ABSTRACT
Middle East respiratory syndrome coronavirus (MERS-CoV) utilizes dipeptidyl peptidase 4 (DPP4) as an entry receptor. While bat, camel, and human DPP4 support MERS-CoV infection, several DPP4 orthologs, including mouse, ferret, hamster, and guinea pig DPP4, do not. Previous work revealed that glycosylation of mouse DPP4 plays a role in blocking MERS-CoV infection. Here, we tested whether glycosylation also acts as a determinant of permissivity for ferret, hamster, and guinea pig DPP4. We found that, while glycosylation plays an important role in these orthologs, additional sequence and structural determinants impact their ability to act as functional receptors for MERS-CoV. These results provide insight into DPP4 species-specific differences impacting MERS-CoV host range and better inform our understanding of virus-receptor interactions associated with disease emergence and host susceptibility.
IMPORTANCE MERS-CoV is a recently emerged zoonotic virus that is still circulating in the human population with an ∼35% mortality rate. With no available vaccines or therapeutics, the study of MERS-CoV pathogenesis is crucial for its control and prevention. However, in vivo studies are limited because MERS-CoV cannot infect wild-type mice due to incompatibilities between the virus spike and the mouse host cell receptor, mouse DPP4 (mDPP4). Specifically, mDPP4 has a nonconserved glycosylation site that acts as a barrier to MERS-CoV infection. Thus, one mouse model strategy has been to modify the mouse genome to remove this glycosylation site. Here, we investigated whether glycosylation acts as a barrier to infection for other nonpermissive small-animal species, namely, ferret, guinea pig, and hamster. Understanding the virus-receptor interactions for these DPP4 orthologs will help in the development of additional animal models while also revealing species-specific differences impacting MERS-CoV host range.
KEYWORDS: MERS-coronavirus, DPP4, orthologs, host range expansion, animal models, glycosylation, host range
INTRODUCTION
Coronaviruses are a diverse family of viruses that infect a wide range of hosts, including both mammalian and avian species. Phylogenetic studies suggest that over the last 800 years, several zoonotic coronaviruses have expanded their host range into humans, resulting in four antigenically distinct strains that are still circulating in the human population (1, 2). Whereas most human coronaviruses cause only mild symptoms in healthy adults, two recent emergence events have resulted in severe disease in humans. Severe acute respiratory syndrome coronavirus (SARS-CoV) emerged from its zoonotic reservoir in 2003 and infected over 8,000 people with a 9% mortality rate before being controlled by public health measures (3). Although SARS-CoV is no longer circulating in the human population, several SARS-like coronaviruses persist in bats, can use the human receptor for entry, and are poised for emergence (4, 5). Middle East respiratory syndrome coronavirus (MERS-CoV) emerged from its zoonotic reservoir in 2012 and continues to cause human infection. As of July 2017, MERS-CoV has infected 2,040 people with an ∼35% mortality rate (6). The exact origins of MERS-CoV remain unclear. However, most data suggest that the virus originated in bats and spread into human populations by the use of camels as an intermediate host species (7, 8). Understanding how MERS-CoV and other coronaviruses evolve and spread will allow us to combat MERS-CoV infection while also developing approaches for dealing with future pandemic coronavirus strains.
One of the key determinants of viral host range is the interaction between the virus spike protein and the host cell receptor. For MERS-CoV, the host cell receptor is dipeptidyl peptidase IV (DPPIV) (9), a ubiquitously expressed cell surface protein that functions in immune homeostasis. Interestingly, whereas MERS-CoV can utilize bat DPP4 (bDPP4), camel DPP4 (cDPP4), and human DPP4 (hDPP4) molecules for entry (10–12), it is unable to infect cells using the DPP4 molecules from traditional small-animal models, including mice, ferrets, guinea pigs, and hamsters (13–17). The inability of MERS-CoV to infect these species in vivo and in vitro is primarily due to spike-receptor incompatibilities and not due to other species-specific host cell factors (13, 18, 19). Because MERS-CoV is unable to infect traditional small-animal models, our ability to study MERS-CoV pathogenesis is restricted, as is the ability to evaluate drugs and vaccine efficacy. To overcome this obstacle, several mouse models have recently been developed in order to study MERS-CoV pathogenesis in vivo. These include a model employing adeno-associated virus delivery of hDPP4 (18), overexpression or knock-in of hDPP4 (20–23), and the use of gene editing to generate mice carrying a chimeric mouse DPP4 (mDPP4) gene (24). These mouse models greatly improve our ability to study MERS-CoV infection, motivating us to investigate the potential for alternative species to act as models for MERS-CoV infection. Importantly, the development of vaccines and therapeutics relies on positive results from more than one animal species. For MERS-CoV, the disparate findings reported for nonhuman primate systems, including the rhesus macaque and common marmoset (25–28), emphasize the need for additional animal models.
In our previous studies, we found that receptor incompatibilities between MERS-CoV and mDPP4 can be alleviated with just two amino acid substitutions (13). Introducing mutations A288L and T330R (residues are numbered relative to mDPP4) allows mDPP4 to act as a functional receptor for MERS-CoV infection. The A288L mutation helps stabilize a hydrophobic core in the MERS-CoV receptor binding domain (RBD) shown to be important for proper binding to DPP4 (29). Conversely, the T330R mutation knocks out a nonconserved glycosylation site present in mDPP4. This glycosylation site is a substantial barrier to MERS-CoV infection, with mDPP4 acting as a functional receptor only when the glycan is absent (19). Taking the data together, these two mutations informed the development of the aforementioned chimeric mDPP4 mouse model (24) and improved our understanding of the biochemical mechanisms that govern the interactions between DPP4 and the MERS-CoV RBD.
Here we investigated whether glycosylation acts as a broader determinant of permissivity using alternative small-animal models, including the ferret, hamster, and guinea pig. Overall, we found that, while glycosylation of these DPP4 orthologs plays an important role in blocking MERS-CoV infection, additional species-specific determinants contribute to the inability of each of these receptors to support MERS-CoV infection. Further investigation is required to identify these determinants and establish whether they lie at the level of the sequence or the structure. Improving our understanding of the interactions between DPP4 and the MERS-CoV RBD can help inform the development of alternative small-animal models as well as help reveal how coronaviruses are able to emerge into novel species.
RESULTS
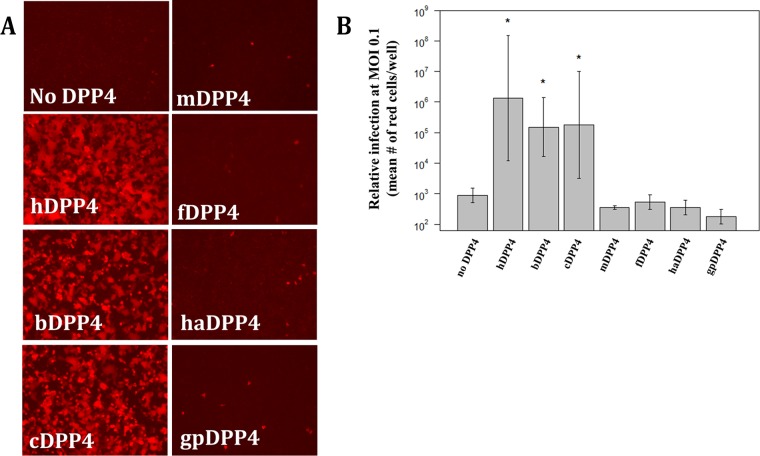
Susceptibility to MERS-CoV varies among a number of host species, making detailed analyses of orthologous DPP4 receptors essential for elucidating the fundamental mechanisms that govern coronavirus species specificity. DPP4 orthologs are classified as permissive (human, bat, and camel) or nonpermissive (mouse, ferret, hamster, and guinea pig) (Fig. 1A). Based on mean fluorescent cell count values, the hDPP4, bDPP4, and cDPP4 orthologs support higher levels of infection than the nonpermissive orthologs (P < 0.05 [Student's t test]) (Fig. 1B). Orthologs categorized as nonpermissive have levels of infection that are not significantly different from those seen when no DPP4 is present (Fig. 1B). Our previous work found that mDPP4 could support MERS-CoV infection with just two amino acid substitutions (13) and that knocking out a glycosylation site present on blade IV of mDPP4 was crucial for this result (19). Here, we investigated alternative nonpermissive DPP4 orthologs to determine whether they can act as backbones to support MERS-CoV infection and, if so, what determinants are responsible for blocking infection in the wild-type molecules.
FIG 1.
