Abstract
A genome-scale genetic interaction map was constructed by examining 5.4 million gene-gene pairs for synthetic genetic interactions, generating quantitative genetic interaction profiles for ~75% of all genes in the budding yeast, Saccharomyces cerevisiae. A network based on genetic interaction profiles reveals a functional map of the cell in which genes of similar biological processes cluster together in coherent subsets, and highly correlated profiles delineate specific pathways to define gene function. The global network identifies functional cross-connections between all bioprocesses, mapping a cellular wiring diagram of pleiotropy. Genetic interaction degree correlated with a number of different gene attributes, which may be informative about genetic network hubs in other organisms. We also demonstrate that extensive and unbiased mapping of the genetic landscape provides a key for interpretation of chemical-genetic interactions and drug target identification.
The relation between an organism's genotype and its phenotype are governed by myriad genetic interactions (1). Although a complex genetic landscape has long been anticipated (2), exploration of genetic interactions on a genome-wide level has been limited. Systematic deletion analysis in the budding yeast, Saccharomyces cerevisiae, demonstrates that the majority of its ~6000 genes are individually dispensable, with only a relatively small subset (~20%) required for viability (1), which suggests the evolution of extensive buffering against genetic perturbations (3). Genome-scale screens for genetic interactions that affect the fitness of a cell or organism can chart the genetic network underlying functional redundancy (1). In particular, synthetic genetic array (SGA) methodology (4) enables the systematic mapping of synthetic lethal genetic interactions through an automated form of genetic analysis that produces high-density arrays of double mutants (5). Here, we report construction of a functionally unbiased genetic interaction map for a eukaryotic cell.
Genome-scale, quantitative analysis of genetic interactions
We consider a digenic interaction as a double mutant that shows a significant deviation in fitness compared with the expected multiplicative effect of combining two single mutants (6). Negative interactions refer to a more severe fitness defect than expected, with the extreme case being synthetic lethality; positive interactions refer to double mutants with a less severe fitness defect than expected. To quantitatively score genetic interactions in large-scale SGA screens, we developed a model to estimate fitness defects directly from double-mutant colony sizes (7, 8) (fig. S1A). We screened 1712 S. cerevisiae query genes, including 334 conditional or hypomorphic alleles of essential genes, for a total of ~5.4 million gene pairs spanning all biological processes (fig. S1, B and C) (7, 8). These queries were selected randomly with respect to function; however, preference was given to mutants exhibiting fitness defects (7, 8). Comparing fitness estimates of single mutants with their corresponding double-mutant phenotypes identified ~170,000 interactions, a threefold increase over all previously reported genetic interaction data (fig. S1, D and E). Our data captured ~35% of previously reported negative genetic interactions (7, 8) (fig. S1D) and exhibited significant correlation (r = 0.89) (fig. S1F) with genetic interactions identified by high-resolution liquid growth profiles (7–9), which confirmed the accuracy of our measurements (fig. S1F). Thus, our approach enabled assembly of a quantitative fitness-based profile of genetic interactions on a genome-wide scale.
We determined false-negative and false-positive rates at a defined confidence threshold (|ε| > 0.08, P < 0.05) (fig. S2A) (7, 8) and used this filtered data set for all analyses. Data evaluation, by several different measures (7, 8), indicated that interactions that corresponded to specific confidence levels were functionally informative (fig. S2, B and C). In particular, enrichment for Gene Ontology (GO) coannotated gene pairs was correlated with the significance and magnitude of genetic interaction (fig. S2B), as well as with genetic profile similarity (fig. S2C) (7, 8). Notably, we found about twice as many negative interactions as positive genetic interactions (fig. S1B). Moreover, negative genetic interactions tended to be more informative for identifying physical interactions and GO coannotated gene pairs than positive interactions (fig. S2C).
