Abstract
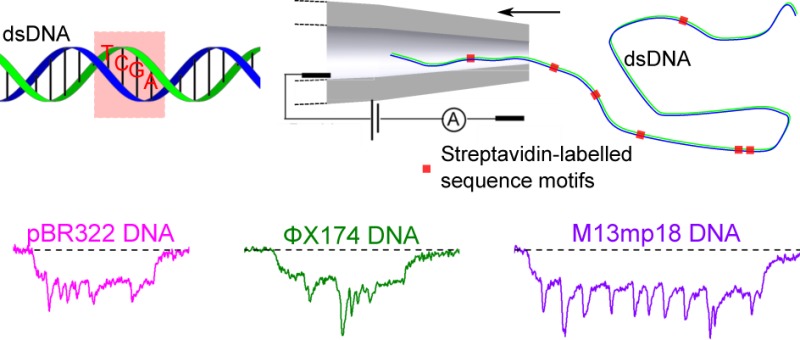
Nanopore sensors show great potential for rapid, single-molecule determination of DNA sequence information. Here, we develop an ionic current-based method for determining the positions of short sequence motifs in double-stranded DNA molecules with solid-state nanopores. Using the DNA-methyltransferase M.TaqI and a biotinylated S-adenosyl-l-methionine cofactor analogue we create covalently attached biotin labels at 5′-TCGA-3′ sequence motifs. Monovalent streptavidin is then added to bind to the biotinylated sites giving rise to additional current blockade signals when the DNA passes through a conical quartz nanopore. We determine the relationship between translocation time and position along the DNA contour and find a minimum resolvable distance between two labeled sites of ∼200 bp. We then characterize a variety of DNA molecules by determining the positions of bound streptavidin and show that two short genomes can be simultaneously detected in a mixture. Our method provides a simple, generic single-molecule detection platform enabling DNA characterization in an electrical format suited for portable devices for potential diagnostic applications.
Keywords: DNA detection, nanopore sensing, genome mapping, single-molecule detection, S-adenosyl-l-methionine analogue, DNA methyltransferase
Precise determination of DNA sequence information has underpinned many recent advances in biomedicine. Alongside direct determination of the sequence, accurate mapping of the position of specific base pair motifs has become an important tool, for instance, in reconstructing large genomes.1 Mapping of short sequence motifs can also be used for species characterization by comparing given maps with databases of known organisms.2 Until now, optical methods have been the primary technique employed for genome mapping.3 In these methods the DNA is first stretched by shear flows or nanochannel confinement followed by determination of the positions of specific sequences by fluorescent labeling.3−7
Nanopores offer another attractive route to DNA identification through genome mapping with the potential for integration into small, portable devices.8,9 Nanopores present a distinct advantage when compared to the requirements for optical microscopy in terms of equipment size and complexity. The principle of the sensing method is that single molecules can be characterized when they pass through a nanosized pore and block the flow of ions. The 1D threading of a long polymer such as DNA through such a nanoscale pore naturally leads to the possibility of reading sequence information. Synthetic or solid-state nanopores have potential advantages such as robustness and amenability to wafer scale integration which makes them attractive alternatives to biological nanopores.10 Singer et al.11,12 showed that bound peptide nucleic acids (PNA) sequences could be detected for targeted identification of a single viral genome using ∼4 nm diameter solid-state nanopores. Recent developments in solid-state nanopore technology have shown that the positions of single proteins13−16 and DNA hairpins17 can also be detected on double-stranded DNA.
In this paper, we build on these developments to demonstrate a generic solid-state nanopore method capable of mapping genomic DNA. We modify short sequence motifs with a biotin residue by using a DNA-methyltransferase (MTase) and a synthetic cofactor analogue.18 The biotin sites are then labeled by tight binding of monovalent streptavidin19 thereby creating additional, sequence specific blockades as the DNA passes through the solid-state nanopore. We demonstrate the feasibility of our method by mapping a variety of DNA plasmid and bacteriophage genomes and find a minimum resolvable distance between sequence motifs of ∼200 bp. Our results show the potential application of solid-state nanopores for DNA identification.
