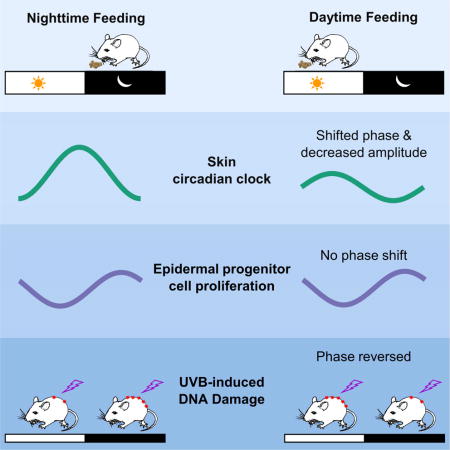
Summary
The epidermis is a highly regenerative barrier protecting organisms from environmental insults, including ultraviolet radiation, the main cause of skin cancer and skin aging. Here we show that time-restricted feeding (RF) shifts the phase and alters the amplitude of the skin circadian clock, and affects the expression of approximately 10% of the skin transcriptome. Furthermore, a large number of skin-expressed genes are acutely regulated by food intake. While the circadian clock is required for daily rhythms in DNA synthesis in epidermal progenitor cells, RF-induced shifts in clock phase do not alter the phase of DNA synthesis. Yet, RF alters both the diurnal sensitivity to UVB-induced DNA damage, and the expression of the key DNA repair gene Xpa. Together, our findings indicate an unexpected regulation of skin function by time of feeding and emphasize a link between circadian rhythm, food intake, and skin health.
Keywords: skin, circadian clock, time-restricted feeding, cell cycle, metabolism, DNA damage, aging
Graphical abstract
Introduction
Acting as a strong barrier to physical, chemical and pathogen insults from the exterior, and to water loss from the interior, the epidermis, the outermost layer of the skin, is a stratified epithelium. Its homeostasis is balanced by stem-cell progeny production in the basal layer and loss of cells through terminal differentiation culminating in shedding of corneocytes at the skin’s surface (Blanpain and Fuchs, 2009). Skin biology research focuses largely on the responses to various forms of external injury, including UV irradiation, a major cause of DNA-damage, accelerated skin ageing, and cancer (Armstrong and Kricker, 2001; Fisher et al., 2002). Recent work, however, has unearthed an important role for the circadian clock in regulating skin function (Plikus et al., 2015). This raises the intriguing possibility that signals that influence circadian clocks, such as the time of feeding, could act as a regulator of skin function; the clocks in some peripheral tissues, especially metabolic organs, including the liver, can be entrained by time-restricted feeding (RF) (Damiola et al., 2000; Hara et al., 2001; Izumo et al., 2014; Kuroda et al., 2012; Stokkan et al., 2001).
The hierarchically organized mammalian circadian clock comprises the central clock, located in the suprachiasmatic nucleus (SCN), and peripheral clocks, possessed by almost all cells (Dibner et al., 2010; Mohawk et al., 2012). Entrained by the day-night cycle, the central clock synchronizes the phases of peripheral clocks, thus coordinating the locomotor and metabolic activity of the animal with the Earth’s rotation. At a molecular level, the central and peripheral clocks are transcription-translation feedback loops wherein the heterodimeric CLOCK/BMAL1 transcription complex activates a large number of genes. These include PERs and CRYs that form heterodimers to inhibit CLOCK/BMAL1 activity, thus establishing an oscillating transcriptional output with a 24-hour periodicity (Dibner et al., 2010; Lowrey and Takahashi, 2011; Mohawk et al., 2012). The direct and indirect targets of the circadian clock encode key regulators of many, if not most, biological processes, including metabolism (Bass, 2012; Lamia et al., 2011), cell proliferation (Levi et al., 2007; Masri et al., 2013), and response to therapeutic treatment (Levi et al., 2007; Levi and Schibler, 2007).
Previous studies showed important roles for the circadian clock in skin biology (Plikus et al., 2015). The clock is highly active in the progenitor cells of the secondary hair germ where it plays a role in the initiation of the hair growth cycle (Lin et al., 2009). It also contributes to heterogeneity in hair follicle stem cells by regulating the sensitivity to activation signals (Janich et al., 2011). In addition, in the matrix of growing hair follicles, the clock determines diurnal variation in cell division which affects the sensitivity of hair follicles to external gamma radiation (Plikus et al., 2013). The circadian clock also gates the response to UVB-radiation in the skin (Gaddameedhi et al., 2011; Geyfman et al., 2012), at least in part by controlling the expression of Xpa, a rate limiting enzyme involved in the repair of UVB-induced DNA damage (Gaddameedhi et al., 2011; Khang et al., 2010). In fact, the skin is most sensitive to UVB-induced tumor induction at night when expression of Xpa is lowest (Gaddameedhi et al., 2011). Studies also showed a link between the circadian clock and skin aging as Bmal1-deleted mice exhibit accelerated skin aging (Janich et al., 2011; Kondratov et al., 2006), perhaps related to excessive reactive oxygen species (ROS) generation. In the interfollicular epidermal progenitor cells, the clock is required for a prominent diurnal variation in DNA synthesis (Gaddameedhi et al., 2011; Geyfman et al., 2012). While the function of these diurnal rhythms in epidermal progenitor cell DNA synthesis remains unknown, transcriptome studies from yeast to mammals have suggested that the circadian clock may coordinate the timing of different cellular processes (Geyfman et al., 2012; Gillette and Sejnowski, 2005; Jouffe et al., 2013; Panda et al., 2002); in the case of epidermal progenitor cells, its role may be to synchronize intermediary metabolism and the cell cycle, thus minimizing cellular damage from oxidative phosphorylation-generated ROS (Stringari et al., 2015). The circadian clock, then, may be a mediator of the long-appreciated, yet incompletely understood cross-talk between metabolism and the cell cycle (Buchakjian and Kornbluth, 2010; Fritz and Fajas, 2010; Laporte et al., 2011).
Despite the circadian clock’s multiple roles in skin biology, other than the SCN, little is known of the factors that entrain the skin circadian clock. Restriction of food intake to defined time periods is known to change the phase of the circadian clock and gene expression programs, especially in primary metabolic organs such as the liver (Adamovich et al., 2014; Damiola et al., 2000; Kuroda et al., 2012; Stokkan et al., 2001). But not all peripheral tissues are entrained by RF (Izumo et al., 2014), and the effect of RF on the skin has not been investigated. Hence, we examined whether RF can entrain the circadian clock in skin and affect skin function. We discovered that RF can shift the circadian clock of skin, but that the phase of the skin circadian clock is not as tightly coupled to feeding time as that of the liver. We found RF-schedule-specific changes in the skin transcriptome, including changes in the expression of multiple metabolic genes and the nucleotide excision repair factor, Xpa. Although the phase of the cell cycle was insensitive to changes in circadian clock phase, RF decreased overall progenitor proliferation rates, and day-time RF reversed diurnal rhythm of epidermal sensitivity to UVB-induced DNA damage. This study points to unexpected influences of time of feeding on the biology of skin, suggesting that time of feeding may affect UVB-induced conditions such as skin cancer and premature aging.
Results
RF entrains the skin circadian clock in a manner distinct from that of the liver
To determine whether RF can shift the phase of the skin circadian clock, we administered five different feeding schedules (Figure 1A). The ad libitum (AD) group of mice had unlimited access to food. The early daytime (ED) feeding group had access to food from zeitgeber time 0 (ZT0) for 4 hours. The midday (MD) feeding group had access to food from ZT5 for 4 hours. The early nighttime (EN) feeding group had access to food starting at ZT12. Finally, the long daytime (LD) feeding group had access to food from ZT3 for 8 hours. These feeding lengths and times were modeled on previous RF studies (Damiola et al., 2000; Stokkan et al., 2001).
Figure 1. Entrainment of peripheral circadian clocks by time-restricted feeding.