Permissivity of DPP4 orthologs to MERS-CoV. (A) Seven DPP4 orthologs were tested for their ability to support infection by rMERS-CoV-RFP. DPP4 constructs were transfected into HEK 293T cells and infected at an MOI of 5 at ∼20 h posttransfection. Cells were imaged for fluorescence at ∼24 hpi. hDPP4, human DPP4; cDPP4, camel DPP4; bDPP4, bat DPP4; mDPP4, mouse DPP4; fDPP4, ferret DPP4; haDPP4, hamster DPP4; gpDPP4, guinea pig DPP4. (B) Mean fluorescent cell counts of MERS-CoV infection utilizing various DPP4 orthologs. Cells were infected at an MOI of 0.1 and the numbers of infected cells counted at ∼72 hpi. Each DPP4 ortholog was measured in triplicate. Only hDPP4, bDPP4, and cDPP4 had levels of infection significantly higher than those seen in the absence of DPP4 (*, P < 0.05 [Student's t test]). All DPP4 orthologs had significantly lower levels of infection than hDPP4 (P < 0.05 [Student's t test]). The levels of infection seen between bDPP4 and cDPP4 were not significantly different. Error bars indicate mean values ± 1 standard deviation.
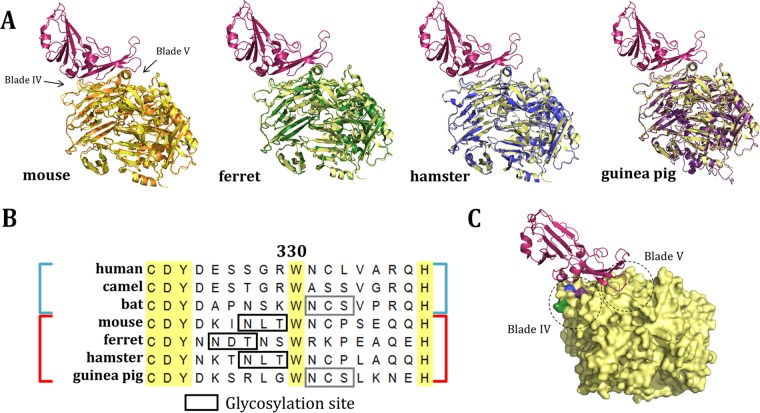
To assess the potential for ferret DPP4 (fDPP4), hamster DPP4 (haDPP4), and guinea pig DPP4 (gpDPP4) to act as functional receptors for MERS-CoV, we compared the overall structures of these molecules. As the crystal structures for these orthologs have not yet been solved, we generated homology models using I-TASSER (30). Each structure is predicted to have a backbone topology highly similar to that of hDPP4 (Fig. 2A). The root mean square deviation (RMSD) scores obtained for mDPP4, fDPP4, haDPP4, and gpDPP4 aligned to hDPP4 (PDB code 4L72) are 0.644, 0.616, 0.378, and 0.604, respectively, with a lower number indicating greater structural homology. These values can be compared to amino acid sequence identity values of 85%, 88%, 85%, and 87%, respectively. The high sequence similarities and predicted structural similarities between hDPP4 and these orthologs suggest that they can likely act as backbones to support MERS-CoV infection, consistent with previous DPP4 ortholog work (15, 17).
FIG 2.
Sequence and structural comparison of nonpermissive DPP4 orthologs. (A) Structural comparison of threaded molecules (30) of mDPP4 (orange), fDPP4 (green), haDPP4 (blue), and gpDPP4 (purple) overlaid on hDPP4 (yellow) complexed with the MERS-CoV RBD (red) (PDB code 4L72). (B) Sequence alignment of permissive (human, camel, bat; blue) and nonpermissive (mouse, ferret, hamster, guinea pig; red) DPP4 amino acid sequences. Residue 330 is numbered relative to mDPP4. Boxes represent glycosylation sites that are either unique to nonpermissive species (black) or shared with a permissive species (gray). (C) hDPP4 (yellow) complexed with the MERS-CoV RBD (red) (PDB code 4L72). Residues aligning to the ferret (green), hamster and mouse (blue), and guinea pig (purple) glycosylation sites are highlighted. Dashed-line circles indicate the regions of the DPP4 molecule that correspond to blades IV and V.
To gain an intuition of the specific differences between permissive and nonpermissive DPP4 orthologs, we can assess their relatedness through phylogenetics. Unfortunately, analysis of the DPP4 gene tree does not reveal a pattern of permissivity on a global sequence scale (i.e., permissive and nonpermissive species do not form distinct monophyletic groups) (31, 32). However, it is possible that a signature may exist at a local scale. Our previous work in mDPP4 revealed that glycosylation on blade IV of the DPP4 molecule can act as a barrier to MERS-CoV infection (19). Thus, we investigated whether glycosylation (designated by motif NXS and motif NXT) acts as a broader determinant of DPP4 ortholog permissivity. A total of eight N-linked glycans are known to be present in the extracellular domain of hDPP4 (29). Without solved crystal structures for DPP4 orthologs, it is unknown how many glycans are truly present on the surface of these molecules. However, predictive software (33) estimates that there are 7 to 11 putative glycosylation sites for each of the DPP4 orthologs analyzed in this study (Table 1). Aligning the sequences of these orthologs species revealed the presence of putative glycosylation sites on blade IV of fDPP4, haDPP4, and gpDPP4 in a region that is crucial for interactions with the MERS-CoV RBD (Fig. 2B; Table 1). Interestingly, haDPP4 encodes a glycosylation site at the residues aligning to the site identified in our previous mDPP4 studies. In contrast, fDPP4 encodes a glycosylation site slightly upstream whereas gpDPP4 encodes a glycosylation site slightly downstream from the mDPP4 site (Fig. 2B). All three glycosylation sites lie on blade IV, within a region of DPP4 that interacts with the MERS-CoV RBD (Fig. 2C). Of note, the downstream gpDPP4 glycosylation site is also present within the permissive bDPP4 receptor used in this study (from Pipistrellus pipistrellus), causing us to hypothesize that host restriction might be mediated by differences in the sequence or structure of gpDPP4 that are independent of this glycosylation site.
TABLE 1.
Known and putative N-linked glycans in DPP4 orthologsa
| Ortholog | Residue | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| hDPP4 | 85 | 92 | 150 | 219 | 229 | 281 | 321 | 520 | |||||||||
| bDPP4 | 82 | 89 | 147 | 216 | 226 | 272 | 315 | 332 | 393 | 432 | 490 | ||||||
| mDPP4 | 83 | 90 | 144 | 223 | 315 | 328 | 514 | ||||||||||
| fDPP4 | 84 | 91 | 178 | 228 | 280 | 320 | 437 | 519 | |||||||||
| haDPP4 | 83 | 90 | 148 | 227 | 275 | 319 | 332 | 518 | |||||||||
| gpDPP4 | 32 | 295 | 302 | 360 | 439 | 491 | 531 | 548 | 730 | ||||||||
Numbers indicate the residues known (hDPP4) or predicted (all other DPP4 orthologs) (33) to have N-linked glycans for each DPP4 molecule. All residues are numbered relative to the given DPP4 molecule, with columns representing residues that align. Among the eight N-linked glycans in hDPP4, only four are conserved across all orthologs. Bold font indicates sites that align to within two residues of a contact residue in hDPP4 (see reference 29).
