Abstract
Key points
Glomus cells in the carotid body (CB) and chromaffin cells in the adrenal medulla (AM) are essential for reflex cardiorespiratory adaptation to hypoxia. However, the mechanisms whereby these cells detect changes in O2 tension are poorly understood.
The metabolic properties of acute O2‐sensing cells have been investigated by comparing the transcriptomes of CB and AM cells, which are O2‐sensitive, with superior cervical ganglion neurons, which are practically O2‐insensitive.
In O2‐sensitive cells, we found a characteristic prolyl hydroxylase 3 down‐regulation and hypoxia inducible factor 2α up‐regulation, as well as overexpression of genes coding for three atypical mitochondrial electron transport subunits and pyruvate carboxylase, an enzyme that replenishes tricarboxylic acid cycle intermediates.
In agreement with this observation, the inhibition of succinate dehydrogenase impairs CB acute O2 sensing. The responsiveness of peripheral chemoreceptor cells to acute hypoxia depends on a ‘signature metabolic profile’.
Abstract
Acute O2 sensing is a fundamental property of cells in the peripheral chemoreceptors, e.g. glomus cells in the carotid body (CB) and chromaffin cells in the adrenal medulla (AM), and is necessary for adaptation to hypoxia. These cells contain O2‐sensitive ion channels, which mediate membrane depolarization and transmitter release upon exposure to hypoxia. However, the mechanisms underlying the detection of changes in O2 tension by cells are still poorly understood. Recently, we suggested that CB glomus cells have specific metabolic features that favour the accumulation of reduced quinone and the production of mitochondrial NADH and reactive oxygen species during hypoxia. These signals alter membrane ion channel activity. To investigate the metabolic profile characteristic of acute O2‐sensing cells, we used adult mice to compare the transcriptomes of three cell types derived from common sympathoadrenal progenitors, but exhibiting variable responsiveness to acute hypoxia: CB and AM cells, which are O2‐sensitive (glomus cells > chromaffin cells), and superior cervical ganglion neurons, which are practically O2‐insensitive. In the O2‐sensitive cells, we found a characteristic mRNA expression pattern of prolyl hydroxylase 3/hypoxia inducible factor 2α and up‐regulation of several genes, in particular three atypical mitochondrial electron transport subunits and some ion channels. In addition, we found that pyruvate carboxylase, an enzyme fundamental to tricarboxylic acid cycle anaplerosis, is overexpressed in CB glomus cells. We also observed that the inhibition of succinate dehydrogenase impairs CB acute O2 sensing. Our data suggest that responsiveness to acute hypoxia depends on a ‘signature metabolic profile’ in chemoreceptor cells.
Keywords: acute oxygen sensing, carotid body, gene expression, peripheral chemoreceptors
Key points
Glomus cells in the carotid body (CB) and chromaffin cells in the adrenal medulla (AM) are essential for reflex cardiorespiratory adaptation to hypoxia. However, the mechanisms whereby these cells detect changes in O2 tension are poorly understood.
The metabolic properties of acute O2‐sensing cells have been investigated by comparing the transcriptomes of CB and AM cells, which are O2‐sensitive, with superior cervical ganglion neurons, which are practically O2‐insensitive.
In O2‐sensitive cells, we found a characteristic prolyl hydroxylase 3 down‐regulation and hypoxia inducible factor 2α up‐regulation, as well as overexpression of genes coding for three atypical mitochondrial electron transport subunits and pyruvate carboxylase, an enzyme that replenishes tricarboxylic acid cycle intermediates.
In agreement with this observation, the inhibition of succinate dehydrogenase impairs CB acute O2 sensing. The responsiveness of peripheral chemoreceptor cells to acute hypoxia depends on a ‘signature metabolic profile’.
Abbreviations
- 7‐AAD
7‐aminoactinomycin D
- Acacb
acetyl‐CoA carboxylase b
- Acly
ATP citrate lyase
- AM
adrenal medulla
- Cacna1d/Cav1.3
calcium channel, voltage‐dependent, L type, alpha 1D subunit
- Cacna1h/Cav3.2
calcium channel, voltage‐dependent, T type, alpha 1H subunit
- CB
carotid body
- Chga
chromogranin A
- Cox4i2
cytochrome c oxidase subunit IV isoform 2
- Cox8b
cytochrome c oxidase subunit VIIIb
- DAPI
4’,6’‐diamidino‐2‐phenylindole
- DMEM
Dulbecco's modified Eagle's medium
- DMM
dimethyl malonate
- ETC
electron transport chain
- ETF
electron transport flavin/quinone oxidoreductase
- FFA
free fatty acid
- Gdf10
growth differentiation factor 10
- Gdnf
glial cell line derived neurotrophic factor
- GFP
green fluorescent protein
- Gls
glutaminase
- Hif1β/Arnt
aryl hydrocarbon receptor nuclear translocator
- Hif2α/Epas1
hypoxia inducible factor 2α/endothelial PAS domain protein 1
- Hif2β/Arnt2
aryl hydrocarbon receptor nuclear translocator 2
- HVR
hypoxic ventilatory response
- Idh1
isocitrate dehydrogenase 1 (NADP+), soluble
- Idh3
isocitrate dehydrogenase 3 (NAD+)
- Igfbd3
insulin‐like growth factor binding protein 3
- Kcnh5
potassium voltage‐gated channel, subfamily H (eag‐related), member 5
- Kcnh7
potassium voltage‐gated channel, subfamily H (eag‐related), member 7
- Kcnip3
Kv channel interacting protein 3, calsenilin
- Kcnj3
potassium inwardly rectifying channel, subfamily J, member 3
- Kcnma1
potassium large conductance calcium‐activated channel, subfamily M, alpha member 1
- Kcnmb1
potassium large conductance calcium‐activated channel, subfamily M, beta member 1
- Kcnmb2
potassium large conductance calcium‐activated channel, subfamily M, beta member 2
- Kcnn2
potassium intermediate/small conductance calcium‐activated channel, subfamily N, member 2
- Kcnq3
potassium voltage‐gated channel, subfamily Q, member 3
- Kcnq5
potassium voltage‐gated channel, subfamily Q, member 5
- Kcnt2
potassium channel, subfamily T, member 2
- Kv/Kcn
potassium voltage gated channel
- Kv3/Kcnc
potassium voltage gated channel, Shaw‐related subfamily
- Kv4/Kcnd
potassium voltage‐gated channel, Shal‐related family
- L‐15
Leibowitz medium
- Ldh
lactate dehydrogenase
- MCI, MCII, MCIII, MCIV
mitochondrial complex I, II, III, IV, respectively
- Ndufa4l2
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4‐like 2
- Ntrk1
neurotrophic tyrosine kinase receptor
- Olfr78
olfactory receptor 78
- Pcx
pyruvate carboxylase
- Pdha1
pyruvate dehydrogenase E1 alpha 1
- Pdk4
pyruvate dehydrogenase kinase, isoenzyme 4
- pFDR
P values adjusted with the false discovery rate
- Phd3/Egln3
prolyl hydroxylase 3/egl‐9 family prolyl hydroxylase 3
- Pnmt
phenylethanolamine‐N‐methyltransferase
- Ppia
peptidylpropyl isomerase A
- QH2
ubiquinol/reduced ubiquinone
- Rgs5
regulator of g‐protein signalling 5
- RIN
RNA integrity number
- ROS
reactive oxygen species
- SCG
superior cervical ganglion
- Scn7a
sodium channel, voltage‐gated, type VII, alpha
- Scn9a/Nav1.7
sodium channel, voltage‐gated, type IX, alpha
- SDHD
succinate dehydrogenase complex, subunit D, integral membrane protein
- Slc1a5
solute carrier family 1 (neutral amino acid transporter), member 5
- Slc18a1
solute carrier family 18 (vesicular monoamine), member 1
- Slc7a5
solute carrier family 7 (cationic amino acid transporter, y+ system), member 5
- SST‐RMA
signal space transformation‐robust multiarray average
- Task1/Kcnk3
potassium channel, subfamily K, member 3
- Task3/Kcnk9
potassium channel, subfamily K, member 9
- TCA
tri‐carboxylic acid
- TH
tyrosine hydroxylase
- Trpc5
transient receptor potential cation channel, subfamily C, member 5
- Ucp2
uncoupling protein 2
- Vegfa
vascular endothelial growth factor A
- Vegfc
vascular endothelial growth factor C
Introduction
Acute oxygen (O2) sensing is essential for individuals to survive in environmental or pathological conditions that result in low O2 tension () in the blood (hypoxaemia). The carotid body (CB), strategically located at the carotid bifurcation, is the main arterial chemoreceptor that responds to hypoxia by triggering fast (in seconds) adaptive cardiorespiratory reflexes (hyperventilation and sympathetic activation) to compensate for the decrease in (for a recent review see López‐Barneo et al. 2016a). Together with other O2‐sensitive organs, the CB forms part of the homeostatic acute O2 sensing system (Weir et al. 2005). It has close developmental and functional links with the adrenal medulla (AM), which is innervated by sympathetic nerves and, although less potently than the CB, also has, particularly in the neonate, intrinsic, non‐neurogenic O2 sensitivity (Adams et al. 1996; Mochizuki‐Oda et al. 1997; Mojet et al. 1997; Thompson et al. 1997, 2002; García‐Fernández et al. 2007a). Recently, the physiology of the CB–AM axis has attracted medical interest due to the fact that its over‐activation can contribute to the exaggerated sympathetic outflow underlying hypertension and other comorbidities associated with highly prevalent human diseases (McBryde et al. 2013; Ribeiro et al. 2013; Marcus et al. 2014; del Río et al. 2016).
The mechanisms of acute O2 sensing have been studied in greatest detail in the CB, which is composed of clusters of O2‐sensitive glomus cells. These neurosecretory, presynaptic‐like elements release transmitters that activate sensory fibres, which impinge upon brainstem neurons involved in the control of respiration and autonomic function. Glomus cells contain a variety of K+ channels, which are inhibited during hypoxia to produce depolarization and Ca2+‐dependent secretory vesicle exocytosis (see Lopez‐Barneo et al. 2016a). O2‐regulated K+ channels have also been described in AM cells as well as in other cells of the acute O2‐sensing system (for reviews see Weir et al. 2005; Nurse et al. 2009). However, the precise molecular processes underlying the detection of changes in O2 by chemoreceptor cells, and the nature of the signals that link O2 sensing to membrane ion channels have remained unclear and a matter of debate (Peers, 2015; for an updated review see López‐Barneo et al. 2016b). Recently, we have shown that ablation of the mouse Ndufs2 gene, which encodes a component of the ubiquinone biding site in mitochondrial complex I (MCI) (see Baradaran et al. 2013), results in selective abolition of both the hypoxic ventilatory response (HVR) and sensitivity to hypoxia in single glomus and AM chromaffin cells. The data suggest that normoxic peripheral chemoreceptor cells possess special metabolic features, which result in the accumulation of reduced ubiquinone (QH2). Slow‐down of the mitochondrial electron transport chain (ETC) during hypoxia may further increase the QH2 pool, thereby enhancing the production of reactive oxygen species (ROS) and reduced pyridine nucleotides in MCI to signal membrane ion channels (Fernández‐Agüera et al. 2015; Gao et al. 2017).
To advance our knowledge of the metabolic specifications characteristic to acute O2‐sensing cells, we performed a comparative analysis of the gene expression profile in the mouse CB, AM and superior cervical ganglion (SCG). A previous gene expression study, carried out on the CB and AM from mice subjected to either normoxia or sustained hypoxia, focused on ion channels and considered the AM as an O2‐insensitive tissue, the gene expression profile of which was used for background subtraction (Ganfornina et al. 2005). In another study, the gene expression profile of the CB from two different mouse strains with variable responsiveness to hypoxia was analysed. This work reported differences in genes encoding ion channels or related to neurotransmitter metabolism, synaptic vesicles and the development of neural crest‐derived cells (Balbir et al. 2007). More recently, the human CB transcriptome has been studied with attention to the expression of channels and receptors relevant for anaesthesia and the up‐regulation of CB genes involved in the inflammatory response (Fagerlund et al. 2010; Mkrtchian et al. 2012). While the current investigation was in progress, a transcriptomic analysis, comparing mouse CB with olfactory or vomeronasal sensory neurons using single‐cell RNA sequencing, was reported (Zhou et al. 2016). This study identified abundant G protein‐coupled receptor signalling, various types of ion channels and hypoxia inducible factor 2α (Hif2α) in neonatal [postnatal day 4 (P4)–P5] glomus cells. In addition, two atypical mitochondrial ETC subunits were among the most specifically expressed genes identified in CB cells (Zhou et al. 2016). In the current study, we used adult mice to compare the transcriptomes of three cell types with the same embryological origin (neural crest‐derived sympathoadrenal progenitors) but variable responsiveness to acute hypoxia: CB glomus cells and AM chromaffin cells, which are O2‐sensitive (glomus cells > chromaffin cells), and SCG neurons, which are O2‐insensitive. We expected this experimental approach to facilitate the identification of genes relevant to acute O2 sensing in comparison with genes related to other cellular functions, developmental specifications or age. Our results reveal a characteristic mRNA expression pattern of prolyl hydroxylase 3 (Phd3)/Hif2α in O2‐sensitive cells and expand previous studies regarding the abundance of atypical mitochondrial ETC subunits in these cells. In addition, we found that several metabolic enzymes, in particular pyruvate carboxylase (Pcx), which is fundamental to tricarboxylic acid (TCA) cycle anaplerosis (see Owen et al. 2002), are differentially expressed in CB chemoreceptor cells compared to the SCG. Finally, we show that pharmacological or genetic inhibition of succinate dehydrogenase impairs CB acute O2 sensing. Our data support the concept that responsiveness to acute hypoxia depends on a ‘signature metabolic profile’ in peripheral chemoreceptor cells.
Methods
Ethical approval
All procedures were approved by the Institutional Committee of the University of Seville for Animal Care and Use (2012PI/LB02 and 22‐09‐15‐332). Handling of the animals was conducted in accordance with the European Community Council directives 86/609/EEC, and 2010/63/EU for the Care and Use of Laboratory Animals. The experiments comply with the principles of animal research established by the Journal of Physiology (Grundy, 2015).
Animals
TH‐GFP transgenic mice were originally obtained from GENSAT (RRID: MMRRC_000292‐UNC) on a mixed background (Gong et al. 2003) and back‐crossed to C57/B6 background in our laboratory. This line was generated by random insertion of a bacterial artificial chromosome containing regulatory sequences of tyrosine hydroxylase (TH) expression followed by EGFP reporter gene. TH‐SDHD mice, in which the mitochondrial complex II (MCII) subunit D (Sdhd) was deleted in TH+ cells, were generated previously in our laboratory (Díaz‐Castro et al. 2012; Platero‐Luengo et al. 2014). Transgenic and wild‐type (C57/B6) mice were housed at regulated temperature (22 ± 1°C) in a 12 h light/dark cycle with ad libitum access to food and drink. Both male and female mice were used in the current study. Mice were killed via intraperitoneal administration of a lethal dose of sodium thiopental (120–150 mg kg−1) before tissue dissection. Dissected tissues were either fast‐frozen with liquid N2 and stored at −80 °C for RNA isolation, or processed for immunohistochemical analysis, cell sorting, or functional analyses, as described below.
Microarray analysis
Total RNA was isolated from CB, AM and SCG of wild‐type adult (∼2 months old) mice using RNeasy Micro kit (Qiagen, Valencia, CA, USA). Due to the small tissue size, each CB replicate was pooled from 10 mice, whereas each AM and SCG replicate was pooled from three mice to obtain sufficient RNA. The RNA quality was determined using an Agilent 2100 Bioanalyzer (Agilent, Santa Clara, CA, USA). RNA samples with RNA integrity number (RIN) ≥ 7.8 were further processed for microarray analysis.
RNA was amplified and labelled using the GeneChip WT PLUS Reagent Kit (Affymetrix, Santa Clara, CA, USA). Amplification was performed with 50 ng of total RNA input following procedures described in the WT PLUS Reagent Kit user manual. The amplified cDNA was quantified, fragmented and labelled in preparation for hybridization to GeneChip Mouse Transcriptome 1.0 Array (Affymetrix) using 5.5 μg of single‐stranded cDNA product and following protocols outlined in the user manual. Washing, staining (GeneChip Fluidics Station 450, Affymetrix) and scanning (GeneChip Scanner 3000, Affymetrix) were performed following protocols outlined in the user manual for cartridge arrays. Data were processed for gene‐level background subtraction, normalization and signal summarization (SST‐RMA, signal space transformation‐robust multi‐array average) using an Affymetrix Expression Console. Gene‐level differential expression analysis was then performed using Transcriptome Analysis Console 3.0 (Affymetrix). One‐way between‐subject (unpaired) ANOVA was used and P values adjusted with the false discovery rate (pFDR) were calculated. Gene expression was considered different between groups with pFDR <0.05 and fold change >2 or <−2. In addition, after the SST‐RMA normalization, data were analysed with Bioconductor to determine the similarity of gene expression profiles among samples using hierarchical clustering analysis, to evaluate the difference of gene expression patterns among the samples using principal component analysis, and to visualize differentially expressed genes using a volcano plot (Huber et al. 2015). Microarray raw data are deposited in the NCBI GEO database (accession number: GSE99593).
Flow cytometry
Freshly dissected CB and AM from TH‐GFP mice were quickly placed in ice‐cooled modified Tyrode solution (in mm: 148 NaCl, 2 KCl, 3 MgCl2, 10 Hepes, 10 glucose, pH 7.4) for enzymatic dispersion. Dispersion of CB glomus cells and AM chromaffin cells was performed following the procedures described by our laboratory (Piruat, et al. 2004; Muñoz‐Cabello et al. 2005; Levitsky and López‐Barneo, 2009). SCGs dissected from TH‐GFP mice were collected in ice‐cooled Leibowitz medium (L‐15), followed by enzymatic cell dispersion as described previously (Alberola‐Die et al. 2013). Dispersed cells from each tissue were incubated with 7‐aminoactinomycin D (7‐AAD, 1:200 dilution, BD Biosciences, Franklin Lakes, NJ, USA) in FACS buffer (L‐15 supplemented with 1% penicillin/streptomycin, 0.2% bovine serum albumin, 10 mm Hepes, 5 mm EDTA) to label the non‐viable cells. Green fluorescent protein positive (GFP+) cells were sorted using a FACSJazz cell sorter (BD Biosciences). GFP+ cells were either collected in PBS for immunocytochemical analysis or in Buffer RLT (Qiagen) and immediately frozen at −80 °C for RNA isolation.
