Abstract
Monoubiquitylation of histone H2B (H2Bub1) is catalyzed mainly by the RNF20/RNF40 complex and erased by multiple deubiquitylating enzymes (DUBs). H2Bub1 influences many aspects of chromatin function, including transcription regulation and DNA repair. Cancer cells often display reduced levels of H2Bub1, and this reduction may contribute to cancer progression. The let-7 family of microRNAs comprises multiple members with reported tumor suppressive features, whose expression is frequently downregulated in cancer. We now report that let-7b and let-7c can positively regulate cellular H2Bub1 levels. Overexpression of let-7b and let-7c in a variety of non-transformed and cancer-derived cell lines results in H2Bub1 elevation. The positive effect of let-7b and let-7c on H2Bub1 levels is achieved through targeting of multiple mRNAs, coding for distinct components of the H2B deubiquitylation machinery. Specifically, let-7b and let-7c bind directly and inhibit the mRNAs encoding the DUBs USP42 and USP44, and also the mRNA encoding the adapter protein ATXN7L3, which is part of the DUB module of the SAGA complex. RNF20 knockdown strongly reduces H2Bub1 levels and increases the migration of non-transformed mammary epithelial cells and breast cancer-derived cells. Remarkably, overexpression of let-7b, which partly counteracts the effect of RNF20 knockdown on H2Bub1 levels, also reverses the pro-migratory effect of RNF20 knockdown. Likewise, ATXN7L3 knockdown also increases H2Bub1 levels and reduces cell migration, and this anti-migratory effect is abolished by simultaneous knockdown of RNF20. Together, our findings uncover a novel function of let-7 microRNAs as regulators of H2B ubiquitylation, suggesting an additional mechanism whereby these microRNAs can exert their tumor suppressive effects.
Keywords: H2B monoubiquitylation, let-7, deubiquitylating enzymes, RNF20, cancer
Introduction
Eukaryotic chromatin is organized in nucleosomes, where DNA is wrapped around the histone core. Histone tails undergo multiple post-translational modifications (PTMs), including methylation, acetylation, phosphorylation, SUMOylation, ubiquitylation and more, which regulate chromatin-associated processes such as gene expression, DNA replication and DNA repair45, 98. Not surprisingly, histone PTMs are often perturbed in cancer, and mutations, aberrant expression or aberrant functionality of numerous histone modifying enzymes have been observed in a growing number of human cancer types5.
One PTM that has recently emerged as frequently altered in cancerous cells and tissues is monoubiquitylation of histone H2B (H2Bub1) on Lys120. H2Bub1 is catalyzed predominantly by the E3 ubiquitin-ligase complex RNF20/RNF4036, 44, 99, and can be erased by multiple deubiquitylating enzymes (DUBs), including USP22, USP7, USP44 and others3, 23, 80, 91, 96. H2Bub1 has been implicated in a variety of processes, including transcription initiation and elongation32, 61, regulation of gene expression55, 67, DNA damage response and repair11, 42, 58, 59, regulation of chromatin structure9, 22 and stem cell differentiation23, 43. Multiple observations suggest that H2Bub1 deregulation may favor tumorigenesis13, 39. Thus, H2Bub1 is downregulated in the cancerous tissue, compared to the normal counterpart, in lung, breast, colorectal and gastric cancer79, 85. Moreover, H2Bub1 decreases during progression of breast cancer and colorectal cancer63, 75, and H2Bub1 loss is associated with worse prognosis in colorectal and gastric cancer54, 85, 86. RNF20 gene mutations were observed, albeit at low frequency, in different cancer types including colorectal, head and neck, and ovarian cancer, as well as melanoma1, 6, 71, 87. Furthermore, the RNF20 promoter is frequently hypermethylated in breast cancer67. Conversely, the gene encoding USP22, a major H2Bub1 DUB, is part of a gene signature associated with tumor aggressiveness27, and its expression correlates with poor prognosis in many cancer types, including breast cancer, lung adenocarcinoma and hepatocellular carcinoma34, 74, 93. Similarly, USP44 is overexpressed in T-cell leukemia92.
In cultured cells, downregulation of RNF20 and H2Bub1 impairs the expression of the p53 tumor suppressor, and promotes the expression of proto-oncogenes such as c-MYC67, 68. Furthermore, RNF20 can act as a transcriptional co-activator for p5344, 90. Loss of RNF20 promotes cell migration and anchorage-independent growth67, and enhances the activation of NF-κB in response to pro-inflammatory signals75. Indeed, decreased H2Bub1 in RNF20+/− mice promotes inflammation-associated colorectal cancer, in conjunction with increased expression of pro-inflammatory NF-κB target genes75. Collectively, these observation have led to RNF20 being considered a putative tumor suppressor, and H2Bub1 being viewed as a tumor suppressive chromatin modification. It should be noted, however, that this impact of RNF20 and H2Bub1 on cancer is not universal, but rather context-dependent; in fact, RNF20 and H2Bub1 can actually exert tumor-supportive effects in several human malignancies8, 76, 81.
MicroRNAs (miRs) are small non-coding RNAs that post-transcriptionally regulate the expression of target genes by inhibiting the translation and/or promoting the degradation of target mRNAs. MicroRNAs are involved in many cellular processes7, 60. Importantly, deregulation of miRNAs can impact cancer initiation and progression, and many miRNAs may promote cancer (oncomiRs) or suppress it (tumor-suppressor miRs) by targeting relevant genes implicated in tumorigenesis62. miRNAs can target the transcripts of chromatin modifier genes21, 26, 57, 65, representing a powerful mechanism whereby they can modulate global chromatin-associated processes. Indeed, perturbation of miRNA-mediated regulation of chromatin remodelers has been implicated in carcinogenesis and correlated with disease prognosis10, 38, 53, 69, 77, 94.
Given the increasing evidence that maintenance of proper H2Bub1 levels may contribute to tumor suppression, we sought to determine whether the enzymatic machinery involved in H2Bub1 homeostasis is also regulated by miRNAs, particularly those implicated in cancer, and whether this might impact cancer-related processes. We now report that members of the let-7 family of miRNAs, particularly let-7b and let-7c, play a positive role in maintaining H2Bub1 through direct targeting of multiple components of the H2B deubiquitylation machinery. This novel activity of let-7 miRNAs may contribute to their extensively documented tumor suppressor capabilities.
Results
let-7b and let-7c are predicted to target negative regulators of H2Bub1
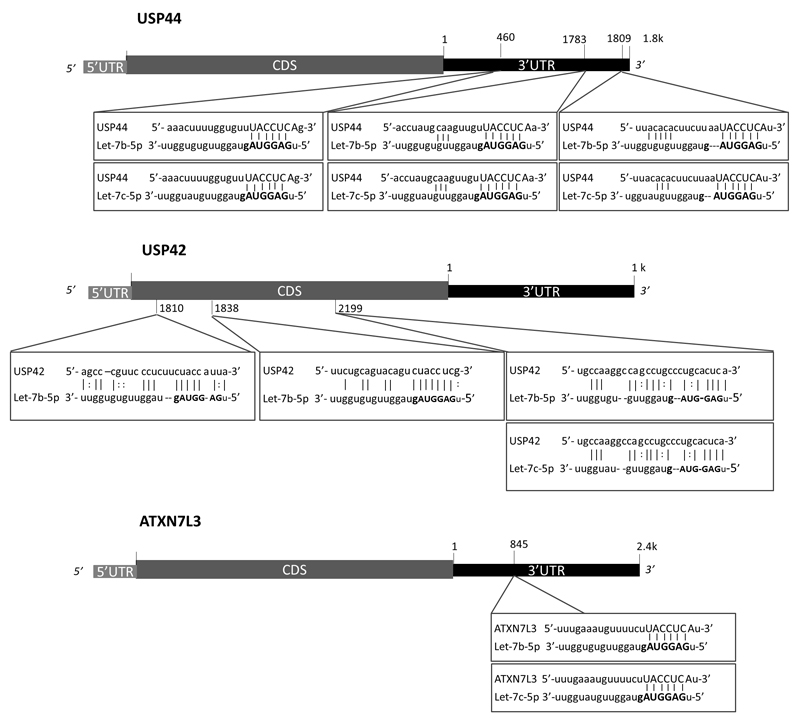
To identify microRNAs that might affect H2Bub1 homeostasis, we performed a bioinformatic screen using the Mirwalk 2.0 database18, 19, which allows simultaneous retrieval of information on miRNA-gene interaction predictions from multiple programs. This analysis suggested that several members of the let-7 miRNA family might target negative regulators of H2Bub1. The let-7 family has well-documented tumor suppressive activities12, 50, 83 and was shown to target multiple oncogenes, including Ras, c-MYC and HMGA284; indeed, let-7 miRNAs are often downregulated in various cancers2, 37, 66, 73, 88, 89. Specifically, let-7b is predicted to target USP44, USP42 and ATXN7L3 [Fig 1]. USP44 and USP42 are deubiquitylating enzymes (DUBs) reported to act on H2Bub123, 33, while ATXN7L3 is part of the DUB module of the SAGA transcriptional co-activator complex, which removes H2Bub1 through the activity of USP2249, 96. According to TargetScan (Release 7.1, June 2016)51, the 3’ UTR of USP44 mRNA is predicted to have three conserved 7mer-A1 target sites for let-7b-5p, and likewise for let-7c-5p, whereas ATXN7L3 mRNA is predicted to harbor one such site [Fig 1]. Furthermore, according to the RNA22 database (v.2.0)56, USP42 mRNA is predicted to have three binding sites for let-7b-5p and one for let-7c-5p within its coding sequence [Fig 1]. Although most documented miRNA interactions occur within the 3’ UTR of target genes, there is increasing evidence that binding within the coding sequence can also exert inhibitory effects31, 64. Additionally, the RNA22 database predicts one binding site for let-7b-5p in the 3’ UTR of USP42 mRNA, albeit with a statistically insignificant p-value [not shown]. Overall, this in silico analysis suggested a possible role for let-7 miRNAs in regulation of H2B monoubiquitylation.
