Abstract
Persistent insomnia is among the most frequent complaints in general practice. To identify genetic factors for insomnia complaints, we performed a genome-wide association study (GWAS) and a genome-wide gene-association study (GWGAS) in 113,006 individuals. We identify three loci and seven genes of which one locus and five genes are supported by joint analysis with an independent sample (n=7,565). Our top association (MEIS1, P<5×10-8) has previously been implicated in Restless Legs Syndrome (RLS). Additional analyses favor the hypothesis that MEIS1 shows pleiotropy for insomnia and RLS, and that the observed association with insomnia complaints cannot be explained only by the presence of an RLS subgroup. Sex-specific analyses suggested different genetic architectures across sexes in addition to common genetic factors. We show substantial positive genetic overlap with internalizing and metabolic traits and negative overlap with subjective well-being and educational attainment. These findings provide novel insight into the genetic architecture of insomnia.
Insomnia Disorder (ID) is the second-most prevalent mental disorder1 with prevalence estimates ranging from 10% (adults) to 22% (elderly)2,3, and is characterized by lasting problems falling asleep or waking up in the night or early morning, with subjective repercussions for daytime functioning. It is the primary risk factor for depression4 and contributes to the risks of cardiovascular disease5–9, type 2 diabetes10 and obesity11. Heritability estimates of 38% (males) and 59% (females)12 suggest a substantial role for genetic factors. In contrast to presumed neurobiological mechanisms involved in most mental disorders, it has been suggested that insomnia merely involves reversible maladaptive learning of sleep-related cognitions and behaviors13,14. Indeed, interventions that address these are at least partly effective in about two-thirds of cases15, but ameliorate complaints only by about 50%16, often resulting in a persistent course17. Family and twin studies suggest the involvement of genetic factors in the etiology of insomnia18–21. However, few, and mainly underpowered (n < 5,000) linkage and association studies of insomnia-related phenotypes have been conducted, and recent larger studies used non-validated proxy measures for ID22–24. For example, the habitual duration or the timing of sleep25,26 do not discriminate reliably between cases and controls of ID27, and therefore genes found for these phenotypes are not indicative of biological mechanisms of insomnia. The most recent GWAS for sleep disturbance traits reported several novel loci24. However, this study did not provide information on the predictive accuracy of the included traits for ID, making it difficult to interpret their findings in terms of clinical relevance. In addition, their top finding (MEIS1) is a known risk factor for Restless Legs Syndrome (RLS), and it is still unclear whether the shared associations in this gene reflect causality, partial mediation or pleiotropy24.
Here we report a GWAS using the UK Biobank sample28 (see URLs) including 113,006 individuals (mean age 56.92, sd = 7.94) to identify genetic risk factors for insomnia complaints (Online Methods, Supplementary Table 1). The discriminative value of insomnia complaints for identifying ID cases versus controls was validated in an independent cohort (n = 1,918). In addition, we report extensively on the possible mechanisms of action explaining the shared genetic signal in MEIS1 for insomnia complaints and RLS.
Results
Insomnia complaints is predictive of Insomnia Disorder
ID was assessed by using a single question on trouble falling asleep and waking up in the middle of the night (Supplementary Note, Supplementary Fig. 1). Individuals that answered ‘usually’ were scored as cases and individuals reporting ‘never/rarely’ or ‘sometimes’ were scored as controls. We note that this operationalization differs from24 where the same question in the UK Biobank sample was used but where cases were defined as scoring ‘usually’ and controls as scoring ‘never/rarely’ (Supplementary Note). We validated the predictive utility of this question for ID in an independent sample of 1,918 participants (845 insomniacs and 1,073 controls) of the Netherlands Sleep Registry29 (NSR, see URLs) (Supplementary Note). The equivalent of the UK Biobank question (using our response category cut-off) in the NSR had a sensitivity of 98%, and a specificity of 96% in discriminating questionnaire-defined insomnia ID cases from non-affected controls (χ2 = 1356.45, P < 0.0001), or discriminating ID cases from cases with exclusively RLS (sensitivity 0.96, specificity 0.97, χ2 = 639.06, P < 0.0001; Supplementary Note, Supplementary Table 2). It did not discriminate RLS from controls well, with a sensitivity of only 0.43, and a specificity of 0.74 (χ2 = 1.28, P = 0.26; Supplementary Fig. 1 and 2, Supplementary Table 2), nor any cases versus controls of 19 possibly related disease categories (Supplementary Note, Supplementary Fig. 3). Further strong support for the validity of the UK Biobank insomnia phenotype was provided by an accuracy of correct classification of 91% of clinical ID in NSR participants diagnosed by a structured interview (Supplementary Note). Moreover, sleep and mood characteristics of individuals who reported difficulties falling and staying asleep closely resembled the corresponding profile of NSR participants with ID, but not the profile of those with RLS (Supplementary Note, Supplementary Fig. 4). These findings show that the UK Biobank insomnia phenotype is predictive of ID, with little confounding by comorbidity. We will refer to this classification as 'insomnia complaints'.
Implicated genes and functional mechanisms
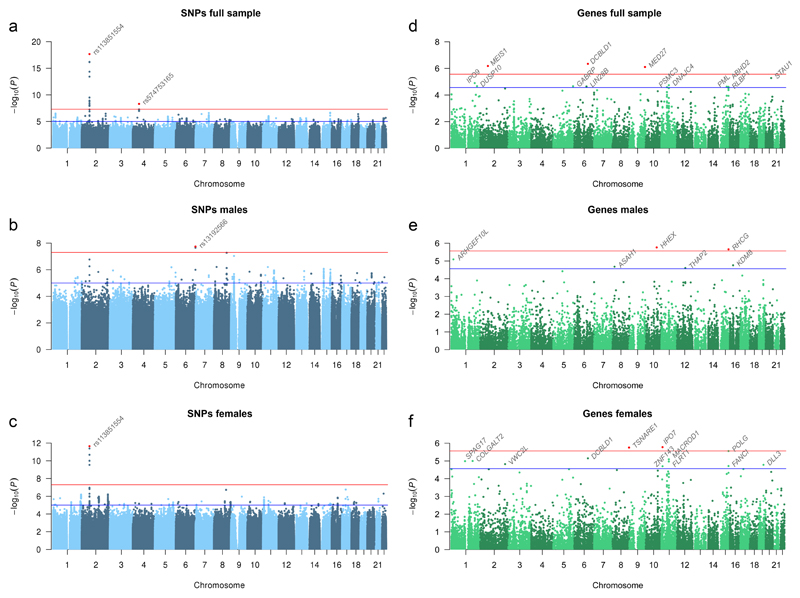
Prevalence of insomnia complaints was 29% in the UK Biobank sample (Supplementary Table 1, and further descriptives in Supplementary Table 3), in keeping with previous estimates for people with advanced age in the UK30 and elsewhere31,32. Females (33%) had a higher prevalence than males (24%). The sex odds ratio of 1.37 matches the previously published meta-analytic estimate of 1.412. GWAS was performed on all individuals of European descent, and standard quality-control procedures included correction for population stratification and filtering on minor allele frequency and imputation quality (Online Methods). The GWAS included 12,444,916 SNPs. The pooled-sex analysis identified two genome-wide significant loci (Fig. 1a; Table 1), implicating two genes: MEIS1 on chromosome 2 and SCFD2 on chromosome 4 (Supplementary Fig. 5a/b). Sex-specific analyses revealed MEIS1 for females as well, including the same significant SNPs found in the pooled-sex analysis, and an additional locus for males, WDR27 on chromosome 6 (Fig. 1b/c; Table 1; Supplementary Fig. 5c/d). Both MEIS1 and WDR27 have been identified by the recent GWAS for sleep disturbance24 traits using the UK Biobank data as well, using a slightly different insomnia phenotype (Supplementary Note, Supplementary Figs. 6 and 7). Our top SNPs in both genes were the same as in this study with similar association signals (MEIS1: rs113851554, P=9.11×10-19; WDR27: rs13192566, P=3.17×10-8).
Figure 1. Manhattan plots for SNP associations with insomnia complaints.
Association results for the frequency of experiencing trouble falling asleep or waking up in the middle of the night in 113,006 individuals of European descent in the UK Biobank study, of whom those experiencing these complaints usually (cases, n = 32,384) were contrasted with those experiencing these complaints never, rarely or sometimes only (controls, n = 80,622). (a) Manhattan plot of the GWAS including all individuals, (b) males only (12,863 cases and 40,776 controls), and (c) females only (19,521 cases and 39,846 controls). Negative log10-transformed P values for each SNP (y axis) are plotted by chromosomal position (x axis). The red and blue lines represent the thresholds for genome-wide statistical significant associations (P = 5 × 10−8) and suggestive associations (P = 1 × 10−5), respectively. Red dots represent top SNPs. (d) Manhattan plot of the gene analysis including all individuals, (e) males only, and (f) females only. Here, each dot represents a gene, and the red and blue lines represent the thresholds for gene-wide statistical significant associations (P = 2.72 × 10−6) and suggestive associations (P = 2.72 × 10−5), respectively.