Real‐time quantitative PCR
To validate the results of microarray analysis, total RNA was isolated using an RNeasy Micro kit (Qiagen) from CB, AM, and SCG of wild‐type adult mice, which were different from those used for microarray analysis. Each CB replicate was pooled from 15 mice, whereas each AM and SCG replicate was pooled from four mice. In addition, total RNA was also isolated from GFP+ cells of each tissue from TH‐GFP mice, which were sorted by flow cytometry as mentioned above. The RNA quality was determined using an Agilent 2100 Bioanalyzer (Agilent) and cRNA was amplified using either a GeneChip WT PLUS Reagent Kit (Affymetrix) from whole tissues or GeneChip WT Pico kit (Affymetrix) from GFP+ cells.
In total, 500 ng of cRNA was copied to cDNA using a QuantiTect Reverse Transcription Kit (Qiagen) in a final volume of 20 μl. Taqman mouse Endogenous Control Array (Applied Biosystems, Carlsbad, CA, USA) was used in a Viia7 Real‐Time PCR system (Applied Biosystems) to select a housekeeping gene among three tissues. Real‐time quantitative PCRs were performed in a 7500 Fast Real Time PCR System (Life Technologies). PCRs were performed in duplicate in a total volume of 20 μl containing 1–4 μl of cDNA solution and 1 μl of Taqman probe of the specific gene (ThermoFisher Scientific Inc., Waltham, MA, USA). Peptidylprolyl isomerase A (Ppia) was also estimated in each sample to normalize the amount of cRNA input in order to perform relative quantifications.
Immunohistological analysis
GFP+ cells sorted by flow cytometry were plated on poly‐l‐lysine‐treated coverslips and incubated for 2 h in either Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum, 1% penicillin/streptomycin, 1% l‐glutamine and 84 mU ml−1 insulin for CB and SCG, or DMEM supplemented with 10% fetal bovine serum, 2 mm l‐glutamine, 1% penicillin/streptomycin for AM. For immunohistochemical studies, mice were perfused first with PBS and then with 4% paraformaldehyde in PBS before scarification and tissue dissection. Carotid bifurcation and adrenal gland were fixed with 4% paraformaldehyde in PBS for 2 h, cryoprotected overnight with 30% sucrose in PBS and embedded in OCT (Tissue‐Tek). Tissue sections of 10 μm were cut with a cryostat (Leica, Wetzlar, Germany). Cells and tissue sections were incubated with primary antibodies overnight at 4°C: Cox4i2 (1:100 dilution, 11463‐1‐1AP, Proteintech, Chicago, IL, USA); Ndufa4l2 (1:50 dilution, 16480‐1‐AP, Proteintech); Pcx (1:100 dilution, ab115579, Abcam, Cambridge, MA, USA); TH (1:5000 dilution, NB300‐109, Novus Biological, Inc., Littleton, CO, USA); or TH (1:100 dilution, AB1542, Millipore, Billerica, MA, USA). This was followed by incubation with fluorescent secondary antibodies: Alexa 568 or Alexa 488 (1:500 dilution, A11057, A11011, A11008, A11015, Invitrogen, Carlsbad, CA, USA). Nuclei were labelled with 4’,6’‐diamidino‐2‐phenylindole (DAPI) staining. Immunofluorescence images were obtained using Nikon A1R+ confocal microscopy (Nikon).
Amperometric recording of catecholamine secretion
Dissected CBs were placed in ice‐cooled modified Tyrode solution and CB slices were prepared as described previously (Ortega‐Sáenz et al. 2010). Briefly, CB slices (150 μm) were sectioned using a vibratome (VT1000, Leica). After a brief enzymatic digestion, the slices were incubated in DMEM supplemented with 10% fetal bovine serum, 1% penicillin/streptomycin, 1% l‐glutamine, 1.2 U ml−1 erythropoietin and 84 mU ml−1 insulin at 37°C in 5% CO2 for 24 h. In some experiments, 2 mm dimethyl malonate (DMM) was added 12 h before recording. Catecholamine secretion was recorded perfusing CB slices continuously with a recording solution (in mm: 117 NaCl, 4.5 KCl, 23 NaHCO3, 1 MgCl2, 2.5 CaCl2, 5 glucose and 5 sucrose, pH 7.4) using an upright microscope (Axioscope, Zeiss, Oberkochen, Germany). The ‘normoxic’ solution was bubbled with 5% CO2, 20% O2 and 75 % N2 ( = 150 mmHg). The ‘hypoxic’ solution was bubbled with 5% CO2 and 95% N2 ( ∼10–20 mmHg). In the high K+ solution, NaCl was replaced by KCl equimolarly. DMM (2 mm) was added to the recording solution when necessary. All experiments were carried out at ∼36°C. Secretory events were recorded with a 10 μm polarized carbon fibre electrode. Amperometric currents were recorded with an EPC‐8 patch‐clamp amplifier (HEKA Electronics, Lambrecht/Pfaltz, Germany). Data acquisition and analysis were carried out with an ITC‐16 interface (Instrutech Corporation, Longmont, CO, USA) and PULSE/PULSEFIT software (HEKA Electronics). The secretion rate (fC min–1) was calculated as the amount of charge transferred to the recording electrode during a given period of time and cumulative secretion was calculated as the integral on time of the amperometric recording.
Cytosolic Ca2+ measurements by microfluorimetry
CB glomus cells were dispersed as described previously (Piruat et al. 2004), seeded on glass poly‐l‐lysine‐treated glass coverslips, and kept in DMEM supplemented with 10% fetal bovine serum, 1% penicillin/streptomycin, 1% l‐glutamine and 84 mU ml−1 insulin. To measure changes in cytosolic Ca2+ concentration, glomus cells were first incubated with 4 μm 2‐AM Fura (F1225, Thermo Scientific) in DMEM for 30 min at 37°C in a 5% CO2 incubator. Loaded cells were then recorded using an inverted microscope (Eclipse Ti, Nikon) equipped with epifluorescence and photometry (Fernández‐Agüera et al. 2015). Alternating excitation wavelengths of 340 and 380 nm were used, and background fluorescence was subtracted before calculating the ratio of fluorescence intensity between 340 and 380 nm. Data were digitized at a sampling interval of 1 s. All the experiments were performed at 36°C.
Statistics
Data were presented as mean ± SEM with the number (n) of experiments indicated and analysed using Sigmaplot v12.0. Normality was tested with the Shapiro–Wilk test. When necessary, a log transformation was performed to normalize the data distribution prior to parametric analyses using a t test. A P value <0.05 was considered statistically significant. The statistical analysis of the microarray data is described in the ‘Microarray analysis’ section.
Results
Gene expression profiles in the CB, AM and SCG
Gene expression profiles were compared among cells in the CB, AM, and SCG from adult mice using Affymetrix GeneChip Mouse Transcriptome Assay 1.0 Array, which includes >23000 protein coding genes and >55000 non‐protein coding genes, such as non‐coding RNA, pseudogenes or rRNA. Hierarchical clustering analysis was first performed to analyse the similarity of the gene expression profiles between the samples. As shown in Fig. 1 A, replicates from the same tissue were clustered together and appeared distant from replicates of other tissues. Principal component analysis was performed to determine the difference in gene expression patterns among the samples, revealing that the three tissues were clearly separated with two principal components (Fig. 1 B). These data demonstrate the existence of a global difference in gene expression among CB, AM and SCG cells.
Figure 1. Microarray analysis of transcriptomes of the carotid body (CB), adrenal medulla (AM), and superior cervical ganglion (SCG) from adult mice.
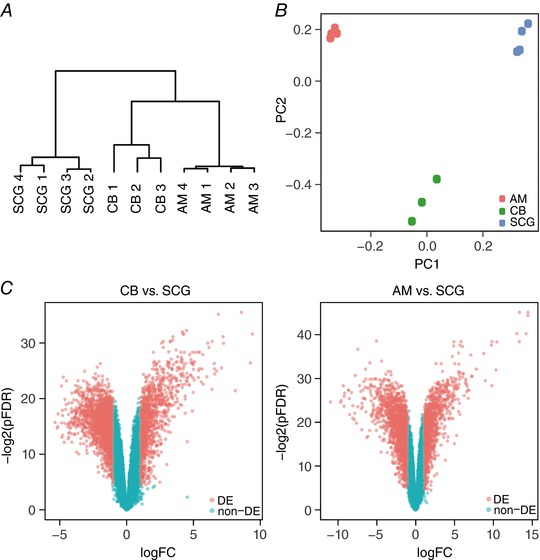
A, hierarchical clustering analysis. Numbers indicate separate tissue samples. B, principal component (PC) analysis. C, volcano plots demonstrating differentially expressed genes (DE, light red, fold change > 2, pFDR < 0.05) compared to genes without differential expression (non‐DE, light blue). Left, CB versus SCG; right, AM versus SCG.
Differential gene expression between the CB and SCG
Gene expression in the CB compared to the SCG was studied based on cutoff values of fold change >2 and pFDR <0.05. Up‐ and down‐regulated genes can be visualized in Fig. 1 C (light red). The genes with the largest changes in expression level (top 20 up‐regulated and top 20 down‐regulated genes) are listed in Table 1. Among the most differentially expressed genes, a number related to G‐protein signalling [e.g. regulator of g‐protein signalling 5 (Rgs5)] were up‐regulated in the CB. Genes related to trophic factors were also up‐regulated in the CB [growth differentiation factor 10 (Gdf10) and insulin‐like growth factor binding protein (Igfbd3)] or the SCG [neurotrophic tyrosine kinase receptor (Ntrk1)]. Notably, several cyto/chemokines and extracellular matrix proteins were overexpressed in the CB, which may be related to the abundance of immune cells in this organ (Mkrtchian et al. 2012) and the organization of various cell types in the CB glomeruli. In this unbiased general analysis, three genes putatively involved in O2 sensing were among the most highly expressed in the CB: NADH dehydrogenase 1 alpha subcomplex, 4‐like 2 (Ndufa4l2), endothelial PAS domain protein 1 (Hif2α) and the Kcnk9 K+ channel (Task3) (see below).
Table 1.
Top 20 up‐ and down‐regulated genes in the CB versus the SCG of adult mice by microarray analysis
| Gene symbol | Description | Fold change (linear) | pFDR |
|---|---|---|---|
| Up‐regulated | |||
| Gdf10 | growth differentiation factor 10 | 760 | 1.49E‐03 |
| Ndufa4l2 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4‐like 2 | 721 | 3.90E‐04 |
| Mgp | matrix Gla protein | 485 | 4.55E‐03 |
| Acta2 | actin, alpha 2, smooth muscle, aorta | 388 | 2.20E‐04 |
| Dpt | dermatopontin | 164 | 1.26E‐03 |
| Igfbp3 | insulin‐like growth factor binding protein 3 | 160 | 2.29E‐04 |
| Rgs5 | regulator of G‐protein signalling 5 | 142 | 8.02E‐04 |
| Cfh | complement component factor h | 119 | 2.29E‐04 |
| Ccl21a | chemokine (C‐C motif) ligand 21A (serine) | 104 | 1.39E‐03 |
| Epas1(Hif2a) | endothelial PAS domain protein 1 | 102 | 2.49E‐03 |
| Cpa3 | carboxypeptidase A3, mast cell | 100 | 1.39E‐03 |
| Myh11 | myosin, heavy polypeptide 11, smooth muscle | 98 | 6.71E‐04 |
| Itga8 | integrin alpha 8 | 96 | 2.74E‐03 |
| Cytl1 | cytokine‐like 1 | 85 | 4.35E‐03 |
| Kcnk9(Task3) | potassium channel, subfamily K, member 9 | 83 | 2.19E‐03 |
| Ccl21a/Ccl21b/Ccl21c | chemokine (C‐C motif) ligand, 21A (serine), 21B (leucine), 21C (leucine) | 81 | 1.49E‐03 |
| Slc9a2 | solute carrier family 9 (sodium/hydrogen exchanger), member 2 | 71 | 8.22E‐04 |
| Adm | adrenomedullin | 64 | 1.99E‐03 |
| Myl9 | myosin, light polypeptide 9, regulatory | 62 | 2.23E‐03 |
| Mcpt4 | mast cell protease 4 | 62 | 3.36E‐03 |
| Down‐regulated | |||
| Cxcr4 | chemokine (C‐X‐C motif) receptor 4 | −41 | 7.12E‐03 |
| Napb | N‐ethylmaleimide sensitive fusion protein attachment protein beta | −39 | 2.34E‐02 |
| Tubb4a | tubulin, beta 4A class IVA | −38 | 4.66E‐02 |
| Mapk11 | mitogen‐activated protein kinase 11 | −36 | 1.29E‐02 |
| Tmem179 | transmembrane protein 179 | −35 | 1.06E‐02 |
| Htr3a | 5‐hydroxytryptamine (serotonin) receptor 3A | −35 | 2.19E‐02 |
| Ntrk1 | neurotrophic tyrosine kinase, receptor, type 1 | −30 | 3.42E‐02 |
| Nrip3 | nuclear receptor interacting protein 3 | −29 | 4.13E‐03 |
| Sult4a1 | sulfotransferase family 4A, member 1 | −29 | 1.03E‐02 |
| Htr3b | 5‐hydroxytryptamine (serotonin) receptor 3B | −28 | 1.83E‐02 |
| Nefl | neurofilament, light polypeptide | −27 | 3.10E‐02 |
| Vwc2l | von Willebrand factor C domain‐containing protein 2‐like | −27 | 3.00E‐03 |
| Maob | monoamine oxidase B | −27 | 1.71E‐02 |
| Gpr158 | G protein‐coupled receptor 158 | −25 | 1.64E‐02 |
| Ehd3 | EH‐domain containing 3 | −25 | 3.06E‐02 |
| Ttll1 | tubulin tyrosine ligase‐like 1 | −24 | 2.16E‐02 |
| Cyp2j12 | cytochrome P450, family 2, subfamily j, polypeptide 12 | −24 | 1.01E‐02 |
| Stk32a | serine/threonine kinase 32A | −23 | 9.87E‐03 |
| Slc6a15 | solute carrier family 6 (neurotransmitter transporter), member 15 | −22 | 7.08E‐03 |
| Maoa | monoamine oxidase A | −21 | 1.92E‐02 |
pFDR, P value adjusted with the false discovery rate.
We focused our analysis on genes implicated in the O2‐sensing pathway or related to cellular functions that could be relevant to acute responsiveness to hypoxia. In addition to the up‐regulation of Hif2α, we found significantly increased expression of the constitutively active Hifβ subunit (Arnt2) and Hif‐dependent angiogenic genes (Vegfa and Vegfc). Among the Phd enzymes, which hydroxylate Hifα protein isoforms for degradation by the proteasome, we found selective mRNA down‐regulation of Phd3 (Egln3) (Table 2). As MCI integrity seems to be essential for acute O2 sensing by peripheral chemoreceptors (Fernández‐Agüera et al. 2015), we also studied the expression of genes encoding ETC subunits. The level of mRNA expression of most of the ETC subunits was similar between the CB and SCG. However, several genes, which code for subunits of MCI to MCIV, were slightly down‐regulated in the CB compared to the SCG (Table 2). In contrast, the mRNAs of three ETC subunits were markedly overexpressed in CB cells: the Ndufa4l2 subunit and the genes encoding for cytochrome c oxidase subunit IV isoform 2 (Cox4i2) and cytochrome c oxidase subunit VIIIb (Cox8b).
Table 2.
Hypoxia‐related differential gene expression in the CB compared to the SCG of adult mice by microarray analysis*
| Gene symbol | Description | Fold change (linear) | pFDR |
|---|---|---|---|
| Phd/Hif pathway and targets | |||
| Epas1(Hif2a) | endothelial PAS domain protein 1 | 102.0 | 2.49E‐03 |
| Arnt2 | aryl hydrocarbon receptor nuclear translocator 2 | 2.9 | 1.47E‐02 |
| Egln3(Phd3) | egl‐9 family hypoxia‐inducible factor 3 | −4.1 | 1.89E‐02 |
| Vegfa | vascular endothelial growth factor A | 4.2 | 1.86E‐02 |
| Vegfc | vascular endothelial growth factor C | 3.9 | 7.34E‐03 |
| Mitochondria | |||
| Ndufa4l2 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4‐like 2 | 720.9 | 3.90E‐04 |
| Ndufa8 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 8 | −2.1 | 4.01E‐02 |
| Ndufa9 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 9 | −3.4 | 5.17E‐03 |
| Ndufa10 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex 10 | −2.4 | 1.28E‐02 |
| Ndufaf7 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex assembly factor 7 | −3.1 | 1.20E‐02 |
| Ndufb6 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 6 | −2.1 | 3.16E‐02 |
| Ndufs8 | NADH dehydrogenase (ubiquinone) Fe‐S protein 8 | −3.9 | 1.09E‐02 |
| Sdhd | succinate dehydrogenase complex, subunit D, integral membrane protein | ‐4.0 | 1.03E‐02 |
| Uqcrc1 | ubiquinol‐cytochrome c reductase core protein 1 | −2.6 | 1.07E‐02 |
| Cyb561 | cytochrome b‐561 | −5.0 | 2.78E‐02 |
| Cyb561d2 | cytochrome b‐561 domain containing 2 | −2.7 | 3.33E‐03 |
| Cyb5b | cytochrome b5 type B | −5.0 | 6.07E‐03 |
| Cyb5d1 | cytochrome b5 domain containing 1 | −3.6 | 3.94E‐02 |
| Cyba | cytochrome b‐245, alpha polypeptide | 3.2 | 1.49E‐02 |
| Cox4i2 | cytochrome c oxidase subunit IV isoform 2 | 10.1 | 6.03E‐03 |
| Cox8b | cytochrome c oxidase subunit VIIIb | 11.8 | 1.30E‐02 |
| Cox5a | cytochrome c oxidase subunit Va | −2.2 | 4.54E‐02 |
| Coa3 | cytochrome C oxidase assembly factor 3 | −3.0 | 2.21E‐02 |
| Cox15 | cytochrome c oxidase assembly protein 15 | −3.6 | 1.79E‐02 |
| Slc25a27 | solute carrier family 25, member 27 | −2.8 | 2.33E‐03 |
| TCA cycle/anaplerosis/biotin‐related | |||
| Pcx | pyruvate carboxylase | 4.9 | 1.31E‐02 |
| Pdha1 | pyruvate dehydrogenase E1 alpha 1 | −2.5 | 1.90E‐02 |
| Pdk4 | pyruvate dehydrogenase kinase, isoenzyme 4 | 9.1 | 3.90E‐03 |
| Clybl | citrate lyase beta like | −2.7 | 9.23E‐03 |
| Acacb | acetyl‐Coenzyme A carboxylase beta | 2.7 | 9.57E‐03 |
| Slc7a5 | solute carrier family 7 (cationic amino acid transporter, y+ system), member 5 | −3.6 | 2.24E‐02 |
| Idh1 | isocitrate dehydrogenase 1 (NADP+), soluble | −3.5 | 1.34E‐02 |
| Idh3a | isocitrate dehydrogenase 3 (NAD+) alpha | −3.1 | 5.50E‐03 |
| Idh3b | isocitrate dehydrogenase 3 (NAD+) beta | −4.6 | 3.83E‐02 |
| Ogdhl | oxoglutarate dehydrogenase‐like | −2.6 | 2.09E‐02 |
| Ogfod1 | 2‐oxoglutarate and iron‐dependent oxygenase domain containing 1 | −3.2 | 9.12E‐03 |
| Mdh1 | malate dehydrogenase 1, NAD (soluble) | −2.4 | 8.89E‐03 |
| Others | |||
| Gdnf | glial cell line derived neurotrophic factor | 2.1 | 9.36E‐03 |
| Pparg | peroxisome proliferator activated receptor gamma | 2.4 | 8.63E‐03 |
*pFDR (P value adjusted with the false discovery rate) < 0.05 and fold change >2 or <−2.