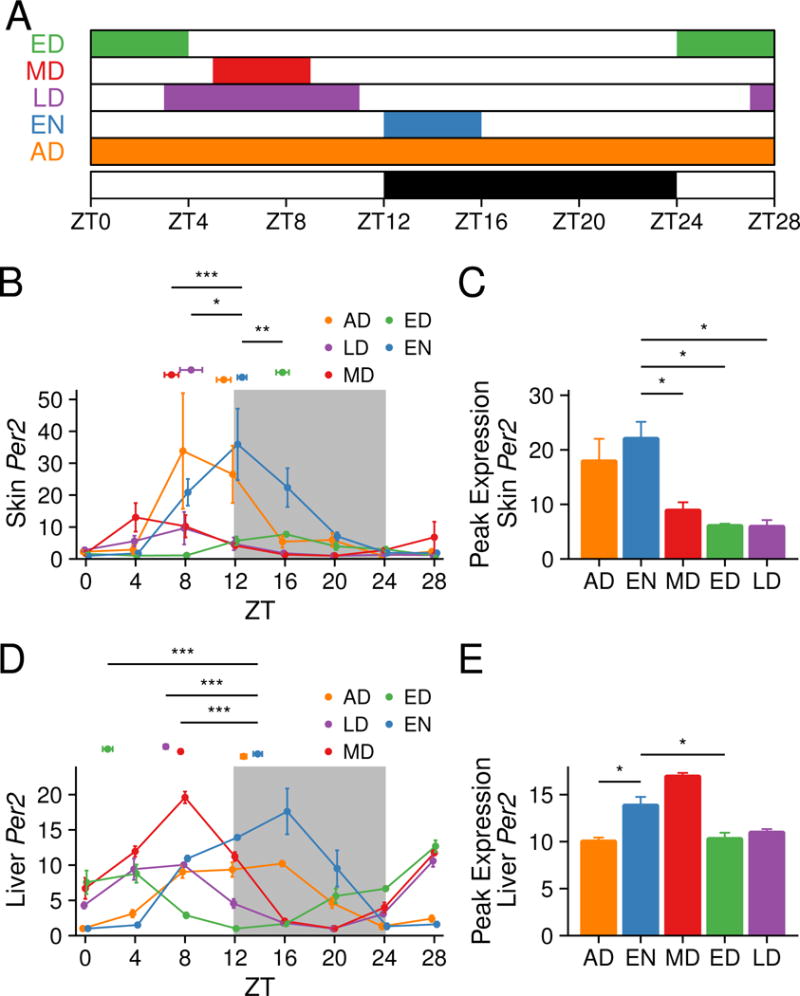
(A) Schematic showing the RF schedules. Colored boxes indicate timing of food access: AD, ad libitum; ED, early day; MD, mid-day; EN, early night; and LD, long day. Day and night are indicated below with white and black bars. The sampling time points are indicated by ZT on the x-axis. Per2 gene expression in the skin (B–C) and liver (D–E) as measured by qPCR. (B, D) The values represent mean ± SEM, N = 3–5. The peak time, a proxy for the circadian phase, is shown above the curves as mean ± SEM, N = 4. Watson-Williams test was used to compare the peak times. (C, E) The peak expression values are shown as mean ± SEM, N = 4. Statistical significance comparing peak expression values was determined by Welch’s t-test, shown as *p < 0.05, **p < 0.01, *** p < 0.001. See also Figure S2.
To evaluate the effect of RF schedules on body weight and food intake, we recorded food intake daily and body weight immediately prior to food availability every other day for 21–26 days for the AD, EN, MD and ED groups (Figure S1A–B). Both MD and ED mice ate significantly less than the AD mice during the first two days of R F, but by the end of the experiment there was no significant difference in food intake across the groups (Figure S1A). All groups weighed approximately the same prior to the beginning of the RF (data not shown), but throughout the RF experiment, body weight was significantly affected by RF schedule: AD weighed more than all RF groups, ED and EN had approximately equal body weight, and MD weighed consistently less than all other groups (Figure S1B). We performed two RF experiments as described in the Methods section, with similar results for body weight and food intake in both experiments (data not shown). We also measured skin compartment width (epidermis; dermis, including dermal fat layer; and muscle layer) by histology and found no significant changes except that dermis width was decreased by about 16–17% in EN and MD compared to AD (Figure S1C–F).
After implementing these RF schedules for 18 to 21 days, we harvested skin and liver from cohorts of mice every 4 hours for 28 hours starting at ZT0. We then determined the circadian clock phase by analyzing the peak time of skin mRNA expression of Per2, a commonly used indicator of the circadian phase. The phase of Per2 in mice fed during the night (EN) was equivalent to that of AD (Figure 1B). Using AD as a reference, we found that MD induced a phase advance on average of 4.19 ± 0.43 hours; in contrast, ED caused a phase delay on average of 4.72 ± 0.38 hours (Figure 1C, Figure S2A). The phase of Per2, then, was almost 9 hours apart for MD and ED, the groups with the most widely separated phases (Figure 1B). The magnitude of phase advances was the same in LD and MD (Figure S2A). We also found that the amplitude of Per2 was significantly lower in day-fed mice compared to EN (Figure 1C). Using either AD or EN as a reference, the phase shift of Per2 in ED was significantly different compared to LD and MD (Figure S2A). By contrast, in the liver, the phase of Per2 expression in all feeding groups was tightly linked to the time of initiation of food intake (Figure 1D, Figure S2B) and the peak expression of neither ED nor LD were significantly different from EN (Figure 1E).
To determine if the shift in the phase of the circadian clock was consistent in the skin and liver, we compared the phase shift of Per2 in the RF groups relative to AD or EN, and found that, while MD exhibited the same phase advance in skin and liver, the phase shifts for ED and LD were different in these organs (Figure S2C). Expression results for core clock genes Dbp (Figure S2D–F) and Per1 (Figure S2G–I) matched the Per2 results, indicating a true phase shift of the core clock machinery. We also studied the phase and peak expression of Per2 in isolated epidermis of EN and ED, finding that they are similar to that in whole skin (Figure S2J–K and Figure 1B–C).
Collectively, these data demonstrate that time of feeding influences the phase and peak expression of the skin circadian clock in manner distinct from that of the liver. While feeding appears to be a direct zeitgeber for the liver with Per2 expression having a constant relation to feeding time, there is a less direct relationship between the initiation of feeding and the phase of the skin circadian clock.
Diverse but functionally similar diurnal transcriptomes under different RF schedules
To define the diurnal transcriptome of skin under different RF schedules, we performed RNA-Seq on telogen skin collected every 4 hours for 28 hours under the three 4-hour RF schedules: ED, MD and EN (Figure 1A and Table S1). We selected mice from these RF schedules for further study in order to minimize effects of differences in caloric intake (see Methods section). EN mice were selected as the control because their circadian phase and time of feeding is similar to that of AD mice (Figure 1A–B); AD mice feed mainly during the early night (Vollmers et al., 2009; Yoon et al., 2012). Analysis of ED and MD mice allows us to observe the effect of maximum diurnal phase shifts as their circadian phases are nearly 9 hours apart in skin (Figure 1B, Figure S2A).
To quantify RNA expression levels, we separately counted the reads in exons, introns and antisense strands of all genes in the UCSC canonical gene set (Figure S3A). Using a combination of four algorithms (Figure S3B), we identified 2,997, 3,198 and 2,546 exon transcripts as diurnally oscillating (out of 22,583 detected) in the EN, ED and MD groups, respectively (Figure 2A, Table S2). Surprisingly, for each feeding group, there were a large number of genes with diurnally oscillating expression unique to that group, and only 147 genes were common to all three feeding groups (Figure 2A, Table S2). These common genes include the core clock regulators Clock, Cry1/2, Per2/3, Dbp, and Npas2 (Figure 2B, Table S2). The overall shift direction of the diurnal transcriptome common between any two RF schedules was consistent with the shift of the circadian clock (Figure 1B, Figure 2B–E, Table S2). Specifically, compared to EN, the phase of most diurnal genes in MD advanced while the phase of most diurnal genes in ED delayed (Figure 2C–E, Table S2). Whereas the specific genes overlapping between any two groups differ, their functional categories were conserved, including cell death, redox regulation, cell cycle and circadian clock (Figure 2B–E, Table S2).
Figure 2. Diurnal transcriptome of skin under time-restricted feeding.