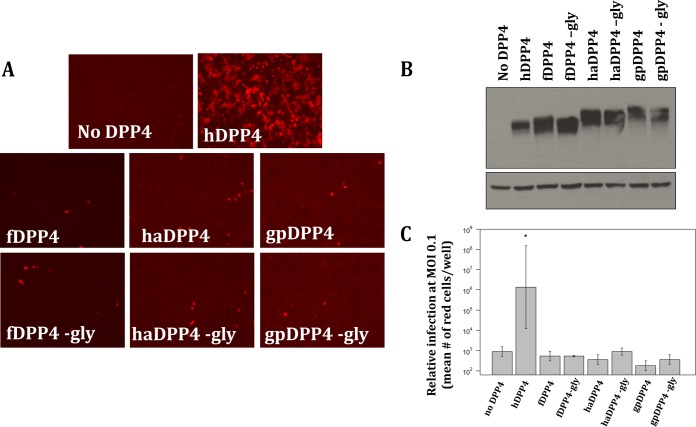
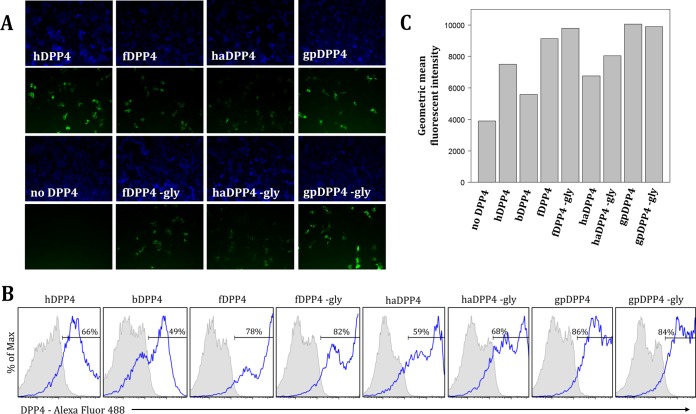
To investigate whether the putative glycosylation sites identified in fDPP4, haDPP4, and gpDPP4 play a role in hindering MERS-CoV infection, we assessed receptor permissivity for glycosylation knockout mutants. Each DPP4 ortholog knockout includes a mutation that changes the N of the glycosylation NXT (or NXS) motif to an alanine, designated by “−gly” (the specific mutations are fDPP4 N331A, haDPP4 N332A, and gpDPP4 N548A, with residues numbered relative to its own sequence). Results showed that removing glycosylation from these three DPP4 orthologs, confirmed by an ∼2.5-kDa downward shift in protein mobility via Western blot analysis (Fig. 3B), did not result in an increase in infection (Fig. 3A). In fact, the levels of infection supported by all three glycosylation knockout molecules were not significantly different from those seen with the respective wild-type DPP4 molecules (P < 0.05 [Student's t test]) (Fig. 3C). Immunofluorescence assay (IFA) and flow cytometry results confirmed that the DPP4 variants were expressed at the surface of the cell (Fig. 4), ruling out the possibility of nonpermissivity resulting from a lack of proper expression of the DPP4 ortholog glycosylation knockouts.
FIG 3.
DPP4 ortholog glycosylation knockout mutants. (A) Neither wild-type nor glycosylation knockout (−gly) DPP4 molecules for ferret (fDPP4), hamster (haDPP4), or guinea pig (gpDPP4) support infection by MERS-CoV. (B) Successful removal of glycosylation is supported by an ∼2.5-kDa downward shift seen via Western blotting. The top blot represents DPP4, and the bottom blot represents β-actin as a control. (C) Fluorescent cell counts from MERS-CoV infection utilizing various DPP4 orthologs and their respective glycosylation knockout mutants. Cells were infected at an MOI of 0.1, and numbers of infected cells were counted at 72 hpi. The level of each DPP4 ortholog was measured in triplicate. Only hDPP4 had levels of infection significantly higher than those seen in the absence of DPP4 (*, P < 0.05 [Student's t test]). The remaining DPP4 orthologs and glycosylation knockouts had infection levels that were not significantly different from those seen in the absence of DPP4. Error bars indicate mean values ± 1 standard deviation.
FIG 4.
DPP4 and mutant variants are expressed on the surface of cells as evidenced by results of immunofluorescence assay (A) and flow cytometry (B and C). (A) Cells were transfected with each DPP4 ortholog, fixed, and probed with primary goat anti-DPP4 polyclonal antibody (R&D Systems) at 1:50 and secondary donkey anti-goat Alexa Fluor 488 (Life Technologies) at 1:500. Cells were imaged at a magnification of ×20 for DAPI (300 ms exposure) and DPP4 (1.5 s exposure). (B) DPP4 expression frequencies (blue outlined histogram) by DPP4 construct after subtraction of background from replicate wells stained with secondary donkey anti-goat IgG (H+L) Alexa Fluor 488 antibody only (gray-shaded histogram). Percentage values represent averages of results across two duplicate wells. Max, maximum. (C) Geometric mean fluorescence intensity of the DPP4-positive populations for each DPP4 construct.
Although removing the blade IV glycosylation site on its own was not enough to confer permissivity to fDPP4, haDPP4, and gpDPP4 (Fig. 3A), we have previously shown in mDPP4 that a second determinant acts in conjunction with the blade IV glycosylation site to impact the interactions between DPP4 and the MERS-CoV RBD (13, 19). Specifically, introducing select human amino acids into the mDPP4 sequence on blade V (A288L) acts to support MERS-CoV infection in combination with the blade IV glycosylation knockout (N328A). In the case of haDPP4, the L288 amino acid identity is conserved between the hamster and human sequences, suggesting that different determinants in haDPP4 and mDPP4 are responsible for blocking MERS-CoV infection. This is consistent with prior studies which identified several amino acid differences between hDPP4 and haDPP4 in blade V that were required to support MERS-CoV infection (17). Therefore, we asked whether specific changes in blade V of haDPP4 could allow it to act as a functional MERS-CoV receptor in combination with the blade IV glycosylation knockout mutation. We found that three mutations on blade V (E289A, V291M, and T293I [residue numbering relative to haDPP4]) conferred MERS-CoV receptor function to haDPP4 when introduced in conjunction with the blade IV glycosylation knockout mutation (Fig. 5A). Two of the identified mutations overlap previous work, while a third site knocked out the same blade IV glycosylation site but did so by mutating a different residue of the glycosylation site motif (Table 2). Testing combinations of these mutations in sets that included fewer than the four identified here, including testing haDPP4 E289A, V291M, and T293I [denoted haDPP4 (289)] in a glycosylation-positive background, did not result in an increase in infection (data not shown). These results demonstrate that the presence of the blade IV glycosylation site plays an important role in regulating the MERS-CoV receptor function of haDPP4. However, species-specific differences in blade V of haDPP4, which differ from those found in mDPP4, are also responsible for haDPP4's inability to act as a functional receptor for MERS-CoV infection.
FIG 5.
Many amino acid changes are required to make fDPP4 and haDPP4 permissive to MERS-CoV infection. (A) Removing glycosylation alone did not confer permissivity to haDPP4. However, combining three amino acid changes on blade V (starting at residue 289) with the glycosylation knockout mutant on blade IV (N332A) resulted in high levels of MERS-CoV infection. Sequences show the alignment between hDPP4 and haDPP4, with the black boxes indicating the amino acids that were swapped from hDPP4 into haDPP4. (B) Removing glycosylation alone did not confer permissivity to fDPP4. However, introducing a set of amino acid changes on blade V (starting at residue 278) and blade IV (starting at residue 330) allowed fDPP4 to support MERS-CoV infection [fDPP4 (278) (330)]. Sequences show the alignment between hDPP4 and fDPP4, with the black boxes indicating the amino acids that were swapped from hDPP4 into fDPP4. Note that fDPP4 −gly is a negative control and includes only the single point mutation N331A. (C) Western blot analysis of fDPP4 and haDPP4 and designated variants for DPP4 and β-actin expression. Successful glycosylation knockout is indicated by a downward shift of ∼2.5 kDa. (D) Fluorescent cell counts of MERS-CoV infection utilizing DPP4 orthologs. Cells were infected at an MOI of 0.1, and numbers of red cells were counted at 72 hpi. Each DPP4 ortholog was measured in triplicate. hDPP4, fDPP4 (278) (330), and haDPP4 (289), −gly had levels of infection that were significantly greater than those seen in the absence of DPP4 (P < 0.05 [Student's t test]). fDPP4 −gly and haDPP4 −gly infection levels were not significantly different from those seen in the absence of DPP4. Error bars indicate mean values ± 1 standard deviation.
TABLE 2.
Residues identified to be important for MERS-CoV permissivitya
| Ortholog | Important residue |
Reference(s) or source | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Blade V | Blade VI | ||||||||||
| Human DPP4 | 267 | 294 | 295 | 336 | 34 | ||||||
| Mouse DPP4 | 294 | 336* | 13, 24 | ||||||||
| Hamster DPP4 | 291 | 295 | 336* | 341 | 346 | 16, 17 | |||||
| 293 | 295 | 297 | 334* | This study | |||||||
| Ferret DPP4 | 279–295 | 331–341 | This study | ||||||||
| 246–503 | 15 | ||||||||||
Data represent the residues that have been identified as important for mediating permissivity to MERS-CoV among various DPP4 orthologs in vitro. All residue numbering is relative to the aligning residue in hDPP4. Residues were either on blade IV or on blade V. Residues that knock out a glycosylation site in the mouse or hamster DPP4 molecules are indicated by an asterisk (*).
