SUMMARY
Zika virus (ZIKV) directly infects neural progenitors and impairs their proliferation. How ZIKV interacts with the host molecular machinery to impact neurogenesis in vivo is not well understood. Here, by systematically introducing individual proteins encoded by ZIKV into the embryonic mouse cortex, we show that expression of ZIKV-NS2A, but not Dengue virus (DENV)-NS2A, leads to reduced proliferation and premature differentiation of radial glial cells, and aberrant positioning of newborn neurons. Mechanistically, in vitro mapping of protein-interactomes and biochemical analysis suggest interactions between ZIKA-NS2A and multiple adherens junction complex (AJ) components. Functionally, ZIKV-NS2A, but not DENV-NS2A, destabilizes the AJ complex, resulting in impaired AJ formation and aberrant radial glial fiber scaffolding in the embryonic mouse cortex. Similarly, ZIKA-NS2A, but not DENV-NS2A, reduces radial glial cell proliferation and causes AJ deficits in human forebrain organoids. Together, our results reveal pathogenic mechanisms underlying ZIKV infection in the developing mammalian brain.
eTOC
Zika virus infects neural stem cells and causes microcephaly. In this study, Yoon et al. showed that NS2A protein encoded by Zika virus, but not by Dengue virus, impairs proliferation of radial glial cells in both embryonic mouse cortex and human forebrain organoids. Mechanistically, ZIKV-NS2A disrupts adherens junction formation.
INTRODUCTION
Zika virus (ZIKV) belongs to the flavivirus genus in the Flaviviridae family, which includes many significant pathogens, such as dengue virus (DENV), yellow fever virus, West Nile virus, and Japanese encephalitis virus (Lindenbach et al., 2007; Ming et al., 2016). In the wake of the recent ZIKV outbreak, the greatest concern has been the link between ZIKV infection during pregnancy and congenital neurodevelopmental birth defects, such as microcephaly (Rasmussen et al., 2016). Since the World Health Organization declared a Public Health Emergency of International Concern (Heymann et al., 2016), tremendous progress has been made in both clinical and basic ZIKV research (Li et al., 2016b; Ming et al., 2016). ZIKV was found in microcephalic brains of fetuses from women infected with ZIKV during pregnancy (Driggers et al., 2016; Mlakar et al., 2016) and ZIKV has been shown to directly infect cortical neural progenitors in various experimental model systems, including human induced pluripotent stem cell (iPSC)-derived and fetal brain tissue-derived neural progenitors in monolayer, 3D neurosphere and brain organoid cultures, and in mice (Li et al., 2016b; Ming et al., 2016). At the cellular level, productive infection of neural progenitors by ZIKV delays cell cycle progression and increases cell death (Ming et al., 2016). At the molecular level, ZIKV infection leads to dysregulation of many signaling pathways (Wen et al., 2017). For example, ZIKV infection of human fetal neurospheres in culture inhibits the Akt-mTOR pathway, leading to defective neurogenesis and aberrant activation of autophagy (Liang et al., 2016). How ZIKV directly interacts with the host machinery to impact neurogenesis in the developing mammalian cortical cortex in vivo remains unknown.
The ZIKV genome consists of a positive-sense, single-stranded RNA approximately 11,000 nucleotides in length, encoding a single open reading frame (ORF) (Garcia-Blanco et al., 2016). Translation of the long ORF produces a large polyprotein with over 3,000 amino acid residues, which is then cleaved by both viral and host proteases to produce three structural proteins (C, prM, and E) and seven nonstructural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B and NS5; Figure S1A) (Garcia-Blanco et al., 2016). Recent in vitro studies have shown that ZIKV-NS4A and ZIKV-NS4B inhibit neural progenitor growth (Liang et al., 2016). Here we took an unbiased and systematic approach to screen for individual ZIKV protein components that may impact embryonic mouse cortical neurogenesis in vivo, followed by mechanistic analyses. We further extended our analysis to human embryonic cortical development using forebrain organoids derived from human iPSCs (Qian et al., 2016).
RESULTS
Reduced proliferation and premature differentiation of radial glial cells upon ZIKV-NS2A expression in the developing mouse cortex
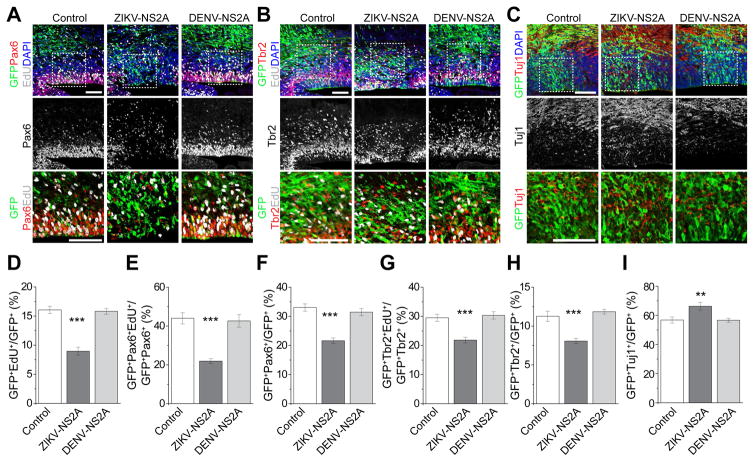
We cloned each ORF of the ZIKV genome into an expression vector (Table S1) and co-expressed individual ZIKV proteins and GFP in E14.5 embryonic mouse cortex via in utero electroporation (Yoon et al., 2014). For the initial screen we pulsed animals with EdU at E17.5 for 2 hr and examined the percentage of EdU+ cells among GFP+Pax6+ radial glial cells (RGCs) as the proliferation index (Figure S1B). Among all ZIKV encoded proteins, ZIKV-NS2A expression resulted in the most dramatic reduction in the proliferation index, whereas ZIKV-C had a mild effect (Figure 1A and S1C).
Figure 1. ZIKV-NS2A, but not DENV-NS2A, dysregulates radial glial cells in the embryonic mouse cortex.
Embryonic mouse cortex was electroporated at E14.5 to express GFP, GFP and ZIKV-NS2A, or GFP and DENV-NS2A, followed by EdU labeling 2 hr before analysis at E17.5. Sample confocal images of immunostaining for Pax6 (A), Tbr2 (B), Tuj1 (C) and GFP, and staining for EdU and DAPI are shown. Scale bars: 100 μm (A, B, C). The regions in white boxes (top panels) are shown at a higher magnification (bottom panels). Quantifications are also shown (D–I). Values represent mean + SEM (n = 5–7 sections from 3–4 animals; ***: P < 0.001; **: P < 0.01; One-way ANOVA).
Also see Figure S1 and S2.
We next focused on ZIKV-NS2A for detailed analyses given that it produced the largest effect in vivo. ZIKV-NS2A exhibits 95.6–99.9% identity at the protein level among different ZIKV strains, suggesting a highly conserved protein (Figure S2). Quantitative analysis showed that ZIKV-NS2A expression led to reductions in the percentage of EdU+ cells among all GFP+ cells and among GFP+Pax6+ RGCs compared to GFP expression alone, indicating deficits in RGC proliferation (Figure 1A, D, E). In addition, the percentage of GFP+Pax6+ RGCs among all GFP+ cells was reduced upon expression of ZIKV-NS2A, suggesting depletion of RGCs in the developing mouse cortex (Figure 1F). RGCs give rise to Tbr2+ intermediate neural progenitor cells (IPCs), which differentiate into neurons (Gotz and Huttner, 2005). We found that ZIKV-NS2A expression also reduced percentages of EdU+ cells among GFP+Tbr2+ IPCs, and of GFP+Tbr2+ IPCs among all GFP+ cells (Figure 1B, G, H). These results are reminiscent of the impact of ZIKV infection on neurogenesis in the embryonic mouse cortex in vivo (Brault et al., 2016; Cugola et al., 2016; Li et al., 2016a; Wu et al., 2016). In addition to reduced proliferation of RGCs and IPCs, we also found a significant increase in the percentage of GFP+Tuj1+ immature neurons among all GFP+ cells, indicating premature differentiation of RGCs into neurons (Figure 1C, I), which is similar to a recent finding of the impact of direct ZIKV infection in cultured human neural progenitors and brain organoids (Gabriel et al., 2017).
DENV, a member of the flaviviridae family closely related to ZIKV, has not been linked to either microcephaly or deficits in neural progenitor proliferation (Brault et al., 2016; Garcez et al., 2016). For comparison, we examined the functional outcome of DENV-NS2A expression. DENV-NS2A shares 24.8% identity at the protein level with ZIKV-NS2A (Figure S2) and presumably plays similar roles in viral replication and assembly for DENV. Upon in utero electroporation to express DENV-NS2A, we did not observe any significant differences in RGC and IPC proliferation or differentiation, compared to GFP expression alone (Figure 1). Therefore, ZIKV-NS2A, but not DENV-NS2A, regulates radial glial neural stem cell properties in the embryonic mouse brain in vivo.
