ABSTRACT
Erf is a gene for a ubiquitously expressed Ets DNA-binding domain-containing transcriptional repressor. Erf haploinsufficiency causes craniosynostosis in humans and mice, while its absence in mice leads to failed chorioallantoic fusion and death at embryonic day 10.5 (E10.5). In this study, we show that Erf is required in all three waves of embryonic hematopoiesis. Mice lacking Erf in the embryo proper exhibited severe anemia and died around embryonic day 14.5. Erf epiblast-specific knockout embryos had reduced numbers of circulating blood cells from E9.5 onwards, with the development of severe anemia by E14.5. Elimination of Erf resulted in both reduced and more immature primitive erythroblasts at E9.5 to E10.5. Reduced definitive erythroid colony-forming activity was found in the bloodstream of E10.5 embryos and in the fetal liver at E11.5 to E13.5. Finally, elimination of Erf resulted in impaired repopulation ability, indicating that Erf is necessary for hematopoietic stem cell maintenance or differentiation. We conclude that Erf is required for both primitive and erythromyeloid progenitor waves of hematopoietic stem cell (HSC)-independent hematopoiesis as well as for the normal function of HSCs.
KEYWORDS: Erf, mouse, embryo, anemia, primitive wave, definitive wave, HSCs, hemogenic endothelium, differentiation, fetal liver, developmental biology, Erf, hematopoiesis
INTRODUCTION
The ontogeny of the hematopoietic system is a complex process with multiple waves of hemopoietic potential emerging from different anatomical sites (1). The first hematopoietic progenitors that emerge in the yolk sac at embryonic day 7.5 (E7.5) generate the first circulating blood cells of erythroid, megakaryocyte, and macrophage lineages (2–4). Shortly after, at E8.25, a second wave of hematopoietic progenitors, termed erythro-myeloid progenitors (EMPs), emerges in the yolk sac and colonizes the fetal liver at E10.5 (5–8). A third wave of hematopoietic potential, consisting of adult repopulating hematopoietic stem cells (HSCs), emerges from large arterial vessels beginning at E10.5, and colonizes the fetal liver but ultimately engrafts the bone marrow to provide lifelong postnatal blood cell production (9–12).
Several ets genes have been shown to play a crucial role in distinct steps of hematopoiesis. PU.1 regulates the HSCs to myeloid commitment (13, 14). Other transcription factors of the ETS family have also been shown to regulate hematopoiesis. Etv2 regulates the emergence of blood and endothelial cells activating the Flk1 gene that is critical for initiation of hemangioblast formation (15–20) and that is also required for the formation of hemogenic endothelium at the onset of yolk sac hematopoiesis (21). Erg is a critical regulator for HSC maintenance during embryonic development (22). Fev regulates the number of Runx1-expressing cells by transcriptionally activating Erk2 to enhance extracellular signal-regulated kinase (ERK) signaling (23), while PU.1 synergizes with Runx1 and determines its hematopoiesis-specific expression (24).
Erf is a ubiquitously expressed ets family gene, and it has been suggested that it enhances erythroid differentiation. Erf is regulated by Erk1/2 phosphorylation and nucleocytoplasmic shuttling. In its nonphosphorylated nuclear form, Erf blocks cell proliferation-arresting cells at the G0/G1 phase in a cell-type-specific manner; it can suppress Ets- and Ras-induced tumorigenicity in fibroblasts, cell lines derived from Ewing's sarcomas harboring the EWS-FLI1 rearrangement, and the epithelial-to-mesenchymal transition via semaphorin 7a inhibition (25–30). Erf-null embryos fail to undergo chorioallantoic fusion and labyrinth development because of a block in chorion trophoblast stem cell (Ch-TSC) differentiation, resulting in fetal demise at E10.5 (31). Finally, Erf haploinsufficiency causes craniosynostosis in humans and in mice, identifying Erf as a novel regulator of osteogenesis within the RAS-ERK signaling pathway (32).
Here, we demonstrate that elimination of Erf in the murine embryo proper at E5 leads to severe anemia and fetal death at E14.5. Employing histological, cellular, molecular, and in vivo repopulation assays, we show that Erf is required for the timely or quantitative production of both primitive and definitive yolk sac-derived hematopoiesis, as well as the production or maintenance of HSCs. In addition, Erf is required for the efficient maturation of erythroid precursors into mature erythrocytes. Thus, our data indicate that Erf is required throughout hematopoietic development for the homeostasis of this complex system.
RESULTS
Erfed/ed embryos die in utero due to severe anemia.
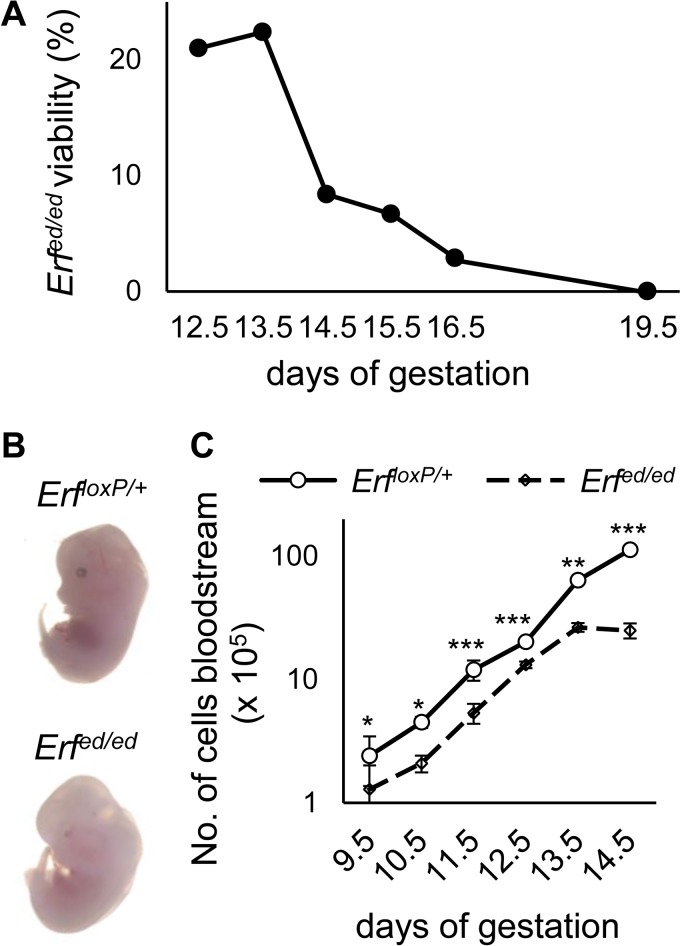
We have previously shown that elimination of Erf in mice leads to lethality at E10.5 because of failed chorioallantoic fusion and labyrinth development (31), while Erf haploinsufficiency leads to defective suture development and craniosynostosis (32). To investigate other developmental processes that Erf may regulate and bypass the placental defect, we eliminated Erf in the epiblast at E5, crossing ErfloxP/loxP mice with mice expressing the Cre recombinase under the control of the Meox2 gene (33) in order to delete Erf in the embryos but not in the placenta (epiblast-derived Erf [Erfed/ed]). Elimination of Erf in the epiblast resulted in embryonic death around E14.5 (Fig. 1A). The embryos were pale, indicative of anemia (Fig. 1B). Erfed/ed embryos contained approximately 50% of normal circulating primitive erythroblasts from E9.5 through E12.5. The blood levels of Erfed/ed fetuses subsequently dropped precipitously by E14.5 to less than 5% of the normal level (Fig. 1C). A few Erf knockout fetuses exhibited somewhat higher levels of circulating blood at E14.5, consistent with the occasional survival until E15.5 or E16.5. The mutant embryos did not appear to have defects in other tissues (data not shown), and overall embryo size was comparable to that of Erf-expressing embryo (see Fig. S1 in the supplemental material). The anemia and fetal demise did not appear to be of placental origin, as placental development appeared unaffected in the Erfed/ed embryos (Fig. S2), but rather appeared to result from a defect in the embryo proper, where Erf elimination exceeded 90% (Fig. S3A to C).
FIG 1.
