Abstract
The CAGI-4 Hopkins clinical panel challenge was an attempt to assess state-of-the-art methods for clinical phenotype prediction from DNA sequence. Participants were provided with exonic sequences of 83 genes for 106 patients from the Johns Hopkins DNA Diagnostic Laboratory. Five groups participated in the challenge, predicting both the probability that each patient had each of the 14 possible classes of disease, as well as one or more causal variants. In cases where the Hopkins laboratory reported a variant, at least one predictor correctly identified the disease class in 36 of the 43 patients (84%). Even in cases where the Hopkins laboratory did not find a variant, at least one predictor correctly identified the class in 39 of the 63 patients (62%). Each prediction group correctly diagnosed at least one patient that was not successfully diagnosed by any other group. We discuss the causal variant predictions by different groups and their implications for further development of methods to assess variants of unknown significance. Our results suggest that clinically relevant variants may be missed when physicians order small panels targeted on a specific phenotype. We also quantify the false-positive rate of DNA-guided analysis in the absence of prior phenotypic indication.
Keywords: CAGI, genetic testing, phenotype prediction, variant interpretation
1 INTRODUCTION
DNA sequencing tests are increasingly being used in medical practice to confirm or assign clinical diagnoses (Katsanis & Katsanis, 2013). However, the interpretation and classification of novel sequence variants identified in a patient remains difficult, even for well-studied disorders like cystic fibrosis (Sosnay et al., 2017). Improved computational methods may aid in the interpretation of sequence variants and, when used in conjunction with clinical data, could increase the confidence of a diagnosis (Schulz, Tormey, & Torres, 2015). Until recently, genetic testing was limited to genes associated with a specific clinical phenotype. However, recent technological advances have made it feasible to sequence large gene panels, exomes, and genomes (Lee et al., 2014, Posey et al., 2016, Vassy et al., 2014). As the number of genes sequenced per patient increases, the number of novel, rare, and unclassified variants also increases. Clinical molecular geneticists must determine which variants, if any, are likely to contribute to the patient’s clinical presentation. The current gold standards for assessing a variant’s pathogenicity are segregation of the variant with the clinical phenotype in multiple pedigrees, and functional assays demonstrating a detrimental effect of that specific nucleotide change. In most instances, when a novel genetic variant is identified, there is no rapid and reliable method to assess its pathogenicity. Predictive software tools are interrogated, but none are considered strong evidence to assert a novel variant’s pathogenicity (Richards et al., 2015). The shift toward analyzing large datasets has led to a need for high-throughput methods to aid in variant classification and also for computation tools to help better interrogate the increasing number of variants of uncertain clinical significance.
Crowdsourced data analysis challenges such as the 4th Critical Assessment of Genome Interpretation (CAGI-4) have emerged as a framework to compare predictive methods and assess the overall state of particular analysis areas (Saez-Rodriguez et al., 2016). In the CAGI-4 Hopkins Clinical Panel challenge, participants were asked to develop or use existing computational methods to analyze data from a next-generation sequencing (NGS) panel in order to match a patient’s genotype to their clinical phenotype in the absence of additional clinical information. The Johns Hopkins DNA Diagnostic Laboratory (henceforth, Hopkins), a CLIA- and CAP-certified lab that specializes in clinical molecular testing for rare, inherited disorders, provided data for this challenge. The Hopkins lab offers testing for approximately 50 phenotypes and disorders, totaling 3,500 tests annually. They offer NGS-based tests targeted for ~20 specific phenotypes. The same NGS capture probe set is used for all panels and only the requested genes are analyzed in each patient. Hopkins provided CAGI-4 organizers with the VCF files for the entire NGS panel for 106 patients with a range of clinical presentations. The genetic disorders associated with variants in the 83 genes on the panel were grouped into 14 “disease classes,” which include lung disorders, peroxisomal disorders, aneurysm disorders, and craniofacial disorders (Table 1; Supp. Table S4). The goal of the challenge was for the participants to match each patient to a disease class based on informatics analysis of the sequence data. A further part of the challenge was to predict the specific gene and variant(s) that is/are the underlying cause of disease.
TABLE 1.
A summary of the 14 disease classes in the CAGI-4 Hopkins clinical panel challenge
| Disease class | Description |
|---|---|
| Cystic fibrosis and CF-related disorders | Classic cystic fibrosis consists of progressive lung disease, exocrine pancreatic insufficiency, and male infertility. |
| Diffuse lung disease | Diffuse lung disease is an umbrella term encompassing multiple lung disease phenotypes. |
| Primary ciliary dyskinesia | Primary ciliary dyskinesia is a genetically heterogeneous group of disorders resulting from dysfunction in different parts of the cilia. |
| Peroxisomal beta-oxidation defects | The majority of patients with peroxisomal beta-oxidation defects have liver disease, brain malformations, developmental retardation, sensory deficits, and dysmorphic craniofacial features. |
| Rhizomelic chondrodysplasia punctata | Symptoms of rhizomelic chondroplasia punctata include proximal shortening of the limbs, cataracts, severe intellectual disability, seizures, and calcific stippling of cartilage. |
| Zellweger spectrum disorders | Zellweger spectrum disorders consist of Zellweger syndrome (cerebro-hepato-renal syndrome; most severe phenotype), neonatal adrenoleukodystrophy (intermediate phenotype), and infantile Refsum disease (mildest phenotype). |
| Loeys-Dietz syndrome | Loeys-Dietz syndrome is a connective tissue disorder that predisposes individuals to aortic aneurysms. |
| Marfan syndrome | Marfan syndrome is an inherited connective tissue disorder that affects the skeletal, ocular, and cardiovascular systems. |
| Thoracic aortic aneurysm and dissection | Thoracic aortic aneurysm and dissection is a cardiovascular disease characterized by dilation of the aorta, which leads to aortic aneurysms (most commonly in the ascending aorta) and aortic dissection. |
| Ataxia telangiectasia | Ataxia-telangiectasia is a disorder of childhood onset progressive cerebellar ataxia and occulocutaneous telangiectasias. |
| Liddle syndrome | Liddle syndrome is a rare genetic disorder characterized by early onset high blood pressure (hypertension) and low blood potassium (hypokalemia). |
| Pseudohypoaldosteronism type 1 | Pseudohypoaldosteronism type 1 is a salt-wasting disease with onset during infancy. |
| Telomere shortening disorders | Telomere shortening disorders represent a spectrum of phenotypes that result from mutations in genes involved in telomere maintenance protein complexes. |
| Treacher Collins and related syndromes | Treacher Collins syndrome is a rare disorder affecting craniofacial development. |
2 MATERIALS AND METHODS
2.1 Sequencing, variant calling, and analysis by the Hopkins lab
Gene sequences were captured using one of the two custom probe sets (Agilent SureSelectXT Target Enrichment Kit, Agilent Technologies, Santa Clara, CA) and sequenced by a NGS platform (Illumina MiSeq, 2 × 100 nt reads, Illumina, San Diego, CA). The NGS panels were used to test assessed exons and exon-adjacent sequences for 64 or 83 loci (Supp. Tables S4 and S5). Sequences were aligned to the human reference genome (GRCh37/hg19) using the Burrows-Wheeler Aligner. Sequence variants were called individually for each patient to produce two Variant Call Format (VCF) files, one for single-nucleotide variants (SNVs; GATK UnifiedGenotyper, v2.7-4, Broad Institute, Cambridge, MA) and one for insertion–deletion variants (InDels; GATK HaplotypeCaller, v2.7-4, Broad Institute, Cambridge, MA). Deidentified VCF files were provided to the CAGI-4 organizers. Note that the CAGI-4 organizers combined individual VCF files for each patient into a single VCF, resulting in potentially misleading data in the INFO and FILTER fields of the file. The panel of 83 genes was sequenced in 96 of the 106 patients; for the other 10 patients, a partially overlapping list of 64 genes was sequenced (Supp. Table S5). Although the whole NGS panel was sequenced in all patients, only the genes selected on the patient’s test requisition form were analyzed by the lab (n = 1–24 genes/patient). For more information on the specific NGS tests offered by the lab, refer to the Hopkins lab Website (http://www.hopkinsmedicine.org/dnadiagnostic/tests/). The Hopkins lab included variants in the genes they analyzed, which were classified as variants of uncertain significance (VUS), likely pathogenic, and pathogenic as an answer key. The disease class of each patient was also provided in the answer key and reflects the test selected by the patient’s physician on the test requisition form. Approximately 20 phenotypes that Hopkins tested were narrowed down to 14 disease classes in order to simplify the challenge (Supp. Table S1). Some disease classes were not represented by any patients and were included as red herrings (Supp. Fig. S9).
