ABSTRACT
Hepatitis E virus (HEV), a single-stranded positive-sense RNA virus, generally causes self-limiting acute viral hepatitis, although chronic HEV infection has recently become a significant clinical problem in immunocompromised individuals, especially in solid-organ transplant recipients. Innate immunity, via the type I interferon (IFN) response, plays an important role during the initial stages of a viral infection. IFN-stimulated gene 15 (ISG15), an IFN-induced ubiquitin-like protein, is known to have an immunomodulatory role and can have a direct antiviral effect on a wide spectrum of virus families. In the present study, we investigated the antiviral effect as well as the potential immunomodulatory role of ISG15 during HEV replication. The results revealed that HEV induced high levels of ISG15 production both in vitro (Huh7-S10-3 liver cells) and in vivo (liver tissues from HEV-infected pigs); however, ISG15 is not required for virus replication. We also demonstrated that ISG15 silencing potentiates enhanced type I IFN-mediated signaling, resulting in an increase in the type I IFN-mediated antiviral effect during HEV replication. This observed enhanced type I IFN signaling correlated with an increase in IFN-stimulated gene expression levels during HEV replication. Furthermore, we showed that PKR and OAS1 played important roles in the ISG15-mediated type I IFN sensitivity of HEV. Taken together, the results from this study suggest that ISG15 plays an important immunomodulatory role and regulates HEV sensitivity to exogenous type I IFN.
IMPORTANCE Hepatitis E virus (HEV) infection typically causes self-limiting acute viral hepatitis. However, chronic HEV infection has recently become a significant clinical problem in immunocompromised patients. Pegylated interferon (IFN) has been used to treat chronic HEV infection in solid-organ transplant patients with some success. However, the mechanism behind the type I IFN-mediated antiviral effect against HEV remains unclear. This report demonstrates that ISG15 induced by HEV replication in Huh7-S10-3 human liver cells plays an immunomodulatory role by negatively regulating type I IFN signaling and, thus, HEV sensitivity to type I IFN. Our results also show that PKR and OAS1 play important roles in the ISG15-mediated type I IFN sensitivity of HEV.
KEYWORDS: ISG15, hepatitis E virus (HEV), type I IFN signaling, immunomodulation
INTRODUCTION
Hepatitis E virus (HEV) is a positive-sense single-stranded RNA virus approximately 7.2 kb in size belonging to the family Hepeviridae, which consists of two genera (Orthohepevirus and Piscihepevirus) and 5 species (1). Within the orthohepevirus A species, there exist at least 7 genotypes: genotypes 1 and 2 are restricted to humans, genotypes 3 and 4 infect humans and several other animal species, genotypes 5 and 6 infect wild boars, and genotype 7 infects camel (1–3). It is recognized that the genotype 3 and 4 HEV strains are zoonotic; pigs and other species serve as the reservoirs (3–5).
The genome of HEV encodes three proteins. ORF1 is a nonstructural protein involved in virus replication, ORF2 is the capsid protein involved in virus assembly and receptor binding, and ORF3 is involved in virus egress. HEV virions were initially identified as nonenveloped viral particles; however, quasi-enveloped HEV virions have recently been reported (6). HEV replication occurs in cytoplasm (7) and uses a clathrin-dependent pathway (8) as well as an endosomal pathway (9, 10) for cellular entry. Endosomal acidification has been shown to be important for HEV infectivity (9). A recent study has also shown that multivesicular bodies and the exosomal pathway play an important role in virus egress (11). Therefore, it is postulated that cloaking the viral capsid protein within the host-derived membrane enables HEV to avoid triggering the host immune response (10).
The disease caused by HEV, hepatitis E, is of global public health importance. It is estimated that there are approximately 20 million HEV infections each year, resulting in 3.3 million symptomatic cases and more than 56,000 hepatitis E-related deaths annually (12). HEV is mainly transmitted via the fecal-oral route through contaminated water or food. Hepatitis E is usually a self-limiting acute disease; however, chronic HEV infection has recently become a major clinical problem in immunocompromised patients, especially in organ transplant recipients (13) and in patients with HIV infection, leukemia, or lymphoma (14). The chronic HEV infections are almost exclusively caused by the zoonotic genotype 3 HEV strains (14). Although the overall mortality rate associated with HEV infection is less than 1%, HEV can cause fulminant hepatitis in infected pregnant women, with a mortality rate of up to 20% to 25% (12, 15). Currently, there is no specific treatment for HEV infection, although broad-spectrum antivirals such as ribavirin and pegylated interferon (IFN) have been used to treat chronic HEV infection in solid-organ transplant patients, which sometime can result in severe side effects, including graft rejection (16).
The mechanism behind type I IFN-mediated antiviral activity against HEV remains unclear. HEV is known to modulate the type I IFN induction pathway. Studies have shown that HEV ORF3 enhances type I IFN induction (17), while HEV ORF1 has been shown to inhibit IFN induction (18). The interferon alpha 2a (IFN-α2a) subtype is known to exert the strongest antiviral effect against HEV in vitro (19). It has been reported that type I IFN mediates a dose-dependent reduction in the viral RNA level and that HEV ORF3 inhibits type I IFN signaling in A549 cells (20). HEV has been shown to be less susceptible to type I IFN-mediated antiviral effects than the hepatitis C virus (HCV) in vitro (19, 21). Therefore, it is important to delineate the mechanism of interactions between type I IFN and HEV to enable a better understanding of HEV pathobiology as well as development of a better prognostic use of type I IFN during HEV infection.
Type I IFNs include a multigene family of secreted cytokines. Type I IFN signaling induces expression of various IFN-stimulated genes (ISGs) via the JAK-STAT pathway to establish an antiviral state. ISG15 is a type I IFN-induced ubiquitin-like protein with pleotropic functions (22). ISG15 conjugation of target proteins (i.e., ISGylation) is a sequential process involving UBE1L (E1), UBE2L6 (E2), and HERC5 (E3) ligase (23). An increase in ISG15 mRNA levels has been reported in patients chronically infected with hepatitis viruses, including HCV (24) and HEV (25). It has been reported that HEV induces ISG15 in A549 cells and hepatic cells (26, 27), as well as in HEV-infected chimpanzee liver biopsy tissues (28). However, the role of ISG15 in viral infection remains controversial (22), as some studies have reported that ISG15 acts in an antiviral manner against human immunodeficiency virus (HIV) (29), Ebola virus (30), HCV (31), and influenza virus (32), whereas other studies showed that ISG15 also acts as a proviral during HCV infection (33, 34). More recently, ISG15 was reported to have an immunomodulatory effect by acting as a negative regulator of type I IFN signaling, thus regulating the antiviral response during viral infection (35, 36). An increased level of ISGs is suspected to favor the persistence of HEV infection in clinical cases (25).
Therefore, in the present study we investigated the role of ISG15 in HEV replication and determined whether ISG15 plays any potential role in type I IFN-mediated antiviral activity during HEV replication. Our results show that HEV induces ISG15 at both the mRNA level and the protein level and that ISG15 has an immunomodulatory role during HEV replication rather than a direct antiviral effect.
RESULTS
HEV induces ISG15 both in vitro and in vivo.
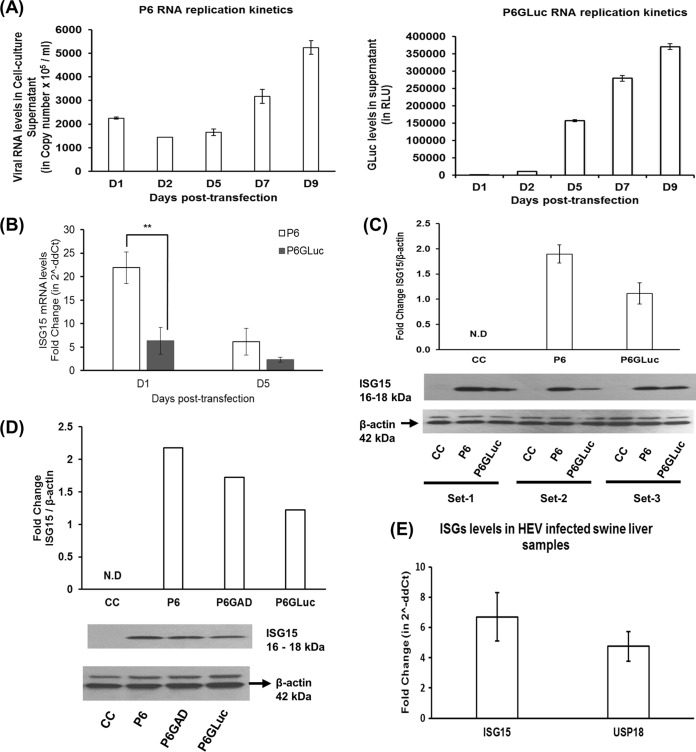
ISG15 is a 17-kDa type I IFN-induced ubiquitin-like protein and is known to modulate virus replication, including HCV replication (31, 34). To better understand the role of ISG15 in HEV replication, we first determined whether HEV could induce ISG15. Throughout this study, we used a cell culture-adapted strain (Kernow P6) of genotype 3 HEV (37). The Huh7-S10-3 liver cells used in the study support the replication of both the P6 HEV infectious cDNA clone (designated “HEV P6”; Fig. 1A) and HEV replicon P6 encoding the Gaussia luciferase (GLuc) clone (designated “HEV P6GLuc”; Fig. 1A). P6GLuc was originally constructed by replacing the 5′ terminal region of ORF2 with the in-frame GLuc reporter gene (37). Although the HEV-P6GLuc replicon mimics viral replication and serves as a convenient tool to monitor viral protein translation and virus replication, due to its lack of ORF2 expression the HEV P6GLuc replicon lacks viral assembly, maturation, and/or budding. Therefore, in this study we also employed an HEV infection system, the HEV P6 infectious cDNA clone, to further verify the results obtained from the HEV P6GLuc replicon system. We observed increases in HEV RNA copy numbers and GLuc levels in supernatant from 5 days posttransfection (dpt) (D5); therefore, samples were collected at that time point for all further experiments.