Double-stranded DNA (dsDNA) translocations through a solid-state nanopore show no detectable sequence information due to the uniform cross-section of a DNA double-strand. Therefore, we sought a method for creating labels on double-stranded DNA that would increase the ionic current blockade at specific sequence motifs. A variety of methods have been demonstrated for labeling specific internal sequences on a double-stranded DNA molecule.20,21 Synthetic PNA binds to a double-helix at highly specific sites ∼8–15 bp in length.11,12,22 Similarly nicking endonucleases can be used to create nicks in DNA for subsequent labeling by nick translation with fluorophore-labeled deoxyribonucleotides.5 Also different variants of Cas9 can be used for fluorescence labeling or to create additional ionic current blockade in solid-state nanopore measurements.23,24 The length of the recognition sites of PNA and Cas9 varies from 8–20 bp. This means that labeling is usually targeted for identification of a specific genome because sequence motifs of this length do not repeat frequently enough to create unique fingerprints for different DNA samples. We sought a method that would label shorter sequence motifs and therefore create a way to easily barcode many DNA molecules based on the distribution of these short sequences along the polymer.
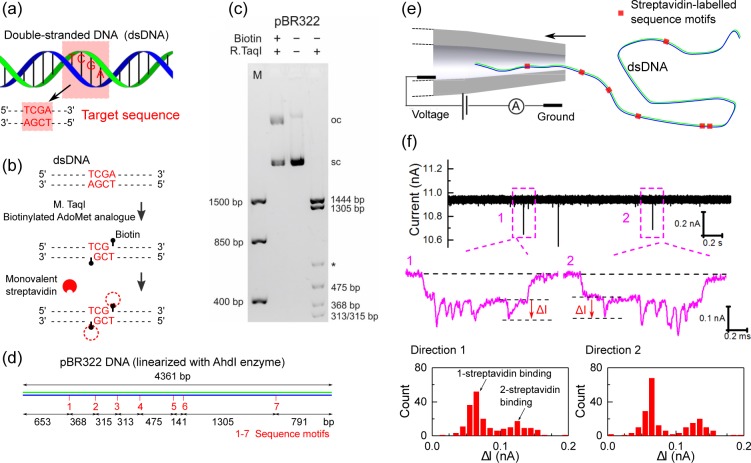
A robust method for targeted labeling of DNA positions is “methyltransferase-directed Transfer of Activated Groups” (mTAG).25 In this technique a DNA MTase is tricked into covalently adding a functional group by replacing the methyl group of the natural cofactor S-adenosyl-l-methionine (AdoMet) with other chemical entities. The mTAG-method has been shown to have a yield close to 100% and can be performed in a simple one-step reaction.2,25Figure 1a,b shows schematics of the DNA MTase-based strategy we employed here. We used the DNA MTase M.TaqI26 which recognizes the four base pair sequence 5′-TCGA-3′ to covalently label the N6-atom of the adenine base of the recognition sequence with a biotin residue.
Figure 1.
Labeling DNA at specific short sequence motifs and translocating them through a glass nanopore. (a) Schematic representation of the sequence (5′-TCGA-3′, shown in the red square) on dsDNA. (b) The labeling procedure: M.TaqI is used to transfer biotin from a biotinylated cofactor analogue to the target adenine within its recognition sequence. Monovalent streptavidin is added which binds to biotin and creates an additional detectable current change during the DNA translocation through a nanopore. (c) Analysis of plasmid modification by agarose gel electrophoresis. Biotinylated and purified pBR322 (Biotin + ) is protected against cleavage by R.TaqI (R.TaqI + ) indicating complete labeling of all 5′-TCGA-3′ sites. Only bands for supercoiled DNA (sc) and open-circular DNA (oc, nicked DNA) are observed as with untreated pBR322 (Biotin −; R.TaqI −). In contrast, treatment of unmodified pBR322 (Biotin −) with R.TaqI (R.TaqI + ) leads to the formation of expected fragments (the asterisk indicates a fragment resulting from partial protection of a hemimethylated M.TaqI site overlapping with a Dam site: 5′-TCGATC-3′). (d) Schematic of the positions of 5′-TCGA-3′ sequence motifs in pBR322 DNA. (e) A streptavidin-labeled DNA molecule is driven into a glass nanopore by a potential difference. (f) An example of ionic current trace at 600 mV and close-up of two selected events. The histograms show the magnitude of the current change at the seventh site for the two directions and are calculated from 200 events.
