Summary
Background
Topical retinoids are effective in retarding skin ageing and restoring homeostasis in skin conditions such as psoriasis. However their adverse effects (AEs), which include irritation (retinoid dermatitis), photosensitivity and teratogenicity, limit their use and patient compliance. Development of retinoid analogues with minimal AEs would allow a broader and more compliant use.
Aim
To synthesise a novel molecule, bakuchiol salicylate (bakusylan), with a modulatory gene expression profile similar to retinoids, using as reference three prescription retinoids: tretinoin, tazarotene and adapalene.
Methods
We hypothesized that because bakuchiol salicylate has a structure entirely different from existing retinoids, there would be at least a partial uncoupling of AEs from the skin normalizing activity of this retinoid. This hypothesis was tested at the transcriptional level in psoriatic cytokine-treated cultures of keratinocytes and organotypic skin substitutes, using DNA microarrays and custom PCR arrays.
Results
Evaluation of the gene expression profile of bakuchiol salicylate revealed elimination of several components of the retinoid-like proinflammatory response and teratogenic signature, without a substantial loss of normalizing potential. A possible mechanism of action, consisting of keratinocyte desensitization to psoriatic cytokine signalling through the inhibition of the signal transducer and regulator of transcription (STAT)1/3/interferon inflammatory signal transduction axis was also identified.
Conclusion
Bipartite materials obtained by merging two skin-active entities with specific, complementary bioactivities, such as bakuchiol and salicylic acid, may yield a new class of functional retinoids.
Introduction
Topical retinoids are effective in delaying skin ageing processes and restoring homeostasis in immune-mediated skin diseases. However, the normalizing effects of retinoids, such as keratinocyte differentiation1 and inhibition of cytokine-induced gene expression2–3 carry a price of adverse effects (AEs), which have an impact on tolerance and compliance. Among the most common AEs are irritation and dryness (retinoid dermatitis), which can actually aggravate some symptoms of targeted skin conditions.4–6 In an attempt to discover retinoid-like compounds with minimal AEs, we screened libraries of natural compounds from plants traditionally used for the treatment of skin conditions for retinoid functionality. This collaborative project yielded one product candidate, a meroterpene and resveratrol structural analogue7 called bakuchiol.8,9 In the current study, we report the synthesis of the salicylic acid ester of that molecule, called bakusylan, and its normalizing properties in two in vitro psoriasiform-surrogate models.
Psoriasis is characterized by cytokine-triggered alteration of epidermal homeostasis, resulting in a shift from cell differentiation to hyperproliferation and inflammation. T helper (Th)1 and Th17 subsets of T cells play a key role in this process by releasing cytokines such as tumour necrosis factor (TNF)-α, interleukin (IL)-17A and IL-22, which promote inflammatory response in keratinocytes, leading to further T-cell infiltration and parakeratosis.10–13 Therefore, our psoriasis-surrogate models to test bakusylan were prepared by treating keratinocyte and skin substitutes with psoriasiform cytokines TNF-α, IL-17A and IL-22.
Methods
Synthesis of bakusylan
Bakuchiol salicylate was synthesized with salicylic acid (Sigma-Aldrich, St Louis, MO, USA) and bakuchiol (Sytheon, Boonton, NJ, USA and Abcam, Cambridge, MA, USA; two different suppliers were used to ensure reproducibility) by the modified Steglich esterification method using dichloromethane as solvent, with dicyclohexylcarbodiimide as condensing agent and 4-N,N-dimethylaminopyridine as catalyst. After solvent evaporation and removal of dicyclourea by filtration, the obtained residue was loaded onto a silicon dioxide (high-purity Fluka silica gel, pore size 60 Å, 70–230 mesh) column, and eluted with toluene/cyclohexane (1/1 v/v). The solvent was evaporated under vacuum, and the oily residue was analysed by infrared detection (Satellite FT-IR spectrophotometer; Mattson Instruments, Madison, WI, USA), nuclear magnetic resonance (200 MHz; Gemini; Varian, Palo Alto, CA, USA), diode array spectrophotometry (HP-8452A; Agilent Technologies, Palo Alto, CA, USA) and high-performance liquid chromatography (HPLC) (series 1100; Agilent Technologies) equipped with an injector (Rheodyne® 7725), binary pump, diode array–ultraviolet detection modules, C18 column (Xterra MS Waters; Milford, MA, USA) and ChemStation software. Samples dissolved in ethanol (10 μL) were eluted with 0.1% formic acid/acetonitrile gradient at 0.3 mL/min.
Psoriasis-surrogate human epidermal keratinocytes
Epidermal keratinocyte progenitors from stratum basale (Zen-Bio, RTP, NC, USA) were incubated with CnT-PCT media supplemented with a mixture of IL-17A, IL-22 and TNF-α (100, 100 and 10 ng/mL, respectively; all BioLegend, San Diego, CA, USA), for 24 h, then the test materials were added and the cells incubated in the presence of the cytokines for another 24 h.
Psoriasis-surrogate human epidermal substitutes
Immature epidermal skin substitutes (MatTek, Ashland, MA, USA) were cultured for their final differentiation time (3 days) in media containing IL-17A/IL-22/TNFα (100/100/10 ng/mL, respectively). After the first 24 h of cytokine treatment, the test materials were added and the incubations were pursued in the presence of the cytokines for another 24 h.
