Abstract
Malignant glioma is the most common brain cancer with dismal outcomes. Individual variation of the patients’ survival times is remarkable. Here, we investigated the transcriptome and promoter methylation differences between patients of malignant glioma with short (less than one year) and the patients with long (more than three years) survival in CGGA (Chinese Glioma Genome Atlas), and validated the differences in TCGA (The Cancer Genome Atlas) to identify the genes whose expression levels showed high concordance with prognosis of glioma patients, as well as played an important role in malignant progression. The gene coding a key enzyme in genetic material synthesis, dCMP deaminase (DCTD), was found to be significantly correlated with overall survival and high level of DCTD mRNA indicated shorter survival of the patients with malignant glioma in different databases. Our finding revealed DCTD as an efficient prognostic factor for malignant glioma. As DCTD inhibitor gemcitabine has been proposed as an adjuvant therapy for malignant glioma, our finding also suggests a therapeutic value of gemcitabine for the patients with high expression level of DCTD.
Introduction
Glioma is the most common primary intracranial tumor, accounting for 46% of all intracranial tumors, and 2% of all adult cancers1. The WHO classification of central nervous system tumors (2007, fourth edition) divided diffuse glioma into WHO II, III and IV grades2. High-grade diffuse gliomas (WHO grade III and IV) and low-grade diffuse gliomas (WHO grade II) vary widely in tumor pathological morphology (such as collagen fiber content and morphological diversity), tumor development and prognosis of patients. Patients with glioblastoma (GBM, WHO grade IV), the most invasive glioma3, have the poorest prognosis, with a median overall survival of only 12–14 months, and a 5-year survival rate of only 9%4. The five-year survival rate for WHO grade III gliomas is 30%. The low-grade gliomas (WHO II grade) have a five-year survival rate of as high as 50%5. Clinically, WHO III and IV grade gliomas, which were characterized by strong invasion and significantly short survival are collectively mentioned as malignant glioma6.
The current standard treatment for malignant glioma is surgical resection followed by radiotherapy combined with concurrent and/or adjuvant temozolomide (TMZ) chemotherapy7–9. However, a large number of clinical studies have indicated that only approximately 9% of malignant glioma patients who received standard treatment could survive more than 5 years4. Although many studies have been conducted to improve the treatment of malignant glioma and to facilitate the increase of patients’ survival time, there have been no newly found effective treatments. Since the year of 2015, immunotherapeutic approaches have made remarkable progresses in hematopoietic tumors. However, the immunotherapeutic clinical trials in solid tumors including glioma are far from satisfying10. Antibody-drugs are currently explored to cure glioma while quite a number of challenges still exists11. Sustaining lag in the therapeutic approaches development is due to the limited understanding of the extremely complex networks of genomic alterations and molecular regulations controlling the initiation and development of malignant gliomas.
It is noteworthy that although the prognosis of malignant glioma patients remains generally poor, individual variation of the patients’ survival times is remarkable. Significantly, different outcomes reflect the intrinsic different expression level of the critical oncogenic genes or genomic alteration. Exploration of the intrinsic differences between malignant glioma with long survival and those with short survival may help us to reveal efficient predictive factors of survival time and potential therapeutic targets.
To explore efficient prognostic factor and effective therapeutic targets, we analyzed mRNA expression and methylation datasets to screen overall survival (OS)-correlated genes by shuttling between datasets from TCGA (The Cancer Genome Atlas) and CGGA (Chinese Glioma Genome Atlas). Seven genes including DCTD (dCMP deaminase) passed the filtering criteria. The prognostic efficiency of DCTD expression level was validated in another two databases and the oncogenic features of DCTD were revealed and verified in four independent databases.
