ABSTRACT
Astrovirus VA1/HMO-C (VA1; mamastrovirus 9) is a recently discovered astrovirus genotype that is divergent from the classic human astroviruses (mamastrovirus 1). The gastrointestinal tract is presumed to be the primary site of infection and pathogenicity for astroviruses. However, VA1 has been independently detected in brain tissue of five cases of human encephalitis. Studies of the pathogenicity of VA1 are currently impossible because there are no reported cell culture systems or in vivo models that support VA1 infection. Here, we describe successful propagation of VA1 in multiple human cell lines. The initial inoculum, a filtered clinical stool sample from the index gastroenteritis case cluster that led to the discovery of VA1, was first passaged in Vero cells. Serial blind passage in Caco-2 cells yielded increasing copies of VA1 RNA, and multistep growth curves demonstrated a >100-fold increase in VA1 RNA 72 h after inoculation. The full-length genomic and subgenomic RNA strands were detected by Northern blotting, and crystalline lattices of viral particles of ∼26-nm diameter were observed by electron microscopy in infected Caco-2 cells. Unlike other human astrovirus cell culture systems, which require addition of exogenous trypsin for continued propagation, VA1 could be propagated equally well with or without the addition of trypsin. Furthermore, VA1 was sensitive to the type I interferon (IFN-I) response, as VA1 RNA levels were reduced by pretreatment of Caco-2 cells with IFN-β1a. The ability to propagate VA1 in cell culture will facilitate studies of the neurotropism and neuropathogenesis of VA1.
IMPORTANCE Astroviruses are an emerging cause of central nervous system infections in mammals, and astrovirus VA1/HMO-C is the most prevalent astrovirus in cases of human encephalitis. This virus has not been previously propagated, preventing elucidation of the biology of this virus. We describe the first cell culture system for VA1, a key step necessary for the study of its ability to cause disease.
KEYWORDS: astrovirus, astrovirus VA1, cell culture, electron microscopy, encephalitis, pathogenesis, subgenomic RNA, viral propagation
INTRODUCTION
Initially identified in 1975, members of the single-stranded, positive-sense RNA viral family Astroviridae have been frequently detected in vertebrate stool samples (1, 2). In humans, the first identified astrovirus species was mamastrovirus 1 (classic human astrovirus), and eight serotypes of this virus have been described (2–4). Most humans are exposed to the classic human astroviruses with seroprevalence as high as 94% to specific serotypes (5–9). The classic human astroviruses predominantly cause a self-limited gastrointestinal illness and have been characterized as the third- to fifth-most-common viral etiology of diarrhea and gastroenteritis in humans (10–14).
Astroviruses contain three open reading frames (ORFs) and based on conserved genomic elements are hypothesized to share common mechanisms of replication (2). Astroviruses encode a slippery sequence and stem-loop secondary RNA structure that enables ribosomal frameshifting to express a polyprotein from ORF1a and ORF1b (15–17). In addition, astroviruses transcribe a subgenomic, positive-sense RNA strand (18–20) that encodes the capsid protein (ORF2). The subgenomic RNA strand is produced during viral replication but is not incorporated into the mature astrovirus virion (19).
Application of molecular methods, including high-throughput sequencing and consensus PCR assays, has identified several novel astroviruses in human stool specimens, including the prototype species of mamastrovirus 6 (representative astrovirus strain MLB1), mamastrovirus 8 (representative astrovirus strain VA2/HMO-A), and mamastrovirus 9 (representative astrovirus strain VA1/HMO-C [VA1]) (21–28). Humans are frequently exposed to these novel astroviruses, as seroprevalence in adults ranges from 65 to 100% (29, 30). Based on their detection in stool samples, the novel human astroviruses are presumed to cause gastrointestinal diseases similar to those produced by the classic human astroviruses (23). However, in the limited number of case-control studies performed to date, the novel human astroviruses have not been clearly associated as etiological agents of diarrhea (28, 31).
Recent studies have identified a novel central nervous system (CNS) tropism for astroviruses. VA1 is the most frequently identified genotype of astrovirus in cases of human encephalitis, with a total of five independently published cases to date (32–36). In these cases, all patients were immunocompromised, including three hematopoietic stem cell transplant recipients and two patients with X-linked agammaglobulinemia (32–36). VA1 sequences were detected in brain tissue biopsy specimens of all five cases by unbiased next-generation sequencing (32–36). These data were corroborated in two of the cases by immunohistochemistry and in another case by in situ hybridization, localizing VA1 to neurons and astrocytes (32–34). Death occurred in four of five patients, demonstrating a high mortality rate in the reported VA1-associated cases of encephalitis (32–36).
Other astroviruses have also been recently detected in the CNS. In humans, cases of meningoencephalitis associated with human astrovirus 4, astrovirus MLB1, and MLB2 have been described (37–39). In minks, an astrovirus genotype is the causative agent of shaking mink syndrome, a neurological disorder of minks characterized by tremors and seizures (40, 41). Bovine astroviruses have been identified as a significant cause of encephalitis in cattle, as these viruses have been detected in 34% of previously unexplained cases of encephalitis (42–47). An additional astrovirus genotype has also been identified as a cause of encephalitis in sheep (48). Porcine astroviruses are associated with a congenital tremor syndrome in newborn piglets, while the “white chicks” hatchery disease is caused by a chicken astrovirus that also has been detected in brain tissue (49, 50). Together, these examples demonstrate a previously unrecognized neurotropism of multiple astroviruses.
While cell culture models for classic human astroviruses and several other astroviruses exist, no cell culture model for VA1 has been described to date in the literature (18, 51–55). Multiple cell lines are broadly permissive to most serotypes of classic human astroviruses, including Caco-2, HT-29, SK-CO-1, HEK293, MA-104, Cos-1, and Vero cell lines (52). Additional human cell lines, including A549, HEp-2, and HeLa, are selectively permissive for replication of some of the different classic human astrovirus serotypes, but only minimal levels of replication have been reported in nonprimate-derived cell lineages (52). Propagation in cell culture is generally thought to be trypsin dependent for classic human, porcine, bovine, and feline astroviruses (51–55). Proteolytic cleavage of the classic human astrovirus capsid by trypsin has been identified as a maturation step necessary for infectivity of the virus in cell culture for classic human astroviruses 1 and 8 (56–58).
Currently, only limited data regarding the host innate immune response to astrovirus infection exist. Classic human astroviruses induce a beta interferon (IFN-β) response in cell culture, and exogenous addition of type I interferons to cell culture reduces replication of classic human astroviruses (59, 60). Furthermore, addition of a human neutralizing antibody to IFN-β increases replication (59, 60). In vivo, loss of function of the interferon α/β receptor or STAT1 increases murine astrovirus replication (61).
We describe here the first cell culture models for propagation of VA1, a necessary tool that will facilitate studies of VA1 tropism, pathogenesis, and immune control. We demonstrate that multiple human cell lines, including Caco2, HEK293T, and A549 support robust VA1 replication, thus enabling detailed studies of VA1.
RESULTS
qRT-PCR assay.
To develop a quantitative reverse transcription-PCR (qRT-PCR) assay capable of detecting all VA1 genotypes, we performed a multiple-sequence alignment of all published VA1 strains to identify highly conserved regions of the genome. Primers and probe were selected from a region that includes the 5′ end of ORF2 and the internal promoter sequence (nucleotide positions 4134 to 4272 of the reference strain [NC_013060.1]). The nucleotide sequences of the primer and probe binding sites were 100% conserved between the VA1 strains. We generated a standard curve for the VA1 qRT-PCR assay using in vitro-transcribed RNA (Fig. 1A). The optimized qRT-PCR assay was sensitive to five copies of target of RNA per reaction mixture and had an efficiency of 98.9%.
FIG 1.