Figure 1.
let-7b and let-7c miRNAs are predicted to target negative regulators of H2Bub1. Shown are schematic representations of the USP44, USP42 and ATXN7L3 genes, with the predicted let-7b and let-7c target sites. miRNA seed sequences are highlighted in bold. Target site predictions for ATXN7L3 and USP44 are from TargetScan (Release 7.1, June 2016); predictions for the coding sequence of USP42 are according to RNA22 v2.0.
let-7b and let-7c positively regulate H2Bub1
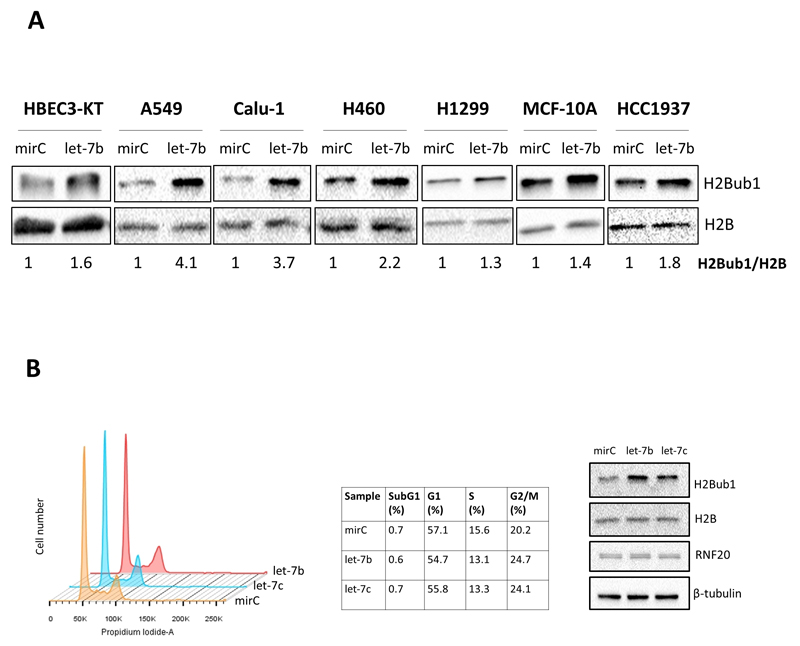
The above in silico predictions raised the possibility that, by downregulating the H2B deubiquitylation machinery, let-7b and let-7c might positively regulate H2Bub1 levels. To address this possibility, we transiently overexpressed a let-7b-5p mimic in a panel of epithelial cell lines, including the non-transformed lung-derived HBEC3-KT cells and the lung cancer-derived Calu-1, H1299, A549, H460 cells, as well as the non-transformed breast-derived MCF-10A cells and the breast cancer-derived HCC1937 cells. Indeed, in all cell types, let-7b mimic overexpression resulted in elevated H2Bub1 [Fig 2A]. Likewise, overexpression of let-7c-5p mimic in Calu-1 and H1299 cells also increased H2Bub1 levels [Fig S1A]. Overexpression of let-7b or let-7c in Calu-1 cells caused a mild reduction in the S phase fraction and a mild increase in the G2/M fraction [Fig 2B]; although one can not exclude completely the possibility that this cell cycle change may also affect indirectly H2B ubiquitylation, its scope is too limited to account for the major increase in H2Bub1 levels (3.7 fold, Fig. 2A).Thus, let-7b and let-7c positively regulate histone H2B monoubiquitylation.
Figure 2.
A) let-7b overexpression increases H2Bub1 in non-transformed and cancer-derived cell lines of lung and breast epithelial origin. Cell cultures were transfected with 25 nM (final concentration) non-targeting microRNA control mimic (mirC) or let-7b-5p mimic for 48 hours, and cell extracts were subjected to Western blot analysis with the indicated antibodies. Numbers indicate quantification (Image Lab Software) of the H2Bub1 band, normalized to total H2B. The experiment was repeated at least 3 times for each cell line. B) Effect of let-7b and let-7c overexpression on cell cycle in Calu-1 cells. Calu-1 cells were transfected with 25 nM mirC, let-7b-5p or let-7c-5p for 48 hours, fixed, stained with propidium iodide, and subjected to FACS–based cell cycle analysis (left and middle panels). The right panel displays Western blot analysis of transfected cell extracts. The experiment was repeated twice.
let-7b reverses the effect of H2Bub1 depletion on cell migration
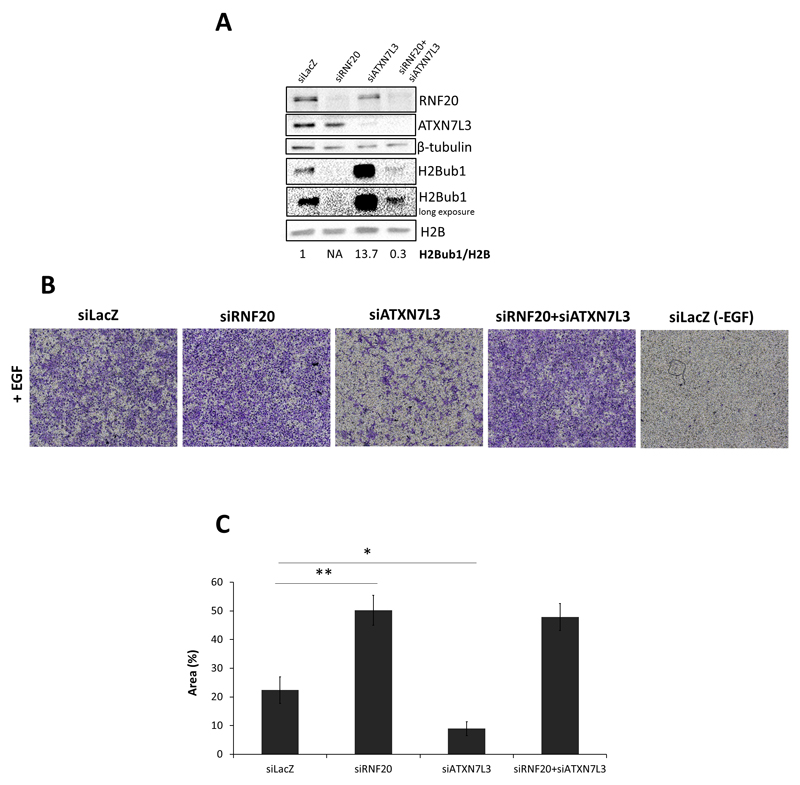
Both downregulation of let-7 family microRNAs and decreased H2Bub1 levels are frequently observed in cancer, and both have been associated with cancer progression13, 39, 83. Therefore, we sought to investigate whether upregulation of H2Bub1 by let-7b has an impact on cancer-related cellular properties. As reported previously, depletion of H2Bub1, through RNF20 knockdown (KD), promotes MCF-10A cell migration67. Indeed, transient siRNA-mediated RNF20 KD, which resulted in a prominent decrease in H2Bub1 [Fig 3A], strongly promoted MCF-10A cell migration towards EGF [Fig 3B,C]. In this model, let-7b overexpression alone did not have a significant effect on migration [Fig 3B,C]. This may not be surprising, as these non-transformed cells display rather low basal migration rates; moreover, let-7b levels are high in these cells48, and hence it is possible that further augmentation of let-7b does not affect migration in basal conditions. Concurrent overexpression of let-7b in RNF20-depleted cells partly restored their H2Bub1 levels [Fig 3A]. Importantly, it also reversed the pro-migratory effect of RNF20 depletion [Fig 3B,C]. Essentially similar results were obtained also in HCC1937 breast cancer cells [Fig 3D-F]; note that basal migration rates were higher in these cancer cells than in the non-transformed MCF-10A cells.