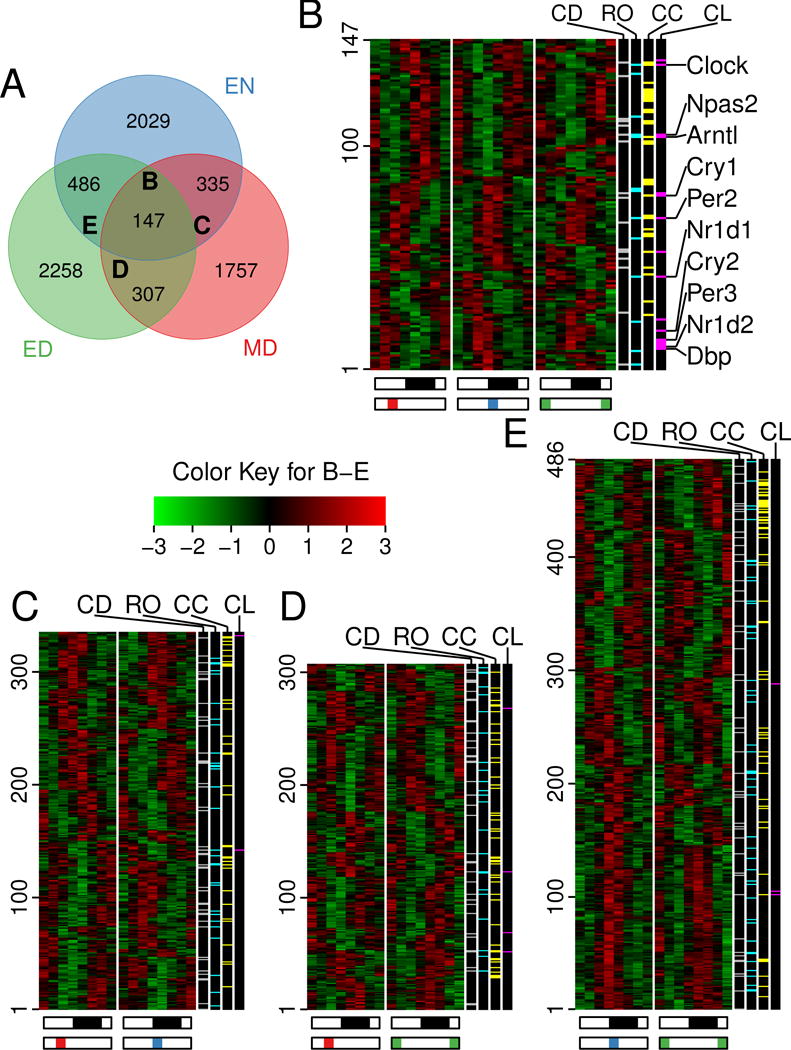
(A) Venn diagram depicting the overlap of diurnal exons expressed in skin of mice after time-restricted feeding. Out of 7,317 total diurnally oscillating exon transcripts, 147 are common within all 3 RF schedules. Exons for core circadian clock genes (such as Clock, Npas2, and Arntl) maintained diurnal rhythm in all RF schedules (core clock gene examples listed in panel B, to the right). (B–E) Heatmaps of the diurnal exons common under all (B) or 2 (C, D and E) of the RF schedules (corresponding to the B–E labels in (A)). Note that in B, most genes in MD are phase advanced while those in ED are phase delayed compared to EN. The white-black bars below indicate day and night, respectively. Colored bars under the heat maps indicate the time of food availability. The color key for the heat maps show the Z-score of expression value, where red is highly expressed and green is minimally expressed. The colored lines in black bars to the right of the heat maps indicate genes annotated to cell death (CD), reduction-oxidation (RO), cell cycle (CC), and circadian clock (CL) biological processes. See also Table S1–2 and Figure S3.
We also identified diurnal intron (Figure S3C, Table S2) and antisense (Figure S3D, Table S2) transcripts. As transcription affects both intron and exon components of transcripts, the comparison of phase shifts between introns and exons in a gene gives insights into the extent to which the phases of diurnal genes and biological processes under different RF schedules are controlled at a transcriptional or a post-transcriptional level. Overall, we found that the phase shift in peak expression in exons and introns of a gene was correlated (Pearson R= 0.68) suggesting that transcriptional regulation is an important mechanism underlying food entrainment in the skin (Figure 3A; Figure S3E–G, Table S2). This mode of regulation was reflected in our gene ontology (GO) analysis showing that the circadian clock genes are clearly regulated transcriptionally (Figure 3B). However, there were a number of genes that are exceptions to this general rule as exemplified by cytokinesis genes, which appear to be regulated post-transcriptionally (Figure 3B).
Figure 3. Functional and regulatory aspects of the skin diurnal transcriptome.
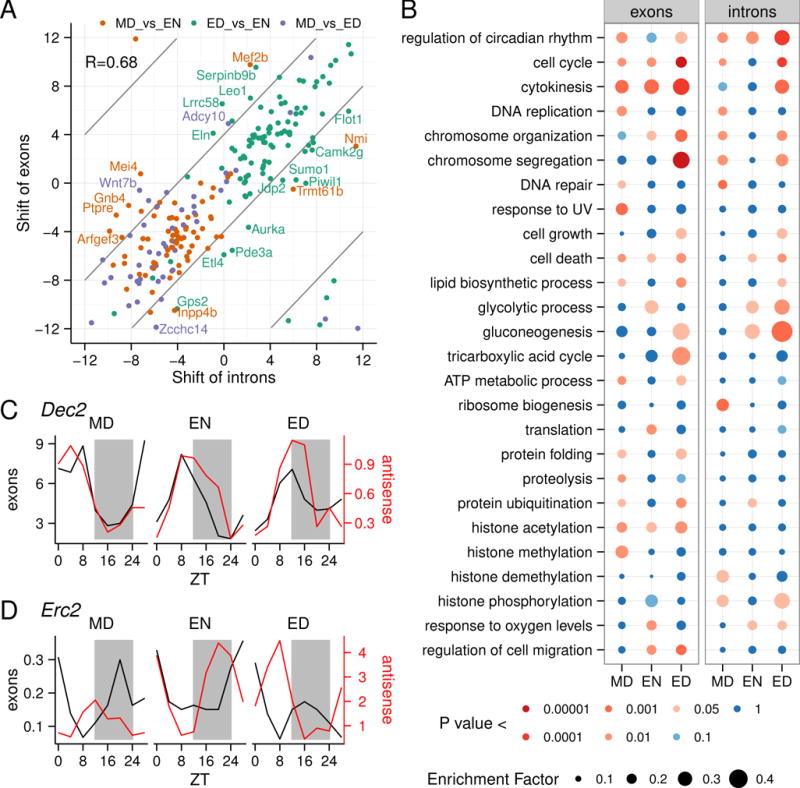
(A) Linear correlation of the time-shift of exon and intron expression under different RF schedules. Examples of genes where the differential timing of intron and exon expression is greater than 4 hours are indicated with gene symbols. (B) Enrichment of biological processes was determined for genes with diurnally expressed introns and exons by Fisher’s exact test. Rows are Gene ontology (GO) terms and columns are RF schedules. Enrichment factor was determined as the ratio of diurnal genes in a term over all genes in a term; this is represented by circle size. The color of the circles indicates p-value. This data suggests that genes annotated to “regulation of circadian rhythm” are mainly regulated at a transcriptional level, while genes annotated to “cell cycle”, “cell death” and “histone acetylation” are mainly regulated post-transcriptionally. (C and D) The expression of the exon and antisense transcripts for two diurnally expressed genes, Dec2 and Erc2, featuring diurnally expressed antisense transcripts in all three RF schedules. For Dec2, the exon and antisense oscillation is in phase, while for Erc2, the exon and antisense transcripts have antiphasic oscillations.
We found only 37 antisense transcripts with diurnal expression in all three feeding schedules (Figure S3D). Some of these, including the clock regulator Dec2 (also known as Bhlhe41), show expression in phase with the exons (Figure 3C) while others, including Erc2, which is involved in regulation of neurotransmitter release, show antiphasic expression to the exons (Figure 3D).
Together, these data indicate that RF shifts the phase of most diurnal genes in skin by transcriptional mechanisms. The RF-affected genes, while participating in similar functions, are largely unique to each RF schedule. In addition, we identified two antisense transcripts that may have a regulatory relationship with the gene from which they are encoded.
Food intake acutely regulates a portion of the skin transcriptome
We next considered the possibility that food intake could acutely affect skin gene expression. To search for such effects in the data, we rearranged the RNA-seq reads across all three feeding schedules according to the feeding time (Figure 4A), generating feeding time (FT) series (FT0-FT4-FT8) where FT0 represents the initiation of feeding and FT4 and FT8 represent times 4 and 8 hours after the start of feeding.
Figure 4. Food intake alters the skin transcriptome.