Given our findings with haDPP4, we then went on to assess whether the same principles applied to fDPP4. Specifically, we tested whether additional changes in blade V would act in concert with the blade IV glycosylation knockout mutation to enhance the MERS-CoV receptor function. Previous studies found that swapping fDPP4 amino acids 246 to 503 with hDPP4 amino acids 247 to 504 allowed fDPP4 to support MERS-CoV infection, but no single mutations were identified that could recapitulate the susceptible phenotype (15). Because the mDPP4 and haDPP4 data suggest that changes on both blade IV and blade V of DPP4 are likely required to confer infection, we mutated residues on blade V and tried them in combination with the blade IV glycosylation knockout mutation (N331A [residue numbering relative to fDPP4]). We found that no set of mutations on blade V conferred permissiveness to MERS-CoV when made only with the blade IV glycosylation knockout mutant (data not shown). Given this result, we expanded our search and made additional mutations on blade IV of fDPP4. By identifying residues not present in permissive DPP4 orthologs, we generated a chimeric fDPP4 that changed 10 residues on blade IV (including the glycosylation knockout mutation [N331A]) and 9 residues on blade V to the equivalent human amino acid identities, indicated by their starting residues of 330 and 278, respectively (numbering relative to the fDPP4 sequence) (Fig. 5B). The combination of these 19 amino acid changes on blades IV and V resulted in a significant increase in infection (P < 0.05 [Student's t test]) (Fig. 5B and D). Western blot analysis confirmed high expression of each DPP4 variant. The larger downward shift seen for the fDPP4 (278) receptor was likely due to the large number of amino acid changes made on blade V (Fig. 5C). Combinations of individual mutations from these blocks did not result in an increase in infection (data not shown). These data reveal that, in contrast to the results seen with mDPP4 and haDPP4, additional determinants in blade IV contribute to the ability of fDPP4 to act as a functional MERS-CoV receptor.
In addition to the determinants of haDPP4 and fDPP4, we investigated those of gpDPP4. Whereas haDPP4 and fDPP4 do not share their glycosylation sites with permissive molecules, gpDPP4 shares its downstream glycosylation site with bDPP4 (Fig. 2B). Because of this, we knocked out the blade IV glycosylation site in bDPP4 to determine (i) whether it is truly glycosylated and (ii) its impact on the permissivity of bDPP4. The results showed that removing glycosylation from bDPP4 caused no statistically significant change in its ability to support MERS-CoV infection (Fig. 6A and D). Western blot analysis confirmed that the bDPP4 glycosylation motif is a true glycosylation site when expressed in human cells, as evidenced by a downward shift in the glycosylation knockout protein band (Fig. 6B). Additionally, flow cytometry and IFA confirmed surface expression of each variant tested in this assay (Fig. 4B and C and 6C). Low apparent expression of bDPP4 was likely due to inefficient binding of the anti-hDPP4 antibody to bDPP4, an observation that is supported by previous work (35). Unfortunately, the gpDPP4 plasmid was not stable, and the gene was repeatedly lost during site-directed mutagenesis. This instability prevented further investigation of the determinants responsible for gpDPP4's inability to support MERS-CoV infection. Therefore, future studies are needed to determine whether changes on both blade IV and blade V or on just blade IV of gpDPP4 can confer permissivity in combination with the blade IV glycosylation knockout mutation such as was shown with fDPP4 and haDPP4.
FIG 6.
Bat and guinea pig DPP4 share the same glycosylation site downstream of the site identified to be important in mDPP4 (Fig. 2B). (A) Removal of the glycosylation site from bDPP4 showed no decrease in infection, while removal of glycosylation from gpDPP4 resulted in no increase in infection. (B) Western blot analysis of bDPP4 and gpDPP4 and their respective glycosylation knockout mutants for DPP4 and β-actin expression. Successful glycosylation knockout is indicated by a downward shift of ∼2.5 kDa. (C) DPP4 and mutant variants are expressed on the surface of cells, visible by immunofluorescence. Cells were transfected with each DPP4 ortholog, fixed, and probed with primary goat anti-DPP4 polyclonal antibody (R&D Systems) at 1:50 and secondary donkey anti-goat Alexa Fluor 488 (Life Technologies) at 1:500. Cells were imaged at a magnification of ×20 for DAPI (300-ms exposure) and DPP4 (1.5-s exposure). (D) Fluorescent cell counts of MERS-CoV infection utilizing various DPP4 orthologs. Cells were infected at an MOI of 0.1, and numbers of infected cells were counted at 72 hpi. Each DPP4 ortholog was measured in triplicate. hDPP4, bDPP4, and bDPP4 −gly had levels of infection that are significantly higher than those seen in the absence of DPP4 (P < 0.05 [Student's t test]). gpDPP4 and gpDPP4 −gly infection levels were not significantly different from those seen in the absence of DPP4. Error bars indicate mean values ± 1 standard deviation.
DISCUSSION
The interactions between the virus spike and host cell receptor are crucial for mediating infection and for acting as a barrier for nonpermissive species. However, viruses frequently overcome species barriers to expand their host range. Glycosylation plays an important role in mediating permissivity for many viruses, in the context both of the virus spike protein and of the host cell receptor. For example, glycosylation of the H1N1 influenza virus hemagglutinin protein is highly conserved and has been shown to be crucial for host cell receptor binding specificity (36, 37). Conversely, glycosylation of the parvovirus host cell receptor (transferrin receptor type-1) in canines was found to confer resistance to infection by previously circulating feline parvovirus strains (38). In fact, dogs did not become susceptible to parvovirus until a lineage (now canine parvovirus) was able to overcome this glycosylation through specific mutations in the capsid protein (38–40).
Glycosylation has previously been identified as a key determinant of coronavirus host range. For example, some lineages of group 1 coronaviruses use aminopeptidase N (APN) as their host cell receptor and APN orthologs have different glycosylation profiles that influence the species specificity of coronavirus infection. Porcine and feline APN orthologs are glycosylated and can support infection by porcine and feline coronaviruses, respectively (41). However, adding a glycosylation site into human APN near residue 290 can abrogate its ability to support human coronavirus 229E (HCoV-229E) infection (41). Similarly, removal of the aligning glycosylation site in mouse APN can allow it to act as a functional receptor for HCoV-229E (42). As another example, SARS-CoV utilizes angiotensin-converting enzyme 2 (ACE2) as its functional receptor. Whereas rat ACE2 is not permissive to SARS-CoV, introduction of a glycosylation site combined with a point mutation allows rat ACE2 to support SARS-CoV infection (43). Taken together, these results highlight glycosylation as a key determinant of receptor species specificity for numerous coronaviruses.
Here we investigated the extent to which glycosylation of DPP4 can act to mediate the host range of MERS-CoV. Our previous work revealed the importance of glycosylation in blocking MERS-CoV infection in the context of mDPP4 (19). Here, we found that glycosylation is not the only determinant that mediates MERS-CoV infection in the nonpermissive orthologs fDPP4, haDPP4, and gpDPP4. This is particularly surprising in the case of haDPP4; this protein not only has a glycosylation site in the same location as mDPP4 (Fig. 2B) but also has the same amino acid identity as hDPP4 at the secondary blade V residue (288) that was identified to be important for mDPP4. Instead, we found that three amino acid substitutions in blade V of haDPP4 affect the molecule's ability to mediate MERS-CoV infection when coupled with the blade IV glycosylation knockout mutation (Fig. 5A). For fDPP4, we found that extensive changes on both blade IV and blade V were required to support MERS-CoV infection (Fig. 5B), indicating that there are likely species-specific determinants that are not shared with mDPP4 and haDPP4. Finally, the blade IV glycosylation site in gpDPP4 is conserved with the permissive bDPP4 ortholog (Fig. 2B). We found that removing this glycosylation site in bDPP4 did not affect its ability to support MERS-CoV infection (Fig. 6A). This suggests that the impact of the bDPP4 blade IV glycosylation site on receptor permissivity may be species specific and that further research should be performed to elucidate the additional changes that might allow gpDPP4 to act as a functional receptor for MERS-CoV. These changes can then be made independently and in combination with the blade IV glycosylation site to determine its impact within the gpDPP4 backbone. Overall, we found that glycosylation in DPP4 orthologs is a substantial barrier to MERS-CoV infection, particularly in combination with species-specific changes in blade V or blade IV or both. The biochemical mechanism of these additional determinants can be investigated in future studies.