CB glomus cells (and, to a lesser degree, AM chromaffin cells) contain large amounts of biotin, a coenzyme of carboxylases (Ortega‐Sáenz et al. 2016). In addition, CB cells contain high levels of succinate, which could be involved in acute O2 sensing (Fernández‐Agüera et al. 2015). These facts led us to explore the status of genes involved in pyruvate metabolism and the TCA cycle. Notably, our analysis showed a clear induction of Pcx with decreased expression of a pyruvate dehydrogenase subunit (Pdha1), suggesting pyruvate‐mediated TCA anaplerosis (see Owen et al. 2002). We also found increased mRNA expression of pyruvate dehydrogenase kinase (Pdk4 isoform), which phosphorylates Pdha1 and inhibits the conversion of pyruvate to acetyl‐CoA (Table 2). However, Pdk4 overexpression was not confirmed in real‐time quantitative PCR analyses of sorted tyrosine hydroxylase‐positive (TH+) cells (see below). In contrast, the mRNA level of citrate lyase (Acly), a classical cataplerotic enzyme that converts cytosolic citrate to acetyl‐CoA and oxaloacetate (see Owen et al. 2002), was down‐regulated in sorted CB cells (see below). Acetyl‐CoA carboxylase b (Acacb) was slightly up‐regulated in CB cells, whereas several mitochondrial and cytosolic isoforms of isocitrate dehydrogenase (Idh1 and Idh3) were down‐regulated in CB samples. In parallel, we observed a decrease in the mRNA expression of the cationic amino acid transporter (Slc7a5), which could be related to decreased glutamate uptake and cytosolic production of α‐ketoglutarate (Table 2). Among other relevant genes, we found that the neurotrophic factor Gdnf, which is necessary for the maintenance of glomus cells (Villadiego et al. 2005; Pascual et al. 2008), was significantly overexpressed in the CB (Table 2).
Peripheral chemoreceptor cells contain several subtypes of O2‐regulated K+ channels, which play a central role in cellular responsiveness to hypoxia. In addition, other channel types mediate the cell excitability and Ca2+ influx that are necessary for neurotransmitter release. The ion channels which were differentially expressed in the CB relative to the SCG are listed in Table 3. Task3 (Kcnk9, see above) was the most highly up‐regulated K+ channel gene in CB cells, although Task1 (Kcnk3) and maxi‐K+ β‐subunits (Kcnmb1 and Kcnmb2) were also induced. CB cells expressed a broad variety of voltage‐gated K+ channel α‐subunits, including members of the Kv3 (Kcnc) and Kv4 (Kcnd) families, which are regulated by hypoxia in mouse glomus cells (Pérez‐García et al. 2004). However, several subclasses of Kv (Kcn) α‐subunits were markedly down‐regulated in the CB (see below). The robust up‐regulation of a Na+‐activated K+ channel (Kcnt2) seen in the microarray analysis was not validated by quantitative PCR studies (see below). Interestingly, calsenilin (Kcnip3), a K+ channel‐interacting Ca2+‐binding protein involved in the regulation of gene expression and cell excitability (Spreafico et al. 2001), was up‐regulated in CB tissue. Kcnq3 channels, which mediate the acetyl choline‐activated M‐current characteristic of sympathetic neurons (see Brown & Passmore, 2009), were markedly up‐regulated in SCG tissue (Table 3), further supporting the proposition that the differences in the microarray expression profile reflect genes that are differentially expressed in CB versus SCG cells. Similar to Kv channels, most Na+ and Ca2+ channel subunits were up‐regulated in the SCG, which is compatible with the need for a high density of voltage‐gated ion channels to support the electrical excitability of large SCG sympathetic neurons in comparison with small CB glomus cells (Table 3). Notable exceptions to this general trend were the CB expression of Cacna1h (T‐type Ca2+ channel α‐1H subunit), which is induced by hypoxia in an HIF2α‐dependent manner (del Toro et al. 2003; Carabelli et al. 2007), and Scn7a, an atypical Na+ channel activated by extracellular Na+ that is involved in the regulation of salt intake behaviour (Hiyama et al. 2002). CB overexpression of Scn7a, however, was not confirmed in our PCR validation studies. Interestingly, the Scn9a gene, which encodes the Na+ channel α‐subunit (Nav1.7) that is involved in transmission of pain sensation (Cox et al. 2006), was markedly down‐regulated in the CB relative to the SCG. In agreement with the single‐cell sequencing study of Zhou et al. (2016), the gene encoding the cation‐permeable transient receptor potential channel Trpc5 was among the most highly expressed in CB cells.
Table 3.
Differential expression of ion channel genes in the CB compared to the SCG of adult mice by microarray analysis*
| Gene symbol | Description | Fold change (linear) | pFDR |
|---|---|---|---|
| Potassium channels | |||
| Kcnab1 | potassium voltage‐gated channel, shaker‐related subfamily, beta member 1 | −5.0 | 1.09E‐02 |
| Kcnab2 | potassium voltage‐gated channel, shaker‐related subfamily, beta member 2 | −5.3 | 6.56E‐03 |
| Kcnb2 | potassium voltage gated channel, Shab‐related subfamily, member 2 | −4.0 | 6.99E‐03 |
| Kcnc4 | potassium voltage gated channel, Shaw‐related subfamily, member 4 | −10.3 | 1.66E‐02 |
| Kcnd1 | potassium voltage‐gated channel, Shal‐related family, member 1 | −2.7 | 1.64E‐02 |
| Kcnd2 | potassium voltage‐gated channel, Shal‐related family, member 2 | −11.6 | 6.27E‐03 |
| Kcne4 | potassium voltage‐gated channel, Isk‐related subfamily, gene 4 | 2.3 | 1.88E‐02 |
| Kcnh1 | potassium voltage‐gated channel, subfamily H (eag‐related), member 1 | −6.7 | 4.35E‐03 |
| Kcnh5 | potassium voltage‐gated channel, subfamily H (eag‐related), member 5 | −13.3 | 3.20E‐03 |
| Kcnh6 | potassium voltage‐gated channel, subfamily H (eag‐related), member 6 | −2.5 | 3.48E‐02 |
| Kcnh7 | potassium voltage‐gated channel, subfamily H (eag‐related), member 7 | −4.1 | 1.20E‐02 |
| Kcnip3 | Kv channel interacting protein 3, calsenilin | 5.7 | 1.54E‐03 |
| Kcnip4 | Kv channel interacting protein 4 | −4.8 | 3.64E‐02 |
| Kcnj3 | potassium inwardly rectifying channel, subfamily J, member 3 | −7.0 | 6.51E‐03 |
| Kcnj10 | potassium inwardly rectifying channel, subfamily J, member 10 | −2.9 | 2.30E‐02 |
| Kcnk3(Task1) | potassium channel, subfamily K, member 3 | 2.5 | 2.17E‐02 |
| Kcnk9(Task3) | potassium channel, subfamily K, member 9 | 83.5 | 2.19E‐03 |
| Kcnk10 | potassium channel, subfamily K, member 10 | −3.7 | 5.80E‐03 |
| Kcnk18 | potassium channel, subfamily K, member 18 | −6.3 | 9.87E‐03 |
| Kcnma1 | potassium large conductance calcium‐activated channel, subfamily M, alpha member 1 | −2.1 | 3.64E‐02 |
| Kcnmb1 | potassium large conductance calcium‐activated channel, subfamily M, beta member 1 | 4.6 | 5.68E‐03 |
| Kcnmb2 | potassium large conductance calcium‐activated channel, subfamily M, beta member 2 | 3.1 | 1.45E‐02 |
| Kcnmb4 | potassium large conductance calcium‐activated channel, subfamily M, beta member 4 | −2.3 | 1.96E‐02 |
| Kcnn3 | potassium intermediate/small conductance calcium‐activated channel, subfamily N, member 3 | −3.8 | 1.87E‐02 |
| Kcnq2 | potassium voltage‐gated channel, subfamily Q, member 2 | −3.5 | 1.01E‐02 |
| Kcnq3 | potassium voltage‐gated channel, subfamily Q, member 3 | −20.1 | 1.40E‐02 |
| Kcnq5 | potassium voltage‐gated channel, subfamily Q, member 5 | −5.6 | 1.75E‐02 |
| Kcnt1 | potassium channel, subfamily T, member 1 | −4.7 | 4.81E‐03 |
| Kcnt2 | potassium channel, subfamily T, member 2 | 7.4 | 2.03E‐03 |
| Kctd9 | potassium channel tetramerization domain containing 9 | −2.3 | 9.29E‐03 |
| Calcium channels | |||
| Cacna1a | calcium channel, voltage‐dependent, P/Q type, alpha 1A subunit | −3.0 | 1.40E‐02 |
| Cacna1b | calcium channel, voltage‐dependent, N type, alpha 1B subunit | −5.5 | 4.54E‐02 |
| Cacna1c | calcium channel, voltage‐dependent, L type, alpha 1C subunit | 2.2 | 3.95E‐03 |
| Cacna1h | calcium channel, voltage‐dependent, T type, alpha 1H subunit | 2.7 | 7.70E‐04 |
| Cacna1i | calcium channel, voltage‐dependent, alpha 1I subunit | 2.6 | 9.87E‐03 |
| Cacna2d3 | calcium channel, voltage‐dependent, alpha2/delta subunit 3 | −4.1 | 1.38E‐02 |
| Cacnb1 | calcium channel, voltage‐dependent, beta 1 subunit | −2.2 | 2.01E‐02 |
| Cacnb3 | calcium channel, voltage‐dependent, beta 3 subunit | −6.4 | 1.07E‐02 |
| Cacnb4 | calcium channel, voltage‐dependent, beta 4 subunit | −3.6 | 1.63E‐03 |
| Cacng2 | calcium channel, voltage‐dependent, gamma subunit 2 | −6.1 | 1.29E‐02 |
| Cacng3 | calcium channel, voltage‐dependent, gamma subunit 3 | −12.2 | 1.05E‐02 |
| Sodium channels | |||
| Scn1a | sodium channel, voltage‐gated, type I, alpha | −8.8 | 6.04E‐03 |
| Scn2a1 | sodium channel, voltage‐gated, type II, alpha 1 | −4.0 | 4.71E‐03 |
| Scn2b | sodium channel, voltage‐gated, type II, beta | −11.6 | 5.63E‐03 |
| Scn3a | sodium channel, voltage‐gated, type III, alpha | −4.9 | 1.98E‐02 |
| Scn3b | sodium channel, voltage‐gated, type III, beta | −7.7 | 1.23E‐02 |
| Scn7a | sodium channel, voltage‐gated, type VII, alpha | 7.5 | 4.71E‐03 |
| Scn9a | sodium channel, voltage‐gated, type IX, alpha | −14.9 | 1.38E‐02 |
| Trp channels | |||
| Trpc5 | transient receptor potential cation channel, subfamily C, member 5 | 30.1 | 2.74E‐03 |
| Trpc6 | transient receptor potential cation channel, subfamily C, member 6 | −3.0 | 2.08E‐02 |
| Trpc7 | transient receptor potential cation channel, subfamily C, member 7 | −4.1 | 1.01E‐02 |
| Trpv2 | transient receptor potential cation channel, subfamily V, member 2 | −2.3 | 9.96E‐03 |
| Trpv4 | transient receptor potential cation channel, subfamily V, member 4 | 2.2 | 4.47E‐03 |
*pFDR (P value adjusted with the false discovery rate) < 0.05 and fold change >2 or <−2.
Differential gene expression between the AM and SCG
Up‐ and down‐regulated AM genes can be visualized in Fig. 1 C (light red). The genes with the largest changes in expression level (top 20 up‐regulated and down‐regulated genes) are listed in Table 4. Among the most differentially up‐regulated genes in the AM were some related to steroid metabolism, which suggests contamination from adrenocortical cells that in the mouse can be embedded in the AM region. However, within this group we also found several highly induced genes, such as phenylethanolamine‐N‐methyltransferase (Pnmt), vesicle monoamine transporter 1 (Slc18a1) and chromogranin A (Chga), which are characteristic of AM chromaffin cells. In addition, this unbiased analysis revealed genes relevant to CB acute O2 sensing (mitochondrial subunit Ndufa4l2 and Task3 and Task1 K+ channels; see Table 1), which were also highly expressed in AM cells (Table 4). Moreover, a number of genes that were more highly expressed in the SCG than the CB, such as Htr3a, Htr3b, Napb, Nefl, Tubb2a and Nrip3 (see Table 1), appeared in the list of the top 20 down‐regulated genes in AM cells (Table 4).
Table 4.
Top 20 up‐ and down‐regulated genes in the AM versus the SCG of adult mice by microarray analysis
| Gene symbol | Description | Fold change (linear) | pFDR |
|---|---|---|---|
| Up‐regulated | |||
| Cyp11b1 | cytochrome P450, family 11, subfamily b, polypeptide 1 | 22442 | 3.01E‐07 |
| Srd5a2 | steroid 5 alpha‐reductase 2 | 21977 | 2.07E‐07 |
| Cyp11a1 | cytochrome P450, family 11, subfamily a, polypeptide 1 | 19438 | 4.11E‐07 |
| Hsd3b1 | hydroxy‐delta‐5‐steroid dehydrogenase, 3 beta‐ and steroid delta‐isomerase 1 | 10373 | 2.00E‐06 |
| Star | steroidogenic acute regulatory protein | 9955 | 2.07E‐07 |
| Cyp21a1 | cytochrome P450, family 21, subfamily a, polypeptide 1 | 8609 | 1.00E‐06 |
| Adh1 | alcohol dehydrogenase 1 (class I) | 1340 | 2.00E‐06 |
| Kcnk9(Task3) | potassium channel, subfamily K, member 9 | 1244 | 1.60E‐05 |
| Abcb1b | ATP‐binding cassette, sub‐family B (MDR/TAP), member 1B | 966 | 2.00E‐06 |
| Akr1cl | aldo‐keto reductase family 1, member C‐like | 913 | 2.00E‐06 |
| Akr1b7 | aldo‐keto reductase family 1, member B7 | 850 | 5.10E‐05 |
| Pnmt | phenylethanolamine‐N‐methyltransferase | 500 | 3.70E‐05 |
| Dlk1 | delta‐like 1 homolog (Drosophila) | 438 | 5.90E‐07 |
| Ndufa4l2 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4‐like 2 | 233 | 4.00E‐06 |
| Slc18a1 | solute carrier family 18 (vesicular monoamine), member 1 | 156 | 2.40E‐05 |
| Scarb1 | scavenger receptor class B, member 1 | 148 | 4.00E‐06 |
| Chga | chromogranin A | 116 | 3.90E‐05 |
| Gdf10 | growth differentiation factor 10 | 114 | 8.00E‐05 |
| Tacr2 | tachykinin receptor 2 | 106 | 7.30E‐05 |
| Kcnk3(Task1) | potassium channel, subfamily K, member 3 | 105 | 3.20E‐05 |
| Down‐regulated | |||
| Htr3a | 5‐hydroxytryptamine (serotonin) receptor 3A | −1920 | 3.46E‐07 |
| Tubb3 | tubulin, beta 3 class III | −831 | 8.00E‐06 |
| Htr3b | 5‐hydroxytryptamine (serotonin) receptor 3B | −828 | 2.00E‐06 |
| Sv2c | synaptic vesicle glycoprotein 2c | −627 | 1.00E‐05 |
| Tspan8 | tetraspanin 8 | −624 | 2.00E‐06 |
| Prph | peripherin | −622 | 2.00E‐05 |
| Ddah1 | dimethylarginine dimethylaminohydrolase 1 | −598 | 2.00E‐06 |
| Tubb2b | tubulin, beta 2B class IIB | −378 | 2.00E‐05 |
| Ret | ret proto‐oncogene | −313 | 1.90E‐05 |
| Sncg | synuclein, gamma | −289 | 2.40E‐05 |
| Avil | advillin | −276 | 3.20E‐05 |
| Napb | N‐ethylmaleimide sensitive fusion protein attachment protein beta | −257 | 2.00E‐06 |
| Ngfr | nerve growth factor receptor (TNFR superfamily, member 16) | −244 | 3.00E‐06 |
| Ppp1r1c | protein phosphatase 1, regulatory (inhibitor) subunit 1C | −212 | 1.00E‐06 |
| Nefl | neurofilament, light polypeptide | −195 | 1.60E‐05 |
| Fxyd7 | FXYD domain‐containing ion transport regulator 7 | −188 | 2.00E‐06 |
| Tubb2a | tubulin, beta 2A class IIA | −173 | 2.60E‐05 |
| Nrip3 | nuclear receptor interacting protein 3 | −156 | 2.00E‐06 |
| Areg | amphiregulin | −146 | 1.60E‐05 |
| Rab6b | RAB6B, member RAS oncogene family | −134 | 1.00E‐05 |
pFDR, P value adjusted with the false discovery rate.
When we focused our analysis on genes related to O2 sensing or potentially relevant to acute responsiveness to hypoxia, we found a qualitative profile similar to that previously revealed in the comparison between CB and SCG cells. We observed high expression of Hif2α and Hifβ (Arnt) isoforms as well as down‐regulation of Phd3 in the AM versus SCG. Moreover, in parallel with a slight decrease in the expression of some subunits of mitochondrial ETC complexes, the three atypical mitochondrial subunits up‐regulated in the CB (Ndufa4l2, Cox4i2 and Cox8b) were also enriched in AM cells (Table 5). In addition, our analysis identified a clear induction of Pcx, with down‐regulation of Pdha1 and decreased expression of the neutral amino acid transporter (Slc1a5), which can also transport glutamine (van Geldermalsen et al. 2016). The data further revealed a clear decrease in lactate dehydrogenase (Ldh) and glutaminase (Gls) expression in AM cells, which was not detected in CB tissue.
Table 5.
