ABSTRACT
Human respiratory syncytial virus (RSV) is the leading etiologic agent of lower respiratory tract infections in children, but no licensed vaccine exists. Previously, we developed two parainfluenza virus 5 (PIV5)-based RSV vaccine candidates that protect mice against RSV challenge. PIV5 was engineered to express either the RSV fusion protein (F) or the RSV major attachment glycoprotein (G) between the hemagglutinin-neuraminidase (HN) and RNA-dependent RNA polymerase (L) genes of the PIV5 genome [PIV5-RSV-F (HN-L) and PIV5-RSV-G (HN-L), respectively]. To investigate the stability of the vaccine candidates in vitro, they were passaged in Vero cells at high and low multiplicities of infection (MOIs) for 11 generations and the genome sequences, growth kinetics, and protein expression of the resulting viruses were compared with those of the parent viruses. Sporadic mutations were detected in the consensus sequences of the viruses after high-MOI passages, and mutation rates increased under low-MOI-passage conditions. None of the mutations abolished antigen expression. Increased numbers of mutations correlated with increased growth rates in vitro, indicating that the viruses evolved through the course of serial passages. We also examined the in vivo stability of the vaccine candidates after a single passage in African green monkeys. No mutations were detected in the consensus sequences of viruses collected from the bronchoalveolar lavage (BAL) fluid of the animals. In vivo, mutations in RSV G and PIV5 L were found in individual isolates of PIV5-RSV-G (HN-L), but plaque isolates of PIV5-RSV-F (HN-L) had no mutations. To improve upon the PIV5-RSV-F (HN-L) candidate, additional vaccine candidates were generated in which the gene for RSV F was inserted into earlier positions in the PIV5 genome. These insertions did not negatively impact the sequence stability of the vaccine candidates. The results suggest that the RSV F and G gene insertions are stable in the PIV5 genome. However, the function of the foreign gene insertion may need to be considered when designing PIV5-based vaccines.
IMPORTANCE The genetic stability of live viral vaccines is important for safety and efficacy. PIV5 is a promising live viral vector and has been used to develop vaccines. In this work, we examined the genetic stability of a PIV5-based RSV vaccine in vitro and in vivo. We found that insertions of foreign genes, such as the RSV F and G genes, were stably maintained in the PIV5 genome and there was no mutation that abolished the expression of RSV F or G. Interestingly, the function of the inserted gene may have an impact on PIV5 genome stability.
KEYWORDS: PIV5, vaccine, genome stability, respiratory syncytial virus
INTRODUCTION
Human respiratory syncytial virus (RSV) is a leading cause of viral bronchiolitis and hospitalizations in infants and young children. Globally, an estimated 33.8 million cases of RSV-associated acute lower respiratory infection occur annually in children under the age of 5 years, resulting in approximately 3 million hospitalizations (1). In the United States, greater than 60% of children are infected with RSV by the age of 1 year, and nearly all children are infected by the age of 2 years (2). Furthermore, severe infection during infancy has been implicated in the development of childhood asthma and wheezing (3, 4). Since natural RSV infection does not confer long-lasting immunity, reinfection, although usually mild in healthy individuals, can occur throughout life. Therefore, RSV infection can still cause serious complications in immunocompromised and elderly individuals (5). For these reasons, vaccines are needed for both young and aged populations.
While many attempts to develop an RSV vaccine have been made for over 40 years, no licensed vaccine currently exists (6). The most advanced pediatric RSV vaccine candidates to date have been live attenuated or vectored vaccines. One live attenuated candidate, rA2 cp248/404/1030ΔSH, was immunogenic and well tolerated in RSV-naive, 1- to 2-month-old infants. However, 34% of the virus isolates recovered from postvaccination nasal washes demonstrated a partial loss of the temperature-sensitive (ts) phenotype. Specifically, the loss of the ts248 or ts1030 mutation was observed, with 83% of the reversions occurring at ts1030 (7). Further examination in in vitro studies showed that the two mutations occurred as a result of selective pressure at 35°C, similar to the maximum temperature of the respiratory tract. MEDI-534, a live-vectored RSV vaccine candidate, also encountered obstacles related to genome stability. MEDI-534 is a chimeric, recombinant vaccine consisting of a bovine parainfluenza virus 3 (bPIV3) backbone engineered to express the human parainfluenza virus 3 (hPIV3) fusion protein, hPIV3 hemagglutinin-neuraminidase (HN), and the RSV fusion protein (F). In a phase 1 study conducted in seronegative children ages 6 to 24 months, all subjects seroconverted in response to hPIV3, but only 50% seroresponded to RSV (8). Sequence analysis of postvaccination nasal wash samples showed mutations in the poly(A) sequence downstream of the bPIV3 nucleocapsid (N) gene as well as in the F open reading frame. These variant subpopulations existed at low levels in the administered vaccine, and the mutations were implicated in the downregulation of F expression and the subsequent reduction in the antibody response against F (9). Therefore, genome stability is important for live attenuated and live viral vector-based vaccine candidates.
Parainfluenza virus 5 (PIV5) is a nonsegmented, negative-sense RNA virus of the genus Rubulavirus in the Paramyxoviridae family (10). Our previous work has shown that PIV5 is safe and efficacious as a vaccine vector and is able to overcome host preexisting immunity (11). PIV5-based vaccine candidates against influenza virus and rabies virus have conferred protection against infection in various animal models (12–18). Furthermore, in the canine model of H3N2 influenza virus infection, PIV5 expressing H3 was able to generate protective hemagglutination inhibition (HAI) titers in PIV5-immunized animals (11). Recently, we developed PIV5-based RSV vaccine candidates expressing either RSV F or the major attachment glycoprotein (G) between the HN and RNA-dependent RNA polymerase (L) genes of PIV5. We showed that the vaccine candidates conferred potent immunity against RSV challenge in mice. Both candidates induced RSV antigen-specific antibodies and reduced RSV lung titers with no evidence of enhanced disease (19).
The genome structure of PIV5 is stable, unlike the genomes of positive-strand RNA viruses (20). Recombinant PIV5 expressing green fluorescent protein (GFP) maintained reporter gene expression for more than 10 generations (the duration of the experiment) (21). Sequence variation is also low among PIV5 isolates, and the PIV5 genome remains stable through high-multiplicity-of-infection (MOI) passages in tissue culture cells (22). In this work, we determined the stability of our vaccine candidates after multiple passages in cell culture and a single passage in African green monkeys.
RESULTS
Recombinant PIV5-based RSV vaccine viruses retained inserted genes through multiple passages in vitro.
