Abstract
A total of 333 fecal specimens from horses in southwestern China were genotyped based on analysis of the small subunit rRNA (SSU rRNA) gene. Cryptosporidium hominis and Cryptosporidium andersoni were identified in 2 and 4 stool specimens, respectively. The identification of C. hominis was confirmed by sequence analysis of the 70‐kDa heat shock protein (HSP70) and oocyst wall protein (COWP) genes. Subtyping analysis of the 60‐kDa glycoprotein (GP60) gene sequence of C. hominis revealed a new rare subtype Id, named IdA15; only three Id isolates have been reported in humans to date. Multilocus sequence typing (MLST) analysis indicated that the C. andersoni subtype was A6, A5, A2, and A1 at the four minisatellite loci (MS1, MS2, MS3, and MS16, respectively). This is the first report to identify the presence of C. andersoni and C. hominis in horses in southwestern China and the first to identify a rare zoonotic subtype Id of C. hominis in horses. These findings suggest that infected horses may act as potential reservoirs of Cryptosporidium to transmit infections to humans.
Keywords: GP60, multilocus sequence typing, transmission
CRYPTOSPORIDIUM spp. are common protozoan parasites that cause diarrhea in a wide variety of vertebrate animals. Although Cryptosporidium was initially discovered by Tyzzer in 1907, it was not recognized as an important parasite for the next 50 yr (Tyzzer 1907). Subsequently, Cryptosporidium was also identified in turkeys, calves and humans with diarrhea, and received more attention around the world (Current et al. 1983; Panciera et al. 1970; Slavin 1955). Cryptosporidium attracted great public health attention in the large waterborne outbreak in Milwaukee (U.S.A.) in 1993, where over 400,000 people were infected (Kenzie et al. 1995). In young calves, high morbidity caused by Cryptosporidium can lead to substantial production losses to the livestock industry (Graaf et al. 1999).
Most animals become infected through direct contact with an infected host, or through indirect ingestion of oocyst‐contaminated water or foods (Cacciò 2005; Fayer et al. 2000). Although clinical presentations of cryptosporidiosis are often self‐limiting or sometimes asymptomatic in healthy individuals, fatal and chronic diarrhea can occur in immunocompromised or immunosuppressed individuals (Petri et al. 2008). Recent molecular epidemiologic surveys of cryptosporidiosis from different host species have identified at least 30 valid species and more than 70 genotypes, with new genotypes continually being found (Ryan et al. 2014, 2015). Of these, Cryptosporidium hominis is primarily a human pathogen (Xiao et al. 2001), and Cryptosporidium andersoni is the major species responsible for bovine cryptosporidiosis (Liu et al. 2015).
In China, horses are commonly used for work, entertainment, and competitions, usually living in close association with humans. Cryptosporidiosis was initially described in immune‐deficient Arabian foals by Snyder et al. (1978). To date, nine Cryptosporidium species/genotypes have been identified in equines in more than eight countries (Liu et al. 2015). However, only limited reports about horse cryptosporidiosis are available in China, and the zoonotic transmission of Cryptosporidium between humans and horses remains unknown. Thus, the present study was conducted to identify Cryptosporidium species/genotypes in horses from southwestern China, and to assess the zoonotic potential of cryptosporidiosis from horses to humans.
Materials and Methods
Ethics statement
The present study protocol was reviewed and approved by the Research Ethics Committee and the Animal Ethical Committee of Sichuan Agricultural University. Permission was obtained from farm managers before the collection of fecal specimens from horses.
Specimen collection
In total, 333 fecal samples from horses of different ages (3 mo to 16 yr) were randomly collected on five farms with different management systems in the Sichuan and Yunnan provinces of southwestern China between August 2015 and April 2016. In total, 100 horses from Farm 1 (48 horses) and Farm 2 (52 horses), which are both equestrian clubs, were kept in stables; 56 horses from Farm 3 were kept in pastures; 12 horses from Farm 4, mainly used for experimental research, were kept in stables; and 165 horses from Farm 5 were used for breeding in pastures and stables. Each sample was collected using a sterile disposable latex glove immediately after the horse defecated onto the ground, and was then placed into an individual sterile plastic container. No obvious clinical signs were observed in any of the sampled horses. Samples were maintained at 4 °C until DNA extraction.
