Abstract
Multifunctional adapter and chaperone protein Daxx participates in the regulation of a number of mainly transcription-related processes. Most notably in a complex with chromatin-remodelling ATPase ATRX, Daxx serves as a histone H3.3 chaperone at telomeric regions and certain genes. In this report we document that Daxx interacts with another chromatin-remodelling, ATPase Brg1. We confirm the Daxx-Brg1 association both in vitro and in cells and show that Daxx interacts with Brg1 in high-molecular-weight complexes. Ectopic co-expression of Daxx with Brg1 and PML could shift disperse nuclear localisation of Brg1 into PML bodies. Mapping the Daxx-Brg1 interaction revealed that Daxx preferentially binds the region between Brg1 N-terminal QLQ and HSA domains, but also weakly interacts with its C-terminal part. Brg1 interacted with both the central and N-terminal parts of Daxx. SiRNA-mediated down-regulation of Daxx in SW13 adrenal carcinoma cells markedly enhanced expression of Brg1-activated genes CD44 or SCEL, suggesting that Daxx either directly through Brg1 and/or indirectly via other factors is a negative regulator of their transcription. Our findings point to Brg1 as another chromatin-remodelling protein that might similarly, as ATRX, target Daxx to specific chromatin regions where it can carry out its chromatin- and transcription-regulating functions.
Keywords: Adapter, Transcriptional repression, Chromatin remodelling, Interaction
Highlights
-
•
Chromatin-remodelling ATPase Brg1 was uncovered as a novel Daxx-interacting protein.
-
•
Their interaction occurs in high molecular weight complexes in a multifocal manner.
-
•
Daxx expression attenuates transactivation of some Brg1-activated genes in SW13 cells.
1. Introduction
Death-domain associated protein 6 (Daxx) is a multifunctional, promyelotic leukaemia (PML)-interacting adapter protein, which is vital for mouse embryogenesis [1], [2] and participates in many processes including apoptosis, protein stability and regulation of transcription [3], [4], [5], [6], [7].
Being predominantly a transcriptional repressor, Daxx bridges many transcription factors including nuclear factor –kB (NF-κB) or nuclear receptors [8], [9] to histone deacetylases, such as histone deacetylase 2 (HDAC2), and thus mediates transcriptional repression of a number of genes [5]. Through its association with DNA methyltransferase DNMT1, Daxx can enhance methylation and thus transcriptional suppression of RelB target genes or the tumour suppressor RASSF1A [4], [10]. Daxx is also required for the epigenetic silencing of the integrated retroviral DNA [11], or the repression of human cytomegalovirus (HCMV) immediate-early genes (IEG) [12].
The most recently revealed function of Daxx is connected with chromatin remodelling, namely with the regulation of HIRA- and the replication-independent deposition of the histone variant H3.3 [13], [14], [15], [16]. Daxx, through interaction with the chromatin-remodelling ATPase Alpha thalassaemia/mental retardation syndrome X-linked (ATRX), mediates H3.3 deposition either at telomeres, pericentromeric heterochromatin or in some actively transcribed genes. Daxx interaction with chromatin- and PML-associated protein DEK [17] is required for proper loading of H3.3 on the telomeres [18] and the Daxx/ATRX complex, through interaction with Suv39h methyltransferase suppresses via H3K9 trimethylation expression of a number of genes during mouse embryogenesis [19]. ATRX-Daxx-dependent reposition of the histone H3.3 variant is associated with repression of adenoviral or CMV promoters [20], [21], but interestingly during early EBV infection, tegument protein BNRF1 apparently replaces ATRX, and via reprogramming Daxx-mediated H3.3 loading thus allowing late gene expression [22]. In contrast, ATRX-Daxx-mediated H3.3 loading enhanced transcriptional activation of neural immediate early genes Bdnf or Egr2 [23]. In addition to telomeres and transcribed genes, Daxx also participates in the distribution of centromeric H3 variant CenH3 at centromers, which is connected with enhanced cellular resistance to DNA damage [24]. Recently Elsasser et al. uncovered that ATRX-Daxx-mediated H3.3 loading contributes to the endogenous retroviral elements in embryonic stem cells [25]. ATRX-Daxx acts as a chaperone in anti-tumour protection – their mutations were found in pancreatic neuroendocrine tumours and paediatric glioblastomas and were associated with a more aggressive phenotype and reduced survival [26], [27], [28]. The possible ATRX-Daxx tumour suppressive function could be related to its negative modulation of alternative lengthening of telomeres [29].
Brg1 and its sibling Brm belong, similarly as ATRX, to the family of chromatin-remodelling ATPases. These evolutionarily conserved enzymes function in multiprotein complexes and participate in the regulation of chromatin structure, influencing transcription, DNA replication, DNA repair and other chromatin-related processes [30]. The importance of Brg1 is underscored by the fact that Brg1 null mouse embryos die several days after implantation, as Brg1 regulates the self-renewal potential of stem cells through the expression of the core pluripotency-related genes [31], [32]. Among other Brg1-regulated genes are cytokine response genes, nuclear receptors, genes involved in differentiation (e. g. homeotic genes, Wnt signalling pathway), cell cycle modulation (regulated by p53, E2Fs and pRB) and many others [33], [34], [35], [36]. Brg1 is also involved in carcinogenesis both as a tumour suppressor and as a tumour promoting gene [37].
In this report we aimed to get a better understanding of the role and function of Daxx in cell physiology and we searched for and analysed novel Daxx-interacting proteins. Among others we found the chromatin-remodelling ATPase Brg1 as a novel Daxx-interacting protein and analysed some functional consequences of their interaction. Our data document that Daxx participates in the transcriptional suppression of the expression of several Brg1-regulated genes in SW13 cells. However, whether Daxx/Brg1 can have wider functional consequences, such as H3.3 loading and regulation of gene expression, remains to be investigated.
2. Materials and methods
2.1. Cell cultures
SW13, HeLa and HEK293T cells (all ATCC) were cultivated in DMEM, and supplemented with 10% FBS and antibiotics (penicillin+streptomycin). MCF10a (ATCC) cells were cultivated in MCF10a medium (ATCC).
2.2. Plasmids and oligonucleotides
Lists of used plasmids and oligonucleotides are shown in Supplementary Tables 1 and 2.
2.3. Antibodies
The following antibodies were used for western-blotting, immunoprecipitation and immunocytochemistry: Daxx02 and Daxx03 (mouse IgG1s), anti-hDaxx pAbU (rabbit polyclonal antibody) – all produced in our laboratory; H-88 and G-7 (hBrg1), E-1 (hBrm), H-76 (hBAF155), TU-01 (α-tubulin) and C-11 (actin) – all from Santa Cruz Biotechnologies; 1B1B2 (histone 3) – from Cell Signalling Technology; M2 (FLAG epitope), 9E10 (Myc epitope)-both from Sigma Aldrich, and AFP01 (α-fetoprotein, AFP) – mouse IgG1 kindly provided by V. Hořejší.
