Abstract
We established a method to generate integration from extrachromosomal arrays with the CRISPR/Cas9 system. Multi-copy transgenes were integrated into the defined loci of chromosomes by this method, while a multi-copy transgene is integrated into random loci by previous methods, such as UV- and gamma-irradiation. The effects of a combination of sgRNAs, which define the cleavage sites in extrachromosomes and chromosomes, and the copy number of potential cleavable sequences were examined. The relative copy number of cleavable sequences in extrachromosomes affects the frequency of fertile F1 transgenic animals. The expression levels of the reporter gene were almost proportional to the copy numbers of the integrated sequences at the same integration site. The technique is applicable to the transgenic strains abundantly stored and shared among the C. elegans community, particularly when researchers use sgRNAs against common plasmid sequences such as β-lactamase.
Keywords: C. elegans, CRISPR/Cas9, Transgenic integration
Highlights
-
•
A method to generate multi-copy integration lines with the CRISPR/cas9 system.
-
•
Transgenes were integrated into defined loci in the chromosomes of C. elegans.
-
•
Positive/negative selection was applicable to the CRISPR/Cas9 integration system.
1. Introduction
To perform molecular genetic analyses using a model organism, techniques for generating transgenic animals are essential. In C.elegans transgenic animals are often obtained using DNA microinjection into the hermaphrodite gonad, which generates multi-copy extrachromosomal (Ex) arrays [1], [2]. Extrachromosomal arrays can then be integrated into random loci of chromosomes by gamma-ray or ultraviolet irradiation [3], [4], so that the integrated arrays contain a high copy-number of transgenes. A technique termed Mos1-mediated single-copy insertion (MosSCI) is often the method of choice to insert a single-copy transgene into a defined location in the chromosomes of C. elegans [5], [6]. Extrachromosomal arrays can also be integrated into random loci of chromosomes by using UV/TMP, benzimidazole and temperature selection [7], [8]. In these cases, integrated transgenes contain single/low-copy transgenes and are useful for genes with low expression levels. Recently, the CRISPR/Cas9 system, which can induce double-strand breaks at specific loci in genomic DNA using specific sgRNAs, has been successfully applied to C. elegans [9]. The system has been widely used to produce small indels [9]. In addition, the CRISPR/Cas9 system allows homologous recombination-mediated insertion of sequence tags such as GFP [10], [11], [12].
In the present study, we developed a technique using the CRISPR/Cas9 system to produce integrations from extrachromosomal arrays. This method allows the insertion of multi-copy transgenes at desired loci in chromosomes. In addition, we also demonstrate transgene integration of a relatively small number of copies using the CRISPR/Cas9 system together with benzimidazole and temperature selection.
2. Materials and methods
2.1. Nematode strains
C. elegans strain N2 worms were used as the wild-type animals. Worms were grown at 20 °C under well-fed conditions using the standard methods [13]. Strains carrying the following mutations were obtained from a UV/TMP-mutagenized library as described previously [14] and identified by PCR amplification with primers spanning the deletion region of ben-1(tm234)III and vps-45(tm246)X, as described previously [14], [15]. The primers used for PCR genotyping were as follows: tm234_1stround, 5′-ACGTGGGAATGGAACCATGT-3′, 5′-TCTCCATTTCCTCTTCCTCC-3′; tm234_2ndround, 5′-CTCCGGACATTGTAACGGAA-3′, 5′-CCCTCCATTTGAAAGAGTCC-3′; tm246, 5′-CGCAATTGGATACTACTTGT-3′, 5′-TCTCCTGCTCTACTTCTGCT-3′. The mutants were backcrossed four times with N2 before use.
2.2. Plasmid construction
We used site-directed mutagenesis to insert the guide sequences into a Cas9-sgRNA (single guide RNA) expression vector (pDD162) containing both sgRNA and Cas9 protein expression units, obtained through Addgene [11]. dpy-3 was targeted for Cas9 cleavage using the guide sequence (TCACCGTCCAGTCTGCTAC) inserted into pDD162. ben-1 was targeted for Cas9 cleavage using the guide sequence (AAGCAACTGCAGAGGAAGA) inserted into pDD162. The β-lactamase (ampicillin resistance) gene, which is contained in commonly used plasmids, was targeted for Cas9 cleavage using the guide sequence (TTAATAGACTGGATGGAGG) inserted into pDD162. We searched “CRISPR Design” (http://crispr.mit.edu) for potential off-target sequences [16]. The guide sequence of dpy-3 had 1 candidate of off-target sites (0 are in genes, 4 mismatches (MMs), score of off-target: 0.0). The guide sequence of ben-1 had 32 candidates of off-target sites (0 are in genes, 4 of 3MMs and 28 of 4MMs, max score of off-target: 1.5). The guide sequence of the β-lactamase gene had 7 candidates of off-target sites (0 are in genes, 7 of 4MMs, max score of off-target: 0.8). To generate a dpy-3 genome fragment plasmid, pPD95.79_dpy-3, a part of the dpy-3 genome (491 bp), was amplified by PCR using the primers 5′-CGACTCTAGAGGATCCGAACAGTGAAGTAGACTTCTGCAAG-3′ and 5′- CGCTCAGTTGGAATTCCATCTTGTCCTGATGTACCGGCAT-3′ with the N2 genome as a template. The fragment was inserted into the plasmid pPD95.79 using the EcoRI and BamHI sites.
The plasmids for transgenic markers, myo-2p::Venus and dpy-7p::DsRed, were generated as outlined previously [17]. The plasmids, pGEMT_ben-1(+), pFX_HBG_Lw_dpy-30p::NLS::GFP and eft-3p::vps-45 were constructed as described in a previous study [7].
