ABSTRACT
Global histone hyperacetylation is suggested to play a critical role for replacement of histones by transition proteins and protamines to compact the genome during spermiogenesis. However, the underlying mechanisms for hyperacetylation-mediated histone replacement remains poorly understood. Here, we report that EPC1 and TIP60, two critical components of the mammalian nucleosome acetyltransferase of H4 (NuA4) complexes, are coexpressed in male germ cells. Strikingly, genetic ablation of either Epc1 or Tip60 disrupts hyperacetylation and impairs histone replacement, in turn causing aberrant spermatid development. Taking these observations together, we reveal an essential role of the NuA4 complexes for histone hyperacetylation and subsequent compaction of the spermatid genome.
KEYWORDS: EPC1, TIP60, histone acetylation, spermiogenesis, spermatids, mouse
INTRODUCTION
Gametogenesis is a critical step for transmitting both genetic and epigenetic information to the next generation and is regulated in an asymmetric manner between males and females. In mammals, this asymmetry is partly represented by mechanisms to convey the respective haploid genomes by using either nucleoprotamines or nucleosomes. Indeed, most of the sperm genome is intensively compacted by protamines (PRMs) while in oocytes the genome retains nucleosomes. Global replacement of histones by PRMs occurring only in male germ cells during spermiogenesis (the postmeiotic phase of spermatogenesis) plays a role in establishing such asymmetry.
During spermiogenesis, male germ cells undergo sequential changes in cell morphology and condensation of the nucleus as represented by the stepwise emergence of round, elongating, condensing, and condensed spermatids (1). Spermatid development in mice is arbitrarily divided into 16 steps based on cell shapes and structures of the nuclei and acrosome, as schematically summarized in Fig. 1A. Spermatids of steps 1 to 8, which are also designated round spermatids (RSs), possess sphere-shaped cells and nuclei. The acrosome is recognized as early as step 3 as a vesicle and develops to form a cap-like structure covering a prospective apical hemisphere of the nucleus by step 8. Elongating spermatids (ESs) of steps 9 to 11 exhibit elongation of the nucleus and concomitant extension of the acrosome along the dorsal surface of the nucleus. Replacement of histones by transition proteins (TNPs) and subsequently by PRMs occurs in condensing spermatids of steps 12 to 14. In steps 15 and 16 condensed spermatids (spermatozoa) exhibit a typical hook type head morphology and are ready to be released into the lumen of seminiferous tubules. In the RSs of steps 1 to 8, histones function as the major basic nuclear proteins and play a role to maintain active transcriptional states (2, 3). Upon the commencement of nuclear elongation, both somatic and testis-specific histones are replaced first with TNPs and later with PRMs to achieve a spermatozoon-specific nuclear structure (4, 5). Histone hyperacetylation temporally precedes histone replacement and emerges in step 8 RSs, just prior to the beginning of nuclear elongation. Although the hyperacetylation marks are retained in ESs, these mostly disappear in the condensing spermatids (6).
FIG 1.
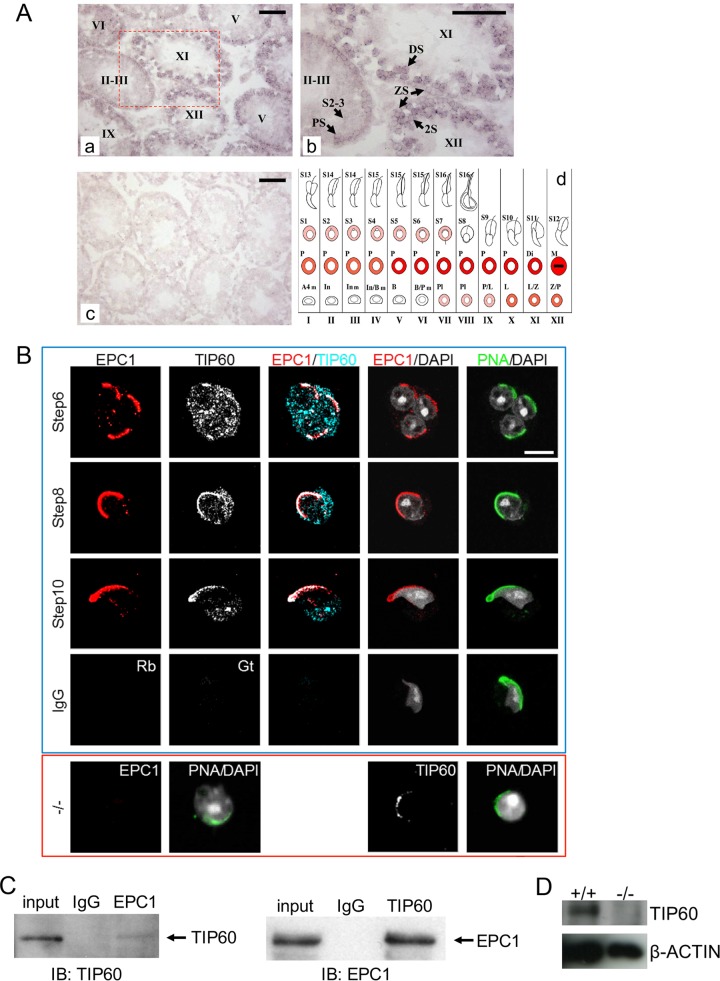
Colocalization and physical interaction of EPC1 and TIP60 in male germ cells. (A) The expression of Epc1 in adult mouse (about 8 weeks old) testis revealed by in situ hybridization. Digoxigenin-labeled Epc1 antisense (a and b) and sense (c; a negative control) probes were used. Stages of the seminiferous tubules were determined by staining the adjacent sections with PAS and hematoxylin and are indicated by roman numerals in panels a, b, and d. Panel b represents an enlarged view for the boxed region in panel a. Epc1 was transcribed in a stage-specific manner and mainly in germ cells in the seminiferous tubules. Epc1 transcription was first observed in preleptotene spermatocytes. The signal was especially strong in primary spermatocytes after the mid-pachytene stage and then appeared to decline in round spermatids. PS, pachytene spermatocyte; ZS, zygotene spermatocyte; DS, diplotene spermatocyte; 2S, secondary spermatocyte; S2-3, spermatid steps 2 and 3. Scale bars, 50 μm. Panel d is a schematic summary of Epc1 expression during spermatogenesis. The relative Epc1 expression level at the respective developmental stage is represented by the intensity of the red. The following abbreviations were used to indicate developmental stages: A4, A4 spermatogonia; In, intermediate spermatobonia; B, B-type spermatogonia; Pl, preleptotene spermatocyte; L, leptotene spermatocyte; Z, zygotene spermatocyte; P, pachytene spermatocyte; Di, diplotene spermatocyte; M, metaphase; S1 to S16, step 1 to 16 spermatids. (B) Colocalization of EPC1 and TIP60 in round and elongating spermatids at steps 6, 8, and 10. Immunofluorescence staining was performed using surface-spread slides of wild-type germ cells to visualize the distribution of EPC1 and TIP60 in samples shown in the top (blue-lined) box. Fluorescein isothiocyanate (FITC)-conjugated PNA was used to demarcate the acrosome for spermatid staging. There is a specific accumulation of both EPC1 and TIP60 at the apical polar region from the round spermatids of step 6 to elongating spermatids. IgG fractions (IgG) from normal rabbit (Rb) and goat (Gt) serum were used as negative controls. For the images shown in the lower (red-lined) box, we also used Epc1-KO spermatids (−/−) to show specificity of the anti-EPC1 (using a step 6 round spermatid) and examine the impact of EPC1 loss on TIP60 distribution (using a step 7 round spermatid). Scale bar, 5 μm. (C) Physical interaction of EPC1 and TIP60 in male germ cells. Whole-cell lysates from wild-type adult male germ cells (input) were immunoprecipitated by control IgG, anti-EPC1 (left), or anti-TIP60 (right) and subjected to immunoblotting with anti-TIP60 or anti-EPC1. (D) Reduced TIP60 levels in Epc1-KO male germ cells. Whole-cell lysates of wild type (+/+) or Epc1-KO (−/−) germ cells were subjected to immunoblotting.
From an evolutionary point of view, histone hyperacetylation in spermatids is closely linked to histone replacement by TNPs and PRMs. Histone hyperacetylation has been observed in species, including roosters, trout, rodents, and humans, all of which exhibit replacement of histones by PRMs (7–12), while in other vertebrates such as carp and winter flounder, histones remain underacetylated throughout spermiogenesis and are not replaced by PRMs (13). Consistent with these views, biochemical studies have shown that hyperacetylated histones are more amenable to replacement by PRMs than underacetylated ones (14, 15). Collectively, these previous insights indicate that histone hyperacetylation could be a critical switch for a transition from nucleosomes to nucleoprotamines. This model is further supported by the recent discovery of a testis-specific bromodomain protein, BRDT (bromodomain, testis-specific), which recognizes acetylated histones and contributes to the deposition of TNPs and PRMs (16). A plethora of evidence gained from previous studies indicates a role of histone acetylation for destabilization and remodeling of nucleosomes (17–19). Thus, histone hyperacetylation mediated by specific histone acetyltransferases (HATs) during spermiogenesis could also facilitate histone replacement per se or augment the accessibility of factors involved in this process. Although recent studies have identified a testis-specific HAT, CDYL (chromodomain protein, Y-chromosome-like), expressed in ESs (20, 21), the detailed molecular mechanism that regulates histone hyperacetylation during spermiogenesis is poorly understood.