2.4. Yeast two-hybrid screening for proteins interacting with Daxx 1-595
The pGBKT7-Daxx 1-595 construct was used for screening a HeLa cDNA library, cloned into the prey vector pGAD-GH (Clontech) for potential Daxx-interacting proteins. The screening and post-screening analyses were carried out according to Matchmaker 3 Two-Hybrid System manual (Clontech).
2.5. Transient transfections
HEK293T cells were transfected using polyethylene imine (Sigma Aldrich) according to [38]. SW13, HeLa cells were transfected with GeneCellin (BioCellChallenge) and siRNA was transfected into SW13 cells using Lipofectamine RNAiMAX (Invitrogen) and the standard protocol (Life Technologies). Used siRNA: siDaxx #1 (GGAGTTGGATCTCTCAGAA, according to [39]) and siDaxx B+C+D (a mixture of siGENOME siRNAs from Dharmacon: J-004420-07, J-004420-06, J-004420-05)
2.6. Purification of recombinant protein and in vitro pulldown assay
Combined His6+GST-tagged Daxx and its variants were produced in E. coli BL21 (DE3) from pET42b-based expression plasmids and purified with TALON (Clontech) and Glutathion-Sepharose beads (GE Healthcare). For the pull-down assays, these beads were incubated with in vitro translated 35S-labelled Brg1, prepared by TNT Quick Coupled Transcription/Translation kit (Promega) using TRAN 35S Label (MP Radiochemicals). The bound proteins were eluted with the sample buffer, subjected to SDS-electrophoresis and visualised on a Phosphor Imaging Plate in BAS 5000 instrument (FujiFilm).
2.7. Cell lysis and immunoprecipitation
In non-detergent lysis, the cellular pellets (~50 million cells) were resuspended in 3 volumes of hypotonic buffer (10 mM HEPES pH 7.5, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT; EDTA-free Complete protease inhibitors, Roche), incubated on ice, lysed by pushing through a 25×G needle and centrifuged (13,500 rpm, 30 s). The supernatants (cytosolic extracts) were mixed with 1.5 volume of hypertonic buffer (20 mM HEPES pH 7.5, 1.5 mM MgCl2, 420 mM NaCl, 0.2 mM EDTA pH 8.0, 25% glycerol, 0.5 mM DTT; Complete). The pellets (nuclei) were resuspended in 1.5 volume of hypertonic buffer, shortly sonicated, incubated on ice for 30 min and centrifuged ( 13,500 rpm, 10 min). These supernatants (nuclear extracts) were diluted 3-times with hypotonic buffer and both cytosolic and nuclear extracts were used for immunoprecipitations.
For the detergent lysis, the cellular pellets (~7 million cells) were resuspended in 3 volumes of lysis buffer (50 mM HEPES pH 7.5, 250 mM NaCl, 0,5% Nonidet P-40, 2% glycerol; Complete), shortly sonicated, incubated at 4 °C for 30 min and centrifuged (13,500 rpm, 10 min). The supernatants were diluted with 1.5 volumes of water containing 1.5 mM DTT and used for immunoprecipitations.
For the immunoprecipitations, the lysates were incubated with 3–5 μg of appropriate antibodies at 4 °C for 15 h, centrifuged ( 13,500 rpm, 10 min) and the immunocomplexes were pulled-down by 50 μl of Protein A/G agarose beads (SC Biotechnologies) on a rotation wheel at 4 °C for 90 min. After washing, the bound proteins were eluted with the sample buffer, heat-denatured and analysed by Western blotting.
2.8. Confocal microscopy
Transfected HeLa cells grown on glass cover-slips in a 24-wells plate were fixed with 3% paraformaldehyde in PBS for 20 min, permeabilised with 0.5% Triton X-100 for 20 min and finally blocked with 10% human serum in PBS for 1 h. These cells were incubated with the primary antibodies and diluted in PBS for 1 h, then with the secondary antibodies (goat-anti-mouse with conjugated Alexa 647 and goat-anti-rabbit with conjugated Rhodamine Red-X, Jackson Biotechnologies), diluted in PBS (1:500) for 1 h and finally with 1.25 μM DAPI for 5 min. The cover-slips were mounted on slides in ProLong Gold anti-fade reagent (Invitrogen) and analysed using a Leica TCS SP5 confocal microscope.
2.9. Preparation of adenoviruses
Recombinant adenoviruses expressing either EGFP or EGFP-tagged Brg1 (kindly provided by R. Bremner) were amplified in HEK293T cells and purified by Adeno-X purification kit (Clontech). Purified adenoviruses were stored in aliquots in the storage buffer (1×GTS,) at −80 °C. One aliquot of each adenovirus was titrated and in the experiments the recombinant adenoviruses were used at MOI=1–2.
3. Transfections, transductions, isolation of RNA, reverse transcription, qRT-PCR and statistical analysis
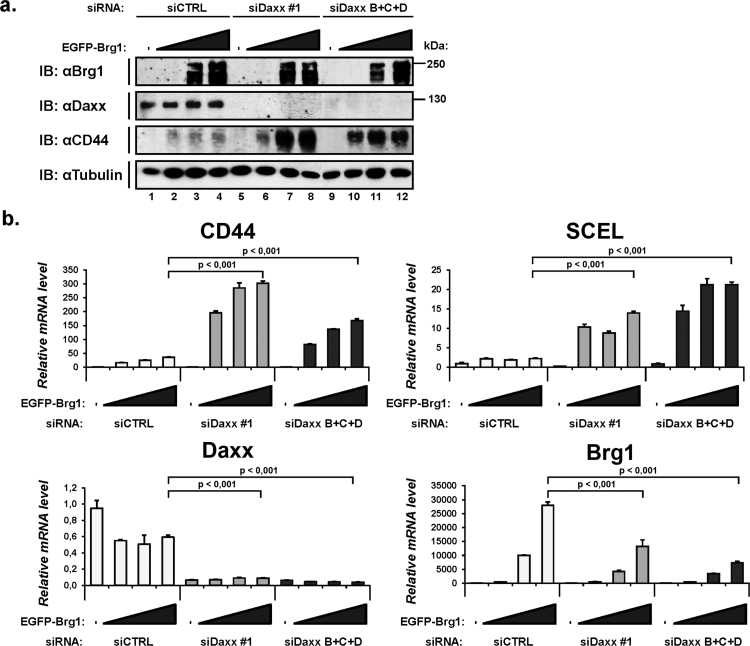
SW13 cells grown on 12-wells plates were twice transfected with siRNAs in two consecutive days using RNAiMax. Cells were then transduced with 0.05, 0.1 or 0.15 μl of adenoviruses, expressing either EGFP or EGFP-Brg1. 48 h post-transduction, the cells were lysed for both protein (SDS lysis buffer) and RNA (RNeasy Plus Mini kit, QIAGEN) analyses. One μg of extracted total RNA was reverse transcribed with RevertAid (Fermentas) and used for quantitative real-time PCR (LightCycler 480 SYBR Green I master-mix and LightCycler 480 instrument, Roche). The sequences of primers are shown in Supplementary Table 2. For the statistical analysis in Fig. 4 we used paired Student's t-test.