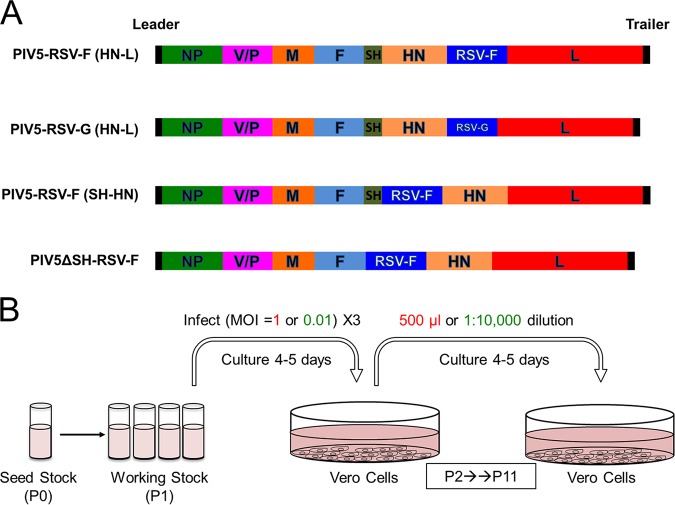
Two recombinant vaccine viruses were previously generated by inserting the coding sequence of RSV F or RSV G between the HN and L genes of PIV5 (Fig. 1A) (19). To determine the stability of the recombinant viruses through serial passages in cell culture, the vaccine viruses were passaged at a high MOI or a low MOI every 4 to 5 days in Vero cells, which have been approved for use for vaccine production by WHO and FDA. A total of 11 passages were performed (Fig. 1B).
FIG 1.
In vitro passage of PIV5-based RSV vaccine constructs. (A) Schematic of PIV5-vectored RSV vaccine constructs. NP, nucleoprotein; V, V protein; P, phosphoprotein; M, matrix protein; F, fusion protein; SH, small hydrophobic protein; HN, hemagglutinin-neuraminidase protein; L, RNA-dependent RNA polymerase; RSV F, respiratory syncytial virus fusion protein; RSV G, respiratory syncytial virus G attachment protein. (B) Vero cells were infected with PIV5-RSV-F (HN-L), PIV5-RSV-G (HN-L), PIV5-RSV-F (SH-HN), or PIV5ΔSH-RSV-F at an MOI of 1 PFU per cell (high MOI) or 0.01 PFU per cell (low MOI). For high-MOI-passage conditions, 500 μl of infected cell culture supernatant was used to infect fresh Vero cells every 4 to 5 days, for a total of 11 passages. For low-MOI-passage conditions, the cell culture supernatant was diluted 1:10,000 and 2.5 ml was used for infection of fresh Vero cells.
Full-genome sequencing of PIV5-RSV-F (HN-L) at high-MOI passage 0 (P0) and passage 11 (P11) showed no differences between the consensus sequence of the initial stock virus and that of the viruses at P11 in three out of the four replicates. High-MOI replicate 4 had a thymine-to-guanine variant in the leader sequence at nucleotide position 26 (Table 1).
TABLE 1.
Comparison of P0 and P11 virus sequences (in vitro passage)
| Virus | Passage, MOIa | Replicate | Mutation | Mutation location | Presence of: |
|||
|---|---|---|---|---|---|---|---|---|
| Population consensus sequence | Plaque isolate | Full genome | Insertion only | |||||
| PIV5-RSV-F (HN-L) | P11, high MOI | 1 | None | • | • | |||
| 2 | None | • | • | |||||
| 3 | None | • | • | |||||
| 4 | ntb T26 → nt G | Leader | • | • | ||||
| P11, low MOI | 1 | A154V | V/P | • | • | |||
| 1 | T372S | HN | • | • | ||||
| 1 | V56M | RSV F | • | • | ||||
| 2 | N306K | P | • | • | ||||
| 3 | E303K | P | • | • | ||||
| 3 | P256H | HN | • | • | ||||
| P0 (3/23) | 1 | V56M | RSV F | • | • | |||
| 1 | T174A | RSV F | • | • | ||||
| 1 | F114S/Y117H | RSV F | • | • | ||||
| P11, high MOI (6/23) | 1 | Mixed N569L | RSV F | • | • | |||
| 1 | Mixed L95L (silent) | RSV F | • | • | ||||
| 1 | Mixed I76N | RSV F | • | • | ||||
| 1 | Mixed F572F (silent) | RSV F | • | • | ||||
| 1 | Mixed K461 stop/mixed F572F (silent) | RSV F | • | • | ||||
| PIV5-RSV-G (HN-L) | P11, high MOI | 1 | K78E (mixed) | V/P | • | • | ||
| 1 | nt 4292C → nt A | 3′ UTR of M | • | • | ||||
| 2 | K78E | V/P | • | • | ||||
| 2 | nt 4292C → nt A | 3′ UTR of M | • | • | ||||
| 3 | K78E | V/P | • | • | ||||
| 3 | nt 4292C → nt A | 3′ UTR of M | • | • | ||||
| 4 | K78E | V/P | • | • | ||||
| 4 | nt 4292C → nt A | 3′ UTR f M | • | • | ||||
| P11, low MOI | 1 | L50P | V/P | • | • | |||
| 1 | T63T (silent) | V/P | • | • | ||||
| 1 | I85T | V/P | • | • | ||||
| 1 | L103L (silent) | V/P | • | • | ||||
| 1 | Y127H | V/P | • | • | ||||
| 1 | F135P | V/P | • | • | ||||
| 1 | P152P (silent) | V/P | • | • | ||||
| 1 | Y175H | V | • | • | ||||
| 1 | S175S (silent) | P | • | • | ||||
| 2 | D315N | P | • | • | ||||
| 2 | T222I | M | • | • | ||||
| 3 | K78E | V/P | • | • | ||||
| 3 | S26T | M | • | • | ||||
| 3 | nt 4292C → nt A | 3′ UTR of M | • | • | ||||
| 3 | V1667A | L | • | • | ||||
| P0 (1/23) | 1 | I243I (silent) | RSV G | • | • | |||
| P11, high MOI (0/24) | 1 | None | • | • | ||||
| PIV5-RSV-F (SH-HN) | P11, high MOI | 1 | P158L | V/P | • | • | ||
| 2 | None | • | • | |||||
| 3 | None | • | • | |||||
| P11, low MOI | 1 | P152S | V/P | • | • | |||
| 1 | N1767D | L | • | • | ||||
| 2 | nt 136C → nt T | Leader | • | • | ||||
| 2 | V330F | P | • | • | ||||
| 2 | S316A | M | • | • | ||||
| 2 | T1017I | L | • | • | ||||
| 3 | I169T | M | • | • | ||||
| PIV5ΔSH-RSV-F | P11, high MOI | 1 | None | • | • | |||
| 2 | Q258K | PIV5 F | • | • | ||||
| 3 | nt 26T → nt G | Leader | • | • | ||||
| 3 | N1763D | L | • | • | ||||
| P11, low MOI | 1 | E303K | P | • | • | |||
| 2 | R80Q | V/P | • | • | ||||
| 2 | L1766S | L | • | • | ||||
| 3 | I169T | M | • | • | ||||
| 3 | V310V (silent) | M | • | • | ||||
| 3 | T1462I | L | • | • | ||||
Values in parentheses represent the number of isolates with the mutation/total number of isolates tested.
nt, nucleotide.