A functional map of the cell
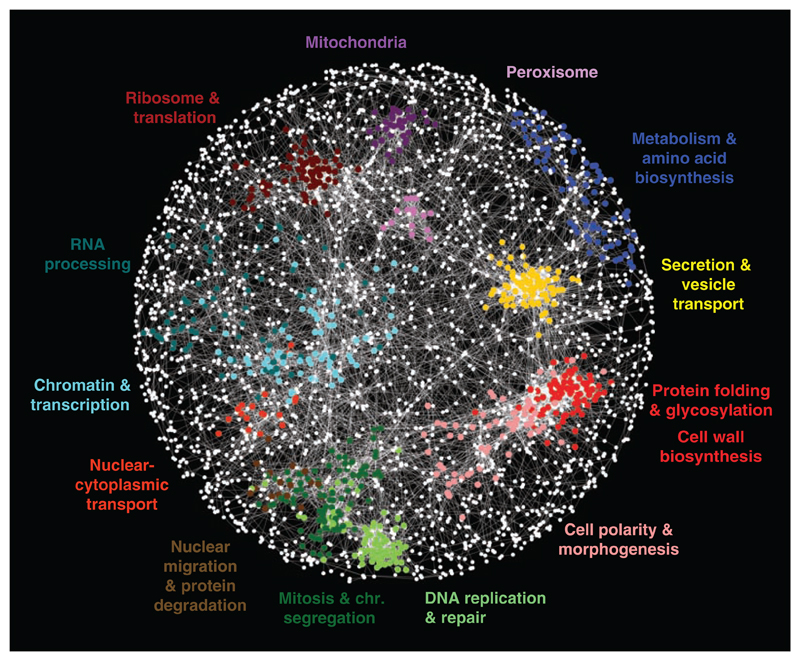
Genes belonging to the same pathway or biological process tend to share similar profiles of genetic interactions (5). We exploited this property to construct a global network, grouping genes with similar interaction patterns together: Nodes in this network represent genes, and edges connect gene pairs that share common sets of genetic interactions or similar interaction profiles (Fig. 1). This network highlights genetic relations between diverse biological processes and the inherent functional organization of the cell. Genes displaying tightly correlated profiles form discernible clusters corresponding to distinct bioprocesses, and the relative distance between distinct clusters appears to reflect shared functionality (Fig. 1). For example, the role of the microtubule cytoskeleton in bridging nuclear chromosomal- and actin cytoskeleton–based functions is illustrated by the close proximity and relative positioning of clusters corresponding to genes annotated with roles in cell polarity and morphogenesis, mitosis and chromosome segregation, and DNA replication and repair (Fig. 1). Despite screening only ~30% of the genome as query genes, we recovered genetic interactions for ~75% of the genome because partial genetic interaction profiles were generated for nearly all nonessential genes in the genome. Our data were able to precisely predict known gene functions (GO biological process annotations), as well as or better than all other genome-scale data sets (fig. S2D), and assigned a substantial amount of unique functional information for the genes not captured by previous genetic interaction studies (fig. S2D).
Fig. 1.
A correlation-based network connecting genes with similar genetic interaction profiles. Genetic profile similarities were measured for all gene pairs by computing Pearson correlation coefficients (PCCs) from the complete genetic interaction matrix. Gene pairs whose profile similarity exceeded a PCC > 0.2 threshold were connected in the network and laid out using an edge-weighted, spring-embedded, network layout algorithm (7, 8). Genes sharing similar patterns of genetic interactions are proximal to each other; less-similar genes are positioned farther apart. Colored regions indicate sets of genes enriched for GO biological processes summarized by the indicated terms.
Predicting function and relations
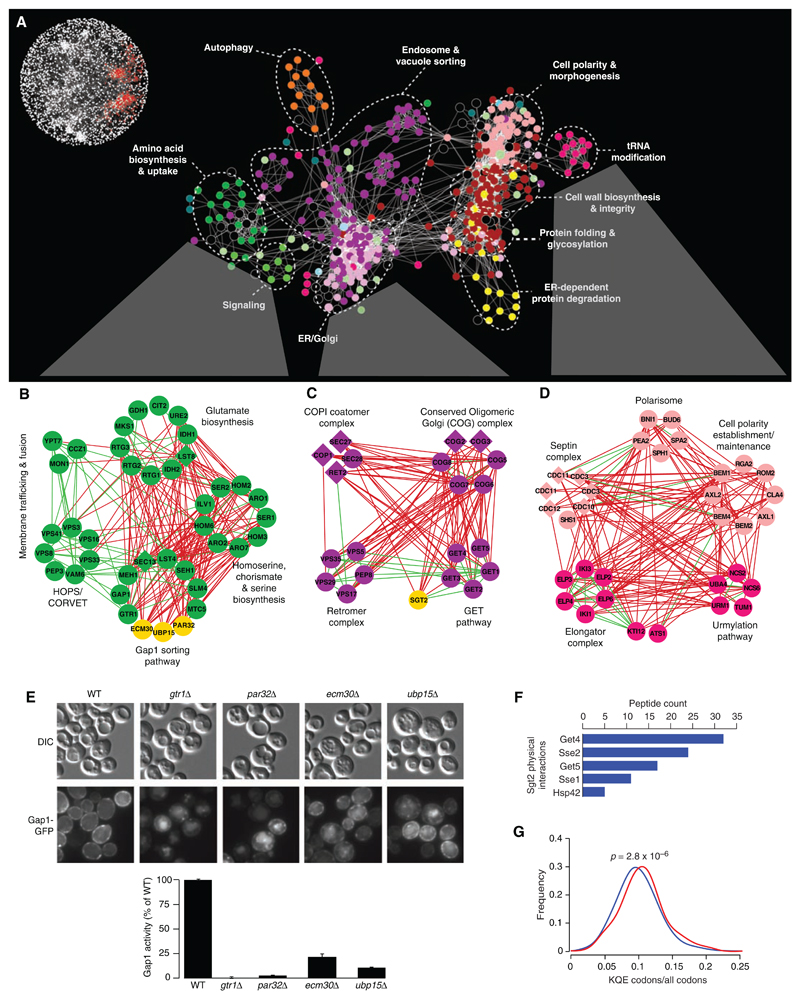
Although complex, the genetic interaction network contains functional information at multiple levels of resolution. The interrogation of the genetic map at higher resolution enabled the dissection of broad biological processes into distinct, yet interdependent, gene cohorts (Fig. 2) [supporting data file S8 (8)]. In even more detail, we can also visualize networks in which genes are connected by edges that correspond to genetic interactions directly. Indeed, gene clusters that are correlated by negative (red) and positive (green) genetic interactions reveal network organization reflecting biological pathways and/or protein complexes and their functional integration with one another (Fig. 2, B to D). The genetic interactions occurring between different pathways and complexes were often monochromatic, as predicted previously (10), such that they were composed almost exclusively of a single type of genetic interaction, either all negative or all positive.