Aberrant neuronal positioning upon ZIKV-NS2A expression in the developing mouse cortex
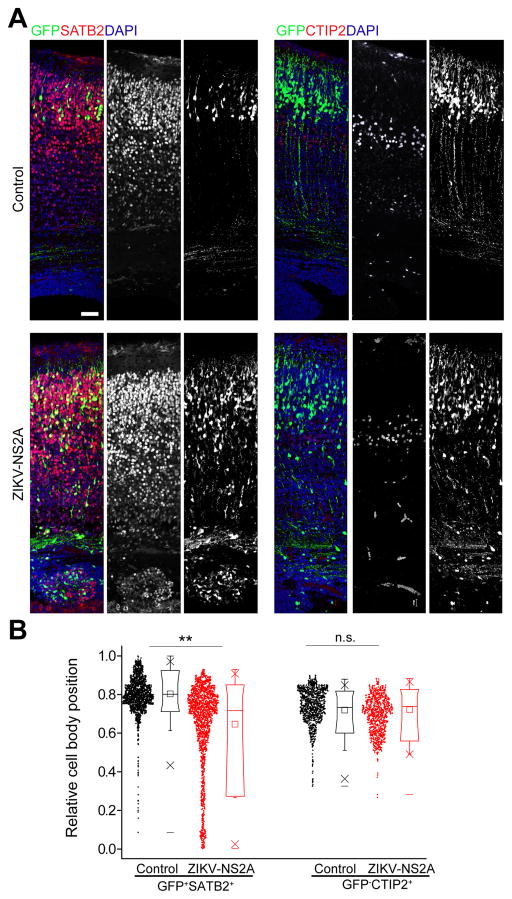
To investigate the impact of ZIKV-NS2A expression on later processes of cortical development, we performed electroporation at E14.5 and examined cortical layer formation at E19.5. At this stage, the majority of transfected cells (GFP+) became SATB2+ upper layer neurons, while most early-born CTIP2+ lower layer neurons were GFP− (Figure 2A). We found significant deficits in the positioning of GFP+SATB2+ neurons upon ZIKV-NS2A expression, many of which failed to migrate to the upper cortical layers and exhibited a much broader distribution (Figure 2). On the other hand, the early-born GFP−CTIP2+ neurons were largely unaffected (Figure 2). These results revealed that expression of ZIKV-NS2A during embryonic cortical development also disrupts positioning of newborn neurons and cortical layer organization in vivo.
Figure 2. ZIKV-NS2A, but not DENV-NS2A, expression leads to aberrant localization of neurons in the developing mouse cortex.
(A) Embryonic mouse cortex was electroporated at E14.5 to express GFP, or GFP and ZIKV-NS2A, followed by analysis at E19.5. Sample confocal images of immunostaining for SATB2, CTIP2 and GFP, and staining for DAPI are shown. Scale bar: 50 μm.
(B) Scatter plots and summary of cell body position of GFP+SATB2+ neurons and GFP−CTIP2+ neurons in the mouse cortex at E19.5. The distance of each cell to the apical surface was normalized to the total thickness of the neocortex. Each dot represents one neuron. The box plots show the medians (line), means (square), interquartile ranges (box; 25–75%), and extremes of the distribution (whisker; 99%: upper crosshatch; 1%: lower crosshatch) (n = 8 sections from 4 animals for each condition; ** P < 0.01; n.s. P > 0.1; Student’s t-test).
Interaction between ZIKV-NS2A and adherens junction complex components
To investigate how ZIKV-NS2A directly interacts with the host protein machinery to impact neural stem cell behavior, we began with an unbiased and systematic approach to generate testable hypotheses. We screened human proteins that can bind to recombinant ZIKV-NS2A protein or DENV-NS2A protein with protein microarray. Among 20,240 full-length recombinant human proteins spotted on the protein microarray (Jeong et al., 2012), which covers over 95% of protein encoding genes in the human genome, 143 and 47 proteins were identified for ZIKV-NS2A and DENV-NS2A, respectively (Figure 3A and Table S2). A total of 45 proteins were shared between the two homologous NS2A proteins. Gene Ontology (GO) analysis of 143 ZIKV-NS2A interacting proteins revealed enrichment for multiple pathways, including extracellular exosome, cytoplasmic stress granule, and focal adhesion (Figure S3A and Table S3). Based on known protein interaction networks, 83 ZIKV-NS2A interacting proteins can be visualized in a connected network (P < 4.91 × 10−13), whereas the remaining 60 proteins are singletons (Figure 3A and Table S4). Within the interaction network, 8 proteins (NME2, ARPC3, HSPB1, PABPC1, PTK2/FAK, VASP, PLEK and SMAD7) have been reported to be related to cell adhesion (P = 0.03). Among singletons, NUMBL is also adhesion-related (Rasin et al., 2007). Notably, 7 out of these 9 cell adhesion related proteins are ZIKV-specific in this in vitro assay (Figure 3A and S3A).
Figure 3. Protein-protein interactomes of ZIKV-NS2A and DENV-NS2A across the human proteome.
(A) 143 and 47 direct ZIKV-NS2A and DENV-NS2A interacting proteins were identified in vitro using a protein array containing 20,240 full-length human proteins. Among 143 ZIKV-NS2A interacting proteins, 83 proteins can be visualized in a connected network based on existing literatures (P < 4.91 × 10−13; STRING analysis), whereas the remaining 60 proteins are singletons. ZIKV-NS2A-specific interacting proteins are coded in gray and adhesion-related proteins are highlighted with circles.
(B) Sample western blot images of co-IP analysis of HEK293 cells expressing GFP, ZIKV-NS2A and GFP, or DENV and GFP, and analyzed for adherens junction complex components. Histone H3 served as a negative control for Co-IP. Also see Figure S3.
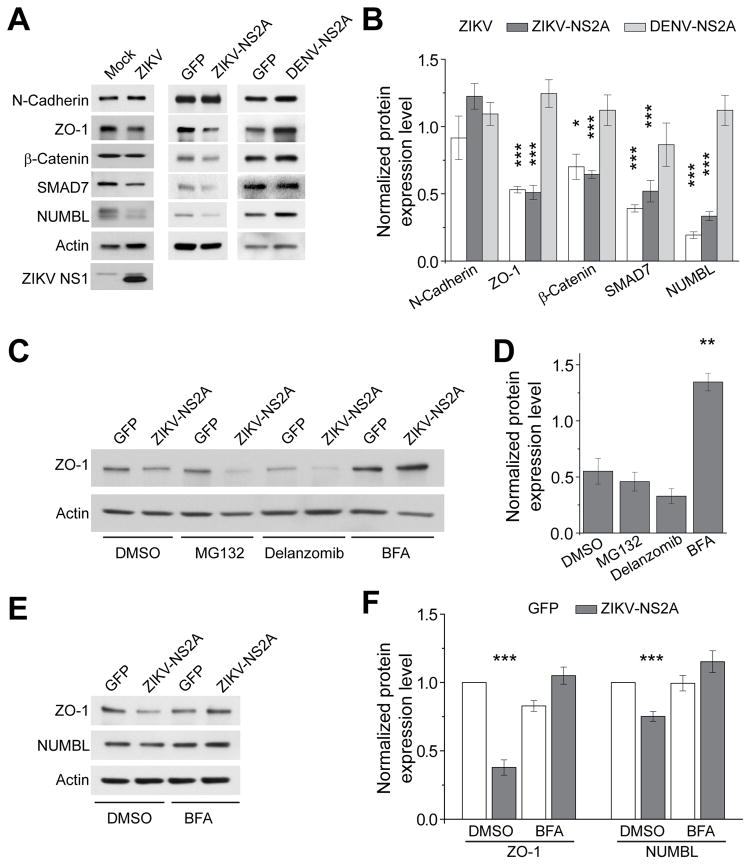
The majority of these adhesion-related proteins (PTK2, VASP, NUMBL, SMAD7, ARPC3) are linked to adherens junctions (AJ), which anchor RGCs and regulate their properties (Stocker and Chenn, 2015). To confirm that ZIKV-NS2A interacts with AJ complex components in a cellular context, we expressed ZIKV-NS2A in HEK293 cells and performed co-immunoprecipitation (co-IP) analysis. Indeed, ZIKV-NS2A co-IPed with multiple AJ components, including N-Cadherin, ZO-1, β-Catenin, SMAD7, NUMBL and ARPC3 (Figure 3B). In contrast, these AJ components were not in the complex with DENV-NS2A (Figure 3B). SMAD7 was identified as an interactor of DENV-NS2A on the protein array, but not in the co-IP analysis, which could be due to differences in the sensitivity of the two assays or differences in the biological context. We further confirmed the interaction of ZIKV-NS2A with multiple AJ complex components in cultured neural progenitors derived from E11.5 mouse cortex (Figure S3B).
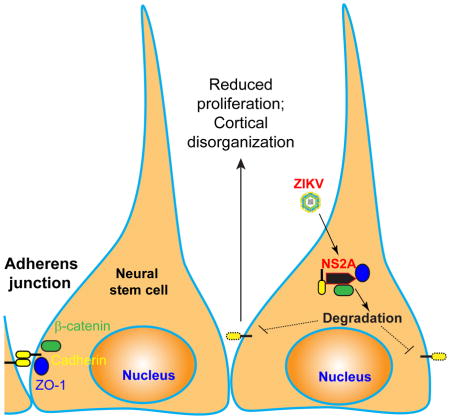
Given that disruption of AJ formation in RGCs via a number of genetic manipulations all leads to premature differentiation and depletion of RGCs in the embryonic mouse cortex (Stocker and Chenn, 2015), which are the same phenotypes we observed for ZIKV-NS2A expression, these results from the in vitro screen raised the possibility that ZIKV-NS2A, but not DENV-NS2A, regulates AJ formation in RGCs in vivo.
Deficits in adherens junction formation upon ZIKV-NS2A expression
To test our hypothesis, we first examined the protein levels of AJ complex components upon ZIKV infection or NS2A expression. Direct ZIKV infection of mouse cortical neural progenitors led to reduced protein levels of several AJ components, including ZO-1, β-Catenin, SMAD7, and NUMBL (Figure 4A–B). A similar reduction in these AJ components at the protein level was observed in mouse cortical neural progenitors expressing ZIKV-NS2A, but not DENV-NS2A (Figure 4A–B). The mRNA levels of most of these genes were not reduced upon ZIKV infection or ZIKV-NS2A expression, suggesting a post-transcriptional regulatory mechanism (Figure S4A–B). It is known that some AJ components are pre-assembled in the endoplasmic reticulum (ER) before they are delivered to the cell membrane to form AJs (Chen et al., 1999). As expected, immunostaining showed that some ZIKV-NS2A and DENV-NS2A were co-localized with an ER marker Calnexin (Figure S4C). In addition, levels of AJ complex components we tested were reduced in the fractionated ER preparation (Figure S4D). To explore cellular pathways that may degrade AJ complex components, we used a pharmacological approach. We found that ZIKV-NS2A-induced reduction of ZO-1 levels in HEK293 cells was prevented by bafilomycin A1 (BFA), an inhibitor for fusion of autophagosomes and lysosomes (Yoshimori et al., 1991), but not by proteasome inhibitors, MG-132, or Delanzomib (Figure 4C–D). Furthermore, BFA treatment prevented ZIKV-NS2A-induced decreases of ZO-1 and NUMBL protein levels in mouse cortical neural progenitors (Figure 4E–F).
