Abstract
Photosynthetic unicellular organisms are considered as promising alternative protein sources. The aim of this study is to understand the extent to which these green sources differ with respect to their gross composition and how these differences affect the final protein isolate. Using mild isolation techniques, proteins were extracted and isolated from four different unicellular sources (Arthrospira (spirulina) maxima, Nannochloropsis gaditana, Tetraselmis impellucida, and Scenedesmus dimorphus). Despite differences in protein contents of the sources (27–62% w/w) and in protein extractability (17–74% w/w), final protein isolates were obtained that had similar protein contents (62–77% w/w) and protein yields (3–9% w/w). Protein solubility as a function of pH was different between the sources and in ionic strength dependency, especially at pH < 4.0. Overall, the characterization and extraction protocol used allows a relatively fast and well-described isolation of purified proteins from novel protein sources.
Keywords: Microalgae, cyanobacteria, single-cell protein, amino acid composition, carbohydrate composition, physicochemical properties
Introduction
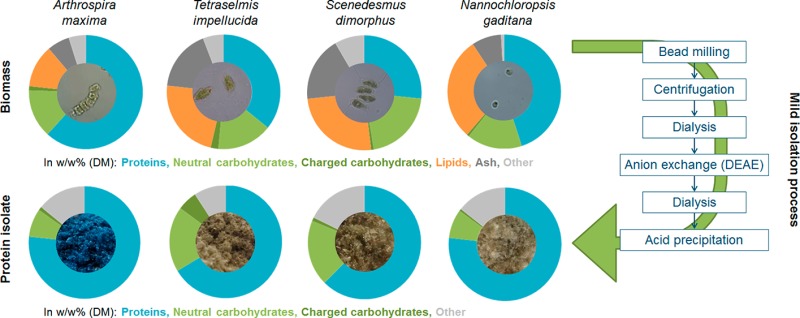
Photosynthetic single-cell organisms (microalgae and cyanobacteria) have received interest as potential alternative protein sources for the food and feed industry. These organisms belong to over 11 phyla and are biologically very diverse, ranging from marine prokaryotes to freshwater eukaryotes.1 Despite the interest, there is a lack of studies describing the detailed chemical composition of these organisms and of protein isolates that can be derived from them. The aim of this study is to understand the extent to which these green sources differ with respect to their gross composition, which is relevant for the feed industry. Second, we aim to understand how these differences affect the final protein isolates, which could be later applied in the food industry. The protein isolates obtained were studied with respect to their chemical composition and techno-functional properties. Four different unicellular sources (Arthrospira (spirulina) maxima, Nannochloropsis gaditana, Tetraselmis impellucida, and Scenedesmus dimorphus), encompassing in total 3 different phyla, were used to extract and further isolate proteins.
The research approach for proteins from these unicellular sources can be expected to develop the same way as the approach that has been developed in the past 50 years for proteins from seeds from leguminous plants, like soy, pea, and lupines. These legumes are biologically related, and studies showed that they contain similar types or classes of proteins. Leguminous proteins include the well-known multimeric vicilin (7S) and legumin (11S) globulin fractions that, in soy, account for >80% of the total proteins.2,3 It is known, however, that differences in nonprotein compounds present in legume seeds, like high contents of starch (e.g., pea) and oil (e.g., soy), necessitate changes in protein isolation procedures.4 In addition, significant differences have been found between the techno-functional properties and the thermostability of protein isolates obtained from various legumes.4 These differences are in part due to impurities caused by differences in the legumes’ biomass composition but are also partly due to differences in the intrinsic molecular properties of the proteins. For example, the multimeric state of leguminous proteins makes them quite distinctly different from, for instance, the monomeric whey proteins. For the study of proteins from unicellular sources, an example should be taken from these past studies on leguminous proteins. Similar to leguminous sources, the gross composition of unicellular green sources like microalgae and cyanobacteria varies greatly (Table 1). Extreme differences in composition between species have been reported, with values for protein and carbohydrate contents ranging from 6% to 72% (w/w dry matter) and from 8% to 64% (w/w dry matter), respectively.5,6 It is important to note that the reported differences within one species, due to differences in growing or harvesting conditions, can be at least as large as the differences between species.6,7 The variation within the composition of the cyanobacterium Arthrospira sp. and the microalgae Nannochloropsis sp., Scenedesmus sp., and Tetraselmis sp. is shown in Table 1. It should be noted that part of this variation may be caused by the different methods used in the literature to measure protein, carbohydrate, and lipid contents.
Table 1. Gross Chemical Composition of Microalgae and Cyanobacteria [% w/w] on a Dry Weight Basis.
| protein |
carbohydrate |
lipid |
ash |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| species | phylum | mean ± SD | rangen | mean ± SD | rangen | mean ± SD | rangen | mean ± SD | rangen | total | ref |
| Arthrospira | Cyanobacteria | ||||||||||
| A. platensis | 35a ± 10 | 21–43 | 36e ± 22 | 11–66 | 7h ± 2 | 4–8 | n.d. | 63–92 | (60) | ||
| 35a ± 7 | 20–43 | 33e ± 18 | 9–66 | 7h ± 2 | 4–13 | n.d. | 59–99 | (60) | |||
| 65b ± 5 | 59–72 | 15e ± 4 | 11–20 | 7I ± 0.4 | 6–7 | n.d. | 85–89 | (6) | |||
| A. maxima | 67b ± 3 | 63–70 | 14e ± 4 | 10–20 | 6I ± 1 | 6–7 | n.d. | 86–90 | (6) | ||
| Arthrospira sp. | 47a | 14e | 11j | 8 | 80 | (27) | |||||
| Nannochloropsis | Ochrophyta | ||||||||||
| N. gaditana | 44a ± 5 | 32–51 | n.d. | n.d. | 27k ± 4 | 20–30 | 8 ± 2 | 5–14 | 66–89 | (61) | |
| 52a,m ± 9 | 37–59 | 21e,m ± 5 | 16–27 | 27k,n ± 6 | 21–36 | n.d. | 100m | (62) | |||
| 41c | 25f | 26l | n.d. | 92 | (63) | ||||||
| Tetraselmis | Chlorophyta | ||||||||||
| T. impellucidao | 36d | 24g | 19I | 15 | 94 | (19) | |||||
| T. chuii | 31d | 12e | 17k | n.d. | 60 | (25) | |||||
| Tetraselmis sp. | 26b | 9e | 14k | 14 | 63 | (26) | |||||
| 30b | 8e | 13k | 17 | 68 | (26) | ||||||
| Scenedesmus | Chlorophyta | ||||||||||
| S. dimorphus | 50a | 6e | 23k | 2 | 83 | (33) | |||||
| S. obliquus | 31a ± 13 | 11–43 | 28e ± 10 | 16–44 | 19j ± 3 | 16–24 | 10 ± 5 | 5–18 | 79–98 | (27) | |
| S. almeriensis | 44c | 25f | 25l | n.d. | 94 | (63) | |||||
Protein content determined by Lowry after alkaline treatment.
Protein content determined by N*6.25.
Protein content determined by combustion with TGA-MS.
Protein content determined by amino acid composition.
Carbohydrate content determined by Dubois.
Carbohydrate content determined by combustion with TGA-MS.
Carbohydrate content determined by acid hydrolysis with HPAEC.
Lipid content determined by sulfo-phospho-vanillin.
Lipid content determined by Soxhlet.
Lipid content determined by Folch.
Lipid content determined by Bligh and Dyer.
Lipid content determined by combustion with TGA-MS.
Expressed as percentage of the organic fraction.
Range due to varying culture conditions.
Tetraselmis sp. used by Schwenzfeier et al. was later confirmed by the supplier to be T. impellucida. N.d. Not determined.
Similar to leguminous seeds, various proteins in microalgae and cyanobacteria are from similar classes and types. This means that they will share certain intrinsic molecular properties (e.g., multimeric state), which are important for their techno-functional properties. For example, all photosynthetic organisms contain a form of the enzyme ribulose-1,5-biphosphate carboxylase/oxygenase (Rubisco) which catalyzes carbon fixation. In microalgae and cyanobacteria, it is present in the so-called form I, which consists of 8 large and 8 small subunits.8 No post-translational modifications and prosthetic groups of Rubisco have been reported in online databases.9 (Uniprot search terms: rbcS/cbbS genes in Arthrospira sp., Nannochloropsis sp., Scenedesmus sp., and Tetraselmis sp. Accession numbers used: D4ZVW5, W6SIC7, K1VV20, A0A023PJK0, and K9ZWI1. Uniprot search terms: rbcL/cbbL genes in Arthrospira sp., Nannochloropsis sp., Scenedesmus sp., and Tetraselmis sp. Accession numbers used: T1RH29, Q3S3D2, B5VXI0, D4ZVW7, Q1KVV0, and K9ZV74.). The Mw of Rubisco’s large subunit over most reported species seems to be quite constant, and in N. gaditana, T. impellucida, S. dimorphus, and A. maxima the Mw is found to be between 52 and 54 kDa.9 (Uniprot search terms: rbcL/cbbL genes in Arthrospira sp., Nannochloropsis sp., Scenedesmus sp., and Tetraselmis sp. Accession numbers used: T1RH29, Q3S3D2, B5VXI0, D4ZVW7, Q1KVV0, and K9ZV74.) The small subunit is more variable in size and structure between species than the large subunit10 and is known to have a Mw range of 10–17 kDa in these genera.9 (Uniprot search terms: rbcS/cbbS genes in Arthrospira sp., Nannochloropsis sp., Scenedesmus sp., and Tetraselmis sp. Accession numbers used: D4ZVW5, W6SIC7, K1VV20, A0A023PJK0, and K9ZWI1.) Additionally, photosynthetic organisms contain various proteins that are active in light harvesting. In microalgae, these proteins are associated with the light harvesting complexes (LHC). The major LHC protein in N. gaditana is the violaxanthin–chlorophyll, a binding protein (VCP), with a Mw of 22 kDa.11,12 Other LHC proteins in N. gaditana also have molecular weights in the 21–32 kDa range.9 (Uniprot search terms: LHC genes in N. gaditana. Accession numbers used: K8YPQ7, W7TX20, W7UAI7, W7T6P5, W7TFG9, W7TZB5, W7TTD7, W7UBF0, W7U2H0, and W7TCK1.) LHC proteins of Tetraselmis sp. and of Scenedesmus have molecular weights of 24–44 and 26–27 kDa, respectively.9 (Uniprot search terms: LHC genes in Tetraselmis sp. Accession numbers used: A0A061RA39, A0A061RJR5, A0A061SK82, A0A061S745, A0A061SA24, A0A061R6B3, A0A061R2N8, A0A061S1P5, A0A061R213, A0A061S9W9, and O22496. Uniprot search terms: LHC genes in Scenedesmus sp. Accession numbers used: A2SY33, A2SY34, A2SY35, and A2SY32.) The LHC proteins of these sources are expected to be multimeric, similar to the LHC-II proteins from spinach. Spinach LHC-II proteins are trimers, where each monomer consists of 10 polypeptide chains each (PDB ID 1RWT).13 These proteins can form supercomplexes with photosystem II via antenna proteins.14 Cyanobacteria do not contain LHCs but synthesize blue pigmented phycocyanins for light harvesting.1 These multimeric phycocyanics have subunits with molecular masses between 15 and 22 kDa.15 Overall, Rubisco and the light harvesting proteins/phycocyanins in the four unicellular sources are all multimeric and have monomeric units in the same size range (15–54 kDa). It is therefore expected that these proteins will behave the same during protein extraction and isolation as a function of ionic strength (association/dissociation of the multimers) and dialysis.
Few studies have been performed on mild protein extraction from microalgae and cyanobacteria. Devi et al. reported an aqueous protein extraction from defatted Arthrospira (Spirulina) platensis with a yield up to 85%.16 Postma et al. also performed a mild extraction of protein and reported a Chlorella vulgaris protein extractability of 32–42%.17 Ursu et al. reported a soluble protein yield of 35% [w/w] from C. vulgaris using high-pressure cell disruption (2700 bar) at pH 7.18 Schwenzfeier et al. reported a T. impellucida protein extractability of 21% [w/w] under mild conditions, with a final protein isolate yield of 7% ([w/w] and protein isolate purity of 64% [w/w].19 Most studies published on protein extraction from microalgae and cyanobacteria, however, involve harsh chemical or physical treatments to disintegrate the cells, which affect the quality of the proteins. By using harsh chemicals (e.g., organic solvents) or physical treatments (e.g., high temperatures), proteins can lose their native tertiary structure or can be hydrolyzed to peptides or amino acids. This will affect the application possibilities in foods, for which techno-functional properties like good solubility, emulsification, and gelling behavior are desired. For example, heating has been shown to reduce protein solubility in alfalfa leaves, whereas acid precipitation can retain protein solubility.20 In this study, the aim was to isolate the proteins in a structure as close to the native structure as possible to provide a baseline observation of the intrinsic properties of the proteins.
