Summary
As a member of subgingival multispecies biofilms, Tannerella forsythia is commonly associated with periodontitis. The bacterium has a characteristic cell surface (S‐) layer modified with a unique O‐glycan. Both the S‐layer and the O‐glycan were analyzed in this study for their role in biofilm formation by employing an in vitro multispecies biofilm model mimicking the situation in the oral cavity. Different T. forsythia strains and mutants with characterized defects in cell surface composition were incorporated into the model, together with nine species of select oral bacteria. The influence of the T. forsythia S‐layer and attached glycan on the bacterial composition of the biofilms was analyzed quantitatively using colony‐forming unit counts and quantitative real‐time polymerase chain reaction, as well as qualitatively by fluorescence in situ hybridization and confocal laser scanning microscopy. This revealed that changes in the T. forsythia cell surface did not affect the quantitative composition of the multispecies consortium, with the exception of Campylobacter rectus cell numbers. The localization of T. forsythia within the bacterial agglomeration varied depending on changes in the S‐layer glycan, and this also affected its aggregation with Porphyromonas gingivalis. This suggests a selective role for the glycosylated T. forsythia S‐layer in the positioning of this species within the biofilm, its co‐localization with P. gingivalis, and the prevalence of C. rectus. These findings might translate into a potential role of T. forsythia cell surface structures in the virulence of this species when interacting with host tissues and the immune system, from within or beyond the biofilm.
Keywords: Campylobacter rectus, cell surface, oral biofilm, periodontal disease, S‐layer glycosylation, Tannerella forsythia
1. INTRODUCTION
To proliferate and persist in their habitat, bacteria tend to live predominately in biofilms, which are highly complex and dynamic, polymicrobial communities providing protection from shear forces and host immune responses.1 In the oral cavity, multispecies biofilms constitute what is known as “dental plaque”.2 In a healthy individual, the oral bacteria exist in a natural balance with their host. However, different factors such as smoking, diabetes, genetic predisposition, or poor dental hygiene can cause the community to become dysbiotic,3, 4 enabling potentially pathogenic bacteria to increase in numbers and cause persistent infections, such as periodontitis.
It has been recognized that periodontitis has a polymicrobial biofilm etiology and is primarily characterized by a shift in the microbial composition and promotion of growth of Gram‐negative anaerobes; among these are the periodontal pathogens Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia.5 These so‐called “red complex” bacteria are able to subvert host immune responses, modulate the infection process within the subgingival pocket, and promote dysbiosis through the expression of virulence factors.6 In the case of P. gingivalis, interbacterial interaction and adhesion to host cells are facilitated through the production of colonization factors such as hemagglutinins and fimbriae.7 The latter also induce the expression of pro‐inflammatory cytokines, such as interleukin‐1 (IL‐1), IL‐6, IL‐8, and tumor necrosis factor‐α (TNF‐α),8 stimulating the immune response during infection. Porphyromonas gingivalis further possesses a set of specialized cell surface cysteine proteinases, the gingipains. They can modulate the host immune response through T‐cell receptor cleavage,9 proteolytic processing of components of the complement system,10 activation of protease‐activated receptors, and inactivation of pro‐ and anti‐inflammatory cytokines.11, 12, 13, 14 The oral spirochete T. denticola is the only motile member of the “red complex” consortium.15 Through the expression of flagellar, chemotactic, and proteolytic factors, T. denticola is able to penetrate and directly interact with the gingival epithelium and underlying connective tissue.16, 17 Here, the principal immunogenic surface antigen of T. denticola, the major sheath protein Msp, facilitates actin remodeling and reorganization in host cells and thereby impairs neutrophil chemotaxis and phagocytic activity.18, 19, 20 Through the action of a surface‐associated protease dentilisin, T. denticola has been shown to modulate host cell immune responses by degradation of IL‐1β, IL‐6, TNF‐α, and monocyte chemoattractant protein 1.21, 22
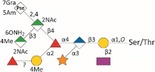
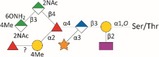
Like T. denticola, T. forsythia is characterized by its fastidious growth requirements and is, especially through its initial recalcitrance to genetic manipulation, a less characterized member of the “red complex” consortium. It has been shown to express several putative virulence factors;23 among them is its characteristic two‐dimensional (2D) crystalline cell surface (S‐) layer.24, 25 Tannerella forsythia is the only member of the “red complex” consortium that possesses an S‐layer fully covering the bacterial cells; this is formed by self‐assembly of the two S‐layer proteins TfsA and TfsB,25 both of which are modified by a unique, complex, branched dekasaccharide that is synthesized by the general protein O‐glycosylation system of the bacterium26 (Table 1). This dekasaccharide is O‐glycosidically bound to multiple serine or threonine residues within a D(S/T)(A/I/L/M/T/V) amino acid target motif present on TfsA and TfsB, but also on several other T. forsythia proteins.26 S‐layer protein glycosylation was shown to be completed in the bacterial periplasm before glycoprotein export via a type IX secretion system27, 28 followed by anchoring of the glycoproteins in the cell envelope and equimolar self‐assembly into the mature S‐layer lattice at the cell surface. Given the nanometer‐scaled periodicity of the 2D S‐layer lattice, this strategy results in a high‐density cell surface display of O‐glycans. This surface glycosylation affects the physicochemical properties of the bacterial cell surface through the introduction of charged sugar residues (for structure of the O‐glycan see Table 1) and modulates bacterial cell hydrophobicity. The prominent cellular location and abundance of the O‐glycan as well as the S‐layer matrix itself make them ideal candidates for influencing interbacterial or bacterium–host interactions as may occur in oral biofilms.
Table 1.
Tannerella forsythia strains and their cell surface mutants cultivated in the subgingival “Zurich biofilm model”
| Strains | Cell surface and glycan properties | Structure of O‐glycan |
|---|---|---|
| ATCC 43037 | ||
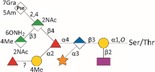
| Wild‐type | wild‐type; S‐layer glycan with terminal Pse residue |
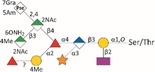
|
| ΔpseC (Tanf_01190) | S‐layer glycan devoid of terminal Pse |
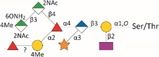
|
| ΔwecC (Tanf_01280) | S‐layer glycan devoid of trisaccharide branch containing Pse and two ManNAcA residues |
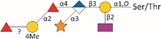
|
| ΔtfsAB (Tanf_03370; Tanf_03375) | S‐layer deficient mutant; this mutant may expose R‐type lipopolysaccharide or O‐glycans from outer membrane glycoproteins |
|
| ΔpseC comp (Tanf_01190) | reconstituted mutant ΔpseC |
|
| UB4 | ||
| Wild‐type | wild‐type; S‐layer glycan with terminal Leg residue |

|
| ΔlegC (TFUB4_00900) | S‐layer glycan devoid of terminal Leg |
|
| ΔlegC comp (TFUB4_00900) | reconstituted mutant ΔlegC |

|
 galactose;
galactose;  xylose;
xylose;  nonulosonic acid; Gra N‐glyceroyl;
nonulosonic acid; Gra N‐glyceroyl;  glucuronic acid;
glucuronic acid;  digitoxose; NAc N‐acetyl; Me O‐methyl;
digitoxose; NAc N‐acetyl; Me O‐methyl;  fucose;
fucose;  mannosaminuronic acid; Am acetamidino; Gc glycolyl
mannosaminuronic acid; Am acetamidino; Gc glycolyl
In biofilms, the physical properties of the bacterial cell surface come into play, as initial attachment by planktonic bacteria to a substrate is primarily influenced by factors such as surface charge, hydrophobicity or electrostatic interactions, whereas the formation of a stable biofilm is facilitated by specialized surface components such as flagella, fimbriae, or pili and the production of an exopolysaccharide matrix.29, 30 The oral bacterium Streptococcus sanguis, for instance, has been shown to largely depend on hydrophobic effect interactions for adhesion to the salivary pellicle.31, 32 Streptococcus parasanguinis, another early colonizer of the dental surface, requires glycosylation of the fimbria‐associated adhesin Fap1 for the formation of stable biofilms.33, 34 In Campylobacter spp., loss of flagellum glycosylation negatively affects the bacterium's ability to form microcolonies and, subsequently, biofilms.35, 36 In Campylobacter jejuni, the flagellum is heavily glycosylated by the addition of O‐linked pseudaminic acid (Pse) and legionaminic acid (Leg),37, 38 a feature that has been shown to orchestrate the bacterium's virulence potential.36
We recently found evidence that the T. forsythia ATCC 43037 wild‐type strain carries a modified Pse residue as a terminal constituent of the S‐layer O‐glycan,26 whereas in the clinical isolate T. forsythia UB4, this residue is present as its stereoisomer, Leg39 (Table 1, see Supplementary material, Fig. S1). Pse (5,7‐diacetamido‐3,5,7,9‐tetradeoxy‐l‐glycero‐l‐manno‐non‐2‐ulosonic acid) as well as Leg (5,7‐diacetamido‐3,5,7,9‐tetradeoxy‐d‐glycero‐d‐galacto‐non‐2‐ulosonic acid) appear to be unique to bacteria.39 They belong to the class of nonulosonic acids, acidic nine‐carbon (C9) α‐keto sugars, which are best represented by the sialic acid family abundantly displayed on the exterior of mammalian cells functioning in cell–cell communication and adhesion.40
The T. forsythia S‐layer has been described to facilitate adhesion to and invasion of gingival epithelial cells,41 suppress pro‐inflammatory cytokine production,42 and inhibit monospecies biofilm formation;28 however, without dissecting any potential contribution of the O‐glycan attached to the S‐layer. Honma et al. (43) observed an increase in T. forsythia biofilm formation upon deletion of a UDP‐N‐acetyl‐d‐mannosaminuronic dehydrogenase (WecC) – later found to cause a three‐sugar truncation of the T. forsythia O‐glycan26 (compare with Table 1) – when cells were cultivated in an untreated polystyrene culture dish.43 In contrast, deficiency in the O‐glycan's terminal nonulosonic acid in a T. forsythia ATCC 43037 ΔpseC and a T. forsythia UB4 ΔlegC mutant, respectively, decreased biofilm formation on a mucin‐coated surface.39 Although these data together demonstrate the involvement of both S‐layer and attached sugar moieties in monospecies biofilm formation, the question arises to what extent these observations are influenced by the physical properties of the surface provided for cell attachment and, above that, demand an investigation into if and how the described effects translate into a multispecies biofilm that more adequately mirrors the in vivo situation. As part of a multispecies biofilm consortium, T. forsythia needs to interact with numerous other bacteria. How these interactions are mediated and whether they depend on the T. forsythia S‐layer and/or its O‐glycosylation has yet to be elucidated.
