Abstract
Syndecan-4 (SDC4) is a cell-surface proteoglycan associated with cell adhesion, motility, and intracellular signaling. Here, we present that SDC4 functions as a positive regulator of the transforming growth factor (TGF)-β1-induced epithelial to mesenchymal transition (EMT) via Snail in lung adenocarcinoma, A549 cells. TGF-β1 up-regulated the expression of SDC4, accompanied by the induction of EMT. Wound-healing and transwell chemotaxis assay revealed that SDC4 promoted cell migration and invasion. SDC4 knockdown recovered the E-cadherin and decreased vimentin and Snail expression in EMT-induced A549 cells. However, depletion of SDC4 resulted in little change of the Slug protein expression and mesenchymal cell morphology induced by TGF-β1. The double knockdown of SDC-4 and Slug was required for reversal of epithelial morphology; it did not occur from the SDC4 single knockdown. These findings suggest that Snail is a transcriptional factor downstream of SDC4, and SDC4 regulates TGF-β1-induced EMT by cooperating with Slug. Our data provide a novel insight into cellular mechanisms, whereby the cell-surface proteoglycan modulated TGF-β1-induced EMT in lung adenocarcinoma, A549 cells.
Abbreviations: EMT, epithelial to mesenchymal transition; TGF-β, transforming growth factor-β; SDC4, Syndecan-4
Keywords: Syndecan-4, Transforming growth factor-β 1, Epithelial to mesenchymal transition, Snail, Slug
Highlights
-
•
TGF β1 induced to increase SDC-4 expression with cells undergo EMT in A549 cells.
-
•
SDC-4 up-regulation leads to decreased E-cadherin expression via Snail.
-
•
Slug suppresses E-cadherin expression, independent from SDC-4 expression.
-
•
SDC-4 and Slug knockdown conserved an epithelial morphology in TGF-β exposure.
-
•
TGF-β induced β1 and β3 integrin alteration under SDC-4 and Slug knockdown.
1. Introduction
Lung cancer is one of the most commonly diagnosed cancers, and the leading cause of death worldwide. Of these deaths, nearly 60% of patients progress into advanced stages with metastasis [1]. Chemotherapy is an important therapeutic strategy for advanced non-small cell lung cancer (NSCLC). However, most patients treated with chemotherapy frequently acquire the resistance to anti-cancer drugs [2]. Therefore, the mechanisms of the biologic processes that drive metastasis and drug resistance need to be elucidated.
Accumulating evidence suggests that the acquisition of epithelial-to-mesenchymal transition (EMT) is one of the cause of chemo-resistance of NSCLC [3]. Furthermore, EMT is associated with the invasiveness and metastasis [3]. EMT is a complex process, which involves cytoskeletal remodeling and cell–cell and cell–matrix adhesion, leading to the transition from a polarized, epithelial phenotype to a highly motile mesenchymal phenotype [4]. A major hallmark of EMT is the down-regulation of cell–cell adhesion molecule, E-cadherin [5], and the up-regulation of several transcriptional factors such as Snail, Slug and Twist, which repress the transcription of E-cadherin [5].
In particular, it has been well documented that Snail and Slug (a closely related member of the Snail family) regulate several genes involved in cell adhesion and cell junctions [6]. Despite many similarities between Snail and Slug, they have different biological functions via their target genes in cancer cells [7], [8]. However, little is known about the upstream molecules that modulate the expression of Snail and Slug, which is a cause of subsequent occurrence of EMT.
Syndecans (SDCs) are evolutionary conserved transmembrane heparan sulfate proteoglycan. They are composed of four genes (SDC1-4), and act as receptors and co-receptors of cytokines, growth factors and extracellular matrix components. They participate in regulation of cell–cell and cell–extracellular matrix (ECM) adhesion, cell migration, and growth factor activity. Among them, SDC4 is concentrated into focal adhesions together with integrins, which cooperate in generating the signals for the formation of focal adhesion and actin-stress fibers, resulting in the organization of both morphology and cell migration [9]. To date, up-regulation of SDC4 has been identified in the hepatocellular carcinomas and malignant mesotheliomas [10], [11]. Nevertheless, it is not clear whether SDC4 play a role in tumor progression and metastasis including EMT.
In the present study, we investigated the role of SDC4 in the control of EMT elicited by transforming growth factor (TGF)-β1 in human lung adenocarcinoma, A549 cells. We found that SDC4 is implicated in the regulation of TGF-β1-induced EMT via Snail. In addition, both SDC4 and Slug is required for completion of TGF-β1-induced EMT in A549 cells.
2. Materials and methods
2.1. Cell culture and reagents
The human lung adenocarcinoma A549 cell line was obtained from Riken Gene Bank (Tsukuba, Japan) and NCI-H292 cell line was purchased from ATCC. A549 cells were maintained in Dulbecco's modified Eagle's medium (DMEM), and NCI-H292 cells were cultured in RPMI-1640 medium supplemented with 10% fetal bovine serum (FBS) in a humidified atmosphere of 5% CO2 at 37 °C. TGF-β1 was purchased from R&D Systems (Minneapolis, MN, USA).