For this study, protein sources were selected from three different unicellular photosynthetic phyla: one cyanobacterium (Arthrospira maxima), one heterokontophyta (N. gaditana), and two chlorophyta (T. impellucida and S. dimorphus). A mild isolation technique was used to avoid possible negative effects to the structure and conformational state of the proteins.
Materials and Methods
Materials
Nonviable samples of N. gaditana (NAN), S. dimorphus (SCE), and A. (spirulina) maxima (ART) were kindly provided by AlgaSpring (Almere, The Netherlands) as a frozen paste (microalgae) or a dried powder (cyanobacteria). Nonviable T. impellucida (TET, Instant Algae, strain CCMP892) was purchased from Reed Mariculture (Campbell, CA, USA) as a frozen paste. The TET material was the same product that was used in the work by Schwenzfeier et al.19 The growing conditions of the biomass samples were not provided by the suppliers. All samples were stored frozen (−20 °C) prior to use. All chemicals used were of analytical grade and purchased from either Merck (Darmstadt, Germany) or Sigma-Aldrich (St. Louis, MO, USA) unless stated otherwise. All water was obtained from a Milli-Q system (Millipore, Billerica, MA, USA) unless stated otherwise.
Protein Isolation
Protein was isolated from the microalgae and cyanobacteria using the isolation method described before.19 Algae paste or cyanobacteria powder was diluted or dispersed to 12% w/w dry matter in a potassium phosphate buffer with a final pH of 8.0 and a final concentration of 50 mM. The cells were disrupted using an agitation bead mill DYNO-Mill type MULTI LAB (Willy A. Bachofen Maschinenfabrik, Muttenz, Switzerland). The bead milling (recirculation) time was adjusted for each source. The ART, TET, NAN, or SCE samples were recirculated for 20, 30, 45, or 60 min, respectively, per 1 L sample, using a set pump speed of 1.5 L/min and a tube inner diameter of 0.8 cm. These times were used to reach complete cell disruption for each source, as confirmed by microscopic analyses. The 0.3 L grinding chamber was filled with 190 mL (approximately 65% [v/v]) yttria-stabilized zirconia SiLiBeads grinding beads, type ZY Premium, of 0.4–0.6 mm (Sigmund Lindner, Warmensteinach, Germany). Water cooled to 2 °C was recirculated through the cooling jacket of the grinding chamber, and the samples were kept on ice to ensure that the sample temperature at the bead mill outlet never exceeded 21 °C. The bead milled biomass was centrifuged (70 000g, 30 min, 4 °C) with exception of NAN. The NAN sample was first centrifuged at 16 000g (30 min, 4 °C) and then filtered using a Whatmann paper filter, and subsequently, the filtrate was centrifuged at 70 000g (30 min, 4 °C). The protein extractability was defined as the amount of protein in the supernatant (algae juice; AJ) divided by the amount of protein in the corresponding biomass × 100% (i.e., g protein in AJ/100 g protein in the biomass). The AJ of all samples was dialyzed (MWCO 12 000–14 000) against demineralized water and subsequently against a potassium phosphate buffer (“buffer A”, pH 7.6, 35 mM) at 4 °C to remove low Mw peptides and nonproteinaceous nitrogen. Each dialyzed algae juice (AJD) was applied on a glass filter (pore size 2) containing the anion exchange adsorbent Streamline DEAE (GE Healthcare, Uppsala, Sweden) in a volumetric ratio of 2:1. The DEAE was previously washed with an excess of demineralized water and then equilibrated with buffer A in a DEAE:buffer volumetric ratio of 1:2. The eluent was applied three times to ensure maximum protein binding (elution under gravity took 30–60 min). The DEAE was washed with buffer A in a DEAE:buffer volumetric ratio of 1:2. Bound protein was eluted by applying buffer A containing 2 M NaCl in a DEAE:buffer volumetric ratio of 1:2. The eluate was dialyzed (MWCO 12 000–14 000) against demineralized water and subsequently against buffer A at 4 °C, yielding the crude algae soluble protein isolate (CASPI). The CASPI was acidified to pH 3.5 with 1 M HCl and then kept at 4 °C for 1 h. The acidified CASPI was centrifuged at 4700g for 30 min at 4 °C. The pellet was redissolved in water by adjusting the pH to 7.6 with 1 M NaOH, and the algae soluble protein isolate (ASPI) obtained was freeze dried or stored frozen with 0.5 M sucrose. The protein isolation yield was defined as the amount of protein in each ASPI divided by the amount of protein in the corresponding biomass × 100% (i.e., g protein in ASPI/100 g protein in the biomass). At all isolation steps, aliquots of samples were freeze dried as such, and additional aliquots were stored frozen with 0.8 M sucrose for further analyses. ASPIs derived from ART, TET, NAN, and SCE will be further referred to as ASPI-A, ASPI-T, ASPI-N, and ASPI-S, respectively.
Compositional Analyses
All samples were freeze dried prior to analysis except for the aliquots needed for moisture content determination. All analysis results of the freeze-dried samples were expressed on a dry weight basis, assuming a residual moisture content of 10% after freeze drying (which was the typical moisture content measured in the freeze-dried biomass).
Dry Matter Content
Dry matter content of liquid samples was determined gravimetrically in triplicate by drying the samples overnight at 80 °C followed by 3 h at 105 °C.
Ash Content
Ash content was determined gravimetrically in triplicate by burning freeze-dried samples overnight at 550 °C. Ash content was additionally determined on washed biomass. For this, freeze-dried biomass was dispersed in water (6% w/w dry matter), stirred on a magnetic stirrer for 1 h, and subsequently centrifuged (10 min, 4,500 g, 20 °C). The supernatant was discarded, and the pellet was resuspended and centrifuged in the same manner two times. The washed biomass was oven dried (overnight at 80 °C followed by 3 h at 105 °C). Ash content was determined of the dried washed biomass.
Amino Acid Composition
Amino acid composition was determined in duplicate according to ISO method 13903:2005, with the exception of tryptophan. Analysis of tryptophan content was only performed for the biomass and not for the derived fractions. Tryptophan was determined in duplicate by a commercial laboratory (NutriControl, Veghel, The Netherlands). Standard deviations were found to be on average <0.5% of the mean. In the worst case the standard deviation was 11.8%.
Total Protein Content and Nitrogen to Protein Conversion Factors
Total nitrogen content was determined in triplicate with the Dumas method using a Flash EA 1112 N analyzer (Thermo Fisher Scientific, Waltham, MA, USA) and d-methionine for calibration. Nitrogen-to-protein (N-Prot) conversion factors kp and ka were calculated as described previously.19 The first N-Prot factor, kp, was calculated as the ratio between the sum of amino acid residues (total protein content) and total nitrogen content (including nonproteinaceous nitrogen). The second N-Prot factor, ka, was calculated as the ratio of the sum of amino acid residues (total protein content) to nitrogen from recovered amino acids (proteinaceous nitrogen only). Due to acid hydrolysis during amino acid quantification, asparagine (ASN) and glutamine (GLN) cannot be distinguished from (ASP) and glutamic acid (GLU). Therefore, the nitrogen recovered from amino acids was calculated assuming either 100% ASN/GLN or 100% ASP/GLU. Presented protein contents of samples are based on the total nitrogen contents and using the calculated N-Prot factors.
Lipid Content and Fatty Acid Composition
Lipid content was determined gravimetrically in duplicate according to Folch et al.21 Bead-milled biomass (1.5 g) was mixed with dichloromethane:methanol (2:1; 100 mL). The mixture was homogenized by sonication (20 s) and shaken for 2 h (200 rpm, 20 °C). Water (25 mL) was added to reach a methanol:dichloromethane:water ratio of 8:4:3, and the mixture was centrifuged (20 min, 4000g, 20 °C). The upper layer was removed, and the dichloromethane/pellet mixture was stored for 12 h at 4 °C. The mixture was paper filtered and flushed with dichloromethane. The dichloromethane was evaporated in a rotatory evaporator. Fatty acid composition was analyzed in duplicate on bead-milled biomass according to Breuer et al.22 In short, lipids were extracted with chloroform:methanol (ratio 4:5 v/v) followed by transesterification of the fatty acids to fatty acid methyl esters (FAMEs). FAMEs were quantified by GC-FID using a Nukol column, as described by Breuer et al. A triglyceride (C15:0) was used as an internal standard. The GC was calibrated using TraceCERT FAME standards purchased from Supelco (CRM18918, 18913-1AMP, and CRM18920, Supelco, Bellefonte, PA, USA). Annotation % of the fatty acids was calculated by assuming all unidentified GC peaks were unidentified FAMEs (<8% of total F). To quantify the unidentified FAMEs, molecular weights were used of FAMEs with similar retention times (<30 s difference).
Sugar Composition and Total Uronic Acid Content
Neutral carbohydrate composition was determined in triplicate according to the procedure by Englyst and Cummings using inositol as internal standard and a prehydrolysis with H2SO4 (72% w/w).23 Alditol acetates formed were analyzed by gas chromatography (Focus-GC, Thermo Scientific, Waltham, MA, USA) using arabinose, galactose, glucose, fucose, mannose, rhamnose, ribose, and xylose as standards. Total uronic acid content was determined in triplicate according to an automated colorimetric m-hydroxydiphenyl assay based on Ahmed et al.24 using an autoanalyzer (Skalar Analytical B.V., Breda, The Netherlands). Samples were prehydrolyzed as described in the neutral carbohydrate composition method. Adaptations to this method were the concentrations used of sodium tetraborate (23.7 mM) and m-hydroxydiphenyl (0.04% in 0.5% NaOH). Galacturonic acid (0–100 μg/mL) was used for calibration.
SDS-PAGE and Immunoblotting
SDS-PAGE was performed in duplicate under reducing conditions (10 mM β-mercapthoethanol) on a Mini-Protean II system (Bio-Rad Laboratories, Hercules, CA, USA) according to the manufacturer’s protocol. The PageRuler Plus Prestained Protein Ladder (Thermo Fisher Scientific, Waltham, MA, USA) was used as a molecular weight marker. Gels (Mini-Protean TGX) were either stained with Instant Blue coomassie stain (Expedeon, San Diego, CA, USA) or transferred to a 0.2 μm pore-size nitrocellulose membrane (Bio-Rad Laboratories) for immunoblotting. Immunoblot assays were carried out with standard reagents according to the protocol. Rabbit polyclonal antibodies against the large subunit of Rubisco (MBS715138, MyBioSource, San Diego, CA, USA) were detected with polyclonal goat antirabbit immunoglobulins conjugated with horseradish peroxidase (P0448, Dako, Carpinteria, CA, USA) using Clarity Western ECL (Bio-Rad Laboratories) as a substrate. To reduce the influence of coprecipitated soluble proteins in the insoluble fractions of the biomass, freeze-dried aliquots of the pellet fractions were washed with a potassium phosphate buffer (50 mM, pH 8.0) prior to analyzing them with SDS-PAGE.
Protein Solubility
ASPI of each alga or cyanobacterium was dispersed in Milli-Q water, and the pH was adjusted to 8.0. Samples that were not completely soluble were stirred overnight (4 °C). All samples were subsequently centrifuged (10 min, 10 000g, 20 °C), and the supernatant was used for further analyses. In all cases, > 80% of the protein was soluble, and the amount of ASPI dispersed was adapted per source to yield a final concentration of soluble proteins of 5 mg/mL in each supernatant. A buffer of 3.65 mM potassium phosphate, pH 7.6, was adjusted to ionic strengths of I = 0.01, 0.20, and 0.50 M with NaCl. The NaCl concentrations of the buffers were 0, 0.19, and 0.49 M, respectively. The ASPI supernatants were diafiltered with the potassium phosphate buffers of various ionic strengths. Subsequently, the protein solutions were adjusted to pH 2.0 using 1 M HCl, resulting in ionic strengths of I = 0.01 (SD = 0.002), 0.21 (SD = 0.006), and 0.49 (SD < 0.001) M. Using a pH-stat, the pH of the protein solutions was adjusted up to pH 8.5 with unit intervals of 0.5 using 0.2 M NaOH. At each pH, an aliquot of each protein solution was taken for further analyses. Actual pH and NaOH additions were recorded during the pH adjustments. The aliquots were kept at 4 °C for 1 h and subsequently centrifuged (10 min, 10 000g, 4 °C). The protein concentration of the supernatants was determined using the BCA protein assay (Pierce ThermoScientific, Waltham, MA USA). Protein concentrations calculated were corrected for the dilutions by NaOH titration and aliquots taken during the pH adjustments. Due to the pH adjustments, the final ionic strengths were calculated to be I = 0.01 (SD = 0.003), 0.19 (SD = 0.006), and 0.48 (SD = 0.010) M for samples with initial ionic strengths of I = 0.01, 0.20, and 0.50 M, respectively. At each ionic strength the protein solubility at pH 8.0 was set at 100%.
