Abstract
Mouse nucleoplasmin M.NPM2 was recombinantly expressed and the protein consisting of the complete sequence was purified and characterized. Similar to its Xenopus laevis X.NPM2 counterpart, the protein forms stable pentameric complexes and exhibits an almost undistinguishable hydrodynamic ionic strength-dependent unfolding behavior. The interaction of N.PM2 with histones and mouse P1/P2 protamines revealed that these chromosomal proteins bind preferentially to the distal part of the nucleoplasmin pentamer. Moreover, the present work highlights the critical role played by histones H2B and H4 in the association of the histone H2A-H2B dimers and histone octamer with nucleoplasmin.
Highlights
-
•
Characterization of the entire mouse M.NPM2 protein.
-
•
Determination of sites of interaction of M.NPM2 with histones and mouse protamines.
-
•
Use of crosslinking mass spectrometry to determine protein-protein interactions.
-
•
Analysis of the C-terminal NPM2 unfolding.
1. Introduction
The term “chaperone” to describe a protein function was first coined in 1978 by Ron Laskey. It was meant to explain the role of nucleoplasmin, a highly abundant protein in the egg, initially isolated from Xenopus, which is found associated with histones [1]. Yet, despite the almost 40 years since its initial discovery, the molecular details of the nucleoplasmin-histone complex are elusive and remain ill-defined. It was subsequently shown that the nucleoplasmin has additional functions that transcend those of its role as histone storage within its initial description, and evidence has been provided that shows it also plays a critical role in the remodelling of the paternal chromatin immediately after fertilization [2]. This means that it operates both as an assembly and disassembly factor during early embryogenesis [3]. Indeed, the ability of Xenopus nucleoplasmin (henceforth referred to as X.NPM2) to bind to the nuclear sperm arginine-rich protamines from different organisms other than Xenopus has been well documented [4], [5], [6].
The nucleoplasmin family of proteins, can be classified into three main groups: NPM1, NPM2 and NPM3. NPM1 also known with the name of nucleophosmin is mainly localized in the nucleolus and it is present in a wide variety of tissues. It is one of the most extensively studied members of this family due mainly to its dis-regulation in different types of cancers. Despite its important involvement in ribosome biogenesis, NPM1 has also been involved in histone chaperone activity. NPM3 has also been involved in ribosome biogenesis in conjunction with NPM1 and it has been implicated in the remodelling of paternal chromatin organization after fertilization in mammals. NPM2 is present in the nucleus of eggs and oocytes and during the early stages of development and it participates both in histone chaperoning and in the removal of paternal histones after fertilization. From an evolutionary point of view, NPM1 and NPM3 are more closely related and appear to have differentiated later than NPM2. Structurally, protein members of the three groups have a very similar primary and secondary structure which is characterized by the presence of several acidic amino acid clusters, a nuclear localization sequence signal (NLS) and an N-terminal core domain consisting of several beta sheets which are responsible for the highly stable pentameric organization of these proteins (see [7], [8] for extensive reviews).
From a structural point of view, the nucleoplasmin NPM2 monomer contains several highly characteristic acidic amino acid clusters (A1, A2 and A3) (Fig. 1A) that were early hypothesized to be responsible for the interactions of this protein with histones. The protein also contains a nuclear localization signal (NLS). In solution, the protein forms a highly stable pentamer [9] with a characteristic beta-strand barrel organization [10]. The crystallization of the nucleoplasmin pentamer led the authors to hypothesize an interaction between the histone octamer and NPM2 through the lateral side of the pentamer [10], not necessarily dependent on the acidic tracts, in contrast to its chromatin remodelling activity. Protamine removal from sperm nuclei has been shown to be highly dependent on the A1 tract [11].
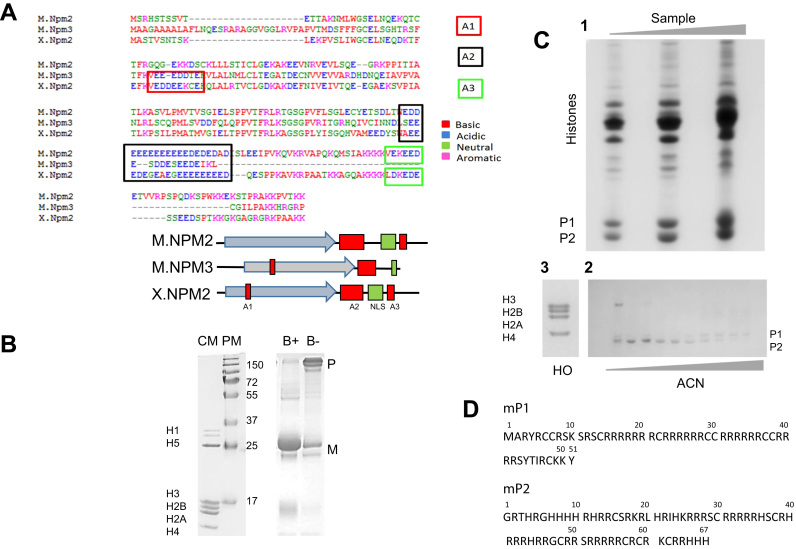
Fig. 1.