FIG 1.
HEV induces ISG15 both in vitro and in vivo. (A) Replication kinetics of viral RNAs from the HEV P6 infectious cDNA clone and HEV P6GLuc replicon in Huh7-S10-3 liver cells. The cells were transfected with capped RNA transcripts from the genotype 3 HEV P6 infectious cDNA clone (HEV P6) or with the HEV P6GLuc replicon (HEV P6GLuc; P6 encoding Gaussia luciferase clone). The culture supernatants were collected at various time points and used to measure viral RNA replication levels by HEV real-time quantitative RT-PCR or by the Gaussia luciferase assay (GLuc assay). RLU, relative light units. (B) ISG15 mRNA expression levels in Huh7-S10-3 liver cells transfected with capped RNA transcripts from the HEV P6 infectious cDNA clone or the HEV P6GLuc replicon at 1 and 5 days posttransfection (dpt) were determined by Sybr green-based quantitative PCR (qPCR). The ISG15-mRNA fold change was calculated using the 2−ΔΔCT method for comparisons to untransfected control cells, and the RPS18 gene was used as the housekeeping gene. (C and D) ISG15 protein expression levels in Huh7-S10-3 cells transfected with capped RNA from the HEV P6 infectious cDNA clone or the HEV P6GLuc replicon (C and D) or the HEV P6 replication-deficient infectious cDNA clone (P6GAD) (D). CC, untransfected cell control. The ISG15 protein expression, at 5 days posttransfection, in cell lysates (40 μg/lane) was analyzed by Western blotting using anti-ISG15 antibody (1:1,000 dilution). Fold change in band density, as calculated via normalization of ISG15 with β-actin (loading control), was measured using ImageJ software (NIH, Bethesda, MD). (E) ISG15 and USP18 mRNA expression levels in swine liver tissue samples obtained from animals experimentally infected with HEV (n = 3) at 3 weeks postinfection. The fold changes in swine ISG15 and USP18 mRNA levels were calculated using the 2−ΔΔCT method compared to the uninfected control animals (n = 2), and the RPS18 gene was used as the housekeeping gene. The data represent means ± standard errors of the means (SEM) of results from three independent transfection experiments (A, B, and C), one experiment representative of two independent transfection experiments (D), and HEV-infected animals (n = 3) compared to uninfected animals (n = 2) (E). **, P ≤ 0.01.
Huh7-S10-3 liver cells were transfected with capped RNA transcripts from genotype 3 HEV P6 or P6GLuc, and the fold changes in ISG15 mRNA and protein levels were estimated using real-time quantitative reverse transcription-PCR (RT-PCR) and Western blot analysis, respectively. We also determined the ISG15 protein levels at 5 dpt in cells transfected with the replication-deficient P6 HEV infectious clone (designated “P6GAD”) (38). We performed negative-strand-specific quantitative RT-PCR to measure replicative HEV RNA levels in HEV P6GAD- or HEV P6-transfected Huh7-S10-3 cells at 5 dpt. Negative-strand viral RNA levels remained undetectable in HEV P6GAD infectious clone-transfected Huh7-S10-3 cells, while negative-strand HEV RNA levels in HEV P6 infectious clone-transfected cells were approximately 1,600 ± 117 RNA copies/μg of total intracellular RNA. This confirms the replication deficiency of the HEV P6GAD clone. In corroboration of previous studies (25, 26), our results showed that the genotype 3 HEV RNA by itself, irrespective of its replication capacity, induced an increase in ISG15 mRNA levels (Fig. 1B) and protein levels (Fig. 1C and D) compared to the untransfected control. Transfection of capped RNA transcripts from full-length infectious cDNA clone HEV P6 in Huh7-S10-3 liver cells resulted in significantly higher levels of ISG15 mRNA (∼21.8-fold ± 3.3-fold increase) than in cells transfected with the capped RNA transcripts from the HEV P6GLuc replicon (∼ 6.3-fold ± 2.8-fold increase) at 1 dpt, and the ISG15 mRNA levels declined at 5 dpt. The enhanced ISG15 mRNA levels seen at 1 dpt were likely due to the residual HEV RNA present posttransfection.
Since our in vitro model with the HEV RNA transfection-based system showed that HEV RNA induced ISG15 expression regardless of its replication property, we further tested if HEV replication could induce ISG15 in vivo by using liver tissues from pigs experimentally infected with HEV. We determined the levels of ISG15 and USP18 (deISGylating enzyme) mRNA expression in swine liver tissue samples (n = 3) collected at 3 weeks postinfection (wpi) from pigs experimentally infected with a genotype 3 strain of HEV in a previous unrelated study. We chose the liver tissue samples from the 3 wpi time point since we observed an increase in viral RNA loads in fecal samples at 3 wpi, and fecal viral shedding gradually became reduced and ceased by 8 wpi (39). Hence, the 3-wpi liver tissue samples were considered to represent an active stage of acute HEV infection and replication in pigs. The results showed that the ISG15 mRNA levels (6.71-fold ± 1.59-fold increase) and USP18 mRNA levels (4.76-fold ± 0.98-fold increase) in the HEV-infected swine liver tissue samples (n = 3) were higher than those in the uninfected control liver tissue samples (n = 2) (Fig. 1E). Therefore, the results from the in vivo study using HEV-infected pig livers further suggested that genotype 3 HEV can potentiate induction of ISG15 and its related gene (i.e., the USP18 gene); thus, our data suggest that genotype 3 HEV induces ISG15 both in vitro and in vivo.
ISG15 is not required for HEV replication.
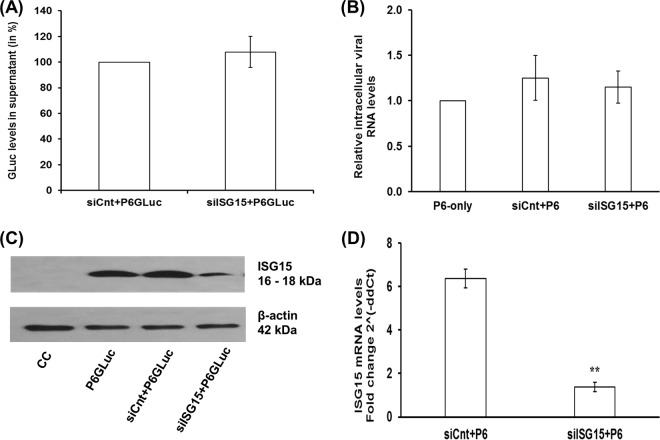
The role of ISG15 during HEV replication is unknown; therefore, we conducted further experiments to determine if ISG15 affects HEV replication. In preliminary standardization experiments, we had seen that transfection of ISG15-targeted small interfering RNA (siRNA) (siISG15) 1 day prior to (D−1) or on the same day as (D0) virus RNA transfection resulted in similar knockdown efficiencies with respect to ISG15 levels when measured at D2 and D5 post-IFN-α treatment (data not shown). Therefore, Huh7-S10-3 liver cells were cotransfected with 20 nM ISG15-targeted siRNA (siISG15) or with control siRNA (siCnt) along with capped RNA transcripts of HEV P6GLuc or P6. At 5 dpt, the levels of GLuc activity in cell culture supernatant and of the intracellular HEV P6 viral RNA were determined. In contrast to a previous study with HCV (31), we found that suppression of ISG15 did not result in enhanced HEV replication as no change was observed either in GLuc activity levels (Fig. 2A) or in the intracellular HEV P6 viral RNA levels (Fig. 2B) compared to control siRNA-transfected samples. Western blot analysis and real-time RT-PCR showed a significant reduction in ISG15 protein (Fig. 2C) and RNA (Fig. 2D) levels in siISG15-treated cells compared to siCnt-treated cells.
FIG 2.