Fig. 4.
Down-regulation of Daxx leads to up-regulation of Brg1-regulated CD44 and SCEL genes. SW13 cell line was transfected with siRNA (control, siDaxx #1 and siDaxx B+C+D, a combination of three distinct siRNAs) and then infected with recombinant adenoviruses (0.05, 0.1 or 0.15 μl) expressing either EGFP or EGFP-tagged Brg1. The cells were then subjected to both (a) Western blotting and (b) qRT-PCR analyses. The statistical significance was assessed by paired Student's t-test.
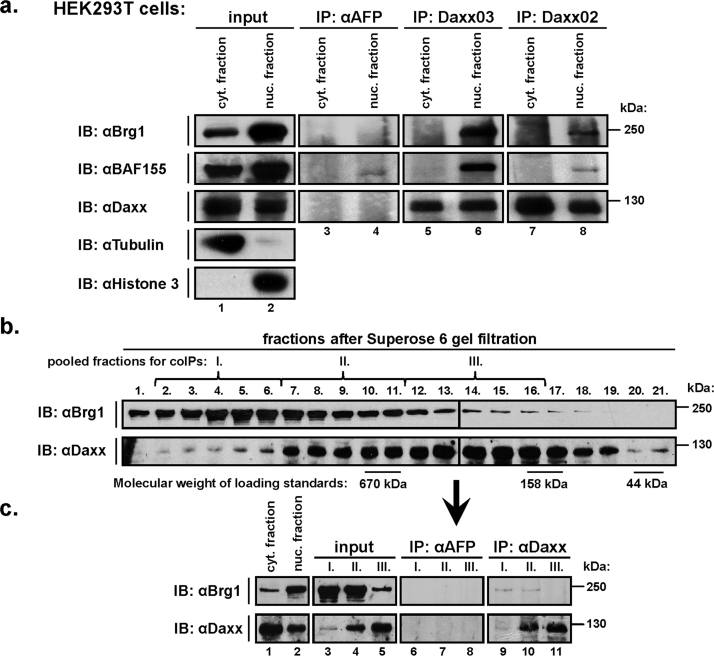
3.1. Gel filtrations
HEK293T cells (3×108) were lysed by “non-detergent lysis”, the nuclear lysates were concentrated to 1.5 ml by ultrafiltration (Amicon Ultra 30 k, Millipore) and applied to a Superose 6 column (GE Healthcare). Pooled fractions were divided into two parts: the first (1/3 of total volume) was concentrated on Microcon-10 (Millipore) to 15 μl, mixed with 15 μl of SDS-sample buffer and analysed by Western blotting; the second part (2/3) was pooled (inputs: fractions 2.-6. to input I., 7.–11. to input II., 12.-16. to input III.), concentrated to 400 μl, and used for immunoprecipitations.
4. Results
4.1. Daxx interacts with the chromatin-remodelling ATPase Brg1 both in vitro and in vivo.
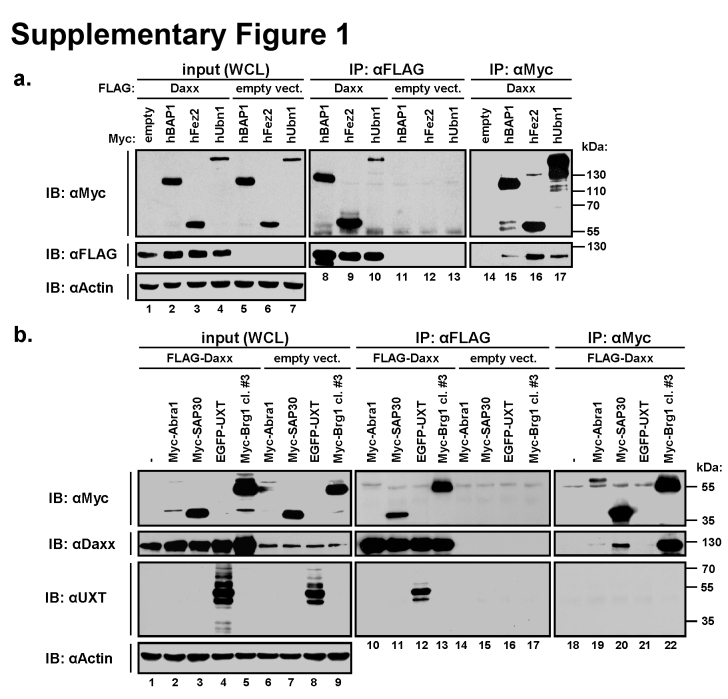
Daxx interacts with a number of mainly nuclear proteins, predominantly through its C-terminal Ser/Pro/Thr-rich (SPT) region or distal SUMO interacting motif (SIM) [40]. To exclude proteins that interact with this promiscuous C-terminal part of Daxx, in our yeast two hybrid (Y2H) screening for novel Daxx-interacting partners, we used as bait the evolutionary and structurally conserved N-terminal [41] and central part of Daxx (amino acids 1-595).The screening provided 24 positive hits (mainly nuclear and chromatin-associated proteins, including previously identified ATRX or DMAP1, see Supplementary Table 3). Y2H re-screening for possible false interactors narrowed down this selection to chromatin-remodelling ATPase ATRX [42], then DMAP1 [43] and seven other proteins. All of these seven novel putative interactors (BAP1, FEZ2, UBN1, SAP30, UXT, BRG1 and ABRA1) co-immunoprecipitated with Daxx from the lysates of co-transfected HEK293T cells (Supplementary Fig. 1). Having in mind the important function of ATRX-Daxx as a novel chromatin-remodelling H3.3 chaperone, we have chosen Brg1 as another chromatin-remodelling ATPase for in detail analysis of its molecular and functional interactions with Daxx.
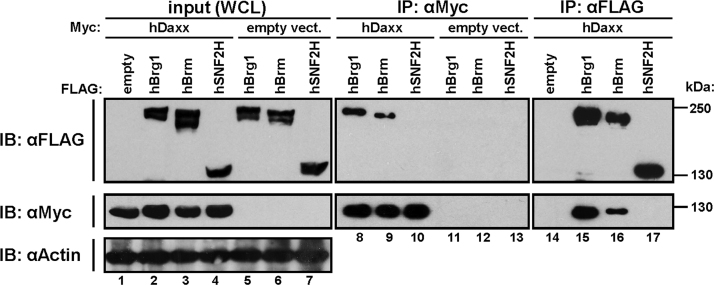
Brg1 or the related Brm, but not unrelated ATPase SNF2H, were co-immunoprecipitated with Daxx from the lysates of co-transfected HEK293T cells (Fig. 1, lanes 8, 9 and 15, 16). Using two different anti-Daxx monoclonal antibodies, we also co-immunoprecipitated the endogenous Brg1 with Daxx from the nuclear lysates of non-transfected HEK293T (Fig. 2a, lanes 6, 8; anti-alpha fetoprotein, AFP antibody was used as non-specific control) or MCF10A (Supplementary Fig. 2, lane 4) cells, indicating that their interaction also occurs under physiological conditions. In addition to Brg1, we also co-precipitated BAF155, a core member of Brg1-containing chromatin-remodelling complexes (Fig. 2a), implying that Daxx interacts with Brg1 bound in a complex. Indeed, the gel filtration analysis showed that a minor part of Daxx is found in fractions containing high molecular weight complexes (1 MDa and higher) (Fig. 2b, lanes 2–6) and only from these fractions is Brg1 efficiently co-immunoprecipitated with Daxx (Fig. 2c, pool I, lane 9).