Figure 4. ZIKV-NS2A, but not DENV-NS2A, expression leads to adherens junction complex component degradation.
(A–B) Sample western blot images of expression levels of adherens junction complex components in mouse neural progenitors infected with ZIKV, expressing ZIKV-NS2A, or expressing DENV-NS2A (A), and quantifications (B). Data were normalized to that of mock infection for the ZIKV infection condition, or to that of GFP expression alone for the ZIKV-NS2A or DENV-NS2A conditions. Values represent mean + SEM (n = 3 cultures; ***: P < 0.001; *: P < 0.05; Student’s t-test).
(C–D) Sample western blot images of expression levels of ZO-1 upon 24 hr treatment of DMSO, MG-132 (20 μM), Delanzomib (60 nM) and BFA (100 nM) in HEK293 cells expressing GFP and ZIKV-NS2A, or GFP alone (C), and quantifications (D). Data were first normalized to actin expression levels and then to the data from expression of GFP alone. Values represent mean + SEM (n = 3 cultures; **: P < 0.01; Student’s t-test).
(E–F) Sample western blot images of expression levels of ZO-1 and NUMBL upon 24 hr treatment of DMSO, or BFA (100 nM) of mouse cortical neural progenitors expressing GFP and ZIKV-NS2A, or GFP alone (E), and quantifications (F). Data were normalized to that of actin. Values represent mean + SEM (n = 3 cultures; ***: P < 0.001; Student’s t-test).
Also see Figure S4.
The above findings raised the possibility that interaction of ZIKV-NS2A with AJ complex components may lead to their depletion, resulting in deficits in AJ formation in RGCs. Indeed, immunostaining and quantification of β-Catenin, PKCλ, or ZO-1 expression revealed significant deficits in AJ formation by RGCs upon expression of ZIKV-NS2A, but not DENV-NS2A (Figure 5A–B). Reduction of ZO-1 expression in Pax6+ and Nestin+ RGCs (Figure S5A–B) suggests that AJ deficits induced by ZIKV-NS2A occur prior to the depletion and premature differentiation of RGCs (Figure 1F, I). ZIKV-NS2A expression also led to disorganized radial glia fiber scaffolding and ventricular protrusions (Figure 5A and S5C–D). Given that newborn neurons migrate along radial fibers of RGCs during embryonic cortical development (Buchman and Tsai, 2007), this deficit may contribute to aberrant cortical layer formation (Figure 2).
Figure 5. ZIKV-NS2A expression and direct ZIKV infection disrupt the formation of adherens junction complex in the embryonic mouse cortex.
(A–B) Embryonic mouse brains were electroporated at E14.5 to express GFP, GFP and ZIKV-NS2A, or GFP and DENV-NS2A, and analyzed at E17.5. Sample confocal images of immunostaining for GFP, β-Catenin (A), or PKCλ and ZO-1 (B), and staining for DAPI under different conditions are shown (left panels). Scale bars: 50 μm. Arrows point to regions with discontinuous AJ formation. Regions in white boxes in (A) are shown at a higher magnification (bottom panels). Quantifications of continuous AJ formation and number of protrusions are also shown (right panels). Values represent mean + SEM (n = 5 sections from 3 animals; ***: P < 0.001; **: P < 0.01; Student’s t-test).
(C–D) ZIKV-SZ strain was injected into lateral ventricles of E13.5/E14.5 mice. Similar to (A–B), sample confocal images of immunostaining for ZIKV, β-Catenin and ZO-1, and staining for DAPI at E18.5 (left panels) and quantifications of continuous AJ formation at E18.5 and ventricular protrusions at P3 (right panels) are shown. Values represent mean + SEM (n = 6 sections from 4 animals; ***: P < 0.001; *: P < 0.05; Student’s t-test).
Also see Figure S5.
Next, we compared the impact of ZIKV-NS2A expression in RGCs to direct ZIKV infection of the embryonic mouse cortex. Following direct injection of the ZIKV-SZ strain (Li et al., 2016a) into the lateral ventricle of E13.5 mouse brain in utero, we observed deficits in AJ formation and radial fiber scaffolding at E18.5 (Figure 5C–D) and ventricular protrusions at P3 (Figure 5C and S5E). Together, these results provide evidence that ZIKV infection and ZIKV-NS2A expression result in similar deficits of AJ formation of RGCs in the embryonic mouse cortex.
Deficits in human cortical neurogenesis upon ZIKV-NS2A expression
Finally, to determine whether ZIKV-NS2A dysregulates embryonic human cortical neurogenesis, we used the recently established human iPSC-derived forebrain organoid model (Qian et al., 2016; Xu et al., 2016). We co-expressed GFP and ZIKV-NS2A, or DENV-NS2A, in ventricular RGCs in day 45 forebrain organoids by electroporation and 3 days later pulsed with EdU (10 μM) for 1 hr (Figure S6A). Expression of ZIKV-NS2A, but not DENV-NS2A, resulted in reduced percentages of EdU+ cells among all GFP+ cells and GFP+PAX6+ cells, and of Ki67+ cells among all GFP+ cells within the ventricular structures, compared to GFP expression alone (Figure 6A and S6B–C). Furthermore, immunostaining and quantification of PKCλ expression revealed disrupted AJ formation in ZIKV-NS2A expressing regions at the ventricular surface (Figure 6B).
Figure 6. Expression of ZIKV-NS2A, but not DENV-NS2A, reduces proliferation and disrupts adherens junction formation of ventricular radial glial cells in human forebrain organoids.
(A) Day 45 forebrain organoids were electroporated to express GFP, GFP and ZIKV-NS2A, or GFP and DENV-NS2A, and analyzed 3 days later (45+3) after pulsing with EdU (10 μM) for 1 hr. Sample confocal images for immunostaining for GFP and PAX6, and staining for EdU and DAPI are shown (top panels). Scale bar: 100 μm. Quantifications of percentages of EdU+GFP+ cells among all GFP+ cells, or GFP+PAX6+EdU+ cells among GFP+PAX6+ cells are also shown (bottom panels). Values represent mean + SEM (n = 10 organoids; ***: P < 0.001; Student’s t-test).
(B) Sample confocal images of immunostaining for GFP, PKCλ, and staining for DAPI are shown (left panels). Scale bar: 100 μm. Arrows point to regions with discontinuous AJ formation. Quantifications of AJ continuity based on PKCλ expression are also shown (right panel). Values represent mean + SEM (n = 9 organoids; **: P < 0.01; Student’s t-test).
(C) Expression of ZIKV-NS2A, but not DENV-NS2A, or GFP alone, led to a loss of typical radial glia morphology of PAX6+GFP+ cells at 7 days after electroporation (45+7). Sample confocal images of immunostaining for GFP and PAX6, and staining for DAPI (left panels; scale bar: 100 μm) and quantification of percentages of GFP+PAX6+ cells with multipolar morphologies are shown. Values represent mean + SEM (n = 7 organoids; **: P < 0.01; Student’s t-test).
Also see Figure S6.
We observed some disruption of ventricular organization upon expression of ZIKV-NS2A, but not DENV-NS2A, at day 3 (45+3; Figure S6B), and more dramatically at day 7 (Figure 6C). Quantitative analysis showed a significantly increased percentage of GFP+PAX6+ cells that lost their radial morphology and instead exhibited multipolar morphology (Figure 6C). These results are reminiscent of a recent observation of AJ formation deficits, ventricular protrusions and disorganized radial glia scaffolding in postmortem forebrain tissue of the first reported ZIKV-infected microcephalic fetus from an infected mother (Onorati et al., 2016).
DISCUSSION
The ZIKV epidemic and its association with microcephaly present a serious public health challenge. Understanding mechanisms underlying ZIKV pathogenesis in the developing mammalian brain may reveal potential targets for anti-ZIKV and neuroprotective therapeutic interventions. Our systematic and functional screen of ZIKV-encoded proteins led to the identification of an in vivo mechanism and a direct link of a ZIKV component to specific host machinery that can explain, at least in part, ZIKV-induced neural stem cell deficits during mammalian cortical neurogenesis in vivo. Our study also revealed additional deficits during cortical development that occurs after the early phases of neurogenesis, specifically, in layer-specific positioning of newborn neurons in the developing cortex. Given the recent clinical findings that babies born to women infected with ZIKV during pregnancy develop significant postnatal neurological deficits, such as epilepsy (Besnard et al., 2016; Moura da Silva et al., 2016; Sarno et al., 2016), our finding has important implications for future studies.