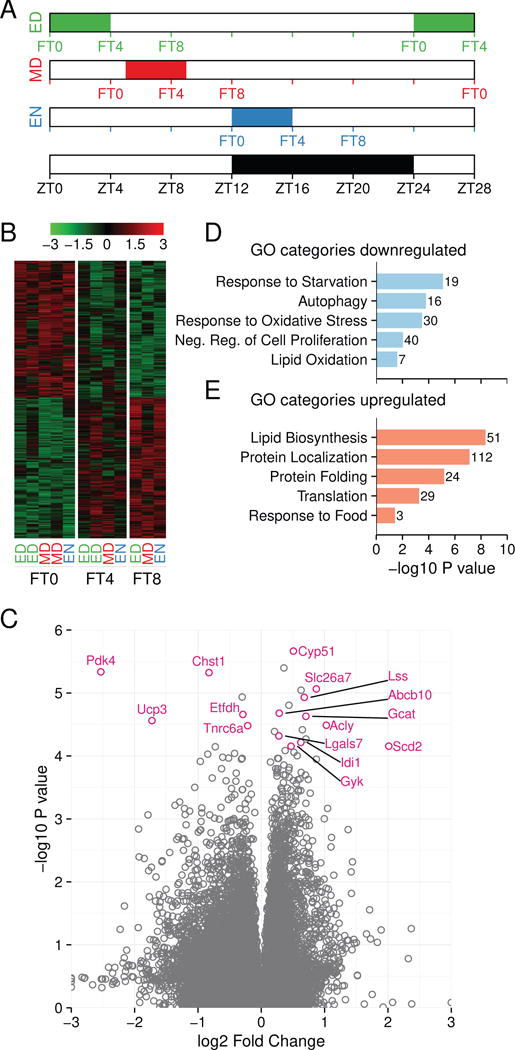
(A) A schematic showing how the RNA-seq reads from data presented in Figure 2 and 3 were regrouped and analyzed based on the timing of food availability. Feeding time zero (FT0) indicates reads immediately before food availability, feeding time 4 (FT4) indicates reads 4 hours after the onset of food availability, and feeding time 8 (FT8) indicates reads 8 hours after the onset of food availability, as described in the Methods section. Feeding groups are indicated below each column. The white-black bar indicates day and night, respectively. (B) 2,026 exons were affected by feeding based on the regrouping described in (A) (one-way ANOVA p < 0.01). Shown is the heat map of food intake-affected genes at FT0, FT4 and FT8. The color key represents the Z-score of expression value with red being highly expressed genes and green being minimally expressed genes. (C) Volcano plot showing feeding-affected exons. Representative, statistically significant metabolic genes are labeled. (D–E) Graphs of representative enriched GO categories for genes down-regulated (D) or up-regulated (E) after feeding. The numbers at the end of the bars refer to the number of genes affected in each category. See also Figure S4 and Table S3.
We identified 2,026 exons differentially regulated by food intake (Figure 4B and Figure S4A, B). Exons showing the most significant change in response to feeding were linked to metabolism (Figure 4C), including Pdk4, Ucp3 and Scd2. The food intake-affected exons fell into two groups: those that decreased (998) after food intake and those that increased (1,028) after food intake (Figure 4B–E, Figure 4SA–B). Genes showing decreased expression after food intake were overrepresented in GO categories including response to starvation, autophagy, response to oxidative stress, negative regulation of cell proliferation, and lipid oxidation (Figure 4D, Table S3) and those showing increased expression included lipid biosynthesis and protein synthesis (Figure 4E, Table S3). These results indicate that the metabolism of skin is oxidative before feeding and becomes anabolic after feeding. We also identified 1,890 introns and 662 antisense transcripts affected by food intake (Figure S4A–B). About half of the food intake-affected genes were identified as having diurnal expression in one or more of the three feeding schedules (Figure S4C), suggesting that food intake contributes to regulation of the diurnal transcriptome in the skin. In conclusion, these data indicate that expression of many genes in the skin, including those involved in oxidative-reductive metabolism and cell proliferation, respond acutely to food intake, and that the metabolic status of skin is determined by feeding.
RF-induced phase shifts of the circadian clock do not affect the diurnal phase of epidermal progenitor cell S-phase
Previous studies showed diurnal changes in epidermal progenitor cell proliferation with 3- to 4-fold greater number of cells in S-phase during the night than during the day (Geyfman et al., 2012). Deletion of Bmal1, either constitutionally or selectively in epidermal cells, obliterates the diurnal variation in cell proliferation, indicating that the circadian clock controls the diurnal variation in epidermal progenitor cell proliferation (Geyfman et al., 2012).
To determine whether the RF-induced phase shifts in the skin’s circadian clock alter the phase of the diurnal variation in epidermal progenitor cell proliferation, we measured the proportion of epidermal progenitor cells in S-phase by BrdU staining over a 28 hour period. The proportion of cells in S-phase is diurnal under each of the five RF schedules. Interestingly, despite the different phases in the circadian clock in different RF schedules, the phase of S-phase was not shifted by RF (Figure 5A). We also evaluated whether RF affects the proliferation rate of interfollicular epidermal progenitor cells, finding that each of the RF schedules resulted in a similar and decreased peak and overall progenitor cell proliferation compared to AD (Figure 5B). Together with previous findings showing that the circadian clock is required for diurnal DNA synthesis rhythms in epidermal progenitor cells (Geyfman et al., 2012), these results indicate that whereas the clock is critical for establishing diurnal variation in progenitor cell proliferation, it alone does not control the phase of the cell cycle.
Figure 5. Effect of time-restricted feeding on epidermal cell proliferation.
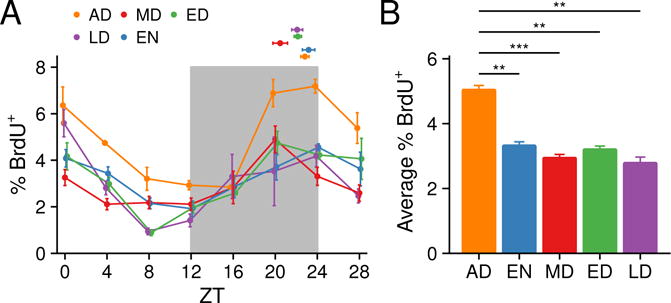
(A) BrdU-positive cells were counted in the epidermis by immunohistochemistry. Values represent mean ± SEM, N = 2–5. The proportion of cells in S-phase is diurnal under each of the five different RF schedules (one-way ANOVA p < 0.01). The dots above the curves indicate the peak time of BrdU incorporation. Values represent mean ± SEM, N = 4. Watson-Williams test was used to compare the peak times. The phase of diurnal rhythms of BrdU incorporation were unchanged among the four RF schedules. (B) The average percentage of BrdU-positive epidermal cells of each RF schedule. Values represent mean ± SEM, N = 4. AD had higher proliferation rate than all RF feeding schedules. Statistical significance was determined by Welch’s t-test, shown as **p < 0.01, and ***p < 0.001.
Daytime feeding shifts skin sensitivity to UVB-induced DNA damage
Previous work in mice showed that sensitivity to UVB-induced DNA damage in the epidermis is diurnal with more damage when UVB is applied during the night than during the day (Gaddameedhi et al., 2011; Geyfman et al., 2012). This diurnal variation depends on the circadian clock as mutations in Bmal1 (Geyfman et al., 2012) and Cry1/Cry2 (Gaddameedhi et al., 2011) obliterate the diurnal variation. To test whether daytime feeding, with its consequent shift in the phase of the clock, modulates the epidermal sensitivity to UVB-induced DNA damage, we applied UVB during the day (ZT9) and night (ZT21) to the shaved backs of AD, EN, ED, and MD mice, collecting the skin 15 minutes after UVB exposure. Consistent with previous studies (Gaddameedhi et al., 2011; Geyfman et al., 2012), mice that ate mainly (AD) or only (EN) at night formed more cyclobutane pyrimidine dimers (CPD) when exposed to UVB during the night than during the day (Figure 6A). By contrast, mice fed during the day (ED and MD) exhibited a reverse pattern, forming more CPD when exposed to UVB during the day than during the night (Figure 6A). Similar trends were observed in a earlier RF experiment with fewer mice in which we measured both CPDs and the second most common UVB-induced lesion, (6-4) Photoproducts ((6-4)PP) (Figure S5A). Thus, while not altering the phase of S-phase in epidermal progenitor cells, daytime RF reverses the diurnal rhythm of sensitivity to UVB-induced DNA damage. In addition, we found that the expression of Xpa, the gene encoding a rate-limiting protein necessary for nucleotide excision repair of UVB-induced DNA (Li et al., 2011; Miyamoto et al., 1992), is dampened in EN, MD, and ED compared to AD. Furthermore, Xpa expression oscillates in a diurnal fashion in AD but not as robustly in the RF schedules (Figure 6B).
Figure 6. Skin sensitivity to UVB-induced DNA damage.