After low-MOI passage, each replicate of PIV5-RSV-F (HN-L) acquired between one and three nonsynonymous changes in the consensus sequence. Among the three low-MOI replicates, three variants arose in the V/phosphoprotein (P)- or P-coding sequences and two variants were acquired in the HN coding sequence. Low-MOI replicate 1 had a V56M variant in the RSV F-coding sequence (Table 1). Overall, low-MOI passage of the virus resulted in increased mutation rates compared with those after high-MOI passage.
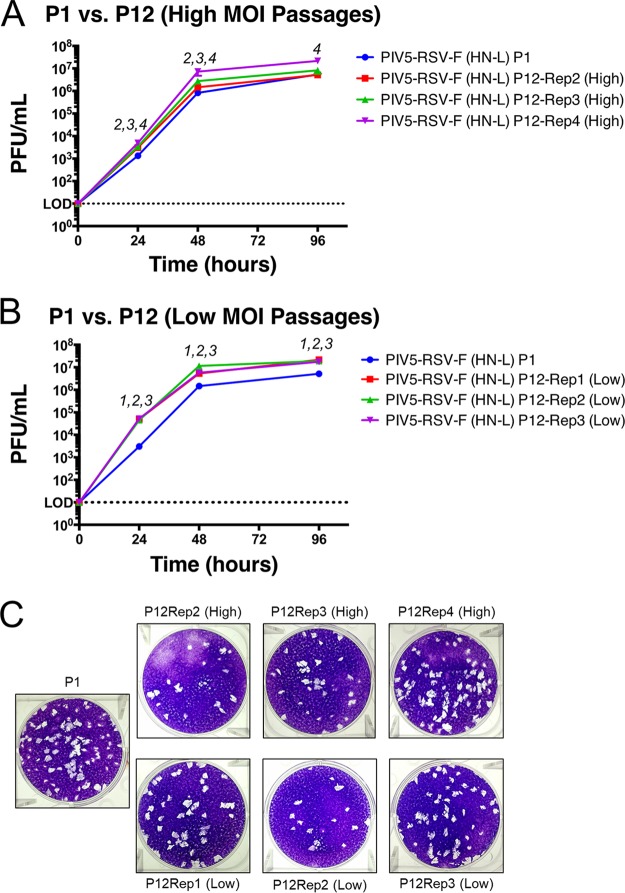
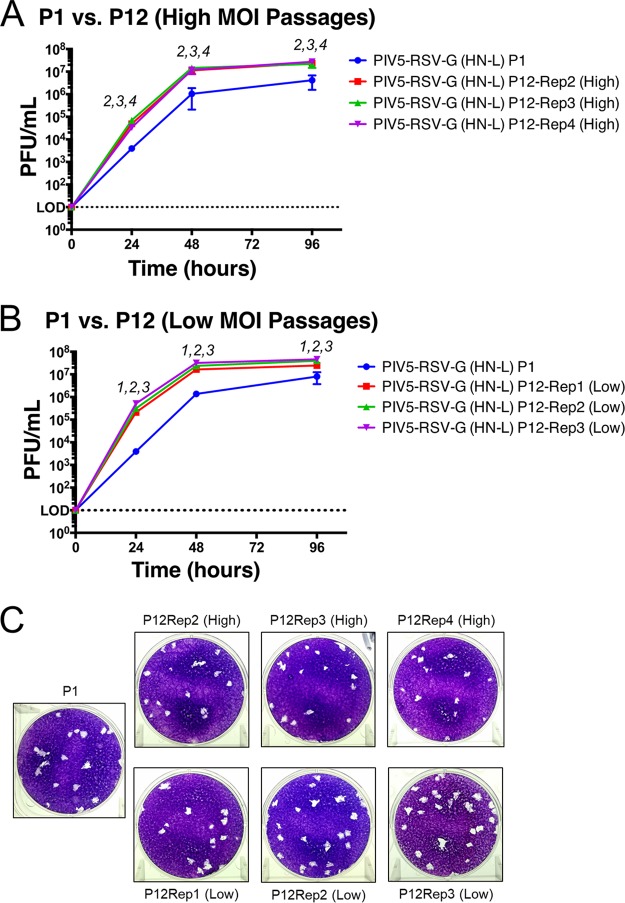
To investigate whether the mutations impacted the growth kinetics of the late-passage viruses, multicycle growth curves were performed to compare the growth of PIV5-RSV-F (HN-L) P1 and P12 viruses (P11 was expanded to generate P12). The high-MOI-passage viruses grew similarly to the parent virus. High-MOI replicate 4 reached significantly higher titers than the P1 virus (0.8 log10), and it was the only high-MOI replicate to have a mutation in the consensus sequence (Fig. 2A). All three low-MOI replicates grew significantly faster and to higher titers than the parent virus. The titers after passage at a low MOI were 1 log10 higher than those after P1 at 24 h postinfection (hpi), and the titer was 0.5 log10 higher at 96 hpi (Fig. 2B). The plaque morphologies of the passaged viruses appeared similar to the plaque morphology of the parent virus (Fig. 2C). The results suggest that the sequence variations acquired through serial passaging increased virus fitness in tissue culture cells.
FIG 2.
Growth kinetics of PIV5-RSV-F (HN-L) at P1 versus P12. Multicycle growth curves of PIV5-RSV-F (HN-L) in Vero cells at P0 and P12 are shown. (A and B) Vero cells were infected with PIV5-RSV-F (HN-L) at an MOI of 0.01 PFU per cell after P1 or P12 at high-MOI passages (A) or low-MOI passages (B). Samples of the tissue culture supernatant were collected at 0, 24, 48, and 96 hpi. Samples of three biological replicates were collected at each time point, and plaque assays were performed in BHK21 cells to determine virus titers. Error bars represent the standard errors of the means. The statistical significance of the difference between virus titers was determined using two-way ANOVA followed by Dunnett's correction for multiple comparisons. The number(s) above each time point indicates which replicate(s) had virus titers significantly different from those at P1 (P ≤ 0.05). (C) Comparison of plaque phenotypes of PIV5-RSV-F (HN-L) after P1 and P12. Representative images of plaques were obtained at 48 hpi. Rep, replicate.
To assess differences within the P0 and P11 virus populations, over 20 plaques were isolated at P0 and P11 and cultured from high-MOI replicate 1 of PIV5-RSV-F (HN-L). The gene insertion, including the upstream and downstream intergenic regions, was sequenced. Interestingly, the sequences of 13% (3 out of 23) of the PIV5-RSV-F (HN-L) P0 isolates differed from the parent sequence, even though P0 virus was grown from a single isolate. The mutations were nucleotide substitutions within the RSV F gene, all of which resulted in amino acid mutations. By P11, the sequences of 26% (6 out of 23) of the PIV5-RSV-F (HN-L) isolates differed from the sequence of the parent virus. However, all of these mutations were mixed bases within the RSV F gene. Of the six mutated isolates, only three isolates (13%) actually sustained amino acid mutations. None of the mixed bases in the P11 viruses matched the mutations in the P0 viruses (Table 1).