Histone hyperacetylation in ESs occurs globally at lysine residues in histone tails (6) and accompanies transient accumulation of double-strand breaks (22, 23). Interestingly, previous studies show that the piccolo nucleosome acetyltransferase of H4 (NuA4) complex formed by Epl1, Esa1, and Yng2 in Saccharomyces cerevisiae plays a role in preferentially mediating global rather than local histone acetylation (24). Importantly, their orthologues, namely, EPC1 (an orthologue of Drosophila Enhancer of Polycomb), TIP60 (KAT5; lysine acetyltransferase 5), and ING3 (inhibitor of growth family, member 3), are conserved in mammals and are components of the mammalian NuA4 complex (25). Given that Tip60 is also transcribed during spermiogenesis (26), we hypothesized that the NuA4 complex could have a role in regulating hyperacetylation, followed by global replacement of histones by PRMs during spermiogenesis.
Consistent with this notion, in this study, we report that EPC1 and TIP60 colocalize at the nuclear periphery near the acrosomes in both RSs and ESs. Furthermore, deletion of Epc1 results in arrest of spermiogenesis predominantly at the transition from RS to ES, coincident with a significant reduction in spermatids exhibiting histone hyperacetylation. Similarly, genetic ablation of Tip60 causes reduced levels of histone acetylation in ESs. Based on these findings, we suggest a crucial involvement of the NuA4-related complexes to mediate histone hyperacetylation in RSs and ESs to promote spermiogenesis in mammals.
RESULTS
The expression of EPC1 and TIP60 during spermiogenesis.
To gain insight into the roles of the mammalian NuA4 complex during spermiogenesis, we first examined the expression of Epc1 in adult testis by in situ hybridization. Epc1 transcription was first observed in preleptotene spermatocytes and peaked after the mid-pachytene stage but declined again in RSs (Fig. 1A). These results revealed that Epc1 as well as Tip60 was expressed during spermiogenesis (26). We then performed multicolor immunofluorescence (IF) analyses to analyze the expression of EPC1 and TIP60 proteins in postmeiotic spermatids. We observed weak EPC1 signals in the cytoplasm and nucleus in RSs of steps 1 to 4 (data not shown) and an apical polar cap-like distribution near the acrosome (demarcated by peanut agglutinin [PNA]) from step 5 onward (Fig. 1B). A similar distribution of EPC1 near the acrosome was seen in ESs (Fig. 1B, step 10). The Epc1 knockout (Epc1-KO) RSs did not show any detectable signals (Fig. 1B, bottom left), verifying the specificity of signals.
In addition, we detected TIP60 in both cytoplasm and nucleus in RSs of steps 1 to 5 (data not shown). Following EPC1 accumulation at the nuclear periphery near the acrosome in step 5 RSs, TIP60 intensively colocalized with EPC1 from step 6 RSs onward (Fig. 1B). This observation implied that the perinuclear distribution of TIP60 follows that of EPC1, which is consistent with previous reports revealing the association of TIP60 with an acrosomal structure protein, Dickkopf-like 1 (DKKL1) (27). We carried out immunoprecipitation (IP) and immunoblotting (IB) analyses and found that EPC1 and TIP60 physically interacted with each other in the testis (Fig. 1C). We further examined whether EPC1 influenced the expression and/or localization of TIP60 by using Epc1-KO RSs and indeed observed a considerable reduction in TIP60 levels although the protein retained its localization near the acrosome (Fig. 1B, bottom right, and D). Taken together, these observations indicate that EPC1 and TIP60 colocalized and interacted with each other at the nuclear periphery near the acrosome in late RSs and ESs. This apical localization is consistent with the previously reported polarity of chromatin condensation and replacement (4, 6). Since histone hyperacetylation starts at around step 8, these observations support a role for EPC1/TIP60 complexes in this process.
Defects in generation of elongating spermatids in Epc1-KO mice.
To examine the role of EPC1 in spermatogenesis, we generated Epc1-KO mice (Fig. 2A and B). Homozygous Epc1-KO mutants were viable but exhibited growth retardation, homeotic transformations of the axis, and sterility in both males and females although the heterozygotes were apparently normal (data not shown). We also noted that the testis weight of adult Epc1-KO mice in reference to their body weight was disproportionally lower than that of the Epc1+/− mice (Fig. 2C).
FIG 2.
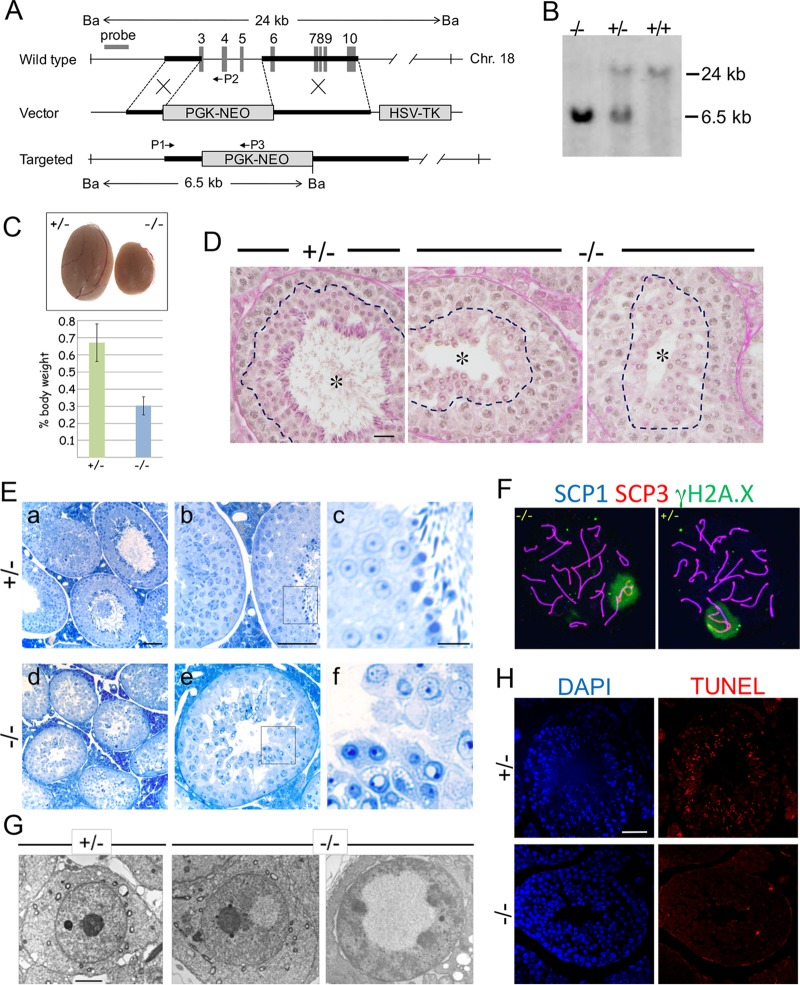
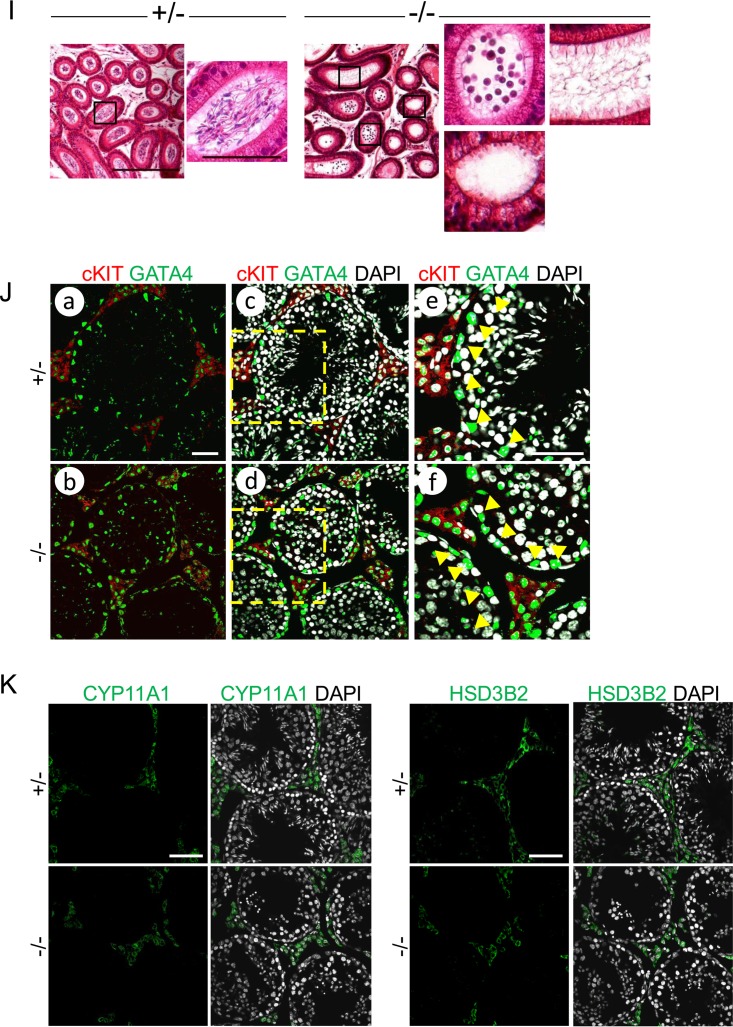
Impaired postmeiotic germ cell maturation of Epc1-KO spermatids. (A) Schematic representation of the strategy to generate an Epc1-KO allele and its genomic configuration. Black boldface bars delineate the 5′ and 3′ homology arms for Epc1 targeting. The targeting vector was designed to replace the genomic region encompassing exons 3 to 6 with a PGK-NEO cassette. Ba, BamHI restriction sites for Southern blot analysis; HSV, herpes simplex virus; TK, thymidine kinase. (B) Confirmation of the knockout mouse genotype by Southern blot analysis. (C) Testes in adult Epc1-KO mice are considerably smaller than those in the heterozygotes. Testes from littermates at 16 weeks after birth are shown (top). Relative testis weights (bottom) as a percentage of body weight of adult Epc1-KO mice (n = 8) or their heterozygous littermates (n = 5) are graphically depicted (P = 5.16E−06). (D) PAS and hematoxylin staining of Epc1+/− (+/−) and Epc1-KO (−/−) testis revealed the presence of the round spermatids and lack of elongating, condensing, or condensed spermatids. Seminiferous tubule lumens are indicated by asterisks. PAS-positive regions are indicated by dotted lines. Scale bar, 50 μm. (E) Toluidine blue staining of testis sections of Epc1-KO mice revealed maturation arrest mainly at the round spermatid stage and lack of elongating spermatids. Panels c and f represent enlarged views of the boxed regions in panels b and e, respectively. Scale bars, 50 μm (a, b, d, and e) and 10 μm (c and f). (F) Synapsis and sex chromosome inactivation in Epc1-KO pachytene spermatocytes. IF staining was performed using surface-spread slides of Epc1-KO (−/−) and Epc1+/− (+/−) germ cells to visualize the distribution of SCP1 (blue), SCP3 (red), and γH2A.X (green). (G) Electron microscopy revealed vacuoles where chromatin is excluded in Epc1-KO round spermatids. Scale bar, 2 μm. (H) Lack of TUNEL-labeled RSs in Epc1-KO (−/−) seminiferous tubules. Note that a considerable number of condensing and condensed spermatids are labeled in Epc1+/− (+/−) tubules. Scale bar, 50 μm. (I) Epc1-KO (−/−) epididymis lumens were empty or contained round spermatids while mature spermatozoa were abundantly observed in the Epc1+/− (+/−) epididymis. Enlarged views of boxed regions in the images at left are shown on the right. Scale bars, 200 μm (left) and 50 μm (right). (J) The expression of GATA4 and c-Kit in Epc1+/− (+/−) and Epc1-KO (−/−) seminiferous tubules, as indicated. Enlarged views of boxed regions in panels c and d are shown in panels e and f, respectively. Note that GATA4-positive Sertoli cells (indicated by arrowheads in panels e and f) are aligned at the basal layer of respective seminiferous tubules, and their distributions are similar between Epc1+/− and Epc1-KO mice. The distribution of Leydig cells demarcated by coexpression of GATA4 (green) and c-Kit (red) is not considerably changed in Epc1-KO mice compared with that in Epc1+/− mice. Scale bars, 50 μm. (K) The expression of CYP11A1 or HSD3B2 in Epc1+/− (+/−) and Epc1-KO (−/−) seminiferous tubules. Note that the expression of CYP11A1 or HSD3B2 in Leydig cells is not considerably changed in Epc1-KO seminiferous tubules compared with that in Epc1+/− seminiferous tubules. Scale bars, 50 μm.