Erfed/ed mice are embryonic lethal and appear anemic. (A) Pregnant mice were sacrificed at the indicated day of gestation, and the genotypes of the embryos were analyzed. The graph indicates the percentages of live Erfed/ed embryos. The data are from 160 litters, with at least 5 litters for each gestation stage (see Table S1 in the supplemental material). A chi-square test showed a statistically important decrease with the expected Mendelian distribution after E13.5. (B) Representative microphotographs of embryos at E13.5 showing apparent anemia and a lack of other gross morphology differences between Erfed/ed embryos and their ErfloxP/+ littermates. (C) Number of blood cells from the yolk sac and embryo proper at the indicated day of gestation. Erfed/ed embryos exhibit a statistically significant decrease throughout E9.5 to E13.5, which is exacerbated at E14.5. Samples are represented in a logarithmic scale. All values are means ± SE from 35 litters with at least 7 litters for each gestation stage (Table S2). Statistical analysis was performed using an unpaired t test with two-tailed distribution. *, P < 0.05; **, P < 0.005; ***, P < 0.0005, for results with ErfloxP/+ mice versus those with Erfed/ed mice.
Erf elimination impairs primitive erythropoiesis.
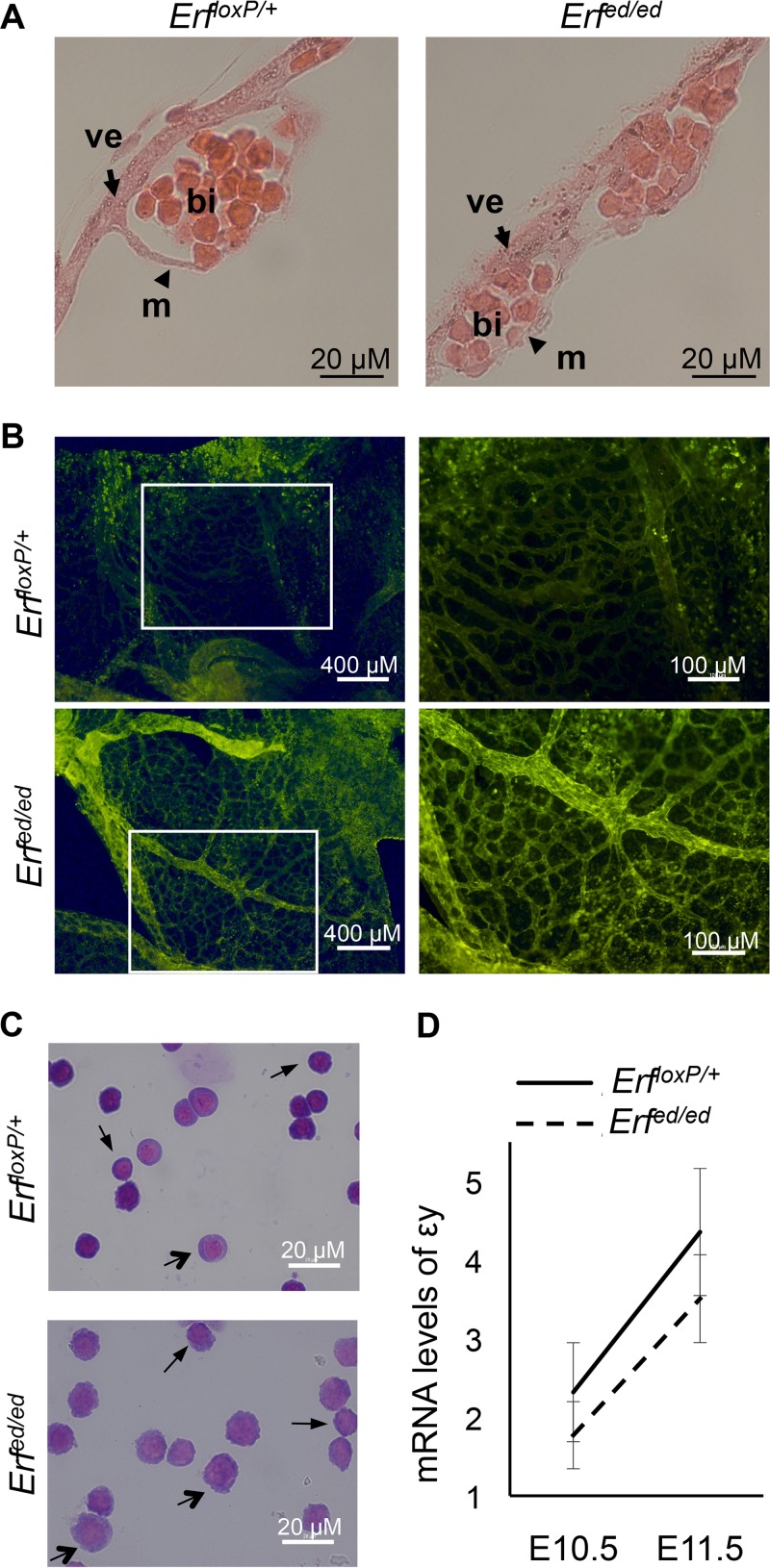
Because of the onset of anemia as early as E9.5, we initially examined erythropoiesis at E9.5 and E10.5, when erythroblasts mature semisynchronously. At E9.5, the Erfed/ed yolk sac revealed normal mesothelium and visceral endoderm junctions, but the blood islands contained fewer and larger primitive erythroblasts (Fig. 2A). In contrast, the yolk sac vasculature of Erfed/ed embryos appeared normal (Fig. 2B). More than 95% of the circulating erythroid cells from E9.5 to E12.5 derive from the primitive wave of hematopoietic progenitors in the yolk sac (4). Thus, the reduced numbers of circulating blood cells would suggest a defect in primitive erythropoiesis. In the absence of Erf, primitive erythroblasts at E10.5 are larger and contain enlarged nuclei with less condensed chromatin, features consistent with a delay in their maturation (Fig. 2C). Globin mRNA levels analyzed by real-time PCR indicate that εy-globin expression in primitive erythroblasts of Erfed/ed embryos was significantly lower than that in their Erf-expressing littermates at both E10.5 and E11.5 (Fig. 2D), consistent with delayed maturation (34). These data indicate that elimination of Erf results in both decreased numbers and delayed maturation of primitive erythroid precursors.
FIG 2.
Erfed/ed yolk sacs exhibit immature primitive erythroid progenitors. (A) Sagittal sections of paraffin-embedded yolk sacs from E9.5 Erfed/ed and ErfloxP/+ embryos stained with H&E. ve, visceral endoderm; m, mesothelium; bi, blood islands. (B) Fluorescent microphotographs of yolk sac from ErfloxP/+ and Meox2-cre; Erfed/ed (Erfed/ed) mice at E10.5 stained with the angiogenesis marker PECAM-1. The boxed areas of the images on the left are magnified on the right. (C) Representative microphotographs of Giemsa-stained peripheral blood cells from E10.5 embryos. Erfed/ed cells exhibit larger primitive precursor cells with less condensed chromatin and larger nuclei than their Erf-expressing littermates. Thin arrows indicate the more immature, larger progenitors while thick arrows indicate the more mature, smaller progenitors in each genotype. (D) εy-globin mRNA levels, normalized to Gapdh mRNA levels, were determined by QPCR in blood cells from E10.5 and E11.5 Erfed/ed and ErfloxP/+ embryos. All values are means ± SE from at least 6 biological samples of each genotype from six litters at E10.5 and from at least 11 biological samples of each genotype from seven litters at E11.5.
Erf elimination impairs yolk sac-derived definitive erythroblasts.
