/jb.100.1.215-219.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hapfelmeier S, Ehrbar K, Stecher B, Barthel M, Kremer M, Hardt WD. Role of the Salmonella pathogenicity island 1 effector proteins SipA, SopB, SopE, and SopE2 in Salmonella enterica subspecies 1 serovar Typhimurium colitis in streptomycin-pretreated mice. Infection and immunity. 2004;72:795–809. doi: 10.1128/IAI.72.2.795-809.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hapfelmeier S, Stecher B, Barthel M, Kremer M, Muller AJ, Heikenwalder M, Stallmach T, Hensel M, Pfeffer K, Akira S, et al. The Salmonella pathogenicity island (SPI)-2 and SPI-1 type III secretion systems allow Salmonella serovar typhimurium to trigger colitis via MyD88-dependent and MyD88-independent mechanisms. J Immunol. 2005;174:1675–1685. doi: 10.4049/jimmunol.174.3.1675. [DOI] [PubMed] [Google Scholar]
- Hensel M, Hinsley AP, Nikolaus T, Sawers G, Berks BC. The genetic basis of tetrathionate respiration in Salmonella typhimurium. Molecular microbiology. 1999;32:275–287. doi: 10.1046/j.1365-2958.1999.01345.x. [DOI] [PubMed] [Google Scholar]
- Hensel M, Shea JE, Waterman SR, Mundy R, Nikolaus T, Banks G, Vazquez-Torres A, Gleeson C, Fang FC, Holden DW. Genes encoding putative effector proteins of the type III secretion system of Salmonella pathogenicity island 2 are required for bacterial virulence and proliferation in macrophages. Molecular microbiology. 1998;30:163–174. doi: 10.1046/j.1365-2958.1998.01047.x. [DOI] [PubMed] [Google Scholar]
- Hoiseth SK, Stocker BA. Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature. 1981;291:238–239. doi: 10.1038/291238a0. [DOI] [PubMed] [Google Scholar]
- Iuchi S, Lin EC. arcA (dye), a global regulatory gene in Escherichia coli mediating repression of enzymes in aerobic pathways. Proceedings of the National Academy of Sciences of the United States of America. 1988;85:1888–1892. doi: 10.1073/pnas.85.6.1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsley RA, Humphries AD, Weening EH, De Zoete MR, Winter S, Papaconstantinopoulou A, Dougan G, Baumler AJ. Molecular and phenotypic analysis of the CS54 island of Salmonella enterica serotype typhimurium: identification of intestinal colonization and persistence determinants. Infection and immunity. 2003;71:629–640. doi: 10.1128/IAI.71.2.629-640.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsley RA, Reissbrodt R, Rabsch W, Ketley JM, Tsolis RM, Everest P, Dougan G, Baumler AJ, Roberts M, Williams PH. Ferrioxamine-mediated Iron(III) utilization by Salmonella enterica. Applied and environmental microbiology. 1999;65:1610–1618. doi: 10.1128/aem.65.4.1610-1618.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koropatkin NM, Martens EC, Gordon JI, Smith TJ. Starch catabolism by a prominent human gut symbiont is directed by the recognition of amylose helices. Structure. 2008;16:1105–1115. doi: 10.1016/j.str.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koropatkin NM, Cameron EA, Martens EC. How glycan metabolism shapes the human gut microbiota. Nature reviews Microbiology. 2012;10:323–335. doi: 10.1038/nrmicro2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroger C, Colgan A, Srikumar S, Handler K, Sivasankaran SK, Hammarlof DL, Canals R, Grissom JE, Conway T, Hokamp K, et al. An infection-relevant transcriptomic compendium for Salmonella enterica Serovar Typhimurium. Cell host & microbe. 2013;14:683–695. doi: 10.1016/j.chom.2013.11.010. [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawes M, Maloy S. MudSacI, a transposon with strong selectable and counterselectable markers: use for rapid mapping of chromosomal mutations in Salmonella typhimurium. Journal of bacteriology. 1995;177:1383–1387. doi: 10.1128/jb.177.5.1383-1387.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawley TD, Bouley DM, Hoy YE, Gerke C, Relman DA, Monack DM. Host transmission of Salmonella enterica serovar Typhimurium is controlled by virulence factors and indigenous intestinal microbiota. Infection and immunity. 2008;76:403–416. doi: 10.1128/IAI.01189-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- Liu JZ, Jellbauer S, Poe AJ, Ton V, Pesciaroli M, Kehl-Fie TE, Restrepo NA, Hosking MP, Edwards RA, Battistoni A, et al. Zinc sequestration by the neutrophil protein calprotectin enhances Salmonella growth in the inflamed gut. Cell host & microbe. 