Based on the analysis of planktonic and monospecies biofilm growth, we employed in this study the subgingival “Zurich biofilm model”44 to investigate how the T. forsythia wild‐type strains ATCC 43037 and UB4 and defined cell surface mutants thereof perform in a multispecies consortium. Through the incorporation of 10 different species of oral bacteria in the biofilm, this in vitro model mimics the natural situation in the oral cavity, whereby several microbial species assemble and grow together in the form of a biofilm, and therefore poses an excellent platform to dissect the role of individual species within the community. In the in vitro model, the selected oral bacteria, including the three “red complex” species out of which T. forsythia was varied, were co‐cultivated to form biofilms on pellicle‐coated hydroxylapatite (HA) disks in saliva and serum‐containing growth medium.12, 45, 46, 47, 48, 49 Tannerella forsythia wild‐type strains and mutants with different cell surface glycosylation patterns as well as an S‐layer‐deficient mutant were introduced in order to monitor their biofilm growth as well as the structural behavior of the biofilm communities as a whole.
In particular, biofilms grown under these conditions were analyzed with the following aims: (i) to numerically determine cell numbers of all individual species within the bacterial consortium and the overall biofilm composition by quantitative real‐time polymerase chain reaction (qPCR) and colony‐forming unit (CFU) counts and (ii) to analyze the localization and distribution of individual species within the microbial structure through fluorescence in situ hybridization (FISH) using species‐specific probes against the 16S rRNA and confocal laser scanning microscopy (CLSM) analysis. This study is intended to be a first characterization of the behavior of T. forsythia strains with varying cell surface composition in a multispecies biofilm setting.
2. METHODS
2.1. Bacterial strains
Tannerella forsythia ATCC 43037 (American Type Culture Collection, Manassas, VA) and T. forsythia UB4 (obtained from Dr. Ashu Sharma, University of Buffalo, NY, USA) wild‐type strains and defined mutants thereof (see below) were grown anaerobically at 37°C for 4‐7 days in brain–heart infusion broth (Oxoid, Basingstoke, UK), supplemented with N‐acetylmuramic acid, horse serum, and 50 μg mL−1 gentamicin as described previously,27 with one passage before biofilm inoculation.
Mutants of T. forsythia ATCC 43037 (JUET0000000050) and T. forsythia UB4 (FMMN0100000051) with characterized defects in their cell surface protein glycosylation, affecting the terminal Pse (ATCC 43037) or Leg (UB4) residue, were available in our laboratory from a previous study.39 Briefly, T. forsythia ATCC 43037 ∆pseC (coding for a dedicated aminotransferase from the Pse biosynthesis pathway) and T. forsythia UB4 ∆legC (coding for a dedicated aminotransferase from the Leg biosynthesis pathway) mutants were constructed by chromosomal insertion of a gene knockout cassette consisting of an erythromycin resistance gene flanked by homologous upstream and downstream regions, ~1000 bp, each. The complementation cassette for T. forsythia mutants consisted of a chloramphenicol resistance gene flanked by a homologous ~1000‐bp upstream region, the gene of interest and a ~1000‐bp downstream region. The T. forsythia ATCC 43037 ∆wecC mutant, which lacks a trisaccharide glycan branch including the Pse residue, was obtained from Dr. Ashu Sharma. In addition to that, the S‐layer‐deficient mutant T. forsythia ATCC 43037 ∆tfsAB 41 was included in this study. This mutant lacks S‐layer glycans due to the absence of the S‐layer, but may expose underlying R‐type lipopolysaccharide52 or even O‐glycans present on outer membrane glycoproteins that become exposed upon removal of the S‐layer.53 All T. forsythia strains and mutants used in this study, together with their cell surface composition, are summarized in Table 1.
2.2. Monospecies biofilm growth of T. forsythia
The monospecies biofilm behavior of all T. forsythia strains and mutants included in this study was analyzed in a microtiter plate assay.39 In brief, bacteria were passaged once before biofilm inoculation at an optical density at 600 nm (OD600) of 0.05 and grown anaerobically for 6 days in 1 mL of half‐concentrated brain–heart infusion medium, with supplements as above,27 in 24‐well polystyrene plates (STARLAB) coated with 5 mg mL−1 mucin (from bovine submaxillary gland; Sigma‐Aldrich, Vienna, Austria) solution (in 0.1 mol L−1 sodium acetate buffer pH 4.5). In each experiment, two wells were used to determine the total cells of each strain and mutant, sterile medium served as negative control. For biofilm quantification, medium and planktonic cells were removed and the wells were washed once with 500 μL of PBS. Subsequently, biofilms were resuspended in 1 mL of PBS and the OD600 of the biofilm cell suspension was measured. Biofilm values were normalized to the corresponding absorbance (OD600) of the total cells. Data represent mean values ±SD of four independent experiments with three replicates each and were analyzed by the unpaired Student's t‐test.
2.3. Multispecies biofilm cultivation
To set‐up the “Zurich subgingival biofilm model”, T. forsythia wild‐type strains and defined cell surface mutants thereof (Table 1) were co‐cultivated with the following organisms: Prevotella intermedia ATCC 25611T (OMZ278), Campylobacter rectus (OMZ388), Veillonella dispar ATCC 17748T (OMZ493), Fusobacterium nucleatum (OMZ598), Streptococcus oralis SK248 (OMZ607), Streptococcus anginosus ATCC 9895 (OMZ871), Actinomyces oris (OMZ745), Porphyromonas gingivalis (OMZ925), and Treponema denticola ATCC 35405 (OMZ661). Each biofilm contained nine standard subgingival species plus one of the eight T. forsythia strains and mutants. Biofilm bacteria were maintained as described previously.44
For biofilm formation, bacterial cultures at an OD600 of 1.0 were mixed at equal volumes and 200 μL of this cell suspension was used to inoculate 1.6 mL of growth medium (60% pooled saliva, 10% fetal bovine serum [Sigma], 30% modified fluid universal medium)54 for biofilm formation on sintered pellicle‐coated HA disks (9 mm in diameter; Clarkson Chromatography Products, South Williamsport, PA) positioned in 24‐well polystyrene tissue‐culture plates. The medium was changed after 16 and 24 hours and disks were dip‐washed in 0.9% NaCl three times a day. After incubating anaerobically at 37°C for 64 hours, biofilms were dip‐washed once more and either harvested by vigorous vortexing for 2 minutes in 0.9% NaCl or fixed for 1 hour at 4°C in 4% paraformaldehyde solution (Merck, Darmstadt, Germany) for FISH.
2.4. Quantitative analysis
Cell numbers were determined by serial dilution plating and CFU counting as well as qPCR on genomic DNA purified from biofilm samples. Cell numbers were taken as a measure for the bacterial growth rate within the biofilm.
For CFU counts, biofilm suspensions were diluted 1:104 and 1:105 in 0.9% NaCl and plated on selective agar plates (Table 2) using a spiral diluter. For the more fastidious strains – i.e. T. denticola, C. rectus, and T. forsythia – cell numbers were determined by qPCR only.
Table 2.
Selective agar plates used for colony‐forming unit counting
| Selective agar plates | Organism |
|---|---|
| Mitis Salivarius Agar (Difco)+1% sodium tellurite solution | Streptococcus anginosus, Streptococcus oralis |
| Columbia Blood Agar (Oxoid)+5% horse blood (Sigma) |
Actinomyces oris, Veillonella dispar
total CFU |
| Fastidious Anaerobe Agar (BAG)+1 mg L−1 erythromycin (Sigma), 4 mg L−1 vancomycin (Sigma), 1 mg L−1 norfloxacin (Sigma) | Fusobacterium nucleatum |
| Columbia Blood Agar+5% horse blood (Sigma), 80 mg L−1 phosphomycin (Sigma) | Prevotella intermedia, Porphyromonas gingivalis |
For qPCR, bacterial genomic DNA was extracted from 500 μl of biofilm suspension using the GenEluteTM Bacterial Genomic DNA Kit (Sigma) and qPCR was performed on an ABI Prism SDS 7000 device (Applied Biosystems, Foster City, CA) according to Ammann et al.47 Each sample was analyzed using species‐specific primers amplifying the 16S rRNA gene.47 For each species, a standard curve was generated and the sample DNA concentration was calculated from the obtained quantification cycle (Cq) values. The abundance of each organism in the biofilm was calculated using the respective theoretical genome weight.47 Cell numbers per biofilm were determined in three independent experiments with three technical replicates for each biofilm. Statistical significance was tested by analysis of variance (Tukey's post‐hoc test for multiple comparisons, P≤.05) using graphpad prism version 7.00 for Windows (GraphPad Software, La Jolla, CA).