2.3. Transgenic lines
Plasmids were injected using the standard C. elegans microinjection procedures [1]. To generate tmEx4252, tmEx4253 and tmEx4277 transgenic animals, the pPD95.79_dpy-3 genome was injected with myo-2p::Venus as an injection marker into N2 worms. The injection ratios of the mixed plasmids and the selection markers are listed in Table 1. We used the same tmEx3199 [eft-3p::vps-45, pGEMT_ben-1(+), pFX_HBG_Lw_dpy-30p::NLS::GFP] transgenic animals that were used in a previous study [7]. Images were taken with a BX-51 microscope (Olympus).
Table 1.
Summary of the multi-copy integration efficiencies into the dpy-3 locus by using the CRISPR/Cas9 system.
| Experiment no. | Parent strain | Cas9 target | No. injecteda | total F1b | No. F1 producing F2 (viability %)c | No. F1 producing Dpy F2d | No. producing Is (Allele name)e | Frequency (Is lines/P0 animals) % |
|---|---|---|---|---|---|---|---|---|
| Exp. 1 | tmEx4252[dpy-3 genome : myo-2p::Venus = 9 : 1] | dpy-3 | 55 | 189 | 97 (51.3%) | 34 | 2 (tmIs1149, tmIs1151) | 3.6 |
| Exp. 2 | tmEx4277[dpy-3 genome : myo-2p::Venus = 1 : 1] | dpy-3 | 45 | 123 | 69 (56.1%) | 15 | 2 (tmIs1155, tmIs1156) | 4.4 |
| Exp. 3 | tmEx4253[dpy-3 genome : myo-2p::Venus = 1 : 9] | dpy-3 | 43 | 94 | 73 (77.6%) | 27 | 4 (tmIs1152, tmIs1154, tmIs1158, tmIs1162) | 9.3 |
| Exp. 4 | tmEx4252[dpy-3 genome : myo-2p::Venus = 9 : 1] | dpy-3, β-lactamase gene | 40 | 35 | 14 (40.0%) | 1 | 1 (tmIs1150) | 2.5 |
| Exp. 5 | tmEx3199[pFX_HBG_Lw_dpy-30p::NLS::GFP, peft-3::vps-45, pGEMT_ben-1(+)] | dpy-3, β-lactamase gene | 50 | 65 | 48 (73.8%) | 8 | 4 (tmIs1157, tmIs1161, tmIs1163, tmIs1164) | 8.0 |
| Exp. 6 | tmEx3199[pFX_HBG_Lw_dpy-30p::NLS::GFP, peft-3::vps-45, pGEMT_ben-1(+)] | dpy-3 | 32 | 114 | 82 (71.9%) | 30 | 0 | 0.0 |
The number of injected parent Ex strains.
The number of Ex marker (myo-2p::Venus or dpy-30p::NLS::GFP)-positive and injection marker (dpy-7p::DsRed or myo-2p::DsRed)-positive F1 animals.
The number of F1 animals producing viable DsRed(+) or DsRed(−) F2 animals. The viability (No. F1 producing F2 / total F1) is described in parentheses.
The number of F1 animals which laid GFP/Venus(+) or GFP/Venus(−) Dpy animals in F2 progeny.
The number and the names of produced Is lines.
2.4. Generating integration lines using the CRISPR/Cas9 system
We injected Cas9-sgRNA plasmids with a selection marker, dpy-7p::DsRed or myo-2p::DsRed, into the extrachromosomal transgenic animals. The target genes for the CRISPR/Cas9 system and injected animals are listed in Table 1 and Supplementary Fig. 2B.
2.5. Quantitative PCR
Genomic DNA was isolated from adult animals using the DNeasy Tissue & Blood kit (QIAGEN). Quantitative PCR was performed in a 7500 Real-time Thermal cycler (Applied Biosystems) using the Power SYBR master mix (Applied Biosystems) with the following parameters: 95 °C for 10 min and 40 cycles of 95 °C for 5 s, 55 °C for 10 s and 72 °C for 34 s. All data were normalized to the ama-1 gene. The primers used are listed in Supplementary Table 1.
2.6. Isolation of integration lines using the CRISPR/Cas9 system and the Dpy phenotype
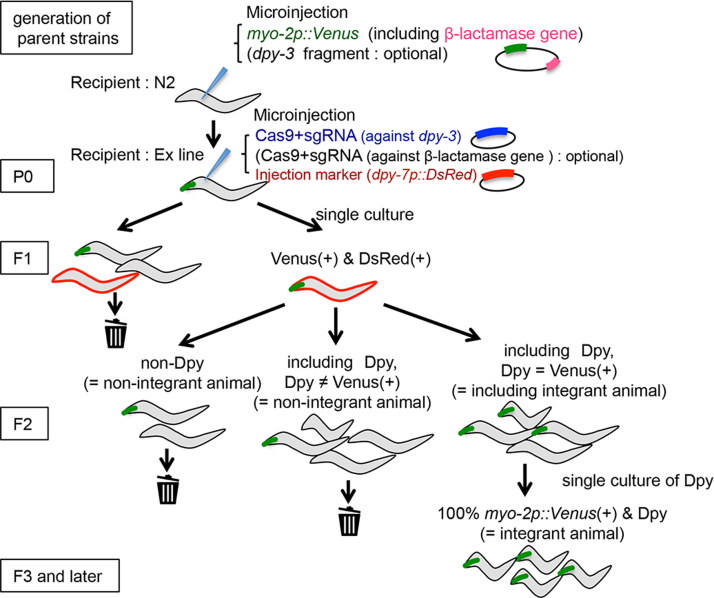
Schematic diagram of the multi-copy integration is shown in the Fig. 1. Ex line marker (Venus/GFP)-positive worms (P0) were injected with the Cas9-sgRNA plasmid(s), against the dpy-3 and/or β-lactamase gene(s), with another injection marker, dpy-7p::DsRed or myo-2p::DsRed. After a few days, both the parent Ex marker- and injection marker-positive F1 worms were singled to new plates. We selected the lines that expressed Venus/GFP in all Dpy animals. The strains harboring inserted parental extrachromosomal arrays into the genomic location of the dpy-3 gene were selected in this process. We selected Dpy worms as insertion (Is) lines that had the parent Ex marker in all the F3 or later progeny. We confirmed that the Dpy phenotype did not segregate with fluorescence, and confirmed that transgenes were integrated at the dpy-3 gene by PCR and sequencing (Supplementary Fig. 1). We performed PCR using purified genomic DNA of each Is strain as a template. We designed primer sets located within the dpy-3 gene, which is not included in the extrachromosomal arrays, and the vectors, which is included only in the extrachromosomal arrays. The primers used in this experiment are listed in Supplementary Table 2. High transmitting Ex lines were produced in this experiment, but it was possible to exclude these Ex lines by our method.