2.2. RNA interference
Small interfering (si) RNAs for SDC4, Snail, Slug and control scramble siRNA were obtained from Sigma-Aldrich (MISSION® siRNA, St. Louis, MO, USA). siRNAs with the following sense and antisense sequences were used: SDC4, 5′-GUAUCUCCAGCUCUGAUUATT-3′ (sense), 5′-UAAUCAGAGCUGGAGAUACTT-3′ (antisense); SNAIL, 5′-GCCUUCAACUGCAAAUACUTT-3′ (sense), 5′-AGUAUUUGCAGUUGAAGGCTT-3′ (antisense); SLUG, 5′-GCAUUUGCAGACAGGUCAATT-3′ (sense), 5′-UUGACCUGUCUGCAAAUGCTT-3′ (antisense); control scramble, 5′-CAGUGAAAUUUAUCCACAATT-3′ (sense), 5′-UUGUGGAUAAAUUUCACUGTT-3′ (antisense). For transient RNA interference, the siRNA were transfected at a concentration of 100 pmol per well with Lipofectamine RNAi MAX (Life Technologies Inc., Carlsbad, CA, USA) according to the manufacturer's protocol. Depletion of the targeted genes was confirmed with Western blot, Dot blot analysis, or the real-time reverse transcriptase PCR (RT-PCR).
2.3. RNA purification and real-time RT-PCR analysis
RNA was isolated with the RNeasy Mini Kit (Qiagen, Valencia, CA, USA). First-strand cDNA was synthesized with the Prime Script RT reagent kit (Takara Bio Inc., Otsu, Japan), and gene expression was quantified with the SYBR Green method of real-time PCR with the StepOne Real-Time PCR System (Life Technologies Inc.). Primer sequences are provided in Supplementary Table S1. Relative messenger (m) RNA levels, after normalization with GAPDH, were assessed with the 2−ΔΔCt method. The experiments were performed in triplicate.
2.4. Western blot and dot blot analysis
Whole cell lysates were prepared by lysing the cells in a buffer containing 50 mM Tris–HCl (pH 7.5), 0.15 M NaCl, 0.1% sodium dodecylsulfate, 1% sodium deoxycholate, 1% Triton X-100, and proteinase and phosphatase inhibitor cocktails (Sigma-Aldrich). Lysates were centrifuged for 10 min at 4 °C and an equal amount of protein 25–50 μg from the supernatants was used for SDS-PAGE and immunoblotting.
For Western blot analysis, the primary antibodies were as follows: E-cadherin (BD Biosciences, San Jose, CA, USA); N-cadherin, Snail and Slug (Cell Signaling Technology); integrin α5, β1, and β3 (BD Biosciences); GAPDH (EMD Millipore, Bedford, MA, USA) and SDC4 (Sigma-Aldrich). For Dot blot analysis, the conditioned medium from A549 cells was collected after 24 h of TGF-β1 stimulation, and blotted onto PVDF membranes. Then the membrane was probed with SDC4-specific antibody and visualized with ECL detection reagent.
2.5. Immunofluorescent staining
Cells grown on a glass slide (Poly-D-Lysine 8-well Culture Slides, BD Biosciences) were fixed in 4% paraformaldehyde and permeabilized with 0.1% Triton X-100. After washing with PBS, the cells were blocked with 1% bovine serum albumin in PBS, and incubated with primary antibodies against SDC4 (1:50) overnight. The cells were then incubated in Alexa Fluor 488-conjugated anti-rabbit IgG (Life Technologies Inc.). For phalloidin staining, the cells were incubated for 20 min at room temperature with Alexa Fluor 594 Phalloidin (Life Technologies Inc.) diluted with PBS and 0.1% BSA. After immunostaining, the slides were stained with 4', 6-diamidino-2-phenylindole (DAPI) and mounted.
2.6. Wound-healing assay
Cells were seeded in triplicate on 24-well culture plates at 2×104 cells/well. A scratch through the central axis of the plate was gently made using 200-μl micropipette tip 48 h after the cells had been transfected with control non-specific, SDC4 or Slug siRNA. The cells were washed with PBS to remove any loose cells, and fresh media were added with or without TGF-β1 (5 ng/ml). The images were obtained immediately after wounding and after 20 h of incubation. The percentage (%) change in restitution was determined by comparing the difference in wound width (n=3).
2.7. Transwell chemotaxis assay
The chemotactic response was assessed with the BD Falcon FluoroBlok system (BD Biosciences) with pore sizes of 8.0 μm in 24-well inserts. Cells (2×104 cells) transfected with control, SDC4 or Slug siRNA were loaded into the inserts in 200 µl of DMEM medium containing 0.5% FBS in the presence or absence of TGF-β1 (5 ng/ml). Lower wells of the plate were filled with 800 µl of DMEM supplemented with 10% FBS as an attractant. After 24 h, the lower side of the membrane were fixed with methanol and mounted on glass slides for DAPI staining. DAPI-positive migrated cells were counted with a fluorescent microscope. Assays were performed in triplicate with 3 separate microscope fields per membrane.
2.8. Cell proliferation assay
Cells were transfected with control or SDC4 siRNA at a concentration of 100 pmol per well. After transfection for 48 h, cells (5×103 cells/well) were detached and re-seeded in a 96-well plate in the presence or absence of TGF-β1. After 72 h incubation, the MTT (Cell Titer 96 assay kit, Promega, Madison, WI) assay was performed following the manufacturer's instruction. All experimental points were set up in 6–12 wells and all experiments were repeated at least 3 times.
3. Statistical analysis
Statistical analysis was carried out using a two-tailed Student's t-Test. P values<0.05 were considered significant.