We initially tested our method with a DNA plasmid of 4361 bp in length (pBR322). This circular plasmid has seven 5′-TCGA-3′ recognition sites (Sites 1–7) for M.TaqI. After biotin labeling, the plasmid was completely protected against cleavage by R.TaqI indicating at least one adenine within the palindromic 5′-TCGA-3′ is labeled with quantitative efficiency (Figure 1c). The biotinylated plasmid was then linearized by digestion with the restriction endonuclease R.AhdI that cuts at a single site (Figure 1d). A 5-fold excess of monovalent streptavidin (over the total number of 5′-TCGA-3′ sites) was then added to bind to the biotinylated sites and thereby create additional ionic blockades.27,28
The sample was added to the reservoir outside a conical glass nanopore (∼14 nm diameter, see the Supporting Information for nanopore fabrication and characterization) and the ionic current was detected upon applying a positive voltage (Figure 1e). Previous studies have shown that this diameter of conical nanopore enables full translocations of DNA and DNA bound with proteins.13,29 Four molar LiCl solution was used as the electrolyte as it is known to slow the DNA translocation relative to other commonly used salts such as NaCl and KCl.13,30,31 A voltage of 600 mV was chosen to strike a balance between the requirements for high capture rate and slow translocation velocity.32 When entering the nanopore, DNA molecules cause a lowering of the current and passage of streptavidin bound to the DNA creates additional current drops. Figure 1f shows an example of a recorded current trace and events caused by translocations of streptavidin-labeled pBR322 DNA. Because of the wide opening of the nanopores used here (∼14 nm diameter) the DNA can translocate not only in a linear form but also in folded conformations. These folded confirmations have been extensively characterized in the literature and it is well-known that the predominant folded conformation has a single hairpin at the beginning.33,34 We wrote a selection algorithm to exclude those events with a fold at the beginning and only the subset of unfolded events (∼30% of the total) were retained for further analysis (see Figures S2 and S3).
The DNA can pass through the pore in two orientations named as direction 1 (D1) and direction 2 (D2), as shown in Figure 1f. For these two directions, we measured the distribution of signal sizes for the peak caused by the seventh 5′-TCGA-3′ site (Figure 1d) along the plasmid. The distributions were bimodal showing that at each 5′-TCGA-3′ site there are either one or two monovalent streptavidin bound. Control experiments with an engineered DNA double strand containing a single biotin site showed only a unimodal distribution thereby indicating the effect was not due to dimerization of streptavidin in 4 M LiCl (see Figure S5 for details).13 Therefore, the observation of two monovalent streptavidin molecules at some sites likely results from the labeling of both adenine residues within the palindromic 5′-TCGA-3′ sequence. The signal for a single streptavidin is ∼60 pA irrespective of the DNA direction (Figure 1f) which is high compared to the typical noise of our nanopores of 6–10 pA RMS in a 50 kHz bandwidth.17
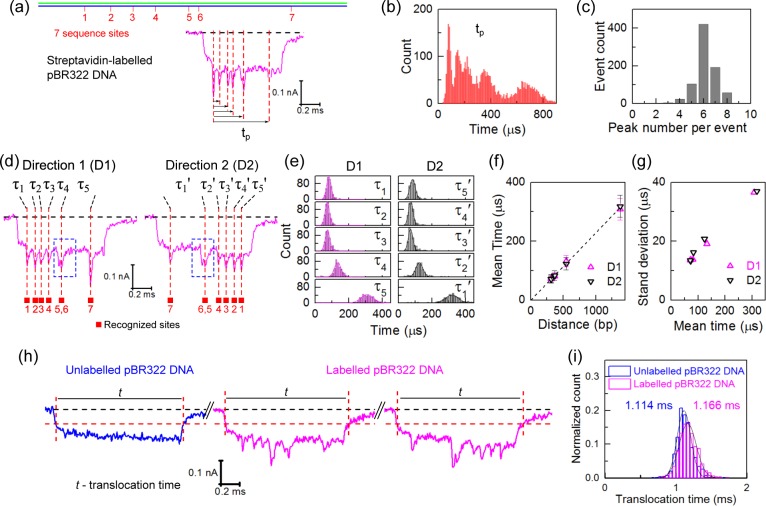
After separating the data set into the two directions, we then used an algorithm to determine the positions of the intraevent peaks caused by the additional current blockade when each streptavidin label passed through the pore (Figure S4). This algorithm first corrects for the baseline DNA translocation and then uses a peak finding routine to determine the time points of each peak in the ionic current as a streptavidin passes through. From visual inspection, streptavidin labels that were separated by >300 bp in the pBR322 DNA were easily resolved whereas the two 5′-TCGA-3′ sites separated by 141 bp mostly showed up as a single peak.
In Figure 2b, we show a histogram of the time points of all peaks measured with respect to the first peak in the translocation (Figure 2a). The data is an aggregate of 812 translocations for direction 1 only. Five peaks are distinguishable in the histogram because the two 5′-TCGA-3′ sites separated by 141 bp were not resolved. The width of each peak in this histogram increases as a function of the time due to fluctuations as the DNA passed through the nanopore. Figure 2c shows a histogram of the number of peaks detected per translocation. Approximately 50% of the translocations were found to have the expected six peaks. Higher or lower numbers of detected peaks can be due to a variety of factors such as errors in the baseline tracking, complex folds, or knots in the DNA creating additional signals or some cases where the two peaks separated by 141 bp were in fact resolved. We note however that unlabeled sites are not thought to be important because the gel characterization of Figure 1c shows that all 5′-TCGA-3′ sites are labeled with at least one biotin.