Table 1. Three genome-wide significant loci associated with insomnia complaints in a GWAS including 113,006 individuals and the sex-specific GWASes.
| FULL |
MALE |
FEMALE |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| rsID | Annotation | Chr | BPa | EA | non-EA | INFO | EAF | OR | 95% CI | P | OR | 95% CI | P | OR | 95% CI | P |
| rs375216017 | MEIS1 intron variant | 2 | 66,728,627 | GT | G | 0.942 | 0.105 | 1.09 | 1.05-1.12 | 1.21E-08 | 1.09 | 1.04-1.14 | 2.27E-04 | 1.09 | 1.04-1.13 | 1.60E-05 |
| rs62144051 | MEIS1 intron variant | 2 | 66,730,783 | G | A | 0.964 | 0.094 | 1.10 | 1.06-1.13 | 3.81E-09** | 1.09 | 1.04-1.14 | 9.21E-04 | 1.10 | 1.06-1.15 | 1.01E-06 |
| rs62144053 | MEIS1 intron variant | 2 | 66,745,864 | A | G | 0.964 | 0.095 | 1.10 | 1.06-1.13 | 1.20E-09** | 1.08 | 1.03-1.13 | 1.79E-03 | 1.11 | 1.07-1.16 | 1.18E-07 |
| rs62144054 | MEIS1 intron variant | 2 | 66,747,480 | A | G | 0.976 | 0.094 | 1.10 | 1.06-1.13 | 1.68E-09** | 1.08 | 1.03-1.13 | 2.45E-03 | 1.11 | 1.07-1.16 | 1.09E-07 |
| rs113851554 | MEIS1 intron variant | 2 | 66,750,564 | T | G | 1 | 0.056 | 1.19 | 1.14-1.24 | 2.14E-18** | 1.18 | 1.11-1.25 | 1.69E-07 | 1.20 | 1.14-1.26 | 2.25E-12** |
| rs182588061 | MEIS1 intron variant | 2 | 66,757,709 | T | G | 0.852 | 0.020 | 1.21 | 1.13-1.28 | 3.18E-10* | 1.21 | 1.10-1.33 | 7.08E-05 | 1.21 | 1.11-1.32 | 1.13E-06 |
| rs139775539 | MEIS1 intron variant | 2 | 66,782,432 | A | AC | 0.961 | 0.048 | 1.19 | 1.14-1.24 | 7.00E-17* | 1.17 | 1.09-1.24 | 2.49E-06 | 1.21 | 1.14-1.28 | 4.08E-12* |
| rs11679120 | MEIS1 intron variant | 2 | 66,785,180 | A | G | 0.960 | 0.047 | 1.19 | 1.14-1.24 | 6.18E-17* | 1.18 | 1.11-1.26 | 5.54E-07 | 1.20 | 1.14-1.27 | 2.09E-11* |
| rs115087496 | MEIS1 intron variant | 2 | 66,793,725 | C | G | 0.950 | 0.047 | 1.18 | 1.13-1.23 | 4.43E-15* | 1.16 | 1.09-1.24 | 7.00E-06 | 1.19 | 1.13-1.26 | 9.91E-11* |
| rs549771308 | MEIS1 intron variant | 2 | 66,795,237 | C | CT | 0.832 | 0.120 | 1.08 | 1.05-1.11 | 9.51E-09** | 1.06 | 1.02-1.11 | 3.14E-03 | 1.09 | 1.05-1.13 | 5.17E-07 |
| rs11693221 | MEIS1 downstream gene variant | 2 | 66,799,986 | T | C | 0.943 | 0.048 | 1.17 | 1.12-1.22 | 3.79E-14** | 1.15 | 1.08-1.23 | 1.92E-05 | 1.19 | 1.12-1.25 | 2.88E-10** |
| rs574753165 | SCFD2 intron variant | 4 | 53,977,261 | G | A | 0.930 | 0.005 | 1.40 | 1.24-1.57 | 4.98E-09* | 1.38 | 1.15-1.65 | 1.85E-04 | 1.42 | 1.21-1.66 | 6.84E-06 |
| rs71554396 | intergenic | 6 | 169,841,072 | GT | G | 0.946 | 0.137 | 0.95 | 0.93-0.98 | 1.43E-03 | 0.89 | 0.85-0.93 | 1.95E-08 | 1.01 | 0.98-1.05 | 5.17E-01 |
| rs13208844 | WDR27 intron variant | 6 | 169,961,603 | G | A | 1 | 0.147 | 0.96 | 0.93-0.98 | 1.50E-03 | 0.89 | 0.85-0.93 | 2.25E-08* | 1.01 | 0.98-1.05 | 5.21E-01 |
| rs13192566 | WDR27 intron variant | 6 | 169,961,635 | C | G | 0.999 | 0.146 | 0.95 | 0.93-0.98 | 1.26E-03 | 0.89 | 0.85-0.93 | 1.80E-08* | 1.01 | 0.98-1.05 | 5.41E-01 |
Summary of the three significant loci present in the full GWAS and/or the male (n = 53,639) and female (n = 59,367) GWAS. All SNPs with P < 5 × 10-8 in one of the analyses are reported for all three analyses. SNP P values and odds ratios were calculated for each GWAS with an additive genetic model using logistic regression adjusted for age, sex in the full GWAS, genotyping array, and principal components. Bold P values are significantly associated with insomnia (P < 5 × 10-8). SNP association results in the deCODE sample and the meta-analysis are reported in Supplementary Tables 11 and 13. Chr, chromosome; BP, base pair; EA, effect allele; EAF. effect allele frequency; OR, odds ratio; CI, confidence interval.
reported on GRCh37.
Genome-wide significant in in the combined analysis of UK Biobank and deCODE.
Stronger association signal in the combined analysis of UK Biobank and deCODE.
Possible functional mechanisms of the identified SNPs and the SNPs in high LD (r2 > 0.6) are reported in the Supplementary Note and Supplementary Table 4. The majority of genome-wide significant SNPs were intronic and unlikely to be deleterious, or part of a regulatory element. However, the SNPs in the locus at chromosome 6 were associated with increased expression of two neighboring genes in blood cells (PHF10, lowest P = 3.65 × 10-13; C6orf120, lowest P = 3.81 × 10-13). One SNP (rs113851554) in the MEIS1 locus showed evidence (P = 1.08 × 10-6, FDR < 0.05) to act as a cis-methylation quantitative trait locus (meQTL). Credible set analysis (Online Methods) of the SNPs in the MEIS1 locus identified 2 variants (rs113851554 and rs182588061) within the 99% confidence set that are plausibly the causal variants (Supplementary Table 5). When incorporating functional annotation, rs113851554 accounted for the full posterior probability, suggesting that the most strongly associated SNP in the MEIS1 locus is also the most likely causal SNP.
SNP heritability
SNP-based heritability was estimated at 0.09 (SE=0.0082) by LD score regression33 (LDSC) and at 0.11 (SE = 0.0093) by BOLT-REML34 (BR) analysis. The sex difference was small with estimates at 0.12 (SE=0.018; LDSC) and 0.11 (SE = 0.02; BR) in males, versus 0.08 (SE = 0.014; LDSC) and 0.09 (SE = 0.02; BR) in females. The quantile-quantile (Q-Q) plots of all SNPs exhibited only mild inflation (λALL = 1.11; λM = 1.06; λF = 1.05; Supplementary Fig. 8) as is expected for a polygenic trait using the current sample size. The intercepts estimated by LDSC of 1.00, 0.99, and 1.00 for sex-combined, males and females respectively, suggested that this mild inflation was unlikely to be due to population stratification.
Gene-based results
A genome-wide gene-association study (GWGAS) as implemented in MAGMA35 (Online Methods) on all individuals identified three genes: MEIS1 (also implicated by the GWAS), DCBLD1 and MED27. Sex-specific GWGAS identified two additional genes (HHEX and RHCG) for males, and two additional genes (IPO7 and TSNARE1) for females (Fig. 1d/e/f, Table 2, Supplementary Table 6). Some of these genes have been associated before with other phenotypes such as diabetes and schizophrenia (Supplementary Table 7; Supplementary Note section 3.1). The top gene MEIS1 encodes a homeobox protein that acts as transcriptional regulator and activator, and it is thought to be important for normal development36. The highest expression levels are found in the female internal reproductive organs, but it is expressed in many other tissues as well, including the brain37. HHEX and MED27 are involved in the regulation of transcription as well. TSNARE1 and SCFD2 (implicated in the GWAS) play a role exocytosis (Supplementary Note, Supplementary Tables 8, 9, Supplementary Figs. 9-12).