2.2 Challenge format
Participants in the Hopkins clinical panel challenge were provided with the two VCF files above, a detailed description of the 14 disease classes given in Table 1, a submission template, a submission validation script, and the gene capture regions used in sequencing the patients (in Browser Extensible Data, or BED format). Participants were also instructed that every patient matched exactly one disease class.
Participants were asked to submit predictions of each patient’s disease class based on their gene panel sequences, along with predicted causal variant(s). Each participant was allowed to submit up to six distinct submissions, in which each submission contained predictions for each patient. For each submission, participants were required to predict the probability that the patient has a referring disease in each of the 14 disease classes in the provided list, as well as the predicted causal variant(s) from the gene panel sequence dataset for every disease class with a nonzero probability. Each predicted probability of disease class also included a mandatory standard deviation (SD) field indicating confidence in the prediction, with low SD indicating high confidence, and high SD indicating low confidence.
2.3 Assessment
Formatting errors in all submissions were corrected to the best of the assessor’s ability, and redundant submissions were removed. Predicted disease classes made in each submission for each patient were assessed against the correct disease class given in the Hopkins answer key, using the metrics described below. The predicted causal variant(s) were also compared with interpretations from the clinical laboratory, but in cases where predictions deviated from the answer key only limited assessment of the predictions could be performed. In their answer key, Hopkins noted which variants they regarded as VUS, likely pathogenic, and pathogenic; however, for purposes of matching participants’ predictions to the answer key, all variants noted by Hopkins for each patient were treated equivalently.
Assessors first calculated the number of correct predictions of disease class made in each submission. For each patient, the predicted disease class was the one assigned the highest probability among all 14 disease classes. Ties (i.e., cases where multiple disease classes were all assigned the highest probability) were handled as described below.
If all 14 probabilities for a patient were equal (e.g., all zeroes), those predictions were not counted in the following three metrics.
In other cases, assessors calculated one metric (nCorrect) in which the number of correct predictions was counted, giving ties full credit; another metric (nCorrecttie) was calculated in which N-way ties were given 1/N credit.
Finally, assessors calculated a third metric (nCorrectvar) in which they counted the number of predictions for which the disease class was correct (giving ties full credit), and for which at least one of the variants submitted in the corresponding column for that disease class matched one of the variants noted by Hopkins.
Assessors also calculated the following metrics for each submission:
avgPCorrect: The average probability assigned by the predictor to the correct disease class. This statistic provides an assessment of predictions that is not dependent on whether the submitter’s highest probability prediction was correct.
avgPCorrectnorm: The average probability assigned by the predictor to the correct disease class, after normalizing all probabilities predicted in each submission for each patient to sum to 1.0 (exception: if all probabilities for a patient were 0, they were not normalized).
avgRank: The average rank assigned by the predictor to the correct disease class. Ties were assigned the average rank of each set of tied predictions; for example, if the two highest probability disease classes had equal rank, both were assigned a rank of 1.5; a three-way tie for second highest probability would be assigned a rank of 3. Note that because there were 14 disease classes, an all-zero prediction would have an avgRank score of 7.5 (i.e., was scored as a 14-way tie).
avgError: The average error in predictions, where the error was measured as the absolute difference between the probability assigned each disease class and zero (if not the correct disease class) or one (if the correct disease class). Like avgPcorrect, avgError assesses predictions independent of their rank, but also includes correct negative predictions.
2.4 Prediction methodology
A summary of each group’s prediction methods is given below.
Group 57 (Jones): The Jones-UCL group made use of one-class support vector machine (SVM) classifiers to automatically assign disease classes according to the supplied exome data. In a normal machine learning experiment, sufficient positive and negative cases are needed to define a hypersurface that separates the two classes. Standard SVMs attempt to define this hypersurface such that the chance of misclassifying new cases is minimized. In some applications, however, only positive or negative cases are readily available, but not both. One-class SVMs (Schölkopf, Platt, Shawe-Taylor, Smola,&Williamson, 2001) have been proposed for problems where either negative or positive case data are unavailable. In this situation, the SVM attempts to identify outliers from a distribution modeled on the available single class of data, and it is assumed that the outliers belong to the alternative class.
In this CAGI challenge, of course, neither negative nor positive training data were readily available. However, the assumption was made that the 1000 Genomes data set (1000 Genomes Project Consortium et al., 2010) could be used as a proxy for negative case data. This is a reasonable assumption if we assume that the diseases in question are relatively rare. To start with, gene variants relating to each disease class were collated using ClinVar (Landrum et al., 2016). Feature sets were generated for each disease class by encoding variant 0/0, 0/1, and 1/1 calls as 0, 1, and 2, respectively, and for each disease-specific feature set, a one-class ν-SVM (using a RBF kernel) was trained. The single parameter ν, which controls both the number of support vectors and the misclassification cost, was optimized for each disease class so as to minimize the number of outliers detected in the 1000 Genome training data. Once trained, the SVM was then applied to the test sample data, and the distance to decision boundary was used as a proxy for classification confidence. The most important variant was identified in each case by systematically removing each variant from the feature set and recalculating the confidence scores.
Group 58 (Tosatto): The analysis started with a manually curated association between the genes of the panel and the 14 clinical phenotypes of interest based on literature review. Sequencing data were annotated with ANNOVAR (Wang, Li, & Hakonarson, 2010), considering for each variant the corresponding affected gene, frequency estimated from the 1000 Genomes Project (1000 Genomes Project Consortium et al., 2013) and predicted pathogenicity score from SIFT (Ng & Henikoff, 2003) and PolyPhen-2 (Adzhubei, Jordan, & Sunyaev, 2013). The method to define association between genetic data and phenotypes was based mainly on two phases. For each individual, variations that are less probable to be disease causing were filtered out, and a probability to be affected based on the analysis of variants was defined. Only coding and splice-site variants that can affect protein function were considered according to the Common Disease-Rare Variant Hypothesis (CDRVH) (El-Fishawy, 2013). Common (MAF > 5%) and/or synonymous SNVs were filtered out. Insertion and deletions were excluded as their impact on protein function is difficult to predict compared to SNVs. Only insertions and deletions (indels) affecting the coding part of a gene and predicted to be “damaging” or known to be pathogenic were considered. Heterozygous indels in genes with autosomal-recessive inheritance, occurring in GC-rich or repeated regions, were filtered out from the disease candidate mutation pool. An empirically derived scoring scheme was implemented to define association between patients and phenotypes, considering both disease inheritance and predicted SNV pathogenicity (Supp. Table S2). Different weights were assigned to different mutation types, that is, a high score for known variants associated with a specific disease (mainly by literature review) and a lower score for mutations not affecting protein function according to predictor output (i.e., tolerated, benign, and unknown). For autosomal-dominant pathologies, only heterozygous variants plus few manually curated homozygous mutations were considered (i.e., the one with the highest probability score). The disease cutoffs were set at different values between submissions, allowing the stringency of the analysis to vary. Both homozygous and compound heterozygous variants were considered for autosomal-recessive conditions. When more than one match per patient occurred, only the most likely was considered (i.e., the one with higher probability score). Different submissions correspond to different sets of weights.
In particular, in the first submission, a slightly lower weight was assigned to variants whose effect is more difficult to assess (i.e., compound heterozygous, homozygous VUS, variants affecting different genes coding for subunits of the same complex) with respect to submission 4.