Figure 2.
Analysis of sequence motif positions in linearized pBR322 DNA. (a) An example event showing the time of each peak (tp) measured with respect to the first peak. (b) Histogram of positions of all peaks measured as defined in (a); only direction 1 translocations are considered (N = 812 events). (c) Histogram of number of peaks detected per translocation in direction 1. (d) Example events observed in nanopore measurements showing the two translocation orientations with sites 1 (Direction 1) and 7 (Direction 2) entering first, respectively. Sites 5 and 6 are separated by only 141 bp so that two separate peaks are not fully resolved (marked by the blue dashed box). (e) Histograms of the translocation times (τ1–τ5 and τ5′−τ1′ for only those with translocations with six peaks detected) between adjacent identification sites in two directions obtained from 422 and 432 events, respectively. (f) Translocation time between peaks as a function of label separation distance. The line shows a least-squares linear fit. The times and error bars are the mean values and standard deviations of the Gaussian fits to the histograms in (e). Sites 5 and 6 are regarded as one and the center between them is used. (g) The standard deviation as a function of mean of the Gaussian fit. (h) Example events measured for a mixture of unlabeled and labeled pBR322 DNA; the red vertical lines denote where the translocation begins and ends which we define at −0.06 nA. (i) Comparison between distributions of translocation times (only unfolded events are considered). The mean translocation time is shown. Experiments shown in (a–g) are conducted with a single pore and (h–i) with a second pore.
Analyzing only those translocations that have six peaks, Figure 2f shows how the mean translocation time (calculated from Gaussian fits to the histograms, Figure 2e) between adjacent peaks varies as a function of the label distance (computed from the known sequence). The data indicates that to a good approximation the intraevent velocity remains constant during a translocation. We recently analyzed the mean intraevent velocity using custom designed DNA rulers that showed a small velocity decrease of ∼5% during the translocation of 7.2 kbp DNA through a nanopore.32 The smaller statistical samples and DNA length used here likely obscures this slight average velocity reduction that is not visible in Figure 2f and for our further analyses in this paper we assume a constant velocity. The velocity did not show a significant change over several hours′ measurement (Figure S6).
Data on the standard deviation of the time interval between peaks as a function of the mean time (both were obtained from the Gaussian fits to the histograms in Figure 2e) is shown in Figure 2g. The standard deviation reflects the uncertainty of position due to natural fluctuations as the DNA threads through the nanopore. These fluctuations create a fundamental physical limit to the accuracy of determining the positions of the labeled sites (Figure 2b). The physical mechanism behind these fluctuations remains to be fully understood but may reflect different conformations of the DNA when it arrives at the pore mouth.32,35,36 Bell et al.32 quantified a standard deviation of ∼500 bp for a 5 kbp distance. This is of the same order of magnitude as the estimated error of ∼1.5 kbp for a 10 kbp distance for optical mapping due to DNA fluctuations in nanochannels.37 This uncertainty caused by fluctuations together with the minimum distance required to resolve two peaks quantifies the genome mapping capability of our system. DNA translocation time and the detection performance were also characterized with different pores (see Figure S7). The percentage of unfolded events that had six peaks ranged from 30 to 50%.
We also performed experiments to determine whether the presence of the protein labels significantly modifies the velocity of the DNA. We mixed together unlabeled pBR322 and labeled pBR322 and measured translocations simultaneously with a single nanopore (Figure 2h). The two populations were separated by determining the number of intraevent peaks for unfolded DNA translocations. Figure 2g shows histograms comparing the translocation times for the labeled and unlabeled DNA (see Figures S8 and S9 for data analysis). The presence of the protein labels might be expected to change the DNA translocation time due to changes in the total charge, increased viscous drag and the possibility of surface interactions between the streptavidin and the nanopore. However, we observe only an increase from 1.1 ± 0.1 ms (μ ± σ from the Gaussian fit) for the unlabeled DNA to 1.2 ± 0.1 ms (μ ± σ) for the labeled DNA indicating that the presence of the labels does not have a significant impact on the DNA velocity (Figure 2g) under our experimental conditions.