ISG15 is not required for HEV replication. Huh7-S10-3 liver cells were cotransfected with 20 nM ISG15-targeted siRNA (siISG15) or control siRNA (siCnt) along with HEV P6GLuc replicon RNA (A and C) or HEV P6 infectious clone viral genomic RNA (B and D). The GLuc activity levels in culture supernatant (A) and the intracellular viral RNA levels (B) were determined by GLuc assay and qPCR, respectively, at 5 dpt. The efficiency of knockdown of ISG15 by siISG15 was determined by measuring ISG15 protein levels in siRNA and HEV P6GLuc replicon RNA cotransfected samples at 5 dpt (C) and ISG15 mRNA levels in siRNA and HEV P6 viral RNA cotransfected samples at 5 dpt (D); fold change compared to untransfected control cells was calculated using the 2−ΔΔCT method, and the RPS18 gene was used as the housekeeping gene. The data represent means ± SEM of results from two independent transfection experiments.
ISG15 silencing enhances the type I IFN-mediated antiviral effect against HEV.
ISG15 is also known to impart an immunomodulatory effect. Recent studies have reported increased resistance to viral infection in ISG15-deficient individuals (36, 40). Thus, we tested if a lack of ISG15 could affect HEV sensitivity to type I IFN, since HEV is known to be less sensitive to type I IFN and to downregulate type I IFN-induced ISG expression levels (19).
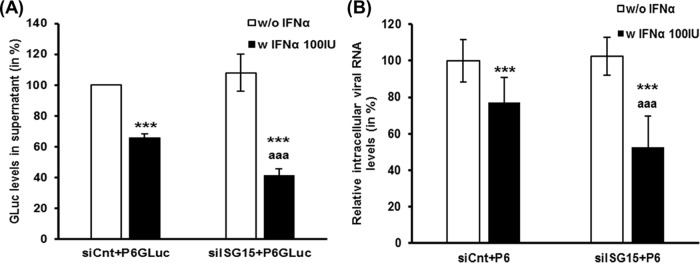
Huh7-S10-3 liver cells were cotransfected with capped RNA transcripts of HEV P6 or HEV P6GLuc along with siISG15 or siCnt. At 24 h posttransfection (hpt), the cells were treated with 100 IU/ml IFN-α or left untreated. The levels of GLuc activity in culture supernatant and of intracellular HEV RNA were determined at 5 dpt. We used a dose of IFN-α (100 IU/ml) throughout the study, as our preliminary experiment (data not shown) and previous studies in HCV and HEV had shown that 100 IU/ml IFN-α imparted a moderate antiviral effect against HEV replication (19).
The results showed that IFN-α treatment resulted in a significant decrease in HEV replication levels compared to IFN-α-untreated samples, as measured both in the HEV P6GLuc replicon system and in the HEV P6 infection system. Loss of ISG15 (i.e., siISG15 plus IFN-α) during IFN-α treatment resulted in a further enhancement of the IFN-α-mediated antiviral effect against HEV. We also observed a significant decrease in GLuc activity levels in culture supernatant (P ≤ 0.001; Fig. 3A) and in intracellular HEV RNA levels (P ≤ 0.01; Fig. 3B) in siISG15-plus-IFN-α-treated cells compared to the corresponding control siRNA-transfected samples (siCnt plus IFN-α).
FIG 3.
ISG15 silencing enhances the type I IFN-mediated antiviral effect against HEV. Huh7-S10-3 liver cells were cotransfected with 20 nM ISG15-targeted siRNA (siISG15) or with control siRNA (siCnt) along with HEV P6GLuc replicon RNA (A) or HEV P6 infectious cDNA clone viral genomic RNA (B) at 24 hpt. The cells were either left untreated (w/o IFN-α) or treated with 100 IU/ml IFN-α (w IFN-α 100IU), and the amounts of HEV RNA were monitored at 5 dpt. The GLuc activity levels in culture supernatants (A) and the intracellular HEV P6 viral RNA levels (B) were normalized to the IFN-α-untreated control levels. The data represent means ± SEM of results of n = 5 (A) and n = 2 (B) independent transfection experiments. ***, P ≤ 0.001 (compared to siCnt w/o IFN-α); aaa, P ≤ 0.001 (compared to siCnt w IFN-α100IU).
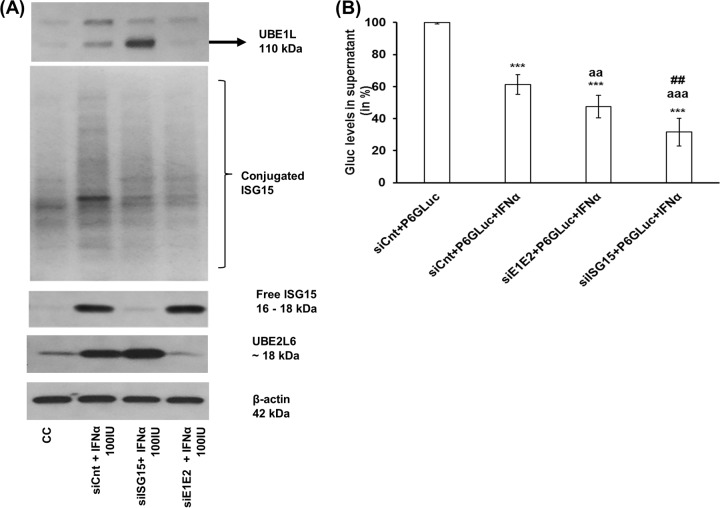
Furthermore, we performed additional experiments to evaluate the contribution of ISGylating enzymes, UBE1L (E1), and UBE2L6 (E2), to the type I IFN-mediated antiviral effect against HEV. Huh7-S10-3 liver cells were cotransfected with capped RNA transcripts of HEV P6GLuc and siRNA, and at 24 hpt the cells were either left untreated or treated with IFN-α (100 IU/ml). The levels of GLuc activity in culture supernatant were estimated at 5 dpt. The results showed that knockdown of UBE1L and UBE2L6 (siE1E2) during IFN-α treatment led to a significant loss of ISGylation of proteins in the cells whereas free ISG15 levels remained similar to those of siCnt-transfected samples (Fig. 4A). Interestingly, we found that the loss of UBE1L and UBE2L6 did not result in an enhanced type I IFN-mediated antiviral effect against HEV P6GLuc as drastic as that of ISG15 knockdown (Fig. 4B).
FIG 4.
Loss of UBE1L and UBE2L6 did not result in an enhanced type I IFN-mediated antiviral effect against HEV. (A) ISG15 protein levels as measured by Western blot analysis in IFN-α-treated cells during ISG15 knockdown (siISG15) or during UBE1L (UBE1L) and UBE2L6 (UBE2L6) double knockdown (siE1E2). Huh7-S10-3 liver cells were transfected with 20 nM siCnt or siISG15 or siE1E2. At 24 hpt, cells were treated with 100 IU/ml IFN-α. Cell lysates were collected at 24 h post-IFN-α treatment to measure conjugated as well as free-form ISG15. The ISG15 protein levels in cell lysates (40 μg/lane) were analyzed by Western blotting using anti-ISG15 antibody (1:750 dilution). The data are representative of the results of one of three independent experiments. (B) GLuc activity levels in culture supernatant at 5 dpt. Cells were cotransfected with siCnt/siISG15/siE1E2 along with HEV P6GLuc replicon RNA; at 24 hpt, the cells were treated with 100 IU/ml IFN-α. The relative GLuc activity levels were determined compared to the IFN-α-untreated control levels. The data represent means ± SEM of n = 4 independent transfection experiments. ***, P ≤ 0.001 (compared to siCnt plus HEV P6GLuc); aaa, P ≤ 0.001; aa, P ≤ 0.01 (compared to siCnt plus P6GLuc plus IFN-α); ##, P ≤ 0.01 (compared to siE1E2 plus HEV P6GLuc plus IFN-α). Statistical analysis was done using ANOVA and a post hoc Tukey test.
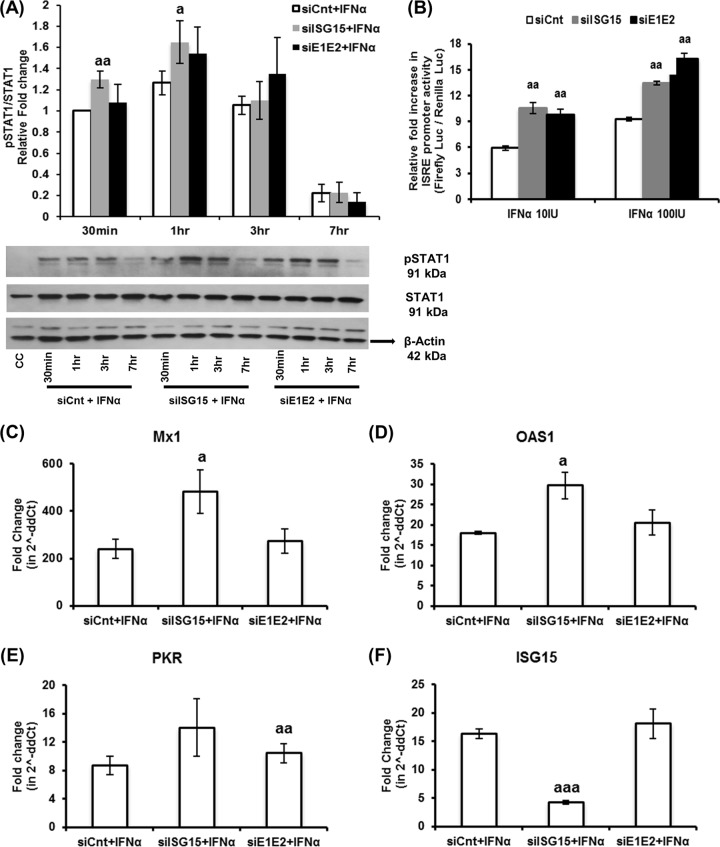
ISG15 regulates type I IFN signaling in Huh7-S10-3 liver cells.
