ABSTRACT
Chronic hepatitis B virus (HBV) infection can lead to liver cirrhosis and hepatocellular carcinoma. HBV reactivation during or after chemotherapy is a potentially fatal complication for cancer patients with chronic HBV infection. Transcription of HBV is a critical intermediate step of the HBV life cycle. However, factors controlling HBV transcription remain largely unknown. Here, we found that different P-TEFb complexes are involved in the transcription of the HBV viral genome. Both BRD4 and the super elongation complex (SEC) bind to the HBV genome. The treatment of bromodomain inhibitor JQ1 stimulates HBV transcription and increases the occupancy of BRD4 on the HBV genome, suggesting the bromodomain-independent recruitment of BRD4 to the HBV genome. JQ1 also leads to the increased binding of SEC to the HBV genome, and SEC is required for JQ1-induced HBV transcription. These findings reveal a novel mechanism by which the HBV genome hijacks the host P-TEFb-containing complexes to promote its own transcription. Our findings also point out an important clinical implication, that is, the potential risk of HBV reactivation during therapy with a BRD4 inhibitor, such as JQ1 or its analogues, which are a potential treatment for acute myeloid leukemia.
KEYWORDS: BRD4, hepatitis B virus, super elongation complex, transcriptional elongation
INTRODUCTION
Around 240 million people worldwide are affected by chronic hepatitis B virus (HBV) infection, which is the most common cause of life-threatening liver diseases like cirrhosis, liver failure, and hepatocellular carcinoma (HCC) (1, 2). HBV is a circular, partially double-stranded, and enveloped hepatotropic DNA virus. It can be transmitted percutaneously, sexually, and perinatally.
The HBV genome is about 3.2 kb in size, containing four viral protein-coding genes and two identified enhancer elements (3–5). Transcription of the HBV genome is an essential step of the HBV replication and infection cycle (6). Upon infection, the capsid-associated relaxed circular DNA (rcDNA) from incoming HBV virion is converted into covalently closed circular DNA (cccDNA), which exists in the host hepatocyte nucleus as an episomal minichromosome and serves as a template for host RNA polymerase II (Pol II) to transcribe all of the viral RNAs, including the pregenomic RNA (pgRNA) (7). The minichromosome structure affects the accessibility of the HBV viral genome to transcription factors and RNA Pol II through the host cellular chromatin modification machinery (8–10). The hepatitis B virus X protein (HBx) is a 17-kDa viral transcriptional coactivator that communicates with the host cellular factors and promotes HBV transcriptional activation (11, 12).
Current antiviral treatments can achieve effective viral suppression in chronic HBV patients, but they rarely lead to the eradication of the viral cccDNA from the infected hepatocytes (13, 14). The HBV viral RNAs are crucial intermediates of the HBV life cycle, and their transcription from cccDNA could be an alternative target for the discovery of new medical interventions in controlling HBV replication in the infected hepatocytes (15, 16). Furthermore, due to the persistence of cccDNA, HBV reactivation occurs frequently in patients with chronic HBV infection when receiving chemotherapeutic and immunosuppressive agents, causing fatal fulminant hepatic failure (FHF) if they do not receive antiviral prophylaxis (17). Therefore, it is important to identify any drugs that may clear HBV and take preventive measures to reduce the risk of such serious complications.
The positive transcription elongation factor b, or P-TEFb, composed of the kinase module CDK9 and its regulatory subunit, cyclin T, is essential in regulating transcription by RNA Pol II (18). P-TEFb exists in multiple complexes in vivo. Previously we identified the super elongation complex (SEC), which is centered on the scaffold AF4/FMR2 family proteins, AFF1 and AFF4, and also contains P-TEFb, the RNA Pol II elongation factor ELL proteins, and the frequent mixed-lineage leukemia (MLL) translocation partners AF9 and ENL (19–21). SEC activates the transcription of MLL target oncogenic genes, stress-responsive genes, and HIV proviral genes through regulating transcriptional elongation checkpoint control (20, 22, 23). Another major active P-TEFb-containing complex is BRD4/P-TEFb (24, 25). BRD4, belonging to the bromodomain and extraterminal (BET) family, mainly interacts with chromatin through recognizing acetylated lysine residues on the histone tails (26). The BRD4 small inhibitor JQ1, which dislodges the bromodomain of BRD4 from the acetylated chromatin, emerges as a potential chemotherapy drug in treating MYC-high cancers (27, 28).
In this paper, we found that both SEC and BRD4 can bind to the HBV genome. SEC is required for the transcription of the HBV genome. However, JQ1 treatment leads to the upregulation of HBV transcription and higher hepatitis B surface antigen (HBsAg) levels in HepG2.2.15 cells. Detailed analyses demonstrated that the recruitment of BRD4 to the HBV genome is increased after JQ1 treatment, which suggests the bromodomain-independent recruitment of BRD4 in this scenario. Furthermore, the recruitment of SEC to the HBV genome is also slightly increased after JQ1 treatment. Inhibition of SEC by either RNA interference (RNAi) or the treatment of the CDK9 inhibitor flavopiridol is able to block HBV upregulation after JQ1 treatment. Thus, our results indicated that the inhibition of BRD4 by JQ1 can result in the reactivation of HBV, and SEC could be a target for the inhibition of HBV pgRNA transcription.