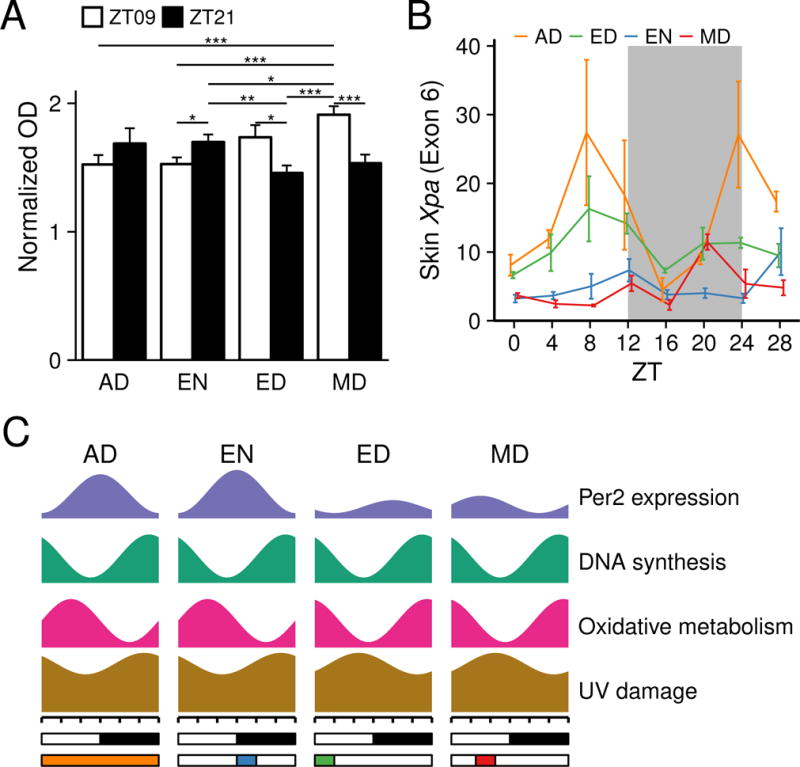
(A) Quantification of CPD photoproducts after UVB exposure at ZT9 and ZT21. The shaved back skins of mice from AD, EN, ED and MD feeding schedules were exposed to single dose of 500 J/m2 UVB. Greater UVB-induced DNA damage is seen in AD and EN mice exposed to UVB at night versus the day, while mice fed during the day (ED and MD) have greater UVB-induced DNA damage when exposed to UVB during the day than during the night. Values represent mean ± SEM, N = 15. Statistical significance was determined by Welch’s t-test, shown as *p < 0.05, **p < 0.01 and ***p < 0.001. (B) Skin Xpa expression is dampened under RF, as detected by qPCR. Values represent mean ± SEM (N = 3–5). Two-way ANOVA shows significance for RF group (p < 0.0001), ZT time (p < 0.002), and RF group X ZT time (p < 0.02). Tukey’s post hoc test shows AD has greater Xpa expression compared to EN (p < 0.0001), and MD (p < 0.0001), while AD is not significantly different than ED (p < 0.056). (C) A schematic summarizing the effects of abnormal feeding time on diurnal rhythms UVB-induced DNA damage, expression of oxidative-reductive metabolic enzymes, DNA synthesis and Per2 expression. See also Figure 1, Figure 2, Figure 5, Figure 6A.
In sum, these results demonstrate that daytime-restricted feeding affects the sensitivity to DNA damage in the skin of mice, and dampens the expression of a key DNA repair factor.
Discussion
This work shows that time of feeding is an important regulator of skin function. As summarized in Figure 6C, we found that: 1) RF shifts the phase of the skin circadian clock, in a pattern distinct from that of the liver; 2) RF alters the expression of many diurnally expressed genes in the skin, including that of the key DNA repair factor Xpa; 3) feeding acutely causes large scale gene expression changes in the skin, most prominently of metabolic genes; 4) day-time RF reverses the time-of-day-dependent sensitivity to UVB-induced DNA damage in the skin. Together, our results indicate that timing of food intake has a more pronounced influence on skin biology than previously recognized, representing a modifiable regulator of skin health.
Studies focusing on the liver, a key organ for organismal metabolism, showed that the phase of the circadian clock is entrained by RF (Damiola et al., 2000; Kuroda et al., 2012; Stokkan et al., 2001) and that a significant portion of the liver transcriptome is affected by the time of feeding (Vollmers et al., 2009). The effect of RF on clocks in peripheral organs not primarily involved in organismal metabolism is less well studied (Izumo et al., 2014; Reznick et al., 2013); in particular, there are no such studies in the skin. Our findings indicate that both the phase and peak expression of the skin circadian clock are distinct from that of the liver. In the skin, day-fed mice (MD, ED, LD) exhibit lower amplitude of the core clock gene Per2 than mice fed during the night (EN), while in the liver, Per2 amplitude does not seem to be sensitive to time of feeding. Furthermore, mice fed during the early daytime (ED) exhibited a 4.7-hour phase delay in Per2 expression compared to AD mice in the skin, while in the liver the phase of Per2 expression is advanced by 10.9-hours, corresponding to the initiation of food intake. These findings suggest that RF controls the phase of the skin circadian clock by different mechanisms than in the liver where feeding appears to be a more direct and dominant cue (Figure 1). These observations suggest that, in addition to regulation of the skin clock by the SCN’s central clock (Tanioka et al., 2009), the skin clock is independently modulated by the timing of food intake. The exact mechanism by which feeding time controls the skin clock likely involves many factors such as physical activity, sleep-wake cycles, metabolism, and circulating hormones.
Comparison of the telogen skin transcriptome under three different RF schedules (ED, MD, EN) reveals that feeding schedule dictates the diurnal expression of approximately 10% of the skin transcriptome (Figure 2). Specifically, RF schedules generate diurnal gene rhythms that are largely unique to each RF schedule while dampening diurnal expression of other genes that are diurnal in the other RF schedules (Figure 2).
Furthermore, while feeding time largely defines the identity of individual genes with oscillating expression, the functional categories annotated to these genes are similar among the three RF schedules. We also found that some diurnally expressed genes are conserved in all the three RF schedules; these genes are enriched for regulators of the circadian clock. The phase shift of the clock-related transcripts under the three RF schedules correlates well with the phase shift of the core circadian clock as indicated by the peak time of Per2 expression (Figure 1). In addition, the shift of expression of exons is linearly correlated with that of introns, indicating that shift of transcriptional activity is an important mechanism underlying the shift of the whole transcriptome (Figure 3). Genes annotated to regulation of circadian rhythm tend to be diurnally expressed in introns and exons while some genes annotated to the cell cycle, especially cytokinesis, tend to be diurnally expressed in exons and not introns. These findings suggest that different diurnal processes may be regulated to varying extents by both transcriptional and/or post-transcriptional mechanisms.
In contrast to exons, there are very few diurnal antisense transcripts. Also, the expression of antisense transcripts is largely uncorrelated with the expression of either introns or exons of the corresponding diurnal genes. We have identified a few antisense genes, however, that exhibit unique diurnal expression in relation to their corresponding genes, indicating a potential regulatory role.
We observed striking changes in skin gene expression directly in response to food intake (Figure 4). This suggests that at least part of the RF schedule-mediated changes in gene expression are a direct feeding effect although we cannot rule out contributions by changes in rhythms of activity level and sleep caused by restricted food availability. An analysis of these gene expression changes indicates that after feeding, cellular metabolism becomes biosynthetic and reductive. Instead of oxidation of fatty acids, the skin transcriptome becomes more characteristic of the synthesis and import of lipids, especially steroids, which are involved in cellular membrane systems. In addition, in response to food intake, genes involved in transcription, translation, and protein folding and localization are up-regulated and those involved in apoptosis are down-regulated. This analysis indicates that components of the global diurnal gene expression program are acutely responsive to food-intake.
In previous studies, we showed that the circadian clock intrinsic to keratinocytes is required for the diurnal fluctuation in the proportion of epidermal progenitors undergoing S-phase (Geyfman et al., 2012). Interestingly, in the current study we find that despite the shift in the phase of the circadian clock by RF, the rhythm of DNA synthesis in epidermal progenitors of the skin did not shift (Figure 5). Together these findings indicate that while an intact clock is required for the diurnal variation in DNA synthesis, the phase of the clock is not the dominant regulator of the phase of the S-phase oscillations in the mouse skin. These findings are in agreement with studies showing that the cell mitotic cycle can be uncoupled from the circadian clock in immortalized rat-1 fibroblasts (Yeom et al., 2010) and Lewis lung carcinoma cells (Pendergast et al., 2010), and a recent study showing that cell division cycles could be gated by WNT-signaling (Matsu-Ura et al., 2016). While a previous study showed that time restricted feeding can shift the daily proliferative rhythm of the digestive tract (Burholt et al., 1985), in the gut, cell proliferation may be mechanically stimulated by food.
Given our finding that oxidative metabolism in the skin is affected by time-restricted feeding, it is likely that a deviation in the time of food intake may lead to asynchrony between oxidative metabolism and DNA replication which normally is coordinated with cell cycle stages in epidermal progenitors (Stringari et al., 2015). We hypothesize that such asynchrony between the timing peak oxidative phosphorylation metabolism and cell division due to unusual feeding times could contribute to increased ROS-mediated DNA damage in progenitor and stem cells, leading to aging and carcinogenesis.