4. Results
4.1. Up-regulation of SDC4 expression in TGF-β1-induced EMT
A549 cells have been frequently used as a model of inducible TGF-β1-mediated EMT in lung cancer. As shown in Fig. 1A, treatment with TGF-β1 induced a spindle-like mesenchymal morphology characteristic of EMT in A549 cells. This morphological change was accompanied by decrease of E-cadherin expression and increased expression of mesenchymal marker, N-cadherin and EMT-related transcriptional factor, Snail and Slug (Fig. 1B, Fig. 1D). These data indicated that A549 cells exhibit phenotype consistent with EMT. In parallel with the occurrence of EMT, TGF-β1 significantly induced the expression of SDC4 at both protein and mRNA levels (Fig. 1C, left panel, Fig. 1D). Immunofluorescence analysis revealed the punctate localization of SDC4 in the TGF-β1–treated A549 cells (Fig. 1C, right panel). Similar results were obtained from the experiments in another human lung adenocarcinoma cell lines, NCI-H292. SDC4 was induced in NCI-H292 cells by the treatment of TGF-β1, accompanied by the changed expression of some of EMT-related genes including E-cadherin, vimentin and Snail (Fig. S4).
Fig. 1.
SDC4 is significantly increased in TGF-β1-induced EMT. A, The images of TGF-β1-induced EMT. Scale bar: 200 μm. A549 cells were exposed in the presence or absence of TGF-β1 (5 ng/ml) for 48 h then images were obtained. B, E-cadherin, N-cadherin and Snail protein expression were determined by Western blot analysis. GAPDH was used as loading control. C, Left: SDC4 mRNA expression was analyzed by real-time RT-PCR using specific primers for SDC4 (Supplementary Table S1). The data were normalized to GAPDH. Each bar represents means ±SEM (n=3). “※” indicates statistically significant (P<0.05). Right: the images of SDC4 expression by immunofluorescent staining. The arrows indicate SDC4 positive cells. Scale bar: 100 μm. D, The effects of SDC4 siRNA on E-cadherin, Vimentin, Snail and Slug expression were determined by Western blot analysis. Cells were seeded in 6-well culture plates at 1×105 cells/well, 48 h after transfection with indicated siRNA(s), then harvest protein samples. GAPDH was used as loading control.
4.2. SDC4 partially contributes to TGF-β1-induced EMT via the up-regulation of Snail
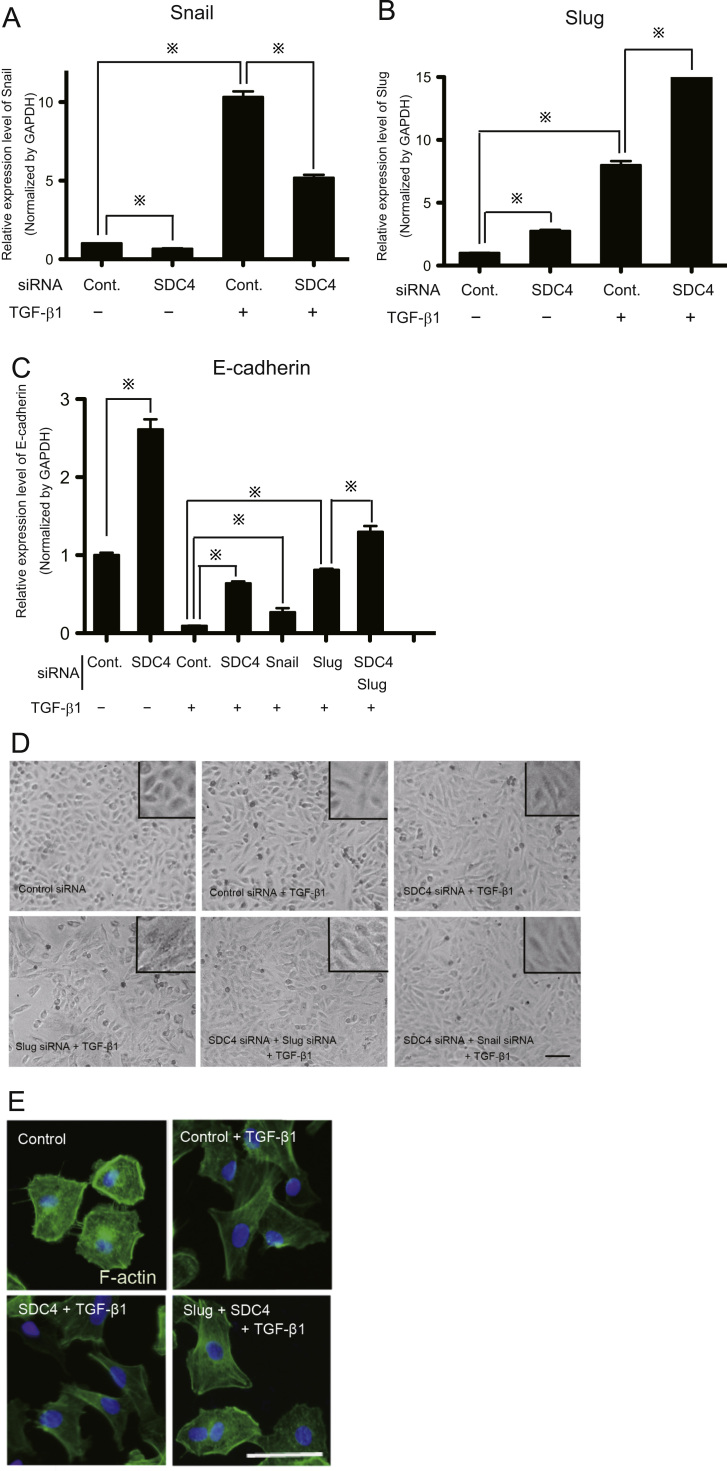
To clarify the role of SDC4 in TGF-β1-induced EMT, we examined the effects of SDC4 siRNA on the EMT phenotype. As shown in Fig. 1D, SDC4 knockdown partially restored the protein expression of E-cadherin, and decreased vimentin. RT-PCR analysis also showed that SDC4 knockdown upregulated the expression of E-cadherin, regardless of whether or not TGF-β1 was present (Fig. 2C). mRNA induction of mesenchymal markers, vimentin and N-cadherin, by TGF-β1 treatment was reduced in the SDC4-siRNA expressed A549 cells (Fig. S6). Furthermore, SDC4 overexpression enhanced the induction of EMT-related genes by the stimulation of TGF-β1 in the both A549 and NCI-H292 cells (Fig. S5). These data show that SDC4 up-regulates the process of TGF-β1-induced EMT. However, the