Results and Discussion
Before describing the protein isolation and the composition of the isolates obtained, it is important to consider the chemical composition of the biomass. This information is relevant for the extraction of the proteins for food applications but may also provide relevant information about the nonprotein compounds, which can be used for other applications such as the aquaculture industry.
Chemical Composition of the Biomass
For all samples 92–99% [w/w] of the total dry matter of the starting material was accounted for (annotated) in the gross compositional analysis (Table 2). Protein contents differed greatly between the four materials, with values of 61.7%, 45.0%, 35.8%, and 26.6% [w/w] measured for, respectively, ART, NAN, TET, and SCE. The total carbohydrate content was found to be quite similar for all sources, ranging between 15.1%and 21.5% [w/w], including between 0.7% and 2.2% [w/w] uronic acids. Total lipid contents ranged between 12.1% and 29.3% [w/w]. These gross composition analysis results fall within the ranges reported in the literature.6,19,25−27 It should be noted that growing and harvesting conditions can greatly influence the chemical composition of algae and cyanobacterial biomass.6,7
Table 2. Gross Chemical Composition of the Starting Materials [% w/w] on a Dry Weight Basis.
| component | A. maxima | N. gaditana | T. impellucida | S. dimorphus |
|---|---|---|---|---|
| proteinsa | 61.7 ± 0.5 | 45.0 ± 0.6 | 34.7 ± 0.1 | 26.6 ± 2.6 |
| carbohydrates | 15.1 ± 0.2 | 16.5 ± 0.2 | 17.9 ± 0.2 | 21.5 ± 0.2 |
| neutral | 13.7 ± 0.2 | 15.8 ± 0.3 | 15.7 ± 0.2 | 20.8 ± 0.3 |
| charged | 1.2 ± <0.1 | 0.6 ± <0.1 | 2.2 ± <0.1 | 0.7 ± <0.1 |
| lipidsb | 12.1 ± 0.2 | 29.3 ± 0.2 | 23.1 ± 0.7 | 25.2 ± 2.1 |
| ashc | 6.3 ± <0.1 | 8.4 ± 0.2 | 17.3 ± <0.1 | 18.2 ± <0.1 |
| total annotated | 95.2 | 99.1 | 93.0 | 91.5 |
On the basis of total amino acid analysis, i.e., including peptides and free amino acids.
Determined as MeOH/CH2Cl2 soluble material.
All measurements were performed on the biomass as such. Ash contents of washed biomass, thus excluding the contribution of extracellular material, were 2.9 ± 0.1%, 3.9 ± 0.0%, 11.8 ± 0.1%, and 16.7 ± 0.01% w/w for A. maxima, N. gaditana, T. impellucida, and S. dimorphus, respectively.
Fatty Acid Composition
The fatty acid composition of unicellular organisms is relevant for the nutritional quality of the sources, especially in the aquaculture industry.28 Specifically, the essential fatty acids and other omega-3 and -6 fatty acids are of relevance for assessing the nutritional quality. The two essential fatty acids were identified in the unicellular sources: linoleic acid (4–32 mol % of FAtot) and α-linolenic acid (13 and 32 mol % of FAtot in TET and SCE, respectively). Other omega-3 and -6 fatty acids present were eicosapentaenoic acid (EPA) and γ-linolenic acid. NAN and TET contained EPA (31 and 3 mol % of FAtot, respectively). ART and TET contained γ-linolenic acid (23 and 4 mol % of FAtot, respectively). No docosahexaenoic acid (DHA) was detected in the samples. Out of the four sources, NAN and TET can be considered interesting sources for aquaculture, because they contain EPA. Overall, all four samples samples contained high amounts of palmitic acid, i.e., palmitic acid accounts for 21–30 mol % of the total amount of fatty acids (FAtot) (Table 3). In addition, The contents and type of fatty acids that formed the majority of the FAtot in these sources, as indicated with an asterisk in Table 3, were similar to literature findings for NAN,29,30 TET,31,32 SCE,31,33 and ART.34,35
Table 3. Fatty Acid Composition of Biomass (% mol of total fatty acids; ± SD)a.
| shorthand | C14:0 | C14:1 cis9 | C16:0 | C16:1 | C16:2 | C16:3 | C16:4 | C18:0 | C18:1 n9 | C18:2 n6 |
|---|---|---|---|---|---|---|---|---|---|---|
| trivial name | myristic acid | myristoleic acid | palmitic acid | palmitoleic acid | hexadecadienoic acid | hexadeca-trienoic acid | hexadecatetraenoic acid | stearic acid | oleic acid | linoleic acid |
| A. maxima | 0.8 ± 0.2 | 0.8 ± 0.4 | 29.9 ± 4.6b | 3.1 ± 1.0 | n.d. | 0.9 ± 0.3 | n.d. | 1.1 ± 0.3 | 5.9 ± 0.5 | 32.2 ± 3.2b |
| N. gaditana | 7.2 ± <0.1 | 1.0 ± <0.1 | 23.5 ± 0.1b | 21.8 ± 0.2b | 0.6 ± <0.1 | 1.3 ± <0.1 | n.d. | n.d. | 4.4 ± <0.1 | 3.9 ± <0.1 |
| T. impellucida | 2.2 ± <0.1 | 1.5 ± <0.1 | 30.1 ± 0.5b | 5.0 ± 0.1 | 2.0 ± <0.1 | 3.2 ± <0.1 | 11.0 ± 0.1b | 0.9 ± <0.1 | 11.2 ± 0.1b | 7.5 ± 0.2b |
| S. dimorphus | 2.0 ± 0.3 | 1.8 ± 0.6 | 20.7 ± 0.3b | 11.1 ± 0.2 | n.d. | 3.4 ± 0.1 | 16.4 ± 0.3b | n.d. | 7.4 ± 0.4 | 5.2 ± 0.3 |
| shorthand | C18:3 n3 | C18:3 n6 | C18:4 | C20:1 | C20:2 n6 | C20:3 n3 | C20:5 n3 | C22:1 | |
|---|---|---|---|---|---|---|---|---|---|
| trivial name | α-linolenic acid | γ-linolenic acid | stearidonic acid | gondoic acid | eicosadienoic acid | eicosatrienoic acid | eicosapentaenoic acid (EPA) | erucic acid | total annotated (% w/w) |
| A. maxima | n.d. | 23.2 ± 1.7b | n.d. | n.d. | 2.2 ± 0.8 | n.d. | n.d. | n.d. | 98.1 |
| N. gaditana | n.d. | n.d. | n.d. | n.d. | n.d. | 5.3 ± <0.1 | 31.0 ± 0.2b | n.d. | 96.9 |
| T. impellucida | 12.6 ± 0.3b | 3.5 ± 0.1 | 3.4 ± 0.1 | 1.6 ± 0.4 | n.d. | 1.2 ± 0.1 | 2.8 ± 0.7 | 0.5 ± 0.2 | 92.1 |
| S. dimorphus | 32.1 ± 0.5b | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | n.d. | 100.0 |
n.d.: Not detected (<0.5 w/w% of total fatty acids).
Major fatty acids (sum up to >70 mol % of total fatty acids of the species).
Carbohydrate Composition
The carbohydrate composition is indicative of the types of oligo- and polysaccharides present in the unicellular sources. Oligo- and polysaccharides can act as fibers in food or feed but can also be copassengers during the isolation of proteins. Glucose and galactose were the major carbohydrate constituents (28–66 and 8–19 mol % of total carbohydrates, respectively) of all four starting materials (Table 4). Charged sugars (uronic acids) accounted for 3–11 mol % of the total carbohydrates in all starting materials. These uronic acids may form complexes with proteins during isolation, as was, for instance, shown in the emulsion properties of an algae protein isolate.36 In the microalgae (NAN, TET, and SCE) mannose was also a major carbohydrate (14–32 mol %), while it was only a minor part of the carbohydrates in ART (2 mol %). The high glucose content was expected, since in all four sources the storage carbohydrates are glucose-based polymers.37−40 In addition, the cell walls of N. gaditana(40,41) and S. dimorphus consist primarily of cellulose.42,43 Although the high mannose contents in NAN, TET, and SCE match literature values,19,44,45 the presence of mannose cannot be explained by storage carbohydrates or by the cell walls of these sources.41,42,46 Another difference between the samples was the rhamnose and ribose content. The rhamnose content was higher in ART and NAN than in TET and SCE (5–6 mol % and <1 mol %, respectively). The highest ribose amount was found in ART (10 mol %), compared to 6, 5, and 2 mol % in NAN, TET, and ART, respectively. Overall, the carbohydrate composition measured in the starting materials was similar to what has been described in literature, with high glucose, galactose, and mannose contents for T. impellucida (30%, 38%, and 7 mol %, respectively),19Nannochloropsis sp. (46, 17, and 34 mol %, respectively)44 and Scenedesmus sp. (38–70, 11–31, and 1–7 mol %, respectively).45 In Arthrospira sp., the major carbohydrate constituents are similar to the present findings and are reported to be glucose and galactose (59–74 and 10–20 mol %, respectively).47 The differences in carbohydrate composition between the sources can affect both the isolation process as well as the techno-functional properties of the proteins isolated.
Table 4. Monocarbohydrate Composition of Total Carbohydrates in Biomass [mol %; ± SD].
| Rha | Fuc | Ara | Xyl | Man | Gal | Glc | Rib | UA | |
|---|---|---|---|---|---|---|---|---|---|
| A. maxima | 5.81 ± 0.05 | 0.83 ± 0.11 | 1.24 ± 0.29 | 3.04 ± 0.12 | 1.85 ± 0.16 | 11.40 ± 0.13 | 59.06 ± 0.27 | 9.65 ± 0.27 | 7.12 ± 0.18 |
| N. gaditana | 4.63 ± 0.16 | 0.88 ± 0.12 | 1.47 ± 0.21 | 1.85 ± 0.09 | 14.29 ± 0.15 | 18.59 ± 0.15 | 48.66 ± 0.10 | 6.10 ± 0.14 | 3.53 ± 0.19 |
| T. impellucida | 0.78 ± 0.02 | 0.20 ± 0.06 | 2.60 ± 0.15 | 1.15 ± 0.10 | 32.41 ± 0.19 | 18.39 ± 0.05 | 28.42 ± 0.17 | 4.94 ± 0.16 | 11.11 ± 0.03 |
| S. dimorphus | 0.84 ± 0.02 | 0.61 ± 0.02 | 0.85 ± 0.10 | 1.54 ± 0.10 | 16.18 ± 0.27 | 7.79 ± 0.02 | 66.54 ± 0.27 | 2.46 ± 0.02 | 3.19 ± 0.08 |
Protein Composition
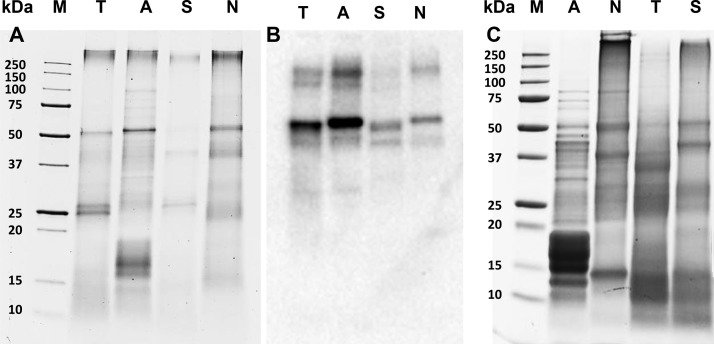
SDS-PAGE analysis showed major bands at ∼50 kDa and at >250 kDa in all sources (Figure 1 A). The ∼50 kDa band corresponds to the large subunit of Rubisco, as shown by immunoblotting (Figure 1B). The >250 kDa proteins are expected to be protein aggregates. Bands between 10 and 17 kDa detected in all sources are expected to represent the small subunit of Rubisco.9 (Uniprot search terms: rbcS/cbbS genes in Arthrospira sp., Nannochloropsis sp., Scenedesmus sp., and Tetraselmis sp. Accession numbers used: D4ZVW5, W6SIC7, K1VV20, A0A023PJK0, and K9ZWI1.) Other major proteins detected varied between sources and were 25–27 (TET), 15–18 (ART), 26 and 38 (SCE) and 15–20, 23, 30, and 39 kDa (NAN). Additionally, these sources contained various proteins that are part of the photosynthetic complex. The TET bands of 25–27 and ∼40 kDa are in the range of light harvesting complex (LHC) proteins reported for this genus (24–44 kDa).9 (Uniprot search terms: LHC genes in Tetraselmis sp. Accession numbers used: A0A061RA39, A0A061RJR5, A0A061SK82, A0A061S745, A0A061SA24, A0A061R6B3, A0A061R2N8, A0A061S1P5, A0A061R213, A0A061S9W9, and O22496.) The intense 15–18 kDa bands in ART match the molecular mass of phycocyanin subunits (15–22 kDa).15 The ∼26 kDa band found in SCE matches the reported presence of a 27 kDa LHC in Scenedesmus sp. (Uniprot search terms: LHC(x) genes in Scenedesmus sp. Accession numbers used: A2SY33, A2SY34, A2SY35, A2SY32).9,48 The ∼23 kDa band detected in NAN is expected to be the 22 kDa violaxanthin–chlorophyll a binding protein (VCP).11,12
Figure 1.