PCR arrays and DNA microarrays
At the end of the incubations described above, total RNA was extracted (NucleoSpin II Kit; Macherey-Nagel, Bethlehem, PA, USA). For PCR, reverse transcription was performed using RT2 First Strand Kit and quantitative PCR was performed (iCycler iQ; BioRad, Hercules, CA, USA), using primers relevant to psoriasis and/or retinoic acid signalling. Gene expression was standardized to the housekeeping gene peptidylprolyl isomerase A (PPIA). All PCR reagents were from Qiagen (Germantown, MD, USA).
For the DNA microarray experiment, gene expression was profiled using an array platform (Human Genome U133 Plus 2.0; Affymetrix, Santa Clara, MD, USA), containing > 470 000 transcripts associated with 20 150 human genes. False discovery rate (FDR), was controlled for by adjusting raw P values derived from linear models using the Benjamini–Hochberg method.14 Enrichment of differentially expressed genes with respect to ordered gene lists was performed using the Wilcoxon rank sum test.15
Statistical analysis
Statistical analysis of DNA microarray results is described in the relevant section above. Elsewhere, two-tailed (paired) t-test was used for estimating statistical significance of differences between comparisons. Differences were considered significant at P < 0.05. Experiments were performed in biological duplicates or greater repeats.
Results
Synthesis of bakusylan
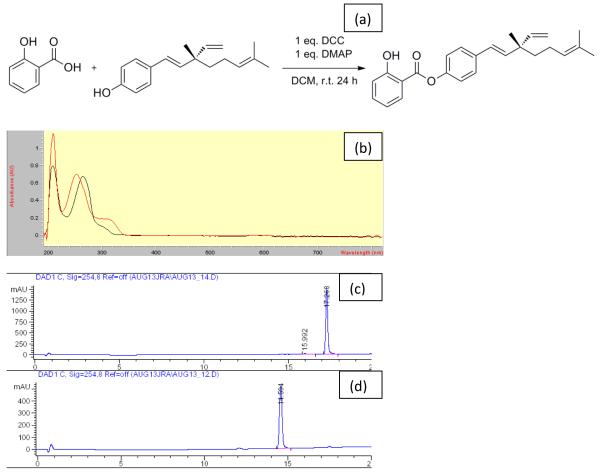
The final product [(E)-4-(3,7-dimethyl-3-vinylocta-1,6-dien-1-yl)phenyl 2-hydroxybenzoate)] eluted from the silica column was a light colourless oil. Its identification yielded the following results: IR (KBr): 3226 (ν O-H), 2967, 2924, 2854 (ν C-H), 1691 (ν C=O), 1615, 1583, 1506, 1484 (ν C=C), 1300 (δ O-H), 1193 (ν C-O), 1156, 1066 (δ C-H), 756 (γ C-H) per cm; 1H NMR (200 MHz, CDCl3): 1.23 [singlet (s) 3H, CH3], 1.24–1.27 (m, 2H, CH2), 1.60 (s, 3H, CH3), 1.70 (s, 3H, CH3), 1.98 (q, 2H, CH2, J = 8 Hz), 5.00–5.13 (m, 3H, 3CH), 5.84–5.98 [multiplet (m) 1H, CH), 6.20 (d, 1H, CH, J = 14 Hz), 6.37 (d, 1H, CH J = 16 Hz), 6.94–7.07 (m, 2H, ArH), 7.15 (d, 2H, ArH, J = 9Hz), 7.44 (d, 2H, ArH, J = 9 Hz), 7.51–7.55 (m, 1H, ArH), 8.08 [doublet of doublets (dd), 1H, ArH, J1 = 6 Hz, J2 = 2H), 10.53 [broad singlet (br s) 1H, OH + D2O exchangeable) ppm; 13C NMR (200 MHz, CDCl3): δ 18.2, 23.7, 26.2, 30.2, 41.7, 43.2, 112.7, 118.3, 120.0, 122.1 (2C), 125.1, 126.6, 127.6 (2C), 130.8, 131.9, 136.8, 137.0 (2C), 139.2, 146.1, 149.4, 162.7, 169.5 ppm. Elemental analysis was consistent with the expected chemical formula [analytical calculations for C25H28O3 (376.20): C, 79.75; H, 7.50; actual result: C, 79.64; H, 7.39]; purity: >99% (Fig. 1a). λmax was established at 250 nm (Fig. 1b). In HPLC the larger, less polar ester eluted later than bakuchiol (compare Fig.1c with Fig. 1d), as predicted by the nonpolar bonding of salicylate with the parent molecule.
Figure 1.
(a) Synthesis of bakuchiol salicylate (right) from salicylic acid (far left) and bakuchiol (left); (b) spectra of bakusylan and bakuchiol, showing absorption peaks at 250 and 260 nm, respectively; (c,d) reverse-phase high-performance liquid chromatography results for (c) bakusylan and (d) bakuchiol.