We next carried out histological analyses of mutant testes during adult spermatogenesis. Consistent with sterility, histological analysis of adult testis sections after periodic acid-Schiff (PAS)–hematoxylin staining revealed a lack of spermatozoa in Epc1-KO testis though RSs were generated (Fig. 2D). This was further tested by using thin sections with toluidine blue staining (Fig. 2E). Seminiferous tubules in the Epc1-KO testis were slightly thinner than those of the heterozygotes (Fig. 2E, panels a and d), and the pachytene spermatocytes exhibited premature detachment from Sertoli cells and the adjacent germ cells and a failure to form a properly layered architecture (Fig. 2E, panels b and e). Despite such morphological alterations in premeiotic stages, we observed that Epc1-KO germ cells developed into the RS stage over meiotic divisions (Fig. 2E, panels c and f; also unpublished observations). Consistently, we observed that synapsis and sex chromosome inactivation occurred normally in Epc1-KO pachytene spermatocytes, as revealed by IF analysis on germ cell spreads (Fig. 2F). We did not, however, find elongating, condensing, or condensed spermatids or mature spermatozoa in Epc1-KO seminiferous tubules, suggesting the presence of developmental defects of Epc1-KO male germ cells prior to or around RS-to-ES transition (Fig. 2E, panel f). Consistent with this, we found that a considerable number of Epc1-KO RSs possessed subnuclear vacuoles that were never seen in Epc1+/− or wild-type RSs (Fig. 2E, panel f). Vacuolation of Epc1-KO RSs was further confirmed by electron microscopic analysis (Fig. 2G). Emergence of vacuoles seems to affect the position or morphology of the nucleolus. These observations suggest a role for EPC1 in the maintenance of RSs and/or their differentiation of ESs.
To test this hypothesis, we checked whether Epc1-KO RSs and/or ESs were eliminated by apoptotic outbursts using terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) analysis (Fig. 2H). We did not, however, find an obvious increase of apoptotic bursts in Epc1-KO RSs. In contrast, condensing and condensed spermatids in Epc1+/− tubules were clearly labeled, likely due to physiological emergence of DNA nicks. Intriguingly, we observed RSs trafficking in the epididymis of Epc1-KO mice while spermatozoa were absent (Fig. 2I). These observations suggest that Epc1-KO RSs failed to differentiate into ESs and were prematurely released into the lumen at the RS stage.
We finally evaluated the impacts of knocking out Epc1 on the development of testicular somatic cells that support spermatogenesis. We used GATA4 and c-Kit as markers to detect Sertoli and Leydig cells in adult testis and did not find obvious changes in their distributions in Epc1-KO seminiferous tubules compared with distributions in Epc1+/− testis (Fig. 2J). We further examined the expression of CYP11A1 (cytochrome P450, family 11, subfamily a, polypeptide 1) and HSD3B2 (hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 2), which are known to mediate steroidogenesis in Leydig cells. We did not detect considerable differences in their expression levels in Leydig cells of Epc1-KO and Epc1+/− testes (Fig. 2K). These observations suggest that EPC1 intrinsically contributes to maintain the developmental potential of RSs or to promote RS differentiation to generate ES.
Activation of ES-related genes in Epc1-KO RSs.
We then investigated how functional properties were altered in Epc1-KO RSs by using a round-spermatid injection (ROSI) technique. We collected 84 Epc1-KO RSs and injected them into oocytes. More than 90% of oocytes survived injection and formed pronuclei, and more than 70% of zygotes developed into 2 cells after 24 h in culture, irrespective of their genotypes (Table 1). After transfer of these 2-cell embryos into the oviducts of recipient females, normal pups were obtained at term in both genotype groups (Fig. 3A), indicating that a considerable fraction of Epc1-KO RSs can support full-term fetal development. However, we also found subtle but considerable differences in the implantation and birth rates between the Epc1-KO and wild-type RSs (Table 1) (P < 0.05). Together these data indicate that EPC1 is dispensable for spermatid development to the RS stage but is required to maintain spermatid functions.
TABLE 1.
Development of ROSI-generated embryos and fetuses from Epc1+/− or Epc1−/− male mice
| Mouse genotype | No. of oocytes cultured | No. (%) of 2-cell embryos | No. of embryos transferred | No. (%) of embryos implanteda | No. (%) of pupsa |
|---|---|---|---|---|---|
| Epc1+/− | 84 | 62 (74) | 62 | 37 (60) A | 20 (32) A |
| Epc1−/− | 84 | 68 (81) | 68 | 20 (29) B | 9 (13) B |
Values with different letters within the same column are significantly different (P < 0.05, Fisher's exact test).
FIG 3.
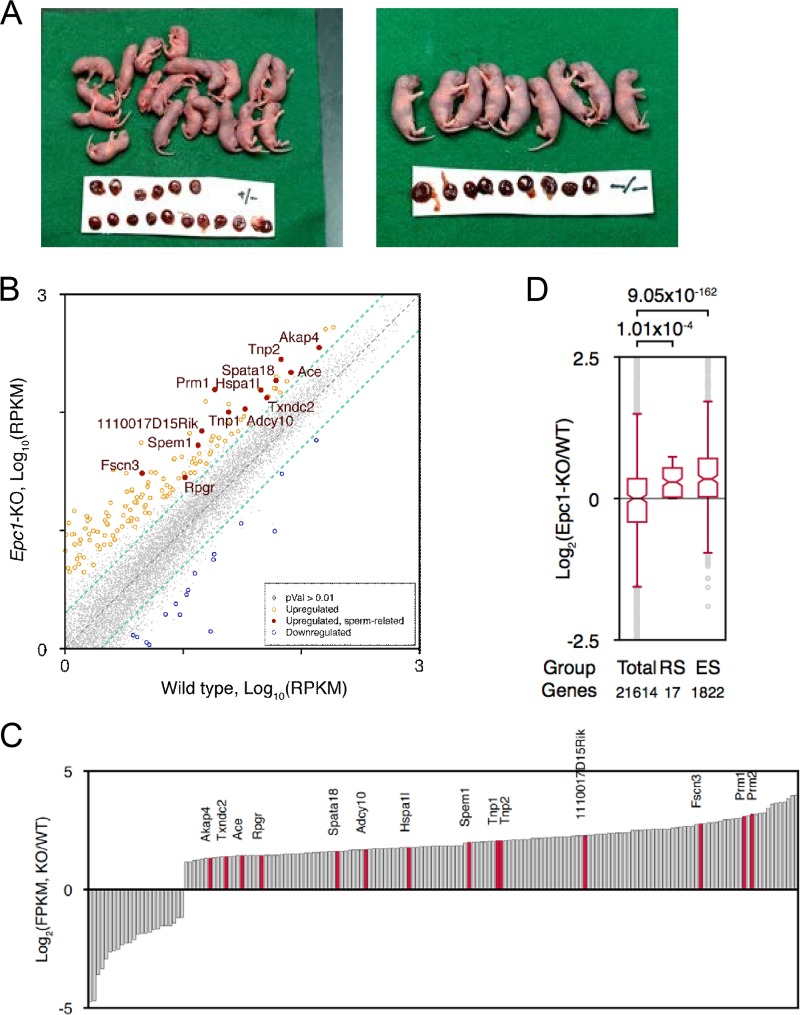
Latent developmental activity of Epc1-KO spermatids. (A) Round spermatid injection (ROSI) analysis of Epc1+/− or Epc1-KO round spermatids. ROSI-mediated perinatal fetuses and placentas recovered by caesarean section at 19.5 days postcoitus are shown. The mean body weight of pups derived from RSs of Epc1-KO mice (1.88 ± 0.04 g) was considerably more than that of Epc1+/− mice (1.74 ± 0.04 g) (P < 0.01). This is speculated to be a secondary result of the difference in the litter size (2.3 from Epc1-KO RSs versus 5.0 from Epc1+/−, on average). (B) A scatter plot diagram comparing gene expression profiles between Epc1-KO and wild-type RSs. Genes expressed 2-fold more or less (P value of <0.01) in Epc1-KO RSs are indicated in orange and red or in blue, respectively. Genes annotated as sperm- or spermatid-related genes by gene ontology are with red circles and gene names. RPKM, reads per kilobase exon per million reads; gray dots, genes not statistically changed. (C) A bar chart representation to show differences of respective gene expression levels between Epc1-KO and wild-type (WT) RSs. (D) A box plot view to show differences of average gene expression levels between Epc1-KO and wild-type RSs of total (21,614 genes), RS-specific (17 genes), and ES-specific (1,822 genes) genes.