(A) SDS-PAGE gels stained with coomassie of bead-milled biomass, under reducing conditions and corresponding Western Blot, detecting rabbit polyclonal antibodies against the large subunit of Rubisco with polyclonal goat antirabbit immunoglobulins (B). (C) SDS-PAGE gels stained with coomassie of ASPIs (C). M = molecular weight marker, T = T. impellucida, A = A. maxima, S = S. dimorphus, and N = N. gaditana.
Amino Acid Composition
Despite differences in the protein composition of the four sources, the overall amino acid profiles of the starting materials were very similar to each other (Table 5). The standard deviations of the mean of 14 out of 18 analyzed amino acids were 4–14% among the various sources. The amino acids that showed the highest deviations between the sources were CYS, PRO, ARG, and TRP. The compositions measured are similar to what has been reported earlier.25 When comparing to common food protein sources, like soy and bovine milk,49−52 the microalgae and cyanobacterium amino acid compositions are more similar to soy proteins than to bovine milk (Figure 2). This is illustrated by the high linear regressions between the determined amino acid compositions and the literature values of microalgae,25 soy,49,50 and bovine milk51,52 with determination coefficients of R2 = 0.89, 0.82, and 0.66, respectively. Compared to bovine milk, the unicellular sources have proportionally one-half the amounts (in w/w% total amino acids) of GLX and proline and more than twice the amounts of GLY and ALA. Compared to soy proteins, the unicellular sources have over 60% more MET, ALA, and TRP and approximately 50% less HIS.
Table 5. Amino Acid Profile of Samples Obtained during the Different Isolation Steps of the Various Unicellular Sources [g/100 g protein] with According Standard Deviationsa.
| CYS | MET | ASX | THR | SER | GLX | GLY | ALA | PRO | |
|---|---|---|---|---|---|---|---|---|---|
| A. maxima | |||||||||
| biomass | 0.81 ± 0.01 | 2.51 ± 0.01 | 10.17 ± 0.04 | 5.11 ± 0.03 | 5.08 ± 0.04 | 14.63 ± 0.06 | 4.49 ± 0.01 | 7.61 ± 0.02 | 4.11 ± 0.09 |
| pellet | 0.77 ± <0.01 | 2.42 ± <0.01 | 10.63 ± 0.01 | 5.21 ± <0.01 | 5.12 ± <0.01 | 13.37 ± 0.04 | 4.68 ± <0.01 | 7.59 ± 0.01 | 4.29 ± 0.04 |
| AJ | 0.72 ± <0.01 | 2.69 ± 0.01 | 10.45 ± 0.01 | 5.39 ± 0.01 | 5.30 ± 0.02 | 13.73 ± 0.04 | 4.56 ± <0.01 | 7.52 ± <0.01 | 3.98 ± 0.03 |
| AJD | 0.87 ± <0.01 | 2.68 ± <0.01 | 10.20 ± <0.01 | 5.33 ± <0.01 | 5.21 ± <0.01 | 14.69 ± 0.01 | 4.65 ± <0.01 | 7.72 ± 0.01 | 3.92 ± 0.04 |
| CASPI | 1.04 ± 0.01 | 2.91 ± 0.02 | 11.23 ± 0.01 | 5.42 ± 0.01 | 5.14 ± <0.01 | 14.98 ± 0.01 | 4.50 ± <0.01 | 7.84 ± 0.03 | 3.62 ± 0.11 |
| ASPI | 1.01 ± <0.01 | 3.03 ± 0.01 | 10.93 ± 0.02 | 5.54 ± <0.01 | 5.27 ± 0.01 | 12.56 ± 0.09 | 4.72 ± <0.01 | 7.95 ± 0.02 | 3.66 ± 0.06 |
| N. gaditana | |||||||||
| biomass | 0.81 ± 0.01 | 2.37 ± 0.01 | 9.75 ± 0.04 | 5.10 ± 0.02 | 4.39 ± 0.01 | 13.21 ± 0.08 | 5.12 ± 0.01 | 7.45 ± 0.01 | 5.57 ± 0.13 |
| pellet | 0.71 ± 0.01 | 2.61 ± 0.02 | 9.96 ± 0.05 | 5.36 ± 0.01 | 4.59 ± 0.01 | 11.77 ± <0.01 | 5.61 ± <0.01 | 7.53 ± 0.01 | 5.38 ± <0.01 |
| AJ | 0.79 ± <0.01 | 2.63 ± <0.01 | 9.66 ± <0.01 | 5.35 ± 0.01 | 4.46 ± 0.01 | 11.81 ± 0.01 | 5.60 ± 0.01 | 7.62 ± 0.01 | 5.40 ± 0.04 |
| AJD | 0.72 ± <0.01 | 2.66 ± <0.01 | 10.00 ± 0.02 | 5.41 ± <0.01 | 4.55 ± <0.01 | 11.86 ± <0.01 | 5.57 ± <0.01 | 7.13 ± 0.01 | 4.82 ± 0.01 |
| CASPI | 1.22 ± 0.01 | 2.56 ± 0.01 | 11.33 ± 0.03 | 5.52 ± <0.01 | 4.58 ± 0.01 | 13.15 ± 0.01 | 5.02 ± <0.01 | 6.64 ± <0.01 | 4.48 ± 0.04 |
| ASPI | 0.97 ± 0.03 | 2.68 ± 0.01 | 10.83 ± 0.02 | 5.50 ± 0.01 | 4.54 ± 0.01 | 12.35 ± 0.01 | 5.43 ± 0.01 | 6.85 ± 0.01 | 4.49 ± 0.05 |
| T. impellucida | |||||||||
| biomass | 1.30 ± 0.01 | 2.71 ± <0.01 | 9.71 ± 0.02 | 4.95 ± 0.02 | 4.23 ± 0.03 | 12.01 ± 0.03 | 5.53 ± <0.01 | 8.78 ± <0.01 | 6.58 ± 0.08 |
| pellet | 1.17 ± 0.01 | 3.01 ± 0.01 | 10.00 ± 0.05 | 5.14 ± <0.01 | 4.66 ± 0.01 | 11.56 ± 0.04 | 5.70 ± <0.01 | 7.75 ± <0.01 | 5.41 ± 0.03 |
| AJ | 1.61 ± 0.01 | 2.53 ± 0.02 | 9.84 ± 0.01 | 4.87 ± <0.01 | 3.93 ± 0.01 | 13.10 ± 0.04 | 5.74 ± 0.01 | 11.00 ± 0.03 | 7.91 ± 0.01 |
| AJD | 1.24 ± <0.01 | 3.07 ± 0.01 | 10.80 ± 0.02 | 5.64 ± 0.01 | 4.83 ± <0.01 | 12.16 ± <0.01 | 5.58 ± 0.01 | 6.84 ± <0.01 | 5.05 ± 0.06 |
| CASPI | 1.79 ± 0.01 | 2.70 ± <0.01 | 12.41 ± <0.01 | 6.14 ± 0.02 | 5.10 ± 0.01 | 14.09 ± <0.01 | 5.19 ± 0.01 | 7.05 ± 0.01 | 4.84 ± 0.07 |
| ASPI | 1.69 ± <0.01 | 3.21 ± 0.01 | 11.37 ± 0.03 | 5.78 ± 0.01 | 4.84 ± <0.01 | 12.15 ± 0.03 | 4.95 ± <0.01 | 6.70 ± <0.01 | 4.63 ± 0.02 |
| S. dimorphus | |||||||||
| biomass | 1.14 ± 0.02 | 2.39 ± 0.01 | 11.10 ± 0.02 | 5.17 ± 0.03 | 4.66 ± 0.03 | 12.51 ± 0.02 | 5.81 ± 0.01 | 7.72 ± 0.03 | 5.41 ± 0.07 |
| pellet | 1.20 ± 0.01 | 2.31 ± 0.01 | 12.75 ± 0.07 | 5.55 ± <0.01 | 4.96 ± 0.02 | 11.83 ± 0.08 | 6.56 ± 0.01 | 7.62 ± 0.02 | 4.98 ± 0.12 |
| AJ | 0.96 ± <0.01 | 2.51 ± <0.01 | 10.89 ± 0.03 | 5.53 ± 0.01 | 4.42 ± <0.01 | 13.59 ± 0.04 | 5.96 ± 0.01 | 9.12 ± <0.01 | 5.15 ± 0.14 |
| AJD | 1.79 ± <0.01 | 2.69 ± 0.01 | 12.07 ± 0.02 | 5.92 ± <0.01 | 4.56 ± <0.01 | 12.61 ± 0.03 | 5.61 ± 0.01 | 7.24 ± 0.01 | 5.11 ± 0.03 |
| CASPI | 2.29 ± 0.02 | 2.53 ± 0.01 | 12.69 ± 0.06 | 5.86 ± <0.01 | 4.48 ± 0.02 | 13.45 ± 0.04 | 5.44 ± <0.01 | 7.11 ± <0.01 | 5.05 ± 0.01 |
| ASPI | 1.98 ± 0.01 | 2.83 ± 0.01 | 11.52 ± 0.02 | 5.73 ± 0.03 | 4.25 ± 0.02 | 11.86 ± 0.28 | 5.32 ± <0.01 | 6.87 ± 0.01 | 4.69 ± 0.05 |
| average biomass | 1.02 ± 0.25 | 2.49 ± 0.16 | 10.18 ± 0.65 | 5.08 ± 0.09 | 4.59 ± 0.37 | 13.09 ± 1.14 | 5.24 ± 0.58 | 7.89 ± 0.60 | 5.41 ± 1.01 |
| literature valuesb | |||||||||
| algae | 0.51 ± 0.08 | 2.15 ± 0.48 | 9.02 ± 0.68 | 5.00 ± 0.61 | 5.57 ± 0.81 | 10.84 ± 0.74 | 6.00 ± 0.32 | 7.62 ± 0.51 | 6.28 ± 2.17 |
| soy beans | 1.48 ± 0.14 | 1.49 ± 0.09 | 10.83 ± 1.15 | 4.21 ± 0.41 | 5.63 ± 0.82 | 15.82 ± 3.40 | 5.60 ± 1.81 | 4.76 ± 0.51 | 5.13 ± 0.42 |
| bovine milk | 0.73 ± 0.02 | 2.51 ± 0.12 | 7.37 ± 0.03 | 4.30 ± <0.01 | 5.22 ± 0.03 | 20.45 ± 0.05 | 1.92 ± 0.07 | 3.25 ± 0.09 | 9.20 ± 0.09 |
| VAL | ILE | LEU | TYR | PHE | HIS | LYS | ARG | TRP | |
|---|---|---|---|---|---|---|---|---|---|
| A. maxima | |||||||||
| biomass | 6.31 ± 0.04 | 5.85 ± 0.02 | 9.29 ± 0.01 | 4.28 ± 0.01 | 4.93 ± 0.02 | 1.82 ± 0.05 | 4.93 ± 0.02 | 6.58 ± 0.05 | 1.50 ± 0.04 |
| pellet | 6.37 ± 0.01 | 5.94 ± <0.01 | 9.70 ± <0.01 | 4.34 ± <0.01 | 5.73 ± 0.02 | 2.10 ± 0.01 | 4.69 ± <0.01 | 7.08 ± 0.01 | n.d. |
| AJ | 6.53 ± 0.02 | 6.32 ± 0.02 | 9.53 ± 0.01 | 4.82 ± <0.01 | 4.63 ± <0.01 | 1.68 ± 0.01 | 5.41 ± <0.01 | 6.72 ± <0.01 | n.d. |
| AJD | 6.36 ± 0.01 | 6.11 ± <0.01 | 9.32 ± <0.01 | 4.63 ± 0.01 | 4.60 ± <0.01 | 1.67 ± 0.02 | 5.38 ± <0.01 | 6.65 ± 0.01 | n.d. |
| CASPI | 5.56 ± 0.01 | 6.41 ± 0.01 | 8.84 ± 0.02 | 5.64 ± 0.03 | 4.08 ± 0.01 | 1.40 ± 0.01 | 4.90 ± 0.01 | 6.50 ± 0.02 | n.d. |
| ASPI | 5.68 ± 0.02 | 6.53 ± 0.01 | 9.23 ± 0.01 | 5.79 ± 0.04 | 4.35 ± <0.01 | 1.45 ± <0.01 | 5.12 ± 0.01 | 7.19 ± 0.01 | n.d. |