Hypoxia‐related differential gene expression in the AM compared to the SCG of adult mice by microarray analysis*
| Gene symbol | Description | Fold change (linear) | pFDR |
|---|---|---|---|
| Phd/Hif pathway and targets | |||
| Hif1a | hypoxia inducible factor 1, alpha subunit | 2.1 | 7.46E‐03 |
| Epas1(Hif2a) | endothelial PAS domain protein 1 | 6.6 | 1.31E‐03 |
| Arnt | aryl hydrocarbon receptor nuclear translocator | 2.8 | 7.19E‐04 |
| Arnt2 | aryl hydrocarbon receptor nuclear translocator 2 | 15.5 | 5.10E‐05 |
| Egln3(Phd3) | egl‐9 family hypoxia inducible factor 3 | −4.6 | 3.44E‐03 |
| Vegfa | vascular endothelial growth factor A | 5.1 | 4.64E‐03 |
| Mitochondria | |||
| Ndufa4 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4 | −4.6 | 3.67E‐03 |
| Ndufa4l2 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4‐like 2 | 232.6 | 4.00E‐06 |
| Ndufaf5 | NADH dehydrogenase (ubiquinone) complex I, assembly factor 5 | −2.0 | 1.29E‐03 |
| Ndufb6 | NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 6 | −2.5 | 6.00E‐03 |
| Ndufs4 | NADH dehydrogenase (ubiquinone) Fe‐S protein 4 | −2.1 | 1.05E‐02 |
| Sdhd | succinate dehydrogenase complex, subunit D, integral membrane protein | −2.6 | 3.54E‐03 |
| Cybb | cytochrome b‐245, beta polypeptide | 3.8 | 4.11E‐04 |
| Cox4i2 | cytochrome c oxidase subunit IV isoform 2 | 2.1 | 1.72E‐03 |
| Cox5a | cytochrome c oxidase subunit Va | −4.4 | 2.58E‐04 |
| Cox5b | cytochrome c oxidase subunit Vb | −2.2 | 1.35E‐02 |
| Cox6b1 | cytochrome c oxidase, subunit VIb polypeptide 1 | −3.1 | 4.10E‐03 |
| Cox7a2 | cytochrome c oxidase subunit VIIa 2 | −2.5 | 1.14E‐02 |
| Cox8b | cytochrome c oxidase subunit VIIIb | 6.4 | 5.48E‐04 |
| Coa3 | cytochrome C oxidase assembly factor 3 | −2.8 | 4.69E‐03 |
| Ucp1 | uncoupling protein 1 (mitochondrial, proton carrier) | −3.7 | 3.11E‐02 |
| Ucp2 | uncoupling protein 2 (mitochondrial, proton carrier) | 10.1 | 5.54E‐04 |
| Ucp3 | uncoupling protein 3 (mitochondrial, proton carrier) | 2.1 | 1.14E‐03 |
| Slc25a27 | solute carrier family 25, member 27 | −5.0 | 7.00E‐05 |
| TCA cycle/anaplerosis/biotin‐related | |||
| Pcx | pyruvate carboxylase | 6.1 | 6.21E‐04 |
| Pdha1 | pyruvate dehydrogenase E1 alpha 1 | −3.0 | 4.32E‐04 |
| Pdk4 | pyruvate dehydrogenase kinase, isoenzyme 4 | −2.6 | 1.12E‐02 |
| Slc1a5 | solute carrier family 1 (neutral amino acid transporter), member 5 | −3.4 | 1.60E‐02 |
| Acaca | acetyl‐coenzyme A carboxylase alpha | −3.7 | 8.01E‐04 |
| Ldha | lactate dehydrogenase A | −2.8 | 2.40E‐03 |
| Ldhb | lactate dehydrogenase B | −9.3 | 7.70E‐05 |
| Gls | glutaminase | −4.4 | 1.24E‐03 |
| Tgm2 | transglutaminase 2, C polypeptide | 2.1 | 4.65E‐03 |
| Aco1 | aconitase 1 | 2.2 | 4.74E‐03 |
| Idh3a | isocitrate dehydrogenase 3 (NAD+) alpha | −2.1 | 1.09E‐03 |
| Suclg2 | succinate‐coenzyme A ligase, GDP‐forming, beta subunit | 3.9 | 1.52E‐04 |
| Others | |||
| Ppara | peroxisome proliferator activated receptor alpha | −2.4 | 2.61E‐04 |
| Ppargc1a | peroxisome proliferative activated receptor, gamma, coactivator 1 alpha | −6.0 | 3.96E‐04 |
| Olfr78/Olfr560 | olfactory receptor 78; olfactory receptor 560 | −2.3 | 9.02E‐03 |
*pFDR (P value adjusted with the false discovery rate) < 0.05 and fold change >2 or <−2.
The general pattern of ion channel gene expression in the AM versus SCG was, despite some notable exceptions, qualitatively similar to that observed in the comparison between the CB and SCG. The K+ channels with more pronounced differential expression were Task3 (Kcnk9) and Task1 (Kcnk3). As in the CB, maxi‐K+ channel subunits (Kcnma1 and Kcnmb2) and calsenilin (Kcnip3) were up‐regulated in AM cells (Table 6). A member of the small/intermediate conductance Ca2+‐activated K+ channel family (Kcnn2), which could mediate part of the O2‐sensitive K+ current (Keating et al. 2001), was also slightly overexpressed in AM cells. Interestingly, the level of expression of the Kcnj11 subunit, which encodes a KATP channel (Kir 6.2) highly relevant to the developmental decrease of hypoxia sensitivity in chromaffin cells (Buttigieg et al. 2009; Salman et al. 2014), was practically the same in AM in comparison with CB or SCG cells (1.09‐fold change AM vs. SCG and 1.06‐fold change CB vs. SCG). As in the CB, we also observed a generalized decrease in the expression of voltage‐gated K+ channel subunits in the AM, particularly in the case of the genes encoding eag‐related (Kcnh5 and Kcnh7) and G protein‐regulated inward rectifier (Kcnj3) channels. K+ channels mediating the M‐current (Kcnq3 and Kcnq5) were down‐regulated in AM cells. Similar to the CB, several subunits of voltage‐gated Ca2+ and Na+ channels were down‐regulated in the AM in comparison with SCG cells. The largest difference was seen in the Scn9a gene, which encodes the Na+ channel α‐subunit (Nav1.7) that mediates pain sensation in the peripheral nervous system (Cox et al. 2006). In contrast, the Cacna1d and Cacna1h genes which encode, respectively, the α‐subunit of low voltage‐activated L‐type Ca2+ channels (Cav1.3) (Vandael et al. 2015) and the O2‐sensitive component of the T‐type current (del Toro et al. 2003; Carabelli et al. 2007) were overexpressed in chromaffin cells. As in the CB, the Trpc5 gene was also highly induced in AM cells.
Table 6.
Differential expression of ion channel genes in the AM compared to the SCG of adult mice by microarray analysis*
| Gene symbol | Description | Fold change (linear) | pFDR |
|---|---|---|---|
| Potassium channels | |||
| Kcna1 | potassium voltage‐gated channel, shaker‐related subfamily, member 1 | −28.4 | 4.27E‐04 |
| Kcna2 | potassium voltage‐gated channel, shaker‐related subfamily, member 2 | −12.3 | 1.60E‐03 |
| Kcna6 | potassium voltage‐gated channel, shaker‐related, subfamily, member 6 | −13.9 | 1.94E‐04 |
| Kcnab1 | potassium voltage‐gated channel, shaker‐related subfamily, beta member 1 | −10.4 | 1.57E‐04 |
| Kcnab2 | potassium voltage‐gated channel, shaker‐related subfamily, beta member 2 | −8.2 | 5.00E‐05 |
| Kcnb2 | potassium voltage gated channel, Shab‐related subfamily, member 2 | −5.0 | 2.81E‐04 |
| Kcnc4 | potassium voltage gated channel, Shaw‐related subfamily, member 4 | −4.5 | 3.40E‐04 |
| Kcnd1 | potassium voltage‐gated channel, Shal‐related family, member 1 | −3.5 | 8.80E‐04 |
| Kcnd2 | potassium voltage‐gated channel, Shal‐related family, member 2 | −21.8 | 1.95E‐04 |
| Kcng4 | potassium voltage‐gated channel, subfamily G, member 4 | −2.6 | 1.16E‐02 |
| Kcnh1 | potassium voltage‐gated channel, subfamily H (eag‐related), member 1 | −11.8 | 4.60E‐05 |
| Kcnh5 | potassium voltage‐gated channel, subfamily H (eag‐related), member 5 | −24.4 | 1.60E‐05 |
| Kcnh7 | potassium voltage‐gated channel, subfamily H (eag‐related), member 7 | −22.7 | 2.40E‐05 |
| Kcnh8 | potassium voltage‐gated channel, subfamily H (eag‐related), member 8 | −6.8 | 7.70E‐05 |
| Kcnip3 | Kv channel interacting protein 3, calsenilin | 16.2 | 5.00E‐05 |
| Kcnip4 | Kv channel interacting protein 4 | −9.2 | 6.94E‐04 |
| Kcnj3 | potassium inwardly rectifying channel, subfamily J, member 3 | −56.3 | 4.50E‐05 |
| Kcnj6 | potassium inwardly rectifying channel, subfamily J, member 6 | 2.2 | 8.71E‐04 |
| Kcnj8 | potassium inwardly rectifying channel, subfamily J, member 8 | 2.3 | 8.72E‐03 |
| Kcnj10 | potassium inwardly rectifying channel, subfamily J, member 10 | −5.9 | 7.14E‐04 |
| Kcnj13 | potassium inwardly rectifying channel, subfamily J, member 13 | −9.4 | 2.20E‐05 |
| Kcnj16 | potassium inwardly rectifying channel, subfamily J, member 16 | −2.9 | 7.69E‐04 |
| Kcnk2 | potassium channel, subfamily K, member 2 | 12.1 | 1.41E‐04 |
| Kcnk3(Task1) | potassium channel, subfamily K, member 3 | 105.4 | 3.20E‐05 |
| Kcnk9(Task3) | potassium channel, subfamily K, member 9 | 1244.1 | 1.60E‐05 |
| Kcnk10 | potassium channel, subfamily K, member 10 | −6.1 | 9.80E‐05 |
| Kcnk18 | potassium channel, subfamily K, member 18 | −12.2 | 5.30E‐04 |
| Kcnma1 | potassium large conductance calcium‐activated channel, subfamily M, alpha member 1 | 3.5 | 8.40E‐05 |
| Kcnmb2 | potassium large conductance calcium‐activated channel, subfamily M, beta member 2 | 6.0 | 5.09E‐04 |
| vKcnmb4 | potassium large conductance calcium‐activated channel, subfamily M, beta member 4 | −3.6 | 9.57E‐04 |
| Kcnn2 | potassium intermediate/small conductance calcium‐activated channel, subfamily N, member 2 | 2.0 | 2.29E‐03 |
| Kcnn3 | potassium intermediate/small conductance calcium‐activated channel, subfamily N, member 3 | −6.1 | 1.19E‐04 |
| Kcnn4 | potassium intermediate/small conductance calcium‐activated channel, subfamily N, member 4 | −3.6 | 1.74E‐04 |
| Kcnq2 | potassium voltage‐gated channel, subfamily Q, member 2 | −3.2 | 4.39E‐04 |
| Kcnq3 | potassium voltage‐gated channel, subfamily Q, member 3 | −16.4 | 3.12E‐04 |
| Kcnq5 | potassium voltage‐gated channel, subfamily Q, member 5 | −19.8 | 8.40E‐05 |
| Kcnt1 | potassium channel, subfamily T, member 1 | −7.7 | 5.00E‐05 |
| Kctd9 | potassium channel tetramerization domain containing 9 | −2.6 | 5.76E‐04 |
| Kctd16 | potassium channel tetramerization domain containing 16 | −2.7 | 6.83E‐04 |
| Calcium channels | |||
| Cacna1a | calcium channel, voltage‐dependent, P/Q type, alpha 1A subunit | −2.6 | 5.76E‐04 |
| Cacna1b | calcium channel, voltage‐dependent, N type, alpha 1B subunit | −6.2 | 3.67E‐04 |
| Cacna1c | calcium channel, voltage‐dependent, L type, alpha 1C subunit | 5.5 | 3.63E‐04 |
| Cacna1d | calcium channel, voltage‐dependent, L type, alpha 1D subunit | 14.3 | 7.00E‐05 |
| Cacna1h | calcium channel, voltage‐dependent, T type, alpha 1H subunit | 6.4 | 8.67E‐04 |
| Cacna2d1 | calcium channel, voltage‐dependent, alpha2/delta subunit 1 | 3.2 | 3.68E‐03 |
| Cacna2d3 | calcium channel, voltage‐dependent, alpha2/delta subunit 3 | −18.5 | 8.50E‐05 |
| Cacnb1 | calcium channel, voltage‐dependent, beta 1 subunit | −2.7 | 2.09E‐03 |
| Cacnb2 | calcium channel, voltage‐dependent, beta 2 subunit | 4.6 | 1.08E‐04 |
| Cacnb3 | calcium channel, voltage‐dependent, beta 3 subunit | −5.8 | 2.90E‐04 |
| Cacnb4 | calcium channel, voltage‐dependent, beta 4 subunit | −6.2 | 1.90E‐05 |
| Cacng2 | calcium channel, voltage‐dependent, gamma subunit 2 | −10.0 | 6.20E‐05 |
| Cacng3 | calcium channel, voltage‐dependent, gamma subunit 3 | −4.9 | 7.60E‐04 |
| Cacng4 | calcium channel, voltage‐dependent, gamma subunit 4 | −2.4 | 4.31E‐04 |
| Sodium channels | |||
| Scn1a | sodium channel, voltage‐gated, type I, alpha | −13.7 | 4.80E‐05 |
| Scn2a1 | sodium channel, voltage‐gated, type II, alpha 1 | −6.0 | 1.96E‐04 |
| Scn2b | sodium channel, voltage‐gated, type II, beta | −13.6 | 9.90E‐05 |
| Scn3a | sodium channel, voltage‐gated, type III, alpha | −2.3 | 4.34E‐03 |
| Scn3b | sodium channel, voltage‐gated, type III, beta | −6.8 | 7.21E‐04 |
| Scn7a | sodium channel, voltage‐gated, type VII, alpha | −6.2 | 4.05E‐04 |
| Scn9a | sodium channel, voltage‐gated, type IX, alpha | −42.2 | 1.30E‐05 |
| Scn11a | sodium channel, voltage‐gated, type XI, alpha | −2.1 | 5.84E‐03 |
| Trp channels | |||
| Trpc1 | transient receptor potential cation channel, subfamily C, member 1 | −2.4 | 4.09E‐03 |
| Trpc5 | transient receptor potential cation channel, subfamily C, member 5 | 32.5 | 4.80E‐05 |
| Trpc6 | transient receptor potential cation channel, subfamily C, member 6 | −3.0 | 2.59E‐03 |
| Trpc7 | transient receptor potential cation channel, subfamily C, member 7 | −3.2 | 1.82E‐03 |
| Trpm3 | transient receptor potential cation channel, subfamily M, member 3 | −2.9 | 4.37E‐03 |
| Trpv2 | transient receptor potential cation channel, subfamily V, member 2 | −3.3 | 3.72E‐04 |
*pFDR (P value adjusted with the false discovery rate) < 0.05 and fold change >2 or <−2.
Expression of selected genes in tissues and sorted TH‐positive cells
To validate our microarray results, real‐time quantitative PCR was performed using a new set of biologically independent samples. As shown in Table 7, similar results were obtained for most of the genes tested in the two comparative analyses (CB vs. SCG and AM vs. SCG), thereby supporting the results of the microarray study. A limitation of using whole tissues is the contamination from non‐neuronal cells that occurs, particularly in the case of the CB, which, in addition to TH+ glomus cells, contains numerous capillaries and other cell types. To circumvent this issue, we used TH‐GFP mice in which the expression of GFP was under the control of the TH promoter. This allowed us to isolate TH+ (GFP+) cells from each tissue (Fig. 2 A). We confirmed that the sorted cells were TH+ by immunofluorescence staining (Fig. 2 B). The percentage of TH+/GFP+ cells counted in random samples was 87% (377/434), 95% (124/131) and 83% (80/97) from the CB, SCG and AM, respectively. Genes analysed by real‐time PCR using the sorted TH+ cells showed similar relative expression to that seen in the analysis of whole tissues (Table 7). The up‐regulation of Hif2α and the ETC subunits Ndufa4l2, Cox4i2 and Cox8b, and the down‐regulation of Phd3 in the CB and, less potently, in the AM versus SCG were validated with high significance in the PCR analyses on isolated TH+ cells. Overexpression of Pcx and down‐regulation of Acly, Slc7a5 and Idh1 were also confirmed in the comparison between TH+ CB and SCG cells. With the exception of Pcx and Cox4i2, this metabolic gene profile was also clearly seen in AM cells (Table 7). All the ion channel‐related genes overexpressed in the CB and AM were also validated with high significance by quantitative PCR of sorted TH+ cells, with the exception of Trpc5 in the AM. In general, high variability among the replicates of sorted TH+ cells from each tissue was observed compared to that of whole tissue analysis, which could explain the lack of statistical significance obtained using TH+ cells. Among other genes tested, uncoupling protein 2 (Ucp2) was up‐regulated in TH+ CB and AM cells, which also validates the microarray data. In addition, the quantitative PCR analysis of whole tissue or sorted TH+ cells showed that the Olfr78 gene, which encodes an atypical olfactory receptor expressed in several tissues outside the nasal epithelium, and particularly in the ganglia of the autonomic nervous system (Weber et al. 2002), was up‐regulated in CB glomus cells relative to sympathetic neurons, but markedly down‐regulated in AM chromaffin cells (Table 7).
Table 7.