DCTD is a key enzyme in genetic material synthesis and taking charge of conversion of deoxycytidylate (dCMP) to deoxy-uridine monophosphate (dUMP). dUMP is a synthetic substrate of thymidylate12. Abnormal expression of DCTD would affect the stability of genetic material synthesis, which is vital important for rapid tumor expansion. We speculate that DCTD acts as a “biosynthetic catalyst” in cancer progression to meet the rapid cell proliferation and active demand for genetic material. This also suggests that the oxycytidine analog antimetabolite gemcitabine, an inhibitor of DCTD, might be an efficient drug for the treatment of patients with high level of DCTD transcription.
Results
Screening for critical genes in glioma origin or development through gene expression and methylation analyses
To search for the critical genes in glioma origin or development, we firstly investigated the global transcriptome differences (target proportion of false discoveries: 0.1, number of permutations: 100, percentile for determining called genes that are false: 90) between patients with WHO grade III and IV glioma surviving for less than 1 year (n = 63) and those surviving for more than 3 years (n = 36) after diagnosis using data from the Chinese Glioma Genome Atlas (CGGA) database (Table 1). Subsequently, the gene promoter methylation levels of these patients were also compared. Therefore, we derived a cluster of genes with hypomethylated promoter and a high level of transcription correlated with short survival time (Cox proportional hazards model, nominal significance level of each univariate test: 0.001). The efficacy of these genes in distinguishing prognosis of all the patients with WHO grade III and IV glioma (except for the screening group, additional patients whose survival time is between 1 to 3 years were included, and the total number of samples was 178) were tested (Cox proportional hazards model, nominal significance level of each univariate test: 0.001). So far, we had identified genes with promoter methylation and transcriptional levels related to patients’ OS and were able to predict the survival time of the malignant glioma patients. Next, we validated the correlation between prognosis and the expression levels of these genes in TCGA microarray database for GBM (n = 476, Cox proportional hazards model, nominal significance level of each univariate test: 0.001). Only seven genes were found to be significantly correlated with the survival of the TCGA GBM patients (Fig. 1). DCTD (dCMP deaminase) encoding critical enzyme in genetic material synthesis was among the 7 filtered genes (Table 2). The hazard ratio for survival of DCTD expression in TCGA microarray (n = 476) was 1.279 and the parametric p-value was 0.0004785 (significance of correlation between expression level and OS generated by BRB Array Tools) (Fig. 1).
Table 1.
Clinical information of the patients in gene expression and methylation analyses.
| Survival <1 year | Survival >3 years | |
|---|---|---|
| Age (year) | 50.0* (13–70) | 41.5* (17–66) |
| Gender | ||
| Male | 40 | 23 |
| Female | 23 | 13 |
| OS (day) | 231.0* (27–363) | 1596.5* (1121–2257) |
| Grade | ||
| WHOIII | 11 | 21 |
| WHOIV | 52 | 15 |
| Histology | ||
| AA | 6 | 3 |
| AO | 1 | 9 |
| AOA | 4 | 9 |
| GBM | 52 | 15 |
*Median value.
Figure 1.

Data analysis pipeline to search for the OS-correlated critically important genes. The differences in the transcriptome between the WHO grade III and IV patients in CGGA database who lived for less than 1 year (n = 63) and those who lived for more than 3 years (n = 36) after diagnosis were analyzed. The level of gene promoter methylation in these patients was also compared. Gene lists derived based on transcriptional level and promoter methylation level was overlapped. The efficacy of the overlapping genes in predication of prognosis of all of the patients with WHO grade III and IV (except for the screening group, additional patients whose survival time is between 1 to 3 years were included, and the total number of samples was 178) glioma were tested. The genes with capability of predicting the survival length of malignant glioma patients were reserved. The prognosis effects of these genes in TCGA microarray data for GBM (n = 476) were tested. Only 7 genes, including DCTD were significantly correlated with the survival length of the TCGA GBM patients.
Table 2.