Our study revealed two potential cellular mechanisms by which ZIKV-NS2A reduces neural stem cell numbers in the developing cortex in vivo, by reducing proliferation and through premature differentiation and depletion of RGCs. Previous studies in monolayer and 3D organoid cultures and in mice have already shown that ZIKV infection leads to reduced proliferation of neural progenitor cells (Ming et al., 2016). A recent study also showed that ZIKV infection of human brain organoids leads to premature differentiation of neural progenitors (Gabriel et al., 2017). Here we identified ZIKV-NS2A protein, but not DENV-NS2A, as capable of mimicking, at least in part, the effect of direct ZIKV infection both in mice in vivo and in human forebrain organoids. Mechanistically, ZIKV-NS2A, but not DENV-NS2A, interacts with AJ complex components and its expression leads to degradation of AJ complex components in neural progenitors and deficits in AJ formation of RGCs in vivo. Deficits in AJ formation have been shown to lead to aberrations in niche signaling that impact cortical neurogenesis (Stocker and Chenn, 2015). Additional deficits in radial glial fiber scaffolding may explain the positional defects of newborn neurons. Notably, two other flaviviruses, West Nile Virus and Japanese Encephalitis Virus, also interact with AJ proteins and target junction proteins for lysosomal degradation in non-neuronal cells via viral proteins different from NS2A (Agrawal et al., 2013; Medigeshi et al., 2009). It will be useful in the future to engineer ZIKVs with NS2A mutations that have high infection capacity but with no impact on neural stem cells to test how our identified mechanism contributes to the overall pathological impact of ZIKV on cortical neurogenesis. Given the tropism of ZIKV towards neural stem cells, these engineered viral vectors can be developed to target and manipulate radial glial cells in both fetal and adult mouse brains in vivo and in human brain organoids (Ming et al., 2016). Our finding does not rule out the possibility that interactions between ZIKV-NS2A and other host molecules may additionally regulate neural stem cell behavior. Our datasets of NS2A protein-protein interaction networks across the human proteome provide a rich resource for future exploration. This global protein interactome may also be useful for understanding ZIKV/DENV replication and assembly to identify antiviral therapeutic targets. It is possible that other ZIKV components, including both ZIKV-encoded proteins (e.g. ZIKV-C) (Liang et al., 2016) and noncoding RNAs (Pylro et al., 2016), may also contribute to the microcephaly phenotype observed in human fetuses and in animal models. Here we provide an experimental paradigm that combines in vivo analysis of mouse cortical neural stem cells and in vitro analysis of human forebrain organoids to address basic mechanisms regulating mammalian cortical neurogenesis under normal and pathological conditions.
STAR METHODS
KEY RESOURCES TABLE
The table highlights the genetically modified organisms and strains, cell lines, reagents, software, and source data essential to reproduce results presented in the manuscript. Depending on the nature of the study, this may include standard laboratory materials (i.e., food chow for metabolism studies), but the Table is not meant to be comprehensive list of all materials and resources used (e.g., essential chemicals such as SDS, sucrose, or standard culture media don’t need to be listed in the Table). Items in the Table must also be reported in the Method Details section within the context of their use. The number of primers and RNA sequences that may be listed in the Table is restricted to no more than ten each. If there are more than ten primers or RNA sequences to report, please provide this information as a supplementary document and reference this file (e.g., See Table S1 for XX) in the Key Resources Table.
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-Pax6 | BioLegend | PRB-278P RRID: AB_2313780 |
| Mouse anti-Pax6 | BD Bioscience | 561664 RRID: AB_10895587 |
| Rabbit anti-Tbr2 | Abcam | ab23345 RRID: AB_778267 |
| Chicken anti-GFP | Aveslab | GFP-1020 RRID: AB_10000240 |
| Mouse anti-Ki67 | BD Bioscience | 550609 RRID: AB_393778 |
| Goat anti-Sox2 | Santa Cruz | sc-17320 RRID: AB_2286684 |
| Chicken anti-Nestin | Aveslab | NES RRID: AB_2314882 |
| Mouse anti-PKCλ | BD Bioscience | 610207 RRID: AB_397606 |
| Mouse anti-N-Cadherin | Innovative Research | 18-0224 RRID: AB_86735 |
| Rabbit anti-Actin | Cytoskeleton | AAN01 RRID: AB_10708070 |
| Mouse anti-β-Catenin | BD Bioscience | 610153 RRID:AB_397554 |
| Rabbit anti-ZO-1 | Thermo Fisher Scientific | 40-2200 RRID:AB_10104693 |
| Rabbit anti- Numbl | Abcam | ab37500 RRID:AB_881766 |
| Rabbit anti-SMAD7 | Thermo Fisher Scientific | 42-0400 RRID:AB_2533512 |
| Rabbit anti-ZIKV NS1 | GeneTex | GTX133307 |
| Goat anti-GFP | Rockland | 600-101-215 RRID:AB_218182 |
| Rabbit anti-Calnexin | Abcam | ab10286 RRID:AB_2069009 |
| Human serum of ZIKV-infected patients | (Li et al., 2016) | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| ProLong Gold Antifade Mountant | Thermo Fisher Scientific | P10144 |
| DAPI | Thermo Fisher Scientific | D1306 RRID: AB_2629482 |
| StemPro Accutase Cell Dissociation Reagent | Thermo Fisher Scientific | A1110501 |
| Matrigel Matrix | Corning | 354277 |
| SimplyBlue SafeStain | Thermo Fisher Scientific | LC6060 |
| Y-27632 | Cellagen Technology | C9127-2s |
| EdU | Thermo Fisher Scientific | A10044 |
| Phosphatase Inhibitor Cocktail | Cell Signaling | 5870 |
| Protease Inhibitor Cocktail | Sigma | P8340 |
| 4× Laemmli Sample Buffer | Bio-Rad | 1610747 |
| A83-01 | Stemcell Technologies | 72022 |
| Dorsomorphin | Stemcell Technologies | 72102 |
| SB431542 | Stemcell Technologies | 72232 |
| CHIR99021 | Stemcell Technologies | 72052 |
| Dulbecco’s Phosphate-Buffered Saline (DPBS) | Corning | 21-031 |
| Dulbecco’s Modification of Eagle’s Medium (DMEM) | Corning | 10-013 |
| DMEM/F-12, HEPES | Gibco | 11330-032 |
| Neurobasal Medium | Gibco | 21103049 |
| KnockOut Serum Replacement | Gibco | 10828028 |
| GlutaMAX Supplement | Gibco | 35050061 |
| MEM Non-Essential Amino Acids Solution | Gibco | 11140050 |
| Penicillin-Streptomycin (10,000 U/mL) | Gibco | 15140122 |
| 2-Mercaptoethanol | Gibco | 21985023 |
| N-2 Supplement | Gibco | 17502048 |
| B-27 Supplement | Gibco | 17504044 |
| Matrigel Growth Factor Reduced (GFR) Basement Membrane Matrix | Corning | 354230 |
| Insulin solution | Sigma-Aldrich | I0516 |
| Fetal Bovine Serum (FBS) | Corning | 35-010 |
| 0.1% Gelatin in Water | Stemcell Technologies | 7903 |
| Advanced DMEM/F-12 | Gibco | 12634010 |
| Human recombinant LIF | Stemcell Technologies | 78055 |
| Compound E | Stemcell Technologies | 73952 |
| MG-132 | Cell Signaling | 2194 |
| Recombinant Human FGF-basic | Peprotech | 100-18B |
| Recombinant Human EGF-basic | R&D Systems | P01133 |
| Polybrene Infection/Transfection Reagent | Thermo Fisher Scientific | TR1003G |
| Bafilomycin A1 | Sigma-Aldrich | B1793 |
| Delanzomib | Kindly gifted by Dr. Wei Zheng (NIH/NCATS) | N/A |
| Critical Commercial Assays | ||
| Click-iT EdU Assay Kits | Thermo Fisher Scientific | C10340 |
| Cy5-NHS ester | GE Healthcare | PA15101 |
| SuperScript III First-Strand Synthesis System | Thermo Fisher Scientific | 18080051 |
| Fast SYBR Green Master Mix | Thermo Fisher Scientific | 4385610 |
| Lipofectamine 2000 | Thermo Fisher Scientific | 11668027 |
| Anti-HA Magnetic Beads | Thermo Fisher Scientific | 88836 |
| Endoplasmic Reticulum Isolation Kit | Sigma-Aldrich | ER0100 |
| RNeasy Mini Kit | Qiagen | 74104 |
| Experimental Models: Cell Lines | ||
| HEK293 | ATCC | CRL-1573, RRID:CVCL_0045 |
| C12 (iPSC from normal human foreskin fibroblasts) | (Wen et al., 2014) | N/A |
| Experimental Models: Organisms/Strains | ||
| Mouse: Crl:CD1(ICR) | Charles River Laboratory | RRID: IMSR_CRL:22 |
| Mouse: Crl:CF188 | Charles River Laboratory | RRID:IMSR_CRL:23 |
| Yeast: Y258 | Kindly gifted by Dr. Michael Snyder (Yale University) | N/A |
| Virus: ZIKV-MR766 | ZeptoMetrix | 0810521CF |
| Virus: ZIKV-SZ | (Li et al., 2016) | N/A |
| Oligonucleotides | ||
| Primers for ZIKV ORF cloning | See Table S1 | N/A |
| Primers for Q-PCR analysis | See Table S1 | N/A |
| Recombinant DNA | ||
| pDONR221 | Thermo Fisher Scientific | 12536017 |
| pCAG-GFP | (Matsuda and Cepko, 2004) | Addgene plasmid: 11150 |
| pEGH-A | (Zhu et al., 2009) | N/A |
| pCWX-R4-DEST-R2-PG | (Giry-Laterriere et al., 2011) | Addgene plasmid: 45957 |
| pENTR-L4-Ubi-L1R | (Giry-Laterriere et al., 2011) | Addgene plasmid: 45959 |
| Software and Algorithms | ||
| Image J | NIH | https://imagej.nih.gov/ij/ |
| Imaris | Bitplane | http://www.bitplane.com/imaris/imaris |
| MATLAB | MathWorks | https://www.mathworks.com/products/matlab.html |
| STRING 10.0 database | (Szklarczyk et al., 2015) | http://string-db.org/ |
| DAVID 6.8 | (Huang da et al., 2009) | https://david.ncifcrf.gov/ |
| Other | ||
| SuperSignal West Dura Extended Duration Substrate | Thermo Fisher Scientific | 34075 |
| Costar 6 Well Clear Flat Bottom Ultra Low Attachment plate | Sigma-Aldrich | CLS3471 |
| Nucleofector Kits for Mouse Neural Stem Cells | Lonza | VAPG-1004 |
| Nucleofector 2b Device | Lonza | AAB-1001 |
| Square wave electroporator | Nepa Gene | CUY21SC |
| Tweezers with platinum disk electrode | Nepa Gene | CUY650-5 |
| Immun-Blot PVDF Membrane | Bio-Rad | 1620177 |
| 12-well Spinning Bioreactor | (Qian et al., 2016) | N/A |
| LSM 800 confocal microscope | Zeiss | LSM 800 |
| Leica CM3050S Research Cryostat | Leica Biosystems | CM3050S |
Please note that ALL references cited in the Key Resources Table must be included in the References list. Please report the information as follows:
REAGENT or RESOURCE: Provide full descriptive name of the item so that it can be identified and linked with its description in the manuscript (e.g., provide version number for software, host source for antibody, strain name). In the Experimental Models section, please include all models used in the paper and describe each line/strain as: model organism: name used for strain/line in paper: genotype. (i.e., Mouse: OXTRfl/fl: B6.129(SJL)-Oxtrtm1.1Wsy/J). In the Biological Samples section, please list all samples obtained from commercial sources or biological repositories. Please note that software mentioned in the Methods Details or Data and Software Availability section needs to be also included in the table. See the sample Table at the end of this document for examples of how to report reagents.