Fig. 1.
Overview of multi-copy integration experiments by using the CRISPR/Cas9 system. An injection marker (myo-2p::Venus including β-lactamase gene) and a portion of the dpy-3 genome sequence (optional) were co-injected into the N2 worms to generate transgenic strains carrying Ex arrays. Ex line marker (Venus)-positive worms (P0) were injected with the Cas9-sgRNA plasmid(s), against the dpy-3 and/or β-lactamase gene(s), with an injection marker (dpy-7p::DsRed). Cas9-sgRNA plasmid(s) were designed to break extrachromosomal arrays and the integration locus. Both the parent Ex marker- and injection marker-positive F1 worms were singled to new plates. The lines that expressed Venus in all Dpy animals were selected as candidates for integrant (Is) strains. F2 Dpy worms in Is candidate strains were singled to new plates. Dpy worms that had the parent Ex marker in all the F3 or later progeny were selected as Is lines.
2.7. Isolation of integration lines using the CRISPR/Cas9 system with the aid of benzimidazole, temperature and PCR selection
Schematic diagram of the integration is shown in the Supplementary Fig. 2A. ben-1(tm234); vps-45(tm246); tmEx3199 [eft-3p::vps-45, pGEMT_ben-1(+), pFX_HBG_Lw_dpy-30p::NLS::GFP] (P0) were injected with the Cas9-sgRNA plasmid, which targeted to the ben-1 gene, and an injection marker, myo-2p::DsRed. After a few days, both the parent Ex marker (dpy-30p::NLS::GFP)- and the injection marker-positive F1 worms were singled to new plates and incubated at 20 °C, while the F3 animals hatched. The F3 animals were selected for integrant strains by benzimidazole and temperature by using a modified version of a previously described method [7], [8]. Briefly, F3 animals were plated onto NGM agar plates containing 10 µg/ml benzimidazole (Wako) (approximately 50 animals/9-cm plate) and were cultured at 20 °C for 7–10 days. F5 animals were plated onto NGM agar plates (approximately 100 animals/9-cm plate) and cultured at 20 °C for 3 days. In this process, the integrant animals carrying the vps-45 transgene but not carrying the complete ben-1 transgene were selected. Surviving animals were singled and examined by PCR. The primers used are listed in Supplementary Table 3. We confirmed that the integration of transgenes at the ben-1 gene by PCR and sequencing (Supplementary Fig. 1). We performed PCR using purified genomic DNA of each Is strain as a template. We designed primer sets located within the ben-1 gene, which is not included in the extrachromosomal arrays, and the vectors, which is included only in the extrachromosomal arrays. The primers used in this experiment are listed in Supplementary Table 2.
3. Results and discussion
3.1. Transgene integration using the CRISPR/Cas9 system
We investigated whether the CRISPR/Cas9 system is able to generate integrated transgenic worms, and how the number of Cas9 cleavage sites in extrachromosomes affects the frequency of isolation of transgenic animals. We first attempted to determine whether a single sgRNA could cleave both extrachromosomal arrays and the genomic target site by using an sgRNA targeting dpy-3 as the simplest experimental design (Fig. 1). Because dpy-3 mutants show a highly penetrant visible and viable recessive phenotype termed “Dpy”, we inferred that integration of an extrachromosomal transgene into the dpy-3 locus would yield Dpy and fluorescence-positive integrant animals. Integrant Dpy lines can be distinguished easily from parental extrachromosomal transgenic animals without the Dpy phenotype. We selected Dpy worms as Is lines that had the parent Ex marker (Venus) in all the F3 or later progeny. To investigate the effect of the number of cleavage sites in extrachromosomal array, we generated 3 parent Ex lines that expressed the myo-2p::Venus together with a portion of the dpy-3 genome sequence (491 bp) at three different concentration ratios (Table 1, exp. 1–3). The dpy-3 genome fragment served as the target for sgRNA against the dpy-3 locus, as shown in Fig. 1 and Table 1.