The 7 filtered genes through gene expression and methylation Analyses.
| Symbol | Parametric p-value | Hazard Ratio | FDR |
|---|---|---|---|
| EFEMP2 | 8e-07 | 1.272 | 0.000108 |
| FBXO17 | 4.67e-05 | 1.279 | 0.0026 |
| PDPN | 5.78e-05 | 1.133 | 0.0026 |
| BICD1 | 0.0001173 | 1.461 | 0.00396 |
| DCTD | 0.0004785 | 1.279 | 0.0129 |
| PTRF | 0.0007646 | 1.203 | 0.0172 |
| MEOX2 | 0.0009865 | 1.085 | 0.019 |
mRNA-level of DCTD could predict OS of the patients with malignant glioma
DCTD expression level was sufficient and efficient to predict the survival time of patients with malignant glioma. We tested the efficiency of DCTD expression level in prognosis of all patients with WHO grade III and IV glioma (except for the screening group, additional patients whose survival time is between 1 to 3 years were included, and the total number of samples was 178) in CGGA mRNA-array data. Half of the patients with relatively higher DCTD expression had marked poor outcomes. The predicative efficiency is also sound in CGGA mRNA-seq data (Fig. 2A). We next validated the efficiency of DCTD as a prognosis indicator in TCGA GBM transcriptional microarray data (n = 512) updated in 2013, which were expanded from the dataset that we used to screen for the above-mentioned 7 genes (Fig. 1). As expected, the efficiency of the DCTD transcriptional level as a prognosis indicator is also ideal (Fig. 2B). The OS indicator role of DCTD was validated in TCGA mRNA-seq (Fig. 2B), GSE16011 and REMBRANDT (Fig. 2C) data. The specificity and sensitivity of DCTD mRNA-level in predication of 5 (Fig. 2D) or 3 (Fig. 2E) years of survival was tested in CGGA and TCGA mRNA-seq data, and compared with “age” and “grade”. The area under curve (AUC) for DCTD transcriptional level in prediction of 5 and 3 years of survival in CGGA are 0.7661 and 0.7196 respectively. Those AUCs in TCGA are 0.7997 and 0.8219. The AUCs for DCTD mRNA-level are all larger than those of “age” in all of the four ROC tests, despite smaller than the AUCs of “grade”.
Figure 2.
The prognosis efficiency of DCTD. (A) The prognosis efficiency of DCTD in all WHO grade III and IV (except for the screening group, additional patients whose survival time is between 1 to 3 years were included, and the total number of samples was 178) patients in CGGA transcriptional microarray data and WHO grade III and IV glioma in CGGA RNA-seq data. (B) The prognosis efficiency of DCTD in GBM (n = 512) from TCGA transcriptional microarray data that were updated in 2013, which was expanded from the dataset that we used to search for the 7 genes and in GBM (n = 393) from TCGA RNA-seq dataset. (C) The prognosis efficiency of DCTD validated in GSE16011 and REMBRANDT datasets. (D) The ROC curves indicating the sensitivity and specificity of predicting 5 years of survival with DCTD-level in CGGA and TCGA database. (E) The ROC curves indicating the sensitivity and specificity of predicting 3 years of survival with DCTD-level in CGGA and TCGA database.
DCTD expression level is correlated with glioma grade and shows a subtype preference
The intensive expression of DCTD in glioma patients with poorer survival suggests oncogenic features of this gene. To further understand whether DCTD plays a role in malignant progression of glioma, we compared the expression levels of DCTD in different WHO grades glioma derived from four datasets. Except for the differentiation between grade II and III in GSE16011 (p = 0.563), DCTD expression increased along with grade progression, and the differentiations are all significant (p < 0.05) (Fig. 3). This suggests that DCTD might play a part in malignant progression of low grade glioma.
Figure 3.
The correlation of DCTD expression level and WHO grade. DCTD expression levels in glioma of WHO grade II-IV in CGGA RNA-seq (A), TCGA RNA-seq (B), GSE16011 (C) and REMBRANDT (D) databases. **p < 0.01.