Fig. 1.
Ectopically expressed Daxx interacts with Brg1 and Brm but not with SNF2H. HEK293T cells were transfected with Myc-Daxx FL or empty vector, together with empty vector or FLAG-Brg1, FLAG-Brm and FLAG-SNF2H. The cell lysates were immunoprecipitated with either anti-Myc or anti-FLAG antibodies and analysed by Western blotting.
Fig. 2.
Endogenous Daxx and Brg1 interact in high molecular weight complexes. HEK293T cells were lysed by non-detergent lysis to separate cytoplasmic and nuclear fractions: (a) immunoprecipitation of both cytoplasmic and nuclear fraction with anti-AFP and anti-Daxx (Daxx03 and Daxx02) antibodies, (b) gel filtration of nuclear lysates on Superose 6 column. Fractions from gel filtration were pooled (fractions 2.–6. to mixture I., 7.–11. to II. and 12.–16. to III.), (c) immunoprecipitated with anti-AFP and anti-Daxx (Daxx03) antibodies, and analysed by Western blotting.
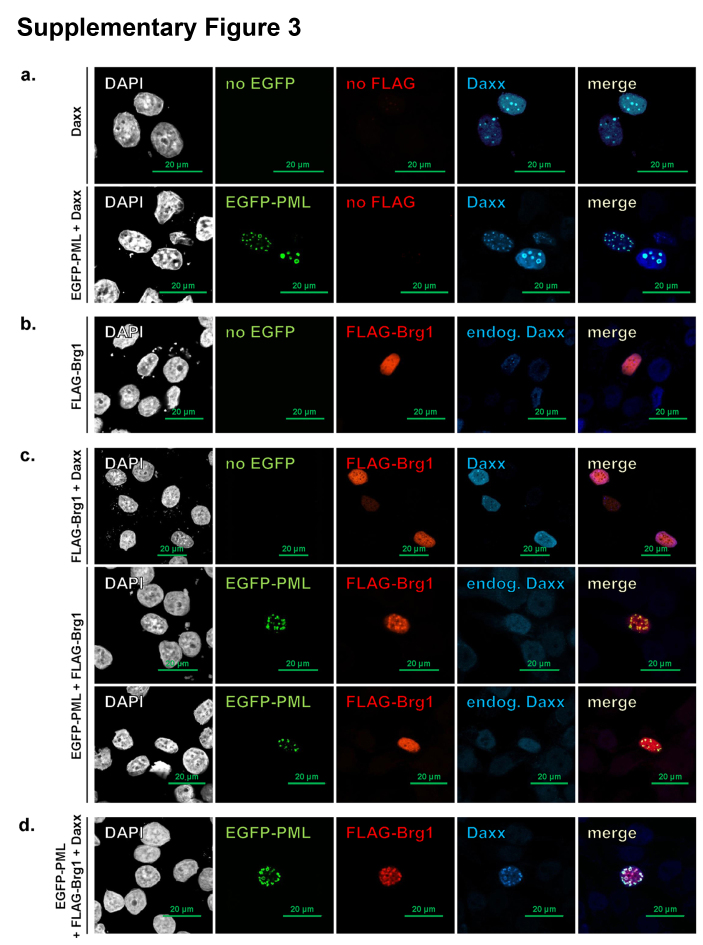
A prominent partner among Daxx-interacting proteins is PML, which shelters Daxx in PML nuclear bodies and could modulate some of the functions of Daxx, such as H3.3 loading [18]. In transfected HeLa cells we confirmed that ectopically expressed Daxx strongly associates with PML bodies, formed by both endogenous and transfected PML (Supplementary Fig. 3a). In contrast, ectopically expressed Brg1 was diffused and did not bind to any nuclear structure (Supplementary Fig. 3b). Co-expression of Daxx or PML with Brg1, also did not largely enhance their association with PML bodies, although in some cells, Brg1/PML co-expression led to weak association of Brg1 with PML bodies, likely mediated by the endogenous Daxx (Supplementary Fig. 3c). However, when all three proteins were expressed at a similar level in transfected HeLa cells, we observed clear co-localization of all of them in PML bodies (Supplementary Fig. 3d), indicating that a significant portion of co-transfected Daxx and Brg1 can interact, and thus confirming the co-immunoprecipitation data (Fig. 1 and Supplementary Fig. 2).