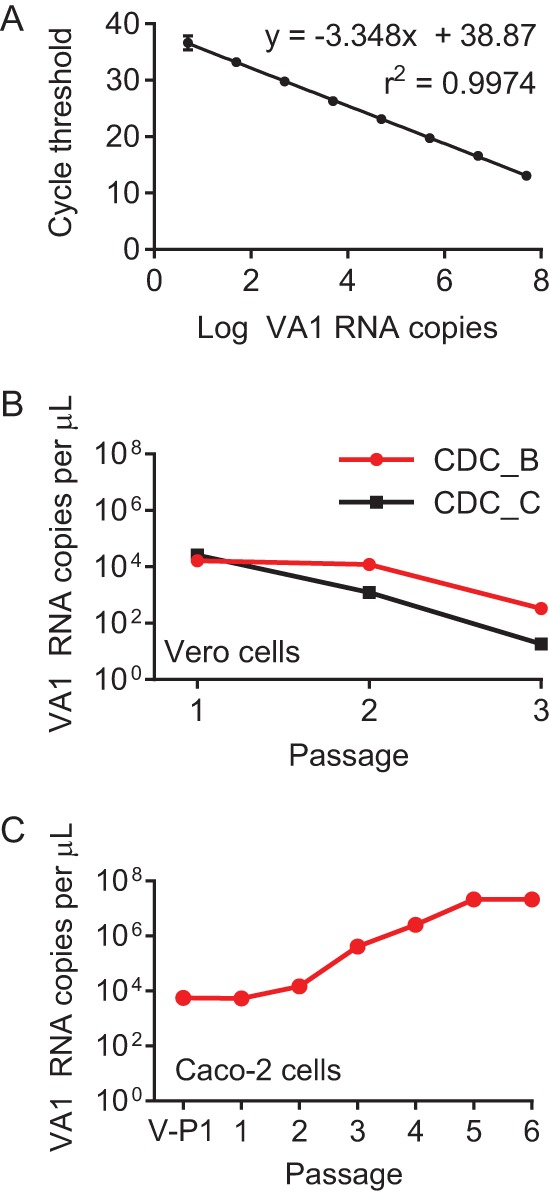
VA1 RNA levels after serial blind passage in cell cultures. (A) qRT-PCR standard curve for quantification of VA1 RNA copies. Serial 10-fold dilutions of VA1 in vitro-transcribed RNA were used as control templates, starting with 5 × 107 copies/reaction mixture. A line of best fit and coefficient of determination were calculated. Error bars represent 1 standard deviation. (B) Vero cell passages. Two stool specimens containing VA1 RNA were inoculated into cell cultures of Vero cells. (C) Caco-2 cell passages. Caco-2 cells were initially inoculated using the Vero passage 1 (V-P1) cell lysate.
Serial passaging of VA1.
We selected two human stool samples that were collected during the originally described outbreak of VA1 gastroenteritis, which also had a high copy number in a previously published qRT-PCR assay (CDC_B and CDC_C samples) (23). After filtration, the resulting stool filtrates were used to inoculate Vero cells. For the CDC_C sample, a steady decrease in VA1 RNA copy number with increasing passage number was observed (Fig. 1B). However, the CDC_B sample demonstrated a stable RNA copy number from passages 1 to 2 (Fig. 1B), suggesting that some VA1 replication may have occurred, given the 40-fold sample dilution with passage. However, RNA levels decreased following the second passage, suggesting that replication was not sustainable in Vero cells under the conditions used (Fig. 1B).
Thus, we tested the ability of Caco-2 cells, a cell line known to be broadly permissive for replication of the classic human astroviruses, to support further replication from the CDC_B V-P1 lysate. In addition, exogenous trypsin was added to our initial passage attempts in Caco-2 cells, as replication of the classic human astroviruses is dependent on addition of trypsin (51, 52). The VA1 copy number was initially stable from passages 1 to 2 in Caco-2 cells (Fig. 1C). Then, after passage 2, the VA1 RNA copy number increased by several log over the next few passages and then plateaued after passage 5 (Fig. 1C). In total, the VA1 RNA copy number increased over 3,000-fold after six serial passages (without accounting for sample dilution at each passage). In contrast, treatment of the C-P4 stock by UV irradiation led to undetectable VA1 RNA after two passages, demonstrating that the virus was sensitive to UV treatment (data not shown). During these passages, no evidence of cytopathic effect was observed.
To determine if trypsin is necessary for the propagation of VA1 in cell culture, we infected Caco-2 cells using the C-P4 lysate with or without addition of exogenous trypsin. We found that after two passages, the VA1 RNA copy numbers of untreated and trypsin-treated cell lysates were not significantly different (passage 1, P = 0.23; passage 2, P = 0.92) (Fig. 2). After passage three, a modest 2-fold decrease was noted in the no-trypsin cell lysate (P = 0.003) (Fig. 2), but this decrease was not sustainable, as there was no significant difference between the trypsin-treated and untreated groups after passage 4 (P = 0.16) (Fig. 2). These results are in stark contrast to previously published cell culture systems with other mammalian astroviruses genotypes, where exogenous trypsin is necessary for successful passage of astroviruses in cell culture (51–55).
FIG 2.
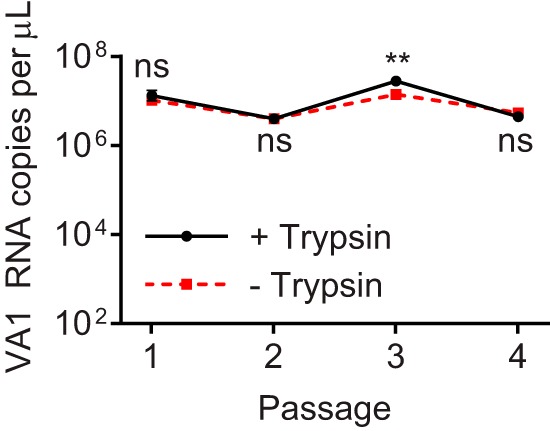
Serial passaging of VA1 in Caco-2 cells in the presence and absence of trypsin. C-P4 aliquots were pretreated and incubated with or without trypsin. Both treatments were passaged four times for 7 days each in Caco-2 cells. Geometric means of each group are shown, and error bars represent 1 geometric standard deviation. **, P ≤ 0.01; ns, not significant (P > 0.05).
To determine the viral titer, we established a 50% tissue culture infectious dose (TCID50) assay using qRT-PCR as a readout for positive infection. The C-P5 VA1 stock had a titer of 1.4 ×106 TCID50/ml. A total of 6,566 of 6,586 nucleotides (nt) of the C-P5 stock were confirmed by Sanger sequencing with 3× coverage. Four genetic variants relative to the published consensus sequence (NC_013060.1) from the original stool sample were detected in the C-P5 stock, occurring at nucleotide positions C499U, A2672G, U5162C, and C5369U. Variants U5162C and C5369U did not result in mutations in the amino acid sequence of ORF2. The C499U variant resulted in a conservative A154V amino acid substitution in ORF1a. The A2672G variant occurred in the overlap region between ORF1a and ORF1b. For ORF1a, the mutation was silent, but in the ORF1b reading frame, this led to a T14A amino acid mutation. When examining the secondary structure of the VA1 genome, the variant occurred within the loop of the stem-loop structure for ribosomal frameshifting of ORF1a and ORF1b.
Because the C499U and A2672G variants resulted in amino acid substitutions, we sequenced these regions in the V-P1 and C-P1 through C-P4 samples to determine when these variants could be first detected. The C499U variant was initially detected in the C-P1 sample, and the A2672G genetic variant was first detected in the V-P1 sample. Based on the detection of these variants in the first two passages of VA1 in cell culture, these variants may represent tissue culture adaptations for propagation of VA1.
VA1 multistep growth curves.
We defined the kinetics of VA1 infection in several cell lines without the addition of exogenous trypsin. Increasing VA1 RNA copies were observed in both the cells and the supernatant fractions of the Caco-2, HEK293T, and A549 lineages but not in BHK-21 cells (Fig. 3A and B). An approximate 10- to 100-fold increase in VA1 RNA copies in the cellular fractions was observed 24 h after inoculation, and the number increased another 10-fold by 96 h. For the supernatant fractions, VA1 RNA increased over time but appeared to accumulate more slowly than in the cellular fraction. In addition, VA1 RNA accumulated more slowly in the supernatant fraction in Caco-2 and HEK293T cells than in A549 cells. This difference between the cellular and supernatant fractions may suggest inefficient viral egress from these cell lines, possibly due to the presence of one or more host restriction factor(s). It is also possible that the viral particles remain tightly associated with the cells. No cytopathic effect was observed in Caco-2, HEK293T, or A549 cells.
FIG 3.
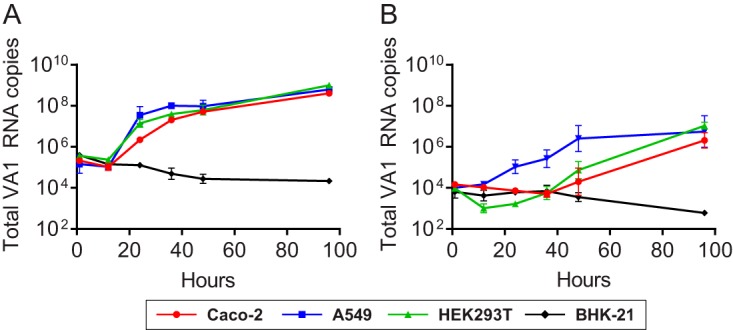
Multistep growth curve of VA1 RNA measured from the cellular and supernatant fractions of Caco-2, HEK293T, A549, and BHK-21 cells. Cells were infected using the C-P5 viral stock, and RNA levels were measured from the cellular fraction (A) or the supernatant fraction (B). The geometric mean of the VA1 RNA in each cell line is shown with error bars representing 1 geometric standard deviation.