Group 59 (Qiagen Bioinformatics): All 106 samples were uploaded to Ingenuity Variant Analysis (QIAGEN- Hereditary Disease Solution) and set up an analysis with all samples to filter low-quality (call quality < 20) and common variants (>0.5% MAF in 1000 Genomes) (1000 Genomes Project Consortium et al., 2010), NHLBI-EVS (NHLBI GO Exome Sequencing Project (ESP), 2015), ExAC (Lek et al., 2016), and Allele Frequency Community (www.allelefrequencycommunity.org), using the confidence and common variant filters, respectively. The Allele Frequency Community is a QIAGEN-hosted allele frequency database, founded by QIAGEN and participating members in 2014. It is a freely accessible “opt-in” community resource designed to facilitate sharing of anonymized, pooled allele frequency statistics among community members. The predicted deleterious filter was used to keep only those variants that are previously published and classified pathogenic or likely pathogenic, using ACMG guidelines, DM variants (pathological mutations reported to be disease causing in the original literature report) present in HGMD, along with other loss of function (frameshift, start/stop loss or gain, splice site) and missense variants. Finally, the biological context filter was applied to find variants linked to each one of the 14 categories, and patient disease category was predicted based on variant–disease connection, using path-to-phenotype evidence.
Group 60 (RSS): Gene phenotype associations were mined from the Hopkins diagnostic panels, OMIM (Hamosh, Scott, Amberger, Bocchini, & McKusick, 2005), and GeneReviews (Pagon et al., 1993). Inheritance mode and penetrance information were extracted from online resources for each gene–phenotype pair.
Variants with low-quality or high-population allele frequencies were filtered out, and the functional impact was annotated with Variant Effect Predictor (McLaren et al., 2010). To estimate the probability that a variant is damaging to protein function, we integrated multiple prediction methods to score all types of variants, for example, missense, nonsense, indels, and intronic variants. The damaging scores were scaled and normalized to reflect the relative deleteriousness, for example, frameshift/nonsense variants would have higher scores than missense variants. We then used the damaging scores to estimate the probability that each individual has a particular phenotype with a probabilistic model, that is, calculated as the probability that at least one associated gene in the individual causes the phenotype. For a particular gene, the probability the gene causes the phenotype was calculated as the probability that the gene is disrupted (taking into account inheritance mode), multiplied by its penetrance score.
The confidence level of the prediction was calculated from the distribution of the estimated probabilities across phenotypes and across individuals. Considering that 14 phenotypes are Mendelian-like diseases, if one individual has high prediction scores across phenotypes, it is more likely to be false positive. Thus, high confidence was assigned to individuals with high variability across phenotypes.
A more detailed description of this group’s prediction methods is included in the Supp. Information.
Group 61 (Moult): The method (implemented in Python) has four modules: variant annotation, QC (quality check), variant prioritization, and probability scoring for the disease. The modules were executed sequentially. Inputs were the two gVCF files and a gene configuration file containing the genes associated with each disease class and their inheritance pattern. The Varant tool (https://doi.org/10.5060/D2F47M2C, http://compbio.berkeley.edu/proj/varant) was used to annotate variants with: region of occurrence in the genome, allele frequency from ExAC (Lek et al., 2016), predicted pathogenicity based on four methods (Adzhubei et al., 2013; Kircher et al., 2014; Kumar, Henikoff, & Ng, 2009; Yue, Melamud, & Moult, 2006) (for missense), and previously reported disease associations in databases (Landrum et al., 2016; Stenson et al., 2003). Three QC analyses were run: (1) variant counts (common vs. rare vs. novel & homozygous vs. heterozygous) per sample, (2) read depth for each gene in each sample was obtained by averaging DP values over all bases in a gene recorded in the gVCF file, and (3) exons with relatively low or no coverage compared with other exons in a gene. The QC-qualified variants per sample were prioritized by first assigning them to one of the three classes, ranked by the likelihood that the variant is causative and further grouping the variants in each class by frequency based on its ExAC MAF (group 1, novel; 2, very rare (MAF < = 0.005); or 3, rare (MAF ≤ 0.01). Class-1 identified variants previously reported in disease databases as pathogenic, Class-2 identified loss of function, splice, and missense variants predicted damaging by in silico prediction tools, and Class-3 identified missense variants (not predicted damaging), UTR, and intronic variants. Variants were further filtered for inheritance model. For each sample, once putative causative variants were found, the process was terminated (e.g., if a suitable variant or variants were found using Class-1, Class-2, and Class-3 were not executed). Finally, a probability score for a sample to have a particular disease was computed based on the type of prioritized variant(s) and inheritance pattern. For the missense variants, the probability model was based on the extent of consensus among the four prediction methods, using a previous HGMD-derived calibration. For other variant types, subjective probability rules were used.
3 RESULTS
3.1 Summary of submissions
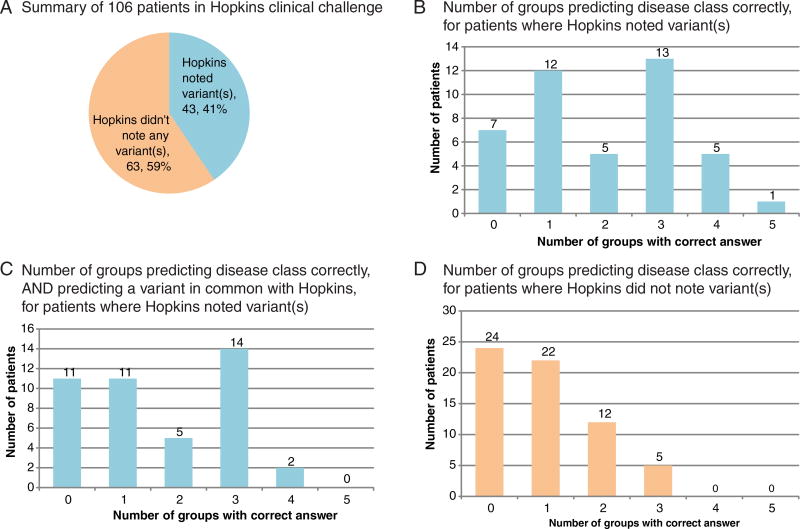
Five groups submitted predictions (with 4, 2, 2, 2, and 1 distinct predictions per group). An overview of the challenge and results is shown in Figure 1. One-hundred six patients in the challenge can be roughly grouped into two difficulty classes: (1) patients for whom Hopkins noted a potentially causal variant in the answer key (43 patients) and (2) patients for whom Hopkins did not note any variants (63 patients) (Fig. 1A). At least one CAGI-4 predicting group correctly predicted the disease class for 36 of the 43 patients who had a reported variant (Fig. 1B). Fewer groups correctly predicted both the disease class and at least one of the variant(s) that Hopkins reported (Fig. 1C). CAGI-4 predictors were not as accurate at predicting disease classes for the remaining 63 patients for whom Hopkins did not note a variant, although at least one group correctly predicted the disease class for the majority of these patients (Fig. 1D). The lower prediction accuracy is perhaps unsurprising given the negative test results for these 63 patients.
FIGURE 1.
Summary of CAGI-4 Hopkins clinical panel challenge and results. A: One-hundred six patients were included in the study. Hopkins noted at least one variant relevant to the disease class for which the patient was referred in 43 cases, and did not note a variant for the remaining 63 cases. Hopkins noted variants of the following classes: variant of uncertain significance, likely pathogenic, or pathogenic. Clinically, Hopkins would have reported 25/43 as positive and 18/43 as uncertain. B: Among the 43 patients for whom Hopkins had noted a variant, at least one CAGI-4 prediction group predicted the correct disease class in 36 cases, and one patient’s disease class was predicted correctly by all five groups. C: Among the 43 patients for whom Hopkins had noted a variant, at least one CAGI-4 prediction group predicted both the correct disease class and a causal variant noted by Hopkins in 32 cases. D: Sixty-three patients for whom Hopkins did not note a variant were more difficult for CAGI-4 groups to predict: 24were not predicted correctly by any group, and only five patients’ disease class was predicted correctly by three groups (none were predicted correctly by four or more groups)
3.2 Numeric assessment summary
Table 2 summarizes our numeric assessment metrics for each nonredundant, submitted prediction, for all patients. Table 3 shows the same statistics for only 43 patients for which Hopkins noted at least one potentially causal variant. The best values for each metric in each table are indicated in bold. Each group’s overall performance is briefly discussed below.