Induction of psoriasis-surrogate phenotypic changes in keratinocyte and organotypic cultures
Various combinations of TNF-α, IL-17A and IL-22 cytokines were tested to induce psoriatic phenotype in human epidermal keratinocyte progenitors (results not shown). The selected concentration (10, 100 or 100 ng/mL, respectively) induced marked shifts in gene expression (Table 1) and cell morphology, consistent with induction of a psoriatic phenotype, after 48 h of exposure [compare Fig. 2a (water control) vs. Fig 2b (cytokine-treated)]. When epidermal skin substitutes were incubated in medium containing the same cytokine cocktail for 48 h, a similar (although not identical) psoriasiform response was observed (Table 1).
Table 1.
Effect of bakusylan and control retinoids on the cytokine-induced modulation of the expression of selected genes in keratinocyte progenitor cell cultures and skin substitutes, assessed using custom PCR arrays.
| Gene symbol |
Cytokines plus:
|
|||||
|---|---|---|---|---|---|---|
| water* | water | BAK | ATRA | ADP | TAZ | |
| S100A7 | NC | 4.0 | NT | NT | NT | NT |
| DEFB4A | 18.9 | 85.8 | −2.0 | NC | NC | NC |
| RARG | −7.5 | −3.2 | 4.1 | 3.4 | NC | 3.3 |
| HPGD | −84.5 | −27.7 | NT | NT | NT | NT |
| CXCL3 | 3.3 | 9.8 | −2.1 | NC | NC | 3.3 |
| IL-8 | 40.7 | 67.2 | −2.6 | 3.7 | 3.4 | NC |
| CTGF | NC | 2 | NT | NT | NT | NT |
| SERPINB1 | NC | 24.0 | −7.5 | NC | 3.3 | 3.0 |
| HAS3 | NC | 5.6 | NT | NT | NT | NT |
| KRT10 | NC | −2950 | NC | −2.5 | NC | NC |
| RARRES1 | −2.6 | NC | NT | NT | NT | NT |
| RBP1 | −4.6 | NC | NT | NT | NT | NT |
| FLG | NC | −247.3 | NT | NT | NT | NT |
| KRT14 | NT | −8 | −2.6 | −4.3 | −2.5 | −2.3 |
| PPARG | NT | −3.3 | −2.3 | NC | NC | 6.9 |
| MMP1 | NT | 1.7 | NC | 3.3 | NC | NC |
| STAT3 | NT | 4.6 | −2.1 | NC | NC | 2.5 |
| LOR | NT | −1783 | NC | NC | −2.2 | −3.3 |
| PTGS2 | NT | 2.6 | NC | NC | NC | −3.9 |
| LAMA3 | NT | NC | NC | NC | NC | 2.1 |
| LAMC2 | NT | NC | 4.1 | NC | NC | NC |
| PTEN | NT | −3.2 | NC | NC | NC | 3.1 |
| AQP3 | NT | −9 | NC | 2.6 | NC | 3.5 |
ADP, adapalene; ATRA, all-trans retinoic acid; BAK, bakulylan; NC, not changed; NT, not tested; TAZ, tazarotene.
This column refers to keratinocytes; the remaining columns to skin substitutes. Experiments were performed in duplicate, P < 0.05. As shown in column 2, in keratinocytes, these cytokines [interleukin (IL)-17A/IL-22/tumour necrosis factor (TNF)-α] triggered a psoriasis-like phenotype in terms of an increase in expression of DEFB4A,21 IL-822 and CXCL3,23,24 and a decrease in HPGD, which is involved in the catabolism of prostaglandins (prostaglandins play an important role in the pathophysiology of psoriasis;19 the decrease in HPGD by IL-17A/IL-22/TNF- α is a novel finding). RARG, RBP1 and RARRES1 are retinoid-responsive genes and were included to monitor mechanistic similarities between the control retinoids and bakusylan. Likewise, in skin substitutes, cytokine-induced gene expression modulation is in agreement with induction of psoriasis-like changes (increase in DEFB4A, CXCL3 and IL-8, and decrease of HPGD). In addition, there were increases in markers of psoriatic epithelium (HAS3, SERPINB1, CTGF24–26) and decreases in markers of epithelial differentiation (KRT10, FLG). In the psoriasiform skin substitutes, bakusylan lowered the expression of six psoriasis-related genes, including STAT3 and the proinflammatory, irritation-related cytokines IL-8 and CXCL3, which, by contrast, were unfavourably upregulated by ATRA and adapalene (ADP) (IL-8) and tazarotene (TAZ) (CXCL3). Note also the unique downregulation of the proinflammatory senescence marker SERPINB1 by bakusylan. This profile indicates a possibly greater therapeutic potential of bakuchiol salicylate compared with the control retinoids.
Figure 2.

(a–e) Morphology of human epidermal keratinocyte progenitor either (a) untreated or (b) treated with (b–e) interleukin (IL)-17A/IL-22/tumour necrosis factor (TNF)-α for 48 h. (b) Note the appearance of a subpopulation of cells with ‘fried egg’ morphology (arrows) in cytokine-treated cells, reminiscent of the proinflammatory senescence-associated secretory phenotype (SASP). (c–e) These gross pathomorphological changes were significantly (P < 0.05) reduced with (c) 10 μg/mL bakusylan (approximately thee-fold) (d) 1.75 μg/mL tazarotene (by 40%) and (e) 0.1 μg/mL adapalene (~approximately thee-fold), as determined by 10 direct microscopic counts of senescent cells (arrows). The highest noncytotoxic doses of the test materials were chosen based upon previous cytotoxicity (MTT) experiments. Test materials were added 24 h after the cytokines, then the cells were incubated with the cytokines and test materials for an additional 24 h. Original magnification: × 40; scale bar, 50 μm.