We next investigated gene expression profiles of Epc1-KO RSs. High-throughput RNA sequencing (RNA-seq) analysis of isolated Epc1-KO and wild-type RSs revealed considerable similarity between them (correlation coefficient, 0.962) but also upregulation of about 160 genes, including sperm- or spermatid-related genes such as Prm1, Prm2, Tnp1, and Tnp2 (Fig. 3B and C). We went on to estimate the average expression levels of genes that are expressed in RSs and ESs in a stage-specific manner. Based on RNA-seq data for male germ cells at different stages during the first wave of spermatogenesis (28), we first defined 28 and 2,390 genes that are more abundantly expressed at RS and ES stages than at premeiotic or other meiotic stages, respectively, and compared these gene expression levels in Epc1-KO RSs against those of the wild type. We indeed observed considerable upregulation of RS- and ES-specific genes in Epc1-KO RSs (Fig. 3D). EPC1 is thus dispensable for transcriptional activation of genes expressed at the ES stage. We thus suggest that defects in ES generation in Epc1-KO mice could involve mechanisms other than activation of ES-specific genes in RSs.
EPC1-dependent histone acetylation is linked to the transition from RS to ES.
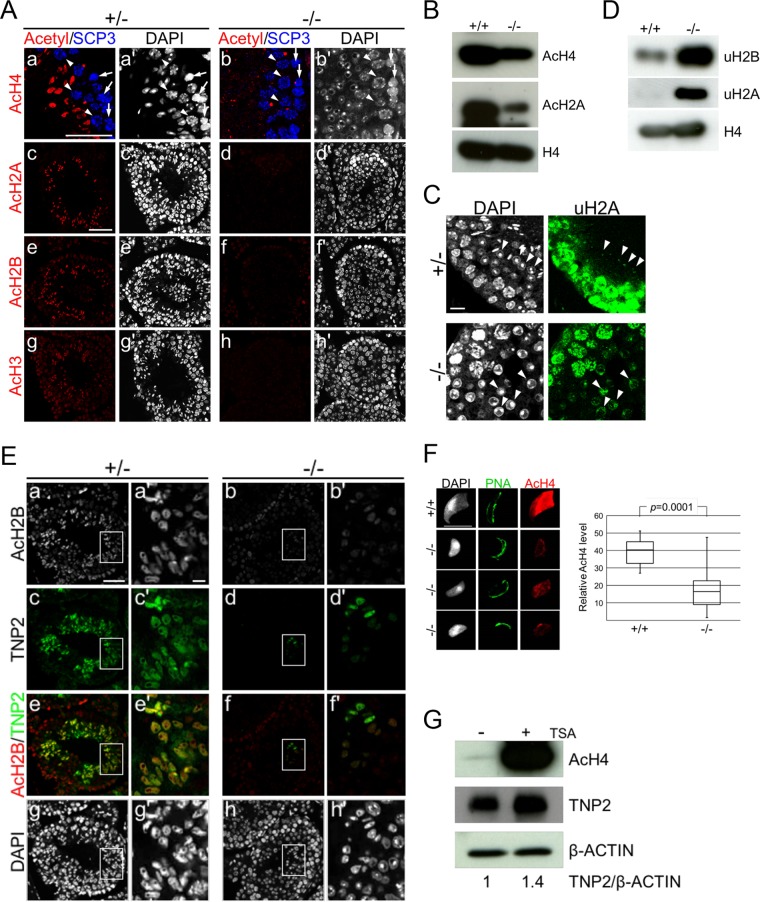
Progression of spermiogenesis from RS to ES has been shown to accompany histone hyperacetylation and subsequent accumulation of TNPs (6). To precisely identify the affected RS maturation step in Epc1-KO spermatids, we investigated the degree of histone acetylation by IF analysis using anti-acetylated histone antibodies, which demarcate step 8 RSs and ESs, together with anti-SCP3 for precise staging of the seminiferous tubules. We particularly focused on stage 9 seminiferous tubules, in which layered alignment of leptotene, pachytene spermatocytes, and ESs was seen in the heterozygotes (Fig. 4A, top row). Here, we observed strong fluorescent signals in ESs using an antibody that recognizes acetylated histone H4K5, -K8, -K12, and -K16. In presumptive stage 9 seminiferous tubules in the Epc1-KO spermatids, we found leptotene and pachytene spermatocytes associated with RSs instead of ESs, and we did not detect hyperacetylated histone H4 in postmeiotic cells (Fig. 4A). As hyperacetylation occurs concurrently at H2A, H2B, and H3 during spermiogenesis, we further examined their acetylation status and observed a considerable reduction in the Epc1-KO spermatids compared to the level of the heterozygotes (Fig. 4A). These results were validated by IB analysis using isolated male germ cells. We indeed observed a clear reduction in histone acetylation (Fig. 4B). Intriguingly, we also found clear increases in H2A and H2B ubiquitination in Epc1-KO cells (Fig. 4C and D), a protein modification that is reported to precede histone hyperacetylation (29, 30). Thus, we propose that EPC1 is mainly required for RS maturation, a stage in which histone is hyperacetylated.
FIG 4.
Impaired histone hyperacetylation and TNP2 incorporation of Epc1-KO spermatids. (A) Defects in histone hyperacetylation in Epc1-KO spermatids. Paraffin sections were indirectly immunostained with anti-acetylated histone H4 (a and b), H2A (c and d), H2B (e and f), or H3 (g and h) antibody in Epc1-KO (−/−) spermatids and littermate heterozygotes (+/−). SCP3 antibody was used for detection of leptotene (arrows) and pachytene (arrowheads) spermatocytes and staging of seminiferous tubules (a, a′, b, and b′). In the presumed stage 9 seminiferous tubules of Epc1-KO mice; leptotene and pachytene spermatocytes were normally layered from basal membrane to seminiferous tubule lumen, but ESs containing hyperacetylated histone were hardly detectable. In addition, a layer of round spermatids was expanded. Scale bars, 50 μm. (B) Reduced histone acetylation levels in Epc1-KO germ cells. Acetylation levels of histone H4 (AcH4) and H2A (AcH2A) in adult germ cells were compared between Epc1-KO (−/−) and wild-type littermates (+/+) by IB analysis. (C) Excess accumulation of RSs demarcated by ubiquitinated histone H2A. Expression of ubiquitinated histone H2A (uH2A) in Epc1-KO seminiferous tubules was examined by IF analysis. Arrowheads indicate RSs. Scale bar, 10 μm. (D) Increased histone ubiquitination levels in Epc1-KO germ cells. Ubiquitination levels of histone H2A (uH2A) and H2B (uH2B) were examined by IB analysis in adult germ cells. (E) Partial restoration of elongating spermatids in Epc1-KO seminiferous tubules. Paraffin sections were immunostained for acetylated histone H2B and transition protein 2 (TNP2) in Epc1-KO mice (−/−) and littermate heterozygotes (+/−). A cluster of Epc1-KO spermatids exhibited a partial restoration of histone H2B acetylation (AcH2B) and a simultaneous incorporation of TNP2 along with an elongating morphology. Enlarged views for the boxed regions in panels a to h are shown in panels a′ to h′, respectively. Scale bars, 50 μm (a) and 5 μm (a′). (F) Partial restoration of histone acetylation levels in Epc1-KO spermatids with an elongating morphology. A spread preparation of spermatids (left) was used to examine morphology, PNA distribution, and histone H4 acetylation levels (AcH4). Representative elongating Epc1-KO spermatids that escaped from maturation arrest are shown. Scale bar, 10 μm. Relative quantification of histone H4 acetylation levels in elongating Epc1-KO (n = 32) and wild-type (n = 20) ESs was performed by using ImageJ, and the results are summarized by box plots (right). The boxes represent the interquartile deviation. (G) The impact of trichostatin A (TSA) treatment on histone H4 acetylation and TNP2 incorporation in Epc1-KO spermatids. Epc1-KO germ cells were cultured for 3 days with (+) or without (−) TSA and subjected to IB analyses for acetylated histone H4, TNP2, and β-actin. TNP2/β-actin ratios based on respective band intensities are shown at the bottom, in which the ratio in untreated Epc1-KO germ cells was set as 1.