We also measured the production of infectious virus in cell lysates using a TCID50 assay at 1 h, 36 h, and 96 h postinfection in Caco-2 cells. The viral titer was below the limit of detection (1.6 × 102 TCID50/ml) at 1 h postinfection, 8.9 × 103 TCID50/ml at 36 h postinfection, and 2.8 × 105 TCID50/ml at 96 h postinfection. These results demonstrated an increase in infectious virus over time.
Detection of VA1 subgenomic RNA.
As additional evidence of replication, we sought to detect the subgenomic RNA that is expected to be transcribed during its replication cycle. The VA1 genome contains the highly conserved promoter sequence present in all known astroviruses for subgenomic RNA synthesis of ORF2 (2). Extrapolating from the published 5′ untranslated region (UTR) of the subgenomic RNA of classic human astrovirus 2 and the annotated VA1 genome (NC_013060.1), VA1 would be hypothesized to produce a subgenomic strand that is at least 2,386 nt [5′ UTR, 11 nt; ORF2, 2,277 nt; 3′ UTR, 98 nt; and poly(A) tail, undefined length] (19). Northern blot analysis of total RNA from Caco-2 cells infected by the C-P5 viral stock yielded two bands (Fig. 4). The first band is >6,000 nt, which presumably represents the full-length VA1 RNA genome [6,586 nt + the poly(A) tail]. A second, fainter band that migrated more slowly than the 2,000-nt marker was detected, consistent with the anticipated size of the subgenomic RNA [2,386 nt + poly(A) tail]. No other significant bands were observed in the VA1-infected Caco-2 cells. The specificity of the probe was confirmed by the absence of any bands in mock-infected Caco-2 cells.
FIG 4.
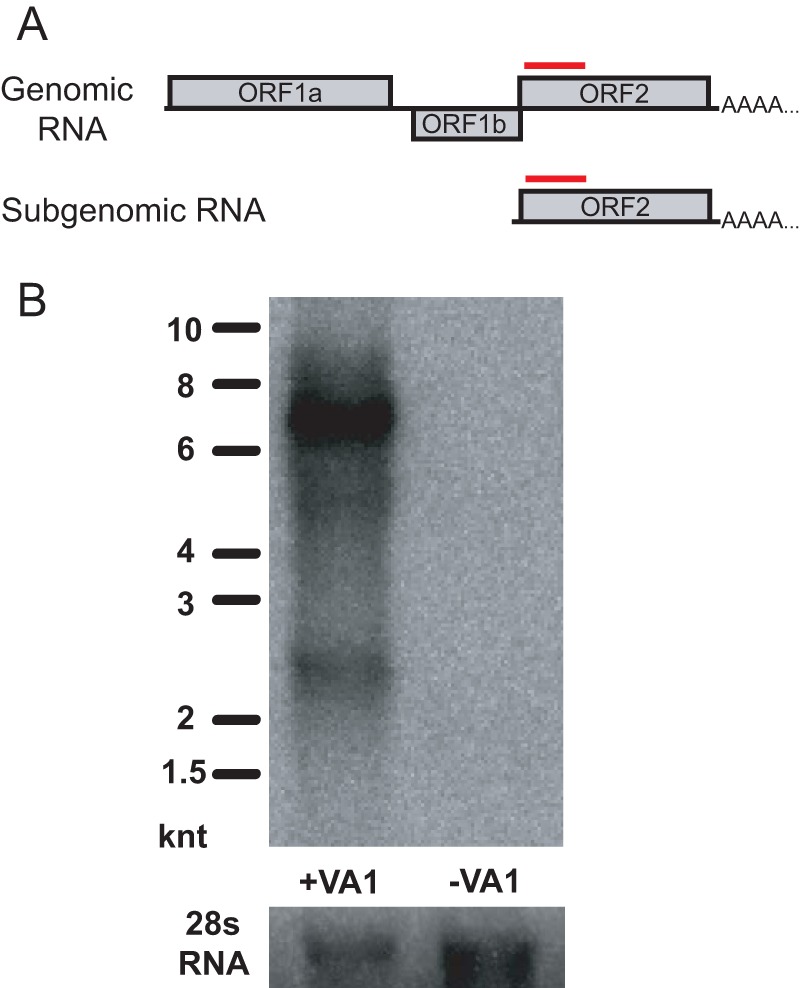
Northern blot analysis of VA1-infected Caco-2 cells. (A) Schematic of the predicted open reading frame structure of the VA1 genomic (6,586 nt in length) and subgenomic RNA (>2,386 nt in length). The red lines denote the region used to generate the radiolabeled probe for Northern blotting. (B) Northern blot of infected and uninfected Caco-2 cells. knt, 1,000 nt. 28s RNA loading controls for each lane are shown in the lower panel.
Detection of VA1 virus-like particles.
Electron microscopy (EM) of infected Caco-2 cells identified crystalline lattices of virus-like particles (Fig. 5a to d). These aggregates were present within the cytosol, and many of these aggregates appeared to be associated with a membrane. The mean size of the virus-like particles was 26.4 nm (standard deviation [SD] = 2.1 nm; n = 10) in diameter, which is comparable to the reported diameters of 28 to 44 nm for serotypes of mamastrovirus 1 (1, 56, 62, 63).
FIG 5.
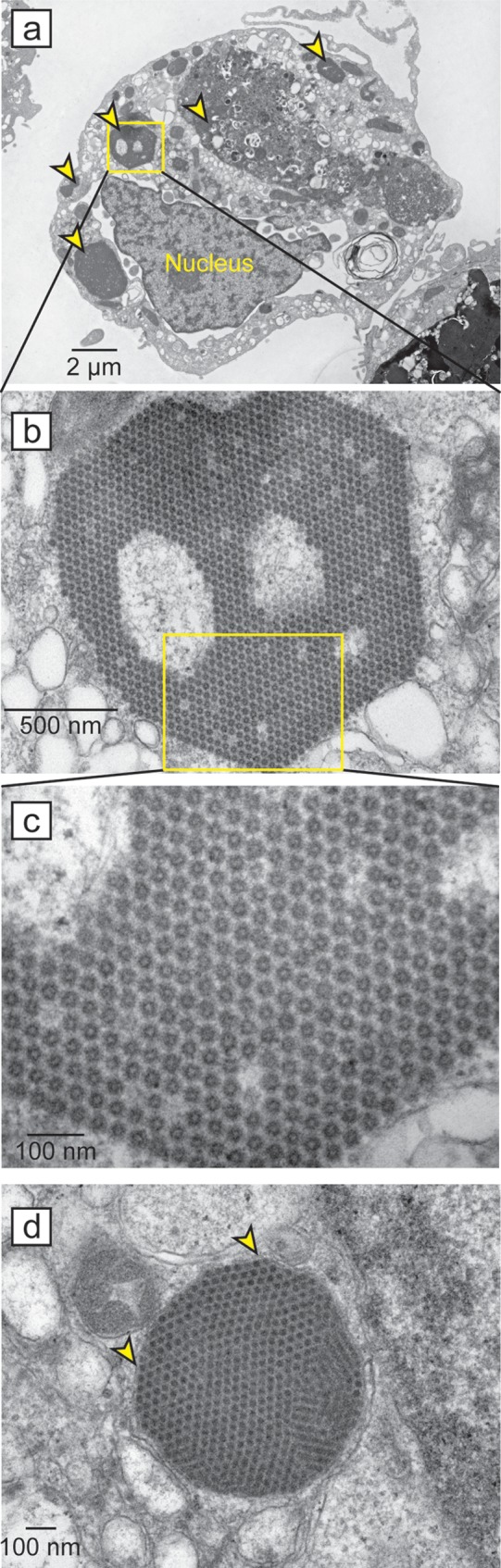
Electron microscopy images of VA1-infected Caco-2 cells. (a to c) Increasing magnifications of a single Caco-2 cell infected with VA1 with the nucleus labeled. Yellow triangles represent regions containing crystalline lattices of virus-like particles. (d) Additional example of a VA1 crystalline lattice contained within a membrane structure (denoted by arrows) in the cytoplasm of an infected Caco-2 cell. Scale bars are shown with each image.
Effect of interferon pretreatment on replication of VA1.