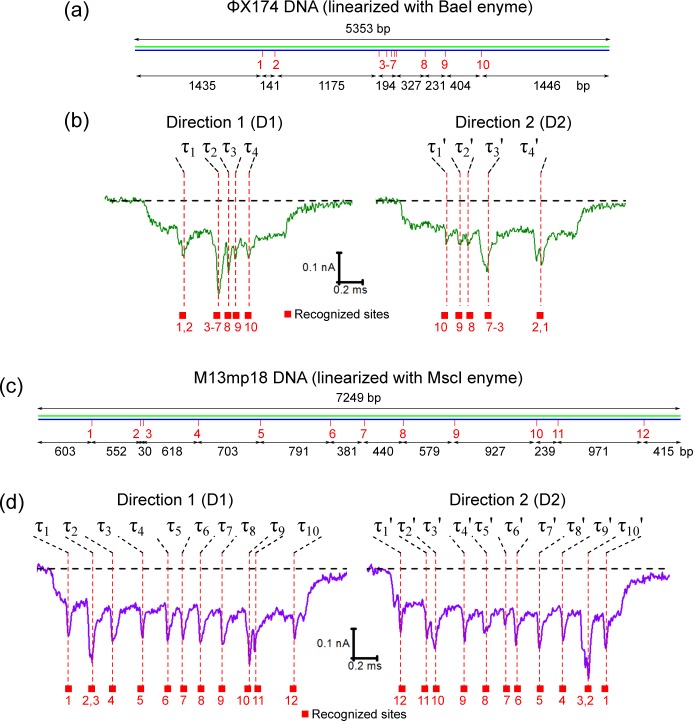
To show that our method can be generally applied to map different genomes, we measured the signals after labeling the DNA of two bacteriophages, ΦX174 DNA (5353 bp after linearization with R.BaeI) and M13mp18 DNA (7249 bp after linearization with R.MscI). These two genomes have 10 and 12 5′-TCGA-3′ sequence motifs, respectively. Figure 3 shows schematics of the labeling of these two genomes and example translocation signals. A detailed analysis on the peak positions is given in the Supporting Information (Figures S10–S15). For the ΦX174 DNA, sites 3–7 are densely distributed on the DNA with an overall distance of 194 bp and cause a wide and deep peak. However, we are able to identify separate peaks with distances of 231 bp (between sites 8 and 9, Figure 3a,b) and 239 bp (between sites 10 and 11, Figure 3c,d). In contrast, the signals are not fully separated for sites 1 and 2 in Figure 3a,b with a smaller distance of 141 bp. Combined with the results for pBR322 DNA in Figure 2, this indicates a minimum resolvable distance between sites of ∼200 bp. This value can be approximately understood based on the known geometry of our nanopores as characterized by scanning electron microscopy.17 The conical shape means the electric field strength 68 nm (∼200 bp) into the pore drops to 37% of its highest value at the pore entrance, according to numerical simulation results calculated with COMSOL Multiphysics 4.4 (Figure S16), so that the baseline has recovered sufficiently for the next peak to be detected. The ∼200 bp resolution is comparable to recent results with super-resolution microscopy.37
Figure 3.
Examples of translocations of ΦX174 and M13mp18 DNA with 10 and 12 labeled sites, respectively. (a) Schematic of the sequence 5′-TCGA-3′ on the ΦX174 DNA linearized by restriction digest with R.BaeI. (b) Example events observed in nanopore measurements. Note that sites 3–7 combine to cause a deep and wide current drop. (c.d) Schematic and example events for M13mp18 DNA (linearized by digestion with R.MscI). The two samples were measured with two different nanopores.