| N. gaditana | |||||||||
| biomass | 5.78 ± <0.01 | 4.90 ± 0.01 | 9.79 ± 0.01 | 3.45 ± 0.01 | 5.67 ± 0.02 | 2.14 ± 0.02 | 6.61 ± 0.02 | 5.60 ± 0.03 | 2.30 ± 0.05 |
| pellet | 6.34 ± 0.01 | 5.03 ± <0.01 | 10.55 ± <0.01 | 3.76 ± <0.01 | 5.94 ± <0.01 | 2.40 ± 0.01 | 6.98 ± <0.01 | 5.48 ± 0.01 | n.d. |
| AJ | 6.28 ± 0.01 | 4.99 ± <0.01 | 9.95 ± <0.01 | 3.98 ± <0.01 | 5.56 ± 0.01 | 2.37 ± 0.01 | 7.62 ± 0.02 | 5.92 ± 0.02 | n.d. |
| AJD | 6.34 ± <0.01 | 5.18 ± <0.01 | 10.36 ± 0.01 | 4.03 ± 0.01 | 5.79 ± 0.01 | 2.41 ± 0.01 | 7.22 ± 0.01 | 5.96 ± 0.01 | n.d. |
| CASPI | 6.19 ± <0.01 | 5.30 ± 0.01 | 9.13 ± 0.01 | 4.67 ± 0.02 | 5.09 ± 0.02 | 2.45 ± 0.03 | 6.74 ± 0.01 | 5.91 ± 0.03 | n.d. |
| ASPI | 6.26 ± 0.01 | 5.37 ± 0.01 | 9.51 ± 0.05 | 4.87 ± 0.14 | 5.31 ± 0.02 | 2.04 ± 0.02 | 6.85 ± 0.02 | 6.15 ± 0.06 | n.d. |
| T. impellucida | |||||||||
| biomass | 5.93 ± 0.02 | 4.51 ± 0.02 | 9.11 ± 0.01 | 3.54 ± 0.02 | 6.19 ± 0.10 | 2.28 ± 0.02 | 6.04 ± <0.01 | 4.40 ± 0.01 | 2.21 ± 0.05 |
| pellet | 6.14 ± 0.01 | 4.91 ± 0.01 | 10.03 ± 0.02 | 4.00 ± 0.05 | 6.94 ± <0.01 | 2.46 ± 0.02 | 5.97 ± 0.04 | 5.14 ± 0.01 | n.d. |
| AJ | 5.93 ± 0.02 | 4.29 ± 0.01 | 8.51 ± 0.01 | 2.98 ± 0.03 | 5.45 ± 0.04 | 2.15 ± <0.01 | 6.65 ± 0.01 | 3.51 ± 0.03 | n.d. |
| AJD | 6.21 ± 0.01 | 4.89 ± <0.01 | 9.72 ± 0.01 | 4.22 ± 0.01 | 6.73 ± <0.01 | 2.41 ± <0.01 | 5.88 ± <0.01 | 4.72 ± 0.01 | n.d. |
| CASPI | 6.65 ± 0.01 | 5.17 ± 0.11 | 9.17 ± 0.01 | 0.23 ± 0.03 | 5.58 ± 0.04 | 2.26 ± 0.02 | 6.54 ± 0.01 | 5.10 ± 0.01 | n.d. |
| ASPI | 6.32 ± <0.01 | 5.18 ± <0.01 | 9.17 ± 0.01 | 4.25 ± 0.02 | 5.68 ± <0.01 | 2.23 ± <0.01 | 6.46 ± <0.01 | 5.39 ± 0.02 | n.d. |
| S. dimorphus | |||||||||
| biomass | 5.80 ± 0.02 | 4.21 ± 0.01 | 9.13 ± 0.02 | 3.34 ± 0.01 | 5.86 ± 0.01 | 2.19 ± 0.02 | 6.03 ± 0.02 | 5.38 ± 0.03 | 2.14 ± 0.03 |
| pellet | 6.02 ± <0.01 | 4.52 ± <0.01 | 9.51 ± 0.01 | 2.20 ± 0.01 | 6.20 ± 0.03 | 1.45 ± 0.01 | 6.75 ± 0.01 | 5.58 ± 0.03 | n.d. |
| AJ | 6.62 ± 0.01 | 4.72 ± <0.01 | 9.03 ± 0.02 | 3.25 ± 0.03 | 5.23 ± 0.01 | 1.80 ± <0.01 | 6.43 ± 0.02 | 4.78 ± <0.01 | n.d. |
| AJD | 6.23 ± 0.01 | 4.67 ± <0.01 | 8.77 ± 0.01 | 3.65 ± <0.01 | 5.67 ± 0.01 | 2.34 ± <0.01 | 5.64 ± <0.01 | 5.41 ± 0.02 | n.d. |
| CASPI | 6.24 ± 0.01 | 4.78 ± <0.01 | 8.49 ± 0.01 | 3.72 ± <0.01 | 5.13 ± <0.01 | 2.10 ± 0.08 | 5.46 ± 0.01 | 5.19 ± 0.01 | n.d. |
| ASPI | 6.27 ± 0.01 | 4.95 ± 0.02 | 9.44 ± 0.03 | 4.04 ± 0.01 | 5.79 ± 0.04 | 2.11 ± 0.02 | 5.69 ± 0.02 | 6.65 ± 0.02 | n.d. |
| average biomass | 5.95 ± 0.24 | 4.87 ± 0.71 | 9.33 ± 0.32 | 3.65 ± 0.42 | 5.66 ± 0.53 | 2.11 ± 0.20 | 5.90 ± 0.70 | 5.49 ± 0.90 | 2.04 ± 0.37 |
| literature valuesb | |||||||||
| algae | 6.22 ± 0.26 | 4.74 ± 0.74 | 8.28 ± 0.55 | 4.40 ± 0.62 | 6.40 ± 0.54 | 1.88 ± 0.26 | 5.93 ± 0.35 | 7.88 ± 2.32 | 1.34 ± 0.26 |
| soy beans | 5.52 ± 0.88 | 4.51 ± 0.08 | 7.06 ± 1.08 | 4.12 ± 0.59 | 5.04 ± 0.20 | 3.78 ± 1.45 | 5.85 ± 0.94 | 8.11 ± 0.21 | 1.07 ± 0.01 |
| bovine milk | 6.13 ± 0.06 | 5.53 ± 0.02 | 9.19 ± 0.17 | 4.80 ± 0.04 | 4.66 ± 0.11 | 2.52 ± 0.01 | 7.50 ± 0.10 | 3.25 ± 0.04 | 1.47 ± 0.17 |
Figure 2.
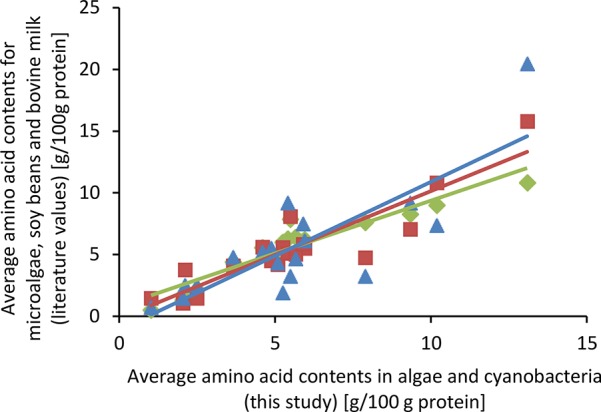
Comparison of average amino acid contents of various microalgae (green),25 soy beans (red),49,50 and bovine milk (blue)51,52 with A. maxima, N. gaditana, T. impellucida, and S. dimorphus (this study). Lines depict linear regressions with determination coefficients of R2 = 0.89, 0.82, and 0.66 for microalgae, soy beans, and bovine milk, respectively.
Nitrogen-to-Protein Conversion Factors
The nitrogen-to-protein conversion kp factors of the four sources ranged from 3.88 to 5.88 (Table 6), indicating that the use of the standard kp factor 6.25 would overestimate the protein contents of these samples up to 1.6 times. The ka factors obtained were much more similar among the four sources than the kp factors, with a lower limit (ASX/GLX = 100% ASP/GLU) ranging between 5.37 and 5.49 and an upper limit (ASX/GLX = 100% ASN/GLN) ranging between 6.30 and 6.37. This shows that the difference in N content between these sources is determined by variations in nonproteinaceous-nitrogen rather than by variations in the amino acid composition (and thus the ka factors). These differences are also reflected in the proteinaceous nitrogen to total nitrogen ratios (NAA/NT). ART was analyzed to have the highest NAA/NT of 80–98% in the biomass and SCE had the lowest NAA/NT of 62–72%.
Table 6. Proteinaceous Nitrogen and Nitrogen-to-Protein Conversion Factors ka and kp at Each Step of the Isolation Procedure of Each of the Unicellular Sources.
| processing step | NAA/NT [%]a,b | N-Prot factor kpc | N-Prot factor kad |
|---|---|---|---|
| A. maxima | |||
| biomasse | 80 < x < 95 | 5.08 | 5.37 < y < 6.35 |
| biomassf | 79 < x < 94 | 5.01 | 5.36 < y < 6.35 |
| pellet | 78 < x < 92 | 4.91 | 5.36 < y < 6.30 |
| algae juice | 78 < x < 92 | 4.97 | 5.38 < y < 6.34 |
| dialyzed algae juice | 70 < x < 83 | 4.44 | 5.36 < y < 6.34 |
| CASPI | 71 < x < 85 | 4.55 | 5.37 < y < 6.42 |
| ASPI | 79 < x < 93 | 5.01 | 5.39 < y < 6.32 |
| N. gaditana | |||
| biomasse | 77 < x < 90 | 4.84 | 5.40 < y < 6.31 |
| biomassf | 75 < x < 88 | 4.73 | 5.38 < y < 6.30 |
| pellet | 71 < x < 82 | 4.45 | 5.42 < y < 6.28 |
| algae juice | 75 < x < 86 | 4.64 | 5.38 < y < 6.22 |
| dialyzed algae juice | 73 < x < 85 | 4.59 | 5.39 < y < 6.25 |
| CASPI | 73 < x < 87 | 4.64 | 5.35 < y < 6.32 |
| ASPI | 80 < x < 93 | 5.03 | 5.38 < y < 6.30 |
| T. impellucida | |||
| biomasse | 70 < x < 82 | 4.48 | 5.49 < y < 6.37 |
| biomassf | 69 < x < 80 | 4.39 | 5.47 < y < 6.36 |
| pellet | 71 < x < 83 | 4.52 | 5.48 < y < 6.35 |
| algae juice | 65 < x < 76 | 4.13 | 5.45 < y < 6.37 |
| dialyzed algae juice | 79 < x < 93 | 5.09 | 5.49 < y < 6.44 |
| CASPI | 82 < x < 98 | 5.16 | 5.29 < y < 6.32 |
| ASPI | 83 < x < 98 | 5.32 | 5.43 < y < 6.39 |
| S. dimorphus | |||
| biomasse | 62 < x < 72 | 3.88 | 5.37 < y < 6.30 |
| biomassf | 60 < x < 71 | 3.78 | 5.35 < y < 6.29 |
| pellet | 60 < x < 71 | 3.77 | 5.32 < y < 6.28 |
| algae juice | 71 < x < 84 | 4.51 | 5.37 < y < 6.34 |
| dialyzed algae juice | 75 < x < 88 | 4.74 | 5.37 < y < 6.35 |
| CASPI | 77 < x < 92 | 4.93 | 5.36 < y < 6.41 |
| ASPI | 80 < x < 94 | 5.03 | 5.36 < y < 6.28 |
Proteinaceous nitrogen (NAA) as proportion of total nitrogen (NT).
Lower limit represents the theoretical value calculated with ASX/GLX = 100% ASP/GLU; upper limit calculated with ASX/GLX = 100% ASN/GLN.
kp values are the average of kp calculated with ASX/GLX = 100% ASN/GLN and kp calculated with ASX/GLX = 100% ASP/GLU. The standard deviations between the values were ≤0.001.
Lower limit represents the theoretical value calculated with ASX/GLX = 100% ASN/GLN; upper limit calculated with ASX/GLX = 100% ASP/GLU.
Values include tryptophan. Since tryptophan was only analyzed in the biomass, the
values are calculated without tryptophan to allow comparison with the other processing steps.