spindle-shaped mesenchymal morphology evoked by TGF-β1 remained in SDC4-knocked down A549 cells (Fig. 2D), indicating that the SDC4 knockdown is insufficient to restore the epithelial morphology. In addition, the SDC4-overexpressing A549 and NCI-H292 cells remained the cobblestone appearance, a characteristic epithelial morphology (Fig. S5A). These results suggest that SDC4 is not involved in the morphological changes accompanied by EMT. Therefore, we investigated how SDC4 knockdown affected the expression of Snail and Slug. The knockdown of SDC4 repressed the expression of Snail at both mRNA and protein levels (Fig. 1D and 2A). In contrast, the expression of Slug protein was not significantly changed by SDC4 knockdown (Fig.1D). Rather, Slug mRNA was increased by SDC4 depletion, as opposed to Snail expression (Fig. 2A and B). In addition, we further addressed the effects of Snail-forced expression in A549 cells (Fig. S2). Single overexpression of Snail, however, did not induce the EMT phenotype. Above findings raise the possibility that SDC4 partially contributes to TGF-β1-induced EMT via the up-regulation of Snail, but additional molecules including Slug is necessary for the completion of EMT in A549 cells.
Fig. 2.
Effects of SDC4 knockdown on the expression of EMT-related genes and cell morphology. A, B, C, Cells were transfected with indicated siRNAs and after 48 h mRNA was isolated with the RNeasy Mini Kit (Qiagen), then real-time RT-PCR for Snail, Slug and E-cadherin was performed by indicated primers in Supplemental Table S1. Each bar represents means ±SEM (n=3). “※” indicates statistically significant (p<0.05). D, The cellular morphological images were observed by optical microscope. A549 cells were transfected by indicated siRNA(s), then exposed TGF-β1 for 48 h. Scale bar: 100 μm. E, Representative cellular images of A549 cells subjected to immunofluorescent staining. F-actin was stained with phalloidin. Images of fluorescent staining were obtained for each field and merged by fluorescent microscope (Nikon Eclipse Ni-E). A549 cells were transfected by indicated siRNA(s), then exposed TGF-β1 for 48 h. The blue color indicates nuclei, stained with DAPI. Scale bar: 50 μm.
4.3. Double knockdown of SDC4 and Slug restores epithelial morphology of A549 cells
This notion mentioned above is supported by the experiments using double knockdown of SDC4 and Slug. In the double-knockdown A549 cells, E-cadherin mRNA increased approximately twice as high as those with either SDC4 or Slug single-knockdown (Fig. 2C). Knockdown of both SDC4 and Slug genes restored the epithelial morphology from spindle shape of A549 elicited by TGF-β1 (Fig. 2D). Moreover, phalloidin staining showed TGF-β1 induced the actin stress fibers formation, a feature of EMT. Single knockdown of SDC4 did not affect this actin remodeling. However, when both SDC4 and Slug were silenced, actin filaments organization completely restored the epithelial morphology (Fig. 2E). These results indicated that the expression of both SDC4 and Slug is necessary for TGF-β1-induced EMT.