As IDH1 mutation is a critical driver and prognosis indicator of glioma4, 13, we further explored the correlation between DCTD transcription level and IDH1 mutation. Both in CGGA (all grades, n = 302) and TCGA (GBM, n = 543) datasets, the patients harboring IDH1 mutation showed much lower expression of DCTD than those with wild-type IDH1 (Fig. 4A,B). The correlation between DCTD expression level and glioma subtype could also reflect the prognosis efficacy of DCTD. DCTD expression levels in the four transcriptional characteristic subtypes were quite different in the CGGA dataset (all grades, n = 302, Fig. 4C). Patients of classical subtype or mesenchymal subtype primarily had strong DCTD expression. In TCGA (GBM, n = 543) data, patients with higher DCTD expression were concentrated in classical, mesenchymal and neural subtypes, whereas patients with lower DCTD expressions primarily belonged to G-CIMP or proneural subtypes (Fig. 4D), which are typically associated with better outcomes14. As validation, the correlations between DCTD transcriptional level and IDH1 mutation or subtype were analyzed in RNA sequencing data derived from CGGA (Fig. 4E) or TCGA (Fig. 4F), as well as in GSE16011 (Fig. 4G). Except for the classical subtype of GSE16011 (the median of DCTD expression in IDH1-mutated glioma is higher than the ones with wild-type IDH1), all the analysis results were corresponding to above conclusions.
Figure 4.
Correlation between DCTD expression level and IDH1 mutation and the subtype preference. (A,B) Correlation of DCTD transcription level and IDH1 mutation in CGGA (A) and TCGA (B) RNA microarray data. (C,D) Correlation of DCTD transcription level and transcriptomic subtype classification in CGGA (C) and TCGA (D) RNA microarray data. (E–G) Correlation of DCTD transcription level and IDH1 mutation in different subtypes of glioma in CGGA RNA-seq (E), TCGA RNA-seq (F, no sample of classical subtype in TCGA seq data harbors IDH1 mutation) data and GSE16011 (G).
DCTD-related genomic alterations and biological processes
To further depict the oncogenic features of DCTD, we obtained an overview of the correlations between DCTD expression level and the genomic or transcriptional alterations contributing to the origin or progression of glioma (Fig. 5). According to the above results, IDH1 mutations occurred more frequently in glioma with lower DCTD expression. The well-known indicator of optimistic outcome, co-deletion of 1p19q gathered in glioma with lower DCTD expression. The incidences of malignant factors including Ki67 high-expression, PTEN mutation, TP53 mutation and EGFR amplification were higher in glioma with higher DCTD expression.
Figure 5.
Correlations of DCTD expressing-level with the classical genomic or transcriptional alterations in glioma. Abbreviation: mut = mutation; expre = expression level; ampli = amplification; L = low; H = high.
To further validate the oncogenic nature of DCTD, we annotated the biological differentiation accompanying with alteration of DCTD expression level (Fig. 6). The differentially expressing genes between glioma with high and low DCTD expression level were separately derived from CGGA or TCGA RNA-sequencing dataset and annotated using the online Database for Annotation, Visualization and Integrated Discovery (DAVID) v6.7. The DCTD-related genes were found more frequently involved in the processes of cell adhesion, immune or inflammatory response and epithelial to mesenchymal transition. Considering the critical role of DCTD in genetic material synthesis, we paid extra attention to the proliferation term and found marked enrichment of the DCTD-related genes in biological processes of positive regulation of cell proliferation.
Figure 6.
Gene ontology analysis for DCTD. Gene ontology analysis for DCTD in CGGA (A) and TCGA (B) RNA sequencing dataset.
Discussion
We revealed a potentially important gene contributing to glioma origin or malignant progression, and as well a promising prognostic indicator. This is the first report elaborating the pathological and biological role of DCTD in glioma. DCTD is a key enzyme in genetic material synthesis, taking charge of conversion of dCMP to dUMP, which is necessary in cancer origin and progression. The critical role of DCTD in genetic material synthesis supports our findings that DCTD-related genes were enriched in cell proliferation process.