2.5. Structural analysis of biofilms
FISH staining was performed according to the protocol established by Thurnheer et al.55 using the probe combinations listed in Table 3. In brief, after fixation, biofilm samples were pre‐hybridized in hybridization buffer (0.9 mol L−1 NaCl, 20 mmol L−1 Tris–HCl, [pH 7.5], 0.01% sodium dodecyl sulfate, formamide (35%‐40%) at 46°C, for 15 minutes, followed by 3 hours of hybridization with specific oligonucleotide probes.45 Samples were washed in wash buffer (20 mmol L−1 Tris–HCl [pH 7.5], 5 mmol L−1 ethylene diaminetetraacetic acid, 0.01% sodium dodecyl sulfate, 46‐70 mmol L−1 NaCl) for 45 minutes at 48°C. For CLSM and image analysis, the samples were counterstained with a mixture of 3 μmol L−1 YoPro‐1 iodide (Invitrogen, Carlsbad, CA) and 15 μmol L−1 Sytox Green (Invitrogen) and embedded in Mowiol56 for confocal microscopy.
Table 3.
Combinations of 16S rRNA probes used for fluorescence in situ hybridization staining of individual bacterial species
| Probes | Target species | FA (%)a | NaCl (mmol L−1)b | Reference |
|---|---|---|---|---|
| Tfor‐997‐Cy3/Pging1006‐2‐prop‐Cy5 | Tannerella forsythia/Porphyromonas gingivalis | 40 | 46 | 44, 75 |
| Tfor‐997‐Cy3/Pging1006 ‐Cy5 | T. forsythia/P. gingivalis | 40 | 46 | 44, 75 |
| Tfor‐997‐Cy3/TrepG1‐679‐Cy5 | T. forsythia/Treponema denticola | 40 | 46 | 75, 76 |
| Tfor‐997‐Cy3/FUS‐664‐Cy5 | T. forsythia/Fusobacterium nucleatum | 40 | 46 | 55, 75 |
| Tfor‐997‐Cy3/CAMP655‐Cy5 | T. forsythia/Campylobacter rectus | 35 | 70 | 44, 75 |
| Tfor‐997‐Cy3/Pging1006‐2‐prop‐Cy5 | T. forsythia/P. gingivalis | 40 | 46 | 44, 75 |
| Tfor‐997‐Cy3/Pging1006 ‐Cy5 | T. forsythia/P. gingivalis | 40 | 46 | 44, 75 |
| Tfor‐997‐Cy3/TrepG1‐679‐Cy5 | T. forsythia/T. denticola | 40 | 46 | 75, 76 |
| Tfor‐997‐Cy3/FUS‐664‐Cy5 | T. forsythia/F. nucleatum | 40 | 46 | 55, 75 |
| Tfor‐997‐Cy3/CAMP655‐Cy5 | T.forsythia/C. rectus | 35 | 70 | 44, 75 |
Formamide concentration used in the hybridization buffer.
NaCl concentration in the wash buffer.
The architecture of the biofilms was analyzed using CLSM. For each of the eight T. forsythia strains and mutants, a minimum of three disks carrying fluorescently labeled biofilms was analyzed using a Leica SP‐5 microscope (Center of Microscopy and Image Analysis of the University of Zürich). Images were captured using a 100× objective and processed with imaris 7.4.0 Software (Bitplane, Zürich, Switzerland). Presented CSLM images (Figures 3 5 6) are snapshots of the biofilm structures present on the HA disks and the depicted structures represent a comprehensive collection of T. forsythia biofilm behavior observed during sampling.
Figure 3.
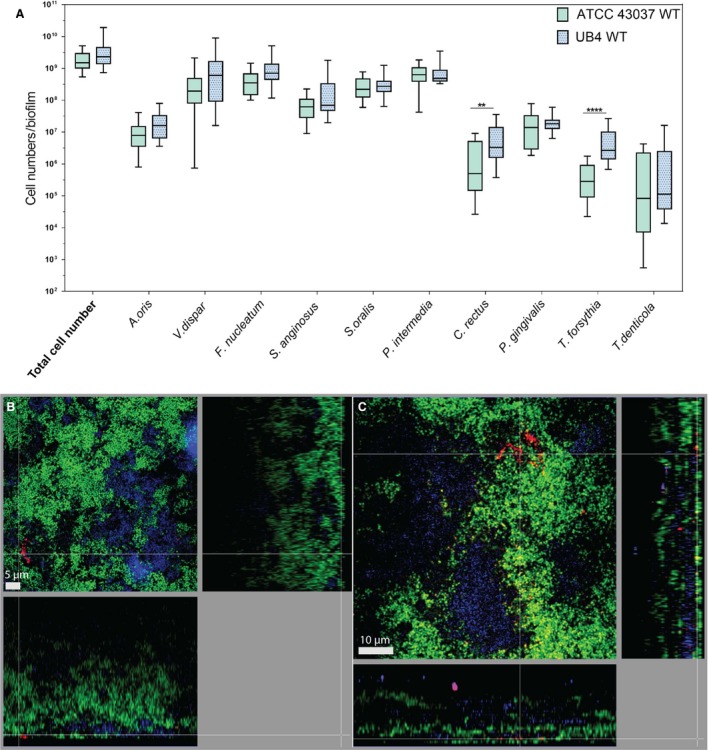
Comparison of 10‐species biofilms with two Tannerella forsythia wild‐type strains. (A) Whiskers boxplots (5th to 95th centile) show bacterial numbers determined by quantitative real‐time PCR from three independent experiments. Asterisk (*) indicates a statistically significant difference (P≤.05) between groups. The two groups represent biofilms with either T. forsythia ATCC 43037 wild‐type or T. forsythia UB4 wild‐type. (B, C) Fluorescence in situ hybridization stainings of fixed biofilms showing the localization of ATCC 43037 wild‐type (B) and UB4 wild‐type (C). Red/yellow: T. forsythia; cyan: Porphyromonas gingivalis, green: non‐hybridized cells (DNA staining YoPro‐1+Sytox). Here a representative area for one disk each is shown with a top view in the left panel and a side view with the biofilm–disk interface directed towards the top view; scale bars 5 μm (B) and 10 μm (C)
Figure 5.
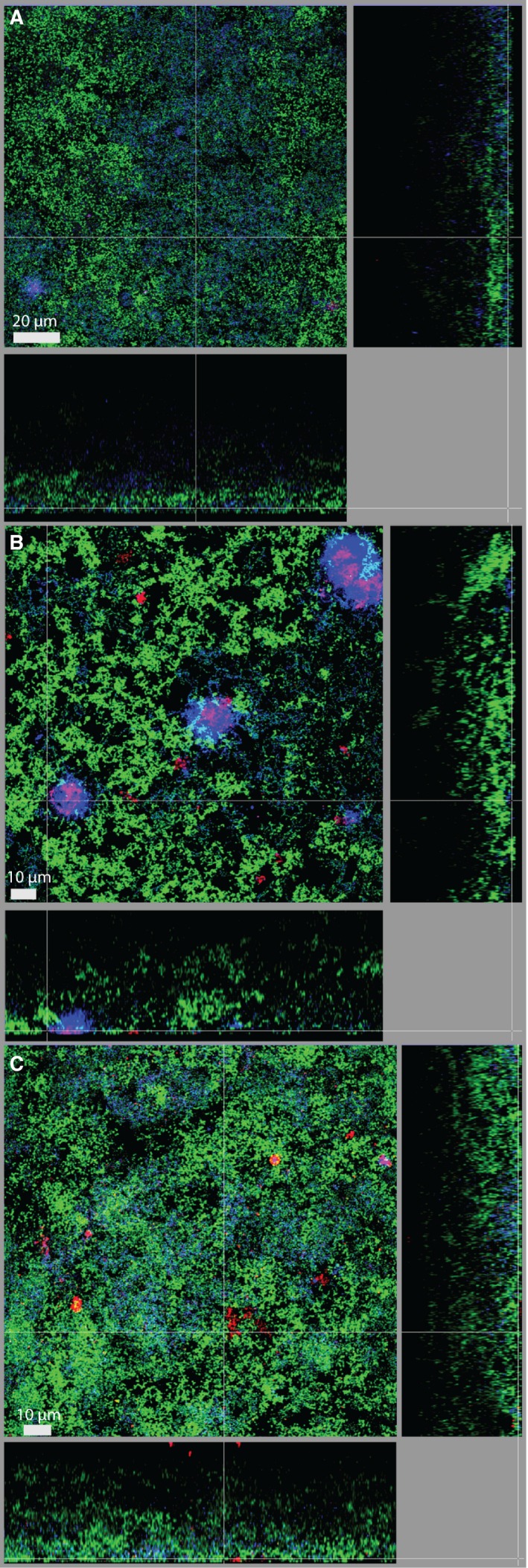
Fluorescence in situ hybridization staining of biofilms harboring Tannerella forsythia ATCC 43037 mutants (A) ∆pseC, (B) ∆wecC, and (C) ∆tfsAB. Red: T. forsythia, cyan: Porphyromonas gingivalis, green: non‐hybridized cells (DNA staining YoPro‐1+Sytox). Scale bars 20 μm (A) and 10 μm (B, C)
Figure 6.