Comparing the data that we have so far on the residues that are important for mediating permissivity in hDPP4, mDPP4, haDPP4, and fDPP4, it is difficult to discern an obvious pattern. The most evident trend is that at least one change is required on both blade IV and blade V of DPP4 (Table 2). This indicates that there are two key points of interaction between DPP4 and the MERS-CoV RBD, which is consistent with previous structural work examining the binding interface of MERS-CoV RBD and hDPP4 (29, 34, 44). We show that these two points of interaction are also important for allowing the virus to utilize a new species receptor. Further understanding of the host range expansion of MERS-CoV will come with identifying which mutations can confer permissivity to DPP4 receptors of currently nonpermissive species. Additionally, solving the crystal structures of nonpermissive DPP4 orthologs will reveal the specific interactions between these molecules and MERS-CoV and help to elucidate what prevents MERS-CoV from successfully utilizing these molecules as functional receptors. The inability to identify a small number of changes that confer permissivity to DPP4 orthologs suggests that extensive remodeling of the MERS-CoV RBD might be required for the virus to infect these nonpermissive species. Access to the crystal structures of these DPP4 orthologs would enable us to model changes in the MERS-CoV RBD that would allow the virus to successfully infect nonpermissive species, thereby enhancing our ability to study MERS-CoV pathogenesis and provide robust small-animal models for evaluating vaccines and therapeutics. As of now, our results indicate that the generation of a transgenic small-animal model in these alternative species would require extensive genomic editing due to the number of mutations needed for each DPP4 to support MERS-CoV infection.
The importance of glycosylation in blocking MERS-CoV infection may vary between species. To gain a better intuition about the extent of glycosylation among DPP4 orthologs, we constructed a phylogenetic tree of a subset of full-length DPP4 protein sequences (Fig. 7). In the tree, shaded colors indicate the general organism group to which each species belongs as follows: blue, reptiles and amphibians; green, avian species; orange, other mammals; red, Chiroptera (bats); purple, ungulates; gray, rodents; pink, primates (Fig. 7). The DPP4 gene tree is slightly discordant with the species tree, notably with respect to the horse and African savanna elephant DPP4 sequences not clustering with those of other ungulate (purple) orthologs. Glycosylation sites that are upstream of (column 1), at the same site as (column 2), or downstream of (column 3) the glycosylation site that is present in mDPP4 (residues 328 to 330) are plotted adjacent to the phylogenetic tree. Permissivity data are indicated in the far right column, with green squares indicating permissive species and red squares indicating nonpermissive species, based on either in vitro or in vivo data (10–17).
FIG 7.

DPP4 protein phylogenetic tree based on amino acid sequences. Shaded colors indicate the group each species falls in. Blue, reptiles and amphibians; green, avian species; orange, other mammals; red, Chiroptera (bats); purple, ungulates; gray, rodents; pink: primates. Colored circles to the right of the species names indicate whether the sequence has a glycosylation site upstream of (first column), at the same site as (second column), or downstream of (third column) the NXT glycosylation site in mDPP4 (residues 332 to 334). Numbers inside the circle designate how many amino acids upstream (or downstream) the N of the NXT or NXS glycosylation site is. For the second column, a circle indicates that there was a glycosylation site aligning to the site present in mDPP4. Squares in the rightmost column indicate permissive (green) or nonpermissive (red) species, as determined from either in vivo or in vitro studies. Numbers indicate bootstrap support values >50.
The small number of known permissive and nonpermissive species makes it difficult to map potential shifts in permissivity on the phylogenetic tree. Because permissive and nonpermissive species do not cluster together on the DPP4 tree (Fig. 7), it is more likely that local differences between the proteins are more important than global similarity. Despite our inability to identify a distinct pattern, a few interesting observations do emerge. First, all nonhuman primates lack glycosylation sites near residue 330 (equivalent to residue 336 in humans) (Fig. 7). Current data would postulate that all nonhuman primates are susceptible to MERS-CoV infection, but testing this hypothesis in vitro would help reveal whether any of these orthologs are nonpermissive. If so, a mechanism other than glycosylation at our location of interest would be responsible for blocking MERS-CoV infection. Second, whereas other permissive DPP4 orthologs (horse DPP4 and common pipistrelle DPP4) have glycosylation sites in this region, of note is the lack of a glycosylation site in the nonpermissive domestic pig, sheep, and cattle DPP4 molecules (Fig. 7). This suggests that glycosylation in this region is not the primary explanation for the lack of support of MERS-CoV infection in these species. One example could be the incompatibility of the host cell proteases which are required for proper cleavage of the MERS-CoV spike prior to cell entry. Previous work has shown that spike cleavage status determines cell tropism (45) and that adaptation to cellular proteases may have played an important role in the cross-species transmission of MERS-CoV from bats to humans (35). Follow-up studies could investigate the mechanism of nonpermissivity in these species specifically and whether or not it lies at the level of the receptor. Third, glycosylation sites in the designated region are prevalent in rodents, other mammals, and avian species (Fig. 7). For example, eight other species have putative glycosylation sites aligning to the glycosylation site identified in mDPP4. The majority of these are present in the rodent (gray) group; however, two are present within the Chiroptera group (black flying fox and large flying fox) (Fig. 7). However, the diversity of glycosylation profiles suggests a lack of the strong conservation of the site seen in mDPP4. Further research to determine whether these other orthologs are permissive can help elucidate whether this region plays a broader role in the MERS-CoV infection susceptibility phenotype. In general, while some trends emerge from the glycosylation profiles across many species (e.g., upstream glycosylation sites in the “other mammals” [orange] group), more data on species permissivity will help determine whether DPP4 phylogenetic relationships can help inform receptor binding dynamics.
Here, we demonstrated that glycosylation is an important barrier to MERS-CoV infection, yet other species-specific determinants are also responsible. Removing the glycosylation sites of fDPP4, haDPP4, and gpDPP4 orthologs did not result in an increase in infection. Rather, a set of 19 and 4 amino acid substitutions in fDPP4 and haDPP4, respectively, were required before a significant increase in infection could be detected. Future work can focus on revealing the importance of these additional substitutions in mediating MERS-CoV infection. By further elucidating the mechanisms by which DPP4 orthologs block MERS-CoV infection, we can better understand the constraints on coronavirus host range expansion and predict coronavirus emergence into new species in the future. Furthermore, this work can inform the development of better animal models for studying MERS-CoV pathogenesis. Our previous work on mDPP4 helped generate a successful transgenic mouse using the clustered regularly interspaced short palindromic repeat (CRISPR)-Cas9 gene editing technique (24). The success of this model emphasizes the importance of understanding receptor-virus interactions and is a potent example of the direct application of these studies.
MATERIALS AND METHODS
Viruses and cells.
MERS-CoV was isolated from a molecular clone as described by Scobey et al. (46). Recombinant MERS-CoV tagged with tomato red fluorescent protein (rMERS-CoV-RFP) was engineered on the basis of the EMC2012 substrain and was shown to infect and replicate similarly to wild-type MERS-CoV (46). Recombinant viruses were passaged once (P1) on Vero cells to generate a working stock of rMERS-CoV-RFP. All recombinant viruses were produced and studied under biosafety level 3 (BSL3) conditions, with staff members wearing Tyvek suits; apron and booties; powered, air-purifying respirators (PAPR); and double gloves as described in the laboratory safety plan.
Measures of rMERS-CoV-RFP infectivity and of receptor protein expression levels, immunofluorescence assays (IFA), and flow cytometry were performed in human embryonic kidney (HEK) 293T cells transfected with cDNAs encoding wild-type or mutant DPP4 orthologs. Cells were seeded at 1 × 106 cells per well in 6-well plates pretreated with 1 ml of 10 μg/ml poly-l-lysine (Sigma)–1× Dulbecco's phosphate-buffered saline (DPBS) (Gibco). All cells were grown at 37°C with 5% CO2.
DPP4 ortholog constructs.
DPP4 orthologs were ordered on plasmids (cDPP4, hDPP4, and mDPP4) or constructed using Gibson assembly (bDPP4, fDPP4, haDPP4, and gpDPP4) (47). The DPP4 orthologs were transferred into 945ΔRRE expression vector, a lentiviral vector derived from pTK945, using the NotI and SpeI restriction sites. DPP4 mutants were generated by overlap PCR mutagenesis and verified by Sanger sequencing. All plasmids were stored at −20°C. The accession numbers for the DPP4 sequences used in this study are as follows: bDPP4, KC249974.1; cDPP4, XM_006176808.2; fDPP4, DQ266376.1; gpDPP4, XM_013142395.1; haDPP4, NM_001310571.1; hDPP4, NM_001935.3; mDPP4, NM_010074.3.
DPP4 ortholog transfections.