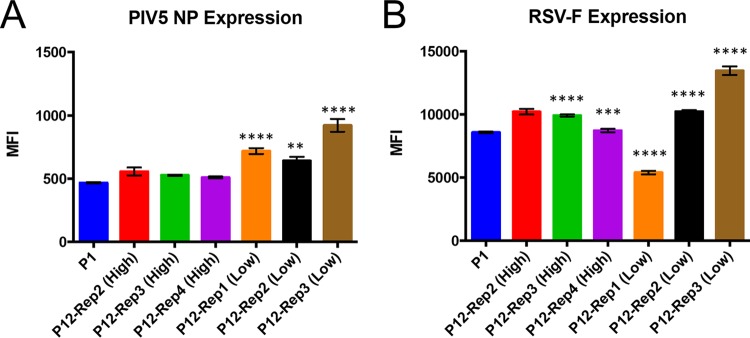
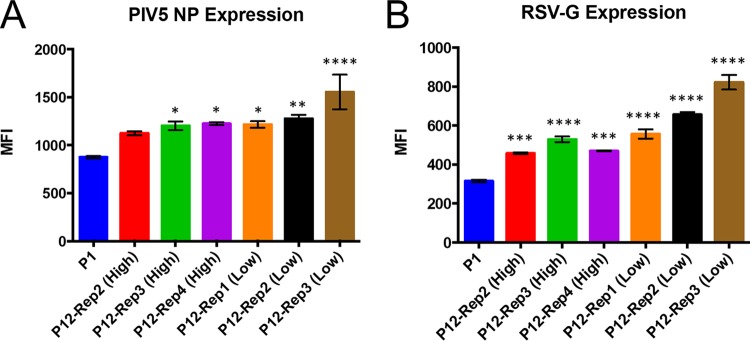
To determine whether changes in the consensus sequence of PIV5-RSV-F (HN-L) affected the expression of the foreign gene (RSV F) insertion, A549 cells were infected with PIV5-RSV-F (HN-L) P1 and P12 isolates, and viral protein expression was measured by flow cytometry. Nucleoprotein (NP) expression in PIV5 was similar to that in P1 virus in the high-MOI passages and significantly increased in the low-MOI passages (Fig. 3A). RSV F expression was increased in two out of the three high-MOI passages and two out of the three low-MOI passages. Low-MOI-passage replicate 1, the only replicate to have a mutation in the RSV F gene, had a 37% reduction in the level of RSV F expression (Fig. 3B). The results indicate that most of the mutations had either no effect on or increased the level of viral protein expression. However, a mutation in the coding sequence of RSV F reduced the level of expression of the foreign gene.
FIG 3.
Viral protein expression by PIV5-RSV-F (HN-L) at P1 versus P12. The levels of viral protein expression by PIV5-RSV-F (HN-L) in infected cells were compared after P1 and P12. A549 cells were infected with PIV5-RSV-F (HN-L) at an MOI of 1 PFU per cell after P1 and P12. After 48 h, the cells were harvested and immunostained with antibodies against PIV5 NP (A) and RSV F (B). Protein expression was measured by flow cytometry. The graphs represent the average mean fluorescence intensity (MFI) for three infection replicates, and error bars represent the standard errors of the means. The statistical significance of the levels of protein expression relative to the level at P1 was determined using one-way ANOVA followed by Dunnett's correction for multiple comparisons. **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001.
Full-genome sequencing of PIV5-RSV-G (HN-L) showed that after all four high-MOI passages the viruses had the same two mutations: one mutation was in the V/P gene, and the other was in the 3′ untranslated region (UTR) of the matrix protein (M). The mutation in the V/P gene caused an amino acid change from lysine to glutamate at amino acid residue 78 in the conserved N terminus. The 3′ UTR of M had a cytosine (C)-to-adenine (A) mutation at nucleotide position 4292 (Table 1). Since the viruses had the same changes after all high-MOI passages, the results suggest that the mutations conferred a fitness advantage in vitro.
After the low-MOI passages, PIV5-RSV-G (HN-L) had between two and nine mutations per replicate. Low-MOI replicate 1 had seven changes in the V/P region; four of these were nonsynonymous, and three of these were synonymous. It also had a nonsynonymous mutation in V and a silent mutation in P. Low-MOI replicate 2 had one mutation in P and one mutation in M. Replicate 3 had four mutations: one in V/P, one in M, one in the 3′ UTR of M, and one in L. This was the only low-MOI replicate in which the same mutations were detected after the high-MOI passages (Table 1). Thus, similar to the findings after the passage of PIV5-RSV-F (HN-L), low-MOI passage of PIV5-RSV-G (HN-L) resulted in increased mutation rates.
At the plaque isolate level in PIV5-RSV-G (HN-L) after P0, the sequences of 4% (1 out of 23) of the isolates were different, with the mutation being a mixed base within the RSV G gene. This mutation was silent. After P11, the sequences of none of the PIV5-RSV-G (HN-L) isolates differed from the parent sequence (Table 1). Thus, the RSV G gene and the flanking intergenic regions appeared to be stable through multiple in vitro passages, even though the genome sustained mutations elsewhere.
Growth curves were performed to compare the growth of PIV5-RSV-G (HN-L) after P1 and P12. All high- and low-MOI-passage viruses grew faster than the P1 virus, with significantly higher viral titers being observed at all time points. The titers of the high-MOI-passage virus were 1 log10 higher than the titer of the P1 virus at 24 hpi and 0.5 log10 higher at 96 hpi (Fig. 4A). The titers of the low-MOI-passage viruses were 1.7 log10 higher at 24 hpi and 0.5 log10 higher at 96 hpi (Fig. 4B). Plaques from the high-MOI-passage viruses appeared slightly smaller than those from the parent virus, while plaques from the low-MOI-passage viruses were similar to those from the parent virus (Fig. 4C). These results indicate that the mutations acquired through high- and low-MOI passage of PIV5-RSV-G (HN-L) increased virus fitness in vitro.
FIG 4.
Growth kinetics of PIV5-RSV-G (HN-L) at P1 versus P12. Multicycle growth curves of PIV5-RSV-G (HN-L) in Vero cells at P0 and P12 are shown. (A and B) Vero cells were infected with PIV5-RSV-G (HN-L) at an MOI of 0.01 PFU per cell after P1 or P12 at high-MOI passages (A) or low-MOI passages (B). Samples of the tissue culture supernatant were collected at 0, 24, 48, and 96 hpi. Samples of three biological replicates were collected at each time point, and plaque assays were performed in BHK21 cells to determine virus titers. Error bars represent the standard errors of the means. The statistical significance of the difference between virus titers was determined using two-way ANOVA followed by Dunnett's correction for multiple comparisons. The number(s) above each time point indicates which replicate(s) had virus titers significantly different from those at P1 (P ≤ 0.05). (C) Comparison of PIV5-RSV-G (HN-L) plaque phenotypes after P1 and P12. Representative images of plaques taken at 48 hpi.