Effect of bakusylan on psoriasis-surrogate human epidermal keratinocytes
To determine whether bakusylan can mitigate the effects of psoriasiform cytokines, it was added at 10 μg/mL to cell culture media 24 h after the cytokines were added, and the cells incubated for a further 24 h. Microscopic count of affected cells showed that bakusylan inhibited the cytokine-induced shift in keratinocyte morphology towards the senescent phenotype by about 60% (Fig. 2c vs. Fig. 2b), as quantified by counting enlarged (by > 2.5 times) cells with disorganized cytoskeletons (Fig. 2b, arrows). This effect was comparable with that of 0.1 μg/mL adapalene (Fig. 2e) and better than 1.75 μg/mL tazarotene (Fig. 2d, 40% inhibition). All compounds were assayed at the highest noncytostatic concentrations.
DNA microarrays revealed a retinoid-like effect of bakuchiol salicylate, as illustrated in Fig. 3. This diagram shows the number of probe sets unique to and common between the comparisons with vehicle control for bakusylan, all-trans retinoic acid (ATRA) and another antipsoriatic compound (compound C) with no analogy to retinoids. While 39 of the 87 (45%) probe sets that were significantly modulated by bakusylan were also modulated by ATRA, only 13 (17%) of the compound C probes were similarly modulated by ATRA, although both compounds have a transcription-modulatory profile consistent with antipsoriatic activity. This result indicates that bakusylan but not compound C has retinoid-like transcription-modulation activity. Interestingly, this similarity appears limited to a subset of ATRA-modulated genes, which make up only about 20% of the genes modulated by the retinoic acid (39 vs. 197).
Figure 3.

Bakusylan is a narrow functional analogue of all-trans retinoic acid (ATRA) in psoriasiform cytokine-treated keratinocytes. This Venn diagram shows the number of probsets modulated uniquely or jointly by three compounds [(a) bakuchiol salicylate, (b) ATRA, (c) retinoid-unrelated compound with antipsoriatic activity] compared with the vehicle control. Whereas 39 of the 87 probe sets (45%) significantly altered by (a) bakusylan were also modulated by(b) ATRA, only 13 (17%) of the compound C-modulated probes were similarly altered by ATRA, although both molecules have antipsoriatic activity. This result indicates that compound A (bakusylan) is much closer functionally to retinoids than compound C (a fumaric acid derivative). Selected probe sets had a fold change of ≥ 2, with the added constraint that the average expression for one of the two groups was ≥ 2,5 preventing large fold changes being selected based on small numbers. This statistical analysis of Affymetrix GeneChip data was based on Human U133 Plus 2.0 microarrays that were processed using the IVT Express kit.
To further investigate this selective retinoid functionality, the biological processes most significantly downregulated by ATRA and bakusylan were compared. This comparison identified a desensitization to cytokine stimulus in general, and to type I interferon in particular, as the most prominent common feature of the two comparators (Table 2), indicating the mechanistic analogy in downregulation of the cytokine-driven inflammatory responses between ATRA and bakusylan. Importantly, this analogy does not extend to morphogenic processes, whose downregulation is associated with the teratogenic effects of retinoids; at least seven morphogenic/developmental pathways were downregulated by ATRA (Table 1) but none by bakusylan. The observed cytokine desensitization seems correlated with the decrease in signal transducer and regulator of transcription 1 (STAT1) (P < 0.01) and STAT1-controlled genes, because bakusylan represses genes with a large number of STAT1 [and interferon-stimulated gene factor 3; ISGF3] motifs in the upstream region (Table 3). These results are consistent with other results (Fig. 3, Table 2), and indicate that bakusylan is a selective transcriptional modulator similar to ATRA, but possibly devoid of teratogenic potential.
Table 2.
Analysis of the GO BP terms most significantly enriched for genes whose expression was decreased by ATRA or bakusylan (DNA microarrays) in psoriasiform cytokine-treated keratinocytes.