We expressed Cas9, the dpy-3-specific sgRNA and a vector driving the expression of dpy-7p::DsRed to label transformed F1 progeny from the parent transgenic worms, tmEx4252, tmEx4277 or tmEx4253. We isolated DsRed- and Venus-positive worms and screened their F2 progeny for Dpy phenotypes (Fig. 1). The integrated transgenic strains in the progeny were easily identified by myo-2p::Venus fluorescence and Dpy (Table 1, column “No. producing Is”). We obtained integrant strains at frequencies of 3.6%, 4.4% and 9.3% from tmEx4252, tmEx4277 and tmEx4253, respectively. Decreasing the number of dpy-3 sequence in extrachromosomal arrays induced higher efficiency of generating integrant strains (Supplementary Table 4, exp. 1–3). Notably, we obtained F2 animals with the Dpy phenotype without Venus fluorescence (Table 1, column “No. F1 producing Dpy F2”). This suggests that not all of the animals with a double-strand break at the dpy-3 locus accepted the extrachromosomal DNA fragments but were instead self-repaired with some changes (small deletion and/or insertion) at the locus, as found in many CRISPR/Cas9-induced knockout strains [9]. Candidates of integrated strains with Dpy and Venus-positive phenotypes did not segregate Dpy and Venus negative nor non-Dpy and Venus-positive in the present experiments. We confirmed that the transgenes were integrated at the dpy-3 locus by PCR and sequencing (Supplementary Fig. 1). Thus, integration into off-target loci is rare if not at all. Practically Venus fluorescence alone is enough to isolate integrated strains. We conclude that extrachromosomal arrays can be integrated into chromosomal loci of interest by using the CRISPR/Cas9 system (Fig. 2). The frequencies of isolation of integrant strains were between 3.6% and 9.3% of P0 strains, which is comparable to or a little higher than the integration method by short wave-length UV irradiation, which yields integrant strains of up to 5% of P0 animals [3]. Our results suggest that a higher concentration of recognition sequences for sgRNAs in extrachromosomal arrays results in a lower frequency of producing transgenic animals. To further examine this hypothesis, we microinjected tmEx4252 transgenic worms with two vectors encoding Cas9, dpy-3-, and β-lactamase-specific sgRNAs together with an injection marker, dpy-7p::DsRed. One integrant strain was isolated from 40 injected P0 worms, suggesting that use of the β-lactamase-specific sgRNA yielded integrated strains at a comparable or slightly lower frequency (2.5%, Table 1, exp. 4) than that of injection of the dpy-3 sgRNA alone (3.6%; Table 1, exp. 1). This result supports the hypothesis that too many fragmented extrachromosomal arrays may reduce the frequency of producing transgenic animals.
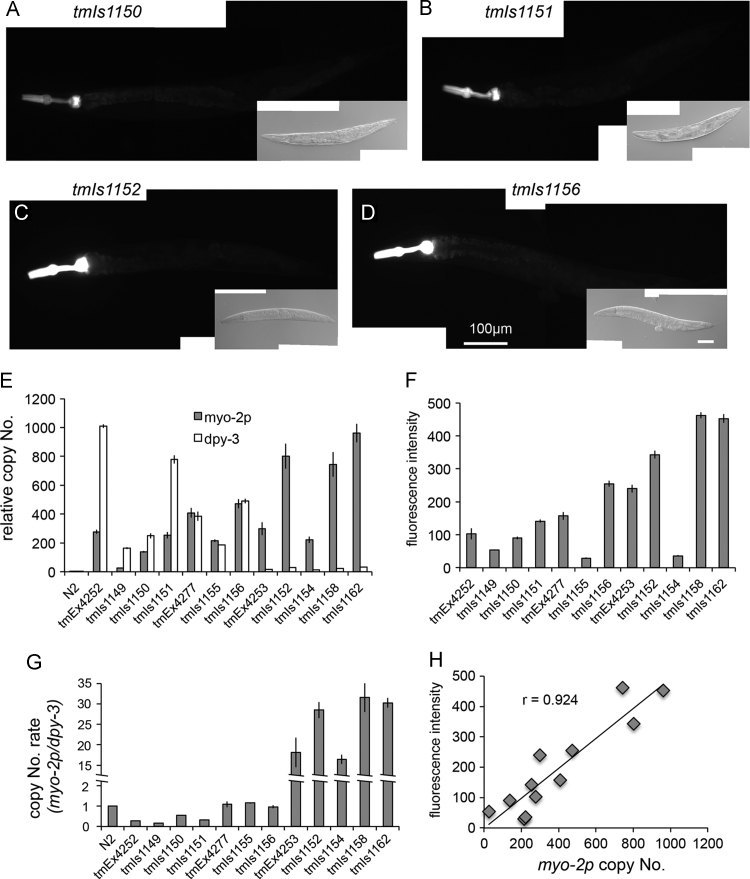
Fig. 2.
Expression of myo-2p::Venus in the multi-copy integrated strains (A-D) Venus protein expressed in pharyngeal muscle cells. Scale bar=100 µm. (400 msec. exposure time) (E) Relative amounts of the myo-2 promoter and the dpy-3 genome as determined by quantitative PCR. The data were normalized to the ama-1 gene. The copy numbers are presented as a ratio to the wild-type N2. Bars represent the mean±SE of three independent experiments. (F) Quantification of the fluorescence in pharyngeal muscle cells. The graph represents the intensity of fluorescence of pharyngeal muscle cells (n>30 animals per strain). Fluorescence intensity was examined using ImageJ (NIH, Bethesda, MD). Bars represent the mean±SE. (G) The copy number ratio of the myo-2 promoter to the dpy-3 genome in the 3 parent Ex lines and the 9 Is lines. Bars represent the mean±SE of three independent experiments. (H) The scatter plot of the number of myo-2 promoter (x-axis) against fluorescence intensity (y-axis) in the 3 Ex lines, tmEx4252, tmEx4253 and tmEx4277, and the 9 Is lines, tmIs1149, tmIs1150, tmIs1151, tmIs1155, tmIs1156, tmIs1152, tmIs1154, tmIs1158 and tmIs1162.