SOURCE: Report the company, manufacturer, or individual that provided the item or where the item can obtained (e.g., stock center or repository). For materials distributed by Addgene, please cite the article describing the plasmid and include “Addgene” as part of the identifier. If an item is from another lab, please include the name of the principal investigator and a citation if it has been previously published. If the material is being reported for the first time in the current paper, please indicate as “this paper.” For software, please provide the company name if it is commercially available or cite the paper in which it has been initially described.
-
IDENTIFIER: Include catalog numbers (entered in the column as “Cat#” followed by the number, e.g., Cat#3879S). Where available, please include unique entities such as RRIDs, Model Organism Database numbers, accession numbers, and PDB or CAS IDs. For antibodies, if applicable and available, please also include the lot number or clone identity. For software or data resources, please include the URL where the resource can be downloaded. Please ensure accuracy of the identifiers, as they are essential for generation of hyperlinks to external sources when available. Please see the Elsevier list of Data Repositories with automated bidirectional linking for details. When listing more than one identifier for the same item, use semicolons to separate them (e.g. Cat#3879S; RRID: AB_2255011). If an identifier is not available, please enter “N/A” in the column.
A NOTE ABOUT RRIDs: We highly recommend using RRIDs as the identifier (in particular for antibodies and organisms, but also for software tools and databases). For more details on how to obtain or generate an RRID for existing or newly generated resources, please visit the RII or search for RRIDs.
Please use the empty table that follows to organize the information in the sections defined by the subheading, skipping sections not relevant to your study. Please do not add subheadings. To add a row, place the cursor at the end of the row above where you would like to add the row, just outside the right border of the table. Then press the ENTER key to add the row. You do not need to delete empty rows. Each entry must be on a separate row; do not list multiple items in a single table cell. Please see the sample table at the end of this document for examples of how reagents should be cited.
TABLE FOR AUTHOR TO COMPLETE
Please upload the completed table as a separate document. Please do not add subheadings to the Key Resources Table. If you wish to make an entry that does not fall into one of the subheadings below, please contact your handling editor. (NOTE: For authors publishing in Current Biology, please note that references within the KRT should be in numbered style, rather than Harvard.)
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact Guo-li Ming (gming@mail.med.upenn.edu). There are no restrictions on any data or materials presented in this paper.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals
All mice were housed with free access to food and water under pathogen-free conditions, in a facility where the temperature and light cycles (12 hour cycle) were controlled. All animal procedures used in this study were performed in accordance with the protocol approved by the Institutional Animal Care and Use Committee of Johns Hopkins University School of Medicine and by the Beijing Institute of Microbiology and Epidemiology Animal Care and Use Committee. Timed-pregnant Crl:CD1(ICR) mice (Charles River Laboratory) at E14.5 were used for in utero electroporation analysis. Pregnant ICR mice at E13.5/E14.5 were used for ZIKV infection.
HEK293 and primary mouse neural progenitor cells
HEK293 cells were obtained from ATCC and maintained in DMEM (Gibco BRL), 10% FBS and Penicillin/Streptomycin (Invitrogen). Mouse NPCs were isolated from CD1 mouse embryonic cortices and cultured in Neurobasal medium (Gibco BRL) containing 20 ng/ml FGF2, 20 ng/ml EGF, 5 mg/ml heparin, 2% B27 (v/v, Gibco BRL), Glutamax (Invitrogen), Penicillin/Streptomycin (Invitrogen) on culture dishes pre-coated with Matrigel matrix (2%, Corning).
Human iPSC lines
The human iPSC line used in the current study (C1, male) was fully characterized (Wen et al., 2014; Yoon et al., 2014). They were cultured in stem cell medium, consisting of DMEM:F12 (Invitrogen) supplemented with 20% Knockout Serum Replacer (Gibco), 1X Non-essential Amino Acids (Invitrogen), 1X Penicillin/Streptomycin (Invitrogen), 1X 2-Mercaptoenthanol (Millipore), 1X Glutamax (Invitrogen), and 10 ng/ml FGF-2 (Peprotech). Culture medium was changed every day. Human iPSCs were passaged every week onto a new plate pre-seeded with irradiated CF1 mouse embryonic fibroblasts (Charles River Laboratory). All studies were performed under approved protocols of Johns Hopkins University School of Medicine.
METHOD DETAILS
DNA constructs
To clone ZIKV- and DENV-encoded ORFs, ZIKV MR766 (African strain) and DENV-1 (Hawaiian strain) were used to infect mosquito cells. One μg of total RNA was converted to cDNA using Superscript III (Thermo Fisher Scientific) for PCR templates. The viral ORFs were constructed by RT-PCR-based cloning from cDNA into the Gateway Entry vector system. Primer sets were designed for amplifying the full-length ORFs and attB1 and attB2 sequences at the 5′-ends of each primer were added to clone PCR amplicon into Gateway Entry vector pDONR221 (Thermo Fisher Scientific) by Gateway recombination (Table S1). Using the expression pEGH-A vector for expression and purification of N-terminal GST fusion protein, a stop codon (TAA) was added between attB2- and the gene-specific reverse primer sequences in all primers. All entry clones were verified to be without any mutations at the amino acid sequences of all viral proteins by comparing to reference genome sequences of each viral strain. To express and purify individual viral proteins, sequence-verified Entry clones were cloned into a yeast expression vector pEGH-A. Verified clones were transformed to yeast strain Y258 that expresses GST fusion proteins under the control of the galactose-inducible GAL1 promoter.
To construct mammalian expression vectors under the control of the human Ubiquitin C promoter, sequence-verified entry clones were cloned into a lentiviral destination vector pCWX-R4-DEST-R2-PG (Addgene plasmid: 45957) by Gateway recombination reactions with pENTR-L4-Ubi-L1R (Addgene plasmid: 45959). To generate HA-tagged ZIKV-NS2A and DENV-NS2A expression vectors for biochemical analysis, the entry clones were amplified by PCR using 5′ primers with attB1 sequence and 3′ primers with HA-attB2 sequence and reinserted into pDONR221 and pCWX-R4-DEST-R2-PG, by the same Gateway recombination, to fuse the HA sequence into the C-terminal of NS2A ORFs. All of the final constructs were sequenced to confirm complete correspondence with original ORF sequences.
In utero electroporation
In utero electroporation was performed as described previously (Yoon et al., 2014). In brief, timed-pregnant CD1 mice (Charles River Laboratory) at E14.5 were anesthetized and the uterine horns were exposed and approximately 1 to 2 μl of plasmid DNA, 0.5 μg/μl pCAG-GFP (Addgene plasmid: 11150) and 2.5 μg/μl a ZIKV ORF expression vector (without HA tag) or an empty lentiviral vector, was injected manually into the lateral ventricles of the embryos using a calibrated micropipette. Five pulses (40 V, 50 ms in duration with a 950 ms interval) were delivered across the uterus with two 5-mm electrode paddles (CUY650-5, Nepa Gene) positioned on either side of the head by a square wave electroporator (CUY21SC, Nepa Gene). After electroporation, the uterus was placed back in the abdominal cavity and the wound was sutured. Mouse embryos were injected with EdU (150 mg/kg of body weight, Thermo Fisher Scientific) 2 hr before sacrifice at E17.5. For the analysis of neuronal positioning, embryos were electroporated at E14.5 and analyzed at E19.5. All animal procedures were performed in accordance with the protocol approved by the Johns Hopkins Institutional Animal Care and Use Committee.
ZIKV infection of embryonic mice and analysis
The ZIKV-SZ strain (GenBank accession no: KU866423) was obtained from Beijing Institute of Microbiology and Epidemiology, China. ZIKV infection of embryonic animals was performed as previously described (Li et al., 2016a). Briefly, pregnant ICR mice at E13.5/E14.5 were anesthetized and dissected to expose the uterine horns, followed by microinjection of ~1 μl of ZIKV (6.5×105 PFU/ml) or PBS into the lateral ventricles of embryos using a calibrated micropipette. After virus injection, the embryos were put back into the abdominal cavity and the incision in the dams was sutured. The brains of infected pups and their littermate controls were collected at E18.5 and P3 for further analysis. The tissues were fixed in 4% PFA overnight, dehydrated in 30% sucrose in PBS and embedded in OCT compound for cryostat sectioning. The brain sections were immunostained with human serum of ZIKV-infected patients (Li et al., 2016a) and antibodies against adherens junction markers (Table S1). The percentage of intact adherens junctions labeled by β–Catenin and ZO-1 along the ventricular surface and the density of ventricular protrusions were evaluated with ImageJ software.