TABLE 2.
Summary of assessment metrics for each nonredundant, submitted prediction, for all patients
| Group | Prediction | nCorrect | nCorrecttie | avgPCorrect | avgPCorrectnorm | avgRank | avgError |
|---|---|---|---|---|---|---|---|
| Jones | 57.1 | 24 | 24 | 0.305 | 0.098 | 5.32 | 0.251 |
| 57.2 | 9 | 9 | 0.239 | 0.068 | 7.66 | 0.287 | |
| 57.3 | 7 | 7 | 0.236 | 0.068 | 7.78 | 0.289 | |
| 57.4 | 7 | 6.5 | 0.426 | 0.074 | 7.1 | 0.42 | |
| Tosatto | 58.1 | 23 | 23 | 0.178 | 0.217 | 6.48 | 0.105 |
| 58.4 | 26 | 25 | 0.223 | 0.227 | 6.15 | 0.107 | |
| Qiagen | 59.1 | 32 | 29.5 | 0.302 | 0.278 | 5.82 | 0.09 |
| 59.2 | 31 | 28.5 | 0.292 | 0.269 | 5.88 | 0.091 | |
| RSS | 60.1 | 12 | 12 | 0.072 | 0.102 | 7.14 | 0.08 |
| 60.2 | 12 | 12 | 0.068 | 0.094 | 7.15 | 0.082 | |
| Moult | 61.1 | 38 | 34.99 | 0.261 | 0.265 | 5.65 | 0.105 |
Notes Predictions are numbered according to the group’s (formerly anonymized) group number (57, Jones; 58, Tosatto; 59, Qiagen Bioinformatics; 60, RSS; 61, Moult) and the group’s submission number (1, most confident prediction; other nonredundant predictions are as numbered by the submitters, up to six per group).
TABLE 3.
Summary of assessment metrics for each nonredundant, submitted prediction, for the 43 patients for which Hopkins noted at least one potentially causal variant
| Group | Prediction | nCorrect | nCorrecttie | nCorrectvar | avgPCorrect | avgPCorrectnorm | avgRank | avgError |
|---|---|---|---|---|---|---|---|---|
| Jones | 57.1 | 5 | 5 | 2 | 0.255 | 0.082 | 6.53 | 0.257 |
| 57.2 | 5 | 5 | 2 | 0.325 | 0.091 | 6.29 | 0.274 | |
| 57.3 | 2 | 2 | 0 | 0.22 | 0.063 | 8.49 | 0.296 | |
| 57.4 | 1 | 1 | 0 | 0.394 | 0.07 | 7.5 | 0.421 | |
| Tosatto | 58.1 | 15 | 15 | 13 | 0.32 | 0.349 | 5.56 | 0.087 |
| 58.4 | 17 | 16 | 16 | 0.38 | 0.339 | 5.16 | 0.094 | |
| Qiagen | 59.1 | 23 | 21 | 19 | 0.535 | 0.488 | 4.24 | 0.065 |
| 59.2 | 22 | 20 | 19 | 0.512 | 0.465 | 4.4 | 0.066 | |
| RSS | 60.1 | 9 | 9 | 8 | 0.149 | 0.193 | 6.41 | 0.073 |
| 60.2 | 9 | 9 | 8 | 0.145 | 0.181 | 6.4 | 0.075 | |
| Moult | 61.1 | 26 | 26 | 25 | 0.5 | 0.512 | 3.78 | 0.07 |
Notes Predictions are numbered as in Table 2.
Table 4 shows a summary of the performance of all predicting groups on each patient. An expanded version of Table 4 with additional columns is provided in Supp.. Information (Supp. Table S6). Tables 5 and 6 summarize the most frequent combinations of groups that predicted the correct disease class for patients (Table 5 ignores causal variant predictions, whereas Table 6 requires each group to predict at least one of the variants noted by Hopkins).
TABLE 4.
Summary of the performance of all predicting groups on each patient
| Patient | nC | nCV | Correct groups | Correct groups, with variant |
Correct predictions | Correct predictions, with variant |
|---|---|---|---|---|---|---|
| P1 | 4 | 4 | 57, 59, 60, 61 | 57, 59, 60, 61 | 59.2, 60.1, 60.2, 61.1, 57.1, 57.3, 59.1 | 59.2, 60.1, 60.2, 61.1, 57.1, 59.1 |
| P2 | 1 | N/A | 57 | N/A | 57.2 | N/A |
| P3 | 0 | N/A | None | N/A | None | N/A |
| P4 | 5 | 3 | 57, 58, 59, 60, 61 | 58, 59, 61 | 59.2, 58.4, 60.1, 60.2, 61.1, 57.1, 58.1, 59.1 | 59.2, 58.4, 61.1, 58.1, 59.1 |
| P5 | 2 | 2 | 60, 61 | 60, 61 | 60.1, 60.2, 61.1 | 60.1, 60.2, 61.1 |
| P6 | 3 | N/A | 57, 59, 61 | N/A | 59.2, 61.1, 57.1, 59.1 | N/A |
| P7 | 0 | N/A | None | N/A | None | N/A |
| P8 | 1 | 1 | 60 | 60 | 60.1, 60.2 | 60.1, 60.2 |
| P9 | 1 | 0 | 57 | None | 57.4, 57.1 | None |
| P10 | 2 | N/A | 57, 58 | N/A | 58.4, 57.1, 58.1 | N/A |
| P11 | 1 | 1 | 61 | 61 | 61.1 | 61.1 |
| P12 | 0 | N/A | None | N/A | None | N/A |
| P13 | 3 | N/A | 57, 58, 60 | N/A | 57.4, 58.4, 60.1, 60.2, 57.2, 58.1 | N/A |
| P14 | 0 | N/A | None | N/A | None | N/A |
| P15 | 0 | N/A | None | N/A | None | N/A |
| P16 | 2 | N/A | 57, 58 | N/A | 58.4, 57.1, 58.1 | N/A |
| P17 | 3 | 3 | 58, 59, 61 | 58, 59, 61 | 59.2, 58.4, 61.1, 58.1, 59.1 | 59.2, 58.4, 61.1, 58.1, 59.1 |
| P18 | 2 | N/A | 57, 58 | N/A | 58.4, 57.1, 58.1 | N/A |
| P19 | 1 | 1 | 60 | 60 | 60.1, 60.2 | 60.1, 60.2 |
| P20 | 1 | N/A | 57 | N/A | 57.1 | N/A |
| P21 | 1 | N/A | 57 | N/A | 57.1 | N/A |
| P22 | 1 | N/A | 59 | N/A | 59.2, 59.1 | N/A |
| P23 | 0 | 0 | None | None | None | None |
| P24 | 4 | 3 | 57, 58, 59, 61 | 58, 59, 61 | 59.2, 58.4, 61.1, 57.2, 58.1, 59.1 | 59.2, 58.4, 61.1, 58.1, 59.1 |
| P25 | 1 | 0 | 57 | None | 57.2 | None |
| P26 | 3 | 3 | 58, 59, 61 | 58, 59, 61 | 59.2, 58.4, 61.1, 59.1 | 59.2, 58.4, 61.1, 59.1 |
| P27 | 3 | 1 | 58, 59, 61 | 59 | 59.2, 58.4, 61.1, 58.1, 59.1 | 59.2, 59.1 |
| P28 | 2 | N/A | 57, 59 | N/A | 59.2, 57.1, 59.1 | N/A |
| P29 | 1 | N/A | 57 | N/A | 57.1 | N/A |
| P30 | 3 | 2 | 58, 59, 61 | 58, 61 | 58.4, 61.1, 58.1, 59.1 | 58.4, 61.1, 58.1 |
| P31 | 1 | N/A | 57 | N/A | 57.1 | N/A |
| P32 | 4 | 3 | 58, 59, 60, 61 | 58, 60, 61 | 59.2, 58.4, 60.1, 60.2, 61.1, 58.1, 59.1 | 58.4, 60.1, 60.2, 61.1, 58.1 |
| P33 | 0 | N/A | N/A | None | N/A | |
| P34 | 3 | 3 | 58, 59, 61 | 58, 59, 61 | 59.2, 58.4, 61.1, 58.1, 59.1 | 59.2, 58.4, 61.1, 58.1, 59.1 |
| P35 | 0 | N/A | None | N/A | None | N/A |