We then examined TNP incorporation in Epc1-KO seminiferous tubules. In heterozygous ESs, TNP2 and hyperacetylated histones were simultaneously detected (Fig. 4E). Consistent with defects in RS maturation into ES, spermatids expressing TNP2 were lost in most Epc1-KO seminiferous tubules (Fig. 4E). However, to our surprise, IF analysis revealed clusters of spermatids incorporating small amounts of TNP2 in some sections (Fig. 4E). Moreover, these TNP2-incorporating spermatids exhibited elongating morphology and more fluorescence intensity for histone acetylation than the surrounding RSs. This observation suggests that a small number of Epc1-KO spermatids can escape the maturation arrest and further develop beyond step 8. We further confirmed the presence of spermatids exhibiting an elongating morphology in Epc1-KO spermatids by using spread spermatids (Fig. 4F). In these Epc1-KO ESs, we observed a considerable level of histone acetylation; however, it was significantly lower than that in wild-type ESs. We used PNA staining to investigate acrosome morphology and found that Epc1-KO ESs possessed acrosome-like structures (Fig. 4F). These observations suggest that, once sufficiently histone acetylated, Epc1-KO spermatids can develop to ESs.
To further examine the role of histone acetylation for spermiogenesis in Epc1-KO spermatids, we artificially raised histone acetylation levels with the histone deacetylase inhibitor trichostatin A (TSA). After 3 days of culture of Epc1-KO testicular cells with or without TSA, we observed a considerable increase in histone acetylation and TNP2 expression (1.4-fold) by IB analysis (Fig. 4G). We further performed IF analysis to assess the frequency of TNP2-positive germ cells and found that this frequency was increased from 3.9% (40 out of 1,026 spermatogenic cells) to 4.9% (65 out of 1,319 spermatogenic cells) by TSA treatment. Although this increase is not statistically significant (P = 0.158, by Fischer's combination test), this suggests that maturation defects in Epc1-KO RSs could be partially restored by forced induction of histone acetylation, which is also suggested by IB results (Fig. 4G). The compensation of the EPC1 loss by TSA suggests that the involvement of EPC1 in promoting RS maturation to generate ESs is via regulation of histone acetylation.
Tip60 deletion affects histone acetylation and the RS-to-ES transition.
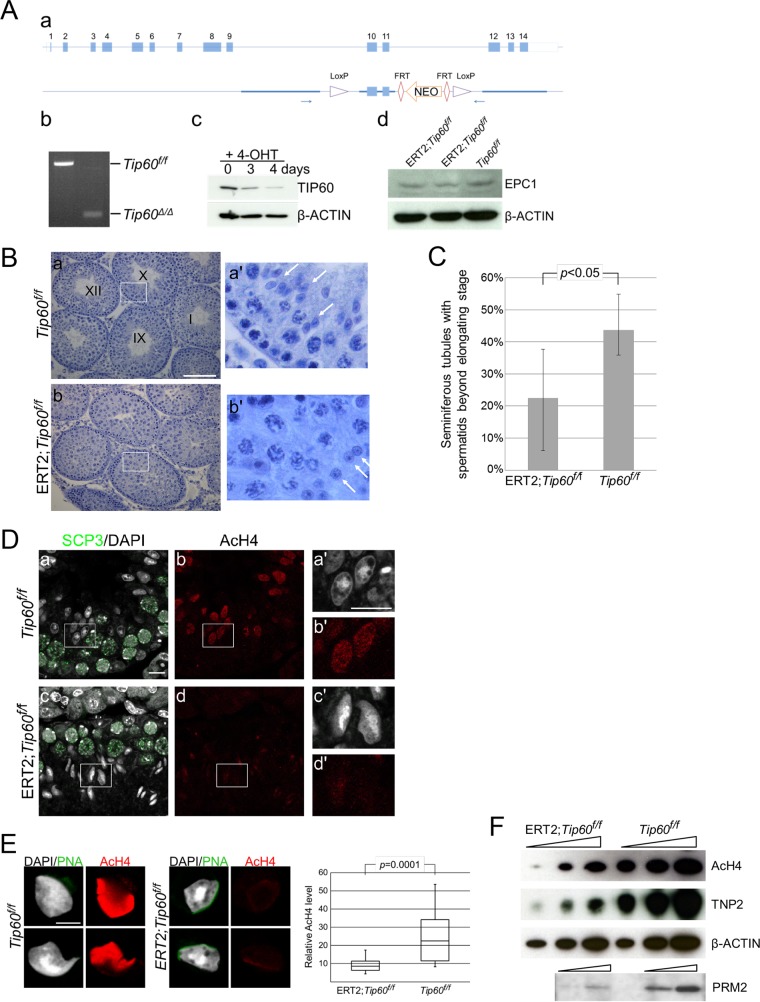
To further examine the contribution of histone acetylation for ES generation, we next investigated the impact of TIP60, which is another component of the NuA4 complexes and potentially acts as an HAT during spermiogenesis. We generated an ERT2-Cre-based conditional knockout allele in which Tip60 could be depleted upon addition of tamoxifen (Fig. 5A). We injected tamoxifen into the peritoneal cavity of ERT2-Cre; Tip60flox/flox pups to induce Tip60 knockout (Tip60-KO) at postnatal day 15 (PND15) and tested germ cell phenotypes at PND35. We found that the testes of the Tip60-KO mice were smaller than those of wild-type mice (data not shown). Histological analysis revealed variegated testicular defects. In the most degenerative tubules, we observed a loss of spermatocytes and RSs and more differentiated cells, while germ cells in the first layer, consisting of spermatogonium and spermatocytes from preleptotene to zygotene stages, were largely intact (data not shown). In moderately affected tubules, germ cell development appeared to be arrested at the RS stage (Fig. 5B). To evaluate the impact of Tip60 deficiency on spermiogenesis, we compared the frequency of seminiferous tubules containing ESs or more mature spermatids between the wild-type and Tip60-KO mice after exclusion of seminiferous tubules exhibiting premeiotic defects in the latter. On average, 43% of wild-type seminiferous tubules contained ESs or more mature spermatids, while only 22% of Tip60-KO seminiferous tubules did (Fig. 5C). This suggests that TIP60 contributes to RS maturation to generate ESs.
FIG 5.
Tip60 deletion affects histone acetylation and ES generation. (A) Generation of the Tip60-KO allele. (a) Schematic representation of the strategy to generate a conditional knockout allele of the Tip60 gene. FRT, Flp recognition target. (b) Cre-mediated deletion of exons 10 and 11 to generate the Tip60-KO (Tip60Δ/Δ) allele revealed by PCR analysis. (c) Reduction of TIP60 protein in Tip60-KO cells. We have established ERT2-Cre; Tip60flox/flox ES cells from blastocysts and induced Tip60 deletion upon 4-hydroxytamoxifen (4-OHT) treatment. (d) EPC1 expression in Tip60-KO germ cells. Whole-cell lysates of germ cells from tamoxifen-injected ERT2-Cre; Tip60flox/flox mice were examined by IB analysis. (B) Morphological changes in moderately affected seminiferous tubules of Tip60-KO testes. (a) Seminiferous tubule stages of a control (Tip60flox/flox cells) are indicated numerically. (b) A section of Tip60-KO (ERT2; Tip60flox/flox) testes with developmental arrest of germ cells are shown. Note that lack of elongating spermatids in the presumed stage 9/10 seminiferous tubules of the Tip60-KO cells. Scale bar, 100 μm. (C) Partial impairment of ES generation in ERT2; Tip60flox/flox mice treated by 4-hydroxytamoxifen. Frequency of seminiferous tubule sections containing ESs or more mature spermatids was calculated from five knockout (ERT2; Tip60flox/flox) and three control (Tip60flox/flox) samples. (D) Reduced acetylation level of histone H4 in Tip60-KO ESs. Stage 9/10 seminiferous tubules from a control and Tip60-KO mouse were immunostained with anti-SCP3 and anti-acetylated histone H4 (AcH4) antibodies. Acetylation levels in elongating spermatids from Tip60-KO mice were lower than those from controls in spite of their similar morphologies. Enlarged views for boxed regions in panels a to d are shown in panels a′ to d′, respectively. Scale bars, 50 μm (a) and 10 μm (a′). (E) Reduced acetylation level of histone H4 in Tip60-KO ESs. (Left) A spread preparation of spermatids was used to examine their morphology and histone H4 acetylation (AcH4) levels (left). Scale bar, 10 μm. Relative quantification for histone H4 acetylation levels in elongating Tip60-KO (n = 22) and control (n = 8) spermatids was performed by using ImageJ (right). (F) Reduction of histone H4 acetylation and TNP2 and PRM2 incorporation in Tip60-KO germ cells. Germ cells from tamoxifen-injected male mice were collected, and their lysates were subjected to IB analysis to examine histone acetylation, TNP2, and PRM2 levels. Tip60f/f, Tip60flox/flox.
We further examined whether changes in spermiogenesis in the Tip60-KO spermatids are accompanied by alterations in histone acetylation status. IF analysis for acetylated histone H4 using apparent stage 9/10 seminiferous tubules revealed considerable reduction of histone acetylation in Tip60-KO spermatids although they retained an elongating morphology (Fig. 5D). Further IF analysis by using surface-spread spermatids confirmed this reduction in elongating spermatids (Fig. 5E). IB analysis indeed revealed a considerable reduction of histone acetylation and TNP2 incorporation in Tip60-KO germ cells (Fig. 5F). Taken together, these results indicate that TIP60 concurrently regulates RS maturation into ESs and histone hyperacetylation, similar to EPC1.