4.4. SDC-4 enhances TGF-β1-stimulated cell migration and proliferation
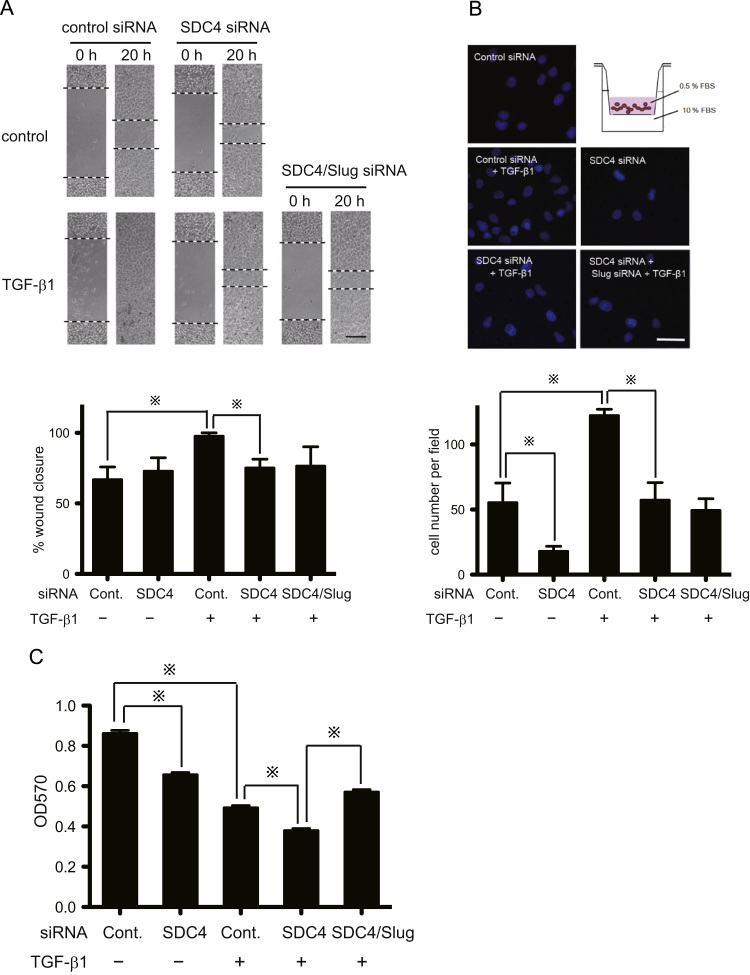
The acquisition of more motile phenotype is important consequences of EMT. In response to TGF-β1, A549 cells exhibited enhanced migratory ability, as shown by the wound-healing and transwell chemotaxis assays (Fig. 3A and B). Knockdown of SDC4 resulted in a decreased migration ability with wound-closure rate of 75.1% compared to control siRNA-transfected cells (97.6% wound closure) in the presence of TGF-β1 (Fig. 3A, lane3, 4). A similar result was observed in the transwell chemotaxis assay. SDC4 knockdown significantly blocked the cellular chemotaxis compared with the control siRNA-treated cells, regardless of whether or not TGF-β1 was present (Fig. 3B). However, we didn't find a synergistic effect of the combination of Slug and SDC4 siRNA on cell motility (Fig. 3A and B). Moreover, SDC4 increased the proliferation of A549 cells as shown in Fig. 3C. These results indicate that SDC4 accelerates the cell migration in the context of EMT.
Fig. 3.
SDC4 enhances TGF-β1 stimulated cellular restitution, chemotaxis and proliferation. A, Scratched restitution assay. Cells were seeded in 24-well culture plates at 2×104 cells/well. Forty-eight hours after transfection with indicated siRNA, a scratch was made using 200-μl micropipette tip. Upper: 0 and 20 h after wounding. Black dotted lines indicate the wound edge. Lower: Percentage (%) change in migration as determined by comparing the difference in wound width (n=3). Each bar represents means ±SEM; ※, p<0.05. Scale bars: 100 μm. B, Transwell chemotaxis assay: The transfected A549 cells (2×104 cells) were loaded into 24-well inserts (8.0 μm-pore size) with DMEM medium containing 0.5% FBS in the presence or absence of TGF-β1 (5 ng/ml). Lower wells of the plate were filled with DMEM with 10% FBS. After 24 h, remove A549 cell on upper-side, then lower side of membrane were stained with DAPI. DAPI-positive migrated cells were counted with a fluorescent microscope. Each bar represents means ±SEM; ※, p<0.05. Scale bars: 50 μm. C, Cellular proliferation assay: The cellular proliferation was assessed with MTT assay. A549 cells were transfected with indicated siRNAs and reseeded into 96-well plate (5×103 cells/well) in the presence or absence of TGF-β1 (5 ng/ml), then after 72 h incubation, MTT assay was performed. Data represent means ±SEM; ※, p<0.05.
4.5. Ectodomain shedding of SDC4 does not affect TGF-β1-induced EMT
SDC4 is known to be shed by proteolytic cleavage, yielding a variety of physiological reactions including cancer progression. Therefore, we examined the effects of the shed SDC4 on EMT. However, the quantity of shed SDC4 was not changed after exposure of TGF-β1 (Fig. S1A). The addition of cell culture supernatant from A549 cells didn’t compensate for the decrease of E-cadherin and Snail mRNA expression by SDC4 siRNA (Fig. S1B). These indicate that the SDC4 ectodomain is not functional for TGF-β1-induced EMT.
4.6. SDC4 is required for retaining the inherent β1 and β3 integrin expression pattern
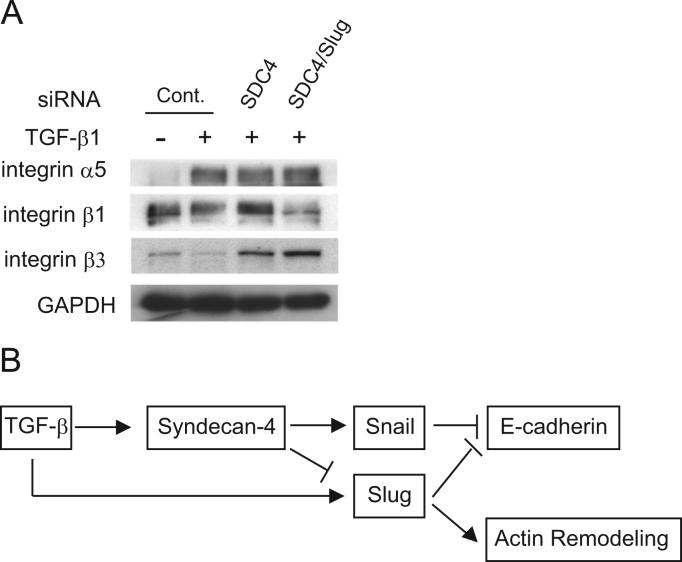
SDC4 acts cooperatively with integrins in the processes of cell spreading, focal adhesion formation and actin stress fiber assembly. Therefore, we investigated the relationship between the expression of SDC4 and integrin in the process of EMT. By treatment with TGF-β1, the expression of α5 integrin was significantly up-regulated, whereas both β1 and β3 integrin expression were down-regulated (Fig. 4A). When SDC4 was knocked down, α5 integrin expression was slightly decreased, while the expression of β1 and β3 integrin was increased (Fig. 4A), suggesting partial reverse to the inherent phenotype of A549 cells. In addition, double knockdown of SDC4 and Slug induced the decreased β1 integrin expression and a further increase of β3 integrin expression (Fig. 4A). These results suggest that the expression of β1 and β3 integrin are independently regulated by SDC4 and Slug, agreeing with the results of actin filaments remodeling shown in Fig. 2E.