3.2. Homologous sequences in extrachromosomal arrays are dispensable for generating integrant strains
There are abundant extrachromosomal transgenic strains shared by the C. elegans research community stored in the CGC (Caenorhabditis Genetics Center; http://www.cbs.umn.edu/research/resources/cgc/strains) and many individual laboratories. Thus, we next investigated whether it is possible to add another sgRNA to target a sequence that is common among most extrachromosomal transgenic animals, to investigate whether such biological materials could be re-used as parent strains for integration into a locus of interest. We used sgRNA against the β-lactamase gene, which is present in all parent strains in the present experiments and is likely to be present in most of the existing extrachromosomal transgenic strains, since the β-lactamase (ampicillin resistance) gene is contained in general plasmid vectors. We attempted to integrate extrachromosomal transgenes by using another strain, tmEx3199, which lacks the dpy-3 sequence in the extrachromosomal arrays. We injected sgRNAs against both the β-lactamase and dpy-3 genes as above and obtained 4 integrant strains from 50 P0 parent animals (8.0%, Table 1, exp. 5). By contrast, we could not generate any Is line by injecting tmEx3199 with sgRNA against only the dpy-3 gene and Cas9 (Table 1, exp. 6, Supplementary Table 4). We conclude that fragmentation of the array, in addition to the cleavage of the genome, is important for efficient integration.
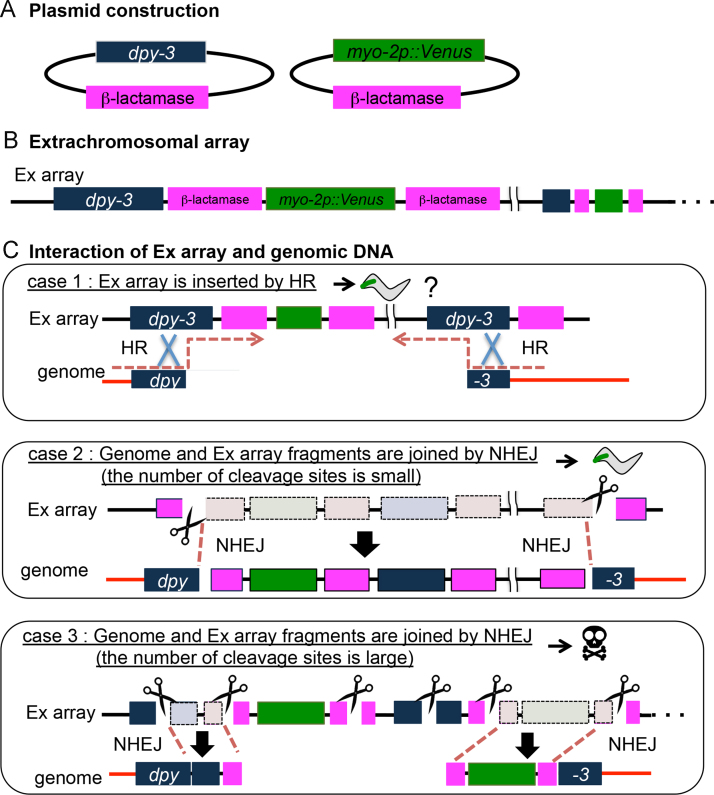
Because the probability of integration with the present method is high enough to generate integrant lines, we next evaluated how the integration processes are affected by the conditions used. The mechanism of this phenomenon is not currently clear but is likely due to DNA repair processes such as homologous recombination (HR) and nonhomologous end-joining (NHEJ) [18] because Cas9 causes double-strand breaks. If the HR system mainly functions in the mechanism of integration, integrated strains should be generated more efficiently by tmEx4252, which has a larger number of homologous sequences (Table 1, exp. 1, Supplementary Table 4), than by tmEx4277 or tmEx4253 (Table 1, exp. 2, 3, Supplementary Table 4). This was not the case. We obtained integrant strains at frequencies of 3.6%, 4.4% and 9.3% from tmEx4252, tmEx4277 and tmEx4253, respectively. These results suggest that the HR system may not play the main role for integration (Fig. 3C, case 1). This idea is compatible with the fact that Cas9 plasmids are injected into the germ cells after meiosis which do not have sister chromosomes. If the NHEJ system mainly functions in the mechanism of integration, even tmEx3199 can generate the integrant strains. This was the case. We obtained integrant strains at a frequency of 8.0% from tmEx3199, which does not have the dpy-3 sequence in the extrachromosomal array (Table 1, exp. 5). This result suggests that the NHEJ system may play the main role for integration (Fig. 3C, case 2).
Fig. 3.
Schematic diagram of Ex array insertion into the genome by the CRIPSR/Cas9 system. (A) Plasmid structure used in the experiment. (B) Schematic diagram of Ex array. (C) In case 1, cleaved dpy-3 sequences in C.elegans genome are ligated by homologous recombination (HR) to the intact extrachromosomal (Ex) array. In case 2, a fragmented Ex array is inserted to the chromosomal break point by nonhomologous end-joining (NHEJ), yielding an ls line. In cases 3, many fragmented Ex arrays are ligated by NHEJ to chromosomal break points but C.elegans genome are not joined each other, yielding aneuploidy.
To investigate the relationship between the numbers of fragmented extrachromosomal array and the efficiency of generating transgenic animals, we compared the results of exp. 1 and exp. 4 in the Table 1 and Supplementary Table 4. Increasing the cleavage sites in extrachromosomal arrays induced higher rates of sterile or larval arrest phenotypes in F1 animals. The same tendency was observed in exp. 1, exp. 2 and exp. 3 (Table 1, exp. 1–3, Supplementary Table 4). Thus, a higher concentration of DNA fragments might result in the ligation of chromosomal break points to the incorrect arms by NHEJ, leading to abortive DNA fragments from extrachromosomal arrays and causing aneuploidy (Fig. 3C, case 3).
Researcher may wonder which types of extrachromosomal arrays should be used. Extrachromosomal arrays with or without homologous sequences to insertion sites may be used. However, it may be easier to use already existing extrachromosomal transgenic animals. The Cas9-sgRNA (for β-lactamase) expression vector is available upon request.