To depict the role of DCTD in malignant progression of glioma, we performed biological functional annotation of the DCTD-related genes. Since the well-known role of DCTD in genetic material synthesis, which is an essential step of cell proliferation and tumor growth, it is unexpected that the DCTD-related genes were mostly noted enriched in the processes of cell adhesion and epithelial to mesenchymal transition. To our knowledge, there is no sound evidence indicating that DCTD acts as a stimulator of migration or invasion of tumor cells. The present study indicates a potential role for DCTD in the invasion capacity of glioma cells and our observations warrants further studies.
Additionally to our proposal that DCTD transcriptional level could have an impact on survival rate for patients with malignant glioma, our findings reveal the potential value of DCTD as a therapeutic target as well. The developments of novel therapeutic approaches continue worldwide. The deoxynucleoside analogue gemcitabine had been considered for combination therapy with radiation in GBM15. Gemcitabine has been routinely used in the treatment of solid tumors, such as non-small-cell lung cancer (NSCLC), breast and ovarian cancer, bladder cancer and pancreatic cancer16. Its active metabolite, gemcitabine triphosphate (dFdCTP) inhibits dCMP deaminase, the protein product of DCTD 17. Although gemcitabine had been proposed as a promising therapy for GBM in consideration of its effect as a radiosensitizer and favorable feasible properties of permeating the blood–tumor barrier, previous trials focused on the treatment of GBM were stopped after phase 0 evaluation15. In the present study, we proposed DCTD as a critically important gene in glioma origin and malignant progression. Since gemcitabine is a ready-made inhibitor of DCTD, we proposed a hypothesis to support the recommission of gemcitabine as an adjuvant therapy for malignant glioma with high DCTD expression (Fig. 7).
Figure 7.
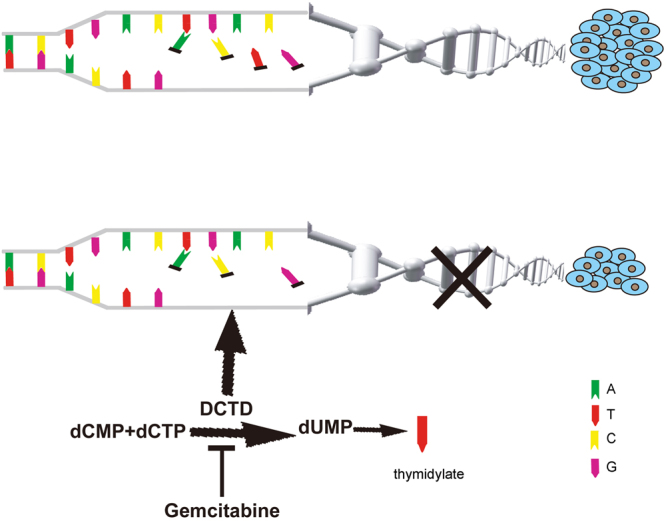
The hypothesis about recommission of gemcitabine as an adjuvant therapy for malignant glioma. The strong proliferation of tumor cells requires active synthesis of genetic material. In the process of genetic material synthesis, the protein product of DCTD, dCMP deaminase, plays an important catalyzing role. The ready-made inhibitor of DCTD, gemcitabine, could suppress the synthesis of dTMP and cause a shortage of genetic material, leading to inhibition of the hyperactive proliferation of tumor cells.
Materials and Methods
Patients and samples
A total of 302 glioma samples of all WHO grades from CGGA were enrolled in this study. This study was approved by the Institutional Review Boards of Beijing Tiantan Hospital, and written informed consent was obtained from all patients. All methods were performed in accordance with the relevant guidelines and regulations of the Institutional Review Boards. The establishment and management of our CGGA databank have been introduced in our previous publications18, 19. Information of Ki67 immunohistochemistry staining20, mutations of IDH1 20, PTEN 21 and TP53 21, deletions of 1p and/or 19q22 and amplifications of EGFR 19 are all derived from CGGA database and the detection methods had been described in our previous publications (the refs 19–22).