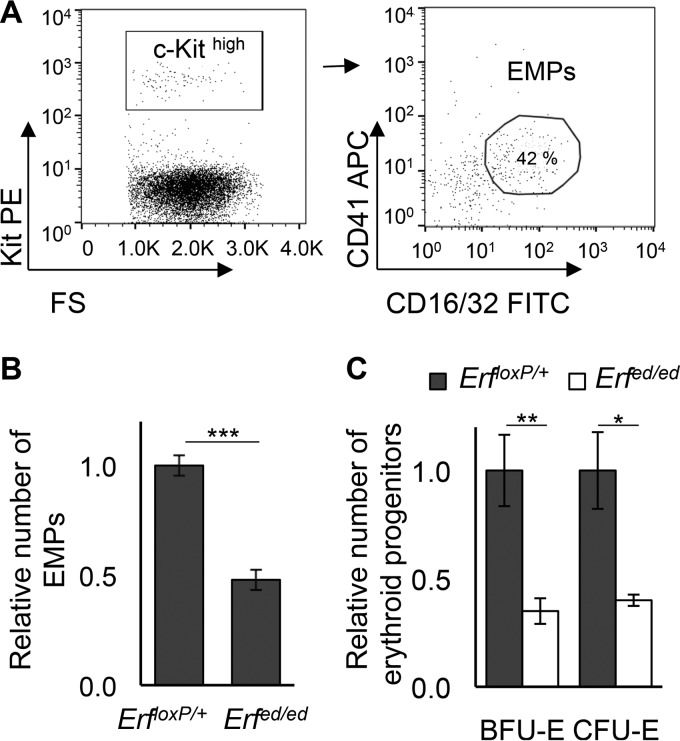
The first circulating definitive erythroid cells that emerge at E11.5 to E12.5 are derived from EMPs/erythroid burst-forming units (BFU-E) that have seeded the fetal liver (35). The profound anemia at E13.5 to E14.5 suggested a defect in EMPs. To explore the effect of Erf elimination on the progenitors of the definitive hematopoietic wave that emerges in the yolk sac, peripheral blood was isolated at E10.5 and CD41+ CD16/32+ EMP cells expressing high levels of c-Kithigh (c-Kithigh) were quantitated by flow cytometry (35) (Fig. 3A). As shown in Fig. 3B, Erfed/ed embryos have half as many phenotypic EMP cells as their Erf-expressing littermates. Consistent with the loss of phenotypic EMPs, the number of definitive erythroid progenitors (BFU-E and erythroid CFU [CFU-E]), defined by their ability to form colonies in semisolid medium, was also significantly reduced at E10.5 (Fig. 3C). Taken together, these data confirm that the yolk sac-derived EMP wave of hematopoietic potential is also impaired by the loss of Erf.
FIG 3.
Erfed/ed embryos have decreased definitive progenitors in bloodstream. (A) Representative flow cytometry analysis of blood from E10.5 embryos stained with anti-c-Kit, anti-CD41, and anti-CD16/32 antibodies. Cells expressing high levels of c-Kit (left panel) that were positive for CD41 and CD16/32 (right panel) were considered the EMP fraction. FS, forward scatter. (B) The numbers of EMP (c-Kithigh CD41+ CD16/32+) cells from E10.5 Erfed/ed and ErfloxP/+ embryonic blood were compared to the average number of the ErfloxP/+ EMP cells of the litter. All values are means ± SE of four biological samples of each genotype from three litters. Statistical analysis was performed using an unpaired t test with two-tailed distribution. (C) Numbers of BFU-E and CFU-E derived from E10.5 Erfed/ed and ErfloxP/+ embryo blood were compared to the average number of the ErfloxP/+ BFU-E/CFU-E of the litter. All values are means ± SE from six biological samples of each genotype from three litters. Statistical analysis was performed using an unpaired t test with two-tailed distribution. *, P < 0.05; **, P < 0.005; ***, P < 0.0005.
Elimination of Erf in hemogenic endothelium recapitulates the Erfed/ed hematopoietic phenotype in definitive hematopoiesis.
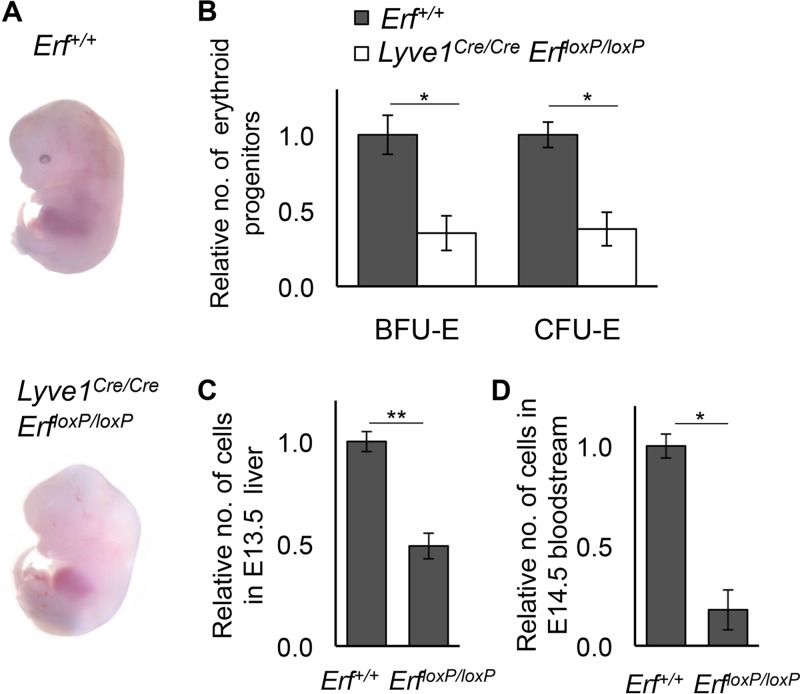
To determine if Erf exerts a cell-autonomous effect on hematopoietic development, we deleted Erf in the hemogenic endothelium, utilizing Lyve1tm1.1(EGFP/cre)Cys mice, which express the Cre recombinase under the control of the Lyve1 gene. Recently, it has been shown that Lyve1 is expressed in hemogenic endothelium and specifically marks EMP cells but not the primitive hematopoietic wave that emerges from the yolk sac (36). Later, Lyve1 is also expressed in blood and lymphatic vessels, lymph nodes, liver, spleen sinuses, and lung and endocardial endothelial cells (37–39). Lyve1tm1.1(EGFP/cre)Cys/tm1.1(EGFP/cre)Cys ErfloxP/loxP (Lyve1Cre/Cre ErfloxP/loxP) mice died in utero and appeared anemic, a phenotype analogous to that of the epiblast-specific Erfed/ed embryos. They were pale (Fig. 4A) and showed reduced BFU-E/CFU-E counts at E10.5 (Fig. 4B), indicating a defect in EMPs before their migration to the liver. At E13.5 the mice exhibited decreased liver cell numbers and severely reduced blood cells in the bloodstream at E14.5 (Fig. 4C and D, respectively). Both the small liver size and the reduced number of circulating blood cells suggest impaired EMP-derived hematopoiesis, similar to that observed in epiblast Erf-null embryos (see below). This would be indicative of a cell-autonomous defect of Erf elimination, specifically in the definitive hematopoietic wave of the yolk sac.
FIG 4.
Elimination of Erf in the definitive yolk sac wave leads to anemia. (A) Representative microphotographs of embryos at E13.5 showing apparent anemia of Lyve1tm1.1(EGFP/cre)Cys/tm1.1(EGFP/cre)Cys; ErfloxP/loxP (Lyve1Cre/Cre ErfloxP/loxP) embryos compared to results in their Erf+/+ littermates. (B) Numbers of BFU-E and CFU-E derived from E10.5 Lyve1Cre/Cre ErfloxP/loxP and Erf+/+ embryo blood were compared to the average numbers of the Erf+/+ BFU-E/CFU-E of the litter. All values are means ± SE from four biological samples from two litters. Statistical analysis was performed using an unpaired t test with two-tailed distribution. *, P < 0.05. (C) Total liver cell counts of Lyve1Cre/Cre ErfloxP/loxP embryos compared to counts in the Erf+/+ littermates at E13.5. All values are means ± SE from two biological samples of each genotype from two litters. Statistical analysis was performed using an unpaired t test with two-tailed distribution. *, P < 0.05; **, P < 0.005. (D) Total cell counts of Lyve1Cre/Cre ErfloxP/loxP embryos compared to counts of the Erf+/+ littermates in E14.5 bloodstream. All values are means ± SE from two biological samples of each genotype from two litters. Statistical analysis was performed using an unpaired t test with two-tailed distribution. *, P < 0.05.
Surprisingly, in contrast to the Lyve1Cre/Cre ErfloxP/loxP embryos, Lyve1Cre/+ ErfloxP/loxP mice were viable, albeit at sub-Mendelian frequencies (Fig. S3D). Analysis, of the Erf expression in fetal livers showed that heterozygous Lyve1-cre embryos have significantly higher expression than the homozygous Lyve1-cre embryos as well as the Meox2-cre embryos (Fig. S3C). This would suggest that the frequency of Erf elimination and/or the level of expression of Erf in EMPs may be critical for the severity of the defect and thus survival.
Erf elimination compromises fetal liver erythropoiesis.