Differential gene expression in the CB, AM and SCG analysed by real‐time quantitative PCR in adult mice
| CB vs. SCG | AM vs. SCG | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A. Microarray | B. Whole tissue | C. TH+ cells | A. Microarray | B. Whole tissue | C. TH+ cells | |||||||
| Gene symbol | Fold change | pFDR | Fold change† | P * | Fold change† | P * | Fold change | pFDR | Fold change† | P * | Fold change# | P * |
| Phd/Hif pathway | ||||||||||||
| Epas1(Hif2a) | 102.0 | 0.00249 | 190.2 ± 38.5 | * | 914.2 ± 270.3 | * | 6.6 | 0.00131 | 6.9 ± 0.7 | * | 16.4 ± 5.2 | * |
| Hif1a | 1.8 | 0.05001 | 6.4 ± 0.6 | * | 1.9 ± 0.5 | 2.1 | 0.00746 | 2.9 ± 0.4 | * | 2.6 ± 0.7 | * | |
| Egln3(Phd3) | −4.1 | 0.01886 | −2.2 ± 0.5 | * | CB NE | −4.6 | 0.00344 | −2.9 ± 0.2 | * | −21.1 ± 16.7 | * | |
| Mitochondrial ETC subunits | ||||||||||||
| Ndufa4l2 | 720.9 | 0.00039 | 1003.8 ± 264.0 | * | 4015.5 ± 849.8 | * | 232.6 | 0.00000 | 257.6 ± 41.1 | * | 512.8 ± 184.5 | * |
| Ndufa4 | −1.9 | 0.21813 | 1.0 ± 0.1 | 1.5 ± 0.2 | −4.6 | 0.00367 | 1.0 ± 0.1 | 1.2 ± 0.1 | ||||
| Cox4i2 | 10.1 | 0.00603 | 360.0 ± 69.1 | * | 1334.4 ± 226.9 | * | 2.1 | 0.00172 | 25.4 ± 5.1 | * | 15.2 ± 12.4 | |
| Cox4i1 | −1.5 | 0.09280 | 1.5 ± 0.2 | * | 1.1 ± 0.1 | −1.7 | 0.01749 | 1.3 ± 0.1 | −1.5 ± 0.2 | |||
| Cox8b | 11.8 | 0.01295 | 512.9 ± 143.9 | * | 4699.9 ± 1024.0 | * | 6.4 | 0.00055 | 255.3 ± 29.4 | * | 8672.1 ± 2019.0 | * |
| Metabolic enzymes and transporters | ||||||||||||
| Pcx | 4.9 | 0.01312 | 20.7 ± 3.8 | * | 23.5 ± 2.8 | * | 6.1 | 0.00062 | 8.7 ± 0.8 | * | −1.3 ± 0.5 | |
| Pdha1 | −2.5 | 0.01900 | −1.4 ± 0.2 | −2.1 ± 1.2 | −3.0 | 0.00043 | −1.2 ± 0.2 | −4.7 ± 1.4 | * | |||
| Slc7a5 | −3.6 | 0.02241 | −2.4 ± 0.4 | * | −3.5 ± 0.6 | * | 1.7 | 0.08917 | 1.9 ± 0.3 | * | −3.9 ± 1.9 | * |
| Idh1 | −3.5 | 0.01335 | −1.8 ± 0.2 | * | −6.7 ± 2.1 | * | 1.6 | 0.07287 | 2.6 ± 0.4 | * | −3.3 ± 0.7 | * |
| Idh3a | −3.1 | 0.00550 | −1.2 ± 0.1 | * | 1.9 ± 0.4 | * | −2.1 | 0.00109 | −1.3 ± 0.1 | * | −1.3 ± 0.4 | |
| Idh3b | −4.6 | 0.03826 | −1.6 ± 0.2 | * | 1.2 ± 0.2 | −1.5 | 0.00257 | −1.2 ± 0.2 | −1.4 ± 0.4 | |||
| Acly | −4.4 | 0.07742 | −1.5 ± 0.3 | −4.6 ± 0.7 | * | 1.5 | 0.19180 | 1.3 ± 0.1 | −4.8 ± 1.1 | * | ||
| Acacb | 2.7 | 0.00957 | 18.9 ± 3.7 | * | ‡ | 1.2 | 0.37025 | 3.8 ± 0.9 | * | 6.3 ± 3.1 | * | |
| Ion channels | ||||||||||||
| Kcnk3(Task1) | 2.5 | 0.02174 | 13.8 ± 2.6 | * | 74.8 ± 12.0 | * | 105.4 | 0.00003 | 23.1 ± 2.4 | * | 19.7 ± 5.4 | * |
| Kcnk9(Task3) | 83.5 | 0.00219 | 2371.4 ± 522.9 | * | 18431.8 ± 5917.5 | * | 1244.1 | 0.00002 | 6527.6 ± 1268.1 | * | 6647.1 ± 1736.0 | * |
| Kcnip3 | 5.7 | 0.00154 | 32.3 ± 3.7 | * | 75.7 ± 10.8 | * | 16.2 | 0.00005 | 18.6 ± 1.3 | * | 30.1 ± 4.5 | * |
| Trpc5 | 30.1 | 0.00274 | 4258.8 ± 628.5 | * | 132974.6 ± 48426.0 | * | 32.5 | 0.00005 | 1014.1 ± 77.6 | * | 1100.1 ± 799.9 | |
| Cacna1h | 2.7 | 0.00077 | 41.6 ± 5.5 | * | 60.9 ± 11.3 | * | 6.4 | 0.00087 | 21.8 ± 2.8 | * | 8.7 ± 3.5 | * |
| Others | ||||||||||||
| Ucp2 | 2.4 | 0.06361 | 7.9 ± 1.3 | * | 10.4 ± 1.5 | * | 10.1 | 0.00055 | 13.1 ± 2.5 | * | 22.1 ± 2.9 | * |
| Olfr78 § | 1.3 | 0.55079 | 8.5 ± 1.6 | * | 5.3 ± 1.0 | * | −2.3 | 0.00902 | −22.6 ± 4.0 | * | −141.4 ± 11.6 | * |
A. For comparison, the results from microarray analysis are also listed. B. Real‐time quantitative PCR analysis using RNA isolated from whole tissue. C. Real‐time PCR analysis using RNA isolated from TH+ cells from each tissue.
* P < 0.05 compared to the SCG.
†Mean ± SEM, n = 3–5 per group.
‡Not available (highly variable, no conclusion).
§Microarray detects Olfr78 and Olfr560, whereas real‐time PCR detects Olfr78 only; CB NE, no expression was detected from CB TH+ cells.
Abbreviations: Epas1(Hif2α), endothelial PAS domain protein 1; Hif1α, hypoxia inducible factor 1, alpha subunit; Egln3(Phd3), egl‐9 family hypoxia inducible factor 3; Ndufa4l2, NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4‐like 2; Ndufa4, NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4; Cox4i2, cytochrome c oxidase subunit IV isoform 2; Cox4i1, cytochrome c oxidase subunit IV isoform 1; Cox8b, cytochrome c oxidase subunit VIIIb; Pcx, pyruvate carboxylase; Pdha1, pyruvate dehydrogenase E1 alpha 1; Slc7a5, solute carrier family 7 (cationic amino acid transporter, y+ system), member 5; Idh1, isocitrate dehydrogenase 1 (NADP+), soluble; Idh3a, isocitrate dehydrogenase 3 (NAD+) alpha; Idh3b, isocitrate dehydrogenase 3 (NAD+) beta; Acly, ATP citrate lyase; Acacb, acetyl‐coenzyme A carboxylase beta; Kcnk3(Task1), potassium channel, subfamily K, member 3; Kcnk9(Task3), potassium channel, subfamily K, member 9; Kcnip3, Kv channel interacting protein 3, calsenilin; Trpc5, transient receptor potential cation channel, subfamily C, member 5; Cacna1h, calcium channel, voltage‐dependent, T type, alpha 1H subunit; Ucp2, uncoupling protein 2 (mitochondrial, proton carrier); Olfr78, olfactory receptor 78.
Figure 2. Sorting of tyrosine hydroxylase (TH) positive cells from adult wild‐type and TH‐GFP mice by flow cytometry.
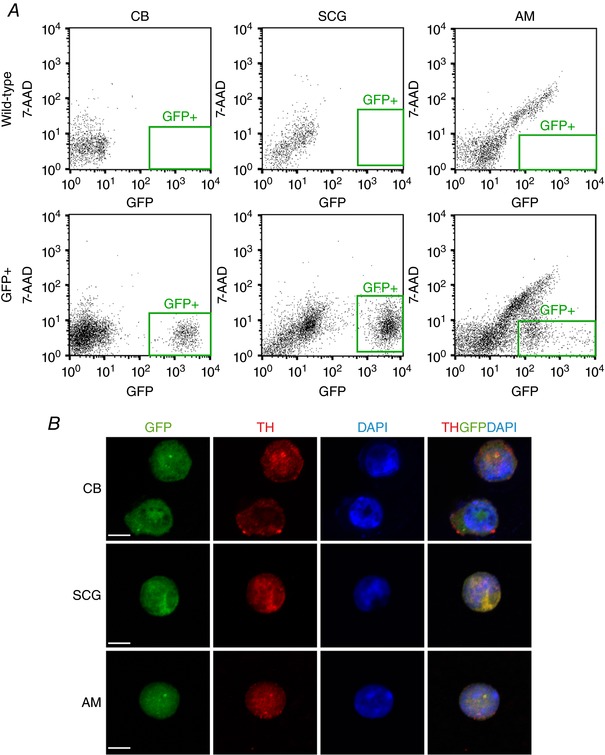
A, isolation of green fluorescent protein positive (GFP+) cells from the carotid body (CB), superior cervical ganglion (SCG) and adrenal medulla (AM) of TH‐GFP mice by flow cytometry. 7‐AAD, 7‐aminoactinomycin D. Plots from wild‐type animals are shown as control. B, immunofluorescent staining demonstrating that the GFP+ cells express TH. Scale bar, 5 μm.
To investigate whether the differential gene expression observed in our microarray analysis was reflected at the protein level, we performed immunofluorescence experiments on carotid bifurcations and adrenal glands. We focused this analysis on the mitochondrial ETC subunits for which antibodies are available (Ndufa4l2 and Cox4i2) and Pcx. As shown in Fig. 3 A, the immunoreactive signal against Ndufa4l2 was higher in CB glomus cells than SCG neurons, as demonstrated by Ndufa4l2/TH co‐localization. A similar result was observed when comparing the AM with the adrenal cortex (Fig. 3 B). Strong immunostaining against the Cox4i2 subunit was observed in both CB and AM TH+ cells (Fig. 4). Pcx was more highly expressed in the CB than the SCG (Fig. 5). However, the immunostaining signal against this protein in the AM was indistinguishable from that in the adrenal cortex (data not shown). This is in agreement with our quantitative PCR data, in which no up‐regulation of Pcx mRNA was observed in sorted AM chromaffin cells (Table 7).
Figure 3. Immunohistochemical analysis of NADH dehydrogenase 1 alpha subcomplex 4‐like 2 (Ndufa4l2) expression in adult mice.
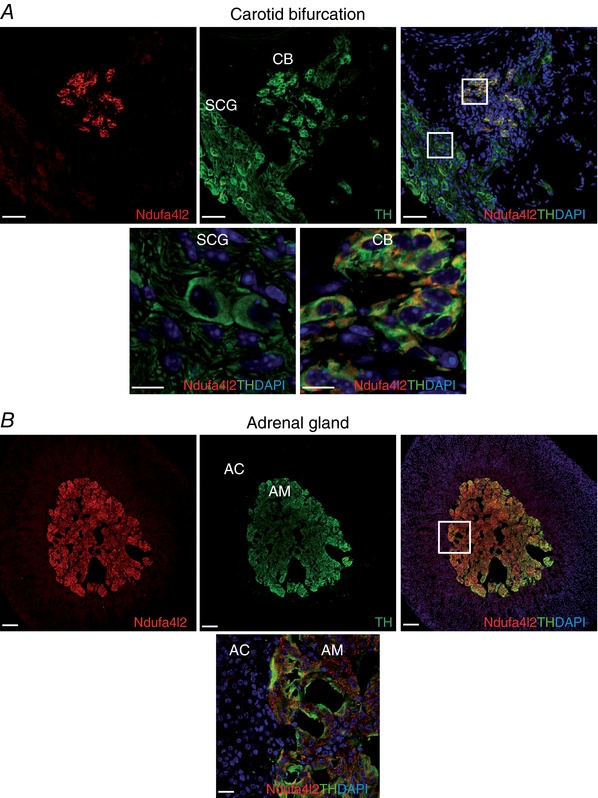
A, upper panels: representative section of the carotid bifurcation demonstrating high Ndufa4l2 immunoreactivity in the carotid body (CB) compared to superior cervical ganglion (SCG) and the co‐localization of Ndufa4l2 (red) and tyrosine hydroxylase (TH, green). Scale bar, 50 μm. Lower panels: magnification of SCG and CB regions indicated in the upper right panel. Scale bar, 10 μm. B, upper panels: representative section of the adrenal gland demonstrating high Ndufa4l2 immunostaining in the adrenal medulla (AM) compared to adrenal cortex (AC) and the co‐localization of Ndufa4l2 (red) and TH (green). Scale bar, 100 μm. Lower panel: magnification of AC and AM regions indicated in the upper right panel. Scale bar, 20 μm.
Figure 4. Immunohistochemical analysis of cytochrome c oxidase subunit IV isoform 2 (Cox4i2) expression in adult mice.
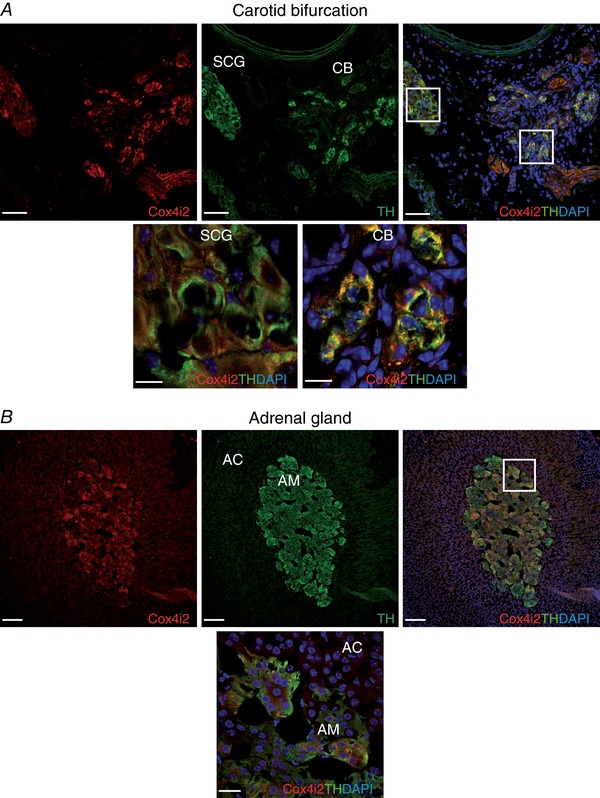
A, upper panels: representative section of the carotid bifurcation demonstrating high Cox4i2 immunoreactivity in the carotid body (CB) compared to superior cervical ganglion (SCG) and the co‐localization of Cox4i2 (red) and tyrosine hydroxylase (TH, green). Scale bar, 50 μm. Lower panels: magnification of SCG and CB regions indicated in the upper right panel. Scale bar, 10 μm. B, upper panels: representative section of the adrenal gland demonstrating high Cox4i2 immunostaining in the adrenal medulla (AM) compared to adrenal cortex (AC) and the co‐localization of Cox4i2 (red) and TH (green). Scale bar, 100 μm. Lower panel: magnification of AC and AM regions indicated in the upper right panel. Scale bar, 20 μm.
Figure 5. Immunohistochemical analysis of pyruvate carboxylase (Pcx) expression in adult mice.
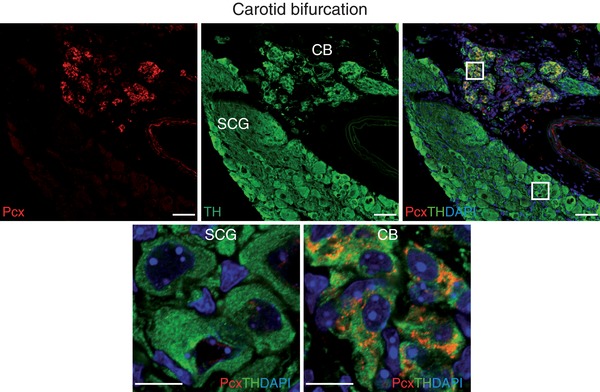
Upper panels: representative section of the carotid bifurcation demonstrating high Pcx immunoreactivity in the carotid body (CB) compared to superior cervical ganglion (SCG) and the co‐localization of Pcx (red) and tyrosine hydroxylase (TH, green). Scale bar, 50 μm. Lower panels. Magnification of SCG and CB regions indicated in the upper right panel. Scale bar, 10 μm.
Impairment of acute O2 sensing by genetic and pharmacological inhibition of succinate dehydrogenase
In a previous study, we suggested that CB glomus cells contain high levels of QH2 due to succinate‐dependent metabolism, and that a further increase in the QH2 pool during hypoxia leads to the generation of ROS and reduced pyridine nucleotides to signal membrane ion channels (Fernández‐Agüera et al. 2015). This proposal, which is compatible with the up‐regulation of Pcx in glomus cells described here (see Discussion), predicts that inhibition of succinate dehydrogenase should decrease acute responsiveness to low . We have generated an MCII‐null mouse (TH‐SDHD) carrying a floxed Sdhd allele that encodes the membrane anchoring subunit D of succinate dehydrogenase. This allele was deleted in CB glomus cells and other catecholaminergic cells by the transgenic expression of a Cre recombinase under the control of the TH promoter (Díaz‐Castro et al. 2012). The analysis of responsiveness to hypoxia in the glomus cells of TH‐SDHD mice is not straightforward because these cells enter a degenerative process that leads to their death (Díaz‐Castro et al. 2012; Platero‐Luengo et al. 2014). However, we were able to demonstrate that responsiveness to hypoxia disappears in Sdhd‐deficient glomus cells before they stop responding to high extracellular K+ or hypoglycaemia (Fig. 6 A and B). These data, which suggest that succinate dehydrogenase activity is required for normal acute O2 sensing, were confirmed by experiments using glomus cells incubated overnight with dimethyl malonate (DMM), a membrane‐permeant competitive inhibitor of succinate dehydrogenase (see Gutman, 1978). DMM‐treated (for ∼12 h) glomus cells showed a drastic decrease in hypoxia‐induced catecholamine secretion, which was partially recovered during washout of malonate from the extracellular solution. Incubation with DMM did not significantly affect the secretory response to high extracellular K+ (Fig. 6 C–E).
Figure 6. Impairment of acute O2 sensing by genetic and pharmacological inhibition of succinate dehydrogenase.