We already showed that a complete loss of ISG15, i.e., of both the conjugated and unconjugated forms of ISG15, led to a substantial increase in the type I IFN-mediated antiviral effect against HEV. Therefore, we conducted further experiments to evaluate type I IFN signaling under those conditions. In agreement with previous studies (35, 40), we observed that loss of ISG15 in Huh7-S10-3 liver cells resulted in enhanced type I IFN signaling as determined by increased pSTAT1 levels (P ≤ 0.05; Fig. 5A) and enhanced luciferase activity in an IFN-stimulated response element (ISRE) promoter assay (P ≤ 0.05; Fig. 5B). Consequently, significant upregulations of type I IFN-induced ISG (i.e., Mx1, OAS1, and PKR) mRNA levels (P ≤ 0.05; Fig. 5C to E) were also observed in these samples. While UBE1L and UBE2L6 silencing did not result in a significant increase in ISG mRNA expression levels compared to that in control samples (Fig. 5C to E), a significant increase in ISRE promoter activity in these samples was observed (Fig. 5B).
FIG 5.
ISG15 regulates type I IFN signaling. (A) Relative pSTAT1 levels. Huh7-S10-3 liver cells were transfected with 20 nM control siRNA (siCnt) or ISG15-siRNA (siISG15) or UBE1L-siRNA plus UBE2L6-siRNA (siE1E2). At 24 hpt, cells were treated with IFN-α (100 IU/ml) for 30 min to 7 h. Cell lysate (20 μg/lane) was analyzed by Western blotting with the indicated antibody, anti-pSTAT1 (1:1,000 dilution), anti-STAT1 (1:1,000 dilution), and anti-β-actin (1:1,000 dilution). Fold change in band intensity was determined using ImageJ software (NIH, Bethesda, MD). The data represent means ± SEM of results from five independent experiments. a, P ≤ 0.05; aa, P ≤ 0.01 (compared with siCnt plus IFN-α at the given time point). (B) IFN-stimulated response element (ISRE) promoter activity levels. Huh7-S10-3 liver cells were cotransfected with siCnt/siISG15/siE1E2 along with pGL4.45[luc2P/ISRE/Hygro] (firefly luciferase) and pGL4.74[hRluc/TK] (renilla luciferase). At 24 hpt, cells were treated with various concentrations of IFN-α. Relative levels of fold induction of the cell-associated firefly luciferase activity, compared to the corresponding untreated cell control levels, at 18 h post-IFN-α treatment were estimated using a dual-luciferase assay kit and were normalized with renilla luciferase expression levels. The data represent means ± SEM of results from triplicate sample experiments. aa, P ≤ 0.01 (compared to siCnt plus IFN-α). (C to F) ISG mRNA levels in siRNA-transfected and IFN-α-treated samples were measured for Mx1 (C), OAS1 (D), PKR (E), and ISG15 (F) using Sybr green qPCR. Fold change in mRNA levels compared to the untransfected control was calculated using the 2−ΔΔCT method, and the RPS18 gene was used as the housekeeping gene. The data represent means ± SEM of results from three independent experiments. a, P ≤ 0.05; aa, P ≤ 0.01; aaa, P ≤ 0.001 (compared to siCnt plus IFN-α).
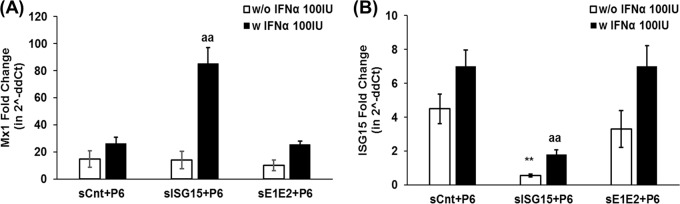
A similar trend was observed in HEV RNA-transfected samples with type I IFN treatment. We found that ISG15 knockdown resulted in enhanced Mx1 levels compared to those in UBE1L and UBE2L6 double-knockdown and control samples at 5 dpt (P ≤ 0.01; Fig. 6). Therefore, we can conclude that the loss of ISG15, but not UBE1L and UBE2L6, results in an enhanced type I IFN signaling-induced ISG expression level, which in turn might enable increased HEV sensitivity to type I IFN. Therefore, the results suggest that ISG15 might play an immunomodulatory role during HEV infection. It is also plausible that HEV upregulates ISG15 to negatively regulate type I IFN signaling for its benefit.
FIG 6.
Loss of ISG15 increases type I IFN-induced Mx1 levels in HEV RNA-transfected Huh7-S10-3 liver cells. (A and B) Mx1 (A) and ISG15 (B) mRNA levels in Huh7-S10-3 liver cells cotransfected with capped RNA from the HEV P6 infectious cDNA clone and siRNA. At 24 hpt, cells were either left untreated (w/o IFN-α 100IU) or treated with 100 IU/ml IFN-α (w IFN-α 100IU). Fold change in mRNA levels compared to the untransfected control levels was calculated using the 2−ΔΔCT method, and the RPS18 gene was used as the housekeeping gene The data represent means ± SEM of results from three independent experiments. **, P ≤ 0.01 (compared to siCnt plus P6); aa, P ≤ 0.01 (compared to siCnt plus P6 plus IFN-α using the Student t test).
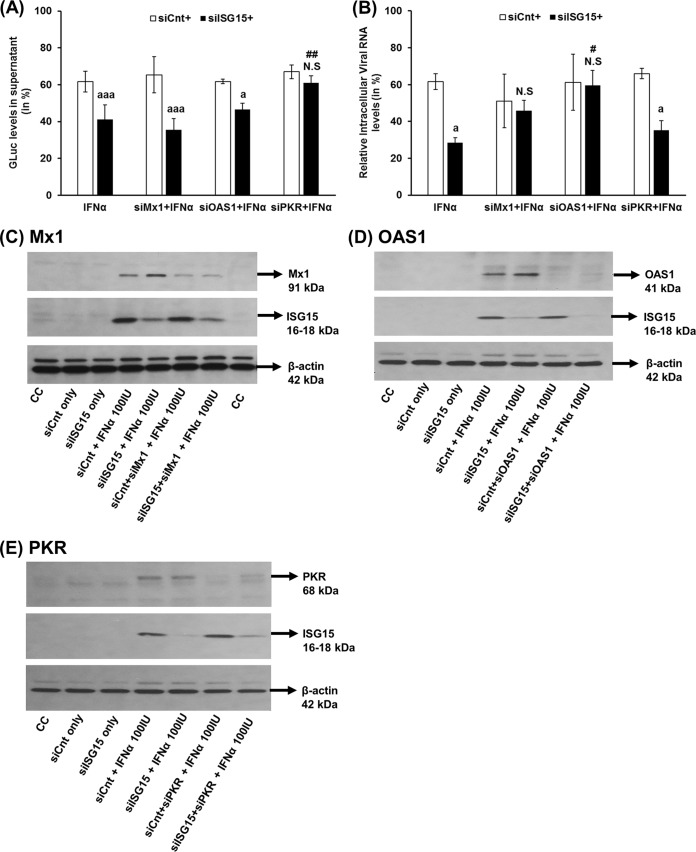
ISG15 silencing mediates an enhanced antiviral effect of IFN-α against HEV via PKR and OAS1.
Since loss of ISG15 led to a significant increase of IFN-α signaling and a significant increase in OAS1, Mx1, and PKR mRNA levels, we decided to further examine if these proteins are involved in the type I IFN-mediated antiviral effect against HEV. We specifically selected these three potential ISGs because type I IFN mediates the antiviral state by targeting various stages of viral replication, and the mRNA levels of these ISGs were significantly upregulated in HEV-infected pig liver tissue samples (data not shown).
Huh7-S10-3 liver cells were cotransfected with a combination of siRNA and capped RNA transcripts of HEV P6GLuc. At 24 hpt, the cells were either treated with IFN-α (100 IU/ml) or left untreated. At 5 dpt, the HEV replication levels were monitored using the GLuc assay. As observed in our previous experiment (Fig. 3A), ISG15 knockdown resulted in a significant increase in the type I IFN-mediated antiviral effect against HEV (Fig. 7A). A double knockdown of PKR and ISG15 resulted in loss of the IFN sensitivity of HEV. The GLuc activity levels in siPKR plus siISG15 plus IFN-α (61.1% ± 3.6%) were similar to those seen with siCnt plus siPKR plus IFN-α (66.9% ± 3.6%), which was statistically significant compared to the results seen with the siISG15-plus-IFN-α samples (41.1% ± 7.9%; P ≤ 0.01). However, we found that loss of PKR by itself did not affect HEV type I IFN sensitivity, as the GLuc activity levels in siCnt plus siPKR plus IFN-α (66.9 ± 3.6%) were similar to that of siCnt plus IFN-α (61.7% ± 5.6%). Additionally, loss of OAS1 or Mx1 in ISG15 knockdown cells did not result in the restoration of GLuc activity to the levels seen with the corresponding control cells. The knockdown efficiency of various siRNAs is shown in Fig. 7C to E.