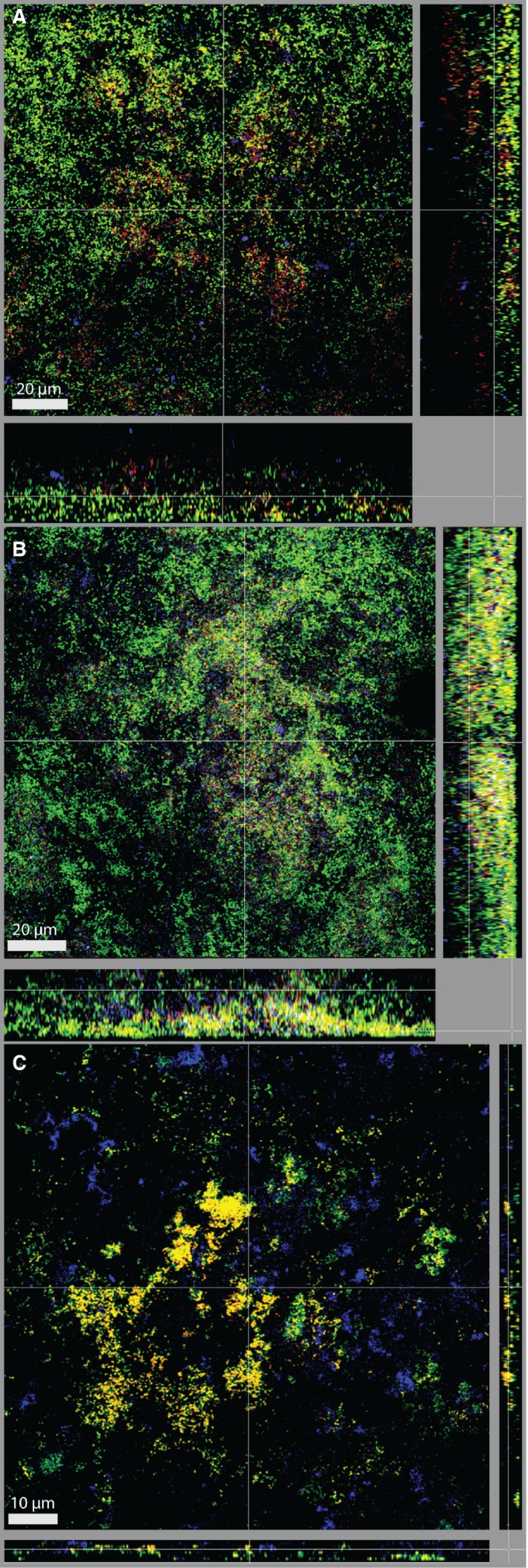
Dual fluorescence in situ hybridization staining of Tannerella forsythia and Campylobacter rectus for biofilms harboring ATCC 43037 wild‐type (A), UB4 wild‐type (B), and ATCC 43037 ∆tfsAB (C). Red/yellow: T. forsythia, cyan: C. rectus; green: non‐hybridized cells (DNA staining YoPro‐1+Sytox). Scale bars 20 μm (A, B) and 15 μm (C)
3. RESULTS
3.1. Monospecies biofilm formation of T. forsythia wild‐type strains and mutants
Based on the observations that deficiency in the protein O‐glycan's terminal nonulosonic acid triggers a decrease in biofilm formation of T. forsythia ATCC 43037 ∆pseC and T. forsythia UB4 ∆legC on a mucin‐coated surface39 and that T. forsythia ATCC 43037 ∆wecC possessing an even more truncated O‐glycan forms more biofilm on untreated plates,43 the biofilm formation capacity of all these strains was compared here in one microtiter plate assay, where the plates were coated with mucin to mimic the native situation on the tooth surface, and biofilm growth was quantified by OD600 measurement of biofilm cells and normalized to the corresponding total cell mass for each strain. In our setting, any manipulation of the cell surface decreased the capacity of the bacteria to form biofilms, as was evident in the absence of the S‐layer (T. forsythia ATCC 43037 ∆tfsAB), of the Pse‐(ManNAcA)2 O‐glycan branch (T. forsythia ATCC 43037 ∆wecC) as well as of the terminal nonulosonic acid alone, i.e. Pse in T. forsythia ATCC 43037 ∆pseC and Leg in T. forsythia UB4 ∆legC (Figure 1). More precisely, biofilms of the ATCC 43037 strain reached an average maximum OD600 of 0.52 ± 0.05 after 6 days of cultivation, whereas biofilm growth of the ∆pseC and ∆tfsAB mutants was reduced by 1.6‐fold, and in the case of the ∆wecC mutant even by five‐fold. Tannerella forsythia UB4 wild‐type biofilms reached an average maximum OD600 of 0.89 ± 0.21 and also here, the biofilm growth was reduced 1.3‐fold in the nonulosonic acid‐deficient mutant ∆legC. In both nonulosonic acid mutants, the growth behavior in the biofilm was restored to the levels of the respective parent strain, with an average maximum OD600 of 0.50 ± 0.06 for ∆pseC comp and 0.84 ± 0.20 for ∆legC comp (Figure 1).
Figure 1.
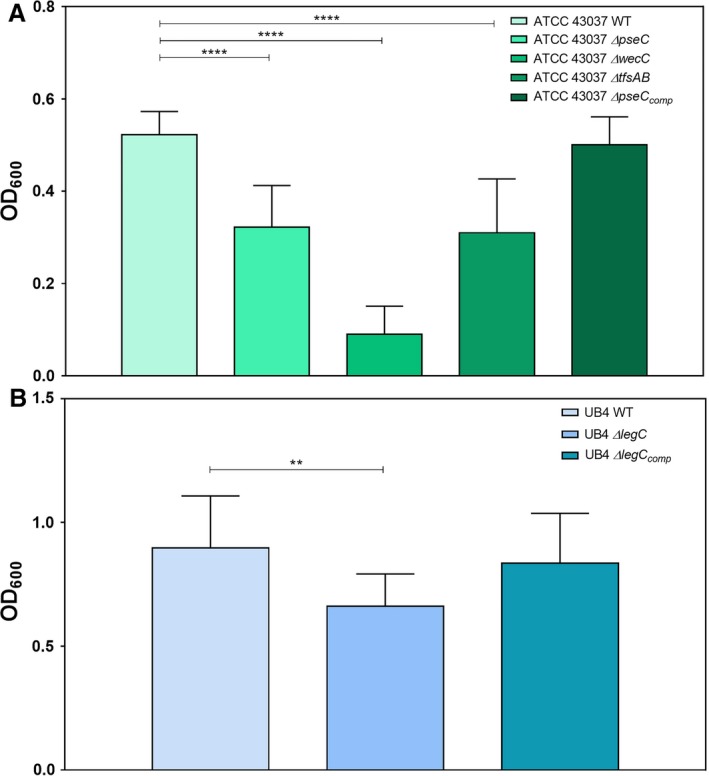
Monospecies biofilm formation of Tannerella forsythia wild‐type and mutant strains. (A) Biofilm formation of T. forsythia ATCC 43037 wild‐type compared with its mutants ATCC 43037 ΔpseC, ΔwecC, ΔtfsAB and the complemented mutant ΔpseC comp. (B) Biofilm formation of T. forsythia UB4 wild‐type compared with its mutant UB4 ΔlegC and the complemented mutant ΔlegC comp. Mean values ±SD of four independent experiments with three replicates, each, are shown. Asterisks (**) indicate significant differences between samples as determined by the unpaired Student's t‐test (P≤.01)
All deletion mutants also showed slower planktonic growth in liquid culture, as concluded from the determination of growth curves and doubling times (see Supplementary material, Fig. S2 and Table S1). These obvious growth defects might result from pleiotropic effects due to the genetic manipulation of the strains rather than from changes of the bacterial cell surface, even though during planktonic growth, both complemented strains performed in a similar way to the parent strain in terms of doubling times, with ∆pseC comp vs ATCC 43037 wild‐type revealing doubling times of 14.99 ± 0.83 hours and 14.41 ± 0.54 hours, and ∆legC comp vs UB4 revealing a slight increase in doubling time (12.28 ± 0.25 hours vs 16.75 ± 3.97 hours) (see Supplementary material, Fig. S2 and Table S1).
3.2. Determination of total biofilm cell numbers in the presence of T. forsythia strains and mutants in the subgingival “Zurich biofilm model”
Total cell numbers in biofilms including nine bacterial species routinely used in the subgingival “Zurich biofilm model” plus one T. forsythia wild‐type strain (T. forsythia ATCC 43037 or UB4) or mutant lacking certain sugar residues (T. forsythia ATCC 43037 ∆pseC, T. forsythia ATCC 43037 ∆wecC, T. forsythia UB4 ∆legC) or the whole S‐layer (T. forsythia ATCC 43037 ∆tfsAB) were analyzed by quantifying the cell numbers of each of the 10 species.
The total cell number per biofilm was not significantly affected, regardless of which T. forsythia strain or mutant had been incorporated into the biofilm (Figure 2). When comparing the total cell number of all biofilm bacteria as determined by strain‐specific qPCR and CFU counts, the latter resulted in lower cell numbers, as only viable cells were enumerable. Both methods, however, provided reproducible results for each of the nine disks that were analyzed for each of the eight T. forsythia strains and mutants included in this study.
Figure 2.