Fig. 2.
Magnification of the functional map better resolves cellular processes. (A) A subnetwork corresponding to a region of the global map described in Fig. 1 is indicated in red (inset). Node color corresponds to a specific biological process: dark green, amino acid biosynthesis and uptake; light green, signaling; light purple, ER-Golgi; dark purple, endosome and vacuole sorting; yellow, ER-dependent protein degradation; red, protein folding and glycosylation, cell wall biosynthesis and integrity; fuchsia, tRNA modification; pink, cell polarity and morphogenesis; orange, autophagy; and black, uncharacterized. Individual genetic interactions contributing to genetic profiles revealed by (A) are illustrated for three specific subnetworks in (B) to (D). (B to D) Subsets of genes belonging to amino acid biosynthesis and uptake, ER-Golgi, and tRNA modification regions of the network were selected, and, in some cases, additional genes were included from the complete network shown in Fig. 1. Nodes are grouped according to profile similarity, and edges represent negative (red) and positive (green) genetic interactions (|ε| > 0.08, P < 0.05). Nonessential (circles) and essential (diamonds) genes are colored according to the biological process indicated in (A), and uncharacterized genes are depicted in yellow. (E) PAR32, ECM30, and UBP15 are required for plasma membrane localization (micrographs) and activity (histogram) of the Gap1 amino acid permease. DIC, differential interference contrast; GFP, green fluorescent protein. (F) Sgt2 physically interacts with components of the GET pathway and members of the Hsp70 chaperone family. Proteins identified with high confidence as specific interactors for tandem affinity purification (TAP)–tagged Sgt2 (Sgt2-TAP) are shown in decreasing order of spectral counts. (G) Distribution of the Elp and Urm modified codon usage among synthetic sick or lethal interaction partners. The fraction of Elp and Urm modified codons (lysine, glutamine, and glutamic acid) relative to all codons was measured for all negative interactors with genes in the Elp or Urm complex (red) relative to the background usage of all genes (blue).
Genetic clusters were used to predict function for uncharacterized genes on the basis of network connectivity (Fig. 2, A to D). Three genes, PAR32, ECM30, and UBP15, had interaction profiles similar to those of members of the Gap1-sorting module (Fig. 2B), and consistent with a role in this process, all three genes led to Gap1 sorting and transport defects when deleted (Fig. 2E). Additional experimental results (fig. S3) (11) suggest that Par32 may function in target of rapamycin (TOR)–dependent regulation of the Gln3, Gat1, Rtg1, and Rtg3 transcription factors (12), whereas Ecm30 forms a stoichiometric complex with the Ubp15 ubiquitin protease (7, 8) that may modulate Gap1 localization, perhaps by controlling its ubiquitination state.
In another example, similar genetic interaction profiles suggested a strong functional relation between the GET pathway and the poorly characterized gene, SGT2 (Fig. 2C). Consistent with a role in endoplasmic reticulum (ER)–dependent membrane targeting (13) or protein folding (14), we found that Sgt2 physically interacts with Get4, Get5, and heat shock 70 (Hsp70) protein family members (Fig. 2F), and, similarly to GET pathway mutants (13), deletion of SGT2 results in mislocalization of the tail-anchored protein, Pex15 (fig. S4).
Deciphering complex regulatory relations from the global genetic network
Because the global genetic interaction map represents a broad functional survey, it should provide insights into the regulatory wiring diagram of the cell. For example, synthetic lethal interactions between genes encoding the elongator (Elp) complex and those of the urmylation (Urm) pathway suggested that the Urm pathway collaborates with the Elp complex in the modification of specific transfer RNAs (tRNAs) (15) (Fig. 2D). In addition to their synthetic lethal relation, Elp and Urm pathway genes shared highly similar genetic interaction profiles; notably, these interactions were enriched for cell polarity and secretion genes (P < 10−3) (Fig. 2D), which reflects a specific cell polarity defect associated with Elp pathway mutants (16).
The elongator tRNA modification machinery has been postulated either to broadly affect the translation of a suite of mRNAs whose genes have cell polarity roles or to selectively influence the activity of a key polarity regulatory gene (17). We were intrigued by the finding that the subset of Elp-Urm negative interactors, as well as cell polarity and secretion genes, in general, encode proteins that are significantly enriched for the amino acids that charge Elp- and Urm-modified tRNAs (Fig. 2G) (7, 8). These findings suggest that Elp and Urm pathways may be biased toward the regulation of a functionally specific subset of cellular proteins. ELP1 is a highly conserved gene whose human ortholog, inhibitor of kappa light polypeptide gene enhancer in B cells, kinase complex–associated protein (IKBKAP), is associated with a neurological disorder, familial dysautonomia, which leads to disruption of cytoskeletal organization when mutated (18, 19). Thus, it is possible that disease manifestation may involve impaired IKBKAP-dependent translation of a set of human genes belonging to a specific functional group.