FIG 7.
ISG15 silencing mediates an enhanced antiviral effect of IFN-α against HEV via PKR and OAS1. Huh7-S10-3 liver cells were cotransfected with HEV RNA and control siRNA (siCnt)/ISG15-siRNA (siISG15), along with siRNA targeted against Mx1 (siMx1) or OAS1 (siOAS1) or PKR (siPKR). At 24 hpt, cells were either left untreated or treated with 100 IU/ml IFN-α. (A) GLuc activity levels in cell culture supernatant at 5 dpt. (B) Intracellular HEV P6 viral RNA levels. The relative changes in GLuc/viral RNA levels were calculated with respect to the corresponding IFN-α-untreated sample. (C to E) Representative Western blot analyses to show the siRNA knockdown efficiency. The Huh7-S10-3 liver cells were cotransfected with control siRNA (siCnt)/ISG15-siRNA (siISG15), along with siRNA targeted against Mx1 (siMx1) (C) or OAS1 (siOAS1) (D) or PKR (siPKR) (E). At 24 hpt, cells were either left untreated or treated with 100 IU/ml IFN-α. After 24 h post-IFN-α treatment, Mx1/PKR/OAS1 protein levels were estimated using Western blot analyses to determine the siRNA knockdown efficiency. The data represent means ± SEM of results from three independent experiments (A) and duplicate cultures (B). a, P ≤ 0.05; aaa, P ≤ 0.001 (compared to siCnt plus IFN-α); #, P ≤ 0.05; ##, P ≤ 0.01 (compared to siISG15 plus IFN-α). N.S, not significant.
To further verify the findings with the replicon system, we performed the same experiment with the HEV P6 infection system to determine if any of these ISGs would affect intracellular HEV RNA levels during type I IFN treatment. Similarly to what was observed in the previous experiment (Fig. 3B), we found that ISG15 knockdown resulted in a significant increase in the type I IFN-mediated antiviral effect against HEV (Fig. 7B). This enhancement in HEV sensitivity to type I IFN is lost during double knockdown of ISG15 with OAS1 or Mx1. In IFN-α-treated samples, loss of OAS1/Mx1 along with ISG15 resulted in an increase in viral RNA levels similar to that seen with the corresponding controls. Intracellular HEV RNA levels in siOAS1 plus siISG15 plus IFN-α (59.53% ± 8.1%) were significantly higher than the levels in siISG15 plus IFN-α (28.43% ± 2.76%; P ≤ 0.05). Interestingly, unlike the HEV P6GLuc system, a double knockdown of ISG15 and PKR during IFN-α treatment (siPKR plus siISG15 plus IFN-α; 35.1% ± 5.2%) in the HEV P6 infection system did not result in a significant increase in the levels of intracellular HEV RNA compared to siISG15 plus IFN-α (28.43% ± 2.76; P = 0.18). Therefore, PKR and OAS1 might play a critical role in ISG15-mediated type I IFN sensitivity to HEV, depending on the study system.
DISCUSSION
HEV is known to be less sensitive to the type I IFN-mediated antiviral effect than HCV (19, 21). High baseline levels of ISG15 have been reported in populations of nonresponders to IFN therapy in patients chronically infected with HCV (34, 41). In the present study, we demonstrated that HEV induces ISG15 and plays an important immunomodulatory role by regulating HEV sensitivity to type I IFN.
High levels of ISG15 expression have been reported in HEV infections of chimpanzee liver tissues (28), in patients with chronic HEV infection (25), and in other persistent hepatic viral infections (24). In vitro studies have also shown that HEV upregulates ISG15 expression levels in A549 and hepatic cells (26, 27), as well as in primary cell cultures of induced pluripotent stem cell-derived hepatocytes (42). Consistent with previous reports, in this study we observed an increase in ISG15 expression at both the mRNA and protein levels in Huh7-S10-3 cells transfected with capped HEV RNA transcripts. Some clones of Huh7 cells such as Huh7.5 cells are known to be RIG-I defective; however, we found that the Huh7-S10-3 cells used in this study expressed RIG-I protein (data not shown). A recent study suggested that an antiviral response can be established independently of type I IFN (43), and an in vitro study has shown an IFN-independent mechanism of a RIG-I-mediated anti-HEV effect (44). Therefore, it is plausible that HEV may use the basal RIG-I mechanism (or another mechanism) to induce ISG15 production in hepatocytes. One of the major drawbacks of studying innate responses to HEV is the lack of an efficient infectious cell culture system. HepG2, Huh7.5, and Huh7-S10-3 hepatic cell lines are known to support differential levels of HEV replication (27). In our previous publications (45, 46), we had used the HepG2 cell line for monitoring HEV infection; however, we found that the HEV infection level in the Huh7-S10-3 infection system is insufficient for ISG15 analyses. Hence, we estimated ISG15 levels in HEV-infected swine liver samples to substantiate our in vitro findings obtained from an in vitro viral RNA transfection-based model but not from in vitro HEV infection of Huh7-S10-3 cells.
The role of ISG15 during a viral infection is complex and is dependent on the host species (22). In vitro studies in human cell lines have shown that ISG15 implements a direct antiviral effect by inhibiting budding of the human pathogens HIV (15) and Ebola virus VP40 (16). However, ISG15-deficient populations do not seem to have impaired antiviral immunity; instead, they display higher levels of expression of ISGs and greater resistance to viral infection (22, 23). The results from this study demonstrate that ISG15 plays an immunomodulatory role rather than a direct antiviral role during HEV replication.
ISG15 is known to mediate a direct antiviral effect via ISGylation (22); thus, we suspected that ISG15 knockdown might affect HEV replication. However, our results showed that the loss of ISG15 alone had no effect on HEV replication. A recent study has shown that overexpression of RIG-I in Huh7.5-P6 HEV cells led to induction of various ISGs (including ISG15); however, type I IFN remained at undetectable levels (44). Therefore, we speculate that the inability to detect any significant changes in HEV levels in siISG15 plus HEV compared to siCnt plus HEV cotransfected Huh7-S10-3 cells was likely due to the ability of HEV to induce an innate response in Huh7 cells independently of type I IFN production.
We further demonstrated here that loss of ISG15 resulted in an improved type I IFN signaling response to exogenous IFN-α treatment and, consequently, enhanced the type I IFN sensitivity of HEV, as observed both in the HEV P6GLuc replicon system and in the HEV P6 infectious cDNA clone system. We also observed that loss of ISGylating enzymes (UBE1L and UBE2L6) did not affect the HEV type I IFN sensitivity. It has been reported that rescuing ISG15-deficient cells with a conjugation-deficient ISG15 mutant can attenuate the type I IFN sensitivity of a cell (36, 40). Free ISG15 is known to stabilize USP18 protein expression levels and thereby to control STAT1 signaling (40). Therefore, it is plausible that the presence of free ISG15 in UBE1L- and UBE2L6-deficient cells resulted in a decrease in levels of IFN-induced ISGs and thus may contribute to the loss of HEV type I IFN sensitivity.
Type I IFN signaling establishes an antiviral state by inducing various ISGs, each of which acts at various stages of viral replication, including early stages post-viral entry, viral transcription, translation, and/or assembly and budding (47). Thus, in this study we tested the roles of OAS1, Mx1, and PKR in the type I IFN-mediated antiviral effect against HEV. OAS1 imparts an antiviral effect via enabling viral double-stranded RNA (dsRNA) degradation in an RNase L-dependent or -independent manner (48, 49), thereby inhibiting viral RNA replication. Mx1 is a GTPase and imparts an antiviral effect at a very early stage of viral replication, although the exact mechanism is not well understood (47). PKR is known to inhibit viral and/or host RNA translation by phosphorylating the α subunit of eukaryotic initiation factor 2 (eIF2A) to prevent viral replication (47). Single nucleotide polymorphisms (SNPs) in PKR and OAS1 have also been suspected to correlate with chronic viral infections (50, 51).