| P36 | 0 | 0 | None | None | None | None |
| P37 | 0 | N/A | None | N/A | None | N/A |
| P38 | 3 | 3 | 58, 60, 61 | 58, 60, 61 | 58.4, 60.1, 60.2, 61.1, 58.1 | 58.4, 60.1, 60.2, 61.1, 58.1 |
| P39 | 1 | 0 | 59 | None | 59.2, 59.1 | None |
| P40 | 0 | N/A | None | N/A | None | N/A |
| P41 | 0 | N/A | None | N/A | None | N/A |
| P42 | 2 | 1 | 59, 61 | 61 | 59.2, 61.1, 59.1 | 61.1 |
| P43 | 2 | N/A | 57, 61 | N/A | 61.1, 57.1 | N/A |
| P44 | 1 | N/A | 59 | N/A | 59.2, 59.1 | N/A |
| P45 | 1 | N/A | 57 | N/A | 57.1 | N/A |
| P46 | 0 | N/A | None | N/A | None | N/A |
| P47 | 1 | 1 | 61 | 61 | 61.1 | 61.1 |
| P48 | 0 | 0 | None | None | None | None |
| P49 | 1 | N/A | 57 | N/A | 57.1 | N/A |
| P50 | 0 | N/A | None | N/A | None | N/A |
| P51 | 1 | N/A | 61 | N/A | 61.1 | N/A |
| P52 | 3 | 3 | 58, 59, 61 | 58, 59, 61 | 59.2, 58.4, 61.1, 59.1 | 59.2, 58.4, 61.1, 59.1 |
| P53 | 2 | N/A | 58, 59 | N/A | 59.2, 58.4, 58.1, 59.1 | N/A |
| P54 | 1 | N/A | 57 | N/A | 57.3 | N/A |
| P55 | 0 | 0 | None | None | None | None |
| P56 | 2 | 1 | 58, 59 | 59 | 59.2, 58.1, 59.1 | 59.2, 59.1 |
| P57 | 1 | 1 | 61 | 61 | 61.1 | 61.1 |
| P58 | 0 | N/A | None | N/A | None | N/A |
| P59 | 0 | 0 | None | None | None | None |
| P60 | 2 | 2 | 59, 61 | 59, 61 | 59.2, 61.1, 59.1 | 59.2, 61.1, 59.1 |
| P61 | 1 | N/A | 57 | N/A | 57.4, 57.3 | N/A |
| P62 | 2 | N/A | 57, 60 | N/A | 60.1, 60.2, 57.1 | N/A |
| P63 | 3 | N/A | 57, 59, 61 | N/A | 59.2, 61.1, 57.1, 59.1 | N/A |
| P64 | 1 | 1 | 60 | 60 | 60.1, 60.2 | 60.1, 60.2 |
| P65 | 3 | N/A | 58, 60, 61 | N/A | 58.4, 60.1, 60.2, 61.1 | N/A |
| P66 | 0 | N/A | None | N/A | None | N/A |
| P67 | 0 | 0 | None | None | None | None |
| P68 | 1 | N/A | 59 | N/A | 59.2, 59.1 | N/A |
| P69 | 0 | 0 | None | None | None | None |
| P70 | 0 | N/A | None | N/A | None | N/A |
| P71 | 0 | N/A | None | N/A | None | N/A |
| P72 | 3 | 2 | 57, 59, 61 | 59, 61 | 59.2, 61.1, 57.2, 59.1 | 59.2, 61.1, 59.1 |
| P73 | 4 | 3 | 57, 58, 59, 61 | 58, 59, 61 | 59.2, 58.4, 61.1, 57.1, 58.1, 59.1 | 59.2, 58.4, 61.1, 58.1, 59.1 |
| P74 | 0 | N/A | None | N/A | None | N/A |
| P75 | 0 | N/A | None | N/A | None | N/A |
| P76 | 0 | N/A | None | N/A | None | N/A |
| P77 | 0 | N/A | None | N/A | None | N/A |
| P78 | 1 | N/A | 61 | N/A | 61.1 | N/A |
| P79 | 0 | N/A | None | N/A | None | N/A |
| P80 | 3 | 3 | 57, 59, 61 | 57, 59, 61 | 59.2, 61.1, 57.1, 57.2, 59.1 | 59.2, 61.1, 57.1, 57.2, 59.1 |
| P81 | 1 | 1 | 58 | 58 | 58.4, 58.1 | 58.4, 58.1 |
| P82 | 1 | N/A | 57 | N/A | 57.2 | N/A |
| P83 | 1 | N/A | 57 | N/A | 57.4, 57.3 | N/A |
| P84 | 4 | 4 | 57, 58, 59, 61 | 57, 58, 59, 61 | 59.2, 58.4, 61.1, 57.2, 58.1, 59.1 | 59.2, 58.4, 61.1, 57.2, 58.1, 59.1 |
| P85 | 2 | N/A | 57, 61 | N/A | 57.4, 61.1 | N/A |
| P86 | 2 | N/A | 58, 59 | N/A | 59.2, 58.4, 58.1, 59.1 | N/A |
| P87 | 3 | N/A | 57, 58, 61 | N/A | 57.4, 58.4, 61.1, 57.1, 58.1, 57.3 | N/A |
| P88 | 1 | N/A | 57 | N/A | 57.1 | N/A |
| P89 | 1 | N/A | 57 | N/A | 57.4, 57.1 | N/A |
| P90 | 2 | N/A | 58, 61 | N/A | 58.4, 61.1, 58.1 | N/A |
| P91 | 1 | N/A | 57 | N/A | 57.2 | N/A |
| P92 | 3 | 3 | 58, 59, 61 | 58, 59, 61 | 59.2, 58.4, 61.1, 59.1 | 59.2, 58.4, 61.1, 59.1 |
| P93 | 1 | 1 | 60 | 60 | 60.1, 60.2 | 60.1, 60.2 |
| P94 | 2 | 2 | 59, 61 | 59, 61 | 59.2, 61.1, 59.1 | 59.2, 61.1, 59.1 |
| P95 | 3 | 3 | 58, 59, 61 | 58, 59, 61 | 59.2, 58.4, 61.1, 58.1, 59.1 | 59.2, 58.4, 61.1, 58.1, 59.1 |
| P96 | 1 | 0 | 57 | None | 57.3 | None |
| P97 | 0 | N/A | None | N/A | None | N/A |
| P98 | 2 | N/A | 57, 61 | N/A | 61.1, 57.1 | N/A |
| P99 | 1 | N/A | 59 | N/A | 59.2, 59.1 | N/A |
| P100 | 0 | N/A | None | N/A | None | N/A |
| P101 | 2 | N/A | 57, 61 | N/A | 61.1, 57.1 | N/A |
| P102 | 1 | N/A | 57 | N/A | 57.3 | N/A |
| P103 | 0 | N/A | None | N/A | None | N/A |
| P104 | 3 | 3 | 58, 59, 61 | 58, 59, 61 | 59.2, 58.4, 61.1, 58.1, 59.1 | 59.2, 58.4, 61.1, 58.1, 59.1 |
| P105 | 3 | 3 | 58, 59, 61 | 58, 59, 61 | 59.2, 58.4, 61.1, 58.1, 59.1 | 59.2, 58.4, 61.1, 58.1, 59.1 |
| P106 | 1 | N/A | 61 | N/A | 61.1 | N/A |
Notes An expanded version of Table 4 with additional columns has been provided in Supp. Information (Supp. Table S6).
nC, number of groups predicting the disease class correctly, among all submissions from each group (counting ties, except in cases where all 14 disease classes were assigned equal probability; nCV, number of groups predicting both the correct disease class and at least one variant noted by Hopkins; correct groups, a list of groups in which the disease class was predicted correctly in at least one submission (counting ties, except in cases where all 14 disease classes were assigned equal probability). Groups are numbered as in Table 2; correct groups, with variant, a list of groups with at least one prediction of the correct disease class, and also at least one variant noted by Hopkins (N/A in this field indicates that Hopkins did not note any variants). Predictions are numbered as in Table 2; correct predictions, with variant, same as above, but indicating individual submission numbers that were correct.