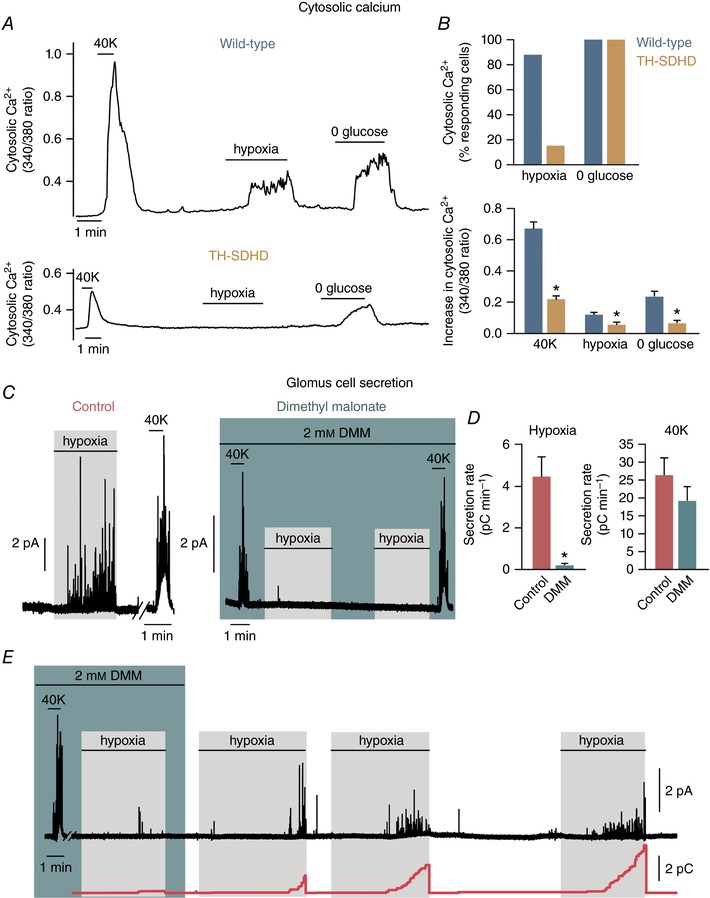
A, representative recordings of the ratiometric increase in cytosolic [Ca2+] elicited in Fura‐2‐loaded dispersed glomus cells from wild‐type and TH‐SDHD animals in response to hypoxia, 0 glucose and 40 mm K+. B, top: percentage of the number of cells that responded to a specific stimulus compared to the number of cells that responded to high potassium (Hypoxia: wild‐type, n = 69 from 7 mice; TH‐SDHD, n = 32 from 9 animals. 0 glucose: wild‐type, n = 19 from 3 mice; TH‐SDHD, n = 14 from 6 animals). Bottom: quantification of the increase in cytosolic [Ca2+] in glomus cells from wild‐type and TH‐SDHD animals that responded to the specific stimulus (40 mm K+: wild‐type, n = 69 from 7 mice; TH‐SDHD, n = 32 from 9 mice. Hypoxia: wild‐type, n = 61 from 7 mice; TH‐SDHD, n = 5 from 2 mice. 0 glucose: wild‐type, n = 19 from 3 mice; TH‐SDHD, n = 14 from 6 mice). C, representative amperometric recordings of responses to hypoxia and 40 mm K+ of glomus cells in wild‐type mouse CB slices incubated overnight with or without 2 mm dimethyl malonate (DMM). D, quantification of the secretion rate elicited by hypoxia (left) and 40 mm K+ (right) in control and DMM‐treated CB glomus cells from wild‐type animals (Hypoxia: control, n = 8 from 6 mice; DMM, n = 7 from 5 animals. 40 mm K+: control, n = 7 from 5 mice; DMM, n = 6 from 4 animals.) E, representative recording demonstrating the recovery of hypoxia‐induced secretory activity of glomus cells in wild‐type mouse slices after the washout of DMM. Cumulative secretion rate (red line) is represented at the bottom. * P < 0.05 compared to wild‐type or control.
Discussion
Differential gene expression profiles in the CB or AM versus SCG
In this study, we analysed the gene expression pattern in three sympathoadrenal tissues (CB, AM and SCG) with variable sensitivity to hypoxia to identify genes that are relevant to acute O2 sensing in peripheral chemoreceptors. Our results reveal several genes potentially related to acute O2 sensing that are modulated (induced or repressed) in the CB (and, to a lesser extent, in the AM) in comparison with the SCG. We then focused on genes that encode proteins related to the Phd/Hif pathway, mitochondrial ETC, metabolic enzymes and transporters, and ion channels. Some of the genes described here were also investigated in previous microarray analyses of CB tissue (Ganfornina et al. 2005; Balbir et al. 2007; Fagerlund et al. 2010; Mkrtchian et al. 2012) and particularly in a recent work that used single CB cell RNA sequencing (Zhou et al. 2016). However, our experimental approach, based on the comparison of adult tissues of the same developmental origin but variable sensitivity to hypoxia, allowed us to focus on genes with a similar qualitative expression pattern in O2‐sensitive tissues (CB and AM) in comparison with O2‐insensitive SCG cells.
Phd3 and Hif2α
Hif2α is one of the most highly expressed genes in CB cells versus SCG neurons, whereas Hif1α is only slightly up‐regulated. This finding confirms previous reports of the constitutive expression of Hif2α in adult mouse CB tissue (Tian et al. 1998) and the large number of Hif2α transcripts in neonatal mouse CB cells (Zhou et al. 2016). In contrast, we found a robust and previously undetected down‐regulation of Phd3 mRNA in CB versus SCG cells. Indeed, Phd3 mRNA expression was negligible in sorted glomus cells. Phd3 has relatively higher influence on Hif2α hydroxylation leading to its degradation (Appelhoff et al. 2004). Therefore, in addition to other functions (see below), down‐regulation of Phd3 probably contributes to the maintenance of a high Hif2α protein level in normoxic CB glomus cells. Hif2α mRNA overexpression and Phd3 down‐regulation were also clearly observed in AM cells. Hif2α has an important role in CB homeostasis (Peng et al. 2011), and is necessary for a normal HVR and CB growth in response to chronic hypoxia (Hodson et al. 2016). On the other hand, the Phd3/Hif2α pathway is involved in the regulation of sympathoadrenal development (Bishop et al. 2008; Macías et al. 2014). The CB and AM are slightly hypertrophied in Phd3‐null animals, and glomus cells exhibit a robust responsiveness to hypoxia (Bishop et al. 2008; Macias et al. 2014; our own unpublished observations). Therefore, our findings suggest that the pattern ‘Phd3 down‐regulation/Hif2α overexpression’ is essential for the metabolic specialization that confers acute O2 sensitivity upon CB and AM cells (see below).
Atypical mitochondrial subunits
We found three atypical mitochondrial ETC subunits, Ndufa4l2, Cox4i2 and Cox8b, the mRNA of which was highly up‐regulated in CB and AM versus SCG cells. The protein levels of Ndufa4l2 and Cox4i2 were also higher in CB and AM chemoreceptor cells in comparison with SCG neurons. High levels of Ndufa4l2 and Cox4i2 transcripts have also been observed in neonatal CB glomus cells (Zhou et al. 2016). It has previously been reported that Ndufa4l2 and Cox4i2 mRNAs are strongly up‐regulated by hypoxia (Fukuda et al. 2007; Brown et al. 2010; Tello et al. 2011; Aras et al. 2013), but, to our knowledge, the regulation of Cox8b expression by hypoxia has not been documented. However, the mouse Cox8b gene contains two putative Hif binding sites in the promoter, one of them near the transcription initiation site (data not shown). Therefore, it is possible that the three atypical mitochondrial subunits are overexpressed in glomus and chromaffin cells due to the constitutively high Hif2α expression in these cells. However, the function of these atypical ETC subunits is unclear and their role in CB or AM cell physiology remains to be determined. Ndufa4l2 is a paralogue of the more ubiquitous subunit Ndufa4, which was thought to be a component of MCI (Carroll et al. 2006), although recent studies have suggested that it is associated with MCIV (Balsa et al. 2012; Kadenbach & Hüttemann, 2015). Ndufa4l2 expression seems to decrease O2 consumption and ROS production by mitochondria (Tello et al. 2011). On the other hand, Cox4i2, the expression of which is restricted to lung and other highly oxygenated tissues, is an isoform of the broadly expressed subunit Cox4i1. In some mammalian cells exposed to hypoxia, Cox4i2 is induced to replace Cox4i1 thereby increasing the efficiency with which MCIV facilitates the transfer of electrons to O2 and decreasing oxidative stress (Fukuda et al. 2007). In lung, Cox4i2 knockout reduces MCIV activity to 50%, despite the presence of the Cox4i1 isoform (Hüttemann et al. 2012). There are three tissue‐specific Cox8 isoforms (a, b and c), although to date no specific function has been identified for any of these (Kadenbach and Hüttemann, 2015). In several studies, however, Cox8b has been associated with thermogenic differentiation (‘browning’) of the white adipose tissue in rodents (Fisher et al. 2012; García et al. 2016). How the combination of the three subunits (Ndufa4l2, Cox4i2 and Cox8b) could render glomus cell mitochondria highly O2‐sensitive is unknown. Cox4i2 and Cox8b are integral proteins with parallel single transmembrane α‐helices that run in close proximity (see Tsukihara et al. 1996). One could therefore speculate that these subunits interact to regulate O2 diffusion through the inner membrane, in order to reach heme a3/CuB, which is buried inside MCIV (Michel et al. 1998). Low O2 accessibility to the catalytic site would make the rate of cytochrome a3/CuB oxidation highly sensitive to decreases in . Together, the data discussed here suggest the existence of mitochondrial specializations that are characteristic of acute O2‐sensing cells. Several decades ago a low affinity cytochrome C oxidase was proposed to exist in the CB, although located in type II rather than O2‐sensitive glomus (type I) cells (Mills and Jöbsis, 1970). More recently, the involvement of mitochondria in acute O2 sensing has been suggested by numerous studies of CB and AM cells (Duchen and Biscoe, 1992a, b; Ortega‐Sáenz et al. 2003; Wyatt & Buckler, 2004; Buttigieg et al. 2008; Buckler & Turner, 2013, Fernández‐Agüera et al. 2015). It is also worth noting that the existence of a mitochondrial based redox acute O2 sensor is a long‐standing hypothesis, which was postulated to explain acute hypoxic pulmonary artery vasoconstriction (Archer et al. 1993; Waypa et al. 2001).
Pyruvate carboxylase and other metabolic enzymes and transporters
Our results demonstrate that Pcx is highly expressed in CB glomus cells (at the level of mRNA and protein) in comparison with SCG neurons. This occurs in parallel with a smaller decrease in pyruvate dehydrogenase (Pdha1) mRNA levels. Therefore, these data suggest that pyruvate is preferentially used by Pcx in glomus cell mitochondria to generate oxaloacetate, which represents a classical anaplerotic reaction that results in replenishment of the pool of TCA cycle intermediates (see Owen et al. 2002). These observations explain the extraordinarily high levels of biotin (a cofactor of carboxylases) recently found in glomus cells (Ortega‐Sáenz et al. 2016) and the need for HCO3 −/CO2 buffers (which are the source of the carbon atoms required by carboxylases) to maintain a robust chemosensory function in CB in vitro preparations (Iturriaga & Lahiri, 1991; our own unpublished observations). Replenishment of TCA cycle intermediates is also compatible with the high levels of succinate in the CB and the postulated accumulation of QH2 in acutely responding O2‐sensitive cells (Fernández‐Agüera et al. 2015). If pyruvate is preferentially used to generate oxaloacetate in glomus cell mitochondria, the main source of acetyl‐CoA necessary for the TCA cycle could be free fatty acid (FFA) β‐oxidation. Although we do not have any direct evidence that FFA catabolism is activated in chemoreceptor cells, this idea is compatible with the existence of abundant adipose tissue in the CB (Ortega‐Sáenz et al. 2013) and the low Phd3 level characteristic of glomus cells, as it is known that Phd3‐dependent hydroxylation of Acacb activates the synthesis of malonyl‐CoA, a potent inhibitor of mitochondrial FFA uptake (German et al. 2016). Enhanced FFA catabolism (facilitated by Phd3 down‐regulation) would also significantly contribute to increasing the QH2 pool in chemoreceptor cells, as each β‐oxidation cycle produces FADH2, which is directly converted to QH2 by the electron transport flavin/quinone oxidoreductase (ETF). Numerous cell types (in particular proliferating cells) rely on glutamine oxidation for TCA cycle anaplerosis. Glutamine is deaminated to glutamate, which is converted into α‐ketoglutarate, a TCA cycle intermediate (Yang et al. 2014) and a substrate of the O2‐regulated prolyl hydroxylases (Epstein et al. 2001). Interestingly, in glomus cells, we observed a significant decrease not only in the expression of the cationic amino acid transporter Slc7a5, but also in the mRNA levels of the soluble isoform of isocytrate dehydrogenase (Idh1), which is necessary for the synthesis of α‐ketoglutarate in the cytosol. These metabolic adaptations, which help to maintain low levels of α‐ketoglutarate, are probably required to ensure low Phd3 activity, as it is known that under conditions in which Phd3 is inhibited (e.g. during hypoxia) the enzyme can be reactivated when cells are exposed to daily α‐ketoglutarate administration (Tennant & Gottlieb, 2010). The metabolic specializations of CB glomus cells, which may be relevant to acute O2 sensing, were also qualitatively present in AM cells with the exception of Pcx which was not up‐regulated in our sorted TH+ AM cells. However, immunocytochemical analyses have directly shown high levels of biotin in the AM, which suggests the presence of high carboxylase activity in chromaffin cells (Ortega‐Sáenz et al. 2016).
Ion channels and other genes
Our comparative microarray analysis has shown that mouse CB and AM cells express a broad variety of calcium‐ and/or voltage‐gated ion channel genes, including some of the subunits which have been proposed to form the O2‐sensitive voltage‐gated K+ channels (Wyatt & Peers, 1995; Pérez‐García et al. 2004). However, we also found that numerous voltage‐dependent K+, Na+ and Ca2+ channel subunits were down‐regulated in the CB or AM in comparison with the SCG. This is probably a consequence of the high density of voltage‐gated ion channels that is needed to sustain the electrical excitability of sympathetic neurons, which have a large somatodendritic arbor and profuse axonal branching. Task3 (Kcnk9) was the most highly up‐regulated K+ channel gene in CB and AM cells, although Task1 (Kcnk3) was also overexpressed. High expression of Task1 has previously been reported in the CB; however, Task3 was not detected in either a human CB microarray study (Mkrtchian et al. 2012) or single neonatal mouse CB cell transcriptomes (Zhou et al. 2016). This suggests that the expression of Task subunits may differ between species or during development. Task‐like channels appear to be the major channels responsible for the hypoxia‐induced depolarization of CB chemoreceptor cells (Buckler et al. 2000; Kim et al. 2009; Kobayashi & Yamamoto 2010). In addition, Task1/Task3 heteromers have been proposed to be the channels that mediate the O2‐sensitive background current in adult rat (Kim et al. 2009) and mouse (Turner & Buckler, 2013) glomus cells. Whereas Task1‐deficient glomus cells have normal electrical parameters, cells from double Task1/Task3 knockout mice exhibit a clear depolarization, thereby supporting the role of Task3 channels in setting the resting potential of glomus cells (Ortega‐Sáenz et al. 2010). However, it must be noted that responsiveness to hypoxia is fully maintained in cells from Task1‐ or Task1/Task3‐null mice. Therefore, Task1 and Task3 channels do not seem to be indispensable for acute O2 sensing, with other channels appearing to mediate sensitivity to changes in in their absence (Ortega‐Sáenz et al. 2010). Two additional ion channel genes potentially related to acute responsiveness to hypoxia were overexpressed in glomus and chromaffin cells: Cacna1h (T‐type Cav3.2) and Trpc5. Ca2+ currents mediated by T‐type channels have previously been recorded from mouse CB cells (Ortega‐Sáenz et al. 2010), and it has recently been suggested that the Cav3.2 channel subtype mediates the CB over‐activation induced by chronic intermittent hypoxia (Makarenko et al. 2016). On the other hand, Cav3.2 mRNA is up‐regulated by chronic hypoxia in an Hif2α‐dependent manner in PC12 and chromaffin cells (del Toro et al. 2003; Carabelli et al. 2007). Cav3.2 expression is high in neonatal AM chromaffin cells but, as this occurs with acute sensitivity to hypoxia, it decreases with postnatal maturation. However, adult chromaffin cell sensitivity to hypoxia increases after AM denervation in parallel with the re‐appearance of T‐type Ca2+ channels (Levitsky and López‐Barneo, 2009). Together, these data suggest that Cav3.2 channels may play a fundamental role in facilitating the responsiveness of peripheral chemoreceptor cells to hypoxia. Similarly, the strong mRNA up‐regulation of Trpc5 channels in CB and AM cells suggests that they may also have an important role in the physiology of peripheral chemoreceptors. The existence of a significant background cationic conductance has been proposed to explain the relatively depolarized resting potential of glomus cells with respect to the K+ equilibrium potential (Carpenter & Peers, 2001; García‐Fernández et al. 2007b). In addition, it has been suggested that cationic currents participate in hypoxic activation of glomus (Kang et al. 2014) and chromaffin (Inoue et al. 1998) cells. Moreover, a cationic conductance, possibly mediated by Trp channels, is activated by hypoglycaemia in glomus cells (García‐Fernández et al. 2007b). As there are several subtypes of Trp channels in the CB (Buniel et al. 2003), Trpc homo‐ or heteromers could mediate the cationic currents mentioned above. Trpc channels are promiscuously activated by stretch, phospholipids and other variables (Beech, 2007). Interestingly, they are also inhibited by anaesthetics such as chloroform, propofol or halothane (Bahnasi et al. 2008), which could help to explain the strong respiratory depression induced by these drugs (Teppema et al. 2002).
Among other genes studied, we found that Ucp2 was systematically up‐regulated in CB and AM cells. Ucp2 and other uncoupling proteins are associated with thermogenesis and appear to be co‐expressed with Cox8b mRNA in some studies of adipose tissue thermogenic differentiation (Wu et al. 1999; Fisher et al. 2012). However, the significance of these observations in the context of CB physiology is currently unknown. Although it has been reported that Ucp2 knockout mice present pseudohypoxic pulmonary vascular remodelling and hypertension (Dromparis et al. 2013), preliminary experiments performed in this mouse model indicate that CB responsiveness to hypoxia is not significantly affected (our own unpublished observations). Olfr78 is an atypical olfactory receptor that is expressed outside the nasal mucosa and particularly in CB glomus cells (Chang et al. 2015; Zhou et al. 2016). In our analyses, Olfr78 mRNA was up‐regulated in the CB relative to the SCG but markedly down‐regulated in the AM. It has been reported that Olfr78 is a lactate receptor that is required for acute O2 sensing, given that Olfr78‐deficient mice lose the HVR and glomus cell responsiveness to hypoxia. Based on these data, an endocrine model of acute O2 sensing has been proposed in which lactate released from tissues during hypoxia activates glomus cells to produce hyperventilation (Chang et al. 2015). However, this model is incompatible with abundant data indicating that acute O2 sensitivity is a cell‐autonomous phenomenon that can be observed in isolated cells bathed in lactate‐free solutions (see López‐Barneo et al. 2016a, b). In addition, two independent groups have found that, in contrast to the findings of Chang et al. (2015), Olfr78 is a poor lactate receptor (Aisenberg et al. 2016; Zhou et al. 2016). The fact that Olfr78 mRNA expression is much higher in the SCG than in the AM argues against any direct involvement of this receptor in acute O2 sensing. However, ongoing experiments in several laboratories using various strains of Olfr78‐null mice should clarify the role of Olfr78 in CB physiology.