In this study, we showed that PKR and OAS1 are critical in ISG15-mediated enhanced HEV sensitivity to type I IFN. We demonstrated that loss of ISG15, along with PKR, during type I IFN treatment resulted in a significant increase in GLuc levels in the HEV P6GLuc replicon system. Therefore, we suspected that loss of PKR would result in an increased level of viral protein and that this might in turn increase the intracellular viral genomic RNA replication levels. However, we did not observe a significant increase in intracellular viral RNA levels in the HEV P6 infection system, while loss of OAS1 resulted in an increase in intracellular HEV viral RNA levels in ISG15 knockdown and type I IFN-treated cells. It has been shown that, in rabies virus, loss of PKR increases viral protein levels without affecting viral mRNA levels (52). Therefore, we speculate that ISG15 may regulate the type I IFN-mediated antiviral effect against HEV in a multistep process. Both PKR and OAS1 appear to be crucial for ISG15-mediated type I IFN sensitivity to HEV, depending on the HEV study system used. Therefore, it is plausible that PKR is a critical regulator of HEV translation and that OAS1 controls HEV RNA stability in a mutually independent manner. Clearly, further studies are warranted to delineate the potential correlation of these ISGs during acute versus chronic HEV infection.
In conclusion, the results from this study suggest that ISG15 may play an important immunomodulatory role in HEV replication and that it regulates HEV sensitivity to exogenous type I IFN. Loss of ISG15 resulted in enhanced HEV type I IFN sensitivity and IFN-induced ISGs levels during HEV replication. The results also suggest that PKR and OAS1 are crucial for the ISG15-mediated IFN sensitivity of HEV, depending on the HEV study system used. Therefore, care needs to be taken in studying the IFN mechanism during HEV infection. Development of more-robust HEV infection cell culture systems is needed to enable better studies of HEV innate response interactions, as such a robust culture system would mimic ex vivo conditions during HEV replication more closely than the currently used HEV transfection-based model. Understanding the expression profile of negative regulators and the subsequent mechanism involved in the regulation of type I IFN signaling pathway, for a given disease state, would enable us to formulate a better IFN therapy regime.
MATERIALS AND METHODS
Cells, immunological reagents, and plasmids.
The human hepatoma cell line Huh7-S10-3 (a gift of Suzanne U. Emerson, NIAID, NIH, Bethesda, MD) was maintained in Dulbecco's modified Eagle's medium (DMEM) (Gibco, USA) supplemented with 10% fetal bovine serum (Atlanta Biologicals, USA), penicillin (100 IU/ml), and streptomycin (100 μg/ml) under 5% CO2 at 37°C. Recombinant human interferon alpha (IFN-α; catalog no. 11100-1) and antibodies specific for ISG15 (catalog no. 21900-1; 1:1,000 dilution) were purchased from PBL Assay Science (Piscataway, NJ). Mx1 (catalog no. sc-271399; 1:1,000 dilution), β-actin (catalog no. sc-1616; 1:1,000 dilution), and STAT1 (catalog no. sc-345; 1:1,000 dilution) were all purchased from Santa Cruz Biotech (Santa Cruz, CA). UBE2L6 (catalog no. ab109086; 1:1,000 dilution) was purchased from AbCam (Cambridge, MA) and pSTAT1 (catalog no. 7649S; 1:1,000 dilution) from Cell Signaling Technologies (Danvers, MA). The plasmids used for the luciferase promoter assay included pGL4.45[luc2P/ISRE/Hygro] and pGL4.74[hRluc/TK], and both were purchased from Promega (Madison, WI).
Transfection with HEV infectious cDNA clone and HEV replicon clone.
The genotype 3 Kernow-C1 P6 HEV infectious cDNA clone (designated “HEV P6”) or the HEV P6GLuc replicon (designated “HEV P6GLuc”) (37) was linearized with MluI. Capped RNA transcripts from HEV P6 or HEV P6GLuc were transcribed in the presence of an m(7)G cap analog using a mMESSAGE mMACHINE T7 Ultra kit (Ambion, Grand Island, NY) per the manufacturer's protocol. The Huh7-S10-3 human liver cells were transfected with capped HEV-P6 RNA or HEV-P6GLuc RNA using DMRIE-C reagent (Invitrogen, Grand Island, NY) according to the manufacturer's protocol.
siRNA-mediated knockdown.
siRNAs targeted against ISG15, UBEL1, PKR, Mx1, and OAS1 were purchased from Santa Cruz Biotech (Santa Cruz, CA), and UBE2L6 was purchased from SignalChem (Richmond, Canada). Huh7-S10-3 liver cells were transiently transfected with 20 nM siRNA using Lipofectamine 2000 or DMRIE-C reagent (Invitrogen, Grand Island, NY).
Dual-luciferase reporter assay.
To monitor type I IFN signaling, Huh7-S10-3 liver cells were transiently cotransfected with 100 ng of pGL4.45[luc2P/ISRE/Hygro] (firefly luciferase) and 5 ng of pGL4.74[hRluc/TK] (renilla luciferase) along with 20 nM siRNA using Lipofectamine 2000. After 24 hpt, the cells were stimulated with IFN-α for an additional 18 h. Cell lysate were prepared and assayed for luciferase using a dual-luciferase assay kit (Promega, Madison, WI). The firefly luciferase activity values were normalized using renilla luciferase values, and the relative fold changes in luciferase activity in IFN-treated samples compared to IFN-untreated control were calculated.
Western blot analysis.
Cells were lysed using radioimmunoprecipitation assay (RIPA) buffer (Millipore, Billerica, MA) containing protease and phosphatase inhibitor (Thermo, Grand Island, NY). Equal amounts (20 to 40 μg) of protein were subjected to SDS-PAGE and transferred to a polyvinylidene difluoride (PVDF) membrane. The membrane was blocked using 5% bovine serum albumin (BSA; Fisher, Grand Island, NY) diluted in phosphate-buffered saline plus 0.1% Tween 20 (PBST) for 1 h and then incubated overnight with specific primary antibody at 4°C. The membranes were then washed with PBST and probed with corresponding secondary horseradish peroxidase (HRP) antibody. The membranes were then developed using ECL substrate (Santa Cruz Biotech, CA). The relative levels of band intensity were determined with ImageJ software (NIH, Bethesda, MD).
Real-time quantitative RT-PCR for quantification of HEV RNA.
The levels of HEV RNA in cell culture supernatant and the levels of intracellular HEV RNA were determined using real-time quantitative RT-PCR as described previously (45). Briefly, viral RNAs were extracted from cell culture supernatant and cell lysates using Tri Reagent (Molecular Research Center, Cincinnati, OH) according to the manufacturer's protocol. Quantitative real-time RT-PCR was performed using a SensiFAST real-time PCR kit (Bioline, USA) and HEV ORF2-specific primers and probe (Table 1).
TABLE 1.
Oligonucleotide primer sequences used for qRT-PCR in the studya
| Primer ID | Primer sequence (5′–3′) |
|---|---|
| ISG15-Fwd | GTGGACAAATGCGACGAACC |
| ISG15-Rev | TCGAAGGTCAGCCAGAACAG |
| OAS1-Fwd | GGAGACCCAAAGGGTTGGAG |
| OAS1-Rev | GTGTGCTGGGTCAGCAGAAT |
| Mx1-Fwd | AAGAGCCGGCTGTGGATATG |
| Mx1-Rev | TTTGGACTTGGCGGTTCTGT |
| PKR-Fwd | ACGTGTGAGTCCCAAAGCAA |
| PKR-Rev | AGGTCAAATCTGGGTGCCAA |
| RPS18-Fwd | TGATCCCTGAAAAGTTCCAGCA |
| RPS18-Rev | CTTCGGCCCACACCCTTAAT |
| Swine-ISG15-Fwd | TGAAGATGCTGGGAGGCAAG |
| Swine-ISG15-Rev | CACCCCATCCTGAAGCACAT |
| Swine-USP18-Fwd | TCCAGCCCAGGGAGTTGT |
| Swine-USP18-Rev | CTGTCCGCAGATTTTTGATG |
| Swine-RSP18-Fwd | CATCGACCTCACCAAGAGGG |
| Swine-RSP18-Rev | CCTGGCTGTACTTCCCATCC |
| HEV-Fwd | GGTGGTTTCTGGGGTGAC |
| HEV-Rev | AGGGGTTGGTTGGATGAA |
| HEV-Probe | 5′FAM-TGATTCTCAGCCCTTCGC3′BHQ |
BHQ, black hole quencher; FAM, 6-carboxyfluorescein; ID, identifier; qRT-PCR, quantitative reverse transcription-PCR.
Real time RT-PCR for ISG quantification.
Total cellular RNAs from Huh7-S10-3 liver cells and from swine liver tissues were isolated using Tri Reagent (Molecular Research Center, Cincinnati, OH) per the manufacturer's protocol. Approximately 1 μg of total RNA was reverse transcribed into cDNA using a High-Capacity cDNA RT kit (Applied Biosystems, Grand Island, NY) with random primers. The mRNA levels of ISG15, OAS1, PKR, Mx1, USP18, and RPS18 (housekeeping control) were determined using SYBR master mix (Applied Biosystems, Grand Island, NY) with gene-specific primer sets (Table 1) and a Bio-Rad IQ5 system. The PCR amplification conditions included 95°C for 2 min (1 cycle) and 35 cycles of 95°C for 10 s, 60°C for 30 s, and 72°C for 30 s. Fold changes in mRNA levels were normalized against the housekeeping gene and calculated using the threshold cycle (2−ΔΔCT) method.
Statistical analysis.