| BPs decreased by optimal nontoxic dose of: | |||||
|---|---|---|---|---|---|
| ATRA (0.1 μg/mL) | Bakusylan (10 μg/mL) | ||||
| GOBPID | P value | BP Term | GOBPID | P value | BP Term |
| GO:0060337 | 4.02 × 10−12 | Type I IFN-mediated signalling pathway* |
GO:0006695 | 3.89 × 10−13 | Cholesterol biosynthesis process |
| GO:0034340 | 5.82 × 10−12 | Response to type I IFN* | GO:0046165 | 2.6 × 10−11 | Alcohol biosynthesis process |
| GO:0051607 | 6.25 × 10−10 | Defence response to virus | GO:1901615 | 4.2 × 10−10 | Organic hydroxy compound metabolic process |
| GO:0043901 | 1.98 × 10−6 | Negative regulation of multiorganism process |
GO:0006694 | 5.07 × 10−10 | Steroid biosynthesis process |
| GO:0006955 | 8.51 × 10−6 | Immune response* | GO:0060337 | 8.58E-09 | Type I IFN-mediated signalling pathway |
| GO:0045071 | 1.09 × 10−5 | Negative regulation of viral genome replication |
GO:0034340 | 9.73E-09 | Response to type I IFN* |
| GO:0060333 | 2.19 × 10−5 | IFN-γ-mediated signalling pathway* |
GO:0008299 | 4.09E-08 | Isoprenoid biosynthesis process |
| GO:0032501 | 3.02 × 10−5 | Multicellular organismal process | GO:0006639 | 9.16E-07 | Acylglycerol metabolism process |
| GO:0010033 | 4.64 × 10−5 | Response to organic substance | GO:0051607 | 2.09 × 10−6 | Defence response to virus |
| GO:0035457 | 8.19 × 10−5 | Cellular response to IFN-alpha* | GO:0046460 | 2.52 × 10−6 | Neutral lipid biosynthesis process |
| GO:0051270 | 8.3 × 10−5 | Regulation of cellular component movement |
GO:0051707 | 8.35 × 10−6 | Response to other organism |
| GO:0070887 | 8.46 × 10−5 | Cellular response to chemical stimulus |
GO:0019432 | 2.33 × 10−6 | Triglyceride biosynthesis process |
| GO:0016525 | 9.33 × 10−5 | Negative regulation of angiogenesis |
GO:0044255 | 3.93 × 10−5 | Cellular lipid metabolic process |
| GO:0002688 | 1.3 × 10−4 | Regulation of leucocyte chemotaxis* |
GO:0044710 | 4.04 × 10−5 | Single-organism metabolism |
| GO:0034341 | 1.96 × 10−4 | Response to IFN-γ* | GO:0006955 | 4.4 × 10−5 | Immune response* |
| GO:0065007 | 2.44 × 10−4 | Biological regulation | GO:0010499 | 1.37 × 10−4 | Proteasomal ubiquitin- independent protein catabolism |
| GO:0050792 | 2.49 × 10−4 | Regulation of viral process | GO:0071616 | 3.55 × 10−4 | Acyl-coenzyme A biosynthesis process |
| GO:0060585 | 2.64 × 10−4 | Positive regulation of prostaglandin-endoperoxide synthase activity |
GO:0048661 | 3.93 × 10−4 | Positive regulation of smooth muscle cell proliferation |
| GO:1901342 | 2.67 × 10−4 | Regulation of vasculature development† |
GO:0009991 | 4.87 × 10−4 | Response to extracellular stimulus |
| GO:0072358 | 2.73 × 10−4 | Cardiovascular system development† |
GO:0016042 | 7.47 × 10−4 | Lipid catabolic process |
| GO:0048729 | 3.1 × 10−4 | Tissue morphogenesis† | GO:0045445 | 8.58 × 10−4 | Myoblast differentiation |
| GO:0001655 | 3.83 × 10−4 | Urogenital system development† | GO:0051591 | 1.07 × 10−3 | Response to cAMP |
| GO:0040012 | 4.69 × 10−4 | Regulation of locomotion | GO:0046503 | 1.41 × 10−3 | Glycerolipid catabolic process |
| GO:0048705 | 7.47 × 10−4 | Skeletal system morphogenesis† | GO:0019433 | 1.47 × 10−3 | Triglyceride catabolic process |
| GO:0000462 | 7.73 × 10−4 | Maturation of SSU-rRNA from tricistronic rRNA transcript (SSU- rRNA, 5.8S rRNA) |
GO:0046461 | 1.47 × 10−3 | Neutral lipid catabolic process |
| GO:0048646 | 7.78 × 10−4 | Anatomical structure formation involved in morphogenesis† |
GO:0045017 | 1.771× 10−3 | Glycerolipid biosynthesis process |
| GO:0060700 | 7.83 × 10−4 | Regulation of ribonuclease activity | GO:0043330 | 1.93 × 10−3 | Response to exogenous double-stranded RNA |
| GO:0008544 | 8.03 × 10−4 | Epidermis development† | GO:0035383 | 1.99 × 10−3 | Thioester metabolic process |
| GO:0071345 | 9.1 × 10−4 | Cellular response to cytokine stimulus* |
GO:0071345 | 2.0 × 10−3 | Cellular response to cytokine stimulus* |
ATRA, all-trans retinoic acid; BP, biological process; GO, Gene Ontology; IFN, interferon; rRNA, ribosomal RNA; SSU, small subunit.
Four biological processes are common to ATRA and bakusylan, indicating a mechanistic analogy in downregulating the cytokine-driven inflammatory responses between the two compounds. Importantly, this analogy does not extend to morphogenic processes, whose downregulation is associated with teratogenic effects of ATRA; at least seven morphogenic/developmental pathways were downregulated by ATRA† and none by bakuchiol salicylate. These results suggest that bakusylan has a functional similarity to ATRA with possibly no teratogenic potential.
Table 3.