Table 2. Genome-wide significant genes associated with insomnia complaints in a GWGAS including 113,006 individuals and the sex-specific GWGASes.
| FULL |
MALE |
FEMALE |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene Symbol | Entrez ID | Chr | Start BPa | Stop BPa | N SNPs | P | P corr | N SNPs | P | P corr | N SNPs | P | P corr |
| DCBLD1 | 285761 | 6 | 117,801,803 | 117,892,021 | 384 | 4.54E-07 | 0.0083 | 385 | 0.0856 | 1 | 386 | 7.12E-06 | 0.131 |
| MEIS1 | 4211 | 2 | 66,660,257 | 66,800,891 | 426 | 6.60E-07 | 0.0121** | 424 | 7.56E-03 | 1 | 428 | 3.58E-04 | 1 |
| MED27 | 9442 | 9 | 134,734,497 | 134,957,274 | 608 | 7.81E-07 | 0.0143* | 608 | 9.56E-03 | 1 | 603 | 5.41E-04 | 1 |
| HHEX | 3087 | 10 | 94,447,681 | 94,456,408 | 19 | 2.49E-03 | 1 | 19 | 1.71E-06 | 0.031** | 19 | 0.5832 | 1 |
| RHCG | 51458 | 15 | 90,013,638 | 90,041,799 | 35 | 1.15E-04 | 1 | 34 | 2.19E-06 | 0.040** | 35 | 0.7115 | 1 |
| IPO7 | 10527 | 11 | 9,404,169 | 9,470,674 | 241 | 7.80E-04 | 1 | 234 | 0.7839 | 1 | 245 | 1.67E-06 | 0.031** |
| TSNARE1 | 203062 | 8 | 143,292,441 | 143,486,543 | 1083 | 0.029503 | 1 | 1074 | 0.9498 | 1 | 1079 | 1.76E-06 | 0.032** |
Gene-based P values are reported for all genes significant after Bonferroni correction (α = 2.72E-06) in at least one of the three analyses (full, males, females). Bold P values are significantly associated with insomnia. Gene association results in the deCODE sample and the meta-analysis are reported in Supplementary Tables 12 and 14. GWGAS, genome-wide gene-association study; Chr, chromosome; BP, base pair; P corr, P corrected for multiple testing.
reported on GRCh37, Including a window of 2,1 kb.
Genome-wide significant in in the combined analysis of UK Biobank and deCODE.
Stronger association signal in the combined analysis of UK Biobank and deCODE.
Joint analysis with independent sample
To examine the robustness of the 3 loci and 7 genes that reached genome-wide significance in the primary analyses, we tested their associations with a well-defined insomnia phenotype in the deCODE sample of n=7,565 (Online Methods, Supplementary Note, Supplementary Tables 10-12), and meta-analyzed these results with the UK Biobank association results, while adhering to the GWAS significance threshold of 5 × 10-8 (Online Methods, Supplementary Tables 13 and 14). The probability to replicate significant SNPs in the deCODE sample was low due to the difference in sample size (Supplementary Note section 4 and Supplementary Table 15), whereas a meta-analysis takes into account standard errors (i.e. sample size) of the observed effects, and allows to evaluate whether discovery P values increase (suggesting no replication) or decrease (supporting a similar effect in the added sample)38. Effects of 11 out of 12 SNPs from the full (both sexes combined) GWAS and all five SNPs from the female GWAS were sign concordant while the three SNPs from the GWAS in males were not. Only SNPs in the MEIS1 locus showed a stronger association signal (six SNPs in the full, and two SNPs in the females GWAS). Six out of seven genes detected in the GWGAS were significant at the genome-wide threshold of 5 × 10-8 in the meta-analysis. The signal for MEIS1, as well as for the four genes associated in the sex-specific analyses, showed a stronger association, and remained below the genome-wide gene-based threshold of association.
The role of MEIS1 in insomnia complaints and RLS
Most strongly associated with insomnia complaints was MEIS1. Winkelmann and colleagues39,40 previously reported an association of multiple SNPs in MEIS1 with RLS, and two of our top SNPs have been associated before with clinically diagnosed RLS patients in a sequencing41 and gene expression study42 of MEIS1. RLS is a prevalent disorder characterized by the urge to move the legs, a symptom which has been suggested to involve deficiencies in the dopaminergic system, while arousals during sleep, which is present in some RLS patients, is related to the glutamate/GABA balance43. The latter has also been implicated in hyperarousal in insomnia44. RLS and insomnia have some form of agitation or restlessness in common: expressed primarily in the cognitive domain in insomnia, and in the sensorimotor domain in RLS. Given the possible phenotypic overlap and reported genetic associations in MEIS1 for RLS, we investigated the mechanisms of action that can explain the shared signal in MEIS1 for insomnia complaints and RLS.
First, we investigated whether the observed associations of MEIS1 with insomnia complaints and RLS are independent. At least two signals in MEIS1 are associated with RLS: one including common SNPs reported in39,40, and a second signal of low-frequency SNPs reported by41,42. The Winkelmann RLS-associated SNPs in MEIS1 are not genome-wide significant in our insomnia complaints GWAS, and are in low LD with our associated SNPs (Supplementary Table 16). Conditioning our top SNP rs113851554 on these SNPs (Online Methods) showed that our top SNP has an effect (P = 2.90 × 10-13) independent from those RLS SNPs in MEIS1. We did not condition on the two SNPs of the second RLS signal in MEIS1 (our top SNP rs11385155442 and rs1169322141) as those SNPs were part of our top findings for insomnia complaints (Table 1).
Second, we applied BUHMBOX45, which provides information on the likelihood that a heterogeneous RLS subgroup exists within our sample that could explain our association results (Online Methods). After establishing sufficient power (0.82 reported by BUHMBOX power calculator based on sample size, RLS SNP effect sizes, and RLS prevalence, Online Methods) to detect heterogeneity when defining RLS genetic structure by the six RLS-associated loci reported in the RLS GWAS40 (Supplementary Table 17), we found no evidence that an RLS subgroup solely drives the reported associations, both when excluding the MEIS1 locus (P = 0.33), and when including this locus (P = 0.36). However, adding our top SNP in MEIS1 for insomnia complaints which was also previously associated with RLS42 (P = 4.80 × 10-12), we did find excessive positive correlation for the RLS loci in a subgroup of the individuals reporting insomnia (P = 0.029), yet this was driven by this single SNP. Note that because of a lack of power (due to the small RLS sample size and smaller effect sizes of the insomnia loci), we could not test the reverse hypothesis that an insomnia subgroup drives the RLS associations.
Third, we performed a genetic risk score analysis (Online Methods) to interpret the BUHMBOX results. This yielded a significant association between insomnia complaints and the RLS associated loci when including the same RLS loci as tested in BUHMBOX (Supplementary Table 17), namely; i) five RLS loci excluding the MEIS1 locus (P = 7.28 × 10-3); ii) six RLS loci including the MEIS1 association of the RLS GWAS (P = 6.23 × 10-4); and iii) six RLS loci including the MEIS1 RLS-associated SNP that was the insomnia complaints top hit as well (P = 5.30 × 10-13). As in the BUHMBOX analysis, including the insomnia complaints top hit strongly increased the association. The results of the BUHMBOX and genetic risk score analyses together are compatible with pleiotropy, but phenotypic overlap between RLS and insomnia complaints might contribute to the association found in the MEIS1 locus.
Fourth, we investigated possible confounding of RLS and insomnia at MEIS1 using conditional phenotypic analysis in data of the Course of Restless Legs Syndrome study (COR; included in40) and the Dortmund Health Study (DHS), containing information on both RLS and insomnia complaints (Online Methods, Supplementary Note). The combined COR+DHS sample included 1,985 individuals with quality-controlled genotypes (Supplementary Table 18). We note that this sample has strong ascertainment biases due to COR (53% of the combined sample) consisting only of patients who are relatively old (65 years on average) and, as members of RLS support groups, tend to have severe RLS. The resulting biases are i) oversampling of insomnia evoked by severe RLS because all individuals with insomnia complaints in COR necessarily have RLS, and ii) overrepresentation of RLS comorbid with insomnia because insomnia increases with age and people seeking help for RLS may be more likely to have comorbid insomnia. Keeping this in mind, MEIS1 was found to be strongly associated with insomnia complaints, supporting our initial finding, and with RLS as expected (Supplementary Tables 19 and 20). Conditioning insomnia complaints on RLS and vice versa reduced the association signals, indicating that phenotypic overlap contributes to the associations of both phenotypes. However, this reduction, which was complete when conditioning insomnia complaints on RLS but incomplete when conditioning RLS on insomnia complaints, cannot exclude pleiotropy of MEIS1 because of the strong ascertainment biases. Up to 83% of all insomnia cases in COR+DHS may belong to the subgroup that is evoked by severe RLS or comorbid with RLS (Supplementary Table 18), explaining why an effect of MEIS1 on insomnia was not visible after conditioning on RLS.
Fifth, we predicted P values for insomnia complaints in the UK Biobank sample under the assumption that RLS alone drives the association. To this end we calculated the expected proportions of RLS individuals in the UK Biobank insomnia cases and controls, using data on age-specific RLS prevalence46, on RLS sensitivity and specificity of the UK biobank question as derived from the NSR and Dortmund Health Study (DHS) sample, and on the reported rs113851554 effect size in RLS cases and controls42 (Supplementary Note). A χ2 test resulted in a predicted association P value of 2 × 10-4 (95% CI 0.056-1.1 × 10-11; based on sampling variances; Supplementary Note). This predicted P value under the assumption that the complete signal was driven by an RLS subgroup was much weaker than the actual P value we observed (2.14 × 10-18), and the latter was outside of the 95% CI of the predicted P value. This finding thus supports the notion that the effect of MEIS1 on insomnia complaints can at most be explained in part by an RLS contamination of the UK Biobank insomnia cases.