TABLE 5.
Frequency with which each combination of groups correctlydiagnosed patients
| Number of patients | Groups predicting correct disease class |
|---|---|
| 31 | No group predicted correct disease |
| 18 | 57 (note: 7 from 57.1) |
| 10 | 58, 59, 61 |
| 6 | 61 |
| 5 | 59 |
| 4 | 60 |
| 4 | 57, 61 |
| 4 | 57, 59, 61 |
| 3 | 59, 61 |
| 3 | 58, 59 |
| 3 | 57, 58, 59, 61 |
| 3 | 57, 58 |
| 2 | 58, 60, 61 |
| 1 | 60, 61 |
| 1 | 58, 61 |
| 1 | 58, 59, 60, 61 |
| 1 | 58 |
| 1 | 57, 60 |
| 1 | 57, 59, 60, 61 |
| 1 | 57, 59 |
| 1 | 57, 58, 61 |
| 1 | 57, 58, 60 |
| 1 | 57, 58, 59, 60, 61 |
Note This table summarizes the number of times each combination of groups correctly diagnosed patients, as shown in the “correct groups” column of Table 4.
TABLE 6.
Frequency with which each combination of groups correctly diagnosed patients, and also noted a Hopkins variant
| Number of patients |
Groups predicting correct disease and variant |
|---|---|
| 63 | (Hopkins did not note any variants) |
| 11 | 58, 59, 61 |
| 11 | (No group predicted disease and variant correctly) |
| 4 | 61 |
| 4 | 60 |
| 3 | 59, 61 |
| 2 | 59 |
| 2 | 58, 60, 61 |
| 1 | 60, 61 |
| 1 | 58, 61 |
| 1 | 58 |
| 1 | 57, 59, 61 |
| 1 | 57, 59, 60, 61 |
| 1 | 57, 58, 59, 61 |
Note: This table summarizes the number of times each combination of groups correctly diagnosed patients and predicted at least one variant noted by Hopkins, as shown in the “correct groups with variant” column of Table 4.
Group 57 (Jones): Group 57’s primary submission (57.1) scored much higher than their other submissions by our metrics. Their method was less accurate than other groups in cases where Hopkins reported a potential causal variant, but it was more accurate at predicting the correct disease class in cases where Hopkins didn’t report a variant. Group 57’s primary submission was also the most accurate among all submissions at rank ordering the disease classes. As seen in Table 5, Group 57 predicted disease classes correctly for 18 patients that no other group predicted correctly, with seven of these cases in their primary submission.
This method was unique in that it did not attempt to mimic a traditional clinical genetics approach. No attempt was made to independently predict the pathogenicity of the ClinVar variants used as features or to correct for linkage disequilibrium, which may explain why the method was able to make correct inferences where no causal variants were reported and why correct inference can arise without reporting the correct variants. A possibility is that some or even a majority of the variants relied on by the classifiers were noncausal variants that simply happen to be in linkage disequilibrium with one or more true causal variants. Thus, the occurrence of these variants were sufficient to identify the sample as a genetic outlier, though not indicating true causation. It is possible that by addressing these issues, the method might be further enhanced to make more accurate predictions relating to true causal variants. It would also be prudent to test this method on a different dataset to rule out the possibility that there is some underlying structure in this dataset that the algorithm is detecting.
Group 58 (Tosatto): As seen in Table 5, most cases that Group 58 predicted correctly were also predicted by at least one other group. However, Group 58 predicted the disease class for one patient (P81) that no other groups predicted; they also assigned 100% probability of the correct disease to that patient, and predicted exactly the same causal variants as noted by Hopkins. Many of the diseases in this challenge result from loss-of-function variants in a given gene, therefore selective filtering of indel variants by Group 58 was questionable. The genes and molecular mechanisms associated with each of the 14 disease classes were not provided as part of the dataset, which increased the difficulty of the matching exercise (Supp. Table S2).
Group 59 (Qiagen): Group 59 had the highest average P values for the correct disease classes, after normalization; they also had some of the best scores in the avgError metric. Group 59 correctly predicted the disease class for five patients that no other groups predicted. Among all the groups, they were the only group for which both P values and SD values were independent and positively correlated with the values they were expected to correlate with (see Accuracy of P and SD Values section below). This challenge was well suited for the Qiagen group, as they specialize in large-scale variant interpretation (Tricarico et al., 2017).
Group 60 (RSS): Due to the misleading fields in the combined VCF files (see the Materials and Methods section on sequencing and variant calling), Group 60 made only 11 high-confidence (P > 0.6) predictions, of which nine were correct. Interestingly, four of these nine cases were not predicted correctly by any other group. Because of the small number of high-confidence predictions, Group 60 had the lowest avgError score among all groups, and the best correlation between assigned P values and correct answers (see Accuracy of P and SD Values section below). After the challenge closed, Group 60 provided the CAGI organizers with a corrected submission, in which the misleading VCF fields were ignored. In this corrected submission (which arrived late and therefore was not formally assessed), Group 60 correctly predicted 38 disease classes. Additional analysis of Group 60’s corrected submission is provided in the Supp. Information. Group 60 adeptly used a series of online clinical genetics resources in their analysis pipeline.
Group 61 (Moult): Group 61 made more correct predictions of both disease class and Hopkins-annotated variants than any other group. For the 43 cases where Hopkins noted variants, Group 61 did especially well, getting 26 disease classes correct, and predicting the best average rank for the correct disease. In 25 of these cases, Group 61 also predicted at least one causal variant that was noted by Hopkins. Group 61 correctly predicted the disease class for six patients that no other groups predicted correctly, and also predicted at least one of the potentially causal variants noted by Hopkins in four of these six cases.
3.3 Accuracy of P and SD values
We expected that predictors’ submitted probabilities for each patient and disease should correlate with the correct disease class for each patient, and we also expected that their submitted SDs on each prediction should correlate with the error in each prediction (i.e., the absolute difference between the P value and either 1 or 0, for cases where the patient does or does not have the disease, respectively). Overall, predictors did better in the first case, and not so well in the second. Only one group (59; Qiagen) had an independent SD model that correlated positively with error. A detailed discussion of the accuracy of P and SD predictions is provided in the Supp. Information.
3.4 Commentary on novel variant predictions
One large limitation in the design of this challenge is that only a subset of the sequence data was clinically analyzed in each patient. This allowed for the possibility of false negatives, where true pathogenic variants may have been present in genes that were not analyzed by the lab. Further, Internal Review Board (IRB) restrictions prevented the data provider from acting as an assessor for the challenge or providing detailed feedback on variant predictions in genes that were not clinically analyzed. In addition, specific variants cannot be listed in the following discussion. In the future, advanced planning is needed to ensure that the appropriate consents and approvals are in place to maximize the use of clinical data. Ideally, a dataset should be fully analyzed by a clinical lab and patients should be specifically asked for consent that their data could be used for research purposes such as the CAGI challenge. This would allow a more critical analysis of the challenge data, would eliminate the possibility of unwanted incidental findings, and would allow more in-depth discussion of challenge results. Clinical data from human patients makes an interesting challenge set, but data from human subjects involve privacy concerns vastly different from that of laboratory model organisms.