RESULTS
P-TEFb-containing complexes occupy the HBV genome.
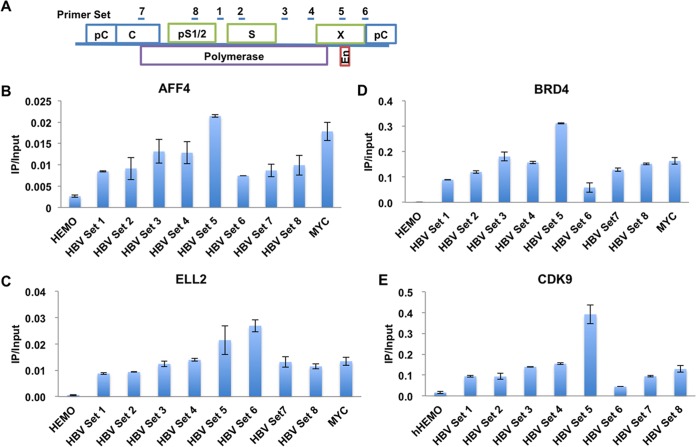
To investigate whether P-TEFb-containing complexes are involved in HBV transcriptional regulation, we first performed chromatin immunoprecipitation (ChIP)-quantitative PCR (qPCR) to assess the interaction of components within the P-TEFb-containing complexes with the HBV genome in HepG2.2.15 cells. The HepG2.2.15 cell line was derived from human hepatoma HepG2 cells, in which a complete HBV genome was stably integrated (29). It has been widely used in HBV studies, since in HepG2.2.15 all HBV viral proteins can be expressed and the viral genome can be replicated. Our analysis indicated that the kinase CDK9, BRD4, and the components of SEC, AFF4 and ELL2, all were detected at the HBV genome by ChIP-qPCR (Fig. 1). The occupancies of BRD4, AFF4, and ELL2 on HBV were comparable to their occupancies at the host endogenous MYC gene, which is a key BRD4 and SEC target gene (Fig. 1B to D).
FIG 1.
Different P-TEFb-containing complexes are recruited to the HBV genome. (A) Cartoon model illustrating the position of the primer pairs used for qPCR after ChIP along the HBV genome. (B to E) The SEC subunits AFF4 (B), ELL2 (C), and BRD4 (D) and the P-TEFb kinase module CDK9 (E) are all recruited to the HBV genome, as detected in HepG2.2.15 cells by ChIP-qPCR assays with specific antibodies. The HEMO gene serves as a negative control for ChIP-qPCR. Error bars represent the standard deviations from three independent measurements.
JQ1 induces HBV expression.
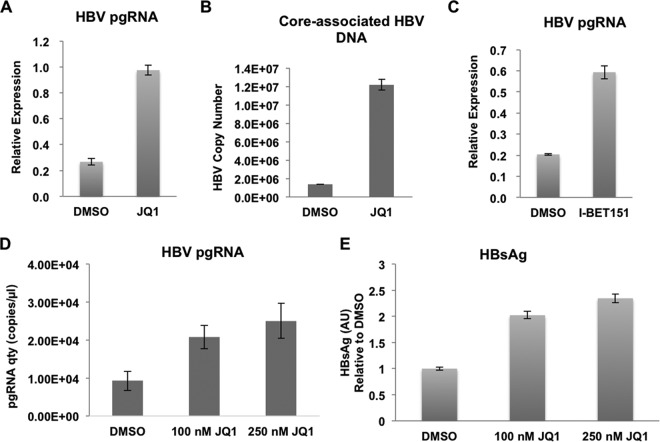
We then sought to determine whether these different P-TEFb-containing complexes occupying the HBV genome also regulate its transcription in the host. BRD4 possesses two bromodomains that bind to acetylated lysine residues, leading to its localization to the hyperacetylated chromatin regions (30). JQ1 is able to specifically displace BRD4 from the acetylated chromatin, disrupting the bromodomain-dependent interaction of BRD4 with chromatin (31, 32). Since BRD4 is present throughout the HBV genome, we first asked whether JQ1 plays a role in HBV transcription by treating HepG2.2.15 cells with JQ1. JQ1 exposure was able to induce HBV expression, as measured by reverse transcription (RT)-qPCR analysis of the viral HBV pgRNA (Fig. 2A). Consequently, the core-associated HBV DNA was also increased after JQ1 treatment (Fig. 2B), suggesting that HBV can be reactivated by bromodomain inhibition. The effect of bromodomain inhibition on HBV reactivation was further validated through treating HepG2.2.15 cells with the JQ1 analogue I-BET151 (Fig. 2C).
FIG 2.