DNA extraction
Approximately 0.5 g of each of the stored fecal specimens was washed three times with distilled water by centrifugation at 2,000 g for 10 min. DNA was extracted from the washed fecal samples, using an EZNA® Stool DNA kit (Omega Biotek, Norcross, GA) according to the manufacturer's recommended protocol. DNA was eluted in 200 μl of the Solution Buffer of the kit and stored at −20 °C until used for polymerase chain reaction (PCR) analysis.
Cryptosporidium genotyping and subtyping
Cryptosporidium species/genotypes were identified in all of the extracted DNA samples by nested PCR amplification of an ∼830‐bp fragment of the small subunit rRNA (SSU rRNA) gene (Xiao et al. 1999) (Table S1). The identification of C. hominis was confirmed by PCR and sequence analysis of the approximately 1,950‐ and 550‐bp fragments for the Cryptosporidium oocyst wall protein (COWP) and 70‐kDa heat shock protein (HSP70) genes, respectively (Sulaiman et al. 2000; Xiao et al. 2000) (Table S1). Subtype identification of C. hominis isolates was performed by sequence analysis of an approximately 800‐bp fragment of the 60‐kDa glycoprotein (GP60) gene, and C. andersoni isolates were determined by amplification at four loci MS1, MS2, MS3, MS16 (fragment lengths of approximately 550, 450, 530, and 590 bp, respectively). Primers and amplification conditions were adopted as previously described (Sulaiman et al. 2005; Feng et al. 2011) (Table S1).
Nucleotide sequencing and analysis
All secondary PCR products were directly sequenced at Life Technologies (Guangzhou, China) using an ABI Big Dye Terminator v3.1 cycle sequencing kit (Applied Biosystems, Carlsbad, CA). The accuracy of the sequences was confirmed by bidirectional sequencing, and a new PCR secondary product was re‐sequenced if necessary. The nucleotide sequences obtained in this study were aligned with the corresponding Cryptosporidium reference sequences downloaded from the GenBank database, using BLAST (http://blast.ncbi.nlm.nih.gov) and ClustalX 1.83 (http://www.clustal.org/) to determine Cryptosporidium species/genotypes and subtypes.
Phylogenetic analysis
Phylogenetic analysis was performed by constructing neighboring‐joining trees of the GP60 genes, using the program Mega 6 (http://www.megasoftware.net/) based on the evolutionary distances calculated by the Kimura‐2‐parameter model. The reliability of the trees was assessed by bootstrap analysis with 1,000 replicates.
Nucleotide sequence accession numbers
The unique partial nucleotide sequences of the SSU rRNA, HSP70, COWP, GP60 genes and the four minisatellite loci (MS1, MS2, MS3, and MS16) obtained in the present study were deposited in the GenBank database under accession numbers KX926452–KX926463 and KY210530–KY210545, respectively.
Results and Discussion
In the present study, six of the 333 (1.8%) fecal specimens were detected to be positive for Cryptosporidium by PCR amplification of the partial SSU rRNA gene, which is consistent with the previously reported low infection rate of Cryptosporidium in horses in Algeria (2.3%), Poland (3.4%), and New York state (5.1%) (Burton et al. 2010; Laatamna et al. 2015; Wagnerová et al. 2015). The age of the Cryptosporidium‐positive horses was between 2 and 3 yr. Analysis of the sequences of the SSU rRNA gene of the six Cryptosporidium‐positive samples revealed the presence of two C. hominis and four C. andersoni isolates. Comparison of the SSU rRNA gene sequences revealed that the two C. hominis isolates were identical to the C. hominis (KF679723) isolate derived from a rhesus macaque in China (Karim et al. 2014), and the three C. andersoni isolates were identical to the C. andersoni (KT175425) isolate derived from wastewater in Iran. However, the remaining sequence of C. andersoni had one nucleotide substitution (T/C at nucleotide 595) when compared to the C. andersoni (KF271453) isolate derived from a human in China. The HSP70 sequence of one isolate had two nucleotide substitutions (T/G and A/T at positions 41 and 879, respectively) compared to the sequence (KF679727) derived from a rhesus macaque in China, but the other isolate was identical to the published C. hominis sequence KM116517 derived from a human in China. Interestingly, the two C. hominis COWP gene sequences obtained in this study were identical to each other, and were also identical to C. hominis isolates obtained from a human (DQ388389) and a monkey (AF266272).