Genetic network connectivity
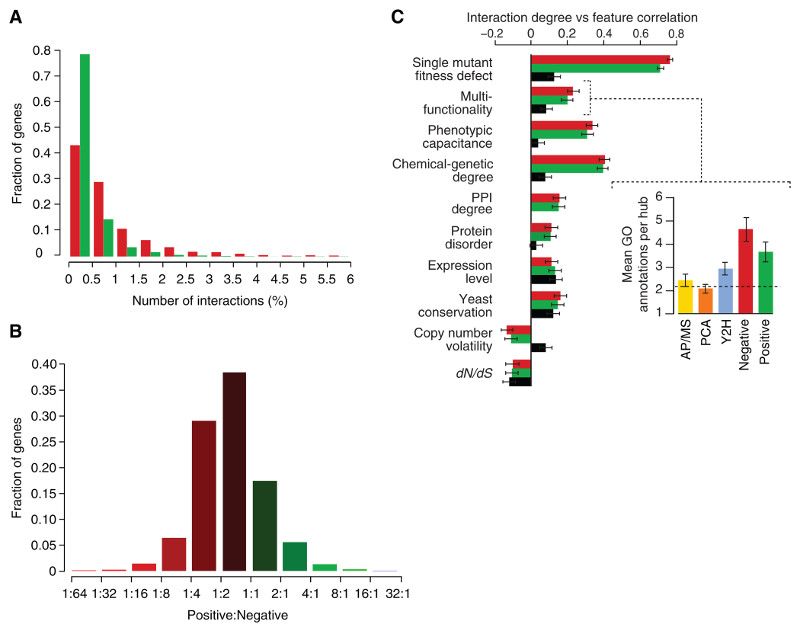
Consistent with the degree distribution of other biological networks (1), the majority of genes have few interactions, whereas a small number are highly connected and serve as network hubs (Fig. 3A). We found subsets of genes that showed a strong bias in their interaction type. About 2% of array genes exhibited more than eight times as many negative interactions as positive ones, whereas a smaller set containing ~1% of all array genes showed four times as many positive as negative interactions (Fig. 3B). Genes displaying this behavior were functionally distinct. Specifically, a bias toward negative interaction was observed for genes required for normal progression of the cell division cycle (P < 10−8), which highlights the central role of checkpoints in maintaining viability in dividing cells. Predominantly positive interactions were indicative of genes involved in translation, ribosomal RNA processing, and mRNA decay (P < 10−5), which may suggest that defects in the translation machinery somehow mask phenotypes that would otherwise be expressed in normal cells.
Fig. 3.
Positive and negative genetic interactions on the basis of a defined confidence threshold (|ε|> 0.08, P < 0.05) (7, 8). (A) The distribution of genetic interaction network degree for negative (red) and positive (green) interactions involving query genes. (B) The ratio of positive to negative interactions for each gene varies across the genome. (C) Pearson correlation between genetic interaction degree (derived from the array mutant strains) and physiological and evolutionary properties was measured for positive (green), negative (red) and protein-protein (black) interaction degree (7, 8). Chemical-genetic degree refers to the number of chemical perturbations to which a gene exhibits hypersensitivity. (Inset) The relation to gene multifunctionality for each of the interaction data sets is illustrated by measuring the average number of annotations to specific biological process GO terms for the top 1% highest degree genes for each interaction type. (7, 8).
Genetic interaction degree, fitness, multifunctionality, and pleiotropy
Genetic interaction hubs show a clear association with several fundamental physiological and evolutionary properties (Fig. 3C), which may be predictive of genetic interactions in other organisms. In particular, we uncovered a strong correlation between genetic interaction degree and single-mutant fitness (r = 0.73). Single mutants with increasingly severe fitness defects tended to exhibit an increased number of both negative and positive interactions (Fig. 3C and fig. S5, A and B) (7, 8). This relation was also observed for essential genes where the average number of interactions involving a temperature-sensitive mutant allele was inversely proportional to allele fitness at a given semipermissive temperature (fig. S5B). The increased connectivity of genes with fitness defects when singly mutated was not due to nonspecific interactions derived from a generally compromised cell or experimental noise; interactions with these genes were found to overlap with known functional relationships just as frequently as other interactions (fig. S5C).
In addition to the correlation with single-mutant fitness defects, genetic interaction hubs showed a high degree of pleiotropy. Specifically, the number of genetic interactions for a particular hub was significantly correlated with the number of distinct annotated functions (multifunctionality) for that gene (Fig. 3C and inset). This connection between network hubs and pleiotropy was further reflected by the rich variation associated with hub mutant phenotypes and increased phenotypic capacitance, the number of different morphological phenotypes linked to a specific gene as defined quantitatively (Fig. 3C) (20). This relation suggests that genetic network hubs play key roles in the integration and execution of morphogenetic programs.
It is noteworthy that these correlations persisted after we controlled for fitness defects of single mutants (fig. S5D). Furthermore, these trends reveal characteristics that distinguish genetic network hubs from hubs on the physical interaction network (Fig. 3C). Notably, the correlation to both fitness and multifunctionality was several fold stronger for genetic interaction degree (Fig. 3C). This likely reflects the ability of genetic perturbation analysis to identify broad phenotypic connections that cannot be captured in networks subject to physical constraints and suggests that large-scale genetic interaction networks will be of wide utility for defining the functional wiring diagrams of cells and organisms.