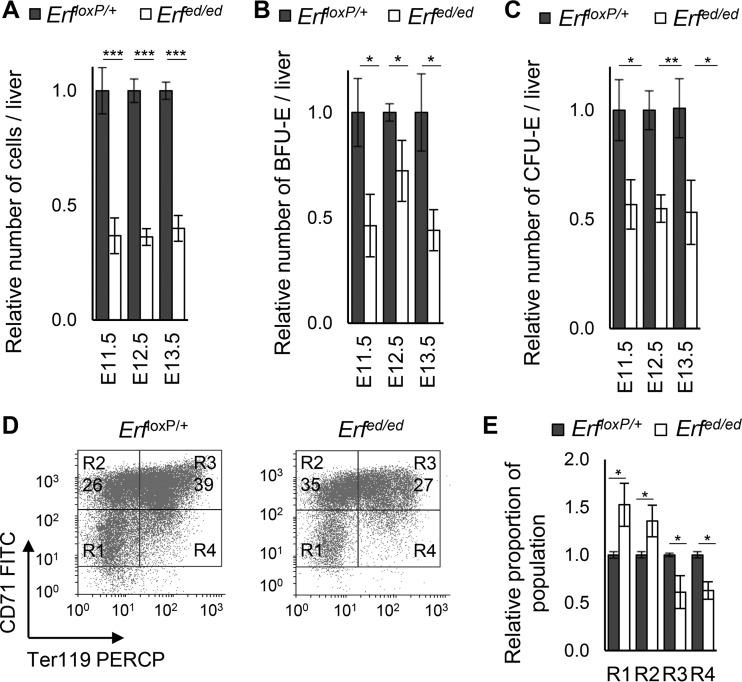
The increased anemia at E13.5 to E14.5 indicated a profound lack of definitive erythroid cells which are produced at this time in the fetal liver and make up the majority of the circulating cells at these time points (5, 40). Analysis of Erfed/ed embryos from E11.5 to E13.5 revealed significantly reduced liver size and liver cell numbers compared to those of their ErfloxP/+ littermates (Fig. 5A and Fig. S4A). To examine if this reduction is due to the colonization of the liver and/or the expansion of the erythroid progenitors in the liver, we analyzed BFU-E and CFU-E numbers at E11.5 to E13.5. Consistent with the reduced number of EMPs in the yolk sac and in the bloodstream, Erfed/ed embryos contained fewer BFU-E and CFU-E in the liver than their ErfloxP/+ littermates (Fig. 5B and C). Interestingly, BFU-E abundance in Erfed/ed embryo livers at E11.5 was comparable to that of their ErfloxP/+ littermates, whereas it was increased at E12.5 to E13.5 (Fig. S4B). The abundance of CFU-E in the Erfed/ed embryo livers was increased compared to that in ErfloxP/+ littermates throughout E11.5 to E13.5 (Fig. S4C). In addition, nonerythroid liver cells appeared unaffected as Erfed/ed embryos had comparable numbers of hepatocytes and stromal cells and comparable rates of liver cell proliferation and apoptosis (Fig. S5; also unpublished data). We further tested the possible contribution of hepatic cells, eliminating Erf specifically in the hepatocytes after crossing the ErfloxP/loxP mice with the Alfp-Cre mice (41). Alfp-Cre; ErfloxP/loxP mice were normal with no hematopoietic defects (not shown). All these data indicate that Erf elimination may also affect erythroid maturation in the fetal liver.
FIG 5.
Erfed/ed embryos have a decreased differentiation rate in the fetal liver. (A) Total number of liver cells. (B) Number of BFU-E per liver at E11.5 to E13.5. (C) Number of CFU-E at E11.5 to E13.5. Values for Erfed/ed and ErfloxP/+ embryos are compared to the average value of the ErfloxP/+ littermates. All values are means ± SE from samples of at least six litters per gestation day (see Table S3 in the supplemental material) for total cells in liver and at least six litters per gestation day (Table S4) for BFU-E/CFU-E counts. Statistical analysis was performed using an unpaired t test with two-tailed distribution. (D) Representative flow cytometry of Erfed/ed E12.5 liver cells stained for Ter119, a marker for early proerythroblasts to mature erythrocytes (R3 to R5), and CD71, a marker for all proerythroblasts, except mature erythrocytes (R1 to R4). (E) Comparison of R1 to R4 populations of E12.5 liver cells, analyzed by flow cytometry as described for panel D. Values for Erfed/ed and ErfloxP/+ cells are compared to the average value of the ErfloxP/+ littermates, and results are expressed as ratios. All values are means ± SE from at least six biological samples of each genotype from at least four litters. Statistical analysis was performed using an unpaired t test with two-tailed distribution. *, P < 0.05; **, P < 0.005; ***, P < 0.0005.
We thus examined erythroid maturation by analyzing the abundance of the populations of region 1 (R1) to R4 in E12.5 fetal liver by CD71 and Ter119 staining and flow cytometry (Fig. 5D). This analysis suggested that, indeed, the early precursor cells and the proerythroblasts (R1), as well as the proerythroblasts and the early basophilic proerythroblasts (R2), are more abundant in Erfed/ed embryo livers, while the early and late basophilic erythroblasts (R3) and the polychromatophilic and orthochromatic erythroblast (R4) populations (42) are less abundant (Fig. 5E). Consistent with the colony assays, these data indicate that in addition to the initial colonization, there is also a defect in the maturation of the erythroid precursors in the fetal liver. This apparently reduced rate of maturation was not due to a defect in cell cycle at E13.5 (Fig. S6) or to the lack of erythroblast contacts with macrophages as they were evident in E12.5 livers (Fig. S7). The defect in maturation was also not evident at E11.5, the earlier stages of liver population by EMPs (Fig. 8). It would thus appear that the maturational defect of the Erfed/ed liver erythroblasts is cell autonomous.
Erf is required for HSC development.
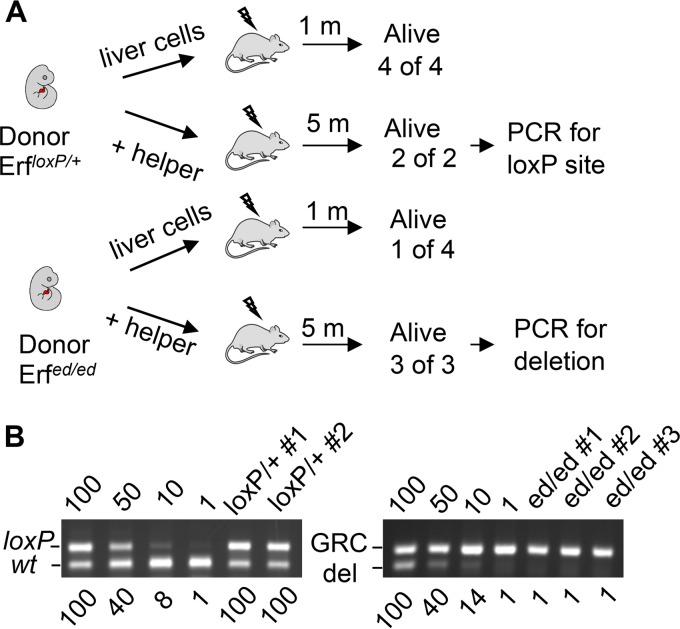
The dramatic decrease in peripheral blood at E14.5 raised the possibility that not only the EMPs but also the HSCs are affected by the Erf loss. Therefore, we examined the role of Erf in hematopoietic stem cell maintenance and differentiation via both noncompetitive and competitive repopulation experiments to evaluate short-term and long-term HSCs. Fifteen to 23 days after the transplantation of E13.5 liver cells in irradiated mice of the same haplotype, 3 of 4 mice repopulated with Erfed/ed cells died in contrast to survival of all of the mice transplanted with Erf-competent cells, indicating a defect in short-term HSCs (Fig. 6A). For the competitive repopulation assays, E12.5 fetal liver cells were coinjected with spleen cells from wild-type (wt) animals in irradiated wt host mice of the same genetic background (Fig. 6A). Donor HSC engraftment was estimated with semiquantitative PCR at 5 months after the injection. Our data suggest that in contrast to the ErfloxP/+ cells that contributed to almost all the hematopoietic cells of the irradiated animals, the Erfed/ed cells had a marginally detectable contribution (Fig. 6B), suggesting that the long-term HSCs may also be affected by the elimination of Erf.
FIG 6.