Suppression of interferon controlled pathways in cytokine-treated keratinocytes.
| Enriched motifs | Motif ID | Esti- mate |
SE | Z stat |
P value | Predicted STAT1/ISGF3 targets downregulated by bakusylan |
|---|---|---|---|---|---|---|
| STAT1: GRAANNGAAAST | STAT_disc3 | 1.61 | 0.20 | 8.01 | 1.2 × 10−15 |
IFI44L, IFIH1, DDX60, DDX60L, EPSTI1, MX2, RSAD2, IFI44, ZNFX1, TRIM14, OAS3, MYOCD, IFIT1, MX1, STAT1, SP100, PNPLA3, ITSN1 |
| STAT1: RGAAANYGAAACT | J9365 | 2.33 | 0.31 | 7.49 | 7 × 10−14 |
DDX60, IFI44L, IFI44, PARP9, TRIM14, IFIH1, IFIT1, STAT1, DDX60L, EPSTI1, MX2 |
| Isgf3g: RAAWCGAAACT | UP00074 | 2.28 | 0.32 | 7.02 | 2.2 × 10−12 |
DDX60L, DDX58, RSAD2, IFI44L, TRIM14, OAS3, DDX60, MX1, STAT1, EPSTI1 |
Bakusylan suppressed interferon-controlled pathways in psoriasiform cytokine-treated keratinocytes by repressing multiple genes (right column) with significant enrichment/over-representation of STAT1 (and ISGF3) motifs (first column) in their upstream region (5 kb). Induction of interferon-mediated pathways associated with STAT1 binding sites is a robust signature in psoriasis lesions.19,20 Enrichment of motifs was calculated using semiparametric generalized additive logistic models.19
We next compared bakusylan-downregulated genes with 301 ordered gene lists16 extracted from microarray datasets deposited in Gene Expression Omnibus (GEO). This comparison identified clinical conditions that similarly led to a decrease (positive association) or an increase (negative association) in the expression of bakusylan-downregulated genes. As shown in Table 4, genes decreased by bakusylan tended to have elevated expression in psoriasis lesions compared with normal skin, whereas genes decreased by bakusylan showed decreased expression in the lesions of patients treated with antipsoriatic drugs. This result supports the concept of bakusylan as a skin-normalizing agent.
Table 4.
GEO database microarray experiments leading to increased or decreased expression of bakusylan-downregulated genes.
| GEO accession (experiment ID) | Gene signature from the GEO database for the given set of experiments |
Correlation with genes downregulated by bakusylan |
|---|---|---|
|
GSE2737, GSE6710, GSE11903, GSE13355, GSE26866, GSE30999, GSE14905, GSE26866, GSE26866 |
Genes upregulated in psoriatic vs normal tissue | Negative* |
| GSE7553, GSE4587, GSE4587 | Genes upregulated in cancerous vs normal tissue | Negative* |
|
GSE32620, GSE32620, GSE24767, GSE20297, GSE7216, GSE36287, GSE1132, GSE7216, GSE12109, GSE36287, GSE2489, GSE32975, GSE36387, GSE20297, GSE9120, GSE25400, GSE7216, GSE20297, GSE24767, GSE28158, GSE37361, GSE37361, GSE24767, GSE32407, GSE32407, GSE24873 |
Genes upregulated in IL1-, IL17-, IL22-, TNF- and/or IFN-treated tissues compared with untreated tissues |
Negative† |
|
GSE16161, GSE32924, GSE5667, GSE5667 |
Genes upregulated in atopic skin vs normal skin | Negative |
|
GSE26487, GSE26523, GSE2822, GSE11903, GSE31652 |
Genes upregulated by antipsoriatic drugs (LY2439821, etanercept, efalizumab, dexamethasone) vs untreated psoriatic tissue |
Positive‡ |
| GSE22298, GSE11792, GSE10433 | Genes upregulated in tissues treated vs retinoids | Positive |
| GSE30768 | Genes upregulated in efalizumab-treated psoriatic skin after relapse vs no relapse |
Negative |
IFN, interferon; IL, interleukin, TNF, tumour necrosis factor; GEO, Gene Expression Omnibus.
Genes upregulated in psoriatic skin in the cited GEO experiments that were downregulated by bakusylan;
genes upregulated by tissue treatment with IL1-, IL17-, IL22-, TNF- and/or IFN in the cited GEO experiments that were downregulated by bakusylan;
genes downregulated in the skin of psoriatic patients treated with antipsoriatic drugs in the cited GEO experiments, which were also downregulated by bakusylan. We screened ordered gene lists derived from GEO to identify experiments with gene signatures that correlated with our own microarray data. Experiments with increased expression of bakusylan-downregulated genes (negative association) and decreased expression of bakusylan-downregulated genes (positive association) were identified. The GEO series accession IDs listed denote experiments showing significant association (P < 0.01; false discovery rate < 0.05). To obtain the enrichment statistic value we used equation (8) from Philippakis et al.15
Confirmation of the normalizing functionality of bakusylan by PCR arrays in organotypic cultures
The narrow-spectrum, retinol-like, cytokine-desensitizing functionality of bakusylan in psoriasis-surrogate keratinocytes determined by DNA microarrays was further compared with that of three current prescription retinoids (tretinoin, tazarotene and adapalene) in psoriasiform skin substitutes, using PCR arrays (Table 1). The result showed that only bakusylan favourably modulated three important genes dysregulated by psoriasiform cytokines: STAT3, IL-8 and CXCL3 (STAT3 is also an important cancer effector).