4.2. Daxx interacts with Brg1 via at least two independent regions.
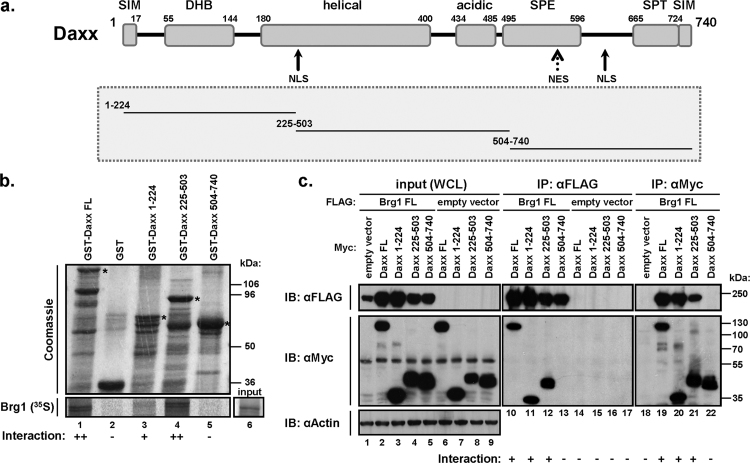
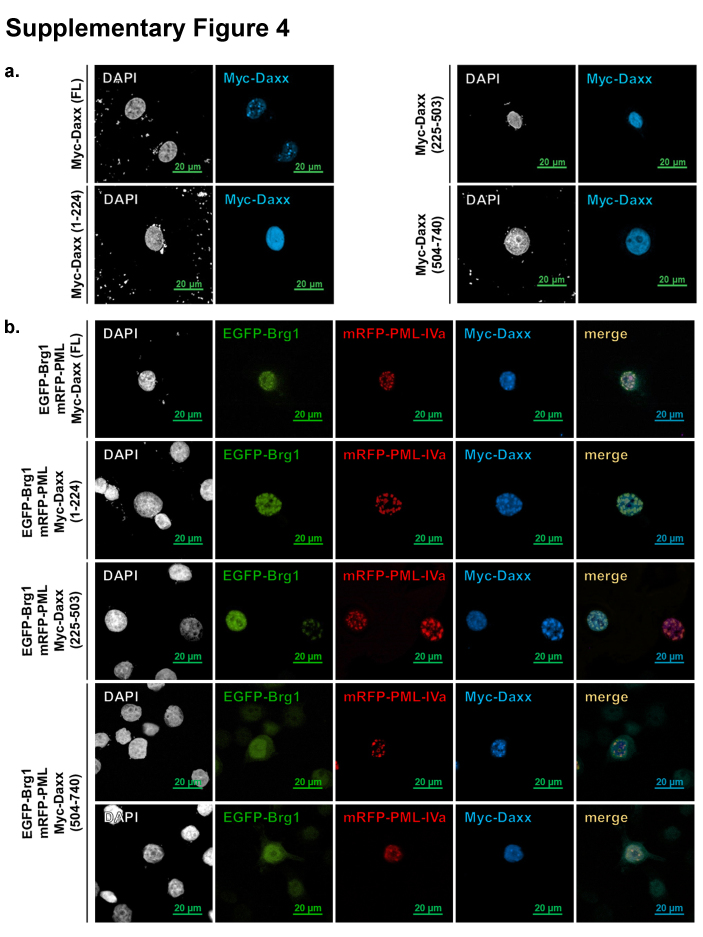
Having confirmed the interaction between Daxx and Brg1 in yeast and mammalian cells, we aimed to determine the interacting regions in both proteins. We chose both in vitro (GST pull-down assay) and in vivo (transient co-transfection of mammalian cells) systems as a methodological approach. For the GST pull-down assays we initially split GST-fused Daxx into three parts – N-terminal, central and C-terminal (Fig. 3a) – and analysed, together with GST-Daxx full length (FL), their interaction with in vitro translated Brg1. The pull-down assays first of all revealed that GST-Daxx FL can directly and reproducibly interact with the full-length Brg1 (Fig. 3b, lane 1 vs. 2 – GST control). We also found that mainly the central part of Daxx, but not its C-terminal SPE- and S/P/T-containing domain strongly interacted with Brg1. However, we also detected slightly weaker but still significant interaction of Brg1 with the N-terminal, DHB domain-containing part of Daxx (Fig. 3b, lanes 3–5). These data were then confirmed by co-immunoprecipitation of expressed Daxx FL or its three parts with co-expressed Brg1 (Fig. 3c). Only Myc-tagged Daxx FL or its N-terminal and central parts but not the C-terminus could form complexes with FLAG-tagged Brg1, that were pulled-down by either anti-Myc or anti-FLAG antibodies (Fig. 3c, compare lanes 10–12 with 13 and 19–21 with 22). Using confocal microscopy of transfected HeLa cells we confirmed that the Daxx deletion variants are localised in the nucleus (Supplementary Fig. 4a). In addition, co-transfection of these deletion variants with Brg1 and PML also confirmed the co-immunoprecipitation data, and revealed that both Brg1-interacting N-terminal (AAs 1-225) and central (AAs 225–503) fragments localise into PML bodies and pull Brg1 into them, while the non-interacting C-terminal Daxx fragment (504–740) does not (Supplementary Fig. 4b).
Fig. 3.
Brg1 interacts with the N-terminal and central parts of Daxx. (a) Human Daxx (SIM-SUMO interacting motif, DHB-Daxx homology bundle, SPE-Ser/Pro/Glu rich, SPT-Ser/Pro/Thr rich) were divided into N-terminal (amino acids 1–224), central (amino acids 225–503) and C-terminal (amino acids 504–740) fragments. (b) GST-tagged Daxx fragments GST-Daxx WT and GST tag alone were incubated with 35S-labelled Brg1 together with Glutathion-Sepharose 4B beads. After the SDS-PAGE, GST-tagged proteins were visualised by Coomassie staining and 35S-labelled Brg1 by autoradiography. (c) HEK293T cells were transfected with FLAG-Brg1 FL or empty vector together with empty vector or Myc-Daxx fragments and Myc-Daxx FL. The cell lysates were immunoprecipitated with either anti-Myc or anti-FLAG antibodies and analysed by Western blotting.
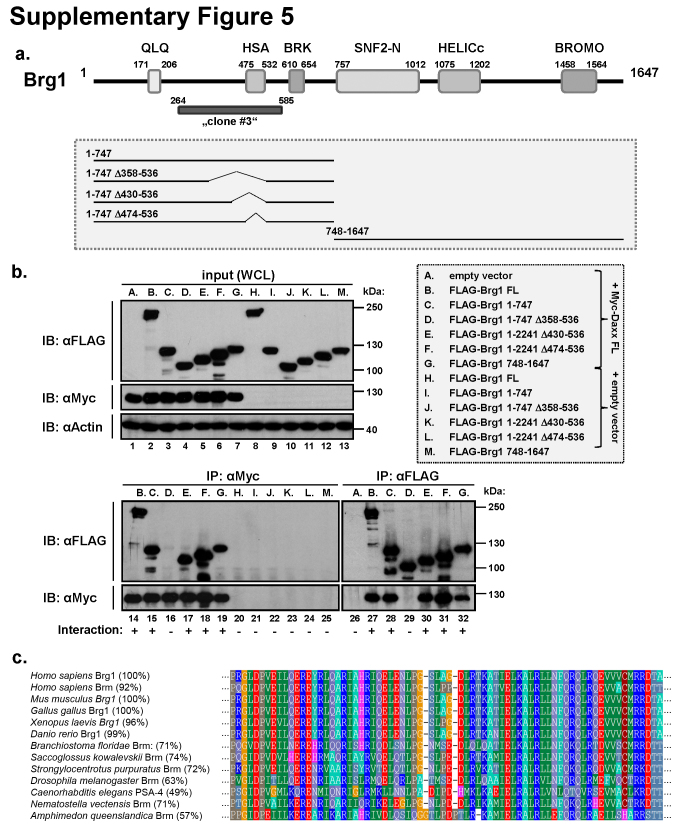
Next we aimed to narrow down the Daxx interaction region on Brg1. The part of Brg1 pulled-down in the yeast two-hybrid screening (clone #3) mapped to the N-terminus of Brg1, containing the HSA domain (Supplementary Fig. 5a). Co-expression of this interaction region containing the N-terminal half of Brg1 with Daxx FL in HEK293T cells followed by immunoprecipitation, indeed confirmed that the Daxx-interacting domain resides in this N-terminal part of Brg1 (Supplementary Fig. 5b, lanes 15 and 28). However, Daxx apparently also co-immunoprecipitated with the second, C-terminal half of Brg1 (Supplementary Fig. 5b, lanes 19 and 32). Thus, as for the Daxx itself, there are also at least two regions mediating interaction between these two proteins. In the N-terminal region of Brg1 we prepared several internal deletion mutants and examined their interaction with co-transfected Daxx (Supplementary Fig. 5b, lanes 16–18 and 29–31). We were able to narrow down the Daxx-interacting region to amino acids 358-430, just N-terminally to the HSA domain (Supplementary Fig. 5b, compare lanes 16 to 17 or 29 to 30). This part of Brg1 also belongs to the phylogenetically most conserved parts of Brg1 (Supplementary Fig. 5c).