Figure 3.
let-7b reverses the effect of RNF20 depletion on cell migration.
A-C) MCF-10A cells were transfected with RNF20 siRNA oligos (siRNF20; final concentration 12.5 nM), let-7b mimic (final concentration 25 nM), or a combination of both; in each case, except for the combination of RNF20 siRNA plus let-7b mimic, the transfection mix was complemented with nM LacZ control siRNA (siLacZ) or non-targeting microRNA control mimic (mirC), as appropriate, to keep the total concentrations of siRNA and miRNA mimics constant. 48 hours later, cells were starved for 7 hours and subsequently collected for Western blot analysis with the indicated antibodies (A), while 50,000 cells were subjected to transwell migration assay towards EGF, as described in Materials and Methods (B). Average migration and SEM from 3 independent experiments is shown in (C); migration is expressed as % of total area stained positive by crystal violet. (*) p-value<0.05, two-tailed Student’s t-test.
D-F) HCC1937 cells were transfected as in (A). 48 hours after transfection, cells were starved for 7 hours and subjected to Western blot analysis (D), while 30,000 cells were subjected to transwell migration assay towards serum (E). Quantitative analysis, based on 2 independent experiments, is shown in (F). Numbers indicate quantification (Image Lab Software) of the H2Bub1 band, normalized to total H2B.
Together, these findings suggest that, in addition to their previously documented modes of action83, let-7b and additional let-7 family members may exert tumor suppressor activities partly also by maintaining H2Bub1 homeostasis.
let-7b directly targets ATXN7L3, USP44 and USP42 mRNA
To experimentally validate the in silico target predictions, we quantified the mRNA levels of ATXN7L3, USP44 and USP42 in MCF-10A and HCC1937 cells after let-7b overexpression; this was performed under the exact experimental conditions employed for the migration assays. Indeed, let-7b overexpression reduced ATXN7L3, USP44 and USP42 mRNA levels in HCC1937 cells [Fig S1C]. let-7b overexpression reduced significantly ATXN7L3 mRNA levels also in MCF-10A cells [Fig. 4A]. However, let-7b did not affect USP42 mRNA levels in MCF-10A cells [not shown], implying that the extent of target downregulation is dependent on the cell context. As recently reported by others47, USP44 mRNA was nearly undetectable in the non-transformed MCF-10A mammary epithelial cells, but was easily detectable in the HCC1937 breast cancer cells [Fig S1B]. Likewise, USP44 mRNA was undetectable in the non-transformed lung-derived HBEC3-KT cells, while being high in the Calu-1 and H1299 lung cancer cell lines [Fig S1B]. Of note, USP44 is highly expressed in embryonic stem cells but decreases during differentiation.23. miRNAs are expected to reduce post-transcriptionally the levels of their direct targets. Therefore, we compared the impact of let-7b overexpression on the levels of the pre-mRNA (indicative of transcription rate) and steady state mRNA of the predicted target transcripts. As seen in Fig. 4B, while let-7b did not downregulate the pre-mRNA of ATXN7L3, USP44 and USP42 in transiently transfected Calu-1 cells, it reduced significantly their steady state mRNA levels, implicating post-transcriptional downregulation. Essentially similar effects were observed with let-7c [Fig. S1D], as well as with let-7b in H1299 cells [Fig S1E]. In agreement, let-7b overexpression also reduced the amount of ATXN7L3 protein in both Calu-1 and H1299 cells [Fig. S2A], as well as in MCF-10A and HCC1937 cells [Fig S2B]. Likewise, let-7b overexpression reduced USP42 protein levels in A549 and HBEC3-KT cells, regardless of whether or not USP42 mRNA was affected [Fig. S2C]. We could not validate the effect of let-7b on USP44 protein levels, because a reproducible USP44 protein signal could not be obtained using several commercially available antibodies [not shown]. Notably, let-7b overexpression did not increase RNF20 protein levels in Calu-1 [Fig. 2B], MCF-10A [Fig. 3A], HCC1937 [Fig. 3D], A549 or HBEC3-KT cells [Fig. S2C], ruling out the possibility that the increase in H2Bub1 is mediated by a secondary effect of let-7b on RNF20 protein levels.
Figure 4.
let-7b targets and downregulates ATXN7L3, USP44 and USP42 mRNA. A) RNA of MCF-10A cells from the migration experiments in Fig 3A-C was subjected to qRT-PCR analysis with primers specific for ATXN7L3. Values were normalized to beta-2 microglobulin (B2M) mRNA as housekeeping control. Graph represents average of 3 independent experiments ± SEM. (*) p-value<0.05, two-tailed Student’s t-test. B) Calu-1 cells were transfected with let-7b mimic or non-targeting microRNA control mimic (mirC) (25 nM final) for 48 hours. RNA was purified and subjected to qRT-PCR analysis with primers specific for the indicated mature mRNAs or pre-mRNAs (see Materials and Methods). Values were normalized to GAPDH mRNA or GAPDH pre-mRNA in the same samples, respectively. Graph represents fold change relative to control, averaged from 5 experiments ±SEM. (*)p-value<0.05; (**)p-value<0.01, two-tailed Student’s t-test. C) Calu-1 cells were transfected with biotinylated miR-control mimic or biotinylated let-7b mimic, and subjected 48 hours later to miRNA pull-down analysis, as described in Materials and Methods. ATXN7L3, USP44 and USP42 mRNA in the pull-down pellets were quantified by qRT-PCR and normalized to RPL8 mRNA (which is not a predicted let-7b target) and to the input. Mean±SEM mRNA fold enrichment with let-7b relative to mirC, calculated from 4 experiments. (*) p-value<0.05; one-tailed Student’s t-test.
To determine whether let-7b targets directly these mRNAs, we performed miRNA pull-down in cells overexpressing a biotinylated let-7b mimic. As seen in Fig 4C, ATXN7L3, USP44 and USP42 mRNA were all significantly enriched by biotinylated let-7b pull-down, as compared to biotinylated non-targeting miRNA mimic control (mirC), in both Calu-1 [Fig 4C] and H1299 [Fig S2D] cells. This strongly supports the conclusion that ATXN7L3, USP44 and USP42 are direct let-7b targets. Hence, by directly targeting mRNAs that encode proteins involved in removal of H2Bub1, let-7b helps maintain high levels of this histone modification.
ATXN7L3 downregulation inhibits cell migration in an RNF20-dependent manner
In agreement with earlier reports4, 49, ATXN7L3 knockdown caused a robust increase in H2Bub1 [Fig 5A]. As seen in Fig. 3, suppression of H2Bub1 by RNF20 knockdown increased cell migration. We therefore asked whether induction of H2Bub1 by ATXN7L3 knockdown might have the opposite effect, namely whether it might inhibit migration. Indeed, transient ATXN7L3 knockdown (siATXN7L3) attenuated significantly EGF-driven MCF-10A cell migration [Fig. 5B,C]. Importantly, concurrent silencing of RNF20, which reversed the positive impact of siATXN7L3 on H2Bub1 levels [Fig 5A], also abrogated almost completely its anti-migratory effect [Fig 5B,C]. These observations suggest that the migration-inhibitory effect of ATXN7L3 depletion relies, to a large extent, on its ability to increase H2Bub1.
Figure 5.
RNF20 depletion alleviates the inhibitory effect of ATXN7L3 depletion on cell migration. A) MCF-10A cells were transfected with each of the indicated siRNA oligos (12.5 nM final) or a combination thereof; LacZ siRNA was used to bring up the total siRNA concentration to 25 nM in each case. 48 hours later, cells were starved for 7 hours and subsequently collected for Western blot analysis with the indicated antibodies (A). Numbers indicate quantification (Image Lab Software) of the H2Bub1 band, normalized to total H2B. NA=not applicable. Transwell migration assay (B) was performed and analyzed as in Fig, 3A-C. Average migration ±SEM from 5 independent experiments is shown in (C).