Although there are several distinguishing characteristics of genetic interaction hubs, we measured a significant correlation (r ~ 0.2) between the genetic and physical interaction degree for any given gene (Fig. 3C). Similar to protein-protein interaction hubs (21–23), we found that genetic network hubs tend to be expressed at higher mRNA levels. In comparison with the whole-genome sequences of 23 different Ascomycota fungi species, we found that genetic interaction degree correlated positively with gene conservation and negatively with copy number volatility, which indicates that they tend to be lost or duplicated less frequently. Genes showing more genetic interactions evolved (dN/dS) more slowly than genes with few interactions (Fig. 3C), which suggests that genetic hubs generally tend to be evolutionarily constrained. However, a subset of genetic interaction hubs appears to behave differently. Despite their tendency to evolve faster (fig. S5F) (24), proteins with higher levels of native disorder tend to exhibit a large number of genetic interactions, which suggests that genes encoding disordered proteins may represent a distinct class of genetic interaction hub.
Distribution of genetic interactions by bioprocess
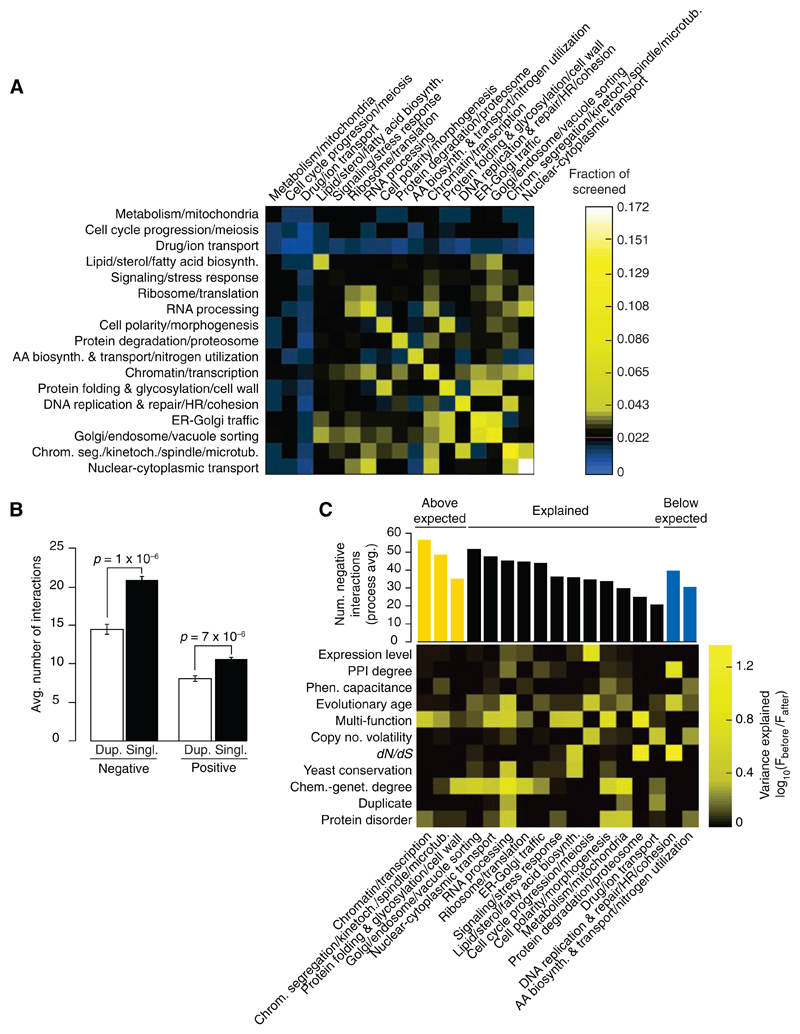
We assessed the distribution of genetic interactions across different cellular processes for both negative (Fig. 4A) and positive (fig. S6A) (7, 8) interactions. The heat map identified functions enriched (yellow) or depleted (blue) for genetic interactions relative to the expected frequency of a random gene set. As expected, genes involved in similar biological processes were enriched for negative interactions; however, we also observed genetic interactions bridging bioprocesses (Fig. 4A). Specifically, genes involved in chromatin, transcription, ER-Golgi transport, and Golgi-endosome transport showed a significant number of interactions that bridge diverse functions, which suggests that many of these genes are interconnected or pleiotropic. These bioprocess-level findings concur with individual gene analyses, which indicated that genes involved in processes related to chromatin structure and transcription (P < 10−14), as well as secretion and vesicle transport (P < 10−9), were among the most highly connected genes in our network. The central role for chromatin- and transcription-related processes identified in the yeast genetic network is consistent with large-scale genetic network mapping in Caenorhabditis elegans (25), and the bridging function for secretory pathway genes emphasizes their role as communication conduits for the cell. In contrast to genetic interactions, protein-protein interactions connect relatively fewer bioprocesses, and thus, although highly informative of local pathway architecture, physical interactions fail to provide a complete picture of multifunctionality or interconnections between cellular processes (fig. S6A). Reduced interactions in particular gene sets, such as meiosis, drug or ion transport, and metabolism or mitochondrial genes (blue in Fig. 4A), may arise because some processes are more buffered than others and require more complex genetic analysis to uncover their interactions (5), whereas others may function only under certain environmental conditions (26).
Fig. 4.