DISCUSSION
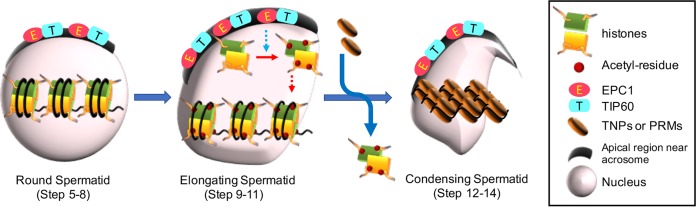
Several lines of genetic evidence presented in this study suggest critical contributions of EPC1 and TIP60 to mediate histone hyperacetylation enabling histone replacement with TNPs and PRMs and maturation of RSs into ESs. Colocalization of EPC1 and TIP60 at the apical side of the nuclear periphery near the acrosome, their physical interaction, and EPC1-dependent loading of TIP60 suggest that these molecules act as constituents of either NuA4- or piccolo NuA4-related multimeric HAT complexes during spermiogenesis. Intriguingly, previous studies reported that linker histone H1T2, which is required for spermatid elongation and DNA condensation, and TNP chaperone HSPA2 also localized at the apical pole (31, 32). Our findings together with those of previous investigators suggest that an apical polarizing activity has a role in organizing the antero-caudal axis of the spermatid nucleus for its maturation. In this case, the nonnucleosomal histones acetylated by NuA4-related complexes containing EPC1 and TIP60 may be transferred and incorporated into chromatin by additional molecules like the histone chaperones (Fig. 6). This could in turn contribute to generate a gradient of nuclear acetylation, with the histones near the apical pole being rapidly hyperacetylated while the ones at the caudal regions would be slowly and moderately acetylated. Such apical polarity might in part explain previous observations indicating that the hyperacetylated histones disappear following an antero-caudal movement in ESs (4, 6), with consequently preferential incorporation of TNPs into chromatin in the anterior region (33).
FIG 6.
A schematic summary of EPC1- and TIP60-dependent histone hyperacetylation during spermatid differentiation. Nuclei and apical regions closely associated with acrosome in round spermatids, elongating spermatids, and condensed spermatids are depicted. EPC1 and TIP60 accumulate at apical regions throughout this differentiation process. In round spermatids of steps 5 to 8, histones are major nuclear proteins but not hyperacetylated. The present study suggests that EPC1/TIP60 complexes likely mediate hyperacetylation of histones in elongating spermatids. Since EPC1 and TIP60 predominantly accumulate at apical regions near the acrosome, we speculate that EPC1/TIP60 complexes mediate acetylation of nonnucleosomal histones (indicated by a dotted blue arrow), which may in turn be incorporated into the chromatin (indicated by a red dotted arrow). Hyperacetylation of histones is shown to facilitate their subsequent replacement with TNPs and/or PRMs during elongating-to-condensing spermatid transition and/or in the elongating-spermatid stage.
This model, however, needs further explanation of how prospective PRM-compacted and nucleosomal genomic domains which were previously described in humans and rodents are distinguished (34, 35). Histone chaperones, remodeling factors, and/or other biochemical activities could be involved in distinguishing these domains and contribute to disseminate histones acetylated by EPC1/TIP60 complexes exclusively to prospective PRM-compacted domains. Intriguingly, RNF8-dependent H2A/B ubiquitination has been reported to be a prerequisite for global histone hyperacetylation and subsequent histone replacement (30). Considerable accumulation of ubiquitinated H2A/B was seen in Epc1-KO RSs (Fig. 4C and D), suggesting that RNF8-dependent H2A/B ubiquitination is a precursor state for selective deposition of acetylated histones by EPC1/TIP60 complexes. However, other models, such as those involving dynamic association of nucleosomes with EPC1/TIP60 at the nuclear periphery, are still not formally excluded. Further studies to test these models are needed.
MATERIALS AND METHODS
Antibodies.
Antibodies against acetylated histone H2A, H2B, and H3 were purchased from Millipore and used for immunoblotting (IB), immunofluorescence (IF), and immunoprecipitation (IP) analyses as follows: 07-376 at 1:2,000 for IB and 1:1,000 for IF; 07-373 at 1:2,000 for IB and 1:1,000 for IF; 06-599 at 1:2,000 for IB and 1:1,000 for IF. Antibodies for ubiquitinated histone H2A and H2B (8240, 1:2,000 for IB and 1:1,000 for IF; 5546, 1:2,000 for IB) were from Cell Signaling Technology. Anti-H4 antibody was from Affinity BioReagents (PA1-84526; 1:2,000 for IB and 1:1,000 for IF). Antibodies for TIP60 (sc-25378; 1:500 for IF), EPC1 (sc-48152; 1:1,000 IB and 1:500 for IF), GATA4 (sc-1237; 1:200 for IF), and TNP2 (sc-21006; 1:2,000 for IB and 1:500 for IF) were purchased from Santa Cruz Biotechnology. Another antibody against TIP60 (DR1041; 1:1,000 for IB and 1:200 for IP) was from Millipore. Antibody for c-Kit (ab112177; 1:100 for IF) was from Abcam. Antibody for CYP11A1 (orb156513; 1:400 for IF) was from Biorbyt. Antibody for HSD3B2 (ABIN2855488; 1:400 for IF) was from Antibodies, Inc. Rabbit anti-EPC1 antiserum was raised against mouse EPC1 peptides (amino acids [aa] 473 to 488 and aa 736 to 750) as previously described (36) and used for IP at 1:200.
Generation of Epc1 and Tip60 mutant alleles.
To generate the targeting construct of Epc1, a 1.5-kb high-fidelity amplified fragment and a 4-kb EcoRI-KpnI fragment from a bacterial artificial chromosome (BAC) clone were subcloned into a pPGK-NEO vector as two homologous arms. The homologous recombination in both arms is expected to replace a genomic region encompassing a part of exon 3 and exons 4 and 5 with a neomycin resistance gene cassette (Fig. 2A, PGK-NEO). The targeted embryonic stem (ES) cell clones were identified by Southern blotting using a 5′ external probe (Fig. 2A and B).
Generation of the Tip60 conditional knockout allele will be described in another paper (K. Yamagata, H. Koseki, and I. Kitabayashi, unpublished data). Briefly, we constructed a targeting vector to induce the deletion of exons 10 and 11 by Cre-mediated recombination (Fig. 5A). Mice carrying the Epc1- and Tip60-targeting constructs were maintained on a mixed genetic background of C57BL/6J and 129/J. All animal experiments were carried out according to the in-house guidelines for the care and use of laboratory animals of the RIKEN, Yokohama Institute, Japan.
In situ hybridization analysis.
In situ hybridization was performed using a digoxigenin (DIG)-labeled full-length Epc1 cRNA probe. Staining procedures were carried out as previously reported (37). Briefly, adult male ICR mice were fixed by transcardiac perfusion with 4% paraformaldehyde in 0.1 M sodium phosphate buffer (pH 7.6) under general anesthesia. Testes were removed and immersed in the same fixative for 4 h at 4°C. Cryostat sections (10 μm in thickness) were prepared and digested with 0.1% pepsin (Dako) in 0.2 M HCl for 5 min at 37°C. Subsequently, sections were hybridized with a cRNA probe (1.5 mg/ml) in hybridization solution containing 10 mM Tris-HCl (pH 7.6), 0.6 M NaCl, 50% formamide, 10% dextran sulfate, 0.025% SDS, Denhardt's solution, 1 mM EDTA, and 0.2 mg/ml yeast tRNA at 50°C for 16 h. Then sections were washed in the 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) containing 50% formamide for 30 min and treated with RNase A (Sigma) (10 mg/ml in a solution of 0.5 M NaCl, 1 mM EDTA, and 10 mM Tris-HCl, pH 7.4) at 37°C for 30 min. The sections were next washed with 2× SSC, and after a final washing in 0.2× SSC at 50°C for 20 min, the hybridized probe was detected with a DIG nucleic acid detection kit (Roche) by using alkaline phosphatase-labeled anti-DIG antibody. Adjacent sections were stained with periodic acid-Schiff (PAS) stain and counterstained with hematoxylin to determine the stages of the seminiferous epithelium.
Terminal deoxynucleotidyl transferase dUTP-biotin nick end labeling (TUNEL) analysis.
Testis sections (fixed with 4% paraformaldehyde [PFA]) were immunostained by using an In situ cell death detection kit (12156792910; Roche) according to the manufacturer's instructions, and nuclei were stained with 4′,6′-diamidino-2-phenylindole (DAPI; blue).
High-resolution histology and transmission electron microscopy analysis.
High-resolution histology and transmission electron microscopy analysis were performed as previously described (38). Briefly, testes were fixed with 2.5% glutaraldehyde in 0.1 M phosphate buffer, postfixed with 1% OsO4, dehydrated, and embedded in Epon 812. One-micrometer sections were cut and stained with 1% toluidine blue for light microscopy analysis. Ultrathin sections were stained with uranyl acetate and lead citrate for transmission electron microscopy analysis.
Immunofluorescence (IF) analysis.
For immunostaining of testis sections, 5-μm paraffin-embedded sections were deparaffinized and rehydrated. Antigen retrieval was performed by boiling slides in 10 mM Tris-HCl (pH 9.0), 1 mM EDTA, and 0.05% Tween 20 for 20 min. Nonspecific binding sites were blocked with 5% normal goat (or donkey) serum in phosphate-buffered serum (PBS) with 0.05% Tween 20 (PBS-T) for 1 h at room temperature. Then sections were incubated with the primary antibodies in 1% bovine serum albumin (BSA)–PBS-T overnight at 4°C in a humidified chamber. After a washing step, Alexa Fluor-conjugated secondary antibodies (Invitrogen) were added for 1 h at room temperature. Finally, slides were mounted in VectaShield with DAPI (Vector Laboratories). The images were captured using a Leica TCS SP2 laser confocal microscope.