Altogether, the above findings strongly argue that let-7b, and additional let-7 family members, can maintain H2Bub1 homeostasis by directly targeting simultaneously multiple negative regulators of this histone modification. Furthermore, they support the conjecture that such positive regulation of H2Bub1 may contribute to the tumor-suppressive activities of the let-7 family of miRNAs.
Discussion
Gradual loss of H2Bub1 is observed in various types of cancer63, 75, and may contribute to both tumor initiation and progression. We now report that members of the let-7 miRNA family, particularly let-7b and let-7c, can positively regulate H2Bub1, by targeting several components of the H2B deubiquitylation machinery. The data presented in our study were all obtained in cell culture models; it will be important to extend our conclusions to in vivo experimental models.
Both let-7b and let-7c have well-documented tumor suppressor activities, and their downregulation has been implicated in many human cancers12, 83. For example, let-7b levels are reduced in lung cancer tissue compared to healthy tissue, and its reduced levels correlate with worse prognosis41. Indeed, let-7b can repress proliferation of lung cancer cell lines and growth of lung cancer cell xenografts in immunodeficient mice20, 40. Furthermore, loss of let-7c was shown to increase migration, invasion, and therapy resistance in lung cancer cell lines14, 82, 95. Likewise let-7b and let-7c are reduced in breast cancer tissue compared to breast benign lesions97 and low levels of let-7b correlate with worse prognosis52. Notably, analysis of the METABRIC dataset15–17 indicates that let-7c is significantly downregulated also in the progression from ductal carcinoma in situ (DCIS) to the IC10 subtype, composed mainly of basal-like triple negative breast tumors29, which tend to display low H2Bub1 levels76. Moreover, let-7c was implicated in inhibition of breast cancer stem cell renewal72.
Metastasis is the major cause of cancer-related deaths, and increased cell migration is a crucial contributor to the metastatic process28. let-7b levels were found to be decreased in metastatic gastric cancer compared to primary gastric cancer, with both expressing less let-7b than normal gastric tissue30, and decreased levels of let-7c were observed in non-small cell lung cancer (NSCLC) patients, in correlation with metastasis, advanced stage and poor survival95. Similarly, let-7b levels are lower in breast cancer patients with lymph node metastases, compared to those without lymph node metastasis35. Furthermore, ectopic let-7b overexpression can reduce migration and invasion of metastatic gastric cancer and breast cancer cell lines30, 35. Repression of cell migration might thus constitute one of the mechanisms through which let-7 miRNAs suppress cancer progression. As shown here, let-7b overexpression can rescue the pro-migratory effect of RNF20 KD-induced H2Bub1 loss. Overall, our findings suggest that maintenance of proper H2Bub1 homeostasis represents an additional mechanism whereby let-7 miRNAs repress cell migration. In view of the connection between cell migration and metastasis, it is tempting to speculate that disruption of H2Bub1 homeostasis in tumors with low let-7 levels might contribute to their increased metastatic potential.
The list of DUBs capable of H2B deubiquitylation is constantly growing24, suggesting the existence of extensive redundancy. Such redundancy might account for the observation that downregulation of a single H2B DUB often fails to yield an increase in global H2Bub1 levels. Thus, in our hands, siRNA-mediated knockdown of USP44 or USP42 alone, or even in combination, did not increase H2Bub1 levels in Calu-1 cells [data not shown]. Similarly, Atanassov et al recently reported that depletion of USP22, a major H2B DUB, not only failed to increase H2Bub1 levels but even caused a mild decrease; this was ascribed to the fact that two newly identified H2B DUBs, USP27X and USP51, compete with USP22 for binding to the SAGA complex adaptor proteins ATXN7L3 and ENY24. Moreover, the binding to ATXN7L3 and ENY2 is necessary for the H2B deubiquitylating activity of USP27X and USP514. We propose that, by targeting simultaneously multiple components of the H2B deubiquitylation machinery, including DUBs as well as ATXN7L3, let-7 miRNAs can overcome this redundancy to effectively maintain appropriate H2Bub1 levels. Moreover, we can not rule out the possibility that let-7 may target additional components of the H2Bub1 regulatory pathway (e.g. additional yet unidentified DUBs), some of which may be cell type-dependent. Conversely, downregulation of let-7 miRNA may favor cancer initiation and progression, at least to some extent, by driving a reduction in H2Bub1 without the need to alter individually the sequences or transcription rates of the genes encoding the multiple components of the H2B ubiquitylation/deubiquitylation machinery.
In vivo miRNA modulation is emerging as a promising potential tool for cancer therapy25. Notably, exogenous delivery of let-7 mimic was shown to impair tumor formation, and induce regression of fully established tumors, in NSCLC mouse xenografts20, 78. Furthermore, let-7 overexpression synergistically enhanced the anti-proliferative effect of Erlotinib in NSCLC cell lines70. Our findings raise the interesting possibility that in vivo administration of let-7 might be particularly efficacious in tumors with constitutively low H2Bub1.
Materials and Methods
Cell culture
Calu-1, H1299 and HCC1937 cell lines were maintained in RPMI 1640 (BI, Kibbutz Beit-Haemek, Israel) supplemented with 10% heat-inactivated fetal bovine serum (GIBCO, Grand Island, NY, USA) and 1% Pen/Strep (BI). A549 and H460 were maintained in DMEM (BI) supplemented with 10% heat-inactivated fetal bovine serum (GIBCO) and 1% Pen/Strep (BI). HBEC3-KT were a generous gift of Vassilis Gourgoulis (University of Athens, Greece), and were maintained in Keratinocytes SFM (GIBCO) supplemented with human recombinant EGF and bovine pituitary extract (GIBCO) according to the manufacturer’s instruction, and gentamicin 5 μg/ml (BI). MCF-10A cells were maintained in DMEM-F12 medium (BI) supplemented with 5% heat-inactivated horse serum (BI) 1% Pen/Strep (BI), 1% L-glutamine (BI), 10 μg/ml insulin (BI), 0.1 μg/ml cholera toxin (Sigma-Aldrich, St. Louis, MO, USA), 0.5 μg/ml hydrocortisone (Sigma-Aldrich) and 10 ng/ml human recombinant EGF (Sigma-Aldrich). All cell lines were grown at 37 °C in a 5% CO2 humidified incubator. Unless otherwise stated, cells were obtained from ATCC (Manassas, VA, USA). All cell lines tested negative for mycoplasma contamination.
Transfections
All transfections were performed using Dharmafect 1 reagent (Dharmacon, GE Healthcare, Lafayette, CO, USA) according to the manufacturer’s protocol. siRNA SMARTpools for RNF20, ATXN7L3 and non-targeting control siLacZ, as well as mature let-7b-5p miR mimic, mature let-7c-5p miR mimic, Negative Control miRNA Mimic, biotinylated-let-7b mimic and biotinylated-miR-Control were all purchased from Dharmacon.
Protein and RNA analysis
Cells were washed twice with ice-cold PBS, harvested and centrifuged at 3,000 rpm for 3 minutes and pellets were flash-frozen in liquid nitrogen and kept at -80° C until further analysis.
For protein analysis cell were lysed using NP40 1% buffer with 0.3% protease inhibitor (Sigma-Aldrich) and 1% phosphatase inhibitor cocktails #2 and #3 (Sigma-Aldrich) followed by centrifugation at 14,000 rpm for 15 min at 4°C. The supernatant was quantified with BCA protein assay (Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s protocol. The lysate was further treated with protein sample buffer (3% SDS, 10% glycerol, 5% β-mercaptoethanol, 62 mM Tris pH6.8) and 3 cycles of vigorous vortexing and boiling at 95°C. Protein lysates were then analyzed by 12.5% SDS-polyacrylamide gel electrophoresis (SDS-PAGE), followed by transfer on nitrocellulose membrane. Membranes were blocked using 5% milk in PBS-T for 30 minutes followed by overnight incubation with primary antibodies at 4°C. After 3 washes in PBS-T membranes were incubated for 45 minutes with HRP-conjugated secondary antibody then washed again 3 times with PBS-T. Finally, membranes were incubated for 1 min with ECL Western blot detection reagent (GE Healthcare) and imaged with Image Lab software on ChemiDoc MP Instrument (Bio-Rad, Hercules, CA, USA). The following antibodies were used: β-tubulin (T7816, Sigma-Aldrich), RNF20 (A300714A, Bethyl, Montgomery, TX, USA), ATXN7L3 (ab99947, Abcam, Cambridge, UK), USP42 (HPA006752, Atlas Antibodies, Bromma, Sweden), H2B (07-371, Millipore, Merck, Billerica, MA, USA) and anti-H2Bub155.