Characterization of the protein interacting partners. (A) Primary structure alignment of mouse NPM2 and NPM3 (M.NPM2/MNPM3) in comparison to Xenopus NPM2 (X.NPM2) and cartoon representation highlighting the position of the three acidic tracts (A1, A2 and A3) and the nuclear localization signal (NLS). The arrow indicates the beta-stranded region. (B) SDS-PAGE analysis of M.NPM2 before (B-) and after boiling (B+) the sample, in comparison to a protein (PM) and a chicken histone marker (CM). M: monomer; P: pentamer. (C) Isolation (1) and purification (2) of the mouse testes sperm nuclear basic proteins (SNBPs) and histone octamers (3). (D) Amino acid sequence of mouse protamines P1 and P2, Reference Sequences: P02319 and NP_032959, respectively.
In solution, the C-terminal non-structured domains of the NPM2 pentamer (C-terminal tails) were shown to be able to adopt an extended organization [5], and the overall interaction of NPM2 with the histones and protamines did not seem to be significantly dependent on their presence [5], [12]. A few recent papers, using high-resolution electron microscopy and three-dimensional image reconstruction, support the notion that the histone H2A-H2B dimer and the entire histone octamer interact with the distal face of the XNPM2 pentamer at a site were the C-terminal tails, which contain the A2 and A3 acidic tracts, protrude from the molecule [13], [14]. In the native physiological settings of the oocyte, the egg, and the embryo, the interactions between the histones in this region appear to be modulated by posttranslational modifications (PTMs), such as phosphorylation and glutamylation of the disordered tails [15], [16]. The differential occurrence of these marks at different stages of development can fine-tune the histone sequestration and deposition [16].
In a further attempt to better understand the interaction between nucleoplasmin, histones and protamines, we have, for the first time, purified the full mouse NPM2 and characterized its interactions with the histone octamer and mouse protamines using crosslinking mass spectrometry [17].
2. Materials and methods
2.1. Gel electrophoresis
SDS-polyacrylamide gel electrophoresis (SDS-PAGE) was carried out according to [18]. Acetic acid-urea (AU)–PAGE were carried out as described elsewhere [19]. Native 12% PAGE was prepared on 40 mM Tris–HCl (pH 7.5), 1 mM EDTA, 20 mM sodium acetate buffer and run in the same buffer.
2.2. Cloning of mouse NPM2
Ovaries from Mus musculus, obtained from the Animal Care Services (University Of Victoria), were flash frozen in liquid nitrogen and stored at −80 °C until use. RNA extraction was performed using the RNeasy® Mini Kit (Qiagen, Valencia, CA). cDNA synthesis was conducted using SuperScript® III RT cDNA Synthesis Kit (Life Technologies, Burlington ON). The full length mouse NPM2 gene was PCR amplified using the following primers:
(5′-GCGCCATATGAGTCGCCACAGCACCA-3′) (forward primer) and (5′GCGCGGATCCTCATTCTTGGTCACTGGCTTC-3′) (reverse primer). The DNA fragment obtained was inserted between the Sap1 and Nde1 sites of a pTXB1 vector, an IMPACT® Kit expression vector containing a self-cleaving intein tag and chitin binding domain (NEB, Ipswich, MA).
2.3. Expression and purification of mouse NPM2
Mouse NPM2 expression was carried out in BL21 (DE3) competent cells (NEB, Ipswich, MA).
After induction with 1 mM IPTG, cells were grown for 3 h at 37 °C. The culture was then centrifuged at 8000 x g for 10 min at 4 °C, and the pellets were re-suspended in one-tenth the volume of 20 mM Tris–HCl (pH 8.5), 500 mM NaCl, 1 mM EDTA containing 1:100 protease inhibitor cocktail (Roche, Indianapolis, IN). The homogenate was lysed with a French Press (SIM Aminco, Rochester, NY) and centrifuged at 8000×g for 10 min. The supernatant was then passed through a 0.45 μm filter, supplemented with 1:50 protease inhibitor cocktail, and loaded onto the IMPACT® Intein chitin resin column (NEB, Ipswich, MA) at 4 °C with a flow rate of 0.5 mL/min, and eluted under the conditions described by the supplier. Fractions containing mouse NPM2 were pooled and dialyzed for 4 h against 50 mM Tris–HCl (pH 8), 100 mM NaCl buffer at 4 °C. The dialysate was concentrated and FPLC fractionated using the ATKA® FPLC System (Amersham Pharmacia Biotech, Piscataway, NJ), using a HiLoad 16/60 Superdex® 200 Size Exclusion Column. Full length Xenopus laevis NPM2 was purified as described previously [12].