3.3. The expression of reporter genes is proportional to the copy number of the construct at the same integration locus
To determine the copy number of insertions, we performed quantitative PCR using purified genomic DNA as a template. We designed primer sets located within the promoter of the myo-2 gene and the dpy-3 gene that were contained in the extrachromosomal array and in the C. elegans genome. Four Ex lines and 13 integrant strains were tested and compared to wild-type N2 (Fig. 2E and Supplementary Fig. 4C). As a result, the myo-2p-to-dpy-3, dpy-30p-to-ben-1 or vps-45-to-ben-1 ratio in each integrant strain was similar to each parent Ex strain (Fig. 2G, Supplementary Fig. 4D).
To compare the copy number of the myo-2 promoter sequences with fluorescence intensity, the integrant strains were photographed under the same exposure time (Fig. 2A–D and F). Fluorescence intensity was highly correlated with the copy number of the myo-2 promoter (correlation coefficient was 0.924; Fig. 2H). This suggests that the expression of transgenic genes in the integrant transgenes is nearly proportional to the copy numbers of the genes among strains with the same integration locus.
3.4. Transgene integration using the CRISPR/Cas9 system, benzimidazole and temperature selection
We previously demonstrated that single/low-copy integrant strains can be isolated using positive (temperature-sensitive lethal phenotype rescue) and negative (benomyl-sensitive phenotype rescue) selection [7], [8]. We next investigated whether a similar selection also works using the CRISPR/Cas9 system to isolate smaller-copy integrations from extrachromosomal arrays. Mutants of ben-1, which encodes a β-tubulin gene of C. elegans, are resistant to an anti-tubulin drug benzimidazole [19]. Wild-type C. elegans worms are severely unhealthy, dumpy, and uncoordinated on benzimidazole-containing selection media [19]. Thus, ben-1 mutants not carrying the ben-1 gene or carrying an incomplete ben-1 gene in the transgene predominantly grow and reproduce on benzimidazole-containing media. Temperature selection was based on rescue of the vps-45 mutant phenotype. The vps-45 mutants are unable to grow and reproduce at 20 °C [15]. vps-45 mutants carrying the vps-45 mini gene grow and reproduce, allowing for easy identification of the transformants. To insert the fragments of extrachromosomal transgenes into the chromosomes, ben-1(tm234); vps-45(tm246); tmEx3199 animals were microinjected with vectors encoding Cas9, a ben-1-specific sgRNA and an injection marker. The progeny of microinjected animals were cultured on benzimidazole-containing NGM plates, and the animals that grew and reproduced on benzimidazole-containing media were cultured on NGM plates at 20 °C. We performed PCR selection to amplify the GFP sequence, using animals that survived this selection as templates (Supplementary Fig. 2A). As a result, 2 integrant strains, tmIs1159 and tmIs1160, were isolated from 54 injected worms (Supplementary Figs. 2B, 3A and B). This method produced a much higher frequency (3.7%) than the previous UV/TMP protocol (0.033%) [7]. We confirmed that the transgenes were integrated at the ben-1 locus in tmIs1159 and tmIs1160 by PCR and sequencing (Supplementary Fig. 1).
To determine the copy number of insertions, we performed quantitative PCR using primer sets for the dpy-30p, vps-45 and ben-1 genes using purified genomic DNA as templates. tmIs1159 and tmIs1160 were tested for the copy number of these sequences and compared to wild-type N2 (Supplementary Fig. 3C and D). tmIs1159 carried approximately 35, 22 and 22 copies of the dpy-30p, vps-45 and ben-1 genes, respectively. tmIs1160 carried approximately 27, 16 and 10 copies of the dpy-30p, vps-45 and ben-1 genes, respectively (Supplementary Fig. 3C). This result indicates that the CRISPR/cas9 system is also useful for positive/negative selection via benzimidazole and temperature selection. tmIs1159 and tmIs1160 contained a slightly larger number of copies than the previously isolated single/low-copy transgenic strains [7], [8]. Because the integrant strains isolated in the present study were resistant to benzimidazole, it is conceivable that the residual ben-1 genes in the integrant strains were disrupted through cleavage by the ben-1 sgRNA and Cas9. Thus, the negative selection by benzimidazole was not as strong as before. We expected that the experimental conditions including the combination of sgRNA against the β-lactamase sequence for extrachromosomes and the dpy-3 sequence for the chromosomal integration site could yield integrants of single/low-copy, as before. GFP Fluorescence was not observed in the germ cells (data not shown), suggesting intermediate-copy number transgenes might be silenced [20].
In conclusion, we developed an efficient method for the locus-specific integration of extrachromosomal arrays into chromosomes using the CRISPR/Cas9 system, while extrachromosomal arrays are integrated into random loci by previous methods. To generate integrant strains efficiently, it is important to cut extrachromosomal arrays in a moderate number of fragments. Although it has been a concern that expression levels are not reproducible depending on the integration site, researchers can now easily compare the expression and functions of various constructs at defined integration loci. Moreover, when using multiple mutants and integrant transgenics, it is apparently beneficial to use transgenic strains with convenient integration sites.
Conflict of interest
The authors declare no conflicts of interests.
Acknowledgments
We thank members of the Mitani laboratory for discussion; and the Caenorhabditis Genetics Center (University of Minnesota, Minneapolis, MN, USA; supported by the National Institutes of Health-National Center for Research Resources) for providing some C. elegans strains. This work was supported partly by a Grant-in-Aid for Scientific Research from JSPS (to S.M., Grant No. 24390053) and by a grant-in-Aid for young scientists from JSPS (to S.Y., Grant No. 25870759).
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.bbrep.2015.11.017.