Finally, we conducted sign concordance and low P value enrichment tests (Online Methods) on the summary statistics of insomnia complaints and RLS (unfortunately the sample of RLS GWAS40 was insufficient to obtain a reliable estimate of genetic correlation). 78% of the independent top SNPs (P = 1 × 10-4) of insomnia complaints were sign-concordant in RLS, while 83% of the top SNPs of RLS were discordant (Supplementary Table 21). The top signals of both studies showed little overlap (Supplementary Tables 22 and 23 and Supplementary Fig. 13). This suggests that besides pleiotropy of some loci there are genetic factors specific to each of the two disorders.
Taken together, the above results suggest that phenotypic overlap between RLS and insomnia can drive some, but not all of the MEIS1 insomnia association. This effect on the association likely has the same effect vice versa (i.e. insomnia complaints confounding RLS). Hence, we conclude that MEIS1 is likely to have pleiotropic effects on both RLS and insomnia.
Overlap with sleep-related phenotypes
Multiple sleep related phenotypes are present in the UK Biobank and multiple loci have been identified for sleep duration24,26, chronotype25,26, and excessive daytime sleepiness24. We performed GWASes on six additional sleep phenotypes in UK Biobank (Supplementary Fig. 14, Supplementary Table 24) and investigated the genetic and phenotypic correlations with insomnia complaints (Supplementary Note). Phenotypically, individuals reporting insomnia complaints have shorter sleep duration, have more trouble getting up, and have more unintentional dozing, but do not show a systematically different chronotype. Furthermore, insomnia complaints showed a significant positive genetic correlation with daytime dozing/sleeping (rg = 0.51, P = 3.25 × 10-4) and napping during the day (rg = 0.42, P = 3.95 × 10-6), and a negative genetic correlation with sleep duration (rg = 0.47, P = 1.97 × 10-16; Supplementary Table 25). The loci identified for insomnia complaints showed no significant association with the six additional sleep phenotypes (Supplementary Table 26). In addition, we investigated possible confounding of other psychiatric, metabolic and social economic traits (Online Methods, Supplementary Note). Adjustment of the significant SNP associations with insomnia complaints by these traits did not show confounding effects (Supplementary Table 27).
Sex-differences in genetic associations
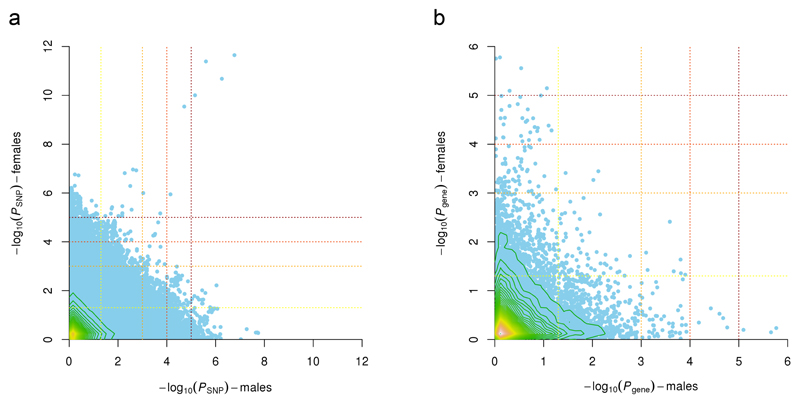
Females have a higher predisposition for insomnia2 than males, which might result from sex-related differences in the genetic architecture. The genetic correlation between sexes was estimated at 0.79 (SE = 0.13) which was just significantly smaller than 1 (one-sided Wald test P = 0.045). This estimate is comparable to e.g. waist circumference for which between-sex genetic heterogeneity is expected, in contrast to height and BMI where no heterogeneity is found47. In keeping with these overall differences, the significant SNP- and gene-based association results also differed between the sexes except for MEIS1 (Fig. 2). Adding sex as an interaction term to the GWAS on the full sample (Online Methods) did not result in genome-wide significant interactions (Supplementary Fig. 15), yet this may also be due to low statistical power for interaction analyses. Sign concordance and low P value enrichment tests of sex-specific results showed little evidence for overlap in the top signals (Online methods, Fig. 2, Supplementary Tables 28-30), suggesting that sex has a role in evoking specific genetic risk factors of insomnia. Whether X-chromosomal loci or sex-specific imprinting48,49 play a role, still needs to be determined. Our finding is in line with sex differences across most sleep variables50, including subjective sleep complaints51, the prevalence2 and heritability12 of insomnia, and physiological signatures of sleep both in the general population52 and within the population that has insomnia53.
Figure 2. Comparison of association results for insomnia complaints in males and females.
(a) SNP and (b) gene associations with insomnia complaints in males plotted against females. Contour lines indicate the density of the data in that region. The lines are colored from green to yellow, indicating increasing data density. Dotted lines indicate the P value thresholds used in the Low P value enrichment tests; from yellow to red P = 0.05, P > 1 × 10-3, P = 1 × 10-4, and P = 1 × 10-5 (note that all SNPs present in both GWASes are displayed, while the enrichment tests were performed on pruned data).
Functional networks
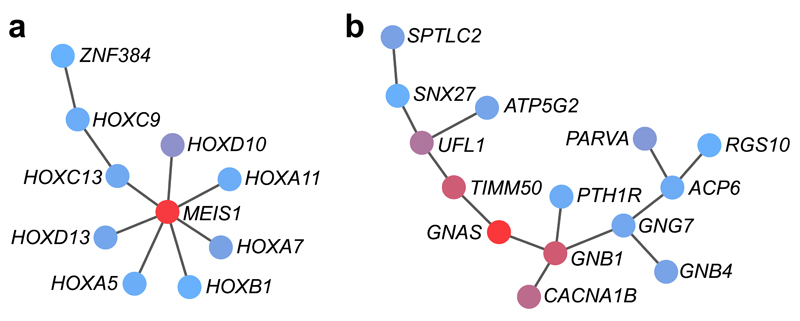
We applied the heat diffusion algorithm HotNet254 (Online Methods) to investigate protein-protein interaction networks enriched with genes most strongly related to insomnia complaints (P < 0.1) in the full and sex-specific GWGASes (Supplementary Note, Supplementary Table 31, Supplementary Figs. 16 and 17). For each input gene, HotNet2 processes a heat diffusion algorithm on the protein-protein interaction network to define a local neighborhood of ‘influence’, followed by a two-stage multiple hypotheses test to identify recurrent subnetworks. As input for HotNet2, we selected genes with P < 0.1 from the GWGAS, thereby considering crosstalk across pathways and network topology. In total, we observed twelve subnetworks of genes for males (P = 0.01, δ = 0.012, k = 7, expected = 8.24) and nine subnetworks for females (P = 0.02, δ = 0.014, k = 7, expected = 5.61; Supplementary Fig. 18). These subnetworks significantly overlapped with known pathways, mostly involved in transcription (Online Methods, Supplementary Table 32). In females, one subnetwork involved MEIS1 (Fig. 3a) together with multiple homeobox genes, encoding a family of transcription factors important for development. Other subnetworks presented candidate genes at ‘hotspots’ that were not detected by GWGAS alone, among them GNAS in the largest of the subnetworks in females (Fig. 3b). GNAS is an imprinted gene that is expressed from the maternal chromosome. It has metabolic functions and modulates REM and NREM sleep states49. This is especially interesting given that stronger maternal transmission21, hypermetabolism55 and instability of those sleep states56 are all characteristic of insomnia. Future studies have to confirm the involvement of the identified subnetworks in insomnia.
Figure 3. Protein-protein-interaction subnetworks identified by the heat diffusion algorithm HotNet2.
The genes most strongly related to insomnia (P < 0.1) in the full and sex-specific GWGASes were used as input to investigate the enrichment of protein-protein interaction networks. Each node (protein) is assigned a score based on the gene P value of the GWGAS. The scores, denoted as “heat” in HotNet2, diffuse across the edges of the network. We report subnetworks that include genes with a wide range of heat scores: red colored nodes send and receive a significant amount of heat, while blue colored nodes do not. (a) Subnetwork identified for females including MEIS1. (b) Subnetwork identified for females including GNAS. Other identified subnetworks for females and males are shown in Supplementary Figure 18.