2.4. Isolation of mouse protamines and chicken erythrocyte core histones
Mouse testes were homogenized in buffer [0.15 M NaCl, 10 mM Tris–HCl (pH 7.5), 0.5% Triton X-100 containing 1:100 protease inhibitor cocktail], and centrifuged at 2,000×g for 10 min at 4 °C. The pellets were re-suspended in [4 M guanidinium hydrochloride, 50 mM Tris–HCl (pH 7.5), 1.25 mM EDTA] and homogenized with 30 strokes of a dounce homogenizer on ice and brought to 52 mM β-mercaptoethanol, then incubated for 90 min at room temperature. 10 μl vinyl pyridine was added and the specimen was incubated for an additional 30 min at room temperature, vortexing every 5 min in the dark. 7 volumes of dH2O was added and the resulting solution was centrifuged at 8000×g for 10 min at 4 °C. Pellets were homogenized in 0.5 N HCl in a dounce on and centrifuged at 8000×g for 10 min at 4 °C. The HCl extract (supernatant) was precipitated six volumes of acetone overnight at −20 °C. The next day, the sample was centrifuged at 8000×g for 10 min at 4 °C and the pellets were re-suspended in half a volume of acetone and centrifuged one more time. The protein pellets thus obtained were dried under vacuum, and the protamine powder was stored at −80 °C. The protamines (P1 and P2) were further purified by reverse-phase high-performance liquid chromatography (RP-HPLC) on a Vydac Protein and Peptide C18 column utilizing 0.1% TFA as the mobile phase and an increasing gradient of ACN from 0% to 60%.
Chicken erythrocyte core histones were prepared as described elsewhere [20].
2.5. NPM2-Chromosomal Protein Interactions
The interactions between mouse NPM2 and core histone octamers or protamines were carried out in native PAGE. Titrations were performed in 240 mM NaCl, 8.8 mM Tris–HCl (pH 7.5), 1.8 mM MgCl2 buffer, and incubated for 30 min at room temperature. The samples were then brought to 5% sucrose and loaded on the gel.
2.6. Analytical ultracentrifuge analysis (AUC)
Sedimentation velocity AUC analyses were performed in a Beckman XL-I analytical ultracentrifuge (Beckman-Coulter Instruments, Brea, CA) using an An-55 Al aluminum rotor and cells with double sector aluminum-filled Epon centerpieces, then analysed as described elsewhere [21].
2.7. Cross linking mass spectrometry (CXMS)
Protamine- and histone-NPM2 complexes, prepared as described in Section 2.5, were extensively dialyzed against 100 mM NaCl, 10 mM PIPES (pH 7.3), and 0.1 mM EDTA buffer for 4 h at 4 °C. A minimum concentration of 30 μg of protein was used for each CXMS experiment. Two samples, NPM2+histone octamers (NPM2-histone run) and NPM2+protamines (NPM2-protamine run) were subjected to CXMS in two independent runs. The pH of each mixture was adjusted to 8.0-8.5 by the addition of 0.2 M Na2HPO4, and proteins were crosslinked with 0.05 mM cyanurobiotindipriopyonylsuccinimide (CBDPS) (Creative Molecules Inc.) for 30 min at 25 °C [17]. Reactions were quenched with ammonium bicarbonate at a final concentration of 30 mM, and cross-linked proteins were then digested with sequencing-grade trypsin (Promega, Madison, WI) overnight at 37 °C at a 10:1 substrate:enzyme ratio. Resulting peptide mixtures were affinity-purified using monomeric avidin-agarose beads (Thermo Scientific, Rockford, IL) and analysed using nano-LC-MALDI-MS/MS via the Eksigent® 1D nano-LC system (AB/Sciex, Dublin, CA), Dionex® Probot spotter (Thermo Scientific, Rockford, IL) and the 4800 MALDI-TOF/TOF mass spectrometer (AB/Sciex, Foster City, CA). Data was analyzed with the DXMSMS Match program [22] of ICC-CLASS [23].
3. Results
3.1. Purification and isolation of full sequence M.NPM2 and chromosomal proteins
Purification of recombinant X.NPM2 is often cumbersome, mainly due to the appearance of truncated forms along the sequence, which are difficult to separate from one another. However, with M.NPM2, we were able to purify the entire protein to higher than 80% purity (Fig. 1B) using a relatively simple two-step protocol involving the use of an intein-conjugated version of the protein, followed by intein cleavage and size-exclusion chromatography. The molecular mass of the protein (23,175 Da) obtained by electron spray ionization mass spectrometry (ESI-MS) matched the molecular mass determined from its amino acid sequence (Fig. 1A). Earlier purification and characterization of a mammalian human NPM2 used a truncated version (M1-D152), which lacked the last 62 C-terminal amino acids containing the acidic tract E3 and the NLS [24].
M.NPM2 was used to determine its interactions between protamines and histones. Mouse testes sperm nuclear basic proteins (SNBPs ) were prepared using a previously described protocol that involves the reduction and alkylation of cysteine residues [25]. The SNBPs obtained allowed for the easy HPLC fractionation of protamines P1/P2 from testes histones (Fig. 1C, 1–2). Protamines have an intrinsically disordered organization [26] that should not be affected by the denaturing conditions of the RP-HPLC purification. A stoichiometric mixture containing equal amounts of P1 and P2 was used in the M.NPM2-protamine binding experiments described in the following section. Histone octamers were obtained from chicken erythrocytes under non-denaturing conditions (Figs. 1C, 3) [20]. Avian histones are compositionally identical to their mammalian histone counterparts. Furthermore, the levels of PTMs in chicken erythrocyte histones are relatively low compared to other tissues or cell types [27].