Examination of PIV5-RSV-G (HN-L) protein expression in infected cells showed that serial passaging of the virus increased the level of overall viral protein expression. Two out of three high-MOI-passage viruses and all low-MOI-passage viruses had increased levels of NP expression (Fig. 5A). The level of RSV G expression was increased in all high- and low-MOI-passage viruses. These results are consistent with the significantly increased virus growth kinetics observed in the growth curves (Fig. 5B).
FIG 5.
Viral protein expression of PIV5-RSV-G (HN-L) at P1 versus P12. The levels of viral protein expression by PIV5-RSV-G (HN-L) in infected cells were compared after P1 and P12. A549 cells were infected with PIV5-RSV-G (HN-L) at an MOI of 1 PFU per cell after P1 and P12. After 48 h, the cells were harvested and immunostained with antibodies against PIV5 NP (A) and RSV G (B). Protein expression was measured by flow cytometry. The graphs represent the average mean fluorescence intensity (MFI) for three infection replicates, and error bars represent the standard errors of the means. The statistical significance of the levels of protein expression relative to the level at P1 was determined using one-way ANOVA followed by Dunnett's correction for multiple comparisons. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001.
The genome of PIV5-RSV-F (HN-L) but not that of PIV5-RSV-G (HN-L) remained stable through in vivo passaging.
Since the stability of the viruses after passage in vitro may not accurately reflect the stability after passage in vivo, we sought to sequence virus isolates that were recovered from an in vivo infection. African green monkeys were inoculated with either PIV5-RSV-F (HN-L) or PIV5-RSV-G (HN-L) after P1, and bronchoalveolar lavage (BAL) fluid wash samples were collected at 5 days postinfection (dpi). Serial dilutions of the BAL fluid wash samples were used for plaque assay in BHK cells. After 5 days, plaques were picked and cultured in Vero cells for 5 days, and the tissue culture supernatant was used for subsequent RNA isolation, reverse transcription (RT)-PCR, and sequencing.
No mutations were found in the consensus sequences of PIV5-RSV-F (HN-L) recovered from the BAL fluid samples from three different animals. None of the six PIV5-RSV-F (HN-L) plaque isolates selected for full-genome sequencing acquired amino acid residue mutations. Isolate 3 from animal A12M032 had a silent mutation in PIV5 F (Table 2). No mutations were observed upon sequencing of the RSV F gene insertion region of six additional isolates, although isolate 6 from animal A12M032 did have a silent mutation in RSV F (Table 2). Thus, the genome of PIV5-RSV-F (HN-L) appeared to be stable after in vivo passage in a nonhuman primate (NHP) model.
TABLE 2.
Comparison of virus sequences after in vivo passagea
| Virus | AGM identifier | Isolate no. | Mutation | Location | Presence of: |
|
|---|---|---|---|---|---|---|
| Full genome | Insertion only | |||||
| PIV5-RSV-F (HN-L) | A12M001 | 1 | None | • | ||
| A12M001 | 2 | None | • | |||
| A12M001 | 3 | None | • | |||
| A12M001 | 4 | None | • | |||
| A12M001 | 5 | None | • | |||
| A12M001 | 6 | None | • | |||
| A12M003 | 1 | None | • | |||
| A12M032 | 1 | None | • | |||
| A12M032 | 3 | V128V (silent) | PIV5 F | • | ||
| A12M032 | 4 | None | • | |||
| A12M032 | 5 | None | • | |||
| A12M032 | 6 | V56V | RSV F | • | ||
| PIV5-RSV-G (HN-L) | A12M004 | 1 | T119T (silent) | RSV G | • | |
| A12M004 | 2 | None | • | |||
| A12M004 | 3 | Q77H | RSV G | • | ||
| A12M004 | 4 | None | • | |||
| A12M004 | 5 | None | • | |||
| A12M004 | 6 | None | • | |||
| A12M015 | 1 | None | • | |||
| A12M015 | 2 | None | • | |||
| A12M015 | 3 | None | • | |||
| A12M015 | 4 | None | • | |||
| A12M015 | 5 | None | • | |||
| A12M015 | 6 | None | • | |||
| A12M008 | 1 | A753S | PIV5 L | • | ||
| A12M008 | 2 | None | • | |||
| A12M008 | 3 | T125A | RSV G | • | ||
| A12M008 | 4 | None | • | |||
| A12M008 | 5 | None | • | |||
Virus was isolated from BAL fluid at 5 dpi. AGM, African green monkey.
The consensus sequences of PIV5-RSV-G (HN-L) recovered from the BAL fluid from three different animals had no mutations. Interestingly, out of six fully sequenced PIV5-RSV-G (HN-L) plaque isolates, one isolate had an A753S mutation in PIV5 L (Table 2). After sequencing of the RSV G gene insertion from 11 additional isolates, 2 isolates were found to have mutations in RSV G (Table 2).
Insertion of RSV F at earlier positions in the PIV5 genome did not increase sequence instability.
In an effort to improve the PIV5-RSV-F (HN-L) vaccine candidate, we sought to increase the level of RSV F expression by inserting the gene at earlier positions in the PIV5 genome. We generated two new candidates in which the RSV F gene was inserted at the small hydrophobic (SH)-HN gene junction [PIV5-RSV-F (SH-HN)] or was inserted in place of SH (PIV5ΔSH-RSV-F) (Fig. 1A) (23). The sequences of the viruses were verified, and then the viruses were passaged in Vero cells as described in Materials and Methods.
The numbers of mutations acquired by the two new vaccine candidates were similar to the numbers acquired by PIV5-RSV-F (HN-L). One out of three high-MOI-passage PIV5-RSV-F (SH-HN) isolates had a mutation in the consensus sequence of the V/P gene. The three low-MOI-passage isolates had between one and four mutations each, and these were located in the leader sequence or the V/P, P, M, or L gene (Table 1). Two out of three high-MOI-passage isolates of PIV5ΔSH-RSV-F acquired one to two mutations, located in PIV5 F (the replicate 2 isolates) or the leader sequence and the L gene (the replicate 3 isolate). The three low-MOI-passage isolates of PIV5ΔSH-RSV-F had between one and three mutations each, and these were located in the V/P, P, M, and L genes (Table 1). Since the numbers of variations acquired through serial passage of PIV5-RSV-F (SH-HN) and PIV5ΔSH-RSV-F were similar to the numbers observed for PIV5-RSV-F (HN-L), the results suggest that that there is no increase in sequence instability when the RSV F gene is inserted at these earlier positions in the PIV5 genome.
DISCUSSION
The encapsidation of the PIV5 RNA genome by NP results in a stable viral genome that retains foreign gene insertions. In this work, we examined the stability of inserts within the PIV5 genome over 11 passages in vitro and a single passage in vivo. At different passages, we determined full-genome consensus sequences as well as the sequences of the foreign gene insertion in individual virus clones. We also investigated viral growth kinetics and protein expression after in vitro passage. The foreign gene insertions were stable, in that there was no loss of the insertions after passages in vitro or in vivo. However, sequence variations emerged in the viruses after high- and low-MOI passage, indicating that the viruses evolved during the serial passages.