In a previous study, C. hominis was first identified in only one horse in Algeria, and a novel GP60 subtype family, Ik, was identified (Laatamna et al. 2015). In Brazil, C. hominis was reported in two foals of 4 and 5 mo of age, respectively, and one subtype IkA20G1 was found (Inácio et al. 2017). In contrast, C. hominis was identified as the dominant parasite (61/87, 70.1%) in horses in China, and two subtypes were found: IkA16G1 and IkA16 (Jian et al. 2016). In the present study, two isolates of C. hominis were successfully amplified at the GP60 locus and were identified for subtypes. DNA sequence analysis showed that the two isolates have a maximum nucleotide identity of 99% to a previously characterized Australian C. hominis subtype, IdA26 (FJ861226). These subtypes have less than 11 nucleotide TCA repeats and five nucleotide substitutions: A/G, G/A, T/C, C/A, and T/C at nucleotides 182, 258, 316, 484, and 835, respectively. The two isolates were determined to belong to the Id subtype based on phylogenetic analysis, and were named IdA15 in accordance with current Cryptosporidium nomenclature conventions (Fig. 1) (Sulaiman et al. 2005). Human infections with subtype Id C. hominis have previously been reported in China, Canada (Ontario and British Columbia), and the Arctic, and no animal‐derived subtype Id isolates have been described to date (Karine et al. 2016; Ong et al. 2008; Trotzwilliams et al. 2006; Wang et al. 2011). The subtype Id of C. hominis has been detected in humans in the area of Tianjin and Henan province in China (Wang et al. 2011). The two isolates found in horses were from Farm 2 in this area. On this farm close contact between people and horses may be responsible for infection of the horses with C. hominis. Whether these horses are truly infected or merely act as mechanical carriers of accidentally ingested oocysts of anthroponotic origin remains to be determined.
Figure 1.
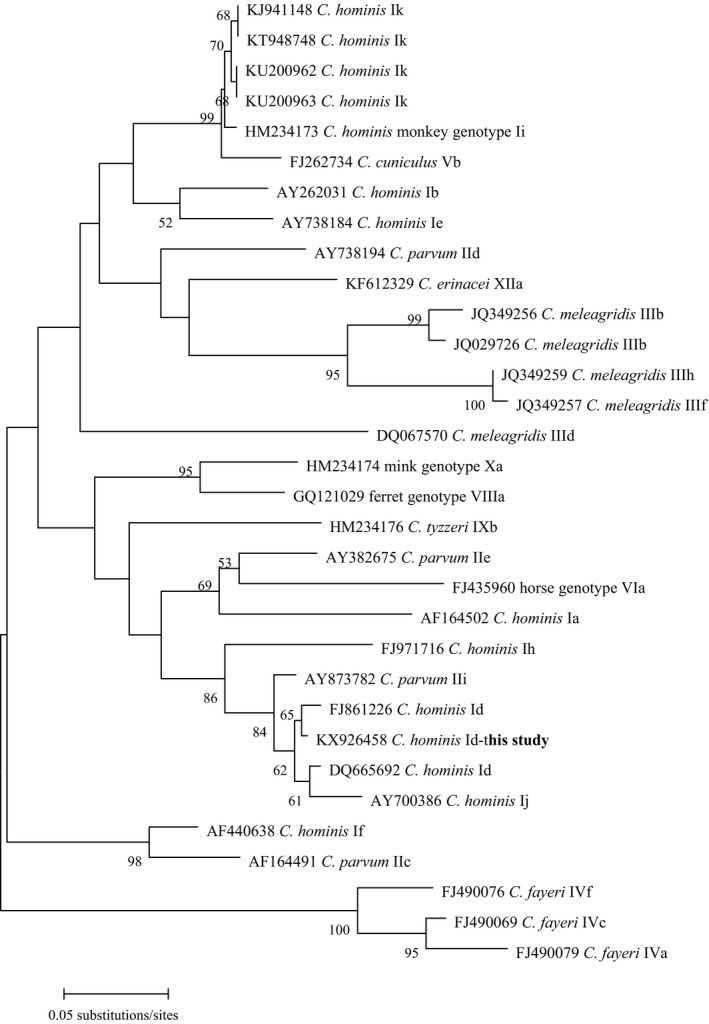
Phylogenetic relationship of 60‐kDa glycoprotein (GP60) nucleotide sequences of the Cryptosporidium equine isolate in this study to multiple subtype families in C. hominis, as inferred by a neighbor‐joining analysis based on evolutionary distances calculated using the Kimura two‐parameter model. The tree was rooted with partial subtypes of C. fayeri. Bootstrap values were obtained using 1,000 pseudo‐replicates, with only values above 50% reported.