Immunohistology and confocal imaging
For immunostaining of tissue sections, brains of embryos were fixed with 4% PFA in PBS overnight at 4°C. Brains were cryoprotected in 30% sucrose in PBS, embedded in OCT compound, and sectioned coronally (20 μm-thickness) on a Leica CM3050S cryostat. For immunostaining of HEK293 cells, cells were fixed with 4% PFA in PBS for 20 min at 4°C. Brain sections and cells were blocked and permeabilized with the blocking solution (5% normal donkey serum, 3% Bovine serum albumin and 0.1% Triton X-100 in PBS) for 1 hr at room temperature, followed by incubation with primary antibodies diluted in the blocking solution at 4°C overnight. Secondary antibodies diluted in blocking solution were applied to the sections for 1 hr at room temperature. Nuclei were visualized by incubating for 10 min with 0.1 mg/ml 4,6-diamidino-2-phenylindole (DAPI; Sigma-Aldrich) in PBS. Stained sections were mounted with ProLong Gold anti-fade reagents (Thermo Fisher Scientific) and analyzed. All the antibodies used are listed in KEY RESOURCE TABLE.
NS2A in vitro binding assay
NS2A proteins were expressed as GST fusions in budding yeast, and purified using glutathione sepharose affinity chromatography as described previously (Hu et al., 2013). On day one, each yeast strain containing the NS2A construct was inoculated in SC-URA media including glucose at 30°C, 200 rpm, overnight. On day two, primary seed cultures were inoculated in two 16 mL of SC-URA media including raffinose and incubated at 30°C, 90 rpm, overnight. On day three, the expression of individual viral proteins was induced by adding final 2% galactose to yeast cultures when the culture reached O.D. 0.9. Induced yeast cells were harvested after 6 hr incubation and stored at −80°C until the protein purification. For the initial protein purification step, Zirconia beads and lysis buffer including protease inhibitor cocktail (Roche) and reducing agent were immediately added to frozen pellets and yeast cells were mechanically lysed. Supernatant was incubated with glutathione beads in fresh plates for 2 hr at 4°C. The mixtures of glutathione beads and individual viral proteins were washed each three times under both high (500 mM NaCl)- and low (100 mM NaCl) - salt washing buffers including a protease inhibitor and reducing agent. For quality control of eluted proteins, all purified proteins were examined by SimplyBlue stain or anti-GST Western blot analysis.
Each purified ZIKV-2A protein and DENV-NS2A protein was fluorescently labeled with Cy5-NHS ester using a commercial kit (GE Life Science) and diluted to a final concentration of 10 ng/μL in 200 μL of 1 X TBST with 2% BSA. HuProt arrays (version III), comprised of 20,240 individually purified full-length human proteins (Jeong et al., 2012), were first blocked with 1 X TBST with 2% BSA at room temperature for 2 hr. Each labeled NS2A protein was incubated on the blocked HuProt arrays in duplicate at RT for 1 hr. After three 15-min washes with 1 X TBST, the HuProt arrays were briefly rinsed with water and dried. After scanning the HuProt arrays with a microarray scanner (GenePix 4000B), the NS2A binding signals were acquired and analyzed using the GenePix software.
GenePix 6.1 was used to align the spot-calling grid. For each protein spot, the median values of foreground (Fij) and background (Bij) intensities at site (i,j) on the microarray were extracted, respectively. The binding intensity (Rij) of each protein spot was defined as Fij/Bij. Since each protein was printed in duplicate on each microarray, Rij was averaged for the duplicate as R’ij. Using a similar method as in our previous study (Hu et al., 2013), the Z-score of each probe was calculated based on the distribution of R’ij,
N is the mean signal value for all proteins on the protein array. A stringent cutoff (Z ≥ 10) was used to determine the positive hit list (Table S2).
For protein-protein interaction analysis, the functional protein association network was constructed using STRING 10.0 database (Szklarczyk et al., 2015) (http://string-db.org/) (Table S3). The protein-protein interactions were obtained with the default parameters (confidence score 0.4). The P value of association enrichment was also given by the database taking all proteins in the protein microarray as background. The association network was generated using Cytoscape 3.2.1(Shannon et al., 2003).
Gene ontology enrichment analyses for NS2A binding proteins were performed using DAVID 6.8 (https://david.ncifcrf.gov/) (Huang da et al., 2009). Compared to all proteins in the protein microarray, enriched terms for NS2A binding proteins (P value < 0.05) were listed (Table S4).
Cell culture, transfection and infection
HEK293 cells were cultured in DMEM containing 10% FBS (Hyclone, Logan, UT, USA), 4 mM L-glutamine (Gibco BRL), 100 IU/ml penicillin (Gibco BRL) and 100 μg/ml streptomycin (Gibco BRL). For co-immunoprecipitation experiments, HEK293 cells were transfected with control, ZIKV-NS2A-HA, and DENV-NS2A-HA expressing constructs with Lipofectamine 2000 (Thermo Fisher Scientific) and collected after 48 hr.
Mouse neural progenitors were isolated from E11.5 CD1 mouse embryos and cultured in Neurobasal medium (Gibco BRL) containing 20 ng/ml FGF2, 20 ng/ml EGF, 5 mg/ml heparin, 2% B27 (v/v, Gibco BRL), and 4 mM L- glutamine as previously described (Currle et al., 2007). High titer lentivirus was produced from HEK293 cells and used to infect mouse neural progenitors in the presence of 4 μg/ml polybrene (Milipore) (Ma et al., 2008). Mouse neural progenitor lysates were collected 4 days after control, ZIKV-NS2A- or DENV-NS2A-expressing lentivirus for Western blotting analysis. For co-IP analysis, mouse neural progenitors were electroporated with empty lentiviral control vector or pCWX-ZIKV-NS2A-HA by Mouse neural stem cell nucleofector kit (Lonza) and Nucleofector 2b Device (Lonza), and collected 3 days later. Mouse neural progenitors were infected with ZIKV MR766 at MOI 0.08 for 64 hr and collected for Western blotting and quantitative PCR analysis (Tang et al., 2016).
Co-IP and Western blot analysis
For co-IP analysis, HEK293 cells were homogenized in the lysis buffer containing Phosphate-buffered saline (pH 7.4), 1.5% Triton-X 100, 1 mM Na3VO4, 1 mM NaF, 1 mM DTT, and protease inhibitor cocktails (Sigma-Aldrich). The lysates were incubated for 15 min on ice, sonicated and centrifuged for 15 min at 15,000 × g 4°C. The super natants were collected and immunoprecipitated with anti-HA magnetic beads (Thermo Fisher Scientific) overnight at 4°C. The beads were thoroughly washed with lysis buffer, boiled with Laemmli Sample Buffer (Bio-Rad) and subjected to Western blot analysis. For Western blotting, samples were separated by 4–20% SDS-PAGE, transferred to PVDF membranes (Bio-Rad), incubated with primary and secondary antibodies and visualized with SuperSignal West Dura Chemiluminescent Substrate (Thermo Fisher Scientific). Primary antibodies are listed in KEY RESOURCE TABLE. Quantification of bands was performed using ImageJ software.
Rough endoplasmic reticulum (RER) fractions were isolated from mouse neural progenitors after 4 days of control, ZIKV-NS2A- and DENV-NS2A-expressing lentivirus infection, using Endoplasmic Reticulum Isolation Kit (Sigma-Aldrich) according to the manufacturer’s protocol. Two 100 mm dishes of mouse neural progenitors were homogenized and RER-enriched microsomes were precipitated by calcium chloride, solubilized by Laemmli Sample Buffer (Bio-Rad) and subjected to Western blot analysis.
RNA preparation and quantitative PCR
For gene expression analysis, the total RNA fraction was immediately isolated from cultured mouse neural progenitor samples with RNeasy Mini Kit (Qiagen), treated with DNaseI and reverse-transcribed into the first-strand cDNA with SuperScript III (Thermo Fisher Scientific) (Su et al., 2017). cDNAs were used for SYBR-green based quantitative real-time PCR to measure the expression level of target genes with the comparative CT method (ABI) (Su et al., 2017). The primers used for quantitative PCR are listed in Table S1.
Forebrain organoid culture
Human iPSC lines from healthy subjects used in the current study have been fully characterized (Chiang et al., 2011; Wen et al., 2014). Protocols for generation of forebrain organoids using the SpinΩ bioreactor were detailed previously (Qian et al., 2016). Briefly, human iPSCs were cultured in stem cell medium, consisting of DMEM:F12 (Invitrogen) supplemented with 20% Knockout Serum Replacer (Gibco), 1X Non-essential Amino Acids(Invitrogen), 1X Penicillin/Streptomycin (Invitrogen), 1X 2-Mercaptoethanol (Millipore), 1X Glutamax (Invitrogen), and 10 ng/ml FGF-2 (Peprotech) on irradiated CF1 mouse embryonic fibroblasts (Charles River). On day 1, iPSC colonies were detached by treatment of 1 mg/ml Collagenase Type IV (Invitrogen) for 1 hr and transferred to an Ultra-Low attachment 6-well plate (Corning Costar), containing 3 ml of stem cell medium (without FGF-2), plus 2 μM Dorsomorphine (Sigma) and 2 μM A83-01 (Tocris). On days 5–6, half of the medium was replaced with the induction medium consisting of DMEM:F12, 1X N2 Supplement (Invitrogen), 1X Penicillin/Streptomycin, 1X Non-essential Amino Acids, 1X Glutamax, 1 μM CHIR99021 (Cellagentech), and 1 μM SB-431542 (Cellagentech). On day 7, organoids were embedded in Matrigel (Corning) and continued to grow in the induction medium for 6 more days. On day 14, embedded organoids were mechanically dissociated from Matrigel and transferred to each well of a 12-well spinning bioreactor (SpinΩ) containing differentiation medium, consisting of DMEM:F12, 1X N2 and B27 Supplements (Invitrogen), 1X Penicillin/Streptomycin, 1X 2-Mercaptoethanol, 1X Non-essential Amino Acids, and 2.5 μg/ml Insulin (Sigma).