Statistical comparisons were performed using JMP software, and analysis of variance (ANOVA) was used with a post hoc Tukey honestly significant difference (HSD) test or the Student t test for multiple comparisons. P values of ≤0.05 were considered statistically significant.
ACKNOWLEDGMENTS
We thank Melissa Markis for her technical support in flow cytometry analysis. We also thank Scott P. Kenney and C. Lynn Heffron for their expert assistance in this project.
This study was supported by grants from the National Institutes of Health (R01AI050611 and R01AI074667).
REFERENCES
- 1.Smith DB, Simmonds P, International Committee on Taxonomy of Viruses Hepeviridae Study Group, Jameel S, Emerson SU, Harrison TJ, Meng XJ, Okamoto H, Van der Poel WH, Purdy MA. 2014. Consensus proposals for classification of the family Hepeviridae. J Gen Virol 95:2223–2232. doi: 10.1099/vir.0.068429-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thiry D, Mauroy A, Pavio N, Purdy MA, Rose N, Thiry E, de Oliveira-Filho EF. 2017. Hepatitis E virus and related viruses in animals. Transbound Emerg Dis 64:37–52. doi: 10.1111/tbed.12351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Purdy MA, Khudyakov YE. 2011. The molecular epidemiology of hepatitis E virus infection. Virus Res 161:31–39. doi: 10.1016/j.virusres.2011.04.030. [DOI] [PubMed] [Google Scholar]
- 4.Meng XJ. 2016. Expanding host range and cross-species infection of hepatitis E virus. PLoS Pathog 12:e1005695. doi: 10.1371/journal.ppat.1005695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Takahashi M, Nishizawa T, Nagashima S, Jirintai S, Kawakami M, Sonoda Y, Suzuki T, Yamamoto S, Shigemoto K, Ashida K, Sato Y, Okamoto H. 2014. Molecular characterization of a novel hepatitis E virus (HEV) strain obtained from a wild boar in Japan that is highly divergent from the previously recognized HEV strains. Virus Res 180:59–69. doi: 10.1016/j.virusres.2013.12.014. [DOI] [PubMed] [Google Scholar]
- 6.Okamoto H. 2011. Hepatitis E virus cell culture models. Virus Res 161:65–77. doi: 10.1016/j.virusres.2011.01.015. [DOI] [PubMed] [Google Scholar]
- 7.Cao D, Meng XJ. 2012. Molecular biology and replication of hepatitis E virus. Emerg Microbes Infect 1:e17. doi: 10.1038/emi.2012.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kapur N, Thakral D, Durgapal H, Panda SK. 2012. Hepatitis E virus enters liver cells through receptor-dependent clathrin-mediated endocytosis. J Viral Hepat 19:436–448. doi: 10.1111/j.1365-2893.2011.01559.x. [DOI] [PubMed] [Google Scholar]
- 9.Yin X, Ambardekar C, Lu Y, Feng Z. 2016. Distinct entry mechanisms for nonenveloped and quasi-enveloped hepatitis E viruses. J Virol 90:4232–4242. doi: 10.1128/JVI.02804-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yin X, Li X, Feng Z. 18 August 2016. Role of envelopment in the HEV life cycle. Viruses doi: 10.3390/v8080229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nagashima S, Jirintai S, Takahashi M, Kobayashi T, Tanggis Nishizawa T, Kouki T, Yashiro T, Okamoto H. 2014. Hepatitis E virus egress depends on the exosomal pathway, with secretory exosomes derived from multivesicular bodies. J Gen Virol 95:2166–2175. doi: 10.1099/vir.0.066910-0. [DOI] [PubMed] [Google Scholar]
- 12.Lozano R, Naghavi M, Foreman K, Lim S, Shibuya K, Aboyans V, Abraham J, Adair T, Aggarwal R, Ahn SY, Alvarado M, Anderson HR, Anderson LM, Andrews KG, Atkinson C, Baddour LM, Barker-Collo S, Bartels DH, Bell ML, Benjamin EJ, Bennett D, Bhalla K, Bikbov B, Bin Abdulhak A, Birbeck G, Blyth F, Bolliger I, Boufous S, Bucello C, Burch M, Burney P, Carapetis J, Chen H, Chou D, Chugh SS, Coffeng LE, Colan SD, Colquhoun S, Colson KE, Condon J, Connor MD, Cooper LT, Corriere M, Cortinovis M, de Vaccaro KC, Couser W, Cowie BC, Criqui MH, Cross M, et al. 2012. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet 380:2095–2128. doi: 10.1016/S0140-6736(12)61728-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Borg BB, Feng Z, Earl TM, Anderson CD. 2016. Hepatitis E in post-liver transplantation: is it time to routinely consider it? Clin Transplant 30:975–979. doi: 10.1111/ctr.12777. [DOI] [PubMed] [Google Scholar]
- 14.Kamar N, Rostaing L, Izopet J. 2013. Hepatitis E virus infection in immunosuppressed patients: natural history and therapy. Semin Liver Dis 33:62–70. doi: 10.1055/s-0033-1338115. [DOI] [PubMed] [Google Scholar]
- 15.Geng Y, Wang Y. 2016. Epidemiology of hepatitis E. Adv Exp Med Biol 948:39–59. [DOI] [PubMed] [Google Scholar]
- 16.Hui W, Wei L, Li Z, Guo X. 2016. Treatment of hepatitis E. Adv Exp Med Biol 948:211–221. [DOI] [PubMed] [Google Scholar]
- 17.Nan Y, Ma Z, Wang R, Yu Y, Kannan H, Fredericksen B, Zhang YJ. 2014. Enhancement of interferon induction by ORF3 product of hepatitis E virus. J Virol 88:8696–8705. doi: 10.1128/JVI.01228-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nan Y, Yu Y, Ma Z, Khattar SK, Fredericksen B, Zhang YJ. 2014. Hepatitis E virus inhibits type I interferon induction by ORF1 products. J Virol 88:11924–11932. doi: 10.1128/JVI.01935-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Todt D, Francois C, Anggakusuma Behrendt P, Engelmann M, Knegendorf L, Vieyres G, Wedemeyer H, Hartmann R, Pietschmann T, Duverlie G, Steinmann E. 2016. Antiviral activities of different interferon types and subtypes against hepatitis E virus replication. Antimicrob Agents Chemother 60:2132–2139. doi: 10.1128/AAC.02427-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dong C, Zafrullah M, Mixson-Hayden T, Dai X, Liang J, Meng J, Kamili S. 2012. Suppression of interferon-alpha signaling by hepatitis E virus. Hepatology 55:1324–1332. doi: 10.1002/hep.25530. [DOI] [PubMed] [Google Scholar]
- 21.Zhou X, Xu L, Wang W, Watashi K, Wang Y, Sprengers D, de Ruiter PE, van der Laan LJ, Metselaar HJ, Kamar N, Peppelenbosch MP, Pan Q. 2016. Disparity of basal and therapeutically activated interferon signalling in constraining hepatitis E virus infection. J Viral Hepat 23:294–304. doi: 10.1111/jvh.12491. [DOI] [PubMed] [Google Scholar]
- 22.Hermann M, Bogunovic D. 2017. ISG15: in sickness and in health. Trends Immunol 38:79–93. doi: 10.1016/j.it.2016.11.001. [DOI] [PubMed] [Google Scholar]
- 23.Zhang D, Zhang DE. 2011. Interferon-stimulated gene 15 and the protein ISGylation system. J Interferon Cytokine Res 31:119–130. doi: 10.1089/jir.2010.0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.MacQuillan GC, Mamotte C, Reed WD, Jeffrey GP, Allan JE. 2003. Upregulation of endogenous intrahepatic interferon stimulated genes during chronic hepatitis C virus infection. J Med Virol 70:219–227. doi: 10.1002/jmv.10381. [DOI] [PubMed] [Google Scholar]
- 25.Moal V, Textoris J, Ben Amara A, Mehraj V, Berland Y, Colson P, Mege JL. 2013. Chronic hepatitis E virus infection is specifically associated with an interferon-related transcriptional program. J Infect Dis 207:125–132. doi: 10.1093/infdis/jis632. [DOI] [PubMed] [Google Scholar]