DPP4 orthologs were transfected into HEK 293T cells, as these cells express little if any endogenous hDPP4 (13). Transfections followed a 2× BES [N,N-Bis(2-hydroxyethyl)-2-aminoethanesulfonic acid, N,N-Bis(2-hydroxyethyl)taurine] protocol. For each DPP4 ortholog, 15 μg of the construct was mixed with 15 μg of pcDNA3.1, 125 μl of 1 M CaCl2, and distilled water (dH2O) to reach a final volume of 500 μl. Then, 500 μl of 2× BES was added to each sample in a dropwise manner, with the reaction mixture subjected to vortex mixing between drops. Samples were incubated at room temperature for 45 min. During this incubation, medium was removed from 6-well plates seeded with 1 × 106 HEK 293T cells/well and 1 ml of fetal bovine serum (FBS)-free medium was added to each well. After the 45-min incubation, 200 μl of each transfection reaction mixture was added (dropwise and in concentric circles) to individual wells. Plates were incubated at 37°C for 2 h before 150 μl of FBS was added to each well, and the cultures were incubated overnight at 37°C. To measure transfection efficiencies, Venus protein-tagged 945 plasmid was transfected in parallel with each experiment (with efficiencies typically between ∼80% and 100%).
Infection of DPP4-transfected cells by MERS-CoV.
At ∼20 h posttransfection, the medium was removed and each well was infected with 200 μl of rMERS-CoV-RFP at a multiplicity of infection (MOI) of 5 for 45 min at 37°C. Next, 2 ml of 3% FBS medium was added to each well and the plates were incubated at 37°C for 24 h. Cells were visualized at 24 h postinfection in the BSL3 containment facility using an Olympus IX51 microscope at a magnification of ×10 and a wavelength of 541 nm. Each well was imaged using a Hamamatsu Orca-Flash4.0 LT camera. Successful virus infection was indicated by the presence of red fluorescence.
Fluorescent cell count assay.
For quantification of levels of MERS-CoV infection, transfected cells were infected as described above, except that an MOI of 0.1 was used. After the 45-min incubation period, virus samples were removed and the cells were washed twice with 1 ml 1× DPBS. Next, 2 ml of 3% FBS medium was added to each well and the infection reaction mixtures were incubated at 37°C for 72 h. At 72 h postinfection, the number of red fluorescent cells was determined. Briefly, three randomly selected microscope fields were counted for each well. The average number of red cells was determined from these three fields and multiplied by a factor of 537 to approximate the number of red cells in the entire well. Infection in the presence of each DPP4 ortholog was measured in triplicate as described; thus, 9 fields were measured to determine the statistical significance of the results of comparisons between two DPP4 treatments using Student's t test.
Protein analysis and Western blotting.
Cells were transfected with DPP4 constructs as described above. At 48 h posttransfection, medium was removed from each well and cells were washed once with 2 ml 1× DPBS. Next, 200 μl of ice-cold AV lysis buffer (20 mM Tris-HCl, 150 mM NaCl, 0.5% deoxycholine, 1% IGEPAL, 0.1% SDS) was added to each well. Cells were incubated at room temperature for 10 min with the lysis buffer. Lysates were then transferred to 1.5-ml microcentrifuge tubes and centrifuged at maximum speed for 2 min. Supernatants were transferred to new tubes, and 200 μl of a 10 mM EDTA–0.9% SDS stock was added to each tube. Lysates were stored at −80°C.
The protein concentration was measured for each sample using a Bradford assay. Briefly, an aliquot of each sample was diluted 1:5 and 10 μl was added to a 96-well plate in triplicate. Bovine serum albumin (BSA) was used as a standard, with 10 μl of a dilution series added to the 96-well plate in triplicate. A 200-μl volume of Coomassie Plus protein assay reagent (ThermoScientific) was added to each well, and the plate was incubated at room temperature in the dark for 10 min. Absorbance was measured at a wavelength of 595 nm, and the protein concentration of each sample was determined by comparison to the BSA dilution standard curve.
Proteins were separated using either a 6% (for DPP4) or an 8% (for actin) polyacrylamide gel. For each sample, 15 μg of the protein lysate was mixed with 6× loading dye and heated to 95°C for 10 min. Samples were loaded into the gel and separated by electrophoresis in 1× running buffer (3.03 g Tris-HCl, 14.4 g glycine, 1 g SDS, 1 liter dH2O) at 55 mA for ∼2 h. Proteins were transferred onto an Immun-Blot polyvinylidene difluoride (PVDF) membrane (Bio-Rad) at 15 V for 45 min using 1× dry transfer buffer (5.82 g Tris base, 2.92 g glycine, 100 ml methanol, 1 liter dH2O). The membranes were blocked in 1× phosphate-buffered saline–Tween 20 (PBST)–5% milk at room temperature for 1 h with shaking. Membranes were then treated with primary antibody using goat anti-DPP4 antibody (R&D Systems) (1:1,000)–1× PBST or goat anti-actin antibody (Santa Cruz Biotechnology) (1:1,000)–1× PBST and incubated at 4°C overnight with shaking. Membranes were washed three times with 1× PBST (10 min per wash) before treatment with secondary antibody. Both the DPP4 and the actin blots used rabbit anti-goat horseradish peroxidase (HRP) (1:10,000)–1× PBST–5% milk and were incubated at room temperature for 1 h with shaking. Membranes were washed three times with 1× PBST (10 min per wash) and developed using Amersham ECL Western blotting detection reagents (GE Healthcare). Membranes were incubated at room temperature for 10 min before imaging was performed using Amersham Hyperfilm ECL (GE Healthcare).
Immunofluorescence assay (IFA).
For IFA analyses, transfections were performed in 50-mm-diameter glass-bottom dishes seeded with 2 × 106 HEK 293T cells. At 24 h posttransfection, cells were washed once with 1× DPBS and then fixed with 2 ml 2% paraformaldehyde (PFA)–1× DPBS for 30 min at room temperature. Cells were washed once with 2 ml of 100 mM glycine–1× DPBS and incubated in 2 ml of 100 mM glycine–1× DPBS for 15 min at room temperature. Cells were washed twice with 1× DPBS before blocking was performed. A 200-μl volume of 3% BSA–10% normal donkey sera–IFA wash (0.05% Tween 20–1× DPBS) was added to each plate, and the reaction mixture was incubated at room temperature for 1 h with shaking. Next, cells were incubated with goat anti-DPP4 polyclonal antibody (R&D Systems) (1:50)–3% BSA–IFA wash at room temperature for 1 h with shaking. Cells were washed three times with IFA wash and then incubated with secondary donkey anti-goat Alexa Fluor 488 (Life Technologies) (1:500) for 1 h at room temperature in the dark with shaking. Cells were again washed three times with IFA wash. Three drops of ProLong Gold antifade reagent (Invitrogen) were added to the cells on each plate before imaging was performed at ×20 magnification. Green fluorescence indicated DPP4 staining, while blue fluorescence indicated DAPI (4′,6-diamidino-2-phenylindole) staining for nuclei.
Flow cytometry analysis.
Transfections were performed as detailed above. At ∼20 h posttransfection, medium was removed and cells were washed with 1× DPBS. Cells were removed from plates using nonenzymatic cell dissociation buffer (Gibco). Cells were then resuspended in complete media and centrifuged (all spins were performed at 500 × g for 5 min). Supernatants were removed, and cells were washed three times with 1× DPBS. Cells were counted and resuspended at a concentration of 5 × 106 cells/ml. A 200-μl volume of cell suspension was added to each well of a 96-well plate (Corning) in duplicate, plates were spun to pellet cells, and the supernatant was discarded. Cells were suspended in 100 μl flow cytometry staining buffer (eBiosciences)–8 μg/ml goat anti-DPP4 polyclonal antibody (R&D Systems) and incubated at 4°C for 1 h. Cells were washed twice with staining buffer and then incubated with 1 μl/well of fluorescein isothiocyanate (FITC)-conjugated donkey anti-goat secondary antibody (Life Technologies)–100 μl staining buffer for 1 h at 4°C. Cells were washed twice with staining buffer and resuspended in 150 μl staining buffer. At least 10,000 events per well were acquired on a Guava easyCyte HT flow cytometer (Millipore) for each read. The background signal of secondary antibody only (no primary antibody) was used to subtract the background. Data were analyzed using FlowJo software version 10.3 (Tree Star).
Phylogenetic analysis.
Amino acid sequences were retrieved from GenBank and aligned using MAFFT (48). The phylogenetic tree was generated using maximum likelihood and 100 bootstrap replicates with the PhyML package. Smart model selection (SMS) was implemented to determine the best model parameters (49). The tree was visualized using EvolView (50). Only bootstrap values of ≥50 are displayed.