4.3. Daxx negatively affects expression of Brg1-regulated genes in SW13 cells.
Brg1, as a part of the Swi/Snf chromatin remodelling complex, actively participates in the regulation of transcription of a number of genes. As one of the important functions of Daxx is also the regulation of transcription, we focused on evaluating the Daxx role in Brg1-dependent, transcription-related processes. SW13 adrenocortical carcinoma cells lack the Brg1 expression and its re-introduction into these cells leads to transactivation of a number of genes including p21, CSF1, SPARC or CD44 [44]. Transduction of SW13 cells with the Brg1-expressing adenovirus did increase the expression of CD44, Sciellin (SCEL), SPARC or CSF1 in a dose-dependent manner (Fig. 4 and not shown). SiRNA-mediated down-regulation of Daxx expression prior to the transduction led to a boost of Brg1-mediated expression of CD44 and SCEL, both at mRNA and protein levels (Fig. 4). Intriguingly, Daxx downregulation also led to the suppression of Brg1 mRNA expression (Fig. 4b) but these changes were only limitedly reflected in Brg1 protein expression (Fig. 4a). Nevertheless despite apparently lower expression of Brg1 in siDaxx transfected cells than in control cells, the expression of its targets CD44 and SCEL significantly increased. This Daxx down-regulation-mediated enhancement of transcription was reproducible with several various Daxx siRNA, and points to an inhibitory rather than activator role of Daxx in transactivation of at least some of Brg1-dependent genes.
5. Discussion
The so-far known Daxx transcription-related functions are mainly repressive, mediated by bridging various chromatin-modifying factors, such as histone deacetylases or DNA-methyltransferases, to transcription regulatory regions [3]. Recently, Daxx bound to chromatin-remodelling ATPase ATRX was also recognised as a histone H3.3 chaperone, bringing H3.3 to telomeres or some actively transcribed genes [13]. However, the proper cellular role of Daxx, e.g. during the embryogenesis (Daxx knockout is lethal at E9.5), still remains elusive.
In this study we point to Brg1, another chromatin-remodelling ATPase with multiple roles in transcription and DNA repair [30], as a novel Daxx-interacting partner. We identified their interaction in a yeast two-hybrid screen and confirmed it by multiple in vitro and in vivo interaction assays, such as GST-pulldown or co-immunoprecipitation of both transfected and endogenous proteins. Moreover, we also co-immunoprecipitated Daxx and Brg1 from gel filtration fractions containing high-molecular-weight complexes (around 1–1.5 MDa) and, under some circumstances (co-expression of Brg1, Daxx and PML), we also observed their co-localization in PML bodies. Importantly, similar high molecular weight complexes and co-localization in PML bodies were also shown for ATRX and its interaction with Daxx [42]. ATRX serves as a DNA-binding anchor, essential for Daxx-mediated positioning of histone H3.3 [13], [45] and for Daxx-mediated repression of CMV-promoter-regulated inducible transgene array [20]. However, the ATRX-Daxx H3.3 loading complex could also contribute to the activation of immediate early genes, such as Bdnf, in activated murine neurons [23]. Our data support rather a repressive role of Daxx in Brg1-dependent transcription of CD44 and SCEL genes in adrenal carcinoma SW13 cells. These cells lack Brg1 expression and its re-introduction leads to enhanced transcription of CD44 and several other genes. Our data document that siRNA-mediated down-regulation of Daxx, greatly enhanced expression of these Brg1-dependent genes, suggesting a direct suppressive role of Daxx, possibly connected to histone deacetylases or its chaperone-like function. This repressive impact of Daxx on the transcription of Brg1-regulated genes might also be independent or only partly dependent on its interaction with Brg1, and further experiments will be required to elucidate the role of Daxx-Brg1 interaction in this process.
We also addressed and mapped the regions responsible for the interaction between Daxx and Brg1. On Daxx, the interaction surface is likely to be broad within its N-terminal and central regions. Interestingly, previously found Daxx-interacting proteins involved in the epigenetic regulation of ATRX and DMAP1 transcription also interact with the N-terminal/central part of Daxx [42], [43], [46], which is also the only region on Daxx, structured into a-helical bundle domain [41]. The part of Brg1 to which we mapped the major interaction with Daxx, i.e. the region between QLQ and HSA domains, also mediates the interaction between Brg1 and BAF250a/ARID1A and it is apparently required for glucocorticoid receptor-dependent transactivation [47].
In summary, we identified chromatin remodelling ATPase Brg1 as a new Daxx-interacting protein and thus strengthened the Daxx connection to the epigenetic regulation of transcription. As both Brg1 and also recently Daxx, are being considered as tumour suppressors, their presumable functional interaction might have an impact on the regulation of chromatin status or gene expression in cancer cells and thus also affect tumour progression.
Acknowledgements
Supported by the AS CR institutional grant RVO68378050, GAAV grant A5052304 to LA and Czech Science Foundation grant no. P305/11/P683 to HH. We thank to Frances Zatrepalkova for the English proofreading the manuscript.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.bbrep.2015.12.012.
Appendix A. Supplementary material
Fig. S1.
Fig. S2.
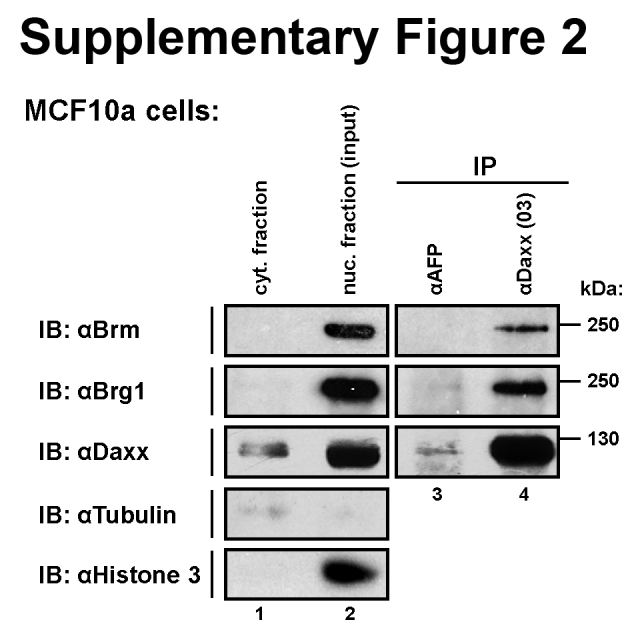
Fig. S3.
Fig. S4.
Fig. S5.