- 26.Devhare PB, Chatterjee SN, Arankalle VA, Lole KS. 2013. Analysis of antiviral response in human epithelial cells infected with hepatitis E virus. PLoS One 8:e63793. doi: 10.1371/journal.pone.0063793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Devhare PB, Desai S, Lole KS. 2016. Innate immune responses in human hepatocyte-derived cell lines alter genotype 1 hepatitis E virus replication efficiencies. Sci Rep 6:26827. doi: 10.1038/srep26827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu C, Boon D, McDonald SL, Myers TG, Tomioka K, Nguyen H, Engle RE, Govindarajan S, Emerson SU, Purcell RH. 2010. Pathogenesis of hepatitis E virus and hepatitis C virus in chimpanzees: similarities and differences. J Virol 84:11264–11278. doi: 10.1128/JVI.01205-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Okumura A, Lu G, Pitha-Rowe I, Pitha PM. 2006. Innate antiviral response targets HIV-1 release by the induction of ubiquitin-like protein ISG15. Proc Natl Acad Sci U S A 103:1440–1445. doi: 10.1073/pnas.0510518103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Okumura A, Pitha PM, Harty RN. 2008. ISG15 inhibits Ebola VP40 VLP budding in an L-domain-dependent manner by blocking Nedd4 ligase activity. Proc Natl Acad Sci U S A 105:3974–3979. doi: 10.1073/pnas.0710629105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Domingues P, Bamford CG, Boutell C, McLauchlan J. 2015. Inhibition of hepatitis C virus RNA replication by ISG15 does not require its conjugation to protein substrates by the HERC5 E3 ligase. J Gen Virol 96:3236–3242. doi: 10.1099/jgv.0.000283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hsiang TY, Zhao C, Krug RM. 2009. Interferon-induced ISG15 conjugation inhibits influenza A virus gene expression and replication in human cells. J Virol 83:5971–5977. doi: 10.1128/JVI.01667-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen L, Sun J, Meng L, Heathcote J, Edwards AM, McGilvray ID. 2010. ISG15, a ubiquitin-like interferon-stimulated gene, promotes hepatitis C virus production in vitro: implications for chronic infection and response to treatment. J Gen Virol 91:382–388. doi: 10.1099/vir.0.015388-0. [DOI] [PubMed] [Google Scholar]
- 34.Broering R, Zhang X, Kottilil S, Trippler M, Jiang M, Lu M, Gerken G, Schlaak JF. 2010. The interferon stimulated gene 15 functions as a proviral factor for the hepatitis C virus and as a regulator of the IFN response. Gut 59:1111–1119. doi: 10.1136/gut.2009.195545. [DOI] [PubMed] [Google Scholar]
- 35.Chua PK, McCown MF, Rajyaguru S, Kular S, Varma R, Symons J, Chiu SS, Cammack N, Najera I. 2009. Modulation of alpha interferon anti-hepatitis C virus activity by ISG15. J Gen Virol 90:2929–2939. doi: 10.1099/vir.0.013128-0. [DOI] [PubMed] [Google Scholar]
- 36.Speer SD, Li Z, Buta S, Payelle-Brogard B, Qian L, Vigant F, Rubino E, Gardner TJ, Wedeking T, Hermann M, Duehr J, Sanal O, Tezcan I, Mansouri N, Tabarsi P, Mansouri D, Francois-Newton V, Daussy CF, Rodriguez MR, Lenschow DJ, Freiberg AN, Tortorella D, Piehler J, Lee B, Garcia-Sastre A, Pellegrini S, Bogunovic D. 2016. ISG15 deficiency and increased viral resistance in humans but not mice. Nat Commun 7:11496. doi: 10.1038/ncomms11496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shukla P, Nguyen HT, Faulk K, Mather K, Torian U, Engle RE, Emerson SU. 2012. Adaptation of a genotype 3 hepatitis E virus to efficient growth in cell culture depends on an inserted human gene segment acquired by recombination. J Virol 86:5697–5707. doi: 10.1128/JVI.00146-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Emerson SU, Nguyen H, Graff J, Stephany DA, Brockington A, Purcell RH. 2004. In vitro replication of hepatitis E virus (HEV) genomes and of an HEV replicon expressing green fluorescent protein. J Virol 78:4838–4846. doi: 10.1128/JVI.78.9.4838-4846.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cao D, Cao QM, Subramaniam S, Yugo DM, Heffron CL, Rogers AJ, Kenney SP, Tian D, Matzinger SR, Overend C, Catanzaro N, LeRoith T, Wang H, Piñeyro P, Lindstrom N, Clark-Deener S, Yuan L, Meng XJ. 19 June 2017. Pig model mimicking chronic hepatitis E virus infection in immunocompromised patients to assess immune correlates during chronicity. Proc Natl Acad Sci U S A doi: 10.1073/pnas.1705446114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang X, Bogunovic D, Payelle-Brogard B, Francois-Newton V, Speer SD, Yuan C, Volpi S, Li Z, Sanal O, Mansouri D, Tezcan I, Rice GI, Chen C, Mansouri N, Mahdaviani SA, Itan Y, Boisson B, Okada S, Zeng L, Wang X, Jiang H, Liu W, Han T, Liu D, Ma T, Wang B, Liu M, Liu JY, Wang QK, Yalnizoglu D, Radoshevich L, Uze G, Gros P, Rozenberg F, Zhang SY, Jouanguy E, Bustamante J, Garcia-Sastre A, Abel L, Lebon P, Notarangelo LD, Crow YJ, Boisson-Dupuis S, Casanova JL, Pellegrini S. 2015. Human intracellular ISG15 prevents interferon-alpha/beta over-amplification and auto-inflammation. Nature 517:89–93. doi: 10.1038/nature13801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Katsounas A, Hubbard JJ, Wang CH, Zhang X, Dou D, Shivakumar B, Winter S, Schlaak JF, Lempicki RA, Masur H, Polis M, Kottilil S, Osinusi A. 2013. High interferon-stimulated gene ISG-15 expression affects HCV treatment outcome in patients co-infected with HIV and HCV. J Med Virol 85:959–963. doi: 10.1002/jmv.23576. [DOI] [PubMed] [Google Scholar]
- 42.Helsen N, Debing Y, Paeshuyse J, Dallmeier K, Boon R, Coll M, Sancho-Bru P, Claes C, Neyts J, Verfaillie CM. 2016. Stem cell-derived hepatocytes: a novel model for hepatitis E virus replication. J Hepatol 64:565–573. doi: 10.1016/j.jhep.2015.11.013. [DOI] [PubMed] [Google Scholar]
- 43.Paludan SR. 2016. Innate antiviral defenses independent of inducible IFNalpha/beta production. Trends Immunol 37:588–596. doi: 10.1016/j.it.2016.06.003. [DOI] [PubMed] [Google Scholar]
- 44.Xu L, Wang W, Li Y, Zhou X, Yin Y, Wang Y, de Man RA, van der Laan LJ, Huang F, Kamar N, Peppelenbosch MP, Pan Q. 3 May 2017. RIG-I is a key antiviral interferon-stimulated gene against hepatitis E virus regardless of interferon production. Hepatology doi: 10.1002/hep.29105. [DOI] [PubMed] [Google Scholar]
- 45.Kenney SP, Wentworth JL, Heffron CL, Meng XJ. 2015. Replacement of the hepatitis E virus ORF3 protein PxxP motif with heterologous late domain motifs affects virus release via interaction with TSG101. Virology 486:198–208. doi: 10.1016/j.virol.2015.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kenney SP, Meng XJ. 2015. The lysine residues within the human ribosomal protein S17 sequence naturally inserted into the viral nonstructural protein of a unique strain of hepatitis E virus are important for enhanced virus replication. J Virol 89:3793–3803. doi: 10.1128/JVI.03582-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schneider WM, Chevillotte MD, Rice CM. 2014. Interferon-stimulated genes: a complex web of host defenses. Annu Rev Immunol 32:513–545. doi: 10.1146/annurev-immunol-032713-120231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kristiansen H, Gad HH, Eskildsen-Larsen S, Despres P, Hartmann R. 2011. The oligoadenylate synthetase family: an ancient protein family with multiple antiviral activities. J Interferon Cytokine Res 31:41–47. doi: 10.1089/jir.2010.0107. [DOI] [PubMed] [Google Scholar]
- 49.Kristiansen H, Scherer CA, McVean M, Iadonato SP, Vends S, Thavachelvam K, Steffensen TB, Horan KA, Kuri T, Weber F, Paludan SR, Hartmann R. 2010. Extracellular 2′–5′ oligoadenylate synthetase stimulates RNase L-independent antiviral activity: a novel mechanism of virus-induced innate immunity. J Virol 84:11898–11904. doi: 10.1128/JVI.01003-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Knapp S, Yee LJ, Frodsham AJ, Hennig BJ, Hellier S, Zhang L, Wright M, Chiaramonte M, Graves M, Thomas HC, Hill AV, Thursz MR. 2003. Polymorphisms in interferon-induced genes and the outcome of hepatitis C virus infection: roles of MxA, OAS-1 and PKR. Genes Immun 4:411–419. [DOI] [PubMed] [Google Scholar]
- 51.García-Álvarez M, Berenguer J, Jiménez-Sousa MA, Pineda-Tenor D, Aldámiz-Echevarria T, Tejerina F, Diez C, Vázquez-Morón S, Resino S. 2017. Mx1, OAS1 and OAS2 polymorphisms are associated with the severity of liver disease in HIV/HCV-coinfected patients: a cross-sectional study. Sci Rep 7:41516. doi: 10.1038/srep41516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nikolic J, Civas A, Lama Z, Lagaudriere-Gesbert C, Blondel D. 2016. Rabies virus infection induces the formation of stress granules closely connected to the viral factories. PLoS Pathog 12:e1005942. doi: 10.1371/journal.ppat.1005942. [DOI] [PMC free article] [PubMed] [Google Scholar]