Statistics.
All quantitative data are presented as means ± 1 standard deviation. All statistical analyses were performed using R programming language version 3.1.2.
ACKNOWLEDGMENTS
We thank members of the Heise and Baric laboratories for reviews of the manuscript. We thank Boyd Yount for his continued BSL3 advice and support and Todd Vision for his advice on phylogenetic analyses.
K.M.P. was supported by an NSF GRFP. This work was supported by grants NIH HHSN272201000019I-HHSN27200003-Task A57, U19 AI109761, AI098728, and AI110700.
REFERENCES
- 1.Pfefferle S, Oppong S, Drexler JF, Gloza-Rausch F, Ipsen A, Seebens A, Muller MA, Annan A, Vallo P, Adu-Sarkodie Y, Kruppa TF, Drosten C. 2009. Distant relatives of severe acute respiratory syndrome coronavirus and close relatives of human coronavirus 229E in bats, Ghana. Emerg Infect Dis 15:1377–1384. doi: 10.3201/eid1509.090224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huynh J, Li S, Yount B, Smith A, Sturges L, Olsen JC, Nagel J, Johnson JB, Agnihothram SS, Gates JE, Frieman MB, Baric RS, Donaldson EF. 2012. Evidence supporting a zoonotic origin of human coronavirus strain NL63. J Virol 86:12816–12825. doi: 10.1128/JVI.00906-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cherry JD. 2004. The chronology of the 2002–2003 SARS mini pandemic. Paediatr Respir Rev 5:262–269. doi: 10.1016/j.prrv.2004.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Menachery VD, Yount BL, Debbink K, Agnihothram SS, Gralinski LE, Plante JA, Graham RL, Scobey T, Ge XY, Donaldson EF, Randell SH, Lanzavecchia A, Marasco WA, Shi ZL, Baric RS. 2015. A SARS-like cluster of circulating bat coronaviruses shows potential for human emergence. Nat Med 21:1508–1513. doi: 10.1038/nm.3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Menachery VD, Yount BL, Sims AC, Debbink K, Agnihothram SS, Gralinksi LE, Graham RL, Scobey T, Plante JA, Royal SR, Swanstrom J, Sheahan TP, Pickles RJ, Corti D, Randell SH, Lanzavecchia A, Marasco WA, Baric RS. 2016. SARS-like WIV1-CoV poised for human emergence. Proc Natl Acad Sci U S A 113:3048–3053. doi: 10.1073/pnas.1517719113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.WHO (World Health Organization). 2017. Middle East respiratory syndrome coronavirus (MERS-CoV). WHO, Geneva, Switzerland: http://www.who.int/emergencies/mers-cov/en/ Accessed 27 Mar 2017. [Google Scholar]
- 7.Reusken CBEM, Haagmans BL, Müller MA, Gutierrez C, Godeke GJ, Meyer B, Muth D, Raj VS, Smits-De Vries L, Corman VM, Drexler JF, Smits SL, El Tahir YE, De Sousa R, van Beek J, Nowotny N, van Maanen K, Hidalgo-Hermoso E, Bosch BJ, Rottier P, Osterhaus A, Gortázar-Schmidt C, Drosten C, Koopmans MP. 2013. Middle East respiratory syndrome coronavirus neutralising serum antibodies in dromedary camels: a comparative serological study. Lancet Infect Dis 13:859–866. doi: 10.1016/S1473-3099(13)70164-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gossner C, Danielson N, Gervelmeyer A, Berthe F, Faye B, Kaasik Aaslav K, Adlhoch C, Zeller H, Penttinen P, Coulombier D. 2016. Human-dromedary camel interactions and the risk of acquiring zoonotic Middle East respiratory syndrome coronavirus infection. Zoonoses Public Health 63:1–9. doi: 10.1111/zph.12171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raj VS, Mou H, Smits SL, Dekkers DH, Muller MA, Annan A, Vallo P, Adu-Sarkodie Y, Kruppa TF, Drosten C. 2013. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature 495:251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Wit E, Prescott J, Baseler L, Bushmaker T, Thomas T, Lackemeyer MG, Martellaro C, Milne-Price S, Haddock E, Haagmans BL, Feldmann H, Munster VJ. 2013. The Middle East respiratory syndrome coronavirus (MERS-CoV) does not replicate in Syrian hamsters. PLoS One 8:e69127. doi: 10.1371/journal.pone.0069127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barlan A, Zhao J, Sarkar MK, Li K, McCray PB Jr, Perlman S, Gallagher T. 2014. Receptor variation and susceptibility to Middle East respiratory syndrome coronavirus infection. J Virol 88:4953–4961. doi: 10.1128/JVI.00161-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eckerle I, Corman VM, Muller MA, Lenk M, Ulrich RG, Drosten C. 2014. Replicative capacity of MERS coronavirus in livestock cell lines. Emerg Infect Dis 20:276–279. doi: 10.3201/eid2002.131182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cockrell AS, Peck KM, Yount BL, Agnihothram SS, Scobey T, Curnes NR, Baric RS, Heise MT. 2014. Mouse dipeptidyl peptidase 4 is not a functional receptor for Middle East respiratory syndrome coronavirus infection. J Virol 88:5195–5199. doi: 10.1128/JVI.03764-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Coleman CM, Matthews KL, Goicochea L, Frieman MB. 2014. Wild-type and innate immune-deficient mice are not susceptible to the Middle East respiratory syndrome coronavirus. J Gen Virol 95:408–412. doi: 10.1099/vir.0.060640-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Raj VS, Smits SL, Provacia LB, van den Brand JM, Wiersma L, Ouwendijk WJ, Bestebroer TM, Spronken MI, van Amerongen G, Rottier PJ, Fouchier RA, Bosch BJ, Osterhaus AD, Haagmans BL. 2014. Adenosine deaminase acts as a natural antagonist for dipeptidyl peptidase 4-mediated entry of the Middle East respiratory syndrome coronavirus. J Virol 88:1834–1838. doi: 10.1128/JVI.02935-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Doremalen N, Miazgowicz KL, Milne-Price S, Bushmaker T, Robertson S, Scott D, Kinne J, McLellan JS, Zhu J, Munster VJ. 2014. Host species restriction of Middle East respiratory syndrome coronavirus through its receptor, dipeptidyl peptidase 4. J Virol 88:9220–9232. doi: 10.1128/JVI.00676-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Doremalen N, Miazgowicz KL, Munster VJ. 2016. Mapping the specific amino acid residues that make hamster DPP4 functional as a receptor for Middle East respiratory syndrome coronavirus. J Virol 90:5499–5502. doi: 10.1128/JVI.03267-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao JC, Li K, Wohlford-Lenane C, Agnihothram SS, Fett C, Zhao J, Gale MJ Jr, Baric RS, Enjuanes L, Gallagher T, McCray PB Jr, Perlman S. 2014. Rapid generation of a mouse model for Middle East respiratory syndrome. Proc Natl Acad Sci U S A 111:4970–4975. doi: 10.1073/pnas.1323279111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peck KM, Cockrell AS, Yount BL, Scobey T, Baric RS, Heise MT. 2015. Glycosylation of mouse DPP4 plays a role in inhibiting Middle East respiratory syndrome coronavirus infection. J Virol 89:4696–4699. doi: 10.1128/JVI.03445-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Agrawal AS, Garron T, Tao X, Peng BH, Wakamiya M, Chan TS, Couch RB, Tseng CT. 2015. Generation of transgenic mouse model of Middle East respiratory syndrome-coronavirus infection and disease. J Virol 89:3659–3670. doi: 10.1128/JVI.03427-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pascal KE, Coleman CM, Mujica AO, Kamat V, Badithe A, Fairhurst J, Hunt C, Strein J, Berrebi A, Sisk JM, Matthews KL, Babb R, Chen G, Lai KM, Huang TT, Olson W, Yancopoulos GD, Stahl N, Frieman MB, Kyratsous CA. 2015. Pre- and postexposure efficacy of fully human antibodies against Spike protein in a novel humanized mouse model of MERS-CoV infection. Proc Natl Acad Sci U S A 112:8738–8743. doi: 10.1073/pnas.1510830112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li K, Wohlford-Lenane C, Perlman S, Zhao J, Jewell AK, Reznikov LR, Gibson-Corley KN, Meyerholz DK, McCray PB Jr. 2016. Middle East respiratory syndrome coronavirus causes multiple organ damage and lethal disease in mice transgenic for human dipeptidyl peptidase 4. J Infect Dis 213:712–722. doi: 10.1093/infdis/jiv499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Coleman CM, Sisk JM, Halasz G, Zhong J, Beck SE, Matthews KL, Venkataraman T, Rajagopalan S, Kyratsous CA, Frieman MB. 2017. CD8+ T cells and macrophages regulate pathogenesis in a mouse model of Middle East respiratory syndrome. J Virol 91:e01825-16. doi: 10.1128/JVI.01825-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cockrell AS, Yount BL, Scobey T, Jensen K, Douglas M, Beall A, Tang XC, Marasco WA, Heise MT, Baric RS. 2016. A mouse model for MERS coronavirus-induced acute respiratory distress syndrome. Nat Microbiol 2:16226. doi: 10.1038/nmicrobiol.2016.