2012;11:227–239. doi: 10.1016/j.chom.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez CA, Winter SE, Rivera-Chavez F, Xavier MN, Poon V, Nuccio SP, Tsolis RM, Baumler AJ. Phage-mediated acquisition of a type III secreted effector protein boosts growth of salmonella by nitrate respiration. mBio. 2012;3 doi: 10.1128/mBio.00143-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupp C, Robertson ML, Wickham ME, Sekirov I, Champion OL, Gaynor EC, Finlay BB. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell host & microbe. 2007;2:119–129. doi: 10.1016/j.chom.2007.06.010. [DOI] [PubMed] [Google Scholar]
- Macy J, Probst I, Gottschalk G. Evidence for cytochrome involvement in fumarate reduction and adenosine 5′-triphosphate synthesis by Bacteroides fragilis grown in the presence of hemin. Journal of bacteriology. 1975;123:436–442. doi: 10.1128/jb.123.2.436-442.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier L, Vyas R, Cordova CD, Lindsay H, Schmidt TS, Brugiroux S, Periaswamy B, Bauer R, Sturm A, Schreiber F, et al. Microbiota-derived hydrogen fuels Salmonella typhimurium invasion of the gut ecosystem. Cell host & microbe. 2013;14:641–651. doi: 10.1016/j.chom.2013.11.002. [DOI] [PubMed] [Google Scholar]
- Martens EC, Kelly AG, Tauzin AS, Brumer H. The devil lies in the details: how variations in polysaccharide fine-structure impact the physiology and evolution of gut microbes. J Mol Biol. 2014;426:3851–3865. doi: 10.1016/j.jmb.2014.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer-Severs GJ, van Santen E. Short-chain fatty acids and succinate in feces of healthy human volunteers and their correlation with anaerobe cultural counts. Scand J Gastroenterol. 1987;22:672–676. doi: 10.3109/00365528709011141. [DOI] [PubMed] [Google Scholar]
- Mercado-Lubo R, Gauger EJ, Leatham MP, Conway T, Cohen PS. A Salmonella enterica serovar typhimurium succinate dehydrogenase/fumarate reductase double mutant is avirulent and immunogenic in BALB/c mice. Infection and immunity. 2008;76:1128–1134. doi: 10.1128/IAI.01226-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercado-Lubo R, Leatham MP, Conway T, Cohen PS. Salmonella enterica serovar Typhimurium mutants unable to convert malate to pyruvate and oxaloacetate are avirulent and immunogenic in BALB/c mice. Infection and immunity. 2009;77:1397–1405. doi: 10.1128/IAI.01335-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller VL, Mekalanos JJ. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol. 1988;170:2575–2583. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overbergh L, Giulietti A, Valckx D, Decallonne R, Bouillon R, Mathieu C. The use of real-time reverse transcriptase PCR for the quantification of cytokine gene expression. J Biomol Tech. 2003;14:33–43. [PMC free article] [PubMed] [Google Scholar]
- Pal D, Venkova-Canova T, Srivastava P, Chattoraj DK. Multipartite regulation of rctB, the replication initiator gene of Vibrio cholerae chromosome II. Journal of bacteriology. 2005;187:7167–7175. doi: 10.1128/JB.187.21.7167-7175.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrenoud A, Sauer U. Impact of global transcriptional regulation by ArcA, ArcB, Cra, Crp, Cya, Fnr, and Mlc on glucose catabolism in Escherichia coli. Journal of bacteriology. 2005;187:3171–3179. doi: 10.1128/JB.187.9.3171-3179.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porwollik S, Santiviago CA, Cheng P, Long F, Desai P, Fredlund J, Srikumar S, Silva CA, Chu W, Chen X, et al. Defined single-gene and multi-gene deletion mutant collections in Salmonella enterica sv Typhimurium. PLoS One. 2014;9:e99820. doi: 10.1371/journal.pone.0099820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prohl C, Wackwitz B, Vlad D, Unden G. Functional citric acid cycle in an arcA mutant of Escherichia coli during growth with nitrate under anoxic conditions. Archives of microbiology. 1998;170:1–7. doi: 10.1007/s002030050608. [DOI] [PubMed] [Google Scholar]