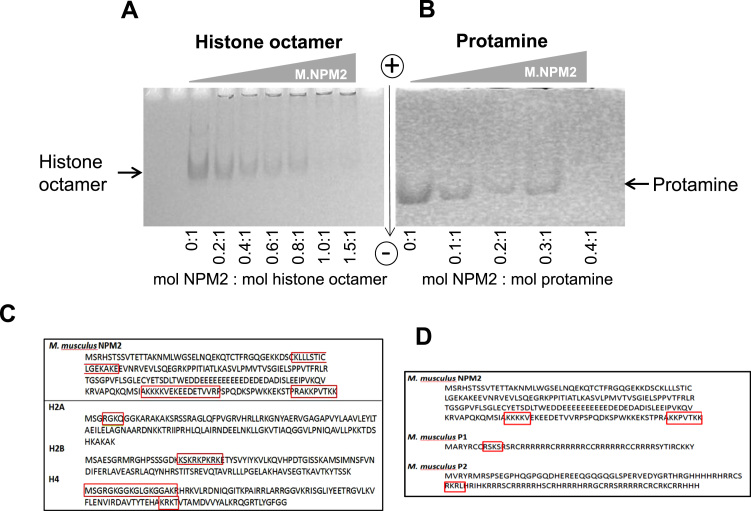
Fig. 2.
Native PAGE of a titration of histone octamers (A) and mouse protamines P1/P2 (B) with increasing amounts of M.NPM2. ). In this type of analysis, NPM2: histone/protamine complexes display a complex ‘shift’ and are unable to enter the gel. (C) Amino acid sequence of mouse NPM2 and core histones H2A, H2B and H4, with interaction sites determined through CXMS data (Supp. Table 1a) highlighted in red. Three peptide sequences in NPM2 represent strong cross-linking candidates with sequences in H2A, H2B and H4 (highlighted in red). Gallus gallus H2A NCBI Reference Sequence: AAC60008.1, G. gallus H2B NCBI Reference Sequence: AAC60000.1, G. gallus H4 NCBI Reference Sequence: NP_001032932.1. (D) Amino acid sequence of mouse NPM2, P1 and P2, with possible interaction sites determined through CXMS data (Supp. Table 1a) highlighted in red. Three peptide sequences within M.NPM2 represent possible cross-linking candidates. M. musculus NPM2 NCBI Reference Sequence: NP_851990.2; M. musculus P1 NCBI Reference Sequence: NP_038665.1; M. musculus P2 NCBI Reference Sequence: P07978.1.
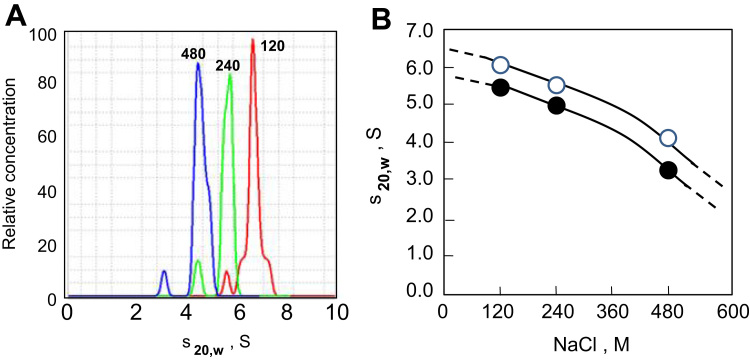
Fig. 3.
(A) Sedimentation velocity analysis of X.NPM2 and M.NPM2 at different sodium chloride concentrations. (A) Plots of the relative sample concentration vs. the sedimentation coefficient at three different NaCl concentrations (120, 240, and 480 mM). Data was obtained using the histogram envelope analysis from the UltraScan software [31]. (B) Sodium chloride dependence of the sedimentation coefficient of X.NPM2 (open circles) and M.NPM2 (black circles). S20,w=sedimentation coefficient corrected to standard conditions (water and 20 °C). S=Svedberg units.
3.2. Interaction between M.NPM2, histone octamers and protamines
The interaction between histone octamers and Xenopus X.NPM2 has been quite extensively studied [10], [12], [14], [16]. Only one similar study is available for a C-terminally truncated version of its mouse counterpart [24]. As shown in Fig. 2A, M.NPM2 saturation takes place upon the binding of a whole histone octamer, as had been previously reported for X.NPM2 [12].
The interaction between Xenopus nucleoplasmin and protamines has been also studied, albeit to a lesser extent [2], [5], [6], [11]. In contrast to core histones, saturation takes place at a ratio of 3 mouse P1+P2:M.NPM2, which is also very similar to what was observed in the interaction between X.NMP2 and fish protamines [5].
Cross linking mass spectrometry (CXMS) analysis (Fig. 3) revealed three sites of interaction with histones: The two strongest ones were located in the C-terminal unstructured regions, and the weakest one in the lateral face of the nucleoplasmin pentamer (see Fig. 2C and supplementary Table 1). One of the C-terminal sites overlaps with the acidic tract A3 (see Fig. 1A and Fig. 4). These sites interact with the N-terminal tails of H2A, H2B and H4. In this later case, an additional site of interaction was found in the region corresponding to the loop L2 of the histone fold domain, and which represents the interaction site with the higher score for this histone. No interactions could be detected for histone H3 (Fig. 2C and supplementary Table 1). Interestingly, the few sites of interaction we were able to detect with this technique revealed that protamines were found to interact at two similar locations, as histones in the C-terminal domains of M.NPM2 (supplementary Table I), with no other interacting sites along nucleoplasmin (Fig. 2D).