RNA was extracted using miRNeasy mini kit (Qiagen, Hilden, Germany), and reverse-transcribed using MMLV reverse transcriptase (Promega, Fitchburg, WI, USA), random hexamer primers (Applied Biosystems, Thermo Fisher Scientific) and nucleotide mix (Larova, Jena, Germany). Quantitative real-time PCR was performed using Platinum Sybr Green qPCR mix (Invitrogen, Thermo Fisher Scientific) in a StepOnePlus Instrument (Applied Biosystems, Thermo Fisher Scientific). Primer sequences for qRT-PCR are listed in Table 1.
Table 1.
qRT-PCR primer sequences
| Gene | Forward Primer 5’-3’ | Reverse Primer 5’-3’ |
|---|---|---|
| GAPDH (mature mRNA) | ACCCACTCCTCCACCTTTGA | CTGTTGCTGTAGCCAAATTCGT |
| RNF20 (mature mRNA) | GAACAGCGACTCAACCGACA | GGAATTCACCCGTTCTAGGACTT |
| USP44 (mature mRNA) | ATGCCACCTACCTCAGGTTCT | TCTTCTCTCGGTTATTACGTCCTG |
| USP42 (mature mRNA) | CCTACACACCACCTCTTGCC | GGGTAATATGTGCTTGCATTG |
| ATXN7L3 (mature mRNA) | GTCTTTGTCTGGCCTGGATAAC | GGTGTACCTCAAAGCAGAATCC |
| GAPDH (pre-mRNA) | CATGCCTTCTTGCCTCTTGT | GTTGAGGTCAATGAAGGGGTC |
| USP44 (pre-mRNA) | GGTTCACTCAGCTTTTCCCTG | TTGGAGGAAAACCTTCTACGC |
| USP42 (pre-mRNA) | GCCTTGATGTGTGTATATGGTTGTA | TGCCTGGGTAATATGTGCTTG |
| ATXN7L3 (pre-mRNA) | GTCTTTGTCTGGCCTGGATAAC | TCTTCCTGGATCCTTGGCTT |
| RPL8 | GCGGACGGAGCTGTTCATT | TGTTGAGCTGGGCCTTCTTG |
| B2M | GGCATTCCTGAAGCTGAC | TCTTTGGAGTACGCTGGATAG |
Cell cycle analysis
Cells were trypsinized, washed once with ice-cold PBS, and fixed in 70% ethanol for at least 30 min at -20°C. Fixed cells were resuspended in PBS containing 50 μg/ml propidium iodide and 50 μg/ml RNase A, and analyzed in an LSRII flow cytometer (Becton Dickinson, Franklin Lakes, NJ, USA).
microRNA pulldown analysis
The biotinylated-miR pulldown assay protocol was adapted from Lal et al46. 1x106 cells were transfected with 100 nM biotinylated-miR-control mimic or biotinylated-let-7b mimic. After 48h, cells were trypsinized and resuspended in 1 ml lysis buffer [(20 mM Tris pH 7.5, 100 mM KCl (Ambion, Grand Island, NY, USA), 5 mM MgCl2 (Ambion), 0.3% NP40)], supplemented with 100 U RNAsein (Promega) and protease inhibitor cocktail (Sigma-Aldrich), and incubated on ice for 20 min. The lysate was then centrifuged at 10,000xg for 15 min at 4 °C. Streptavidin-coated magnetic beads (Invitrogen, Thermo Fisher Scientific) were washed 3 times with coupling solution (5 mM Tris pH 7.5, 0.5 mM EDTA, 1 M NaCl) once with solution A (0.1 M NaOH, 0.05 NaCl) and once with solution B (0.1 NaCl). The beads were then blocked at 4 °C for 1 hour in lysis buffer supplemented with 1 mg/ml yeast tRNA (Ambion) and 1 mg/ml BSA (Ambion), then washed twice with lysis buffer. After centrifugation, 10% of the cellular lysate was frozen at -80 °C to be used as input, and the remaining lysate was loaded on the beads and incubated for 4 hours at 4 °C. Beads were then washed 5 times with lysis buffer, and the RNA bound to the beads as well as the input RNA was purified with the miRNeasy kit (Qiagen). Specific pulled-down mRNAs were quantified by qRT-PCR, and for each sample values were normalized to RPL8 mRNA and to the input.
Transwell migration assays
Transwell migration was assayed using 6.5 mm inserts with 8.0 μm pore polycarbonate membrane (Corning, Sigma-Aldrich) in 24 well plate. 48h after siRNA or miRNA mimic transfection, cells were starved for EGF or serum for 7 hours and then trypsinized, counted, and 50,000 MCF-10A cells or 30,000 HCC1937 cells were seeded in the upper part of the insert. Cells were allowed to migrate for 16 hours to the lower chamber, containing medium with or without attractant. After 16 hours, inserts were washed in PBS, fixed in 100% methanol for 5 minutes, incubated in crystal violet solution for 30 minutes, washed briefly in distilled water, and the inner part was scraped gently with a cotton swab to remove non-migrated cells. The membranes were then imaged by microscopy and migrated cells were quantified using ImageJ software (NIH, version 1.49v, June 2015).
Statistical Analysis
For all the experiments described we performed independent biological replicates as detailed in the figure legends. All values represent mean ± SEM averaged from independent experiments. Statistical significance was assessed by Student's t-test as appropriate. p-value<0.05 was considered significant.
Supplementary Information
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc)
Acknowledgements
We thank Ofra Golani from the Bioinformatics Unit of the Weizmann Institute for expert help with image analysis, Yonit Hoffman and Gali Brand for help with data analysis, Sharath Chandra Arandkar and Ohad Tarcic for valuable discussions. This study was supported in part by the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation, grant 293438 (RUBICAN) from the European Research Council, a Center of Excellence grant from the Israel Science Foundation, the Robert Bosch Foundation (project 11.5.8000.0094), and the Moross Integrated Cancer Center. M.O. is incumbent of the Andre Lwoff chair in molecular biology.
Footnotes
Conflict of Interest
The authors declare no conflict of interest.