- Pudlo NA, Urs K, Kumar SS, German JB, Mills DA, Martens EC. Symbiotic Human Gut Bacteria with Variable Metabolic Priorities for Host Mucosal Glycans. mBio. 2015;6:e01282–01215. doi: 10.1128/mBio.01282-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabin RS, Stewart V. Dual response regulators (NarL and NarP) interact with dual sensors (NarX and NarQ) to control nitrate- and nitrite-regulated gene expression in Escherichia coli K-12. Journal of bacteriology. 1993;175:3259–3268. doi: 10.1128/jb.175.11.3259-3268.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffatellu M, George MD, Akiyama Y, Hornsby MJ, Nuccio SP, Paixao TA, Butler BP, Chu H, Santos RL, Berger T, et al. Lipocalin-2 resistance confers an advantage to Salmonella enterica serotype Typhimurium for growth and survival in the inflamed intestine. Cell host & microbe. 2009;5:476–486. doi: 10.1016/j.chom.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson AR, Payne EC, Younger N, Karlinsey JE, Thomas VC, Becker LA, Navarre WW, Castor ME, Libby SJ, Fang FC. Multiple targets of nitric oxide in the tricarboxylic acid cycle of Salmonella enterica serovar typhimurium. Cell host & microbe. 2011;10:33–43. doi: 10.1016/j.chom.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Chavez F, Winter SE, Lopez CA, Xavier MN, Winter MG, Nuccio SP, Russell JM, Laughlin RC, Lawhon SD, Sterzenbach T, et al. Salmonella uses energy taxis to benefit from intestinal inflammation. PLoS pathogens. 2013;9:e1003267. doi: 10.1371/journal.ppat.1003267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Chavez F, Zhang LF, Faber F, Lopez CA, Byndloss MX, Olsan EE, Xu G, Velazquez EM, Lebrilla CB, Winter SE, et al. Depletion of Butyrate-Producing Clostridia from the Gut Microbiota Drives an Aerobic Luminal Expansion of Salmonella. Cell host & microbe. 2016;19:443–454. doi: 10.1016/j.chom.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogowski A, Briggs JA, Mortimer JC, Tryfona T, Terrapon N, Lowe EC, Basle A, Morland C, Day AM, Zheng H, et al. Glycan complexity dictates microbial resource allocation in the large intestine. Nat Commun. 2015;6:7481. doi: 10.1038/ncomms8481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinstein R, Howard AV, Wrong OM. In vivo dialysis of faeces as a method of stool analysis. IV. The organic anion component. Clin Sci. 1969;37:549–564. [PubMed] [Google Scholar]
- Schell MA, Karmirantzou M, Snel B, Vilanova D, Berger B, Pessi G, Zwahlen MC, Desiere F, Bork P, Delley M, et al. The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:14422–14427. doi: 10.1073/pnas.212527599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmieger H. Phage P22-mutants with increased or decreased transduction abilities. Mol Gen Genet. 1972;119:75–88. doi: 10.1007/BF00270447. [DOI] [PubMed] [Google Scholar]
- Schwarz WH, Zverlov VV, Bahl H. Extracellular glycosyl hydrolases from clostridia. Adv Appl Microbiol. 2004;56:215–261. doi: 10.1016/S0065-2164(04)56007-0. [DOI] [PubMed] [Google Scholar]
- Shimizu T, Ohtani K, Hirakawa H, Ohshima K, Yamashita A, Shiba T, Ogasawara N, Hattori M, Kuhara S, Hayashi H. Complete genome sequence of Clostridium perfringens, an anaerobic flesh-eater. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:996–1001. doi: 10.1073/pnas.022493799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon R, Priefer U, Puhler A. A Broad Host Range Mobilization System for In Vivo Genetic Engineering: Transposon Mutagenesis in Gram Negative Bacteria. Nature Biotechnology. 1983;1:784–791. [Google Scholar]
- Stecher B, Barthel M, Schlumberger MC, Haberli L, Rabsch W, Kremer M, Hardt WD. Motility allows S. Typhimurium to benefit from the mucosal defence. Cell Microbiol. 2008;10:1166–1180. doi: 10.1111/j.1462-5822.2008.01118.x. [DOI] [PubMed] [Google Scholar]