Bromodomain inhibitor JQ1 activates HBV transcription and replication. (A) Treatment of HepG2.2.15 cells by JQ1 promotes HBV transcription. The levels of HBV pgRNA were assessed by RT-qPCR and normalized to the internal control GAPDH mRNA levels before and after JQ1 exposure. (B) HBV core-associated DNAs were extracted from culture medium treated with DMSO and JQ1. The DNA levels were then quantified by standard curve qPCR analysis. (C) Treatment of HepG2.2.15 cells by I-BET151 promotes HBV transcription. The levels of HBV pgRNA were assessed by RT-qPCR and normalized to the internal control GAPDH mRNA levels before and after JQ1 exposure. (D) JQ1 treatment also upregulates the transcription from the nonintegrated form of the HBV genome. Huh7 cells transiently transfected with HBV were treated with different doses (0, 100, and 250 nM) of JQ1. The levels of HBV pgRNA were then assessed by RT-qPCR and normalized to the internal control GAPDH mRNA levels. Error bars represent standard deviations from three independent measurements. (E) The surface antigen of HBV, HBsAg, is upregulated by JQ1 treatment in Huh7 cells, which were treated as described for panel B. HBsAg levels in cell culture supernatant before and after different doses of JQ1 treatment were quantified by ELISA.
In order to mimic the situation of free nonintegrated HBV, we performed transient transfection of Huh7 cells with HBV and then treated them with different concentrations (0, 100, and 250 nM) of JQ1. Consistent with the situation of free nonintegrated HBV, HBV pgRNA was upregulated by JQ1 in Huh7 cells transiently transfected with HBV in a dose-dependent manner (Fig. 2D). We also determined the effect of JQ1 on the HBV replication level by an enzyme-linked immunosorbent assay (ELISA) analysis of HBsAg levels in the culture medium. The HBsAg level was also upregulated by JQ1, suggesting that JQ1 could lead nonintegrated HBV to synthesize and secrete more HBV viral particles in the host (Fig. 2E).
JQ1 leads to the accumulation of BRD4 on the HBV genome.
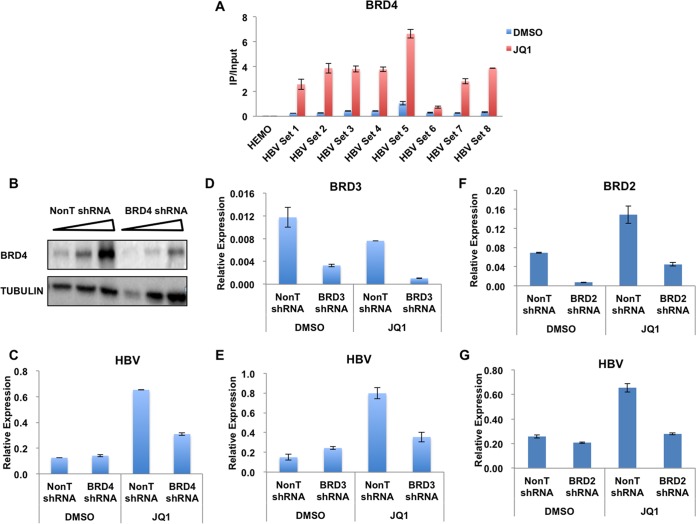
We have demonstrated so far that BRD4 is enriched on the HBV genome, while the treatment of the bromodomain inhibitor JQ1 leads to the upregulation of HBV transcription. These findings pointed to two possibilities: (i) the recruitment of BRD4 to the HBV genome is bromodomain independent, and (ii) BRD4 binds to the HBV genome in a bromodomain-dependent manner and inhibits the transcription of HBV in the host. To test our hypotheses, we first performed BRD4 ChIP in HepG2.2.15 cells, followed by qPCR in the absence or presence of JQ1. The occupancy of BRD4 throughout the HBV genome was significantly increased after JQ1 treatment (Fig. 3A), suggesting the bromodomain-independent binding of BRD4 to the HBV genome.
FIG 3.
JQ1 treatment increases the occupancy of BRD4 and SEC on the HBV genome. (A) HepG2.2.15 cells were treated with 250 nM JQ1 for 48 h. Nontreated and treated cells were used in ChIP assays with specific antibodies, followed by qPCR amplification using the primer pairs spanning the HBV genome, as illustrated in Fig. 1A. The occupancy of BRD4 is significantly enhanced after JQ1 treatment. The HEMO gene serves as a negative control for ChIP-qPCR. Error bars represent the standard deviations from three independent measurements. (B to G) BRD2, BRD3, and BRD4 were individually knocked down to examine their effects on HBV transcription upon JQ1 treatment. The levels of HBV pgRNA were then assessed by RT-qPCR and normalized to the internal control GAPDH mRNA levels. Error bars represent the standard deviations from three independent measurements.
To further examine whether the increase of BRD4 occupancy at the HBV genome leads to the upregulation of HBV transcription, we specifically depleted BRD4 by using lentivirus-mediated short hairpin RNA (shRNA) (Fig. 3B). As the results indicated, although BRD4 was not required for the basal transcription of HBV, the knockdown of BRD4 compromised JQ1-mediated HBV reactivation (Fig. 3C). JQ1 also can block the recognition of other bromodomain-containing proteins, especially the BRD4-interacting proteins BRD2 and BRD3, with the hyperacetylated chromatin. Thus, we also examined the requirement of BRD2 and BRD3 for JQ1-mediated HBV reactivation. Consistent with the above-described findings, the depletion of both BRD2 and BRD3 reduced the effect of JQ1 on HBV upregulation. Taken together, our data suggest that in addition to BRD4, BRD2 and BRD3 also function in mediating the HBV reactivation by JQ1 (Fig. 3D to G).
SEC is required for both basal and JQ1-induced HBV transcription.