The CAGI-4 Hopkins clinical panel challenge gives us an opportunity to test state-of-the-art genetic analysis pipelines on a subset of the data that would be obtained from complete exome sequencing of patients, and to explore potential advantages and disadvantages of genomics-driven approaches to clinical testing versus the phenotype-driven approach currently employed by Hopkins. In some cases, multiple groups reported the same causal variant for a case where Hopkins did not identify a variant. Since Hopkins only analyzed the genes ordered by the physician, it is possible that there were true pathogenic variants identified in the challenge that were not included on the answer key, such cases are elaborated on below. In order to explore the potential complication of false positives in the genomics-driven approach, we also examined cases in which CAGI-4 predictors consistently predicted the wrong disease class along with the same causal variants. Several of these cases are described below:
Patient P7: Groups 57 (submission 4), 58, 59, and 61 all predicted Telomere Shortening Disorders, and the latter three groups consistently noted a missense variant in TERT. The patient’s diagnosis was cystic fibrosis and CF-related disorders, and Hopkins did not note any reportable variants and did not analyze the TERT gene. The TERT variant is described in the literature; it leads to telomere shortening and is involved in bone marrow failure. Telomere shortening due to mutations in TERT is known to be involved in pulmonary fibrosis. Clinical presentation of pulmonary fibrosis is very different from cystic fibrosis. This TERT variant is annotated in ClinVar as involved in pulmonary fibrosis, but literature support for this phenotype is unclear. The variant is found in 120 ExAC participants including two homozygotes.
Patient P36: Groups 57 (submission 2), 58, 59, and 61 all predicted Liddle syndrome, with the same missense variant in SCNN1G. The patient’s diagnosis was diffuse lung disease. The SCNN1G variant is a known pathogenic variant observed in two independent patients with bronchiectasis. The predictors presumably predicted Liddle syndrome because the same gene is involved in that disorder. Hopkins did not note a reportable variant for this patient and the SCNN1G gene was not analyzed.
Patient P37: Groups 57 (submission 2), 58, 59, and 61 all predicted Marfan syndrome with the same variant, a missense variant in FBN1. The patient’s diagnosis was diffuse lung disease. FBN1 is involved in Marfan syndrome and in other cardiac phenotypes. A subgroup of Marfan patients develop lung emphysema, which is possibly a reason for the predictions. The missense variant is a known low-frequency polymorphism annotated as “benign” in ClinVar, so this is likely a false-positive prediction. Hopkins did not note any variants for this patient and did not analyze the FBN1 gene.
Patient P14: Groups 57 (submissions 3 and 4), 58, 59, and 61 all predicted cystic fibrosis and CF-related disorders, along with one to two out of four variants in CFTR. The patient’s diagnosis was diffuse lung disease, and Hopkins did not analyze the CFTR gene. All the predicted CFTR variants have previously been reported. One is a common polymorphism, and seems unlikely to contribute to disease. Another is intronic, and it is not clear whether it may be involved in splicing. The remaining two CFTR variants were rare missense variants. One missense variant is seen in ExAC 739 times including once in the homozygous state, and there is no information on its pathogenicity reported in the literature or public databases. The second missense variant is seen in ExAC 623 times including once in the homozygous state, and there is conflicting evidence reported in the literature regarding its pathogenicity. The latter two variants appear to be too common to be causal in this case, but as mentioned above, CF studies may be included in ExAC.
4 DISCUSSION
Overall, we found that current state-of-the-art computational prediction methods do a reasonable job of predicting clinical phenotype from genotype, even when blinded to clinical diagnoses. At the same time, current genotype-driven prediction methodologies generate false positives and false negatives at a rate unacceptable for clinical use. In cases where the Hopkins lab reported a variant, predictors did relatively well, with at least one group correctly identifying the disease class in 36 of 43 patients (84%), and at least one group identifying the correct disease class and variant in 33 of 43 cases (77%). In cases where the Hopkins lab did not find a reportable variant in the genes they analyzed, at least one group correctly matching the disease class in 39 of 63 patients (62%). In the latter cases, methods based on machine learning (SVM) technology appeared to be most effective at correctly identifying the disease. Interestingly, despite the ability to correctly match genotype to phenotype, the SVM-based method could not correctly identify the pathogenic variant. It is unclear what is happening in cases where groups correctly identify the disease class, but not the causal variant. In retrospect, it would have been helpful to include a list of gene–disease associations as well as modes of inheritance to the predictors to aid in the matching process. Different groups performed better depending on which metric was used; there was no clear “winner” that dominated performance across all metrics. Indeed, every group predicted at least one patient’s disease class correctly that no other group predicted correctly. This result suggests that a “meta-predictor” or a human clinical expert with access to all groups’ results might improve on the performance of each individual group.
Currently, clinical genetic testing is almost entirely phenotype-driven: given a clinical diagnosis, laboratories analyze variants in genes known to be relevant to the diagnosed disease. This is partially due to the historic technical limitations on genetic testing, for example, sequencing costs limited the number of genes for which data could be obtained. The standards for reporting variants to the patient are also currently conservative, in part because common, benign polymorphic variants have caused many false positives in past genetic analyses (Manrai et al., 2016, Walsh et al., 2017). However, as whole-exome and whole-genome sequencing become more economical, the phenotype-driven paradigm may be replaced by a genomics-driven approach, in which all rare, putatively functional variants in a patient’s genome are first identified, then evaluated based on the plausibility that they may be pathogenic. The genomics-driven approach has the potential for higher sensitivity, due to more genes being analyzed, and also has the potential to diagnose diseases not identified by the referring physician. However, the main tradeoff compared with phenotype-driven approaches is a potentially higher false-positive rate.
MultipleCAGI-4 groups in the Hopkins challenge were in consensus in identifying several possible causative variants that were not identified by the current panel testing paradigm. They also identified several other variants that were likely to be false positives. Distinguishing these two possibilities and identifying which variants to report to the patient is a topic that requires further research. The American College of Medical Genetics and Genomics has published guidelines for the interpretation of sequence variants in order to help codify variant assessment (Richards et al., 2015). However, even when adhering to these guidelines, there are still elements of variant interpretation that are subjective and vary between labs (Amendola et al., 2016, Garber et al., 2016). Given large databases of “control” exomes (i.e., without a known phenotype), researchers could develop statistical models to predict whether particular variants are in fact causative (Lek et al., 2016). Such models could inform the development of new statistically justified reporting standards based on, e.g., particular thresholds on the probability that the prediction of a causal variant is a false positive.
This challenge was designed to reflect the range of cases seen in the Hopkins diagnostic lab (Fig. 1A). This includes a high percentage of cases for which no likely pathogenic variant was identified, despite the patient presenting with a clinical phenotype. Even for clinical exome sequencing, roughly 70% of cases are negative (Lee et al., 2014, Posey et al., 2016). Negative cases proved especially challenging to participants, as “phenotype not discernable” was not listed as a matching option. Despite the fact that no pathogenic variants were identified by the Hopkins lab, most groups were able to make a disease prediction and to identify putative pathogenic alleles in these negative cases. Indeed, the reason data from all 83 genes was included in the challenge was to highlight the difficulty in interpreting a large data set of rare variants that are unrelated to the patient’s phenotype. The presence of negative cases in the data set reflects clinical practice and cautions on the overinterpretation of rare variants.
Unlike prior prediction challenges, where the activity of an enzyme had been quantitatively measured in the laboratory, there was no definitive answer key for this challenge. The predictors were asked to match sequencing data to a phenotype, and many groups did so by first identifying a causative variant. Only in a minority of cases (~23%in this dataset) could it be said with high confidence that a variant was likely contributing to disease in a patient. When a clinical laboratory reports a variant as pathogenic, this is often because the variant has previously been reported in patients with the same phenotype or the nucleotide change introduces a premature termination codon in a gene where loss-of-function variants cause disease (Richards et al., 2015). Thus, with a foundation in clinical genetics and access to online resources, one could identify a large proportion of the “pathogenic” variants in this dataset. However, many of the variants detected in the clinical laboratory are rare missense or synonymous variants that have not previously been reported in the literature; these are almost always classified as variants of uncertain clinical significance. It is for these VUS, which are difficult to interpret and for which there is no answer key, that better assessment tools are needed.