For spread preparations on slides, stage-specific segments of wild-type seminiferous tubules were collected in PBS as described previously (39). For collection of Epc1−/− (Epc1-KO) germ cells, the decapsulated testes were minced briefly, and then germ cells were released by pipetting and washed twice with PBS. Cell pellets were resuspended in 4% paraformaldehyde–PBS-T and then spread onto aminopropyltriethoxy silane (APES)-coated slides in a humidified chamber for 20 min at room temperature. For immunostaining, germ cells were permeabilized with 0.3% Triton X-100–PBS for 20 min, and downstream treatments were the same as described above.
Immunoprecipitation (IP).
Testicular cells were collected as described above and resuspended in 1% NP-40 buffer containing 50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1 mM dithiothreitol (DTT), and protease inhibitor cocktail (Roche). A brief sonication was carried out with a hand-held sonicator at 50% output for 10 s three times to disrupt cells, and then chromatin was digested with an enzymatic shearing cocktail (Active Motif) for 90 min on ice. After centrifugation at 12,000 × g for 10 min, the supernatant was precleared by incubation with protein G Dynabeads for 30 min at 4°C. Subsequently, antibodies against TIP60 or EPC1 and IgG as a negative control were added and incubated at 4°C overnight. Immune complexes were captured with protein G Dynabeads and washed with PBS-T four times. Immunoprecipitated materials were eluted by boiling in SDS sample buffer and subjected to IB analysis.
ROSI.
Round spermatid injection (ROSI) was performed using a piezodriven micromanipulator (Prime Tech, Ltd., Ibaraki, Japan) as described previously (40–42). Briefly, male germ cell suspensions containing RSs were mixed with 10% polyvinylpyrrolidone in HEPES–Chatot-Ziomek-Bavister (CZB) medium in a microinjection chamber. Before injection of RS nuclei, oocytes were activated by treatment with Ca2+-free CZB medium containing 2.5 mM SrCl2 for 20 min at 37°C (43). Oocytes reaching telophase II at 40 to 90 min after onset of the activation treatment were each injected with an RS nucleus. They were kept in HEPES-CZB medium at room temperature (24°C) for ∼10 min before culture in CZB medium at 37°C under 5% CO2 in air. Embryos that reached the 2-cell stage by 24 h of culture in CZB medium were transferred into the oviducts of pseudopregnant ICR strain females (8 to 12 weeks old) on the day after mating (day 0.5). On day 19.5, the number of live, term fetuses per uterus was examined by Caesarean section.
Sorting of round spermatids.
Testes from adult C57BL/6J and Epc1-KO mice were decapsulated in erythrocyte lysing buffer (Sigma) and briefly washed twice in PBS. Tubule masses were minced into small pieces using fine scissors, and testicular cells were released by pipetting and then filtered through nylon mesh (pore sizes of 100, 50, and 30 μm, sequentially). Next, germ cells were treated with cell dissociation buffer (Gibco) for 1 min and washed twice with Dulbecco's modified Eagle's medium (DMEM; Sigma) containing 1% BSA. Finally, testicular cells were resuspended at a concentration of 2 × 107 cells/ml and Vybrant DyeCycle ruby stain (Invitrogen) was added for 30 min to define DNA content; then germ cells were sorted using a FACSAria (Becton, Dickinson, and Company) instrument.
RNA sequencing and data analysis.
RNA was extracted from sorted RSs using an RNeasy kit (Qiagen), and then libraries were generated using a NEBNext RNA library prep kit for Illumina and sequenced on a HiSeq 1500 according to the manufacturer's instructions. Sequences were aligned to the mouse genome (mm9) using the tophat2 (version 2.0.9) and cufflinks (version 2.1.0) programs. Expression changes (degree and statistical significance) were calculated by cuffdiff (version 2.1.0).
Signature genes that represent developmental stages of spermatids were determined by using RNA-seq data retrieved from a public database (NCBI GEO accession number GSE39970) (28). Gene expression profiles of testicular germ cells at postnatal days (PNDs) 7, 14, 17, 21, and 28 were compared with each other to select genes specific for spermatogonia, early spermatocytes, late spermatocytes, RSs, and ESs, respectively. At each PND genes were selected that were expressed at significantly higher levels (q value of <0.05; logarithm of fold change of >1.0) than at the other stages (see Table S1 in the supplemental material). Since the difference between results for PND14 and those for PND17 was very small, we identified spermatocyte-specific genes using only PND14 and not PND17. We identified 1,989 genes predominantly expressed in spermatogonia, 68 genes in spermatocytes, 28 genes in RSs, and 2,390 genes in ESs.
Culture of Epc1-KO testicular cells.
Testicular cells from Epc1-KO mice were collected and washed twice with DMEM containing 10% fetal calf serum (FCS; Gibco), 50 IU/liter follicle stimulation hormone (Sigma), and 1 μM testosterone enanthate (Sigma). Then, the mixture of germ cells was cultured with or without 100 μg/liter trichostatin A (Sigma) for 3 days at 33°C in a water-saturated atmosphere with 5% CO2.
Tamoxifen injection.
Tamoxifen (Sigma) was dissolved in corn oil at 10 mg/ml. PND15 mice were intraperitoneally administered with tamoxifen at a concentration of 10 mg/kg body weight.
Statistical analysis.
Student's t tests were used for all statistical analyses. A P value of <0.05 was considered to be statistically significant. In box plots, the box corresponds to the central 50% of data. Mean and standard deviations were plotted.
Accession number(s)
All raw data used in this study are available in the NCBI Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo) under accession number GSE89640.
Supplementary Material
ACKNOWLEDGMENTS
We thank the members of the Koseki laboratory for daily discussions.
We declare that we have no competing interests.
Y.D., K.-I.I., and H.K. designed the study, performed most of experiments, and wrote the manuscript. T.A.E. and O.O. performed informatics analyses. M.M., Y.T., C.I., K.T., and K.O. performed high-resolution histological analyses. K.H. generated anti-EPC1 antibody. N.O., K.I., and A.O. performed round spermatid injection. K.Y. and I.K. generated the Tip60 conditional allele.
This work was supported by Grants-in-Aid for Scientific Research and for Scientific Research (to K.I. and H.K.) from the Ministry of Education, Culture, Sports, Science and Technology of Japan and CREST (to H.K.) programs of Japan Science and Technology Agency.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/MCB.00082-17.
REFERENCES
- 1.Pogany GC, Corzett M, Weston S, Balhorn R. 1981. DNA and protein content of mouse sperm. Implications regarding sperm chromatin structure. Exp Cell Res 136:127–136. [DOI] [PubMed] [Google Scholar]
- 2.Hecht NB. 1998. Molecular mechanisms of male germ cell differentiation. Bioessays 20:555–561. doi:. [DOI] [PubMed] [Google Scholar]
- 3.Steger K. 2001. Haploid spermatids exhibit translationally repressed mRNAs. Anat Embryol (Berl) 203:323–334. doi: 10.1007/s004290100176. [DOI] [PubMed] [Google Scholar]
- 4.Meistrich ML, Mohapatra B, Shirley CR, Zhao M. 2003. Roles of transition nuclear proteins in spermiogenesis. Chromosoma 111:483–488. doi: 10.1007/s00412-002-0227-z. [DOI] [PubMed] [Google Scholar]
- 5.Lewis JD, Saperas N, Song Y, Zamora MJ, Chiva M, Ausió J. 2004. Histone H1 and the origin of protamines. Proc Natl Acad Sci U S A 101:4148–4152. doi: 10.1073/pnas.0308721101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hazzouri M, Pivot-Pajot C, Faure AK, Usson Y, Pelletier R, Sèle B, Khochbin S, Rousseaux S. 2000. Regulated hyperacetylation of core histones during mouse spermatogenesis: involvement of histone deacetylases. Eur J Cell Biol 79:950–960. doi: 10.1078/0171-9335-00123. [DOI] [PubMed] [Google Scholar]
- 7.Christensen ME, Rattner JB, Dixon GH. 1984. Hyperacetylation of histone H4 promotes chromatin decondensation prior to histone replacement by protamines during spermatogenesis in rainbow trout. Nucleic Acids Res 12:4575–4592. doi: 10.1093/nar/12.11.4575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grimes SR Jr, Henderson N. 1984. Hyperacetylation of histone H4 in rat testis spermatids. Exp Cell Res 152:91–97. doi: 10.1016/0014-4827(84)90232-5. [DOI] [PubMed] [Google Scholar]
- 9.Oliva R, Mezquita C. 1982. Histone H4 hyperacetylation and rapid turnover of its acetyl groups in transcriptionally inactive rooster testis spermatids. Nucleic Acids Res 10:8049–8059. doi: 10.1093/nar/10.24.8049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meistrich ML, Trostle-Weige PK, Lin R, Bhatnagar YM, Allis CD. 1992. Highly acetylated H4 is associated with histone displacement in rat spermatids. Mol Reprod Dev 31:170–181. doi: 10.1002/mrd.1080310303. [DOI] [PubMed] [Google Scholar]