Caco-2 cells pretreated with IFN-β1a for 12 h were infected with VA1 using a multiplicity of infection (MOI) of 0.01 (Fig. 6). At 96 h postinfection, a dose-dependent effect of IFN-β1a on VA1 RNA copy abundance was detected (one-way analysis of variance [ANOVA] F(4,15) = 21.03, P < 0.0001), with a 20-fold decrease in VA1 RNA copy number at the maximum dose of 1,000 U/ml compared to no treatment (Dunnett's multiple-comparison test, P = 0.0001). This result supports the role of the type I interferon response in limiting the replication of VA1.
FIG 6.
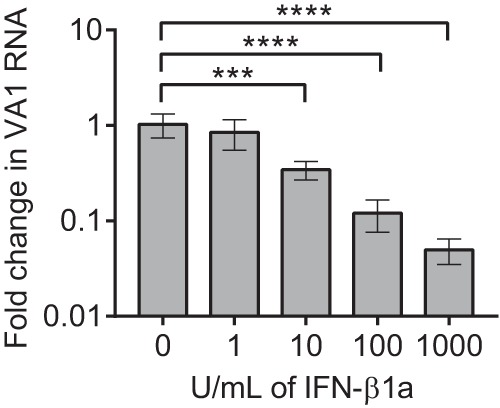
Effect of interferon on replication of VA1. Caco-2 cells were pretreated with increasing doses of IFN-β1a and then inoculated with VA1 in the presence of interferon. RNA was measured 96 h after inoculation. VA1 RNA was normalized to 18s RNA expression and is shown as mean fold change relative to treatment with 0 U/ml of IFN-β1a. Error bars represent 1 standard deviation. ***, P ≤ 0.001; ****, P ≤ 0.0001.
DISCUSSION
Our results demonstrate bona fide replication of VA1 in cell culture, confirmed by multiple lines of evidence. The data include detection of viral subgenomic RNA, multiple log increases in viral RNA in multistep growth curves, and visualization by EM of virus-like particles in infected cells. To our knowledge, this is the first report of propagation of any the newly identified, nonclassic human astroviruses (mamastroviruses 6, 8, and 9). The lack of any cell culture or in vivo model of infection has been a significant limiting factor in the study of VA1 and VA clade astroviruses. The establishment of a culture system for VA1 fulfills the first step of Koch's postulates and provides a necessary reagent for efforts to complete Koch's postulates.
Interestingly, the C-P5 isolate of VA1 can be propagated in cell culture without the addition of exogenous trypsin. In all other previously published mammalian astrovirus cell culture systems, the addition of exogenous trypsin is necessary for continued viral propagation. For the classic human astroviruses 1 and 8, trypsin significantly enhances infectivity by contributing to proteolytic activation of the capsid (56–58). Currently, it is unclear what proteolytic processing of the VA1 capsid, if any, occurs in vivo. Based on the ability to passage VA1 in the absence of exogenous trypsin, VA1 may simply have a capsid that does not need to be proteolytically cleaved to be infectious. Alternatively, endogenous proteases produced by Caco-2, HEK293T, and A549 cell lines may be sufficient for activation of the VA1 capsid.
If proteolytic cleavage of the capsid by trypsin is important for astrovirus infectivity in vivo, it may be a factor that contributes to the tissue tropism of astroviruses. Trypsin is predominantly expressed in the gastrointestinal tract of humans, with weaker expression in other tissues, including the brain (64). To date, the classic human astroviruses have been detected almost exclusively from the gastrointestinal tract. Replication of the classic human astroviruses could be largely restricted to tissues with high expression of trypsin. If VA1 does not require proteolytic activation, VA1 may have a broader spectrum of tissues that it can infect in vivo, including the CNS. In support of this general concept is one report of chicken astroviruses isolated from diverse tissue specimens (respiratory tract, leukocytes, and small intestine) that could be passaged in tissue culture without the addition of trypsin (65). In addition, a mink astrovirus can cause shaking mink syndrome after intracranial injection into unaffected minks without trypsin treatment (40, 41). A single case report of classic human astrovirus 4 RNA being detected in other tissue sites obtained postmortem, including cerebrospinal fluid, has been published (37). To our knowledge, the trypsin dependence of astrovirus 4 has not been explicitly examined. Moreover, in that study, the initial culturing experiments that led to isolation of human astrovirus 4 were done in the absence of trypsin treatment (37). Thus, it is possible that some of the classic human astroviruses may not be as dependent on trypsin as astroviruses 1 and 8.
Sequencing of the C-P5 stock identified two nonsynonymous mutations relative to the reference VA1 amino acid sequence. The C499U variant occurs near a coiled-coil region of the VA1 ORF1A-encoded protein that is of unknown function. While an alanine-to-valine substitution is often presumed to be a conservative mutation, C499U was first detected in the C-P1 sample and might represent an adaptive mutation for propagation of VA1 in cell culture. The A2672G variant occurs in the overlap region between ORF1a and ORF1b, altering the amino acid code of ORF1b. The location of this variant is within the loop of the stem-loop structure for ribosomal frameshifting. A previous study of classic human astrovirus 1 found no effect on ribosomal frameshifting when the nucleotides of the loop were mutated to a reverse complement sequence (15). The A2672G variant was first detected after the first passage in Vero cells and is also a candidate adaptive mutation for cell culture. The genetic variants could also be simply stochastic mutations with no effect on adaptation to cell culture. The effects of these genetic variants on propagation of VA1 might be examined in the future by development of a VA1 reverse genetic system, as has been described for the classic human astroviruses (66, 67).
Treatment of Caco-2 cells with exogenous IFN-β1a reduced replication of VA1 20-fold. Our results are similar to those reported in two previous publications, whereby addition of exogenous type I interferon to Caco-2 cells reduced the replication of classic human astroviruses approximately 10-fold (59, 60). The specific interferon-stimulated genes that mediate the antiviral effect on replication of astroviruses in general are currently unknown.
The establishment of a cell culture system for VA1 infection opens many new frontiers in the study of VA1. Astroviruses have only recently been recognized as a neurotropic family of RNA viruses, and they may have unique mechanisms that mediate neuroinvasion and neuroinfection. Based on the clinical cases of VA1 encephalitis occurring in immunocompromised hosts, the adaptive immune system could be important in controlling VA1 infection. Future work will focus on developing cell culture and in vivo models of VA1 CNS infection. In addition, with the ability to perform antibody neutralization against VA1, it will be possible to identify critical antigens for vaccine development as well as protective antibodies. The VA1 cell culture model may also facilitate high-throughput pharmaceutical screens for antiviral molecules with the ultimate goal of reducing the high mortality rate associated with VA1 CNS infection.
MATERIALS AND METHODS
qRT-PCR.
To identify conserved regions for primer design, we generated a multiple-sequence alignment of all available VA1 genomes using MUSCLE (NC_013060.1, GQ891990.1, GQ415662.1, KJ920196.1, KJ920197.1, KM358468.1, KM401565.1) (68). The primers and probe used in the assay include forward primer AJ0071, 5′-CTAGTGGTGGGGAAGAAC-3′, reverse primer AJ0072, 5′-CCTTGGCTATTTGCTTTGC-3′, and TaqMan probe AJ0073, 5′-/56-FAM/CCATGACTT/ZEN/TGCTTTGGACCTCCC/3IABkFQ/-3′ (Integrated DNA Technologies).
As a positive control, plasmid DW639 harboring a 536-bp fragment of the VA1 genome that encompassed the primer/probe region was used as the template for in vitro RNA transcription using the Megascript T7 kit (ThermoFisher). RNA was quantified by NanoDrop 1000 (ThermoScientific), and 10-fold serial dilutions of RNA were made. Quantitative reverse transcription-PCR (qRT-PCR) was performed using the manufacturer's instructions for TaqMan Fast Virus 1-Step master mix (Applied Biosystems) on a ViiA 7 Real Time PCR system (Applied Biosystems) with an annealing temperature of 55°C. To define a standard curve, 10-fold serial dilutions of the VA1 RNA were analyzed in triplicate. A plot of RNA copy number and cycle threshold (CT) number was created, and a linear regression was calculated from the data points. The VA1 RNA copy number was calculated using the linear regression model from Prism 7 (GraphPad).
Cell culture.
All cell lines and infection steps were carried out at 37°C with 5% CO2. The African green monkey kidney epithelial cell line (Vero), undifferentiated human intestinal epithelial cell line (Caco-2), human embryonic kidney cell line (HEK293T), human lung epithelial cell line (A549), and baby hamster kidney fibroblast cell line (BHK-21) were all cultured in growth medium consisting of Dulbecco's modified Eagle medium (DMEM) with l-glutamine supplemented with 10% fetal bovine serum (FBS; Gibco) and 1% of 10,000 units/ml of penicillin and streptomycin (Gibco).