Appendix A. Supplementary material
Supplementary material Supplementary Fig. 1 Analysis of the boundary sequences between genome and the transgene in Is lines. (A) Schematic diagram of PCR. Primer 1 and Primer 3 are located in the genome, but not included in the extrachromosomal arrays. Primer 2 and Primer 4 are located in the vectors, and included only in the extrachromosomal arrays but not in the wild-type genome. The PCR fragments were analyzed by the Sanger sequencing method. (B, C) The boundary sequences between genome and the transgene in Is lines. The primers used in this experiment are listed in Supplementary Table 2. Sequences of sgRNAs, vectors and PAM sequences are indicated in red, blue and green letters, respectively. Inserted sequences of unknown origin are presented with underscored letters. Numbers in parentheses indicate the lengths of nucleotide sequences that are not shown. (B) Wild-type of the dpy-3 genome sequence is shown in the top row. The boundary sequences of the dpy-3 locus in each Is line are shown in subsequent rows. Sequences of the dpy-3 genome are indicated in black letters. (C) Wild-type of the ben-1 genome sequence is shown in the top row. The boundary sequences of the ben-1 locus in tmIs1159 and tmIs1160 are shown in subsequent rows. Sequences of the ben-1 genome are indicated in black letters. Lower case letters in (C) indicate the ben-1 sequence from the Ex arrays that was inserted in the reverse direction. In tmIs1159, the vector sequence was detected at about 6 kb upstream from the ben-1 integration site. We found the tandem repeats from the Ex arrays including the ben-1 sequence in this 6 kb insertion.
Supplementary material Supplementary Fig. 2 Overview of intermediate-copy integration experiments by the CRISPR/Cas9 system, benzimidazole and temperature selection. (A) Schematic diagram of intermediate-copy integration and selection. (B) Summary of intermediate-copy integration efficiency. Three of five-benzimidazole selected strains were not real integrant lines, but Ex transgenic worms without ben-1 rescue.
Supplementary material Supplementary Fig. 4 Expression of dpy-30p::NLS::GFP in the intermediate-copy integrated strains. (A, B) GFP protein was expressed ubiquitously. Scale bar = 100 µm. (600 msec. exposure time) (C) Relative amounts of the dpy-30 promoter and the vps-45 and ben-1 genes as determined by quantitative PCR. The data were normalized to the ama-1 gene. The copy numbers are presented as a ratio to the wild-type N2. Bars represent the mean±SE of three independent experiments. (D) The copy number ratios of the dpy-30 promoter to the ben-1 gene and the vps-45 gene to the ben-1 gene in tmEx3199, tmIs1159 and tmIs1160. Bars represent the mean±SE of three independent experiments.
Supplementary material Supplementary Fig. 4 Expression of dpy-30p::NLS::GFP in the multi-copy integrated strains. (A, B) The GFP protein was expressed ubiquitously. Scale bar=100 µm. (400 msec. exposure time) (C) The relative amount of the dpy-30 promoter and the vps-45 and ben-1 genes as determined by quantitative PCR. The data were normalized to the ama-1 gene. The copy numbers are presented as a ratio to the wild-type N2. Bars represent the mean±SE of three independent experiments. (D) The copy number ratios of the dpy-30 promoter to the ben-1 gene and the vps-45 gene to the ben-1 gene in tmEx3199 and the 4 Is lines. Bars represent the mean±SE of three independent experiments.
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
References
- 1.Mello C.C., Kramer J.M., Stinchcomb D., Ambros V. Efficient gene transfer in C.elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mello C., Fire A. DNA transformation. Methods Cell. Biol. 1995;48:451–482. [PubMed] [Google Scholar]
- 3.Mitani S. Genetic regulation of mec-3 gene expression implicated in the specification of the mechanosensory neuron cell types in Caenorhabditis elegans. Dev. Growth Differ. 1995;37:551–557. doi: 10.1046/j.1440-169X.1995.t01-4-00010.x. [DOI] [PubMed] [Google Scholar]
- 4.Way J.C., Wang L., Run J.Q., Wang A. The mec-3 gene contains cis-acting elements mediating positive and negative regulation in cells produced by asymmetric cell division in Caenorhabditis elegans. Genes. Dev. 1991;5:2199–2211. doi: 10.1101/gad.5.12a.2199. [DOI] [PubMed] [Google Scholar]
- 5.Frøkjaer-Jensen C., Davis M.W., Hopkins C.E., Newman B.J., Thummel J.M., Olesen S.P., Grunnet M., Jorgensen E.M. Single-copy insertion of transgenes in Caenorhabditis elegans. Nat. Genet. 2008;40:1375–1383. doi: 10.1038/ng.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zeiser E., Frøkjær-Jensen C., Jorgensen E., Ahringer J. MosSCI and gateway compatible plasmid toolkit for constitutive and inducible expression of transgenes in the C. elegans germline. PLoS One. 2011;6:e20082. doi: 10.1371/journal.pone.0020082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kage-Nakadai E., Imae R., Yoshina S., Mitani S. Methods for single/low-copy integration by ultraviolet and trimethylpsoralen treatment in Caenorhabditis elegans. Methods. 2014;68:397–402. doi: 10.1016/j.ymeth.2014.02.036. [DOI] [PubMed] [Google Scholar]