- Stecher B, Robbiani R, Walker AW, Westendorf AM, Barthel M, Kremer M, Chaffron S, Macpherson AJ, Buer J, Parkhill J, et al. Salmonella enterica serovar typhimurium exploits inflammation to compete with the intestinal microbiota. PLoS biology. 2007;5:2177–2189. doi: 10.1371/journal.pbio.0050244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steeb B, Claudi B, Burton NA, Tienz P, Schmidt A, Farhan H, Maze A, Bumann D. Parallel exploitation of diverse host nutrients enhances Salmonella virulence. PLoS pathogens. 2013;9:e1003301. doi: 10.1371/journal.ppat.1003301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojiljkovic I, Baumler AJ, Heffron F. Ethanolamine utilization in Salmonella typhimurium: nucleotide sequence, protein expression, and mutational analysis of the cchA cchB eutE eutJ eutG eutH gene cluster. Journal of bacteriology. 1995;177:1357–1366. doi: 10.1128/jb.177.5.1357-1366.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchawa Yimga M, Leatham MP, Allen JH, Laux DC, Conway T, Cohen PS. Role of gluconeogenesis and the tricarboxylic acid cycle in the virulence of Salmonella enterica serovar Typhimurium in BALB/c mice. Infection and immunity. 2006;74:1130–1140. doi: 10.1128/IAI.74.2.1130-1140.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiennimitr P, Winter SE, Winter MG, Xavier MN, Tolstikov V, Huseby DL, Sterzenbach T, Tsolis RM, Roth JR, Baumler AJ. Intestinal inflammation allows Salmonella to use ethanolamine to compete with the microbiota. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:17480–17485. doi: 10.1073/pnas.1107857108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turton LJ, Drucker DB, Ganguli LA. Effect of glucose concentration in the growth medium upon neutral and acidic fermentation end-products of Clostridium bifermentans, Clostridium sporogenes and peptostreptococcus anaerobius. J Med Microbiol. 1983;16:61–67. doi: 10.1099/00222615-16-1-61. [DOI] [PubMed] [Google Scholar]
- Unden G, Kleefeld A. C4-Dicarboxylate Degradation in Aerobic and Anaerobic Growth. EcoSal Plus. 2004;1 doi: 10.1128/ecosalplus.3.4.5. [DOI] [PubMed] [Google Scholar]
- Vazquez-Baeza Y, Pirrung M, Gonzalez A, Knight R. EMPeror: a tool for visualizing high-throughput microbial community data. Gigascience. 2013;2:16. doi: 10.1186/2047-217X-2-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RP, Raffatellu M, Chessa D, Winter SE, Tukel C, Baumler AJ. The Vi-capsule prevents Toll-like receptor 4 recognition of Salmonella. Cell Microbiol. 2008;10:876–890. doi: 10.1111/j.1462-5822.2007.01090.x. [DOI] [PubMed] [Google Scholar]
- Wimpenny JW, Cole JA. The regulation of metabolism in facultative bacteria. 3. The effect of nitrate. Biochim Biophys Acta. 1967;148:233–242. doi: 10.1016/0304-4165(67)90298-x. [DOI] [PubMed] [Google Scholar]
- Winter SE, Thiennimitr P, Nuccio SP, Haneda T, Winter MG, Wilson RP, Russell JM, Henry T, Tran QT, Lawhon SD, et al. Contribution of flagellin pattern recognition to intestinal inflammation during Salmonella enterica serotype typhimurium infection. Infection and immunity. 2009;77:1904–1916. doi: 10.1128/IAI.01341-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter SE, Thiennimitr P, Winter MG, Butler BP, Huseby DL, Crawford RW, Russell JM, Bevins CL, Adams LG, Tsolis RM, et al. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature. 2010;467:426–429. doi: 10.1038/nature09415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter SE, Winter MG, Poon V, Keestra AM, Sterzenbach T, Faber F, Costa LF, Cassou F, Costa EA, Alves GE, et al. Salmonella enterica Serovar Typhi conceals the invasion-associated type three secretion system from the innate immune system by gene regulation. PLoS pathogens. 2014;10:e1004207. doi: 10.1371/journal.ppat.1004207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter SE, Winter MG, Xavier MN, Thiennimitr P, Poon V, Keestra AM, Laughlin RC, Gomez G, Wu J, Lawhon SD, et al. Host-derived nitrate boosts growth of E. coli in the inflamed gut. Science. 2013;339:708–711. doi: 10.1126/science.1232467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynosky-Dolfi MA, Snyder AG, Philip NH, Doonan PJ, Poffenberger MC, Avizonis D, Zwack EE, Riblett AM, Hu B, Strowig T, et al. Oxidative metabolism enables Salmonella evasion of the NLRP3 inflammasome. J Exp Med. 2014;211:653–668. doi: 10.1084/jem.20130627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Bjursell MK, Himrod J, Deng S, Carmichael LK, Chiang HC, Hooper LV, Gordon JI. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science. 2003;299:2074–2076. doi: 10.1126/science.1080029. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.