Protein Extraction
Large differences were found in the protein extractability between the four sources. After bead milling and centrifugation, 17%, 41%, 58%, and 74% [w/w] of the total protein in the biomass was extracted for SCE, TET, NAN, and ART, respectively (Table 7). The unicellular sources have different types of cell walls, which affected the duration of bead milling needed to disrupt the cells. Scenedesmus sp. and N. gaditana cell walls are mainly composed of cellulose, with an outer hydrophobic algaenan layer.41−43 Cell walls of Tetraselmis sp. consist of various carbohydrate acids and neutral carbohydrates.38,46 The cell walls of cyanobacteria (including Arthrospira sp.) are mainly composed of peptidoglycans.53 It is generally assumed that peptidoglycan cell walls of cyanobacteria are less robust than the cellulose cell walls of microalgae. Indeed, more time was needed to break the cells of SCE and NAN than of ART (60 and 45 min compared to 20 min, respectively, per liter of sample (12% w/w dry matter)). Cell walls of TET were less recalcitrant than the cellulose-based cell walls of SCE and NAN but more recalcitrant than ART cell walls (30 min bead milling/L at 12% dry matter). All samples were bead milled until most cells were disrupted (as verified by light microscopy). No relation could be found between protein extractability of the bead-milled samples and the cell wall recalcitrance to disintegration by bead milling.
Table 7. Protein Yield and Protein Content of Samples at Each Processing Step, on a Dry Matter Basis (± SD).
|
A. maxima |
N. gaditana |
T. impellucida |
S. dimorphus |
|||||
|---|---|---|---|---|---|---|---|---|
| proteinaceous material [w/w%] | proteinaceous yield [%] | proteinaceous material [w/w%] | proteinaceous yield [%] | proteinaceous material [w/w%] | proteinaceous yield [%] | proteinaceous material [w/w%] | proteinaceous yield [%] | |
| biomass | 61.74 ± 0.51 | 100.00 | 44.99 ± 0.61 | 100.00 | 35.75 ± 1.90 | 100.00 | 29.08 ± 4.99 | 100.00 |
| pellet | 59.54 ± 0.51 | 26.31 ± 2.24 | 40.86 ± 0.15 | 41.83 ± 15.24 | 34.85 ± 2.48 | 60.09 ± 6.49 | 31.16 ± 2.55 | 82.75 ± 10.19 |
| AJ | 63.90 ± 0.89 | 73.69 ± 11.85 | 43.08 ± 0.42 | 58.17 ± 19.90 | 34.68 ± 1.27 | 41.06 ± 5.99 | 18.79 ± 14.75 | 17.29 ± 10.07 |
| AJD | 60.77 ± 1.00 | 35.91 ± 9.16 | 50.39 ± 0.89 | 46.24 ± 13.40 | 52.09 ± 4.79 | 26.85 ± 5.35 | 17.85 ± 9.09 | 12.38 ± 2.08 |
| CASPI | 66.57 ± 2.60 | 7.92 ± 0.23 | 67.67 ± 10.18 | 10.79 ± 2.18 | 61.92 ± 3.54 | 8.60 ± 2.56 | 18.62 ± 4.07 | 3.89 ± 2.75 |
| ASPI | 76.68 ± 0.07 | 6.16 ± 1.14 | 76.79 ± 2.56 | 8.79 ± 1.60 | 66.37 ± 6.60 | 6.25 ± 2.38 | 62.47 ± 5.31 | 3.24 ± 1.31 |
Part of the proteinaceous material extracted was found to be low molecular weight (LMW; <12–14 kDa) peptides or free amino acids, based on the lower protein yield after dialysis of the algae juice (AJD): 12%, 27%, 48%, and 36% [w/w] in SCE, TET, NAN, and ART, respectively. This means that 21–51% of the soluble proteinaceous material was of LMW, with ART having the highest LMW fraction. The high molecular weight protein fraction contains the (intact) proteins of interest and was therefore used for further isolation. Previous publications on algal protein extractability showed a similar protein extractability for T. impellucida (21% [w/w])19 and a 30% [w/w] protein extractability for Nannochloropsis sp.54 No data was found on protein extractability from Arthrospira and Scenedesmus species. The proportion of LMW proteinaceous material in the extracts (12–36%) was similar to what was reported by Schwenzfeier et al., who showed that 38% [w/w] of the extracted proteins of T. impellucida is of low Mw.19 The final protein isolates, obtained after AEC and acid precipitation, had protein contents of 62–77% [w/w], corresponding to protein yields of 3–9% [w/w]. The differences in protein content between the starting materials was thus reduced during the isolation process. The yields are not high, but the aim of the method was to obtain representative fractions of the soluble part of the proteins. It should, however, be noted that the soluble proteins may represent only part of all proteins in the algae and that the proteins represent only part of the total nitrogen. The processing step in which most of the solubilized protein was lost for all sources is the AEC step, where 68–78% [w/w] of the protein in AJD was not bound to the DEAE. The combination of the AEC step and the acid precipitation step increased the protein purity the most (with 26–250%). The TET results were very similar to the work by Schwenzfeier et al., in which a T. impellucida protein isolate was obtained with a protein content of 64% [w/w] and a protein yield of 7% [w/w].19
Protein and Amino Acid Distribution upon Extraction
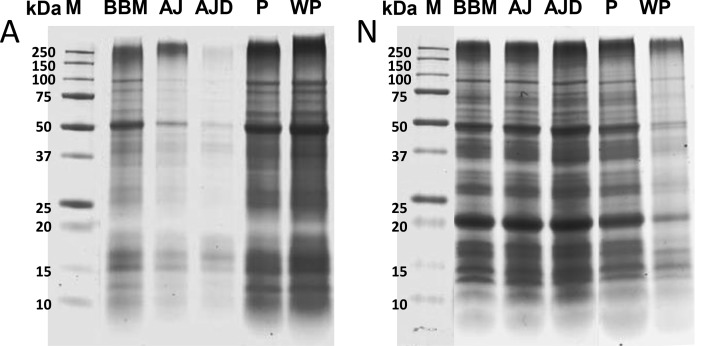
The soluble and insoluble fractions obtained after extraction of proteins from SCE, TET, NAN, and ART had identical protein compositions (Figure 3). The identical protein composition was not caused by coprecipitation of soluble proteins in the pellets: pellets washed with a potassium phosphate buffer still had the same protein compositions. The similarity between insoluble and soluble protein upon extraction also shows in the amino acid compositions (Table 5), which are the same for the pellet and algae juice fractions (the average SD of the mean is 1.3%). On the basis of the SDS-PAGE and AA results it was concluded that the majority of proteins in the insoluble fraction are essentially the same proteins as in the soluble fractions. This indicates that the insoluble fraction (which is often referred to as hydrophobic proteins or cell wall bound proteins55) does not necessarily have to consist of different proteins than those that are obtained in the extract.
Figure 3.
Coomassie stained SDS-PAGE gels with various fractions of A. maxima (A) and N. gaditana (N) under reducing conditions. BBM = bead milled biomass, AJ = algae juice (nondialyzed), AJD = algae juice (dialyzed), P = insoluble fraction of the biomass, WP = washed pellet, and M = protein molecular weight marker.
Protein Isolate Characterization
Chemical Composition
The final ASPIs contained 62–77% [w/w] protein and 9–24% [w/w] carbohydrates (Table 8). These components formed the majority of the ASPIs, representing 82–87% of the total dry weight. The isolates thus had higher protein contents than the biomass they were isolated from. Compared to the biomass, the total carbohydrate content is lower in ASPI-N and -A and is increased in ASPI-T and -S. The ratios of uronic acids (charged carbohydrates) to protein (UA:P) of the isolates are 0.01, 0.01, 0.09, and 0.02 for ASPI -A, -N, -T, and -S. This means that for SCE, NAN, and ART the UA:P in the ASPI was about a factor of 2 lower than in the biomass, while for TET it was a factor of 1.4 higher. Previously, the presence of the charged carbohydrates in ASPI from T. impellucida was linked to higher stability of emulsions against flocculation around the pI.36 The monocarbohydrate constituents of the total carbohydrates in the ASPIs are different than those of the corresponding biomass (Table 9). Overall, carbohydrates containing rhamnose, arabinose, and xylose represent a larger fraction (% mol) of the total carbohydrates in all ASPIs than in their initial biomass. Ribose is coisolated in the protein isolation process of ART, TET, and SCE and is the major carbohydrate constituent in the associated ASPIs (25–67 % mol of total carbohydrates). In contrast, the ribose fraction of ASPI-N carbohydrates is similar to that of the NAN biomass (6% vs 8% mol of total carbohydrates). Likewise, mannose and galactose appear to be more coisolated in NAN and TET, respectively, than in the other samples. The glucose fraction of the total carbohydrates decreases during the protein isolation of ART, NAN, and TET (from 49–67% to 2–10% mol) but remains constant in SCE.
Table 8. Gross Chemical Composition of Protein Isolates [% w/w] on a Dry Weight Basis.
| component | A. maxima | N. gaditana | T. impellucida | S. dimorphus |
|---|---|---|---|---|
| proteins | 76.7 ± 0.1 | 76.8 ± 2.6 | 66.4 ± 6.6 | 62.5 ± 5.3 |
| carbohydrates | 9.2 ± 0.4 | 8.9 ± 0.3 | 24.4 ± 0.4 | 19.8 ± 1.1 |
| neutral | 8.1 ± 0.3 | 8.3 ± 0.4 | 18.7 ± 0.5 | 18.8 ± 1.2 |
| charged | 1.0 ± <0.1 | 0.6 ± <0.1 | 5.7 ± 0.1 | 1.0 ± 0.1 |
| total annotated | 85.8 | 85.7 | 90.8 | 82.2 |
Table 9. Monocarbohydrate Composition of Total Carbohydrates in Protein Isolates [mol %; ± SD]a.
| Rha | Fuc | Ara | Xyl | Man | Gal | Glc | Rib | UA | |
|---|---|---|---|---|---|---|---|---|---|
| A. maxima | 14.60 ± 0.01 | 2.64 ± 0.03 | 6.22 ± 0.01 | 4.95 ± 0.07 | 1.04 ± 0.03 | 2.90 ± 0.03 | 1.78 ± 0.25 | 67.30 ± 0.02 | 9.68 ± 0.03 |
| N. gaditana | 15.56 ± 0.06 | 8.47 ± 0.09 | 10.00 ± 0.01 | 10.51 ± 0.01 | 26.94 ± 0.13 | 14.82 ± 0.03 | 10.11 ± 0.06 | 8.21 ± 0.01 | 6.50 ± 0.02 |
| T. impellucida | 9.30 ± 0.30 | n.d. | 18.21 ± 0.10 | 7.47 ± 0.33 | 5.04 ± 0.30 | 35.52 ± 0.16 | 4.25 ± 1.20 | 24.75 ± 0.11 | 6.58 ± 0.11 |
| S. dimorphus | 2.07 ± 0.07 | 1.12 ± 0.01 | 3.44 ± 0.07 | 2.60 ± 0.07 | 5.87 ± 0.08 | 5.23 ± 0.12 | 53.47 ± 0.76 | 32.46 ± 0.22 | 4.85 ± 0.07 |
n.d.: Not detected.
Protein and Amino Acid Composition
The protein composition of the cyanobacterial ASPI-A was different from those of the three microalgal ASPIs (-N, -T, and -S); the latter three are quite similar to each other (Figure 3). The microalgal ASPIs had a diverse protein composition (Figure 1 C), whereas ASPI-A contained one dominant group of proteins (15–18 kDa). On the basis of the intense blue color of the isolate and the dominance of the 15–18 kDa bands, this shows that the phycocyanins present in the biomass were predominantly retained during the isolation process. The ∼50 kDa protein that is considered to be Rubisco’s large subunit was much less pronounced in ASPI-A compared to the other ASPIs. The protein composition of ASPI-A was more homogeneous than that of the ART biomass. The most intense bands detected in ASPI-N represent proteins of 15, 37, and 50 kDa and proteins of large Mw (>250 kDa). Compared to the NAN biomass, the 15 kDa band (attributed to the small subunit of Rubisco) was more pronounced in the isolate. ASPI-T mostly contained 10–15, 25–30, 35–37, and 50 kDa proteins. Compared to the TET biomass, the bands of 10–15 kDa were more pronounced and the large subunit Rubisco is less pronounced. The protein composition of ASPI-S was comparable to that of ASPI-N and ASPI-T, with major bands at <15, 25–30, 37, and 50 kDa. Like in ASPI-N, large Mw proteins (>250 kDa) were present in the isolate. Additionally, glycoprotein analysis with PAS staining revealed the presence of glycoproteins of ≥250 kDa in all ASPIs (data not shown). Overall, a shared property of the ASPIs is the presence of proteins that are subunits of multimeric proteins (i.e., Rubisco and phycocyanins), which may lead to similar techno-functionalities. Additionally, ASPI -N, -T, and -S are more similar to each other than ASPI-A, based on their more diverse protein composition and presence of Rubisco.