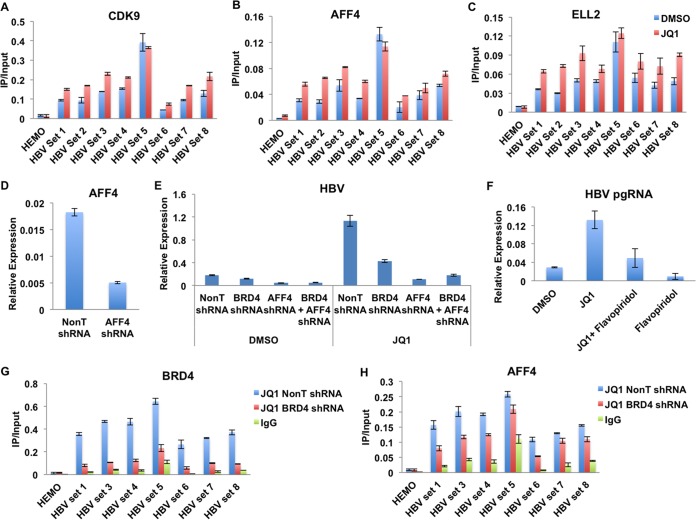
Previously, we demonstrated that SEC is required for the transcriptional induction of immediate-early genes in response to external stimuli (20). SEC also can be recruited by the Tat protein to the HIV promoter and regulates Tat-mediated HIV transactivation. Here, we found that SEC is localized at the HBV genome (Fig. 1). The treatment of JQ1 further leads to the slightly increased occupancies of the kinase subunit CDK9 and other SEC components, AFF4 and ELL2, across the HBV genome (Fig. 4A to C).
FIG 4.
SEC is required for JQ1-mediated HBV induction. (A to C) JQ1 treatment slightly enhances the occupancies of SEC subunits CDK9, AFF4, and ELL2. (D) Knockdown of AFF4 in HepG2.2.15 cells by lentivirus-mediated shRNA targeting AFF4. (E) Knockdown of AFF4 in HepG2.2.15 cells inhibits basal and JQ1-induced HBV pgRNA transcription. Control and AFF4-depleted cells were treated with DMSO, for the vehicle control, and JQ1. The expression levels were normalized to the internal control GAPDH mRNA levels. Error bars represent the standard deviations from three independent measurements. (F) The CDK9 inhibitor flavopiridol inhibits basal and JQ1-induced HBV pgRNA transcription. pgRNA levels were measured by RT-qPCR and normalized to GAPDH mRNA levels. Error bars represent the standard deviations from three independent measurements. (G and H) BRD4 and AFF4 enrichment analysis in BRD4-depleted cells. The HepG2.2.15 cells were treated with BRD4- and AFF4-specific shRNAs for 2 days, followed by 48 h of JQ1 treatment. IgG serves as the nonrelated antibody control. The HEMO gene serves as a negative control for ChIP-qPCR. Error bars represent the standard deviations from three independent measurements.
In order to investigate whether SEC functions in HBV transcription, we performed shRNA-mediated knockdown of the central component of SEC, AFF4, in HepG2.2.15 cells. Knockdown efficiency, measured by RT-qPCR, shows that AFF4 RNAi can lead to around 80% suppression of AFF4 (Fig. 4D). Transcription of both the basal and JQ1-induced pgRNA was compromised after the depletion of AFF4 in HepG2.2.15 cells, which suggests that SEC is a major factor in regulating the transcription of HBV (Fig. 4E). We also examined the inhibitory effects of the CDK9 inhibitor flavopiridol in HBV transcription. The results further support the hypothesis that the inhibition of CDK9 can significantly compromise HBV pgRNA transcription and also HBV reactivation by JQ1 (Fig. 4F).
It has been suggested that BRD4 and SEC function synergistically in controlling gene expression, like that of the MYC gene (22). Here, we observed the colocalization of BRD4 and AFF4, the central component of SEC, to the HBV genome. In addition, JQ1 treatment leads to the concomitant increased occupancies of these two factors, and both BRD4 and AFF4 are required for JQ1-induced HBV upregulation. We then asked whether AFF4/SEC is directly recruited to the HBV genome by BRD4 under this condition. We examined AFF4 occupancy after the knockdown of BRD4. The depletion of BRD4 significantly reduced BRD4 occupancy at the HBV genome (Fig. 4G). However, AFF4 occupancy was only slightly affected (Fig. 4H). In summary, these data indicated that SEC and BRD4 function differently in regulating HBV transcription.
DISCUSSION
Transcription from HBV cccDNA to pgRNA is a critical step of HBV replication. Here, we identified a novel mechanism by which different P-TEFb-containing complexes coordinately regulate the transcription of HBV. Both BRD4 and SEC are highly enriched in the HBV genome. BRD4 is recruited to the HBV genome in a bromodomain-independent way. The bromodomain small inhibitor JQ1 treatment leads to the increased occupancy of BRD4 and also SEC components, as well as upregulation of HBV transcription. We also demonstrated that SEC is required for both basal and JQ1-induced HBV transcription. Our studies point out that the potentially fatal complication of HBV reactivation can be induced in inactive HBV carriers who receive the chemotherapy drug JQ1 or its analogs. Meanwhile, our studies also suggest a therapeutic strategy to reduce HBV transcription through inhibiting the activity of SEC.