Transcriptomic subtype classification
The samples were classified into four transcriptional characteristic subtypes according to the method reported by Brennan et al.23. The mRNA levels were analyzed according to the Proneural-Neural-Classical-Mesenchymal classes using the signatures published in Verhaak et al.24, and the single sample Gene Set Enrichment Analysis algorithm (ssGSEA).
Statistical analysis
Median Absolute Deviation (MAD) was calculated using Matlab. Probes targeting genes that showed the highest variable expression were selected for further analysis. Significance Analysis of Microarray (SAM) was performed using BRB Array Tools developed by Dr. Richard Simon and his team to gain the genes whose transcriptional levels or promoter methylation levels were significantly different between the patients lived for less than 1 year (n = 63) and those lived for more than 3 years (n = 36) after diagnosis using data from the Chinese Glioma Genome Atlas (CGGA). FDR was set as 0.1. Then the gene lists derived based on transcriptional level and promoter methylation level was overlapped. Therefore, we derived a cluster of genes with hypomethylated promoter and a high level of transcription correlated with short survival time. Survival analysis (Cox proportional hazards model) was used to estimate the efficacy of these genes for distinguishing prognosis of all patients with WHO grade III and IV glioma in CGGA (except for the screening group, additional patients whose survival time is between 1 to 3 years were included, and the total number of samples was 178) by BRB Array Tools (Version: 4.3.1, Stable, June 2013). Efficacy of these genes in distinguishing prognosis of GBMs in TCGA dataset was also tested. The genes that could efficiently (nominal significance level of each univariate test was both set as 0.001) indicate OS in CGGA and TCGA datasets were overlapped. Finally, only 7 genes were filtered out.
The diagrams describing the correlations between the expression level of DCTD with OS, grades, IDH1 mutant and subtype were generated using R programming language.
Transcriptome differences between patients with DCTD expression higher than median value and those with DCTD expression level lower than median value were obtained from CGGA or TCGA databases and gene ontology analysis of the DCTD expression level-related genes was performed using online Database for Annotation, Visualization and Integrated Discovery (DAVID) v6.7 (https://david.ncifcrf.gov/).
Electronic supplementary material
Acknowledgements
We appreciate the patients who have participated in CGGA and this study. This study was supported by funds from the National Nature Science Foundation of China (grant NO. 81502495); The National Key Research and Development Plan (No. 2016YFC0902500); Capital Medical Development Research Fund (2016-1-1072); Youth Innovation Fund Of Beijing Neurosurgical Institute (2014009).
Author Contributions
H.M.H. and Z.W. provided equal contributions to the design of the study, data analysis and planned and wrote the manuscript. F.Z., K.Y.W., R.Y.H., H.Y.W., F.Y., T.Y.L. and H.H. participated in data downloading and preliminary analysis. T.J. and M.Y.L. critically revised the manuscript. All of the authors have read and approved the final manuscript.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Huimin Hu and Zheng Wang contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-11962-y.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Louis DN. Molecular pathology of malignant gliomas. Annu Rev Pathol. 2006;1:97–117. doi: 10.1146/annurev.pathol.1.110304.100043. [DOI] [PubMed] [Google Scholar]
- 2.Louis DN, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang M, et al. L1 stimulation of human glioma cell motility correlates with FAK activation. J Neurooncol. 2011;105:27–44. doi: 10.1007/s11060-011-0557-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiang T, et al. CGCG clinical practice guidelines for the management of adult diffuse gliomas. Cancer Lett. 2016;375:263–273. doi: 10.1016/j.canlet.2016.01.024. [DOI] [PubMed] [Google Scholar]