The PIV5-RSV-F (HN-L) vaccine candidate retained the RSV F gene insert over 11 high-MOI and low-MOI passages in vitro. Under high-MOI-passage conditions, the consensus sequence was stable, with only one mutation in the leader sequence of one passage replicate isolate being found. The insertion of the RSV F gene into different positions of the PIV5 genome did not negatively impact the sequence stability of the vaccine candidates, as the mutation rates of PIV5-RSV-F (SH-HN) and PIV5ΔSH-RSV-F were similar to those of PIV5-RSV-F (HN-L). In contrast, high-MOI passage of PIV5-RSV-G (HN-L) consistently resulted in the emergence of two mutations in the consensus sequence: a K78E mutation in V/P and a C-to-A substitution at nucleotide position 4292 in the 3′ UTR of M. For all of the vaccine candidates, low-MOI-passage conditions drove the emergence of more variations throughout the genome consensus sequence, several of which were concentrated in the V/P and P genes. Passage replicate isolates with increased mutation rates also had increased growth potentials, indicating that the vaccine viruses evolved during in vitro passage and the mutations conferred increased virus fitness in Vero cells.
Examination of the PIV5 vaccine viruses showed that mutations within clonal populations of each virus exist. For PIV5-RSV-F (HN-L), 3 out of 23 clones contained mutations within the RSV F gene at P0. By P11, 6 out of 23 clones contained mutations as a mixture at a given site. An RSV F V56M variant detected in a plaque isolate of the P0 stock later emerged in the consensus sequence of low-MOI-passage replicate 1 virus, suggesting that the variant became fixed in the population. Cells infected with this virus had a 37% reduction in the level of RSV F expression. Although the V56M mutation occurred in the majority of viruses in the population, it did not abolish RSV F expression, as cells infected with virus with this mutation expressed RSV F at 63% of the levels at which it was expressed in cells infected with P1 virus. Since this variant emerged in only one out of four passage replicates, outgrowth in the population may have been a random event. The RSV F V56M variant was not detected in the BAL fluid of the nonhuman primates, but a V56V silent mutation was detected in a BAL fluid plaque isolate, possibly indicating instability at that position. For the purpose of vaccine production, the V56 codon may need to be stabilized and stringent quality control will be necessary to monitor the sequences of virus populations within the vaccine lots.
While no amino acid changes were detected within the RSV G gene of PIV5-RSV-G (HN-L) at the consensus or clonal level after in vitro passage, mutations emerged within the PIV5 backbone. All high-MOI-passage isolates and one low-MOI-passage isolate of PIV5-RSV-G (HN-L) had the K78E mutation in V/P and the C-to-A nucleotide substitution at position 4292 in the 3′ UTR of M. The K78E mutation is within a previously reported RNA binding region of the V/P gene (24). The mutation in the 3′ UTR of the M gene may affect the expression of M as well as F, the downstream gene. The length of the RSV G gene within the PIV5 genome is approximately 900 nucleotides, which is about half the length of RSV F. The absence of mutations within RSV G after in vitro passage is consistent with its smaller size and with RNA viruses being a quasispecies. However, the insertion of the G gene, despite its smaller size, resulted in mutations within the PIV5 backbone. We speculate that the emergence of mutations within the PIV5-RSV-G (HN-L) genome may indicate that the RSV G gene impacted the stability of the PIV5 genome. The RSV G protein binds to cell surface heparin sulfate and serves as an attachment protein for RSV (25). It is therefore possible that the functionality of RSV G affects the replication of PIV5-RSV-G (HN-L) in cultured cells.
We also investigated the stability of the vaccine candidates after a single in vivo passage in African green monkeys. No mutations were found in the consensus sequence of PIV5-RSV-F (HN-L) recovered from BAL fluid 5 days after infection. Further examination of individual plaque isolates also revealed no mutations. The PIV5-RSV-G (HN-L) consensus sequence appeared to be stable after passage, but 3 out of 17 isolates had mutations in various parts of the genome. One isolate sustained an A753S mutation in L, while 2 isolates had mutations in RSV G. The A753S mutation was located in the polymerase domain of L, but residue 753 is not known to be associated with catalytic activity (26). Both mutations in G were found in a hypervariable mucin-like region, with the T125A mutation being in a potential O-linked glycosylation site (27, 28). Thus, while PIV5-RSV-F (HN-L) remained stable through in vivo passage, PIV5-RSV-G (HN-L) acquired mutations in both the PIV5 backbone and the RSV G gene insertion. Together with the in vitro data, these results suggest that the RSV G gene may affect the stability of the PIV5-RSV-G (HN-L) candidate.
When considering further vaccine design using PIV5 as a vector, it is important to consider the functions of the antigens whose genes are inserted and their potential impact on viral genome stability. Previously, we generated a PIV5-based H5N1 vaccine using the gene for the hemagglutinin (HA) of an H5N1 influenza virus that lacks the cleavage site, rendering the HA molecule inactive but maintaining the immunogenicity of the protein (13, 17). A similar approach may be considered for improving the stability of PIV5-based RSV G vaccines.
MATERIALS AND METHODS
Cells and viruses.
Vero and BHK21 cells were maintained in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS) plus 100 IU/ml penicillin and 100 μg/ml streptomycin (1% P/S; Mediatech Inc., Manassas, VA, USA). Cells were passaged 1 day prior to infection and achieved approximately 90% confluence by the following day.
PIV5-RSV-F (HN-L), PIV5-RSV-G (HN-L), PIV5-RSV-F (SH-HN), and PIV5ΔSH-RSV-F were generated (19) and grown in MDBK cells as previously described (21).
Serial passage of recombinant viruses.
Vero cells in 6-cm dishes were infected with PIV5-RSV-F (HN-L), PIV5-RSV-G (HN-L), PIV5-RSV-F (SH-HN), or PIV5ΔSH-RSV-F at an MOI of 1 or 0.01 PFU per cell for high- and low-MOI passages, respectively. For high-MOI passages, 500 μl of cell supernatant was used to inoculate fresh 6-cm dishes of Vero cells every 4 to 5 days. For low-MOI passages, fresh Vero cells were inoculated with a 1:10,000 dilution of virus from the previous passage every 4 to 5 days. The remaining cell supernatant was supplemented with 1% bovine serum albumin (BSA) and stored at −80°C. Eleven passages were performed.
Initially, the high-MOI passages were performed with one replicate, which was designated replicate 1. The experiment was then repeated with three replicates, designated replicates 2 through 4. All low-MOI passages were performed in triplicate, with the isolates being designated replicates 1 through 3.
Growth curves.
Vero cells in 6-well plates were infected with P0 or P12 virus at an MOI of 0.01 PFU per cell and incubated for 1 to 2 h at 37°C in 5% CO2. The inocula were aspirated and replaced with 2 ml of DMEM containing 2% FBS and 1% P/S. One-hundred-microliter samples of supernatant were collected at 0, 24, 48, and 96 h postinfection (hpi). Each sample was supplemented with 10% sucrose-phosphate-glutamate (SPG) buffer, snap-frozen in liquid nitrogen, and stored at −80°C immediately after collection. Virus titers were determined by plaque assay as described by Chen et al. (16).