Comparison of colony‐forming unit (CFU) counting and quantitative polymerase chain reaction (qPCR) for Tannerella forsythia wild‐type strains and mutants in the subgingival “Zurich biofilm”. Total bacteria for 10‐species biofilms with different T. forsythia strains and mutants enumerated by CFU counts (red boxes) and qPCR (blue boxes) for three independent experiments with three technical replicates, each, are shown (Whiskers boxplots 5th to 95th centile)
3.3. Influence of T. forsythia wild‐type strains on composition and structure of the subgingival “Zurich biofilm model”
First, the multispecies biofilm behavior of the T. forsythia wild‐type strains ATCC 43037 and UB4 was compared with regard to bacterial growth and localization in the 10‐species biofilm.
For quantitative analysis, the cell number of each individual species in the biofilm was determined by qPCR after 64 hours of incubation. The total cell number of all species, except for C. rectus OMZ388 (see below), was not affected by the incorporation of the different T. forsythia strains, but there was a clear difference in the biofilm growth of the T. forsythia strains (Figure 3A). In accordance with the results observed in monospecies biofilms (Figure 1), also in the 10‐species consortium, T. forsythia UB4 seemed to perform better with mean cell numbers higher by 11.9‐fold when compared with the mean cell numbers of T. forsythia ATCC 43037 (Figure 3A), as determined by qPCR. This coincided with C. rectus OMZ388 to be found at significantly higher levels (3.6‐fold) in biofilms containing strain UB4 as determined by analysis of variance (P≤.01).
The influence of the two T. forsythia wild‐type strains on the biofilm structure and their localization were determined by CLSM. Tannerella forsythia ATCC 43037 tended to be primarily localized at the outer biofilm surface in the form of clearly visible cell clusters (Figure 3B). In contrast, T. forsythia UB4 was found in the form of microcolonies as well as singly dispersed close to the biofilm surface and in small clusters in deeper layers of the biofilm (Figure 3C).
3.4. Analysis of T. forsythia cell surface mutants in the subgingival biofilm
3.4.1. Quantitative analysis
In order to assess to what extent the difference in biofilm growth of T. forsythia strains ATCC 43037 and UB4 (see above) was influenced by their cell surface composition, defined mutants of either strain (Table 1) were incorporated into the biofilm and their cell numbers were again determined via qPCR.
Contrary to their performance in monospecies biofilms (Figure 1) and their slower planktonic growth (see Supplementary material, Fig. S2) in the 10‐species consortium, the T. forsythia ATCC 43037 mutants behaved in a very similar way to the parent strain, and neither the lack of the terminal Pse residue (∆pseC) nor the lack of the trisaccharide branch (∆wecC) of the S‐layer O‐glycan significantly affected the growth of T. forsythia or the other species of the biofilm (Figure 4A). The same was observed for the reconstituted strain ATCC 43037 ∆pseC comp. Interestingly, the absence of the S‐layer in the ∆tfsAB mutant, although not affecting growth of T. forsythia itself, led to a strong increase in the growth of C. rectus OMZ388 in the biofilm (Figure 4A) when compared with biofilms with T. forsythia ATCC 43037 wild‐type or ∆pseC, indicating that the loss of the S‐layer causes a growth benefit for C. rectus OMZ388 in these biofilms. As a control, the complemented mutant T. forsythia ATCC 43037 ∆pseC comp reverted C. rectus OMZ388 cell numbers back to the wild‐type level (with a non‐significant reduction of C. rectus cell numbers).
Figure 4.
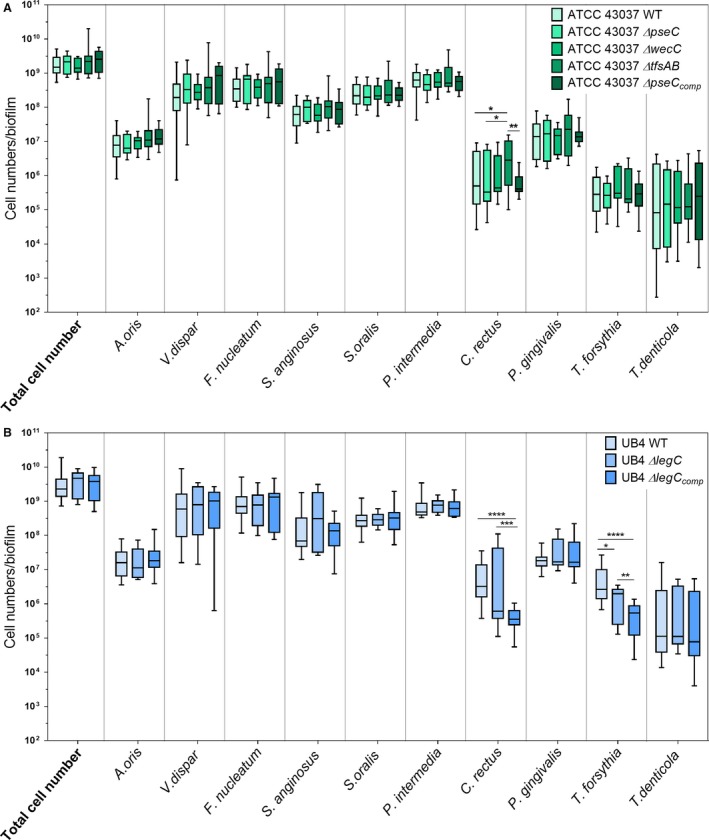
Box plots showing cell numbers of all species determined by quantitative real‐time PCR for biofilms with Tannerella forsythia ATCC 43037 wild‐type or mutants (∆pseC, ∆wecC, ∆tfsAB, ∆pseC comp) (A) and UB4 wild‐type or mutants (∆legC, ∆legC comp), respectively (B). Data derived from three independent experiments were plotted on a logarithmic scale. Asterisk (*) indicates significant differences (P≤.05) between the groups
In contrast to the almost identical performance of T. forsythia ATCC 43037 wild‐type and mutants in the multispecies biofilm, genetic manipulation of T. forsythia UB4 (i.e. T. forsythia UB4 ∆legC and ∆legC comp) resulted in a decrease in cell numbers in multispecies biofilms. The cell number of T. forsythia UB4 ∆legC was significantly decreased in the biofilm when compared with the parent strain, a fact that had already been observed in monospecies biofilms (Figure 1). Contrary to its behavior in monospecies biofilms, in the multispecies community, the reconstituted strain ∆legC comp could not restore the parent phenotype (Figure 4B).
As described before, at the high levels of T. forsythia UB4 wild‐type that developed in the biofilm, cell numbers of C. rectus OMZ388 were elevated in comparison with biofilms harboring T. forsythia ATCC 43037. In biofilms containing the ∆legC mutant this effect was less pronounced, with C. rectus OMZ388 mean cell numbers being significantly decreased by 1.4‐fold when compared with biofilms with UB4 wild‐type (P≤.001) (Figure 4B). In the presence of the complemented strain ∆legC comp, the growth of C. rectus OMZ388 was significantly reduced when compared with biofilms harboring UB4 wild‐type or ∆legC (P≤.001) (Figure 4B). Given that in monospecies biofilm experiments as well as during planktonic growth, UB4 ∆legC comp was shown to behave in the same way as its parent strain (Figure 1, see Supplementary material, Fig. S2), its impaired growth in the multispecies community suggests that the modification of this gene locus has a pleiotropic effect causing a growth defect in the environment of the multispecies biofilm.
3.4.2. Evaluation of the biofilm structure by CLSM
Since changes in the cell surface composition of T. forsythia did not affect the numeric composition of the 10‐species biofilms, FISH staining and CLSM analysis were performed for a qualitative evaluation of the biofilm structure.
Similar to the T. forsythia ATCC 43037 wild‐type, both the ATCC 43037 ∆pseC and the ATCC 43037 ∆wecC mutants were detected at the biofilm surface. Whereas ∆pseC was also found singly dispersed and in pronounced cell clusters at the biofilm surface, with only very few cells being detected (Figure 5A), ∆wecC formed dense superficial clusters (Figure 5B). The S‐layer‐deficient mutant T. forsythia ATCC 43037 ∆tfsAB was observed as small microcolonies scattered along the surface as well as in the form of single cells dispersed in the upper layers of the biofilm (Figure 5C).
Tannerella forsythia ATCC 43037 ∆wecC formed clearly distinguishable aggregates with P. gingivalis OMZ925, an effect that was not observed for the other T. forsythia strains and mutants analyzed. In contrast to biofilms incorporating other T. forsythia strains and mutants, in ∆wecC biofilms, cells appeared to grow less dense, as seen by YoPro‐1+Sytox staining of non‐hybridized bacteria. Porphyromonas gingivalis OMZ925 appeared to have changed its localization, being detected predominantly at the biofilm surface. This potentially direct interaction of P. gingivalis OMZ925 with the ∆wecC mutant was followed up in co‐aggregation studies (see Supplementary material, Fig. S3B). Porphyromonas gingivalis OMZ925 coaggregated with all T. forsythia strains at different levels. Significant differences between wild‐type and mutant strains or a distinct affinity of P. gingivalis OMZ925 for the ∆wecC mutant could not be observed in these assays. The truncation of the O‐glycan, however, was found to affect the autoaggregation of T. forsythia (see Supplementary material, Fig. S3A). This could be observed as a strong decrease of the OD600 of the cell suspensions and higher percentage of autoaggregation of ATCC 43037 ∆pseC (38.1%) compared with T. forsythia ATCC 43037 wild‐type (1.5%) but also when compared with ∆wecC (18.1%), ∆tfsAB (4.8%) and ∆pseC comp (4.7%).