We also show that feeding at non-physiological times can alter the skin’s susceptibility to UVB-induced DNA damage. Mice that eat during the normal time (night) have greater sensitivity to UVB-induced DNA damage at night compared to during the day, consistent with previous studies (Figure S5; Geyfman et al., 2012; Gaddameedhi et al., 2011). This rhythm in DNA damage is attributed to the increased number of progenitors undergoing S-phase at night (Geyfman et al., 2012) and the diurnal variation in the efficacy of DNA excision repair mediated by the key nucleotide excision repair factor Xpa (Gaddameedhi et al., 2011). We found that daytime-fed mice had reversed diurnal rhythm of sensitivity to UVB-induced DNA damage with greater sensitivity during the day than during the night (Figure 6A). Since phase of S-phase is not altered by RF, other factors may contribute to the RF-altered rhythm in the susceptibility to DNA damage. In fact, the expression of the key repair gene Xpa is diurnal, peaking during the day in AD mice (Figure 6B) (Gaddameedhi et al., 2011; Khang et al., 2010), but exhibiting dampened and less rhythmic expression under RF schedules (Figure 6B).
In conclusion, our findings show that time-restricted feeding (EN, ED, MD) decreases the proportion of cells in S-phase, and dampens expression of DNA repair factor Xpa. In addition to these changes, daytime-restricted feeding (ED and MD) also shifts the phase of core clock genes and oxidative metabolism genes, and reverses the rhythm of sensitivity to UVB-induced DNA damage. By disrupting the natural expression and diurnal variation of such important processes in the skin, abnormally-timed food intake may contribute to the development of skin pathologies involving sun damage, skin aging and skin cancer.
Experimental Procedures
Animals
Twenty-three day old male C57BL/6NCrl mice (Charles River Laboratories, strain code 027) were acclimated to the mouse facility for 1 to 2 weeks before starting the RF experiments. The mice were housed under 12h:12h light/dark cycles (Light on at 6:30am, ZT0, and off at 6:30pm, ZT12) with free access to water during the whole experiment, and free access to food before initiation of the RF experiments. Regular chow was used in all experiments. The RF schedules were carried out for 18 to 21 days for the RNA-seq, qPCR and cell proliferation experiments (the first RF experiment) and 21 to 26 days for the DNA damage and body weight/food intake data (the second RF experiment). In the first RF experiment, EN mice had free access to food from ZT12 for 4 hours during the first 8 days, but after that were given the same amount food as was eaten by ED mice. Slight modifications to the RF schedules in the second RF experiment were as follows: The mice were weighed every other day immediately prior to daily food availability. Each day, ED and MD mice were given access to 20g of chow for 4 hours and leftover food was measured. The average food consumed per mouse in ED and MD was given to EN mice beginning at ZT12. Because EN mice ate all the food they received during the first 4 hours of food availability, EN food intake (Figure S1A) was represented as the food weight they received. In both experiments, skin was sampled at the end of the experiment during the second telogen when mice were at around postnatal day 54; the telogen state of the skin was verified by histology. All procedures were approved by the Institutional Animal Care and Use Committee of the University of California, Irvine (approval number 2001-2239).
Cell proliferation assays
Four hours prior to sampling, 50 μg/g BrdU was injected intraperitoneally. The histological staining for BrdU was performed as previously described (Geyfman et al., 2012). BrdU-positive and BrdU-negative cells were quantified with ImageJ with the observer blind to experimental conditions.
Quantitative PCR
Total RNA from whole dorsal back skin was purified with TRIzol Reagent (Life Technologies), cDNA was synthesized with iScript cDNA Synthesis Kit (Bio-Rad) and qPCR was performed with SsoFast EvaGreen/Probes Supermix (Bio-Rad). The Taqman probes for Per2 (Mm00478113.m1) and the Xpa probe detecting exons 2–3 (Mm00457111_m1) were from Applied Biosystems. Primers for Per1 and Dbp were from PrimerBank (Wang et al., 2012) (ID: 26349399a1 for Per1 and 8393240a1 for Dbp). The Xpa primer sequences for the last exon (exon 6) of Xpa are (5′-3′) ATTGCGGCGAGCAATAAGAAG and ACAAGGTACAAGTCTTGCGGT, from Eurofins.
Statistics on qPCR and cell proliferation
The biological replicates of each time point of each RF schedule were assigned to 4 sets of artificial time series by random permutation and the peak time, mean and peak expression of each time series was calculated with the cosinor function of psych package (Revelle, 2016). The peak value was calculated as mean × (1+amplitude). The phase shift between any two RF schedules was calculated using the peak time of the curves in the same set, so that 4 replicates of the peak time, average value, peak value and peak shift were produced at each permutation. The calculation of the mean and standard deviation of peak time as well as the Watson-Williams test of difference between peak time means and between the shifts was done with the circular package (Agostinelli and Lund, 2013). The calculation and test were done for 100 permutations and the average of the mean, SEM and the p-values were presented.
Stranded total RNA sequencing
Total RNA from mouse whole back skin with RNA integrity number (RIN) higher than 6.5 were pooled, treated with RNase-Free DNase (Qiagen cat. No. 79254), and cleaned with RNeasy Mini Elute Cleanup Kit (Qiagen). Library was constructed with TruSeq Stranded Total RNA Sample Prep Kit (Illumina) and sequenced with Hiseq 2500 (Illumina).
Analysis of RNA-Seq data
RNA-Seq short reads were mapped to mouse genome mm10 by Tophat2 (Kim et al., 2013), and rRNA reads were removed from the bam file via RSeQC (Wang et al., 2012). The reads with mapping quality less than 5 were removed with SAMtools (Li et al., 2009) (Table S1). Then the exon and intron gene reads and the reads for the antisense region of the whole gene body were counted with HOMER (Heinz et al., 2010). Transcripts were annotated using the UCSC known canonical gene set (31872 total; 22526 with introns). The reads were normalized into RPKM (Reads Per Kilobase per Million) by CQN (Hansen et al., 2012). The data analysis thereafter was performed with R (Ihaka and Gentleman, 1996; R Core Team, 2014), if not differently specified.
Identification of diurnal genes
Circadian genes identified in each of four algorithms, ARSER (Yang and Su, 2010), Lomb-Scargle (Glynn et al., 2006; Lomb, 1976), JTK-Cycle (Hughes et al., 2010) and sinusoid curve fitting (Geyfman et al., 2012), were combined to generate one circadian gene list. The exon reads from EN were ranked by the p-value of each method and compared with the genes circadian in telogen skin (Geyfman et al., 2012), or circadian in at least seven other tissues (Yan et al., 2008). The ratio of reference genes (positive found ratio, PFR) was found within a sliding window of 100 genes moving from the top (most significant p-values) for each method. Then, the PFRs of each method’s list were smoothed using the loess function in R base. A p-value cutoff was chosen for each method so that the smoothed PFR was high enough to be significant compared to a random ordering of genes, using a permutation test with a p-value cutoff of p<0.01. These cutoffs were applied to all transcripts for each method, and transcripts identified by at least one of these methods were incorporated into the final circadian list. Because each algorithm has pros and cons (Deckard et al., 2013), combining the different methods using PFR to determine the cutoffs in a consistent way yielded a more comprehensive circadian list. The peak time predicted by Lomb-Scargle method (Glynn et al., 2006; Lomb, 1976) was adopted for the phase shift analysis.
Identification of feeding affected genes
Whole skin RNA-seq exon reads from EN, ED and MD were re-grouped according to the feeding time (FT) (FT0 being the onset of feeding, FT4 being 4 hours after the onset of feeding, FT8 being 8 hours after the onset of feeding, for each particular RF schedule) and then averaged. Feeding-affected genes were identified by comparing RNA-seq expression values at FT0 (ZT4 and ZT28 of MD, ZT12 for EN, ZT0 and ZT24 for ED), FT4 (ZT8 of MD, ZT16 for EN, ZT4 and ZT28 for ED) and FT8 (ZT12 of MD, ZT20 for EN, ZT8 for ED) by one-way ANOVA. The higher ratio out of the FT8/FT0 and FT4/FT0 ratios was used to represent the fold change in gene expression after feeding.
Functional enrichment analysis
Gene ontology annotations were downloaded on Oct 1st 2014 from EBI (ftp://ftp.ebi.ac.uk/pub/databases/GO/goa/MOUSE/gene_association.goa_mouse.gz) and MGI (ftp://ftp.informatics.jax.org/pub/reports/gene_association.mgi). Annotations were combined to get a more comprehensive list of gene sets. Fisher’s exact test was employed for enrichment test and the IEA annotations were included.