Signature gene‐expression profile in chemoreceptor cells and the mechanism of acute O2 sensing
Together, the data available on the gene expression profile of chemoreceptor cells versus O2‐insensitive SCG neurons suggest that a mix of genes encoding mitochondrial subunits, metabolic enzymes/transporters and ion channels is characteristic of cells that are acutely responding to decreases in . Our study suggests that down‐regulation of Phd3 and up‐regulation of Hif2α, Ndufa4l2, Cox4i2, Cox8b and Pcx confer CB glomus cells with their special sensitivity to hypoxia. Other genes that may also contribute to the ‘acute O2‐sensing signature metabolic profile’ of chemoreceptor cells are Pdha1, Idh1, Acacb and Slc7a5. The absence of frank Pcx overexpression is probably one of the reasons why AM cells are less O2‐sensitive than glomus cells. In addition to these ten ‘metabolic’ genes, four other genes which encode ion channels (Task3, Task1, Trp5, Cacna1h) are characteristic of CB and AM O2‐sensitive cells. Although it is possible that none of these genes is absolutely required for acute O2 sensing, their concerted action could result in a metabolic status that renders the cells sensitive to hypoxia. These data fit quite well with the MCI signalling model of acute O2 sensing, which is based on high succinate content and QH2 accumulation in glomus cells (Fernández‐Agüera et al. 2015), as well as the abundant evidence suggesting that special mitochondrial properties could contribute to hypoxia responsiveness in glomus cells (Mill & Jöbsis, 1970; Duchen & Biscoe, 1992a, b; Ortega‐Sáenz et al. 2003; Wyatt & Buckler, 2004; Buckler & Turner, 2013, Fernández‐Agüera et al. 2015).
The metabolic features of chemoreceptor cells, in particular glomus cells, are schematically summarized in Fig. 7. Phd3 has a central position in this scheme, as its low level of expression probably permits Hif2α stabilization and the subsequent induction of the genes that encode the three atypical mitochondrial subunits (Ndufa4l2, Cox4i2 and possibly Cox8b). The suppression of Phd3 activity, which is favoured by maintaining a low level of cytosolic α‐ketoglutarate, also decreases malonyl‐CoA synthesis and therefore enables FFA‐dependent mitochondrial metabolism and the generation of both acetyl‐CoA for the TCA cycle and large amounts of QH2. In parallel, the high levels of Pcx and relative down‐regulation of Pdh provide abundant oxaloacetate to replenish TCA cycle intermediates and to further increase the QH2 pool through the activity of MCI and MCII. The presence of Cox4i2, Cox8b and Ndufa4l2 could make cytochrome c oxidase activity highly sensitive to decreases in , such that even relatively mild hypoxia causing accumulation of an extra amount of QH2 leads to the slow‐down or even reversal of MCI and the production of molecules (reduced pyridine nucleotides and ROS) that signal via ion channels in the plasma membrane (see Fernández‐Agüera et al. 2015).
Figure 7. Model of the metabolic features of chemoreceptor glomus cells relevant to O2 sensing based on their gene expression profile.
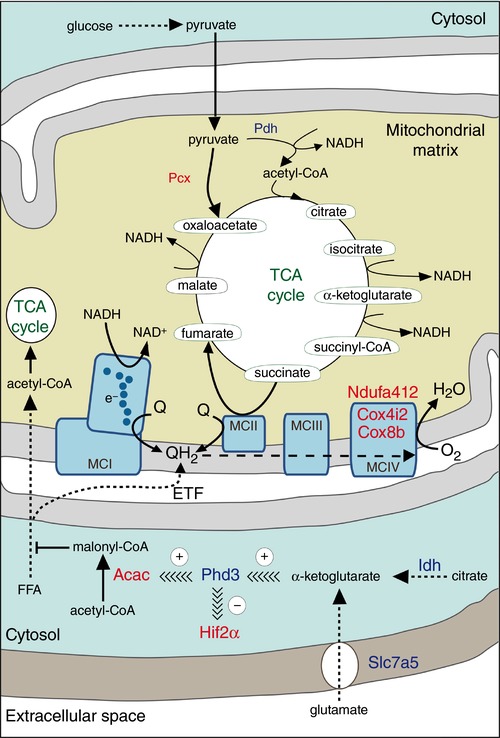
Proteins (or subunits of an enzyme complex) highly expressed in the CB in comparison to the SCG, as suggested by the microarray analysis, are highlighted in red, whereas proteins (or subunits of an enzyme complex) with relatively low expression in the CB are represented in dark blue. Arrows attached to continuous lines indicate one‐step chemical reactions, whereas arrows attached to discontinuous lines represent multistep chemical modifications. Acac, acetyl‐coenzyme A carboxylase; Cox4i2, cytochrome c oxidase subunit IV isoform 2; Cox8b, cytochrome c oxidase subunit VIIIb; ETF, electron transport flavin/quinone oxidoreductase; FFA, free fatty acid; Hif2α, endothelial PAS domain protein 1 (Epas1); Idh, isocitrate dehydrogenase; MCI, MCII, MCIII, MCIV, mitochondrial complex I, II, III, IV, respectively; Ndufa4l2, NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4‐like 2; Pcx, pyruvate carboxylase; Pdh, pyruvate dehydrogenase; Phd3, egl‐9 family prolyl hydroxylase 3 (Egln3); Q, ubiquinone; QH2, ubiquinol/reduced ubiquinone; Slc7a5, solute carrier family 7 (cationic amino acid transporter, y+ system), member 5; TCA, tri‐carboxylic acid. See text for detailed explanation.
For the past three decades, the identification of the ‘O2 sensor’ in glomus cells has been a matter of much discussion and investigation. However, numerous studies, in many cases using genetically modified animals with selective ablation of specific genes, have suggested that, in line with the model proposed here, O2 sensing is a multifactorial process that depends on the biophysical and metabolic properties of chemoreceptor cells rather than on the function of a specific O2‐sensing molecule (for recent reviews see Lopez‐Barneo et al. 2016a, b). Interestingly, recent work on mice with ablation of MCI genes (Fernandez‐Agüera et al. 2015) and the gene expression analyses discussed here suggest the compatibility of the two classical ‘models’ of CB acute O2 sensing: the ‘membrane model’, based on the modulation of ion channels by hypoxia and the ‘metabolic hypothesis’ which claims a fundamental role for mitochondria in this process. A major advantage of the scheme in Fig. 7 is that it allows predictions that can be experimentally tested. It has already been shown that pharmacological or genetic abolition of MCI selectively abolishes responsiveness to hypoxia in glomus and chromaffin cells (Ortega‐Saenz et al. 2003; Thompson et al. 2007; Fernández‐Agüera et al. 2015). Similarly, it is also known that CB cells are highly dependent on MCII for survival (Díaz‐Castro et al. 2012; Platero‐Luengo et al. 2014) and that genetic or pharmacological MCII dysfunction causes inhibition of responsiveness to hypoxia in pulmonary myocytes (this paper; see also Paddenberg et al. 2012). It can therefore be expected that future experiments, focused on the genes discussed here, will provide further understanding of the mechanisms of acute O2 sensing by chemoreceptor cells, and their modifications in variable pathophysiological or developmental conditions.
Additional information
Competing interests
The authors have no conflicts of interest to declare
Author contributions
L.G., V.B.‐H., P.G.‐F., I.A.‐M. and P.O.‐S. performed the experiments and participated in the interpretation of data. L.G., P.O.‐S and J.L.‐B. designed the figures and wrote the first draft of the paper. J.L.B. wrote the final draft of the paper and supervised the project. All authors read and approved the final paper.
Funding
This research was supported by the Botín Foundation, the Spanish Ministry of Economy and Innovation (SAF2012‐39343, SAF2016‐74990‐R) and the European Research Council (ERC Advanced Grant PRJ201502629).
Acknowledgements
We wish to thank Elizabeth Pintado for insightful comments and suggestions throughout this work. We also thank Alberto Pascual and Clara Ortega‐de San Luis for their help in microarray analysis. We are indebted to Konstantin L. Levitsky, Francisco J. Morón‐Civanto, Juan Antonio Cordero‐Varela and Mª José Castro‐Pérez for their technical support, and other services provided by the core facilities of Instituto de Biomedicina de Sevilla. The TH‐GFP mouse was obtained from the Gene Expression Nervous System Atlas (GENSAT) Project, NINDS Contracts N01NS02331 and HHSN271200723701C to The Rockefeller University (New York, NY, USA).
Linked articles This article is highlighted by a Perspective by Nurse. To read this Perspective, visit https://doi.org/10.1113/JP274880.
This is an Editor's Choice article from the 15 September 2017 issue.
Contributor Information
Lin Gao, Email: lgao-ibis@us.es.
José López‐Barneo, Email: lbarneo@us.es.
References
- Adams MB, Simonetta G & McMillen IC (1996). The non‐neurogenic catecholamine response of the fetal adrenal to hypoxia is dependent on activation of voltage sensitive Ca2+ channels. Brain Res Dev Brain Res 94, 182–189. [DOI] [PubMed] [Google Scholar]
- Aisenberg WH, Huang J, Zhu W, Rajkumar P, Cruz R, Santhanam L, Natarajan N, Yong HM, De Santiago B, Oh JJ, Yoon AR, Panettieri RA, Homann O, Sullivan JK, Liggett SB, Pluznick JL & An SS (2016). Defining an olfactory receptor function in airway smooth muscle cells. Sci Rep 6, 38231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberola‐Die A, Reboreda A, Lamas JA & Morales A (2013). Lidocaine effects on acetylcholine‐elicited currents from mouse superior cervical ganglion neurons. Neurosci Res 75, 198–203. [DOI] [PubMed] [Google Scholar]
- Appelhoff RJ, Tian YM, Raval RR, Turley H, Harris AL, Pugh CW, Ratcliffe PJ & Gleadle JM (2004). Differential function of the prolyl hydroxylases PHD1, PHD2, and PHD3 in the regulation of hypoxia‐inducible factor. J Biol Chem 279, 38458–38465. [DOI] [PubMed] [Google Scholar]
- Aras S, Pak O, Sommer N, Finley R, Jr , Huttemann M, Weissmann N & Grossman LI (2013). Oxygen‐dependent expression of cytochrome c oxidase subunit 4‐2 gene expression is mediated by transcription factors RBPJ, CXXC5 and CHCHD2. Nucleic Acids Res 41, 2255–2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archer SL, Huang J, Henry T, Peterson D & Weir EK (1993). A redox‐based O2 sensor in rat pulmonary vasculature. Circ Res 73, 1100–1112. [DOI] [PubMed] [Google Scholar]
- Bahnasi YM, Wright HM, Milligan CJ, Dedman AM, Zeng F, Hopkins PM, Bateson AN & Beech DJ (2008). Modulation of TRPC5 cation channels by halothane, chloroform and propofol. Br J Pharmacol 153, 1505–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balbir A, Lee H, Okumura M, Biswal S, Fitzgerald RS & Shirahata M (2007). A search for genes that may confer divergent morphology and function in the carotid body between two strains of mice. Am J Physiol Lung Cell Mol Physiol 292, L704–715. [DOI] [PubMed] [Google Scholar]
- Balsa E, Marco R, Perales‐Clemente E, Szklarczyk R, Calvo E, Landazuri MO & Enriquez JA (2012). NDUFA4 is a subunit of complex IV of the mammalian electron transport chain. Cell Metab 16, 378–386. [DOI] [PubMed] [Google Scholar]
- Baradaran R, Berrisford JM, Minhas GS & Sazanov LA (2013). Crystal structure of the entire respiratory complex I. Nature 494, 443–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beech DJ (2007). Bipolar phospholipid sensing by TRPC5 calcium channel. Biochem Soc Trans 35, 101–104. [DOI] [PubMed] [Google Scholar]
- Bishop T, Gallagher D, Pascual A, Lygate CA, de Bono JP, Nicholls LG, Ortega‐Saenz P, Oster H, Wijeyekoon B, Sutherland AI, Grosfeld A, Aragones J, Schneider M, van Geyte K, Teixeira D, Diez‐Juan A, Lopez‐Barneo J, Channon KM, Maxwell PH, Pugh CW, Davies AM, Carmeliet P & Ratcliffe PJ (2008). Abnormal sympathoadrenal development and systemic hypotension in PHD3–/– mice. Mol Cell Biol 28, 3386–3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown ST, Buttigieg J & Nurse CA (2010). Divergent roles of reactive oxygen species in the responses of perinatal adrenal chromaffin cells to hypoxic challenges. Respir Physiol Neurobiol 174, 252–258. [DOI] [PubMed] [Google Scholar]
- Brown DA & Passmore GM (2009). Neural KCNQ (Kv7) channels. Br J Pharmacol 156, 1185–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckler KJ & Turner PJ (2013). Oxygen sensitivity of mitochondrial function in rat arterial chemoreceptor cells. J Physiol 591, 3549–3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckler KJ, Williams BA & Honore E (2000). An oxygen‐, acid‐ and anaesthetic‐sensitive TASK‐like background potassium channel in rat arterial chemoreceptor cells. J Physiol 525, 135–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buniel MC, Schilling WP & Kunze DL (2003). Distribution of transient receptor potential channels in the rat carotid chemosensory pathway. J Comp Neurol 464, 404–413. [DOI] [PubMed] [Google Scholar]
- Buttigieg J, Brown ST, Lowe M, Zhang M & Nurse CA (2008). Functional mitochondria are required for O2 but not CO2 sensing in immortalized adrenomedullary chromaffin cells. Am J Physiol Cell Physiol 294, C945–956. [DOI] [PubMed] [Google Scholar]
- Buttigieg J, Brown S, Holloway AC & Nurse CA (2009). Chronic nicotine blunts hypoxic sensitivity in perinatal rat adrenal chromaffin cells via upregulation of KATP channels: role of α7 nicotinic acetylcholine receptor and hypoxia‐inducible factor‐2α. J Neurosci 29, 7137–7147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carabelli V, Marcantoni A, Comunanza V, de Luca A, Diaz J, Borges R & Carbone E (2007). Chronic hypoxia up‐regulates α1H T‐type channels and low‐threshold catecholamine secretion in rat chromaffin cells. J Physiol 584, 149–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter E & Peers C (2001). A standing Na+ conductance in rat carotid body type I cells. Neuroreport 12, 1421–1425. [DOI] [PubMed] [Google Scholar]
- Carroll J, Fearnley IM, Skehel JM, Shannon RJ, Hirst J & Walker JE (2006). Bovine complex I is a complex of 45 different subunits. J Biol Chem 281, 32724–32727. [DOI] [PubMed] [Google Scholar]
- Chang AJ, Ortega FE, Riegler J, Madison DV & Krasnow MA (2015). Oxygen regulation of breathing through an olfactory receptor activated by lactate. Nature 527, 240–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox JJ, Reimann F, Nicholas AK, Thornton G, Roberts E, Springell K, Karbani G, Jafri H, Mannan J, Raashid Y, Al‐Gazali L, Hamamy H, Valente EM, Gorman S, Williams R, McHale DP, Wood JN, Gribble FM & Woods CG (2006). An SCN9A channelopathy causes congenital inability to experience pain. Nature 444, 894–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Rio R, Andrade DC, Lucero C, Arias P & Iturriaga R (2016). Carotid body ablation abrogates hypertension and autonomic alterations induced by intermittent hypoxia in rats. Hypertension 68, 436–445. [DOI] [PubMed] [Google Scholar]
- Del Toro R, Levitsky KL, Lopez‐Barneo J & Chiara MD (2003). Induction of T‐type calcium channel gene expression by chronic hypoxia. J Biol Chem 278, 22316–22324. [DOI] [PubMed] [Google Scholar]
- Diaz‐Castro B, Pintado CO, Garcia‐Flores P, Lopez‐Barneo J & Piruat JI (2012). Differential impairment of catecholaminergic cell maturation and survival by genetic mitochondrial complex II dysfunction. Mol Cell Biol 32, 3347–3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dromparis P, Paulin R, Sutendra G, Qi AC, Bonnet S & Michelakis ED (2013). Uncoupling protein 2 deficiency mimics the effects of hypoxia and endoplasmic reticulum stress on mitochondria and triggers pseudohypoxic pulmonary vascular remodeling and pulmonary hypertension. Circ Res 113, 126–136. [DOI] [PubMed] [Google Scholar]
- Duchen MR & Biscoe TJ (1992a). Mitochondrial function in type I cells isolated from rabbit arterial chemoreceptors. J Physiol 450, 13–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchen MR & Biscoe TJ (1992b). Relative mitochondrial membrane potential and [Ca2+]i in type I cells isolated from the rabbit carotid body. J Physiol 450, 33–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein AC, Gleadle JM, McNeill LA, Hewitson KS, O'Rourke J, Mole DR, Mukherji M, Metzen E, Wilson MI, Dhanda A, Tian YM, Masson N, Hamilton DL, Jaakkola P, Barstead R, Hodgkin J, Maxwell PH, Pugh CW, Schofield CJ & Ratcliffe PJ (2001). C. elegans EGL‐9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell 107, 43–54. [DOI] [PubMed] [Google Scholar]
- Fagerlund MJ, Kahlin J, Ebberyd A, Schulte G, Mkrtchian S & Eriksson LI (2010). The human carotid body: expression of oxygen sensing and signaling genes of relevance for anesthesia. Anesthesiology 113, 1270–1279. [DOI] [PubMed] [Google Scholar]
- Fernandez‐Aguera MC, Gao L, Gonzalez‐Rodriguez P, Pintado CO, Arias‐Mayenco I, Garcia‐Flores P, Garcia‐Perganeda A, Pascual A, Ortega‐Saenz P & Lopez‐Barneo J (2015). Oxygen sensing by arterial chemoreceptors depends on mitochondrial complex I signaling. Cell Metab 22, 825–837. [DOI] [PubMed] [Google Scholar]
- Fisher FM, Kleiner S, Douris N, Fox EC, Mepani RJ, Verdeguer F, Wu J, Kharitonenkov A, Flier JS, Maratos‐Flier E & Spiegelman BM (2012). FGF21 regulates PGC‐1α and browning of white adipose tissues in adaptive thermogenesis. Genes Dev 26, 271–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda R, Zhang H, Kim JW, Shimoda L, Dang CV & Semenza GL (2007). HIF‐1 regulates cytochrome oxidase subunits to optimize efficiency of respiration in hypoxic cells. Cell 129, 111–122. [DOI] [PubMed] [Google Scholar]
- Ganfornina MD, Perez‐Garcia MT, Gutierrez G, Miguel‐Velado E, Lopez‐Lopez JR, Marin A, Sanchez D & Gonzalez C (2005). Comparative gene expression profile of mouse carotid body and adrenal medulla under physiological hypoxia. J Physiol 566, 491–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L, Gonzalez‐Rodriguez P, Ortega‐Saenz P & Lopez‐Barneo J (2017). Redox signaling in acute oxygen sensing. Redox Biol 12, 908–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia‐Fernandez M, Mejias R & Lopez‐Barneo J (2007a). Developmental changes of chromaffin cell secretory response to hypoxia studied in thin adrenal slices. Pflugers Arch 454, 93–100. [DOI] [PubMed] [Google Scholar]