Given the demonstrated growth‐promoting effect of T. forsythia UB4 wild‐type (Figure 3A) and T. forsythia ATCC 43037 ∆tfsAB (Figure 3A) on C. rectus OMZ388, dual FISH stainings were performed to determine a possible coaggregation of these species (Figure 6). In the section of the biofilm shown in the CLSM images, a high number of T. forsythia cells was detected for both ATCC 43037 and UB4 wild‐type strains. Both were present as single cells throughout the whole biofilm structure as well as in small clusters close to the biofilm surface in the case of the ATCC 43037 wild‐type (Figure 6A) and in deeper layers for the UB4 wild‐type (Figure 6B), as had been found before (Figure 3B, C). The S‐layer mutant ∆tfsAB was present in the form of clusters in close proximity to the HA‐disc surface in a relatively thin section of the biofilm (Figure 6B). Campylobacter rectus OMZ388 cells appeared in the form of irregularly interspersed microcolonies in all layers of the biofilm.
Although it was obvious that, compared with the ATCC 43037 wild‐type strain (Figure 6A), C. rectus OMZ388 cell numbers in the biofilm were elevated in the presence of the UB4 wild‐type strain (Figure 6B) and the ATCC 43037 S‐layer mutant ∆tfsAB (Figure 6C), co‐localization, which would be a prerequisite of a direct interaction between the bacteria, could not be observed. Also, co‐aggregation assays did not show a direct interaction between C. rectus OMZ388 and the ATCC 43037 S‐layer mutant or UB4 wild‐type strain (see Supplementary material, Fig. S3C). Here, as for P. gingivalis OMZ925, aggregation was elevated only with the nonulosonic‐acid‐deficient strains T. forsythia ATCC 43037 ∆pseC (14.3%) and T. forsythia UB4 ∆legC (9.5%) when compared with ATCC 43037 wild‐type (4.8%) and UB4 wild‐type (4.1%) (see Supplementary material, Fig. S3C).
4. DISCUSSION
The purpose of this study was to analyze different T. forsythia wild‐type strains and selected mutants thereof with defined differences in cell surface composition with regard to their behavior in a multispecies biofilm community. First we showed that the monospecies biofilm lifestyle of T. forsythia was clearly influenced by its S‐layer and attached O‐glycan. Alteration of the T. forsythia cell surface composition significantly reduced the capability of the bacterium to form monospecies biofilms as evidenced previously with the nonulosonic‐acid‐deficient mutants ATCC 43037 ∆pseC and T. forsythia UB4 ∆legC.39 In this study, this effect was confirmed for a mutant with an even more truncated O‐glycan ATCC 43037 ∆wecC as well as for the S‐layer‐deficient mutant ATCC43037 ∆tfsAB (Figure 1). For the nonulosonic‐acid‐deficient mutants, biofilm formation could be fully restored in the complemented mutants, suggesting a direct correlation between loss of the terminal sugar residue and reduced biofilm formation. These data were derived from biofilm experiments using mucin‐coated polystyrene plates and are contradictory to previous findings by others, where on untreated polystyrene, biofilm formation was enhanced for the ATCC 43037 ∆wecC mutant.57 In their natural habitat, mucin provides an initial adhesion site and nutrient source for bacteria and fosters biofilm growth.58 Mucin coating introduces highly hydrophilic properties to the otherwise hydrophobic polystyrene surface.59 Bacterial adhesion and interaction is influenced by hydrophobic interactions as well as steric forces and charge effects.60 The decrease of biofilm formation of strains that lack one (∆pseC, ∆legC) or more (∆wecC) charged sugar residues on a hydrophilic surface documented in this study vs the previously observed opposite effect on a hydrophobic surface57 shows that biofilm behavior is decisively influenced by the properties of the surface provided for attachment.
In this study, we investigated polymicrobial biofilms that approximate the native situation in the oral cavity much better than a planktonic or monospecies biofilm culture and, therefore, constitute an ideal system to examine the growth performance of individual species and strains. When introduced into in vitro 10‐species subgingival biofilms, alteration of the bacterial cell surface composition as present in the defined mutants did not impair the growth behavior of T. forsythia in terms of cell numbers per biofilm (Figure 4A). Interestingly, at the wild‐type level, T. forsythia UB4 occurred in higher numbers than T. forsythia ATCC 43037 (Figures 1, 3A, 4A), which may indicate a better adaptation of T. forsythia UB4 to the niche.
Comparison of the planktonic growth of both T. forsythia ATCC 43037 and UB4 wild‐type strains supported these observations, as UB4 was found to have a shorter generation time and grew to a higher OD600 before reaching the stationary phase in comparison with strain ATCC 43037 (see Supplementary material, Fig. S2). To our knowledge the data presented here constitute a first description of the different growth characteristics of these two T. forsythia strains in biofilm settings as well as in planktonic form. A preliminary bioinformatic analysis of the genomes of different T. forsythia isolates available in databases reflects the variability in the genetic make‐up for either Leg or Pse biosynthesis.39 Considering the differences in biofilm behavior of T. forsythia ATCC 43037 and UB4 wild‐type and that Leg is a better mimic of the biologically important sialic acid than Pse, this might suggest that the presence of either nonulosonic acid could reflect the adaptation of T. forsythia strains to different oral microenvironments.
Whereas in the multispecies biofilms the overall cell numbers remained relatively constant, the distribution of T. forsythia changed depending on the bacterium's cell surface composition. Neither the S‐layer nor its glycosylation seemed to be required for the bacterium to establish itself in the multispecies community. However, changes thereof influenced T. forsythia's autoaggregation, which was enhanced upon truncation of the O‐glycan in the mutants T. forsythia ATCC 43037 ∆pseC, T. forsythia ATCC 43037 ∆wecC, and T. forsythia UB4 ∆legC (see Supplementary material, Fig. S3). Alteration of the cell surface might, therefore, change the way that cells interact with each other within the microcolonies and multispecies cell aggregations. As cell surface glycosylation affected biofilm formation on a mucin‐coated surface (Figure 1), it is tempting to speculate that the decreased ability to adhere to the heavily sialylated salivary glycoprotein mucin in a monospecies biofilm setting is mirrored in the multispecies consortium in a way that T. forsythia cell surface mutants might exhibit an altered capability to adhere to sialic‐acid‐like structures present on other oral bacteria, such as streptococci or Campylobacter species37, 38 and, thereby, vary their localization within the multispecies consortium.
From the other bacterial species in the multispecies biofilm, C. rectus OMZ388 seems to be strongly affected by the T. forsythia cell surface composition. Upon presence of the T. forsythia ATCC 43073 ∆tfsAB mutant, which is deficient for the S‐layer and, hence, also the attached O‐glycans, C. rectus OMZ388 was increased in its cell numbers per biofilm (Figures 3A, 4A, 6C). Hence, it is conceivable that in the native multispecies situation, the glycosylated S‐layer as an entity (ATCC 43037) might have a regulatory role in keeping C. rectus cell numbers below a certain threshold. In fact, a previous proteomic analysis of T. forsythia biofilms identified the two S‐layer proteins TfsA and TfsB to be upregulated in comparison with the planktonic cells,61 which underlines the importance of the S‐layer for the biofilm lifestyle of the bacterium. The causative factors and underlying mechanism for the increased growth of C. rectus OMZ388 in biofilms harboring the T. forsythia ATCC 43037 ΔtfsAB mutant still await further investigation. Structural analysis of these biofilms and coaggregation assays performed so far suggest that the observed growth effect is independent of a direct interaction between the two species (Figure 6, and see Supplementary material, Fig. S3). Campylobacter rectus is often associated with periodontal disease5, 62 where it occurs in elevated numbers in the deep periodontal pockets.63, 64 Campylobacter spp. have long been described as of clinical relevance.65, 66 They are Gram‐negative, microaerophilic bacteria whose motility is conferred by a single polar, glycosylated flagellum.67 Even though little has been described about C. rectus flagellar glycosylation, the bacterium possesses the genetic make‐up for Pse biosynthesis (V. Friedrich, M. L. Braun, S. Bloch, C. Schäffer, unpublished observation) and, interestingly, also covers its cells with a 2D S‐layer.67, 68, 69
For P. gingivalis and T. forsythia, a direct synergistic interaction has been described previously, albeit for another strain.70 In the biofilms analyzed in this study, P. gingivalis OMZ925 seemed to strongly co‐localize with T. forsythia ATCC 43037 ∆wecC (Figure 4B) but was not affected in its growth by the T. forsythia cell surface composition (Figure 4). Coaggregation of P. gingivalis OMZ925 with T. forsythia did not differ significantly between T. forsythia wild‐type strains ATCC 43037 and UB4 and their respective mutants and a preferential direct interaction of P. gingivalis OMZ925 with T. forsythia ATCC 43037 ∆wecC could not be observed. Porphyromonas gingivalis outer membrane vesicles enhance attachment to and invasion of epithelial cells by T. forsythia,71 co‐infection of T. forsythia and P. gingivalis increases abscess formation in a mouse model,72 and T. forsythia cell extracts have been shown to have a growth‐promoting effect on P. gingivalis.73 In support of the synergistic interaction between the two species, Bao et al. described, in the very same experimental model as used here, reduced growth of T. forsythia in multispecies biofilms containing a P. gingivalis Lys‐gingipain‐deficient strain.12 However, despite this observation, the molecular mechanism of coaggregation between the two pathogens is still unclear.74
In conclusion, the present study shows that the growth of T. forsythia in an in vitro multispecies biofilm, as represented by the “Zurich biofilm model”, does not depend on the bacterium's cell surface composition. Deletion of one or more sugars (T. forsythia ATCC 43073 ∆pseC, ∆wecC, T. forsythia UB4 ∆legC) has a disadvantageous effect on neither the biofilm growth of T. forsythia, nor on overall cell numbers in the biofilm. Tannerella forsythia is able to establish itself in the multispecies consortium even without an S‐layer (T. forsythia ATCC 43073 ∆tfsAB). These findings suggest that the glycosylated S‐layer of T. forsythia does not play a crucial role in regulating the bacterium's growth in a multispecies biofilm. Nevertheless, we observed that it affected the bacterium's localization in the biofilm, the interaction with C. rectus, for which the glycosylated S‐layer has a growth retarding effect, and its co‐localization with P. gingivalis, which is increased upon a three‐sugar truncation of the O‐glycan in the T. forsythia ATCC 43037 ∆wecC mutant. Hence, changes in the S‐layer and surface glycosylation of T. forsythia might actually contribute to the bacterium's virulence potential by promoting structural arrangements within in the biofilm. Whether this contributes to the immune evasion of the biofilm‐associated species needs to be tested in functional interaction assays with host cells.