Forebrain organoid electroporation
On day 45, forebrain organoids were transferred into PBS solution in a 10 cm petri dish for electroporation. A mixture of 0.5 μl of plasmid DNA and 0.05% Fast green was injected into the ventricle-like cavity of neural tube structures in the forebrain organoid using a calibrated micropipette. About 3–4 locations on one side of each forebrain organoid were targeted by the injection. The DNA-injected side of the organoid was placed toward the positive electrode in the middle of 5 mm gap of electrode paddles (CUY650-5, Nepa Gene). Five pulses (40 V, 50 ms in duration with a 950 ms interval) were delivered by a square wave electroporator (CUY21SC, Nepa Gene). After electroporation, organoids were transferred back to SpinΩ bioreactor for continued culturing. On day 48 (45+3) or day 52 (45+7), organoids were pulsed by 10 μM EdU (ThermoFisher) for 1 hr by directly adding EdU into culture media and fixed for immunostaining analysis (Qian et al., 2016).
QUANTIFICATION AND STATISTICAL ANALYSIS
No statistical methods were used to pre-determine sample sizes but our sample sizes are similar to those generally employed previously (Qian et al., 2016; Yoon et al., 2014). No randomization or blinding was employed. Data in figure panels reflect several independent experiments performed on different days. Different statistical tests were performed as listed in figure legends.
Analyses of mouse cortical neurogenesis
For quantitative analysis of electroporated neocortices, only GFP+ cells localized within the dorsolateral cortex were examined. 3×3 tiled images were obtained to cover the electroporated region of each coronal section with a 20× or 40× objective by scanning microscope (Zeiss LSM 800) and compared with equivalent sections in littermate counterparts. Quantifications were performed using Imaris software (Bitplane). In detail, for quantification of cell proliferation and cell fate, GFP+ cells were marked, and GFP+Pax6+, GFP+Tbr2+, GFP+EdU+, GFP+Pax6+EdU+, and GFP+Tuj1+ cells were defined and counted based on the intensity of Pax6, Tbr2, Tuj1 and EdU immunofluorescence in GFP+ cells measured with the same criteria among different groups. For quantifications of β-Catenin, PKCλ, or ZO-1 expression and distribution, the total length of the GFP+ apical membrane and the total length of the β-Catenin+, PKCλ+, or ZO-1+ apical membrane on the ventricular surface were measured by ImageJ software and % of intact expression of the total electroporated region was calculated. For quantification of the number of ventricular protrusions, bulged cell clusters more than 15 μm away from the ventricular surface in DAPI stained images were defined and counted as ventricular protrusions. The average number of ventricular protrusions in 100 μm length of the GFP+ apical membrane was calculated. All quantifications were performed with 4–7 brain sections from at least 3 animals. Data are presented as the mean ± SEM and statistical significance was assessed using unpaired Student’s t-test and One-way ANOVA. For distribution plots of neuronal positioning, the distances between GFP+SATB2+ or GFP−CTIP2+ cells and the ventricular surface were calculated and plotted after dividing each distance by the total thickness of the neocortex. For statistical analysis, the normalized mean distance to the ventricular surface was determined for each section, an equal number of sections was used for each condition and the mean of mean distances was compared for each condition using unpaired Student’s t-test. Quantification was performed with eight brain sections from four animals.
Statistical analysis on forebrain organoids
For quantification after electroporation, randomly selected ventricular structures with electroporated “fan-shaped” regions facing towards the pial surface, but not the interior of organoid, were imaged by confocal microscope (Zeiss LSM 800). Among electroporated cells labelled by GFP, EdU+, Ki67+ or PAX6+ nuclei were counted using ImageJ software, and the effect from expression of different constructs (GFP, GFP and ZIKV-NS2A, or GFP and DENV-NS2A) was evaluated by the percentage of EdU+ or Ki67+ cells among total GFP+ or GFP+ PAX6+ cells. For analysis of AJ continuity, the length of PKCλ laminated ventricular surface was measured using ImageJ software, and divided by the total length of ventricular surface in the electroporated region. A ratio above 80% was defined as continuous AJ. For analysis of cell morphology, GFP+PAX6+ cells with 3 or more processes were defined as having multi-polar morphology, as opposed to bi-polar radial glia cells with a basal and an apical process. The effect from the expression of different constructs was evaluated by the percentage of multi-polar cells among total GFP+PAX6+ cells. Multiple organoids were quantified and Student’s t-test was used for statistical analysis.
Supplementary Material
Highlights.
ZIKV-NS2A, but not DENV-NS2A, depletes RGCs in the embryonic mouse cortex
ZIKV-NS2A expression causes mis-positioning of newborn neurons in the mouse cortex
ZIKA-NS2A interacts and depletes adherens junction (AJ) complex proteins
ZIKV-NS2A impairs RGC proliferation and AJ formation in human forebrain organoids
Acknowledgments
We thank members of Ming and Song laboratories for suggestions, L. Liu and Y. Cai for technical support, J. Schnoll for manuscript editing, and Dr. Park for help with statistical analysis. This work was supported by National Institutes of Health grants R35NS097370 and R01MH105128 (G-l.M.), U19AI131130 (G-l.M., H.T, P.J., & Z.W.), R37NS047344 (H.S.), U19MH106434 (G-l.M., J.Q., & H.S.), P01NS097206 (H.S., J.Q., & P.J.), R01GM111514 (H.Z & J.Q.), R25NS065729 and K12NS098482 (C.H)., and AI119530 (H.T.), Simons Foundation (SFARI Grant 308988 to H.S.), Maryland Stem Cell Research Fund (MSCRF; G-l.M. & H.S.), start-up fund from Emory (Z.W.), start-up fund from IGDB of CAS (Q.W)., China NSF (31430037) and MOST “973” program (2014CB942801 and 2012YQ03026006 to Y.W. & Z.X.), and Zika seed funding from Florida State University (H.T.). K-J.Y. was supported by a postdoctoral fellowship from MSCRF and by a Young Investigator Award from Brain & Behavior Research Foundation. C.V. was partially supported by a NSF fellowship and T32GM007445.
Footnotes
AUTHOR CONTRIBUTIONS: K-J. Y. led the project and was involved in all aspects of the study. G.S., J.P., H.S.R., J.Q., and H.Z. performed protein array analysis. X.Q. and F.J. performed human organoid study. E.L. and H.T. performed viral infection. D.X., C.L., L.Y., C-F. Q., Q.W., and Z.X. performed mouse ZIKV infection experiments, N-S.K., C.H. F.Z., L.Z., W-K. H., F.R.R., C.V., K.K., S.K., J.Y., Y.C., S.L., Z.W., K.C., and P.J. contributed to additional data collection and writing. K-J.Y., H.J.S. and G-l.M designed the project, analyzed the data and wrote the paper.
COMPETING FINANCIAL INTERESTS: The authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agrawal T, Sharvani V, Nair D, Medigeshi GR. Japanese encephalitis virus disrupts cell-cell junctions and affects the epithelial permeability barrier functions. PLoS One. 2013;8:e69465. doi: 10.1371/journal.pone.0069465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besnard M, Eyrolle-Guignot D, Guillemette-Artur P, Lastere S, Bost-Bezeaud F, Marcelis L, Abadie V, Garel C, Moutard ML, Jouannic JM, et al. Congenital cerebral malformations and dysfunction in fetuses and newborns following the 2013 to 2014 Zika virus epidemic in French Polynesia. Euro Surveill. 2016:21. doi: 10.2807/1560-7917.ES.2016.21.13.30181. [DOI] [PubMed] [Google Scholar]
- Brault JB, Khou C, Basset J, Coquand L, Fraisier V, Frenkiel MP, Goud B, Manuguerra JC, Pardigon N, Baffet AD. Comparative Analysis Between Flaviviruses Reveals Specific Neural Stem Cell Tropism for Zika Virus in the Mouse Developing Neocortex. EBioMedicine. 2016;10:71–76. doi: 10.1016/j.ebiom.2016.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchman JJ, Tsai LH. Spindle regulation in neural precursors of flies and mammals. Nat Rev Neurosci. 2007;8:89–100. doi: 10.1038/nrn2058. [DOI] [PubMed] [Google Scholar]
- Chen YT, Stewart DB, Nelson WJ. Coupling assembly of the E-cadherin/beta-catenin complex to efficient endoplasmic reticulum exit and basal-lateral membrane targeting of E-cadherin in polarized MDCK cells. J Cell Biol. 1999;144:687–699. doi: 10.1083/jcb.144.4.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang CH, Su Y, Wen Z, Yoritomo N, Ross CA, Margolis RL, Song H, Ming GL. Integration-free induced pluripotent stem cells derived from schizophrenia patients with a DISC1 mutation. Mol Psychiatry. 2011;16:358–360. doi: 10.1038/mp.2011.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cugola FR, Fernandes IR, Russo FB, Freitas BC, Dias JL, Guimaraes KP, Benazzato C, Almeida N, Pignatari GC, Romero S, et al. The Brazilian Zika virus strain causes birth defects in experimental models. Nature. 2016;534:267–271. doi: 10.1038/nature18296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currle DS, Hu JS, Kolski-Andreaco A, Monuki ES. Culture of mouse neural stem cell precursors. J Vis Exp. 2007:152. doi: 10.3791/152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driggers RW, Ho CY, Korhonen EM, Kuivanen S, Jaaskelainen AJ, Smura T, Rosenberg A, Hill DA, DeBiasi RL, Vezina G, et al. Zika Virus Infection with Prolonged Maternal Viremia and Fetal Brain Abnormalities. N Engl J Med. 2016;374:2142–2151. doi: 10.1056/NEJMoa1601824. [DOI] [PubMed] [Google Scholar]