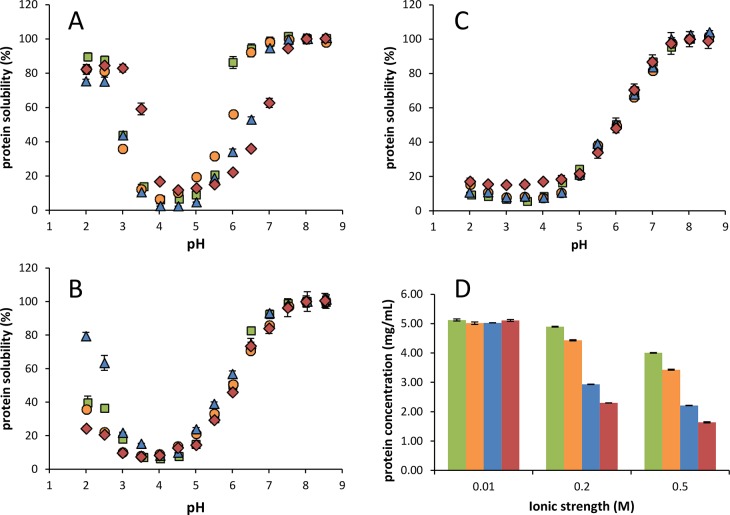
Solubility
Despite the differences in protein composition, the protein isolates of ART, NAN, TET, and SCE displayed similar pH dependent solubility (Figure 4). At low ionic strength (I = 0.01), the proteins were completely soluble at pH > 6.5 (ASPI-T and ASPI-S) or at pH > 7.0 (ASPI-A and ASPI-N) and least soluble at pH 4.0–4.5. This point of lowest solubility is close to the computed pI based on amino acid compositions, which were calculated to be 4.98, 5.16, 4.94, and 4.98 for ASPI-A, -N, -T, and -S respectively. For these calculations, GLU/GLN and ASP/ASN ratios of Rubisco from N. gaditana and Tetraselmis suecica (1.0:2.2 and 1.0:1.4, respectively) were used (based on ref (9). (Accession numbers used: A0A023PJK0, Q3S3D2, and K9ZV74.) These pI values are lower than the theoretical pI of Rubisco (5.88–8.00).56 (Accession numbers used: 4MKV, 1WDD, 1RLC, 1RLD, 1RBL, 1RSC, 1BWV, 1BXN, 1EJ7, 1IWA, 2YBV, 3AXM, 3AXK, 3ZXW, 4F0M, 4F0K, 4F0H, 1UPM, 1UPP, 1IR1, AA1, 1RCX, 1RXO, 1RBO, 1RCO, 8RUC, 1AUS, 2VDH, 2VDI, 2V67, 2V68, 2 V63, 2 V69, 2V6A, 1UW9, 1UWA, 1IR2, and1GK8.) These differences are expected to be partially due to the presence of other proteins apart from Rubisco, as shown in the SDS-PAGE profiles. Additionally, the presence of protein-bound uronic acids contribute to the overall charge and solubility of the ASPIs. Using a pKa value of 3.3 for uronic acids,57 the uronic acids present in the ASPIs were calculated to decrease the pI by 0.24–0.52 pH units.
Figure 4.
Protein solubility (starting concentration 5 mg protein/mL) as a function of pH of ASPI-T (green), ASPI-S (orange), ASPI-A (blue), and ASPI-N (red) at different ionic strengths (I = 0.05 (A), 0.2 (B), and 0.5 M (C). Solubility is expressed relative to pH 8.0 (=set as 100% soluble) (A–C) and the amount of solubilized protein at pH 8.0 as affected by ionic strength (D). Error bars indicate standard deviations.
Solubility increased again at pH values below the theoretical pI; below pH 3.0, >80% of all ASPIs was soluble. It should be noted that all isolates were obtained using a similar isolation procedure, which would select proteins with similar solubility at the pH used for extraction and precipitation (pH 8.0 and 3.5). The point of minimum solubility of the ASPIs is lower than some conventional vegetal protein sources, including soy glycinin (pH 4.7–6.2),58 and more comparable to that of sunflower helianthinin (pH 4.0–5.5).59 Values reported for unicellular proteins (from Arthrospira platensis,16Nannochloropsis sp.,54 and T. impellucida(19)) are very similar and are in the range of pH 3.0–4.0.
At pH 8, protein solubility of all ASPIs was found to be dependent on ionic strength (Figure 4 D). When increasing the ionic strength to I = 0.2 and 0.5 M, the protein solubility decreased. At low pH (<pH 4.5), this decrease was more apparent (Figure 4 A–C). Protein solubility of ASPI-T and ASPI-S was least dependent on ionic strength, since at pH 7.6 I = 0.2 M 85% [w/w] protein was in solution, whereas 38–39% [w/w] protein of the other ASPIs was in solution under these conditions. At I = 0.5 M, the solubility at pH 7.6 was lower than that at I = 0.2 M for all ASPIs (38–69% w/w), apart from ASPI-T (85% w/w). At low pH (≤4.0) and high ionic strength (I = 0.5 M), protein solubility was considerably decreased (4–10% [w/w]) for all ASPIs. The ionic strength dependence of the ASPIs was different from previously reported solubility profiles of a T. impellucida ASPI,19 which show a low ionic strength dependency at ionic strengths of 0.03–0.5 M19 This difference in solubility between the two T. impellucida isolates is thought to be due to batch-to-batch variations in the microalgae. The behavior of the ASPIs was similar to that of sunflower helianthinin, of which the protein solubility at lower pH range decreases drastically at I = 0.25 M as compared to I = 0.03 M.59
The aim of this study was to make a first step in the description of the differences in gross composition of various types of unicellular biomass and to understand how these differences affect the final protein isolate. A single isolation method was used in this study as a tool to isolate proteins from four different unicellular photosynthetic sources. The current protocol was not aimed at optimizing protein isolation yield. It aimed at enabling a relatively fast isolation of purified proteins from various novel protein sources in order to compare the proteins’ characteristics. The key findings of this study were that in spite of the different chemical compositions of the unicellular sources used, protein isolates were obtained with comparable purity (62–77% [w/w] protein) and proteinaceous yield (3–9% [w/w]). Additionally, protein solubility as a function of pH of the ASPIs was similar at low ionic strength (I = 10 mM). At higher ionic strengths (I = 0.2–0.5 M) differences in protein solubility between the sources were observed, especially at pH < 4.0. Overall, this study showed that the isolation method applied can yield protein isolates that have similar protein purity and solubility, regardless of the chemical composition and protein composition of the starting algal or cyanobacterial biomass.
Acknowledgments
We are grateful to the Animal Nutrition group from Wageningen University & Research for performing the amino acid analyses and to Bioprocess Engineering from Wageningen University & Research for their contribution to the FAME analyses.
This research was executed within a project of the Protein Innovation Program of the Dutch Technology Foundation STW, which is part of NWO, the Dutch National Science Foundation. The program is also partly funded by the Dutch Ministry of Economic Affairs. The project was entitled “Proteins from green sources for use in both food and fish feed”, project number STW 12637. As of January 2017 STW continues its activities as NWO Applied and Engineering Sciences, NWO domain TTW.
The authors declare no competing financial interest.
References
- Barsanti L.; Gualtieri P.. Algae: Anatomy, Biochemistry, and Biotechnology, 2nd ed.; CRC Press: Boca Raton, FL, USA, 2014. [Google Scholar]
- Derbyshire E.; Wright D. J.; Boulter D. Legumin and vicilin, storage proteins of legume seeds. Phytochemistry 1976, 15, 3–24. 10.1016/S0031-9422(00)89046-9. [DOI] [Google Scholar]
- Saio K.; Kamiya M.; Watanabe T. Food processing characteristics of soybean 11S and 7S proteins. Agric. Biol. Chem. 1969, 33, 1301–1308. 10.1271/bbb1961.33.1301. [DOI] [Google Scholar]
- Gueguen J. Legume seed protein extraction, processing, and end product characteristics. Qual. Plant. - Plant Foods Hum. Nutr. 1983, 32, 267–303. 10.1007/BF01091191. [DOI] [Google Scholar]
- Becker E. W. Micro-algae as a source of protein. Biotechnol. Adv. 2007, 25, 207–210. 10.1016/j.biotechadv.2006.11.002. [DOI] [PubMed] [Google Scholar]
- De Oliveira M. A. C. L.; Monteiro M. P. C.; Robbs P. G.; Leite S. G. F. Growth and chemical composition of Spirulina maxima and Spirulina platensis biomass at different temperatures. Aquacult. Int. 1999, 7, 261–275. 10.1023/A:1009233230706. [DOI] [Google Scholar]
- Olofsson M.; Lamela T.; Nilsson E.; Bergé J. P.; del Pino V.; Uronen P.; Legrand C. Seasonal variation of lipids and fatty acids of the microalgae Nannochloropsis oculata grown in outdoor large-scale photobioreactors. Energies 2012, 5, 1577–1592. 10.3390/en5051577. [DOI] [Google Scholar]
- Tabita F. R.; Satagopan S.; Hanson T. E.; Kreel N. E.; Scott S. S. Distinct form I, II, III, and IV Rubisco proteins from the three kingdoms of life provide clues about Rubisco evolution and structure/function relationships. J. Exp. Bot. 2008, 59, 1515–1524. 10.1093/jxb/erm361. [DOI] [PubMed] [Google Scholar]
- UniProtKB, UniProt: the universal protein knowledgebase. Nucleic Acids Res. 2017, 45, D158–D169. 10.1093/nar/gkw1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson I.; Backlund A. Structure and function of Rubisco. Plant Physiol. Biochem. 2008, 46, 275–291. 10.1016/j.plaphy.2008.01.001. [DOI] [PubMed] [Google Scholar]
- Basso S.; Simionato D.; Gerotto C.; Segalla A.; Giacometti G. M.; Morosinotto T. Characterization of the photosynthetic apparatus of the Eustigmatophycean Nannochloropsis gaditana: Evidence of convergent evolution in the supramolecular organization of photosystem I. Biochim. Biophys. Acta, Bioenerg. 2014, 1837, 306–314. 10.1016/j.bbabio.2013.11.019. [DOI] [PubMed] [Google Scholar]
- Carbonera D.; Agostini A.; Di Valentin M.; Gerotto C.; Basso S.; Giacometti G. M.; Morosinotto T. Photoprotective sites in the violaxanthin–chlorophyll a binding Protein (VCP) from Nannochloropsis gaditana. Biochim. Biophys. Acta, Bioenerg. 2014, 1837, 1235–1246. 10.1016/j.bbabio.2014.03.014. [DOI] [PubMed] [Google Scholar]
- Liu Z.; Yan H.; Wang K.; Kuang T.; Zhang J.; Gui L.; An X.; Chang W. Crystal structure of spinach major light-harvesting complex at 2.72 Å resolution. Nature 2004, 428, 287–292. 10.1038/nature02373. [DOI] [PubMed] [Google Scholar]
- Wei X.; Su X.; Cao P.; Liu X.; Chang W.; Li M.; Zhang X.; Liu Z. Structure of spinach photosystem II–LHCII supercomplex at 3.2 Å resolution. Nature 2016, 534, 69–74. 10.1038/nature18020. [DOI] [PubMed] [Google Scholar]
- Larkum T.; Howe C. J.. Chapter IV: Light-harvesting proteins, subChapter F: Phycobiliproteins and associated proteins; Molecular Aspects of Light-harvesting Processes in Algae. Academic Press: San Diego, 1997. [Google Scholar]
- Devi M. A.; Subbulakshmi G.; Devi K. M.; Venkataraman L. Studies on the proteins of mass-cultivated, blue-green alga (Spirulina platensis). J. Agric. Food Chem. 1981, 29, 522–525. 10.1021/jf00105a022. [DOI] [PubMed] [Google Scholar]
- Postma P.; Miron T.; Olivieri G.; Barbosa M.; Wijffels R.; Eppink M. Mild disintegration of the green microalgae Chlorella vulgaris using bead milling. Bioresour. Technol. 2015, 184, 297–304. 10.1016/j.biortech.2014.09.033. [DOI] [PubMed] [Google Scholar]
- Ursu A.-V.; Marcati A.; Sayd T.; Sante-Lhoutellier V.; Djelveh G.; Michaud P. Extraction, fractionation and functional properties of proteins from the microalgae Chlorella vulgaris. Bioresour. Technol. 2014, 157, 134–139. 10.1016/j.biortech.2014.01.071. [DOI] [PubMed] [Google Scholar]
- Schwenzfeier A.; Wierenga P. A.; Gruppen H. Isolation and characterization of soluble protein from the green microalgae Tetraselmis sp. Bioresour. Technol. 2011, 102, 9121–9127. 10.1016/j.biortech.2011.07.046. [DOI] [PubMed] [Google Scholar]
- Betschart A. A. Nitrogen solubility of alfalfa protein concentrate as influenced by various factors. J. Food Sci. 1974, 39, 1110–1115. 10.1111/j.1365-2621.1974.tb07329.x. [DOI] [Google Scholar]