- Garcia‐Fernandez M, Ortega‐Saenz P, Castellano A & Lopez‐Barneo J (2007b). Mechanisms of low‐glucose sensitivity in carotid body glomus cells. Diabetes 56, 2893–2900. [DOI] [PubMed] [Google Scholar]
- Garcia RA, Roemmich JN & Claycombe KJ (2016). Evaluation of markers of beige adipocytes in white adipose tissue of the mouse. Nutr Metab (Lond) 13, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- German NJ, Yoon H, Yusuf RZ, Murphy JP, Finley LW, Laurent G, Haas W, Satterstrom FK, Guarnerio J, Zaganjor E, Santos D, Pandolfi PP, Beck AH, Gygi SP, Scadden DT, Kaelin WG, Jr & Haigis MC (2016). PHD3 loss in cancer enables metabolic reliance on fatty acid oxidation via deactivation of ACC2. Mol Cell 63, 1006–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong S, Zheng C, Doughty ML, Losos K, Didkovsky N, Schambra UB, Nowak NJ, Joyner A, Leblanc G, Hatten ME & Heintz N (2003). A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature 425, 917–925. [DOI] [PubMed] [Google Scholar]
- Grundy D (2015). Principles and standards for reporting animal experiments in the Journal of Physiology and Experimental Physiology . J Physiol 593, 2547–2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutman M (1978). Modulation of mitochondrial succinate dehydrogenase activity, mechanism and function. Mol Cell Biochem 20, 41–60. [DOI] [PubMed] [Google Scholar]
- Hiyama TY, Watanabe E, Ono K, Inenaga K, Tamkun MM, Yoshida S & Noda M (2002). Nax channel involved in CNS sodium‐level sensing. Nat Neurosci 5, 511–512. [DOI] [PubMed] [Google Scholar]
- Hodson EJ, Nicholls LG, Turner PJ, Llyr R, Fielding JW, Douglas G, Ratnayaka I, Robbins PA, Pugh CW, Buckler KJ, Ratcliffe PJ & Bishop T (2016). Regulation of ventilatory sensitivity and carotid body proliferation in hypoxia by the PHD2/HIF‐2 pathway. J Physiol 594, 1179–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber W, Carey VJ, Gentleman R, Anders S, Carlson M, Carvalho BS, Bravo HC, Davis S, Gatto L, Girke T, Gottardo R, Hahne F, Hansen KD, Irizarry RA, Lawrence M, Love MI, MacDonald J, Obenchain V, Oles AK, Pages H, Reyes A, Shannon P, Smyth GK, Tenenbaum D, Waldron L & Morgan M (2015). Orchestrating high‐throughput genomic analysis with Bioconductor. Nat Methods 12, 115–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttemann M, Lee I, Gao X, Pecina P, Pecinova A, Liu J, Aras S, Sommer N, Sanderson TH, Tost M, Neff F, Aguilar‐Pimentel JA, Becker L, Naton B, Rathkolb B, Rozman J, Favor J, Hans W, Prehn C, Puk O, Schrewe A, Sun M, Hofler H, Adamski J, Bekeredjian R, Graw J, Adler T, Busch DH, Klingenspor M, Klopstock T, Ollert M, Wolf E, Fuchs H, Gailus‐Durner V, Hrabe de Angelis M, Weissmann N, Doan JW, Bassett DJ & Grossman LI (2012). Cytochrome c oxidase subunit 4 isoform 2‐knockout mice show reduced enzyme activity, airway hyporeactivity, and lung pathology. FASEB J 26, 3916–3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue M, Fujishiro N & Imanaga I (1998). Hypoxia and cyanide induce depolarization and catecholamine release in dispersed guinea‐pig chromaffin cells. J Physiol 507, 807–818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iturriaga R & Lahiri S (1991). Carotid body chemoreception in the absence and presence of CO2‐HCO3 . Brain Res 568, 253–260. [DOI] [PubMed] [Google Scholar]
- Kadenbach B & Huttemann M (2015). The subunit composition and function of mammalian cytochrome c oxidase. Mitochondrion 24, 64–76. [DOI] [PubMed] [Google Scholar]
- Kang D, Wang J, Hogan JO, Vennekens R, Freichel M, White C & Kim D (2014). Increase in cytosolic Ca2+ produced by hypoxia and other depolarizing stimuli activates a non‐selective cation channel in chemoreceptor cells of rat carotid body. J Physiol 592, 1975–1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keating DJ, Rychkov GY & Roberts ML (2001). Oxygen sensitivity in the sheep adrenal medulla: role of SK channels. Am J Physiol Cell Physiol 281, C1434–1441. [DOI] [PubMed] [Google Scholar]
- Kim D, Cavanaugh EJ, Kim I & Carroll JL (2009). Heteromeric TASK‐1/TASK‐3 is the major oxygen‐sensitive background K+ channel in rat carotid body glomus cells. J Physiol 587, 2963–2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi N & Yamamoto Y (2010). Hypoxic responses of arterial chemoreceptors in rabbits are primarily mediated by leak K channels. Adv Exp Med Biol 669, 195–199. [DOI] [PubMed] [Google Scholar]
- Levitsky KL & Lopez‐Barneo J (2009). Developmental change of T‐type Ca2+ channel expression and its role in rat chromaffin cell responsiveness to acute hypoxia. J Physiol 587, 1917–1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez‐Barneo J, Ortega‐Saenz P, Gonzalez‐Rodriguez P, Fernandez‐Aguera MC, Macias D, Pardal R & Gao L (2016a). Oxygen‐sensing by arterial chemoreceptors: mechanisms and medical translation. Mol Aspects Med 47–48, 90–108. [DOI] [PubMed] [Google Scholar]
- Lopez‐Barneo J, Gonzalez‐Rodriguez P, Gao L, Fernandez‐Aguera MC, Pardal R & Ortega‐Saenz P (2016b). Oxygen sensing by the carotid body: mechanisms and role in adaptation to hypoxia. Am J Physiol Cell Physiol 310, C629–642. [DOI] [PubMed] [Google Scholar]
- Macias D, Fernandez‐Aguera MC, Bonilla‐Henao V & Lopez‐Barneo J (2014). Deletion of the von Hippel‐Lindau gene causes sympathoadrenal cell death and impairs chemoreceptor‐mediated adaptation to hypoxia. EMBO Mol Med 6, 1577–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarenko VV, Ahmmed GU, Peng YJ, Khan SA, Nanduri J, Kumar GK, Fox AP & Prabhakar NR (2016). CaV3.2 T‐type Ca2+ channels mediate the augmented calcium influx in carotid body glomus cells by chronic intermittent hypoxia. J Neurophysiol 115, 345–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus NJ, Del Rio R, Schultz EP, Xia XH & Schultz HD (2014). Carotid body denervation improves autonomic and cardiac function and attenuates disordered breathing in congestive heart failure. J Physiol 592, 391–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBryde FD, Abdala AP, Hendy EB, Pijacka W, Marvar P, Moraes DJ, Sobotka PA & Paton JF (2013). The carotid body as a putative therapeutic target for the treatment of neurogenic hypertension. Nat Commun 4, 2395. [DOI] [PubMed] [Google Scholar]
- Michel H, Behr J, Harrenga A & Kannt A (1998). Cytochrome c oxidase: structure and spectroscopy. Annu Rev Biophys Biomol Struct 27, 329–356. [DOI] [PubMed] [Google Scholar]
- Mills E & Jobsis FF (1970). Simultaneous measurement of cytochrome a3 reduction and chemoreceptor afferent activity in the carotid body. Nature 225, 1147–1149. [DOI] [PubMed] [Google Scholar]
- Mkrtchian S, Kahlin J, Ebberyd A, Gonzalez C, Sanchez D, Balbir A, Kostuk EW, Shirahata M, Fagerlund MJ & Eriksson LI (2012). The human carotid body transcriptome with focus on oxygen sensing and inflammation–a comparative analysis. J Physiol 590, 3807–3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki‐Oda N, Takeuchi Y, Matsumura K, Oosawa Y & Watanabe Y (1997). Hypoxia‐induced catecholamine release and intracellular Ca2+ increase via suppression of K+ channels in cultured rat adrenal chromaffin cells. J Neurochem 69, 377–387. [DOI] [PubMed] [Google Scholar]
- Mojet MH, Mills E & Duchen MR (1997). Hypoxia‐induced catecholamine secretion in isolated newborn rat adrenal chromaffin cells is mimicked by inhibition of mitochondrial respiration. J Physiol 504, 175–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz‐Cabello AM, Toledo‐Aral JJ, Lopez‐Barneo J & Echevarria M (2005). Rat adrenal chromaffin cells are neonatal CO2 sensors. J Neurosci 25, 6631–6640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurse CA, Buttigieg J, Brown S & Holloway AC (2009). Regulation of oxygen sensitivity in adrenal chromaffin cells. Ann N Y Acad Sci 1177, 132–139. [DOI] [PubMed] [Google Scholar]
- Ortega‐Saenz P, Garcia‐Fernandez M, Pardal R, Alvarez E & Lopez‐Barneo J (2003). Studies on glomus cell sensitivity to hypoxia in carotid body slices. Adv Exp Med Biol 536, 65–73. [DOI] [PubMed] [Google Scholar]
- Ortega‐Saenz P, Levitsky KL, Marcos‐Almaraz MT, Bonilla‐Henao V, Pascual A & Lopez‐Barneo J (2010). Carotid body chemosensory responses in mice deficient of TASK channels. J Gen Physiol 135, 379–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega‐Saenz P, Macias D, Levitsky KL, Rodriguez‐Gomez JA, Gonzalez‐Rodriguez P, Bonilla‐Henao V, Arias‐Mayenco I & Lopez‐Barneo J (2016). Selective accumulation of biotin in arterial chemoreceptors: requirement for carotid body exocytotic dopamine secretion. J Physiol 594, 7229–7248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega‐Saenz P, Pardal R, Levitsky K, Villadiego J, Munoz‐Manchado AB, Duran R, Bonilla‐Henao V, Arias‐Mayenco I, Sobrino V, Ordonez A, Oliver M, Toledo‐Aral JJ & Lopez‐Barneo J (2013). Cellular properties and chemosensory responses of the human carotid body. J Physiol 591, 6157–6173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen OE, Kalhan SC & Hanson RW (2002). The key role of anaplerosis and cataplerosis for citric acid cycle function. J Biol Chem 277, 30409–30412. [DOI] [PubMed] [Google Scholar]
- Paddenberg R, Tiefenbach M, Faulhammer P, Goldenberg A, Gries B, Pfeil U, Lips KS, Piruat JI, Lopez‐Barneo J, Schermuly RT, Weissmann N & Kummer W (2012). Mitochondrial complex II is essential for hypoxia‐induced pulmonary vasoconstriction of intra‐ but not of pre‐acinar arteries. Cardiovasc Res 93, 702–710. [DOI] [PubMed] [Google Scholar]
- Pascual A, Hidalgo‐Figueroa M, Piruat JI, Pintado CO, Gomez‐Diaz R & Lopez‐Barneo J (2008). Absolute requirement of GDNF for adult catecholaminergic neuron survival. Nat Neurosci 11, 755–761. [DOI] [PubMed] [Google Scholar]
- Peers C (2015). Acute oxygen sensing–inching ever closer to an elusive mechanism. Cell Metab 22, 753–754. [DOI] [PubMed] [Google Scholar]
- Peng YJ, Nanduri J, Khan SA, Yuan G, Wang N, Kinsman B, Vaddi DR, Kumar GK, Garcia JA, Semenza GL & Prabhakar NR (2011). Hypoxia‐inducible factor 2α (HIF‐2α) heterozygous‐null mice exhibit exaggerated carotid body sensitivity to hypoxia, breathing instability, and hypertension. Proc Natl Acad Sci USA 108, 3065–3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez‐Garcia MT, Colinas O, Miguel‐Velado E, Moreno‐Dominguez A & Lopez‐Lopez JR (2004). Characterization of the Kv channels of mouse carotid body chemoreceptor cells and their role in oxygen sensing. J Physiol 557, 457–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piruat JI, Pintado CO, Ortega‐Saenz P, Roche M & Lopez‐Barneo J (2004). The mitochondrial SDHD gene is required for early embryogenesis, and its partial deficiency results in persistent carotid body glomus cell activation with full responsiveness to hypoxia. Mol Cell Biol 24, 10933–10940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platero‐Luengo A, Gonzalez‐Granero S, Duran R, Diaz‐Castro B, Piruat JI, Garcia‐Verdugo JM, Pardal R & Lopez‐Barneo J (2014). An O2‐sensitive glomus cell‐stem cell synapse induces carotid body growth in chronic hypoxia. Cell 156, 291–303. [DOI] [PubMed] [Google Scholar]
- Ribeiro MJ, Sacramento JF, Gonzalez C, Guarino MP, Monteiro EC & Conde SV (2013). Carotid body denervation prevents the development of insulin resistance and hypertension induced by hypercaloric diets. Diabetes 62, 2905–2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salman S, Holloway AC & Nurse CA (2014). Chronic opioids regulate KATP channel subunit Kir6.2 and carbonic anhydrase I and II expression in rat adrenal chromaffin cells via HIF‐2α and protein kinase A. Am J Physiol Cell Physiol 307, C266–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spreafico F, Barski JJ, Farina C & Meyer M (2001). Mouse DREAM/calsenilin/KChIP3: gene structure, coding potential, and expression. Mol Cell Neurosci 17, 1–16. [DOI] [PubMed] [Google Scholar]
- Tello D, Balsa E, Acosta‐Iborra B, Fuertes‐Yebra E, Elorza A, Ordonez A, Corral‐Escariz M, Soro I, Lopez‐Bernardo E, Perales‐Clemente E, Martinez‐Ruiz A, Enriquez JA, Aragones J, Cadenas S & Landazuri MO (2011). Induction of the mitochondrial NDUFA4L2 protein by HIF‐1α decreases oxygen consumption by inhibiting Complex I activity. Cell Metab 14, 768–779. [DOI] [PubMed] [Google Scholar]
- Tennant DA & Gottlieb E (2010). HIF prolyl hydroxylase‐3 mediates alpha‐ketoglutarate‐induced apoptosis and tumor suppression. J Mol Med (Berl) 88, 839–849. [DOI] [PubMed] [Google Scholar]
- Teppema LJ, Nieuwenhuijs D, Sarton E, Romberg R, Olievier CN, Ward DS & Dahan A (2002). Antioxidants prevent depression of the acute hypoxic ventilatory response by subanaesthetic halothane in men. J Physiol 544, 931–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson RJ, Buttigieg J, Zhang M & Nurse CA (2007). A rotenone‐sensitive site and H2O2 are key components of hypoxia‐sensing in neonatal rat adrenomedullary chromaffin cells. Neuroscience 145, 130–141. [DOI] [PubMed] [Google Scholar]
- Thompson RJ, Farragher SM, Cutz E, Nurse CA (2002). Developmental regulation of O2 sensing in neonatal adrenal chromaffin cells from wild‐type and NADPH‐oxidase‐deficient mice. Pflugers Arch 444, 539–548. [DOI] [PubMed] [Google Scholar]
- Thompson RJ, Jackson A & Nurse CA (1997). Developmental loss of hypoxic chemosensitivity in rat adrenomedullary chromaffin cells. J Physiol 498, 503–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian H, Hammer RE, Matsumoto AM, Russell DW & McKnight SL (1998). The hypoxia‐responsive transcription factor EPAS1 is essential for catecholamine homeostasis and protection against heart failure during embryonic development. Genes Dev 12, 3320–3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukihara T, Aoyama H, Yamashita E, Tomizaki T, Yamaguchi H, Shinzawa‐Itoh K, Nakashima R, Yaono R & Yoshikawa S (1996). The whole structure of the 13‐subunit oxidized cytochrome c oxidase at 2.8 A. Science 272, 1136–1144. [DOI] [PubMed] [Google Scholar]
- Turner PJ & Buckler KJ (2013). Oxygen and mitochondrial inhibitors modulate both monomeric and heteromeric TASK‐1 and TASK‐3 channels in mouse carotid body type‐1 cells. J Physiol 591, 5977–5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Geldermalsen M, Wang Q, Nagarajah R, Marshall AD, Thoeng A, Gao D, Ritchie W, Feng Y, Bailey CG, Deng N, Harvey K, Beith JM, Selinger CI, O'Toole SA, Rasko JE & Holst J (2016). ASCT2/SLC1A5 controls glutamine uptake and tumour growth in triple‐negative basal‐like breast cancer. Oncogene 35, 3201–3208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandael DH, Marcantoni A & Carbone E (2015). Cav1.3 channels as key regulators of neuron‐like firings and catecholamine release in chromaffin cells. Curr Mol Pharmacol 8, 149–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villadiego J, Mendez‐Ferrer S, Valdes‐Sanchez T, Silos‐Santiago I, Farinas I, Lopez‐Barneo J & Toledo‐Aral JJ (2005). Selective glial cell line‐derived neurotrophic factor production in adult dopaminergic carotid body cells in situ and after intrastriatal transplantation. J Neurosci 25, 4091–4098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waypa GB, Chandel NS & Schumacker PT (2001). Model for hypoxic pulmonary vasoconstriction involving mitochondrial oxygen sensing. Circ Res 88, 1259–1266. [DOI] [PubMed] [Google Scholar]
- Weber M, Pehl U, Breer H & Strotmann J (2002). Olfactory receptor expressed in ganglia of the autonomic nervous system. J Neurosci Res 68, 176–184. [DOI] [PubMed] [Google Scholar]
- Weir EK, Lopez‐Barneo J, Buckler KJ & Archer SL (2005). Acute oxygen‐sensing mechanisms. N Engl J Med 353, 2042–2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Puigserver P, Andersson U, Zhang C, Adelmant G, Mootha V, Troy A, Cinti S, Lowell B, Scarpulla RC & Spiegelman BM (1999). Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC‐1. Cell 98, 115–124. [DOI] [PubMed] [Google Scholar]
- Wyatt CN & Buckler KJ (2004). The effect of mitochondrial inhibitors on membrane currents in isolated neonatal rat carotid body type I cells. J Physiol 556, 175–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyatt CN & Peers C (1995). Ca2+‐activated K+ channels in isolated type I cells of the neonatal rat carotid body. J Physiol 483, 559–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Ko B, Hensley CT, Jiang L, Wasti AT, Kim J, Sudderth J, Calvaruso MA, Lumata L, Mitsche M, Rutter J, Merritt ME & DeBerardinis RJ (2014). Glutamine oxidation maintains the TCA cycle and cell survival during impaired mitochondrial pyruvate transport. Mol Cell 56, 414–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Chien MS, Kaleem S & Matsunami H (2016). Single cell transcriptome analysis of mouse carotid body glomus cells. J Physiol 594, 4225–4251. [DOI] [PMC free article] [PubMed] [Google Scholar]