CONFLICT OF INTEREST
The authors declare no conflict of interest related to this study.
Supporting information
ACKNOWLEDGEMENTS
The authors thank Manuela Flury and Helga Lüthi‐Schaller (Division of Oral Microbiology and Immunology, Institute of Oral Biology, University of Zürich) for excellent technical assistance. We thank the Centre of Microscopy and Image Analysis (ZMB) of the University of Zürich for their support with confocal microscopy.
Bloch S, Thurnheer T, Murakami Y, Belibasakis GN, Schäffer C. Behavior of two Tannerella forsythia strains and their cell surface mutants in multispecies oral biofilms. Mol Oral Microbiol. 2017;32:404–418. https://doi.org/10.1111/omi.12182
Funding information
Financial support came from the Austrian Science Fund FWF, projects P26836‐B22 and P24317‐B22 (to CS) and the Doctoral Programme “Biomolecular Technology of Proteins” W1224.
REFERENCES
- 1. Marsh PD. Dental plaque: Biological significance of a biofilm and community life‐style. J Clin Periodontol. 2005;32:7‐15. [DOI] [PubMed] [Google Scholar]
- 2. Marsh PD. Dental plaque as a microbial biofilm. Caries Res. 2004;38:204‐211. [DOI] [PubMed] [Google Scholar]
- 3. Van Dyke TE, Sheilesh D. Risk factors for periodontitis. J Int Acad Periodontol. 2005;7:3‐7. [PMC free article] [PubMed] [Google Scholar]
- 4. Hajishengallis G, Lamont RJ. Beyond the red complex and into more complexity: The polymicrobial synergy and dysbiosis (PSD) model of periodontal disease etiology. Mol Oral Microbiol. 2012;27:409‐419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL Jr. Microbial complexes in subgingival plaque. J Clin Periodontol. 1998;25:134‐144. [DOI] [PubMed] [Google Scholar]
- 6. Hajishengallis G. Immunomicrobial pathogenesis of periodontitis: Keystones, pathobionts, and host response. Trends Immunol. 2014;35:3‐11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lamont RJ, Jenkinson HF. Life below the gum line: Pathogenic mechanisms of Porphyromonas gingivalis . Microbiol Mol Biol Rev. 1998;62:1244‐1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bostanci N, Belibasakis GN. Porphyromonas gingivalis: An invasive and evasive opportunistic oral pathogen. FEMS Microbiol Lett. 2012;333:1‐9. [DOI] [PubMed] [Google Scholar]
- 9. Kitamura Y, Matono S, Aida Y, Hirofuji T, Maeda K. Gingipains in the culture supernatant of Porphyromonas gingivalis cleave CD4 and CD8 on human T cells. J Periodontal Res. 2002;37:464‐468. [DOI] [PubMed] [Google Scholar]
- 10. Wingrove JA, DiScipio RG, Chen Z, Potempa J, Travis J, Hugli TE. Activation of complement components C3 and C5 by a cysteine proteinase (gingipain‐1) from Porphyromonas (Bacteroides) gingivalis . J Biol Chem. 1992;267:18902‐18907. [PubMed] [Google Scholar]
- 11. Lourbakos A, Potempa J, Travis J, et al. Arginine‐specific protease from Porphyromonas gingivalis activates protease‐activated receptors on human oral epithelial cells and induces interleukin‐6 secretion. Infect Immun. 2001;69:5121‐5130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bao K, Belibasakis GN, Thurnheer T, Aduse‐Opoku J, Curtis MA, Bostanci N. Role of Porphyromonas gingivalis gingipains in multi‐species biofilm formation. BMC Microbiol. 2014;14:258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. O'Brien‐Simpson NM, Pathirana RD, Walker GD, Reynolds EC. Porphyromonas gingivalis RgpA‐Kgp proteinase‐adhesin complexes penetrate gingival tissue and induce proinflammatory cytokines or apoptosis in a concentration‐dependent manner. Infect Immun. 2009;77:1246‐1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Belibasakis GN, Bostanci N, Reddi D. Regulation of protease‐activated receptor‐2 expression in gingival fibroblasts and Jurkat T cells by Porphyromonas gingivalis . Cell Biol Int. 2010;34:2872‐2892. [DOI] [PubMed] [Google Scholar]
- 15. Dashper SG, Seers CA, Tan KH, Reynolds EC. Virulence factors of the oral spirochete Treponema denticola . J Dent Res. 2011;90:691‐703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lux R, Sim JH, Tsai JP, Shi W. Construction and characterization of a cheA mutant of Treponema denticola . J Bacteriol. 2002;184:3130‐3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Grenier D, Uitto VJ, McBride BC. Cellular location of a Treponema denticola chymotrypsinlike protease and importance of the protease in migration through the basement membrane. Infect Immun. 1990;58:347‐351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Batista da Silva AP, Lee W, Bajenova E, McCulloch CA, Ellen RP. The major outer sheath protein of Treponema denticola inhibits the binding step of collagen phagocytosis in fibroblasts. Cell Microbiol. 2004;6:485‐498. [DOI] [PubMed] [Google Scholar]
- 19. Amin M, Grove DA, Kapus A, Glogauer M, Ellen RP. An actin‐stabilizing peptide conjugate deduced from the major outer sheath protein of the bacterium Treponema denticola . Cell Motil Cytoskeleton. 2007;64:662‐674. [DOI] [PubMed] [Google Scholar]
- 20. Puthengady TB, Sun CX, Bajenova E, Ellen RP, Glogauer M. Modulation of human neutrophil functions in vitro by Treponema denticola major outer sheath protein. Infect Immun. 2006;74:1954‐1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Miyamoto M, Ishihara K, Okuda K. The Treponema denticola surface protease dentilisin degrades interleukin‐1 beta (IL‐1 beta), IL‐6, and tumor necrosis factor alpha. Infect Immun. 2006;74:2462‐2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Okuda T, Kimizuka R, Miyamoto M, et al. Treponema denticola induces interleukin‐8 and macrophage chemoattractant protein 1 production in human umbilical vein epithelial cells. Microbes Infect. 2007;9:907‐913. [DOI] [PubMed] [Google Scholar]
- 23. Sharma A. Virulence mechanisms of Tannerella forsythia . Periodontol 2000. 2010;54:106‐116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sabet M, Lee SW, Nauman RK, Sims T, Um H‐S. The surface (S‐) layer is a virulence factor of Bacteroides forsythus . Microbiology. 2003;149(Pt 12):3617‐3627. [DOI] [PubMed] [Google Scholar]
- 25. Sekot G, Posch G, Oh YJ, et al. Analysis of the cell surface layer ultrastructure of the oral pathogen Tannerella forsythia . Arch Microbiol. 2012;194:525‐539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Posch G, Pabst M, Brecker L, Altmann F, Messner P, Schäffer C. Characterization and scope of S‐layer protein O‐glycosylation in Tannerella forsythia . J Biol Chem. 2011;286:38714‐38724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tomek MB, Neumann L, Nimeth I, et al. The S‐layer proteins of Tannerella forsythia are secreted via a type IX secretion system that is decoupled from protein O‐glycosylation. Mol Oral Microbiol. 2014;29:307‐320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Narita Y, Sato K, Yukitake H, et al. Lack of a surface layer in Tannerella forsythia mutants deficient in the type IX secretion system. Microbiology. 2014;160(Pt 10):2295‐2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Dunne WM Jr. Bacterial adhesion: Seen any good biofilms lately? Clin Microbiol Rev. 2002;15:155‐166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Garrett TR, Bhakoo M, Zhang ZB. Bacterial adhesion and biofilms on surfaces. Prog Nat Sci. 2008;18:1049‐1056. [Google Scholar]
- 31. Nesbitt WE, Doyle RJ, Taylor KG. Hydrophobic interactions and the adherence of Streptococcus sanguis to hydroxylapatite. Infect Immun. 1982;38:637‐644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nesbitt WE, Doyle RJ, Taylor KG, Staat RH, Arnold RR. Positive coooperativity in the binding of Streptococcus sanguis to hydroxylapatite. Infect Immun. 1982;35:157‐165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wu H, Zeng M, Fives‐Taylor P. The glycan moieties and the N‐terminal polypeptide backbone of a fimbria‐associated adhesin, Fap1, play distinct roles in the biofilm development of Streptococcus parasanguinis . Infect Immun. 2007;75:2181‐2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Liang X, Chen YY, Ruiz T, Wu H. New cell surface protein involved in biofilm formation by Streptococcus parasanguinis . Infect Immun. 2011;79:3239‐3248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Guerry P, Ewing CP, Schirm M, et al. Changes in flagellin glycosylation affect Campylobacter autoagglutination and virulence. Mol Microbiol. 2006;60:299‐311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Guerry P. Campylobacter flagella: Not just for motility. Trends Microbiol. 2007;15:456‐461. [DOI] [PubMed] [Google Scholar]