(A) Frequency of synthetic lethal/sick (negative) genetic interactions within and across biological processes. The fraction of screened gene pairs exhibiting negative interactions was measured for 17 broadly defined functional gene sets (7, 8). A color was assigned to each process-process element reflecting the fraction of interaction (blue, below the frequency of random pairs; black, statistically indistinguishable from the random background of interactions; and yellow, above the frequency of random pairs), with the diagonal representing within-process interactions. The red line in the color scale bar indicates random background. (B) Genetic interaction frequency of duplicate genes. T bars, SEM. (C) Gene-specific factors explaining the variation in number of negative interactions across biological processes. (Top) The average number of interactions across each process with the color indicating processes that have more interactions than expected (yellow, P < 0.05); processes whose interaction degree is explained by the factors indicated on the y axis; and those with fewer interactions than expected (blue, P < 0.05). The influence of each gene-specific factor in explaining the number of interactions observed was measured by plotting the ratio of F statistics of the bioprocess factor before and after incorporating the additional gene-specific factor. This ratio is indicated by the corresponding column in the heat map (7, 8). (AA, amino acids; chrom. seg., chromosome segregation; HR, homologous recombination; kinetoch., kinetochore)
Because variation was observed in the average number of genetic interactions for genes across different bioprocesses, we tested whether gene-specific properties (Fig. 3C) were predictive of this variation. For example, we found that gene duplicates exhibited fewer interactions when surveyed across the entire genome (Fig. 4B) (7, 8), and therefore, we asked if bioprocesses with relatively few genetic interactions could be explained by specific factors, such as a high percentage of duplicated genes. An analysis of covariance (ANCOVA) (Fig. 4C) (7, 8) showed that a linear model including the gene-specific properties predictive of genetic interaction hubs (Fig. 3C) was sufficient to explain the number of negative (12 out of 17) (Fig. 4C) and positive (13 out of 17) (fig. S6B) genetic interactions for the majority of bioprocesses. For example, the relatively few genetic interactions seen for genes with roles in drug and ion transport are explained by a combination of a high rate of gene duplication (~50 to 60%) and copy number volatility among genes annotated to this process. This is consistent with the tendency of genes encoding protein pumps to undergo numerous duplication events (27), which confirms that extensive redundancy associated with large gene families complicates the identification of digenic interactions. Three bioprocesses had significantly more negative interactions than predicted (Fig. 4C) (P < 0.05), including those that show functional enrichment for genetic interaction hubs (Fig. 4A). Conversely, DNA replication and repair and amino acid biosynthesis showed significantly fewer negative interactions than predicted (P < 0.05), which suggested that either more genetic interactions remain to be found for these genes under different environmental conditions or that these genes are more buffered and thus are inherently less connected on the digenic network.
Overlap between the genetic and the protein-protein interaction networks
We observed genetic interactions overlapping with 10 to 20% of protein-protein interaction pairs, depending on the physical interaction mapping methodology (fig. S7), which is significantly higher than expected randomly (~3%). Considering the global yeast physical interaction network as defined by affinity purification–mass spectrometry (28, 29), yeast two-hybrid protocol (30), or protein-fragment complementation assay (PCA) (31), roughly an equivalent number of physical interactions overlapped with negative and positive genetic interaction pairs: ~7% of protein-protein interacting pairs shared a negative genetic interaction, whereas ~5% shared a positive interaction. Conversely, considering our genetic interaction network, only a small fraction of gene pairs that show a genetic interaction (0.4% negative and 0.5% positive) are also physically linked. These findings suggest that the vast majority of both positive and negative interactions occurs between, rather than within, complexes and pathways, connecting those that presumably work together or buffer one another, respectively.
Navigating from genetic to chemical-genetic interaction networks
The set of ~4700 viable yeast deletion mutants has been exposed to hundreds of different chemical compounds (26). We quantified the chemical-genetic degree for each gene by counting the number of chemical (environmental) perturbations for which the corresponding gene deletion mutant showed hypersensitivity. We found a significant correlation (r = 0.4, P < 10−5) between genetic interaction and chemical-genetic degree (Fig. 3C). These observations suggest that hubs on a chemical-genetic network are predictive of hubs on the genetic interaction network and can be used to link environmental capacitance and genetic robustness. Furthermore, our data suggest that the same genes buffer the cell against both environmental and genetic insults. It is not known whether natural selection favors genetic robustness (32), but the positive correlation between genetic interaction degree and environmental capacitance suggests that genetic and environmental robustness may coevolve (33).