Erfed/ed embryos have reduced HSCs in liver transplant recipients. (A) Outline of repopulation experiments of sublethally irradiated mice with E13.5 liver cells or competitive repopulation experiments with E12.5 liver donor cells (+ helper) from Erfed/ed and ErfloxP/+ embryos assayed 1 or 5 months (m) after irradiation. (B) Detection of the repopulating cells in the competitive repopulation experiment 5 months after irradiation by semiquantitative PCR. Transplanted Erf-expressing cells are detected by the presence of the 170-bp band (loxP; left panel) while Erf-null cells are detected by the expression of the 178-bp band (del; right panel) generated after the excision of the Erf gene. The 101-bp wt Erf band and the 206-bp band from the GRCm38.p4 locus (GRC) were used as PCR controls. Lanes 100, 50, 10, and 1 are quantitation controls containing 100%, 50%, 10%, 1% ErfloxP/+ or Erfed/ed DNA mixed with Erf+/+ DNA. The numbers below indicate the percentage of the test band compared to the 100% sample.
DISCUSSION
In this study, we demonstrate for first time that elimination of Erf in the murine embryo proper at E5 is detrimental to embryonic erythropoiesis and leads to embryonic death at around E14.5 due to severe anemia. Our data indicate that Erf elimination decreases, but does not block, the primitive and definitive yolk sac-derived erythropoiesis waves and decreases the efficiency of fetal HSCs that support adult hematopoiesis.
A wave of primitive erythroid progenitors emerges from the yolk sac at E7.5 to E9.0 (5), providing the majority of circulating blood cells until E12.5 (4). The reduced numbers of blood cells at E9.5 and onwards in the Erfed/ed embryos indicate a defect in the production and/or differentiation of these cells. The immature cell morphology and the decreased expression of εy-globin suggest that Erfed/ed may affect the onset or differentiation rate of these cells.
The second, EMP, wave emerging from the yolk sac at E8.25 and colonizing the fetal liver (5) is also reduced in the absence of Erf. Both the number of EMPs assessed by flow cytometry and the number of BFU-E and CFU-E, circulating in the bloodstream of E10.5 embryos are reduced in Erfed/ed embryos. Erfed/ed embryos show additional differentiation defects in definitive erythropoiesis after EMPs have homed to the fetal liver. Consistent with the decrease in the number of EMPs, total cells in the liver, as well as erythroid precursor cells, are decreased in the absence of Erf. However, both BFU-E/CFU-E colony assays and flow cytometry of liver cells indicate increased proportions of R1 and R2 proerythroblasts and reduced proportions of the mature R3 and R4 populations. These data suggest an additional role for Erf in R2-R3 differentiation. The apparent defect in the production and differentiation of the definitive erythroid lineage in the absence of Erf appears to be a cell-autonomous event, as evidenced by the elimination of Erf in the Lyve1-expressing hemogenic endothelium cells of the yolk sac. Similar to the epiblast-deleted Erfed/ed embryos, the Lyve1Cre/Cre ErfloxP/loxP embryos appear anemic, have reduced BFU-E/CFU-E counts in the bloodstream at E10.5, and have reduced numbers of total cells and erythroid precursors in the liver at E14.5.
Finally, Erf appears to also have a crucial role in the differentiation of HSCs emerging from the aorta-gonad-mesonephros (AGM) region at E10.5, migrating to the liver and ultimately constituting the adult HSCs in the bone marrow. Transplantation of Erfed/ed liver cells into irradiated mice shows that recipients could not live beyond 4 weeks, suggesting a defect in short-term HSCs. Addition of spleen helper cells from wt mice to assess long-term HSCs indicates a marginal contribution from the Erfed/ed cells at 5 months after transplantation. These data allude to a defect in HSC maintenance or differentiation ability that may have implications in adult pathologies and needs further examination.
In spite of the wide effect of Erf on hematopoiesis, it remains unclear if there is a common underlying mechanism or if there are distinct downstream effectors involved in the different cell types affected. The fact that in the absence of Erf we have quantitative differences rather than a block in any specific differentiation step or the cell cycle makes determination of possible mechanisms even more challenging. Indeed, transcriptional analysis of E13.5 livers, before the collapse of peripheral blood levels, failed to reveal statistically significant differences in genes or pathways between wt and Erf-null cells (unpublished data). This would be consistent with a quantitative rather than an instructive effect. This conclusion is also supported by the apparent difference in viability of the Lyve1-Cre heterozygous and homozygous animals in which the observed difference may be the result of the extent of the deletion. In addition, our unpublished data indicate a minimal but statistically significant sub-Mendelian frequency of the viable ErfloxP/− animals, suggesting an effect because of the timing and/or the extent of Erf expression.
Erf is a member of the ETS family of transcription factors and an effector in the RTK/Ras/Erk pathway, both of which have been involved in diverse facets of hematopoiesis. Erk has been reported to negatively affect fetal hemopoiesis, leading to tight junctions between endothelial cells in the AGM region of zebrafish, repressing the emergence of HSCs, and promoting the arterial endothelial identity (43). Such an effect would be consistent with the apparent decrease of the HSCs observed in the absence of Erf and the reduced numbers of EMPs and primitive cells that emerge from the hemogenic endothelium in the yolk sac (44). In addition, oncogenic H-Ras blocks the terminal differentiation of CFU-E progenitors in vivo (45), consistent with the differentiation defect of the R2 population observed in Erfed/ed livers.
ETS transcription factors recognize a common DNA-binding motif (46) and may antagonize each other on a variety of targets. Erf, a transcriptional repressor may ensure the absence of spurious activation by other Ets family proteins, leading to the proper differentiation programs and homeostasis. Overexpression of PU-1 is reported to block the differentiation of BFU-E/CFU-E to mature erythrocytes (47). The archetypal fusion Gag-Myb-Ets oncoprotein, encoded by E26 retrovirus, induces acute leukemia and can block both myeloid and erythroid differentiation at the stage of BFU-E/CFU-E (48). Overactivation of Etv2 leads to the upregulation of Fli1 (49), which promotes the formation of the vascular system (50) and the upregulation of PU.1. Fli1 was identified as a proto-oncogene in erythroleukemias induced by retroviral integration (51). Finally, Erf itself has been reported to promote erythroid differentiation (26) and may antagonize Ets1, which may also have a role in this process (52). Thus, elimination of Erf may affect the entire regulatory network of ETS factors in hematopoiesis.
Another plausible hypothesis on the diverse effects of Erf in hematopoiesis is its interplay with the RUNX factors. Runx1 is expressed in HSCs and downstream myeloid cells, but its expression is decreased in maturing erythroid cells (53). Cooperation of Runx1 with PU.1 is critical for the commitment to myeloid lineages (13). Runx2 is highly expressed in HSCs and is decreased as they mature (54). Runx1 transcription appears to be regulated via ETS factor binding to one of its enhancers (24), while Runx2 may also be regulated by Erf or other ETS proteins (32). Finally, we have recently shown that the Runx-Erf interplay on osteogenic targets may be important for bone development and craniosynostosis (32). It is conceivable that a similar interplay may also be relevant in hematopoietic development.
Taken together, our data strongly suggest an important role of Erf in all three waves primitive, EMP, and HSC hematopoiesis during ontogeny. Erf activity is regulated by phosphorylation and nucleocytoplasmic shuttling and thus could be an appealing target for intervention. However, further experiments are needed to elucidate its precise mechanism and the stage of action during hematopoietic differentiation, particularly in HSC differentiation.
MATERIALS AND METHODS
Generation of conditional Erf−/− mice.
Elimination of Erf in epiblast-derived tissues (Erfed/ed) was achieved by the intercross of Meox2tm1(cre)Sor/J (stock no. 003755; Jackson Laboratory) mice with ErfloxP/loxP mice (32) to generate Meox2tm1(cre)Sor/J; Erf+/− mice and by crossing these mice with ErfloxP/loxP mice to totally eliminate Erf in the embryo proper. Lyve1tm1.1(EGFP/cre)Cys/J mice (stock no. 012601; Jackson Laboratory) were crossed with the ErfloxP/loxP mice to eliminate Erf in the hemogenic endothelium (39) and finally in the definitive yolk sac-derived wave (36). The liver-specific Alfp-Cre; ErfloxP/loxP mice were used to eliminate Erf in hepatic cells (41). Mice were maintained in the Institute of Molecular Biology and Biotechnology (IMBB) colony. All experiments were approved by the General Directorate of Veterinary Services, Region Crete (permit numbers EL 91BIO-02 and EL91-BIOexp-02; project license no. 27289 to G. Mavrothalassitis). All efforts were undertaken to minimize animal suffering and use according to Greek and European Union guidelines.