Flow cytometry.
A549 cells in 6-well dishes were infected with P0 and P12 viruses at an MOI of 1 PFU per cell. Forty-eight hours later, the cells were detached using 10 mM EDTA in Dulbecco's phosphate-buffered saline (DPBS). The cells were washed with DPBS and then fixed and permeabilized in BD Cytofix/Cytoperm solution for 20 min at 4°C. PIV5-RSV-F (HN-L)-infected cells were immunostained with palivizumab conjugated to allophycocyanin and a primary monoclonal antibody against NP (antibody NP214, made in-house), followed by staining with goat anti-mouse IgG conjugated to Alexa Fluor 488. PIV5-RSV-G (HN-L)-infected cells were immunostained with an anti-G monoclonal antibody (antibody RSV1C2; Abcam) conjugated to Alexa Fluor 488 and an anti-NP monoclonal antibody (antibody NP214) conjugated to allophycocyanin. Mock-infected cells served as negative controls for staining.
NHP infection and BAL fluid collection.
Animal experiments were performed according to protocols approved by the Merck Institutional Animal Care and Use Committee. African green monkeys (Chlorocebus sabaeus) seronegative for PIV5 and seropositive for RSV were anesthetized with tiletamine-zolazepam (Telazol; 4 to 6 mg/kg of body weight) supplemented with ketamine (5 mg/kg), if needed. Either PIV5-RSV-F (HN-L) (P1) or PIV5-RSV-G (HN-L) (P1) (106 PFU) was administered intranasally in a 1-ml volume and was distributed equally over both nares.
Bronchoalveolar lavage (BAL) fluid samples were collected at 3, 5, 7, 10, and 14 dpi. Briefly, 5 ml of Hanks' balanced salt solution (HBSS) was delivered into the lung and then aspirated using a tracheal tube and syringe. BAL fluid samples were mixed with 10% SPG buffer and 1% gelatin, flash-frozen in a dry-ice ethanol bath, and stored at −80°C.
PIV5-RSV-F (HN-L) and PIV5-RSV-G (HN-L) plaque purification.
Viruses from passages 0 and 11 of PIV5-RSV-F (HN-L) and PIV5-RSV-G (HN-L) were diluted by a factor of 104 to 106 in DMEM containing 1% BSA. Fifty microliters of each dilution was used to infect 6-well plates of BHK21 cells as described above. After 5 days, plaques were selected and cultured in Vero cells with DMEM containing 2% FBS and 1% P/S. After 4 to 5 days, cell supernatants were harvested, supplemented with 1% BSA, and stored at −80°C.
For plaque purification of PIV5-RSV-F (HN-L) and PIV5-RSV-G (HN-L) from NHP samples, BAL fluid wash samples from 5 dpi were diluted by a factor of 2 to 2 × 105 in DMEM containing 1% BSA. Four hundred microliters of each dilution was used to infect BHK21 cells, and plaques were cultured as stated above.
RT-PCR and sequencing.
Viral RNA was extracted from cell culture supernatants using a QIAamp viral RNA minikit (Qiagen Inc., Valencia, CA). Reverse transcription (RT) was performed with SuperScript III reverse transcriptase (Life Technologies) and random hexamers (Promega, Madison, WI). The cDNA templates and 19 genome-specific primer pairs were used to amplify overlapping fragments of the viral genome by PCR. These primers were also used to sequence the PCR products.
For NHP plaque isolates, viral RNA was extracted as stated above. Genome-specific primers and 5 μl of RNA were used for one-step RT-PCR (Life Technologies).
Sequence analysis was performed using Sequencher (version 5.1) sequence analysis software (Gene Codes Corporation, Ann Arbor, MI). Primer sequences are available upon request.
Statistical analysis.
GraphPad Prism (version 5.04) software for Windows was used for statistical analysis (GraphPad Software, La Jolla, CA). Two-way analysis of variance (ANOVA) followed by Dunnett's correction for multiple comparisons was used to compare the virus titers at different time points on the growth curves after P0 and P12. One-way ANOVA followed by Dunnett's correction for multiple comparisons was used to compare the levels of protein expression by P1 and P12 viruses.
ACKNOWLEDGMENTS
We thank all members of Biao He's laboratory for their helpful discussions and technical assistance. We also thank the teams at Merck & Co., Inc., and the New Iberia Research Center for their assistance with the nonhuman primate studies.
This work was supported by an endowment from the Fred C. Davison Distinguished University Chair in Veterinary Medicine to B.H.
Footnotes
For a companion article on this topic, see https://doi.org/10.1128/JVI.00560-17.
REFERENCES
- 1.Nair H, Nokes DJ, Gessner BD, Dherani M, Madhi SA, Singleton RJ, O'Brien KL, Roca A, Wright PF, Bruce N, Chandran A, Theodoratou E, Sutanto A, Sedyaningsih ER, Ngama M, Munywoki PK, Kartasasmita C, Simões EA, Rudan I, Weber MW, Campbell H. 2010. Global burden of acute lower respiratory infections due to respiratory syncytial virus in young children: a systematic review and meta-analysis. Lancet 375:1545–1555. doi: 10.1016/S0140-6736(10)60206-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Glezen WP, Taber LH, Frank AL, Kasel JA. 1986. Risk of primary infection and reinfection with respiratory syncytial virus. Am J Dis Child 140:543–546. [DOI] [PubMed] [Google Scholar]
- 3.Escobar GJ, Ragins A, Li SX, Prager L, Masaquel AS, Kipnis P. 2010. Recurrent wheezing in the third year of life among children born at 32 weeks' gestation or later: relationship to laboratory-confirmed, medically attended infection with respiratory syncytial virus during the first year of life. Arch Pediatr Adolesc Med 164:915–922. doi: 10.1001/archpediatrics.2010.177. [DOI] [PubMed] [Google Scholar]
- 4.Wu P, Hartert TV. 2011. Evidence for a causal relationship between respiratory syncytial virus infection and asthma. Expert Rev Anti Infect Ther 9:731–745. doi: 10.1586/eri.11.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Falsey AR, Walsh EE. 2000. Respiratory syncytial virus infection in adults. Clin Microbiol Rev 13:371–384. doi: 10.1128/CMR.13.3.371-384.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anderson LJ, Dormitzer PR, Nokes DJ, Rappuoli R, Roca A, Graham BS. 2013. Strategic priorities for respiratory syncytial virus (RSV) vaccine development. Vaccine 31(Suppl 2):B209–B215. doi: 10.1016/j.vaccine.2012.11.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karron RA, Wright PF, Belshe RB, Thumar B, Casey R, Newman F, Polack FP, Randolph VB, Deatly A, Hackell J, Gruber W, Murphy BR, Collins PL. 2005. Identification of a recombinant live attenuated respiratory syncytial virus vaccine candidate that is highly attenuated in infants. J Infect Dis 191:1093–1104. doi: 10.1086/427813. [DOI] [PubMed] [Google Scholar]