- Folch J.; Lees M.; Stanley G. H. S. A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 1957, 226, 497–509. [PubMed] [Google Scholar]
- Breuer G.; Evers W. A. C.; de Vree J. H.; Kleinegris D. M. M.; Martens D. E.; Wijffels R.; H; Lamers P. P. Analysis of fatty acid content and composition in microalgae. J. Visualized Exp. 2013, e50628. 10.3791/50628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Englyst H. N.; Cummings J. H. Simplified method for the measurement of total non-starch polysaccharides by gas-liquid chromatography of constituent sugars as alditol acetates. Analyst 1984, 109, 937–942. 10.1039/an9840900937. [DOI] [PubMed] [Google Scholar]
- Ahmed A. E. R.; Labavitch J. M. A simplified method for accurate determination of cell wall uronide content. J. Food Biochem. 1978, 1, 361–365. 10.1111/j.1745-4514.1978.tb00193.x. [DOI] [Google Scholar]
- Brown M. R. The amino-acid and sugar composition of 16 species of microalgae used in mariculture. J. Exp. Mar. Biol. Ecol. 1991, 145, 79–99. 10.1016/0022-0981(91)90007-J. [DOI] [Google Scholar]
- Renaud S. M.; Thinh L.-V.; Parry D. L. The gross chemical composition and fatty acid composition of 18 species of tropical Australian microalgae for possible use in mariculture. Aquaculture 1999, 170, 147–159. 10.1016/S0044-8486(98)00399-8. [DOI] [Google Scholar]
- Biller P.; Ross A. B. Pyrolysis GC-MS as a novel analysis technique to determine the biochemical composition of microalgae. Algal Res. 2014, 6, 91–97. 10.1016/j.algal.2014.09.009. [DOI] [Google Scholar]
- Miller M. R.; Nichols P. D.; Carter C. G. n-3 Oil sources for use in aquaculture--alternatives to the unsustainable harvest of wild fish. Nutr. Res. Rev. 2008, 21, 85–96. 10.1017/S0954422408102414. [DOI] [PubMed] [Google Scholar]
- Hita Peña E.; Robles Medina A.; Jiménez Callejón M. J.; Macías Sánchez M. D.; Esteban Cerdán L.; González Moreno P. A.; Molina Grima E. Extraction of free fatty acids from wet Nannochloropsis gaditana biomass for biodiesel production. Renewable Energy 2015, 75, 366–373. 10.1016/j.renene.2014.10.016. [DOI] [Google Scholar]
- Taleb A.; Pruvost J.; Legrand J.; Marec H.; Le-Gouic B.; Mirabella B.; Legeret B.; Bouvet S.; Peltier G.; Li-Beisson Y.; Taha S.; Takache H. Development and validation of a screening procedure of microalgae for biodiesel production: Application to the genus of marine microalgae Nannochloropsis. Bioresour. Technol. 2015, 177, 224–232. 10.1016/j.biortech.2014.11.068. [DOI] [PubMed] [Google Scholar]
- Custódio L.; Soares F.; Pereira H.; Barreira L.; Vizetto-Duarte C.; Rodrigues M. J.; Rauter A. P.; Alberício F.; Varela J. Fatty acid composition and biological activities of Isochrysis galbana T-ISO, Tetraselmis sp. and Scenedesmus sp.: Possible application in the pharmaceutical and functional food industries. J. Appl. Phycol. 2014, 26, 151–161. 10.1007/s10811-013-0098-0. [DOI] [Google Scholar]
- Huerlimann R.; de Nys R.; Heimann K. Growth, lipid content, productivity, and fatty acid composition of tropical microalgae for scale-up production. Biotechnol. Bioeng. 2010, 107, 245–257. 10.1002/bit.22809. [DOI] [PubMed] [Google Scholar]
- Renaud S. M.; Parry D. L.; Thinh L.-V. Microalgae for use in tropical aquaculture I: Gross chemical and fatty acid composition of twelve species of microalgae from the Northern Territory, Australia. J. Appl. Phycol. 1994, 6, 337–345. 10.1007/BF02181948. [DOI] [Google Scholar]
- Chaiklahan R.; Chirasuwan N.; Loha V.; Bunnag B. Lipid and fatty acids extraction from the cyanobacterium. ScienceAsia 2008, 34, 299–305. 10.2306/scienceasia1513-1874.2008.34.299. [DOI] [Google Scholar]
- Baunillo K. E.; Tan R. S.; Barros H. R.; Luque R. Investigations on microalgal oil production from Arthrospira platensis: towards more sustainable biodiesel production. RSC Adv. 2012, 2, 11267–11272. 10.1039/c2ra21796a. [DOI] [Google Scholar]
- Schwenzfeier A.; Wierenga P. A.; Eppink M. H.; Gruppen H. Effect of charged polysaccharides on the techno-functional properties of fractions obtained from algae soluble protein isolate. Food Hydrocolloids 2014, 35, 9–18. 10.1016/j.foodhyd.2013.07.019. [DOI] [Google Scholar]
- Corteggiani Carpinelli E.; Telatin A.; Vitulo N.; Forcato C.; D’Angelo M.; Schiavon R.; Vezzi A.; Giacometti G. M.; Morosinotto T.; Valle G. Chromosome scale genome assembly and transcriptome profiling of Nannochloropsis gaditana in nitrogen depletion. Mol. Plant 2014, 7, 323–335. 10.1093/mp/sst120. [DOI] [PubMed] [Google Scholar]
- Kermanshahi-pour A.; Sommer T. J.; Anastas P. T.; Zimmerman J. B. Enzymatic and acid hydrolysis of Tetraselmis suecica for polysaccharide characterization. Bioresour. Technol. 2014, 173, 415–421. 10.1016/j.biortech.2014.09.048. [DOI] [PubMed] [Google Scholar]
- Wang L.; Li Y.; Sommerfeld M.; Hu Q. A flexible culture process for production of the green microalga Scenedesmus dimorphus rich in protein, carbohydrate or lipid. Bioresour. Technol. 2013, 129, 289–295. 10.1016/j.biortech.2012.10.062. [DOI] [PubMed] [Google Scholar]
- Aikawa S.; Izumi Y.; Matsuda F.; Hasunuma T.; Chang J.-S.; Kondo A. Synergistic enhancement of glycogen production in Arthrospira platensis by optimization of light intensity and nitrate supply. Bioresour. Technol. 2012, 108, 211–215. 10.1016/j.biortech.2012.01.004. [DOI] [PubMed] [Google Scholar]
- Scholz M. J.; Weiss T. L.; Jinkerson R. E.; Jing J.; Roth R.; Goodenough U.; Posewitz M. C.; Gerken H. G. Ultrastructure and composition of the Nannochloropsis gaditana cell wall. Eukaryotic Cell 2014, 13, 1450–1464. 10.1128/EC.00183-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickett-Heaps J. D.; Staehelin L. A. The ultrastructure of Scenedesmus (Chlorophyceae). II. Cell division and colony formation. J. Phycol. 1975, 11, 186–202. 10.1111/j.1529-8817.1975.tb02766.x. [DOI] [Google Scholar]
- Allard B.; Templier J. Comparison of neutral lipid profile of various trilaminar outer cell wall (TLS)-containing microalgae with emphasis on algaenan occurrence. Phytochemistry 2000, 54, 369–380. 10.1016/S0031-9422(00)00135-7. [DOI] [PubMed] [Google Scholar]
- Templeton D. W.; Quinn M.; Van Wychen S.; Hyman D.; Laurens L. M. L. Separation and quantification of microalgal carbohydrates. Journal of Chromatography A 2012, 1270, 225–234. 10.1016/j.chroma.2012.10.034. [DOI] [PubMed] [Google Scholar]
- Shamala T. R.; Drawert F.; Leupold G. Studies on Scenedesmus acutus growth II. Effect of autotrophic and mixotrophic growth on the amino acid and the carbohydrate composition of Scenedesmus acutus. Biotechnol. Bioeng. 1982, 24, 1301–1317. 10.1002/bit.260240606. [DOI] [PubMed] [Google Scholar]
- Becker B.; Melkonian M.; Kamerling J. P., The cell wall (theca) of Tetraselmis striata (Chlorophyta): Macromolecular composition and structural elements of the complex polysaccharides. 1998, 34, 779–787.
- Ortega-Calvo J. J.; Mazuelos C.; Hermosin B.; Saiz-Jimenez C. Chemical composition of Spirulina and eukaryotic algae food products marketed in Spain. J. Appl. Phycol. 1993, 5, 425–435. 10.1007/BF02182735. [DOI] [Google Scholar]
- Hermsmeier D.; Schulz R.; Senger H. Formation of light-harvesting complexes of photosystem II in Scenedesmus. Planta 1994, 193, 398–405. 10.1007/BF00201819. [DOI] [Google Scholar]
- Cervantes-Pahm S.; Stein H. Effect of dietary soybean oil and soybean protein concentration on the concentration of digestible amino acids in soybean products fed to growing pigs. J. Anim. Sci. 2008, 86, 1841–1849. 10.2527/jas.2007-0721. [DOI] [PubMed] [Google Scholar]
- Panthee D.; Pantalone V.; Saxton A.; West D.; Sams C. Genomic regions associated with amino acid composition in soybean. Mol. Breed. 2006, 17, 79–89. 10.1007/s11032-005-2519-5. [DOI] [Google Scholar]
- Belitz H.; Grosch W.; Schieberle P.. Food Chemistry, 3rd ed.; Springer: Berlin Heidelberg, 2004. [Google Scholar]
- Williams A.; Bishop D. R.; Cockburn J.; Scott K. Composition of ewe’s milk. J. Dairy Res. 1976, 43, 325–329. 10.1017/S0022029900015892. [DOI] [PubMed] [Google Scholar]
- Venkataraman L. V.Spirulina platensis (Arthrospira): Physiology, cell biology and biotechnology; Kluwer Academic Publishers, 1997; Vol. 9, p 6. [Google Scholar]
- Gerde J. A.; Wang T.; Yao L.; Jung S.; Johnson L. A.; Lamsal B. Optimizing protein isolation from defatted and non-defatted Nannochloropsis microalgae biomass. Algal Res. 2013, 2, 145–153. 10.1016/j.algal.2013.02.001. [DOI] [Google Scholar]
- Tamayo Tenorio A.; Boom R. M.; van der Goot A. J. Understanding leaf membrane protein extraction to develop a food-grade process. Food Chem. 2017, 217, 234–243. 10.1016/j.foodchem.2016.08.093. [DOI] [PubMed] [Google Scholar]
- Berman H. M.; Westbrook J.; Feng Z.; Gilliland G.; Bhat T. N.; Weissig H.; Shindyalov I. N.; Bourne P. E. The protein data bank. Nucleic Acids Res. 2000, 28, 235–242. 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohn R.; Kovac P. Dissociation constants of D-galacturonic and D-glucuronic acid and their O-methyl derivatives. Chem. Zvesti 1978, 32, 478–485. [Google Scholar]
- Lakemond C. M.; de Jongh H. H.; Hessing M.; Gruppen H.; Voragen A. G. Soy glycinin: influence of pH and ionic strength on solubility and molecular structure at ambient temperatures. J. Agric. Food Chem. 2000, 48, 1985–1990. 10.1021/jf9908695. [DOI] [PubMed] [Google Scholar]
- González-Pérez S.; Vereijken J. M.; Merck K. B.; van Koningsveld G. A.; Gruppen H.; Voragen A. G. Conformational states of sunflower (Helianthus annuus) helianthinin: effect of heat and pH. J. Agric. Food Chem. 2004, 52, 6770–6778. 10.1021/jf049612j. [DOI] [PubMed] [Google Scholar]
- Markou G.; Chatzipavlidis I.; Georgakakis D. Carbohydrates production and bio-flocculation characteristics in cultures of Arthrospira (Spirulina) platensis. BioEnergy Res. 2012, 5, 915–925. 10.1007/s12155-012-9205-3. [DOI] [Google Scholar]
- Camacho-Rodríguez J.; González-Céspedes A. M.; Cerón-García M. C.; Fernández-Sevilla J. M.; Acién-Fernández F. G.; Molina-Grima E. Appl. Microbiol. Biotechnol. 2014, 98, 2429–2440. 10.1007/s00253-013-5413-9. [DOI] [PubMed] [Google Scholar]
- Ferreira M.; Coutinho P.; Seixas P.; Fábregas J.; Otero A. Enriching rotifers with “premium” microalgae. Mar. Biotechnol. 2009, 11, 585–595. 10.1007/s10126-008-9174-x. [DOI] [PubMed] [Google Scholar]
- López-González D.; Fernandez-Lopez M.; Valverde J. L.; Sanchez-Silva L. Kinetic analysis and thermal characterization of the microalgae combustion process by thermal analysis coupled to mass spectrometry. Appl. Energy 2014, 114, 227–237. 10.1016/j.apenergy.2013.09.055. [DOI] [Google Scholar]