It has been reported that JQ1 can activate HIV latency in resting CD4+ T cells and Jurkat T cell-based latency models (33–35). Consistent with the role of JQ1 in blocking bromodomain-acetylated histone interactions, JQ1 dislodges BRD4 from the HIV promoter and thus allows Tat to recruit SEC to activate HIV transcription (36). In the current study, we found that JQ1 can also reactivate HBV transcription. Unlike HIV, JQ1 enhances the occupancy of BRD4 at the HBV genome. It is very important to test whether JQ1 can activate latent HBV in the future.
How can JQ1 lead to enhanced occupancy of BRD4 at the HBV genome? A possible explanation is that JQ1 treatment leads to elevated levels of chromatin-free BRD4, which is then relocated to the HBV genome. It suggests that the recruitment of BRD4 to the HBV genome is independent of its bromodomains. A recent study in triple-negative breast cancer revealed a hyperphosphorylation form of BRD4 might be responsible for the bromodomain-independent roles of BRD4 (32). However, it is unclear whether the interrupted interaction of BRD4 with acetylated histones by JQ1 could actually lead to the increased phosphorylation of BRD4. More studies in papillomavirus also showed that the viral protein E2 could directly interact with BRD4 and then recruit it to the viral genome in a bromodomain-independent manner (37, 38). These findings suggest the context-dependent recruitment mechanisms of BRD4.
Besides the BRD4–P-TEFb complex, we found the SEC–P-TEFb complex also localizes at the HBV genome. SEC is required for both the basal expression and JQ1-mediated reactivation of HBV pgRNA suggesting that the transcriptional elongation step plays critical roles in controlling HBV transcription. It has been reported that the treatment of another bromodomain inhibitor, I-BET151, in myeloid leukemia cells can dissociate both BRD4 and SEC components from cancer-related genes, such as MYC, BCL2, and CDK6 (28). Although we did observe the concomitant increase of BRD4 and AFF4 occupancies across the HBV genome after JQ1 treatment, BRD4 does not seem to be a major recruiter for AFF4 at the HBV genome. Our data indicated a dominant role of the SEC–P-TEFb complex in controlling HBV transcription.
MATERIALS AND METHODS
Cell culture.
HepG2.2.15, Huh7, and 293T cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS). The cells were maintained under 5% CO2 at 37°C. JQ1 and IBET151 were dissolved in dimethyl sulfoxide (DMSO) for a stock solution and added to the culture medium fresh on the day of the experiment.
Antibodies.
Antibodies for AFF4, ELL2, and CDK9 were described previously (19). A fragment of human BRD4 (amino acids 1110 to 1270) was expressed as a His tag fusion protein in pET-16b, purified on nitrilotriacetic acid (NTA)-agarose according to Qiagen's protocol, and sent to Pocono Rabbit Farm & Laboratory, Inc., for immunization into rabbits.
Lentivirus-mediated RNAi.
AFF4, BRD2, BRD3, and BRD4 shRNA constructs were purchased from Open Biosystems. The nontargeting shRNA construct (SHC002) was purchased from Sigma. Lentiviral particle preparation and infection were performed as previously described. Briefly, around 70% confluent 293T cells in 150-mm tissue culture plates were cotransfected with 8 μg of the shRNA construct or nontargeting control shRNA, 6 μg of psPAX2 packaging plasmids, and 2 μg of pMD2.G envelope plasmids using X-tremeGENE HP (Roche). The medium was replaced with fresh DMEM supplemented with 10% FBS after 16 h of transfection. The lentiviral supernatants were collected 48 and 72 h after the transfection and filtered through 0.45-μm filters. The HepG2.2.15 cells were infected with filtered lentiviral supernatant with Polybrene (Sigma) at a concentration of 8 μg/ml. Twenty-four hours after infection, the HepG2.2.15 cells were subjected to selection with 2 μg/ml of puromycin for an additional 48 h.
Preparation of full-length HBV DNA.
Linear HBV monomers were prepared as previously described (39). Briefly, plasmid DNAs (full-length HBV genome in TOPO-pCR2.1) were purified using a PureYield plasmid miniprep system (Promega, Madison, WI). After that, linear HBV monomers were released by cleavage with 2 U of BspQI per μg of DNA (New England BioLabs) at 50°C for at least 3 h. Digested DNA was gel purified using a QIAquick gel extraction kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. The DNA was eluted in 50 μl distilled water. The concentration and quality (optical density at 260 nm [OD260]/OD280) of the purified DNA were quantified using a NanoDrop ND-1000 UV-visible spectrophotometer (Thermo Scientific, USA).
Transfection of HBV DNA.
A total of 1 × 105 Huh7 cells were seeded into each well of a 12-well plate and allowed to recover for 24 h before transfection. The well should be 40 to 80% confluent at the day of transfection. Aliquots of 0.375 μg of HBV DNA and 0.125 μg of pSEAP2-Control vector (Clontech) were cotransfected into Huh7 cells using Effectene transfection reagent (Qiagen, Hilden, Germany) at a 1:10 DNA/Effectene ratio. The media were changed to DMEM with 0.5% FBS at 24 h posttransfection. At 48 h posttransfection, the cells were treated with different doses (0, 100, and 250 nM) of JQ1, and the cells were harvested 2 days later. The experiment was done in triplicate.