- 11.Sonnack V, Failing K, Bergmann M, Steger K. 2002. Expression of hyperacetylated histone H4 during normal and impaired human spermatogenesis. Andrologia 34:384–390. doi: 10.1046/j.1439-0272.2002.00524.x. [DOI] [PubMed] [Google Scholar]
- 12.Faure AK, Pivot-Pajot C, Kerjean A, Hazzouri M, Pelletier R, Péoc'h M, Sèle B, Khochbin S, Rousseaux S. 2003. Misregulation of histone acetylation in Sertoli cell-only syndrome and testicular cancer. Mol Hum Reprod 9:757–763. doi: 10.1093/molehr/gag101. [DOI] [PubMed] [Google Scholar]
- 13.Kennedy BP, Davies PL. 1980. Acid-soluble nuclear proteins of the testis during spermatogenesis in the winter flounder. Loss of the high mobility group proteins. J Biol Chem 255:2533–2539. [PubMed] [Google Scholar]
- 14.Oliva R, Mezquita C. 1986. Marked differences in the ability of distinct protamines to disassemble nucleosomal core particles in vitro. Biochemistry 25:6508–6511. doi: 10.1021/bi00369a025. [DOI] [PubMed] [Google Scholar]
- 15.Oliva R, Bazett-Jones D, Mezquita C, Dixon GH. 1987. Factors affecting nucleosome disassembly by protamines in vitro. Histone hyperacetylation and chromatin structure, time dependence, and the size of the sperm nuclear proteins. J Biol Chem 262:17016–17025. [PubMed] [Google Scholar]
- 16.Gaucher J, Boussouar F, Montellier E, Curtet S, Buchou T, Bertrand S, Hery P, Jounier S, Depaux A, Vitte AL, Guardiola P, Pernet K, Debernardi A, Lopez F, Holota H, Imbert J, Wolgemuth DJ, Gérard M, Rousseaux S, Khochbin S. 2012. Bromodomain-dependent stage-specific male genome programming by Brdt. EMBO J 31:3809–3820. doi: 10.1038/emboj.2012.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tse C, Sera T, Wolffe AP, Hansen JC. 1998. Disruption of higher-order folding by core histone acetylation dramatically enhances transcription of nucleosomal arrays by RNA polymerase III. Mol Cell Biol 18:4629–4638. doi: 10.1128/MCB.18.8.4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL. 2006. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science 311:844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- 19.Kan P, Caterino T, Hayes J. 2009. The H4 tail domain participates in intra- and internucleosome interactions with protein and DNA during folding and oligomerization of nucleosome arrays. Mol Cell Biol 29:538–546. doi: 10.1128/MCB.01343-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lahn BT, Tang ZL, Zhou J, Barndt RJ, Parvinen M, Allis CD, Page DC. 2002. Previously uncharacterized histone acetyltransferases implicated in mammalian spermatogenesis. Proc Natl Acad Sci U S A 99:8707–8712. doi: 10.1073/pnas.082248899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Caron C, Pivot-Pajot C, van Grunsven LA, Col E, Lestrat C, Rousseaux S, Khochbin S. 2003. Cdyl: a new transcriptional co-repressor. EMBO Rep 4:877–882. doi: 10.1038/sj.embor.embor917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marcon L, Boissonneault G. 2004. Transient DNA strand breaks during mouse and human spermiogenesis new insights in stage specificity and link to chromatin remodeling. Biol Reprod 70:910–918. doi: 10.1095/biolreprod.103.022541. [DOI] [PubMed] [Google Scholar]
- 23.Leduc F, Maquennehan V, Nkoma GB, Boissonneault G. 2008. DNA damage response during chromatin remodeling in elongating spermatids of mice. Biol Reprod 78:324–332. doi: 10.1095/biolreprod.107.064162. [DOI] [PubMed] [Google Scholar]
- 24.Boudreault AA, Cronier D, Selleck W, Lacoste N, Utley RT, Allard S, Savard J, Lane WS, Tan S, Côté J. 2003. Yeast enhancer of Polycomb defines global Esa1-dependent acetylation of chromatin. Genes Dev 17:1415–1428. doi: 10.1101/gad.1056603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Doyon Y, Selleck W, Lane WS, Tan S, Côté J. 2004. Structural and functional conservation of the NuA4 histone acetyltransferase complex from yeast to humans. Mol Cell Biol 24:1884–1896. doi: 10.1128/MCB.24.5.1884-1896.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shima JE, McLean DJ, McCarrey JR, Griswold MD. 2004. The murine testicular transcriptome: characterizing gene expression in the testis during the progression of spermatogenesis. Biol Reprod 71:319–330. doi: 10.1095/biolreprod.103.026880. [DOI] [PubMed] [Google Scholar]
- 27.Reynard LN, Cocquet J, Burgoyne PS. 2009. The multi-copy mouse gene Sycp3-like Y-linked (Sly) encodes an abundant spermatid protein that interacts with a histone acetyltransferase and an acrosomal protein. Biol Reprod 81:250–257. doi: 10.1095/biolreprod.108.075382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laiho A, Kotaja N, Gyenesei A, Sironen A. 2013. Transcriptome profiling of the murine testis during the first wave of spermatogenesis. PLoS One 8:e61558. doi: 10.1371/journal.pone.0061558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baarends WM, Hoogerbrugge TW, Roest HP, Ooms M, Vreeburg J, Hoeijmakers JHJ, Grootegoed JA. 1999. Histone ubiquitination and chromatin remodeling in mouse spermatogenesis. Dev Biol 207:322–333. doi: 10.1006/dbio.1998.9155. [DOI] [PubMed] [Google Scholar]
- 30.Lu LY, Wu J, Ye L, Gavrilina GB, Saunders TL, Yu X. 2010. RNF8-dependent histone modifications regulate nucleosome removal during spermatogenesis. Dev Cell 18:371–384. doi: 10.1016/j.devcel.2010.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martianov I, Brancorsini S, Catena R, Gansmuller A, Kotaja N, Parvinen M, Sassone-Corsi P, Davidson I. 2005. Polar nuclear localization of H1T2, a histone H1 variant, required for spermatid elongation and DNA condensation during spermiogenesis. Proc Natl Acad Sci U S A 102:2808–2813. doi: 10.1073/pnas.0406060102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Govin J, Caron C, Escoffier E, Ferro M, Kuhn L, Rousseaux S, Eddy EM, Garin J, Khochbin S. 2006. Post-meiotic shifts in HSPA2/HSP70.2 chaperone activity during mouse spermatogenesis. J Biol Chem 281:37888–37892. doi: 10.1074/jbc.M608147200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao M, Shirley CR, Mounsey S, Meistrich ML. 2004. Nucleoprotein transitions during spermiogenesis in mice with transition nuclear protein Tnp1 and Tnp2 mutations. Biol Reprod 71:1016–1025. doi: 10.1095/biolreprod.104.028191. [DOI] [PubMed] [Google Scholar]
- 34.Hammoud SS, Nix DA, Zhang H, Purwar J, Carrell DT, Cairns BR. 2009. Distinctive chromatin in human sperm packages genes for embryo development. Nature 460:473–478. doi: 10.1038/nature08162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Erkek S, Hisano M, Liang CY, Gill M, Murr R, Dieker J, Schübeler D, van der Vlag J, Stadler MB, Peters AH. 2013. Molecular determinants of nucleosome retention at CpG-rich sequences in mouse spermatozoa. Nat Struct Mol Biol 20:868–875. doi: 10.1038/nsmb.2599. [DOI] [PubMed] [Google Scholar]
- 36.Attwooll C, Oddi S, Cartwright P, Prosperini E, Agger K, Steensgaard P, Wagener C, Sardet C, Moroni MC, Helin K. 2005. A novel repressive E2F6 complex containing the polycomb group protein, EPC1, that interacts with EZH2 in a proliferation-specific manner. J Biol Chem 280:1199–1208. doi: 10.1074/jbc.M412509200. [DOI] [PubMed] [Google Scholar]
- 37.Hattori K, Uchino S, Isosaka T, Maekawa M, Iyo M, Sato T, Kohsaka S, Yagi T, Yuasa S. 2006. Fyn is required for haloperidol-induced catalepsy in mice. J Biol Chem 281:7129–7135. doi: 10.1074/jbc.M511608200. [DOI] [PubMed] [Google Scholar]
- 38.Kawa S, Ito C, Toyama Y, Maekawa M, Tezuka T, Nakamura T, Nakazawa T, Yokoyama K, Yoshida N, Toshimori K, Yamamoto T. 2006. Azoospermia in mice with targeted disruption of the Brek/Lmtk2 (brain-enriched kinase/lemur tyrosine kinase 2) gene. Proc Natl Acad Sci U S A 103:19344–19349. doi: 10.1073/pnas.0603603103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kotaja N, Kimmins S, Brancorsini S, Hentsch D, Vonesch JL, Davidson I, Parvinen M, Sassone-Corsi P. 2004. Preparation, isolation and characterization of stage-specific spermatogenic cells for cellular and molecular analysis. Nat Methods 1:249–254. doi: 10.1038/nmeth1204-249. [DOI] [PubMed] [Google Scholar]
- 40.Kimura Y, Yanagimachi R. 1995. Mouse oocytes injected with testicular spermatozoa or round spermatids can develop into normal offspring. Development 121:2397–2405. [DOI] [PubMed] [Google Scholar]
- 41.Ogonuki N, Mori M, Shinmen A, Inoue K, Mochida K, Ohta A, Ogura A. 2010. The effect on intracytoplasmic sperm injection outcome of genotype, male germ cell stage and freeze-thawing in mice. PLoS One 5:e11062. doi: 10.1371/journal.pone.0011062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ogura A, Yanagimachi R. 1993. Round spermatid nuclei injected into hamster oocytes from pronuclei and participate in syngamy. Biol Reprod 48:219–225. doi: 10.1095/biolreprod48.2.219. [DOI] [PubMed] [Google Scholar]
- 43.Chatot CL, Ziomek CA, Bavister BD, Lewis JL, Torres I. 1989. An improved culture medium supports development of random-bred 1-cell mouse embryos in vitro. J Reprod Fertil 86:679–688. doi: 10.1530/jrf.0.0860679. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.