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Wit E, Rasmussen AL, Falzarano D, Bushmaker T, Feldmann F, Brining DL, Fischer ER, Martellaro C, Okumura A, Chang J, Scott D, Benecke AG, Katze MG, Feldmann H, Munster VJ. 2013. Middle East respiratory syndrome coronavirus (MERS-CoV) causes transient lower respiratory tract infection in rhesus macaques. Proc Natl Acad Sci U S A 110:16598–16603. doi: 10.1073/pnas.1310744110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Falzarano D, de Wit E, Rasmussen AL, Feldmann F, Okumara Scott ADP, Brining D, Bushmaker T, Martellaro C, Baseler L, Benecke AG, Katze MG, Munster VJ, Feldmann H. 2013. Treatment with interferon-a2b and ribavirin improves outcome in MERS-CoV-infected rhesus macaques. Nat Med 19:1313–1317. doi: 10.1038/nm.3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Falzarano D, de Wit E, Feldmann F, Rasmussen AL, Okumara A, Peng X, Thomas MJ, van Doremalen N, Haddock E, Nagy L, LaCasse R, Liu T, Zhu J, McLellan JS, Scott DP, Katze MG, Feldmann H, Munster VJ. 2014. Infection with MERS-CoV causes lethal pneumonia in the common marmoset. PLoS Pathog 10:e1004250. doi: 10.1371/journal.ppat.1004250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chan JFW, Yao Y, Yeung ML, Deng W, Bao L, Jia L, Li F, Xiao C, Gao H, Yu P, Cai JP, Chu H, Zhou J, Chen H, Qin C, Yuen KY. 2015. Treatment with lopinavir/ritonavir or interferon-B1b improves outcome of MERS-CoV infection in a nonhuman primate model of common marmoset. J Infect Dis 212:1904–1913. doi: 10.1093/infdis/jiv392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang N, Shi X, Jiang L, Zhang S, Wang D, Tong P, Guo D, Fu L, Cui Y, Liu X, Arledge KC, Chen YH, Zhang L, Wang X. 2013. Structure of MERS-CoV spike receptor-binding domain complexed with human receptor DPP4. Cell Res 23:986–993. doi: 10.1038/cr.2013.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Y. 2008. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cui J, Eden JS, Holmes EC, Wang LF. 2013. Adaptive evolution of bat dipeptidyl peptidase 4 (dpp4): implications for the origin and emergence of Middle East respiratory syndrome coronavirus. Virol J 10:304. doi: 10.1186/1743-422X-10-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peck KM, Burch CL, Heise MT, Baric RS. 2015b. Coronavirus host range expansion and Middle East respiratory syndrome coronavirus emergence: biochemical mechanisms and evolutionary perspectives. Annu Rev Virol 2:95–117. [DOI] [PubMed] [Google Scholar]
- 33.Gupta R, Jung E, Brunak S. 2004. NetNGlyc 1.0 server. Center for biological sequence analysis, Technical University of Denmark. http://www.cbs.dtu.dk/services/NetNGlyc.
- 34.Song W, Wang Y, Wang N, Wang D, Guo J, Fu L, Shi X. 2014. Identification of residues on human receptor DPP4 critical for MERS-CoV binding and entry. Virology 471–473:49–53. doi: 10.1016/j.virol.2014.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang CC, Chen JR, Tseng YC, Hsu CH, Hung YF, Chen SW, Chen CM, Khoo KH, Cheng TJ, Cheng YS, Jan JT, Wu CY, Ma C, Wong CH. 2009. Glycans on influenza hemagglutinin affect receptor binding and immune response. Proc Natl Acad Sci U S A 106:18137–18142. doi: 10.1073/pnas.0909696106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang Y, Du L, Liu C, Wang L, Ma C, Tang J, Baric RS, Jiang S, Li F. 2014. Receptor usage and cell entry of bat coronavirus HKU4 provide insight into bat-to-human transmission of MERS-Coronavirus. Proc Natl Acad Sci U S A 111:12516–12521. doi: 10.1073/pnas.1405889111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jayaraman A, Koh X, Li J, Raman R, Viswanathan K, Shriver Z, Sasisekharan R. 2012. Glycosylation at Asn91 of H1N1 haemagglutinin affects binding to glycan receptors. Biochem J 444:429–435. doi: 10.1042/BJ20112101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kaelber JT, Demogines A, Harbison CE, Allison AB, Goodman LB, Ortega AN, Sawyer SL, Parrish CR. 2012. Evolutionary reconstructions of the trasnferrin receptor of Caniforms supports canine parvovirus being a re-emerged and not a novel pathogen in dogs. PLoS Pathog 8:e1002666. doi: 10.1371/journal.ppat.1002666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goodman LB, Lyi SM, Johnson NC, Cifuente JO, Hafenstein SL, Parrish CR. 2010. Binding site on the transferrin receptor for the parvovirus capsid and effects of altered affinity on cell uptake and infection. J Virol 84:4969–4978. doi: 10.1128/JVI.02623-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Allison AB, Organtini LJ, Zhang S, Hafenstein SL, Holmes EC, Parrish CR. 2015. Single mutations in the VP2 300 loop region of the three-fold spike of the carnivore parvovirus capsid can determine host range. J Virol 90:753–767. doi: 10.1128/JVI.02636-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wentworth DE, Holmes KV. 2001. Molecular determinants of species specificity in the coronavirus receptor aminopeptidase N (CD13): influence of N-linked glycosylation. J Virol 75:9741–9752. doi: 10.1128/JVI.75.20.9741-9752.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tusell SM, Schittone SA, Holmes KV. 2007. Mutational analysis of aminopeptidase N, a receptor for several group 1 coronaviruses, identifies key determinants of viral host range. J Virol 81:1261–1273. doi: 10.1128/JVI.01510-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li W, Zhang C, Sui J, Kuhn JH, Moore MJ, Luo S, Wong S, Huang I, Xu K, Vasilieva N, Murakami A, He Y, Marasco WA, Guan Y, Choe H, Farzan M. 2005. Receptor and viral determinants of SARS-coronavirus adaptation to human ACE2. EMBO J 24:1634–1643. doi: 10.1038/sj.emboj.7600640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lu G, Hu Y, Wang Q, Qi J, Gao F, Li Y, Zhang Y, Zhang W, Yuan Y, Bao J, Zhang B, Shi Y, Yan J, Gao GF. 2013. Molecular basis of binding between novel human coronavirus MERS-CoV and its receptor CD26. Nature 500:227–231. doi: 10.1038/nature12328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Park JE, Li K, Barlan A, Fehr AR, Perlman S, McCray PB Jr, Gallagher T. 2016. Proteolytic processing of Middle East respiratory syndrome coronavirus spikes expands virus tropism. Proc Natl Acad Sci U S A 113:12262–12267. doi: 10.1073/pnas.1608147113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Scobey T, Yount BL, Sims AC, Donaldson EF, Agnihothram SS, Menachery VD, Graham RL, Swanstrom J, Bove PF, Kim JD, Grego S, Randell SH, Baric RS. 2013. Reverse genetics with a full-length infectious cDNA of the Middle East respiratory syndrome coronavirus. Proc Natl Acad Sci U S A 110:16157–16162. doi: 10.1073/pnas.1311542110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA III, Smith HO. 2009. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods 6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 48.Katoh K, Misawa K, Kuma K, Miyata T. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 50.Zhang H, Gao S, Lercher MJ, Hu S, Chen WH. 2012. EvolView, an online tool for visualizing, annotating and managing phylogenetic trees. Nucleic Acids Res 40:W569–W72. doi: 10.1093/nar/gks576. [DOI] [PMC free article] [PubMed] [Google Scholar]