RT-qPCR.
For RT-qPCR, total RNA was isolated with the RNeasy kit (Qiagen), treated with DNase I (NEB), and repurified with RNeasy. cDNAs were synthesized with a high-capacity RNA-to-cDNA kit from Applied Biosystems. The expression levels of HBV pgRNA and AFF4 were measured with Fast SYBR green master mix (Thermo Fisher) on a StepOnePlus (Applied Biosystems). HBV pgRNA forward and reverse primers were 5′-CTT TTG GAG TGT GGA TTC GC-3′ and 5′-GCG AGG GAG TTC TTC TTC TA-3′, respectively (Sigma-Aldrich). Expression relative to that of the housekeeping gene encoding glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was calculated.
ChIP assay.
A total of 5 × 107 cells were used per ChIP assay according to the previously described protocol (19). Briefly, cells were cross-linked with 1% paraformaldehyde for 10 min at room temperature; cross-linking was quenched by glycine. Fixed chromatin was sonicated and immunoprecipitated with specific antibodies.
HBsAg ELISA.
The amount of HBsAg secreted into the culture medium was detected using a Monolisa HBsAg ultra assay (Bio-Rad Laboratories). The samples were diluted 3× using fresh culture medium before the assay. First, the control sera and the samples were loaded into the wells of the microplate. Fifty microliters of the conjugate solution containing mouse monoclonal anti-HBs antibodies and goat polyclonal anti-HBs antibodies bound to the peroxidase was added. The microplate was covered with an adhesive film and incubated for 90 min at 37°C. At the end of the incubation, the unbound conjugate was removed and the wells were washed with 1× washing solution a minimum of 5 times. One hundred microliters of development solution containing tetramethyl benzidine, citric acid, and sodium acetate solution, pH 4.0, as well as 0.015% H2O2 and 4% DMSO was added to each well. The plate was incubated in the dark for 30 min at room temperature, and the reaction was stopped by addition of 100 μl stopping solution (1 N sulfuric acid solution). The optical density was read at 450 nm using 650 nm as the reference wavelength. A serial dilution of the positive control was used to generate the relative standard curve.
Extraction of core-associated HBV DNA.
Culture medium was collected and briefly centrifuged at 800 × g for 5 min to pellet any suspended cell debris. The supernatant was transferred to a clean tube with 10% polyethylene glycol solution and 1.75 M sodium chloride. The solution was incubated overnight at 4°C to allow the virus particles to precipitate. The next day, the solution was centrifuged at 7,000 × g for 10 min at 4°C to pellet the precipitated virions. The virus pellet was resuspended in sterile water and then treated with DNase I (10 U per 500 μl culture medium) for 30 min at 37°C to remove any DNA not enclosed in a capsid. The reaction was stopped by adding 25 mM EDTA. A QIAamp DNA minikit was then used to extract the HBV DNA contained within the purified viral particles, eluted in water, and then quantified via standard curve qPCR.
ACKNOWLEDGMENTS
We thank Toby Wai Kiat Chin for generating BRD4 antibody. We are grateful to James E. Bradner for providing JQ1 for this study.
This work was supported by funds provided by the Singapore A*STAR IMCB core fund (MR05010), the NMRC TCR flagship program (Eradication of HBV TCR Program, NMRC/TCR/014-NUHS/2015), and the BMRC YIG fund (BMRC024), as well as the Natural Science Foundation of Jiangsu Province of China (BK20160026), all to C.L.
REFERENCES
- 1.Lavanchy D. 2004. Hepatitis B virus epidemiology, disease burden, treatment, and current and emerging prevention and control measures. J Viral Hepat 11:97–107. doi: 10.1046/j.1365-2893.2003.00487.x. [DOI] [PubMed] [Google Scholar]
- 2.Dienstag JL. 2008. Hepatitis B virus infection. N Engl J Med 359:1486–1500. doi: 10.1056/NEJMra0801644. [DOI] [PubMed] [Google Scholar]
- 3.Seeger C, Mason WS. 2015. Molecular biology of hepatitis B virus infection. Virology 479-480:672–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kaplan PM, Greenman RL, Gerin JL, Purcell RH, Robinson WS. 1973. DNA polymerase associated with human hepatitis B antigen. J Virol 12:995–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Delius H, Gough NM, Cameron CH, Murray K. 1983. Structure of the hepatitis B virus genome. J Virol 47:337–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sureau C, Romet-Lemonne JL, Mullins JI, Essex M. 1986. Production of hepatitis B virus by a differentiated human hepatoma cell line after transfection with cloned circular HBV DNA. Cell 47:37–47. doi: 10.1016/0092-8674(86)90364-8. [DOI] [PubMed] [Google Scholar]