Tissue collection and histological analysis.
Timed-pregnant female mice were sacrificed at specified days of gestation by cervical dislocation according to institutional guidelines. The uteri were isolated from the peritoneum, and placentas were removed using watchmaker's forceps (number 5). The remaining whole yolk sacs were rinsed in phosphate-buffered saline (PBS) to remove vestiges of maternal blood and placed in Iscove's modified Dulbecco's medium (IMDM) (catalog no. 12440053; ThermoFisher) with 2% fetal bovine serum (FBS) (50115; Biochrom). Blood was collected from separated embryos. Fetal livers were isolated from embryos not subjected to bleeding and dissociated by gentle pipetting in IMDM with 2% FBS. Cells were counted on a hemocytometer. Blood collected on slides was left to dry, fixed with methanol for 10 min, stained with Giemsa (Merck) for 2 min, washed with excess water, dried, and mounted for observation.
For hematoxylin-eosin (H&E) staining, yolk sacs were fixed with 4% paraformaldehyde for 1 h at room temperature, dehydrated gradually with an ethanol series of 70%, 90%, and 100% for 30 min at room temperature, cleared in xylol for 30 min, and impregnated with paraffin at 58°C twice for 30 min each time. Paraffin sections (5 μm) were stained with hematoxylin-eosin. For immunostaining, yolk sacs were fixed with 4% paraformaldehyde (PFA) for 1 h at room temperature, washed extensively with PBS, stained with rat anti-platelet endothelial cell adhesion molecule 1 (anti-PECAM-1) (catalog no. 553370; Pharmingen) overnight at 4°C and then incubated with anti-rat fluorescein isothiocyanate (FITC)-conjugated antibody (F0382; Sigma) in a 1:50 dilution for 1 h at room temperature.
Hematopoietic colony assays.
Erythroid CFU (CFU-E) and burst-forming units (BFU-E) were analyzed from bloodstream and fetal livers at E10.5 and at E11.5 to E13.5, respectively. Fifty thousand cells were plated in 1 ml of 1.2% serum-free methylcellulose (catalog no. H4100; Stem Cell Technologies) supplemented with 37% FBS, 1.25% (wt/vol) bovine serum albumin (BSA), 0.25 μM β-mercaptoethanol, 0.01% sodium bicarbonate, 10 ng/ml interleukin-3 (IL-3) (213-13; Peprotech), 10 ng/ml IL-6 (216-16; Peprotech), 1 ng/ml granulocyte-macrophage colony-stimulating factor (GM-CSF) (315-03; Peprotech), and 2 U/ml erythropoietin (gift from Marieke von Lindern). Additionally, 100 ng/ml stem cell factor (SCF; supernatant of CHO producer cells) was added in colonies from E10.5 blood. The cells were cultured in 35-mm dishes at 37°C with 5% CO2 and 100% humidity. The numbers of colonies were scored after 3 days for CFU-E and after 7 days for BFU-E.
Flow cytometric analysis.
Single-cell suspensions were prepared from E11.5 to E13.5 livers and stained for 15 min at 4°C in PBS–1% fetal bovine serum with biotinylated rat anti-Ter119 antibody (catalog no. 116203; Biolegend), a marker for all stages of differentiation from early proerythroblasts to mature erythrocytes, streptavidin-peridinin chlorophyll protein (PerCP) (405213; Biolegend), and FITC-coupled rat anti-CD71, a marker for all erythroblasts except mature erythrocytes (113805; Biolegend). Blood cells were stained with FITC-coupled anti-CD16/32 (101305; Biolegend), phycoerythrin (PE)-coupled anti-c-Kit (105807; Biolegend), and allophycocyanin (APC)-coupled anti-CD41 (133913; Biolegend), markers of EMPs. Stained cells were analyzed in a MoFloT high-performance cell sorter with FlowJo, version 10, software.
Real-time QPCR.
Total RNA was extracted from peripheral blood and fetal livers at E10.5 to E11.5, using TRIzol reagent (catalog no. 15596018; Invitrogen) according to the manufacturer's instructions. mRNA was subjected to reverse transcription using a SuperScript first-strand synthesis kit (11904-018; Invitrogen). Expression levels of β-major globin, detected mainly in definitive blood cells (forward [Fw], 5′-CACAAACCCCAGAAACAGACA-3′; reverse [Rv], 5′-CTGACAGATGCTCTCTTGGG-3′), βΗ1-globin (Fw, 5′-CTCAAGGAGACCTTTGCTCA-3′; Rv: 5′-AGTCCCCATGGAGTCAAAGA-3′), and εy-globin, detected in primitive erythroid cells (Fw, 5′-GGAGAGTCCATTAAGAACCTAGACAA-3′; Rv, 5′-CTGTGAATTCATTGCCGAAGTAC-3′), were examined with real-time PCRs. Reactions were performed using 5 ng of total cDNA using 2× Brilliant III SYBR green quantitative PCR (QPCR) master mix (600882-51; Stratagene) in an Applied Biosystems StepOne Plus real-time PCR machine. All expression levels were normalized to Gapdh levels (Fw, 5′-CCAGTATGACTCCACTCACG-3′; Rv, 5′-GACTCCACGACATACTCAGC-3′) in the same cDNA. To distinguish Alfp-Cre-homozygous from -heterozygous mice, quantitative real-time PCR was performed with a general Cre primer (Fw, 5′-GCGGTCTGGCAGTAAAAACTATC-3′; Rv, 5′GTGAAACAGCATTGCTGTCACTT-3′) in 10 ng of DNA.
Reconstitution analysis.
Mice at 6 weeks of age were lethally irradiated with 950 rads. Five hundred thousand fetal liver cells at E13.5 were injected intravenously into the lethally irradiated recipient Erf+/+ mice. For the competitive repopulation studies, 4 × 105 fetal liver cells from E12.5 embryos were coinjected intravenously with 2 × 105 spleen cells into lethally irradiated recipient Erf+/+ mice. The animals were monitored twice a week for viability and general health. All mice were of mixed C57B6/SV129 background. The engraftment of donor cells was analyzed after 5 months. DNA from blood was extracted with a FlexiGene DNA kit (51204; Qiagen), and PCR for the detection of the loxP allele or the deletion of Erf was performed with the primer pair m11671F (5′-ACGCCACAGCCCACCTCTCC-3′) and 11771R (5′-CAGCAAAAGCTCAGGGAGTG-3′) and the pair 4021F (5′-GCACTGCTAGCTCTGAATGG-3′) and 11771R (5′-CAGCAAAAGCTCAGGGAGTG-3′), respectively.
Statistical analysis.
Statistical analysis was carried out using a two-tailed unpaired t test with Excel 2016. For all graphs, data are presented as means ± standard errors (SE). Pearson's chi-square test was used to evaluate the actual versus the expected frequency of the genotypes. The P values indicating statistical significance are given in the figure legends.
Supplementary Material
ACKNOWLEDGMENTS
We thank C. Kourouniotis and the IMBB animal facility staff for expert animal support, Z. Vlata for flow cytometry analysis, and Marieke von Lindern (Sanquin, Amsterdam, Netherlands) for help with cell cultures and materials. We thank Kathleen McGrath for advice regarding the flow cytometric analysis of EMPs and Anne Koniski for advice regarding erythroid colony assays.
Support was provided by Greek Ministry of Education grant Synergasia 09SYN-11-902 to G.M. and National Institutes of Health (USA) grant R01 HL130670 to J.P.
I.P. designed and performed experiments and wrote the manuscript, J.P. designed experiments and wrote the manuscript, and G.M. conceived the project, designed experiments, and wrote the manuscript.
We have no conflicts of interest to declare.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/MCB.00183-17.