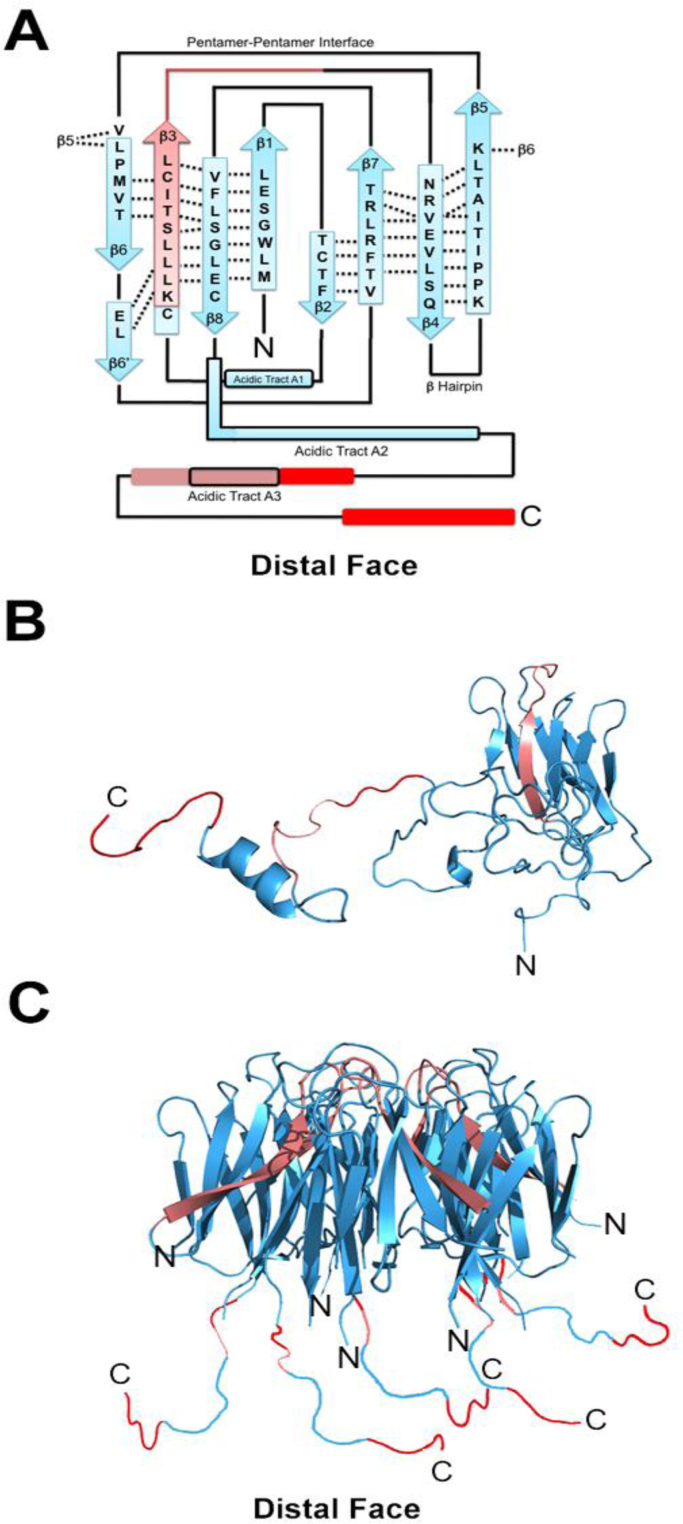
Fig. 4.
Schematic representation and 3D structure of mouse NPM2 with highlighted chromosomal protein interaction sites as determined by CXMS. The intensity of the interactions is depicted in red (see Supp. Table 1). (A) Schematic representation of the secondary structure organization of the NPM2 monomer. Acidic tracts are highlighted in black boxes. Note: Acidic tract A1 denotes the region where the A1 tract is seen in other species (but not in mouse) (see Fig.1). Beta barrels β1-β8 are also shown by wide arrows and intra-sheet hydrogen bonds are represented by dotted black lines. (B) Tertiary structure of the M.NPM2 monomer created using PyMOL. (C) Tertiary structure organization of the M.NPM2 pentamer created using PyMOL.
3.3. X.NPM2 and M.N.PM2 exhibit a very similar hydrodynamic conformation and salt-dependent unfolding
As seen in Fig. 3, both X.NPM2 and M.NPM2 exhibit a very similar hydrodynamic behaviour, with a sedimentation coefficient value of 6.5 S for X.NPM2 and 6.0 S for M.NPM2, which correspond to a pentameric organization [5], [12]. The slightly lower sedimentation coefficient values of the mammalian counterpart are indicative of a slightly more compact organization within the pentameric structure, which may be the result of the different molecular structural changes exhibited by the Xenopus and mouse nucleoplasmin that result in a smaller crystallization unit cell in the latter [24]. The pentaneric association of M.NPM2 can also be clearly visualized in Fig. 1B. The β-barrel organization of the nucleoplasmin pentamer [10] confers the macromolecular complex with high stability, which in the absence of thermal denaturation, allows it to retain its pentameric organization even in the presence of ionic detergents such as SDS [5].