Discussion
Psoriasis is a chronic immune-mediated disease causing harmful dysregulation of the epidermis, and is a significant health problem as it affects 1–3% of the world population.17 It is also a good study model for identifying novel functional retinoids, because of its sensitivity to the normalizing effects of this class of compounds.1–3
When tested in two in vitro psoriasis-surrogate models, bakusylan showed an improved transcription-normalizing profile compared with control retinoids, without a teratogenic retinoid signature. Bakusylan not only downregulated psoriasiform gene expression but also inhibited the expression of IL-8 and CXLC3 in skin substitutes. These proinflammatory cytokines may be responsible for some of irritant AEs of retinoids. Genes whose expression was decreased by bakusylan correlated significantly with the genes whose expression is increased in psoriatic skin and negatively correlated with genes whose expression is increased by treatment with antipsoriatic drugs. This demonstrates at least partial uncoupling of the pro-irritant side effects of this retinoid from its skin-normalizing effects.
The psoriasis-surrogate keratinocyte model also allowed us to address the mechanism of action of this bakuchiol ester in an inflammatory context. One of the key signatures in psoriasis is induction of interferon (IFN)-mediated pathways associated with STAT1 binding sites.18,19 Inhibition of interferon expression by ATRA has been reported previously.20 In the current study, we found that inhibition of type I interferon signal transduction is a common feature between ATRA and bakusylan, and is correlated with a decrease in STAT1 and STAT1-controlled genes. Furthermore, recent analyses suggest that this IFN–STAT1 pathway is activated in a number of different skin diseases and thus the use of bakusylan would not be limited to psoriasis.
Taken together, these results point to a retinoid-like anti-inflammatory potential of bakuchiol salicylate, which would make it suitable for dermatological and skin-care applications. As the effects of retinoids are also exerted on the sebaceous glands, follicular keratinization and certain cancers, it would be interesting to determine whether the functional transcriptional similarity of bakusylan extends in these circumstances as well. Importantly, the narrow activity spectrum of this ester does not include proinflammatory effects or blockage of morphogenic processes, such as cardiovascular and epidermal development, which are common to vitamin A derivatives. AEs are the ‘glass ceiling’ for retinoids, impeding their wider use, and elimination of these AEswould allow a more widespread and compliant utilization of retinoids, unlocking the full functional potential of this class of skin-active compounds. The bipartite materials obtained by merging two skin-active chemicals with complementary bioactivities, such as bakuchiol and salicylic acid, may herald the next generation of functional retinoids.
What's already known about this topic?
Retinoids are important therapies for psoriasis.
Teratogenicity and skin irritation are the limiting factors for retinoid utilization in psoriasis.
What does this study add?
The new compound described here, called bakusylan, is an ester of two skin-active molecules: salicylic acid and the meroterpene bakuchiol
Using DNA microarrays and PCR arrays, bakusylan (bakuchiol salicylate) was found to be a narrow transcriptional analogue of retinol.
This narrow retinol functionality appears to at least partially uncouple the AEs of functionality from the therapeutic effects of this retinoid.
A mechanism of action of bakuchiol salicylate, consisting of keratinocyte desensitization to cytokines through the inhibition of the STAT1/3/interferon inflammatory signalling pathway, is proposed.
Acknowledgements
This study was supported by an SBIR grant from the National Institutes of Health, National Institute of Arthritis, Musculoskeletal and Skin Diseases (NIAMS) # 1 R43 AR062935-01A1.
Footnotes
Conflict of interest: the authors declare that they have no conflicts of interest.
References
- 1.Schroeder M, Zouboulis CC. All-trans-retinoic acid and 13-cis-retinoic acid: pharmacokinetics and biological activity in different cell culture models of human keratinocytes. Horm Metab Res. 2007;39:136–40. doi: 10.1055/s-2007-961813. [DOI] [PubMed] [Google Scholar]
- 2.Saurat JH. Retinoids and psoriasis: novel issues in retinoid pharmacology and implications for psoriasis treatment. J Am Acad Dermatol. 1999;41:S2–6. doi: 10.1016/s0190-9622(99)70358-0. [DOI] [PubMed] [Google Scholar]
- 3.Tjabringa G, Bergers M, van Rens D, et al. Development and validation of human psoriatic skin equivalents. J. Am J Pathol. 2008;173:815–23. doi: 10.2353/ajpath.2008.080173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yeung H, Wan J, Van Voorhees AS, et al. Patient-reported reasons for the discontinuation of commonly used treatments for moderate to severe psoriasis. J Am Acad Dermatol. 2013;68:64–72. doi: 10.1016/j.jaad.2012.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Song X, Xu A, Pan W, et al. Nicotinamide attenuates aquaporin 3 overexpression induced by retinoic acid through inhibition of EGFR/ERK in cultured human skin keratinocytes. Int J Mol Med. 2008;22:229–36. [PubMed] [Google Scholar]