UVB induced DNA damage assays
Two days before the end of the RF experiment, mice were anesthetized by intraperitoneal injection of ketamine (100 mg/kg) and xylazine (10 mg/kg) and then their back skin was shaved using electric clippers during a time that would not interfere with their feeding schedules. A single dose of 500 J/m2 was applied to the mice 15 minutes before sampling as described previously (Geyfman et al., 2012), at ZT9 or ZT19. Whole back skin DNA was isolated with QIAamp DNA Mini Kit (Qiagen cat. No. 51306). ELISA was employed to detect CPDs and (6-4)PP and with antibodies from Cosmo Bio Co. (NMDND001 for CPDs and NMDND002 for (6-4)PPs) following the manufacture’s protocol.
Skin histology measurement
The dorsal skin of the mice used in the second RF experiment was collected, fixed in 10% formalin for 48 hours, ethanol dehydrated and embedded in paraffin. After sectioning, dewaxing and rehydrating, staining with hemotocylin was performed. Twenty-fold magnified mosaic images were acquired on a Keyence microscope, with at least 1,000 μm skin length per image. Histological measurements were performed by a blinded researcher using ImageJ software. At least 2 images per mouse were analyzed and 15 measurements for each skin compartment per image were averaged.
Supplementary Material
Table S2. Genes with diurnal transcripts, Related to Figure 2
List of all genes detected by RNA-seq. The number of algorithms in which a given transcript type from a given feeding group was identified to be diurnal is indicated in columns C–K. Genes significantly associated with the gene ontology categories of cell death, redox, cell cycle and circadian clock are identified by a + in columns L–O.
Table S3. Gene ontology enrichment of genes affected by food intake, Related to Figure 4
Functional enrichment analysis was performed on feeding affected genes identified by RNA-seq. Columns A–B list BP terms, column C indicates whether a gene was up- or down-regulated after feeding, column D indicates the significance of gene expression change after initiation of food intake, and column E lists the feeding-affected genes associated with the respective GO term. GO categories and enrichment was determined as described in the “Functional enrichment analysis” Methods section.
Acknowledgments
This study was supported by the Irving Weinstein Foundation and National Institutes of Health Grant AR 56439 (to B.A.); China Scholarship Council Grant No.2011635103 (to H.W.); NSF Graduate Research Fellowship DGE-1321846 (to E.V.S). J.S.T. is an Investigator in the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
H.W., E.V.S., M.G., V.K., N.L., J.T. and B.A. conceived and designed the experiments. H.W., E.V.S. and M.G. performed the experiments. H.W. E.V.S., Q.L., M.S., A.I., and B.A. analyzed the data and H.W. made the figures with suggestions from all the authors. H.W., E.V.S. and B.A. wrote the manuscript with input from all of the authors.
Accession Numbers
RNA-Seq data is archived in a publicly accessible database at Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) with accession number GEO: GSE83855.
References
- Adamovich Y, Rousso-Noori L, Zwighaft Z, Neufeld-Cohen A, Golik M, KrautCohen J, Wang M, Han X, Asher G. Circadian clocks and feeding time regulate the oscillations and levels of hepatic triglycerides. CELL METAB. 2014;19:319–330. doi: 10.1016/j.cmet.2013.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agostinelli C, Lund U. R package ‘circular’: Circular Statistics (version 0.4-7) 2013 URL https://r-forge.r-project.org/projects/circular/
- Armstrong BK, Kricker A. The epidemiology of UV induced skin cancer. J Photochem Photobiol B. 2001;63:8–18. doi: 10.1016/s1011-1344(01)00198-1. [DOI] [PubMed] [Google Scholar]
- Bass J. Circadian topology of metabolism. NATURE. 2012;491:348–356. doi: 10.1038/nature11704. [DOI] [PubMed] [Google Scholar]
- Blanpain C, Fuchs E. Epidermal homeostasis: a balancing act of stem cells in the skin. Nat Rev Mol Cell Biol. 2009;10:207–217. doi: 10.1038/nrm2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchakjian MR, Kornbluth S. The engine driving the ship: metabolic steering of cell proliferation and death. Nat Rev Mol Cell Biol. 2010;11:715–727. doi: 10.1038/nrm2972. [DOI] [PubMed] [Google Scholar]
- Burholt DR, Etzel SL, Schenken LL, Kovacs CJ. Digestive tract cell proliferation and food consumption patterns of Ha/ICR mice. Cell Tissue Kinet. 1985;18:369–386. doi: 10.1111/j.1365-2184.1985.tb00668.x. [DOI] [PubMed] [Google Scholar]
- Damiola F, Le Minh N, Preitner N, Kornmann B, Fleury-Olela F, Schibler U. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev. 2000;14:2950–2961. doi: 10.1101/gad.183500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibner C, Schibler U, Albrecht U. The mammalian circadian timing system: organization and coordination of central and peripheral clocks. ANNU REV PHYSIOL. 2010;72:517–549. doi: 10.1146/annurev-physiol-021909-135821. [DOI] [PubMed] [Google Scholar]
- Fisher GJ, Kang S, Varani J, Bata-Csorgo Z, Wan Y, Datta S, Voorhees JJ. Mechanisms of photoaging and chronological skin aging. Arch Dermatol. 2002;138:1462–1470. doi: 10.1001/archderm.138.11.1462. [DOI] [PubMed] [Google Scholar]
- Fritz V, Fajas L. Metabolism and proliferation share common regulatory pathways in cancer cells. ONCOGENE. 2010;29:4369–4377. doi: 10.1038/onc.2010.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaddameedhi S, Selby CP, Kaufmann WK, Smart RC, Sancar A. Control of skin cancer by the circadian rhythm. Proc Natl Acad Sci U S A. 2011;108:18790–18795. doi: 10.1073/pnas.1115249108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyfman M, Kumar V, Liu Q, Ruiz R, Gordon W, Espitia F, Cam E, Millar SE, Smyth P, Ihler A, et al. Brain and muscle Arnt-like protein-1 (BMAL1) controls circadian cell proliferation and susceptibility to UVB-induced DNA damage in the epidermis. Proc Natl Acad Sci U S A. 2012;109:11758–11763. doi: 10.1073/pnas.1209592109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillette MU, Sejnowski TJ. Physiology. Biological clocks coordinately keep life on time. SCIENCE. 2005;309:1196–1198. doi: 10.1126/science.1111420. [DOI] [PubMed] [Google Scholar]
- Glynn EF, Chen J, Mushegian AR. Detecting periodic patterns in unevenly spaced gene expression time series using Lomb-Scargle periodograms. BIOINFORMATICS. 2006;22:310–316. doi: 10.1093/bioinformatics/bti789. [DOI] [PubMed] [Google Scholar]
- Hansen KD, Irizarry RA, Wu Z. Removing technical variability in RNA-seq data using conditional quantile normalization. BIOSTATISTICS. 2012;13:204–216. doi: 10.1093/biostatistics/kxr054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara R, Wan K, Wakamatsu H, Aida R, Moriya T, Akiyama M, Shibata S. Restricted feeding entrains liver clock without participation of the suprachiasmatic nucleus. GENES CELLS. 2001;6:269–278. doi: 10.1046/j.1365-2443.2001.00419.x. [DOI] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. MOL CELL. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes ME, Hogenesch JB, Kornacker K. JTK_CYCLE: an efficient nonparametric algorithm for detecting rhythmic components in genome-scale data sets. J Biol Rhythms. 2010;25:372–380. doi: 10.1177/0748730410379711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihaka R, Gentleman R. R: A Language for Data Analysis and Graphics. J COMPUT GRAPH STAT. 1996;5:299–314. [Google Scholar]
- Izumo M, Pejchal M, Schook AC, Lange RP, Walisser JA, Sato TR, Wang X, Bradfield CA, Takahashi JS. Differential effects of light and feeding on circadian organization of peripheral clocks in a forebrain Bmal1 mutant. ELIFE 3. 2014 doi: 10.7554/eLife.04617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janich P, Pascual G, Merlos-Suarez A, Batlle E, Ripperger J, Albrecht U, Cheng HY, Obrietan K, Di Croce L, Benitah SA. The circadian molecular clock creates epidermal stem cell heterogeneity. NATURE. 2011;480:209–214. doi: 10.1038/nature10649. [DOI] [PubMed] [Google Scholar]