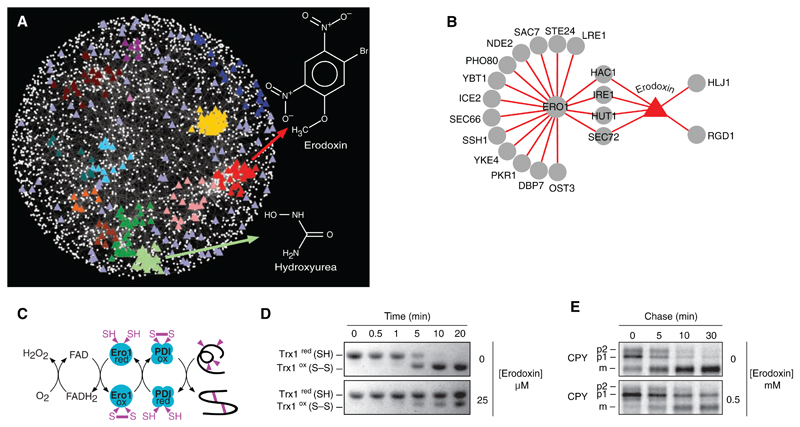
Because chemical perturbations mimic genetic perturbations, the genetic network should be useful for predicting the cellular targets of bioactive molecules (34). We identified genetic interaction profiles that are significantly correlated to a chemical-genetic profile of a particular compound (7, 8, 26, 34) and showed that compounds often clustered to dense regions of the genetic network indicative of specific bioprocesses (Fig. 5A). For example, hydroxyurea, a compound that inhibits ribonucleotide reductase and blocks DNA synthesis, clusters with the gene cohort annotated with roles in DNA replication and repair (Fig. 5A). These results demonstrate that clustering of chemical-genetic and genetic interaction profiles complements haploinsufficiency profiling, which has the potential to identify drug targets directly (26). We used this network approach to examine the previously uncharacterized compound, 0428-0027, which we have subsequently named erodoxin (Fig. 5A). Erodoxin clustered with genes associated with protein folding, glycosylation, and cell wall biosynthesis functions (Fig. 5A) because the erodoxin chemical-genetic profile most closely resembled the genetic interaction profile of ERO1 (Fig. 5B and fig. S8A), an essential gene involved in oxidative protein folding (Fig. 5C) (35). Two additional lines of evidence suggested that Ero1 is the target of erodoxin. First, ero1Δ/+ and fad1Δ/+ heterozygotes were the most hypersensitive mutants identified from haploinsufficency profiling (fig. S8B) (7, 8). Second, we found that erodoxin leads to inhibition of Trx1 oxidation (Fig. 5D) and delayed carboxy peptidase Y (CPY) processing (Fig. 5E), which suggests that it inhibits Ero1 activity both in vitro and in vivo.
Fig. 5.
(A) A chemical-genetic interaction map is shown in which colored triangles represent chemical compounds and white nodes correspond to genes. Compounds were positioned on the map by highlighting the gene node whose genetic interaction profile most closely resembles the chemical genetic profile of the compound derived from three sources (7, 8). Compounds tightly correlated to genes positioned within functional clusters (Fig. 1) were colored accordingly to the color of the cluster as in Fig. 1. The chemical-genetic profile of hydroxyurea clustered with genes involved in DNA replication and repair, whereas that of erodoxin clustered with genes involved in protein folding, glycosylation, and cell wall biosynthesis. Compounds positioned outside functional clusters are colored light purple. (B) Network displaying overlap between ERO1 negative genetic interactions and genes resulting in growth inhibition when deleted in the presence of erodoxin. (C) ERO1-dependent pathway for oxidative protein-folding pathway. (D) Erodoxin inhibits Ero1-dependent oxidation of Trx1 in vitro. (E) Erodoxin inhibits CPY processing to the vacuolar form in vivo. ER (p1), Golgi (p2), and vacuolar (m) forms of CPY are indicated.
Exploring the universe of genetic interactions
Unbiased, systematic, and quantitative analysis of digenic loss-of-function perturbations assigns a rich phenotypic profile to each gene and enables construction of a functional map of the cell, organizing genes and higher-order bioprocesses according to their related roles (Fig. 1). The functional connections defined by genetic interactions complement the information derived from networks based upon physical interactions, which links previously uncharacterized genes to specific pathways and complexes and reveals connections between pathways and complexes. The global mapping of genetic networks is becoming feasible in more complex cells and metazoans because of the growing availability of whole-genome sequences and large-scale sets of gene-knockdown reagents (1). Although negative genetic interactions can be conserved from yeast to worms and from yeast to human cells, the extent to which individual genetic interactions are conserved over large evolutionary distances remains unclear (1). The conservation of the genetic map may also occur at various levels of resolution. For example, overall network topology (Fig. 1) and properties (Fig. 3C) may be more highly conserved than particular genetic interactions because they reflect the fundamental architecture of the cell. The ability to integrate genetic and chemical-genetic perturbation data offers the potential to link bioactive compounds to their targets (Fig. 5), to identify genetic interaction hubs through chemical perturbations (Fig. 3C), to design synthetic lethal therapies for targeting genetically defined tumors (36), and to understand the mechanistic basis of drug synergy (37). Finally, genetic interaction maps provide a model for understanding the link between genotype and phenotype and for outlining the general principles of complex genetic interaction networks, which play a key role in governing inherited phenotypes, including human disease (3).
Supplementary Material
Editor’s Summary.
Making Connections
Genetic interaction profiles highlight cross-connections between bioprocesses, providing a global view of cellular pleiotropy, and enable the prediction of genetic network hubs. Costanzo et al. (p. 425) performed a pairwise fitness screen covering approximately one-third of all potential genetic interactions in yeast, examining 5.4 million gene-gene pairs and generating quantitative profiles for ~ 75% of the genome. Of the pairwise interactions tested, about 3% of the genes investigated interact under the conditions tested. On the basis of these data, a reference map for the yeast genetic network was created.