Cryptosporidium andersoni was considered the major species causing bovine cryptosporidiosis in previous studies (Liu et al. 2015; Zhao et al. 2015), but it has been identified in various hosts in China, including humans, golden takins, bactrian camels, cattle, sheep, goats, and yaks (Liu et al. 2014a,b; Qi et al. 2015; Zhao et al. 2015). Each C. andersoni‐positive isolate was successfully amplified and sequenced at the four minisatellite loci: MS1, MS2, MS3, and MS16. Multiple‐sequence alignment analysis of the four C. andersoni‐positive sequences showed A6, A5, A2, and A1 at the four loci, respectively. This MLST subtype was first reported in Bactrian camels in the Henan province, and has also been reported in Bactrian camels in the Sichuan province in China (Wang et al. 2012).
In summary, this is the first report of C. hominis and C. andersoni in horses in southwestern China, as well as the first report of the rare zoonotic subtype Id of C. hominis and the MLST subtypes A6, A5, A2, and A1 of C. andersoni in horses overall. Cryptosporidium andersoni has been identified in other animals, and the MLST subtypes A6, A5, A2, and A1 have also been reported in other animals in southwestern China. It is therefore likely that the horse‐derived C. andersoni resulted from cross‐species transmission. Importantly, the presence of the C. hominis Id subtype in both humans and horses indicates that it may present a threat to public health.
Supporting information
Table S1. Gene locus and primer sequences used in this study, annealing temperatures used in the PCR and expected sizes of the PCR products.
Acknowledgments
This study was financially supported by the Chengdu Giant Panda Breeding Research Foundation (CPF2014‐14; CPF2015‐4).
Lei Deng, Wei Li, Zhijun Zhong and Chao Gong contributed equally to this work.
Contributor Information
Kongju Wu, Email: 646401864@qq.com.
Guangneng Peng, Email: pgn.sicau@163.com.
Literature Cited
- Burton, A. J. , Nydam, D. V. , Dearen, T. K. , Mitchell, K. , Bowman, D. D. & Xiao, L. 2010. The prevalence of Cryptosporidium, and identification of the Cryptosporidium horse genotype in foals in New York State. Vet. Parasitol., 174:139–144. [DOI] [PubMed] [Google Scholar]
- Cacciò, S. M. 2005. Molecular epidemiology of human cryptosporidiosis. Parassitologia, 47:185–192. [PubMed] [Google Scholar]
- Current, W. L. , Reese, N. C. , Ernst, J. V. , Bailey, W. S. , Heyman, M. B. & Weinstein, W. M. 1983. Human cryptosporidiosis in immunocompetent and immunodeficient persons. Studies of an outbreak and experimental transmission. N. Engl. J. Med., 308:1252–1257. [DOI] [PubMed] [Google Scholar]
- Fayer, R. , Morgan, U. & Upton, S. J. 2000. Epidemiology of Cryptosporidium: transmission, detection and identification. Int. J. Parasitol., 30:1305–1322. [DOI] [PubMed] [Google Scholar]
- Feng, Y. , Yang, W. , Ryan, U. , Zhang, L. , Kváč, M. , Koudela, B. , Modrý, D. , Li, N. , Fayer, R. & Xiao, L. 2011. Development of a multilocus sequence tool for typing Cryptosporidium muris and Cryptosporidium andersoni . J. Clin. Microbiol., 49:34–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graaf, D. C. D. , Vanopdenbosch, E. , Ortega‐Mora, L. M. , Abbassi, H. & Peeters, J. E. 1999. A review of the importance of cryptosporidiosis in farm animals. Int. J. Parasitol., 29:1269–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inácio, S. V. , Widmer, G. , de Brito, R. L. , Zucatto, A. S. , de Aquino, M. C. , Oliveira, B. C. , Nakamura, A. A. , Neto, L. D. , Carvalho, J. G. , Gomes, J. F. , Meireles, M. V. & Bresciani, K. D. 2017. First description of Cryptosporidium hominis GP60 genotype IkA20G1 and Cryptosporidium parvum GP60 genotypes IIaA18G3R1 and IIaA15G2R1 in foals in Brazil. Vet. Parasitol., 233:48–51. [DOI] [PubMed] [Google Scholar]