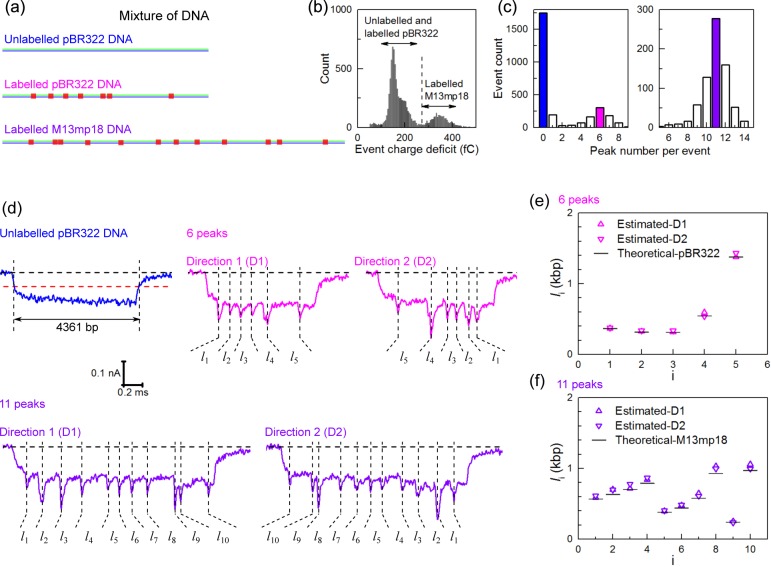
Finally we investigated the potential for simultaneously mapping two DNA molecules present at the same time in the reservoir. We selected labeled pBR322 and labeled M13mp18 as the two genomes to be mapped. We also added unlabeled pBR322 to the sample mixture (Figure 4a) as an independent calibration for determining the velocity of bare DNA for the pore. We added this calibration because it is well established that the DNA velocity can vary slightly from one solid-state nanopore to the next due to differences in geometry and a calibration sample therefore allows us to accurately infer the time-distance relationship for a particular nanopore assuming a constant velocity.13Figure 4b shows a histogram of event charge deficit (ECD), equal to the total charge excluded during a single translocation, for 9244 total events (including folded confirmations). It has previously been shown that ECD gives a measure of DNA length.29 In Figure 4b, the left group of events are caused by the unlabeled and labeled pBR322 DNA and the right group of events are caused by the labeled M13mp18 DNA. Theoretically, unlabeled pBR322 DNA, labeled pBR322 DNA, and labeled M13mp18 DNA would create 0, 6, and 11 peaks, respectively, if all labels with less than 200 bp separation created one peak. We selected events with 0 in the left group as a calibration and events with 6 peaks in the left group and 11 peaks in the right (Figure 4c) to be analyzed. Example events present are shown in Figure 4d and further details of data analysis are shown in the Supporting Information (Figures S17–S19). We estimated the distances between adjacent sites for the two labeled DNA samples based on the velocity computed for unlabeled pBR322 DNA, as shown in Figure 4e,f and Tables S1 and S2 (utilizing the constant velocity assumption). The values are in good agreement with expected ones with the largest error of the estimated mean value around 10% (Tables S1 and S2).
Figure 4.
Detection of multiple DNA molecules in a mixture. (a) The pBR322 DNA, protein-labeled pBR322 DNA (with 7 TCGA sites) and protein-labeled M13mp18 DNA (with 12 TCGA sites) were mixed together. (b) Histogram of the event charge deficit that is approximately proportional to DNA length and shows two populations of pBR322 DNA and M13mp18 DNA. (c) Detected peak number per event for the unfolded events in the left group and right group respectively shown in (b). There are 1752 events with zero peaks and 198 events with one peak showing a false positive peak detection rate of approximately 10% for unlabeled DNA. (d) Example events caused by the mixture. (e) Analysis of distances measured for translocation containing six peaks. The data was transformed from time measurements to distance using the velocity of the unlabeled pBR322 DNA and the value shown is the mean of a Gaussian fit to the distribution. The expected distances are based on the known sequence of pBR322 are also shown. (f) Same analysis as (e) but for only the translocations that show 11 peaks, which is compared with the known separations for M13mp18.
In summary, we have demonstrated a solid-state nanopore platform for mapping genomic DNA by detecting streptavidin labeled on short sequence motifs. Our method shows that labels as close as ∼200 bp can be observed separately. We extensively characterized the relationship between translocation time and distance between labels and showed the ability to simultaneously map two genomes. We envisage several potential avenues for further improvement of the current implementation. Fast voltage switching protocols could enable multiple reads of each individual molecule to reduce the error in localizing each peak.38 This could also help for reading longer DNA molecules because a current difficulty is that longer DNA molecules give fewer unfolded translocations. The event throughput could also be increased by membrane tethering39 or dielectrophoretic concentration.40 We might also use DNA MTases that recognize different sequence motifs in combination with individually addressable nanopores to measure samples at different label densities. The ability to resolve closely separated labels could also be improved by using nanopores in thin 2D membranes because these pores have electric field sensing zones as small as several nanometers.
Overall our solid-state nanopore-based method for determining the positions of short sequence motifs by recording electrical signals can be used for DNA characterization with relatively few labeling and purification steps. In contrast to previous experiments with targeted Cas9 or PNA binding,11,12,24 the short sequence identification enables a generic platform for mapping sequence sites in different DNA molecules with a single workflow. The barcodes generated from labeling short sequence motifs could be correlated with genome databases to enable identification of the different samples present in solution.2 This technique could also straightforwardly be combined with recent developments in specific protein sensing with solid-state nanopores13,17 to provide both DNA and protein detection in a single measurement. The amenability of solid-state nanopores to integration in small chips also suggests potential for portable diagnostic devices.