References
- 1.Integrated genomic analyses of ovarian carcinoma. Nature. 2011;474:609–615. doi: 10.1038/nature10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akao Y, Nakagawa Y, Naoe T. let-7 microRNA functions as a potential growth suppressor in human colon cancer cells. Biological & pharmaceutical bulletin. 2006;29:903–906. doi: 10.1248/bpb.29.903. [DOI] [PubMed] [Google Scholar]
- 3.Atanassov BS, Koutelou E, Dent SY. The role of deubiquitinating enzymes in chromatin regulation. FEBS Lett. 2011;585:2016–2023. doi: 10.1016/j.febslet.2010.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Atanassov Boyko S, Mohan Ryan D, Lan X, Kuang X, Lu Y, Lin K, et al. ATXN7L3 and ENY2 Coordinate Activity of Multiple H2B Deubiquitinases Important for Cellular Proliferation and Tumor Growth. Molecular Cell. 2016;62:558–571. doi: 10.1016/j.molcel.2016.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Audia JE, Campbell RM. Histone Modifications and Cancer. Cold Spring Harbor perspectives in biology. 2016;8:a019521. doi: 10.1101/cshperspect.a019521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barber TD, McManus K, Yuen KWY, Reis M, Parmigiani G, Shen D, et al. Chromatid cohesion defects may underlie chromosome instability in human colorectal cancers. Proceedings of the National Academy of Sciences. 2008;105:3443–3448. doi: 10.1073/pnas.0712384105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beermann J, Piccoli MT, Viereck J, Thum T. Non-coding RNAs in Development and Disease: Background, Mechanisms, and Therapeutic Approaches. Physiological reviews. 2016;96:1297–1325. doi: 10.1152/physrev.00041.2015. [DOI] [PubMed] [Google Scholar]
- 8.Blank M, Tang Y, Yamashita M, Burkett SS, Cheng SY, Zhang YE. A tumor suppressor function of Smurf2 associated with controlling chromatin landscape and genome stability through RNF20. Nature medicine. 2012;18:227–234. doi: 10.1038/nm.2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chandrasekharan MB, Huang F, Sun Z-W. Ubiquitination of histone H2B regulates chromatin dynamics by enhancing nucleosome stability. Proceedings of the National Academy of Sciences. 2009;106:16686–16691. doi: 10.1073/pnas.0907862106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen D-l, Zhang D-s, Lu Y-x, Chen L-z, Zeng Z-l, He M-m, et al. microRNA-217 inhibits tumor progression and metastasis by downregulating EZH2 and predicts favorable prognosis in gastric cancer. Oncotarget. 2015;6:10868–10879. doi: 10.18632/oncotarget.3451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chernikova SB, Razorenova OV, Higgins JP, Sishc BJ, Nicolau M, Dorth JA, et al. Deficiency in Mammalian Histone H2B Ubiquitin Ligase Bre1 (Rnf20/Rnf40) Leads to Replication Stress and Chromosomal Instability. Cancer Research. 2012;72:2111. doi: 10.1158/0008-5472.CAN-11-2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chiu SC, Chung HY, Cho DY, Chan TM, Liu MC, Huang HM, et al. Therapeutic potential of microRNA let-7: tumor suppression or impeding normal stemness. Cell transplantation. 2014;23:459–469. doi: 10.3727/096368914X678418. [DOI] [PubMed] [Google Scholar]
- 13.Cole AJ, Clifton-Bligh R, Marsh DJ. Histone H2B monoubiquitination: roles to play in human malignancy. Endocrine-related cancer. 2015;22:T19–33. doi: 10.1530/ERC-14-0185. [DOI] [PubMed] [Google Scholar]
- 14.Cui S-Y, Huang J-Y, Chen Y-T, Song H-Z, Feng B, Huang G-C, et al. Let-7c Governs the Acquisition of Chemo- or Radioresistance and Epithelial-to-Mesenchymal Transition Phenotypes in Docetaxel-Resistant Lung Adenocarcinoma. Molecular Cancer Research. 2013;11:699. doi: 10.1158/1541-7786.MCR-13-0019-T. [DOI] [PubMed] [Google Scholar]
- 15.Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dawson S-J, Rueda OM, Aparicio S, Caldas C. A new genome-driven integrated classification of breast cancer and its implications. The EMBO Journal. 2013;32:617–628. doi: 10.1038/emboj.2013.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dvinge H, Git A, Graf S, Salmon-Divon M, Curtis C, Sottoriva A, et al. The shaping and functional consequences of the microRNA landscape in breast cancer. Nature. 2013;497:378–382. doi: 10.1038/nature12108. [DOI] [PubMed] [Google Scholar]
- 18.Dweep H, Sticht C, Pandey P, Gretz N. miRWalk – Database: Prediction of possible miRNA binding sites by “walking” the genes of three genomes. Journal of Biomedical Informatics. 2011;44:839–847. doi: 10.1016/j.jbi.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 19.Dweep H, Gretz N. miRWalk2.0: a comprehensive atlas of microRNA-target interactions. Nat Meth. 2015;12:697–697. doi: 10.1038/nmeth.3485. [DOI] [PubMed] [Google Scholar]
- 20.Esquela-Kerscher A, Trang P, Wiggins JF, Patrawala L, Cheng A, Ford L, et al. The let-7 microRNA reduces tumor growth in mouse models of lung cancer. Cell Cycle. 2008;7:759–764. doi: 10.4161/cc.7.6.5834. [DOI] [PubMed] [Google Scholar]
- 21.Fabbri M, Garzon R, Cimmino A, Liu Z, Zanesi N, Callegari E, et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proceedings of the National Academy of Sciences. 2007;104:15805–15810. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fierz B, Chatterjee C, McGinty RK, Bar-Dagan M, Raleigh DP, Muir TW. Histone H2B ubiquitylation disrupts local and higher-order chromatin compaction. Nat Chem Biol. 2011;7:113–119. doi: 10.1038/nchembio.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fuchs G, Shema E, Vesterman R, Kotler E, Wolchinsky Z, Wilder S, et al. RNF20 and USP44 Regulate Stem Cell Differentiation by Modulating H2B Monoubiquitylation. Molecular Cell. 2012;46:662–673. doi: 10.1016/j.molcel.2012.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fuchs G, Oren M. Writing and reading H2B monoubiquitylation. Biochimica et biophysica acta. 2014;1839:694–701. doi: 10.1016/j.bbagrm.2014.01.002. [DOI] [PubMed] [Google Scholar]
- 25.Gambari R, Brognara E, Spandidos DA, Fabbri E. Targeting oncomiRNAs and mimicking tumor suppressor miRNAs: Nuew trends in the development of miRNA therapeutic strategies in oncology (Review) Int J Oncol. 2016;49:5–32. doi: 10.3892/ijo.2016.3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garcia AI, Buisson M, Bertrand P, Rimokh R, Rouleau E, Lopez BS, et al. Down-regulation of BRCA1 expression by miR-146a and miR-146b-5p in triple negative sporadic breast cancers. EMBO Mol Med. 2011;3:279–290. doi: 10.1002/emmm.201100136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Glinsky GV, Berezovska O, Glinskii AB. Microarray analysis identifies a death-from-cancer signature predicting therapy failure in patients with multiple types of cancer. The Journal of Clinical Investigation. 115:1503–1521. doi: 10.1172/JCI23412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guan X. Cancer metastases: challenges and opportunities. Acta Pharmaceutica Sinica B. 2015;5:402–418. doi: 10.1016/j.apsb.2015.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haakensen VD, Nygaard V, Greger L, Aure MR, Fromm B, Bukholm IRK, et al. Subtype-specific micro-RNA expression signatures in breast cancer progression. International Journal of Cancer. 2016;139:1117–1128. doi: 10.1002/ijc.30142. [DOI] [PubMed] [Google Scholar]
- 30.Han X, Chen Y, Yao N, Liu H, Wang Z. MicroRNA let-7b suppresses human gastric cancer malignancy by targeting ING1. Cancer Gene Ther. 2015;22:122–129. doi: 10.1038/cgt.2014.75. [DOI] [PubMed] [Google Scholar]
- 31.Hausser J, Syed AP, Bilen B, Zavolan M. Analysis of CDS-located miRNA target sites suggests that they can effectively inhibit translation. Genome Research. 2013;23:604–615. doi: 10.1101/gr.139758.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Henry KW, Wyce A, Lo W-S, Duggan LJ, Emre NCT, Kao C-F, et al. Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes & Development. 2003;17:2648–2663. doi: 10.1101/gad.1144003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hock AK, Vigneron AM, Vousden KH. Ubiquitin-specific Peptidase 42 (USP42) Functions to Deubiquitylate Histones and Regulate Transcriptional Activity. Journal of Biological Chemistry. 2014;289:34862–34870. doi: 10.1074/jbc.M114.589267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu J, Yang D, Zhang H, Liu W, Zhao Y, Lu H, et al. USP22 promotes tumor progression and induces epithelial–mesenchymal transition in lung adenocarcinoma. Lung Cancer. 2015;88:239–245. doi: 10.1016/j.lungcan.2015.02.019. [DOI] [PubMed] [Google Scholar]