Both nucleoplasmin complexes exhibit an identical ionic strength–dependent change of their sedimentation coefficients with a significant drop as the salt concentration increases. This is indicative of their long C-terminal intrinsically disordered domains becoming unfolded as a result of the charge neutralization that releases the interaction of these regions with the pentameric core [5], [15], a process that in the physiological setting is likely to be enhanced by phosphorylation [15], [16], [28].
4. Discussion
4.1. The interaction of M.NPM2 with histones
The results shown in supplementary Table 1 (see also Fig.2) show that the strongest interactions between M.NPM2 occur between histones H2B and H4 at sites in M.NPM2 located at the C-terminal tail of nucleoplasmin at the distal face of the pentamer (Fig. 4). Interestingly, the two strongest interacting sites detected here for H2B (KSKRKPKRKE) and H4 (KRKT) fall at the interface between the histone tail of H2B and the histone fold, and at a region within the histone fold domain in the case of histone H4. This observation provides support to the notion that stereospecific interactions between histones and nucleoplasmin play an important role in the formation of the complexes, in addition to the electrostatic ones [12]. Interestingly, these stronger interaction sites overlap with the sites of interaction previously determined by Ramos et al. using [14]. The weakest sites of interaction determined here for the N-terminal tails of H2A and H4 could not be detected in [14] probably due to the fact that these represent weaker interaction sites which would be harder to detect by the limited proteolysis approach used [14].
The lack of strong interacting sites for H2A and H3 suggest that these histones play a lesser role in the formation of the nucleoplasmin complexes with either the H2A-H2B dimer or the entire histone octamer complex. It is possible that H2B represents the main anchoring site for the H2A-H2B dimer. As described in the introduction, nucleoplasmin was initially found associated with H2A-H2B dimers in the Xenopus egg [29], but its prevalence in the early stages of development immediately after fertilization [30] in the absence of histone synthesis have suggested a potential involvement in its interaction with the whole histone octamer.
4.2. The interaction of M.NPM2 with mouse protamines
The interaction between the sea bass fish (Dicentrarchus labrax) protamine and Xenopus nucleoplasmin, and the potential involvement of the main X.NPM2 polyglutamic tract, has already been carefully analysed. The amino acid sequence of D. labrax is: PR4QASRPVR5TR2STAER5V2R4. This very simple sequence shares the recurrent presence of arginine clusters with mouse P1/P2 protamines. The very similar M.NPM2 protamine saturation ratio observed for mammalian (mouse) protamines and X.NMP2:fish protamine suggest that the interaction may be primarily electrostatically driven, and reaches an approximate saturation value of 50-60 electrostatic R/E-D, which represent about only half of the E-D residues contributed by the main acidic tract A2 and A3 in the pentamer (Fig. 1A). Interestingly, whilst these tracts appear to play an essential role in protamine binding in fish, they were not found to be indispensable for sperm chromatin decondensation [5].
The results presented here for the protamine-M.NPM2 interaction should be viewed as preliminary. As described in the materials and methods, the novel crosslinking method of isotopically (deuterium) labeled amine-reactive cross-linkers using CBDPS [17] used here relies on the use of trypsin after cross-linking the protein complexes, where the digested cross-linked peptides are avidin-fractionated, HPLC separated, and the peaks subjected to mass spectrometric analysis. Special care is taken in this procedure in regards to cross-linking specificity. This is achieved by titration with different concentrations of the cross-linker and sequential dilution of the sample. Only the persistent cross-linked bands are analysed. This allows for unequivocal determination of the cross-linked sites and amino acid sequence of the domains involved. However, the massive presence of arginine clusters complicates this type of analysis, as it results in peptide overdigestion. Nevertheless, the few sites identified here target the interactions of P1/P2 protamines to the C-terminal domains of M.NPM2 at the distal face of the nucleoplasmin pentamer (Fig. 2D and Fig. 4).
4.3. Binding of M.NPM2 with somatic and sperm chromosomal proteins preferentially occurs through the distal face of nucleoplasmin
Fig. 4 schematically summarizes the results described above. In Fig. 4A–B, the strongest sites of interaction between histones and protamines with M.NPM2 are indicated in red. Weaker sites are indicated in faint red. As observed in the 3D crystallographic image shown in Fig. 4D, most of the interactions observed here take place in the distal face of the nucleoplasmin pentamer, with fewer observed in the lateral face. In this model, the C-terminal regions of NPM2 extend to participate in the binding through electrostatic interactions with the basic amino acids of the chromosomal proteins, similar to what is observed in the ionic competition with monovalent salts (Fig. 3).