- 7.Liu B, Wen X, Huang C, Wei Y. 2013. Unraveling the complexity of hepatitis B virus: from molecular understanding to therapeutic strategy in 50 years. Int J Biochem Cell Biol 45:1987–1996. doi: 10.1016/j.biocel.2013.06.017. [DOI] [PubMed] [Google Scholar]
- 8.Bock CT, Schwinn S, Locarnini S, Fyfe J, Manns MP, Trautwein C, Zentgraf H. 2001. Structural organization of the hepatitis B virus minichromosome. J Mol Biol 307:183–196. doi: 10.1006/jmbi.2000.4481. [DOI] [PubMed] [Google Scholar]
- 9.Newbold JE, Xin H, Tencza M, Sherman G, Dean J, Bowden S, Locarnini S. 1995. The covalently closed duplex form of the hepadnavirus genome exists in situ as a heterogeneous population of viral minichromosomes. J Virol 69:3350–3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pollicino T, Belloni L, Raffa G, Pediconi N, Squadrito G, Raimondo G, Levrero M. 2006. Hepatitis B virus replication is regulated by the acetylation status of hepatitis B virus cccDNA-bound H3 and H4 histones. Gastroenterology 130:823–837. doi: 10.1053/j.gastro.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 11.Belloni L, Pollicino T, De Nicola F, Guerrieri F, Raffa G, Fanciulli M, Raimondo G, Levrero M. 2009. Nuclear HBx binds the HBV minichromosome and modifies the epigenetic regulation of cccDNA function. Proc Natl Acad Sci U S A 106:19975–19979. doi: 10.1073/pnas.0908365106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lucifora J, Arzberger S, Durantel D, Belloni L, Strubin M, Levrero M, Zoulim F, Hantz O, Protzer U. 2011. Hepatitis B virus X protein is essential to initiate and maintain virus replication after infection. J Hepatol 55:996–1003. doi: 10.1016/j.jhep.2011.02.015. [DOI] [PubMed] [Google Scholar]
- 13.Liaw YF, Kao JH, Piratvisuth T, Chan HL, Chien RN, Liu CJ, Gane E, Locarnini S, Lim SG, Han KH, Amarapurkar D, Cooksley G, Jafri W, Mohamed R, Hou JL, Chuang WL, Lesmana LA, Sollano JD, Suh DJ, Omata M. 2012. Asian-Pacific consensus statement on the management of chronic hepatitis B: a 2012 update. Hepatol Int 6:531–561. doi: 10.1007/s12072-012-9365-4. [DOI] [PubMed] [Google Scholar]
- 14.Zoulim F, Lebosse F, Levrero M. 2016. Current treatments for chronic hepatitis B virus infections. Curr Opin Virol 18:109–116. doi: 10.1016/j.coviro.2016.06.004. [DOI] [PubMed] [Google Scholar]
- 15.Ahmed M, Wang F, Levin A, Le C, Eltayebi Y, Houghton M, Tyrrell L, Barakat K. 2015. Targeting the Achilles heel of the hepatitis B virus: a review of current treatments against covalently closed circular DNA. Drug Discov Today 20:548–561. doi: 10.1016/j.drudis.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 16.Summers J, Mason WS. 1982. Replication of the genome of a hepatitis B–like virus by reverse transcription of an RNA intermediate. Cell 29:403–415. doi: 10.1016/0092-8674(82)90157-X. [DOI] [PubMed] [Google Scholar]
- 17.Bozza C, Cinausero M, Iacono D, Puglisi F. 2016. Hepatitis B and cancer: a practical guide for the oncologist. Crit Rev Oncol Hematol 98:137–146. doi: 10.1016/j.critrevonc.2015.10.017. [DOI] [PubMed] [Google Scholar]
- 18.Peterlin BM, Price DH. 2006. Controlling the elongation phase of transcription with P-TEFb. Mol Cell 23:297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 19.Lin C, Smith ER, Takahashi H, Lai KC, Martin-Brown S, Florens L, Washburn MP, Conaway JW, Conaway RC, Shilatifard A. 2010. AFF4, a component of the ELL/P-TEFb elongation complex and a shared subunit of MLL chimeras, can link transcription elongation to leukemia. Mol Cell 37:429–437. doi: 10.1016/j.molcel.2010.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin C, Garrett AS, De Kumar B, Smith ER, Gogol M, Seidel C, Krumlauf R, Shilatifard A. 2011. Dynamic transcriptional events in embryonic stem cells mediated by the super elongation complex (SEC). Genes Dev 25:1486–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luo Z, Lin C, Guest E, Garrett AS, Mohaghegh N, Swanson S, Marshall S, Florens L, Washburn MP, Shilatifard A. 2012. The super elongation complex family of RNA polymerase II elongation factors: gene target specificity and transcriptional output. Mol Cell Biol 32:2608–2617. doi: 10.1128/MCB.00182-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luo Z, Lin C, Shilatifard A. 2012. The super elongation complex (SEC) family in transcriptional control. Nat Rev Mol Cell Biol 13:543–547. doi: 10.1038/nrm3417. [DOI] [PubMed] [Google Scholar]
- 23.Mohan M, Lin C, Guest E, Shilatifard A. 2010. Licensed to elongate: a molecular mechanism for MLL-based leukaemogenesis. Nat Rev Cancer 10:721–728. doi: 10.1038/nrc2915. [DOI] [PubMed] [Google Scholar]