- 8.Kage-Nakadai E., Kobuna H., Funatsu O., Otori M., Gengyo-Ando K., Yoshina S., Hori S., Mitani S. Single/low-copy integration of transgenes in Caenorhabditis elegans using an ultraviolet trimethylpsoralen method. BMC Biotechnol. 2012;12:1. doi: 10.1186/1472-6750-12-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Friedland A.E., Tzur Y.B., Esvelt K.M., Colaiácovo M.P., Church G.M., Calarco J.A. Heritable genome editing in C. elegans via a CRISPR-Cas9 system. Nat. Methods. 2013;10:741–743. doi: 10.1038/nmeth.2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen C., Fenk L.A., de Bono M. Efficient genome editing in Caenorhabditis elegans by CRISPR-targeted homologous recombination. Nucleic Acids Res. 2013;41:e193. doi: 10.1093/nar/gkt805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dickinson D.J., Ward J.D., Reiner D.J., Goldstein B. Engineering the Caenorhabditis elegans genome using Cas9-triggered homologous recombination. Nat. Methods. 2013;10:1028–1034. doi: 10.1038/nmeth.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tzur Y.B., Friedland A.E., Nadarajan S., Church G.M., Calarco J.A., Colaiácovo M.P. Heritable custom genomic modifications in Caenorhabditis elegans via a CRISPR-Cas9 system. Genetics. 2013;195:1181–1185. doi: 10.1534/genetics.113.156075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gengyo-Ando K., Mitani S. Characterization of mutations induced by ethyl methanesulfonate, UV, and trimethylpsoralen in the nematode Caenorhabditis elegans. Biochem. Biophys. Res. Commun. 2000;269:64–69. doi: 10.1006/bbrc.2000.2260. [DOI] [PubMed] [Google Scholar]
- 15.Gengyo-Ando K., Kuroyanagi H., Kobayashi T., Murate M., Fujimoto K., Okabe S., Mitani S. The SM protein VPS-45 is required for RAB-5-dependent endocytic transport in Caenorhabditis elegans. EMBO Rep. 2007;8:152–157. doi: 10.1038/sj.embor.7400882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang W., Bikard D., Cox D., Zhang F., Marraffini L.A. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat. Biotechnol. 2013;31:233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gengyo-Ando K., Yoshina S., Inoue H., Mitani S. An efficient transgenic system by TA cloning vectors and RNAi for C. elegans. Biochem. Biophys. Res. Commun. 2006;349:1345–1350. doi: 10.1016/j.bbrc.2006.08.183. [DOI] [PubMed] [Google Scholar]
- 18.A.J.B. Alberts, J. Lewis, M. Raff, K. Roberts, P. Walter, Molecular biology of the cell, 5th edition, Biochemistry and Molecular Biology Education, 2008, pp. 317–318.
- 19.Driscoll M., Dean E., Reilly E., Bergholz E., Chalfie M. Genetic and molecular analysis of a Caenorhabditis elegans beta-tubulin that conveys benzimidazole sensitivity. J. Cell. Biol. 1989;109:2993–3003. doi: 10.1083/jcb.109.6.2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kelly W.G., Xu S., Montgomery M.K., Fire A. Distinct requirements for somatic and germline expression of a generally expressed Caernorhabditis elegans gene. Genetics. 1997;146:227–238. doi: 10.1093/genetics/146.1.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material Supplementary Fig. 1 Analysis of the boundary sequences between genome and the transgene in Is lines. (A) Schematic diagram of PCR. Primer 1 and Primer 3 are located in the genome, but not included in the extrachromosomal arrays. Primer 2 and Primer 4 are located in the vectors, and included only in the extrachromosomal arrays but not in the wild-type genome. The PCR fragments were analyzed by the Sanger sequencing method. (B, C) The boundary sequences between genome and the transgene in Is lines. The primers used in this experiment are listed in Supplementary Table 2. Sequences of sgRNAs, vectors and PAM sequences are indicated in red, blue and green letters, respectively. Inserted sequences of unknown origin are presented with underscored letters. Numbers in parentheses indicate the lengths of nucleotide sequences that are not shown. (B) Wild-type of the dpy-3 genome sequence is shown in the top row. The boundary sequences of the dpy-3 locus in each Is line are shown in subsequent rows. Sequences of the dpy-3 genome are indicated in black letters. (C) Wild-type of the ben-1 genome sequence is shown in the top row. The boundary sequences of the ben-1 locus in tmIs1159 and tmIs1160 are shown in subsequent rows. Sequences of the ben-1 genome are indicated in black letters. Lower case letters in (C) indicate the ben-1 sequence from the Ex arrays that was inserted in the reverse direction. In tmIs1159, the vector sequence was detected at about 6 kb upstream from the ben-1 integration site. We found the tandem repeats from the Ex arrays including the ben-1 sequence in this 6 kb insertion.
Supplementary material Supplementary Fig. 2 Overview of intermediate-copy integration experiments by the CRISPR/Cas9 system, benzimidazole and temperature selection. (A) Schematic diagram of intermediate-copy integration and selection. (B) Summary of intermediate-copy integration efficiency. Three of five-benzimidazole selected strains were not real integrant lines, but Ex transgenic worms without ben-1 rescue.
Supplementary material Supplementary Fig. 4 Expression of dpy-30p::NLS::GFP in the intermediate-copy integrated strains. (A, B) GFP protein was expressed ubiquitously. Scale bar = 100 µm. (600 msec. exposure time) (C) Relative amounts of the dpy-30 promoter and the vps-45 and ben-1 genes as determined by quantitative PCR. The data were normalized to the ama-1 gene. The copy numbers are presented as a ratio to the wild-type N2. Bars represent the mean±SE of three independent experiments. (D) The copy number ratios of the dpy-30 promoter to the ben-1 gene and the vps-45 gene to the ben-1 gene in tmEx3199, tmIs1159 and tmIs1160. Bars represent the mean±SE of three independent experiments.
Supplementary material Supplementary Fig. 4 Expression of dpy-30p::NLS::GFP in the multi-copy integrated strains. (A, B) The GFP protein was expressed ubiquitously. Scale bar=100 µm. (400 msec. exposure time) (C) The relative amount of the dpy-30 promoter and the vps-45 and ben-1 genes as determined by quantitative PCR. The data were normalized to the ama-1 gene. The copy numbers are presented as a ratio to the wild-type N2. Bars represent the mean±SE of three independent experiments. (D) The copy number ratios of the dpy-30 promoter to the ben-1 gene and the vps-45 gene to the ben-1 gene in tmEx3199 and the 4 Is lines. Bars represent the mean±SE of three independent experiments.
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material