The roles or differential involvement of M.NPM2 and M.NPM3 in mammalian oogenesis and sperm decondensation after fertilization still remain quite obscure [8]. In future studies, it will be of interest to extend the present research to the analysis of the interactions between histones and protamines with M.NPM3, which lacks acidic tract A3. In this regard, molecular swapping experiments between the two proteins, where A1 is incorporated into M.NPM2 or A3 deleted from M.NPM2, may turn out to be very informative.
Acknowledgments
This work was supported by a Natural Sciences and Engineering Research Council of Canada (NSERC) 46399-2012 grant to J.A. CHB would like to thank for support from the the Genomic Innovations Network (GIN) from Genome Canada and Genome British Columbia, and the National Sciences and Engineering Research Council of Canada for RGPIN 346194-2013 grant support. We are also thankful to Abby Truman for careful editing and proofreading of this manuscript.
Footnotes
Transparency document associated with this article can be found in the online version at 10.1016/j.bbrep.2016.04.002.
Appendix A. Supplementary material
Supplementary material
.
References
- 1.Laskey R.A., Honda B.M., Mills A.D., Finch J.T. Nucleosomes are assembled by an acidic protein which binds histones and transfers them to DNA. Nature. 1978;275:416–420. doi: 10.1038/275416a0. [DOI] [PubMed] [Google Scholar]
- 2.Philpott A., Leno G.H., Laskey R.A. Sperm decondensation in Xenopus egg cytoplasm is mediated by nucleoplasmin. Cell. 1991;65:569–578. doi: 10.1016/0092-8674(91)90089-h. [DOI] [PubMed] [Google Scholar]
- 3.Laskey R.A., Mills A.D., Philpott A., Leno G.H., Dilworth S.M., Dingwall C. The role of nucleoplasmin in chromatin assembly and disassembly. Philos. Trans. R. Soc. Lond. Ser. B: Biol. Sci. 1993;339:263–269. doi: 10.1098/rstb.1993.0024. (discussion 268-9) [DOI] [PubMed] [Google Scholar]
- 4.Katagiri C., Ohsumi K. Remodeling of sperm chromatin induced in egg extracts of amphibians. Int. J. Dev. Biol. 1994;38:209–216. [PubMed] [Google Scholar]
- 5.Prieto C., Saperas N., Arnan C., Hills M.H., Wang X., Chiva M., Aligue R., Subirana J.A., Ausió J. Nucleoplasmin interaction with protamines. Involvement of the polyglutamic tract. Biochemistry. 2002;41:7802–7810. doi: 10.1021/bi020120e. [DOI] [PubMed] [Google Scholar]
- 6.Saperas N., Chiva M., Aligu, Itoh T., Katagiri C., Subirana J.A., Ausió J. Physicochemical and functional comparison of Xenopus laevis nucleoplasmin obtained from oocytes and from overexpression in bacteria. Arch. Biochem. Biophys. 1999;361:135–141. doi: 10.1006/abbi.1998.0965. [DOI] [PubMed] [Google Scholar]
- 7.Frehlick L.J., Eirin-Lopez J.M., Ausio J. New insights into the nucleophosmin/nucleoplasmin family of nuclear chaperones. Bioessays. 2007;29:49–59. doi: 10.1002/bies.20512. [DOI] [PubMed] [Google Scholar]
- 8.Finn R.M., Ellard K., Eirin-Lopez J.M., Ausio J. Vertebrate nucleoplasmin and NASP: egg histone storage proteins with multiple chaperone activities. FASEB J. 2012;26:4788–4804. doi: 10.1096/fj.12-216663. [DOI] [PubMed] [Google Scholar]
- 9.Earnshaw W.C., Honda B.M., Laskey R.A., Thomas J.O. Assembly of nucleosomes: the reaction involving X. laevis nucleoplasmin. Cell. 1980;21:373–383. doi: 10.1016/0092-8674(80)90474-2. [DOI] [PubMed] [Google Scholar]
- 10.Dutta S., Akey I.V., Dingwall C., Hartman K.L., Laue T., Nolte R.T., Head J.F., Akey C.W. The crystal structure of nucleoplasmin-core: implications for histone binding and nucleosome assembly. Mol. Cell. 2001;8:841–853. doi: 10.1016/s1097-2765(01)00354-9. [DOI] [PubMed] [Google Scholar]
- 11.Salvany L., Chiva M., Arnan C., Ausio J., Subirana J.A., Saperas N. Mutation of the small acidic tract A1 drastically reduces nucleoplasmin activity. FEBS Lett. 2004;576:353–357. doi: 10.1016/j.febslet.2004.07.095. [DOI] [PubMed] [Google Scholar]
- 12.Arnan C., Saperas N., Prieto C., Chiva M., Ausio J. Interaction of nucleoplasmin with core histones. J. Biol. Chem. 2003;278:31319–31324. doi: 10.1074/jbc.M305560200. [DOI] [PubMed] [Google Scholar]