- Jian, F. , Liu, A. , Wang, R. , Zhang, S. , Qi, M. , Zhao, W. , Shi, Y. , Wang, J. , Wei, J. & Zhang, L. 2016. Common occurrence of Cryptosporidium hominis in horses and donkeys. Infect. Genet. Evol., 43:261–266. [DOI] [PubMed] [Google Scholar]
- Karim, M. R. , Wang, R. , He, X. , Zhang, L. , Li, J. , Rume, F. I. , Dong, H. , Qi, M. , Jian, F. , Zhang, S. , Sun, M. , Yang, G. , Zou, F. , Ning, C. & Xiao, L. 2014. Multilocus sequence typing of Enterocytozoon bieneusi in nonhuman primates in China. Vet. Parasitol., 200:13–23. [DOI] [PubMed] [Google Scholar]
- Karine, T. , Asma, I. , Brent, D. , Réjean, D. , Benoît, L. , Philippe, C. , Lyne, C. , Momar, N. , Jean‐François, P. & Yansouni, C. P. 2016. Cryptosporidium hominis is a newly recognized pathogen in the Arctic region of Nunavik, Canada: molecular characterization of an outbreak. PLoS Negl. Trop Dis., 10(4):e0004534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenzie, W. R. M. , Schell, W. L. , Blair, K. A. , Addiss, D. G. , Dan, E. P. , Hoxie, N. J. , Kazmierczak, J. J. & Davis, J. P. 1995. Massive outbreak of waterborne Cryptosporidium infection in Milwaukee, Wisconsin: recurrence of illness and risk of secondary transmission. Clin. Infect. Dis., 21:57–62. [DOI] [PubMed] [Google Scholar]
- Laatamna, A. E. , Wagnerová, P. , Sak, B. , Květoňová, D. , Xiao, L. , Rost, M. , Mcevoy, J. , Saadi, A. R. , Aissi, M. & Kváč, M. 2015. Microsporidia and Cryptosporidium in horses and donkeys in Algeria: detection of a novel Cryptosporidium hominis subtype family (Ik) in a horse. Vet. Parasitol., 208:135–142. [DOI] [PubMed] [Google Scholar]
- Liu, A. , Jia, Z. , Zhao, J. , Wei, Z. , Wang, R. & Zhang, L. 2015. The first report of Cryptosporidium andersoni in horses with diarrhea and multilocus subtype analysis. Parasit. Vectors, 8:1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H. , Shen, Y. , Yin, J. , Yuan, Z. , Jiang, Y. , Xu, Y. , Pan, W. , Hu, Y. & Cao, J. 2014a. Prevalence and genetic characterization of Cryptosporidium, Enterocytozoon, Giardia and Cyclospora in diarrheal outpatients in China. BMC Infect. Dis., 14:290–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, X. , Zhou, X. , Zhong, Z. , Deng, J. , Chen, W. , Cao, S. , Fu, H. , Zuo, Z. , Hu, Y. & Peng, G. 2014b. Multilocus genotype and subtype analysis of Cryptosporidium andersoni derived from a Bactrian camel (Camelus bactrianus) in China. Parasitol. Res., 113:2129–2136. [DOI] [PubMed] [Google Scholar]
- Ong, C. S. , Chow, S. , Gustafson, R. , Plohman, C. , Parker, R. , Isaac‐Renton, J. L. & Fyfe, M. W. 2008. Rare Cryptosporidium hominis subtype associated with aquatic center use. Emerg. Infect. Dis., 14:1323–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panciera, R. J. , Thomassen, R. W. & Garner, F. M. 1970. Cryptosporidial infection in a calf. Vet. Pathol., 8:479–484. [Google Scholar]
- Petri, W. A. , Miller, M. , Binder, H. J. , Levine, M. M. , Dillingham, R. & Guerrant, R. L. 2008. Enteric infections, diarrhea, and their impact on function and development. J. Clin. Invest., 118:1277–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi, M. , Wang, H. , Jing, B. , Wang, D. , Wang, R. & Zhang, L. 2015. Occurrence and molecular identification of Cryptosporidium spp. in dairy calves in Xinjiang, Northwestern China. Vet. Parasitol., 212:404–407. [DOI] [PubMed] [Google Scholar]
- Ryan, U. , Fayer, R. & Xiao, L. 2014. Cryptosporidium species in humans and animals: current understanding and research needs. Parasitology, 141:1–19. [DOI] [PubMed] [Google Scholar]