REFERENCES
- 1.Dzierzak E, Speck NA. 2008. Of lineage and legacy: the development of mammalian hematopoietic stem cells. Nat Immunol 9:129–136. doi: 10.1038/ni1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Palis J, Malik J, McGrath KE, Kingsley PD. 2010. Primitive erythropoiesis in the mammalian embryo. Int J Dev Biol 54:1011–1018. doi: 10.1387/ijdb.093056jp. [DOI] [PubMed] [Google Scholar]
- 3.Kingsley PD, Malik J, Fantauzzo KA, Palis J. 2004. Yolk sac-derived primitive erythroblasts enucleate during mammalian embryogenesis. Blood 104:19–25. doi: 10.1182/blood-2003-12-4162. [DOI] [PubMed] [Google Scholar]
- 4.Fraser ST, Isern J, Baron MH. 2007. Maturation and enucleation of primitive erythroblasts during mouse embryogenesis is accompanied by changes in cell-surface antigen expression. Blood 109:343–352. doi: 10.1182/blood-2006-03-006569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Palis J, Robertson S, Kennedy M, Wall C, Keller G. 1999. Development of erythroid and myeloid progenitors in the yolk sac and embryo proper of the mouse. Development 126:5073–5084. [DOI] [PubMed] [Google Scholar]
- 6.Bertrand JY, Jalil A, Klaine M, Jung S, Cumano A, Godin I. 2005. Three pathways to mature macrophages in the early mouse yolk sac. Blood 106:3004–3011. doi: 10.1182/blood-2005-02-0461. [DOI] [PubMed] [Google Scholar]
- 7.Tober J, Koniski A, McGrath KE, Vemishetti R, Emerson R, de Mesy-Bentley KK, Waugh R, Palis J. 2007. The megakaryocyte lineage originates from hemangioblast precursors and is an integral component both of primitive and of definitive hematopoiesis. Blood 109:1433–1441. doi: 10.1182/blood-2006-06-031898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rich IN, Kubanek B. 1979. The ontogeny of erythropoiesis in the mouse detected by the erythroid colony-forming technique. I. Hepatic and maternal erythropoiesis. J Embryol Exp Morphol 50:57–74. [PubMed] [Google Scholar]
- 9.Muller AM, Medvinsky A, Strouboulis J, Grosveld F, Dzierzak E. 1994. Development of hematopoietic stem cell activity in the mouse embryo. Immunity 1:291–301. doi: 10.1016/1074-7613(94)90081-7. [DOI] [PubMed] [Google Scholar]
- 10.Godin I, Dieterlen-Lievre F, Cumano A. 1995. Emergence of multipotent hemopoietic cells in the yolk sac and paraaortic splanchnopleura in mouse embryos, beginning at 8.5 days postcoitus. Proc Natl Acad Sci U S A 92:773–777. doi: 10.1073/pnas.92.3.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Medvinsky A, Dzierzak E. 1996. Definitive hematopoiesis is autonomously initiated by the AGM region. Cell 86:897–906. doi: 10.1016/S0092-8674(00)80165-8. [DOI] [PubMed] [Google Scholar]
- 12.de Bruijn MF, Speck NA, Peeters MC, Dzierzak E. 2000. Definitive hematopoietic stem cells first develop within the major arterial regions of the mouse embryo. EMBO J 19:2465–2474. doi: 10.1093/emboj/19.11.2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Imperato MR, Cauchy P, Obier N, Bonifer C. 2015. The RUNX1-PU.1 axis in the control of hematopoiesis. Int J Hematol 101:319–329. doi: 10.1007/s12185-015-1762-8. [DOI] [PubMed] [Google Scholar]
- 14.Scott EW, Simon MC, Anastasi J, Singh H. 1994. Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science 265:1573–1577. doi: 10.1126/science.8079170. [DOI] [PubMed] [Google Scholar]
- 15.Liu F, Patient R. 2008. Genome-wide analysis of the zebrafish ETS family identifies three genes required for hemangioblast differentiation or angiogenesis. Circ Res 103:1147–1154. doi: 10.1161/CIRCRESAHA.108.179713. [DOI] [PubMed] [Google Scholar]
- 16.Lee D, Park C, Lee H, Lugus JJ, Kim SH, Arentson E, Chung YS, Gomez G, Kyba M, Lin S, Janknecht R, Lim DS, Choi K. 2008. ER71 acts downstream of BMP, Notch, and Wnt signaling in blood and vessel progenitor specification. Cell Stem Cell 2:497–507. doi: 10.1016/j.stem.2008.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sumanas S, Gomez G, Zhao Y, Park C, Choi K, Lin S. 2008. Interplay among Etsrp/ER71, Scl, and Alk8 signaling controls endothelial and myeloid cell formation. Blood 111:4500–4510. doi: 10.1182/blood-2007-09-110569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kataoka H, Hayashi M, Nakagawa R, Tanaka Y, Izumi N, Nishikawa S, Jakt ML, Tarui H, Nishikawa S. 2011. Etv2/ER71 induces vascular mesoderm from Flk1+ PDGFRα+ primitive mesoderm. Blood 118:6975–6986. doi: 10.1182/blood-2011-05-352658. [DOI] [PubMed] [Google Scholar]
- 19.Wareing S, Mazan A, Pearson S, Gottgens B, Lacaud G, Kouskoff V. 2012. The Flk1-Cre-mediated deletion of ETV2 defines its narrow temporal requirement during embryonic hematopoietic development. Stem Cells 30:1521–1531. doi: 10.1002/stem.1115. [DOI] [PubMed] [Google Scholar]
- 20.Okano K, Hibi A, Miyaoka T, Inoue T, Sugimoto H, Tsuchiya K, Akiba T, Nitta K. 2012. Inhibitory effects of the transcription factor Ets-1 on the expression of type I collagen in TGF-β1-stimulated renal epithelial cells. Mol Cell Biochem 369:247–254. doi: 10.1007/s11010-012-1388-6. [DOI] [PubMed] [Google Scholar]
- 21.Wareing S, Eliades A, Lacaud G, Kouskoff V. 2012. ETV2 expression marks blood and endothelium precursors, including hemogenic endothelium, at the onset of blood development. Dev Dyn 241:1454–1464. doi: 10.1002/dvdy.23825. [DOI] [PubMed] [Google Scholar]
- 22.Taoudi S, Bee T, Hilton A, Knezevic K, Scott J, Willson TA, Collin C, Thomas T, Voss AK, Kile BT, Alexander WS, Pimanda JE, Hilton DJ. 2011. ERG dependence distinguishes developmental control of hematopoietic stem cell maintenance from hematopoietic specification. Genes Dev 25:251–262. doi: 10.1101/gad.2009211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang L, Liu T, Xu L, Gao Y, Wei Y, Duan C, Chen GQ, Lin S, Patient R, Zhang B, Hong D, Liu F. 2013. Fev regulates hematopoietic stem cell development via ERK signaling. Blood 122:367–375. doi: 10.1182/blood-2012-10-462655. [DOI] [PubMed] [Google Scholar]
- 24.Nottingham WT, Jarratt A, Burgess M, Speck CL, Cheng JF, Prabhakar S, Rubin EM, Li PS, Sloane-Stanley J, Kong ASJ, de Bruijn MF. 2007. Runx1-mediated hematopoietic stem-cell emergence is controlled by a Gata/Ets/SCL-regulated enhancer. Blood 110:4188–4197. doi: 10.1182/blood-2007-07-100883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Allegra M, Zaragkoulias A, Vorgia E, Ioannou M, Litos G, Beug H, Mavrothalassitis G. 2012. Semaphorin-7a reverses the ERF-induced inhibition of EMT in Ras-dependent mouse mammary epithelial cells. Mol Biol Cell 23:3873–3881. doi: 10.1091/mbc.E12-04-0276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Athanasiou M, Blair DG, Mavrothalassitis G. 2003. ERF, an ETS-related transcriptional repressor, can induce erythroid differentiation. Anticancer Res 23:2143–2153. [PubMed] [Google Scholar]
- 27.Athanasiou M, LeGallic L, Watson DK, Blair DG, Mavrothalassitis G. 2000. Suppression of the Ewing's sarcoma phenotype by FLI1/ERF repressor hybrids. Cancer Gene Ther 7:1188–1195. doi: 10.1038/sj.cgt.7700220. [DOI] [PubMed] [Google Scholar]