- Jouffe C, Cretenet G, Symul L, Martin E, Atger F, Naef F, Gachon F. The circadian clock coordinates ribosome biogenesis. PLOS BIOL. 2013;11:e1001455. doi: 10.1371/journal.pbio.1001455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khang T, Lindsey-Boltz A, Reardon J, Sancar A. Circadian control of XPA and excision repair of cisplatin-DNA damage by cryptochrome and HERC2 ubiquitin ligase. PNAS. 2010;107:4890–4895. doi: 10.1073/pnas.0915085107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. GENOME BIOL. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondratov RV, Kondratova AA, Gorbacheva VY, Vykhovanets OV, Antoch MP. Early aging and age-related pathologies in mice deficient in BMAL1, the core component of the circadian clock. Genes Dev. 2006;20:1868–1873. doi: 10.1101/gad.1432206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda H, Tahara Y, Saito K, Ohnishi N, Kubo Y, Seo Y, Otsuka M, Fuse Y, Ohura Y, Hirao A, et al. Meal frequency patterns determine the phase of mouse peripheral circadian clocks. Sci Rep. 2012;2:711. doi: 10.1038/srep00711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamia KA, Papp SJ, Yu RT, Barish GD, Uhlenhaut NH, Jonker JW, Downes M, Evans RM. Cryptochromes mediate rhythmic repression of the glucocorticoid receptor. NATURE. 2011;480:552–556. doi: 10.1038/nature10700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laporte D, Lebaudy A, Sahin A, Pinson B, Ceschin J, Daignan-Fornier B, Sagot I. Metabolic status rather than cell cycle signals control quiescence entry and exit. J CELL BIOL. 2011;192:949–957. doi: 10.1083/jcb.201009028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levi F, Filipski E, Iurisci I, Li XM, Innominato P. Cross-talks between circadian timing system and cell division cycle determine cancer biology and therapeutics. Cold Spring Harb Symp Quant Biol. 2007;72:465–475. doi: 10.1101/sqb.2007.72.030. [DOI] [PubMed] [Google Scholar]
- Levi F, Schibler U. Circadian rhythms: mechanisms and therapeutic implications. Annu Rev Pharmacol Toxicol. 2007;47:593–628. doi: 10.1146/annurev.pharmtox.47.120505.105208. [DOI] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The Sequence Alignment/Map format and SAMtools. BIOINFORMATICS. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Musich PR, Serrano MA, Dong Z, Zou Y. XPA-mediated regulation of global nucleotide excision repair by ATR Is p53-dependent and occurs primarily in S-phase. PLOS ONE. 2011;6:e28326. doi: 10.1371/journal.pone.0028326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin KK, Kumar V, Geyfman M, Chudova D, Ihler AT, Smyth P, Paus R, Takahashi JS, Andersen B. Circadian clock genes contribute to the regulation of hair follicle cycling. PLOS GENET. 2009;5:e1000573. doi: 10.1371/journal.pgen.1000573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomb NR. Least-squares frequency analysis of unequally spaced data. 1976;39:447–462. [Google Scholar]
- Lowrey PL, Takahashi JS. Genetics of circadian rhythms in Mammalian model organisms. ADV GENET. 2011;74:175–230. doi: 10.1016/B978-0-12-387690-4.00006-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masri S, Cervantes M, Sassone-Corsi P. The circadian clock and cell cycle: interconnected biological circuits. CURR OPIN CELL BIOL. 2013;25:730–734. doi: 10.1016/j.ceb.2013.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsu-Ura T, Dovzhenok A, Aihara E, Rood J, Le H, Ren Y, Rosselot AE, Zhang T, Lee C, Obrietan K, et al. Intercellular Coupling of the Cell Cycle and Circadian Clock in Adult Stem Cell Culture. MOL CELL. 2016;64:900–912. doi: 10.1016/j.molcel.2016.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto I, Miura N, Niwa H, Miyazaki J, Tanaka K. Mutational analysis of the structure and function of the xeroderma pigmentosum group A complementing protein. Identification of essential domains for nuclear localization and DNA excision repair. J BIOL CHEM. 1992;267:12182–12187. [PubMed] [Google Scholar]
- Mohawk JA, Green CB, Takahashi JS. Central and peripheral circadian clocks in mammals. ANNU REV NEUROSCI. 2012;35:445–462. doi: 10.1146/annurev-neuro-060909-153128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, Schultz PG, Kay SA, Takahashi JS, Hogenesch JB. Coordinated transcription of key pathways in the mouse by the circadian clock. CELL. 2002;109:307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- Pendergast JS, Yeom M, Reyes BA, Ohmiya Y, Yamazaki S. Disconnected circadian and cell cycles in a tumor-driven cell line. Commun Integr Biol. 2010;3:536–539. doi: 10.4161/cib.3.6.12841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plikus MV, Van Spyk EN, Pham K, Geyfman M, Kumar V, Takahashi JS, Andersen B. The circadian clock in skin: implications for adult stem cells, tissue regeneration, cancer, aging, and immunity. J Biol Rhythms. 2015;30:163–182. doi: 10.1177/0748730414563537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plikus MV, Vollmers C, de la Cruz D, Chaix A, Ramos R, Panda S, Chuong CM. Local circadian clock gates cell cycle progression of transient amplifying cells during regenerative hair cycling. Proc Natl Acad Sci U S A. 2013;110:E2106–E2115. doi: 10.1073/pnas.1215935110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing. R foundation for Statistical Computing; 2014. Available at https://www.r-project.org/ [Google Scholar]
- Revelle W. psych: Procedures for Psychological, Psychometric, and Personality Research. 2016 Available at https://CRAN.R-project.org/package=psych.
- Reznick J, Preston E, Wilks DL, Beale SM, Turner N, Cooney GJ. Altered feeding differentially regulates circadian rhythms and energy metabolism in liver and muscle of rats. Biochim Biophys Acta. 2013;1832:228–238. doi: 10.1016/j.bbadis.2012.08.010. [DOI] [PubMed] [Google Scholar]
- Stokkan KA, Yamazaki S, Tei H, Sakaki Y, Menaker M. Entrainment of the circadian clock in the liver by feeding. SCIENCE. 2001;291:490–493. doi: 10.1126/science.291.5503.490. [DOI] [PubMed] [Google Scholar]
- Stringari C, Wang H, Geyfman M, Crosignani V, Kumar V, Takahashi JS, Andersen B, Gratton E. In Vivo single-cell detection of metabolic oscillations in stem cells. CELL REP. 2015;10:1–7. doi: 10.1016/j.celrep.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanioka M, Yamada H, Doi M, Bando H, Yamaguchi Y, Nishigori C, Okamura H. Molecular clocks in mouse skin. J INVEST DERMATOL. 2009;129:1225–1231. doi: 10.1038/jid.2008.345. [DOI] [PubMed] [Google Scholar]
- Vollmers C, Gill S, DiTacchio L, Pulivarthy SR, Le HD, Panda S. Time of feeding and the intrinsic circadian clock drive rhythms in hepatic gene expression. Proc Natl Acad Sci U S A. 2009;106:21453–21458. doi: 10.1073/pnas.0909591106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Wang S, Li W. RSeQC: quality control of RNA-seq experiments. BIOINFORMATICS. 2012;28:2184–2185. doi: 10.1093/bioinformatics/bts356. [DOI] [PubMed] [Google Scholar]
- Wang X, Spandidos A, Wang H, Seed B. PrimerBank: a PCR primer database for quantitative gene expression analysis, 2012 update. NUCLEIC ACIDS RES. 2012;40:D1144–D1149. doi: 10.1093/nar/gkr1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Wang H, Liu Y, Shao C. Analysis of gene regulatory networks in the mammalian circadian rhythm. PLOS COMPUT BIOL. 2008;4:e1000193. doi: 10.1371/journal.pcbi.1000193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang R, Su Z. Analyzing circadian expression data by harmonic regression based on autoregressive spectral estimation. BIOINFORMATICS. 2010;26:i168–i174. doi: 10.1093/bioinformatics/btq189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeom M, Pendergast JS, Ohmiya Y, Yamazaki S. Circadian-independent cell mitosis in immortalized fibroblasts. Proc Natl Acad Sci U S A. 2010;107:9665–9670. doi: 10.1073/pnas.0914078107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon JA, Han DH, Noh JY, Kim MH, Son GH, Kim K, Kim CJ, Pak YK, Cho S. Meal time shift disturbs circadian rhythmicity along with metabolic and behavioral alterations in mice. PLOS ONE. 2012;7:e44053. doi: 10.1371/journal.pone.0044053. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S2. Genes with diurnal transcripts, Related to Figure 2
List of all genes detected by RNA-seq. The number of algorithms in which a given transcript type from a given feeding group was identified to be diurnal is indicated in columns C–K. Genes significantly associated with the gene ontology categories of cell death, redox, cell cycle and circadian clock are identified by a + in columns L–O.
Table S3. Gene ontology enrichment of genes affected by food intake, Related to Figure 4
Functional enrichment analysis was performed on feeding affected genes identified by RNA-seq. Columns A–B list BP terms, column C indicates whether a gene was up- or down-regulated after feeding, column D indicates the significance of gene expression change after initiation of food intake, and column E lists the feeding-affected genes associated with the respective GO term. GO categories and enrichment was determined as described in the “Functional enrichment analysis” Methods section.