- Ryan, U. , Paparini, A. , Tong, K. , Yang, R. , Gibson‐Kueh, S. , O'Hara, A. , Lymbery, A. & Xiao, L. 2015. Cryptosporidium huwi n. sp. (Apicomplexa: Eimeriidae) from the guppy (Poecilia reticulata). Exp. Parasitol., 150:31–35. [DOI] [PubMed] [Google Scholar]
- Slavin, D. 1955. Cryptosporidium meleagridis (sp. nov.). J. Comp. Pathol. Ther., 65:262–266. [DOI] [PubMed] [Google Scholar]
- Snyder, S. P. , England, J. J. & Mcchesney, A. E. 1978. Cryptosporidiosis in immunodeficient Arabian foals. Vet. Pathol., 15:12–17. [DOI] [PubMed] [Google Scholar]
- Sulaiman, I. M. , Hira, P. R. , Zhou, L. , Alali, F. M. , Alshelahi, F. A. , Shweiki, H. M. , Iqbal, J. , Khalid, N. & Xiao, L. 2005. Unique endemicity of cryptosporidiosis in children in Kuwait. J. Clin. Microbiol., 43:2805–2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulaiman, I. M. , Morgan, U. M. , Thompson, R. C. A. , Lal, A. A. & Xiao, L. 2000. Phylogenetic relationships of Cryptosporidium parasites based on the 70‐kilodalton heat shock protein (HSP70) gene. Appl. Environ. Microbiol., 66:2385–2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotzwilliams, L. A. , Martin, D. S. , Gatei, W. , Cama, V. , Peregrine, A. S. , Martin, S. W. , Nydam, D. V. , Jamieson, F. & Xiao, L. 2006. Genotype and subtype analyses of Cryptosporidium isolates from dairy calves and humans in Ontario. Parasitol. Res., 99:346–352. [DOI] [PubMed] [Google Scholar]
- Tyzzer, E. E. 1907. A sporozoan fund in the peptic glands of the common mouse. Proc. Soc. Exp. Biol. Med., 56:12–13. [Google Scholar]
- Wagnerová, P. , Sak, B. , Mcevoy, J. , Rost, M. , Matysiak, A. P. , Ježková, J. & Kváč, M. 2015. Genetic diversity of Cryptosporidium spp. including novel identification of the Cryptosporidium muris and Cryptosporidium tyzzeri in horses in the Czech Republic and Poland. Parasitol. Res., 114:1619–1624. [DOI] [PubMed] [Google Scholar]
- Wang, R. , Jian, F. , Zhang, L. , Ning, C. , Liu, A. , Zhao, J. , Feng, Y. , Qi, M. , Wang, H. & Lv, C. 2012. Multilocus sequence subtyping and genetic structure of Cryptosporidium muris and Cryptosporidium andersoni . PLoS ONE, 7:e43782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, R. , Zhang, X. , Zhu, H. , Zhang, L. , Feng, Y. , Jian, F. , Ning, C. , Qi, M. , Zhou, Y. & Fu, K. 2011. Genetic characterizations of Cryptosporidium spp. and Giardia duodenalis in humans in Henan, China. Exp. Parasitol., 127:42–45. [DOI] [PubMed] [Google Scholar]
- Xiao, L. , Bern, C. , Limor, J. , Sulaiman, I. , Roberts, J. , Checkley, W. , Cabrera, L. , Gilman, R. H. & Lal, A. A. 2001. Identification of 5 types of Cryptosporidium parasites in children in Lima, Peru. J. Infect. Dis., 183:492–497. [DOI] [PubMed] [Google Scholar]
- Xiao, L. , Escalante, L. , Yang, C. , Sulaiman, I. , Escalante, A. A. , Montali, R. J. , Fayer, R. & Lal, A. A. 1999. Phylogenetic analysis of Cryptosporidium parasites based on the small‐subunit rRNA gene locus. Appl. Environ. Microbiol., 65:1578–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, L. H. , Limor, J. , Morgan, U. M. , Sulaiman, I. M. , Thompson, R. C. A. & Lal, A. A. 2000. Sequence differences in the diagnostic target region of the oocyst wall protein gene of Cryptosporidium parasites. Appl. Environ. Microbiol., 66:5499–5502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, G. H. , Du, S. Z. , Wang, H. B. , Hu, X. F. , Deng, M. J. , Yu, S. K. , Zhang, L. X. & Zhu, X. Q. 2015. First report of zoonotic Cryptosporidium spp., Giardia intestinalis and Enterocytozoon bieneusi in golden takins (Budorcas taxicolor bedfordi). Infect. Genet. Evol., 34:394–401. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Gene locus and primer sequences used in this study, annealing temperatures used in the PCR and expected sizes of the PCR products.