- 5.Goodenberger ML, Jenkins RB. Genetics of adult glioma. Cancer Genet. 2012;205:613–621. doi: 10.1016/j.cancergen.2012.10.009. [DOI] [PubMed] [Google Scholar]
- 6.Stupp R, Tonn JC, Brada M, Pentheroudakis G. High-grade malignant glioma: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2010;21(Suppl 5):v190–v193. doi: 10.1093/annonc/mdq187. [DOI] [PubMed] [Google Scholar]
- 7.Stupp R, et al. Promising survival for patients with newly diagnosed glioblastoma multiforme treated with concomitant radiation plus temozolomide followed by adjuvant temozolomide. J Clin Oncol. 2002;20:1375–1382. doi: 10.1200/JCO.2002.20.5.1375. [DOI] [PubMed] [Google Scholar]
- 8.Walker MD, et al. Randomized comparisons of radiotherapy and nitrosoureas for the treatment of malignant glioma after surgery. N Engl J Med. 1980;303:1323–1329. doi: 10.1056/NEJM198012043032303. [DOI] [PubMed] [Google Scholar]
- 9.Li MY, et al. Low c-Met expression levels are prognostic for and predict the benefits of temozolomide chemotherapy in malignant gliomas. Sci Rep. 2016;6:21141. doi: 10.1038/srep21141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weller M, et al. Vaccine-based immunotherapeutic approaches to gliomas and beyond. Nat Rev Neurol. 2017;13:363–374. doi: 10.1038/nrneurol.2017.64. [DOI] [PubMed] [Google Scholar]
- 11.Gan, H. K. et al. Antibody-drug conjugates in glioblastoma therapy: the right drugs to the right cells. Nat Rev Clin Oncol. 10.1038/nrclinonc.2017.95. Published online 4 Jul (2017). [DOI] [PubMed]
- 12.Weiner KX, et al. Chromosomal location and structural organization of the human deoxycytidylate deaminase gene. J Biol Chem. 1995;270:18727–18729. doi: 10.1074/jbc.270.32.18727. [DOI] [PubMed] [Google Scholar]
- 13.Yan H, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Noushmehr H, et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17:510–522. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sigmond J, et al. Gemcitabine uptake in glioblastoma multiforme: potential as a radiosensitizer. Ann Oncol. 2009;20:182–187. doi: 10.1093/annonc/mdn543. [DOI] [PubMed] [Google Scholar]
- 16.Toschi L, Finocchiaro G, Bartolini S, Gioia V, Cappuzzo F. Role of gemcitabine in cancer therapy. Future Oncol. 2005;1:7–17. doi: 10.1517/14796694.1.1.7. [DOI] [PubMed] [Google Scholar]
- 17.Heinemann V, et al. Cellular elimination of 2′,2′-difluorodeoxycytidine 5′-triphosphate: a mechanism of self-potentiation. Cancer Res. 1992;52:533–539. [PubMed] [Google Scholar]
- 18.Hu H, et al. Genome-wide transcriptional analyses of Chinese patients reveal cell migration is attenuated in IDH1-mutant glioblastomas. Cancer Lett. 2015;357:566–574. doi: 10.1016/j.canlet.2014.12.018. [DOI] [PubMed] [Google Scholar]
- 19.Liu Y, et al. Multidimensional analysis of gene expression reveals TGFB1I1-induced EMT contributes to malignant progression of astrocytomas. Oncotarget. 2014;5:12593–12606. doi: 10.18632/oncotarget.2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cai J, et al. ATRX mRNA expression combined with IDH1/2 mutational status and Ki-67 expression refines the molecular classification of astrocytic tumors: evidence from the whole transcriptome sequencing of 169 samples samples. Oncotarget. 2014;5:2551–2561. doi: 10.18632/oncotarget.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Y, et al. Integrated analysis identified genes associated with a favorable prognosis in oligodendrogliomas. Genes Chromosomes Cancer. 2016;55:169–176. doi: 10.1002/gcc.22323. [DOI] [PubMed] [Google Scholar]
- 22.Yang P, et al. Radiation combined with temozolomide contraindicated for young adults diagnosed with anaplastic glioma. Oncotarget. 2016;7:80091–80100. doi: 10.18632/oncotarget.11756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brennan CW, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Verhaak RG, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.