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material
References
- 1.Ishov A.M., Sotnikov A.G., Negorev D., Vladimirova O.V., Neff N., Kamitani T., Yeh E.T., Strauss J.F., (III.), Maul G.G. PML is critical for ND10 formation and recruits the PML-interacting protein Daxx to this nuclear structure when modified by SUMO-1. J. Cell Biol. 1999;147:221–234. doi: 10.1083/jcb.147.2.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Michaelson J.S., Bader D., Kuo F., Kozak C., Leder P. Loss of Daxx, a promiscuously interacting protein, results in extensive apoptosis in early mouse development. Genes Dev. 1999;13:1918–1923. doi: 10.1101/gad.13.15.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Salomoni P., Khelifi A.F. Daxx: death or survival protein? Trends Cell Biol. 2006;16:97–104. doi: 10.1016/j.tcb.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 4.Puto L.A., Reed J.C. Daxx represses RelB target promoters via DNA methyltransferase recruitment and DNA hypermethylation. Genes Dev. 2008;22:998–1010. doi: 10.1101/gad.1632208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morozov V.M., Massoll N.A., Vladimirova O.V. Regulation of c-met expression by transcription repressor Daxx. Oncogene. 2008;27:2177–2186. doi: 10.1038/sj.onc.1210865. [DOI] [PubMed] [Google Scholar]
- 6.Chang C.-C., Lin D.-Y., Fang H.-I., Chen R.-H., Shih H.-M. Daxx mediates the SUMO-dependent transcriptional repression of Smad4. J. Biol. Chem. 2005;35:1397–1400. doi: 10.1074/jbc.M409161200. [DOI] [PubMed] [Google Scholar]
- 7.Song M.S., Salmena L., Carracedo A., Egia A., Lo-Coco F., Teruya-Feldstein J., Pandolfi P.P. The deubiquitinylation and localization of PTEN are regulated by a HAUSP-PML network. Nature. 2008;455:813–817. doi: 10.1038/nature07290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Croxton R., Puto L.A., de Belle I., Thomas M., Torii S., Hanaii F., Cuddy M., Reed J.C. Daxx represses expression of a subset of antiapoptotic genes regulated by nuclear factor-kappaB. Cancer Res. 2006;66:9026–9035. doi: 10.1158/0008-5472.CAN-06-1047. [DOI] [PubMed] [Google Scholar]
- 9.Lin D.-Y., Fang H.-I., Ma A.-H., Huang Y.-S., Pu Y.-S., Jenster G., Kung H.-J., Shih H.-M. Negative modulation of androgen receptor transcriptional activity by Daxx. Mol. Cell Biol. 2004;24:10529–10541. doi: 10.1128/MCB.24.24.10529-10541.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang H., He J., Li J. Methylation of RASSF1A gene promoter is regulated by p53 and DAXX. FASEB J. 2013;27:232–242. doi: 10.1096/fj.12-215491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shalginskikh N., Poleshko A., Skalka A.M. Retroviral DNA methylation and epigenetic repression are mediated by the antiviral host protein Daxx. J. Virol. 2013;87:2137–2150. doi: 10.1128/JVI.02026-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cantrell S.R., Bresnahan W.A. Human cytomegalovirus (HCMV) UL82 gene product (pp71) relieves hDaxx-mediated repression of HCMV replication. J. Virol. 2006;80:6188–6191. doi: 10.1128/JVI.02676-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drané P., Ouararhni K., Depaux A., Shuaib M., Hamiche A. The death-associated protein DAXX is a novel histone chaperone involved in the replication-independent deposition of H3.3. Genes Dev. 2010;24:1253–1265. doi: 10.1101/gad.566910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goldberg A.D., Banaszynski L.A., Noh K.M., Lewis P.W., Elsaesser S.J., Stadler S., Dewell S., Law M., Guo X., Li X., Wen D., Chapgier A., DeKelver R.C., Miller J.C., Lee Y.L., Boydston E.A., Holmes M.C., Gregory P.D., Greally J.M., Rafii S., Yang C., Scambler P.J., Garrick D., Gibbons R.J., Higgs D.R., Cristea I.M., Urnov F.D., Zheng D., Allis C.D. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell. 2010;140:678–691. doi: 10.1016/j.cell.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elsaesser S.J., Allis C.D. HIRA and Daxx constitute two independent histone H3.3-containing predeposition complexes. Cold Spring Harbor Symp. Quant. Biol. 2010;75:27–34. doi: 10.1101/sqb.2010.75.008. [DOI] [PubMed] [Google Scholar]
- 16.Salomoni P. The PML-interacting protein DAXX: histone loading gets into the picture. Front Oncol. 2013;3:152. doi: 10.3389/fonc.2013.00152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hollenbach A.D., McPherson C.J., Mientjes E.J., Iyengar R., Grosveld G. Daxx and histone deacetylase II associate with chromatin through an interaction with core histones and the chromatin-associated protein Dek. J. Cell Sci. 2002;115:3319–3330. doi: 10.1242/jcs.115.16.3319. [DOI] [PubMed] [Google Scholar]
- 18.Ivanauskiene K., Delbarre E., McGhie J.D. The PML-associated protein DEK regulates the balance of H3.3 loading on chromatin and is important for telomere integrity. Genome Res. 2014;24:1584–1594. doi: 10.1101/gr.173831.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He Q., Kim H., Huang R. The Daxx/Atrx complex protects tandem repetitive elements during DNA hypomethylation by promoting H3K9 trimethylation. Cell Stem Cell. 2015;17:273–286. doi: 10.1016/j.stem.2015.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Newhart A., Rafalska-Metcalf I.U., Yang T. Single-cell analysis of Daxx and ATRX-dependent transcriptional repression. J. Cell Sci. 2012;125:5489–5501. doi: 10.1242/jcs.110148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schreiner S., Burck C., Glass M. Control of human adenovirus type 5 gene expression by cellular Daxx/ATRX chromatin-associated complexes. Nucleic Acids Res. 2013;41:3532–3550. doi: 10.1093/nar/gkt064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsai K., Chan L., Gibeault R. Viral reprogramming of the Daxx histone H3.3 chaperone during early Epstein-Barr virus infection. J. Virol. 2014;88:14350–14363. doi: 10.1128/JVI.01895-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Michod D., Bartesaghi S., Khelifi A. Calcium-dependent dephosphorylation of the histone chaperone DAXX regulates H3.3 loading and transcription upon neuronal activation. Neuron. 2012;74:122–135. doi: 10.1016/j.neuron.2012.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lacoste N., Woolfe A., Tachiwana H. Mislocalization of the centromeric histone variant CenH3/CENP-A in human cells depends on the chaperone DAXX. Mol. Cell. 2014;53:631–644. doi: 10.1016/j.molcel.2014.01.018. [DOI] [PubMed] [Google Scholar]