Genetic overlap with other traits
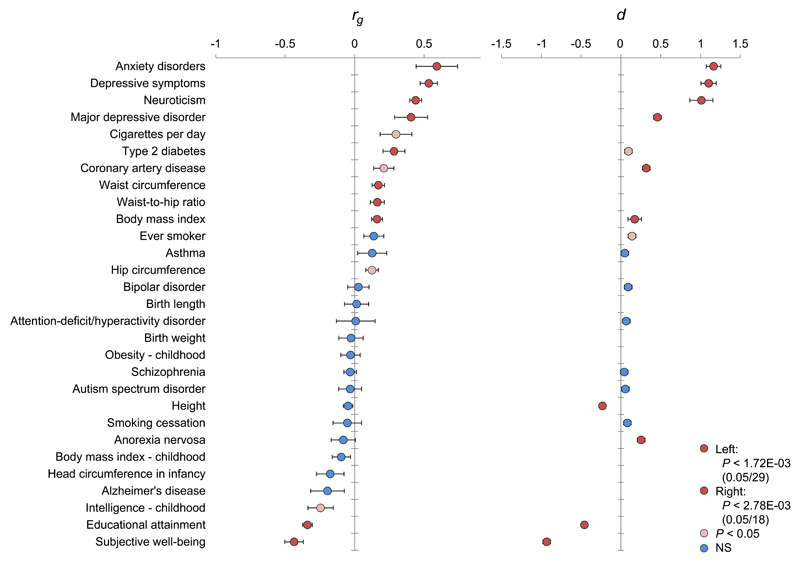
Insomnia implies an increased risk for major health problems, notably in the domains of cardiovascular diseases5–9, obesity11, and psychiatric disorders like depression4. We assessed the genetic correlation of insomnia by whole-genome LD score regression33 against 29 traits from these domains and additional anthropometric and life style traits (Online Methods). Significant genetic correlations (conservatively adjusted for multiple testing: P < 1.72 × 10-3 [= 0.05/29]) were observed between insomnia complaints and ten other traits (Fig. 4, left panel; Supplementary Table 33). Strong positive genetic correlations were observed with anxiety (rg = 0.59, P = 7.14 × 10-5), depressive symptoms (rg = 0.53, P = 1.03 × 10-17), neuroticism (rg = 0.44, P = 1.20 × 10-25), and Major Depressive Disorder (rg = 0.41, P = 6.50 × 10-4). Other positive yet weaker genetic correlations were observed with metabolic traits, i.e. Type II Diabetes, waist circumference, waist-to-hip ratio, and body mass index. Strong but negative genetic correlations were observed with subjective well-being (rg = -0.44, P = 5.64 × 10-11) and educational attainment (rg = -0.34, P = 1.81 × 10-22). Of the 29 traits, 18 have also been assessed in the NSR, allowing to investigate the phenotypic differences of these phenotypes in insomnia cases versus controls. We found that the profile of the magnitudes (d) of phenotypic group differences (Online Methods) was strikingly similar to the profile of genetic correlations (rank correlation 0.82; Fig. 4 right panel and Supplementary Table 34), providing further evidence for a link between the above mentioned traits and insomnia.
Figure 4. Genetic and phenotypic overlap between insomnia complaints and other traits and disorders.
Left bar chart: Genetic correlations (rg) between the frequency of experiencing trouble falling asleep or waking up in the middle of the night and various other traits and diseases. LD Score regression tested genome-wide SNP associations for these insomnia complaints against similar data for 29 other anthropometric and cardiovascular traits and neuropsychiatric outcomes (Supplementary Table 33). Error bars represent standard errors on these estimates. Red bars represent the traits that showed a significant genetic correlation after correction for multiple testing (P < 1.72 × 10−3), pink bars the traits that showed nominal association (P < 0.05), and blue bars the traits that did not show a significant genetic association. Right bar chart: The genetic correlations profile was strikingly similar to phenotypic overlap of insomnia with the same subject-characteristics assessed in an independent sample. Of the 29 disorders, traits and characteristics, 18 had been assessed in the NSR as well. Group differences between the 1,073 individuals without insomnia complaints and the 845 likely to have ID were evaluated using t-tests or χ2-tests (Supplementary Table 34). The profile of the magnitudes (d) of phenotypic group differences strongly resembled the genetic correlations profile.
Discussion
We conducted a large scale GWAS and GWGAS on insomnia complaints, using a measure that reliably discriminates ID from non-affected controls. We identified five novel genes (MEIS1, HHEX, RHCG, IPO7 and TSNARE1) and one locus (including MEIS1) that are associated with insomnia complaints and that were supported by joint analysis with an independent sample. These findings partly (i.e. two genes) overlapped with those reported in a recently published GWAS24 using a slightly different operationalization of the phenotype, which was less discriminative of clinical ID cases versus controls. Our top association was with MEIS1, which was also found in24, and has previously been implicated in RLS. Our extensive analyses now show that both residual phenotypic overlap and pleiotropy are relevant for the involvement of MEIS1 in insomnia as well as RLS.
We also provided evidence of sex-specific genetic effects and showed genetic overlap with several psychiatric and metabolic disorders. These findings provide starting points for subsequent functional analyses to unravel the molecular neurobiological mechanisms underlying the vulnerability to ID.
Online Methods
UK Biobank sample
We used data provided by the UK Biobank Study28 (see URLs). UK Biobank is a major national health resource including >500,000 participants, with the aim of improving the prevention, diagnosis and treatment of a wide range of serious and life-threatening illnesses – including cancer, heart diseases, stroke, diabetes, arthritis, osteoporosis, eye disorders, depression and forms of dementia. All participants provided written informed consent; the UK Biobank received ethical approval from the National Research Ethics Service Committee North West–Haydock (reference 11/NW/0382), and all study procedures were performed in accordance with the World Medical Association Declaration of Helsinki ethical principles for medical research. The current study was conducted under the UK Biobank application number 16406.
The study design of the UK Biobank has been described elsewhere28,57. Briefly, in 2006-2010 about 9.2 million invitation letters to participate in the study were sent to all people aged 40-69 years who were registered with the National Health Service and living up to ~25 miles from one of the 22 study assessment centers. A total of 503,325 participants were recruited into the study28. Apart from registry based phenotypic information, extensive self-reported baseline data were collected by questionnaire, in addition to anthropometric assessments, and DNA collection. For the present study we focused on insomnia, which was measured as experiencing trouble falling asleep or waking up in the middle of the night (Supplementary Note section 1.1).
SNP analysis of the UK Biobank sample
We used imputed genetic data from UK Biobank (May 2015 release) including ~73 million genetic variants in 152,249 individuals. Details on the data are provided elsewhere (see URLs). In summary, the first ~50,000 samples were genotyped on the UK BiLEVE custom array, and the remaining ~100,000 samples were genotypes on the UK Biobank Axiom array. After standard quality control of the SNPs and samples which was centrally performed by UK Biobank, the dataset comprised 641,018 autosomal SNPs in 152,256 samples for phasing and imputation. Imputation was performed with a reference panel that included the UK10K haplotype panel and the 1000 Genomes Project Phase 3 reference panel.
For the analyses in our study, we selected Caucasians only. After removal of related individuals, discordant sex, withdrawn consent, and missing phenotype data, 113,006 individuals remained for analysis (Supplementary Table 1). This is the largest available GWAS sample for insomnia to date. Previous power analyses showed that a samples size of n = 40,000 already allows for high power (>90%) detection of SNPs with small effect sizes explaining only 0.1% of the variance58, indicating sufficient statistical power of detecting SNPs associated with insomnia in our sample.
Association tests were performed in SNPTEST59 (see URLs), using a logistic regression including the covariates age, sex (for the full sample), genotyping array, and the top five genetically determined principal components and additional ones out of ten extra that were associated with the phenotype (tested by logistic regression). SNPs with imputation quality <0.8 (based on the total sample, and on the Caucasians only) and MAF<0.001 were excluded after the association analysis, resulting in 12,444,916 SNPs, 12,428,592 SNPs, and 12,432,937 SNPs for the full, male and female analyses respectively.
Gene analysis
We used all 19,427 protein-coding genes from the NCBI 37.3 gene definitions as basis for a genome-wide gene-association study (GWGAS) in MAGMA35 (see URLs). We annotated all SNPs of our association analysis to these genes, resulting in 18,355 genes that were covered by at least one SNP. We included a window around the genes of 2 kb before transcription start site and 1 kb after transcription stop site. Gene-association test were performed taking LD between SNPs into account. We applied a stringent Bonferroni correction to account for multiple testing.
A GWGAS can identify genes in which multiple genetic variants show a mild effect that is not sufficiently strong to be picked up by GWAS. On the other hand, while a GWAS analysis can indicate a significant locus including a gene, it is possible that this gene is not identified in the GWGAS because the genes can harbor many more SNPs that did not show an association signal, and the GWGAS takes all SNPs within the gene into account.
SNP analysis of the deCODE sample
The Icelandic GWAS dataset used in the current study is based on whole genome sequencing (WGS), chip genotyping and long range phasing of Icelandic population samples60. In brief, we whole genome sequenced 15,220 Icelanders using Illumina technology (Illumina, San Diego, CA, USA) to an average depth of at least 34X, resulting in the identification of some 94 million variants. Using imputation assisted by long-range haplotype phasing61 and after removing variants with imputation information content below 0.8 as well as with an imputed minor allele frequency below 0.01%, we successfully inferred the genotypes of 32,463,443 variants in 434,571 Icelanders, of whom 151,677 had been genotyped using the Illumina chip genotyping platform. The remaining 282,894 Icelanders are first- and second degree relatives of the chip-typed individuals and are imputed by aid of genealogic information. Of the 3,774 cases and 3,791 controls used in this study, 3,671 and 3,697 were directly genotyped, respectively.
Logistic regression was used to test for associations between variants and insomnia, assuming a multiplicative model, treating disease status as the response and expected genotype counts from imputation as covariates. For the Icelandic cohort this was done using software developed at deCODE genetics60. Testing was performed using the likelihood ratio statistic and population stratification was adjusted for by including county of birth as covariates.
To account for inflation in test statistics due to cryptic relatedness and stratification within the case and control sample sets, we applied the method of LD score regression33. With a set of 1.1 million variants we regressed the χ2 statistics from our GWAS scan against LD score and used the intercept as a correction factor. The estimated correction factors were 1.059, 1.025 and 1.036 for the analysis including all, males only and females only, respectively. All P values were adjusted using these correction factors.