References and Notes
- 1.Dixon SJ, Costanzo M, Baryshnikova A, Andrews B, Boone C. Annu Rev Genet. 2009;43:601. doi: 10.1146/annurev.genet.39.073003.114751. [DOI] [PubMed] [Google Scholar]
- 2.Waddington CH. The Strategy of the Gene. Allen & Unwin; London: 1957. [Google Scholar]
- 3.Hartwell L. Science. 2004;303:774. doi: 10.1126/science.1094731. [DOI] [PubMed] [Google Scholar]
- 4.Tong AH, et al. Science. 2001;294:2364. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- 5.Tong AH, et al. Science. 2004;303:808. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- 6.Mani R, St Onge RP, Hartman JL, 4th, Giaever G, Roth FP. Proc Natl Acad Sci USA. 2008;105:3461. doi: 10.1073/pnas.0712255105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Materials and methods are available as supporting material on Science Online
- 8.See supplementary information at http://drygin.ccbr.utoronto.ca/~costanzo2009.
- 9.St Onge RP, et al. Nat Genet. 2007;39:199. doi: 10.1038/ng1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Segrè D, Deluna A, Church GM, Kishony R. Nat Genet. 2005;37:77. doi: 10.1038/ng1489. [DOI] [PubMed] [Google Scholar]
- 11.Huber A, et al. Genes Dev. 2009;23:1929. doi: 10.1101/gad.532109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen EJ, Kaiser CA. J Cell Biol. 2003;161:333. doi: 10.1083/jcb.200210141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jonikas MC, et al. Science. 2009;323:1693. doi: 10.1126/science.1167983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Metzger MB, Michaelis S. Mol Biol Cell. 2009;20:1006. doi: 10.1091/mbc.E08-02-0140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leidel S, et al. Nature. 2009;458:228. doi: 10.1038/nature07643. [DOI] [PubMed] [Google Scholar]
- 16.Rahl PB, Chen CZ, Collins RN. Mol Cell. 2005;17:841. doi: 10.1016/j.molcel.2005.02.018. [DOI] [PubMed] [Google Scholar]
- 17.Esberg A, Huang B, Johansson MJ, Byström AS. Mol Cell. 2006;24:139. doi: 10.1016/j.molcel.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 18.Naumanen T, Johansen LD, Coffey ET, Kallunki T. Cell Adh Migr. 2008;2:236. doi: 10.4161/cam.2.4.6630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johansen LD, et al. J Cell Sci. 2008;121:854. doi: 10.1242/jcs.013722. [DOI] [PubMed] [Google Scholar]
- 20.Levy SF, Siegal ML, Levchenko A. PLoS Biol. 2008;6:e264. doi: 10.1371/journal.pbio.0060264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim PM, Lu LJ, Xia Y, Gerstein MB. Science. 2006;314:1938. doi: 10.1126/science.1136174. [DOI] [PubMed] [Google Scholar]
- 22.Fraser HB, Wall DP, Hirsh AE. BMC Evol Biol. 2003;3:11. doi: 10.1186/1471-2148-3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pál C, Papp B, Hurst LD. Genetics. 2001;158:927. doi: 10.1093/genetics/158.2.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim PM, Sboner A, Xia Y, Gerstein M. Mol Syst Biol. 2008;4:179. doi: 10.1038/msb.2008.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lehner B, Crombie C, Tischler J, Fortunato A, Fraser AG. Nat Genet. 2006;38:896. doi: 10.1038/ng1844. [DOI] [PubMed] [Google Scholar]
- 26.Hillenmeyer ME, et al. Science. 2008;320:362. doi: 10.1126/science.1150021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dunham MJ, et al. Proc Natl Acad Sci USA. 2002;99:16144. doi: 10.1073/pnas.242624799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gavin AC, et al. Nature. 2006;440:631. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- 29.Krogan NJ, et al. Nature. 2006;440:637. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 30.Yu H, et al. Science. 2008;322:104. [Google Scholar]
- 31.Tarassov K, et al. Science. 2008;320:1465. doi: 10.1126/science.1153878. [DOI] [PubMed] [Google Scholar]
- 32.de Visser JA, et al. Evolution. 2003;57:1959. doi: 10.1111/j.0014-3820.2003.tb00377.x. [DOI] [PubMed] [Google Scholar]
- 33.Meiklejohn CD, Hartl DL. Trends Ecol Evol. 2002;17:468. [Google Scholar]
- 34.Parsons AB, et al. Nat Biotechnol. 2004;22:62. doi: 10.1038/nbt919. [DOI] [PubMed] [Google Scholar]
- 35.Sevier CS, et al. Cell. 2007;129:333. doi: 10.1016/j.cell.2007.02.039. [DOI] [PubMed] [Google Scholar]
- 36.Fong PC, et al. N Engl J Med. 2009;361:123. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- 37.Lehár J, Stockwell BR, Giaever G, Nislow C. Nat Chem Biol. 2008;4:674. doi: 10.1038/nchembio.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.We thank S. Dixon, T. Hughes, P. Jorgensen, and M. Tyers for critical comments. Supported by Genome Canada through the Ontario Genomics Institute (2004-OGI-3-01) and the Canadian Institutes of Health Research (GSP-41567) (C.B., B.A.), the University of Minnesota Biomedical Informatics and Computational Biology program (J.B., R.D.), and a seed grant from the Minnesota Supercomputing Institute (J.B., B.V.).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.