- 24.Jang MK, Mochizuki K, Zhou M, Jeong HS, Brady JN, Ozato K. 2005. The bromodomain protein Brd4 is a positive regulatory component of P-TEFb and stimulates RNA polymerase II-dependent transcription. Mol Cell 19:523–534. doi: 10.1016/j.molcel.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 25.Yang Z, Yik JH, Chen R, He N, Jang MK, Ozato K, Zhou Q. 2005. Recruitment of P-TEFb for stimulation of transcriptional elongation by the bromodomain protein Brd4. Mol Cell 19:535–545. doi: 10.1016/j.molcel.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 26.Loven J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, Bradner JE, Lee TI, Young RA. 2013. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 153:320–334. doi: 10.1016/j.cell.2013.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zuber J, Shi J, Wang E, Rappaport AR, Herrmann H, Sison EA, Magoon D, Qi J, Blatt K, Wunderlich M, Taylor MJ, Johns C, Chicas A, Mulloy JC, Kogan SC, Brown P, Valent P, Bradner JE, Lowe SW, Vakoc CR. 2011. RNAi screen identifies Brd4 as a therapeutic target in acute myeloid leukaemia. Nature 478:524–528. doi: 10.1038/nature10334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dawson MA, Prinjha RK, Dittmann A, Giotopoulos G, Bantscheff M, Chan WI, Robson SC, Chung CW, Hopf C, Savitski MM, Huthmacher C, Gudgin E, Lugo D, Beinke S, Chapman TD, Roberts EJ, Soden PE, Auger KR, Mirguet O, Doehner K, Delwel R, Burnett AK, Jeffrey P, Drewes G, Lee K, Huntly BJ, Kouzarides T. 2011. Inhibition of BET recruitment to chromatin as an effective treatment for MLL-fusion leukaemia. Nature 478:529–533. doi: 10.1038/nature10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sells MA, Chen ML, Acs G. 1987. Production of hepatitis B virus particles in Hep G2 cells transfected with cloned hepatitis B virus DNA. Proc Natl Acad Sci U S A 84:1005–1009. doi: 10.1073/pnas.84.4.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dey A, Chitsaz F, Abbasi A, Misteli T, Ozato K. 2003. The double bromodomain protein Brd4 binds to acetylated chromatin during interphase and mitosis. Proc Natl Acad Sci U S A 100:8758–8763. doi: 10.1073/pnas.1433065100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schweiger MR, Ottinger M, You J, Howley PM. 2007. Brd4-independent transcriptional repression function of the papillomavirus e2 proteins. J Virol 81:9612–9622. doi: 10.1128/JVI.00447-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shu S, Lin CY, He HH, Witwicki RM, Tabassum DP, Roberts JM, Janiszewska M, Huh SJ, Liang Y, Ryan J, Doherty E, Mohammed H, Guo H, Stover DG, Ekram MB, Peluffo G, Brown J, D'Santos C, Krop IE, Dillon D, McKeown M, Ott C, Qi J, Ni M, Rao PK, Duarte M, Wu SY, Chiang CM, Anders L, Young RA, Winer EP, Letai A, Barry WT, Carroll JS, Long HW, Brown M, Liu XS, Meyer CA, Bradner JE, Polyak K. 2016. Response and resistance to BET bromodomain inhibitors in triple-negative breast cancer. Nature 529:413–417. doi: 10.1038/nature16508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Banerjee C, Archin N, Michaels D, Belkina AC, Denis GV, Bradner J, Sebastiani P, Margolis DM, Montano M. 2012. BET bromodomain inhibition as a novel strategy for reactivation of HIV-1. J Leukoc Biol 92:1147–1154. doi: 10.1189/jlb.0312165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bartholomeeusen K, Xiang Y, Fujinaga K, Peterlin BM. 2012. Bromodomain and extra-terminal (BET) bromodomain inhibition activate transcription via transient release of positive transcription elongation factor b (P-TEFb) from 7SK small nuclear ribonucleoprotein. J Biol Chem 287:36609–36616. doi: 10.1074/jbc.M112.410746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu J, Gaiha GD, John SP, Pertel T, Chin CR, Gao G, Qu H, Walker BD, Elledge SJ, Brass AL. 2012. Reactivation of latent HIV-1 by inhibition of BRD4. Cell Rep 2:807–816. doi: 10.1016/j.celrep.2012.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Z, Guo J, Wu Y, Zhou Q. 2013. The BET bromodomain inhibitor JQ1 activates HIV latency through antagonizing Brd4 inhibition of Tat-transactivation. Nucleic Acids Res 41:277–287. doi: 10.1093/nar/gks976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schweiger MR, You J, Howley PM. 2006. Bromodomain protein 4 mediates the papillomavirus E2 transcriptional activation function. J Virol 80:4276–4285. doi: 10.1128/JVI.80.9.4276-4285.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McPhillips MG, Oliveira JG, Spindler JE, Mitra R, McBride AA. 2006. Brd4 is required for e2-mediated transcriptional activation but not genome partitioning of all papillomaviruses. J Virol 80:9530–9543. doi: 10.1128/JVI.01105-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gunther S, Li BC, Miska S, Kruger DH, Meisel H, Will H. 1995. A novel method for efficient amplification of whole hepatitis B virus genomes permits rapid functional analysis and reveals deletion mutants in immunosuppressed patients. J Virol 69:5437–5444. [DOI] [PMC free article] [PubMed] [Google Scholar]