Genotyping and association analyses of the ‘Course of Restless Legs Syndrome’ and Dortmund Health Study samples
1,051 Dortmund Health Study (DHS) participants were genotyped using the Illumina HumanOmni chip 2.5-4v1 and the GenomeStudio Genotype module. 1,057 ‘Course of Restless Legs Syndrome’ (COR) study participants were genotyped using the Affymetrix Axiom CEU array and the Axiom GT1 algorithm. Genome-wide imputation of autosomal SNPs according to phase 3 of the 1000 Genomes Project was performed at the Sanger Imputation Service (see URLs). Quality control before imputation removed variants with genotyping rate of less than 95%, minor allele frequency of less than 10 × 10-3, or strong deviation from Hardy-Weinberg equilibrium (P < 10 × 10-8), excluded individuals with sex-mismatch or genotyping rate of less than 95%, and finally selected a maximal set of unrelated individual (Pi hat > 0.16). Variants were recoded before imputation according to human genome build 37 information on position, strand orientation, and major alleles, as reported by genotyping chip strand files (see URLs) and dbSNP (see URLs). Recoded datasets were merged by PLINK software62 (see URLs) and subjected to multidimensional scaling (MDS) on 10 dimensions which involved outlier detection (>4 SD from the population mean) and provided covariates besides age and sex for the association analyses. For the latter, the imputed genotypes were merged, yielding data on 1,985 individuals after quality control and outlier removal. For 1,772 of them, the three-level insomnia severity score was available. In case of individual SNPs, association analysis applied an additive model and linear or logistic regression as implemented in PLINK. For the gene analysis, MAGMA was used with the same phenotypes and covariates, using the same method in MAGMA as the GWGAS.
Credible set analysis
For the MEIS1 locus, we defined a credible set of SNPs that could plausibly be considered as causal using PAINTOR (Probabilistic Annotation INTegratOR)63. PAINTOR uses a multivariate normal approximation to connect the LD structure of the SNPs in the locus to the P value. Data on functional annotation is integrated through an empirical Bayes prior, which results in a prior probability of a variant to be causal that is governed by its score on the functional classes. The SNPs scoring high on certain functional annotations are up-weighted, while the SNPs scoring low on a certain functional annotation are down-weighted. We included functional annotations reported in Supplementary Note section 2.1 that contained information to prioritize the SNPs in the MEIS1 locus: deleteriousness (continuous score), regulatory function (continuous score), meQTL (yes/no), and mean chromatin state over different tissues (continuous score).
Conditional analyses
We performed 2 types of conditional analyses for our SNPs associated with insomnia complaints: i) conditioning our top SNP rs113851554 in MEIS1 upon three SNPs (rs6710341, rs12469063 and rs2300478) representing the association signals detected in RLS GWASes39,40; ii) conditioning all SNPs significantly associated with insomnia complaints upon other traits and characteristics related to insomnia that were available in the UK Biobank study: waist-to-hip ratio, body mass index (BMI), Townsend deprivation index, years of education, depressive symptoms and neuroticism. Analyses were performed in SNPTEST59 using the logistic regression model including covariates as described in the SNP analysis section. Additive effects of the SNPs in i) were added to the regression model with the -condition on flag. The phenotypes in ii) we added to the other covariates with the -cov_names flag.
Genotype × sex interaction analysis
To investigate possible sex effects on insomnia complaints, we performed an association analysis adding sex as an interaction term in the original insomnia complaints GWAS using PLINK62 (--linear interaction). We analyzed the same SNPs included in the main GWAS of this study and included the same covariates (sex, age, array, principal components).
BUHMBOX
We applied Breaking Up Heterogeneous Mixture Based On Cross-locus correlations (BUHMBOX45; see URLs) to test if a heterogeneous subgroup that shows genetic characteristics of RLS is present in our UK Biobank insomnia sample, that should otherwise be homogeneous (i.e. is the sharing of risk alleles by insomnia and RLS driven by all individuals or a subset of individuals). We used the top SNPs of the six associated loci with RLS40 with their risk alleles and allele frequencies (Supplementary Table 17) to define the genetic architecture of RLS. We first ran the BUHMBOX power calculator including 1000 simulations experiments including the UK Biobank sample size, risk allele frequencies and odds ratio’s of the RLS SNPs (Supplementary Table 17), and proportion of expected RLS patients in the insomnia complaints group (0.107). BUHMBOX tests if the RLS risk alleles have higher allele frequencies only in a subset of insomnia cases (when pleiotropy would exist, the RLS risk alleles are expected to have higher allele frequencies across the total sample of insomnia cases). If the RLS risk alleles are enriched in one subgroup of insomnia cases, the expected correlations of number of risk alleles between the loci will be consistently positive. The pairwise correlations are combined in one statistic to test for excessive positive correlations.
Genetic risk score analysis
We used the top SNPs of the six associated loci with RLS (same as for BUHMBOX) as input for the genetic risk score analysis. For each individual, we calculated the GRS by summing the risk allele dosage (0, 1 or 2) multiplied by the effect size (log[OR]) of the six top SNPs. An association analysis was performed between this genetic risk score and the UK Biobank insomnia phenotype using a logistic regression including the genetic principal components as covariates.
Sign concordance test
As input for the sign concordance test, we used independent SNPs that we defined by pruning the data with PLINK62 (--indep-pairwise 1000 100 0.1; see URLs). For the analysis with the RLS and insomnia complaints data, we first removed all SNPs with the allele combinations A/T and C/G to exclude strand ambiguity. In addition, all SNPs with non-matching alleles were removed. Sign concordance between two datasets was tested by a two-sided binominal test for a probability of 0.5, for SNP selections below six different P value thresholds (1, 0.5, 0.05, 1 × 10-3, 1 × 10-4, 1 × 10-5).
Low P value enrichment tests
The pruned data used as input for the sign concordance test (see above) was used for the low P value enrichment test as well. Enrichment of low P values between two datasets was tested with a two-sided Fisher’s exact tests on the cross tabs of the SNPs below and over four different P value thresholds (0.05, 1 × 10-3, 1 × 10-4, 1 × 10-5). In addition, because the RLS and insomnia complaints summary statistics are from samples that substantially differ in size (which is influencing the P values), we performed the analysis for seven different ranked P value thresholds as well (50, 100, 200, 400, 800, 1600, 3200).
Meta-analysis
Meta-analysis of the SNPs in UK Biobank and deCODE was performed in METAL64 (see URLs). The analysis was based on P values, taking sample size and direction of effect into account. Meta-analysis of the genes in UK Biobank and deCODE was performed in MAGMA35 (see URLs), which uses the Stouffer’s weighted Z-transform method.
HotNet2 analysis
We applied the HotNet2 algorithm54 to identify networks of genes that are related to insomnia. HotNet2 is based on a heat diffusion model. The key advantage of HotNet2 compared to conventional methods is the possibility to detect genes in connected subnetworks with associations to the phenotype stronger than expected by chance. Conventional gene enrichment or gene set analyses are limited by the rigid “in or out” definition of a gene set which does not allow for crosstalk between pathways that are represented by different gene sets. To depict an entire network topology, conventional enrichment tools therefore need to define a large number of gene sets resulting in a loss of statistical power due to a high level of multiple testing.
As input for the HotNet2 analysis we selected all genes from our GWGAS results with a P value ≤ 0.1 (2335, 2101, and 2077 genes for full, female, and male analyses respectively). The -log10(P value) was defined as input gene score. HotNet2 was performed based on protein-protein interactions reported by iRefIndex65. For four delta thresholds (minimum edge weight) that were automatically chosen by HotNet2, the significance of N subnetworks at k (the minimum number of proteins in a subnetwork) were reported based on a 100 times permuted influence matrix.
Next, we performed an enrichment analysis of the identified subnetworks by calculating a P value for the fraction of genes that overlaps with pre-defined pathways using the hypergeometric test. We selected all canonical pathways (n = 1,330) and Gene Ontology (GO) pathways (n = 1,454) from the molecular signature database (MSigDB v5.166, see URLs). A pathway was considered statistically significant when the hypergeometric test showed P ≤ 0.05 after correcting for multiple testing using the Benjamini and Hochberg method.
Genetic correlations
Genetic correlations (rg) were calculated between i) insomnia complaints and six other sleep related phenotypes present in UK Biobank; ii) insomnia complaints in males and females; iii) insomnia complaints and 29 other traits for which summary statistics from GWAS were publicly available (Supplementary Table 33), using LD score regression33 (see URLs). We used pre-computed LD scores that were provided by LD score regression, which were calculated using the European panel of 1000 Genomes. No constraining of the intercept was applied. A conservative Bonferroni-corrected P value threshold of 1.72 × 10-3 was used in iii to define significant associations.
Phenotypic group differences between individuals with and without insomnia
For 18 disorders, traits and characteristics measures in the Netherlands Sleep Registry (NSR)29, group differences between 1,073 individuals without insomnia complaints and 845 likely to have ID were evaluated using t-tests (continuous phenotype) or χ2-tests (dichotomous phenotypes).