For infection, cells were grown to 80 to 90% confluence and washed with phosphate-buffered saline (PBS) prior to inoculation with stool filtrate or cell lysates. Incubation of the VA1 inoculum with the cells was carried out for 1 h unless noted otherwise. The medium was then aspirated, cells were washed with PBS, and the cells were incubated. RNA was extracted using TRIzol (ThermoFisher) in Direct-zol mini-prep columns (Zymo Research). VA1 RNA copies were determined by qRT-PCR.
Serial passaging of VA1 in cell culture.
Stool samples positive for VA1 by PCR (CDC_B and CDC_C) from the originally described outbreak of VA1-associated gastroenteritis in Virginia, USA, were used as the initial inoculum (23). A total of 200 μg of stool was diluted 1:6 in PBS and filtered through a 0.45-μm filter, creating a passage 0 (P0) viral filtrate. For passage in cell culture, Vero cells grown in a 6-well plate were inoculated with either 75 μl of CDC_B P0 or 50 μl of CDC_C P0 diluted in 1 ml of DMEM per well. Cells were incubated for 2 h and washed with PBS, and 2 ml growth medium was added to each well. No exogenous trypsin was added at any step. The Vero cells were then incubated for 7 days and monitored daily for cytopathic effect. After 1 week, cells were freeze-thawed three times, creating passage 1 (V-P1) cell lysate. A 50-μl volume of cell lysate was used as the inoculum for another round of passage under the same infection conditions to generate V-P2 and iterated further to generate additional passages. For RNA extraction, 100 μl of cell lysate was extracted using the COBAS Ampliprep instrument (Roche).
The V-P1 CDC_B cell lysate was also passaged into Caco-2 cells grown in a 6-well plate. A 50-μl volume of the V-P1 CDC_B cell lysate was pretreated with 200 μg/ml of trypsin (Gibco) for 1 h at 37°C. The lysate-trypsin mixture was then diluted in 1 ml of DMEM and added to Caco-2 cells. Cells were incubated for 1 h, medium was aspirated, and the cells were washed. A total of 2 ml of DMEM supplemented with 3.3 μg/ml of trypsin was added to the wells, and the cells were incubated for 24 h. After 24 h, 220 μl of FBS was added to the cells, and the cells were incubated for 6 days. Cells were then lysed by freeze-thawing three times, thus generating a Caco-2-P1 (C-P1) cell lysate. Subsequent passages were repeated using 50 μl of cell lysate as the inoculum for each sequential passage. A total of 100 μl of cell lysate was used for RNA extraction.
A 50-μl aliquot of C-P4 was treated with 50 mJ/cm2 of UV light using the Spectrolink XL-1000 UV cross-linker (Spectroline). The sample was incubated with trypsin and serially passaged twice in Caco-2 cells. To determine the effects of exogenous trypsin, 50-μl aliquots of C-P4 lysate were passaged four times with or without the addition of trypsin at any step. A total of three replicates for each trypsin treatment group were completed, and results were compared using a two-tailed Student's t test. P levels of <0.05 were considered significant.
Preparation of VA1 virus stock.
Using a 150-μl aliquot of C-P4 cell lysate, Caco-2 cells in a T75 flask were infected to create a large-scale viral stock (C-P5 stock). No trypsin was added in any of the steps for the creation of the viral stock. Cells were incubated for 1 week, and cells were lysed by freeze-thawing three times. The viral stock was used for all subsequent Sanger sequencing, growth curve assays, Northern blotting, electron microscopy, and the effect of interferon on VA1 replication.
Quantification of infectious virus by TCID50 assay.
The titers (TCID50) of the C-P5 stock and cell lysate samples obtained 1 h, 36 h, and 96 h after infection were determined by infecting Caco-2 cells with serial 10-fold dilutions of each cell lysate in quadruplicate. Cells were incubated for 72 h, and cell extracts were collected in TRIzol for RNA extraction. The TCID50 was calculated by the Spearman-Karber method, and wells were considered infected if VA1 RNA was detected by the qRT-PCR assay (CT < 30) (69). The CT values of uninfected wells were all >37. The TCID50 titer was then used to calculate the MOI for all experiments using the C-P5 stock.
Sequencing of C-P5 VA1 stock.
The VA1 genome consensus sequence in the C-P5 stock was determined in six overlapping RT-PCR amplicons (Table 1). cDNA was synthesized in a 1-h incubation using Thermoscript (ThermoFisher) with 10 pmol of the VA1 fragment-specific reverse complement primer (Table 1) using an incubation temperature of 55°C for all fragments, except for fragment 5, which was incubated at 60°C. The cDNA was then treated with RNase H (ThermoFisher) at 37°C for 20 min. High-fidelity PCR was performed on the six cDNA products using PfuUltra II Fusion HS DNA polymerase (Agilent Genetics) with the corresponding VA1-specific primer set for each fragment (Table 1). The reaction conditions included 95°C for 1 min and then 27 cycles of 95°C for 20 s, annealing temperature of 53°C for all fragments except fragment 5 (59°C) for 20 s, and 72°C for 1 min, followed by a final extension phase at 72°C for 3 min. PCR products were purified using Purelink PCR Micro kit (Invitrogen) and cloned into pCR4 TOPO plasmids (Invitrogen). Plasmids were then Sanger sequenced. A total of three replicates of each fragment of the genome were amplified, cloned, and sequenced.
TABLE 1.
Primers used for sequencing of the C-P5 viral stocka
| Fragment no. | Name of primer | Direction | Sequence (5′→3′) | Nucleotide positions |
|---|---|---|---|---|
| 1 | AJ0117 | Forward | CCAAATTTGTTGGCTGTGCC | 1–20 |
| SF165 | Reverse | TATCACTTTCACATACTCAGCATCAC | 1072–1097 | |
| 2 | SF166 | Forward | CTAATATGGAGGTCACTTCAATGT | 943–966 |
| SF167 | Reverse | ATCTGACCTTTAAGGGCATCCTCTA | 1891–1915 | |
| 3 | SF168 | Forward | CTGATTGAGGATGGCACCGT | 1608–1627 |
| SF169 | Reverse | ATCAATTGGCTCATCATAATCACT | 2798–2821 | |
| 4 | SF170 | Forward | CTATCATAAGTGAGTTGGATGAATTATG | 2539–2566 |
| SF171 | Reverse | TAACTCTGTCCACATAATTGTCACAT | 3799–3824 | |
| 5 | SF172 | Forward | CTCTGGACAAATATCCACAACTA | 3628–3650 |
| AJ0125 | Reverse | CAGCACCCTCATCCACAATCTG | 5139–5160 | |
| 6 | SF174 | Forward | CAAGGCATATGAGTGAAAGGTTC | 5074–5096 |
| Poly(T) primer | Reverse | GGCCACGCGTCGACTAGTACTTTTTTTTTTTTTTTTT | NA |
Nucleotide positions are based on the reference VA1 genome (NC_013060.1). NA, not applicable.
We analyzed the virus stocks from all passages for the presence of the two genetic variants that altered the amino acid sequence. PCR primers that spanned the mutation sites were designed. Amplicons were generated using One-step RT-PCR kits (Qiagen) and primer pairs SF164 (5′-CATGGCAGTGTCAAAGCACGGAT-3′) with AJ0118 (5′-AATGCACCCAACACTGAAAGG-3′) and SF170 with SF169 (Table 1). The reaction conditions included 55°C for 45 min, 95°C for 15 min, and then 40 cycles of 94°C for 30 s, annealing temperature of 55°C for 30 s, and 72°C for 1 min, followed by a final extension phase at 72°C for 10 min. Amplicons were gel extracted using the Qiaquick gel extraction kit (Qiagen) and cloned into pCR4 TOPO plasmids. Plasmids were then Sanger sequenced.
Multistep growth curves.
Caco-2, HEK293T, A549, or BHK-21 cells were grown in 24-well plates. Cells were inoculated with 1.25 μl (MOI, 0.01) of the C-P5 stock diluted in 250 μl of DMEM. The supernatant and cell fractions were harvested 1, 12, 24, 36, 48, and 96 h after inoculation. A 100-μl volume of the supernatant or the complete cell fraction was harvested for RNA extraction. A total of three replicates at each time point were completed.
Northern blotting.