- 13.Ramos I., Martin-Benito J., Finn R., Bretana L., Aloria K., Arizmendi J.M., Ausio J., Muga A., Valpuesta J.M., Prado A. Nucleoplasmin binds histone H2A-H2B dimers through its distal face. J. Biol. Chem. 2010;285:33771–33778. doi: 10.1074/jbc.M110.150664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ramos I., Fernandez-Rivero N., Arranz R., Aloria K., Finn R., Arizmendi J.M., Ausio J., Valpuesta J.M., Muga A., Prado A. The intrinsically disordered distal face of nucleoplasmin recognizes distinct oligomerization states of histones. Nucleic Acids Res. 2014;42:1311–1325. doi: 10.1093/nar/gkt899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Banuelos S., Omaetxebarria M.J., Ramos I., Larsen M.R., Arregi I., Jensen O.N., Arizmendi J.M., Prado A., Muga A. Phosphorylation of both nucleoplasmin domains is required for activation of its chromatin decondensation activity. J. Biol. Chem. 2007;282:21213–21221. doi: 10.1074/jbc.M702842200. [DOI] [PubMed] [Google Scholar]
- 16.Onikubo T., Nicklay J.J., Xing L., Warren C., Anson B., Wang W.L., Burgos E.S., Ruff S.E., Shabanowitz J., Cheng R.H., Hunt D.F., Shechter D. Developmentally Regulated Post-translational Modification of Nucleoplasmin Controls Histone Sequestration and Deposition. Cell Rep. 2015 doi: 10.1016/j.celrep.2015.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.E.V., Petrotchenko, J.J., Serpa, C.H., Borchers (2010) An isotopically coded CID-cleavable biotinylated cross-linker for structural proteomics, Mol Cell Proteomics. 10, M110 001420. [DOI] [PMC free article] [PubMed]
- 18.Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 19.Ausió J. Presence of a highly specific histone H1-like protein in the chromatin of the sperm of the bivalve mollusks. Mol. Cell. Biochem. 1992;115:163–172. doi: 10.1007/BF00230327. [DOI] [PubMed] [Google Scholar]
- 20.Ausió J., Dong F., van Holde K.E. Use of selectively trypsinized nucleosome core particles to analyze the role of the histone “tails” in the stabilization of the nucleosome. J. Mol. Biol. 1989;206:451–463. doi: 10.1016/0022-2836(89)90493-2. [DOI] [PubMed] [Google Scholar]
- 21.Ausió J., Moore S.C. Reconstitution of chromatin complexes from high-performance liquid chromatography-purified histones. Methods (San Diego, Calif) 1998;15:333–342. doi: 10.1006/meth.1998.0637. [DOI] [PubMed] [Google Scholar]
- 22.Petrotchenko E.V., Makepeace K.A., Borchers C.H. DXMSMS match program for automated analysis of LC–MS/MS data obtained using isotopically coded cid-cleavable cross-linking reagents. Curr. Protoc. Bioinform. 2014;48 doi: 10.1002/0471250953.bi0818s48. (8 18 1-8 18 19) [DOI] [PubMed] [Google Scholar]
- 23.Petrotchenko E.V., Borchers C.H. ICC-CLASS: isotopically-coded cleavable crosslinking analysis software suite. BMC Bioinform. 2010;11:64. doi: 10.1186/1471-2105-11-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O., Platonova, I.V., Akey, J.F., Head, C.W., Akey (2011) Crystal Structure and Function of Human Nucleoplasmin (Npm2): A Histone Chaperone in Oocytes and Embryos, Biochemistry. [DOI] [PMC free article] [PubMed]
- 25.Ausio J., Gonzalez-Romero R., Woodcock C.L. Comparative structure of vertebrate sperm chromatin. J. Struct. Biol. 2014;188:142–155. doi: 10.1016/j.jsb.2014.09.004. [DOI] [PubMed] [Google Scholar]
- 26.Roque A., Ponte I., Suau P. Secondary structure of protamine in sperm nuclei: an infrared spectroscopy study. BMC Struct. Biol. 2012;11:14. doi: 10.1186/1472-6807-11-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peterson, C.L., Hansen, J.C. (2008) Chicken erythrocyte histone octamer preparation, CSH protocols. 2008, pdb prot5112 [DOI] [PubMed]
- 28.Banuelos S., Hierro A., Arizmendi J.M., Montoya G., Prado A., Muga A. Activation mechanism of the nuclear chaperone nucleoplasmin: role of the core domain. J. Mol. Biol. 2003;334:585–593. doi: 10.1016/j.jmb.2003.09.067. [DOI] [PubMed] [Google Scholar]
- 29.Dilworth S.M., Black S.J., Laskey R.A. Two complexes that contain histones are required for nucleosome assembly in vitro: role of nucleoplasmin and N1 in Xenopus egg extracts. Cell. 1987;51:1009–1018. doi: 10.1016/0092-8674(87)90587-3. [DOI] [PubMed] [Google Scholar]
- 30.Litvin J., King M.L. Expression and segregation of nucleoplasmin during development in Xenopus. Development (Cambridge, England) 1988;102:9–21. doi: 10.1242/dev.102.1.9. [DOI] [PubMed] [Google Scholar]
- 31.Demeler B. In: UltrScan A Comprehensive Data Analysis Software Package for Analytical Ultracentrifugation Experiments. Modern Analytical Ultracentrifugation: Techniques and Methods. In. Scott D.J., Hardfing S.E., Rowe A.J., editors. Royal Society of Chemistry; UK: 2005. pp. 210–229. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material