- 8.Bernstein DI, Malkin E, Abughali N, Falloon J, Yi T, Dubovsky F. 2012. Phase 1 study of the safety and immunogenicity of a live, attenuated respiratory syncytial virus and parainfluenza virus type 3 vaccine in seronegative children. Pediatr Infect Dis J 31:109–114. doi: 10.1097/INF.0b013e31823386f1. [DOI] [PubMed] [Google Scholar]
- 9.Yang CF, Wang CK, Malkin E, Schickli JH, Shambaugh C, Zuo F, Galinski MS, Dubovsky F, Tang RS. 2013. Implication of respiratory syncytial virus (RSV) F transgene sequence heterogeneity observed in phase 1 evaluation of MEDI-534, a live attenuated parainfluenza type 3 vectored RSV vaccine. Vaccine 31:2822–2827. doi: 10.1016/j.vaccine.2013.04.006. [DOI] [PubMed] [Google Scholar]
- 10.Lamb R, Kolakofsky D. 2001. Paramyxoviridae: the viruses and their replication. In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE (ed), Fields virology, 4th ed Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 11.Chen Z, Xu P, Salyards GW, Harvey SB, Rada B, Fu ZF, He B. 2012. Evaluating a parainfluenza virus 5-based vaccine in a host with pre-existing immunity against parainfluenza virus 5. PLoS One 7:e50144. doi: 10.1371/journal.pone.0050144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Z, Mooney AJ, Gabbard JD, Gao X, Xu P, Place RJ, Hogan RJ, Tompkins SM, He B. 2013. Recombinant parainfluenza virus 5 expressing hemagglutinin of influenza A virus H5N1 protected mice against lethal highly pathogenic avian influenza H5N1 challenge. J Virol 87:354–362. doi: 10.1128/JVI.02321-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Z, Gabbard JD, Mooney A, Gao X, Chen Z, Place RJ, Tompkins SM, He B. 2013. Single-dose vaccination of a recombinant parainfluenza virus 5 expressing NP from H5N1 virus provides broad immunity against influenza A viruses. J Virol 87:5985–5993. doi: 10.1128/JVI.00120-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Z, Gabbard JD, Mooney A, Chen Z, Tompkins SM, He B. 2013. Efficacy of parainfluenza virus 5 mutants expressing hemagglutinin from H5N1 influenza A virus in mice. J Virol 87:9604–9609. doi: 10.1128/JVI.01289-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li Z, Gabbard J, Johnson S, Dlugolenski D, Phan S, Tompkins M, He B. 2015. Efficacy of a parainfluenza virus 5 (PIV5)-based H7N9 vaccine in mice and guinea pigs: antibody titer towards HA was not a good indicator for protection. PLoS One 10:e0120355. doi: 10.1371/journal.pone.0120355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen Z, Zhou M, Gao X, Zhang G, Ren G, Gnanadurai CW, Fu ZF, He B. 2013. A novel rabies vaccine based on a recombinant parainfluenza virus 5 expressing rabies virus glycoprotein. J Virol 87:2986–2993. doi: 10.1128/JVI.02886-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mooney AJ, Li Z, Gabbard JD, He B, Tompkins SM. 2013. Recombinant parainfluenza virus 5 vaccine encoding the influenza virus hemagglutinin protects against H5N1 highly pathogenic avian influenza virus infection following intranasal or intramuscular vaccination of BALB/c mice. J Virol 87:363–371. doi: 10.1128/JVI.02330-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tompkins SM, Lin Y, Leser GP, Kramer KA, Haas DL, Howerth EW, Xu J, Kennett MJ, Durbin RK, Durbin JE, Tripp R, Lamb RA, He B. 2007. Recombinant parainfluenza virus 5 (PIV5) expressing the influenza A virus hemagglutinin provides immunity in mice to influenza A virus challenge. Virology 362:139–150. doi: 10.1016/j.virol.2006.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Phan SI, Chen Z, Xu P, Li Z, Gao X, Foster SL, Teng MN, Tripp RA, Sakamoto K, He B. 2014. A respiratory syncytial virus (RSV) vaccine based on parainfluenza virus 5 (PIV5). Vaccine 32:3050–3057. doi: 10.1016/j.vaccine.2014.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mueller S, Wimmer E. 1998. Expression of foreign proteins by poliovirus polyprotein fusion: analysis of genetic stability reveals rapid deletions and formation of cardioviruslike open reading frames. J Virol 72:20–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.He B, Paterson RG, Ward CD, Lamb RA. 1997. Recovery of infectious SV5 from cloned DNA and expression of a foreign gene. Virology 237:249–260. doi: 10.1006/viro.1997.8801. [DOI] [PubMed] [Google Scholar]
- 22.Rima BK, Gatherer D, Young DF, Norsted H, Randall RE, Davison AJ. 2014. Stability of the parainfluenza virus 5 genome revealed by deep sequencing of strains isolated from different hosts and following passage in cell culture. J Virol 88:3826–3836. doi: 10.1128/JVI.03351-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Phan SI, Zengel JR, Wei H, Li Z, Wang D, He B. 2017. Parainfluenza virus 5 expressing wild-type or prefusion respiratory syncytial virus (RSV) fusion protein protects mice and cotton rats from RSV challenge. J Virol 91:e00560-17. doi: 10.1128/JVI.00560-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin GY, Paterson RG, Lamb RA. 1997. The RNA binding region of the paramyxovirus SV5 V and P proteins. Virology 238:460–469. doi: 10.1006/viro.1997.8866. [DOI] [PubMed] [Google Scholar]
- 25.Bourgeois C, Bour JB, Lidholt K, Gauthray C, Pothier P. 1998. Heparin-like structures on respiratory syncytial virus are involved in its infectivity in vitro. J Virol 72:7221–7227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Poch O, Blumberg BM, Bougueleret L, Tordo N. 1990. Sequence comparison of five polymerases (L proteins) of unsegmented negative-strand RNA viruses: theoretical assignment of functional domains. J Gen Virol 71(Pt 5):1153–1162. [DOI] [PubMed] [Google Scholar]
- 27.Johnson PR, Spriggs MK, Olmsted RA, Collins PL. 1987. The G glycoprotein of human respiratory syncytial viruses of subgroups A and B: extensive sequence divergence between antigenically related proteins. Proc Natl Acad Sci U S A 84:5625–5629. doi: 10.1073/pnas.84.16.5625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tan L, Coenjaerts FEJ, Houspie L, Viveen MC, van Bleek GM, Wiertz EJHJ, Martin DP, Lemey P. 2013. The comparative genomics of human respiratory syncytial virus subgroups A and B: genetic variability and molecular evolutionary dynamics. J Virol 87:8213–8226. doi: 10.1128/JVI.03278-12. [DOI] [PMC free article] [PubMed] [Google Scholar]