Fig. 4.
SDC4 are required for retaining the inherent β1 and β3 integrin expression pattern. A, α5, β1, and β3 integrin expressions were determined by Western-blot analysis. Forty-eight hours after transfection with indicated siRNA, A549 cells were exposed with or without TGF-β1 (5 ng/ml) for 48 h. GAPDH was used as loading control. B, Schema of simplified signal transduction pathways, regulating E-cadherin expression via SDC4, Snail and Slug, and consequent actin remodeling.
5. Discussion
In this study, we demonstrated a correlation between the up-regulation of SDC4 expression and TGF-β1-induced EMT in human lung adenocarcinoma, A549 cells. SDC4 is an upstream molecule of Snail, and subsequently modulates the expression of EMT-related genes and promotes cell migration. However, SDC4 didn't upregulate Slug expression in EMT. Both SDC4 and Slug expression are required for the TGF-β-induced EMT in A549 cells.
SDC4 is a focal adhesion component in a range of cell types, adherent to several different matrix molecules, activating protein kinase C-alpha, focal adhesion kinase (FAK), and small GTPase Rho to promote cell adhesion and motility. The SDC4 overexpressing cells showed larger and denser focal adhesions, and correlated to stronger attachment and decreased cell motility [12], whereas SDC4 null cells are deficient in phosphorylated FAK and show impaired cell motility [13], [14]. E-cadherin is a mediator cell–cell adhesion in epithelial tissue, and loss of E-cadherin can promote invasive and metastatic behavior in many epithelial tumors [15]. Our study revealed that the SDC4 knockdown significantly inhibited Snail expression and induced E-cadherin. Conversely, Kato et al. reported that loss of syndecan-1 resulted in the mesenchymal phenotype accompanied by the decrease of E-cadherin [16]. Therefore, this is the first report that SDC4 is involved in TGF-β1-induced EMT via Snail signaling.
Snail and Slug are key regulators of TGF-β1-induced EMT in a variety of cancers. Several studies have shown that the induction of both Snail and Slug in the response to TGF-β is mediated by common transcriptional factor, Smad3, which binds to the promoter and activates its transcription of these genes [17], [18], [19]. In our study, however, SDC4 raised the expression of Snail and repressed Slug (Fig. 1D, 2A and B). Similar results have been reported in estradiol/estrogen α-induced EMT in breast cancer cell lines [20]. In that paper, the transcription of Slug gene was modulated by the epigenetic histone modification or phosphoinositide 3-kinase/protein kinase B signaling [20]. Furthermore, in pancreatic cancer, Snail and Slug have different functions on cell motility and Rho signaling [21]. Snail, but not Slug, promotes cell migration by β1 integrin [21]. Thus, molecular mechanisms by which the transcription of Snail and Slug is inversely regulated in TGF-β-induced EMT in A549 cells need further elucidation.
Silencing of SDC4 upregulated the E-cadherin expression even in the absence of TGF-β1 (Fig. 2C), though the Snail level was not changed significantly (Fig. 2A). One interpretation of these results is that SDC4 regulates E-cadherin expression via other repressors of E-cadherin expression (e.g. ZEB1/2, Twist1/2 and E47/TCF3 etc) than Snail under the condition without TGF-β1. SDC4 is a multifunctional proteoglycan, which functions as a co-receptor for several growth factors, an independent receptor for FGF or PDGF, a physical connector to extra cellular matrix, and a regulator of Wnt signaling. Therefore, SDC4 might control the expression of E-cadherin through the various and complex signaling in basal condition.
The shed SDC4 ectodomain by proteolytic cleavage is also an important regulatory mechanism for altering pathophysiological conditions, including the processes of tumor development, progression, and metastasis [25–29]. Our study shows that exposure to the conditioned media from A549 cells, including the shed SDC4 ectodomain, is insufficient to disrupt expression of the EMT markers E-cadherin and Snail in response to TGF-β1 in SDC4-attenuated A549 cells (Fig. S1). These findings suggest that the cytoplasmic domain of SDC4 is required for TGF-β–induced EMT.
SDC4 functions as a co-receptor for chemokine and growth factor including TGF-β, vascular endothelial growth factor and fibroblast growth factors [22], [23]. SDC4 facilitates binding of such growth factors to their receptors through the heparan sulfate chains, and enhances the signal evoked by these receptors [24]. Our data showed that TGF-β induced SDC4 expression, followed by the progression of EMT. Possibly, the induced SDC4 protein might further promote EMT by the acceleration of binding TGF-β to the receptor in a positive feedback manner.