A CAGI challenge focused on the interpretation of variants of uncertain clinical significance would be more relevant to current clinical genetics practice. A clinical lab may upgrade a variant’s classification from “uncertain” to “pathogenic” based on new clinical information, segregation of a variant within a family, or identification of the variant in multiple unrelated individuals. Many molecular diagnostic labs maintain internal variant databases; such databases could be mined to curate a challenge set of “uncertain” variants for which there is unpublished data to support pathogenicity. In this proposed challenge, participants would have to correctly identify these “pathogenic” variants from a set of “uncertain” variants (for which there was unpublished data that they were not likely to contribute to disease). This would more directly test the challengers’ ability to predict pathogenicity without relying on allele frequency or online databases and without requiring knowledge of gene–disease associations. Assessment of the challenge would benefit from having fully vetted data and a clear answer key. This type of challenge, while still lacking a phenotype component, would more accurately mirror the clinical challenge of interpreting rare variants. Obtaining this data set would also invite communication between clinical testing labs (both academic and commercial) and the research community.
In this vein, the development of a clinically useful variant assessment tool will require collaboration between clinical geneticists and data scientists. Discussions resulting from the Hopkins clinical challenge demonstrated that although most participants incorporated genetic principles into their pipelines, they approached variant interpretation in a very different manner than a clinical laboratory. In future challenges, it would be interesting to pair an informatics group with a clinical group as a challenge team, particularly for whole-exome sequencing challenges. Ideally, the back-and-forth between clinical and informatics groups would produce a method that could outperform that of either group alone. Diverse collaborations at CAGI could help bridge the communication gap between fields and pave the way for the development of better tools.
Supplementary Material
Acknowledgments
We acknowledge Steven E. Brenner and Roger Hoskins for their important roles in organizing CAGI-4.
Funding Information
This work was supported by the National Institutes of Health (U41 GH007446) through the U.S. Department of Energy under Contract No. DE-AC02-05CH11231. The CAGI experiment coordination is supported by NIHU41 GH007446 and the CAGI conference by NIH R13 HG006650.
Footnotes
Additional Supporting Information may be found online in the supporting information tab for this article.
References
- 1000 Genomes Project Consortium. Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, McVean GA. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1000 Genomes Project Consortium. Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, McVean GA. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adzhubei I, Jordan DM, Sunyaev SR. Predicting functional effect of human missense mutations using PolyPhen-2. Current Protocols in Human Genetics. 2013;(Unit 7.20) doi: 10.1002/0471142905.hg0720s76. Chapter 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amendola LM, Jarvik GP, Leo MC, McLaughlin HM, Akkari Y, Amaral MD, Rehm HL. Performance of ACMG-AMP variant-interpretation guidelines among nine laboratories in the Clinical Sequencing Exploratory Research Consortium. The American Journal of Human Genetics. 2016;98:1067–1076. doi: 10.1016/j.ajhg.2016.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Fishawy DP. Common disease-rare variant hypothesis. In: Volkmar FR, editor. Encyclopedia of autism spectrum disorders. New York: Springer; 2013. pp. 720–722. [Google Scholar]
- Garber KB, Vincent LM, Alexander JJ, Bean LJ, Bale S, Hegde M. Reassessment of genomic sequence variation to harmonize interpretation for personalized medicine. The American Journal of Human Genetics. 2016;99:1140–1149. doi: 10.1016/j.ajhg.2016.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamosh A, Scott AF, Amberger JS, Bocchini CA, McKusick VA. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Research. 2005;33:D514–D517. doi: 10.1093/nar/gki033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsanis SH, Katsanis N. Molecular genetic testing and the future of clinical genomics. Nature Review Genetics. 2013;14:415–426. doi: 10.1038/nrg3493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nature Genetics. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nature Protocols. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- Landrum MJ, Lee JM, Benson M, Brown G, Chao C, Chitipiralla S, Maglott DR. ClinVar: Public archive of interpretations of clinically relevant variants. Nucleic Acids Research. 2016;44:D862–D868. doi: 10.1093/nar/gkv1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Deignan JL, Dorrani N, Strom SP, Kantarci S, Quintero-Rivera F, Nelson SF. Clinical exome sequencing for genetic identification of rare Mendelian disorders. JAMA. 2014;312:1880–1887. doi: 10.1001/jama.2014.14604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lek M, Karczewski K, Minikel E, Samocha K, Banks E, Fennell T Exome Aggregation Consortium. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manrai AK, Funke BH, Rehm HL, Olesen MS, Maron BA, Szolovits P, Kohane IS. Genetic misdiagnoses and the potential for health disparities. The New England Journal of Medicine. 2016;375:655–665. doi: 10.1056/NEJMsa1507092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren W, Pritchard B, Rios D, Chen Y, Flicek P, Cunningham F. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics. 2010;26:2069–2070. doi: 10.1093/bioinformatics/btq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Research. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NHLBI GO Exome Sequencing Project (ESP) 2015 Retrieved from http://evs.gs.washington.edu/EVS/
- Pagon RA, Adam MP, Ardinger HH, Wallace SE, Amemiya A, Bean LJ, Stephens K, editors. Gene Reviews(®) Seattle, Washington: University of Washington, Seattle; 1993. [Google Scholar]
- Posey JE, Rosenfeld JA, James RA, Bainbridge M, Niu Z, Wang X, Plon SE. Molecular diagnostic experience of whole-exome sequencing in adult patients. Genetics in Medicine. 2016;18:678–685. doi: 10.1038/gim.2015.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J ACMG Laboratory Quality Assurance Committee. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genetics in Medicine. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saez-Rodriguez J, Costello JC, Friend SH, Kellen MR, Mangravite L, Meyer P, Stolovitzky G. Crowdsourcing biomedical research: leveraging communities as innovation engines. Nat Rev Genet. 2016;17:470–486. doi: 10.1038/nrg.2016.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schölkopf B, Platt JC, Shawe-Taylor J, Smola AJ, Williamson RC. Estimating the support of a high-dimensional distribution. Neural Computation. 2001;13:1443–1471. doi: 10.1162/089976601750264965. [DOI] [PubMed] [Google Scholar]
- Schulz WL, Tormey CA, Torres R. Computational approach to annotating variants of unknown significance in clinical next generation sequencing. Laboratory Medicine. 2015;46:285–289. doi: 10.1309/LMWZH57BRWOPR5RQ. [DOI] [PubMed] [Google Scholar]
- Sosnay PR, Salinas DB, White TB, Ren CL, Farrell PM, Raraigh KS, Castellani C. Applying cystic fibrosis transmembrane conductance regulator genetics and CFTR2 data to facilitate diagnoses. Journal of Pediatrics. 2017;181S:S27–S232. doi: 10.1016/j.jpeds.2016.09.063. [DOI] [PubMed] [Google Scholar]
- Stenson PD, Ball EV, Mort M, Phillips AD, Shiel JA, Thomas NST, Cooper DN. Human Gene Mutation Database (HGMD): 2003 update. Human Mutation. 2003;21:577–581. doi: 10.1002/humu.10212. [DOI] [PubMed] [Google Scholar]
- Tricarico R, Kasela M, Mareni C, Thompson BA, Drouet A, Staderini L InSiGHT Variant Interpretation Committee. Assessment of the InSiGHT interpretation criteria for the clinical classification of 24 MLH1 and MSH2 gene variants. Human Mutation. 2017;38:64–77. doi: 10.1002/humu.23117. [DOI] [PubMed] [Google Scholar]
- Vassy JL, Lautenbach DM, McLaughlin HM, Kong SW, Christensen KD, Krier J MedSeq Project. The MedSeq Project: A randomized trial of integrating whole genome sequencing into clinical medicine. Trials. 2014;15:85. doi: 10.1186/1745-6215-15-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh R, Thomson KL, Ware JS, Funke BH, Woodley J, McGuire KJ, Watkins H. Reassessment of Mendelian gene pathogenicity using 7,855 cardiomyopathy cases and 60,706 reference samples. Genetics in Medicine. 2017;19:192–203. doi: 10.1038/gim.2016.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Li M, Hakonarson H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Research. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue P, Melamud E, Moult J. SNPs3D: Candidate gene and SNP selection for association studies. BMC Bioinformatics. 2006;7:166. doi: 10.1186/1471-2105-7-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.