- 25.Elsasser S.J., Noh K.M., Diaz N. Histone H3.3 is required for endogenous retroviral element silencing in embryonic stem cells. Nature. 2015;522:240–244. doi: 10.1038/nature14345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schwartzentruber J., Korshunov A., Liu X.Y. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482:226–231. doi: 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]
- 27.Jiao Y., Shi C., Edil B.H., de Wilde R.F., Klimstra D.S., Maitra A., Schulick R.D., Tang L.H., Wolfgang C.L., Choti M.A., Velculescu V.E., Diaz L.A., Jr., Vogelstein B., Kinzler K.W., Hruban R.H., Papadopoulos N. DAXX/ATRX, MEN1, and mTOR pathway genes are frequently altered in pancreatic neuroendocrine tumors. Science. 2011;331:1199–1203. doi: 10.1126/science.1200609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marinoni I., Kurrer A.S., Vassella E. Loss of DAXX and ATRX are associated with chromosome instability and reduced survival of patients with pancreatic neuroendocrine tumors. Gastroenterology. 2014;146 doi: 10.1053/j.gastro.2013.10.020. 453-460. [DOI] [PubMed] [Google Scholar]
- 29.Clynes D., Jelinska C., Xella B. Suppression of the alternative lengthening of telomere pathway by the chromatin remodelling factor ATRX. Nat. Commun. 2015;6:7538. doi: 10.1038/ncomms8538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hargreaves D.C., Crabtree G.R. ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell Res. 2011;21:396–420. doi: 10.1038/cr.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bultman S., Gebuhr T., Yee D., La Mantia C., Nicholson J., Gilliam A., Randazzo F., Metzger D., Chambon P., Crabtree G., Magnuson T. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol. Cell. 2000;6:1287–1295. doi: 10.1016/s1097-2765(00)00127-1. [DOI] [PubMed] [Google Scholar]
- 32.Kidder B.L., Palmer S., Knott J.G. SWI/SNF-Brg1 regulates self-renewal and occupies core pluripotency-related genes in embryonic stem cells. Stem Cells. 2009;27:317–328. doi: 10.1634/stemcells.2008-0710. [DOI] [PubMed] [Google Scholar]
- 33.Barker N., Hurlstone A., Musisi H., Miles A., Bienz M., Clevers H. The chromatin remodelling factor Brg-1 interacts with beta-catenin to promote target gene activation. EMBO J. 2001;20:4935–4943. doi: 10.1093/emboj/20.17.4935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dunaief J.L., Strober B.E., Guha S., Khavari P.A., Alin K., Luban J., Begemann M., Crabtree G.R., Goff S.P. The retinoblastoma protein and BRG1 form a complex and cooperate to induce cell cycle arrest. Cell. 1994;79:119–130. doi: 10.1016/0092-8674(94)90405-7. [DOI] [PubMed] [Google Scholar]
- 35.Huang M., Qian F., Hu Y., Ang C., Li Z., Wen Z. Chromatin-remodelling factor BRG1 selectively activates a subset of interferon-alpha-inducible genes. Nat. Cell Biol. 2002;4:774–781. doi: 10.1038/ncb855. [DOI] [PubMed] [Google Scholar]
- 36.Lee D., Kim J.W., Seo T., Hwang S.G., Choi E.J., Choe J. SWI/SNF complex interacts with tumor suppressor p53 and is necessary for the activation of p53-mediated transcription. J. Biol. Chem. 2002;277:22330–22337. doi: 10.1074/jbc.M111987200. [DOI] [PubMed] [Google Scholar]
- 37.Reisman D., Glaros S., Thompson E.A. The SWI/SNF complex and cancer. Oncogene. 2009;28:1653–1668. doi: 10.1038/onc.2009.4. [DOI] [PubMed] [Google Scholar]
- 38.Boussif O., Lezoualc’h F., Zanta M.A., Mergny M.D., Scherman D., Demeneix B., Behr J.P. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc. Natl. Acad. Sci. USA. 1995;92:7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Michaelson J.S., Leder P. RNAi reveals anti-apoptotic and transcriptionally repressive activities of DAXX. J. Cell Sci. 2003;116:345–352. doi: 10.1242/jcs.00234. [DOI] [PubMed] [Google Scholar]
- 40.Lin D.Y., Huang Y.S., Jeng J.C. Role of SUMO-interacting motif in Daxx SUMO modification, subnuclear localization, and repression of sumoylated transcription factors. Mol. Cell. 2006;24:341–354. doi: 10.1016/j.molcel.2006.10.019. [DOI] [PubMed] [Google Scholar]
- 41.Escobar-Cabrera E., Lau D.K., Giovinazzi S. Structural characterization of the DAXX N-terminal helical bundle domain and its complex with Rassf1C. Structure. 2010;18:1642–1653. doi: 10.1016/j.str.2010.09.016. [DOI] [PubMed] [Google Scholar]
- 42.Xue Y., Gibbons R., Yan Z., Yang D., McDowell T.L., Sechi S., Qin J., Zhou S., Higgs D., Wang W. The ATRX syndrome protein forms a chromatin-remodeling complex with Daxx and localizes in promyelocytic leukemia nuclear bodies. Proc. Natl. Acad. Sci. USA. 2003;100:10635–10640. doi: 10.1073/pnas.1937626100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Muromoto R., Sugiyama K., Takachi A., Imoto S., Sato N., Yamamoto T., Oritani K., Shimoda K., Matsuda T. Physical and Functional Interactions between Daxx and DNA Methyltransferase 1-Associated Protein DMAP1. J. Immunol. 2004;172:2985–2993. doi: 10.4049/jimmunol.172.5.2985. [DOI] [PubMed] [Google Scholar]
- 44.Yamamichi-Nishina M., Ito T., Mizutani T. SW13 cells can transition between two distinct subtypes by switching expression of BRG1 and Brm genes at the post-transcriptional level. J. Biol. Chem. 2003;278:7422–7430. doi: 10.1074/jbc.M208458200. [DOI] [PubMed] [Google Scholar]
- 45.Lewis P.W., Elsaesser S.J., Noh K.M., Stadler S.C., Allis C.D. Daxx is an H3.3-specific histone chaperone and cooperates with ATRX in replication-independent chromatin assembly at telomeres. Proc. Natl. Acad. Sci. USA. 2010;107:14075–14080. doi: 10.1073/pnas.1008850107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ishov A.M., Vladimirova O.V., Maul G.G. Heterochromatin and ND10 are cell-cycle regulated and phosphorylation-dependent alternate nuclear sites of the transcription repressor Daxx and SWI/SNF protein ATRX. J. Cell Sci. 2004;117:3807–3820. doi: 10.1242/jcs.01230. [DOI] [PubMed] [Google Scholar]
- 47.Trotter K.W., Fan H.Y., Ivey M.L., Kingston R.E., Archer T.K. The HSA domain of BRG1 mediates critical interactions required for glucocorticoid receptor-dependent transcriptional activation in vivo. Mol. Cell Biol. 2008;28:1413–1426. doi: 10.1128/MCB.01301-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material
Supplementary Material
Supplementary Material
Supplementary Material