In addition, SDC4 is involved in focal adhesion formation and actin stress fibers by cooperating with integrins in a Rho– and protein kinase Cα–dependent manner [25]. A recent study showed that TGF-β signaling enhances Smad3 binding to the β1 integrin promoter, triggering an up-regulation of β1 integrin gene expression [26]. Another study also reported that TGF-β induced to increase both mRNA and protein of β3 integrin subunit in human lung fibroblasts via Src-, and p38 MAPK-dependent pathway [27]. Generally, raised expression of α5β1 and αvβ3 integrin is favorable for cell movement via the attachment to extracellular matrix (ECM) such as fibronectin and vitronectin. However, in human lung adenocarcinoma, A549 cells, TGF-β induced the α5 integrin, but reduced β1/β3 integrin slightly (Fig. 4A). Additionally, SDC4 knockdown led to further increase of both β1 and β3 integrin, although its knockdown inhibited EMT (Fig. 1, Fig. 4). This finding is contrary to previous reports described above [24], [25]. SDC4 is known to interact with ECM identically to integrins and a full cell-adhesion to ECM requires engagement of both types of receptors. Therefore, the increased expression of β1/β3 integrin by SDC4 knockdown may have resulted from a compensation for the attenuated binding between SDC4 and ECM.
SDC4 knockdown could not change the mesenchymal morphology associated with TGF-β1-induced EMT (Fig. 2D and E). Double knockdown of SDC4 and Slug make it possible to revert to the epithelial morphology. Furthermore, single knockdown of SDC4 repressed the cellular restitution and chemotaxis, while double knockdown of SDC4 and Snail exhibited no synergistic effects on the cell migration (Fig. 3). These data suggest that Snail is a regulator of cell motility downstream of SDC4, and Slug is a modulator of the cytoskeletal changes and actin remodeling in TGF-β1-induced EMT in A549 cells. Moreover, single overexpression of Snail could not induce any EMT phenotypes in A549 cells (Fig. S2), suggesting that the progression of TGF-β1-induced EMT is required for more additional factors, e.g. Zeb and Twist family genes.
In conclusion, we have identified a novel role for SDC4 and a new regulatory mechanism during TGF-β1-mediated EMT in lung adenocarcinoma, A549 cells. A549 cells are the typical model of TGF-β-induced EMT. It is necessary to generalize the involvement of SDC4 in EMT using other NSCLC cell lines. Further studies about the regulatory mechanisms of TGF-β-induced EMT via SDC4 should be carried out, thereby disclosing the aspects of cell motility, cell adhesion, and actin filament remodeling in cancer cell invasion and metastasis.
Acknowledgments
This work was supported in part by MEXT-Supported Program for the Strategic Research Foundation at Private Universities, 2012–2014, from the Ministry of Education, Culture, Sports, Science and Technology of Japan. We would like to thank the members of the institute of molecular oncology and Dr. Stephanie Constantin for helpful discussions, and Mr. Andrew Bourger and Gerald O’rourke for correction of the English.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.bbrep.2015.11.021.
Appendix A. Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
References
- 1.Mountain C.F., Dresler C.M. Regional lymph node classification for lung cancer staging. Chest. 1997;111:1718–1723. doi: 10.1378/chest.111.6.1718. [DOI] [PubMed] [Google Scholar]
- 2.Mok T.S., Ramalingam S.S. Maintenance therapy in nonsmall-cell lung cancer: a new treatment paradigm. Cancer. 2009;115:5143–5154. doi: 10.1002/cncr.24563. [DOI] [PubMed] [Google Scholar]
- 3.Massague J. TGFβ in Cancer. Cell. 2008;134:215–230. doi: 10.1016/j.cell.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huber M.A., Kraut N., Beug H. Molecular requirements for epithelial-mesenchymal transition during tumor progression. Curr. Opin. Cell. Biol. 2005;17:548–558. doi: 10.1016/j.ceb.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 5.Thiery J.P., Acloque H., Huang R.Y., Nieto M.A. Epithelial–mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 6.Hajra K.M., Chen D.Y., Fearon E.R. The SLUG zinc-finger protein represses E-cadherin in breast cancer. Cancer Res. 2002;62:1613–1618. [PubMed] [Google Scholar]
- 7.Shih J.Y., Yang P.C. The EMT regulator slug and lung carcinogenesis. Carcinogenesis. 2011;32:1299–1304. doi: 10.1093/carcin/bgr110. [DOI] [PubMed] [Google Scholar]
- 8.Bernfield M., Kokenyesi R., Kato M., Hinkes M.T., Spring J., Gallo R.L., Lose E.J. Biology of the syndecans: a family of transmembrane heparan sulfate proteoglycans. Annu. Rev. Cell. Biol. 1992;8:365–393. doi: 10.1146/annurev.cb.08.110192.002053. [DOI] [PubMed] [Google Scholar]
- 9.Woods A., Couchman J.R. Syndecan-4 and focal adhesion function. Curr. Opin. Cell. Biol. 2001;13:578–583. doi: 10.1016/s0955-0674(00)00254-4. [DOI] [PubMed] [Google Scholar]