Caco-2 cells in a T75 flask were incubated with either 150 μl of the C-P5 stock or PBS (mock) diluted in 7.5 ml of DMEM for 1 h. After incubation with the inoculum, the cells were washed, growth medium was added, and the cells were incubated for 48 h prior to total cellular RNA extraction. One microgram of total cellular RNA was run on a 1% denaturing formaldehyde-MOPS (morpholinepropanesulfonic acid) agarose gel. RNA was transferred from the gel to a nylon membrane (Amersham Hybond-XL; GE Healthcare) using a vertical transfer apparatus (Schleicher and Schuell TurboBlotter Transfer System) for 3 h using 10× saline-sodium citrate (SSC; 1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) transfer buffer. The membrane was baked at 70°C for 20 min and cross-linked using the Spectrolink XL-1000 UV cross-linker (Spectroline) per the manufacturer's optimal UV exposure. The blot was stained with 0.02% methylene blue to visualize 28S rRNA bands. The membrane was destained and prehybridized with Rapid-hyb buffer (GE Healthcare Amersham) for 30 min at 65°C.
A region of ORF2 of the reference VA1 genome (NC_013060.1; nucleotide positions 4282 to 5014) was selected for probe generation. A 732-bp PCR product using primers AJ0094 (5′-AACAGGAGAAGAAGGAACC-3′) and AJ0081 (5′-GCATTGGTAGTTTCAGTCACC-3′) was generated using Accuprime Taq Polymerase (ThermoFisher) with an annealing temperature of 55°C. PCR products were run on a 1% agarose gel, and PCR products were extracted using the QIAquick gel extraction kit (Qiagen). Radiolabeled double-stranded DNA probes were generated by random priming. A total of 50 ng of the AJ0094-AJ0081 PCR product was incubated with [α-32P]-dCTP, dATP, dGTP, and dTTP, random hexamers, and DNA polymerase I large Klenow fragment (New England BioLabs) for 15 min at room temperature. The radiolabeled probe was purified using the Qiaquick nucleotide removal kit (Qiagen). The probe was quantified using a Beckman LS6000IC liquid scintillation counter.
A total of 5 ×106 CPM of probe was added to the hybridization buffer and incubated with the blot for 3 h at 65°C. The membrane was washed with increasingly stringent washes at room temperature, first with 2× SSC + 0.1% SDS for 5 min (twice), and then 1× SSC + 0.1% SDS for 10 min (twice) and 0.1× SSC + 0.1% SDS for 5 min (4 times). The membrane was finally washed with 0.1× SSC + 0.1% SDS at 42°C for 15 min (twice) and then at 65°C for 15 min (twice). The blot was then rinsed with 2× SSC, and the membrane was exposed to a phosphoimager plate for autoradiography.
EM.
Caco-2 cells infected with C-P5 viral stock for 1 week were trypsinized and fixed in 2% paraformaldehyde–2.5% glutaraldehyde (Polysciences Inc.) in 100 mM sodium cacodylate buffer, pH 7.2, for 1 h at room temperature. The cells were then washed with sodium cacodylate buffer and postfixed in 1% osmium tetroxide (Polysciences Inc.) for 1 h. The sample was then rinsed extensively in distilled water (dH2O) prior to en bloc staining with 1% aqueous uranyl acetate (Ted Pella Inc.) for 1 h. Following several rinses in dH2O, the sample was dehydrated in a graded series of ethanol and embedded in Eponate 12 resin (Ted Pella Inc.). Sections of 95 nm were cut with a Leica Ultracut UCT ultramicrotome (Leica Microsystems Inc.), stained with uranyl acetate and lead citrate, and viewed on a JEOL 1200 EX transmission electron microscope (JEOL USA Inc.) equipped with an AMT 8-megapixel digital camera and AMT Image Capture Engine V602 software (Advanced Microscopy Techniques). The diameter of the virus-like particles was measured using ImageJ.
Effects of pretreatment of IFN-β1a on VA1 replication.
Caco-2 cells were pretreated with 10-fold dilutions of a 1,000 U/ml stock of IFN-β1a (PBL Assay Science) for 12 h. Cells were then inoculated with an MOI of 0.01 of the C-P5 stock in the presence of IFN-β1a. The cells were then washed and incubated with IFN-β1a for 96 h, and RNA was extracted. VA1 RNA and 18S RNA were measured by qRT-PCR (Integrated DNA Technologies), and the relative RNA was quantified using the 2−ΔΔCT method. Groups were compared using a one-way ANOVA in Prism 7 (GraphPad). A post hoc analysis comparing no-treatment groups with each IFN-β1a dose group was completed using Dunnett's multiple-comparison test. P values of <0.05 were considered significant.
Accession number(s).
The sequence of the C-P5 stock has been deposited in GenBank under accession number KY933670.
ACKNOWLEDGMENTS
We thank Wandy Beatty and the Washington University School of Medicine Molecular Microbiology Imaging Facility for their support in obtaining the electron microscopy images.
A.B.J. is supported in part by the NIH under training grant NIH T32 AI106688. L.R.H. is supported in part by the Children's Discovery Institute (MD-FR-2013-292). D.W. holds an Investigator in the Pathogenesis of Infectious Disease award from the Burroughs Wellcome Fund. This project was supported in part by NIH grant R21 NS101371.
REFERENCES
- 1.Madeley CR, Cosgrove BP. 1975. Letter: viruses in infantile gastroenteritis. Lancet ii:124. [DOI] [PubMed] [Google Scholar]
- 2.Bosch A, Pinto RM, Guix S. 2014. Human astroviruses. Clin Microbiol Rev 27:1048–1074. doi: 10.1128/CMR.00013-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kurtz JB, Lee TW. 1984. Human astrovirus serotypes. Lancet ii:1405. [DOI] [PubMed] [Google Scholar]
- 4.Lee TW, Kurtz JB. 1982. Human astrovirus serotypes. J Hyg (Lond) 89:539–540. doi: 10.1017/S0022172400071114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koopmans MP, Bijen MH, Monroe SS, Vinje J. 1998. Age-stratified seroprevalence of neutralizing antibodies to astrovirus types 1 to 7 in humans in The Netherlands. Clin Diagn Lab Immunol 5:33–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mitchell DK, Matson DO, Cubitt WD, Jackson LJ, Willcocks MM, Pickering LK, Carter MJ. 1999. Prevalence of antibodies to astrovirus types 1 and 3 in children and adolescents in Norfolk, Virginia. Pediatr Infect Dis J 18:249–254. doi: 10.1097/00006454-199903000-00008. [DOI] [PubMed] [Google Scholar]
- 7.Kriston S, Willcocks MM, Carter MJ, Cubitt WD. 1996. Seroprevalence of astrovirus types 1 and 6 in London, determined using recombinant virus antigen. Epidemiol Infect 117:159–164. doi: 10.1017/S0950268800001266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kurtz J, Lee T. 1978. Astrovirus gastroenteritis age distribution of antibody. Med Microbiol Immunol 166:227–230. doi: 10.1007/BF02121154. [DOI] [PubMed] [Google Scholar]
- 9.Midthun K, Greenberg HB, Kurtz JB, Gary GW, Lin FY, Kapikian AZ. 1993. Characterization and seroepidemiology of a type 5 astrovirus associated with an outbreak of gastroenteritis in Marin County, California. J Clin Microbiol 31:955–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kirkwood CD, Clark R, Bogdanovic-Sakran N, Bishop RF. 2005. A 5-year study of the prevalence and genetic diversity of human caliciviruses associated with sporadic cases of acute gastroenteritis in young children admitted to hospital in Melbourne, Australia (1998-2002). J Med Virol 77:96–101. doi: 10.1002/jmv.20419. [DOI] [PubMed] [Google Scholar]