- 35.Hu X, Guo J, Zheng L, Li C, Zheng TM, Tanyi JL, et al. The heterochronic microRNA let-7 inhibits cell motility by regulating the genes in the actin cytoskeleton pathway in breast cancer. Molecular cancer research : MCR. 2013;11:240–250. doi: 10.1158/1541-7786.MCR-12-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hwang WW, Venkatasubrahmanyam S, Ianculescu AG, Tong A, Boone C, Madhani HD. A Conserved RING Finger Protein Required for Histone H2B Monoubiquitination and Cell Size Control. Molecular Cell. 2003;11:261–266. doi: 10.1016/s1097-2765(02)00826-2. [DOI] [PubMed] [Google Scholar]
- 37.Iorio MV, Ferracin M, Liu C-G, Veronese A, Spizzo R, Sabbioni S, et al. MicroRNA Gene Expression Deregulation in Human Breast Cancer. Cancer Research. 2005;65:7065. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 38.Jansen MPHM, Reijm EA, Sieuwerts AM, Ruigrok-Ritstier K, Look MP, Rodríguez-González FG, et al. High miR-26a and low CDC2 levels associate with decreased EZH2 expression and with favorable outcome on tamoxifen in metastatic breast cancer. Breast Cancer Research and Treatment. 2012;133:937–947. doi: 10.1007/s10549-011-1877-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johnsen SA. The enigmatic role of H2Bub1 in cancer. FEBS Letters. 2012;586:1592–1601. doi: 10.1016/j.febslet.2012.04.002. [DOI] [PubMed] [Google Scholar]
- 40.Johnson CD, Esquela-Kerscher A, Stefani G, Byrom M, Kelnar K, Ovcharenko D, et al. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res. 2007;67:7713–7722. doi: 10.1158/0008-5472.CAN-07-1083. [DOI] [PubMed] [Google Scholar]
- 41.Jusufović E, Rijavec M, Keser D, Korošec P, Sodja E, Iljazović E, et al. let-7b and miR-126 Are Down-Regulated in Tumor Tissue and Correlate with Microvessel Density and Survival Outcomes in Non–Small–Cell Lung Cancer. PLoS ONE. 2012;7:e45577. doi: 10.1371/journal.pone.0045577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kari V, Shchebet A, Neumann H, Johnsen SA. The H2B ubiquitin ligase RNF40 cooperates with SUPT16H to induce dynamic changes in chromatin structure during DNA double-strand break repair. Cell Cycle. 2011;10:3495–3504. doi: 10.4161/cc.10.20.17769. [DOI] [PubMed] [Google Scholar]
- 43.Karpiuk O, Najafova Z, Kramer F, Hennion M, Galonska C, König A, et al. The Histone H2B Monoubiquitination Regulatory Pathway Is Required for Differentiation of Multipotent Stem Cells. Molecular Cell. 2012;46:705–713. doi: 10.1016/j.molcel.2012.05.022. [DOI] [PubMed] [Google Scholar]
- 44.Kim J, Hake SB, Roeder RG. The Human Homolog of Yeast BRE1 Functions as a Transcriptional Coactivator through Direct Activator Interactions. Molecular Cell. 2005;20:759–770. doi: 10.1016/j.molcel.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 45.Kouzarides T. Chromatin Modifications and Their Function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 46.Lal A, Thomas MP, Altschuler G, Navarro F, O'Day E, Li XL, et al. Capture of MicroRNA–Bound mRNAs Identifies the Tumor Suppressor miR-34a as a Regulator of Growth Factor Signaling. PLoS Genet. 2011;7 doi: 10.1371/journal.pgen.1002363. e1002363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lan X, Atanassov BS, Li W, Zhang Y, Florens L, Mohan RD, et al. USP44 IS AN INTEGRAL COMPONENT OF N-COR THAT CONTRIBUTES TO GENE REPRESSION BY DEUBIQUITINATING HISTONE H2B. Cell reports. 2016;17:2382–2393. doi: 10.1016/j.celrep.2016.10.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, et al. A Mammalian microRNA Expression Atlas Based on Small RNA Library Sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lang G, Bonnet J, Umlauf D, Karmodiya K, Koffler J, Stierle M, et al. The Tightly Controlled Deubiquitination Activity of the Human SAGA Complex Differentially Modifies Distinct Gene Regulatory Elements. Molecular and Cellular Biology. 2011;31:3734–3744. doi: 10.1128/MCB.05231-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee H, Han S, Kwon CS, Lee D. Biogenesis and regulation of the let-7 miRNAs and their functional implications. Protein & cell. 2016;7:100–113. doi: 10.1007/s13238-015-0212-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 52.Ma L, Li G-z, Wu Z-s, Meng G. Prognostic significance of let-7b expression in breast cancer and correlation to its target gene of BSG expression. Medical Oncology. 2013;31:773. doi: 10.1007/s12032-013-0773-7. [DOI] [PubMed] [Google Scholar]
- 53.Majid S, Dar AA, Saini S, Shahryari V, Arora S, Zaman MS, et al. miRNA-34b Inhibits Prostate Cancer through Demethylation, Active Chromatin Modifications, and AKT Pathways. Clinical Cancer Research. 2013;19:73. doi: 10.1158/1078-0432.CCR-12-2952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Melling N, Grimm N, Simon R, Stahl P, Bokemeyer C, Terracciano L, et al. Loss of H2Bub1 Expression is Linked to Poor Prognosis in Nodal Negative Colorectal Cancers. Pathology & Oncology Research. 2016;22:95–102. doi: 10.1007/s12253-015-9977-9. [DOI] [PubMed] [Google Scholar]
- 55.Minsky N, Shema E, Field Y, Schuster M, Segal E, Oren M. Monoubiquitinated H2B is associated with the transcribed region of highly expressed genes in human cells. Nat Cell Biol. 2008;10:483–488. doi: 10.1038/ncb1712. [DOI] [PubMed] [Google Scholar]
- 56.Miranda KC, Huynh T, Tay Y, Ang Y-S, Tam W-L, Thomson AM, et al. A Pattern-Based Method for the Identification of MicroRNA Binding Sites and Their Corresponding Heteroduplexes. Cell. 2006;126:1203–1217. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 57.Moskwa P, Buffa FM, Pan Y, Panchakshari R, Gottipati P, Muschel RJ, et al. miR-182-Mediated Downregulation of BRCA1 Impacts DNA Repair and Sensitivity to PARP Inhibitors. Molecular Cell. 2011;41:210–220. doi: 10.1016/j.molcel.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Moyal L, Lerenthal Y, Gana-Weisz M, Mass G, So S, Wang S-Y, et al. Requirement of ATM-Dependent Monoubiquitylation of Histone H2B for Timely Repair of DNA Double-Strand Breaks. Molecular Cell. 2011;41:529–542. doi: 10.1016/j.molcel.2011.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nakamura K, Kato A, Kobayashi J, Yanagihara H, Sakamoto S, Oliveira Douglas VNP, et al. Regulation of Homologous Recombination by RNF20-Dependent H2B Ubiquitination. Molecular Cell. 2011;41:515–528. doi: 10.1016/j.molcel.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 60.Olive V, Minella AC, He L. Outside the coding genome, mammalian microRNAs confer structural and functional complexity. Science signaling. 2015;8 doi: 10.1126/scisignal.2005813. re2-re2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pavri R, Zhu B, Li G, Trojer P, Mandal S, Shilatifard A, et al. Histone H2B Monoubiquitination Functions Cooperatively with FACT to Regulate Elongation by RNA Polymerase II. Cell. 2006;125:703–717. doi: 10.1016/j.cell.2006.04.029. [DOI] [PubMed] [Google Scholar]
- 62.Peng Y, Croce CM. The role of MicroRNAs in human cancer. Signal Transduction And Targeted Therapy. 2016;1:15004. doi: 10.1038/sigtrans.2015.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Prenzel T, Begus-Nahrmann Y, Kramer F, Hennion M, Hsu C, Gorsler T, et al. Estrogen-Dependent Gene Transcription in Human Breast Cancer Cells Relies upon Proteasome-Dependent Monoubiquitination of Histone H2B. Cancer Research. 2011;71:5739. doi: 10.1158/0008-5472.CAN-11-1896. [DOI] [PubMed] [Google Scholar]
- 64.Reczko M, Maragkakis M, Alexiou P, Grosse I, Hatzigeorgiou AG. Functional microRNA targets in protein coding sequences. Bioinformatics. 2012;28:771–776. doi: 10.1093/bioinformatics/bts043. [DOI] [PubMed] [Google Scholar]
- 65.Sander S, Bullinger L, Klapproth K, Fiedler K, Kestler HA, Barth TFE, et al. MYC stimulates EZH2 expression by repression of its negative regulator miR-26a. Blood. 2008;112:4202. doi: 10.1182/blood-2008-03-147645. [DOI] [PubMed] [Google Scholar]
- 66.Schultz J, Lorenz P, Gross G, Ibrahim S, Kunz M. MicroRNA let-7b targets important cell cycle molecules in malignant melanoma cells and interferes with anchorage-independent growth. Cell Res. 2008;18:549–557. doi: 10.1038/cr.2008.45. [DOI] [PubMed] [Google Scholar]