Supplementary Material
Acknowledgements
This work was funded by The Netherlands Organization for Scientific Research (NWO Brain & Cognition 433-09-228, NWO VICI 453-14-005 and 453-07-001, 645-000-003) and by the European Research Council (ERC-ADG-2014-671084 INSOMNIA). The analyses were carried out on the Genetic Cluster Computer, which is financed by the Netherlands Scientific Organization (NWO: 480-05-003), by the VU University, Amsterdam, The Netherlands, and by the Dutch Brain Foundation, and is hosted by the Dutch National Computing and Networking Services SurfSARA. This research has been conducted using the UK Biobank Resource under Application Number 16406. We thank the participants and researchers who collected and contributed to the data. We thank the participants of the Netherlands Sleep Registry for providing extensive phenotypic data. We thank participants who provided samples and data for the Icelandic study and our valued colleagues who contributed to data collection and phenotypic characterization of clinical samples, genotyping and analysis of genome sequences data. We also thank the EU-RLS consortium and the Cooperative Research in the Region of Augsburg (KORA) for providing the RLS summary statistics. KORA was initiated and financed by the Helmholtz Zentrum München, which is funded by the German Federal Ministry of Education and Research and by the State of Bavaria. The collection of sociodemographic and clinical data in the Dortmund Health Study (DHS) was supported by the German Migraine & Headache Society (DMKG) and by unrestricted grants of equal share from Almirall, Astra Zeneca, Berlin Chemie, Boehringer, Boots Health Care, Glaxo-Smith-Kline, Janssen Cilag, McNeil Pharma, MSD Sharp & Dohme and Pfizer to the University of Muenster. Blood collection in the Dortmund Health Study was done through funds from the Institute of Epidemiology and Social Medicine University of Muenster. Genotyping for the Human Omni Chip was supported by the German Ministry of Education and Research (BMBF, grant no. 01ER0816). Researchers interested in using DHS data are required to sign and follow the terms of a Cooperation Agreement that includes a number of clauses designed to ensure protection of privacy and compliance with relevant laws. The 'Course-of-Restless-Legs-Syndrome' (COR) Study was supported by unrestricted grants to the University of Muenster from the German Restless Legs Patient Organisation (RLS e.V. Deutsche Restless Legs Vereinigung), the Swiss RLS Patient Association (Schweizerische Restless Legs Selbsthilfegruppe), and from a consortium formed by Boeringer Ingelheim Pharma, Mundipharma Research, Neurobiotec, Roche Pharma, UCB (Germany + Switzerland) and Vifor Pharma. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Researchers interested in using COR data are required to sign and follow the terms of a Cooperation Agreement that includes a number of clauses designed to ensure protection of privacy and compliance with relevant laws. For further information on DHS and COR, contact K. Berger (bergerk@uni-muenster.de). Acknowledgements for data contributed by other consortia that are used for secondary analyses are presented in the Supplementary Note.
Footnotes
URLs. UK Biobank, www.ukbiobank.ac.uk; Netherlands Sleep Registry, http://www.sleepregistry.nl; UK Biobank genotyping and QC, http://biobank.ctsu.ox.ac.uk/crystal/refer.cgi?id=155580; SNPTEST, https://mathgen.stats.ox.ac.uk/genetics_software/snptest/snptest.html; MAGMA, http://ctg.cncr.nl/software/magma; Sanger Imputation Service, https://imputation.sanger.ac.uk/; Genotyping chip strand files, www.well.ox.ac.uk/~wrayner/strand/; dbSNP data, ftp://ftp.ncbi.nih.gov/snp/; PLINK, https://www.cog-genomics.org/plink2; BUHMBOX, http://software.broadinstitute.org/mpg/buhmbox/; METAL, http://genome.sph.umich.edu/wiki/METAL_Program; MSigDB, http://software.broadinstitute.org/gsea/msigdb/; LD score regression, https://github.com/bulik/ldsc.
Author Contributions: D.P. and E.J.W.V.S conceived the study. A.R.H., and D.P. performed the analyses. T.F.B., K.D., B.H.W.t.L, R.W. and E.J.W.V.S. recruited participants of the NSR and collected and analyzed data for the phenotypic validation. C.d.L., S.Sn., K.W., and E.T. performed secondary analyses. S.St. prepared the UK Biobank data for analyses and wrote a pipeline to facilitate efficient data processing. G.T. and I.J. performed the deCODE analyses. K.O. performed the COR and DHS analyses. A.R.H., K.O., E.J.W.V.S. and D.P. wrote the paper. All authors discussed the results and commented on the paper. K.O., E.J.W.V.S. and D.P. contributed equally to this work.
Competing Financial Interests: Authors affiliated with deCODE genetics / Amgen Inc. declare competing financial interests as employees. The other authors declare no competing financial interests.
Data Availability.
Summary statistics from our insomnia GWASes are available for download at http://ctg.cncr.nl/software/summary_statistics. The data generated in the secondary analyses of this study are included in this published article (Supplementary Tables). The genotype data analyzed during the current study was provided by the UK Biobank Study (see URLs), received under the UK Biobank application number 16406. The genotype data from deCODE, DHS and COR was obtained through the PIs of these studies.
References
- 1.Wittchen HU, et al. The size and burden of mental disorders and other disorders of the brain in Europe 2010. Eur Neuropsychopharmacol. 2011;21:655–679. doi: 10.1016/j.euroneuro.2011.07.018. [DOI] [PubMed] [Google Scholar]
- 2.Zhang B, Wing Y-K. Sex differences in insomnia: a meta-analysis. Sleep. 2006;29:85–93. doi: 10.1093/sleep/29.1.85. [DOI] [PubMed] [Google Scholar]
- 3.Morin CM, et al. Insomnia disorder. Nat Rev Dis Prim. 2015;1:15026. doi: 10.1038/nrdp.2015.26. [DOI] [PubMed] [Google Scholar]
- 4.Baglioni C, et al. Insomnia as a predictor of depression: A meta-analytic evaluation of longitudinal epidemiological studies. Journal of Affective Disorders. 2011;135:10–19. doi: 10.1016/j.jad.2011.01.011. [DOI] [PubMed] [Google Scholar]
- 5.Palagini L, et al. Sleep Loss and Hypertension: A Systematic Review. Curr Pharm Des. 2013;19:2409–2419. doi: 10.2174/1381612811319130009. [DOI] [PubMed] [Google Scholar]
- 6.Mallon L, Broman JE, Hetta J. Sleep complaints predict coronary artery disease mortality in males: A 12-year follow-up study of a middle-aged Swedish population. J Intern Med. 2002;251:207–216. doi: 10.1046/j.1365-2796.2002.00941.x. [DOI] [PubMed] [Google Scholar]
- 7.Nilsson PM, Nilsson JÅ, Hedblad B, Berglund G. Sleep disturbance in association with elevated pulse rate for prediction of mortality - Consequences of mental strain? J Intern Med. 2001;250:521–529. doi: 10.1046/j.1365-2796.2001.00913.x. [DOI] [PubMed] [Google Scholar]
- 8.Schwartz SW, et al. Are sleep complaints an independent risk factor for Myocardial infarction? Ann Epidemiol. 1998;8:384–392. doi: 10.1016/s1047-2797(97)00238-x. [DOI] [PubMed] [Google Scholar]
- 9.Clark A, Lange T, Hallqvist J, Jennum P, Rod NH. Sleep impairment and prognosis of acute myocardial infarction: a prospective cohort study. Sleep. 2014;37:851–8. doi: 10.5665/sleep.3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cappuccio FP, D’Elia L, Strazzullo P, Miller MA. Quantity and quality of sleep and incidence of type 2 diabetes: A systematic review and meta-analysis. Diabetes Care. 2010;33:414–420. doi: 10.2337/dc09-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hargens TA, Kaleth AS, Edwards ES, Butner KL. Association between sleep disorders, obesity, and exercise: A review. Nature and Science of Sleep. 2013;5:27–35. doi: 10.2147/NSS.S34838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lind MJ, Aggen SH, Kirkpatrick RM, Kendler KS, Amstater AB. A Longitudinal Twin Study of Insomnia Symptoms in Adults. Sleep. 2015;38:1423–1430. doi: 10.5665/sleep.4982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bootzin RR, Epstein DR. Understanding and Treating Insomnia. Annu Rev Clin Psychol. 2011;7:435–458. doi: 10.1146/annurev.clinpsy.3.022806.091516. [DOI] [PubMed] [Google Scholar]
- 14.Bettolo A. L’insonnia e sua importanza clinica: le sue cause predisponenti e determinanti, i suoi caratteri nelle diverse malattie, Ia sua importanza prognostica ed il suo trattamento generale e speciale. Studium. 1931;21:54–65. [Google Scholar]
- 15.Morin CM, et al. Cognitive behavioral therapy, singly and combined with medication, for persistent insomnia: a randomized controlled trial. JAMA. 2009;301:2005–2015. doi: 10.1001/jama.2009.682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harvey AG, Tang NKY. Cognitive behaviour therapy for primary insomnia: can we rest yet? Sleep Med Rev. 2003;7:237–262. doi: 10.1053/smrv.2002.0266. [DOI] [PubMed] [Google Scholar]
- 17.Morin CM, et al. The natural history of insomnia: A population-based 3-year longitudinal study. Arch Intern Med. 2009;169:447–53. doi: 10.1001/archinternmed.2008.610. [DOI] [PubMed] [Google Scholar]
- 18.Bastien CH, Morin CM. Familial incidence of insomnia. J Sleep Res. 2000;9:49–54. doi: 10.1046/j.1365-2869.2000.00182.x. [DOI] [PubMed] [Google Scholar]