- 6.Hecker D, Worsley J, Yueh G, et al. Interactions between tazarotene and ultraviolet light. J Am Acad Dermatol. 1999;41:927–30. doi: 10.1016/s0190-9622(99)70248-3. [DOI] [PubMed] [Google Scholar]
- 7.Chen Z, Jin K, Gao L, et al. Anti-tumor effects of bakuchiol, an analogue of resveratrol, on human lung adenocarcinoma A549 cell line. Eur J Pharmacol. 2010;643:170. doi: 10.1016/j.ejphar.2010.06.025. [DOI] [PubMed] [Google Scholar]
- 8.Chaudhuri RK, Bojanowski K. Bakuchiol: a retinol-like functional compound revealed by gene expression profiling and clinically proven to have anti-aging effects. Int J Cosmet Sci. 2014;36:221–30. doi: 10.1111/ics.12117. [DOI] [PubMed] [Google Scholar]
- 9.Chaudhuri RK. Bakuchiol. A retinol-like functional compound, modulating multiple retinol and non-retinol targets. In: Sivamani RK, Jagdeo J, Elsner P, Maibach HI, editors. Cosmeceuticals and Active Cosmetics. 3rd edn 2015. pp. 1–18. [Google Scholar]
- 10.Tokura Y, Mori T, Hino R. Psoriasis and other Th17-mediated skin diseases. J UOEH. 2010;32:317–28. doi: 10.7888/juoeh.32.317. [DOI] [PubMed] [Google Scholar]
- 11.Van Belle AB, de Heusch M, Lemaire MM, et al. IL-22 is required for imiquimod-induced psoriasiform skin inflammation in mice. J Immunol. 2012;188:462–9. doi: 10.4049/jimmunol.1102224. [DOI] [PubMed] [Google Scholar]
- 12.Hänsel A, Günther C, Ingwersen J, et al. Human slan (6-sulfo LacNAc) dendritic cells are inflammatory dermal dendritic cells in psoriasis and drive strong TH17/TH1 T-cell responses. J Allergy Clin Immunol. 2011;127:787–94. doi: 10.1016/j.jaci.2010.12.009. [DOI] [PubMed] [Google Scholar]
- 13.Kim J, Krueger JG. The immunopathogenesis of psoriasis. Dermatol Clin. 2015;33:13–23. doi: 10.1016/j.det.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 14.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol. 1995;B 57:289–300. [Google Scholar]
- 15.Philippakis AA, Busser BW, Gisselbrecht SS, et al. Expression-guided in silico evaluation of candidate cis regulatory codes for Drosophila muscle founder cells. PLoS Comput Biol. 2006;2:e53. doi: 10.1371/journal.pcbi.0020053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Swindell WR, Remmer HA, Sarkar MK, et al. Proteogenomic analysis of psoriasis reveals discordant and concordant changes in mRNA and protein abundance. Genome Med. 2015;7:86. doi: 10.1186/s13073-015-0208-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.WHO Global Report on Psoriasi. 2016 ISBN 978 92 4 156518 9; http://apps.who.int/iris/bitstream/10665/204417/1/9789241565189_eng.pdf.
- 18.Swindell WR, Johnston A, Xing X, et al. Modulation of epidermal transcription of circuits in psoriasis: new links between inflammation and hyperproliferation. PLoS One. 2013;8:e79253. doi: 10.1371/journal.pone.0079253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Swindell WR, Johnston A, Voorhees JJ, et al. Dissecting the psoriasis transcriptome: inflammatory- and cytokine-driven gene expression in lesions from 163 patients. BMC Genomics. 2013;14:527. doi: 10.1186/1471-2164-14-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang HK, Hou WS. Retinoic acid modulates interferon-γ production by hepatic natural kilter T cells via phosphatase 2A and the extracellular signal-regulated kinase pathway. J Interferon Cytokine Res. 2015;35:200–12. doi: 10.1089/jir.2014.0098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Simanski M, Rademacher F, Schroder L, et al. IL-17A and IFN-γ synergistically induce RNase 7 expression via STAT3 in primary keratinocytes. PLoS One. 2013;8:e59531. doi: 10.1371/journal.pone.0059531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arican O, Aral M, Sasmaz S, et al. Serum levels of TNF-alpha, IFN-gamma, IL-6, IL-8, IL-12, IL-17, and IL-18 in patients with active psoriasis and correlation with disease severity. Mediators Inflamm. 2005;2005:273–9. doi: 10.1155/MI.2005.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van Belle AB, de Heusch M, Lemaire MM, Hendrickx E, Warnier G, Dunussi-Joannopoulos K, Fouser LA, Renauld JC, Dumoutier L. IL022 is required for imiquimod-induced psoriasiform skin inflammation in mice. J Immunol. 2012;188:462–9. doi: 10.4049/jimmunol.1102224. [DOI] [PubMed] [Google Scholar]
- 24.Lundin A, Engstrom-Laurent A, Michaelsson G, Tengblad A. High levels of hyaluronate in suction blister fluid from active psoriatic lesions. Br J Dermatol. 1987;116:335–40. doi: 10.1111/j.1365-2133.1987.tb05847.x. [DOI] [PubMed] [Google Scholar]
- 25.Manczinger M, Kemeny L. Novel factors in the pathogenesis of psoriasis and potential drug candidates are found with systems biology approach. PLoS One. 2013;8:e80751. doi: 10.1371/journal.pone.0080751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao P, Hou L, Farley K, Sundrud MS, Remold-O’Donnel E. SerpinB1 regulates homeostatic expansion of IL-17+ γδ and CD4+ Th17 cells. J Leukoc Biol. 2014;95:521–30. doi: 10.1189/jlb.0613331. [DOI] [PMC free article] [PubMed] [Google Scholar]