- 11.Buss SN, Leber A, Chapin K, Fey PD, Bankowski MJ, Jones MK, Rogatcheva M, Kanack KJ, Bourzac KM. 2015. Multicenter evaluation of the BioFire FilmArray gastrointestinal panel for etiologic diagnosis of infectious gastroenteritis. J Clin Microbiol 53:915–925. doi: 10.1128/JCM.02674-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Platts-Mills JA, Babji S, Bodhidatta L, Gratz J, Haque R, Havt A, McCormick BJ, McGrath M, Olortegui MP, Samie A, Shakoor S, Mondal D, Lima IF, Hariraju D, Rayamajhi BB, Qureshi S, Kabir F, Yori PP, Mufamadi B, Amour C, Carreon JD, Richard SA, Lang D, Bessong P, Mduma E, Ahmed T, Lima AA, Mason CJ, Zaidi AK, Bhutta ZA, Kosek M, Guerrant RL, Gottlieb M, Miller M, Kang G, Houpt ER, MAL-ED Network Investigators. 2015. Pathogen-specific burdens of community diarrhoea in developing countries: a multisite birth cohort study (MAL-ED). Lancet Glob Health 3:e564–575. doi: 10.1016/S2214-109X(15)00151-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vu DL, Bosch A, Pinto RM, Guix S. 2017. Epidemiology of classic and novel human astrovirus: gastroenteritis and beyond. Viruses 9(2):E33. doi: 10.3390/v9020033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cunliffe NA, Booth JA, Elliot C, Lowe SJ, Sopwith W, Kitchin N, Nakagomi O, Nakagomi T, Hart CA, Regan M. 2010. Healthcare-associated viral gastroenteritis among children in a large pediatric hospital, United Kingdom. Emerg Infect Dis 16:55–62. doi: 10.3201/eid1601.090401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marczinke B, Bloys AJ, Brown TD, Willcocks MM, Carter MJ, Brierley I. 1994. The human astrovirus RNA-dependent RNA polymerase coding region is expressed by ribosomal frameshifting. J Virol 68:5588–5595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lewis TL, Matsui SM. 1995. An astrovirus frameshift signal induces ribosomal frameshifting in vitro. Arch Virol 140:1127–1135. doi: 10.1007/BF01315421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lewis TL, Matsui SM. 1996. Astrovirus ribosomal frameshifting in an infection-transfection transient expression system. J Virol 70:2869–2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Monroe SS, Stine SE, Gorelkin L, Herrmann JE, Blacklow NR, Glass RI. 1991. Temporal synthesis of proteins and RNAs during human astrovirus infection of cultured cells. J Virol 65:641–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Monroe SS, Jiang B, Stine SE, Koopmans M, Glass RI. 1993. Subgenomic RNA sequence of human astrovirus supports classification of Astroviridae as a new family of RNA viruses. J Virol 67:3611–3614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koci MD, Seal BS, Schultz-Cherry S. 2000. Molecular characterization of an avian astrovirus. J Virol 74:6173–6177. doi: 10.1128/JVI.74.13.6173-6177.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Finkbeiner SR, Allred AF, Tarr PI, Klein EJ, Kirkwood CD, Wang D. 2008. Metagenomic analysis of human diarrhea: viral detection and discovery. PLoS Pathog 4:e1000011. doi: 10.1371/journal.ppat.1000011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Finkbeiner SR, Kirkwood CD, Wang D. 2008. Complete genome sequence of a highly divergent astrovirus isolated from a child with acute diarrhea. Virol J 5:117. doi: 10.1186/1743-422X-5-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Finkbeiner SR, Li Y, Ruone S, Conrardy C, Gregoricus N, Toney D, Virgin HW, Anderson LJ, Vinje J, Wang D, Tong S. 2009. Identification of a novel astrovirus (astrovirus VA1) associated with an outbreak of acute gastroenteritis. J Virol 83:10836–10839. doi: 10.1128/JVI.00998-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Finkbeiner SR, Holtz LR, Jiang Y, Rajendran P, Franz CJ, Zhao G, Kang G, Wang D. 2009. Human stool contains a previously unrecognized diversity of novel astroviruses. Virol J 6:161. doi: 10.1186/1743-422X-6-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kapoor A, Li L, Victoria J, Oderinde B, Mason C, Pandey P, Zaidi SZ, Delwart E. 2009. Multiple novel astrovirus species in human stool. J Gen Virol 90:2965–2972. doi: 10.1099/vir.0.014449-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jiang H, Holtz LR, Bauer I, Franz CJ, Zhao G, Bodhidatta L, Shrestha SK, Kang G, Wang D. 2013. Comparison of novel MLB-clade, VA-clade and classic human astroviruses highlights constrained evolution of the classic human astrovirus nonstructural genes. Virology 436:8–14. doi: 10.1016/j.virol.2012.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Phan TG, Nordgren J, Ouermi D, Simpore J, Nitiema LW, Deng X, Delwart E. 2014. New astrovirus in human feces from Burkina Faso. J Clin Virol 60:161–164. doi: 10.1016/j.jcv.2014.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meyer CT, Bauer IK, Antonio M, Adeyemi M, Saha D, Oundo JO, Ochieng JB, Omore R, Stine OC, Wang D, Holtz LR. 2015. Prevalence of classic, MLB-clade and VA-clade astroviruses in Kenya and The Gambia. Virol J 12:78. doi: 10.1186/s12985-015-0299-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holtz LR, Bauer IK, Jiang H, Belshe R, Freiden P, Schultz-Cherry SL, Wang D. 2014. Seroepidemiology of astrovirus MLB1. Clin Vaccine Immunol 21:908–911. doi: 10.1128/CVI.00100-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burbelo PD, Ching KH, Esper F, Iadarola MJ, Delwart E, Lipkin WI, Kapoor A. 2011. Serological studies confirm the novel astrovirus HMOAstV-C as a highly prevalent human infectious agent. PLoS One 6:e22576. doi: 10.1371/journal.pone.0022576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Holtz LR, Bauer IK, Rajendran P, Kang G, Wang D. 2011. Astrovirus MLB1 is not associated with diarrhea in a cohort of Indian children. PLoS One 6:e28647. doi: 10.1371/journal.pone.0028647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Quan PL, Wagner TA, Briese T, Torgerson TR, Hornig M, Tashmukhamedova A, Firth C, Palacios G, Baisre-De-Leon A, Paddock CD, Hutchison SK, Egholm M, Zaki SR, Goldman JE, Ochs HD, Lipkin WI. 2010. Astrovirus encephalitis in boy with X-linked agammaglobulinemia. Emerg Infect Dis 16:918–925. doi: 10.3201/eid1606.091536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Naccache SN, Peggs KS, Mattes FM, Phadke R, Garson JA, Grant P, Samayoa E, Federman S, Miller S, Lunn MP, Gant V, Chiu CY. 2015. Diagnosis of neuroinvasive astrovirus infection in an immunocompromised adult with encephalitis by unbiased next-generation sequencing. Clin Infect Dis 60:919–923. doi: 10.1093/cid/ciu912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brown JR, Morfopoulou S, Hubb J, Emmett WA, Ip W, Shah D, Brooks T, Paine SM, Anderson G, Virasami A, Tong CY, Clark DA, Plagnol V, Jacques TS, Qasim W, Hubank M, Breuer J. 2015. Astrovirus VA1/HMO-C: an increasingly recognized neurotropic pathogen in immunocompromised patients. Clin Infect Dis 60:881–888. doi: 10.1093/cid/ciu940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fremond ML, Perot P, Muth E, Cros G, Dumarest M, Mahlaoui N, Seilhean D, Desguerre I, Hebert C, Corre-Catelin N, Neven B, Lecuit M, Blanche S, Picard C, Eloit M. 2015. Next-generation sequencing for diagnosis and tailored therapy: a case report of astrovirus-associated progressive encephalitis. J Pediatric Infect Dis Soc 4(3):e53–e57. doi: 10.1093/jpids/piv040. [DOI] [PubMed] [Google Scholar]
- 36.Lum SH, Turner A, Guiver M, Bonney D, Martland T, Davies E, Newbould M, Brown J, Morfopoulou S, Breuer J, Wynn R. 2016. An emerging opportunistic infection: fatal astrovirus (VA1/HMO-C) encephalitis in a pediatric stem cell transplant recipient. Transpl Infect Dis 18:960–964. doi: 10.1111/tid.12607. [DOI] [PubMed] [Google Scholar]
- 37.Wunderli W, Meerbach A, Gungor T, Berger C, Greiner O, Caduff R, Trkola A, Bossart W, Gerlach D, Schibler M, Cordey S, McKee TA, Van Belle S, Kaiser L, Tapparel C. 2011. Astrovirus infection in hospitalized infants with severe combined immunodeficiency after allogeneic hematopoietic stem cell transplantation. PLoS One 6:e27483. doi: 10.1371/journal.pone.0027483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sato M, Kuroda M, Kasai M, Matsui H, Fukuyama T, Katano H, Tanaka-Taya K. 2016. Acute encephalopathy in an immunocompromised boy with astrovirus-MLB1 infection detected by next generation sequencing. J Clin Virol 78:66–70. doi: 10.1016/j.jcv.2016.03.010. [DOI] [PubMed] [Google Scholar]