Acknowledgments
We thank the lab of Mark Howarth at Oxford University for kindly providing monovalent streptavidin and K. Glensk for preparing M.TaqI.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.nanolett.7b01009.
Details on materials and methods including the chemicals, sequence-specific biotinylation of plasmid DNA, glass nanopore fabrication, setup, and nanopore measurements. Data analysis for translocation events caused by different samples, simulation of the electric filed with details shown in Supplementary Figures S1–S19 and Tables S1–S2 (PDF)
Author Present Address
∥ (N.A.W.B.) Department of Chemistry, University of Oxford, Oxford, OX1 3TA, United Kingdom.
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
K.C. acknowledges funding from the China Scholarship Council (201506210147). N.A.W.B. and U.F.K. acknowledge funding from an ERC consolidator grant (Designerpores 647144). E.W. acknowledge financial support from the German-Israeli Foundation for Scientific Research and Development (I-1196-195.9/2012).
The authors declare no competing financial interest.
Supplementary Material
References
- Friedrich S. M.; Zec H. C.; Wang T. H. Lab Chip 2016, 16, 790–811. 10.1039/C5LC01294E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunwald A.; Dahan M.; Giesbertz A.; Nilsson A.; Nyberg L. K.; Weinhold E.; Ambjornsson T.; Westerlund F.; Ebenstein Y. Nucleic Acids Res. 2015, 43, e117. 10.1093/nar/gkv563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neely R. K.; Deen J.; Hofkens J. Biopolymers 2011, 95, 298–311. 10.1002/bip.21579. [DOI] [PubMed] [Google Scholar]
- Doerks T.; Copley R. R.; Schultz J.; Ponting C. P.; Bork P. Genome Res. 2002, 12, 47–56. 10.1101/gr.203201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao M.; Phong A.; Ha C.; Chan T. F.; Cai D.; Leung L.; Wan E.; Kistler A. L.; DeRisi J. L.; Selvin P. R.; Kwok P. Y. Nucleic Acids Res. 2007, 35, e16. 10.1093/nar/gkl1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howorka S.; Siwy Z. ACS Nano 2016, 10, 9768–9771. 10.1021/acsnano.6b07041. [DOI] [PubMed] [Google Scholar]
- McCaffrey J.; Sibert J.; Zhang B.; Zhang Y.; Hu W.; Riethman H.; Xiao M. Nucleic Acids Res. 2016, 44, e11. 10.1093/nar/gkv878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howorka S.; Siwy Z. Chem. Soc. Rev. 2009, 38, 2360–2384. 10.1039/b813796j. [DOI] [PubMed] [Google Scholar]
- Miles B. N.; Ivanov A. P.; Wilson K. A.; Dogan F.; Japrung D.; Edel J. B. Chem. Soc. Rev. 2013, 42, 15–28. 10.1039/C2CS35286A. [DOI] [PubMed] [Google Scholar]
- Tahvildari R.; Beamish E.; Tabard-Cossa V.; Godin M. Lab Chip 2015, 15, 1407–1411. 10.1039/C4LC01366B. [DOI] [PubMed] [Google Scholar]
- Singer A.; Wanunu M.; Morrison W.; Kuhn H.; Frank-Kamenetskii M.; Meller A. Nano Lett. 2010, 10, 738–742. 10.1021/nl100058y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer A.; Rapireddy S.; Ly D. H.; Meller A. Nano Lett. 2012, 12, 1722–1728. 10.1021/nl300372a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell N. A.; Keyser U. F. J. Am. Chem. Soc. 2015, 137, 2035–2041. 10.1021/ja512521w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulushev R. D.; Marion S.; Radenovic A. Nano Lett. 2015, 15, 7118–7125. 10.1021/acs.nanolett.5b03264. [DOI] [PubMed] [Google Scholar]
- Plesa C.; Ruitenberg J. W.; Witteveen M. J.; Dekker C. Nano Lett. 2015, 15, 3153–3158. 10.1021/acs.nanolett.5b00249. [DOI] [PubMed] [Google Scholar]
- Yu J.-S.; Lim M.-C.; Huynh D. T. N.; Kim H.-J.; Kim H.-M.; Kim Y.-R.; Kim K.-B. ACS Nano 2015, 9, 5289–5298. 10.1021/acsnano.5b00784. [DOI] [PubMed] [Google Scholar]
- Bell N. A.; Keyser U. F. Nat. Nanotechnol. 2016, 11, 645–651. 10.1038/nnano.2016.50. [DOI] [PubMed] [Google Scholar]