- 67.Shema E, Tirosh I, Aylon Y, Huang J, Ye C, Moskovits N, et al. The histone H2B-specific ubiquitin ligase RNF20/hBRE1 acts as a putative tumor suppressor through selective regulation of gene expression. Genes & Development. 2008;22:2664–2676. doi: 10.1101/gad.1703008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Shema E, Kim J, Roeder RG, Oren M. RNF20 inhibits TFIIS-facilitated transcriptional elongation to suppress pro-oncogenic gene expression. Molecular cell. 2011;42:477–488. doi: 10.1016/j.molcel.2011.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Song QC, Shi ZB, Zhang YT, Ji L, Wang KZ, Duan DP, et al. Downregulation of microRNA-26a is associated with metastatic potential and the poor prognosis of osteosarcoma patients. Oncol Rep. 2014;31:1263–1270. doi: 10.3892/or.2014.2989. [DOI] [PubMed] [Google Scholar]
- 70.Stahlhut C, Slack FJ. Combinatorial Action of MicroRNAs let-7 and miR-34 Effectively Synergizes with Erlotinib to Suppress Non-small Cell Lung Cancer Cell Proliferation. Cell Cycle. 2015;14:2171–2180. doi: 10.1080/15384101.2014.1003008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, et al. The Mutational Landscape of Head and Neck Squamous Cell Carcinoma. Science. 2011;333:1157. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sun X, Xu C, Tang SC, Wang J, Wang H, Wang P, et al. Let-7c blocks estrogen-activated Wnt signaling in induction of self-renewal of breast cancer stem cells. Cancer Gene Ther. 2016;23:83–89. doi: 10.1038/cgt.2016.3. [DOI] [PubMed] [Google Scholar]
- 73.Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 74.Tang B, Tang F, Li B, Yuan S, Xu Q, Tomlinson S, et al. High USP22 expression indicates poor prognosis in hepatocellular carcinoma. Oncotarget. 2015;6:12654–12667. doi: 10.18632/oncotarget.3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tarcic O, Pateras Ioannis S, Cooks T, Shema E, Kanterman J, Ashkenazi H, et al. RNF20 Links Histone H2B Ubiquitylation with Inflammation and Inflammation-Associated Cancer. Cell Reports. 2016;14:1462–1476. doi: 10.1016/j.celrep.2016.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tarcic O, Granit RZ, Pateras IS, Masury H, Maly B, Zwang Y, et al. RNF20 and histone H2B ubiquitylation exert opposing effects in Basal-Like versus luminal breast cancer. Cell Death Differ. 2017;24:694–704. doi: 10.1038/cdd.2016.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tommasi S, Pinto R, Danza K, Pilato B, Palumbo O, Micale L, et al. miR-151-5p, targeting chromatin remodeler SMARCA5, as a marker for the BRCAness phenotype. Oncotarget. 2016:3. doi: 10.18632/oncotarget.10345. Advance Online Publications. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Trang P, Medina PP, Wiggins JF, Ruffino L, Kelnar K, Omotola M, et al. Regression of murine lung tumors by the let-7 microRNA. Oncogene. 2010;29:1580–1587. doi: 10.1038/onc.2009.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Urasaki Y, Heath L, Xu CW. Coupling of Glucose Deprivation with Impaired Histone H2B Monoubiquitination in Tumors. PLoS ONE. 2012;7:e36775. doi: 10.1371/journal.pone.0036775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.van der Knaap JA, Kumar BRP, Moshkin YM, Langenberg K, Krijgsveld J, Heck AJR, et al. GMP Synthetase Stimulates Histone H2B Deubiquitylation by the Epigenetic Silencer USP7. Molecular Cell. 2005;17:695–707. doi: 10.1016/j.molcel.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 81.Wang E, Kawaoka S, Yu M, Shi J, Ni T, Yang W, et al. Histone H2B ubiquitin ligase RNF20 is required for MLL-rearranged leukemia. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:3901–3906. doi: 10.1073/pnas.1301045110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang P-Y, Sun Y-X, Zhang S, Pang M, Zhang H-H, Gao S-Y, et al. Let-7c inhibits A549 cell proliferation through oncogenic TRIB2 related factors. FEBS Letters. 2013;587:2675–2681. doi: 10.1016/j.febslet.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 83.Wang T, Wang G, Hao D, Liu X, Wang D, Ning N, et al. Aberrant regulation of the LIN28A/LIN28B and let-7 loop in human malignant tumors and its effects on the hallmarks of cancer. Molecular Cancer. 2015;14:125. doi: 10.1186/s12943-015-0402-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang X, Cao LEI, Wang Y, Wang X, Liu N, You Y. Regulation of let-7 and its target oncogenes (Review) Oncology Letters. 2012;3:955–960. doi: 10.3892/ol.2012.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang Z-J, Yang J-L, Wang Y-P, Lou J-Y, Chen J, Liu C, et al. Decreased histone H2B monoubiquitination in malignant gastric carcinoma. World Journal of Gastroenterology : WJG. 2013;19:8099–8107. doi: 10.3748/wjg.v19.i44.8099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang Z, Zhu L, Guo T, Wang Y, Yang J. Decreased H2B monoubiquitination and overexpression of ubiquitin-specific protease enzyme 22 in malignant colon carcinoma. Human Pathology. 2015;46:1006–1014. doi: 10.1016/j.humpath.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 87.Wei X, Walia V, Lin JC, Teer JK, Prickett TD, Gartner J, et al. Exome sequencing identifies GRIN2A as frequently mutated in melanoma. Nat Genet. 2011;43:442–446. doi: 10.1038/ng.810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yang H, Kong W, He L, Zhao JJ, O'Donnell JD, Wang J, et al. MicroRNA expression profiling in human ovarian cancer: miR-214 induces cell survival and cisplatin resistance by targeting PTEN. Cancer Res. 2008;68:425–433. doi: 10.1158/0008-5472.CAN-07-2488. [DOI] [PubMed] [Google Scholar]
- 89.Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong C, et al. let-7 Regulates Self Renewal and Tumorigenicity of Breast Cancer Cells. Cell. 2007;131:1109–1123. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
- 90.Zhang F, Yu X. WAC, a Functional Partner of RNF20/40, Regulates Histone H2B Ubiquitination and Gene Transcription. Molecular Cell. 2011;41:384–397. doi: 10.1016/j.molcel.2011.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhang X-Y, Varthi M, Sykes SM, Phillips C, Warzecha C, Zhu W, et al. The Putative Cancer Stem Cell Marker USP22 Is a Subunit of the Human SAGA Complex Required for Activated Transcription and Cell-Cycle Progression. Molecular Cell. 2008;29:102–111. doi: 10.1016/j.molcel.2007.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhang Y, van Deursen J, Galardy PJ. Overexpression of Ubiquitin Specific Protease 44 (USP44) Induces Chromosomal Instability and Is Frequently Observed in Human T-Cell Leukemia. PLoS ONE. 2011;6:e23389. doi: 10.1371/journal.pone.0023389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zhang Y, Yao L, Zhang X, Ji H, Wang L, Sun S, et al. Elevated expression of USP22 in correlation with poor prognosis in patients with invasive breast cancer. Journal of Cancer Research and Clinical Oncology. 2011;137:1245–1253. doi: 10.1007/s00432-011-0998-9. [DOI] [PubMed] [Google Scholar]
- 94.Zhang Z, Tang H, Wang Z, Zhang B, Liu W, Lu H, et al. MiR-185 targets the DNA methyltransferases 1 and regulates global DNA methylation in human glioma. Mol Cancer. 2011;10:124. doi: 10.1186/1476-4598-10-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhao B, Han H, Chen J, Zhang Z, Li S, Fang F, et al. MicroRNA let-7c inhibits migration and invasion of human non-small cell lung cancer by targeting ITGB3 and MAP4K3. Cancer Letters. 2014;342:43–51. doi: 10.1016/j.canlet.2013.08.030. [DOI] [PubMed] [Google Scholar]
- 96.Zhao Y, Lang G, Ito S, Bonnet J, Metzger E, Sawatsubashi S, et al. A TFTC/STAGA Module Mediates Histone H2A and H2B Deubiquitination, Coactivates Nuclear Receptors, and Counteracts Heterochromatin Silencing. Molecular Cell. 2008;29:92–101. doi: 10.1016/j.molcel.2007.12.011. [DOI] [PubMed] [Google Scholar]
- 97.Zhao Y, Deng C, Wang J, Xiao J, Gatalica Z, Recker RR, et al. Let-7 family miRNAs regulate estrogen receptor alpha signaling in estrogen receptor positive breast cancer. Breast Cancer Research and Treatment. 2011;127:69–80. doi: 10.1007/s10549-010-0972-2. [DOI] [PubMed] [Google Scholar]
- 98.Zhao Y, Garcia BA. Comprehensive Catalog of Currently Documented Histone Modifications. Cold Spring Harbor perspectives in biology. 2015;7:a025064. doi: 10.1101/cshperspect.a025064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhu B, Zheng Y, Pham A-D, Mandal SS, Erdjument-Bromage H, Tempst P, et al. Monoubiquitination of Human Histone H2B: The Factors Involved and Their Roles in HOX Gene Regulation. Molecular Cell. 2005;20:601–611. doi: 10.1016/j.molcel.2005.09.025. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.