- 37. Thibault P, Logan SM, Kelly JF, et al. Identification of the carbohydrate moieties and glycosylation motifs in Campylobacter jejuni flagellin. J Biol Chem. 2001;276:34862‐34870. [DOI] [PubMed] [Google Scholar]
- 38. Angata T, Varki A. Chemical diversity in the sialic acids and related alpha‐keto acids: An evolutionary perspective. Chem Rev. 2002;102:439‐469. [DOI] [PubMed] [Google Scholar]
- 39. Friedrich V, Janesch B, Windwarder M, et al. Tannerella forsythia strains display different cell‐surface nonulosonic acids: Biosynthetic pathway characterization and first insight into biological implications. Glycobiology. 2017;27:342‐357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Varki A. Diversity in the sialic acids. Glycobiology. 1992;2:25‐40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sakakibara J, Nagano K, Murakami Y, et al. Loss of adherence ability to human gingival epithelial cells in S‐layer protein‐deficient mutants of Tannerella forsythensis . Microbiology. 2007;153(Pt 3):866‐876. [DOI] [PubMed] [Google Scholar]
- 42. Sekot G, Posch G, Messner P, et al. Potential of the Tannerella forsythia S‐layer to delay the immune response. J Dent Res. 2011;90:109‐114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Honma K, Inagaki S, Okuda K, Kuramitsu HK, Sharma A. Role of a Tannerella forsythia exopolysaccharide synthesis operon in biofilm development. Microb Pathog. 2007;42:156‐166. [DOI] [PubMed] [Google Scholar]
- 44. Guggenheim B, Gmür R, Galicia JC, et al. In vitro modeling of host–parasite interactions: The ‘subgingival’ biofilm challenge of primary human epithelial cells. BMC Microbiol. 2009;9:280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ammann TW, Gmür R, Thurnheer T. Advancement of the 10‐species subgingival Zurich biofilm model by examining different nutritional conditions and defining the structure of the in vitro biofilms. BMC Microbiol. 2012;12:227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ammann TW, Belibasakis GN, Thurnheer T. Impact of early colonizers on in vitro subgingival biofilm formation. PLoS ONE. 2013;8:e83090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ammann TW, Bostanci N, Belibasakis GN, Thurnheer T. Validation of a quantitative real‐time PCR assay and comparison with fluorescence microscopy and selective agar plate counting for species‐specific quantification of an in vitro subgingival biofilm model. J Periodontal Res. 2013;48:517‐526. [DOI] [PubMed] [Google Scholar]
- 48. Belibasakis GN, Thurnheer T. Validation of antibiotic efficacy on in vitro subgingival biofilms. J Periodontol. 2014;85:343‐348. [DOI] [PubMed] [Google Scholar]
- 49. Thurnheer T, Bostanci N, Belibasakis GN. Microbial dynamics during conversion from supragingival to subgingival biofilms in an in vitro model. Mol Oral Microbiol. 2016;31:125‐135. [DOI] [PubMed] [Google Scholar]
- 50. Friedrich V, Pabinger S, Chen T, Messner P, Dewhirst FE, Schäffer C. Draft genome sequence of Tannerella forsythia type strain ATCC 43037. Genome Announc. 2015;3:e00660‐00615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Stafford GP, Chaudhuri RR, Haraszthy V, et al. Draft genome sequences of three clinical isolates of Tannerella forsythia isolated from subgingival plaque from periodontitis patients in the United States. Genome Announc. 2016;4:e01268‐01316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Posch G, Andrukhov O, Vinogradov E, et al. Structure and immunogenicity of the rough‐type lipopolysaccharide from the periodontal pathogen Tannerella forsythia . Clin Vaccine Immunol. 2013;20:945‐953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Friedrich V, Gruber C, Nimeth I, et al. Outer membrane vesicles of Tannerella forsythia: Biogenesis, composition, and virulence. Mol Oral Microbiol. 2015;30:451‐473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Gmür R, Guggenheim B. Antigenic heterogeneity of Bacteroides intermedius as recognized by monoclonal antibodies. Infect Immun. 1983;42:459‐470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Thurnheer T, Gmür R, Guggenheim B. Multiplex FISH analysis of a six‐species bacterial biofilm. J Microbiol Methods. 2004;56:37‐47. [DOI] [PubMed] [Google Scholar]
- 56. Thurnheer T, van der Ploeg JR, Giertsen E, Guggenheim B. Effects of Streptococcus mutans gtfC deficiency on mixed oral biofilms in vitro . Caries Res. 2006;40:163‐171. [DOI] [PubMed] [Google Scholar]
- 57. Honma K, Mishima E, Inagaki S, Sharma A. The OxyR homologue in Tannerella forsythia regulates expression of oxidative stress responses and biofilm formation. Microbiology. 2009;155(Pt 6):1912‐1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Derrien M, van Passel MW, van de Bovenkamp JH, Schipper RG, de Vos WM, Dekker J. Mucin‐bacterial interactions in the human oral cavity and digestive tract. Gut Microbes. 2010;1:254‐268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Crouzier T, Jang H, Ahn J, Stocker R, Ribbeck K. Cell patterning with mucin biopolymers. Biomacromol. 2013;14:3010‐3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Renner LD, Weibel DB. Physicochemical regulation of biofilm formation. MRS Bull. 2011;36:347‐355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Pham TK, Roy S, Noirel J, Douglas I, Wright PC, Stafford GP. A quantitative proteomic analysis of biofilm adaptation by the periodontal pathogen Tannerella forsythia . Proteomics. 2010;10:3130‐3141. [DOI] [PubMed] [Google Scholar]
- 62. Macuch PJ, Tanner AC. Campylobacter species in health, gingivitis, and periodontitis. J Dent Res. 2000;79:785‐792. [DOI] [PubMed] [Google Scholar]
- 63. Dzink JL, Tanner AC, Haffajee AD, Socransky SS. Gram negative species associated with active destructive periodontal lesions. J Clin Periodontol. 1985;12:648‐659. [DOI] [PubMed] [Google Scholar]
- 64. Ihara H, Miura T, Kato T, et al. Detection of Campylobacter rectus in periodontitis sites by monoclonal antibodies. J Periodontal Res. 2003;38:64‐72. [DOI] [PubMed] [Google Scholar]
- 65. Lee S, Lee J, Ha J, et al. Clinical relevance of infections with zoonotic and human oral species of Campylobacter . J Microbiol. 2016;54:459‐467. [DOI] [PubMed] [Google Scholar]
- 66. Man SM. The clinical importance of emerging Campylobacter species. Nat Rev Gastroenterol Hepatol. 2011;8:669‐685. [DOI] [PubMed] [Google Scholar]
- 67. Bolton DJ. Campylobacter virulence and survival factors. Food Microbiol. 2015;48:99‐108. [DOI] [PubMed] [Google Scholar]
- 68. Dokland T, Olsen I, Farrants G, Johansen BV. Three‐dimensional structure of the surface layer of Wolinella recta . Oral Microbiol Immunol. 1990;5:162‐165. [DOI] [PubMed] [Google Scholar]
- 69. Thompson SA. Campylobacter surface‐layers (S‐layers) and immune evasion. Ann Periodontol. 2002;7:43‐53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Yao ES, Lamont RJ, Leu SP, Weinberg A. Interbacterial binding among strains of pathogenic and commensal oral bacterial species. Oral Microbiol Immunol. 1996;11:35‐41. [DOI] [PubMed] [Google Scholar]
- 71. Inagaki S, Onishi S, Kuramitsu HK, Sharma A. Porphyromonas gingivalis vesicles enhance attachment, and the leucine‐rich repeat BspA protein is required for invasion of epithelial cells by Tannerella forsythia . Infect Immun. 2006;74:5023‐5028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Yoneda M, Hirofuji T, Anan H, et al. Mixed infection of Porphyromonas gingivalis and Bacteroides forsythus in a murine abscess model: Involvement of gingipains in a synergistic effect. J Periodontal Res. 2001;36:237‐243. [DOI] [PubMed] [Google Scholar]
- 73. Yoneda M, Yoshikane T, Motooka N, et al. Stimulation of growth of Porphyromonas gingivalis by cell extracts from Tannerella forsythia . J Periodontal Res. 2005;40:105‐109. [DOI] [PubMed] [Google Scholar]
- 74. Zhu WD, Lee SW. Surface interactions between two of the main periodontal pathogens: Porphyromonas gingivalis and Tannerella forsythia . J Periodontal Implant Sci. 2016;46:2‐9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Züger J, Lüthi‐Schaller H, Gmür R. Uncultivated Tannerella BU045 and BU063 are slim segmented filamentous rods of high prevalence but low abundance in inflammatory disease‐associated dental plaques. Microbiology. 2007;153(Pt 11):3809‐3816. [DOI] [PubMed] [Google Scholar]
- 76. Zijnge V, van Leeuwen MB, Degener JE, et al. Oral biofilm architecture on natural teeth. PLoS ONE. 2010;5:e9321. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