- 28.Le Gallic L, Sgouras D, Beal G Jr, Mavrothalassitis G. 1999. Transcriptional repressor ERF is a Ras/mitogen-activated protein kinase target that regulates cellular proliferation. Mol Cell Biol 19:4121–4133. doi: 10.1128/MCB.19.6.4121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sgouras DN, Athanasiou MA, Beal GJ Jr, Fisher RJ, Blair DG, Mavrothalassitis GJ. 1995. ERF: an ETS domain protein with strong transcriptional repressor activity, can suppress ETS-associated tumorigenesis and is regulated by phosphorylation during cell cycle and mitogenic stimulation. EMBO J 14:4781–4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Verykokakis M, Papadaki C, Vorgia E, Le Gallic L, Mavrothalassitis G. 2007. The RAS-dependent ERF control of cell proliferation and differentiation is mediated by c-Myc repression. J Biol Chem 282:30285–30294. doi: 10.1074/jbc.M704428200. [DOI] [PubMed] [Google Scholar]
- 31.Papadaki C, Alexiou M, Cecena G, Verykokakis M, Bilitou A, Cross JC, Oshima RG, Mavrothalassitis G. 2007. Transcriptional repressor Erf determines extraembryonic ectoderm differentiation. Mol Cell Biol 27:5201–5213. doi: 10.1128/MCB.02237-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Twigg SR, Vorgia E, McGowan SJ, Peraki I, Fenwick AL, Sharma VP, Allegra M, Zaragkoulias A, Sadighi Akha E, Knight SJ, Lord H, Lester T, Izatt L, Lampe AK, Mohammed SN, Stewart FJ, Verloes A, Wilson LC, Healy C, Sharpe PT, Hammond P, Hughes J, Taylor S, Johnson D, Wall SA, Mavrothalassitis G, Wilkie AO. 2013. Reduced dosage of ERF causes complex craniosynostosis in humans and mice and links ERK1/2 signaling to regulation of osteogenesis. Nat Genet 45:308–313. doi: 10.1038/ng.2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tallquist MD, Soriano P. 2000. Epiblast-restricted Cre expression in MORE mice: a tool to distinguish embryonic vs. extra-embryonic gene function. Genesis 26:113–115. doi:. [DOI] [PubMed] [Google Scholar]
- 34.Kingsley PD, Malik J, Emerson RL, Bushnell TP, McGrath KE, Bloedorn LA, Bulger M, Palis J. 2006. “Maturational” globin switching in primary primitive erythroid cells. Blood 107:1665–1672. doi: 10.1182/blood-2005-08-3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McGrath KE, Frame JM, Fegan KH, Bowen JR, Conway SJ, Catherman SC, Kingsley PD, Koniski AD, Palis J. 2015. Distinct sources of hematopoietic progenitors emerge before HSCs and provide functional blood cells in the mammalian embryo. Cell Rep 11:1892–1904. doi: 10.1016/j.celrep.2015.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee LK, Ghorbanian Y, Wang W, Wang Y, Kim YJ, Weissman IL, Inlay MA, Mikkola HK. 2016. LYVE1 marks the divergence of yolk sac definitive hemogenic endothelium from the primitive erythroid lineage. Cell Rep 17:2286–2298. doi: 10.1016/j.celrep.2016.10.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goode DK, Obier N, Vijayabaskar MS, Lie ALM, Lilly AJ, Hannah R, Lichtinger M, Batta K, Florkowska M, Patel R, Challinor M, Wallace K, Gilmour J, Assi SA, Cauchy P, Hoogenkamp M, Westhead DR, Lacaud G, Kouskoff V, Gottgens B, Bonifer C. 2016. Dynamic gene regulatory networks drive hematopoietic specification and differentiation. Dev Cell 36:572–587. doi: 10.1016/j.devcel.2016.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dellinger MT, Meadows SM, Wynne K, Cleaver O, Brekken RA. 2013. Vascular endothelial growth factor receptor-2 promotes the development of the lymphatic vasculature. PLoS One 8:e74686. doi: 10.1371/journal.pone.0074686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gordon EJ, Gale NW, Harvey NL. 2008. Expression of the hyaluronan receptor LYVE-1 is not restricted to the lymphatic vasculature; LYVE-1 is also expressed on embryonic blood vessels. Dev Dyn 237:1901–1909. doi: 10.1002/dvdy.21605. [DOI] [PubMed] [Google Scholar]
- 40.Houssaint E. 1981. Differentiation of the mouse hepatic primordium. II. Extrinsic origin of the haemopoietic cell line. Cell Differ 10:243–252. [DOI] [PubMed] [Google Scholar]
- 41.Kellendonk C, Opherk C, Anlag K, Schutz G, Tronche F. 2000. Hepatocyte-specific expression of Cre recombinase. Genesis 26:151–153. doi:. [DOI] [PubMed] [Google Scholar]
- 42.Sieff CA, Yang J, Merida-Long LB, Lodish HF. 2010. Pathogenesis of the erythroid failure in Diamond Blackfan anaemia. Br J Haematol 148:611–622. doi: 10.1111/j.1365-2141.2009.07993.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang C, Lv J, He Q, Wang S, Gao Y, Meng A, Yang X, Liu F. 2014. Inhibition of endothelial ERK signalling by Smad1/5 is essential for haematopoietic stem cell emergence. Nat Commun 5:3431. doi: 10.1038/ncomms4431. [DOI] [PubMed] [Google Scholar]
- 44.Padron-Barthe L, Temino S, Villa del Campo C, Carramolino L, Isern J, Torres M. 2014. Clonal analysis identifies hemogenic endothelium as the source of the blood-endothelial common lineage in the mouse embryo. Blood 124:2523–2532. doi: 10.1182/blood-2013-12-545939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang J, Socolovsky M, Gross AW, Lodish HF. 2003. Role of Ras signaling in erythroid differentiation of mouse fetal liver cells: functional analysis by a flow cytometry-based novel culture system. Blood 102:3938–3946. doi: 10.1182/blood-2003-05-1479. [DOI] [PubMed] [Google Scholar]
- 46.Hollenhorst PC, McIntosh LP, Graves BJ. 2011. Genomic and biochemical insights into the specificity of ETS transcription factors. Annu Rev Biochem 80:437–471. doi: 10.1146/annurev.biochem.79.081507.103945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schuetze S, Stenberg PE, Kabat D. 1993. The Ets-related transcription factor PU.1 immortalizes erythroblasts. Mol Cell Biol 13:5670–5678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rascle A, Ferrand N, Gandrillon O, Samarut J. 1996. Myb-Ets fusion oncoprotein inhibits thyroid hormone receptor/c-ErbA and retinoic acid receptor functions: a novel mechanism of action for leukemogenic transformation by E26 avian retrovirus. Mol Cell Biol 16:6338–6351. doi: 10.1128/MCB.16.11.6338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wong KS, Proulx K, Rost MS, Sumanas S. 2009. Identification of vasculature-specific genes by microarray analysis of Etsrp/Etv2 overexpressing zebrafish embryos. Dev Dyn 238:1836–1850. doi: 10.1002/dvdy.21990. [DOI] [PubMed] [Google Scholar]
- 50.Brown LA, Rodaway AR, Schilling TF, Jowett T, Ingham PW, Patient RK, Sharrocks AD. 2000. Insights into early vasculogenesis revealed by expression of the ETS-domain transcription factor Fli-1 in wild-type and mutant zebrafish embryos. Mech Dev 90:237–252. doi: 10.1016/S0925-4773(99)00256-7. [DOI] [PubMed] [Google Scholar]
- 51.Ben-David Y, Giddens EB, Letwin K, Bernstein A. 1991. Erythroleukemia induction by Friend murine leukemia virus: insertional activation of a new member of the ets gene family, Fli-1, closely linked to c-ets-1. Genes Dev 5:908–918. doi: 10.1101/gad.5.6.908. [DOI] [PubMed] [Google Scholar]
- 52.Clausen PA, Athanasiou M, Chen Z, Dunn KJ, Zhang Q, Lautenberger JA, Mavrothalassitis G, Blair DG. 1997. ETS-1 induces increased expression of erythroid markers in the pluripotent erythroleukemic cell lines K562 and HEL. Leukemia 11:1224–1233. doi: 10.1038/sj.leu.2400735. [DOI] [PubMed] [Google Scholar]
- 53.North TE, Stacy T, Matheny CJ, Speck NA, de Bruijn MF. 2004. Runx1 is expressed in adult mouse hematopoietic stem cells and differentiating myeloid and lymphoid cells, but not in maturing erythroid cells. Stem Cells 22:158–168. doi: 10.1634/stemcells.22-2-158. [DOI] [PubMed] [Google Scholar]
- 54.Kuo YH, Zaidi SK, Gornostaeva S, Komori T, Stein GS, Castilla LH. 2009. Runx2 induces acute myeloid leukemia in cooperation with Cbfβ-SMMHC in mice. Blood 113:3323–3332. doi: 10.1182/blood-2008-06-162248. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.