- 10.Gulyas M., Hjerpe A. Proteoglycans and WT1 as markers for distinguishing adenocarcinoma, epithelioid mesothelioma, and benign mesothelium. J. Pathol. 2003;199:479–487. doi: 10.1002/path.1312. [DOI] [PubMed] [Google Scholar]
- 11.Roskams T., De Vos R., David G., Van Damme B., Desmet V. Heparan sulphate proteoglycan expression in human primary liver tumours. J. Pathol. 1998;185:290–297. doi: 10.1002/(SICI)1096-9896(199807)185:3<290::AID-PATH91>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 12.Longley R.L., Woods A., Fleetwood A., Cowling G.J., Gallagher J.T., Couchman J.R. Control of morphology, cytoskeleton and migration by syndecan-4. J. Cell. Sci. 1999;112(Pt 20):3421–3431. doi: 10.1242/jcs.112.20.3421. [DOI] [PubMed] [Google Scholar]
- 13.Woods A. Syndecans: transmembrane modulators of adhesion and matrix assembly. J. Clin. Invest. 2001;107:935–941. doi: 10.1172/JCI12802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cary L.A., Chang J.F., Guan J.L. Stimulation of cell migration by overexpression of focal adhesion kinase and its association with Src and Fyn. J. Cell. Sci. 1996;109:1787–1794. doi: 10.1242/jcs.109.7.1787. [DOI] [PubMed] [Google Scholar]
- 15.Birchmeier W., Behrens J. Cadherin expression in carcinomas: role in the formation of cell junctions and the prevention of invasiveness. Biochim. Et. Biophys. acta. 1994;1198:11–26. doi: 10.1016/0304-419x(94)90003-5. [DOI] [PubMed] [Google Scholar]
- 16.Kato M., Saunders S., Nguyen H., Bernfield M. Loss of cell surface syndecan-1 causes epithelia to transform into anchorage-independent mesenchyme-like cells. Mol. Biol. Cell. 1995;6:559–576. doi: 10.1091/mbc.6.5.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoot K.E., Lighthall J., Han G., Lu S.L., Li A., Ju W., Kulesz-Martin M., Bottinger E., Wang X.J. Keratinocyte-specific Smad2 ablation results in increased epithelial–mesenchymal transition during skin cancer formation and progression. J. Clin. Invest. 2008;118:2722–2732. doi: 10.1172/JCI33713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cho H.J., Baek K.E., Saika S., Jeong M.J., Yoo J. Snail is required for transforming growth factor-β-induced epithelial-mesenchymal transition by activating PI3 kinase/Akt signal pathway. Biochem. Biophys. Res. Commun. 2007;353:337–343. doi: 10.1016/j.bbrc.2006.12.035. [DOI] [PubMed] [Google Scholar]
- 19.Morita T., Mayanagi T., Sobue K. Dual roles of myocardin-related transcription factors in epithelial mesenchymal transition via slug induction and actin remodeling. J. Cell. Biol. 2007;179:1027–1042. doi: 10.1083/jcb.200708174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ye Y., Xiao Y., Wang W., Yearsley K., Gao J.X., Shetuni B., Barsky S.H. ERalpha signaling through slug regulates E-cadherin and EMT. Oncogene. 2010;29:1451–1462. doi: 10.1038/onc.2009.433. [DOI] [PubMed] [Google Scholar]
- 21.Shields M.A., Krantz S.B., Bentrem D.J., Dangi-Garimella S., Munshi H.G. Interplay between β1-integrin and Rho signaling regulates differential scattering and motility of pancreatic cancer cells by snail and Slug proteins. J. Biol. Chem. 2012;287:6218–6229. doi: 10.1074/jbc.M111.308940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang D., Liang J., Campanella G.S., Guo R., Yu S., Xie T., Liu N., Jung Y., Homer R., Meltzer E.B., Li Y., Tager A.M., Goetinck P.F., Luster A.D., Noble P.W. Inhibition of pulmonary fibrosis in mice by CXCL10 requires glycosaminoglycan binding and syndecan-4. J. Clin. Invest. 2010;120:2049–2057. doi: 10.1172/JCI38644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ishiguro K., Kojima T., Muramatsu T. Syndecan-4 as a molecule involved in defense mechanisms. Glycoconj. J. 2002;19:315–318. doi: 10.1023/A:1025308702966. [DOI] [PubMed] [Google Scholar]
- 24.Tkachenko E., Rhodes J.M., Simons M. Syndecans: new kids on the signaling block. Circ. Res. 2005;96:488–500. doi: 10.1161/01.RES.0000159708.71142.c8. [DOI] [PubMed] [Google Scholar]
- 25.Saoncella S., Echtermeyer F., Denhez F., Nowlen J.K., Mosher D.F., Robinson S.D., Hynes R.O., Goetinck P.F. Syndecan-4 signals cooperatively with integrins in a Rho-dependent manner in the assembly of focal adhesions and actin stress fibers. Proc. Natl. Acad. Sci. USA. 1999;96:2805–2810. doi: 10.1073/pnas.96.6.2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yeh Y.C., Wei W.C., Wang Y.K., Lin S.C., Sung J.M., Tang M.J. Transforming growth factor-β1 induces Smad3-dependent β1 integrin gene expression in epithelial-to-mesenchymal transition during chronic tubulointerstitial fibrosis. Am. J. Pathol. 2010;177:1743–1754. doi: 10.2353/ajpath.2010.091183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pechkovsky D.V., Scaffidi A.K., Hackett T.L., Ballard J., Shaheen F., Thompson P.J., Thannickal V.J., Knight D.A. Transforming growth factor β1 induces αvβ3 integrin expression in human lung fibroblasts via a beta3 integrin-, c-Src-, and p38 MAPK-dependent pathway. J. Biol. Chem. 2008;283:12898–12908. doi: 10.1074/jbc.M708226200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material
Supplementary material