- Hanz G. M.; Jung B.; Giesbertz A.; Juhasz M.; Weinhold E. J. Visualized Exp. 2014, 10.3791/52014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howarth M.; Chinnapen D. J.; Gerrow K.; Dorrestein P. C.; Grandy M. R.; Kelleher N. L.; El-Husseini A.; Ting A. Y. Nat. Methods 2006, 3, 267–273. 10.1038/nmeth861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottfried A.; Weinhold E. Biochem. Soc. Trans. 2011, 39, 623–628. 10.1042/BST0390623. [DOI] [PubMed] [Google Scholar]
- Zohar H.; Muller S. J. Nanoscale 2011, 3, 3027–3039. 10.1039/c1nr10280j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray A.; Nordén B. FASEB J. 2000, 14, 1041–1060. [DOI] [PubMed] [Google Scholar]
- Deng W.; Shi X.; Tjian R.; Lionnet T.; Singer R. H. Proc. Natl. Acad. Sci. U. S. A. 2015, 112, 11870–11875. 10.1073/pnas.1515692112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulushev R. D.; Marion S.; Petrova E.; Davis S. J.; Maerkl S. J.; Radenovic A. Nano Lett. 2016, 16, 7882–7890. 10.1021/acs.nanolett.6b04165. [DOI] [PubMed] [Google Scholar]
- Lukinavičius G.; Lapienė V.; Staševskij Z.; Dalhoff C.; Weinhold E.; Klimašauskas S. J. Am. Chem. Soc. 2007, 129, 2758–2759. 10.1021/ja0691876. [DOI] [PubMed] [Google Scholar]
- Goedecke K.; Pignot M.; Goody R. S.; Scheidig A. J.; Weinhold E. Nat. Struct. Biol. 2001, 8, 121–125. 10.1038/84104. [DOI] [PubMed] [Google Scholar]
- Carlsen A. T.; Zahid O. K.; Ruzicka J. A.; Taylor E. W.; Hall A. R. Nano Lett. 2014, 14, 5488–5492. 10.1021/nl501340d. [DOI] [PubMed] [Google Scholar]
- Kong J.; Bell N. A.; Keyser U. F. Nano Lett. 2016, 16, 3557–3562. 10.1021/acs.nanolett.6b00627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell N. A.; Muthukumar M.; Keyser U. F. Phys. Rev. E: Stat. Phys., Plasmas, Fluids, Relat. Interdiscip. Top. 2016, 93, 022401. 10.1103/PhysRevE.93.022401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalczyk S. W.; Wells D. B.; Aksimentiev A.; Dekker C. Nano Lett. 2012, 12, 1038–1044. 10.1021/nl204273h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K.; Bell N. A.; Kong J.; Tian Y.; Keyser U. F. Biophys. J. 2017, 112, 674–682. 10.1016/j.bpj.2016.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell N. A.; Keyser U. F.. 2016, arXiv:1607.04612. Accessed on 01/01/17.
- Storm A. J.; Chen J. H.; Zandbergen H. W.; Dekker C. Phys. Rev. E 2005, 71, 051903. 10.1103/PhysRevE.71.051903. [DOI] [PubMed] [Google Scholar]
- Mihovilovic M.; Hagerty N.; Stein D. Phys. Rev. Lett. 2013, 110, 028102. 10.1103/PhysRevLett.110.028102. [DOI] [PubMed] [Google Scholar]
- Chen P.; Gu J.; Brandin E.; Kim Y.-R.; Wang Q.; Branton D. Nano Lett. 2004, 4, 2293–2298. 10.1021/nl048654j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu B.; Albertorio F.; Hoogerheide D. P.; Golovchenko J. A. Biophys. J. 2011, 101, 70–79. 10.1016/j.bpj.2011.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffet J.; Kobo A.; Su T.; Grunwald A.; Green O.; Nilsson A. N.; Eisenberg E.; Ambjörnsson T.; Westerlund F.; Weinhold E.; et al. ACS Nano 2016, 10, 9823. 10.1021/acsnano.6b05398. [DOI] [PubMed] [Google Scholar]
- Gershow M.; Golovchenko J. A. Nat. Nanotechnol. 2007, 2, 775–779. 10.1038/nnano.2007.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusko E. C.; Johnson J. M.; Majd S.; Prangkio P.; Rollings R. C.; Li J.; Yang J.; Mayer M. Nat. Nanotechnol. 2011, 6, 253–260. 10.1038/nnano.2011.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedman K. J.; Otto L. M.; Ivanov A. P.; Barik A.; Oh S. H.; Edel J. B. Nat. Commun. 2016, 7, 10217. 10.1038/ncomms10217. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.