- 39.Cordey S, Vu DL, Schibler M, L'Huillier AG, Brito F, Docquier M, Posfay-Barbe KM, Petty TJ, Turin L, Zdobnov EM, Kaiser L. 2016. Astrovirus MLB2, a new gastroenteric virus associated with meningitis and disseminated infection. Emerg Infect Dis 22:846–853. doi: 10.3201/eid2205.151807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Blomstrom AL, Widen F, Hammer AS, Belak S, Berg M. 2010. Detection of a novel astrovirus in brain tissue of mink suffering from shaking mink syndrome by use of viral metagenomics. J Clin Microbiol 48:4392–4396. doi: 10.1128/JCM.01040-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gavier-Widen D, Brojer C, Dietz HH, Englund L, Hammer AS, Hedlund KO, Hard af Segerstad C, Nilsson K, Nowotny N, Puurula V, Thoren P, Uhlhorn H, Weissenbock H, Agren E, Klingeborn B. 2004. Investigations into shaking mink syndrome: an encephalomyelitis of unknown cause in farmed mink (Mustela vison) kits in Scandinavia. J Vet Diagn Invest 16:305–312. doi: 10.1177/104063870401600408. [DOI] [PubMed] [Google Scholar]
- 42.Selimovic-Hamza S, Boujon CL, Hilbe M, Oevermann A, Seuberlich T. 2017. Frequency and pathological phenotype of bovine astrovirus CH13/NeuroS1 infection in neurologically-diseased cattle: towards assessment of causality. Viruses 9(1):E12. doi: 10.3390/v9010012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schlottau K, Schulze C, Bilk S, Hanke D, Hoper D, Beer M, Hoffmann B. 2016. Detection of a novel bovine astrovirus in a cow with encephalitis. Transbound Emerg Dis 63:253–259. doi: 10.1111/tbed.12493. [DOI] [PubMed] [Google Scholar]
- 44.Bouzalas IG, Wuthrich D, Selimovic-Hamza S, Drogemuller C, Bruggmann R, Seuberlich T. 2016. Full-genome based molecular characterization of encephalitis-associated bovine astroviruses. Infect Genet Evol 44:162–168. doi: 10.1016/j.meegid.2016.06.052. [DOI] [PubMed] [Google Scholar]
- 45.Wuthrich D, Boujon CL, Truchet L, Selimovic-Hamza S, Oevermann A, Bouzalas IG, Bruggmann R, Seuberlich T. 2016. Exploring the virome of cattle with non-suppurative encephalitis of unknown etiology by metagenomics. Virology 493:22–30. doi: 10.1016/j.virol.2016.03.009. [DOI] [PubMed] [Google Scholar]
- 46.Bouzalas IG, Wuthrich D, Walland J, Drogemuller C, Zurbriggen A, Vandevelde M, Oevermann A, Bruggmann R, Seuberlich T. 2014. Neurotropic astrovirus in cattle with nonsuppurative encephalitis in Europe. J Clin Microbiol 52:3318–3324. doi: 10.1128/JCM.01195-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li L, Diab S, McGraw S, Barr B, Traslavina R, Higgins R, Talbot T, Blanchard P, Rimoldi G, Fahsbender E, Page B, Phan TG, Wang C, Deng X, Pesavento P, Delwart E. 2013. Divergent astrovirus associated with neurologic disease in cattle. Emerg Infect Dis 19:1385–1392. doi: 10.3201/eid1909.130682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pfaff F, Schlottau K, Scholes S, Courtenay A, Hoffmann B, Hoper D, Beer M. 2017. A novel astrovirus associated with encephalitis and ganglionitis in domestic sheep. Transbound Emerg Dis 64:677–682. doi: 10.1111/tbed.12623. [DOI] [PubMed] [Google Scholar]
- 49.Blomstrom AL, Ley C, Jacobson M. 2014. Astrovirus as a possible cause of congenital tremor type AII in piglets? Acta Vet Scand 56:82. doi: 10.1186/s13028-014-0082-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nunez LF, Santander Parra SH, Carranza C, Astolfi-Ferreira CS, Buim MR, Piantino Ferreira AJ. 2016. Detection and molecular characterization of chicken astrovirus associated with chicks that have an unusual condition known as “white chicks” in Brazil. Poult Sci 95:1262–1270. doi: 10.3382/ps/pew062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee TW, Kurtz JB. 1981. Serial propagation of astrovirus in tissue culture with the aid of trypsin. J Gen Virol 57:421–424. doi: 10.1099/0022-1317-57-2-421. [DOI] [PubMed] [Google Scholar]
- 52.Brinker JP, Blacklow NR, Herrmann JE. 2000. Human astrovirus isolation and propagation in multiple cell lines. Arch Virol 145:1847–1856. doi: 10.1007/s007050070060. [DOI] [PubMed] [Google Scholar]
- 53.Shimizu M, Shirai J, Narita M, Yamane T. 1990. Cytopathic astrovirus isolated from porcine acute gastroenteritis in an established cell line derived from porcine embryonic kidney. J Clin Microbiol 28:201–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aroonprasert D, Fagerland JA, Kelso NE, Zheng S, Woode GN. 1989. Cultivation and partial characterization of bovine astrovirus. Vet Microbiol 19:113–125. doi: 10.1016/0378-1135(89)90077-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Harbour DA, Ashley CR, Williams PD, Gruffydd-Jones TJ. 1987. Natural and experimental astrovirus infection of cats. Vet Rec 120:555–557. doi: 10.1136/vr.120.23.555. [DOI] [PubMed] [Google Scholar]
- 56.Dryden KA, Tihova M, Nowotny N, Matsui SM, Mendez E, Yeager M. 2012. Immature and mature human astrovirus: structure, conformational changes, and similarities to hepatitis E virus. J Mol Biol 422:650–658. doi: 10.1016/j.jmb.2012.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mendez E, Fernandez-Luna T, Lopez S, Mendez-Toss M, Arias CF. 2002. Proteolytic processing of a serotype 8 human astrovirus ORF2 polyprotein. J Virol 76:7996–8002. doi: 10.1128/JVI.76.16.7996-8002.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bass DM, Qiu S. 2000. Proteolytic processing of the astrovirus capsid. J Virol 74:1810–1814. doi: 10.1128/JVI.74.4.1810-1814.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Marvin SA, Huerta CT, Sharp B, Freiden P, Cline TD, Schultz-Cherry S. 2015. Type I interferon response limits astrovirus replication and protects against increased barrier permeability in vitro and in vivo. J Virol 90:1988–1996. doi: 10.1128/JVI.02367-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Guix S, Perez-Bosque A, Miro L, Moreto M, Bosch A, Pinto RM. 2015. Type I interferon response is delayed in human astrovirus infections. PLoS One 10:e0123087. doi: 10.1371/journal.pone.0123087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yokoyama CC, Loh J, Zhao G, Stappenbeck TS, Wang D, Huang HV, Virgin HW, Thackray LB. 2012. Adaptive immunity restricts replication of novel murine astroviruses. J Virol 86:12262–12270. doi: 10.1128/JVI.02018-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Risco C, Carrascosa JL, Pedregosa AM, Humphrey CD, Sanchez-Fauquier A. 1995. Ultrastructure of human astrovirus serotype 2. J Gen Virol 76(Part 8):2075–2080. doi: 10.1099/0022-1317-76-8-2075. [DOI] [PubMed] [Google Scholar]
- 63.Caul EO, Appleton H. 1982. The electron microscopical and physical characteristics of small round human fecal viruses: an interim scheme for classification. J Med Virol 9:257–265. doi: 10.1002/jmv.1890090403. [DOI] [PubMed] [Google Scholar]
- 64.Koshikawa N, Hasegawa S, Nagashima Y, Mitsuhashi K, Tsubota Y, Miyata S, Miyagi Y, Yasumitsu H, Miyazaki K. 1998. Expression of trypsin by epithelial cells of various tissues, leukocytes, and neurons in human and mouse. Am J Pathol 153:937–944. doi: 10.1016/S0002-9440(10)65635-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Baxendale W, Mebatsion T. 2004. The isolation and characterisation of astroviruses from chickens. Avian Pathol 33:364–370. doi: 10.1080/0307945042000220426. [DOI] [PubMed] [Google Scholar]
- 66.Geigenmuller U, Ginzton NH, Matsui SM. 1997. Construction of a genome-length cDNA clone for human astrovirus serotype 1 and synthesis of infectious RNA transcripts. J Virol 71:1713–1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Velazquez-Moctezuma R, Banos-Lara Mdel R, Acevedo Y, Mendez E. 2012. Alternative cell lines to improve the rescue of infectious human astrovirus from a cDNA clone. J Virol Methods 179:295–302. doi: 10.1016/j.jviromet.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 68.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hierholzer JC, Killington RA. 1996. Virus isolation and quantitation, p 25–46. In Mahy BW, Kangro HO (ed), Virology methods manual, 1st ed Academic Press, Cambridge, MA. [Google Scholar]