- Gabriel E, Ramani A, Karow U, Gottardo M, Natarajan K, Gooi LM, Goranci-Buzhala G, Krut O, Peters F, Nikolic M, et al. Recent Zika Virus Isolates Induce Premature Differentiation of Neural Progenitors in Human Brain Organoids. Cell Stem Cell. 2017;20:397–406. e395. doi: 10.1016/j.stem.2016.12.005. [DOI] [PubMed] [Google Scholar]
- Garcez PP, Loiola EC, Madeiro da Costa R, Higa LM, Trindade P, Delvecchio R, Nascimento JM, Brindeiro R, Tanuri A, Rehen SK. Zika virus impairs growth in human neurospheres and brain organoids. Science. 2016;352:816–818. doi: 10.1126/science.aaf6116. [DOI] [PubMed] [Google Scholar]
- Garcia-Blanco MA, Vasudevan SG, Bradrick SS, Nicchitta C. Flavivirus RNA transactions from viral entry to genome replication. Antiviral Res. 2016;134:244–249. doi: 10.1016/j.antiviral.2016.09.010. [DOI] [PubMed] [Google Scholar]
- Gotz M, Huttner WB. The cell biology of neurogenesis. Nature reviews Molecular cell biology. 2005;6:777–788. doi: 10.1038/nrm1739. [DOI] [PubMed] [Google Scholar]
- Heymann DL, Hodgson A, Sall AA, Freedman DO, Staples JE, Althabe F, Baruah K, Mahmud G, Kandun N, Vasconcelos PF, et al. Zika virus and microcephaly: why is this situation a PHEIC? Lancet. 2016;387:719–721. doi: 10.1016/S0140-6736(16)00320-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S, Wan J, Su Y, Song Q, Zeng Y, Nguyen HN, Shin J, Cox E, Rho HS, Woodard C, et al. DNA methylation presents distinct binding sites for human transcription factors. eLife. 2013;2:e00726. doi: 10.7554/eLife.00726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Jeong JS, Jiang L, Albino E, Marrero J, Rho HS, Hu J, Hu S, Vera C, Bayron-Poueymiroy D, Rivera-Pacheco ZA, et al. Rapid identification of monospecific monoclonal antibodies using a human proteome microarray. Mol Cell Proteomics. 2012;11:O111016253. doi: 10.1074/mcp.O111.016253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Xu D, Ye Q, Hong S, Jiang Y, Liu X, Zhang N, Shi L, Qin CF, Xu Z. Zika Virus Disrupts Neural Progenitor Development and Leads to Microcephaly in Mice. Cell Stem Cell. 2016a;19:120–126. doi: 10.1016/j.stem.2016.04.017. [DOI] [PubMed] [Google Scholar]
- Li H, Saucedo-Cuevas L, Shresta S, Gleeson JG. The Neurobiology of Zika Virus. Neuron. 2016b;92:949–958. doi: 10.1016/j.neuron.2016.11.031. [DOI] [PubMed] [Google Scholar]
- Liang Q, Luo Z, Zeng J, Chen W, Foo SS, Lee SA, Ge J, Wang S, Goldman SA, Zlokovic BV, et al. Zika virus NS4A and NS4B proteins deregulate Akt-mTOR signaling in human fetal neural stem cells to inhibit neurogenesis and induce autophagy. Cell Stem Cell. 2016;19:663–671. doi: 10.1016/j.stem.2016.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindenbach BD, Thiel HJ, Rice CM. Flaviviridae: The viruses and their replication. In: Knipe DM, Howley PM, editors. Fields Virology. Philadelphia, PA, USA: Lippincott Williams and Wilkins; 2007. pp. 1101–1152. [Google Scholar]
- Ma DK, Chiang CH, Ponnusamy K, Ming GL, Song H. G9a and Jhdm2a regulate embryonic stem cell fusion-induced reprogramming of adult neural stem cells. Stem cells. 2008;26:2131–2141. doi: 10.1634/stemcells.2008-0388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medigeshi GR, Hirsch AJ, Brien JD, Uhrlaub JL, Mason PW, Wiley C, Nikolich-Zugich J, Nelson JA. West nile virus capsid degradation of claudin proteins disrupts epithelial barrier function. J Virol. 2009;83:6125–6134. doi: 10.1128/JVI.02617-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ming GL, Tang H, Song H. Advances in Zika Virus Research: Stem Cell Models, Challenges, and Opportunities. Cell Stem Cell. 2016;19:690–702. doi: 10.1016/j.stem.2016.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlakar J, Korva M, Tul N, Popovic M, Poljsak-Prijatelj M, Mraz J, Kolenc M, Resman Rus K, Vesnaver Vipotnik T, Fabjan Vodusek V, et al. Zika Virus Associated with Microcephaly. N Engl J Med. 2016;374:951–958. doi: 10.1056/NEJMoa1600651. [DOI] [PubMed] [Google Scholar]
- Moura da Silva AA, Ganz JS, Sousa PD, Doriqui MJ, Ribeiro MR, Branco MD, Queiroz RC, Pacheco MJ, Vieira da Costa FR, Silva FS, et al. Early Growth and Neurologic Outcomes of Infants with Probable Congenital Zika Virus Syndrome. Emerg Infect Dis. 2016;22:1953–1956. doi: 10.3201/eid2211.160956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onorati M, Li Z, Liu F, Sousa AM, Nakagawa N, Li M, Dell’Anno MT, Gulden FO, Pochareddy S, Tebbenkamp AT, et al. Zika virus disrupts phospho-TBK1 localization and mitosis in human neuroepithelial stem cells and radial glia. Cell Rep. 2016;16:2576–2592. doi: 10.1016/j.celrep.2016.08.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pylro VS, Oliveira FS, Morais DK, Cuadros-Orellana S, Pais FS, Medeiros JD, Geraldo JA, Gilbert J, Volpini AC, Fernandes GR. ZIKV - CDB: A Collaborative Database to Guide Research Linking SncRNAs and ZIKA Virus Disease Symptoms. PLoS Negl Trop Dis. 2016;10:e0004817. doi: 10.1371/journal.pntd.0004817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian X, Nguyen HN, Song MM, Hadiono C, Ogden SC, Hammack C, Yao B, Hamersky GR, Jacob F, Zhong C, et al. Brain-Region-Specific Organoids Using Mini-bioreactors for Modeling ZIKV Exposure. Cell. 2016;165:1238–1254. doi: 10.1016/j.cell.2016.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasin MR, Gazula VR, Breunig JJ, Kwan KY, Johnson MB, Liu-Chen S, Li HS, Jan LY, Jan YN, Rakic P, et al. Numb and Numbl are required for maintenance of cadherin-based adhesion and polarity of neural progenitors. Nat Neurosci. 2007;10:819–827. doi: 10.1038/nn1924. [DOI] [PubMed] [Google Scholar]
- Rasmussen SA, Jamieson DJ, Honein MA, Petersen LR. Zika Virus and Birth Defects--Reviewing the Evidence for Causality. N Engl J Med. 2016;374:1981–1987. doi: 10.1056/NEJMsr1604338. [DOI] [PubMed] [Google Scholar]
- Sarno M, Aquino M, Pimentel K, Cabral R, Costa G, Bastos F, Brites C. Progressive lesions of Central Nervous System in microcephalic fetuses with suspected congenital Zika virus syndrome. Ultrasound Obstet Gynecol. 2016 doi: 10.1002/uog.17303. [DOI] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome research. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocker AM, Chenn A. The role of adherens junctions in the developing neocortex. Cell Adh Migr. 2015;9:167–174. doi: 10.1080/19336918.2015.1027478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y, Shin J, Zhong C, Wang S, Roychowdhury P, Lim J, Kim D, Ming GL, Song H. Neuronal activity modifies the chromatin accessibility landscape in the adult brain. Nat Neurosci. 2017;20:476–483. doi: 10.1038/nn.4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H, Hammack C, Ogden SC, Wen Z, Qian X, Li Y, Yao B, Shin J, Zhang F, Lee EM, et al. Zika Virus Infects Human Cortical Neural Progenitors and Attenuates Their Growth. Cell Stem Cell. 2016;18:587–590. doi: 10.1016/j.stem.2016.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Z, Nguyen HN, Guo Z, Lalli MA, Wang X, Su Y, Kim NS, Yoon KJ, Shin J, Zhang C, et al. Synaptic dysregulation in a human iPS cell model of mental disorders. Nature. 2014;515:414–418. doi: 10.1038/nature13716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Z, Song H, Ming GL. How does Zika virus cause microcephaly? Genes Dev. 2017;31:849–861. doi: 10.1101/gad.298216.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu KY, Zuo GL, Li XF, Ye Q, Deng YQ, Huang XY, Cao WC, Qin CF, Luo ZG. Vertical transmission of Zika virus targeting the radial glial cells affects cortex development of offspring mice. Cell research. 2016;26:645–654. doi: 10.1038/cr.2016.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Lee EM, Wen Z, Cheng Y, Huang WK, Qian X, Tcw J, Kouznetsova J, Ogden SC, Hammack C, et al. Identification of small-molecule inhibitors of Zika virus infection and induced neural cell death via a drug repurposing screen. Nat Med. 2016;22:1101–1107. doi: 10.1038/nm.4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon KJ, Nguyen HN, Ursini G, Zhang F, Kim NS, Wen Z, Makri G, Nauen D, Shin JH, Park Y, et al. Modeling a genetic risk for schizophrenia in iPSCs and mice reveals neural stem cell deficits associated with adherens junctions and polarity. Cell Stem Cell. 2014;15:79–91. doi: 10.1016/j.stem.2014.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimori T, Yamamoto A, Moriyama Y, Futai M, Tashiro Y. Bafilomycin A1, a specific inhibitor of vacuolar-type H(+)-ATPase, inhibits acidification and protein degradation in lysosomes of cultured cells. J Biol Chem. 1991;266:17707–17712. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.