- 19.Dauvilliers Y, et al. Family studies in insomnia. J Psychosom Res. 2005;58:271–278. doi: 10.1016/j.jpsychores.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 20.Beaulieu-Bonneau S, LeBlanc M, Mérette C, Dauvilliers Y, Morin CM. Family history of insomnia in a population-based sample. Sleep. 2007;30:1739–45. doi: 10.1093/sleep/30.12.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wing YK, et al. Familial aggregation and heritability of insomnia in a community-based study. Sleep Med. 2012;13:985–990. doi: 10.1016/j.sleep.2012.04.013. [DOI] [PubMed] [Google Scholar]
- 22.Byrne EM, et al. A genome-wide association study of sleep habits and insomnia. Am J Med Genet B Neuropsychiatr Genet. 2013;162B:439–51. doi: 10.1002/ajmg.b.32168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Amin N, et al. Genetic variants in RBFOX3 are associated with sleep latency. Eur J Hum Genet. 2016 doi: 10.1038/ejhg.2016.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lane JM, et al. Genome-wide association analyses of sleep disturbance traits identify new loci and highlight shared genetics with neuropsychiatric and metabolic traits. Nat Genet. 2017;49:274–281. doi: 10.1038/ng.3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lane JM, et al. Genome-wide association analysis identifies novel loci for chronotype in 100,420 individuals from the UK Biobank. Nat Commun. 2016;7:10889. doi: 10.1038/ncomms10889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jones SE, et al. Genome-wide association analyses in 128,266 individuals identifies new morningness and sleep duration loci. PLoS Genet. 2016;12:e1006125. doi: 10.1371/journal.pgen.1006125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vgontzas AN, et al. Persistent insomnia: the role of objective short sleep duration and mental health. Sleep. 2012;35:61–68. doi: 10.5665/sleep.1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sudlow C, et al. UK Biobank: An Open Access Resource for Identifying the Causes of a Wide Range of Complex Diseases of Middle and Old Age. PLoS Med. 2015;12:e1001779. doi: 10.1371/journal.pmed.1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Benjamins JS, et al. Insomnia heterogeneity: characteristics to consider for data-driven multivariate subtyping. Sleep Med Rev. 2016 doi: 10.1016/j.smrv.2016.10.005. [DOI] [PubMed] [Google Scholar]
- 30.Morphy H, Dunn KM, Lewis M, Boardman HF, Croft PR. Epidemiology of insomnia: a longitudinal study in a UK population. Sleep. 2007;30:274–280. [PubMed] [Google Scholar]
- 31.Paparrigopoulos T, et al. Insomnia and its correlates in a representative sample of the Greek population. BMC Public Health. 2010;10:531. doi: 10.1186/1471-2458-10-531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cho YW, et al. Epidemiology of Insomnia in Korean Adults: Prevalence and Associated Factors. J Clin Neurol. 2009;5:20–23. doi: 10.3988/jcn.2009.5.1.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bulik-Sullivan BK, et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47:291–295. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Loh P-R, et al. Contrasting genetic architectures of schizophrenia and other complex diseases using fast variance-components analysis. Nat Genet. 2015;47:1385–92. doi: 10.1038/ng.3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.de Leeuw CA, Mooij JM, Heskes T, Posthuma D. MAGMA: Generalized Gene-Set Analysis of GWAS Data. PLOS Comput Biol. 2015;11:e1004219. doi: 10.1371/journal.pcbi.1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cai M, et al. Dual actions of Meis1 inhibit erythroid progenitor development and sustain general hematopoietic cell proliferation. Blood. 2012;120:335–346. doi: 10.1182/blood-2012-01-403139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.GTEx Consortium. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Skol AD, Scott LJ, Abecasis GR, Boehnke M. Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat Genet. 2006;38:209–213. doi: 10.1038/ng1706. [DOI] [PubMed] [Google Scholar]
- 39.Winkelmann J, et al. Genome-wide association study of restless legs syndrome identifies common variants in three genomic regions. Nat Genet. 2007;39:1000–1006. doi: 10.1038/ng2099. [DOI] [PubMed] [Google Scholar]
- 40.Winkelmann J, et al. Genome-Wide association study identifies novel restless legs syndrome susceptibility loci on 2p14 and 16q12.1. PLoS Genet. 2011;7:e1002171. doi: 10.1371/journal.pgen.1002171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schulte EC, et al. Targeted resequencing and systematic in vivo functional testing identifies rare variants in MEIS1 as significant contributors to restless legs syndrome. Am J Hum Genet. 2014;95:85–95. doi: 10.1016/j.ajhg.2014.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xiong L, et al. MEIS1 intronic risk haplotype associated with restless legs syndrome affects its mRNA and protein expression levels. Hum Mol Genet. 2009;18:1065–1074. doi: 10.1093/hmg/ddn443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Allen RP, Barker PB, Horská A, Earley CJ. Thalamic glutamate/glutamine in restless legs syndrome: Increased and related to disturbed sleep. Neurology. 2013;80:2028–2034. doi: 10.1212/WNL.0b013e318294b3f6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Spiegelhalder K, et al. Magnetic Resonance Spectroscopy in Patients with Insomnia: A Repeated Measurement Study. PLoS One. 2016;11:e0156771. doi: 10.1371/journal.pone.0156771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Han B, et al. A method to decipher pleiotropy by detecting underlying heterogeneity driven by hidden subgroups applied to autoimmune and neuropsychiatric diseases. Nat Genet. 2016;48:803–810. doi: 10.1038/ng.3572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ohayon MM, O’Hara R, Vitiello MV. Epidemiology of restless legs syndrome: a synthesis of the literature. Sleep Med Rev. 2012;16:283–295. doi: 10.1016/j.smrv.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang J, et al. Genome-wide genetic homogeneity between sexes and populations for human height and body mass index. Hum Mol Genet. 2015;24:7445–7449. doi: 10.1093/hmg/ddv443. [DOI] [PubMed] [Google Scholar]
- 48.Tucci V. Genomic Imprinting: A New Epigenetic Perspective of Sleep Regulation. PLoS Genet. 2016;12:e1006004. doi: 10.1371/journal.pgen.1006004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lassi G, et al. Loss of Gnas imprinting differentially affects REM/NREM sleep and cognition in mice. PLoS Genet. 2012;8:e1002706. doi: 10.1371/journal.pgen.1002706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dijk D-J. Sleep of aging women and men: back to basics. Sleep. 2006;29:12–13. doi: 10.1093/sleep/29.1.12. [DOI] [PubMed] [Google Scholar]
- 51.Ursin R, Bjorvatn B, Holsten F. Sleep duration, subjective sleep need, and sleep habits of 40- to 45-year-olds in the Hordaland Health Study. Sleep. 2005;28:1260–1269. doi: 10.1093/sleep/28.10.1260. [DOI] [PubMed] [Google Scholar]
- 52.Redline S, et al. The effects of age, sex, ethnicity, and sleep-disordered breathing on sleep architecture. Arch Intern Med. 2004;164:406–418. doi: 10.1001/archinte.164.4.406. [DOI] [PubMed] [Google Scholar]
- 53.Buysse DJ, et al. EEG spectral analysis in primary insomnia: NREM period effects and sex differences. Sleep. 2008;31:1673–1682. doi: 10.1093/sleep/31.12.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Leiserson MDM, et al. Pan-cancer network analysis identifies combinations of rare somatic mutations across pathways and protein complexes. Nat Genet. 2014;47:106–114. doi: 10.1038/ng.3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bonnet MH, Arand DL. 24-Hour metabolic rate in insomniacs and matched normal sleepers. Sleep. 1995;18:581–588. doi: 10.1093/sleep/18.7.581. [DOI] [PubMed] [Google Scholar]
- 56.Feige B, et al. The microstructure of sleep in primary insomnia: An overview and extension. Int J Psychophysiol. 2013;89:171–180. doi: 10.1016/j.ijpsycho.2013.04.002. [DOI] [PubMed] [Google Scholar]
- 57.Allen NE, Sudlow C, Peakman T, Collins R. UK Biobank Data: Come and Get It. Sci Transl Med. 2014;6:224ed4. doi: 10.1126/scitranslmed.3008601. [DOI] [PubMed] [Google Scholar]
- 58.Visscher PM. Sizing up human height variation. Nat Genet. 2008;40:489–490. doi: 10.1038/ng0508-489. [DOI] [PubMed] [Google Scholar]
- 59.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 60.Gudbjartsson DF, et al. Large-scale whole-genome sequencing of the Icelandic population. Nat Genet. 2015;47:435–444. doi: 10.1038/ng.3247. [DOI] [PubMed] [Google Scholar]
- 61.Kong A, et al. Detection of sharing by descent, long-range phasing and haplotype imputation. Nat Genet. 2008;40:1068–1075. doi: 10.1038/ng.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kichaev G, et al. Integrating functional data to prioritize causal variants in statistical fine-mapping studies. PLoS Genet. 2014;10:e1004722. doi: 10.1371/journal.pgen.1004722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Willer CJ, Li Y, Abecasis GR. METAL: Fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Razick S, Magklaras G, Donaldson IM. iRefIndex: a consolidated protein interaction database with provenance. BMC Bioinformatics. 2008;9:405. doi: 10.1186/1471-2105-9-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.